Abstract
The risk of severe irinotecan-induced neutropenia has been shown to be related to the UGT1 variant UGT1A1*28, which increases exposure to the potent metabolite SN-38. Our goal was to identify a novel UGT1 marker(s) using 28 haplotype-tagged single nucleotide polymorphisms genotyped by mass spectrometry. By characterizing the UGT1 sequence from a cohort of 167 Canadian metastatic colorectal cancer (mCRC) patients and a validation cohort of 250 Italian mCRC patients, we found rs11563250G, located in the intergenic region downstream of UGT1, to be significantly associated with reduced risk of severe neutropenia (odds ratio (OR)=0.21; p=0.043 and OR=0.27; p=0.036, respectively, and OR=0.31 when combined; p=0.001), which remained significant upon correction for multiple testing in the combined cohort (p=0.041). For the two-marker haplotype rs11563250G and UGT1A1*1 (rs8175347 TA6), the OR was of 0.17 (p=0.0004). Genetic testing of this marker may identify patients who might benefit from increased irinotecan dosing.
Keywords: Colorectal cancer, Genetic variants, Irinotecan, Neutropenia, UGT1
INTRODUCTION
Irinotecan is a chemotherapeutic agent used in combination with folinic acid (leucovorin), and 5-fluorouracil as a first-line treatment of metastatic colorectal cancer (mCRC) in a regimen denoted FOLFIRI. Irinotecan exerts its cytotoxicity by inhibiting topoisomerase I during DNA replication through its active metabolite SN-38. As an anticancer agent with a narrow therapeutic index, dose management of irinotecan is necessary to minimize associated toxicities, i.e., neutropenia and diarrhea. Hepatic and extrahepatic phase II UDP-glucuronosyltransferase (UGT) drug-metabolizing enzymes, i.e., UGT1A1, UGT1A7, and UGT1A9, convert SN-38 into an inactive form SN-38 glucuronide (SN-38G).1 Neutropenia is the most significant dose-limiting toxicity associated with irinotecan treatment and is directly related to the plasma SN-38 concentration, which, in addition to UGT1 activity, depends on biliary excretion and the activities of several transporter genes.2–6 In patients, germline information concerning the UGT1 pathway may help to optimize the chemotherapeutic agent dose and type of therapy.7–9 Several reports of distinct irinotecan-reaction profiles associated with common UGT1 variants have highlighted the relevance of characterizing UGT1 variants.2, 4, 5, 10, 11
Most tested biomarkers for UGT1 variants have been shown to be useful tools to identify patients more likely to experience severe neutropenia related to irinotecan-containing regimens. In particular, the variant UGT1A1*28, which contains seven, instead of six TA repeats in its promoter A(TA)nTAA region, is associated with significantly decreased glucuronidation activity, which results in reduced SN-38 clearance 12. This reduced clearance is consistently associated, in a dose-dependent manner, with an increased risk of severe neutropenia in patients homozygous for this allele.2, 13–15 More recently, it was established that UGT1 haplotypes, e.g., combination of variants in UGT1A1, UGT1A6, UGT1A7, and UGT1A9, are also associated with an increased risk of severe neutropenia.11, 16, 17 These findings demonstrate that in addition to the well-established UGT1A1 rs8175347 TATA box promoter variant, other UGT1 variants might be involved in irinotecan-induced toxicities. Through haplotyping, our group recently found that the presence of the variant rs8330 in the 3’–untranslated region (3’UTR) of the UGT1 locus improves the ability to predict the risk of severe irinotecan-induced neutropenia, which suggests variance in this region common to all UGT1A transcripts, may also participate in the toxic effect of irinotecan.16. Furthermore, the clinical relevance of rs8330 was recently demonstrated for acetaminophen-induced acute liver failure associated with modification of the exon 5a/5b splice variants mRNA ratio.18. These findings, therefore, support the contribution of variants across UGT1 in irinotecan pharmacogenetics and the putative role of the 3’-region of the gene in the overall glucuronidation capacity and subsequent risk of severe toxicity.
The study reported herein aimed to examine the genetic association across the UGT1 locus with the risk of developing severe neutropenia in mCRC patients treated with FOLFIRI-based regimens using a haplotype-tagging SNP (htSNP) strategy to maximize gene coverage and discover novel markers. We initially studied a prospective cohort of mCRC patients recruited in Canada (n = 167) treated with FOLFIRI-based regimens (discovery cohort) and replicated the main findings of that study in a similar, but independent, cohort of 250 Italian patients (validation cohort). The most significant and replicated finding of this work is the discovery that the variant allele rs11563250G in the 3’-flanking region of UGT1 is associated with a substantially reduced risk of irinotecan-induced neutropenia in both populations. This new marker may help refine our ability to predict the risk of severe neutropenia, optimize irinotecan dosage, and personalize treatment to improve clinical outcomes.
MATERIALS AND METHODS
Patient cohorts and liver samples.
One hundred and sixty-seven Eastern Canadian mCRC patients were recruited and then begun on a FOLFIRI regimen. All patients received a FOLFIRI regimen that included 180 mg irinotecan/m2 intravenously with 69 patients also receiving a co-treatment, i.e., bevacizumab, an experimental drug, or a placebo. Specific treatment modalities and eligibility criteria have been published.16 Participants provided written consent for genetic analysis. Each local research ethics committee approved the research protocol. Table 1 summarizes patient demographics (age and sex) and clinical information (treatment, toxicity, tumor site). The replication cohort consisted of 250 Northeastern Italian mCRC patients that are receiving a FOLFIRI treatment of the same dose and delivery method as described.9, 11 The severity of neutropenia was evaluated according to the National Cancer Institute Common Terminology Criteria for Adverse Events version 3.0. We studied 48 livers to assess the relationship between severe irinotecan-induced neutropenia and UGT1 genotypes (see below for the genotyping procedure). UGT1A1 expression levels and rates of bilirubin and SN-38 glucuronidation for these liver samples have been reported.19, 20
Table 1.
