Abstract
Advances in high-throughput DNA sequencing have revolutionized the discovery of variants in the human genome; however, interpreting the phenotypic effects of those variants is still a challenge. While several computational approaches to predict variant impact are available, their accuracy is limited and further improvement is needed. Here, we introduce ClinPred, an efficient tool for identifying disease-relevant nonsynonymous variants. Our predictor incorporates two machine learning algorithms that use existing pathogenicity scores and, notably, benefits from inclusion of normal population allele frequency from the gnomAD database as an input feature. Another major strength of our approach is the use of ClinVar—a rapidly growing database that allows selection of confidently annotated disease-causing variants—as a training set. Compared to other methods, ClinPred showed superior accuracy for predicting pathogenicity, achieving the highest area under the curve (AUC) score and increasing both the specificity and sensitivity in different test datasets. It also obtained the best performance according to various other metrics. Moreover, ClinPred performance remained robust with respect to disease type (cancer or rare disease) and mechanism (gain or loss of function). Importantly, we observed that adding allele frequency as a predictive feature—as opposed to setting fixed allele frequency cutoffs—boosts the performance of prediction. We provide pre-computed ClinPred scores for all possible human missense variants in the exome to facilitate its use by the community.
Keywords: pathogenicity prediction, whole-exome sequencing, rare disease, cancer, diagnostic, computational biology, machine learning, variant interpretation, predictive modeling
Introduction
Immense progress in high-throughput sequencing technologies provides new opportunities for identifying genetic determinants of disease. “Next generation” sequencing is now firmly established in diagnostic and research laboratories. Although recent advances in these technologies make them affordable, interpreting the effect of discovered variants remains a serious challenge. Since the human exome on average contains around 20,000 single-nucleotide variants, as compared with the reference,1 it is crucial to accurately predict deleteriousness of genomic changes, especially nonsynonymous single-nucleotide variants (nsSNVs). Distinguishing pathogenic amino acid changes from background polymorphisms is essential for efficient use of these technologies in personalized medicine. Experimental validation of the pathogenicity of large numbers of variants is not feasible because it is expensive and time consuming. Consequently, many algorithms have been developed to predict the potential impact of a variant on protein structure and/or function. These methods use different properties of the variant, such as relationship to local protein structure, evolutionary conservation, and/or physiochemical and biochemical properties of amino acids.
While the current programs provide positive predictive power, their results are often in disagreement with each other,2, 3 and there are currently no guidelines as to which predictions are the most reliable. It is believed that individual methods have complementary strengths, depending on their specific features and computational algorithms.3, 4, 5 Hence, recently, new “ensemble” predictors have combined individual predictors in order to achieve higher classification accuracy. Existing ensemble prediction tools apply machine learning algorithms and have been trained on known pathogenic and neutral nsSNVs mostly from HGMD or UniProt databases. While those databases provide important information about variants associated with diseases, they have known limitations. In a study by Dorschner et al., 239 unique variants classified as disease causing in HGMD were re-reviewed manually and only 16 unique AD variants and 1 AR variant pair were consistent with a pathogenic or likely pathogenic category.6 In another study, Bell et al. found that 27% of annotations for recessive disease-causing genes were annotated incorrectly or are common polymorphisms.7 These incorrect annotations resulted from varying and inconsistent levels of evidence applied by different laboratories to determine causation. To improve functional annotation of human variation, the more recently developed ClinVar8 database recommends that submitters use American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) guidelines9 for clinical interpretation of variants. Since its release in 2013, ClinVar has grown rapidly and has become the powerful resource representing current understanding of the relationship between genotypes and medically important phenotypes.10
In this article, we develop a machine learning approach trained on the most up-to-date and highest quality data, aimed to facilitate more accurate and reliable prediction of variants’ relevance to genetic disease. Our classifier, ClinPred, combines random forest and gradient boosting models. As predictive features, we combine commonly used and recently developed individual prediction tool scores, as well as allele frequencies (AFs) of the variant in different populations from the gnomAD database. The first innovative aspect of our model is that it trains on variants from the ClinVar database. The second innovative aspect of our approach is the use of AFs in different populations as features, rather than filtering variants based on arbitrary AF cutoffs, as is the case with most currently used approaches. We also assemble large independent test sets to evaluate ClinPred on cancer and rare disease data and to compare its performance with other existing methods. Finally, we provide pre-computed ClinPred scores for all possible human variants through our website to facilitate its use in general practice.
Material and Methods
Training Dataset
ClinVar database dated January 2016 was downloaded and nonsynonymous variants that were categorized as (1) benign or likely benign and (2) pathogenic or likely pathogenic were selected as our negative (benign) and positive (pathogenic) labels, respectively. All variants with conflicting interpretations in the clinical significance reports were excluded. We restricted training data to the high-confidence variants with review status of “criteria provided” from submitter or “reviewed by expert panel,” resulting in 32,910 variants. Subsequently, the variants added to ClinVar prior to January 2013 were eliminated to minimize overlap with training data of the component features of our predictors and the tools that are being compared. Since the training data of PolyPhen-2 and CADD overlap with our training set, to prevent type 1 circularity, any variant existing in their training data was excluded from our dataset. Only missense variants were retained, resulting in the 11,082 variants, with 7,059 labeled as benign and 4,023 labeled as pathogenic.
Test Datasets
We assembled nine test datasets. The first independent test dataset (ClinVarTest) was constructed from missense variants that were added to ClinVar database after January 2016 to minimize any overlap with our features’ as well as other available deleteriousness prediction tools’ training data. Any variant that was last evaluated before 2016 was excluded from this data. To further investigate generalizability of our model with respect to data collection method, we constructed our second and third datasets from different sources.
The second distinct database was compromised of pathogenic variants in the mutagenetix database (see Web Resources). This is a database of phenotypes and mutations produced through random germline mutagenesis induced with N-ethyl-N-nitrosourea (ENU) in mice. Phenotypic mutations that are responsible for a particular phenotype were obtained from the mutagenetix database. UCSC genome browser LiftOver tool was applied to convert genome coordinates and annotation from mouse to human GRCH37. Only variants that cause the same amino acid changes in human and mouse were kept. We obtained our neutral SNVs for the second test data from VariSNP database, which is the benchmark database for neutral SNVs.11 In order to prevent type II circularity that arises when all the variants in a gene are labeled either pathogenic or benign,12 we retained only genes that contained both benign (VariSNP) and pathogenic (mutagenetix) variants, to create the MouseVariSNP test data.
