Abstract
Epitope identification is essential for developing effective antibodies that can detect and neutralize bioactive proteins. Computational prediction is a valuable and time-saving alternative for experimental identification. Current computational methods for epitope prediction are underused and undervalued due to their high false positive rate. In this work, we targeted common properties of linear B-cell epitopes identified in an individual protein class (metalloendopeptidases) and introduced an alternative method to reduce the false positive rate and increase accuracy, proposing to restrict predictive models to a single specific protein class. For this purpose, curated epitope sequences from metalloendopeptidases were transformed into frame-shifted Kmers (3 to 15 amino acid residues long). These Kmers were decomposed into a matrix of biochemical attributes and used to train a decision tree classifier. The resulting prediction model showed a lower false positive rate and greater area under the curve when compared to state-of-the-art methods. Our predictions were used for synthesizing peptides mimicking the predicted epitopes for immunization of mice. A predicted linear epitope that was previously undetected by an experimental immunoassay was able to induce neutralizing-antibody production in mice. Therefore, we present an improved prediction alternative and show that computationally identified epitopes can go undetected during experimental mapping.
Introduction
Correct epitope identification is essential for developing vaccines and selecting high-affinity antibodies for immunotherapy and immunodiagnostics1. Experimental epitope identification is an expensive procedure and comprises several challenges. These challenges include antibody production to identify antigenic regions in a target protein, adequate animal models, and further epitope validation through crystallography. Besides, high-affinity antibody methods and immunoassays can contradict each other on which region is a better target. On the other hand, computational approaches can help to guide experimental assays and improve precision by selecting specific regions with high probability of being effective epitopes2.
Attempts to predict B-cell epitopes started in the 70 s3. They focused on amino acid properties within a sequence, such as hydrophobicity, hydrophilicity, or antigenicity, to identify propensities and patterns4,5. Exhaustive benchmark procedures for (and rigorous statistical analysis of) the biochemical properties that influence epitopes have revealed that single-scale amino acid profiles cannot be used to reliably predict epitope localization6,7. Current classification techniques involve a combination of attributes to increase the information gain. These methods are sensitive to data quality and are often subject to under- or over-representation of attributes. This approach can produce false negatives and positives, which—despite good accuracy (area under the curve [AUC] of 0.7)—lead to misidentification of epitopes7.
The experimental methods that accompany computational identification have limitations. Often, different wet-lab techniques are in disagreement on the important regions in a protein, thus resulting in highly heterogeneous epitope composition8,9. The removal of this experimental noise to train proper classifiers has been attempted by combining attributes, but the results have not been significantly better than those obtained with a few physicochemical attributes4,10,11.
It is generally accepted that most epitopes are conformational12, but even though some algorithms focus on structural properties to target these epitopes, prediction has not improved13,14. A key factor for achieving greater success in separating epitopes from the background is a reduction in both computational and experimental bias8,15. Public databases compiled from validated information16 and statistical analyses17 are essential for building adequate computational models designed for epitope prediction18,19. Moreover, it should be considered that immunological attributes are strongly related to animal models and evolutionary traits of a protein20–22.
Restricting the problem to a specific antigen group may lead to more precise epitope prediction because of the increase in information quality and a reduction in the noise from other protein groups. For testing this hypothesis, we focused on metalloendopeptidases carefully curated by means of available information to predict epitopes16,23. This article describes a methodology—based on a decision tree classifier—for identifying epitopes in a single protein class and for producing neutralizing antibodies against target proteins. The proteins used to experimentally validate our hypothesis belong to the venom of three snake species: Bothrops atrox, Bothrops asper, and Bothrops leucurus. Snakes from the Bothrops genus cause more than 80% of yearly snakebite accidents in Brazil24, thus being medically significant25,26. These venoms exert proteolytic action with well-known biological effects such as hemorrhage that can be evaluated in vitro and in vivo27,28. These characteristics allow for studying the neutralizing-antibody capabilities on the basis of our computational and experimental results, thereby, unveiling antigenic and immunogenic protein properties.
Results
Kmer classification rules allow for reducing false positives from nonepitope residues
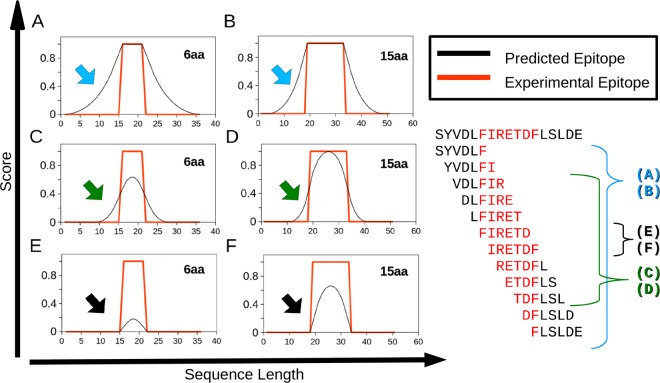
Our manually curated dataset is available in Supplementary Material 1. It contains 40 epitopes, as described in the Methods section. This dataset was transformed into a matrix summarizing 101,115 elements representing Kmers of 3 to 15 amino acid residues (aa), each described by 33 attributes. When a Kmer maps 50% or more of its length onto an epitope or nonepitope, it is assigned to that respective class by our approach. This method produced fewer false positives than did other labeling methods, as illustrated in Fig. 1. Red lines indicate a true epitope, while black lines represent computational prediction. The example shows Kmers of 6 and 15 aa under three selection conditions: when only 1 aa is required to label the Kmers as an epitope (A and B), when the 50% or more rule used in our method is applied (C and D), and when the exact epitope must be matched to regard a Kmer as an epitope (E and F). It must be highlighted that the method proposed here is less sensitive for small epitopes (under 5 aa), but it can be later corrected by the SMOTE algorithm during the decision tree classifier training.
Figure 1.
Selection of Kmers as epitopes. The graphs illustrate how the selection of positive Kmers to be considered epitopes alters the rate of false positives based on compositional rules. As an example, Kmers of 6 and 15 aa were employed. The X-axis shows the amino acid sequence position. The Y-axis shows the probability of an amino acid residue to be a part of an epitope. Red lines represent a true epitope. Black lines represent computational prediction. (A) and (B) illustrate a prediction where at least one amino acid residue must be predicted as an epitope to label a Kmer as positive (C) and (D) show that when 50% of the amino acids must be predicted as an epitope to label a Kmer as positive (E) and (F) shows when all the amino acid residues from of a Kmer must be predicted as an epitope to label a Kmer as positive. The arrows indicate the portion of potentially false positives in each prediction method. On the right side of the figure, there is an example of prediction of an epitope marked red within the sequence SYVDLFIRETDFLSLDE by means of a 6 aa Kmer and the three approaches illustrated in the graphs.
Charged amino acid residues contribute to B-cell epitope prediction with a decision tree model
We used a decision tree classifier for predicting epitopes of three metalloendopeptidases from Bothrops snake venoms (Bap1, Atr-I, and Leuc-a). Epitopes derived from these proteins were not present in our classifier training dataset. The classification tree is presented in Supplementary material 2. A comparison with random-forest attribute analysis highlighted the importance (for the aliphatic index) of the percentages of Arg (R), His (H), Lys (K), Glu (E), Asp (N), Pro (P), and Trp (W). Table 1 summarizes this comparison. The highlighted attributes represent the first nodes of the decision tree model. Decreased Gini values are an inequity measure between epitope and nonepitope classes. The lower values represent the best attributes across a million trees growth with random forest.
