Abstract
Since their discovery, the study of maternal mRNAs has led to the identification of mechanisms underlying their spatiotemporal regulation within the context of oogenesis and early embryogenesis. Following synthesis in the oocyte, maternal mRNAs are translationally silenced and sequestered into storage in cytoplasmic granules. At the same time, their unique distribution patterns throughout the oocyte and embryo are tightly controlled and connected to their functions in downstream embryonic processes. At certain points in oogenesis and early embryogenesis, maternal mRNAs are translationally activated to perform their functions in a timely manner. The cytoplasmic polyadenylation machinery is responsible for the translational activation of maternal mRNAs, and its role in initiating the maternal to zygotic transition events has recently come to light. Here, we summarize the current knowledge on maternal mRNA regulation, with particular focus on cytoplasmic polyadenylation as a mechanism for translational regulation.
Keywords: cytoplasmic polyadenylation, maternal mRNA, maternal to zygotic transition, translational regulation
Abbreviations
CPE, cytoplasmic polyadenylation element
CPEB, CPE‐binding protein
GV, germinal vesicle
MBT, mid‐blastula transition
MPF, maturation promoting factor
MZT, maternal‐to‐zygotic transition
PAS, polyadenylation site
SCA, subcortical aggregates
snRNA, small nuclear RNA
In the early 1950s, Alexander Neyfakh observed that different doses of X‐ray irradiation cause different effects in the fertilized eggs of the loach (Misgurnus fossilis) (Fig. 1A). A lower dose caused damage to the nucleus and triggered the arrest of development at late blastula stage. A higher dose of irradiation caused damage to the cytoplasm followed by an almost immediate developmental arrest 1. Neyfakh's observation became the basis for the idea of a separate genetic function of the nucleus and cytoplasm, where the former (defined as the ‘morphogenetic function’) required only starting from late blastula, and the latter being indispensable from the very first steps of development. This idea, which had been validated by X‐ray irradiation studies in fish, amphibians, echinoderms, worms, and molluscs 2, was in line with the observations that embryos resulting from interspecies hybrids of fishes, amphibians, and echinoderms stop developing at approximately the same stage of late blastula 3, 4, presumably due to the discrepancy between their cytoplasmic and nuclear genetic programs.
Figure 1.
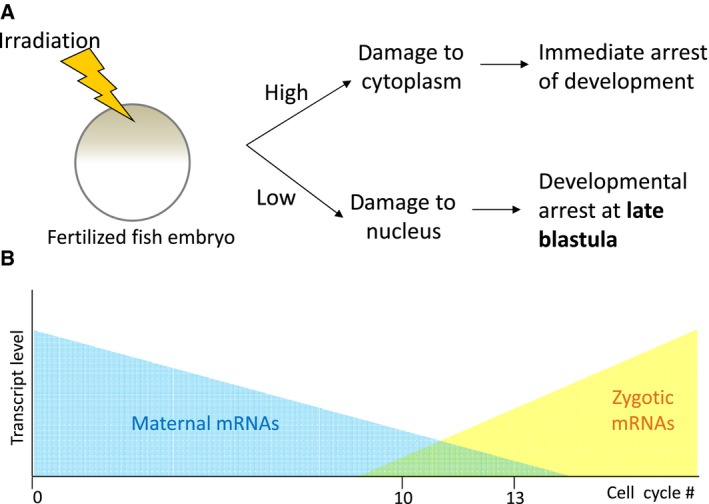
Maternal mRNAs control development up to the point when zygotic genome is activated during the MZT occurring at late blastula in fishes and amphibians. (A) Neyfakh's experiment with irradiated loach embryos show different phenotypes depending on the doses of radiation which affects either cytoplasm or nucleus. The delayed developmental arrest resulting upon nuclear damage gave rise to the understanding of the morphogenetic function of the nuclei, which is responsible for development from late blastula onward. (B) Current understanding at the molecular level established that maternal mRNAs present at high levels prior to MZT drives development up to this point, before it is taken over by zygotic genes expressed from here onward.
Depending on the size of an egg and the amount of cytoplasm deposited into an egg by the mother in the species studied, fertilization is followed by a period of transcriptional quiescence of varying length when the genome of the embryo which is contained in the nucleus is not yet expressed. During this period of transcriptional silence, development is driven by cytoplasmic factors which largely consist of maternally deposited mRNAs. At a certain point in embryogenesis, maternal mRNAs are degraded and the zygotic genome becomes active as manifested by an activation of transcription 5, 6, thus developmental control is passed on from maternal components to that of zygotic ones (Fig. 1B). As it turned to be, Neyfakh's ‘morphogenetic function of the nucleus’ consists of the zygotic genome activation (ZGA, i.e., expression of the genome of the embryo proper) and is a crucial part of the mid‐blastula transition (MBT), which also includes the asynchronization of the cell cycle 7 and acquisition of cell motility 8, 9. In fish and frogs, the occurrence of MBT was shown to be dependent on the nuclear to cytoplasmic ratio which increases with each cell division, causing a titration of maternal suppressors which, at a critical point, allows the activation of zygotic transcription 8, 10, 11. In parallel, some studies in insects and mammals led to the development of the maternal‐to‐zygotic transition (MZT) concept, which is equivalent to that of MBT in fish and amphibians. Later on, the concept of MZT was extended to other changes at the molecular as well as cellular level. These include the clearance of maternal mRNAs 12, 13, 14, 15, 16, 17, 18 and major epigenetic rearrangements 19, 20, 21. The consequence of these events culminates in the initiation of gastrulation, during which cells migrate and form the three germ layers.
Despite an absence of transcriptional input from the embryonic genome up to the point of MBT/MZT, developmental processes dynamically progress throughout early embryogenesis. This is possible thanks to the maternally deposited factors, which include a large pool of maternal mRNAs. In order to ensure progression through different developmental milestones, the activity of maternal mRNAs is confined to a precise time and space. In this Review, we will describe how the maternal mRNAs are spatiotemporally organized in the oocyte and early embryo and how their activity is regulated during development.
The genesis of maternal mRNAs
Before we discuss the regulation of maternal mRNAs, it is necessary to understand their origin. The process of oogenesis in all model organisms studied includes two meiotic arrests. The first arrest occurs during prophase I, which is also known as the period of oocyte growth. During this phase, the oocyte accumulates maternal components required for subsequent maturation and embryonic development. In some organisms such as Drosophila, maternal mRNAs are transcribed by cells supporting the oocyte, called nurse cells 22. These maternal RNAs are subsequently transferred to the oocyte by microtubules, cytoskeletal networks, and cytoplasmic streaming 23, 24, 25. During the period of oocyte growth in vertebrates, maternal RNAs are transcribed by the oocyte proper. From this time onward, the oocyte undergoes dynamic changes which include its final maturation, activation, and the earliest steps of embryonic development following its fertilization. These dynamic changes depend on maternal mRNAs activated according to precise timetable to confer functions required.
To prevent premature activation of developmental program, maternal mRNAs transcribed during the oocyte growth undergo translational repression. Their poly(A) tails are shortened immediately after their export into the cytoplasm. Injecting a tPA RNA with a long poly(A) tail into an immature oocyte causes its deadenylation, suggesting that the RNA‐silencing mechanism of oocyte depends on deadenylation 26. The immature oocytes of Xenopus and other species contain translationally dormant mRNAs with short poly(A) tails of about 20–40 nucleotides 27, 28, 29, 30, 31, 32, 33. These are stored for activation at appropriate developmental time points.
Spatial organization of maternal mRNAs in the oocyte and early embryo
All these studies raised a number of important questions, which need to be answered to understand the role of maternal RNAs in more detail. Where do translationally silent maternal mRNAs reside in the egg? What keeps them sequestered from the pervasive translation or degradation machinery in the cytoplasm? And finally, what mechanism ensures precise spatial and temporal activation of maternal mRNAs?
Many maternal mRNAs are spatially localized in the oocyte and early embryos. In Drosophila, more than 70% of the early embryonic transcriptome were found to be subcellularly localized 34. Localization of transcripts generally serves to confine their activity and function to the appropriate time and space. In parallel, it also facilitates interactions with important protein regulators. In Drosophila, localization of maternal mRNAs in the oocyte is known to be crucial to establish the first embryonic axis 35, while in germ cells, localization of oskar and nanos mRNAs to the posterior of the germ cell progenitor is necessary to select whether cell will become an oocyte or a nurse cell. As another example, the zebrafish mos mRNA is localized to the animal pole, where Zorba protein, required for its translational activation during oocyte maturation, is present 36. During development, differential localization of transcripts into subcellular or subembryonic domains is especially relevant in order to confer asymmetry, which will in turn define embryonic patterning. In early Xenopus oocyte, xdazl and nanos1 are localized to the vegetal cortex; in later stage oocytes, vg1 and vegt are transported to vegetal cortex and subsequently inherited during cell cleavage by vegetal blastomeres. Similarly, many maternal mRNAs are localized in zebrafish oocytes. These include the homologs of previously mentioned Xenopus mRNAs such as dazl and dvr1 37, 38.
The localization of maternal mRNAs in the fertilized egg and early embryo is driven by several different mechanisms 39, 40. On fertilization in Xenopus, the maternal transcripts of xCr1, a coreceptor of the Nodal signaling, are translationally activated in selected cells while repressed in others. This results in the accumulation of xCR1 specifically in the animal and marginal cells and its absence in vegetal cells 41. Localization of mRNAs in the early embryo can occur through cell division. In the early mouse embryo, the cell fate decision between cells of the inner cell mass (ICM) and trophoectoderm occurs due to localization of Cdx2 mRNA at the apical side of blastomeres at the 8‐ to 16‐cell stage and inherited by daughter cells upon division. Maternally inherited magoh transcripts encode a core component of the splicing‐dependent multiprotein exon junction complex deposited at the pre‐mRNA splice junctions. This complex includes several other maternal components (Rbm8A, Casc3, Eif4a3). As a component of exon junction complex, Magoh may influence downstream processes, including nuclear mRNA export, subcellular mRNA localization, translation efficiency, nonsense‐mediated mRNA decay and through its interaction with Pym1 inhibits apoptosis by regulating splicing of BCL2L1/Bcl‐X as well as some other apoptotic factors 42, 43, 44. Magoh is known for its role in germ plasm assembly and germline development in Drosophila and zebrafish 38. It is noteworthy that despite its uniform distribution in the 1‐ and 2‐cell stage zebrafish embryo and during late embryogenesis, these transcripts become transiently enriched in the central four blastomeres during 8–16 cell stage 45. This is of interest since these blastomeres contribute to the ectoderm giving rise to the neural tube and epidermis 46, 47. The functional implications of this phenomenon remain to be understood. All we know for now is that a deficiency of one of the members of the exon junction complex, EIF4A3, have been linked to craniofacial abnormalities with learning and language disabilities in human suggestive of defects in neural crest derivatives. In support of this role of EIF4A3, the zebrafish morphants show extensive apoptosis early on and craniofacial phenotype later 48. It remains to be seen how specific is the morphant phenotype and whether an accumulation of magoh transcripts in blastomeres giving rise to ectodermal derivatives have any connection to physiological apoptosis in the developing neural tube 49.
