Abstract
Enrichment of calf diets with exogenous butyrate has shown promise as a promotor of calf growth and intestinal development. However, the impact of dietary derived butyrate on the gut microbiota and their potential role, in turn, as mediators of its effect on calf growth and development is not known. Here, the effects of butyrate supplementation on rumen and hindgut microbiota and fermentation profiles were assessed in 16 Holstein-Friesian bull calves randomly assigned to one of two groups: Control (CON) fed conventional milk replacer or Sodium-Butyrate (SB – added to milk replacer) from days 7 to 56 of life. In the colon, total short chain fatty acid (SCFA), propionate and acetate concentrations were increased by SB (P < 0.05). 16S rRNA gene amplicon sequencing showed cecal abundance of butyrate producers Butyrivibrio and Shuttleworthia were decreased by SB (P < 0.05), while that of the propionate producer Phascolarctobacterium was higher (P < 0.05). Mogibacterium is associated with impaired gut health and was reduced in the cecum of SB calves (P < 0.05). These data show that the beneficial effects of SB on growth and performance occur in tandem with changes in the abundance of important SCFA producing and health-associated bacteria in the hindgut in milk-fed calves.
Introduction
The digestive physiology of calves changes dramatically in the first weeks and months of life, and the transition from a nominal monogastric to functional ruminant is fraught with challenges1,2. The occurrence of gastrointestinal disorders in this period is a source of substantial economic loss in dairy production systems, responsible for around 10% of calf mortality3. With rising concerns surrounding the prophylactic and growth-promoting use of antibiotics in livestock production promotion4, there is much interest in the development of synthetic and natural alternatives to promote bovine intestinal health and development in early life.
The gastrointestinal tract (GIT) microbiota of ruminants and other production animals is well established as a key feature underscoring animal health, development and productivity5. In adult cattle, the rumen microbiota is the predominant feed-degrading microbial community. However, up to 20% of milk solids may pass to the hindgut for digestion during the milk feeding phase, placing elevated importance on the hindgut microbiota in this period6. Short chain fatty acids (SCFAs) are organic acids produced throughout the intestinal tract by microbial fermentation, and are vital in the stimulation of intestinal growth and development7,8. The antimicrobial properties of SCFAs and their natural presence in the mammalian digestive tract suggested that SCFA-derived feed additives may be an alternative to conventional antimicrobials in livestock production9. Among the most prominent of the luminal SCFAs, butyrate has been investigated for its effectiveness in enhancing animal growth and intestinal integrity and development in young livestock, with promising results10,11. Butyrate is the primary energy source for rumen epithelial cells and colonocytes, which are important mediators of water, mineral, and nutrient absorption12. Butyrate inclusion in both milk replacer and solid feed has been shown to have beneficial effects on both intestinal development and animal growth in young livestock9,10,13,14.
Enteric disorders in calves are associated with microbial dysbiosis in the gut15, and thus the health-promoting effects of exogenous butyrate may be underpinned by modulation of the GIT microbiota. There is evidence that encapsulated butyrate can reduce enteric pathogen colonisation in swine and poultry13,16,17, and direct infusion of butyrate into the mature cow rumen caused significant changes to the resident microbiota18. However, there are little data concerning the effect of long-term supplementation of butyrate on GIT microbial communities in pre-weaned calves. Given the established impact of butyrate on animal growth and intestinal development, we hypothesised that provision of a butyrate-fortified milk replacer impacts microbial communities throughout the GIT while improving host performance. Therefore, the objectives of this study were to assess microbial composition and fermentation in the rumen and hindgut at weaning in dairy calves offered milk replacer enriched with butyrate.
Materials and Methods
Ethical statement
All procedures involving animals were approved by University College Dublin, animal research ethics committee (UCD AREC), under licence from the Irish Department of Health and Children in accordance with the Cruelty to Animals Act (Ireland 1897) and European Community Directive 86/609/EC.
Animal study
Forty-four male Holstein-Friesian calves (13 ± 5 days of age) were obtained from one dairy farm, and were housed on one research farm for use in this study. Calves were blocked according to age and body weight and were randomly assigned to one of two treatment groups; CON (fed unaltered milk replacer, n = 22) or SB (encapsulated sodium butyrate included in milk replacer at 4 g/kg of DM daily, n = 22). Calves were placed on a standard 56-day calf rearing program upon arrival at the research farm, with milk replacer (12.5% solids; Crude Protein 23% and Crude Fat 20%; BlossomTM, Volac, UK) offered at 6 L/day via an automatic feeder (Forester Tecknik, KFA3-MA3). Concentrates (rolled barley 26.5%; soya bean meal 25%; maize 15%; beet pulp 12.5%; soya hulls 12.5%; molasses 5%; minerals & vitamins 2.5%; vegetable oil 1%; Nutriad, Belgium) and water were offered on an ad libitum basis throughout the experimental period. Detailed dietary composition is presented in Supplementary Table S1. All calves were in good health throughout the experimental period. Calves were weaned over a 7-day period (D49–56) via gradual reduction in the allocation of milk replacer. On D56, eight animals from each group were randomly selected for euthanasia using an intravenous overdose of pentobarbital sodium (DolethalTM, Vetoquinol UK, 1.4 ml/kg live body-weight). Death was confirmed by lack of an ocular response and heartbeat. Digesta samples from the rumen, cecum, and colon were collected, immediately snap frozen on liquid nitrogen, and stored at −80 °C pending molecular analysis. A further digesta sample was collected from both the rumen and colon (representative of the total hindgut SCFA profile, as previously shown19) for SCFA analysis. These samples were passed through four layers of cheesecloth and stored in H2SO4 at −80 °C prior to SCFA analysis using gas chromatography.
Microbial DNA extraction
Frozen digesta from the cecum, colon and rumen was ground under liquid nitrogen to a fine powder. Total DNA was extracted using the RBB + C method as previously described20; approximately 250 mg of ground frozen sample was subjected to repeated bead beating followed by column purification with a QIAGEN DNeasy Stool Kit (Qiagen, UK). DNA quantity and purity were assessed by two consecutive readings at A260 and A280nm on a Nanodrop 1000 spectrophotometer, and visualisation with UV light in a 0.8% agarose gel. Samples with DNA purity values <1.6 were re-extracted, as were samples of low concentration (<100 ng/µl). Mean quantity and purity values in each gastrointestinal region are provided in Supplementary Data S5.
Microbial profiling using amplicon sequencing
Amplicons of the V4 hyper-variable region of the 16S rRNA gene were prepared using Illumina Nextera chemistry, as previously reported21. DNA concentrations recorded on the Nanodrop were used to normalise each sample to a concentration of 100 ng/µl with molecular water. A 25 µl PCR reaction using 20 ng of DNA, and KAPA Hi-Fi PCR (New England Biolabs Inc.) mix was prepared using 515 F/806 R primers22 to simultaneously characterise bacterial and archaeal members using the following cycle programme: 95 °C for 3 minutes, and 25 cycles of: 95 °C for 30 sec, 55 °C for 30 sec, 72 °C for 30 sec, with a final elongation step of 72 °C for 30 seconds. Amplicons were purified using the QIAGEN QIAquick PCR Purification Kit. A second PCR step was performed to add Illumina dual indices and NexteraTM adapters to the purified fragments (Illumina, San Diego, CA, USA). Following another column purification, the barcoded amplicon products were combined into two pools in equimolar quantities to ensure adequate sequencing coverage. Each pool was subjected to gel (QIAquick Gel Extraction Kit, Qiagen) and column purification (QIAquick Purification Kit, Qiagen) to remove primer dimers and any residual agarose. Purified pools were quantified by qPCR using the KAPA SYBR FAST Universal kit with Illumina Primer Premix (New England Biolabs Inc.). Pools were then diluted and denatured according to the Illumina MiSeq library preparation guide. A 6 pM amplicon library was spiked with 30% denatured and diluted PhiX Illumina control library (version 3, 12.5 pM), and subjected to sequencing on the Illumina MiSeq platform with one pool per run.
