Abstract
Noncoding RNAs are emerging as potent and multifunctional regulators in all biological processes. In parallel, a rapidly growing number of studies has unravelled associations between aberrant noncoding RNA expression and human diseases. These associations have been extensively reviewed, often with the focus on a particular microRNA (miRNA) (family) or a selected disease/pathology. In this Mini‐Review, we highlight a selection of studies in order to demonstrate the wide‐scale involvement of miRNAs and long noncoding RNAs in the pathophysiology of three types of diseases: cancer, cardiovascular and neurological disorders. This research is opening new avenues to novel therapeutic approaches.
Keywords: long noncoding RNA, microRNA, noncoding RNA
Abbreviations
AD, Alzheimer's disease
ALS, amyotrophic lateral sclerosis
APP, amyloid precursor protein
BDNF, brain‐derived neurotrophic factor
CARL, cardiac apoptosis‐related lncRNA
CLL, chronic lymphocytic leukaemia
CRP, C‐reactive protein
EC, endothelial cell
FTLD, frontotemporal lobar degeneration
HCC, hepatocellular carcinoma
HD, Huntington's disease
lncRNAs, long noncoding RNAs
miRNA, microRNA
MPTP, 1‐methyl‐4‐phenyl‐1,2,3,6‐tetrahydropyridine
NATs, natural antisense transcripts
ncRNA, noncoding RNA
NSCLC, nonsmall cell lung cancer
PD, Parkinson's disease
SMA, spinal muscular atrophy
Completion of the Human Genome Project has revealed that protein‐coding genes comprise only about 1.5% of the human genome. In fact, two large‐scale consortia, the Encyclopedia of DNA elements (ENCODE) and the Functional Annotation of the Mammalian Genome (FANTOM) have shown that the majority of genome is transcribed and produces a wide spectrum of noncoding RNA species (ncRNAs) 1, 2, 3, 4. Consequently, it is now believed that the degree of complexity of a species correlates better with the number of ncRNAs than with the number of protein‐coding genes 5. Furthermore, the availability of this data has shown that mutations within the noncoding genome are major determinants of human diseases, for example cancer 6.
Noncoding RNAs can be classified, according to their size: short RNAs are < 200 nucleotides (nts) in length and include small interfering RNAs (siRNAs), piwi‐interacting RNAs (piRNAs) and microRNAs (miRNAs) 7, 8; Long noncoding RNAs (lncRNAs) are longer than 200 nts and may comprise thousands of nucleotides 9. Thanks to their major contributions in so many cellular processes, the study of ncRNAs has evolved into a rather inspiring scientific field.
The discovery of miRNAs dates back to 1993, when two laboratories independently reported that a small noncoding RNA transcript lin‐4 from Caenorhabditis elegans regulates lin‐14 through its 3′ untranslated region (3′UTR) 10, 11. At the time of their discovery, it was unclear whether miRNAs were an odd RNA species or ‘emissaries from an unexplored RNA world’ 12. The intense research which followed showed that miRNAs are key regulatory elements of gene expression and essential mediators in a wide range of cellular processes in both health and disease.
The biogenesis of miRNAs (Fig. 1) has been reviewed in detail elsewhere 7. Briefly, miRNAs are expressed as mono‐cistronic primary transcripts or as clusters from polycistronic primary transcripts. MiRNA genes are located in defined transcriptional units or in intergenic regions. Intragenic miRNAs can be found in introns or exons of coding genes (host genes) in the sense orientation. Intragenic miRNAs and their host genes are frequently co‐ordinately expressed, since they share the same promoter 13. Their transcription is driven by RNA Polymerase II (Pol II) producing primary transcripts – called pri‐miRNAs – which are 5′‐capped, spliced and polyadenylated 14. The pri‐miRNA is cleaved at the stem of the hairpin structure by the RNaseII endonuclease III Drosha, together with DGCR8/Pasha proteins resulting in the release of a 60–70 nt hairpin structure, known as the precursor‐miRNA (pre‐miRNA). Pre‐miRNAs are then transported to the cytoplasm by the RanGTP‐dependent nuclear transporter exportin‐5 (XPO5), where they are subsequently processed by an endonuclease cytoplasmic RNase III enzyme Dicer to yield the mature miRNA of 18–25 nt length embedded in an imperfect duplex which is incorporated into the RNA‐Induced Silencing Complex (RISC), together with an Argonaute (Ago) core protein component. One strand of the miRNA duplex (the ‘passenger’ strand) is removed, whereas the other remains bound to Ago as the mature miRNA ‘guide’ strand responsible for guiding RISC to the target mRNAs 8.
Figure 1.
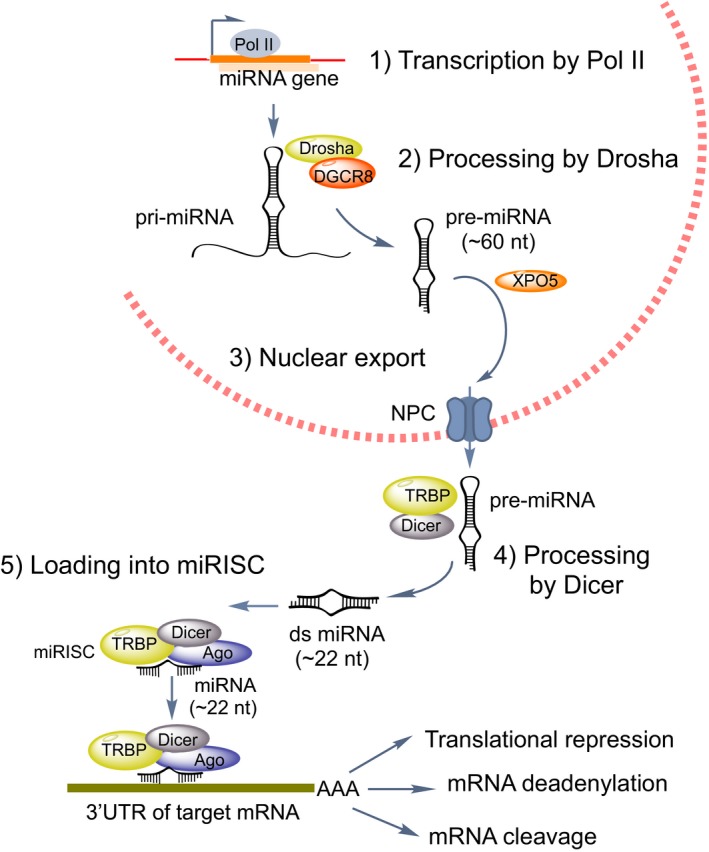
The individual steps of miRNA biogenesis.
MiRNAs attenuate the expression of their target genes by hybridizing, either completely or partially, to complementary binding sites located in the 3ʹUTR of target mRNAs. This leads to mRNA degradation and/or translational inhibition 15. In mammals, miRNAs promote mRNA destabilization, by recruiting the CCR4‐NOT deadenylase complex onto target mRNAs leading to deadenylation. Additionally, miRNAs can mediate translational repression, through various mechanisms, including the recruitment of downstream translational repressors 16.
Bioinformatic predictions suggest that human miRNAs regulate over 60% of transcripts. Given that a single miRNA can regulate the expression of over one hundred mRNAs 8, and each mRNA can be targeted by several miRNAs, miRNAs are highly versatile players in regulatory networks. Furthermore, RNAs containing binding sites for a certain miRNA can attenuate their activity by acting as ‘decoys’ or ‘sponges’, thereby influencing the expression of its other target RNAs 17. The roles of miRNAs also extend beyond suppression of gene expression, as they have also been reported to induce translation of targeted mRNAs 18.
Long noncoding RNAs are a large and diverse class of transcribed RNAs that lack functional open reading frames, though exceptions have been described 19. They are transcribed by RNA Pol II, and are 5′‐capped, spliced and polyadenylated 20. LncRNAs can fold into a variety of secondary structures which facilitate their interactions with DNA, RNA and proteins 21. LncRNAs can be divided into different classes based upon their genomic location: long intergenic noncoding RNAs (lincRNAs) genes are located between coding or noncoding genes. Some lncRNAs are located in the introns of protein‐coding genes. Natural antisense transcripts (NATs) are transcribed from the opposite strand of a coding gene but their transcription start site resides downstream relative to that of the host gene, and these transcripts often overlap with the sequence of the corresponding mRNA.
Long noncoding RNAs function through heterogeneous mechanisms (Fig. 2), conferring additional layers of regulation upon gene expression during for example cell proliferation, cell cycle, metabolism, apoptosis, differentiation and maintenance of pluripotency 22. They also participate in chromatin modification and structure by acting as molecular scaffolds, interacting with components of the epigenetic machinery, such as histone‐modifying enzymes and DNA methyltransferases, and thus mediating their recruitment to DNA loci 23. Additionally, lncRNAs can impact the transcription of other genes, by promoting or preventing the binding of transcription factors and transcriptional mediators to promoters 24, 25. LncRNAs are involved in the regulation of RNA processing, such as RNA splicing 26, or mRNA decay 27.
Figure 2.
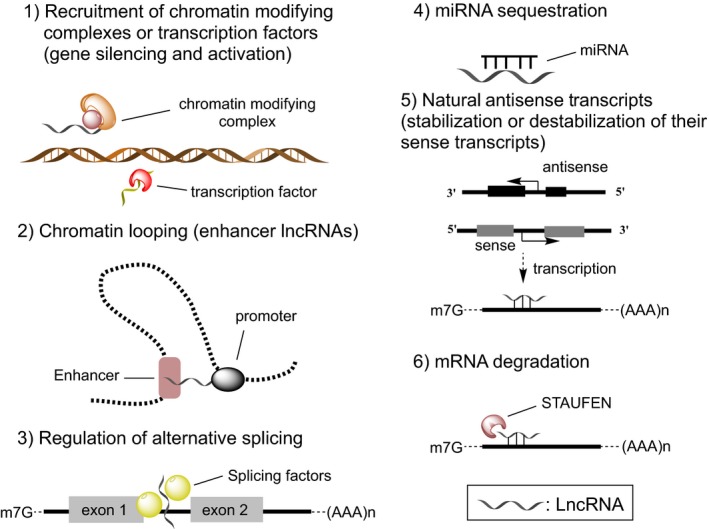
LncRNAs show a wide variety of functions.