Table 1.
Valuable attributes from random-forest and decision tree classifiers.
| Attribute | Decreased Gini | Decreased Accuracy |
|---|---|---|
| Positive charged RHK | 75,28 | 37.50* |
| Negative charged DE | 79,12 | 52,71 |
| Uncharged STNQ | 101,78 | 63,27 |
| Special CGP | 100,9 | 43.43* |
| Hydrophobic AVILMFW | 92,84 | 43.16* |
| gravy | 198,59 | 60,82 |
| Aliphatic index (5) | 118,75 | 45.49* |
| % Atoms of C | 234,22 | 76,8 |
| % Atoms of H | 212,17 | 58,34 |
| % Atoms of N | 233,88 | 59,07 |
| % Atoms of O (12) | 198,23 | 68,39 |
| % Atoms of S (1) | 166,32 | 67,65 |
| % Arg (4) | 72,98 | 39.25* |
| % His (2) | 68.85* | 53,73 |
| % Lys (3) | 36.46* | 42.84* |
| % Asp | 80,68 | 56,97 |
| % Glu (6) | 66.12* | 52,07 |
| % Serine | 107,13 | 76,88 |
| % Thr (11) | 94,8 | 73,62 |
| % Asn (9) | 74,87 | 47.39* |
| % Gln | 73,67 | 55,23 |
| % Cys | 77,22 | 45.50* |
| % Gly | 98,59 | 52,81 |
| % Pro(10) | 58.36* | 39.87* |
| % Ala | 86,08 | 65,12 |
| % Val | 48.48* | 38.31* |
| % Ile | 68.87* | 40.67* |
| % Leu | 74,26 | 42.55* |
| % Met | 87,91 | 61,82 |
| % Phe | 48.65* | 39.62* |
| % Tyr | 64.44* | 56,46 |
| % Trp(8) | 57.03* | 51,84 |
| Isoelectric point (7) | 183,09 | 54,9 |
The attributes in bold represent the first nodes of the decision tree and (*) indicate the best attributes from random-forest.
Experimental and computational B-cell epitope mapping
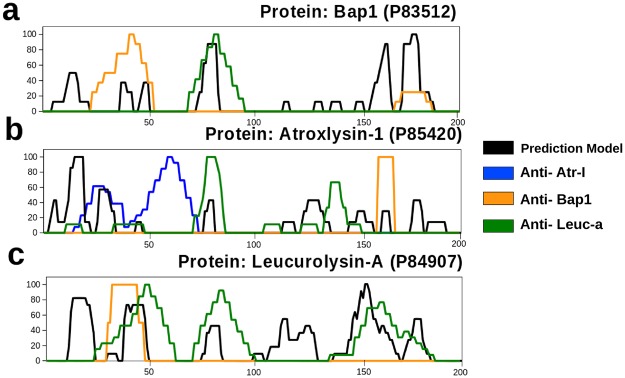
To compare our computational prediction method with experimental approaches, we used SPOT immunoblotting to map epitopes within metalloendopeptidases Atr-I, Leuc-a, and Bap1, using specific antibodies developed against each protein. Each protein was probed with all three antibodies: anti-Atr-I, anti-Leuc-a, and anti-Bap1. We identified two epitopic regions for Atr-I (aa 19–39 and aa 46–75), shown as blue lines (Fig. 2b), three regions for Leuc-a, highlighted as green lines (Fig. 2c), and two regions for Bap1, presented as orange lines (Fig. 2a). The local alignment showed that Atr-I shares a sequence identity of 55.45% and 50% with Bap1 and Leuc-a, respectively, while Leuc-a and Bap1 share 78.22% identity (Table 2). Besides, SPOT-identified epitopes presented similar position within the protein sequences.
Figure 2.
Predicted and experimental epitope overlapping. The X-axis shows amino acid residue position. The Y-axis represents the experimental and predicted epitope score values from 0 to 100. Black lines represent the epitopes predicted by our model. The blue, orange, and green lines represent epitope mapping by SPOT-Immunoblotting using anti-Atr-I, anti-Bap1, and anti-Leuc-a antibodies respectively. Letters (a–c) represent the mapping of epitopes within the individual proteins, Bap1, Atr-I, and Leuc-a from Bothrops asper, B. atrox, and B. leucurus, respectively. The overlapping positions between black and colored lines represent successful predictions, while overlapping between colored lines indicates a cross-reaction.
Table 2.
Clustal Omega Identity matrix between metalloproteinases.
| Atr-I | Leuc-a | Bap1 | |
|---|---|---|---|
| Atr-I | 100 | 50.5 | 55.45 |
| Leuc-a | 50.5 | 100 | 78.22 |
| Bap1 | 55.45 | 78.22 | 100 |
We predicted eight epitopic regions for Atr-I (positions 4–6, 12–20, 28–35, 79–83, 127–136, 153–156, 165–168 and 180–183), six epitopic regions for Bap1 (13–18, 39–44, 49–52, 77–86, 161–169, and 176–188), and six epitopic regions for Leuc-a (11–24, 38–48, 78–84, 114–130, 147–168, and 176–183; Table 3). These predicted regions successfully matched experimental results (Fig. 2). The experimental mapping by SPOT yielded two epitopes in Bap1 (orange lines), two in Atr-I (blue lines), and three in Leuc-a (green lines). These regions were identified by means of specific sera against each of these toxins. Cross-reaction was also observed when we employed different antibodies against each protein. The anti-Leuc-a antibody, when used against the Bap1 spot membrane, recognized a region different from that recognized by anti-Bap1 serum. On the other hand, anti-Atr-I sera did not recognize any epitope from Bap1. Furthermore, the anti-Leuc-a antibody when used against Atr-I, recognized two central regions different from those recognized by anti-Atr-I or anti-Bap1 sera. Anti-Leuc-a identified a single epitope region close to the C-terminal Atr-1 segment. Anti-Atr-I antibodies only identified epitopes in Atr-I, while the other two polyclonal antisera showed cross-reactivity.
Table 3.
Epitopes discovered computationally (A), experimentally (B) and cross reactive regions (C).
| Start | Sequence | End |
|---|---|---|
| A Computationally predicted epitopes | ||
| Atroxlysine-I | uniprot P85420 | |
| 4 | QQR | 6 |
| 11 | FIVVDHGMF | 19 |
| 27 | DKIRRRIH | 34 |
| 78 | FGEWR | 82 |
| 126 | IQDHSEQDLM | 135 |
| 152 | HDTG | 155 |
| 164 | CIMS | 167 |
| 179 | SDCS | 182 |
| Bap | uniprot P83512 | |
| 13 | VVADHG | 18 |
| 39 | NTVGF | 44 |
| 49 | DVHA | 52 |
| 77 | KSFGEWRERD | 86 |
| 161 | GAKSCIMAS | 169 |
| 176 | SYEFSDCSQNQYE | 188 |
| Leucurolysin | uniprot P84907 | |
| 11 | VVADHGMFKKYN | 24 |
| 38 | NTVNGFFRSMN | 48 |
| 78 | FGEWRER | 84 |
| 114 | AGMCDLSQSVAVVMDHS | 130 |
| 147 | NLGMRHDGNQCHCNAPSCIMAD | 168 |
| 176 | FEFSDCSQ | 183 |
| B Experimentally mapped epitopes | ||
| Atroxlysine-I | uniprot P85420 | |
| 19 | FMKYNGNSDKIRRRIHQMVNI | 39 |
| 46 | TMYIDILLTGVEIWSNKDLINVQPAAPQTL | 75 |
| C Cross reactive regions | ||
| Atroxlysine-I | uniprot P85420 | |
| 78 | FGEWRKTDLLN | 88 |
| 137 | AITMAHELGHN | 147 |
| 163 | SCIMSPVL | 167 |
Receiver operating characteristic analysis shows an improvement in accuracy and a reduction in false positives
Three state-of-the-art prediction methods and our model were compared by the ROC curve analysis with default cutoffs that each software suggested. We also used cutoffs that maximized AUC and Precision (Table 4). We obtained mean AUC for the three proteins (Bap1, Leuc-a, and Atr-I) of 0.5407 followed by ABCpred (0.5382), TEPRF (0.5297), and BepiPred (0.4450). All the measurements showed that our predictor had better performance, accuracy, precision, true positive rate, and AUC while always having the lowest false positive rate (Table 4). The false positive rate was found to be significantly lower (0.3266) when compared to ABCpred (0.5752), BepiPred (0.3961), and TEPRF (0.5121).