Sequence data quality control and pre-processing
Demultiplexed paired end reads were trimmed and filtered to remove low quality reads and bases (Phred quality score threshold of 20), and simultaneously merged together using the BBTools suite23. The resulting merged reads were then size selected to retain only reads ± 2 standard deviations from the mean read length, to minimise spurious OTU creation. Finally, merged pairs were combined into a single file for downstream processing using the Quantitative Insights Into Molecular Ecology (QIIME v.1.9) tool24.
Bioinformatic Analysis
Operational taxonomic unit (OTU) identification using a similarity level of 97% was carried out using the open reference picking method implemented in QIIME24. A representative sequence from each identified OTU was then aligned against the reference Greengenes database (v.13_8)25. A graphical representation of the phylogenetic trees created in QIIME was made using the Interactive Tree of Life software26. The raw and unfiltered OTU table created in QIIME was imported into R to create a Phyloseq class object27. α-diversity was computed by first randomly subsampling (rarefying) the OTU table to the lowest read number, to reduce bias due to differential sequencing depth. The Shannon and Chao1 metrics were used to assess diversity and evenness of the rumen and hindgut microbiota. β-diversity was calculated in a similar manner, with a Bray Curtis Dissimilarity matrix constructed from the rarefied OTU table. A cluster dendrogram using Ward linkage equilibrium was generated from the same OTU table in R. Principle Coordinate Analysis (PCoA) was performed in Phyloseq and used to visualise these distance matrices in 2-dimensional space. OTUs represented by <2 sequences were filtered and removed. Relative abundances of taxa at the phylum, family, and genus levels were computed in R.
Statistical Analysis
Permutation based Analysis of Variance (PERMANOVA) analysis based on the Bray Curtis Dissimilarity Matrix was carried out in R using the Vegan package to compare microbial structure between groups and GIT region28,29. Taxonomic abundances at the phylum and genus levels were compared across treatments (within GIT compartment) using a Wald parametric test, offered within the DESeq2 Bioconductor package in R30. A false-discovery rate (FDR) threshold of 0.15 was used to determine statistical significance31. Only taxa present at ≥0.01% of all 16S rRNA sequences in either treatment group were considered present. Exploratory investigation of taxonomic profiles revealed two outlier animals (one from each group), and they were removed from subsequent analysis leaving a total of 7 animals in each treatment group.
Results
Animal performance
This experiment was conducted in association with a larger study designed to examine the effect of SB supplementation on the performance, feed efficiency and immune status of artificially reared dairy calves. Briefly, from this perspective, calves supplemented with SB tended (P = 0.08) to have a higher pre-weaning growth rate compared to CON (0.69 versus 0.59 kg/day). At weaning SB calves (80.2 kg) were 3.1 kg heavier than the CON group (76.9 kg) with bodyweight difference detected from day 42 to weaning (P = 0.08). Bodyweight differences between treatments were not evident prior to this (P > 0.1). Total DMI was not different between dietary treatments but pre-weaning SB supplementation tended (P = 0.08) to improve feed efficiency (measured using feed conversion ratio) of the calves (SB; 1.7:1 compared to CON; 2.5:1; P = 0.07). Feed intakes and growth rates are presented in supplementary data.
Fermentation profiles in the rumen and hindgut at weaning
Short chain fatty acid profiles of the rumen and colon contents at weaning are presented in Table 1. Colonic concentrations of total SCFA, propionate, and acetate were higher for SB fed calves (P < 0.05). SB supplementation reduced ruminal butyrate concentration (P < 0.05), but total SCFA concentration was unaffected.
Table 1.
The effect of SB inclusion in milk replacer on Short Chain Fatty Acid (SCFA) profiles in the rumen and colon.
| SCFA Profiles | ||||||
|---|---|---|---|---|---|---|
| Rumen | Colon | |||||
| Item | CON | SB | P-value | CON | SB | P-value |
| Total SCFA Concentrations (mmol/L) | ||||||
| Acetate | 90.56 ± 7.51a | 78.46 ± 1.93 | NS b | 39.48 ± 3.49 | 60.84 ± 6.03 | 0.01 |
| Propionate | 62.43 ± 7.51 | 58.26 ± 1.73 | NS | 10.77 ± 1.20 | 17.06 ± 2.00 | 0.02 |
| Butyrate | 16.21 ± 1.17 | 11.49 ± 0.57 | 0.04 | 3.56 ± 0.40 | 5.01 ± 0.84 | NS |
| Isobutyrate | 0.71 ± 0.37 | 0.33 ± 0.06 | NS | 0.53 ± 0.05 | 0.50 ± 0.09 | NS |
| Valerate | 4.91 ± 0.61 | 3.43 ± 0.09 | NS | 0.75 ± 0.13 | 0.85 ± 0.09 | NS |
| Isovalerate | 1.79 ± 0.28 | 1.05 ± 0.08 | NS | 0.37 ± 0.06 | 0.31 ± 0.06 | NS |
| Total VFA | 176.44 ± 16.02 | 152.82 ± 3.67 | NS | 55.46 ± 4.87 | 84.57 ± 8.60 | 0.02 |
| Molar Proportions of SCFA | ||||||
| Acetate | 0.517 ± 0.01 | 0.513 ± 0.003 | NS | 0.712 ± 0.01 | 0.721 ± 0.01 | NS |
| Propionate | 0.346 ± 0.01 | 0.379 ± 0.004 | NS | 0.192 ± 0.01 | 0.200 ± 0.01 | NS |
| Butyrate | 0.094 ± 0.004 | 0.077 ± 0.004 | NS | 0.064 ± 0.003 | 0.057 ± 0.01 | NS |
| Isobutyrate | 0.003 ± 0.002 | 0.001 ± 0.001 | NS | 0.010 ± 0.001 | 0.007 ± 0.001 | NS |
| Valerate | 0.028 ± 0.003 | 0.023 ± 0.001 | NS | 0.014 ± 0.001 | 0.010 ± 0.001 | NS |
| Isovalerate | 0.011 ± 0.002 | 0.007 ± 0.001 | NS | 0.007 ± 0.001 | 0.004 ± 0.001 | NS |
P-values were obtained using a Monte-Carlo permutational t-test in R.
aMean ± SEM. bNot significant.
Microbial structure and diversity in the rumen and hindgut in response to SB
Amplicon sequencing of rumen and hindgut digesta samples from calves at weaning yielded a total of 10,348,464 high quality reads, with an average of 215,593 ± 75,380 sequences per sample. Taxon abundance was agglomerated at the genus and phylum levels for comparisons across treatments, and relative abundances of all detected taxa are summarised in Supplementary Data S2.