Certain lncRNAs have enhancer‐like properties. Orom et al. 28 demonstrated that depletion of a lncRNA at multiple sites of the human genome leads to a specific decrease in the expression of neighbouring protein‐coding genes. Enhancer‐derived lncRNAs (eRNAs) are described to control contacts between enhancers and the cognate promoter through chromosome looping. Activating ncRNAs (ncRNA‐a) mediate DNA looping and chromatin remodelling via the Mediator complex to establish a stable transcription initiation process 29. LncRNAs can additionally function as decoy RNAs, by binding and titrating away miRNAs 17. These lncRNAs may harbour sites complementary to miRNA sequences thereby sequestering them and preventing them from binding to their targets.
A special class of lncRNAs are the antisense lncRNAs (NATs) that are transcribed from the opposite strand of a protein‐coding gene locus 30. NATs have either positive 31 or negative effects on the levels of its corresponding sense transcript 32. For example BACE1‐AS is transcribed from the β‐secretase‐1 (BACE1) gene in antisense direction: it binds to BACE1 mRNA and protects it from miRNA‐mediated degradation 31. Brain‐derived neurotrophic factor (BDNF), on the other hand, is normally repressed by a conserved noncoding antisense RNA transcript, BDNF‐AS, by recruiting the enhancer of zeste homolog 2 (EZH2) and polycomb repressive complex 2 (PRC2) to the BDNF promoter region 32. Finally, lncRNAs can interact with proteins to modulate protein function, regulate protein – protein/DNA/RNA interactions, or direct their localization within cellular compartments 33.
MiRNAs and long noncoding RNAs in disease
In the sections below, we highlight a nonexhaustive selection of examples that demonstrate the wide‐scale involvement of miRNAs and lncRNAs in the pathophysiology of cancer, cardiovascular and neurological disorders.
MiRNAs and long noncoding RNAs in cancer
MiRNAs play various roles in processes underlying human malignancies, including sustaining proliferation, resistance to apoptosis, angiogenesis, invasion and metastasis. Altered miRNA expression patterns found in cancer have been attributed to genomic abnormalities (deletions, amplifications or mutations) 34, epigenetic modifications 35, dysregulated transcription factors 36 and dysregulation of RNA‐binding proteins (RBPs) which participate in miRNA biogenesis 37. However, categorizing miRNAs inhibitors or drivers of tumorigenesis is sometimes not clear‐cut, since their activity depends upon the expression of their targets in the tissue/cell type in which they are expressed. The expression of certain miRNAs can be of prognostic value in human cancers 38. Furthermore, it was recently shown that miRNAs can be released through exosomes from cancer cells into body fluids including blood, urine, milk, sputum and saliva 39. Pharmaceutical approaches to the modulation of miRNA activities represent an exciting and promising field in cancer therapeutics 40, 41. In the following paragraphs we highlight some of the most prominent examples of ncRNAs with important roles in cancer.
Calin and associates were the first to describe a role of miRNAs in cancer, when they reported that miR‐15 and miR‐16 are dramatically downregulated in the majority (68%) of patients with B‐cell chronic lymphocytic leukaemia (CLL) due to deletions or mutations on the 13q13.4 chromosome 42. Both miR‐15 and miR‐16 induce apoptosis by repressing Bcl‐2, an antiapoptotic protein overexpressed in malignant nondividing B cells and many solid tumours 43. The New Zealand Black (NZB) mouse model of CLL exhibits genetic alterations in the mir‐15a/16‐1 locus, which results in decreased levels of miR‐15a and miR‐16 in lymphoid tissues 44, whereas the restoration of miR‐16 levels in a New Zealand Black–derived malignant B‐1 cell line mitigates the proliferation of malignant B1 cells 45.
More than 50% of human tumours carry loss of function mutations in the tumour suppressor protein TP53 (p53) 46. P53 drives transcription of the miR‐34 family, which activates apoptotic pathways 47. At the same time, miR‐34a promotes p53 expression by targeting the antiageing factor Sirtuin‐1 (SIRT1), a negative regulator of p53 48. Reduced expression of miR‐34 has been observed in many cancer types 49, including human gliomas, with concomitant increased expression of the target oncogenes c‐Met, Notch‐1/2 and cyclin‐dependent kinase 6 (CDK6). MiR‐34a was used in a ‘miRNA replacement therapy’ approach, where a chemically synthesized miRNA ‘mimic’ of miR‐34a and a lipid‐based delivery vehicle were used to block tumour growth in mouse models of nonsmall cell lung cancer (NSCLC) 50. Subsequently, a liposomal formulation of a miR‐34a mimic became the first miRNA to enter a phase I clinical study (http://clinicaltrials.gov/ct2/show/NCT01829971). It was given intravenously in patients with primary liver cancer or other selected solid tumors or hematologic malignancies. However, the trial was halted after immune‐related severe adverse events were reported in some of the patients. MiR‐26a is an example of a miRNA whose expression is lost in hepatocellular carcinoma (HCC). It regulates the cyclins D1 and D2, which control cell cycle arrest, as well as ULK1, a critical initiator of autophagy that promotes apoptosis 51. The administration of chemically synthesized miR‐26a in a mouse model of HCC results in inhibition of cancer cell proliferation, induction of tumour‐specific apoptosis, and a dramatic slow‐down in disease progression 52.
The let‐7 miRNAs represent a large family of miRNAs that plays an important role in stem cell division and cell differentiation 53. Let‐7 family members are downregulated in many types of cancer, including lung cancer, gastric tumours, colon cancer, melanoma, ovarian cancer and Burkitt's lymphoma 54. Let‐7 miRNAs target several oncogenes including K‐RAS, c‐Myc and HMGA2, and therefore are considered as tumour suppressors 53. The oncofoetal RBP Lin28 and its paralogue Lin28b bind to the terminal loops of most let‐7 precursors and block their processing into mature miRNAs 55, 56. Lin28 is a stem cell pluripotency factor and both paralogues are upregulated in many human cancers including glioblastoma, ovarian, gastric, prostate and breast cancer 37. The Lin28/let‐7 axis is not only prominent in cancer: it also regulates glucose metabolism through the let‐7‐mediated repression of multiple components of the insulin‐PI3K‐mTOR pathway 57. Aberrant glucose metabolism is tightly linked to cancer since a switch towards glycolytic metabolism increases the cancer cell's ability to increase biomass (‘Warburg Effect’). A subsequent study has shown that overexpression of either Lin28 or Lin28b in liver cancer cells elevates glucose uptake, lactate production and oxygen consumption, all of which are reversed upon addition of let‐7 mimics 58. The importance of the Lin28/let‐7 axis has spurred efforts to generate inhibitors of this biology with a new class of future anticancer agents 41. The oncogenic potential of Lin28 was also shown when King and associates constitutively expressed LIN28B in colon cancer cells and implanted them into immunocompromised mice. Tumours with constitutive LIN28B expression exhibited increased expression of colonic stem cell markers LGR5 and PROM1, mucinous differentiation and metastasis 59. Transgenic mouse models overexpressing Lin28B from the mouse Vil1 promoter specifically in the intestine, showed let‐7‐dependent intestine hypertrophy. Restoring mature let‐7a levels in the intestine reversed the observed hyperplasia, reducing the cellular transformation in the intestinal epithelium 60. Importantly, inhibition of either LIN28A or LIN28B via siRNAs suppressed established human xenograft tumours in mice 61. A similar effect was observed when the xenograft models were treated with chemically synthesized let‐7a miRNA.
Many miRNAs are found expressed at higher levels in tumours and can be seen as oncogenes. They promote tumour development by inhibiting tumour suppressor genes and/or genes that control cell cycle, cell differentiation and apoptosis. c‐Myc is an important oncogene that transactivates several miRNAs including the miR‐17~92 and miR‐106a~363 clusters 36. miR‐17~92 is a notable oncogenic miRNA cluster comprising six miRNAs that are located at chromosome 13q31, a genomic locus amplified in several types of lymphoma and solid tumours 62. This cluster is highly expressed in embryonic cells 63 and its miRNAs target the E2F transcription factor which controls the transition from G1 to S phase 64. The cluster is also overexpressed in many types of cancer, including B‐cell lymphoma, colon cancer, pancreatic cancer, breast cancer, ovarian cancer and neuroblastoma 65. MiRNAs from miR‐17~92 target Bim, repressing its proapoptotic activity 63 and the cell cycle inhibitors p21CIP1 and p57KIP2 thereby enhancing cancer cell growth 66, whereas miR‐19a and miR‐19b‐1 regulate the tumour suppressor PTEN 63. Xiao and associates generated mice with elevated miR‐17~92 expression in lymphocytes; these developed lymphoproliferative disease and autoimmunity and died prematurely 67.
MiR‐221 and miR‐222 (miR‐221/222) are two highly homologous miRNAs, which are significantly overexpressed in several types of human malignancies 68. For example, elevated expression of miR‐222 has been reported to contribute to pancreatic cancer invasion by targeting the tissue inhibitor of MMP‐2 (TIMP‐2) 69. In human glioma cells, miR‐221/222 inhibits cell apoptosis by targeting the proapoptotic gene PUMA 70. In breast cancer, overexpression of miR‐221/222 promotes epithelial‐to‐mesenchymal transition by negatively regulating the adiponectin receptor 1 71, as well as trichorhinophalangeal 1 (TRPS1) 72, leading to increased cell migration and invasion. PTEN, a prominent tumour suppressor gene, is a confirmed target of miR‐221/222 in the breast cancer cell line MCF‐7 73. MiR‐222 promotes tumour progression in HCC 74 and lentivirus‐mediated silencing of miR‐221 suppresses proliferation of liver cancer cells and growth of hepatoma xenografts in vivo 75.
There is considerable evidence that miR‐21 has oncogenic properties, being involved in regulatory pathways of proliferation, apoptosis and metastatic potential 76. Its targets include PTEN, as well as PDCD4, and BTG2, which play important roles in oncogenic processes 77. Furthermore, it is strongly upregulated in glioblastoma, head and neck carcinoma, ovarian cancer, B‐cell lymphoma and hepatocellular and cervical carcinoma 78. In a study of 540 clinical cancer samples by Volinia et al. 79, miR‐21 was the most consistently upregulated miRNA. Furthermore, mice conditionally expressing miR‐21 via Tet‐Off and Cre‐recombinase technologies developed clinical signs of haematological malignancies. MiR‐21‐overexpressing tumour cells were found to invade the peripheral blood, and other organs. Once miR‐21 expression was switched off, the tumours regressed, partly due to the activation of apoptosis 80.