Table 4.
Comparison statistics of computational B-cell epitope prediction method.
| Method | AUC | Accuracy | TPR | FPR | Precision | Specificity |
|---|---|---|---|---|---|---|
| Default software cutoff statistics | ||||||
| ABCpred | 0,5382 | 0,4384 | 0,6516 | 0,5752 | 0,0906 | 0,4248 |
| Bepipred | 0,4450 | 0,5655 | 0,2860 | 0,3961 | 0,1356 | 0,6039 |
| Labimq | 0,5407 | 0,6175 | 0,4080 | 0,3266 | 0,2333 | 0,6734 |
| TEPRF | 0,5297 | 0,4972 | 0,5714 | 0,5121 | 0,1097 | 0,4879 |
| Cutoffs that maximize precision | ||||||
| ABCpred | 0,5525 | 0,5116 | 0,6275 | 0,5224 | 0,2041 | 0,4776 |
| Bepipred | 0,4825 | 0,5556 | 0,3976 | 0,4326 | 0,2117 | 0,5674 |
| Labimq | 0,5996 | 0,7210 | 0,4052 | 0,2059 | 0,3157 | 0,7941 |
| TEPRF | 0,5878 | 0,3790 | 0,9896 | 0,8140 | 0,2407 | 0,1860 |
| Cutoffs that maximize Area Under the Curve | ||||||
| ABCpred | 0,6542 | 0,6722 | 0,6617 | 0,3534 | 0,1255 | 0,6466 |
| Bepipred | 0,5499 | 0,6650 | 0,3799 | 0,2802 | 0,1443 | 0,7198 |
| Labimq | 0,6306 | 0,7876 | 0,4189 | 0,1577 | 0,2852 | 0,8423 |
| TEPRF | 0,6330 | 0,4912 | 0,8464 | 0,5805 | 0,1742 | 0,4195 |
Experimental validation of epitope prediction by immunization of mice
We selected two Atr-I regions to be synthesized as peptides by Fmoc chemistry for antibody production. One epitope corresponding to the N-terminal region (9VDLFIVVDHGMFMKY23) was identified by our model with a prediction score of 0.49; we also selected a central region (99LTSTDFNGPTIGLAY113), which was not identified by our model. Both regions were not mapped in SPOT experiments. We chose a cutoff of 0.2 to classify a sequence as positive. The peptides were called AtrCPEN (Computationally Positive Experimentally Negative) and AtrCNEN (Computationally Negative and Experimentally Negative), respectively. Their molecular masses were verified by mass spectrometry after synthesis and corresponded to the predicted amidated and acetylated masses (not shown).
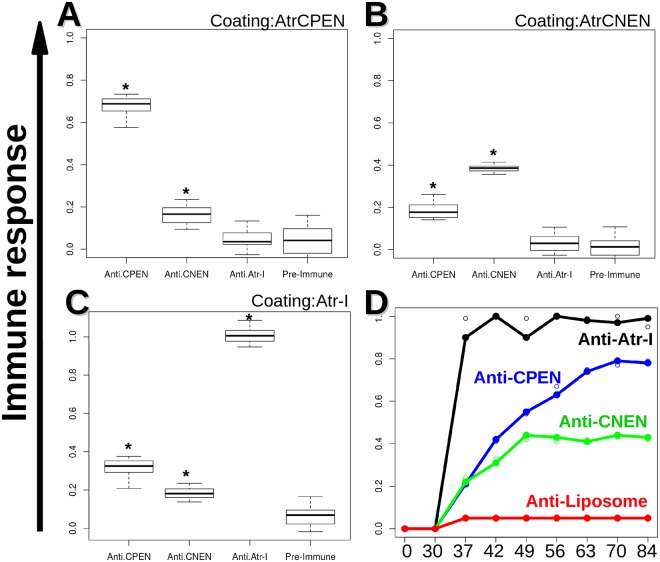
AtrCPEN and AtrCNEN peptides were utilized for immunization of BALB/c mice, after incorporation into liposomes as an adjuvant. An ELISA was conducted to monitor antibody production against AtrCPEN and AtrCNEN. It was possible to detect specific antibody production after the 7th dose (day 63; Fig. 3D). The two synthesized peptides were not recognized by anti-Atr-I serum. By contrast, anti-AtrCPEN (0.3 Abs.) recognition of Atr-I as an antigen was slightly higher than that obtained by Anti-AtrCNEN (0.2 Abs; Fig. 3B). Antipeptide sera (Anti-AtrCPEN and Anti-AtrCNEN) only poorly recognized Atr-I. The antibody responses were compared by the t test showing a p-value lower than 0.05 for all groups (confidence interval: 95%).
Figure 3.
ELISA tests of anti-CPEN, CNEN, and Atr-I sera. (A–C) show the boxplots results for ELISA plates coated individually by CPEN, CNEN, and Atr-I, respectively. |Plates were incubated for 1 h with the respective sera at 37 °C followed by another round of 3× washing before incubation with a respective secondary antibody for 1 h. An OPD substrate was added for ~20 minute incubation, and the reaction was stopped with H2SO4 prior plate reading. The black lines within the boxes correspond to medians. All the samples marked with (*) show to be significantly different for a p < 0.05, when comparing by the t test available in software R. (D) shows antibody binding over 9 doses (x-axis represents days).
To verify whether the produced antibodies against Atr-CPEN and Atr-CNEN had neutralizing properties against Atr-I, we tested the enzymatic activity of Atr-1 over time, in the presence of anti-CPEN and anti-CNEN, using a synthetic substrate, Abz-LVEALYQ, that produces fluorescence when cleaved (Supplementary File 3). This assay showed Atr-I activity reduction by 80–70% for anti-CPEN and 30–20% for anti-CNEN. This result indicated the successful neutralization of Atr-I activity, when incubated with anti-CPEN; this neutralization was significantly stronger than that obtained with the anti-CNEN antibody.