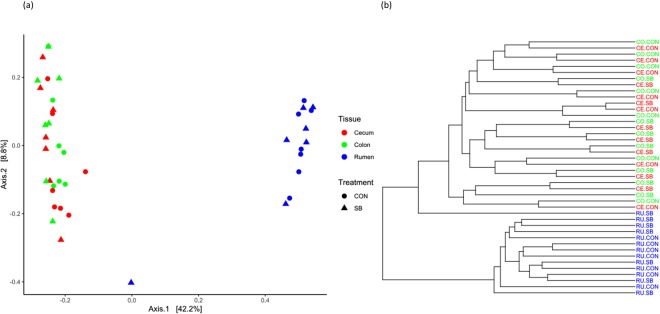
Alpha diversity measured using the Shannon index was not affected by treatment in any region studied, though was higher in both hindgut regions than in the rumen (P < 0.05; Table 2). The Chao1 index of species richness was lower in the rumen of SB animals (P < 0.05), but was similar across treatments in the hindgut (Table 2), and was higher in the colon than both other compartments (P < 0.05; Table 2). Principle coordinate anlaysis (PCoA) and cluster analysis showed some evidence of separation according to treatment, independent of GIT region in the hindgut (Fig. 1a,b), but comparisons using PERMANOVA failed to detect any differences (P > 0.05; Table 3). There was, however, clear separation according to gastrointestinal region, with the rumen community clustering away from both hindgut regions (P < 0.05), while both hindgut regions appeared to harbour a similar microbial community (Fig. 1a,b).
Table 2.
Comparisons of alpha diversity metrics in the rumen, cecum, and colon of calves at weaning.
| α-Diversity | ||||||||
|---|---|---|---|---|---|---|---|---|
| Chao1 | Shannon | |||||||
| Overall | CON | SB | P-value | Overall | CON | SB | P-value | |
| Rumen | 1698.0a | 1887.2 | 1508.7 | 0.01 | 3.6a | 3.7 | 3.6 | 0.15 |
| Cecum | 1728.7a | 1630.3 | 1827.0 | 0.30 | 4.9b | 4.8 | 5.0 | 0.28 |
| Colon | 2849.0b | 2827.3 | 2870.7 | 0.87 | 5.1b | 5.1 | 5.1 | 0.76 |
Significant differences according to gastrointestinal region are denoted with different letters.
Figure 1.
(a) Principle Coordinate Analysis plot and (b) cluster dendrogram generated based on the Bray-Curtis dissimilarity matrix of operational taxonomic units (OTUs) present in the rumen, cecum, and colon.
Table 3.
Comparing microbial communities between treatments and gastrointestinal region in the rumen, cecum and colon.
| β-Diversity | |||||
|---|---|---|---|---|---|
| Treatment | GIT Region | F-value | P-value | ||
| F-value | P-value | ||||
| Rumen | 1.04 | 0.37 | Rumen vs. Cecum | 21.44 | 0.001 |
| Cecum | 1.30 | 0.16 | Rumen vs. Colon | 21.15 | 0.001 |
| Colon | 1.28 | 0.12 | Cecum vs. Colon | 0.82 | 0.700 |
P-values obtained using PERMANOVA analysis based on Bray Curtis similarity matrices.
Microbial composition in the rumen and hindgut in response to SB
Rumen
Among the bacterial phyla detected in the rumen, Bacteroidetes, Firmicutes, and Proteobacteria were predominant, followed by Actinobacteria and Cyanobacteria (Fig. 2), while the remaining minor phyla (<1% 16S rRNA reads) are presented in Fig. 2. Archaea were represented by the Euryarchaetota phylum. 60 genus-level assignements were reported from the rumen, with Prevotella, f.Succivibrionaceae and f.Lachnospiraceae predominant at weaning, regardless of dietary treatment. Notably, only 48.14% of reads recovered from the rumen could not be confidently assigned at the genus level. This is reflected in the high abundances of f.Succinivibrionaceae (f = family level, unassigned at genus level in QIIME), f.Lachnospiraceae and o.Clostridiales in the rumen samples, as well as a further 21 unclassifed genus-level taxa (Supplementary data S2, Fig. 3a). Comparisons of taxon abundance in DESeq2 between SB and CON animals showed no statistically significant effect of dietary treatment on the rumen microbiota at either phylum or genus level following adjustment into FDR.
Figure 2.
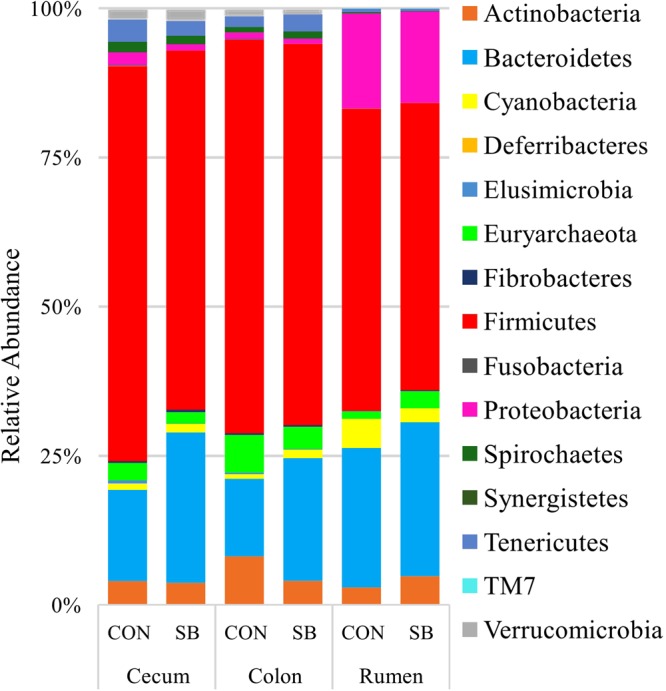
Stacked bar chart of microbial abundances at the phylum level, calculated as a percentage of total 16S rRNA reads within each group.
Figure 3.