The third dataset was comprised of variants from DoCM,13 a database of curated mutations in cancer derived from literature. We retained only missense variants labeled as pathogenic and likely pathogenic to form the DoCM test data. Since this database contains only pathogenic variants, we used this test set to compare sensitivity of different methods.
Next, in order to determine whether the performance differs between gain-of-function and loss-of-function gene products, we constructed four distinct subset datasets from ClinVarTest. Oncogene test data consist of 242 benign and 112 pathogenic variants in genes defined as oncogene based on ONGene database.14 The tumor suppressor gene (TSG) dataset consists of 635 variants (475 benign and 160 pathogenic) based on genes defined as TSG in the TSGene database.15 Gain-of-function (GainFunction) and loss-of-function (LossFunction) datasets were collected based on the gene-disorder relationship as curated by the Orphanet database (see Web Resources). Description of datasets is shown in Table S1. Any variants that existed in our training data and our features’ training data were discarded from all test datasets.
Importantly, to test the application of ClinPred in clinically relevant data, we constructed a dataset comprised of 31 exome case subjects with rare disease obtained from the FORGE Canada, Care4Rare Canada Consortia,16 and collaborators. These samples were considered solved if the variant under consideration was in a known gene and the referring clinician provided feedback that this gene explained the affected individual’s phenotype. Also, in the case of novel disease genes, the variant was considered likely causative for the clinical phenotype in the presence of genetic validation (multiple families with mutations in the same gene and similar phenotype) and/or strong functional evidence. Since all of these data had been published after mid-2015, it has not been used to train any predictor.
Finally, to evaluate how ClinPred matches the results of large-scale functional assay data, we constructed the BRCA1 dataset from “A Database of Functional Classifications of BRCA1 Variants based on Saturation Genome Editing.”17 This test data consist of 437 missense loss-of-function (LoF) and 1,464 functional variants from genome editing in 13 BRCA1 exons that encode critical RING and BRCT domains.
Features
Having collected the high-confidence sets of SNVs, we annotated them with the latest version of ANNOVAR using dbNSFP v.3a to generate the required prediction scores from different component tools. Allele frequencies (AFs) of each variant in different populations were obtained from the gnomAD database: all exome, African/African American (AFR), Latino/Admixed American (AMR), Ashkenazi Jewish (ASJ), East Asian (EAS), Finnish (FIN), Non-Finnish European (NFE), South Asian (SAS), other (OTH). These AFs were assigned zero if the variant was not represented. The potential clinical relevance of each variant is predicted by incorporating AFs and 16 individual prediction scores from SIFT, PolyPhen-2 HDIV, PolyPhen-2 HVAR, LRT, MutationAssessor, PROVEAN, CADD, GERP, DANN, PhastCons, fitCons, PhyloP, and SiPhy.18, 19, 20, 21, 22 These features were selected to provide complementary information, and they either did not require training or their training data are publicly available to allow exclusion from our data and prevent type I circularity.
Model Definition
We applied random forest (cforest) and gradient boosted decision tree (xgboost) models and used the default missing value predictions that these algorithms provide for cases where individual scores for component predictors were not available.
We trained each model using either of balanced or equal weights:
-
•
Equal weights assigns a weight of 1 to each example in the training set. For highly unbalanced datasets (e.g., 90% pathogenic, 10% benign), these models may be trivial/sub-optimal.
-
•
Balanced weights assigns a weight to each example so that the total weight of each class is equal. For example, if there are 900 pathogenic and 100 benign variants, then we assign each pathogenic variant a weight of 1 (total weight = 1∗900 = 900), and each benign variant a weight of 9 (total weight = 9∗100 = 900).
As the results from the balanced weight model was not significantly different from the results from the equal weight model (data not shown), for simplicity, we show only balanced weights results for our final models. The output of each model is a score between 0 and 1, representing the probability of a pathogenic variant. The decision boundary to classify pathogenic or benign is 0.5 (lower for benign, higher for pathogenic). In addition, in order to maximize the sensitivity for detecting pathogenic variants, we defined the higher score of either of these two models (xgboost, cforest) as the ClinPred score.
Comparing the Performance of Individual Predictors
For each input feature and comparator models, we learned a univariate model based on each training set as follows. We learn the sign (−1 if smaller scores are more likely to be pathogenic; 1 otherwise) and threshold (score ∗sign ≤ threshold predicts benign; otherwise pathogenic) that minimizes the number of incorrect predictions using all non-missing features in the training data. This threshold was used to compute evaluation metrics.
To quantitatively compare our models with individual features and other models, we performed 5-fold cross validation on training, ClinVarTest, and MouseVariSNP data. Each dataset was randomly partitioned into five equal sized subsamples. In each round of cross-validation, our models were trained on 80% of training data and tested on 20% of the test data. In order to allow for fair comparison with available methods, the thresholds of other models were learned during the cross validation to maximize accuracy. Thus, the performance of our models was compared to other recent state-of-the-art tools such as VEST3, MetaSVM, MetaLR, M-CAP, fathmm-MKL, Eigen, GenoCanyon, and REVEL.3, 23, 24, 25, 26, 27, 28 Following the guidelines for reporting and using prediction tools, we computed seven evaluation metrics on each test based on the learned threshold described above. These metrics include sensitivity (true positive rate), specificity (1-false positive rate), accuracy, precision, F1 Score, and Matthew correlation coefficient (MCC),29, 30 as well as the area under the test receiver operating characteristic curve (AUC).