Demographic and clinical characteristics of the study populations.
| Canadian cohort (Discovery cohort) |
Italian Cohorta (Validation cohort) |
|||
|---|---|---|---|---|
| Characteristics | N | % | N | % |
| Total number | 167 | 250 | ||
| Gender (male/female) | 110/57 | 162/88 | ||
| Median age (years) | 61.5 | 60.6 | ||
| Primary tumor site | ||||
| Colon | 122 | 73.1 | 179 | 71.6 |
| Rectum | 42 | 25.1 | 71 | 28.4 |
| Unknown | 3 | 1.8 | - | |
| Regimen | ||||
| FOLFIRI | 167 | 250 | ||
| Co-treatment: | ||||
| bevacizumab | 69 | 41.3 | - | |
| Other drug | 6 | 3.6 | - | |
| Toxicity | ||||
| Diarrhea (grade 3–1) | 24 | 14.4 | 21 | 8.4 |
| Neutropenia (grade 3–1) | 28 | 16.8 | 33 | 13.0 |
Genetic analysis, 3’-RACE, and re-sequencing.
Single nucleotide polymorphisms (SNPs) were identified in UGT1A from the CEU population using International HapMap Project information (http://hapmap.ncbi.nlm.nih.gov). To maximize coverage, we included the ±5 kb flanking UGT1. htSNPs were found by Haploview v4.2 (Broad Institute, Cambridge, MA, USA). Markers that had previously been associated with irinotecan-related outcomes in the literature but not listed in the HapMap Project were also included. SNPs that could not be sequenced as the result of poor primer design or because they were located in duplicated regions were replaced with tagged SNPs in complete LD (r2 = 1.0). All selected htSNPs (n = 28) (Supplementary Table S1) were genotyped using an iPLEX matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry (Sequenom, San Diego, CA). Negative controls and a 5% random sample duplicate population were used to ensure the robustness of the assay and genotyping reproducibility.
A 3’-RACE study (Life Technologies, Burlington, ON, CA) was performed as described by the manufacturer using total RNA extracted from two liver samples that had been genotyped as homozygous for the rs11563250 variant (one variant was AA and the other was GG). PCR amplicons were subsequently sequenced on an ABI PRISM 3730XL DNA Analyzer (Applied Biosystems, Foster City, CA). Sequences were analyzed using the Staden package software version 2.0.0b9 (https://sourceforge.net/projects/staden) and compared with the GenBank reference sequence NG_002601. Re-sequencing was performed using germline DNA from the homozygous rs11563250G carrier by PCR amplification of the promoter regions and first exons of UGT1A1, UGT1A7, and UGT1A9, the common exons 2, 3, 4, 5a and 5b, the intron-exon boundaries, and the 3’-UTR regions of exon 5a and exon 5b.
Statistical analysis.
All genetic-association tests were assessed by logistic regression analysis using SPSS 21.0 software (SPSS Inc, Chicago, IL) with independent analyses to account for allelic, dominant, and recessive modes of transmission. ORs were adjusted for age and co-medication, as in our previous study 16, 21. Genetic variants with p < 0.10 were investigated in the replication cohort. The statistically significant threshold was fixed at p ≤ 0.05. Deviation from Hardy-Weinberg Equilibrium values was calculated using the PLINK v1.07 whole genome association analysis toolset.22 Haplotypes and pairwise LDs were inferred using Phase v2.1.1and Haploview v4.2, respectively.23, 24 To account for the false discovery rate associated with the combined cohort analysis, a Bonferroni correction was applied using R software (version 2.15.3). The statistical difference in bilirubin levels between carriers and non-carriers for a given genetic variation was assessed using the Student’s t-test. For studies using the human liver samples, analyses were performed by XLSTAT software version 2014.3.04 (AddinSoft Inc, Brooklyn, NY) using a one-way analysis of variance (ANOVA) with haplotypes, bilirubin-G, SN-38-G, and UGT1A1 expression as variables. A post-hoc Dunnett’s Test was applied with the haplotype 6A set as the reference.
RESULTS
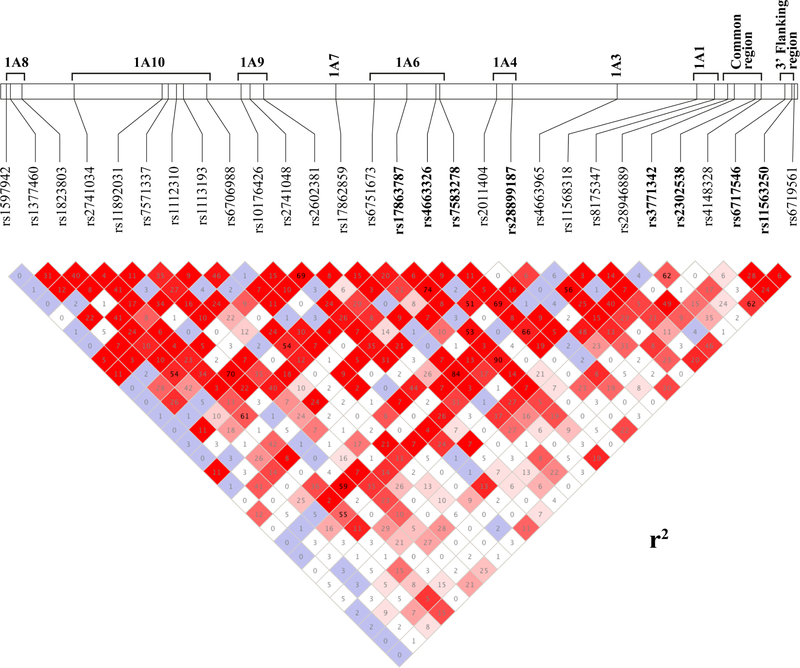
A total of 28 UGT1 htSNPs from the discovery cohort composed of 167 Canadians with mCRC (Table 1) were first assessed for their association with grade 3–4 severe neutropenia. This set of SNPs across the UGT1 gene had never been genotyped in this population. A linkage disequilibrium (LD) map representing these markers, along with the well-established UGT1A1 rs8175347 TATA box promoter variant, is depicted in Figure 1.
Figure 1.
Linkage disequilibrium map of 28 htSNPs genotyped in the discovery cohort. The map illustrates the linkage disequilibrium for the 28 UGT1 htSNPs first assessed in the discovery cohort of 167 Canadian patients and resembles that from the CEU population. The rs8175347 corresponding to the well-known UGT1A1 promoter variant (A(TA)6〉7TAA region) has been included in the LD map but was previously reported for this cohort of patients.16 Values inside each square are those for r2 and are reported as percentages. The colors depict the strength of the LD between each pair of htSNPs.