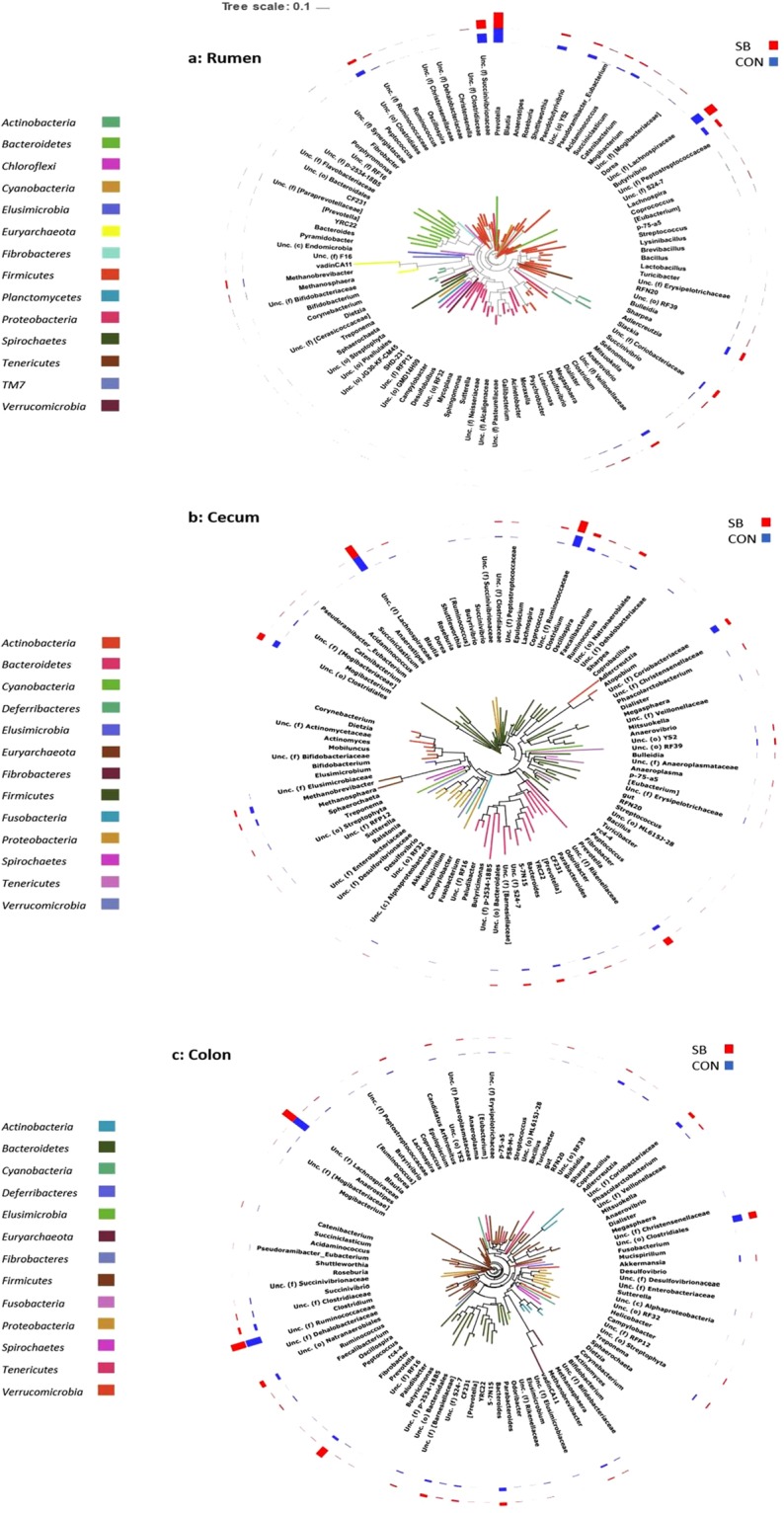
Phylogenetic Trees of (a) rumen, (b) cecum, and (c) colon microbiota. A phylogenetic tree file was built from a multiple sequence alignment generated in QIIME. OTUs were agglomerated at the genus level in R. The trees were visualised using the Interactive Tree of Life (ITOL) software package.
Hindgut
Twelve bacterial phyla and a single archaeal phyla were detected in the hindgut, among which Defferibacteres was unique to the colon (Fig. 2). Like the rumen, a significant proportion of 16S rRNA reads recovered from the cecum and colon could not be resolved taxonomically to the genus level (~59%). 93 and 88 genera were detected in the cecum and colon, respectively. Genera annotated only as f.Lachnospiraceae and f.Ruminococcaceae were the most abundant in both compartments (Fig. 3b,c). There was a minor impact of treatment on composition of the hindgut microbiota. For instance, in the colon, Prevotella was enriched in SB animals (P < 0.05). In the cecum, several taxa were different between treatments; as in the colon Prevotella (4.31–9.48%) was numerically higher in the SB cohort, but this difference was not signficant, possibly due to the large inter-animal variaiton observed (Supplementary Data S2).
An additional 9 genera were different between dietary treatments in the cecum (P < 0.05); Shuttleworthia (0.01 vs. 0.06%), Butyrivibrio (0.13 vs. 0.81), Sharpea (0.32 vs. 1.09%), and Mogibacterium (0.12 vs. 0.26%) were all reduced by SB supplementation (P < 0.05), as well an unidentified member of the f.[Mogibacteriaceae] (0.65 vs. 1.56%) (Fig. 3b). A genus belonging to the Cyanobacterial YS2 order was increased by SB, as were Lachnospira (0.13 vs. 0.06%), Phascolarctobacterium (1.40% vs. 0.66%), and a genus annotated as p-75-af belonging to Erysipelotrichaceae (0.31 vs. 0.11%; P < 0.05). A single genus from the Tenericutes phylum classified only as o. ML615J-28 was also increased in the SB group (0.19 vs. 0.04%; P < 0.05). Additionally, an undetermined genus assigned to the Coriobacteriaceae family was reduced by SB in the cecum (3.96 vs. 8.17%; P < 0.05; Fig. 3b).
Discussion
The beneficial effects of dietary butyrate supplementation (often included in salt form as calcium or sodium butyrate) on animal growth and intestinal development have been demonstrated in calves10,14,32, chickens17 and pigs33. While there is now an established body of evidence supporting the potential of butyrate as a beneficial feed additive, its impact on the gut microbiota is unknown. In adult animals, hindgut fermentation typically provides 5–10% of dietary energy, but this may be elevated during the pre-weaning phase of calf growth, when up to 20% of ingested milk solids may pass to the hindgut6,34. Thus microbial fermentation in the cecum and colon is an important host energy source during this period6. Given that the importance of the hindgut in feed digestion is accentuated during early ruminal development, it is of interest to ascertain what changes may occur in the microbiota and fermentation patterns following SB supplementation. In a previous study, our group showed positive effects on growth and efficiency when dairy calves were supplemented with SB35. Here, we provide evidence that such improved performance is accompanied by changes in microbial composition and fermentation in the hindgut compartments, while the rumen microbiota is mostly unaffected.
SB does not induce substantial changes in the rumen microbiota or fermentation profile
In terms of bacterial composition, the rumen microbiota was unaffected by SB. However, species richness (assessed using the Chao1 estimator) was lower in the SB animals, indicative of a greater number of sparsely abundant OTUs being present the rumen of CON animals than the SB group. Interestingly, we also observed a reduction in ruminal butyrate concentration in the SB cohort. The digestive physiology of the milk-fed calf effectively precludes entry of liquid feed into the reticulorumen via action of the reticular groove36, and so these changes are likely due to an indirect effect of SB on the rumen microbiota, as the exogenous butyrate in the milk replacer did not enter the rumen. Such indirect influences of SB on the rumen have previously been observed; SB-fortified MR significantly improved rumen growth and papillae development compared to calves fed conventional MR37, but we did not observe such effects in the present study where rumen papillae length, width, and perimeter were not affected by SB supplementation (data not shown). Thus, though we observed a reduction in the concentration of ruminal butyrate, this does not appear to have had any detrimental effects on rumen development. It is possible that if the excess dietary butyrate was absorbed in the gut, it may have reduced the requirement for ruminal butyrate in the SB calves. It is also worth noting that many inconsistent results have been reported in the literature when butyrate or its derivatives are used as supplements in livestock diets, as recently reviewed12. Nonetheless, this suggests cross-talk mechanisms may exist between the lower gut and the rumen and warrant further investigation. In studies where SB was included in calf starter, significant development of the rumen epithelium was observed10,14, and future work should also examine changes in the rumen microbiota and fermentation profiles when calves are supplemented with SB in solid feed.