Results
Performance Comparison of Our Models and Individual Component Features
Our two models were superior to all their constituent features and discriminated well between pathogenic and benign variants in ClinVarTest with AUC equal to 0.97 ± 0.004 (mean ± standard deviation in 5-fold CV) for xgboost and cforest (Figure S1). They also showed superior performance in MouseVariSNP with respective AUCs of 0.96 ± 0.01 and 0.96 ± 0.02. Although most features and our models demonstrated little change in AUC score between MouseVariSNP and ClinVarTest, DANN and Siphy29-way attained 11% lower AUC in MouseVariSNP compared to ClinVarTest. Overall, the single features with the highest AUC were AF (gnomAD_exome_ALL), followed by PROVEAN, PolyPhen-HVAR, and CADD (Figure S1). Consistent with other research findings, conservation scores (GERP++, PhastCons, PhyloP, and SiPhy) almost all have lower AUC than functional scores (SIFT, MutationAssessor, PROVEAN, PolyPhen-2 HDIV, and HVAR).23 We further investigated the effect of excluding/including AFs as a feature in the models and found that the inclusion of AFs significantly increases AUC as well as increasing sensitivity and specificity (Figure S2). Finally, we found that combining our models by selecting the higher of the two probability scores improved the AUC to 0.98 ± 0.004 and 0.96 ± 0.01 in ClinVarTest and MouseVariSNP, respectively, while also achieving the best specificity at 95% sensitivity. Hence, we defined this combined model as ClinPred and used it in subsequent tests.
ClinPred in Comparison to Other Ensemble Tools
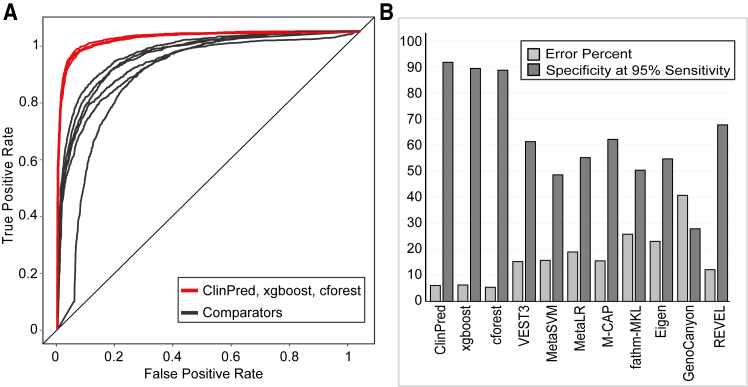
Using the ClinVarTest dataset (Figure 1), ClinPred outperformed other classifiers with the best AUC (0.98 ± 0.004), sensitivity (93.1% ± 3%), and specificity (94.2% ± 0.04%). It had the lowest error rate (6.04%)—the sum of false positives and false negatives over total number of labeled variants—in comparison to other tools (Table S2) where the error rate ranged from 13.2% (REVEL) to 50.3% (M-CAP).
Figure 1.
The Performance of ClinPred Was Compared to Seven Recently Developed Tools using ClinVarTest Data
(A) ClinPred showed increased sensitivity and specificity compared to other methods
(B) Our models had the best specificity at the cut off required to achieve 95% sensitivity. AUC, error percent, and specificity at 95% sensitivity were calculated for 5-fold cross validation and the mean score is shown.
When used on MouseVariSNP, ClinPred again outcompeted other available methods (Table S3). VEST3 was the closest competitor with an AUC of 0.88 ± 0.03. All methods were less accurate in MouseVariSNP than in ClinVarTest; the method with the largest AUC decrease was FATHMM (from 0.78 ± 0.1 to 0.58 ± 0.07) (Figure S1). Although ClinPred achieved the highest specificity in MouseVariSNP, it was followed closely by REVEL. This might be due to type I circularity, considering that VariSNP has overlap with the training set of other tools. On the other hand, as pathogenic variants in MouseVariSNP have the least overlap with the training data used by other tools, sensitivity score is the least biased comparator. ClinPred had the highest sensitivity among tools, detecting 92.79% ± 3.04% of pathogenic variants and VEST3 was the next, achieving 85.26% ± 3.34% sensitivity.
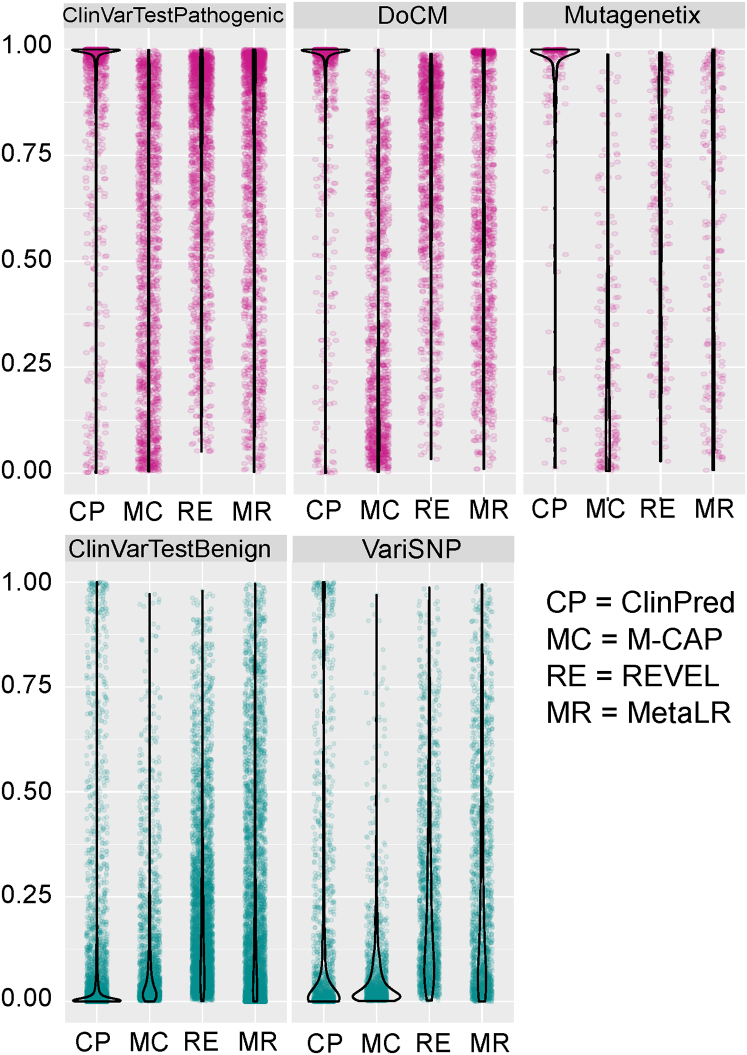
In order to visualize the distribution of scores of recently developed ensemble tools, we plotted raw scores for pathogenic and benign variants in different datasets. As demonstrated in Figure 2, ClinPred scores were highly concentrated near 1 for pathogenic and 0 in the benign variants across datasets. This analysis provides another way to illustrate the ability of ClinPred to differentiate well between benign and pathogenic variants in comparison to other methods.