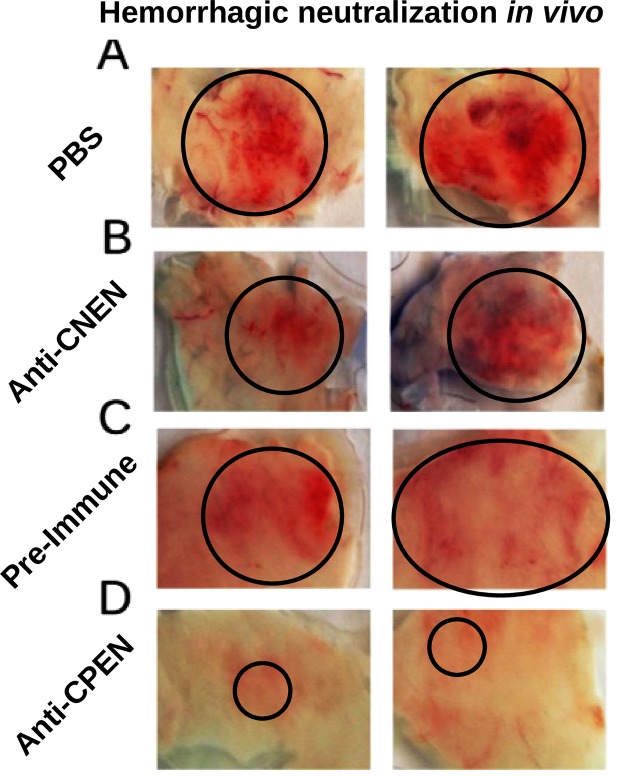
Because Atr-I activity neutralization by anti-CPEN antibodies was observed in vitro, Atr-I-induced hemorrhage neutralization was tested in vivo, in BALB/c mice. They were challenged with a toxin amount corresponding to 1 Minimum Hemorrhage Dose (19 μg of Atr-I in 100 μL), as described by Schneider et al., 2016. The animals challenged with Atr-I mixed with anti-AtrCPEN serum showed a clear reduction in hemorrhage when compared to the positive control group. Negative control with preimmune serum samples and anti-AtrCNEN serum yielded slightly reduced hemorrhage when compared with the positive control, probably owing to other serum components that interfere with the enzymatic metalloendopeptidase activity (Fig. 4).
Figure 4.
Hemorrhagic Atr-I activity neutralization in vivo. Mice were challenged with 1 MHD (Minimal Hemorrhage Dose) of Atr-I diluted either in PBS (A) anti-CNEN serum (B) preimmune serum (4C) or anti-CPEN serum (D). Black circles indicate hemorrhagic areas in animal skin, for each treatment. (A–C) showed a clear hemorrhage area, while serum against CPEN (shown in D) was able to reduce hemorrhage, causing only skin irritation.
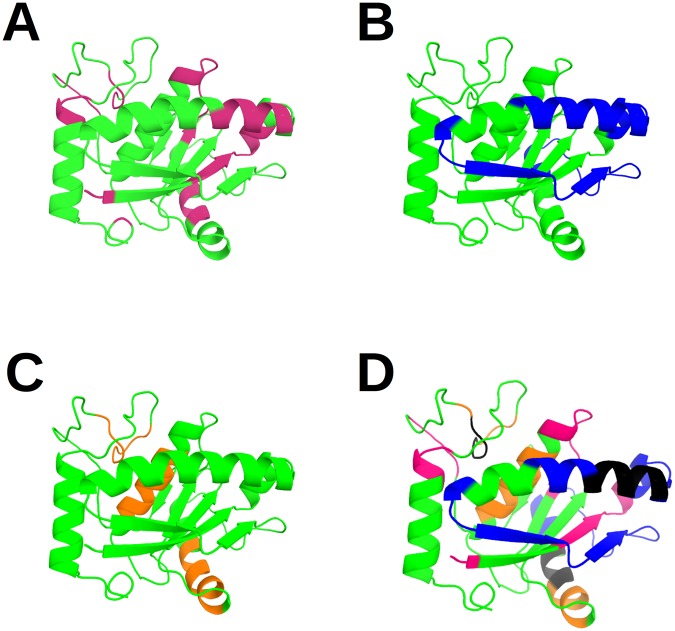
Spatial distribution of epitopes obtained by computational and experimental methods showed an overlapping region, whereas cross-reactive regions did not seem to have any pattern (Fig. 5).
Figure 5.
Localization of predicted epitopes in the Atr-I model. A cartoon view of the structural model of the protein Atr-I is displayed. The residues that belong to the computational prediction are shown in pink (A) while the experimental epitopes and the cross-reactive regions between the different serum samples tested are indicated in blue (anti-Atr1) and orange respectively (anti-Bap1 and anti-Leuc-a) (B,C). The overlap of these methods is presented in (D) where the black regions correspond to the matching computational and experimental predictions.
Discussion
B-cell epitopes are related to a humoral immune response and play a key role in vaccine production and several biotechnological applications, while T-cell epitopes are associated with cell-mediated immunity29,30. The experimental epitope identification is time- and resource-consuming in comparison with other computational techniques2. Computer algorithms to predict B-epitopes by means of an antigen sequence3,4 or structure9,13,31 have been refined over the past decades. These techniques have been accompanied by experimentally characterized datasets comprising both positive epitopes and negative nonepitopes15.
The first challenge for epitope prediction is represented by database construction aimed at organizing the disproportional negative or nonepitope examples16. Another complication is the negative example selection, based on randomly chosen sequences, where no antibody binding is reported22. Collections of both epitopes and nonepitope sequences from experimentally validated data are available, e.g., the Immune Epitope Database (IEDB)16. Nevertheless, experimental nonepitope data still have the potential for being due to possibly flawed result interpretation, a lack of detailed mapping, or simple experimental errors21. Furthermore, this principle applies to all existent experimental and computational methods as demonstrated in our results dealing with the production of neutralizing antibodies having a region undetected by experimental mapping.
The factors that influence the immune response and epitope detection are mostly attributed to genetics19, evolution9, immunological complexity5,18,20, structural conformation32, surface indistinguishability15, and others31. Moreover, we recently explored the differences between epitopes from different antigen classes or families and revealed that these differences can be useful for identification of epitopes23. We took advantage of the antigen sequence from a single protein class (metalloproteinases) to train a decision tree classifier. Furthermore, we validated our hypothesis by comparing our results to those of state-of-the-art predictors and experimental methods for three different proteins, thus showing an accuracy improvement and a reduction in the false positive rate (Table 4). The important attributes for classification included the isoelectric point, lateral chain size, and amino acid residues such as Asn, Gln, Ser, Thr, Lys, or Trp (Table 1), which are described as antigenic2,6,7. Other studies indicate that sequence attributes can be used for analyzing structural33 and sequential epitopes34,35, thus highlighting hydrophilic amino acids because they surround antigenic determinants36.
Classification models are great tools for identifying patterns within complex data and gradually gain importance in computational biology owing to rising information amounts37. Machine-learning approaches have undisputed advantages over simpler methods, such as regression38, but their persistent limitations are the inability to point out relevant characteristics and the necessity of high computational power34. Regression-based methods such as random forest39, decision tree40, and linear regression11 allow researchers to identify these attributes rapidly41,42 (Table 1, Supplementary File 2).
Here, we show an improvement in the performance on in silico epitope prediction (Table 4) and in vivo validation (Figs 2 and 4), especially in terms of the false positive rate, when compared to other methods (BepiPred, ABCpred, and TEPRF). BepiPred employs hidden Markov models to identify propensities in sequence data, despite underperformance of similar approaches, as uncovered by Blythe and Flower6,32. ABCpred is a method that involves recurrent neural networks to analyze fixed length windows of less than 20 amino acids and their biochemical properties43. The random forest approach (TEPRF) takes advantage of two powerful machine-learning techniques: bagging (bootstrap) and random attribute selection. TEPRF yields a large number of false positives that could be explained by the attempt at overpredicting the under-represented groups or experimental epitopes. To overcome these limitations, we increased the negative/positive examples for epitopes by separating the full sequence into Kmers of several lengths (3 to 15 aa) and by correcting the proportions with SMOTE, thus increasing overall performance (Table 4).