Eight novel markers, rs4663326, rs17863787, rs7583278, rs28899187, rs3771342, rs2302538, rs6717546 and rs11563250, were significantly associated with severe neutropenia in the Canadian derivation cohort. Four htSNPs located in the common region (minor alleles rs3771342A and rs2302538G; r2=0.62) or downstream of the 3’-UTR of UGT1 (rs6717546A and rs11563250G; r2=0.28) were significantly associated with a lesser risk of severe neutropenia (OR = 0.22–0.46; p < 0.05). In contrast, carriers of rs17863787G and rs7583278T (r2=0.75) in the first exon of UGT1A6 had an increased risk of severe neutropenia (OR = 2.04 and 1.86; p = 0.019 and 0.039, respectively) (Table 2). In addition, all Canadian carriers of the minor allele for rs4663326G in the first exon UGT1A6 and rs28899187A in the first exon UGT1A4 (r2=0.52) did not experienced severe neutropenia (p < 0.02).
Table 2.
UGT1 htSNPs significantly associated with severe neutropenia (grade 3–4) in the Canadian cohort.
| Neutropenia | Neutropenia | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Regions | htSNPs | Alleles | 0–2 | 3–4 | OR1 (95% CI) | P | Genotypes | 0–2 | 3–4 | OR1 (95% CI) | P |
| 1A8 | rs1597942 | C(Ref) | 267 | 54 | - | 0.5953 | CC (Ref) | 131 | 27 | - | 0.5923 |
| T | 5 | 0 | CT/TT | 5 | 0 | ||||||
| rs1377460 | G(Ref) | 214 | 44 | 0.84(0.40–1.78) | 0.649 | GG (Ref) | 85 | 17 | 1.02(0.43–2.41) | 0.973 | |
| A | 58 | 10 | GA/AA | 51 | 10 | ||||||
| rsl8238032 | C(Ref) | 145 | 34 | 0.67(0.37–1.23) | 0.19 | CC (Ref) | 42 | 13 | 0.48(0.21–1.12) | 0.088 | |
| T | 131 | 20 | CT/TT | 96 | 14 | ||||||
| 1A10 | rs2741034 | A(Ref) | 190 | 31 | 1.63(0.89–2.98) | 0.111 | AA (Ref) | 70 | 10 | 1.72(0.73–4.06) | 0.213 |
| G | 84 | 23 | AG/GG | 67 | 17 | ||||||
| rs11892031 | A(Ref) | 248 | 53 | 0.56(0.16–1.93) | 0.358 | AA(Ref) | 113 | 25 | 0.60(0.17–2.15) | 0.429 | |
| C | 26 | 3 | AC/CC | 24 | 3 | ||||||
| rs7571337 | T(Ref) | 143 | 34 | 0.68(0.37–1.26) | 0.220 | TT/TC (Ref) | 101 | 23 | 0.54(0.17–1.68) | 0.287 | |
| C | 129 | 20 | CC | 35 | 4 | ||||||
| rs1112310 | C(Ref) | 198 | 44 | 0.64(0.31–1.35) | 0.239 | CC (Ref) | 71 | 18 | 0.57(0.24–1.38) | 0.213 | |
| A | 74 | 10 | CA/AA | 65 | 9 | ||||||
| rs1113193 | C(Ref) | 214 | 44 | 0.98(0.48–1.99) | 0.957 | CC (Ref) | 85 | 18 | 0.89(0.38–2.10) | 0.792 | |
| T | 60 | 12 | CT/TT | 52 | 10 | ||||||
| 1A9 | rs6706988 | A(Ref) | 243 | 48 | 1.22(0.53–2.82) | 0.646 | AA (Ref) | 107 | 20 | 1.34(0.53–3.36) | 0.539 |
| G | 33 | 8 | AG/GG | 31 | 8 | ||||||
| rs10176426 | C(Ref) | 251 | 51 | 1.31 (0.46–3.67) | 0.614 | CC (Ref) | 115 | 24 | 1.03(0.32–3.32) | 0.965 | |
| T | 21 | 5 | CT/TT | 21 | 4 | ||||||
| rs2741048 | A(Ref) | 174 | 38 | 0.74(0.39–1.40) | 0.352 | AA (Ref) | 57 | 14 | 0.69(0.30–1.59) | 0.379 | |
| C | 100 | 16 | AC/CC | 80 | 13 | ||||||
| rs26023812 | C(Ref) | 149 | 34 | 0.78(0.43–1.41) | 0.417 | CC/CT (Ref) | 102 | 23 | 0.63(0.22–1.79) | 0.385 | |
| T | 125 | 22 | TT | 35 | 5 | ||||||
| 1A7 | rsl7862859 | G(Ref) | 257 | 54 | 0.52(0.12–2.31) | 0.390 | GG (Ref) | 120 | 26 | 0 54 (0.12–2 51) | 0.434 |
| A | 19 | 2 | GA/AA | 18 | 2 | ||||||
| 1A6 | rs6751673 | G(Ref) | 188 | 44 | 0.58(0.29–1.17) | 0.128 | GG (Ref) | 64 | 17 | 0.57(0.25–1.30) | 0.180 |
| A | 88 | 12 | GA/AA | 74 | 11 | ||||||
| rsl7863787 | T(Ref) | 188 | 28 | 2.04(1.12–3.72) | 0.01 | TT/GT (Ref) | 121 | 19 | 3.16(1.18–8.50) | 0.02 | |
| G | 86 | 26 | GG | 16 | 8 | ||||||
| rs4663326 | A(Ref) | 236 | 56 | - | <0.0013 | AA (Ref) | 100 | 28 | - | <0.0013 | |
| G | 38 | 0 | AG/GG | 37 | 0 | ||||||
| rs7583278 | C(Ref) | 170 | 25 | 1.86(1.03–3.37) | 0.039 | CC/CT (Ref) | 116 | 18 | 2.53(1.00–6.41) | 0.050 | |
| T | 106 | 29 | TT | 22 | 9 | ||||||
| 1A4 | rs2011404 | C(Ref) | 231 | 50 | 0.62(0.25–1.54) | 0.301 | CC (Ref) | 100 | 23 | 0.58(0.20–1.65) | 0.308 |
| T | 43 | 6 | CT/TT | 37 | 5 | ||||||
| rs28899187 | T(Ref) | 247 | 54 | - | 0.0193 | TT (Ref) | 113 | 27 | - | 0.0273 | |