On the list of prominent tumour‐promoting miRNAs is miR‐155, which originates from the B‐cell integration cluster, also known as MIR155HG (miR‐155 host gene). Aberrant expression of miR‐155 has oncogenic potential in several types of haematological malignancies 81. It was recently found that miR‐155 induces resistance to chemotherapeutic agents, which can be reversed by treatment with miR‐155 inhibitors, and that this chemoresistance is dependent on a p53/miR‐155 feedback loop 82. Eμ‐mmu‐miR155 transgenic mice express murine miR‐155 under the control of a VH promoter‐Ig heavy chain Eμ enhancer, which becomes activated at the pro‐B‐cell stage of B‐cell development. These mouse models develop a lymphoproliferative disease, which phenocopies the human form. This study was the first to demonstrate that transgenic overexpression of a single miRNA is sufficient to cause cancer 83.
Long noncoding RNAs have been shown to influence many of the pathways which drive malignant transformation. For instance the lncRNA MALAT1 (also known as NEAT2) is found to be highly expressed in many tumours 84, for example during metastasis in patients with early‐stage NSCLC 85. The elevated expression of MALAT1 is linked to traits such as increased migration, metastasis and clonogenic growth in NSCLC 85, pancreatic 86 and prostate cancer cells 87. Consistent with this, the deletion of MALAT1 in osteosarcoma cell lines inhibited cell proliferation and invasion 84. This lncRNA also promotes the growth and migration of ovarian cancer cells 88. It can bind to active chromatin sites 89 and it co‐localizes with nuclear speckles, where it influences pre‐mRNA splicing 26. MALAT1 is required for G1/S and mitotic progression by modulating the expression and/or pre‐mRNA processing of cell cycle–regulating transcription factors 90.
The Hox transcript antisense intergenic RNA known as HOTAIR is a lncRNA which is transcribed from the HOXC locus. It is considered a biomarker for the prognosis of certain cancers: higher levels of the RNA have been found in colorectal, liver, pancreatic, breast and gastric cancers 91. It forms double stem‐loop structures that bind to lysine‐specific demethylase 1 and PRC2 histone‐modification complexes, which leads to histone H3 tri‐methylation at lysine 27 (H3K27me3) and histone H3 dimethyl Lys4 (H3K4me2) and consequently results in gene silencing. HOTAIR is upregulated in breast cancer and increases cancer invasiveness and metastasis 92.
The lncRNA neuroblastoma associated transcript‐1 (NBAT‐1) was identified as an independent prognostic biomarker, predicting clinical outcome of neuroblastoma patients 93. Loss of NBAT‐1 increases cellular proliferation and invasion. It mediates epigenetic silencing of target genes, through its interaction with the PRC2 repressive chromatin complex.
The lncRNA ANRIL shows increased expression in NSCLC tissues, and this correlates with stages of tumour–node–metastasis and the size of tumours 94. ANRIL is expressed highly in gastric cancers, and higher levels of ANRIL promote proliferation of gastric cancer cells, where it inhibits apoptosis by epigenetic silencing of miR‐99a and miR‐449a transcription 95.
The oncofoetal lncRNA H19 is an important factor in both embryonic development and tumorigenesis. It is upregulated in a series of cancer types, where it reportedly accelerates cellular proliferation rates and increases the resistance of tumour cells to stress 96. Interestingly, H19 transcript has been reported to sequester and inhibit two cancer‐related miRNAs – let‐7 and miR‐106a 97, 98. H19 also serves as a primary miRNA precursor of miR‐675 99, which is considered as oncogenic due to its targeting of the tumour suppressor retinoblastoma protein. The H19 locus belongs to a cluster of imprinted genes that control embryonic and postnatal growth. The H19 gene is located 90 kb distant from the Igf2 gene on chromosome 11p15 in humans and chromosome 7 in mice. The Igf2 locus encodes insulin‐like growth factor‐2 (IGF2), which is a growth‐promoting peptide hormone highly expressed during embryogenesis. H19 and Igf2 genes are reciprocally imprinted from the maternal and paternal alleles respectively. The changes in imprinting of the Igf2‐H19 locus are likely to be involved in tumour formation. In humans, loss of imprinting at this locus are associated with the Beckwith–Wiedemann syndrome (BWS), which is characterized by overgrowth phenotypes in affected children, as well as a predisposition to develop embryonal tumours such as Wilms’ tumour or rhabdomyosarcomas 100. There are inconsistencies between various murine models which aim to define the role of H19 locus in cancer. In some cases, the H19 locus has been suggested to act as a tumour suppressor, and mice bearing a mutation in the Apc gene are murine models for colorectal cancer. When double mutants were generated, lacking both H19 and Apc, they showed an enhanced cancer phenotype compared with their Apc littermates 101. In other cases, H19 has been shown to promote tumour growth in mice. Matouk and associates demonstated that ectopic H19 expression enhances the tumorigenic potential of bladder carcinoma cells in vivo 102.
MiRNAs and long noncoding RNAs in cardiovascular disease
Cardiovascular disease and complications thereof are a leading cause of morbidity and mortality worldwide. The myocardium can undergo remodelling in response to external stressors. However, chronic activation of remodelling processes, such as hypertrophy and fibrosis, can result in multiple cardiovascular diseases, including myocardial infarction, cardiomyopathies and heart failure. Ikeda et al. 103 identified significantly altered miRNA expression profiles in heart disease and showed that patterns of miRNA expression are distinct in different forms of heart disease. A myriad of studies has shown that miRNAs regulate the expression of genes in signalling pathways associated with heart failure, hypertrophy, and ischaemia reperfusion injury. For example, miRNAs have been found to promote or inhibit cardiomyocyte apoptosis, regulate postischaemic neovascularization and control cardiac fibrosis 104. Remarkably when miRNA biogenesis is inhibited through Dicer deletion, dilated cardiomyopathy associated with heart failure is observed in neonates 105, whereas the postnatal myocardium‐specific Dicer deletion drives maladaptive cardiac remodelling 106. Additionally, endothelial knockout of Dicer leads to endothelial dysfunction, revealing a key role for miRNAs in endothelial physiology 107.
Several miRNAs play key roles in vascular development and angiogenesis. For example miR‐24 has a role in cardiac vascularization 108. It is highly expressed in cardiac endothelial cells (ECs) and is significantly upregulated after cardiac ischaemia. Blockage of miR‐24 limits myocardial infarct size of mice, preventing endothelial apoptosis and enhancing vascularity. This miRNA exerts its functions through targeting the endothelium‐enriched transcription factor GATA2 and the p21‐activated kinase PAK4. MiR‐126‐3p is a proangiogenic factor, which is implicated in endothelial gene expression and mediates EC dysfunction as well as atherosclerosis triggered by blood flow changes 109. Overexpression of miR‐126‐3p reduces atherosclerosis 110, whereas its knock‐out causes systemic oedema, multifocal haemorrhages and ruptured blood vessels 111. It is enriched in the apoptotic bodies of dying ECs in a mouse model of atherosclerosis and has an angioprotective role via the CXCL12‐CXCR4 pathway 110.
MiR‐208 is selectively expressed in cardiomyocytes, and is highly expressed in autopsy samples of infarcted heart tissue from patients with myocardial ischaemia 112. In addition, compared to other miRNAs, levels of miR‐208 are high in cardiac tissue of dilated cardiomyopathy patients and it is a strong predictor of clinical outcome 113. In response to cardiac stress such as pressure overload, knockdown of miR‐208 in mice produces no cardiomyocyte hypertrophy and fibrosis 114. The miRNA also plays an important role in cardiac conduction, by regulating the expression of cardiac transcription factors and the gap junction protein connexin 40 (Cx43) 115. The miR‐15 family includes six closely‐related miRNAs that are also increased in myocardial ischaemia 116. Inhibition of miR‐15 family members by antimiR‐oligonucleotides reduces infarct size after ischaemia–reperfusion injury in cardiac tissue of both mice and pigs by de‐repressing the antiapoptotic protein Bcl‐2 and the mitochondrial protecting factor ADP‐ribosylation factor‐like protein 2 116.
Zidar and associates have reported that downregulation of miR‐150 is involved in the pathology of ventricular rupture after myocardial ischaemia 117. Of note, it was recently shown that the cardio‐related lncRNA ZFAS1 can interact directly with miR‐150, acting as a miRNA sponge that induces cardiomyocyte apoptosis in acute myocardial ischaemia via C‐reactive protein (CRP) 118. It regulates adenoreceptor beta 1 and CRP genes, which are associated with heart remodelling 119.
The neurologic‐enriched miRNA miR‐212/132 family becomes activated during heart failure 120. These miRNAs affect cardiac hypertrophy by targeting the anti‐hypertrophic and proautophagic transcription factor forkhead box O3 (FoxO3), leading to induction of the prohypertrophic calcineurin/NFAT signalling pathway 121. Altered levels of miR‐21 are associated with multiple cardiovascular diseases, including proliferative vascular disease, cardiac hypertrophy, heart failure, and ischaemic heart diseases 122. MiR‐21 promotes cardiac fibrosis by regulating genes, such as transforming growth factor β1 receptor III (TβRIII) 123 and matrix metalloprotease‐2 (MMP2) 124, 125. Bang and associates have demonstrated that miR‐21 is transferred through fibroblast derived exosomes, acting as a paracrine mediator of cardiomyocyte hypertrophy 125.
MiR‐1 is most abundantly expressed in heart and plays essential roles in cardiogenesis and in physiological cardiac function. Jayawardena et al. 126 showed that miR‐1 alone is sufficient to induce the fibroblast to cardiomyocyte reprogramming. MiR‐1 targets genes that cluster into several categories, including regulators of cell cycle, cardiac differentiation and the conductive system 127, 128. Cardiac Serca2a, which regulates calcium uptake into the sarcoplasmic reticulum (SR), has also been shown to increase after miR‐1 gene transfer in mice 129. This miRNA attenuates cardiomyocyte hypertrophy in cultured cardiomyocytes and in the intact adult heart by regulation of cardiomyocyte growth responses through modulation of calcium signalling components such as calmodulin 127. Consistent with this, miR‐1 has been found decreased in early‐stage cardiac hypertrophy 130. The miRNA and its primary target Errβ act together to regulate the transition from prenatal to neonatal stages by repressing the cardiac foetal gene program, which is reactivated under pathological conditions 131. Expression of miR‐1 is lost in the myocardium of myotonic dystrophy patients, concomitant with up regulation of its targets Connexin 43 (Cx43) and calcium voltage‐gated channel subunit alpha1C (CAV1.2) may at least partly account for the arrhythmia, which is observed in these patients 128. MiR‐1 is clustered together with miR‐133 on mouse chromosome 2, where they are separated by 9.3 kb, and on mouse chromosome 18, where they are separated by 2.5 kb 132. Although miR‐1 and miR‐133 derive from the same miRNA polycistron and are transcribed together, they have antagonistic effects on muscle development: miR‐1 enhances myogenic differentiation, whereas miR‐133 induces myoblast proliferation 133. MiRNA‐133 is decreased in mouse and human models of cardiac hypertrophy 134 through its regulation of the Ras homolog family member A (RhoA) and cell division control protein 42 homolog (Cdc42). It also plays a role in cardiac fibrosis by controlling the expression of the connective tissue growth factor 135. MiR‐133 affects inotropism by regulating the expression of multiple components of the β1‐adrenergic cascade, including the receptor itself 136.