Sodium butyrate modifies the hindgut microbiota and fermentation profiles in early life
The microbial profiles of the cecum and colon were highly similar. No significant clustering was observed in the PCoA plot according to treatment within either compartment, but finer shifts in the microbial profile were evident in both. In the colon, the proportion of Prevotella was increased by SB, with a similar numerical increase observed in the cecum. Enrichment of Prevotella in the colon and stomach of neonatal piglets has previously been reported following SB supplementation13. Prevotella is established as a primary member of the mammalian gut ecosystem, comprising species capable of fermenting a wide range of non-cellulosic plant polysaccharides and protein38. Prevotella spp. positively correlated with intestinal butyrate concentrations in growing pigs39, and it is possible that excess dietary butyrate reaching the colon conferred a competitive advantage on Prevotella, as they are not notable butyrate producers40, aligning with the significant reduction in known butyrate producing taxa discussed below. Supplementing the diet of neonatal piglets and poultry with SB has previously been reported to reduce the abundance of known gut pathogens (e.g E. coli)41. We did not observe similar effects in our study, which may be attributable to differences in analytical approach (e.g. qPCR for specific scour causing bacteria). We did detect Escherichia in our dataset, but its proportion was very low (<0.005% of total 16S rRNA sequences) and so was not considered in our final analysis. This highlights a limitation of amplicon sequencing surveys, whereby potentially important taxa may be under- or over-represented due to variation in 16S rRNA gene copy number among microbial species42.
We observed most evidence of microbial manipulation through SB supplementation, in the cecum. Most notably, the abundances of several important SCFA producers were changed. Phascolarctobacterium rapidly converts succinate to propionate in the gut43,44. The higher abundance of this genus in the cecum of SB animals may have contributed to improved growth via increased host energy substrate, as propionate is the primary precursor for gluconeogenesis in ruminants44. This, combined with our observation of higher levels of propionate and total SCFA, provides evidence that improved rates of bacterial fermentation in the hindgut may also contribute to SB-driven performance improvements, as well as the increased activation of the IGF-1 pathway previously reported9. Abundances of known butyrate-producing Butyrivibrio and Shuttleworthia were reduced in the cecum under SB supplementation, suggesting that exogenous butyrate suppresses microbial biosynthesis of butyrate in the gut. The reduction of the lactate producer Sharpea may also contribute to lower microbial butyrate as lactate is an intermediate molecule formed by bacterial action in the GIT. Lactate is usually rapidly utilised for SCFA (primarily butyrate) synthesis, as accumulation can lead to harmful acidotic conditions45,46. While the mechanisms and occurrence of ruminal acidosis has been extensively investigated in cattle47–49, there is little knowledge of the prevalence of hindgut acidosis in calves. Lactate was not measured in the present study, but our results suggest that lactate metabolism may be an important intermediary in the response of the gut microbiota to exogenous butyrate, warranting further investigation.
Sodium butyrate supplementation in reduced cecal abundance of taxa associated with lowered gut health and integrity, and elevated inflammation. For instance, Mogibacterium, a known genus of the oral microbiota, was reduced in response to SB supplementation. Whilst the role of Mogibacterium in the gut is not fully understood, previous studies have observed a decreased fecal abundance of this genus in response to beneficial prebiotic supplementation in neonatal piglets50, and mucosal abundance of Mogibacterium was higher in the distal gut of human colorectal cancer patients than healthy controls51. Thus while the dearth of knowledge concerning the characteristics of Mogibacterium spp. in the gut ecosystem make it difficult to speculate as to why SB may affect it, it’s reduction may be indicative of favourable changes in the gut microbiota of calves fed SB. Similarly, the abundance of Actinobacteria was also significantly lower in the cecum of SB calves, driven by a significant reduction in a genus classified only as part of the Coriobacteraceae family (reported as “f__Coriobacteraceae;g.__” in QIIME). There were several other low-abundance genera assigned to Coriobacteraceae (<0.01%), so this is likely an undescribed genus or genera which may have an important role in the maintenance of gut health. Several novel members of this family have been described recently52,53, and further advances in our knowledge of the role of Coriobacteraceae in the gut may resolve the possible role of as-yet undefined Coriobacteraceae species in SB-driven growth improvements. The Coriobacteraceae in the gut have been associated with a suppression in host inflammatory response. Reduced abundance of this family was previously observed in tandem with lower detection of the pro-inflammatory IL-6 in blood plasma54, and so our results may indicate reduced immunogenicity among the cecal microbiota of SB fed calves.
The higher abundance of Cyanobacteria observed in the cecum of SB animals was driven by significant increases of a genus assigned to the YS2 order. This highlights a wider issue concerning 16S rRNA gene investigations of intestinal microbial communities. Although Cyanobacteria have been widely reported as minor contributors to GIT microbial diversity in mammals55–57, the validity of their role in the anaerobic gut ecosystem is questionable, as many species of this phylum are native to marine environments and are notable performers of complex oxygenic photosynthesis58. Recent studies have revealed that the Cyanobacteria found in the gut are genetically dissimilar to their photosynthetic relatives, and likely diverged prior to the latter developing the capability for photosynthesis59,60. Two such novel Cyanobacteria-like lineages have been described in the human GIT to date, the Melainabacteria59, and the Sericytochromatia60, but there is not yet a consensus on the correct nomenclature61. Neither is it known if these novel taxa are also the same Cyanobacteria-derivatives present in the ruminant gut, and this warrants urgent investigation. Regardless, increased abundance of Cyanobacteria has not been previously reported in the gut of SB supplemented calves, suggesting a potential role of the newly described Cyanobacteria groups in the developing intestine, but further work is needed to confirm their role in the ruminant gut ecosystem.
The rumen and hindgut harbour significantly different microbial communities at weaning
While patterns of microbial colonisation in the pre-functioning rumen have been the subject of several investigations recently62–65, there are noticeably fewer published reports concerning the hindgut microbiota of young ruminants. In agreement with the available literature, we found that the rumen and hindgut microbiota differed significantly at weaning55,66. In addition to lower rumen bacterial diversity, SCFA levels were higher in the rumen than in the colon, suggesting that at weaning, the rumen microbiota ferments plant biomass at a greater rate than that of the hindgut. It is likely that the greater range of secondary fermentation products entering the lower gut is the driver of the increased bacterial diversity of the cecum and colon. The bacterial profile of the rumen was similar to that previously reported in young animals and was dominated by Firmicutes and Bacteroidetes. Bacteroidetes have previously been reported as the predominant bacterial phyla in the rumen and hindgut of 3-week old and weaned diary calves64,66, and in the rumen of 6-week old lambs67. Prevotella was the most abundant bacterial genus in the rumen at weaning which is in agreement with published reports66. Our data showed the principal bacterial phylum Firmicutes was dominated by unclassified Succinivibrionaceae in the rumen, but that the hindgut regions harboured higher relative abundances of unclassified genera from the Lachnospiraceae and Ruminococcaceae families while the Succinivibrionaceae members were minor contributors. Succinivibrionaceae has been reported as a member of the core active rumen microbiota in adult cattle68, and is implicated in reduced methane formation in both ruminants and macropods21,69,70. The predominance of Prevotella and Succinivibrionaceae has been previously documented in the rumen of adult dairy cows71, but the high abundance of uncharacterised Succinivibrionaceae in the rumen at weaning has not, to our knowledge, been reported to date. However, caution should be exercised when comparing results of multiple amplicon sequencing surveys, as amplification primer choice can significantly bias results72. Popova, et al.73 and Zhou, et al.7 have previously described the hindgut methanogen populations in lambs and dairy calves, and our findings are largely similar to theirs, with Methanobrevibacter as the predominant genus.