A major point poorly discussed in the literature is that experimental nonepitopes are classified as false positives and are employed as such by predictors30. We demonstrated that one of these epitopes predicted and experimentally mapped as a negative epitope (CPEN) was able to induce neutralizing antibodies (Figs 3 and 4). This finding also means that statistical comparisons for the current classification methods harbor a bias that significantly alters the accuracy of current predictors. This bias could be due to variation in immunological or experimental conditions. Continuous epitopes are powerful diagnostic/treatment tools, despite representing only 10% of all estimated B-cell epitopes14,44,45.
The low accuracy seen during prediction validation across the different algorithms could be due to the statistical bias caused by incomplete experimental results or database failure. Most amino acids identified as epitopes in silico by us were in the proximity of (or were partially included in) experimental epitopes (Fig. 2). This is important because linear epitope sequences can be rapidly produced as soluble peptides for immunization1,29. These peptides are flexible34 and have a higher probability of mimicking epitopes46. Peptides designed on the basis of these epitopes are undoubtedly powerful tools for improving vaccine efficacy38,47.
This study was focused on the snake venom metalloendopeptidase called Atroxlysin-I as a candidate protein for our experiments because it is a well-characterized enzyme. It can enzymatically cleave Xaa-Leu bonds in proteins such as fibrin, fibronectin, type I and IV collagens, and other extracellular-matrix components and can induce hemorrhage. Besides, Atr-I interferes with platelet aggregation in an enzymatically independent manner1.
Of note, in this study, it was shown that a predicted epitope for Atr-I (AtrCPEN), not identified by experimental mapping, can be employed to induce neutralizing antibodies. The region identified (aa 11–19) was expanded to aa 8–22 for synthesis purposes; this approach non significantly reduced the score of our model from 0.6 to 0.5, whereas any sequence with a score above 0.2 was regarded as an epitope (Fig. 2). The sequence chosen to represent a negative prediction had a score of zero, and it was not recognized by experimental procedures. Both peptides induced antibody production with a lower response to Atr-I than its corresponding anti-Atr-I sera (Fig. 3). The lower response associated with peptide-based antibodies has been observed previously14. Atr-I was experimentally found to contain two immunogenic regions in the regions aa 19–39 and aa 46–75, as identified with anti-Atr-I serum obtained elsewhere48. Our prediction match region (positions 27–34) is similar to BepiPred’s (positions 23 to 29) (data not shown), which are closer to the border of the experimentally identified region48. TEPRF and ABCpred select larger protein portions, reflected in a higher false positive rate (Table 4). The experimental epitopes found in Leuc-a, Bap1, and Atr-I contain a common region between residues 28 and 39 (Atr-I), while the two peaks for Atr-I: one, for Bap-1, and the other for Leuc-a, are encapsulated between positions 19 and 64.
The Atr-I structural model revealed that anti-Atr-I sera recognize two regions near the N-terminal portion of Atr-I (Fig. 5). All three analyzed endopeptidases had epitopes located in the region comprised by the first and second sequence portion close to the N-terminal amino acid, as shown by orange, blue, and green color peaks (Fig. 2). This region comprises two α-helices with a loop in between, followed by another loop and a strand. These helix-loop regions were shown to be immunoreactive with anti-Atr-I sera and with cross-reactive sera as well (Fig. 5). The preference of anti-Bap1 sera matched our predictions as well on the first helix-loop with residues 19–39. This structure seems to be conserved among all metalloendopeptidases and could be a source for the development of additional antivenom agents and vaccines against other toxic endopeptidases. This region was erroneously labeled as an experimentally negative region, but we presented evidence to the contrary, and we were able to neutralize the hemorrhagic effect of Atr-I (Fig. 4). The impact of this approach during epitope predictions is clear, and some regions erroneously classified by experimental methods can harbor immunogenic properties (Fig. 2). The other two regions next to aa 65–75 may point to another region that was immunodominant for anti-Leuc-a sera whereas anti-Bap1 showed a preference for a region close to the C-terminal Atr-I segment. These untested regions may be important for other endopeptidases owing to their cross-reactivity and structural identity.
An immunogenic region depends on the system where it is identified and on the parameters of this system49, such as the immunized host type, antigen type, inoculation method, adjuvant presence, and others50. These epitopes can be refined for vaccination51 and their biotechnological applications are well known1,29. Several computational methods are available32, and their results are in agreement within consequences of certain conditions, in which amino acids, hydropathy values, and others are relevant for immunodominance49. These properties escalate to form complex networks and energetic mechanics52 thus making the task of predicting B-cell epitopes a major challenge32. Computational and experimental methods suggest that regions helix-loop, sheet-loop, helix-loop-helix, and helix-loop-sheet are the most likely to result in epitopes, binding, and therefore detection and neutralization of a target protein53. Besides, this study showed an improvement in epitope prediction accuracy by revealing a specific approach to the still complicated task of predicting neutralizing epitopes and vaccine targets.
Conclusion
This work describes development of a classification model based on a protein dataset that belongs to a single antigen class (metalloendopeptidases), but the method can be applied to any protein class. This model successfully predicted linear epitopes that overlap with experimentally determined epitopes on three sample proteins, with better performance than ABCpred, BepiPred, and TEPRF. We also determined which biochemical attributes are important during epitope prediction for this model. Furthermore, antisera raised against these epitope regions were demonstrated to be cross-reactive and will improve the understanding of the immunoreactive regions in metalloendopeptidases. Furthermore, we produced neutralizing antibodies against Atroxlysin-I through immunization with a synthetic peptide. The selected region was based on a predicted positive but experimentally negative epitope. Therefore, it was demonstrated here that computationally positive predictions can serve as a basis for producing peptides capable of raising neutralizing antibodies.
Methods
Ethics statement
The study protocol was approved by the Ethics Committee for Animal Experimentation, Universidade Federal de Minas Gerais (protocol number 200/2010). All the experiments were performed in accordance with Guide for the Care and Use of Laboratory Animals, US National Institutes of Health (NIH Publication No. 85-23, revised 1996).
Animals and venoms
The animals were maintained at Centro de Bioterismo and received water and food under controlled environmental conditions (Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais, Brazil). A venom pool from at least six adult Peruvian B. atrox specimens was donated by the Instituto Nacional de Salud (Lima, Peru). Purified Atr-I was previously obtained in our laboratory, as previously described1,9, ICB-UFMG.
Dataset
The B-cell epitope dataset (Rong and Jianjun, 2011), was used and modified, as previously by us elsewhere23. Briefly, this dataset contains manually curated and selected metalloendopeptidases based on the experimental validation procedures as described (Supplementary Data 1).
Attribute matrix analysis
We produced a matrix based on the properties of Kmers between 3 and 15 aa derived from every protein and epitope in our dataset. The Kmer sizes were chosen according to a common epitope size distribution54. The attributes are the percentages of each of the 20 aa; sequence lengths; hydropathy index; atom percentages of C, H, O, N, and S; the aliphatic chain size index; isoelectric point; and amino acid percentages grouped by hydrophobic, positive, negative, polar, and special amino acids (CGP).
The Kmers were tagged as an epitope or nonepitope if they had 50% or more aa that belonged to the respective class. The attributes were computed by a Biopython package for Python55 and Perl scripts based on Expasy descriptions (Wilkins MR., et al. (1999)). We selected a decision tree model as a data-mining technique for classifying the Kmers according to their attributes. We also chose the ClustalO56 local alignment option for aligning the metalloendopeptidases Bap1 (B. asper), Leuc-a (B. leucurus), and Atr-I (B. atrox).