Next to miRNAs, lncRNAs also play important roles in cardiovascular disease. In fact, data from deep sequencing demonstrated that compared to mRNA and miRNA expression profiles, lncRNA expression profiles are more sensitive to different heart failure aetiologies and that altered lncRNAs reflect increased susceptibility to coronary artery disease, myocardial infarction and heart failure 137. For example, Viereck et al. have recently discovered a new lncRNA – Chast (for ‘cardiac hypertrophy–associated transcript’) – that promotes cardiac remodelling and hypertrophy in mice. Antisense‐mediated degradation of Chast attenuated pathological cardiac remodelling, as it was shown by in vivo gain‐ and loss‐of‐function experiments in mice 138.
Besides its role in cancer, the lncRNA MALAT1 is also linked to cardiovascular disease: silencing of MALAT1 reduces capillary growth in a mouse model of hind limb ischaemia 139 as well as in a rat model of diabetic retinopathy 140. Furthermore, MALAT1‐derived mascRNA (MALAT1‐associated small cytoplasmic RNA) is involved in cardiovascular innate immunity and viral myocarditis 141.
The lncRNA GAS5 (growth arrest–specific 5) is another regulator of hypertension‐related vascular remodelling 142. It is mainly expressed in ECs and vascular smooth muscle cells (VSMCs), and its expression is significantly downregulated in hypertension. GAS5 regulates EC and vascular smooth muscle cell function through β‐catenin signalling. The cardiac apoptosis‐related lncRNA (CARL) has been found to regulate mitochondrial homeostasis and cell death in cardiomyocytes 143. CARL intervenes during the mitochondrial fission process by sequestering miR‐539 and inhibiting the miR‐539‐mediated repression of Prohibitin 143.
Ounzain et al. 144 identified several lncRNAs with potential roles in both cardiac development and pathological cardiac remodelling. One particular novel lncRNA, Novlnc6, is significantly decreased in dilated cardiomyopathy. Knockdown of Novlnc6 in cardiomyocytes results in a concomitant downregulation of BMP10 and NKX2.5, two important mediators of cardiac growth and function.
The lncRNA cardiac hypertrophy related factor, is substantially elevated in response to hypertrophic stimulation by angiotensin II in cardiomyocytes 145. In addition, it is also significantly upregulated in a mouse model of transverse aortic constriction and in human heart failure samples 145. This lncRNA acts as a sponge for miR‐489, de‐repressing Myd88, a direct target of miR‐489 so as to regulate cardiomyocyte hypertrophy.
Finally, the lncRNA myocardial infarction–associated transcript (MIAT) is highly expressed in heart and foetal brain tissue. Polymorphisms in MIAT that were identified by genome‐wide association studies are a risk factor for myocardial infarction 146. MIAT is found at low levels in platelets from patients with myocardial infarction 147, whereas elevated levels of this lncRNA are found in myocardial samples from patients with dilated cardiomyopathy suffering from Chagas disease 148.
MiRNAs and long noncoding RNAs in neurodegenerative disease
Neurodegenerative diseases are hereditary and sporadic conditions which are characterized by progressive dysfunction and the death of neurons. According to the neuronal populations afflicted, these disorders can lead to disturbances in motor, cognitive and/or behavioural performance of affected individuals. They include diseases such as Alzheimer's disease (AD) and other dementias, Parkinson's disease (PD), amyotrophic lateral sclerosis (ALS), spinal muscular atrophy (SMA), Huntington's disease (HD) and others.
MiRNAs in Parkinson's disease
MiRNAs display specific temporal and spatial patterns of expression during embryonic neural development and in adult brain 149. In the central nervous system, they have been shown to participate in a wide range of processes, such as neurodevelopment, brain architecture, neuroplasticity establishment, neurotransmission, etc. Not surprisingly, misregulated miRNAs have been linked to many neurodegenerative and psychiatric disorders. MiRNAs miR‐34b and miR‐34c are decreased in the affected areas of PD patients at an early stage of the disease 150. This miRNA family regulates alpha‐synuclein, a key protein in PD pathogenesis 150. Reduced expression of these miRNAs is associated with mitochondrial abnormalities and increased oxidative stress. MiR‐155 has been shown to mediate immune activation by aggregated α‐synuclein. In a PD mouse model overexpressing α‐SYN (via an adeno‐associated‐virus; AAV2‐SYN), levels of miR‐155 are significantly increased. However, miR‐155 knockout mice models transduced with AAV2‐SYN, exhibit a remarkably decreased proinflammatory response, without a loss of dopaminergic neurons 151. It was recently shown that miR‐30e improves neuronal damage, neuroinflammation and dyskinesia via targeting Nlrp3 expression and inhibiting NLRP3 inflammasome activation in a MPTP‐induced PD mice model 152. MiR‐30e levels were downregulated after MPTP injection, suggesting miR‐30 might also have a role in the pathogenesis of PD. MiR‐124 expression was downregulated in substantia nigra dopaminergic neurons following MPTP administration in mice. A MiR‐124 mimic delivered to the right lateral ventricle in the MPTP mouse model increases the density of tyrosine hydroxylase positive (TH+) neurons and reduced the upregulation of Bim mRNA level and protein level induced by MPTP, leading to reduced apoptosis 153.
MiRNAs in Alzheimer's disease
Expression of the miR‐29a/b‐1 cluster is significantly decreased in the brains of patients suffering from sporadic AD, displaying abnormally high levels of BACE1 protein 154. This miRNA family targets BACE‐1 secretase, which cleaves amyloid precursor protein (APP) and generates toxic Aβ species, thereby contributing to synaptic loss and cognitive decline in AD. MiR‐29 has been also suggested to protect cells from apoptosis by targeting proapoptotic proteins, including BIM, BMF, HRK and PUMA 155. It was recently demonstrated that pre‐miR‐29b encapsulated in polyplexes decreases levels of hBACE1 and Aβ45.156 Levels of miR‐29a are increased by more than two‐fold in cerebrospinal fluid of AD patients, indicating that miR‐29a may be a candidate biomarker for AD 157. MiR‐106b from the miR‐106b~25 cluster is a regulator of Aβ production and clearance through the suppression of ABCA1 expression 158. Suppression of ABCA1 expression by miR‐106b impairs cellular cholesterol efflux and increases the levels of secreted Aβ. MiR‐106b is also aberrantly expressed in a double transgenic mouse model for AD 159. Simvastatin was recently shown to ameliorate the memory decline in AD mouse models via decreased miR‐106b levels 160. Finally, miR‐34a is found over expressed in affected brain regions of AD patients as well as in transgenic AD mice 161. The increased expression of miR‐34a in specific brain regions induces synaptic dysfunction. Its accumulation, along with the interneuronal transfer of miR‐34a‐loaded exosomes, may affect neural networks dedicated to memory. MiR‐34c is also connected to hippocampal memory function. Inhibition of this miRNA rescues memory impairment in AD transgenic mice, with concomitant de‐repression of SIRT1, a confirmed target of miR‐34 162.
MiR‐196 in Huntington's disease
Huntington's disease is an autosomal‐dominant disease that is caused by an expansion of CAG trinucleotide repeats located in the exon 1 region of the huntingtin gene. MiR‐196a has emerged as a protective miRNA in the context of HD. Overexpression of miR‐196a leads to a reduction of mutant huntingtin (HTT) and the formation of pathological aggregates in HD models of human embryonic kidney cells and mouse neuroblastoma cells. In HD transgenic mice overexpressing miR‐196a, suppression of mutant HTT in the brain shows attenuated neuropathological progression, manifested by reduced nuclear, intranuclear and neuropil aggregates as well as late‐stage behavioural phenotypes 163. The effects of miR‐196a might be via its involvement in the ubiquitin–proteasome systems, gliosis, and the CREB pathway.
MiR‐183 in spinal muscular atrophy
MiR‐183 has been shown to contribute to the pathology of SMA via its target mTor. The local axonal translation of mTor is reduced in SMN‐deficient neurons, and this can be restored by inhibition of miR‐183. Importantly, inhibition of miR‐183 expression in the spinal cord of an SMA mouse model prolongs survival and ameliorates motor performance of SMN‐mutant mice 164.
BACE1‐AS in Alzheimer’ s disease
Aberrant lncRNA expression is linked to the onset and progression of several neurodegenerative diseases. For instance BACE1‐AS is a NAT that is transcribed from an intron of the β‐secretase‐1 (BACE1) gene in antisense direction. Its expression is elevated in subjects suffering from AD and in APP transgenic mice 165. Several cell stressors increase BACE1‐AS RNA, which enhances BACE1 mRNA stability, generating additional Abeta 1‐42. It has been postulated that BACE1‐AS prevents translational repression of BACE1 mRNA by miR‐485, by masking the miRNA binding site 31.
MALAT1 and NEAT1_2 in FTLD and ALS and Huntington's disease
TDP‐43 is a nuclear RNA‐binding protein that forms inclusion bodies in frontotemporal lobar degeneration (FTLD) and ALS. The binding of TDP‐43 to MALAT1 and NEAT1_2 lncRNAs is increased in human FTLD brains compared with healthy controls 166. Analyses of human spinal motor neurons in ALS cases shows that NEAT1_2 lncRNA is upregulated during the early stage of ALS pathogenesis. This lncRNA acts as a scaffold for RNAs and RBPs in the nuclei of ALS motor neurons, thereby modulating the functions of ALS‐associated RNA‐binding proteins, such as TDP‐43 and FUS/TLS, during the early phase of ALS 167. NEAT1 levels are also increased in the postmortem brain from patients of HD 168. Gain‐of‐function studies showed that NEAT1 upregulation in HD contributes to the neuroprotective mechanism against neuronal injury.
UCHL1‐AS, MALAT‐1 and HOTAIR in Parkinson's disease
The ubiquitin carboxy‐terminal hydrolase L1 gene (UCHL1) is closely related to brain function and neurodegenerative diseases. An antisense transcript of UCHL1, UCHL1‐AS promotes translation of UCHL1 169, which is strongly attenuated in neurochemical models of PD in vitro and in vivo 169. MALAT1 is highly expressed in neurons 170. It was recently demonstrated that MALAT1 overexpression increases, whereas inhibition decreases alpha‐synuclein expression 171. β‐Asarone, a constituent of Acorus tatarinowii Schott, suppresses the levels of MALAT1 and alpha‐synuclein in the midbrain tissue of PD mice, suggesting that β‐asarone may be a potential therapeutic agent for PD171. HOTAIR is upregulated in a mouse model of PD that is produced by intraperitoneal injection of MPTP, a prodrug to the neurotoxin MPP+. The lncRNA increases the stability of LRRK2 mRNA 172, and thus may interfere with the LRRK2‐associated mitochondrial impairment in PD.