In some cases, certain localization signals are known to be crucial for mRNA localization into appropriate subcellular or subembryonic domains. The evolutionary conserved Nodal signaling acts as a determinant of dorsal cell fate and body axis later on. Squint is one of the Nodal ligands in zebrafish. Along with the Drosophila oskar, Xenopus vegt, the zebrafish squint belongs to a group of the so‐called ‘coding and non‐coding’ RNAs (cncRNAs) acting as key developmental regulators in plants and animals 50. At the 4‐cell stage, the maternal mRNA of squint, a ligand of the Nodal signaling, was found to be localized to two blastomeres giving rise to dorsal cell fates 51. This localization is driven by a structural motif in the form of a hairpin loop at the 3′UTR of the squint transcript 52. This motif is bound by the Ybx1 protein which prevents premature translation of squint 53. Of note, prior to the 16‐cell stage, squint is translationally repressed and is not polyadenylated. It undergoes cytoplasmic polyadenylation from the 16‐cell stage, consistent with a cohort of maternal mRNAs previously described by our group 53, 54, 55, 56. It is still unclear how the squint mRNA is transported to the prospective dorsal blastomeres, although its localization depends on microtubules 51. It is likely that squint transport follows a predominant mode of active translocation mediated by the cytoskeletal network of the cell. Interestingly, a similar structure motif was found in other components of the Nodal signaling pathway, which suggests common translational regulation of factors within the same pathway 57.
Microtubule‐based transport is known to be involved in localization of dorsal determinants in oocytes and early embryos of amphibians and fishes. In the zebrafish, the mRNA and protein of the maternal factors Syntabulin (Sybu) and Wnt8a are initially localized at the vegetal pole of the oocyte (Fig. 2). On egg activation and fertilization, they are translocated toward the future embryonic dorsal, where they activate the canonical Wnt signaling and induce the body axis 58, 59, 60. This transport was shown to be dependent on arrays of microtubules, which upon egg activation is organized at the vegetal pole of the egg in direction of the future embryonic dorsal side 61. Grip2a, another maternal factor with similar dynamics to that of Sybu and Wnt8a, is responsible for the organization of microtubules at the vegetal pole of the activated egg, which represents the initial step in breaking the asymmetry before the long‐range transport of these factors to the dorsal side 62. The failure of microtubule rearrangement in grip2a mutants results in the failure of wnt8a mRNA translocation to the future embryonic dorsal and hence axis induction defects. Surprisingly, a more recently performed loss of function analysis of wnt8a by Hino and colleagues revealed that maternal Wnt8a is likely to be dispensable for the initial dorsal axis determination 63; nevertheless, it cooperates with zygotically expressed Wnt8a in ventrolateral and posterior tissue formation. The same study also identified Wnt6a as an alternative candidate for dorsal determinant which showed similar expression pattern and localization as wnt8a. Among other maternal dorsal determinants located initially at the vegetal pole is Vrtn, whose activity could be suppressed by the putative vegetally localized antagonists. Vrtn binds the bmp2b regulatory sequence and represses its zygotic transcription 64.
Figure 2.
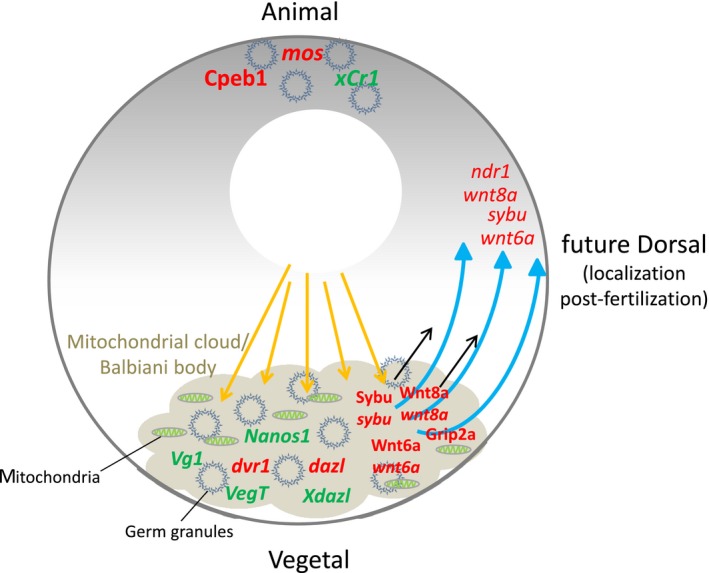
Distribution of key maternal factors in the oocyte. Studies in different organisms have shown that maternal mRNAs are organized in cytoplasmic granules together with several regulatory proteins responsible for their post‐transcriptional processing and thus translational regulation. In fish and amphibians, a large structure known as mitochondrial cloud or Balbiani body is present at the vegetal pole of the oocyte. This structure consists of a large accumulation of mitochondria and cytoplasmic granules (specifically termed germ granules) containing silenced mRNAs. The mitochondrial cloud serves as a vehicle for transporting and localizing maternal factors to the vegetal cortex during oogenesis by means of microtubule network and motor proteins (yellow arrows). At egg activation and fertilization, Sybu and Wnt8 are translocated to the future dorsal axis through microtubule‐mediated transport (blue arrows).
Storage of maternal mRNAs in cytoplasmic granules
Transported mRNAs are often present in RNP complexes with RNA‐binding proteins, most of which serve to regulate mRNA processing and repress their translation 65. These RNP complexes are actively transported along the cell's cytoskeletal network in a directed fashion to their rightful subcellular localization. Once the complex reaches its destination, and upon appropriate biological cues, the RNP complex is remodeled and translational activation of these mRNAs ensues. These RNP complexes are often organized into membrane‐less structures known as cytoplasmic granules.
In metazoan cells, structures known as cytoplasmic granules host specific molecular processes pertaining to the inhibition of mRNA translation or degradation 66, 67, 68, 69. These subcellular structures lack membrane and contain proteins involved in specific RNA metabolism function. The organization of maternal mRNAs into cytoplasmic granules would reasonably facilitate their translational suppression by common factors residing in the granules; at the same time, it would also facilitate collective transport to different subcellular domains and rapid response to developmental cues as the lack of membrane of these granules would allow turnover rate in the order of seconds 70. These granules are indeed dynamic in nature—their formation depends on the steady‐state supply of mRNAs subjected to the particular molecular process of the specific granule type. Interestingly, the types of granules present in a cell are regulated in a precise spatial and temporal manner during development as different combinations of granule types each of which perform distinct functions are present in different cell types, according to specific developmental stage 71.
One of the most common and well‐characterized types of cytoplasmic granules are the processing bodies (P‐bodies) known as sites of mRNA repression, decapping, and degradation. P‐bodies contain a core set of decapping enzymes—DCP1 and DCP2, and other accessory proteins such as the DEAD‐box RNA helicase (Dhh1). In the Caenorhabditis elegans oocyte, a subset of maternal mRNAs is bound to the homolog of Dhh1 (CGH‐1). Binding of mRNA to CGH‐1 causes its stabilization and repression and directs them to a distinct type of RNA granule 72, 73. Unlike P‐bodies, these CGH‐1 granules lack decapping activity and were found to contain only repressed maternal mRNAs and some interactors of CGH‐1 such as the Ataxin2 and PABP homologs. These translational regulators are also found in stress granules—another type of granules known to sequester and transiently suppress the translation of mRNAs during stress 72. Therefore, in the C. elegans oocyte, maternal mRNA association with CGH‐1 and translational regulators nucleates the formation of localized cytoplasmic foci in the form of cytoplasmic granule where translationally repressed maternal mRNAs are stabilized and sequestered.
During oogenesis in Drosophila, different types of germ granules exist at different stages and subcellular locations. These granules differ by mRNA composition. However, a single type of granule may differ in mRNA composition at different developmental stages. The germ granules are typically up to 500 nm large and contain core germ plasm proteins Oskar, Vasa, and Tudor as well as other proteins involved in RNA processing 69, 74. They are also sites of active translation containing large numbers of polysomes 75. Sponge bodies are a type of granules found in the nurse cells and oocyte. It is identified by the presence of Exuperantia which is required for the localization of bicoid RNA to the anterior pole of the oocyte 25. Sponge bodies exhibit dynamic movements between the nurse cells and oocyte and are known to contain different compositions of mRNAs in different subcellular locations, therefore suggesting dynamic exchange of their contents which is coupled to their transport 76. The polar granule is another type of granule found in the late‐stage oocytes. These granules contain materials transported from the nurse cells to the posterior part of the oocyte. Similar to sponge bodies and P‐granules, the polar granules also contain the decapping enzyme Dcp1 and DEAD‐box helicase Me31B 77, suggesting that these structures are closely related. In addition, they also contain Aubergine and Piwi involved in the piRNA pathway 78, 79. Piwi proteins and piRNAs are thought to protect the germ cells from the mutagenic activity of retrotransposons 80.
In the early Drosophila embryo, P‐bodies are sites for translational control of maternal mRNAs required to establish the embryonic axis. By electron microscopy, P‐bodies are identified as electron‐dense structures of about 60–80 nm in diameter‐lacking ribosomes and organized into distinct core and outer region 81. The core contains the Drosophila homolog of the DEAD‐box RNA Helicase Dhh1, Me31B. It contains repressed mRNAs and, therefore, may represent the site of translational repression. The outer region contains translational regulator proteins such as Sqd and Orb which are required for translational activation. Active mRNAs are anchored to the outer regions and translated there. Super‐resolution microscopy revealed that mRNAs within the granules form structured, homotypic clusters of single mRNA species localized in a defined position, whereas proteins are evenly distributed throughout the granule 82. Interestingly, the distribution of mRNA clusters within the granule itself does not seem to correlate with their translational activity or their protection from degradation. The homotypic organization of mRNAs into different subcompartments of the granule may allow storage and quick retrieval of specific mRNAs when needed. Such organization may also facilitate localized interactions between regulatory proteins and their target mRNA by providing high local concentrations of the target as well as the protein itself.
In mouse, P‐bodies are found in early‐stage oocytes arrested at prophase I, where they reside in perinuclear foci as well as at the cell cortex 83. The P‐bodies disappear during the period of oocyte growth, and some of their components including cytoplasmic polyadenylation element (CPE)‐binding protein (CPEB) and DDX6 become localized to the subcortical aggregates (SCA), which contain maternal mRNAs. In contrast to the typical P‐bodies, the SCA lack the decapping enzyme DCP1 implicated in mRNA degradation 84. This may reflect the function of SCA in maternal mRNA storage and repression but not degradation. This is similar to the CGH‐1 granules of C. elegans 72. On oocyte maturation, the SCA disappear, presumably releasing associated maternal mRNAs from repression.