Classification of computational epitopes
The decision tree classifier tends to overpredict the majority class. To avoid this bias, the positive and negative sequence proportion was altered by means of the SMOTE algorithm57 for over-representing the minority class and improving the performance of decision tree models.
A classification model series was tested using KNIME58. The decision tree was selected as the most suitable method because of speed and performance23. The important attributes were compared with two measures (decreased Gini and accuracy) produced by randomForest Package on R, after a million trees produced by means of the same attributes as in our decision tree model. These measures are defined as inequity and inclusion measures, respectively.
Statistical validation
Statistical analysis was carried out in R59, as described elsewhere23 and included 10-fold cross-validation (Krstajic., et al. (2003)). This classification model performance was studied by analyzing a receiver operating characteristic (ROC) curve17, recall, precision, specificity, area under the curve (AUC), and Cohen’s Kappa coefficient39. Mean comparisons between experimental and computational results were conducted by the t test available in the R software.
Cellulose-bound peptide production and immunoassay
Briefly, A Multipep (Intavis) robot was used for automating peptide synthesis of overlapping pentadecapeptides frame-shifted by 3 residues covering the entire amino acid sequence of Atr-I, Leuc-a, and Bap1 on cellulose membranes. Later, these membranes were tested against specific serum samples, as previously described by us26,27,48.
Soluble peptide synthesis
Fmoc amino acids were acquired from Novabiochem or Sigma Aldrich. After epitope prediction, two linear regions were selected for synthesis. The first region was localized near the N-terminal region of Atr-I (11-FIVVDHGMF-19). We increased the predicted sequence by 3 aa for each border, resulting in a final sequence of 15 aa with composition 9-VDLFIVVDHGMFMKY-23 (AtrCPEN). A second region is localized in the central part of the protein 99-LTSTDFNGPTIGLAY-113 (AtrCNEN). AtrCNEN was also 15 aa long and showed no previous immunological response and was undetectable by the computational methods tested in this study. The two peptides were synthesized by the Fmoc chemistry method on an automatic Multiprep robot (Intavis)60. During the synthesis, the growing peptide was immobilized on the Rink Amide resin (Novabiochem). At the end of the synthesis, peptides were released from the resin, and the side chain deprotection was carried out by trifluoroacetic acid treatment (95% TFA, 2.5% triisopropylsilane, and 2.5% water). The molecular masses of synthesis products were analyzed by mass spectrometry (MALDI-TOF, linear mode).
Production antipeptide sera
Two groups of five BALB/c female mice were immunized subcutaneously with AtrCNEN or AtrCPEN. Each peptide was encapsulated into Asolectin liposomes (Sigma Aldrich), as described by us elsewhere27. Aluminum hydroxide (40 μg/μL) was added in a 1:1 (v:w) ratio as an adjuvant. All animal groups received 10 doses during a 3-month protocol with the initial 1-month interval, followed by weekly doses. Experimental groups received 50 μg of an encapsulated peptide per dose per animal. Control animals were immunized with empty liposomes (without an antigen) in aluminum hydroxide (40 µg/µL). One week after the last immunization, the mice were bled to recover the immunized serum.
Antigenic anti-AtrCNEN and anti-AtrCPEN analysis by ELISA
Recognition of the synthesized peptides by BALB/c IgG anti-Atr-I antibody (previously produced and kindly donated by Sanchez et al.26) was tested in Maxisorp plates (Nunc) coated with AtrCNEN, AtrCPEN, (10 μg/mL), or Atr-I (5 μg/mL) overnight at 4 °C in coating buffer (0.05 M Na2CO3, pH 9.6). After blockage for 1 h at 37 °C with a powdered milk solution (2%) in PBS containing Tween 20 (0.1%), IgG Anti-Atr-I, AntiAtrCPEN, or AntiAtrCNEN produced in BALB/c mice was incubated for 1 hour at 37 °C. A goat anti-mouse IgG antibody conjugated to peroxidase (Sigma) served for detecting the reaction followed by addition of the OPD Peroxidase substrate (SIGMAFAST from Sigma-Aldrich).
Methods for computational evaluation and comparison
The immunoassay data from cellulose-bound peptides were employed for calculating a reactivity score for the three out-sample proteins uncharacterized at the time (Bap1, Leuc-a, and Atr-I). The score is based on the aa occurrence numbers in each predicted overlapping reactive peptide and later was scaled to a maximum value of 1. These results were compared with our model, BepiPred, ABCpred, and TEPRF7,43.
Experimental validation
The Atr-I protein and sera against Atr-I Leuc-a and Bap1 were used to experimentally validate the classification model. We selected one region from Atr-I, considered immunogenic only by our predictor and a second region considered immunologically negative by all the tested mapping methods (i.e., computational and experimental). These two peptides were synthesized and used for producing antibodies and were later characterized. In addition, we compared the cross-reactivity of anti-Atr-I, anti-Bap1, and anti-Leuc-a sera (Kindly donated by Schneider, F. et al.61 against each sample protein. Immunoassays with cellulose-bound peptides were conducted. Besides, we compared these results to the predictions. Finally, all the comparisons between experimental and computational methods were expressed in accuracy, precision, recall, AUC, and ROC curves. Furthermore, we compared the epitope spatial distributions using a 3D model built with a combination of methods35,62,63.
An in vitro neutralization assay
The FRET peptide (Abz-LVEALYQ-EDDnp) kindly donated by Schneider et al.61. was used to test the neutralizing activities of antiCPEN and antiCNEN after Abz-LVEALYQ-EDDnp hydrolysis by purified Atr-I also donated by Schenider et al.61, 2016. First, 11 ng of Atr-I was preincubated with 1, 2, or 3 µg of antiCPEN or antiCNEN for 30 min at 37 °C. Then, the substrate was added at a final concentration of 47 mM. Positive controls were set up by preincubating Atr-I alone for 30 min at 37 °C. The residual activity and neutralizing activity were normalized to the positive control. Enzymatic activity was measured by fluorescence on a Sinergy2 (Biotek) instrument (λex = 320 nm and λem = 420 nm) for 30 min at 37 °C as described by Schneider et al.61.
An in vivo neutralizing assay
Atr-I–induced hemorrhage neutralization was tested in 16 BALB\c mice separated into groups of four. One minimum hemorrhagic dose (MHD/kg) of Atr-I (19 μg per mouse of 18–22 g)48 was pre-incubated with 50 μL of either anti-AtrCNEN or anti-AtrCPEN sera for 1 hour at 37 °C. The mixtures were inoculated subcutaneously into four mice per group. As a positive control, Atr-I was injected alone (without antisera). The negative control involved preimmune sera. After 3 hours, the mice were euthanized and their skin was removed for evaluating hemorrhage.
Electronic supplementary material
Proteins used as training for the decision tree
Acknowledgements
The authors thank the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Brazil; Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Brazil; Fundação de Amparo à Pesquisa do Estado de Santa Catarina, Brazil; and Fundação de Amparo à Pesquisa do Estado de Minas Gerais (FAPEMIG), Brazil, for financial support; Centro Nacional de Supercomputação (CESUP) as well as Dr. José Maria Gutiérrez (Instituto Clodomiro Picado), Facultad de Microbiología, Universidad de Costa Rica, for providing the Bap1 protein.