Figure 2.
Comparison of Raw Scores of ClinPred, M-CAP, REVEL, and MetaLR
Violin plots represent the full distribution of scores for pathogenic (pink) and benign (green) variants in different test data.
Using Set Allele Frequency Cutoffs versus Allele Frequency as a Predictor Variable
In most laboratories tasked with analyzing exome data, hard allele frequency cutoffs are used to filter lists of detected variants, and prediction scores are assessed for the remaining variants as part of variant interpretation. As a result, allele frequency has generally not been used explicitly to predict clinical relevance of mutations in previous approaches. This is a sensible approach, since a reasonable estimate of the maximum AF can be based on the mode of inheritance and population frequency of the phenotype. Moreover, this approach is supported by the current ACMG guidelines, where AF is given a higher evidence value than computational predictions.9 However, in many cases, the mode of inheritance may not be evident—for example distinguishing recessive from de novo dominant cases—and population prevalence may not be obvious for non-specific phenotypes. Moreover, our analysis above suggested that population allele frequency is one of the most informative features in our model, and it is likely that it can acquire additional value when used in the machine learning setting alongside other predictor variables. Hence, we investigated whether AF remains an important predictor when the models are used in typical research approaches. We tested our models on datasets filtered according to various AF cutoffs: lower than 0.01, lower than 0.005, and lower than 0.001. In all conditions, ClinPred was superior to other tools, achieving highest AUC, sensitivity, and specificity (Figures S3 and S4). Thus, even when using datasets that are likely to be seen in the research or clinical setting—i.e., filtered using typically applied allele frequency cutoffs—there is still valuable information to be learned from population AF.
Comparing Categorical Scores across Different Tools
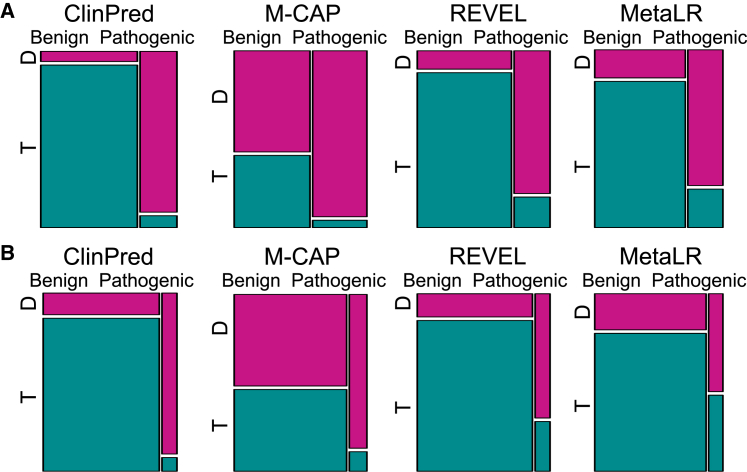
Many current tools provide categorical predictions—pathogenic/damaging versus benign/tolerant—according to the authors’ recommended pathogenicity thresholds. Hence, we compared the categorical predictions across various ensemble tools. As REVEL does not provide categorical scores, any variant with a raw score lower than 0.5 in REVEL was classified as benign while scores greater than or equal to 0.5 were classified as pathogenic. We used the same threshold for ClinPred. We restricted the comparison to variants where scores were available for any tool (excluding missing values). In ClinVarTest, M-CAP had the highest sensitivity, successfully classifying 95.8% pathogenic variants as damaging. However, this came at the cost of very low specificity, with 58% of benign variants misclassified as damaging. ClinPred achieved the second highest sensitivity (93.6%), while maintaining a low false positive rate of 6% (Figure 3; Table 1). When tested on MouseVariSNP, ClinPred had the best performance according to both sensitivity and specificity (Figure 3; Table S4).
Figure 3.
Comparison of ClinPred with Categorical Predictions Available from M-CAP, REVEL, and MetaLR
REVEL and ClinPred scores lower than 0.5 are defined as tolerant and greater than 0.5 as damaging. We show proportions of benign and pathogenic variants that were classified as tolerated (T, green) and damaging (D, pink). ClinPred had the best performance in finding as many pathogenic variants possible while minimizing the number of benign variants that are predicted as damaging both in ClinVarTest (A) and MouseVariSNP (B).
Table 1.
Overview of Performance of ClinPred in Comparison to Categorical Scores of Other Tools in ClinVarTest
| Sensitivity % | Specificity % | FPR | Accuracy | Precision | Error Percent | F1 Score | MCC | |
|---|---|---|---|---|---|---|---|---|
| ClinPred | 93.58 | 94.10 | 0.06 | 0.94 | 0.86 | 6.04 | 0.90 | 0.85 |
| xgboost | 90.75 | 94.65 | 0.05 | 0.94 | 0.87 | 6.42 | 0.89 | 0.84 |
| cforest | 89.06 | 96.59 | 0.03 | 0.95 | 0.91 | 5.49 | 0.90 | 0.86 |
| REVEL | 82.55 | 89.27 | 0.11 | 0.87 | 0.75 | 12.60 | 0.78 | 0.70 |
| M-CAP | 95.79 | 41.62 | 0.58 | 0.64 | 0.54 | 35.79 | 0.69 | 0.42 |
| MetaLR | 77.93 | 83.87 | 0.16 | 0.82 | 0.65 | 17.79 | 0.71 | 0.59 |
| Fathmm_mkl | 96.48 | 43.70 | 0.56 | 0.58 | 0.40 | 41.65 | 0.56 | 0.38 |
Abbreviations: FPR, false positive rate; MCC, Matthews correlation coefficient.
Subsequently, we investigated the performance of our models on the rare variants that are likely to be considered in current clinical testing. As M-CAP scores only rare variants (≤1% allele frequency), we restricted our analysis to the same cutoff. ClinPred maintained its performance as a classifier with lowest error rate in both ClinVarTest and MouseVariSNP restricted to AF ≤ 1% (Figure S5).