In Xenopus oocytes, maternal mRNAs are localized and translationally repressed in the mitochondrial cloud—an electron dense structure containing germinal granules and associated with dense array of mitochondria (Fig. 2). This structure can be found in stage I oocytes at the future vegetal cortex 85, 86. Within the mitochondrial cloud, silenced mRNAs are organized within or around the germinal granules which aggregate at the vegetal pole. The mitochondrial cloud delivers germinal granules and localizes maternal mRNAs to the vegetal cortex by the messenger transport organizer (METRO) pathway during stage I and II of oogenesis. This mechanism works through the entrapment of germ plasm mRNAs by the dense ER network of the mitochondrial cloud and subsequent expansion of the cloud toward the vegetal cortex 87, 88.
A structure termed the Balbiani body analogous to the mitochondrial cloud could also be found in zebrafish 89. This structure resembles the mitochondrial cloud in that it associates with mitochondria as well as germ granules and their associated maternal mRNAs (Fig. 2). The Balbiani body is also known to be involved in transporting and localizing maternal mRNAs to the oocyte vegetal cortex 90, 91. The formation of the Balbiani body is linked to orientation of the chromosomal bouquet, a specific organization assumed by the chromosomes in the nucleus at the onset of meiosis, in a microtubule‐directed process 92. Subsequent to its formation, the Balbiani body associates with the oocyte cortex and releases its mRNPs containing most of the dorsal determinants and germ plasm components, including sybu and wnt8 discussed in previous section 38, 91, 93. The establishment of the Balbiani body at the vegetal pole, therefore, determines the polarity of the oocyte, which is in turn necessary to establish the embryonic axis and specify the germ line 61, 94, 95. To date, very few studies were performed on cytoplasmic granules in zebrafish embryogenesis or for that matter during the embryogenesis of vertebrates. However, it is known that the activation of zygotic transcription results in the formation of distinct types of nuclear granules including those involved in the processing of histone mRNAs and assembly of splicing machinery 96. Given the nature of cytoplasmic bodies in other systems known to be dependent on the presence of mRNA itself, it is plausible that the activation of zygotic transcription represents a trigger for the formation of such granules.
The timely activation of maternal mRNAs at developmental milestones
Up to the point of ZGA, the dynamic processes of oogenesis and embryogenesis rely solely on maternal mRNAs. The absence of transcriptional input from the zygotic genome necessitates regulation of maternal RNAs at the post‐transcriptional level in order to activate different subsets of mRNAs with precise developmental timing. Over the last two decades, several studies have identified translational control by cytoplasmic polyadenylation as one of the mechanisms to regulate the timely activation of maternal mRNAs during the development prior to MZT 28, 33, 97, 98, 99, 100, 101, 102, 103, 104, 105. In the mouse, Xenopus, and zebrafish, maternal mRNAs encoding key drivers of oogenesis and early embryonic development are deadenylated right after their synthesis during oocyte growth and stored to be reactivated at appropriate developmental milestones (Fig. 3). These deposited maternal mRNAs have a short poly(A) tail and are translationally inactive. A portion of the stored maternal mRNAs becomes dramatically adenylated at specific stages of oogenesis as well as upon fertilization, which correlates well with their recruitment into polysomes and translation initiation 29, 30, 56, 106, 107, 108. In Xenopus oocytes, the poly(A) tail could increase by as much as twofold or more, resulting in a corresponding increase of translation efficiency 29, 30, 108.
Figure 3.
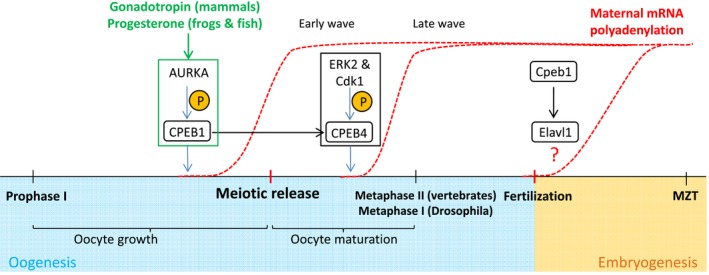
Different waves of maternal mRNA activation throughout oogenesis and early embryogenesis mediated by cytoplasmic polyadenylation. Maternal mRNAs are synthesized throughout the period of oocyte growth and stored in a dormant state. The first wave of activation is stimulated by GH or progesterone, resulting in phosphorylation of CPEB1 by Aurora kinase and the cytoplasmic polyadenylation of several maternal factors required for oocyte maturation. Another late wave is in turn mediated by CPEB4 whose translation was activated during the early wave and is phosphorylated by ERK2 and Cdk1 kinases. Following fertilization, another wave of cytoplasmic polyadenylation occurs on thousands of maternal transcripts. This later wave is required for the embryo to undergo a proper process of MZT. However, the exact molecular mechanism regulating this third wave of cytoplasmic polyadenylation has not yet been worked out.
The initiation of oocyte maturation represents the first point of activation of stored maternal mRNAs. In most vertebrates, upon ovulation, meiosis is reinitiated and arrested a second time in metaphase II 109, whereas in Drosophila, meiotic arrest takes place in metaphase I 110. The point of meiotic release from prophase I is known as oocyte maturation which occurs upon stimulation by hormones—gonadotropin in mouse 73 and progesterone in Xenopus and zebrafish 111, 112 or exposure to a particular signal released by sperm in C. elegans 113, 114. During its maturation, the oocyte undergoes two consecutive M‐phases without an intervening S‐phase. As a result of this maturation, the oocyte can be fertilized, upon which the completion of meiosis ensues and embryonic development initiates. One of the earliest transcripts to be translationally activated upon oocyte maturation is c‐Mos kinase 115. The activity of this kinase is crucial to initiate a subsequent series of cytoplasmic polyadenylation and translation of other mRNAs’ encoding factors essential for oocyte maturation such as Cyclin B1 116, 117, 118, 119. Loss of function of these factors, overexpression, or prevention of their translational activation was shown to prevent oocyte maturation, indicating that their timely activation is essential 119, 120. In the zebrafish and mouse oocytes, cyclin B1 mRNAs are repressed and stored in cytoplasmic RNA granules which disassemble during oocyte maturation. The formation of these granules is required for the translational repression, while their disassembly coincides with translational activation of cyclin B1 mRNAs. Disruption of the granules results in premature translation of Cyclin B1, which negatively affects oocyte maturation 121.
The second event of maternal mRNA activation occurs during embryonic development, which in some species is triggered by fertilization. In zebrafish, a large cohort of maternal mRNA was found to undergo cytoplasmic polyadenylation prior to the MBT 54. This process was shown to correlate with the increase in mRNA association with polysomes. It is essential for developmental progression past MBT/MZT 56.
The molecular machinery of cytoplasmic polyadenylation
Polyadenylation of eukaryotic mRNAs is an inherent part of gene expression. It involves the addition of multiple adenine bases to the 3′ end of the mature mRNA. Polyadenylation of mRNA is tightly linked to its stability and translatability. All mRNAs undergo nuclear polyadenylation following their transcription and maturation 122, 123. This process is dictated by a hexamer motif at the 3′UTR of the mRNA known as the nuclear polyadenylation sequence (AAUAAA) 30. While some mRNAs are translated upon translocation to the cytoplasm, others are deadenylated upon reaching cytoplasm and stored to be translated at a later stage 26. The latter group of mRNA possesses an additional signal in their 3′UTR known as the CPE, a sequence signal at the 3′‐UTR of mRNA molecules, which determines whether the mRNA should undergo such translational regulation 124, 125. This mode of translational regulation is known as cytoplasmic polyadenylation.
The molecular machinery of cytoplasmic polyadenylation is probably best characterized within the context of oocyte maturation and early embryogenesis in Xenopus 106, 107, 126. The CPEB represents a family of RNA‐binding proteins. They bind CPE to regulate translation by nucleating the formation of cytoplasmic polyadenylation complex as well as translational enhancing and repressing complexes 124, 127, 128. All CPEB proteins contain a highly conserved C‐terminal, which consists of two RNA recognition modules responsible for binding target RNAs and a less conserved N‐terminal domain. The latter contains phosphorylation domains and protein–protein interaction domains presumed to be functionally important 129, 130, 131, 132. In the vertebrates, the two classes of CPEBs are represented by CPEB1 and CPEB2‐4 groups 127. The latter group is found to be expressed abundantly in the central nervous system. It is known to function in synaptic plasticity and long‐term memory formation 133, 134, 135. CPEB1 and CPEB4 play a role in oogenesis and are activated through different mechanisms. To be activated, CPEB1 is phosphorylated by the Aurora kinase A (Aurka), whereas CPEB4 by the ERK2 and Cdk1 kinases 136. CPEBs perform a dual role in translational regulation: in their native unphosphorylated form, they repress cap‐dependent translation; in their phosphorylated active form, they activate the translation of mRNAs through promoting cytoplasmic polyadenylation. CPEB1 recognizes the CPE at the 3′UTR of the mRNA molecule. The recognition of target mRNAs by CPEBs occurs in the nucleus. These mRNAs by default have long poly(A) tails. When the mRNAs are exported to the cytoplasm, CPEB nucleates the formation of a repressive complex, which includes the Gld2 poly(A) polymerase and PARN poly(A) ribonuclease. As PARN is more active, the concurrent activity of these two proteins results in the net shortening of poly(A) tail. On appropriate biological stimuli, PARN is expelled from the complex, resulting in elongation of the poly(A) tail due to Gld2 activity 128. The long poly(A) tail is subsequently recognized by the poly(A)‐binding protein (PABP), which recruits the translation initiation complex and initiates mRNA translation 137.
Cytoplasmic polyadenylation mechanism in oogenesis and early embryonic development
In the Xenopus oocyte, a heterodimeric complex of CDC2 and Cyclin B, known as the maturation‐promoting factor (MPF) induces the initial entry into metaphase I from prophase I 116. In oocytes arrested at prophase I, this complex is initially inactive and requires CDC25 for its activation. Progesterone stimulation results in the phosphorylation of Cpeb1 by Aurka (Fig. 3), resulting in the earliest wave of cytoplasmic polyadenylation of mRNAs encoding the components required for activation as well as the assembly of the MPF 125. Importantly, this early wave of polyadenylation also results in the translation of Cpeb4 which is responsible for a second late wave of polyadenylation during the second meiotic division (Fig. 3). Cpeb4 is activated through a distinct mechanism. ERK2 and Cdk1 phosphorylate Cpeb4 at its N‐terminal domain which contains an intrinsically disordered region. This hyperphosphorylation maintains Cpeb4 in a monomeric active state required for cytoplasmic polyadenylation 136. Otherwise, Cpeb4 forms liquid‐like droplets by means of its N‐terminal region, sequestering mRNAs and suppressing their translation.