| A | 23 | 0 | TA/AA | 22 | 0 | ||||||
| 1A3 | rs4663965 | T(Ref) | 145 | 27 | 1.20(0.67–2.15) | 0.537 | TT/TC (Ref) | 106 | 20 | 1.36(0.54–3.42) | 0.509 |
| C | 131 | 29 | CC | 32 | 8 | ||||||
| 1A1 | rsl1568318 | C(Ref) | 258 | 54 | 0.64(0.14–2.90) | 0.564 | CC (Ref) | 121 | 26 | 0.63(0.13–2.92) | 0.553 |
| A | 16 | 2 | CA/AA | 16 | 2 | ||||||
| Common region | rs28946889 | G(Ref) | 208 | 45 | 0.86(0.42–1.77) | 0.677 | GG (Ref) | 80 | 18 | 0.85 (0.36–2.01) | 0.716 |
| T | 60 | 11 | GT/TT | 54 | 10 | ||||||
| rs3771342 | C(Ref) | 237 | 52 | 0.24(0.06–1.03) | 0.054 | CC (Ref) | 99 | 25 | 0.21 (0.05–0.93) | 0.040 | |
| A | 39 | 2 | CA/AA | 39 | 2 | ||||||
| rs2302538 | A(Ref) | 236 | 52 | 0.22 (0.05–0.95) | 0.043 | AA (Ref) | 98 | 25 | 0.19(0.04–0.85) | 0.030 | |
| G | 40 | 2 | AG/GG | 40 | 2 | ||||||
| rs4148328 | C(Ref) | 174 | 35 | 0.95(0.51–1.77) | 0.877 | CC/CT (Ref) | 118 | 24 | 0.85(0.23–3.13) | 0.802 | |
| T | 98 | 19 | TT | 18 | 3 | ||||||
| 3′-Flanking region | rs6717546 | G(Ref) | 171 | 44 | 0.46(0.23–0.91) | 0.026 | GG (Ref) | 54 | 18 | 0.37 (0.16–0.86) | 0.021 |
| A | 103 | 12 | GA/AA | 83 | 10 | ||||||
| rsl1563250 | A(Ref) | 235 | 54 | 0.23(0.05–1.00) | 0.050 | AA (Ref) | 99 | 26 | 0.21 (0.05–0.95) | 0.043 | |
| G | 41 | 2 | AG/GG | 39 | 2 | ||||||
| rs6719561 | C(Ref) | 189 | 39 | 0.93(0.50–1.75) | 0.829 | CC (Ref) | 66 | 13 | 1.03(0.45–2.34) | 0.948 | |
| T | 87 | 17 | CT/TT | 72 | 15 | ||||||
Ref.: reference
OR, odds ratio; 95% CI, 95% confidence interval.
p-Values in bold type are <0.05.
Adjusted OR and p-values.
SNPs that were not found in the Hardy-Weinberg equilibrium (p < 0.05).
P-values obtained by univariate analysis.
Positive markers were subsequently genotyped in the independent Italian cohort (n = 250 mCRC cases; Table 1). As observed in the discovery cohort, the minor 3’-flanking variant rs11563250G was also associated with a decreased occurrence of severe neutropenia in the replication cohort (OR = 0.27; CI 95% 0.08–0.91, p = 0.036). For the combined cohort, the OR value (0.31; p = 0.001) remained significant upon adjustment for multiple testing (p = 0.041). All other associations between htSNPs and risk of severe neutropenia found in the Canadian discovery cohort were not replicated in the Italian validation cohort (p>0.05; Table 3).
Table 3.
Replication of positive UGT1 markers in the Italian cohort in relation to severe neutropenia (grades 3–4).
| Neutrpenia | Neutropenia | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Exons | htSNPs | A11eles | 0–2 | 3–4 | OR1 (95% CI) | P | Genotypes | 0–2 | 3–4 | OR1 (95% CI) | P |
| 1A6 | rs4663326 | A(Ref) | 383 | 55 | 2.62(1.26–5.64) | 0.011 | AA (Ref) | 178 | 22 | 3.21 (1.40–7.36) | 0.006 |
| G | 29 | 11 | AG/GG | 28 | 11 | ||||||
| rsl7863787 | T(Ref) | 276 | 42 | 1.31 (0.74–2.30) | 0.347 | TT | 97 | 14 | 1.30(0.61–2.75) | 0.501 | |
| G | 112 | 22 | GT/GG | 97 | 18 | ||||||
| rs7583278 | C(Ref) | 264 | 42 | 1.02(0.60–1 76) | 0.934 | CC (Ref) | 82 | 13 | 1.02(0.48–2.16) | 0.961 | |
| T | 148 | 24 | CT/TT | 124 | 20 | ||||||
| 1A4 | rs28899187 | T(Ref) | 353 | 57 | 0.95 (0.44–2.01) | 0.884 | TT (Ref) | 185 | 23 | 3.12(1.24–7.89) | 0.016 |
| A | 59 | 9 | TA/AA | 21 | 8 | ||||||
| Common region | rs3771342 | C(Ref) | 391 | 54 | 2.80(0.18–6 65) | 0.020 | CC (Ref) | 168 | 24 | 1.65(0.71–3.84) | 0.244 |
| A | 21 | 8 | CA/AA | 38 | 9 | ||||||
| rs2302538 | A(Ref) | 369 | 57 | 1.35(0.62–2 91) | 0.449 | AA (Ref) | 153 | 25 | 0.93 (0.39–2.18) | 0.859 | |
| G | 43 | 9 | AG/GG | 53 | 8 | ||||||
| 3′-Flanking region | rs6717546 | G(Ref) | 243 | 39 | 1.01 (0.59–1.71) | 0.984 | GG (Ref) | 71 | 11 | 1.06(0.49–2.31) | 0.889 |
| A | 169 | 27 | GA/AA | 135 | 22 | ||||||
| rsl1563250 | A(Ref) | 349 | 63 | 0.26 (0.08–0.87) | 0.028 | AA (Ref) | 150 | 30 | 0.27 (0.08–0.92) | 0.036 | |
| G | 63 | 3 | AG/GG | 56 | 3 | ||||||
Ref.: reference
OR, odds ratio; 95% CI, 95% confidence interval.
p-Values in bold type are <0.05.