Conclusion
The constellations of physiological processes which orchestrate life are subject to intricate control. MiRNAs and lncRNAs have emerged as ubiquitous RNA molecules capable of modulating all cellular processes. In particular, ncRNAs have drawn great attention partly for their putative roles in the pathology of many diseases. In many of the cases highlighted in this Review, in which we have limited the discussion to three types of diseases, the links between ncRNAs and disease pathologies came to light through their aberrant expression in disease cells or tissues. It is noteworthy that some ncRNAs (e.g. miR‐15, miR‐29, miR‐34, ANRIL and MALAT‐1) appear to contribute to more than one pathological mechanisms. In some of these cases, the miRNA‐disease association is sufficiently strong (i.e. possibly causative) that the miRNA represents a potential drug target or a therapeutic entity (e.g. miR‐106 and let‐7 respectively). The availability of potent pharmacological tools for use in animal models of these diseases and/or in clinical trials will ultimately clarify their value in distinct therapeutic applications 173.
Acknowledgements
This work was partly funded by the KrebsForschung Schweiz (grant number: KLS‐3816‐02‐2016). For reasons of limited space, we are unable to cite all of the work contributing to this exciting field.
Edited by Wilhelm Just
References
- 1. Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C et al (2005) The transcriptional landscape of the mammalian genome. Science 309, 1559–1563. [DOI] [PubMed] [Google Scholar]
- 2. Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J et al (2005) Antisense transcription in the mammalian transcriptome. Science 309, 1564–1566. [DOI] [PubMed] [Google Scholar]
- 3. Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H et al (2002) Analysis of the mouse transcriptome based on functional annotation of 60,770 full‐length cDNAs. Nature 420, 563–573. [DOI] [PubMed] [Google Scholar]
- 4. Hangauer MJ, Vaughn IW and McManus MT (2013) Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet 9, e1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Taft RJ, Pheasant M and Mattick JS (2007) The relationship between non‐protein‐coding DNA and eukaryotic complexity. BioEssays 29, 288–299. [DOI] [PubMed] [Google Scholar]
- 6. Melton C, Reuter JA, Spacek DV and Snyder M (2015) Recurrent somatic mutations in regulatory regions of human cancer genomes. Nat Genet 47, 710–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim VN, Han J and Siomi MC (2009) Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10, 126–139. [DOI] [PubMed] [Google Scholar]
- 8. Bartel DP (2018) Metazoan microRNAs. Cell 173, 20–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Batista PJ and Chang HY (2013) Long noncoding RNAs: cellular address codes in development and disease. Cell 152, 1298–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee RC, Feinbaum RL and Ambros V (1993) The C. elegans heterochronic gene lin‐4 encodes small RNAs with antisense complementarity to lin‐14. Cell 75, 843–854. [DOI] [PubMed] [Google Scholar]
- 11. Wightman B, Ha I and Ruvkun G (1993) Posttranscriptional regulation of the heterochronic gene lin‐14 by lin‐4 mediates temporal pattern formation in C. elegans . Cell 75, 855–862. [DOI] [PubMed] [Google Scholar]
- 12. Wickens M and Takayama K (1994) RNA. Deviants–or emissaries. Nature 367, 17–18. [DOI] [PubMed] [Google Scholar]
- 13. Rodriguez A, Griffiths‐Jones S, Ashurst JL and Bradley A (2004) Identification of mammalian microRNA host genes and transcription units. Genome Res 14, 1902–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Carthew RW and Sontheimer EJ (2009) Origins and mechanisms of miRNAs and siRNAs. Cell 136, 642–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pasquinelli AE (2012) MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet 13, 271–282. [DOI] [PubMed] [Google Scholar]
- 16. Iwakawa HO and Tomari Y (2015) The functions of microRNAs: mRNA decay and translational repression. Trends Cell Biol 25, 651–665. [DOI] [PubMed] [Google Scholar]
- 17. Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I (2011) A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147, 358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vasudevan S, Tong Y and Steitz JA (2007) Switching from repression to activation: microRNAs can up‐regulate translation. Science 318, 1931–1934. [DOI] [PubMed] [Google Scholar]
- 19. Wilhelm M, Schlegl J, Hahne H, Gholami AM, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H et al (2014) Mass‐spectrometry‐based draft of the human proteome. Nature 509, 582–587. [DOI] [PubMed] [Google Scholar]
- 20. Quinn JJ and Chang HY (2016) Unique features of long non‐coding RNA biogenesis and function. Nat Rev Genet 17, 47–62. [DOI] [PubMed] [Google Scholar]
- 21. Mercer TR, Dinger ME and Mattick JS (2009) Long non‐coding RNAs: insights into functions. Nat Rev Genet 10, 155–159. [DOI] [PubMed] [Google Scholar]
- 22. Geisler S and Coller J (2013) RNA in unexpected places: long non‐coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol 14, 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian‐Amiri ME, Ru K, Solda G, Simons C et al (2008) Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 18, 1433–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Feng J, Bi C, Clark BS, Mady R, Shah P and Kohtz JD (2006) The Evf‐2 noncoding RNA is transcribed from the Dlx‐5/6 ultraconserved region and functions as a Dlx‐2 transcriptional coactivator. Genes Dev 20, 1470–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ng SY, Bogu GK, Soh BS and Stanton LW (2013) The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell 51, 349–359. [DOI] [PubMed] [Google Scholar]
- 26. Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA et al (2010) The nuclear‐retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39, 925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gong C and Maquat LE (2011) lncRNAs transactivate STAU1‐mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature 470, 284–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q et al (2010) Long noncoding RNAs with enhancer‐like function in human cells. Cell 143, 46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA and Shiekhattar R (2013) Activating RNAs associate with mediator to enhance chromatin architecture and transcription. Nature 494, 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Khorkova O, Myers AJ, Hsiao J and Wahlestedt C (2014) Natural antisense transcripts. Hum Mol Genet 23, R54–R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Faghihi MA, Zhang M, Huang J, Modarresi F, Van der Brug MP, Nalls MA, Cookson MR, St‐Laurent G III and Wahlestedt C (2010) Evidence for natural antisense transcript‐mediated inhibition of microRNA function. Genome Biol 11, R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Modarresi F, Faghihi MA, Lopez‐Toledano MA, Fatemi RP, Magistri M, Brothers SP, van der Brug MP and Wahlestedt C (2012) Inhibition of natural antisense transcripts in vivo results in gene‐specific transcriptional upregulation. Nat Biotechnol 30, 453–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E and Chang HY (2015) Systematic discovery of Xist RNA binding proteins. Cell 161, 404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sevignani C, Calin GA, Nnadi SC, Shimizu M, Davuluri RV, Hyslop T, Demant P, Croce CM and Siracusa LD (2007) MicroRNA genes are frequently located near mouse cancer susceptibility loci. Proc Natl Acad Sci USA 104, 8017–8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Suzuki H, Maruyama R, Yamamoto E and Kai M (2013) Epigenetic alteration and microRNA dysregulation in cancer. Front Genet 4, 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. O'Donnell KA, Wentzel EA, Zeller KI, Dang CV and Mendell JT (2005) c‐Myc‐regulated microRNAs modulate E2F1 expression. Nature 435, 839–843. [DOI] [PubMed] [Google Scholar]
- 37. Viswanathan SR, Powers JT, Einhorn W, Hoshida Y, Ng T, Toffanin S, O'Sullivan M, Lu J, Philips LA, Lockhart VL et al (2009) Lin28 enhances tumorigenesis and is associated with advanced human malignancies. Nat Genet 41, 843–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nair VS, Maeda LS and Ioannidis JP (2012) Clinical outcome prediction by microRNAs in human cancer: a systematic review. J Natl Cancer Inst 104, 528–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tran N (2016) Cancer exosomes as miRNA factories. Trends Cancer 2, 329–331. [DOI] [PubMed] [Google Scholar]
- 40. Rupaimoole R and Slack FJ (2017) MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discovery 16, 203–222. [DOI] [PubMed] [Google Scholar]
- 41. Roos M, Pradere U, Ngondo RP, Behera A, Allegrini S, Civenni G, Zagalak JA, Marchand JR, Menzi M, Towbin H et al (2016) A small‐molecule inhibitor of Lin28. ACS Chem Biol 11, 2773–2781. [DOI] [PubMed] [Google Scholar]
- 42. Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K et al (2002) Frequent deletions and down‐regulation of micro‐ RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 99, 15524–15529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M et al (2005) miR‐15 and miR‐16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA 102, 13944–13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Raveche ES, Salerno E, Scaglione BJ, Manohar V, Abbasi F, Lin YC, Fredrickson T, Landgraf P, Ramachandra S, Huppi K et al (2007) Abnormal microRNA‐16 locus with synteny to human 13q14 linked to CLL in NZB mice. Blood 109, 5079–5086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Salerno E, Scaglione BJ, Coffman FD, Brown BD, Baccarini A, Fernandes H, Marti G and Raveche ES (2009) Correcting miR‐15a/16 genetic defect in New Zealand Black mouse model of CLL enhances drug sensitivity. Mol Cancer Ther 8, 2684–2692. [DOI] [PubMed] [Google Scholar]
- 46. Duffy MJ, Synnott NC and Crown J (1990) (2017) Mutant p53 as a target for cancer treatment. Eur J Cancer 83, 258–265. [DOI] [PubMed] [Google Scholar]