Investigating Generalizability of ClinPred to Different Disease Mechanisms
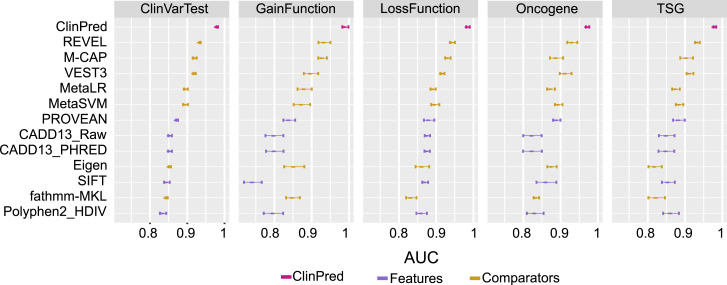
Further, we examined whether our algorithm’s performance differs between mutations resulting in gain or loss of function, in either rare disease or cancer. Similarly, to the first and second test datasets, we calculated AUC and sensitivity for GainFunction, LossFunction, TSG, and Oncogene test data. As demonstrated in Figure 4, ClinPred performance remained robust across all four test datasets. Moreover, ClinPred retains the highest sensitivity to predict pathogenic variants among other tools (Figure S6).
Figure 4.
ClinPred Performance Remained Robust across Distinct Datasets Based on Different Genetic Models and Pathogenic Mechanisms
We show mean AUC and error bars for 5-fold cross validation in all test datasets.
Since the DoCM database consists of only pathogenic variants, we could compile sensitivity scores based only on the categorical predictions provided by the tools (Table S5). ClinPred could successfully predict pathogenic variants in cancer, achieving a sensitivity score equal to 94.02%.
Application of ClinPred to Clinical Data
Finally we evaluated the performance of ClinPred in comparison to commonly used predictors (SIFT, PolyPhen-2, CADD) as well as the recent ensemble predictors MetaSVM, MetaLR, REVEL, M-CAP, and VAAST Variant Prioritizer (VVP)31 in 31 exomes from the FORGE Canada and Care4Rare Canada projects. To compare categorical scores, variants were categorized as pathogenic if they were predicted as pathogenic/probably pathogenic (PolyPhen2) or damaging (SIFT, MetaSVM, MetaLR, M-CAP). Since CADD authors did not provide a categorical score, we defined pathogenic variants according to different CADD_PHRED score cutoffs (more than 10, 15, and 20). We considered any score higher than 0.5 as pathogenic in ClinPred and REVEL, and any score higher than 50 as pathogenic in VVP.
After typical quality filtering,16 an individual’s exome on average harbored 433 non-synonymous variants (AF < 0.05 in ExAC). There were 25 different nonsynonymous variants with strong supporting evidence for being causative in these samples. All studies have been published in peer-reviewed journals or are in press. In this analysis, we defined the sensitivity of a predictor as the number of known causative variants that were predicted as pathogenic, divided by 25 (the total number of known causative variants). Although each exome likely contains other pathogenic variants, in addition to those that cause the disease, we aimed to identify the prediction tools that selected the highest number of the 25 known disease variants, while discarding the highest proportion of the remaining variants.
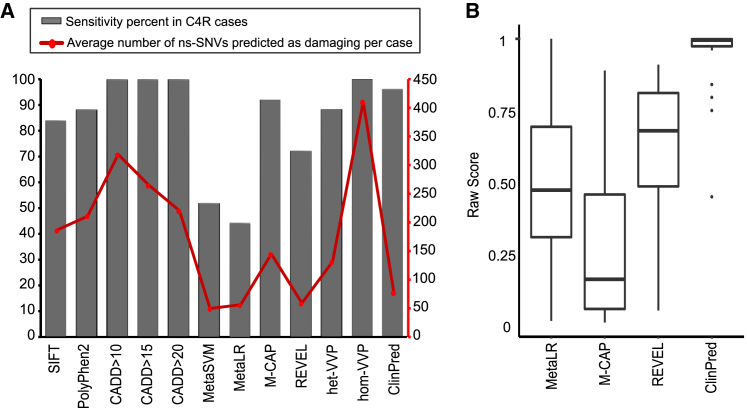
As demonstrated in Figure 5, sensitivity scores among tools ranged from 44% to 100%, with the highest achieved by CADD and VVP for homozygous genotype (hom-VVP). Although CADD and hom-VVP identified all the causative variants as pathogenic, this came with the cost of low specificity: on average 94%, 75%, 60%, and 50% of non-synonymous variants per exome were predicted as pathogenic using hom-VVP and different CADD_PHRED cutoffs (more than 10, 15, and 20, respectively). ClinPred predicted 24/25 = 96% of the causative variants as pathogenic. The only variant missed by ClinPred had a marginal score of 0.449 and was found in late-onset case subject with compound heterozygote variants in the same gene—one frameshift and the other nonsynonymous.32 This nonsynonymous variant is also predicted as benign/neutral by all other predictors except CADD and VVP.
Figure 5.
Illustration of Performance of ClinPred as Compared to Other Tools on Real-Life Clinical Samples from Solved FORGE Canada and Care4Rare Canada Projects
(A) ClinPred reduced the number of nonsynonymous variants predicted as pathogenic and retained high sensitivity.
(B) Raw Scores from MetaLR, M-CAP, REVEL, and ClinPred for any causative variant in these 31 solved FORGE Canada and Care4Rare Canada project cases were shown.
Assessing Concordance between Functional Assay and Computational Prediction Scores
Since ACMG guidelines suggest the result of well-established in vitro or in vivo functional study as evidence for variant interpretation, we examined how our algorithm and other computational prediction methods’ performance matches functional assay data. A recent study on large-scale functional classification of BRCA1 variants provides an excellent opportunity for such comparison.17 While most of the computational methods had the ability to predict LoF variants in the BRCA1 dataset as pathogenic/deleterious (sensitivity ranged from 92.6% to 100%), their performance was poor in predicting functional variants as benign (specificity ranged from 0.1% to 46% with the lowest in M-CAP and the best in MetaSVM). ClinPred predicted 97.5% of LoF variants as pathogenic and 32% of functional variants as benign (Figure S7). The relatively low specificity of computational predictions in the BRCA1 dataset may be at least partly due to the limited sensitivity of the in vitro assays used in that study. Functional scores in the BRCA1 dataset were measured based on cellular fitness in a haploid human cell line, which may not fully reflect the function in the complete organism. Such discrepancy between in vitro and in vivo BRCA1 mutant homologous recombination activity has previously been demonstrated.33, 34 Conversely, computational predictions may be overestimating pathogenicity. At this point, we conclude that the relative strengths of computational predictions and in vitro functional assays warrant further investigation.