Adjusted OR and p-values.
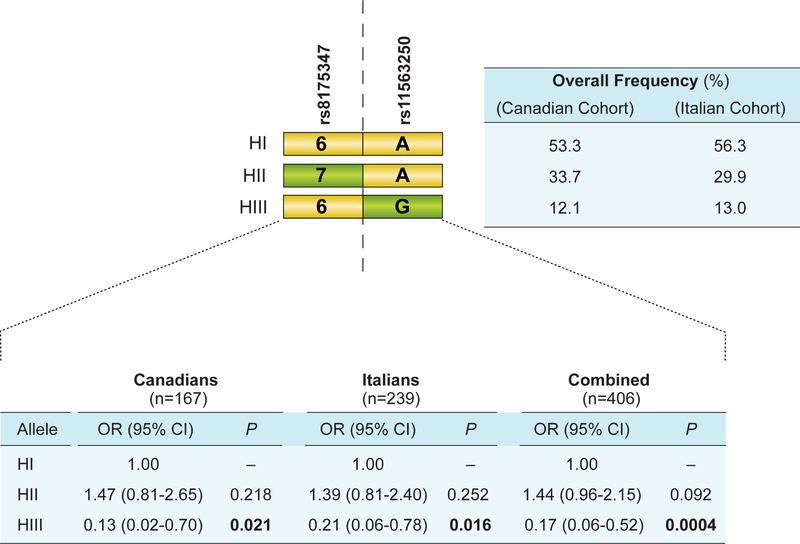
We then sought to evaluate the co-occurrence of the UGT1A1*28 risk allele (rs8175347; −54_−53insTA) and rs11563250 in a two-marker haplotype analysis. Compared with Canadian and Italian patients carrying the reference haplotype I denoted 6A in Figure 2 (UGT1A1*1, containing the reference six TA repeat in its promoter and the major rs11563250A allele), those carrying the UGT1A1*28 risk allele [a seven TA repeat in the promoter] and rs1156250A (haplotype II, 7A) tended to be at greater risk for severe neutropenia (OR = 1.44, p = 0.092; Figure 2). In contrast, haplotype III (6G) (UGT1A1*1 and the rs11563250G corresponding to alleles individually associated with a lower risk of neutropenia) was associated with a significantly decreased risk of severe neutropenia in both populations whether their risk was analyzed individually (Canadian cohort (14.1%), OR = 0.13, p = 0.021; Italian cohort (14.6%), OR = 0.21, p = 0.016) or in combination (OR = 0.17, p = 0.0004).
Figure 2.
Schematic of the two-marker haplotype comprising rs11563250 and the UGT1A1 promoter variant rs8175347. Yellow rectangles represent the reference nucleotide (with respect to the reference sequence, AF297093), whereas olive-green rectangles represent the variant allele. HI-HIII, Haplotype groups I–III; OR, odds ratio; 95% CI, 95% confidence intervals; H1 corresponds to the reference haplotype (OR= 1.0). Frequencies in studied populations are shown.
In a second series of haplotype analysis, we tested a three-marker haplotype incorporating the rs8330 marker previously shown to be associated with reduced risk of severe neutropenia in haplotype analyses,16 Results revealed a comparable association with OR = 0.13 (95% CI= 0.02 – 0.69; p = 0.026) associated with haplotype 6GC (UGT1A1*1, the rs11563250G and the rs8330C) for the Canadian cohort (data not shown). Therefore, the observed protective effect cannot be attributed to this 3’-UTR variation and because there is no LD between rs11563250 and rs8330 in Canadian and Italian populations (r2 < 0.10).
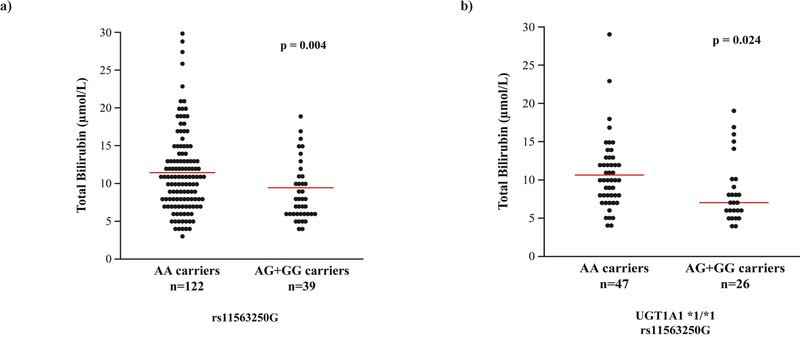
Consistent with the protective effect of rs11563250G, carriers of the G allele (AG + GG) exhibited a 17.5% decrease (p = 0.004) in total bilirubin compared with carriers of the AA genotype (Figure 3a), suggesting that the rs11563250G carriers had elevated UGT1A1 activity. When assessing only UGT1A1*1 carriers, total bilirubin was reduced in carriers of rs11563250G (p = 0.024; Figure 3b). In line with these data, the two-marker haplotype analysis also revealed a trend towards lower levels of unconjugated bilirubin for mCRC carriers with the 6G haplotype compared with those carrying 6A (11.95 vs. 10.06 μmol/L; p = 0.059) in the patients for whom data were available (data not shown). Using a bank of 48 human livers previously studied for UGT1A1 expression levels and rates of bilirubin and SN-38 glucuronidation,19, 20 we also assessed the relationship with HI, HII, and HIII. No significant differences were found for these three endpoints in carriers of HI compared with HIII carriers. As expected, compared with carriers of HI, those with the UGT1A1*28 allele (HII) expressed less UGT1A1 protein and had decreased formation of bilirubin-G and SN-38G (p = 0.0006, 0.003, and 0.0008, respectively; data not shown).
Figure 3.
Total bilirubin levels (μmol/L) in mCRC patients in relation to the presence of rs11563250.a) Data are presented for the discovery cohort patients and b) for the carriers of UGT1A1*1/*1 in that cohort. The p-value significance was determined by comparing the means of the logarithmic-transformed raw bilirubin values using the Student’s t-test. The red bars indicate the mean values for each group.