Besides Cpeb1 which is implicated in oocyte maturation, an additional small CPEB‐like RNA‐binding protein (ElrA) is present at the earliest steps of embryonic development in Xenopus. ElrA recognizes the embryonic‐type CPEs present in maternal mRNAs regulated during embryogenesis 106, 126, 138. It is a member of the ELAV family of RNA‐binding proteins implicated in regulating mRNA stability, translation, and transport in Drosophila and mammalian neurons 139. The zebrafish homolog of ElrA, elavl1, was shown to be under translational regulation through cytoplasmic polyadenylation during pre‐MBT stages 140 Moreover, the translation of elavl1 appeared to be regulated by Zorba, the zebrafish homolog of CPEB1 141.
Previous research from our group revealed that at least 3000 of maternal transcripts undergo progressive increase in poly(A) tail prior to MBT 54. Subsequently, Subtelny and colleagues 108 showed the correlation between poly(A) tail elongation and translation efficiency of maternal mRNAs in several organisms, including the zebrafish. A unifying observation from these studies is that embryonic cytoplasmic polyadenylation seems to occur almost exclusively during the period of transcriptional quiescence 54, 108. Inhibition of this process by genetic mutations or chemical treatment of embryos with 3′ deoxyadenosine (3′dA) has been shown to result in the failure of passing through the MBT milestone in zebrafish 54, 56 and MZT in other organisms 97, 100. The transcripts of at least three different CPEBs—cpeb1/zorba, cpeb4a, and cpeb4b, were maternally supplied 54, 56. In addition, elavl1 is also present at this stage 138. Each of these factors show distinct expression dynamics 54, 56, 138, which suggests their sequential roles in regulating cytoplasmic polyadenylation and translational activation of maternal mRNAs in a timely manner. Zorba transcripts are initially present at high level from egg stage and are gradually degraded as development proceeds, in agreement with its known role in oocyte maturation in Xenopus. The degradation of zorba coincided with the gradual increase in cytoplasmic polyadenylation of cpeb4a, cpeb4b, and elavl1 (in agreement with 140. While the level of elavl1 continues to increase post‐MBT, cpeb4 levels immediately fall following its peak expression at MBT. Moreover, in agreement with their known roles in later developmental processes 133, 134, 135, cpeb2 and cpeb3 were undetectable during the pre‐MBT stages, with cpeb2 starting to be expressed only after MBT 54, 56. The sequential expression patterns of the different CPEBs coincide with the different developmental milestones and thus suggest distinct targets and function at different stages of development.
Mechanistic insights from studies of cytoplasmic polyadenylation and CPEB beyond embryogenesis
Besides its role in oogenesis and embryogenesis pre‐MBT/MZT, cytoplasmic polyadenylation also plays an important role in later stages of life. The best characterized examples of cytoplasmic polyadenylation in somatic cells are known to occur in the central nervous system 133, 134, 135, in synaptic junctions as part of a mechanism for the establishment of long‐term memory. It is known that memory formation is dependent on protein synthesis for the duration of the training stimuli 142, 143, 144. CPEB plays a role in synaptic plasticity and neuronal morphology and hence determines learning and long‐term memory 145, 146, 147, 148. In the neurons, CPEB is localized in RNP granules distributed along the dendritic arbor and localized in the vicinity of synapses 128, 149. The CPEB regulation of localized translation in the synapses has been observed in the sea slug Aplysia 150. Several key evidences support this idea as disruption of CPEB function results in abnormalities in various aspects of neuronal development and function 133, 151, 152.
Although the role of CPEB characterized so far is predominantly within the cytoplasmic domain, CPEBs also have a prominent role in the nucleus. CPEB1 proteins were observed to be shuttling between the cytoplasm and nucleus in cultured cells as well as in Xenopus embryos 153, 154, 155. It is well established that post‐transcriptional mRNA regulation usually starts even before cytoplasmic import, where RNA‐binding proteins regulating translation shuttle between nucleus and cytoplasm. They bind nuclear mRNAs and regulation of translation can occur once they are exported to the cytoplasm. In this sense, the behavior of CPEB1 is in agreement with this concept. In the nucleus of HeLa cells, CPEB1 is associated with nucleolar bodies containing the nuclear export protein Crm1. In the Xenopus oocytes, CPEB1 associates with lampbrush chromosomes. Interestingly, this association is RNAse‐sensitive, showing that CPEB1 binds to nascent‐transcribed mRNAs known to associate with proteins involved in nuclear RNA processing 154. These observations suggest that CPEB1 may play a role in processing of RNAs and translational control. However, despite this knowledge, the exact role of CPEB1 in the nucleus was not known until relatively recently. Bava and colleagues 156 observed that CPEB1 in the nucleus colocalizes with splicing factors. Here, CPEB1 promotes the formation of alternative 3′UTRs of mRNAs: its binding to target mRNAs promotes the usage of a more proximally located alternative polyadenylation site (PAS), resulting in a shorter 3′UTR. In this scenario, CPEB1 recruits the cleavage and polyadenylation specificity factor to a more upstream and weaker PAS. Under normal circumstances, the stronger downstream PAS would be used, thus resulting in a transcript with longer 3′UTR. It is thought that having a shorter 3′UTR would subject the transcript to less translational regulation, as it would contain less regulatory elements. Hence, the presence of CPEB1 would result in more efficient translation of target transcripts. An example of this mechanism could be observed in the liver, where CPEB1 mediates the processing of the 3′UTRs of VEGF and CPEB4 mRNAs. This results in a shorter isoform of both mRNA species lacking translational repression and mRNA destabilizing elements, which results in the overall increase in their translation 157. The consequent increase in CPEB4, in turn, leads to the increase in cytoplasmic polyadenylation of the VEGF mRNA, which stimulates VEGF translation further. In the human liver, CPEB1 and CPEB4 are expressed uniformly at low levels. In patients with cirrhotic liver, however, their expression is strongly increased particularly in hepatocytes at the regenerative foci 157. In addition, CPEBs and VEGF expression is also elevated in the endothelium of new vessels generated in fibrotic areas, which is thought to stimulate endothelial cell recruitment and proliferation necessary for angiogenesis. The loss of function of CPEBs was found to limit the pathologic progression of VEGF‐mediated angiogenesis without compromising physiological function of VEGF, which could thus be explored further as a potential therapeutic target to prevent disease progression.
Extending the morphogenetic function
Besides cytoplasmic polyadenylation, many other means of translational regulation which are not covered in this Review have been characterized in oogenesis and embryogenesis 158. In case of the latter, most of the forms known are suppressive in nature, the end result of which is often the instability of the transcript and its degradation 158, 159, 160. One mechanism which recently came to light is codon optimality 161 which has been demonstrated to regulate maternal mRNA clearance in the zebrafish 162, 163. With the advent of large‐scale analysis of genomes and transcriptomes brought about by high‐throughput next generation sequencing (NGS) technologies, particularly those which enables the profiling of global translation 164, 165, it is reasonable to expect that more and more of such mechanisms will come to light in the near future, which will enable us to gain a more comprehensive understanding of maternal RNA biology. By extension, the routine comparison of large numbers of maternal and zygotic transcripts at the genomic level has driven rapid progress in understanding of the molecular mechanisms of the MBT which had been discovered by Neyfakh in 1950s. Hence, his papers were rediscovered by a new generation of developmental biologists 54, 56, 166. And yet, there was a second part of the MBT story that for now remains hidden in the libraries. One intriguing continuation of the studies on ‘morphogenetic’ function of nuclei (defined as the ability to support embryonic development) has been the ‘vitogenetic’ function (defined as the ability of irradiated embryos to survive without developmental progression 2. In continuation of the ‘vitogenetic’ function story, it was found that the microinjection of a fraction of small nuclear RNA (snRNA) known for their role in pre‐mRNA splicing 167, but not any other RNA extracted during mid‐gastrula embryos into irradiated 1–2 cell stage loach embryos led to a significant increase of zygotic transcription as well as noticeable extension of the period of their survival. In this case, the zygotic transcription has been detected by cultivating the isolated blastoderms at specific stages (7 hpf, note that at optimal temperature, the loach develops approximately twice slower compared to zebrafish) with radioactively labeled precursors of DNA, RNA, and protein for 1 h and measuring radioactivity of the trichloroacetic acid filter precipitate in the liquid scintillation counter. The relative increase of transcription as defined by incorporation of tritiated uridine was higher comparing to replication and protein synthesis as defined by incorporation of labeled thymidine and leucine, correspondingly 168, 169, 170. In other words, an excess of snRNA seems to cause a prolongation of the ‘vitogenetic’ function 171. What could be the reason for this?
It has been shown that, upon fertilization, the Xenopus egg contains a certain amount of snRNA U1, sufficient for 4000–8000 nuclei, that is, roughly corresponding to the number of cells at the MBT. In addition, when zygotic transcription is activated at the twelfth cleavage (4000‐cell stage), RNA polymerase II generates mainly snRNAs. From the twelfth cleavage to gastrulation, U1 RNA increases sevenfold in parallel with increase of cell number. This level of snRNA transcription is much above that present in somatic cells. This suggests much higher U1 transcription or a greater number of active U1 genes in the embryo, suggesting the critical developmental role of snRNA transcripts. Interestingly, microinjection of U1 snRNAs into the cytoplasm of a mature Xenopus oocyte triggers the transport of snRNA‐binding proteins into the nucleus (germinal vesicle, GV) 172. In the sea urchin, the maternal U1 RNA and U1‐specific ribonucleoproteins (snRNP) reside in the GV. On oocyte maturation, they relocate to the cytoplasm where they are maintained during early cleavage and U1 RNA appears in the nuclei of micromeres at the 16‐cell stage. In contrast, the zygotic U1 transcripts are confined to nuclei, while the maternal ones remain cytoplasmic. Being released to the cytoplasm at oocyte maturation, these RNPs (or RNAs) may have been structurally altered 173. Intriguingly, the snRNA themselves are a subject of cytoplasmic polyadenylation 174, 175. It remains to be seen whether the 1988 experimentally induced elongation of the ‘vitogenetic’ function could be due to a strictly limited mass of maternal snRNA or its structural modification involving cytoplasmic polyadenylation. Whatever the answer, it seems that there are multiple fractions of maternal RNAs whose activation during development involves cytoplasmic polyadenylation.