Discussion
Although there are several prediction methods currently available, tools with higher ability to distinguish between pathogenic and neutral variants will be beneficial for future precision medicine. In this study, we use an improved supervised machine learning approach to create ClinPred, a method to efficiently distinguish clinically pathogenic from neutral variants. The first improvement concerns the choice of the most accurate training dataset: we train our predictor on clinically significant variants based on the joint consensus recommendation for the interpretation of sequence variants by the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP). Second, we apply two different machine learning algorithms and include a wide range of recent supervised and unsupervised methods as predictive features. Finally, we identify allele frequency as one of our key predictive features, which improves performance in both the presence and the absence of a predetermined frequency threshold for inclusion of variants in the analysis.
Compared to other methods, ClinPred showed highest sensitivity, improved specificity, and obtained the best performance according to various performance metrics. Our results emphasize that allele frequency is an important factor to be added as a feature in predicting pathogenicity of amino acid change and leads to significant performance improvement. Although traditionally AF is employed to discard benign variants, it is unclear what threshold should be selected at which a variant is considered benign. Many investigators use a cutoff of 5%, which is the upper bound for carrier frequency of most common Mendelian diseases, such as cystic fibrosis. However, in view of rarity of many other phenotypes, researchers often select lower AF cutoffs.35 In designing ClinPred, we did not set any restrictions regarding AFs of either pathogenic or benign variants, and we allowed the algorithms to learn the best use of this feature as a predictor. This contrasts with other methods, where the benign variants are often selected based on certain AFs.3, 26 As examples, M-CAP considers any variant with a mean allele frequency ≤ 1% in ExAC and 1000 Genomes as benign while REVEL selects variants with AFs between 0.1% and 1% across the seven study populations for their benign label. In our approach, we utilized AFs from the largest database available, gnomAD, as one of the predictor variables and allowed the model to learn the optimal parameters, without using a specific threshold. Some other existing methods have also incorporated AFs in their approach. For example, MetaLR, MetaSVM, and Eigen applied AFs from 1000 Genomes database in their model. M-CAP indirectly benefits from AF by using MetaLR and MetaSVM as their feature. The relatively lower level of success in using AFs in those methods may be due to high missing values for AFs in the smaller, less representative databases. As far as we know, Gavin and VVP are the only methods that use AFs from a large database; however, our method is different from them. Gavin applied AFs equal to 0.00346 in ExAC and CADD > 15 as the fixed thresholds for defining variants as pathogenic.36 VVP incorporated population variant frequencies from the WGS portion of gnomAD (15,496 whole genomes) while we not only incorporate gnomAD all-exome AFs (123,136 exome sequences), but also AFs in 8 different populations available in gnomAD: African/African American, Latino/Admixed American, Ashkenazi Jewish, East Asian, Finnish, Non-Finnish European, South Asian, and other ethnicities. We attribute a large part of the increase in performance of ClinPred to allowing our classifier to learn and optimize the use of AFs in making the distinction between pathogenic and benign variants.
We also found that our predictor maintains consistently superior performance across different genetic models and pathogenic mechanisms—for example dominant versus recessive or oncogene versus tumor suppressor classifications. However, it should be noted that this outcome is highly dependent on the types and proportions of variants that are currently present in the disease databases. The currently cataloged pathogenic variants are predominantly highly penetrant monogenic or oncogenic mutations, generally with severe disease phenotypes that are strongly selected against in human populations. As we begin to identify variants responsible for less severe, polygenic, and complex traits, clinically relevant predictors will likely benefit from training on relevant subsets of disease databases. In particular, the use of allele frequency as a feature—even though we found it to be universally beneficial across the currently cataloged disease variants—should optimally be trained on sets of variants most relevant to different inheritance models and severity of diseases.
In addition, our results demonstrate the value of combining different methods that likely provide complementary information as a result of their divergent algorithms and training datasets. The importance of diversifying training datasets is illustrated by comparing PolyPhen-2 HVAR and HDIV scores, where their difference in performance is because they applied different training dataset in spite of the same algorithm. On the other hand, illustrating difference in methodologies, DANN and CADD shared the same training data with different algorithm resulting in disjoint performance.
In our design, we took great care to avoid type I circularity,12 a problem that occurs in supervised machine learning when the training data directly or indirectly overlap with test data. Although in our model we eliminate any such overlap to prevent over-fitting, comparison against other tools is not completely free of bias. First, it is practically impossible to remove from our dataset the neutral variants that were used to train other ensemble tools, such as large number of variants present in the ExAC database. Second, M-CAP, VEST3, and REVEL were trained on private pathogenic datasets which we were unable to access, and which may influence comparison of performance in their favor.
In our illustration of real-life utility with respect to clinical data, the FORGE Canada and Care4Rare Canada case subjects were selected from studies published after mid-2015 to avoid overlap with any of the training data. Applying our predictor as one of the selection criteria for pathogenicity would reduce the list of an average 443 non-synonymous in individual’s exome to an average of 70 variants to be further manually followed up. In most cases, the entire list of selected variants—prioritized by prediction scores—would not have to be examined, as 83% of causative variants ranked within the top 25 candidates. Out of the 25 distinct disease-causing variants, ClinPred misclassified only 1 causative variant as benign. While this illustrates the pitfall of using classifiers such as ClinPred for purely automated filtering, it also suggests to a possible alternative multi-stage protocol, where case subjects can first be scanned using the currently optimized strict score cutoff and, if no definite disease cause is identified, the criteria can be relaxed and re-applied. The only approaches that succeeded in identifying all of the 25 pathogenic variants in this dataset were CADD and hom-VVP, but this came at the cost of specificity, and as a result, their application could narrow down the candidate list to an average of 216 and 409 variants per case for CADD and hom-VVP, respectively. While increasing the threshold of any predictor score results in higher sensitivity, it will jeopardize specificity. In the clinical domain, a test achieving 95% sensitivity with high specificity is generally favorable. Across our test data, ClinPred was able to achieve 95% sensitivity with the best specificity among other tools.