- 47. Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, Meister G and Hermeking H (2007) Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR‐34a is a p53 target that induces apoptosis and G1‐arrest. Cell Cycle 6, 1586–1593. [DOI] [PubMed] [Google Scholar]
- 48. Navarro F and Lieberman J (2015) miR‐34 and p53: new insights into a complex functional relationship. PLoS One 10, e0132767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hermeking H (2012) MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer 12, 613–626. [DOI] [PubMed] [Google Scholar]
- 50. Wiggins JF, Ruffino L, Kelnar K, Omotola M, Patrawala L, Brown D and Bader AG (2010) Development of a lung cancer therapeutic based on the tumor suppressor microRNA‐34. Can Res 70, 5923–5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jin F, Wang Y, Li M, Zhu Y, Liang H, Wang C, Wang F, Zhang CY, Zen K and Li L (2017) MiR‐26 enhances chemosensitivity and promotes apoptosis of hepatocellular carcinoma cells through inhibiting autophagy. Cell Death Dis 8, e2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, Chang TC, Vivekanandan P, Torbenson M, Clark KR et al (2009) Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell 137, 1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Roush S and Slack FJ (2008) The let‐7 family of microRNAs. Trends Cell Biol 18, 505–516. [DOI] [PubMed] [Google Scholar]
- 54. Wang X, Cao L, Wang Y, Wang X, Liu N and You Y (2012) Regulation of let‐7 and its target oncogenes (Review). Oncol Lett 3, 955–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Balzeau J, Menezes MR, Cao S and Hagan JP (2017) The LIN28/let‐7 pathway in cancer. Front Genet 8, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Loughlin FE, Gebert LF, Towbin H, Brunschweiger A, Hall J and Allain FH (2011) Structural basis of pre‐let‐7 miRNA recognition by the zinc knuckles of pluripotency factor Lin28. Nat Struct Mol Biol 19, 84–89. [DOI] [PubMed] [Google Scholar]
- 57. Zhu H, Shyh‐Chang N, Segre AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG et al (2011) The Lin28/let‐7 axis regulates glucose metabolism. Cell 147, 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ma X, Li C, Sun L, Huang D, Li T, He X, Wu G, Yang Z, Zhong X, Song L et al (2014) Lin28/let‐7 axis regulates aerobic glycolysis and cancer progression via PDK1. Nat Commun 5, 5212. [DOI] [PubMed] [Google Scholar]
- 59. King CE, Cuatrecasas M, Castells A, Sepulveda AR, Lee JS and Rustgi AK (2011) LIN28B promotes colon cancer progression and metastasis. Can Res 71, 4260–4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Madison BB, Liu Q, Zhong X, Hahn CM, Lin N, Emmett MJ, Stanger BZ, Lee JS and Rustgi AK (2013) LIN28B promotes growth and tumorigenesis of the intestinal epithelium via Let‐7. Genes Dev 27, 2233–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Piskounova E, Polytarchou C, Thornton JE, LaPierre RJ, Pothoulakis C, Hagan JP, Iliopoulos D and Gregory RI (2011) Lin28A and Lin28B inhibit let‐7 microRNA biogenesis by distinct mechanisms. Cell 147, 1066–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mavrakis KJ, Wolfe AL, Oricchio E, Palomero T, de Keersmaecker K, McJunkin K, Zuber J, James T, Khan AA, Leslie CS et al (2010) Genome‐wide RNA‐mediated interference screen identifies miR‐19 targets in Notch‐induced T‐cell acute lymphoblastic leukaemia. Nat Cell Biol 12, 372–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ventura A, Young AG, Winslow MM, Lintault L, Meissner A, Erkeland SJ, Newman J, Bronson RT, Crowley D, Stone JR et al (2008) Targeted deletion reveals essential and overlapping functions of the miR‐17 through 92 family of miRNA clusters. Cell 132, 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G and Chartrand P (2007) An E2F/miR‐20a autoregulatory feedback loop. J Biol Chem 282, 2135–2143. [DOI] [PubMed] [Google Scholar]
- 65. Fuziwara CS and Kimura ET (2015) Insights into regulation of the miR‐17‐92 cluster of miRNAs in cancer. Front Med 2, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Conkrite K, Sundby M, Mukai S, Thomson JM, Mu D, Hammond SM and MacPherson D (2011) miR‐17~92 cooperates with RB pathway mutations to promote retinoblastoma. Genes Dev 25, 1734–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Xiao C, Srinivasan L, Calado DP, Patterson HC, Zhang B, Wang J, Henderson JM, Kutok JL and Rajewsky K (2008) Lymphoproliferative disease and autoimmunity in mice with increased miR‐17‐92 expression in lymphocytes. Nat Immunol 9, 405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Song J, Ouyang Y, Che J, Li X, Zhao Y, Yang K, Zhao X, Chen Y, Fan C and Yuan W (2017) Potential value of miR‐221/222 as diagnostic, prognostic, and therapeutic biomarkers for diseases. Front Immunol 8, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Xu Q, Li P, Chen X, Zong L, Jiang Z, Nan L, Lei J, Duan W, Zhang D, Li X et al (2015) miR‐221/222 induces pancreatic cancer progression through the regulation of matrix metalloproteinases. Oncotarget 6, 14153–14164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhang CZ, Zhang JX, Zhang AL, Shi ZD, Han L, Jia ZF, Yang WD, Wang GX, Jiang T, You YP et al (2010) MiR‐221 and miR‐222 target PUMA to induce cell survival in glioblastoma. Mol Cancer 9, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hwang MS, Yu N, Stinson SY, Yue P, Newman RJ, Allan BB and Dornan D (2013) miR‐221/222 targets adiponectin receptor 1 to promote the epithelial‐to‐mesenchymal transition in breast cancer. PLoS One 8, e66502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Stinson S, Lackner MR, Adai AT, Yu N, Kim HJ, O'Brien C, Spoerke J, Jhunjhunwala S, Boyd Z, Januario T et al (2011) miR‐221/222 targeting of trichorhinophalangeal 1 (TRPS1) promotes epithelial‐to‐mesenchymal transition in breast cancer. Sci Signal 4, pt5. [DOI] [PubMed] [Google Scholar]
- 73. Li B, Lu Y, Wang H, Han X, Mao J, Li J, Yu L, Wang B, Fan S, Yu X et al (2016) miR‐221/222 enhance the tumorigenicity of human breast cancer stem cells via modulation of PTEN/Akt pathway. Biomed Pharmacother 79, 93–101. [DOI] [PubMed] [Google Scholar]
- 74. Fu X, Wang Q, Chen J, Huang X, Chen X, Cao L, Tan H, Li W, Zhang L, Bi J et al (2011) Clinical significance of miR‐221 and its inverse correlation with p27Kip(1) in hepatocellular carcinoma. Mol Biol Rep 38, 3029–3035. [DOI] [PubMed] [Google Scholar]
- 75. He XX, Guo AY, Xu CR, Chang Y, Xiang GY, Gong J, Dan ZL, Tian DA, Liao JZ and Lin JS (2014) Bioinformatics analysis identifies miR‐221 as a core regulator in hepatocellular carcinoma and its silencing suppresses tumor properties. Oncol Rep 32, 1200–1210. [DOI] [PubMed] [Google Scholar]
- 76. Folini M, Gandellini P, Longoni N, Profumo V, Callari M, Pennati M, Colecchia M, Supino R, Veneroni S, Salvioni R et al (2010) miR‐21: an oncomir on strike in prostate cancer. Mol Cancer 9, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Buscaglia LE and Li Y (2011) Apoptosis and the target genes of microRNA‐21. Chin J Cancer 30, 371–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Feng YH and Tsao CJ (2016) Emerging role of microRNA‐21 in cancer. Biomed Rep 5, 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M et al (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 103, 2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Medina PP, Nolde M and Slack FJ (2010) OncomiR addiction in an in vivo model of microRNA‐21‐induced pre‐B‐cell lymphoma. Nature 467, 86–90. [DOI] [PubMed] [Google Scholar]
- 81. Tili E, Croce CM and Michaille JJ (2009) miR‐155: on the crosstalk between inflammation and cancer. Int Rev Immunol 28, 264–284. [DOI] [PubMed] [Google Scholar]
- 82. Van Roosbroeck K, Fanini F, Setoyama T, Ivan C, Rodriguez‐Aguayo C, Fuentes‐Mattei E, Xiao L, Vannini I, Redis RS, D'Abundo L et al (2017) Combining anti‐Mir‐155 with chemotherapy for the treatment of lung cancers. Clin Cancer Res 23, 2891–2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N and Croce CM (2006) Pre‐B cell proliferation and lymphoblastic leukemia/high‐grade lymphoma in E(mu)‐miR155 transgenic mice. Proc Natl Acad Sci USA 103, 7024–7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Dong Y, Liang G, Yuan B, Yang C, Gao R and Zhou X (2015) MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol 36, 1477–1486. [DOI] [PubMed] [Google Scholar]
- 85. Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E et al (2003) MALAT‐1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early‐stage non‐small cell lung cancer. Oncogene 22, 8031–8041. [DOI] [PubMed] [Google Scholar]
- 86. Li L, Chen H, Gao Y, Wang YW, Zhang GQ, Pan SH, Ji L, Kong R, Wang G, Jia YH et al (2016) Long noncoding RNA MALAT1 promotes aggressive pancreatic cancer proliferation and metastasis via the stimulation of autophagy. Mol Cancer Ther 15, 2232–2243. [DOI] [PubMed] [Google Scholar]
- 87. Ren S, Liu Y, Xu W, Sun Y, Lu J, Wang F, Wei M, Shen J, Hou J, Gao X et al (2013) Long noncoding RNA MALAT‐1 is a new potential therapeutic target for castration resistant prostate cancer. J Urol 190, 2278–2287. [DOI] [PubMed] [Google Scholar]
- 88. Zhou Y, Xu X, Lv H, Wen Q, Li J, Tan L, Li J and Sheng X (2016) The long noncoding RNA MALAT‐1 is highly expressed in ovarian cancer and induces cell growth and migration. PLoS One 11, e0155250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. West JA, Davis CP, Sunwoo H, Simon MD, Sadreyev RI, Wang PI, Tolstorukov MY and Kingston RE (2014) The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol Cell 55, 791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Tripathi V, Shen Z, Chakraborty A, Giri S, Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A et al (2013) Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B‐MYB. PLoS Genet 9, e1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Cai B, Song XQ, Cai JP and Zhang S (2014) HOTAIR: a cancer‐related long non‐coding RNA. Neoplasma 61, 379–391. [DOI] [PubMed] [Google Scholar]