Author Contributions
E.E.G.K. conceived and developed the idea of using data-mining techniques applied to single-protein classes for classification; performed immunization protocols and neutralization assays and wrote most of the manuscript. L.C. oriented, discussed and helped to develop the classification models. F.S.C. produced and purified the antibodies and FRET-peptides which allowed for the comparison and validation of the computational approach. B.T.V. discussed actively all the ideas applied to the study. C.N., B.T.S. and S.A.L. produced the membranes and did most of the experiments on the antisera used on this research. F.M., C.G.D. and L.F. established the basis and initial observations on which this research was built. C.C.O. and R.A.M.A. discussed and guided all the research as well as provided all the necessary equipment. All the authors reviewed and approved the final manuscript.
Data Availability
All the data depicted in the figures are available upon request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Edgar Ernesto Gonzalez Kozlova, Email: eegonzalezk@gmail.com.
Ricardo Andrez Machado-de-Ávila, Email: r_andrez@yahoo.com.br.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-33298-x.
References
- 1.Schneider FS, et al. Use of a synthetic biosensor for neutralizing activity-biased selection of monoclonal antibodies against Atroxlysin-I, an hemorrhagic metalloendopeptidase from Bothrops atrox snake venom. Plos Negl. Trop. Dis. 2014;8(4):e2826. doi: 10.1371/journal.pntd.0002826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansari HR, Raghava GP. Identification of conformational B-cell epitopes in an antigen from its primary sequence. Immunome Res. 2010;10(6):1745–7580. doi: 10.1186/1745-7580-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hopp TP, Woods KR. Prediction of protein antigenic determinants from amino acid sequences. Proc. Natl. Acad. Sci. USA. 1981;78(6):3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kolaskar AS, Tongaonkar PC. A semi-empirical method for prediction of antigenic determinants on protein antigens. FEBS Lett. 1986;276:172–174. doi: 10.1016/0014-5793(90)80535-Q. [DOI] [PubMed] [Google Scholar]
- 5.Moreau V, et al. PEPOP: computational design of immunogenic peptides. BMC Bioinformatics. 1986;30:9–71. doi: 10.1186/1471-2105-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blythe M, Flower D. Benchmarking B cell epitope prediction: Underperformance of existing methods. Protein Sci. 2005;14:246–248. doi: 10.1110/ps.041059505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larsen JE, Lund O, Nielsen M. Improved method for predicting linear B-cell epitopes. Immunome Res. 2006;2:2. doi: 10.1186/1745-7580-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang W, et al. Prediction of conformational B-cell epitopes from 3D structures by random forests with a distance-based feature. BMC Bioinformatics. 2011;17(12):341. doi: 10.1186/1471-2105-12-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin Scott, Cheng Cheng-Wei, Su Emily. Prediction of B-cell epitopes using evolutionary information and propensity scales. BMC Bioinformatics. 2013;14(Suppl 2):S10. doi: 10.1186/1471-2105-14-S2-S10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azoitei ML, et al. Computational design of high-affinity epitope scaffolds by backbone grafting of a linear epitope. J. Mol. Biol. 2012;415(1):175–192. doi: 10.1016/j.jmb.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lian Y, Ge M, Pan X. EPMLR: Sequence-based linear B-cell epitope prediction method using multiple linear regression. BMC Bioinformatics. 2014;15(2):414. doi: 10.1186/s12859-014-0414-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kulp DW, Schief WR. Advances in structure-based vaccine design. Curr. Opin. Virol. 2013;3(3):322–331. doi: 10.1016/j.coviro.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berman Helen M., Battistuz Tammy, Bhat T. N., Bluhm Wolfgang F., Bourne Philip E., Burkhardt Kyle, Feng Zukang, Gilliland Gary L., Iype Lisa, Jain Shri, Fagan Phoebe, Marvin Jessica, Padilla David, Ravichandran Veerasamy, Schneider Bohdan, Thanki Narmada, Weissig Helge, Westbrook John D., Zardecki Christine. The Protein Data Bank. Acta Crystallographica Section D Biological Crystallography. 2002;58(6):899–907. doi: 10.1107/S0907444902003451. [DOI] [PubMed] [Google Scholar]
- 14.Machado-de-Ávila RA, et al. Induction of neutralizing antibodies against mutalysin-II from Lachesis muta muta snake venom elicited by a conformational B-cell epitope predicted by Blue Star Sting Data Base. Immunome Res. 2015;10:083. [Google Scholar]
- 15.Kunik V, Ofran Y. The indistinguishability of epitopes from protein surface is explained by the distinct binding preferences of each of the six antigen-binding loops. Protein Eng. Des. Sel. 2013;26(10):599–609. doi: 10.1093/protein/gzt027. [DOI] [PubMed] [Google Scholar]
- 16.Greenbaum JA, et al. Towards a consensus on datasets and evaluation metrics for developing B-cell epitope prediction tools. J. Mol. Recognit. 2007;20(2):7582. doi: 10.1002/jmr.815. [DOI] [PubMed] [Google Scholar]
- 17.Hajian-Tilaki K. Receiver operating characteristic (ROC) curve analysis for medical diagnostic Test Evaluation. Caspian J. Intern. Med. 2013;4(2):627–635. [PMC free article] [PubMed] [Google Scholar]
- 18.Bremel RD, Homan EJ. An integrated approach to epitope analysis I: Dimensional reduction, visualization and prediction of MHC binding using amino acid principal components and regression approaches. Immunome Res. 2010;6:7. doi: 10.1186/1745-7580-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Resende DM, et al. An assessment on epitope prediction methods for protozoa genomes. BMC Bioinformatics. 2012;13:309. doi: 10.1186/1471-2105-13-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lustrek M, et al. Epitope predictions indicate the presence of two distinct types of epitope-antibody-reactivities determined by epitope profiling of intravenous immunoglobulins. PLoS One. 2013;8:e78605. doi: 10.1371/journal.pone.0078605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kringelum JV, Nielsen M, Padkjær SB, Lund O. Structural analysis of B-cell epitopes in antibody:protein complexes. Mol Immunol. 2013;53(1-2):24–34. doi: 10.1016/j.molimm.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toseland CP, et al. AntiJen: a quantitative immunology database integrating functional, thermodynamic, kinetic, biophysical, and cellular data. Immunome Res. 2005;1(1):4. doi: 10.1186/1745-7580-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozlova E, Viart B, Machado de Avila R, Felicori L, Chavez-Olortegui C. Classification epitopes in groups based on their protein family. BMC Bioinformatics. 2015;16(Suppl 19):S7. doi: 10.1186/1471-2105-16-S19-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ministério da Saúde, Secretaria de Vigilância em Sade, Guia de vigilância epidemiológica & Departamento de Vigilância, E. Braislia, 2010. 816 p (2010).