In summary, we have developed an ensemble classifier for predicting disease relevance of missense SNVs, using a combination of two different machine learning algorithms and incorporating several popular pathogenicity predictors, along with population allele frequencies, as component features. Our classifier is specifically designed to predict pathogenicity of variants that are causative for Mendelian disease. We systematically compared both categorical prediction and raw scores of different commonly used methods, under different AF cutoffs that may typically be used by researchers and clinicians to narrow down lists of variants. ClinPred outperformed all existing ensemble classifiers in distinguishing between disease-relevant pathogenic from neutral variants. Our model generalizes well when applied to variants from various sources not included in its training dataset. It also has high performance both in rare disease and in cancer. We provide pre-computed prediction scores for all possible variants in the human exome with a guide (Table S6) to facilitate interpretation of high-throughput sequencing results. In future developments, the prediction power of our model may be further enhanced by incorporating more components such as specific population genotype frequency, penetrance, disease prevalence, and human phenotype ontology (HPO) terms.37, 38 Furthermore, progress in whole-genome sequencing data will bring the need to accurately predict the effect of non-coding variants. The framework outlined here can help design future predictors for non-coding variants when appropriately large and reliable sources of pathogenic and benign variants become available.
Declaration of Interests
The authors declare no competing interests.
Acknowledgments
The authors first thank the FORGE Canada, Care4Rare Canada Consortium funded by Genome Canada. We would also like to acknowledge Compute Canada and Calcul Québec for providing high performance computing infrastructures.
Published: September 13, 2018
Footnotes
Supplemental Data include seven figures and six tables and can be found with this article online at https://doi.org/10.1016/j.ajhg.2018.08.005.
Contributor Information
Najmeh Alirezaie, Email: najmeh.alirezaie@mail.mcgill.ca.
Jacek Majewski, Email: jacek.majewski@mcgill.ca.
Web Resources
ANNOVAR, http://annovar.openbioinformatics.org/en/latest/
ClinPred, https://sites.google.com/site/clinpred/
ClinVar, https://www.ncbi.nlm.nih.gov/clinvar/
dbNSFP v.2.0, https://sites.google.com/site/jpopgen/dbNSFP
dbSNP, https://www.ncbi.nlm.nih.gov/projects/SNP/
ExAC Browser, http://exac.broadinstitute.org/
GERP++, http://mendel.stanford.edu/SidowLab/downloads/gerp/
Mutagenetix, http://mutagenetix.utsouthwestern.edu
OrphaNet, https://www.orpha.net/
UCSC Genome Browser, https://genome.ucsc.edu
Supplemental Data
References
- 1.Shihab H.A., Gough J., Mort M., Cooper D.N., Day I.N., Gaunt T.R. Ranking non-synonymous single nucleotide polymorphisms based on disease concepts. Hum. Genomics. 2014;8:11. doi: 10.1186/1479-7364-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Q., Liu X., Gibbs R.A., Boerwinkle E., Polychronakos C., Qu H.Q. Gene-specific function prediction for non-synonymous mutations in monogenic diabetes genes. PLoS ONE. 2014;9:e104452. doi: 10.1371/journal.pone.0104452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ioannidis N.M., Rothstein J.H., Pejaver V., Middha S., McDonnell S.K., Baheti S., Musolf A., Li Q., Holzinger E., Karyadi D. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am. J. Hum. Genet. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.González-Pérez A., López-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am. J. Hum. Genet. 2011;88:440–449. doi: 10.1016/j.ajhg.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu X., Jian X., Boerwinkle E. dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum. Mutat. 2011;32:894–899. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dorschner M.O., Amendola L.M., Turner E.H., Robertson P.D., Shirts B.H., Gallego C.J., Bennett R.L., Jones K.L., Tokita M.J., Bennett J.T., National Heart, Lung, and Blood Institute Grand Opportunity Exome Sequencing Project Actionable, pathogenic incidental findings in 1,000 participants’ exomes. Am. J. Hum. Genet. 2013;93:631–640. doi: 10.1016/j.ajhg.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell C.J., Dinwiddie D.L., Miller N.A., Hateley S.L., Ganusova E.E., Mudge J., Langley R.J., Zhang L., Lee C.C., Schilkey F.D. Carrier testing for severe childhood recessive diseases by next-generation sequencing. Sci. Transl. Med. 2011;3:65ra4. doi: 10.1126/scitranslmed.3001756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46(D1):D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J., Grody W.W., Hegde M., Lyon E., Spector E., ACMG Laboratory Quality Assurance Committee Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrison S.M., Riggs E.R., Maglott D.R., Lee J.M., Azzariti D.R., Niehaus A., Ramos E.M., Martin C.L., Landrum M.J., Rehm H.L. Using ClinVar as a resource to support variant interpretation. Curr. Protoc. Hum. Genet. 2016;89:1–23. doi: 10.1002/0471142905.hg0816s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schaafsma G.C., Vihinen M. VariSNP, a benchmark database for variations from dbSNP. Hum. Mutat. 2015;36:161–166. doi: 10.1002/humu.22727. [DOI] [PubMed] [Google Scholar]