- 92. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL et al (2010) Long non‐coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Pandey GK, Mitra S, Subhash S, Hertwig F, Kanduri M, Mishra K, Fransson S, Ganeshram A, Mondal T, Bandaru S et al (2014) The risk‐associated long noncoding RNA NBAT‐1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell 26, 722–737. [DOI] [PubMed] [Google Scholar]
- 94. Nie FQ, Sun M, Yang JS, Xie M, Xu TP, Xia R, Liu YW, Liu XH, Zhang EB, Lu KH et al (2015) Long noncoding RNA ANRIL promotes non‐small cell lung cancer cell proliferation and inhibits apoptosis by silencing KLF2 and P21 expression. Mol Cancer Ther 14, 268–277. [DOI] [PubMed] [Google Scholar]
- 95. Zhang EB, Kong R, Yin DD, You LH, Sun M, Han L, Xu TP, Xia R, Yang JS, De W et al (2014) Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR‐99a/miR‐449a. Oncotarget 5, 2276–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Raveh E, Matouk IJ, Gilon M and Hochberg A (2015) The H19 Long non‐coding RNA in cancer initiation, progression and metastasis – a proposed unifying theory. Mol Cancer 14, 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kallen AN, Zhou XB, Xu J, Qiao C, Ma J, Yan L, Lu L, Liu C, Yi JS, Zhang H et al (2013) The imprinted H19 lncRNA antagonizes let‐7 microRNAs. Mol Cell 52, 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Imig J, Brunschweiger A, Brummer A, Guennewig B, Mittal N, Kishore S, Tsikrika P, Gerber AP, Zavolan M and Hall J (2015) miR‐CLIP capture of a miRNA targetome uncovers a lincRNA H19‐miR‐106a interaction. Nat Chem Biol 11, 107–114. [DOI] [PubMed] [Google Scholar]
- 99. Cai X and Cullen BR (2007) The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 13, 313–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Reik W, Brown KW, Slatter RE, Sartori P, Elliott M and Maher ER (1994) Allelic methylation of H19 and IGF2 in the Beckwith‐Wiedemann syndrome. Hum Mol Genet 3, 1297–1301. [DOI] [PubMed] [Google Scholar]
- 101. Yoshimizu T, Miroglio A, Ripoche MA, Gabory A, Vernucci M, Riccio A, Colnot S, Godard C, Terris B, Jammes H et al (2008) The H19 locus acts in vivo as a tumor suppressor. Proc Natl Acad Sci USA 105, 12417–12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Matouk IJ, DeGroot N, Mezan S, Ayesh S, Abu‐lail R, Hochberg A and Galun E (2007) The H19 non‐coding RNA is essential for human tumor growth. PLoS One 2, e845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Ikeda S, Kong SW, Lu J, Bisping E, Zhang H, Allen PD, Golub TR, Pieske B and Pu WT (2007) Altered microRNA expression in human heart disease. Physiol Genomics 31, 367–373. [DOI] [PubMed] [Google Scholar]
- 104. Boon RA and Dimmeler S (2015) MicroRNAs in myocardial infarction. Nat Rev Cardiol 12, 135–142. [DOI] [PubMed] [Google Scholar]
- 105. Chen JF, Murchison EP, Tang R, Callis TE, Tatsuguchi M, Deng Z, Rojas M, Hammond SM, Schneider MD, Selzman CH et al (2008) Targeted deletion of Dicer in the heart leads to dilated cardiomyopathy and heart failure. Proc Natl Acad Sci USA 105, 2111–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. da Costa Martins PA, Bourajjaj M, Gladka M, Kortland M, van Oort RJ, Pinto YM, Molkentin JD and De Windt LJ (2008) Conditional dicer gene deletion in the postnatal myocardium provokes spontaneous cardiac remodeling. Circulation 118, 1567–1576. [DOI] [PubMed] [Google Scholar]
- 107. Suarez Y, Fernandez‐Hernando C, Pober JS and Sessa WC (2007) Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res 100, 1164–1173. [DOI] [PubMed] [Google Scholar]
- 108. Fiedler J, Jazbutyte V, Kirchmaier BC, Gupta SK, Lorenzen J, Hartmann D, Galuppo P, Kneitz S, Pena JT, Sohn‐Lee C et al (2011) MicroRNA‐24 regulates vascularity after myocardial infarction. Circulation 124, 720–730. [DOI] [PubMed] [Google Scholar]
- 109. Kumar S, Kim CW, Simmons RD and Jo H (2014) Role of flow‐sensitive microRNAs in endothelial dysfunction and atherosclerosis: mechanosensitive athero‐miRs. Arterioscler Thromb Vasc Biol 34, 2206–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, Hristov M, Koppel T, Jahantigh MN, Lutgens E et al (2009) Delivery of microRNA‐126 by apoptotic bodies induces CXCL12‐dependent vascular protection. Science Signaling 2, ra81. [DOI] [PubMed] [Google Scholar]
- 111. Wang S, Aurora AB, Johnson BA, Qi X, McAnally J, Hill JA, Richardson JA, Bassel‐Duby R and Olson EN (2008) The endothelial‐specific microRNA miR‐126 governs vascular integrity and angiogenesis. Dev Cell 15, 261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Bostjancic E, Zidar N, Stajer D and Glavac D (2010) MicroRNAs miR‐1, miR‐133a, miR‐133b and miR‐208 are dysregulated in human myocardial infarction. Cardiology 115, 163–169. [DOI] [PubMed] [Google Scholar]
- 113. Satoh M, Minami Y, Takahashi Y, Tabuchi T and Nakamura M (2010) Expression of microRNA‐208 is associated with adverse clinical outcomes in human dilated cardiomyopathy. J Cardiac Fail 16, 404–410. [DOI] [PubMed] [Google Scholar]
- 114. van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J and Olson EN (2007) Control of stress‐dependent cardiac growth and gene expression by a microRNA. Science 316, 575–579. [DOI] [PubMed] [Google Scholar]
- 115. Callis TE, Pandya K, Seok HY, Tang RH, Tatsuguchi M, Huang ZP, Chen JF, Deng Z, Gunn B, Shumate J et al (2009) MicroRNA‐208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Investig 119, 2772–2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Hullinger TG, Montgomery RL, Seto AG, Dickinson BA, Semus HM, Lynch JM, Dalby CM, Robinson K, Stack C, Latimer PA et al (2012) Inhibition of miR‐15 protects against cardiac ischemic injury. Circ Res 110, 71–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Zidar N, Bostjancic E, Glavac D and Stajer D (2011) MicroRNAs, innate immunity and ventricular rupture in human myocardial infarction. Dis Markers 31, 259–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Wu T, Wu D, Wu Q, Zou B, Huang X, Cheng X, Wu Y, Hong K, Li P, Yang R et al (2017) Knockdown of long non‐coding RNA‐ZFAS1 protects cardiomyocytes against acute myocardial infarction via anti‐apoptosis by regulating miR‐150/CRP. J Cell Biochem 118, 3281–3289. [DOI] [PubMed] [Google Scholar]
- 119. Devaux Y, Vausort M, McCann GP, Zangrando J, Kelly D, Razvi N, Zhang L, Ng LL, Wagner DR and Squire IB (2013) MicroRNA‐150: a novel marker of left ventricular remodeling after acute myocardial infarction. Circ Cardiovasc Genet 6, 290–298. [DOI] [PubMed] [Google Scholar]
- 120. Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, Doevendans PA, Mummery CL, Borlak J, Haverich A et al (2007) MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation 116, 258–267. [DOI] [PubMed] [Google Scholar]
- 121. Ucar A, Gupta SK, Fiedler J, Erikci E, Kardasinski M, Batkai S, Dangwal S, Kumarswamy R, Bang C, Holzmann A et al (2012) The miRNA‐212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat Commun 3, 1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Cheng Y and Zhang C (2010) MicroRNA‐21 in cardiovascular disease. Cardiovasc Transl Res 3, 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Liang H, Zhang C, Ban T, Liu Y, Mei L, Piao X, Zhao D, Lu Y, Chu W and Yang B (2012) A novel reciprocal loop between microRNA‐21 and TGFbetaRIII is involved in cardiac fibrosis. Int J Biochem Cell Biol 44, 2152–2160. [DOI] [PubMed] [Google Scholar]
- 124. Roy S, Khanna S, Hussain SR, Biswas S, Azad A, Rink C, Gnyawali S, Shilo S, Nuovo GJ and Sen CK (2009) MicroRNA expression in response to murine myocardial infarction: miR‐21 regulates fibroblast metalloprotease‐2 via phosphatase and tensin homologue. Cardiovasc Res 82, 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Bang C, Batkai S, Dangwal S, Gupta SK, Foinquinos A, Holzmann A, Just A, Remke J, Zimmer K, Zeug A et al (2014) Cardiac fibroblast‐derived microRNA passenger strand‐enriched exosomes mediate cardiomyocyte hypertrophy. J Clin Investig 124, 2136–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Jayawardena TM, Egemnazarov B, Finch EA, Zhang L, Payne JA, Pandya K, Zhang Z, Rosenberg P, Mirotsou M and Dzau VJ (2012) MicroRNA‐mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res 110, 1465–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Ikeda S, He A, Kong SW, Lu J, Bejar R, Bodyak N, Lee KH, Ma Q, Kang PM, Golub TR et al (2009) MicroRNA‐1 negatively regulates expression of the hypertrophy‐associated calmodulin and Mef2a genes. Mol Cell Biol 29, 2193–2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Rau F, Freyermuth F, Fugier C, Villemin JP, Fischer MC, Jost B, Dembele D, Gourdon G, Nicole A, Duboc D et al (2011) Misregulation of miR‐1 processing is associated with heart defects in myotonic dystrophy. Nat Struct Mol Biol 18, 840–845. [DOI] [PubMed] [Google Scholar]
- 129. Karakikes I, Chaanine AH, Kang S, Mukete BN, Jeong D, Zhang S, Hajjar RJ and Lebeche D (2013) Therapeutic cardiac‐targeted delivery of miR‐1 reverses pressure overload‐induced cardiac hypertrophy and attenuates pathological remodeling. J Am Heart Assoc 2, e000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Bagnall RD, Tsoutsman T, Shephard RE, Ritchie W and Semsarian C (2012) Global microRNA profiling of the mouse ventricles during development of severe hypertrophic cardiomyopathy and heart failure. PLoS One 7, e44744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Wei Y, Peng S, Wu M, Sachidanandam R, Tu Z, Zhang S, Falce C, Sobie EA, Lebeche D and Zhao Y (2014) Multifaceted roles of miR‐1s in repressing the fetal gene program in the heart. Cell Res 24, 278–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Lagos‐Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W and Tuschl T (2002) Identification of tissue‐specific microRNAs from mouse. Curr Biol 12, 735–739. [DOI] [PubMed] [Google Scholar]
- 133. Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL and Wang DZ (2006) The role of microRNA‐1 and microRNA‐133 in skeletal muscle proliferation and differentiation. Nat Genet 38, 228–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang ML, Segnalini P, Gu Y, Dalton ND et al (2007) MicroRNA‐133 controls cardiac hypertrophy. Nat Med 13, 613–618. [DOI] [PubMed] [Google Scholar]