- 25.Gutiérrez JM, Escalante T, Rucavado A, Herrera C. Hemorrhage caused by snake venom metalloproteinases: A journey of discovery and understanding. Toxins (Basel) 2016;8(4):93. doi: 10.3390/toxins8040093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanchez EF, et al. The novel metalloendopeptidase atroxlysin-I from Peruvian Bothrops atrox (Jergn) snake venom acts both on blood vessel ECM and platelets. Arch. Biochem. Biophys. 2010;496(1):9–20. doi: 10.1016/j.abb.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 27.Machado de Avila RA, et al. Mimotopes of mutalysin-II from Lachesis muta snake venom induce hemorrhage inhibitory antibodies upon vaccination of rabbits. Peptides. 2011;32(8):1640–1646. doi: 10.1016/j.peptides.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 28.Ferreira RN, et al. Antibodies against synthetic epitopes inhibit the enzymatic activity of mutalysin II, a metalloendopeptidase from bushmaster snake venom. Toxicon. 2006;48(8):1098–1103. doi: 10.1016/j.toxicon.2006.07.040. [DOI] [PubMed] [Google Scholar]
- 29.Capelli-Peixoto J, et al. Evaluation of the protective potential of a Taenia solium cysticercus mimotope on murine cysticercosis. Vaccine. 2011;29(51):9473–9492. doi: 10.1016/j.vaccine.2011.10.027. [DOI] [PubMed] [Google Scholar]
- 30.Shahsavandi, S., Ebrahimi, M. M., Sadeghi, K. & Mahravani, H. Design of a heterosubtypic epitope-based peptide vaccine fused with hemokinin-1 against influenza viruses. Virol Sin. Apr15 (2015). [DOI] [PMC free article] [PubMed]
- 31.Chen SW, Van Regenmortel MH, Pellequer JL. Structure-activity relationships in peptide-antibody complexes: implications for epitope prediction and development of synthetic peptide vaccines. Curr. Med. Chem. 2009;16(8):953–964. doi: 10.2174/092986709787581914. [DOI] [PubMed] [Google Scholar]
- 32.Sun J, et al. Does difference exist between epitope and non - epitope residues? Analysis of the physicochemical and structural properties on conformational epitopes from B-cell protein antigens. Immunome Res. 2011;7(3):1. [Google Scholar]
- 33.Wilson PC, Andrews SF. Tools to therapeutically harness the human antibody response. Nat. Rev. Immunol. 2012;12(10):709–719. doi: 10.1038/nri3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El-Manzalawy Y, Dobbs D, Honavar V. Predicting linear B-cell epitopes using string kernels. J. Mol. Recognit. 2008;21(4):243–255. doi: 10.1002/jmr.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015;10(6):845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saha S, Bhasin M, Raghava GP. Bcipep: a database of B-cell epitopes. BMC Genomics. 2005;6:79. doi: 10.1186/1471-2164-6-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korber B, LaBute M, Yusim K. Immunoinformatics comes of age. PLoS Comput. Biol. 2006;2(6):e71. doi: 10.1371/journal.pcbi.0020071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rubinstein ND, Mayrose I, Pupko TA. Machine-learning approach for predicting B-cell epitopes. Mol. Immunol. 2009;46(5):840–847. doi: 10.1016/j.molimm.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 39.Breiman L. Random Forests. Mach. Learn. 2001;45(1):5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 40.Chaudhuri R, Kulshreshtha D, Raghunandanan MV, Ramachandran S. Integrative immunoinformatics for Mycobacterial diseases in R platform. Syst. Synth. Biol. 2014;8(1):27–39. doi: 10.1007/s11693-014-9135-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuroda D, Shirai H, Jacobson MP, Nakamura H. Computer-aided antibody design. Protein Eng. Des. Sel. 2012;25(10):507–521. doi: 10.1093/protein/gzs024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao Liang, Li Jinyan. Mining for the antibody-antigen interacting associations that predict the B cell epitopes. BMC Structural Biology. 2010;10(Suppl 1):S6. doi: 10.1186/1472-6807-10-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saha S, Raghava GP. Prediction of continuous B-cell epitopes in an antigen using recurrent neural network. Proteins. 2006;65(1):40–48. doi: 10.1002/prot.21078. [DOI] [PubMed] [Google Scholar]
- 44.Sivalingam GN, Shepherd AJ. An analysis of B-cell epitope discontinuity. Mol. Immunol. 2012;51(3-4):304–309. doi: 10.1016/j.molimm.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stevanovi S. Structural basis of immunogenicity. Transpl. Immunol. 2002;10(2-3):133–136. doi: 10.1016/S0966-3274(02)00059-X. [DOI] [PubMed] [Google Scholar]
- 46.O’Rourke JP, et al. Development of a mimotope vaccine targeting the Staphylococcus aureus quorum sensing pathway. PLoS One. 2014;9(11):e111198. doi: 10.1371/journal.pone.0111198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Legutki JB, Johnston SA. Immunosignatures can predict vaccine efficacy. Proc. Natl. Acad. Sci. USA. 2013;110(46):18614–18619. doi: 10.1073/pnas.1309390110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneider FS, et al. Identification of protective B-cell epitopes of Atroxlysin-I: A metalloendopeptidase from Bothrops atrox snake venom. Vaccine. 2016;34(14):1680–1687. doi: 10.1016/j.vaccine.2016.02.035. [DOI] [PubMed] [Google Scholar]
- 49.Ishii H, et al. Impact of vaccination on cytotoxic T lymphocyte immunodominance and cooperation against simian immunodeficiency virus replication in rhesus macaques. J. Virol. 2012;86(2):738–745. doi: 10.1128/JVI.06226-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwong PD, Mascola JR, Nabel GJ. The changing face of HIV vaccine research. J. Int. AIDS Soc. 2012;15(2):17407. doi: 10.7448/IAS.15.2.17407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Correia BE, et al. Proof of principle for epitope-focused vaccine design. Nature. 2014;507(7491):201–206. doi: 10.1038/nature12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koide A, et al. Exploring the capacity of minimalist protein interfaces: interface energetics and affinity maturation to picomolar KD of a single-domain antibody with a flat paratope. J. Mol. Biol. 2007;373(4):941–953. doi: 10.1016/j.jmb.2007.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sollner J, et al. Analysis and prediction of protective continuous B-cell epitopes on pathogen proteins. Immunome Res. 2008;4:1. doi: 10.1186/1745-7580-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh H, Ansari HR, Raghava GP. Improved method for linear B-cell epitope prediction using antigen’s primary sequence. PLoS One. 2013;8(5):e62216. doi: 10.1371/journal.pone.0062216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cock PJ, et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25(11):1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Larkin MA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 57.Chawla, N. V. L. A., Hall, L. O. & Bowyer, K. SMOTEBoost: Improving Prediction of Minority Class in Boosting. 7th European Conference of Priciples and Practice of Knowledge Discovery in Databases. 2003: pp. 10719 (2003).
- 58.Berthold, M. R. et al. KNIME: The Konstanz Information Miner. Studies in classification, data analysis, and knowledge organization. Springer. ISSN::1431–8814 (2007).
- 59.R Development Core Team R: A language and environment for statistical computing. R Foundation for Statistical Computing. ISBN::3-900051-07-0. (2008).
- 60.Mendes TM, et al. Generation and characterization of a recombinant chimeric protein (rCpLi) consisting of B-cell epitopes of a dermonecrotic protein from Loxosceles intermedia spider venom. Vaccine. 2006;31(25):2749–2755. doi: 10.1016/j.vaccine.2013.03.048. [DOI] [PubMed] [Google Scholar]
- 61.Liu R, Hu J. Prediction of discontinuous B-cell epitopes using logistic regression and structural information. Proteomics Bioinform. 2011;4:010–015. [Google Scholar]
- 62.Grant BJ, Rodrigues AP, ElSawy KM, McCammon JA, Caves LS. Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics. 2006;22(21):2695–2696. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
- 63.Webb B, Sali A. Protein structure modeling with MODELLER. Methods Mol. Biol. 2014;1137:1–15. doi: 10.1007/978-1-4939-0366-5_1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Proteins used as training for the decision tree
Data Availability Statement
All the data depicted in the figures are available upon request.