The understanding of translational regulation mechanism extends beyond early embryonic development and could be extrapolated to different biological aspects. With the development of high‐throughput sequencing techniques, it is now possible to move beyond single‐gene studies and obtain a global view of translation which has often been limited by technical challenges associated with protein analyses. Incorporation of the increasing number of such datasets into the established knowledge of the principles of early embryonic development would be the next step to establish a more comprehensive understanding of translational regulation of maternal mRNAs. One caveat, however, is that this methodology is relatively easy to use for the analysis of transcripts within the temporal aspect, whereas it remained more challenging to use this methodology to resolve developmental processes in space, taking place in 3D environment of oocytes and embryos. This situation changes for the better with the introduction of single‐cell transcriptome analysis. In the zebrafish field, the feasibility of this technique have recently been demonstrated to construct the spatial map of the early zebrafish gastrula based on the expression profiles of individual cells 176 and for lineage tracing of individual cells in the early zebrafish embryo in combination with CRISPR/Cas9 gene editing 177. It is foreseeable in the future that the application of single‐cell‐based techniques, in combination with whole embryo analysis, will open up vast possibilities to study and unravel novel aspects of the regulation of maternal mRNAs in the spatial context.
Funding
CLW is supported by the EU FP7 Grant Fishmed GA No. 316125, Polish National Science Center (NCN) OPUS grant No. 2014/13/B/NZ2/03863, and the FNP First Team grant GA No. First TEAM/2016‐1/8. VK is supported by Polish National Science Center (NCN) OPUS grant No. 2016/21/B/NZ3/00354.
Acknowledgements
We thank members of our laboratories and colleagues for discussions.
Edited by Angel Nebreda
Contributor Information
Cecilia Lanny Winata, Email: cwinata@iimcb.gov.pl.
Vladimir Korzh, Email: vkorzh@iimcb.gov.pl.
References
- 1. Neyfakh AA (1964) Radiation investigation of nucleo‐cytoplasmic interrelations in morphogenesis and biochemical differentiation. Nature 201, 880–884. [DOI] [PubMed] [Google Scholar]
- 2. Neyfakh AA (1961) Comparative study of the morphogenetic function of Nuclei in Animal. Zh Obshch Biol 22, 42–57. [PubMed] [Google Scholar]
- 3. Moore JA (1946) Studies in the development of frog hybrids; embryonic development in the cross Rana pipiens female x Rana sylvatica male. J Exp Zool 101, 173–219. [DOI] [PubMed] [Google Scholar]
- 4. Moore JA (1955) Abnormal combinations of nuclear and cytoplasmic systems in frogs and toads. Adv Genet 7, 139–182. [DOI] [PubMed] [Google Scholar]
- 5. Harvey SA, Sealy I, Kettleborough R, Fenyes F, White R, Stemple D and Smith JC (2013) Identification of the zebrafish maternal and paternal transcriptomes. Development 140, 2703–2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Palfy M, Joseph SR and Vastenhouw NL (2017) The timing of zygotic genome activation. Curr Opin Genet Dev 43, 53–60. [DOI] [PubMed] [Google Scholar]
- 7. Rott NN and Sheveleva GA (1968) Changes in the rate of cell divisions in the course of early development of diploid and haploid loach embryos. J Embryol Exp Morphol 20, 141–150. [PubMed] [Google Scholar]
- 8. Newport J and Kirschner M (1982) A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell 30, 675–686. [DOI] [PubMed] [Google Scholar]
- 9. Signoret J (1971) Contribution a l'etude de la segmentation de l'oeuf d'axolotl: I. Definition de la transition blastuleenne. Ann Embryol Morphol 4, 113–123. [Google Scholar]
- 10. Joseph SR, Palfy M, Hilbert L, Kumar M, Karschau J, Zaburdaev V, Shevchenko A and Vastenhouw NL (2017) Competition between histone and transcription factor binding regulates the onset of transcription in zebrafish embryos. Elife 6, pii: e23326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kane DA and Kimmel CB (1993) The zebrafish midblastula transition. Development 119, 447–456. [DOI] [PubMed] [Google Scholar]
- 12. Clegg KB and Piko L (1982) RNA synthesis and cytoplasmic polyadenylation in the one‐cell mouse embryo. Nature 295, 343–344. [DOI] [PubMed] [Google Scholar]
- 13. Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ and Schier AF (2006) Zebrafish MiR‐430 promotes deadenylation and clearance of maternal mRNAs. Science 312, 75–79. [DOI] [PubMed] [Google Scholar]
- 14. Hamatani T, Carter MG, Sharov AA and Ko MS (2004) Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell 6, 117–131. [DOI] [PubMed] [Google Scholar]
- 15. Piko L and Clegg KB (1982) Quantitative changes in total RNA, total poly(A), and ribosomes in early mouse embryos. Dev Biol 89, 362–378. [DOI] [PubMed] [Google Scholar]
- 16. Stitzel ML and Seydoux G (2007) Regulation of the oocyte‐to‐zygote transition. Science 316, 407–408. [DOI] [PubMed] [Google Scholar]
- 17. Tadros W and Lipshitz HD (2009) The maternal‐to‐zygotic transition: a play in two acts. Development 136, 3033–3042. [DOI] [PubMed] [Google Scholar]
- 18. Walser CB and Lipshitz HD (2011) Transcript clearance during the maternal‐to‐zygotic transition. Curr Opin Genet Dev 21, 431–443. [DOI] [PubMed] [Google Scholar]
- 19. Lindeman LC, Andersen IS, Reiner AH, Li N, Aanes H, Ostrup O, Winata C, Mathavan S, Muller F, Alestrom P et al (2011) Prepatterning of developmental gene expression by modified histones before zygotic genome activation. Dev Cell 21, 993–1004. [DOI] [PubMed] [Google Scholar]
- 20. Lindeman LC, Winata CL, Aanes H, Mathavan S, Alestrom P and Collas P (2010) Chromatin states of developmentally‐regulated genes revealed by DNA and histone methylation patterns in zebrafish embryos. Int J Dev Biol 54, 803–813. [DOI] [PubMed] [Google Scholar]
- 21. Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, Rinn J and Schier AF (2010) Chromatin signature of embryonic pluripotency is established during genome activation. Nature 464, 922–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Spradling AC (1993) Germline cysts: communes that work. Cell 72, 649–651. [DOI] [PubMed] [Google Scholar]
- 23. Cooley L and Theurkauf WE (1994) Cytoskeletal functions during Drosophila oogenesis. Science 266, 590–596. [DOI] [PubMed] [Google Scholar]
- 24. Pokrywka NJ and Stephenson EC (1995) Microtubules are a general component of mRNA localization systems in Drosophila oocytes. Dev Biol 167, 363–370. [DOI] [PubMed] [Google Scholar]
- 25. Wilsch‐Brauninger M, Schwarz H and Nusslein‐Volhard C (1997) A sponge‐like structure involved in the association and transport of maternal products during Drosophila oogenesis. J Cell Biol 139, 817–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huarte J, Stutz A, O'Connell ML, Gubler P, Belin D, Darrow AL, Strickland S and Vassalli JD (1992) Transient translational silencing by reversible mRNA deadenylation. Cell 69, 1021–1030. [DOI] [PubMed] [Google Scholar]
- 27. Ahringer J, Rosenquist TA, Lawson DN and Kimble J (1992) The Caenorhabditis elegans sex determining gene fem‐3 is regulated post‐transcriptionally. EMBO J 11, 2303–2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bachvarova R and Paynton BV (1988) Gene expression during growth and meiotic maturation of mouse oocytes. Prog Clin Biol Res 267, 67–85. [PubMed] [Google Scholar]
- 29. Fox CA, Sheets MD and Wickens MP (1989) Poly(A) addition during maturation of frog oocytes: distinct nuclear and cytoplasmic activities and regulation by the sequence UUUUUAU. Genes Dev 3, 2151–2162. [DOI] [PubMed] [Google Scholar]
- 30. McGrew LL, Dworkin‐Rastl E, Dworkin MB and Richter JD (1989) Poly(A) elongation during Xenopus oocyte maturation is required for translational recruitment and is mediated by a short sequence element. Genes Dev 3, 803–815. [DOI] [PubMed] [Google Scholar]
- 31. Paris J, Osborne HB, Couturier A, Le Guellec R and Philippe M (1988) Changes in the polyadenylation of specific stable RNA during the early development of Xenopus laevis . Gene 72, 169–176. [DOI] [PubMed] [Google Scholar]
- 32. Rosenthal ET and Wilt FH (1986) Patterns of maternal messenger RNA accumulation and adenylation during oogenesis in Urechis caupo . Dev Biol 117, 55–63. [DOI] [PubMed] [Google Scholar]
- 33. Vassalli JD, Huarte J, Belin D, Gubler P, Vassalli A, O'Connell ML, Parton LA, Rickles RJ and Strickland S (1989) Regulated polyadenylation controls mRNA translation during meiotic maturation of mouse oocytes. Genes Dev 3, 2163–2171. [DOI] [PubMed] [Google Scholar]
- 34. Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P and Krause HM (2007) Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131, 174–187. [DOI] [PubMed] [Google Scholar]
- 35. Bastock R and St Johnston D (2008) Drosophila oogenesis. Curr Biol 18, R1082–R1087. [DOI] [PubMed] [Google Scholar]
- 36. Suzuki H, Tsukahara T and Inoue K (2009) Localization of c‐mos mRNA around the animal pole in the zebrafish oocyte with Zor‐1/Zorba. Biosci Trends 3, 96–104. [PubMed] [Google Scholar]
- 37. Howley C and Ho RK (2000) mRNA localization patterns in zebrafish oocytes. Mech Dev 92, 305–309. [DOI] [PubMed] [Google Scholar]
- 38. Marlow FL and Mullins MC (2008) Bucky ball functions in Balbiani body assembly and animal‐vegetal polarity in the oocyte and follicle cell layer in zebrafish. Dev Biol 321, 40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kumano G (2012) Polarizing animal cells via mRNA localization in oogenesis and early development. Dev Growth Differ 54, 1–18. [DOI] [PubMed] [Google Scholar]
- 40. Medioni C, Mowry K and Besse F (2012) Principles and roles of mRNA localization in animal development. Development 139, 3263–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang Y, Forinash KD, McGivern J, Fritz B, Dorey K and Sheets MD (2009) Spatially restricted translation of the xCR1 mRNA in Xenopus embryos. Mol Cell Biol 29, 3791–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fribourg S, Gatfield D, Izaurralde E and Conti E (2003) A novel mode of RBD‐protein recognition in the Y14‐Mago complex. Nat Struct Biol 10, 433–439. [DOI] [PubMed] [Google Scholar]