- 12.Grimm D.G., Azencott C.A., Aicheler F., Gieraths U., MacArthur D.G., Samocha K.E., Cooper D.N., Stenson P.D., Daly M.J., Smoller J.W. The evaluation of tools used to predict the impact of missense variants is hindered by two types of circularity. Hum. Mutat. 2015;36:513–523. doi: 10.1002/humu.22768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ainscough B.J., Griffith M., Coffman A.C., Wagner A.H., Kunisaki J., Choudhary M.N., McMichael J.F., Fulton R.S., Wilson R.K., Griffith O.L., Mardis E.R. DoCM: a database of curated mutations in cancer. Nat. Methods. 2016;13:806–807. doi: 10.1038/nmeth.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y., Sun J., Zhao M. ONGene: A literature-based database for human oncogenes. J. Genet. Genomics. 2017;44:119–121. doi: 10.1016/j.jgg.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 15.Zhao M., Kim P., Mitra R., Zhao J., Zhao Z. TSGene 2.0: an updated literature-based knowledgebase for tumor suppressor genes. Nucleic Acids Res. 2016;44(D1):D1023–D1031. doi: 10.1093/nar/gkv1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beaulieu C.L., Majewski J., Schwartzentruber J., Samuels M.E., Fernandez B.A., Bernier F.P., Brudno M., Knoppers B., Marcadier J., Dyment D., FORGE Canada Consortium FORGE Canada Consortium: outcomes of a 2-year national rare-disease gene-discovery project. Am. J. Hum. Genet. 2014;94:809–817. doi: 10.1016/j.ajhg.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Findlay G.M., Daza R.M., Martin B., Zhang M.D., Leith A.P., Gasperini M., Janizek J.D., Huang X., Starita L.M., Shendure J. Accurate functional classification of thousands of BRCA1 variants with saturation genome editing. bioRxiv. 2018 doi: 10.1038/s41586-018-0461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi Y., Sims G.E., Murphy S., Miller J.R., Chan A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS ONE. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gulko B., Hubisz M.J., Gronau I., Siepel A. A method for calculating probabilities of fitness consequences for point mutations across the human genome. Nat. Genet. 2015;47:276–283. doi: 10.1038/ng.3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quang D., Chen Y., Xie X. DANN: a deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics. 2015;31:761–763. doi: 10.1093/bioinformatics/btu703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reva B., Antipin Y., Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong C., Wei P., Jian X., Gibbs R., Boerwinkle E., Wang K., Liu X. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum. Mol. Genet. 2015;24:2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carter H., Douville C., Stenson P.D., Cooper D.N., Karchin R. Identifying Mendelian disease genes with the variant effect scoring tool. BMC Genomics. 2013;14(Suppl 3):S3. doi: 10.1186/1471-2164-14-S3-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shihab H.A., Rogers M.F., Gough J., Mort M., Cooper D.N., Day I.N., Gaunt T.R., Campbell C. An integrative approach to predicting the functional effects of non-coding and coding sequence variation. Bioinformatics. 2015;31:1536–1543. doi: 10.1093/bioinformatics/btv009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jagadeesh K.A., Wenger A.M., Berger M.J., Guturu H., Stenson P.D., Cooper D.N., Bernstein J.A., Bejerano G. M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat. Genet. 2016;48:1581–1586. doi: 10.1038/ng.3703. [DOI] [PubMed] [Google Scholar]
- 27.Ionita-Laza I., McCallum K., Xu B., Buxbaum J.D. A spectral approach integrating functional genomic annotations for coding and noncoding variants. Nat. Genet. 2016;48:214–220. doi: 10.1038/ng.3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu Q., Hu Y., Sun J., Cheng Y., Cheung K.H., Zhao H. A statistical framework to predict functional non-coding regions in the human genome through integrated analysis of annotation data. Sci. Rep. 2015;5:10576. doi: 10.1038/srep10576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vihinen M. Guidelines for reporting and using prediction tools for genetic variation analysis. Hum. Mutat. 2013;34:275–282. doi: 10.1002/humu.22253. [DOI] [PubMed] [Google Scholar]
- 30.Niroula A., Vihinen M. Variation interpretation predictors: principles, types, performance, and choice. Hum. Mutat. 2016;37:579–597. doi: 10.1002/humu.22987. [DOI] [PubMed] [Google Scholar]
- 31.Flygare S., Hernandez E.J., Phan L., Moore B., Li M., Fejes A., Hu H., Eilbeck K., Huff C., Jorde L. The VAAST Variant Prioritizer (VVP): ultrafast, easy to use whole genome variant prioritization tool. BMC Bioinformatics. 2018;19:57. doi: 10.1186/s12859-018-2056-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoch N.C., Hanzlikova H., Rulten S.L., Tétreault M., Komulainen E., Ju L., Hornyak P., Zeng Z., Gittens W., Rey S.A., Care4Rare Canada Consortium XRCC1 mutation is associated with PARP1 hyperactivation and cerebellar ataxia. Nature. 2017;541:87–91. doi: 10.1038/nature20790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drost R., Bouwman P., Rottenberg S., Boon U., Schut E., Klarenbeek S., Klijn C., van der Heijden I., van der Gulden H., Wientjens E. BRCA1 RING function is essential for tumor suppression but dispensable for therapy resistance. Cancer Cell. 2011;20:797–809. doi: 10.1016/j.ccr.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 34.Millot G.A., Carvalho M.A., Caputo S.M., Vreeswijk M.P., Brown M.A., Webb M., Rouleau E., Neuhausen S.L., Hansen Tv., Galli A., ENIGMA Consortium Functional Assay Working Group A guide for functional analysis of BRCA1 variants of uncertain significance. Hum. Mutat. 2012;33:1526–1537. doi: 10.1002/humu.22150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobayashi Y., Yang S., Nykamp K., Garcia J., Lincoln S.E., Topper S.E. Pathogenic variant burden in the ExAC database: an empirical approach to evaluating population data for clinical variant interpretation. Genome Med. 2017;9:13. doi: 10.1186/s13073-017-0403-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Velde K.J., de Boer E.N., van Diemen C.C., Sikkema-Raddatz B., Abbott K.M., Knopperts A., Franke L., Sijmons R.H., de Koning T.J., Wijmenga C. GAVIN: Gene-Aware Variant INterpretation for medical sequencing. Genome Biol. 2017;18:6. doi: 10.1186/s13059-016-1141-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kernohan K.D., Hartley T., Alirezaie N., Care4Rare Canada Consortium. Robinson P.N., Dyment D.A., Boycott K.M. Evaluation of exome filtering techniques for the analysis of clinically relevant genes. Hum. Mutat. 2018;39:197–201. doi: 10.1002/humu.23374. [DOI] [PubMed] [Google Scholar]
- 38.Eilbeck K., Quinlan A., Yandell M. Settling the score: variant prioritization and Mendelian disease. Nat. Rev. Genet. 2017;18:599–612. doi: 10.1038/nrg.2017.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.