Unclassified genera of the Lachnospiraceae were previously reported as comprising just 5.58% of faecal 16S rRNA sequences 5 days after weaning, in contrast to our observation of high abundance in the cecum and colon66. The same study revealed high abundance of an unclassified Ruminococcaceae genus in the faeces of dairy calves shortly after weaning which is consistent with our results66. Both taxa have been widely reported as important members of the gut microbiota, containing prominent plant polysaccharide hydrolysing species74. Interestingly, visualisation of the phylogenetic tree generated in QIIME shows Prevotella sequences recovered from the rumen appeared to cluster away from the other Bacteroidetes taxa (Fig. 3a), suggesting that at weaning the rumen may contain a phylogenetically distinct cohort of Prevotella spp. compared to that of the hindgut, where Prevotella sequences clustered broadly as expected (Fig. 3b,c). This warrants further investigation, given the ubiquitous and abundant presence of Prevotella in the mammalian digestive tract. Also evident in our dataset is the dominance of undescribed microorganisms in the mammalian GIT. Indeed, among the ten most abundant genus level taxa reported in the hindgut regions, only four (Prevotella, Clostridium, Bacteroides and Ruminococcus) were annotated as a known bacterial genus. This underlines the large number of as-yet uncharacterised bacteria that exist within the mammalian gut, and highlights the inherent difficulties in accurate compositional and functional profiling of the GIT microbiota.
Conclusions
The data presented here and in our companion study35 provide evidence that the improved performance recorded for SB supplemented calves may be mediated through minor changes in the rumen and hindgut microbiota, with a particularly notable response to SB evident in the cecum. However, it is impossible to conclude whether changes in microbial composition are actively contributing to this improved growth and performance, or whether the host phenotype is driving changes in the microbial community. It is possible that the major effects of exogenous butyrate supplementation on the GIT microbiota may occur during the first weeks of life and are not evident at weaning, and indeed previous work has suggested that for maximum impact, butyrate should be supplemented from the first day of life12. The present study may also be limited by the fact that the calves had already undergone a weaning process (between days 49–56) when the samples were collected, and the amount of exogenous butyrate entering the GIT was thus reduced in the week preceding slaughter. It may be advantageous to collect digesta samples throughout the milk-feeding period in future studies, to assess if SB supplementation may facilitate a smoother weaning transition. Nonetheless, considering the significant differences that were still evident one week following the onset of the weaning process, SB supplementation appears to impart persistent changes on gut microbial composition and fermentation in dairy calves, and may be a candidate additive for “microbial programming” of gut microbial communities in early life75. In summary, we conclude that positive trends in growth rate and feed efficiency associated with SB supplementation in early life occur in tandem with changes in bacterial composition and fermentation in the hindgut. More thorough investigations using metagenomic or metatranscriptomic approaches may offer further information as to the mechanisms by which sodium butyrate modulates the gut microbial community in young ruminants.
Electronic supplementary material
Acknowledgements
The assistance of farm staff at UCD Lyons Farm, Clane, Co. Kildare, is gratefully acknowledged. Graduate students and technicians who assisted with sample collection and processing are also thanked. We especially acknowledge the advice and guidance of Dr. Paul Cormican, Teagasc, in analysis of microbial communities, and TrinSeq, Dublin, Ireland, for use of their MiSeq platform.
Author Contributions
E.O’H., S.W., L.L.G., A.K., and D.K. wrote the paper. E.O’H and M.MC.C carried out laboratory analysis and sequencing. E.O’H. performed bioinformatic and statistical analysis. All nucleic acid work was funded by Teagasc (Project Number 6341). Eóin O’Hara was in receipt of a Teagasc Walsh Fellowship (Reference Number WF 2013047).
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Eóin O’Hara, Email: eoin@uaberta.ca.
Sinéad M. Waters, Email: sinead.waters@teagasc.ie
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-33122-6.
References
- 1.Steele Michael A., Penner Greg B., Chaucheyras-Durand Frédérique, Guan Le Luo. Development and physiology of the rumen and the lower gut: Targets for improving gut health 1. Journal of Dairy Science. 2016;99(6):4955–4966. doi: 10.3168/jds.2015-10351. [DOI] [PubMed] [Google Scholar]
- 2.M. Ryle, E. R. Ø. Energy Nutrition in Ruminants. (Springer, Dordrecht, 1992).
- 3.USDA. Dairy 2007, Heifer calf health and management practices on US dairy operations. 122–131 (USDA:APHIS:VS, CEAH. Fort Collins, CO., 2010).
- 4.Van Boeckel TP, et al. Global trends in antimicrobial use in food animals. Proceedings of the National Academy of Sciences. 2015;112:5649–5654. doi: 10.1073/pnas.1503141112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yeoman CJ, White BA. Gastrointestinal tract microbiota and probiotics in production animals. Annual review of animal biosciences. 2014;2:469–486. doi: 10.1146/annurev-animal-022513-114149. [DOI] [PubMed] [Google Scholar]
- 6.Castro JJ, Gomez A, White B, Loften JR, Drackley JK. Changes in the intestinal bacterial community, short-chain fatty acid profile, and intestinal development of preweaned Holstein calves. 2. Effects of gastrointestinal site and age. Journal of dairy science. 2016;99:9703–9715. doi: 10.3168/jds.2016-11007. [DOI] [PubMed] [Google Scholar]
- 7.Zhou M, Chen Y, Griebel PJ, Guan le L. Methanogen prevalence throughout the gastrointestinal tract of pre-weaned dairy calves. Gut microbes. 2014;5:628–638. doi: 10.4161/19490976.2014.969649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Firkins J. L., Yu Z. RUMINANT NUTRITION SYMPOSIUM: How to use data on the rumen microbiome to improve our understanding of ruminant nutrition1,2. Journal of Animal Science. 2015;93(4):1450–1470. doi: 10.2527/jas.2014-8754. [DOI] [PubMed] [Google Scholar]
- 9.Guilloteau P, et al. Sodium-butyrate as a growth promoter in milk replacer formula for young calves. Journal of dairy science. 2009;92:1038–1049. doi: 10.3168/jds.2008-1213. [DOI] [PubMed] [Google Scholar]
- 10.Gorka P, et al. Effect of sodium butyrate supplementation in milk replacer and starter diet on rumen development in calves. Journal of physiology and pharmacology: an official journal of the Polish Physiological Society. 2009;60(Suppl 3):47–53. [PubMed] [Google Scholar]
- 11.Niwińska B, Hanczakowska E, Arciszewski MB, Klebaniuk R. Review: Exogenous butyrate: implications for the functional development of ruminal epithelium and calf performance. Animal. 2017;11:1522–1530. doi: 10.1017/S1751731117000167. [DOI] [PubMed] [Google Scholar]