- 135. Duisters RF, Tijsen AJ, Schroen B, Leenders JJ, Lentink V, van der Made I, Herias V, van Leeuwen RE, Schellings MW, Barenbrug P et al (2009) miR‐133 and miR‐30 regulate connective tissue growth factor: implications for a role of microRNAs in myocardial matrix remodeling. Circ Res 104, 170–178, 6p following 178. [DOI] [PubMed] [Google Scholar]
- 136. Castaldi A, Zaglia T, Di Mauro V, Carullo P, Viggiani G, Borile G, Di Stefano B, Schiattarella GG, Gualazzi MG, Elia L et al (2014) MicroRNA‐133 modulates the beta1‐adrenergic receptor transduction cascade. Circ Res 115, 273–283. [DOI] [PubMed] [Google Scholar]
- 137. Yang KC, Yamada KA, Patel AY, Topkara VK, George I, Cheema FH, Ewald GA, Mann DL and Nerbonne JM (2014) Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation 129, 1009–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Viereck J, Kumarswamy R, Foinquinos A, Xiao K, Avramopoulos P, Kunz M, Dittrich M, Maetzig T, Zimmer K, Remke J et al (2016) Long noncoding RNA Chast promotes cardiac remodeling. Sci Transl Med 8, 326ra22. [DOI] [PubMed] [Google Scholar]
- 139. Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S et al (2014) Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res 114, 1389–1397. [DOI] [PubMed] [Google Scholar]
- 140. Liu JY, Yao J, Li XM, Song YC, Wang XQ, Li YJ, Yan B and Jiang Q (2014) Pathogenic role of lncRNA‐MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis 5, e1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Gast M, Schroen B, Voigt A, Haas J, Kuehl U, Lassner D, Skurk C, Escher F, Wang X, Kratzer A et al (2016) Long noncoding RNA MALAT1‐derived mascRNA is involved in cardiovascular innate immunity. J Mol Cell Biol 8, 178–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Wang YN, Shan K, Yao MD, Yao J, Wang JJ, Li X, Liu B, Zhang YY, Ji Y, Jiang Q et al (2016) Long noncoding RNA‐GAS5: a novel regulator of hypertension‐induced vascular remodeling. Hypertension, 68, 736–748. [DOI] [PubMed] [Google Scholar]
- 143. Wang K, Long B, Zhou LY, Liu F, Zhou QY, Liu CY, Fan YY and Li PF (2014) CARL lncRNA inhibits anoxia‐induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR‐539‐dependent PHB2 downregulation. Nat Commun 5, 3596. [DOI] [PubMed] [Google Scholar]
- 144. Ounzain S, Micheletti R, Beckmann T, Schroen B, Alexanian M, Pezzuto I, Crippa S, Nemir M, Sarre A, Johnson R et al (2015) Genome‐wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart‐specific long non‐coding RNAs. Eur Heart J 36, 353–368a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Wang K, Liu F, Zhou LY, Long B, Yuan SM, Wang Y, Liu CY, Sun T, Zhang XJ and Li PF (2014) The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR‐489. Circ Res 114, 1377–1388. [DOI] [PubMed] [Google Scholar]
- 146. Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, Miyamoto Y, Ikegawa S, Kamatani N, Hori M et al (2006) Identification of a novel non‐coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet 51, 1087–1099. [DOI] [PubMed] [Google Scholar]
- 147. Eicher JD, Chami N, Kacprowski T, Nomura A, Chen MH, Yanek LR, Tajuddin SM, Schick UM, Slater AJ, Pankratz N et al (2016) Platelet‐Related Variants Identified by Exomechip Meta‐analysis in 157,293 Individuals. Am J Hum Genet 99, 40–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Frade AF, Laugier L, Ferreira LR, Baron MA, Benvenuti LA, Teixeira PC, Navarro IC, Cabantous S, Ferreira FM, da Silva Candido D et al (2016) Myocardial infarction‐associated transcript, a long noncoding RNA, is overexpressed during dilated cardiomyopathy due to chronic chagas disease. J Infect Dis 214, 161–165. [DOI] [PubMed] [Google Scholar]
- 149. Ziats MN and Rennert OM (2014) Identification of differentially expressed microRNAs across the developing human brain. Mol Psychiatry 19, 848–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Minones‐Moyano E, Porta S, Escaramis G, Rabionet R, Iraola S, Kagerbauer B, Espinosa‐Parrilla Y, Ferrer I, Estivill X and Marti E (2011) MicroRNA profiling of Parkinson's disease brains identifies early downregulation of miR‐34b/c which modulate mitochondrial function. Hum Mol Genet 20, 3067–3078. [DOI] [PubMed] [Google Scholar]
- 151. Thome AD, Harms AS, Volpicelli‐Daley LA and Standaert DG (2016) microRNA‐155 regulates alpha‐synuclein‐induced inflammatory responses in models of Parkinson disease. J Neurosci 36, 2383–2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Li D, Yang H, Ma J, Luo S, Chen S and Gu Q (2018) MicroRNA‐30e regulates neuroinflammation in MPTP model of Parkinson's disease by targeting Nlrp3. Hum Cell 31, 106–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Wang H, Ye Y, Zhu Z, Mo L, Lin C, Wang Q, Wang H, Gong X, He X, Lu G et al (2016) MiR‐124 regulates apoptosis and autophagy process in MPTP model of Parkinson's disease by targeting to bim. Brain Pathol 26, 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Hebert SS, Horre K, Nicolai L, Papadopoulou AS, Mandemakers W, Silahtaroglu AN, Kauppinen S, Delacourte A and De Strooper B (2008) Loss of microRNA cluster miR‐29a/b‐1 in sporadic Alzheimer's disease correlates with increased BACE1/beta‐secretase expression. Proc Natl Acad Sci USA 105, 6415–6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Kole AJ, Swahari V, Hammond SM and Deshmukh M (2011) miR‐29b is activated during neuronal maturation and targets BH3‐only genes to restrict apoptosis. Genes Dev 25, 125–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Pereira PA, Tomas JF, Queiroz JA, Figueiras AR and Sousa F (2016) Recombinant pre‐miR‐29b for Alzheimer s disease therapeutics. Sci Rep 6, 19946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Muller M, Jakel L, Bruinsma IB, Claassen JA, Kuiperij HB and Verbeek MM (2016) MicroRNA‐29a is a candidate biomarker for Alzheimer's disease in cell‐free cerebrospinal fluid. Mol Neurobiol 53, 2894–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Kim J, Yoon H, Ramirez CM, Lee SM, Hoe HS, Fernandez‐Hernando C and Kim J (2012) MiR‐106b impairs cholesterol efflux and increases Abeta levels by repressing ABCA1 expression. Exp Neurol 235, 476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Wang H, Liu J, Zong Y, Xu Y, Deng W, Zhu H, Liu Y, Ma C, Huang L, Zhang L et al (2010) miR‐106b aberrantly expressed in a double transgenic mouse model for Alzheimer's disease targets TGF‐beta type II receptor. Brain Res 1357, 166–174. [DOI] [PubMed] [Google Scholar]
- 160. Huang W, Li Z, Zhao L and Zhao W (2017) Simvastatin ameliorate memory deficits and inflammation in clinical and mouse model of Alzheimer's disease via modulating the expression of miR‐106b. Biomed Pharmacother 92, 46–57. [DOI] [PubMed] [Google Scholar]
- 161. Sarkar S, Jun S, Rellick S, Quintana DD, Cavendish JZ and Simpkins JW (2016) Expression of microRNA‐34a in Alzheimer's disease brain targets genes linked to synaptic plasticity, energy metabolism, and resting state network activity. Brain Res 1646, 139–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Zovoilis A, Agbemenyah HY, Agis‐Balboa RC, Stilling RM, Edbauer D, Rao P, Farinelli L, Delalle I, Schmitt A, Falkai P et al (2011) microRNA‐34c is a novel target to treat dementias. EMBO J 30, 4299–4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Cheng PH, Li CL, Chang YF, Tsai SJ, Lai YY, Chan AW, Chen CM and Yang SH (2013) miR‐196a ameliorates phenotypes of Huntington disease in cell, transgenic mouse, and induced pluripotent stem cell models. Am J Hum Genet 93, 306–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Kye MJ, Niederst ED, Wertz MH, Goncalves Ido C, Akten B, Dover KZ, Peters M, Riessland M, Neveu P, Wirth B et al (2014) SMN regulates axonal local translation via miR‐183/mTOR pathway. Hum Mol Genet 23, 6318–6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G III, Kenny PJ and Wahlestedt C (2008) Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed‐forward regulation of beta‐secretase. Nat Med 14, 723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Tollervey JR, Curk T, Rogelj B, Briese M, Cereda M, Kayikci M, Konig J, Hortobagyi T, Nishimura AL, Zupunski V et al (2011) Characterizing the RNA targets and position‐dependent splicing regulation by TDP‐43. Nat Neurosci 14, 452–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Nishimoto Y, Nakagawa S, Hirose T, Okano HJ, Takao M, Shibata S, Suyama S, Kuwako K, Imai T, Murayama S et al (2013) The long non‐coding RNA nuclear‐enriched abundant transcript 1_2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol Brain 6, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Sunwoo JS, Lee ST, Im W, Lee M, Byun JI, Jung KH, Park KI, Jung KY, Lee SK, Chu K et al (2017) Altered expression of the long noncoding RNA NEAT1 in Huntington's disease. Mol Neurobiol 54, 1577–1586. [DOI] [PubMed] [Google Scholar]
- 169. Carrieri C, Forrest AR, Santoro C, Persichetti F, Carninci P, Zucchelli S and Gustincich S (2015) Expression analysis of the long non‐coding RNA antisense to Uchl1 (AS Uchl1) during dopaminergic cells’ differentiation in vitro and in neurochemical models of Parkinson's disease. Front Cell Neurosci 9, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Lipovich L, Dachet F, Cai J, Bagla S, Balan K, Jia H and Loeb JA (2012) Activity‐dependent human brain coding/noncoding gene regulatory networks. Genetics 192, 1133–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Zhang QS, Wang ZH, Zhang JL, Duan YL, Li GF and Zheng DL (2016) Beta‐asarone protects against MPTP‐induced Parkinson's disease via regulating long non‐coding RNA MALAT1 and inhibiting alpha‐synuclein protein expression. Biomed Pharmacother 83, 153–159. [DOI] [PubMed] [Google Scholar]
- 172. Wang S, Zhang X, Guo Y, Rong H and Liu T (2017) The long noncoding RNA HOTAIR promotes Parkinson's disease by upregulating LRRK2 expression. Oncotarget 8, 24449–24456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Adams BD, Parsons C, Walker L, Zhang WC and Slack FJ (2017) Targeting noncoding RNAs in disease. J Clin Investig 127, 761–771. [DOI] [PMC free article] [PubMed] [Google Scholar]