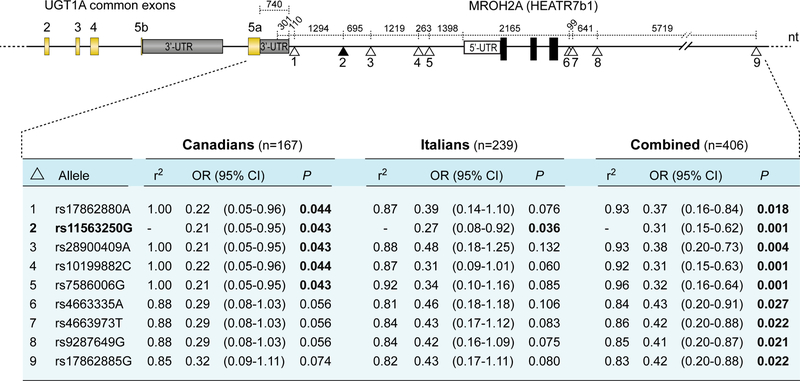
With the aim of identifying additional markers across the UGT1 locus linked to rs11563250G (not in linkage with the UGT1A1 TATA box variant rs8175347; r2=0.35), we genotyped eight additional SNPs found in the International HapMap project and 1000 genomes from the CEU population in strong linkage disequilibrium (r2 ≥ 0.80), that corresponds to a LD block of SNPs is distributed over a 13.6-kb area. Four of these intergenic variants rs17862880, rs28900409, rs10199882, and rs7586006, all located between UGT1 and HEATR7B1 (MROH2A) are almost in complete LD with rs11563250 (r2 ≥ 0.92–1.00) but only the p-value for rs11563250 is significant in the replication cohort (Figure 4). For instance, the rs17826880 variant, which is closest to the 3’-UTR of the UGT1 gene and tightly linked to rs11563250 (r2 = 1.0 for Canadians and r2 = 0.87 for Italians,), is also associated with a reduced risk of neutropenia for the Canadian cohort (OR = 0.22, p = 0.044) but did not reached significance for the replication cohort (OR = 0.39; p = 0.076).
Figure 4.
Schematic showing the positions of the 3’-flanking marker rs11563250 and eight LD markers genotyped in UGT1 from the Canadian and Italian mCRC patients. OR, odds ratio; 95% CI, 95% confidence intervals. R2 values between rs11563250 and each of the eight variants are provided.
Variants in high LD with rs11563250 are all located in the intergenic non-coding DNA region between UGT1 and HEATR7b1 (Figure 4). To determine whether these sequences are present in hepatic UGT1A transcripts, a series of 3’-RACE experiments were conducted using human liver mRNA from homozygous carriers of the rs11563250 A and G alleles. The results indicate that the region encompassing these variants, including the variant rs17826880 closest to the 3’-UTR of exon 5a (rs17826880 is 110 bp downstream this 3’-UTR), is not present in liver mRNAs encoded by the UGT1 locus (data not shown). Furthermore, sequencing of the first exons, promoter regions of the most active UGT1As towards SN-38 (1A1, 1A7, and 1A9),21 the common exons, intron-exon junctions, and the 3’-UTRs of exons 5a and 5b of germline DNA from a homozygous carrier of rs11563250G, did not identify new variants associated with this protective marker.
DISCUSSION
Pharmacogenetic tailoring of irinotecan-based chemotherapy has been the subject of several investigations over the last several years. Despite these efforts, the most reliable predictor of severe neutropenia remains associated with the UGT1A1*28/28 promoter genotype related to decreased SN-38 glucuronidation, greater exposure to SN-38, and an approximately two-fold greater risk of toxicity, which helps to identify patients who would benefit from a reduced irinotecan dose.2 We report herein a novel marker (rs11563250G) located in the 3’-flanking region of UGT1 associated with a better tolerance against severe irinotecan-induced neutropenia in two independent cohorts of mCRC patients, which should allow certain patients to benefit from an increased irinotecan dose and potentially improved benefit from irinotecan-based therapy.
Previous studies advised that patients with a favorable genetic profile might benefit from an increased irinotecan dose to maximize antitumor activity.7–9 Toffoli and collaborators demonstrated that the irinotecan recommended dose of 180 mg /m2 in FOLFIRI regimens is considerably less than the dose that can be tolerated by non-carriers of UGT1A1*28/*28.9 This dose-escalation study established that a dose of 370 or 310 mg/m2 of irinotecan can be safely administered every 2 weeks for mCRC patients, with the *1/*1 or *1/*28 genotype, respectively. In line with this study, Marcuello and colleagues showed that the recommended FOLFIRI dose of 180 mg/m2 irinotecan is ~two-fold lower than the dose that can be tolerated by patients with UGT1A1*1/*1 or *1/*28 (390 mg/m2 and 340 mg/m2, respectively), as part of FOLFIRI treatment.8 More recently, Innocenti and collaborators studied the effect of administering irinotecan once every 3 weeks and demonstrated that the predicted maximum tolerated dose is significantly superior for patients with UGT1A1*1/*1 or *1/*28, i.e., 470 mg/m2 and 390 mg/m2 respectively, compared with the recommended dose of 350 mg/m2.7 According to our findings, the UGT1A1*1 (rs8175347 TA6)/rs11563250G two-marker haplotype significantly improves prediction of a decreased risk for severe neutropenia compared with an assessment of UGT1A1*1 alone, suggesting that patients carrying these two markers are currently being under dosed, further reinforcing the clinical relevance of our findings.7
Consistent with the protective effect of the rs11563250G allele, we found a reduction in total plasma bilirubin, suggesting that this polymorphism, or SNPs in high LD, might be associated with an enhanced glucuronidation capacity. However, we could not assess the relationship between genotype and exposure to active SN-38 and inactive SN-38 glucuronide and, therefore, represents a limitation of the study. However, a genome-wide meta-analysis by Johnson and colleagues also reported an association between rs11563250 and total plasma bilirubin levels (p = 3.7 × 10–8) in three combined cohorts (n = 9,464).25 In line with a protective effect conferred by this allele, a second study found an interaction between a haplotype comprising the intergenic marker rs7586006 tightly linked to rs11563250 and frequent NSAID use that significantly decreased colorectal cancer risk.26
Our data indicate that the rs11563250 variant is located outside the UGT1 3’-UTR region, distant from the first exons of UGT1. In support, our 3’-RACE experiments did not capture 3’-mRNA sequences encompassing rs11563250. The rs11563250 variant is tightly linked to eight other variants (r2 ≥ 0.80). No additional variants located within the UGT1 gene were in significant LD with rs11563250 and sequencing of rs11563250G homozygous carriers did not allow for the identification of additional neutropenia-associated variants. It can be inferred that rs11563250 possesses a functional role in regulating UGT1 expression. However, no significant variation in UGT1A1 expression or bilirubin and SN-38 glucuronidation rates could be detected in our liver samples that contain the rs11563250G allele, possibly a consequence of the small sample size. The rs11563250 variant is within an intergenic zone that corresponds to an open chromatin region at chromosome 2 according to ENCODE project data. It is possible, therefore, that this variant, or closely related variants, display allelic differences in regulatory activity of the UGT1 locus, potentially affecting chromatin folding, epigenetic factors, or is part of cis-regulatory and complex long-range promoter-enhancer communication regulating transcription of UGT1. Such intergenic SNPs, co-localizing with transcription factors that bind to these sequences and act as positive regulators of gene expression, have been reported.27–29
The biological mechanism behind the association of rs11563250G with decreased risk of neutropenia also deserves additional in-depth functional investigations as it may potentially affect the overall conjugation capacity of UGT1A-targeted substrates. In support, a pharmacokinetic study has been undertaken in our laboratory 30 that has found that organ transplant patients receiving mycophenolate mofetil and carriers of rs11563250G present an overall greater mycophenolic acid glucuronide concentration after 2 h of drug administration compared with the levels found in non-carriers, suggesting that increased glucuronidation rates are associated with this allele as mycophenolic acid is a substrate of UGT1A enzymes (unpublished data).