- 12.Bedford Andrea, Gong Joshua. Implications of butyrate and its derivatives for gut health and animal production. Animal Nutrition. 2018;4(2):151–159. doi: 10.1016/j.aninu.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu J, Chen X, Yu S, Su Y, Zhu W. Effects of Early Intervention with Sodium Butyrate on Gut Microbiota and the Expression of Inflammatory Cytokines in Neonatal Piglets. PloS one. 2016;11:e0162461. doi: 10.1371/journal.pone.0162461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorka P, et al. Effect of method of delivery of sodium butyrate on rumen development in newborn calves. Journal of dairy science. 2011;94:5578–5588. doi: 10.3168/jds.2011-4166. [DOI] [PubMed] [Google Scholar]
- 15.Oikonomou G, et al. Fecal Microbial Diversity in Pre-Weaned Dairy Calves as Described by Pyrosequencing of Metagenomic 16S rDNA. Associations of Faecalibacterium Species with Health and Growth. PloS one. 2013;8:e63157. doi: 10.1371/journal.pone.0063157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Czerwinski J, Hojberg O, Smulikowska S, Engberg RM, Mieczkowska A. Effects of sodium butyrate and salinomycin upon intestinal microbiota, mucosal morphology and performance of broiler chickens. Archives of animal nutrition. 2012;66:102–116. doi: 10.1080/1745039X.2012.663668. [DOI] [PubMed] [Google Scholar]
- 17.Hu Z, Guo Y. Effects of dietary sodium butyrate supplementation on the intestinal morphological structure, absorptive function and gut flora in chickens. Animal Feed Science and Technology. 2007;132:240–249. doi: 10.1016/j.anifeedsci.2006.03.017. [DOI] [Google Scholar]
- 18.Li RW, Wu S, Baldwin RL, 6th, Li W, Li C. Perturbation dynamics of the rumen microbiota in response to exogenous butyrate. PloS one. 2012;7:e29392. doi: 10.1371/journal.pone.0029392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elsden SR, et al. Volatile acid in the digesta of ruminants and other animals. The Journal of experimental biology. 1946;22:191–202. doi: 10.1242/jeb.22.3-4.191. [DOI] [PubMed] [Google Scholar]
- 20.Yu Z, Morrison M. Improved extraction of PCR-quality community DNA from digesta and fecal samples. BioTechniques. 2004;36:808–812. doi: 10.2144/04365ST04. [DOI] [PubMed] [Google Scholar]
- 21.McCabe MS, et al. Illumina MiSeq Phylogenetic Amplicon Sequencing Shows a Large Reduction of an Uncharacterised Succinivibrionaceae and an Increase of the Methanobrevibacter gottschalkii Clade in Feed Restricted Cattle. PloS one. 2015;10:e0133234. doi: 10.1371/journal.pone.0133234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bushnell, B. BBtools (no. LBNL-7065E) (2015).
- 24.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guilloteau P, et al. From the gut to the peripheral tissues: the multiple effects of butyrate. Nutrition research reviews. 2010;23:366–384. doi: 10.1017/s0954422410000247. [DOI] [PubMed] [Google Scholar]
- 26.Letunic I, Bork P. Interactive tree of life (iTOL)v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McMurdie PJ, Holmes S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PloS one. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dixon P. VEGAN, a package of R functions for community ecology. Journal of Vegetation Science. 2003;14:927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x. [DOI] [Google Scholar]
- 29.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecology. 2001;26:32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x. [DOI] [Google Scholar]
- 30.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq. 2. Genome biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300. [Google Scholar]
- 32.Guilloteau P, Zabielski R, Blum JW. Gastrointestinal tract and digestion in the young ruminant: ontogenesis, adaptations, consequences and manipulations. Journal of physiology and pharmacology: an official journal of the Polish Physiological Society. 2009;60(Suppl 3):37–46. [PubMed] [Google Scholar]
- 33.Kotunia A, et al. Effect of sodium butyrate on the small intestine development in neonatal piglets fed [correction of feed] by artificial sow. Journal of physiology and pharmacology: an official journal of the Polish Physiological Society. 2004;55(Suppl 2):59–68. [PubMed] [Google Scholar]
- 34.Gressley T. F., Hall M. B., Armentano L. E. RUMINANT NUTRITION SYMPOSIUM: Productivity, digestion, and health responses to hindgut acidosis in ruminants1. Journal of Animal Science. 2011;89(4):1120–1130. doi: 10.2527/jas.2010-3460. [DOI] [PubMed] [Google Scholar]
- 35.Pierce, K. M., Boland, T. M., Kenny, D. A., O’Doherty, J. V., Kelly, A. K. In ADSA-ASAS-CSAS Joint Annual Meeting (2014).
- 36.Black, J. & Sharkey, M. In Mammalia Vol. 34 294 (1970).
- 37.Gorka P, et al. Is rumen development in newborn calves affected by different liquid feeds and small intestine development? Journal of dairy science. 2011;94:3002–3013. doi: 10.3168/jds.2010-3499. [DOI] [PubMed] [Google Scholar]
- 38.Purushe J, et al. Comparative genome analysis of Prevotella ruminicola and Prevotella bryantii: insights into their environmental niche. Microbial ecology. 2010;60:721–729. doi: 10.1007/s00248-010-9692-8. [DOI] [PubMed] [Google Scholar]
- 39.Ivarsson E, Roos S, Liu HY, Lindberg JE. Fermentable non-starch polysaccharides increases the abundance of Bacteroides-Prevotella-Porphyromonas in ileal microbial community of growing pigs. Animal: an international journal of animal bioscience. 2014;8:1777–1787. doi: 10.1017/s1751731114001827. [DOI] [PubMed] [Google Scholar]
- 40.Emerson EL, Weimer PJ. Fermentation of model hemicelluloses by Prevotella strains and Butyrivibrio fibrisolvens in pure culture and in ruminal enrichment cultures. Applied microbiology and biotechnology. 2017;101:4269–4278. doi: 10.1007/s00253-017-8150-7. [DOI] [PubMed] [Google Scholar]
- 41.Xiong, H. et al. Butyrate upregulates endogenous host defense peptides to enhance disease resistance in piglets via histone deacetylase inhibition. Scientific reports6, 27070, 10.1038/srep27070, https://www.nature.com/articles/srep27070#supplementary-information (2016). [DOI] [PMC free article] [PubMed]
- 42.Klappenbach JA, Saxman PR, Cole JR, Schmidt TM. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Research. 2001;29:181–184. doi: 10.1093/nar/29.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watanabe Y, Nagai F, Morotomi M. Characterization of Phascolarctobacterium succinatutens sp. nov., an Asaccharolytic, Succinate-Utilizing Bacterium Isolated from Human Feces. Applied and Environmental Microbiology. 2012;78:511–518. doi: 10.1128/AEM.06035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aschenbach JR, Kristensen NB, Donkin SS, Hammon HM, Penner GB. Gluconeogenesis in dairy cows: the secret of making sweet milk from sour dough. IUBMB life. 2010;62:869–877. doi: 10.1002/iub.400. [DOI] [PubMed] [Google Scholar]
- 45.Flint HJ, Duncan SH, Scott KP, Louis P. Links between diet, gut microbiota composition and gut metabolism. Proceedings of the Nutrition Society. 2014;74:13–22. doi: 10.1017/S0029665114001463. [DOI] [PubMed] [Google Scholar]
- 46.Bourriaud C, et al. Lactate is mainly fermented to butyrate by human intestinal microfloras but inter-individual variation is evident. Journal of applied microbiology. 2005;99:201–212. doi: 10.1111/j.1365-2672.2005.02605.x. [DOI] [PubMed] [Google Scholar]