- 43. Gehring NH, Kunz JB, Neu‐Yilik G, Breit S, Viegas MH, Hentze MW and Kulozik AE (2005) Exon‐junction complex components specify distinct routes of nonsense‐mediated mRNA decay with differential cofactor requirements. Mol Cell 20, 65–75. [DOI] [PubMed] [Google Scholar]
- 44. Michelle L, Cloutier A, Toutant J, Shkreta L, Thibault P, Durand M, Garneau D, Gendron D, Lapointe E, Couture S et al (2012) Proteins associated with the exon junction complex also control the alternative splicing of apoptotic regulators. Mol Cell Biol 32, 954–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pozzoli O, Gilardelli CN, Sordino P, Doniselli S, Lamia CL and Cotelli F (2004) Identification and expression pattern of mago nashi during zebrafish development. Gene Expr Patterns 5, 265–272. [DOI] [PubMed] [Google Scholar]
- 46. Fong SH, Emelyanov A, Teh C and Korzh V (2005) Wnt signalling mediated by Tbx2b regulates cell migration during formation of the neural plate. Development 132, 3587–3596. [DOI] [PubMed] [Google Scholar]
- 47. Helde KA, Wilson ET, Cretekos CJ and Grunwald DJ (1994) Contribution of early cells to the fate map of the zebrafish gastrula. Science 265, 517–520. [DOI] [PubMed] [Google Scholar]
- 48. Favaro FP, Alvizi L, Zechi‐Ceide RM, Bertola D, Felix TM, de Souza J, Raskin S, Twigg SR, Weiner AM, Armas P et al (2014) A noncoding expansion in EIF4A3 causes Richieri‐Costa‐Pereira syndrome, a craniofacial disorder associated with limb defects. Am J Hum Genet 94, 120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Furutani‐Seiki M, Jiang YJ, Brand M, Heisenberg CP, Houart C, Beuchle D, van Eeden FJ, Granato M, Haffter P, Hammerschmidt M et al (1996) Neural degeneration mutants in the zebrafish, Danio rerio . Development 123, 229–239. [DOI] [PubMed] [Google Scholar]
- 50. Sampath K and Ephrussi A (2016) CncRNAs: RNAs with both coding and non‐coding roles in development. Development 143, 1234–1241. [DOI] [PubMed] [Google Scholar]
- 51. Gore AV, Maegawa S, Cheong A, Gilligan PC, Weinberg ES and Sampath K (2005) The zebrafish dorsal axis is apparent at the four‐cell stage. Nature 438, 1030–1035. [DOI] [PubMed] [Google Scholar]
- 52. Gilligan PC, Kumari P, Lim S, Cheong A, Chang A and Sampath K (2011) Conservation defines functional motifs in the squint/nodal‐related 1 RNA dorsal localization element. Nucleic Acids Res 39, 3340–3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kumari P, Gilligan PC, Lim S, Tran LD, Winkler S, Philp R and Sampath K (2013) An essential role for maternal control of Nodal signaling. Elife 2, e00683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Aanes H, Winata CL, Lin CH, Chen JP, Srinivasan KG, Lee SG, Lim AY, Hajan HS, Collas P, Bourque G et al (2011) Zebrafish mRNA sequencing deciphers novelties in transcriptome dynamics during maternal to zygotic transition. Genome Res 21, 1328–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lim S, Kumari P, Gilligan P, Quach HN, Mathavan S and Sampath K (2012) Dorsal activity of maternal squint is mediated by a non‐coding function of the RNA. Development 139, 2903–2915. [DOI] [PubMed] [Google Scholar]
- 56. Winata CL, Lapinski M, Pryszcz L, Vaz C, Bin Ismail MH, Nama S, Hajan HS, Lee SGP, Korzh V, Sampath P et al (2018) Cytoplasmic polyadenylation‐mediated translational control of maternal mRNAs directs maternal‐to‐zygotic transition. Development 145, pii: dev159566. [DOI] [PubMed] [Google Scholar]
- 57. Zaucker A, Nagorska A, Kumari P, Hecker N, Wang Y, Huang S, Cooper L, Sivashanmugam L, VijayKumar S, Brosens J et al (2018) Translational co‐regulation of a ligand and inhibitor by a conserved RNA element. Nucleic Acids Res 46, 104–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Lu FI, Thisse C and Thisse B (2011) Identification and mechanism of regulation of the zebrafish dorsal determinant. Proc Natl Acad Sci USA 108, 15876–15880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Nojima H, Rothhamel S, Shimizu T, Kim CH, Yonemura S, Marlow FL and Hibi M (2010) Syntabulin, a motor protein linker, controls dorsal determination. Development 137, 923–933. [DOI] [PubMed] [Google Scholar]
- 60. Nojima H, Shimizu T, Kim CH, Yabe T, Bae YK, Muraoka O, Hirata T, Chitnis A, Hirano T and Hibi M (2004) Genetic evidence for involvement of maternally derived Wnt canonical signaling in dorsal determination in zebrafish. Mech Dev 121, 371–386. [DOI] [PubMed] [Google Scholar]
- 61. Tran LD, Hino H, Quach H, Lim S, Shindo A, Mimori‐Kiyosue Y, Mione M, Ueno N, Winkler C, Hibi M et al (2012) Dynamic microtubules at the vegetal cortex predict the embryonic axis in zebrafish. Development 139, 3644–3652. [DOI] [PubMed] [Google Scholar]
- 62. Ge X, Grotjahn D, Welch E, Lyman‐Gingerich J, Holguin C, Dimitrova E, Abrams EW, Gupta T, Marlow FL, Yabe T et al (2014) Hecate/Grip2a acts to reorganize the cytoskeleton in the symmetry‐breaking event of embryonic axis induction. PLoS Genet 10, e1004422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hino H, Nakanishi A, Seki R, Aoki T, Yamaha E, Kawahara A, Shimizu T and Hibi M (2018) Roles of maternal wnt8a transcripts in axis formation in zebrafish. Dev Biol 434, 96–107. [DOI] [PubMed] [Google Scholar]
- 64. Shao M, Wang M, Liu YY, Ge YW, Zhang YJ and Shi DL (2017) Vegetally localised Vrtn functions as a novel repressor to modulate bmp2b transcription during dorsoventral patterning in zebrafish. Development 144, 3361–3374. [DOI] [PubMed] [Google Scholar]
- 65. Shav‐Tal Y and Singer RH (2005) RNA localization. J Cell Sci 118, 4077–4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Anderson P and Kedersha N (2009) RNA granules: post‐transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol 10, 430–436. [DOI] [PubMed] [Google Scholar]
- 67. Courchaine EM, Lu A and Neugebauer KM (2016) Droplet organelles? EMBO J 35, 1603–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sengupta MS and Boag PR (2012) Germ granules and the control of mRNA translation. IUBMB Life 64, 586–594. [DOI] [PubMed] [Google Scholar]
- 69. Voronina E, Seydoux G, Sassone‐Corsi P and Nagamori I (2011) RNA granules in germ cells. Cold Spring Harb Perspect Biol 3, pii: a002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Dundr M, Hebert MD, Karpova TS, Stanek D, Xu H, Shpargel KB, Meier UT, Neugebauer KM, Matera AG and Misteli T (2004) In vivo kinetics of Cajal body components. J Cell Biol 164, 831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schisa JA (2012) New insights into the regulation of RNP granule assembly in oocytes. Int Rev Cell Mol Biol 295, 233–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Boag PR, Atalay A, Robida S, Reinke V and Blackwell TK (2008) Protection of specific maternal messenger RNAs by the P body protein CGH‐1 (Dhh1/RCK) during Caenorhabditis elegans oogenesis. J Cell Biol 182, 543–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Noble SL, Allen BL, Goh LK, Nordick K and Evans TC (2008) Maternal mRNAs are regulated by diverse P body‐related mRNP granules during early Caenorhabditis elegans development. J Cell Biol 182, 559–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gao X, Ge L, Shao J, Su C, Zhao H, Saarikettu J, Yao X, Yao Z, Silvennoinen O and Yang J (2010) Tudor‐SN interacts with and co‐localizes with G3BP in stress granules under stress conditions. FEBS Lett 584, 3525–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mahowald AP (1968) Polar granules of Drosophila. II. Ultrastructural changes during early embryogenesis. J Exp Zool 167, 237–261. [DOI] [PubMed] [Google Scholar]
- 76. Snee MJ and Macdonald PM (2009) Dynamic organization and plasticity of sponge bodies. Dev Dyn 238, 918–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Nakamura A, Amikura R, Hanyu K and Kobayashi S (2001) Me31B silences translation of oocyte‐localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development 128, 3233–3242. [DOI] [PubMed] [Google Scholar]
- 78. Thomson T and Lin H (2009) The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol 25, 355–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Thomson T, Liu N, Arkov A, Lehmann R and Lasko P (2008) Isolation of new polar granule components in Drosophila reveals P body and ER associated proteins. Mech Dev 125, 865–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A and Hannon GJ (2008) An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322, 1387–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Weil TT, Parton RM, Herpers B, Soetaert J, Veenendaal T, Xanthakis D, Dobbie IM, Halstead JM, Hayashi R, Rabouille C et al (2012) Drosophila patterning is established by differential association of mRNAs with P bodies. Nat Cell Biol 14, 1305–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Trcek T, Grosch M, York A, Shroff H, Lionnet T and Lehmann R (2015) Drosophila germ granules are structured and contain homotypic mRNA clusters. Nat Commun 6, 7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Flemr M and Svoboda P (2011) Ribonucleoprotein localization in mouse oocytes. Methods 53, 136–141. [DOI] [PubMed] [Google Scholar]
- 84. Flemr M, Ma J, Schultz RM and Svoboda P (2010) P‐body loss is concomitant with formation of a messenger RNA storage domain in mouse oocytes. Biol Reprod 82, 1008–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Heasman J, Quarmby J and Wylie CC (1984) The mitochondrial cloud of Xenopus oocytes: the source of germinal granule material. Dev Biol 105, 458–469. [DOI] [PubMed] [Google Scholar]
- 86. Kloc M, Zearfoss NR and Etkin LD (2002) Mechanisms of subcellular mRNA localization. Cell 108, 533–544. [DOI] [PubMed] [Google Scholar]