In conclusion, we report the identification of an intergenic variant, rs11563250G located in the 3’-flanking region of UGT1, which is associated with better tolerance to irinotecan-induced neutropenia. rs11563250 genotyping may be clinically useful to identify patients who would better tolerate a greater irinotecan dose, especially those individuals with the UGT1A1*1/*1 genotype. We base these conclusions on our study of the two independent cohorts of the 406 FOLFIRI-treated mCRC patients and reiterate the need to validate the presence of a biomarker(s) in independent populations to obtain clinically meaningful findings with translational potential. The strengths of our study include the substantial plausibility of an association(s) given UGT1A enzymes involvement in irinotecan disposition, the extensive coverage of UGT1 htSNPs, replication in an independent population, and correction for multiple testing. Further investigations related to the function of this intergenic variant are required to decipher the molecular mechanism underlying its protective effect and potential role in affecting the metabolism of other substrates of UGT1A enzymes. We conclude that this relatively common variation (12%) influences irinotecan toxicity and should be considered to refine pharmacogenetic testing. The need to genotype the two markers rs11563250 and rs8175347 in the UGT1A1 promoter variant is our major conclusion as it may have clinical consequences in irinotecandosing management, especially in patients who are carriers of rs11563250G and might, therefore, tolerate, and likely benefit, from greater irinotecan dosing to maximize antitumor activity without increasing toxicity. Our study represents another step towards personalized and more precise FOLFIRI-related treatment of mCRC patients.
Supplementary Material
Acknowledgements:
The authors thank all participants in this study and the research nurses from Québec and Ottawa hospitals for their contributions. The Canadian Institutes of Health Research (C.G.; grant MOP-42392) and the Canada Research Chair Program (C.G.) supported this work. S.C. was a recipient of the studentship award ‘Fonds de l’Enseignement et de la Recherche’ from Faculty of Pharmacy at Laval University. I.L. is a recipient of a Canadian Institutes of Health Research Frederick Banting and Charles Best studentship award and a Graduate Scholarship for clinician-scientist from FRQ-S. M.H.C. was supported by the United States National Institutes of Health, National Institute of General Medical Sciences [Grant GM-102130] and the William R. Jones Endowment at Washington State University. E.L. is a recipient of a Canadian Institutes of Health Research clinician-scientist phase II award and Prostate Cancer Canada Rising Star Award (RS2013–55). C.G. holds a Canada Research Chair in Pharmacogenomics (Tier I).
Abbreviations:
- UGT
UDP-glucuronosyltransferase
- PCR
polymerase chain reaction
- UTR
untranscribed region
- SNP
single nucleotide polymorphism
- ECOG
Eastern Cooperative Oncology Group
- LD
linkage disequilibrium
- OR
Odds Ratio
- RACE
RNA ligase-mediated rapid amplification of cDNA end
- htSNP
haplotype-tagging SNP
Footnotes
Conflict of interest disclosure: The authors have no conflicts of interest to disclose.