- 47.Gao X, Oba M. Characteristics of dairy cows with a greater or lower risk of subacute ruminal acidosis: Volatile fatty acid absorption, rumen digestion, and expression of genes in rumen epithelial cells. Journal of dairy science. 2016 doi: 10.3168/jds.2016-11570. [DOI] [PubMed] [Google Scholar]
- 48.Kim YH, et al. Effects of Dietary Forage and Calf Starter Diet on Ruminal pH and Bacteria in Holstein Calves during Weaning Transition. Frontiers in microbiology. 2016;7:1575. doi: 10.3389/fmicb.2016.01575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim YH, et al. Effects of dietary forage and calf starter on ruminal pH and transcriptomic adaptation of the rumen epithelium in Holstein calves during the weaning transition. Physiological genomics. 2016;48:803–809. doi: 10.1152/physiolgenomics.00086.2016. [DOI] [PubMed] [Google Scholar]
- 50.Berding K, et al. Prebiotics and Bioactive Milk Fractions Affect Gut Development, Microbiota, and Neurotransmitter Expression in Piglets. Journal of pediatric gastroenterology and nutrition. 2016;63:688–697. doi: 10.1097/mpg.0000000000001200. [DOI] [PubMed] [Google Scholar]
- 51.Chen W, Liu F, Ling Z, Tong X, Xiang C. Human Intestinal Lumen and Mucosa-Associated Microbiota in Patients with Colorectal Cancer. PLOS ONE. 2012;7:e39743. doi: 10.1371/journal.pone.0039743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kobayashi H, Fu Q, Maeda H, Sato K. Draft Genome Sequence of a Novel Coriobacteriaceae sp. Strain, EMTCatB1, Reconstructed from the Metagenome of a Thermophilic Electromethanogenic Biocathode. Genome announcements. 2017;5:e00022–00017. doi: 10.1128/genomeA.00022-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Looft T, Bayles DO, Alt DP, Stanton TB. Complete Genome Sequence of Coriobacteriaceae Strain 68-1-3, a Novel Mucus-Degrading Isolate from the Swine Intestinal Tract. Genome announcements. 2015;3:e01143–01115. doi: 10.1128/genomeA.01143-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.KEMP P., LANDER D. J. Hydrogenation in vitro of -Linolenic Acid to Stearic Acid by Mixed Cultures of Pure Strains of Rumen Bacteria. Microbiology. 1984;130(3):527–533. doi: 10.1099/00221287-130-3-527. [DOI] [Google Scholar]
- 55.Meale SJ, et al. Weaning age influences the severity of gastrointestinal microbiome shifts in dairy calves. Scientific reports. 2017;7:198. doi: 10.1038/s41598-017-00223-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kittelmann S, et al. Simultaneous amplicon sequencing to explore co-occurrence patterns of bacterial, archaeal and eukaryotic microorganisms in rumen microbial communities. PloS one. 2013;8:e47879. doi: 10.1371/journal.pone.0047879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jenkins T. C., Wallace R. J., Moate P. J., Mosley E. E. BOARD-INVITED REVIEW: Recent advances in biohydrogenation of unsaturated fatty acids within the rumen microbial ecosystem1. Journal of Animal Science. 2008;86(2):397–412. doi: 10.2527/jas.2007-0588. [DOI] [PubMed] [Google Scholar]
- 58.Ley RE, et al. Obesity alters gut microbial ecology. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Veneman JB, et al. Does Dietary Mitigation of Enteric Methane Production Affect Rumen Function and Animal Productivity in Dairy Cows? PloS one. 2015;10:e0140282. doi: 10.1371/journal.pone.0140282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bickerstaffe R., Noakes D. E., Annison E. F. Quantitative aspects of fatty acid biohydrogenation, absorption and transfer into milk fat in the lactating goat, with special reference to thecis- andtrans-isomers of octadecenoate and linoleate. Biochemical Journal. 1972;130(2):607–617. doi: 10.1042/bj1300607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Soo RM, et al. An Expanded Genomic Representation of the Phylum Cyanobacteria. Genome Biology and Evolution. 2014;6:1031–1045. doi: 10.1093/gbe/evu073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jami E, Israel A, Kotser A, Mizrahi I. Exploring the bovine rumen bacterial community from birth to adulthood. The ISME journal. 2013;7:1069–1079. doi: 10.1038/ismej.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rey M, et al. Establishment of ruminal bacterial community in dairy calves from birth to weaning is sequential. Journal of applied microbiology. 2013 doi: 10.1111/jam.12405. [DOI] [PubMed] [Google Scholar]
- 64.Malmuthuge N, Griebel PJ, Guan LL. Taxonomic Identification of Commensal Bacteria Associated with the Mucosa and Digesta throughout the Gastrointestinal Tracts of Preweaned Calves. Applied and environmental microbiology. 2014;80:2021–2028. doi: 10.1128/aem.03864-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Malmuthuge N, Li M, Goonewardene LA, Oba M, Guan LL. Effect of calf starter feeding on gut microbial diversity and expression of genes involved in host immune responses and tight junctions in dairy calves during weaning transition. Journal of dairy science. 2013;96:3189–3200. doi: 10.3168/jds.2012-6200. [DOI] [PubMed] [Google Scholar]
- 66.Meale SJ, et al. Development of Ruminal and Fecal Microbiomes Are Affected by Weaning But Not Weaning Strategy in Dairy Calves. Frontiers in microbiology. 2016;7:582. doi: 10.3389/fmicb.2016.00582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang, W. et al. Effects of early feeding on the host rumen transcriptome and bacterial diversity in lambs. 6, 32479, 10.1038/srep32479, https://www.nature.com/articles/srep32479#supplementary-information (2016). [DOI] [PMC free article] [PubMed]
- 68.Li, F. & Guan, L. L. Metatranscriptomic Profiling Reveals Linkages between the Active Rumen Microbiome and Feed Efficiency in Beef Cattle. Applied and environmental microbiology83, 10.1128/aem.00061-17 (2017). [DOI] [PMC free article] [PubMed]
- 69.Danielsson R, et al. Methane Production in Dairy Cows Correlates with Rumen Methanogenic and Bacterial CommunityStructure. Frontiers in microbiology. 2017;8:226. doi: 10.3389/fmicb.2017.00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pope PB, et al. Isolation of Succinivibrionaceae implicated in low methane emissions from Tammar wallabies. Science. 2011;333:646–648. doi: 10.1126/science.1205760. [DOI] [PubMed] [Google Scholar]
- 71.Dill-McFarland, K. A., Breaker, J. D. & Suen, G. Microbial succession in the gastrointestinal tract of dairy cows from 2 weeks to first lactation. 7, 40864, 10.1038/srep40864, https://www.nature.com/articles/srep40864#supplementary-information (2017). [DOI] [PMC free article] [PubMed]
- 72.Nelson MC, Morrison HG, Benjamino J, Grim SL, Graf J. Analysis, optimization and verification of Illumina-generated 16S rRNA gene amplicon surveys. PloS one. 2014;9:e94249. doi: 10.1371/journal.pone.0094249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Popova M, Morgavi DP, Martin C. Methanogens and Methanogenesis in the Rumens and Ceca of Lambs Fed Two Different High-Grain-Content Diets. Applied and environmental microbiology. 2013;79:1777–1786. doi: 10.1128/AEM.03115-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut microbes. 2012;3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yanez-Ruiz, D. R., Abecia, L. & Newbold, C. J. Manipulating rumen microbiome and fermentation through interventions during early life: a review. Frontiers in microbiology6, 10.3389/fmicb.2015.01133 (2015). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.