- 87. Chang P, Torres J, Lewis RA, Mowry KL, Houliston E and King ML (2004) Localization of RNAs to the mitochondrial cloud in Xenopus oocytes through entrapment and association with endoplasmic reticulum. Mol Biol Cell 15, 4669–4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wilk K, Bilinski S, Dougherty MT and Kloc M (2005) Delivery of germinal granules and localized RNAs via the messenger transport organizer pathway to the vegetal cortex of Xenopus oocytes occurs through directional expansion of the mitochondrial cloud. Int J Dev Biol 49, 17–21. [DOI] [PubMed] [Google Scholar]
- 89. Kloc M, Bilinski S, Dougherty MT, Brey EM and Etkin LD (2004) Formation, architecture and polarity of female germline cyst in Xenopus . Dev Biol 266, 43–61. [DOI] [PubMed] [Google Scholar]
- 90. Knaut H, Steinbeisser H, Schwarz H and Nusslein‐Volhard C (2002) An evolutionary conserved region in the vasa 3′UTR targets RNA translation to the germ cells in the zebrafish. Curr Biol 12, 454–466. [DOI] [PubMed] [Google Scholar]
- 91. Kosaka K, Kawakami K, Sakamoto H and Inoue K (2007) Spatiotemporal localization of germ plasm RNAs during zebrafish oogenesis. Mech Dev 124, 279–289. [DOI] [PubMed] [Google Scholar]
- 92. Elkouby YM, Jamieson‐Lucy A and Mullins MC (2016) Oocyte polarization is coupled to the chromosomal bouquet, a conserved polarized nuclear configuration in meiosis. PLoS Biol 14, e1002335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Houston DW (2013) Regulation of cell polarity and RNA localization in vertebrate oocytes. Int Rev Cell Mol Biol 306, 127–185. [DOI] [PubMed] [Google Scholar]
- 94. Colozza G and De Robertis EM (2014) Maternal syntabulin is required for dorsal axis formation and is a germ plasm component in Xenopus . Differentiation 88, 17–26. [DOI] [PubMed] [Google Scholar]
- 95. Langdon YG and Mullins MC (2011) Maternal and zygotic control of zebrafish dorsoventral axial patterning. Annu Rev Genet 45, 357–377. [DOI] [PubMed] [Google Scholar]
- 96. Heyn P, Salmonowicz H, Rodenfels J and Neugebauer KM (2017) Activation of transcription enforces the formation of distinct nuclear bodies in zebrafish embryos. RNA Biol 14, 752–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Aoki F, Hara KT and Schultz RM (2003) Acquisition of transcriptional competence in the 1‐cell mouse embryo: requirement for recruitment of maternal mRNAs. Mol Reprod Dev 64, 270–274. [DOI] [PubMed] [Google Scholar]
- 98. Evsikov AV, de Vries WN, Peaston AE, Radford EE, Fancher KS, Chen FH, Blake JA, Bult CJ, Latham KE, Solter D et al (2004) Systems biology of the 2‐cell mouse embryo. Cytogenet Genome Res 105, 240–250. [DOI] [PubMed] [Google Scholar]
- 99. Hwang SY, Oh B, Fuchtbauer A, Fuchtbauer EM, Johnson KR, Solter D and Knowles BB (1997) Maid: a maternally transcribed novel gene encoding a potential negative regulator of bHLH proteins in the mouse egg and zygote. Dev Dyn 209, 217–226. [DOI] [PubMed] [Google Scholar]
- 100. Lieberfarb ME, Chu T, Wreden C, Theurkauf W, Gergen JP and Strickland S (1996) Mutations that perturb poly(A)‐dependent maternal mRNA activation block the initiation of development. Development 122, 579–588. [DOI] [PubMed] [Google Scholar]
- 101. Oh B, Hwang S, McLaughlin J, Solter D and Knowles BB (2000) Timely translation during the mouse oocyte‐to‐embryo transition. Development 127, 3795–3803. [DOI] [PubMed] [Google Scholar]
- 102. Paynton BV, Rempel R and Bachvarova R (1988) Changes in state of adenylation and time course of degradation of maternal mRNAs during oocyte maturation and early embryonic development in the mouse. Dev Biol 129, 304–314. [DOI] [PubMed] [Google Scholar]
- 103. Potireddy S, Vassena R, Patel BG and Latham KE (2006) Analysis of polysomal mRNA populations of mouse oocytes and zygotes: dynamic changes in maternal mRNA utilization and function. Dev Biol 298, 155–166. [DOI] [PubMed] [Google Scholar]
- 104. Salles FJ, Darrow AL, O'Connell ML and Strickland S (1992) Isolation of novel murine maternal mRNAs regulated by cytoplasmic polyadenylation. Genes Dev 6, 1202–1212. [DOI] [PubMed] [Google Scholar]
- 105. Salles FJ, Lieberfarb ME, Wreden C, Gergen JP and Strickland S (1994) Coordinate initiation of Drosophila development by regulated polyadenylation of maternal messenger RNAs. Science 266, 1996–1999. [DOI] [PubMed] [Google Scholar]
- 106. Simon R, Tassan JP and Richter JD (1992) Translational control by poly(A) elongation during Xenopus development: differential repression and enhancement by a novel cytoplasmic polyadenylation element. Genes Dev 6, 2580–2591. [DOI] [PubMed] [Google Scholar]
- 107. Stebbins‐Boaz B, Hake LE and Richter JD (1996) CPEB controls the cytoplasmic polyadenylation of cyclin, Cdk2 and c‐mos mRNAs and is necessary for oocyte maturation in Xenopus . EMBO J 15, 2582–2592. [PMC free article] [PubMed] [Google Scholar]
- 108. Subtelny AO, Eichhorn SW, Chen GR, Sive H and Bartel DP (2014) Poly(A)‐tail profiling reveals an embryonic switch in translational control. Nature 508, 66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Nishiyama T, Tachibana K and Kishimoto T (2010) Cytostatic Arrest: Post‐Ovulation Arrest until Fertilization in Metazoan Oocytes, Oogenesis. pp. 357–384. John Wiley & Sons, Ltd., Hoboken, NJ. [Google Scholar]
- 110. Doane WW (1960) Completion of meiosis in uninseminated eggs of Drosophila melanogaster . Science 132, 677–678. [DOI] [PubMed] [Google Scholar]
- 111. Iwabuchi M, Ohsumi K, Yamamoto TM, Sawada W and Kishimoto T (2000) Residual Cdc2 activity remaining at meiosis I exit is essential for meiotic M‐M transition in Xenopus oocyte extracts. EMBO J 19, 4513–4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Nagahama Y (1997) 17 alpha,20 beta‐dihydroxy‐4‐pregnen‐3‐one, a maturation‐inducing hormone in fish oocytes: mechanisms of synthesis and action. Steroids 62, 190–196. [DOI] [PubMed] [Google Scholar]
- 113. Miller MA, Nguyen VQ, Lee MH, Kosinski M, Schedl T, Caprioli RM and Greenstein D (2001) A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science 291, 2144–2147. [DOI] [PubMed] [Google Scholar]
- 114. Tunquist BJ and Maller JL (2003) Under arrest: cytostatic factor (CSF)‐mediated metaphase arrest in vertebrate eggs. Genes Dev 17, 683–710. [DOI] [PubMed] [Google Scholar]
- 115. de Moor CH and Richter JD (1997) The Mos pathway regulates cytoplasmic polyadenylation in Xenopus oocytes. Mol Cell Biol 17, 6419–6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Hochegger H, Klotzbucher A, Kirk J, Howell M, le Guellec K, Fletcher K, Duncan T, Sohail M and Hunt T (2001) New B‐type cyclin synthesis is required between meiosis I and II during Xenopus oocyte maturation. Development 128, 3795–3807. [DOI] [PubMed] [Google Scholar]
- 117. Mendez R, Hake LE, Andresson T, Littlepage LE, Ruderman JV and Richter JD (2000) Phosphorylation of CPE binding factor by Eg2 regulates translation of c‐mos mRNA. Nature 404, 302–307. [DOI] [PubMed] [Google Scholar]
- 118. Sagata N, Daar I, Oskarsson M, Showalter SD and Vande Woude GF (1989) The product of the mos proto‐oncogene as a candidate “initiator” for oocyte maturation. Science 245, 643–646. [DOI] [PubMed] [Google Scholar]
- 119. Sheets MD, Wu M and Wickens M (1995) Polyadenylation of c‐mos mRNA as a control point in Xenopus meiotic maturation. Nature 374, 511–516. [DOI] [PubMed] [Google Scholar]
- 120. Gautier J, Minshull J, Lohka M, Glotzer M, Hunt T and Maller JL (1990) Cyclin is a component of maturation‐promoting factor from Xenopus . Cell 60, 487–494. [DOI] [PubMed] [Google Scholar]
- 121. Kotani T, Yasuda K, Ota R and Yamashita M (2013) Cyclin B1 mRNA translation is temporally controlled through formation and disassembly of RNA granules. J Cell Biol 202, 1041–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Nagaike T, Logan C, Hotta I, Rozenblatt‐Rosen O, Meyerson M and Manley JL (2011) Transcriptional activators enhance polyadenylation of mRNA precursors. Mol Cell 41, 409–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Proudfoot NJ (2011) Ending the message: poly(A) signals then and now. Genes Dev 25, 1770–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Hake LE and Richter JD (1994) CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell 79, 617–627. [DOI] [PubMed] [Google Scholar]
- 125. Pique M, Lopez JM, Foissac S, Guigo R and Mendez R (2008) A combinatorial code for CPE‐mediated translational control. Cell 132, 434–448. [DOI] [PubMed] [Google Scholar]
- 126. Simon R, Wu L and Richter JD (1996) Cytoplasmic polyadenylation of activin receptor mRNA and the control of pattern formation in Xenopus development. Dev Biol 179, 239–250. [DOI] [PubMed] [Google Scholar]
- 127. Ivshina M, Lasko P and Richter JD (2014) Cytoplasmic polyadenylation element binding proteins in development, health, and disease. Annu Rev Cell Dev Biol 30, 393–415. [DOI] [PubMed] [Google Scholar]
- 128. Richter JD (2007) CPEB: a life in translation. Trends Biochem Sci 32, 279–285. [DOI] [PubMed] [Google Scholar]
- 129. Afroz T, Skrisovska L, Belloc E, Guillen‐Boixet J, Mendez R and Allain FH (2014) A fly trap mechanism provides sequence‐specific RNA recognition by CPEB proteins. Genes Dev 28, 1498–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Hake LE, Mendez R and Richter JD (1998) Specificity of RNA binding by CPEB: requirement for RNA recognition motifs and a novel zinc finger. Mol Cell Biol 18, 685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Merkel DJ, Wells SB, Hilburn BC, Elazzouzi F, Perez‐Alvarado GC and Lee BM (2013) The C‐terminal region of cytoplasmic polyadenylation element binding protein is a ZZ domain with potential for protein‐protein interactions. J Mol Biol 425, 2015–2026. [DOI] [PubMed] [Google Scholar]
- 132. Theis M, Si K and Kandel ER (2003) Two previously undescribed members of the mouse CPEB family of genes and their inducible expression in the principal cell layers of the hippocampus. Proc Natl Acad Sci USA 100, 9602–9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Keleman K, Kruttner S, Alenius M and Dickson BJ (2007) Function of the Drosophila CPEB protein Orb2 in long‐term courtship memory. Nat Neurosci 10, 1587–1593. [DOI] [PubMed] [Google Scholar]