References
- 1.Gagne JF, Montminy V, Belanger P, Journault K, Gaucher G, Guillemette C. Common human UGT1A polymorphisms and the altered metabolism of irinotecan active metabolite 7-ethyl-10-hydroxycamptothecin (SN-38). Mol Pharmacol 2002; 62: 608–617. [DOI] [PubMed] [Google Scholar]
- 2.Barbarino JM, Haidar CE, Klein TE, Altman RB. PharmGKB summary: very important pharmacogene information for UGT1A1. Pharmacogenet Genomics 2014; 24: 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Jong FA, Kitzen JJ, de Bruijn P, Verweij J, Loos WJ. Hepatic transport, metabolism and biliary excretion of irinotecan in a cancer patient with an external bile drain. Cancer Biol Ther 2006; 5: 1105–1110. [DOI] [PubMed] [Google Scholar]
- 4.De Mattia E, Toffoli G, Polesel J, D’Andrea M, Corona G, Zagonel V, et al. Pharmacogenetics of ABC and SLC transporters in metastatic colorectal cancer patients receiving first-line FOLFIRI treatment. Pharmacogenet Genomics 2013; 23: 549–557. [DOI] [PubMed] [Google Scholar]
- 5.Marsh S, Hoskins JM. Irinotecan pharmacogenomics. Pharmacogenomics 2010; 11: 1003–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith NF, Figg WD, Sparreboom A. Pharmacogenetics of irinotecan metabolism and transport: an update. Toxicol In Vitro 2006; 20: 163–175. [DOI] [PubMed] [Google Scholar]
- 7.Innocenti F, Schilsky RL, Ramirez J, Janisch L, Undevia S, House LK, et al. Dose-finding and pharmacokinetic study to optimize the dosing of irinotecan according to the UGT1A1 genotype of patients with cancer. J Clin Oncol 2014; 32: 2328–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcuello E, Paez D, Pare L, Salazar J, Sebio A, del Rio E, et al. A genotype-directed phase IIV dose-finding study of irinotecan in combination with fluorouracil/leucovorin as first-line treatment in advanced colorectal cancer. Br J Cancer 2011; 105: 53–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toffoli G, Cecchin E, Gasparini G, D’Andrea M, Azzarello G, Basso U, et al. Genotype-driven phase I study of irinotecan administered in combination with fluorouracil/leucovorin in patients with metastatic colorectal cancer. J Clin Oncol 2010; 28: 866–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guillemette C, Levesque E, Rouleau M. Pharmacogenomics of human uridine diphosphoglucuronosyltransferases and clinical implications. Clin Pharmacol Ther 2014; 96: 324–339. [DOI] [PubMed] [Google Scholar]
- 11.Cecchin E, Innocenti F, D’Andrea M, Corona G, De Mattia E, Biason P, et al. Predictive role of the UGT1A1, UGT1A7, and UGT1A9 genetic variants and their haplotypes on the outcome of metastatic colorectal cancer patients treated with fluorouracil, leucovorin, and irinotecan. J Clin Oncol 2009; 27: 2457–2465. [DOI] [PubMed] [Google Scholar]
- 12.Beutler E, Gelbart T, Demina A. Racial variability in the UDP-glucuronosyltransferase 1 (UGT1A1) promoter: a balanced polymorphism for regulation of bilirubin metabolism? Proc Natl Acad Sci U S A 1998; 95: 8170–8174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu ZY, Yu Q, Pei Q, Guo C. Dose-dependent association between UGT1A1*28 genotype and irinotecan-induced neutropenia: low doses also increase risk. Clin Cancer Res 2010; 16: 3832–3842. [DOI] [PubMed] [Google Scholar]
- 14.Hu ZY, Yu Q, Zhao YS. Dose-dependent association between UGT1A1*28 polymorphism and irinotecan-induced diarrhoea: a meta-analysis. Eur J Cancer 2010; 46: 1856–1865. [DOI] [PubMed] [Google Scholar]
- 15.Dias MM, McKinnon RA, Sorich MJ. Impact of the UGT1A1*28 allele on response to irinotecan: a systematic review and meta-analysis. Pharmacogenomics 2012; 13: 889–899. [DOI] [PubMed] [Google Scholar]
- 16.Levesque E, Belanger AS, Harvey M, Couture F, Jonker D, Innocenti F, et al. Refining the UGT1A haplotype associated with irinotecan-induced hematological toxicity in metastatic colorectal cancer patients treated with 5-fluorouracil/irinotecan-based regimens. J Pharmacol Exp Ther 2013; 345: 95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Innocenti F, Liu W, Chen P, Desai AA, Das S, Ratain MJ. Haplotypes of variants in the UDP-glucuronosyltransferase1A9 and 1A1 genes. Pharmacogenet Genomics 2005; 15: 295–301. [DOI] [PubMed] [Google Scholar]
- 18.Court MH, Freytsis M, Wang X, Peter I, Guillemette C, Hazarika S, et al. The UDPGlucuoronosyltransferase (UGT) 1A polymorphism c.2042C>G (rs8330) is associated with increased human liver acetaminophen glucuronidation, increased UGT1A Exon 5a/5b Splice Variant mRNA ratio, and decreased risk of unintentional acetaminophen-induced acute liver failure. J Pharmacol Exp Ther 2013; 345: 297–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Girard H, Thibaudeau J, Court MH, Fortier LC, Villeneuve L, Caron P, et al. UGT1A1 polymorphisms are important determinants of dietary carcinogen detoxification in the liver. Hepatology 2005; 42: 448–457. [DOI] [PubMed] [Google Scholar]
- 20.Girard H, Villeneuve L, Court MH, Fortier LC, Caron P, Hao Q, et al. The novel UGT1A9 intronic I399 polymorphism appears as a predictor of 7-ethyl-10-hydroxycamptothecin glucuronidation levels in the liver. Drug Metab Dispos 2006; 34: 1220–1228. [DOI] [PubMed] [Google Scholar]
- 21.Toffoli G, Cecchin E, Corona G, Russo A, Buonadonna A, D’Andrea M, et al. The role of UGT1A1*28 polymorphism in the pharmacodynamics and pharmacokinetics of irinotecan in patients with metastatic colorectal cancer. J Clin Oncol 2006; 24: 3061–3068. [DOI] [PubMed] [Google Scholar]
- 22.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet 2003; 73: 1162–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 2001; 68: 978–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson AD, Kavousi M, Smith AV, Chen MH, Dehghan A, Aspelund T, et al. Genome-wide association meta-analysis for total serum bilirubin levels. Hum Mol Genet 2009; 18: 2700–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Angstadt AY, Hartman TJ, Lesko SM, Muscat JE, Zhu J, Gallagher CJ, et al. The effect of UGT1A and UGT2B polymorphisms on colorectal cancer risk: haplotype associations and gene-environment interactions. Genes Chromosomes Cancer 2014; 53: 454–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fogarty MP, Panhuis TM, Vadlamudi S, Buchkovich ML, Mohlke KL. Allele-specific transcriptional activity at type 2 diabetes-associated single nucleotide polymorphisms in regions of pancreatic islet open chromatin at the JAZF1 locus. Diabetes 2013; 62: 1756–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sebio A, Paez D, Salazar J, Berenguer-Llergo A, Pare-Brunet L, Lasa A, et al. Intergenic polymorphisms in the amphiregulin gene region as biomarkers in metastatic colorectal cancer patients treated with anti-EGFR plus irinotecan. Pharmacogenomics J 2014; 14: 256–262. [DOI] [PubMed] [Google Scholar]
- 29.Stadhouders R, Aktuna S, Thongjuea S, Aghajanirefah A, Pourfarzad F, van Ijcken W, et al. HBS1L-MYB intergenic variants modulate fetal hemoglobin via long-range MYB enhancers. J Clin Invest 2014; 124: 1699–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levesque E, Delage R, Benoit-Biancamano MO, Caron P, Bernard O, Couture F, et al. The impact of UGT1A8, UGT1A9, and UGT2B7 genetic polymorphisms on the pharmacokinetic profile of mycophenolic acid after a single oral dose in healthy volunteers. Clin Pharmacol Ther 2007; 81: 392–400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.