- 134. Kruttner S, Stepien B, Noordermeer JN, Mommaas MA, Mechtler K, Dickson BJ and Keleman K (2012) Drosophila CPEB Orb2A mediates memory independent of Its RNA‐binding domain. Neuron 76, 383–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Majumdar A, Cesario WC, White‐Grindley E, Jiang H, Ren F, Khan MR, Li L, Choi EM, Kannan K, Guo F et al (2012) Critical role of amyloid‐like oligomers of Drosophila Orb2 in the persistence of memory. Cell 148, 515–529. [DOI] [PubMed] [Google Scholar]
- 136. Guillen‐Boixet J, Buzon V, Salvatella X and Mendez R (2016) CPEB4 is regulated during cell cycle by ERK2/Cdk1‐mediated phosphorylation and its assembly into liquid‐like droplets. Elife 5, pii: e19298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Darnell JC and Richter JD (2012) Cytoplasmic RNA‐binding proteins and the control of complex brain function. Cold Spring Harb Perspect Biol 4, a012344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. O'Connell ML, Cavallo WC Jr and Firnberg M (2014) The expression of CPEB proteins is sequentially regulated during zebrafish oogenesis and embryogenesis. Mol Reprod Dev 81, 376–387. [DOI] [PubMed] [Google Scholar]
- 139. Wu L, Good PJ and Richter JD (1997) The 36‐kilodalton embryonic‐type cytoplasmic polyadenylation element‐binding protein in Xenopus laevis is ElrA, a member of the ELAV family of RNA‐binding proteins. Mol Cell Biol 17, 6402–6409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Bally‐Cuif L, Schatz WJ and Ho RK (1998) Characterization of the zebrafish Orb/CPEB‐related RNA binding protein and localization of maternal components in the zebrafish oocyte. Mech Dev 77, 31–47. [DOI] [PubMed] [Google Scholar]
- 141. Wagner DS, Dosch R, Mintzer KA, Wiemelt AP and Mullins MC (2004) Maternal control of development at the midblastula transition and beyond: mutants from the zebrafish II. Dev Cell 6, 781–790. [DOI] [PubMed] [Google Scholar]
- 142. Flexner JB, Flexner LB, Stellar E, De La Haba G and Roberts RB (1962) Inhibition of protein synthesis in brain and learning and memory following puromycin. J Neurochem 9, 595–605. [DOI] [PubMed] [Google Scholar]
- 143. Flexner LB and Flexner JB (1968) Intracerebral saline: effect on memory of trained mice treated with puromycin. Science 159, 330–331. [DOI] [PubMed] [Google Scholar]
- 144. Sutton MA and Schuman EM (2006) Dendritic protein synthesis, synaptic plasticity, and memory. Cell 127, 49–58. [DOI] [PubMed] [Google Scholar]
- 145. Alarcon JM, Hodgman R, Theis M, Huang YS, Kandel ER and Richter JD (2004) Selective modulation of some forms of schaffer collateral‐CA1 synaptic plasticity in mice with a disruption of the CPEB‐1 gene. Learn Mem 11, 318–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Berger‐Sweeney J, Zearfoss NR and Richter JD (2006) Reduced extinction of hippocampal‐dependent memories in CPEB knockout mice. Learn Mem 13, 4–7. [DOI] [PubMed] [Google Scholar]
- 147. McEvoy M, Cao G, Montero Llopis P, Kundel M, Jones K, Hofler C, Shin C and Wells DG (2007) Cytoplasmic polyadenylation element binding protein 1‐mediated mRNA translation in Purkinje neurons is required for cerebellar long‐term depression and motor coordination. J Neurosci 27, 6400–6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Miniaci MC, Kim JH, Puthanveettil SV, Si K, Zhu H, Kandel ER and Bailey CH (2008) Sustained CPEB‐dependent local protein synthesis is required to stabilize synaptic growth for persistence of long‐term facilitation in Aplysia. Neuron 59, 1024–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Huang YS, Carson JH, Barbarese E and Richter JD (2003) Facilitation of dendritic mRNA transport by CPEB. Genes Dev 17, 638–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Si K, Giustetto M, Etkin A, Hsu R, Janisiewicz AM, Miniaci MC, Kim JH, Zhu H and Kandel ER (2003) A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse‐specific long‐term facilitation in aplysia. Cell 115, 893–904. [DOI] [PubMed] [Google Scholar]
- 151. Bestman JE and Cline HT (2008) The RNA binding protein CPEB regulates dendrite morphogenesis and neuronal circuit assembly in vivo. Proc Natl Acad Sci USA 105, 20494–20499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Si K, Choi YB, White‐Grindley E, Majumdar A and Kandel ER (2010) Aplysia CPEB can form prion‐like multimers in sensory neurons that contribute to long‐term facilitation. Cell 140, 421–435. [DOI] [PubMed] [Google Scholar]
- 153. Ernoult‐Lange M, Wilczynska A, Harper M, Aigueperse C, Dautry F, Kress M and Weil D (2009) Nucleocytoplasmic traffic of CPEB1 and accumulation in Crm1 nucleolar bodies. Mol Biol Cell 20, 176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Lin CL, Evans V, Shen S, Xing Y and Richter JD (2010) The nuclear experience of CPEB: implications for RNA processing and translational control. RNA 16, 338–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Rouget C, Papin C and Mandart E (2006) Cytoplasmic CstF‐77 protein belongs to a masking complex with cytoplasmic polyadenylation element‐binding protein in Xenopus oocytes. J Biol Chem 281, 28687–28698. [DOI] [PubMed] [Google Scholar]
- 156. Bava FA, Eliscovich C, Ferreira PG, Minana B, Ben‐Dov C, Guigo R, Valcarcel J and Mendez R (2013) CPEB1 coordinates alternative 3′‐UTR formation with translational regulation. Nature 495, 121–125. [DOI] [PubMed] [Google Scholar]
- 157. Calderone V, Gallego J, Fernandez‐Miranda G, Garcia‐Pras E, Maillo C, Berzigotti A, Mejias M, Bava FA, Angulo‐Urarte A, Graupera M et al (2016) Sequential functions of CPEB1 and CPEB4 regulate pathologic expression of vascular endothelial growth factor and angiogenesis in chronic liver disease. Gastroenterology 150, 982–997.e930. [DOI] [PubMed] [Google Scholar]
- 158. Despic V and Neugebauer KM (2018) RNA tales – how embryos read and discard messages from mom. J Cell Sci 131, pii: jcs201996. [DOI] [PubMed] [Google Scholar]
- 159. Eichhorn SW, Subtelny AO, Kronja I, Kwasnieski JC, Orr‐Weaver TL and Bartel DP (2016) mRNA poly(A)‐tail changes specified by deadenylation broadly reshape translation in Drosophila oocytes and early embryos. Elife 5, pii: e16955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Kronja I, Yuan B, Eichhorn SW, Dzeyk K, Krijgsveld J, Bartel DP and Orr‐Weaver TL (2014) Widespread changes in the posttranscriptional landscape at the Drosophila oocyte‐to‐embryo transition. Cell Rep 7, 1495–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Presnyak V, Alhusaini N, Chen YH, Martin S, Morris N, Kline N, Olson S, Weinberg D, Baker KE, Graveley BR et al (2015) Codon optimality is a major determinant of mRNA stability. Cell 160, 1111–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Bazzini AA, Del Viso F, Moreno‐Mateos MA, Johnstone TG, Vejnar CE, Qin Y, Yao J, Khokha MK and Giraldez AJ (2016) Codon identity regulates mRNA stability and translation efficiency during the maternal‐to‐zygotic transition. EMBO J 35, 2087–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Mishima Y and Tomari Y (2016) Codon usage and 3′ UTR length determine maternal mRNA stability in zebrafish. Mol Cell 61, 874–885. [DOI] [PubMed] [Google Scholar]
- 164. Ingolia NT, Ghaemmaghami S, Newman JR and Weissman JS (2009) Genome‐wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Sampath P, Lee QY and Tanavde V (2011) Identifying translationally regulated genes during stem cell differentiation. Curr Protoc Stem Cell Biol Chapter 1, Unit1B.8. [DOI] [PubMed] [Google Scholar]
- 166. Korzh V (2009) Before maternal‐zygotic transition… There was morphogenetic function of nuclei. Zebrafish 6, 295–302. [DOI] [PubMed] [Google Scholar]
- 167. Maniatis T and Reed R (1987) The role of small nuclear ribonucleoprotein particles in pre‐mRNA splicing. Nature 325, 673–678. [DOI] [PubMed] [Google Scholar]
- 168. Burakova TA, Korzh VP and Neifakh AA (1984) Activation of the synthesis of macromolecules by the Snrna injection into loach (Misgurnus fossilis L) embryos. Dokl Akad Nauk SSSR 274, 413–416. [Google Scholar]
- 169. Burakova TA, Korzh VP, Shostak NA and Neifakh AA (1980) Activation of RNA synthesis in loach embryos after injection of a nuclear extract obtained by using 0.35 M NaCl. Mol Biol (Mosk) 14, 922–927. [PubMed] [Google Scholar]
- 170. Korzh V, Burakova T, Mazin A and Neyfakh A (1985) Activation of genetic apparatus of teleost fish embryos after microinjection of snRNA. Biopolim Cell 1, 14–20. [Google Scholar]
- 171. Burakova TA, Korzh VP, Khainovskaia AM and Neifakh AA (1988) Low‐molecular weight nuclear RNAs increase the duration of loach anucleate embryo life. Dokl Akad Nauk SSSR 303, 733–735. [PubMed] [Google Scholar]
- 172. Forbes DJ, Kornberg TB and Kirschner MW (1983) Small nuclear‐RNA transcription and ribonucleoprotein assembly in early Xenopus development. J Cell Biol 97, 62–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Nash MA, Kozak SE, Angerer LM, Angerer RC, Schatten H, Schatten G and Marzluff WF (1987) Sea urchin maternal and embryonic U1 RNAs are spatially segregated in early embryos. J Cell Biol 104, 1133–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Perumal K, Gu J and Reddy R (2000) Evolutionary conservation of post‐transcriptional 3′ end adenylation of small RNAs: S. cerevisiae signal recognition particle RNA and U2 small nuclear RNA are post‐transcriptionally adenylated. Mol Cell Biochem 208, 99–109. [DOI] [PubMed] [Google Scholar]
- 175. Sinha KM, Gu J, Chen Y and Reddy R (1998) Adenylation of small RNAs in human cells. Development of a cell‐free system for accurate adenylation on the 3’‐end of human signal recognition particle RNA. J Biol Chem 273, 6853–6859. [DOI] [PubMed] [Google Scholar]
- 176. Satija R, Farrell JA, Gennert D, Schier AF and Regev A (2015) Spatial reconstruction of single‐cell gene expression data. Nat Biotechnol 33, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. McKenna A, Findlay GM, Gagnon JA, Horwitz MS, Schier AF and Shendure J (2016) Whole‐organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907. [DOI] [PMC free article] [PubMed] [Google Scholar]


