The synthesis and structures of three coordination polymers composed of manganese(II) acetate and pyridine N-oxide complexes are reported. The pyridine N-oxide, 2-methylpyridine N-oxide, and 4-methylpyridine N-oxide complexes form different networks owing to the substituent group effects.
Keywords: crystal structure, manganese(II) acetate, pyridine N-oxide ligand, coordination polymer
Abstract
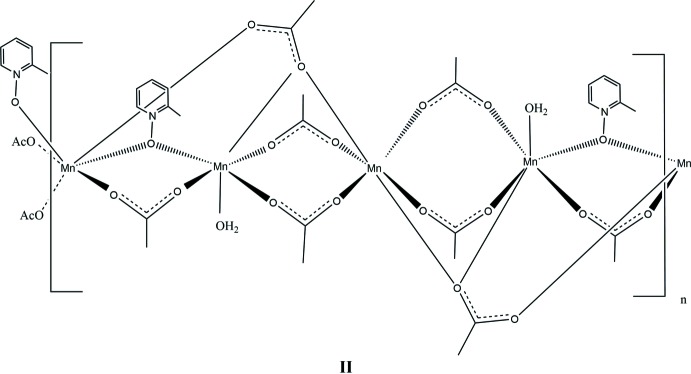
Manganese(II) acetate coordination polymers have been prepared with three derivatives of pyridine N-oxide. The compounds are catena-poly[manganese(II)-μ3-acetato-di-μ2-acetato-[aquamanganese(II)]-μ2-acetato-μ-(pyridine N-oxide)-manganese(II)-μ3-acetato-μ2-acetato-μ-(pyridine N-oxide)-[aquamanganese(II)]-di-μ2-acetato], [Mn4(CH3COO)8(C5H5NO)2(H2O)2]n, (I), catena-poly[[manganese(II)]-μ3-acetato-μ2-acetato-μ-(2-methylpyridine N-oxide)-[aquamanganese(II)]-di-μ2-acetato-manganese(II)-di-μ2-acetato-μ3-acetato-[aquamanganese(II)]-μ2-acetato-μ-(2-methylpyridine N-oxide)], [Mn4(CH3COO)8(C6H7NO)2(H2O)2]n, (II), and catena-poly[[manganese(II)-di-μ2-acetato-μ-(4-methylpyridine N-oxide)] monohydrate], {[Mn(CH3COO)2(C6H7NO)]·H2O}n, (III). Compounds (I) and (II) both have three unique Mn atoms; in both compounds two of them sit on a crystallographic inversion center while the third is on a general position. In compound (III), the single unique Mn atom sits on a general position. Pseudo-octahedral six-coordinate manganese(II) centers are found in all compounds. All of the compounds form chains of Mn atoms bridged by acetate ions and the oxygen atom of the N-oxide in pyridine N-oxide (PNO), 2-methylpyridine N-oxide (2MePNO), or 4-methylpyridine N-oxide (4MePNO). Compound (I) and (II) both exhibit a bound water of solvation. In (I), the water hydrogen bonds to a nearby acetate whereas in (II) the water molecule forms bridging hydrogen bonds between two neighboring acetates. In compound (III) a water molecule of solvation is found in the lattice, not bound to the metal ion but hydrogen bonding to a bridging acetate.
Chemical context
N-Oxides and acetates both have interesting binding modes that facilitate the growth of unique coordination structures. The structures take advantage of the versatility of the acetate ions and the hybridization and dipole at the oxygen atom on the N-oxide. The structures extend to the formation of coordination polymers that have been reported previously (Sarma et al., 2008 ▸, 2009 ▸; Sarma & Baruah, 2011 ▸). A recent report shows the utility of pyridine N-oxide to facilitate coordination polymer formation with both zinc(II) and manganese(II) metal ions with a single bifunctional ligand containing an acetate and N-oxide moiety (Ren et al., 2018 ▸). In a previous paper in this series, we examined the initial utility of aromatic N-oxide ligands to form polymeric structures with manganese(II) chloride (Kang et al., 2017 ▸). Complexes have also been used previously as metal centers for catalytic transformations (Liu et al., 2014 ▸).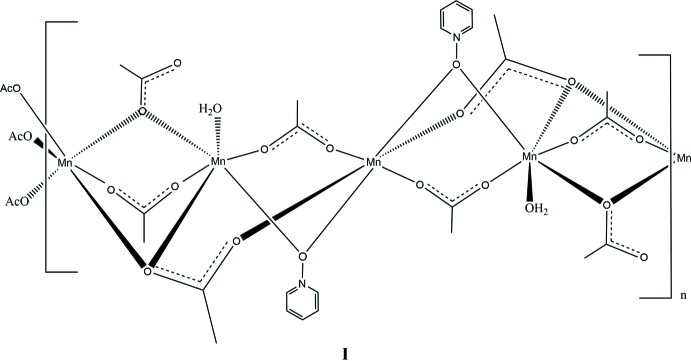
In this contribution, we report the synthesis and solid-state structures of three manganese(II) complexes with the versatile mono- or bidentate bridging ligands acetate and three derivatives of pyridine N-oxide (Figs. 1 ▸–3 ▸ ▸). In this study, each of the ligands pyridine N-oxide, 2-methyl and 4-methyl pyridine N-oxide has an impact on the structures of manganese(II) acetate complexes. All three complexes form coordination polymers with the N-oxide bridging in a μ2-1,1 mode and varying acetate ligation. The study was conducted to investigate the utility of both acetate and substituted pyridine N-oxide to facilitate the growth of unique coordination polymers.
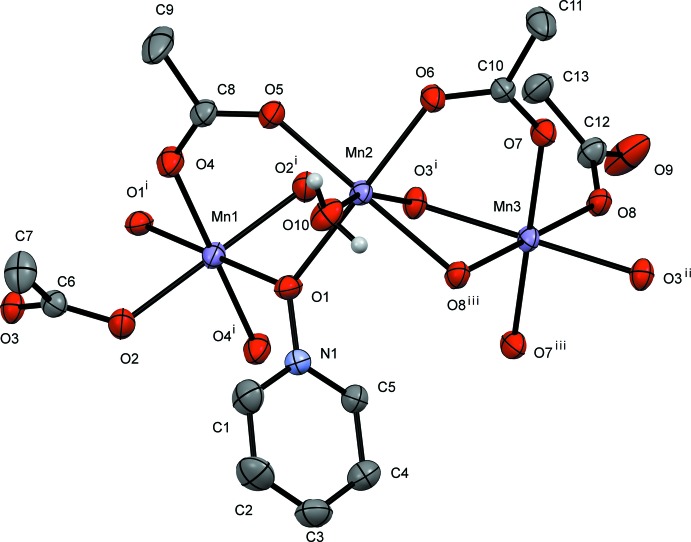
Figure 1.
A view of compound I, showing the atom labeling. Displacement ellipsoids are drawn at the 50% probability level, H atoms not involved in hydrogen bonding have been omitted for clarity. [Symmetry codes: (i) −x + 1, –y + 1, −z + 1; (ii) x + 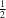 , y −
, y − 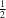 , z; (iii) −x +
, z; (iii) −x +  , −y +
, −y + 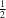 , −z + 1.]
, −z + 1.]
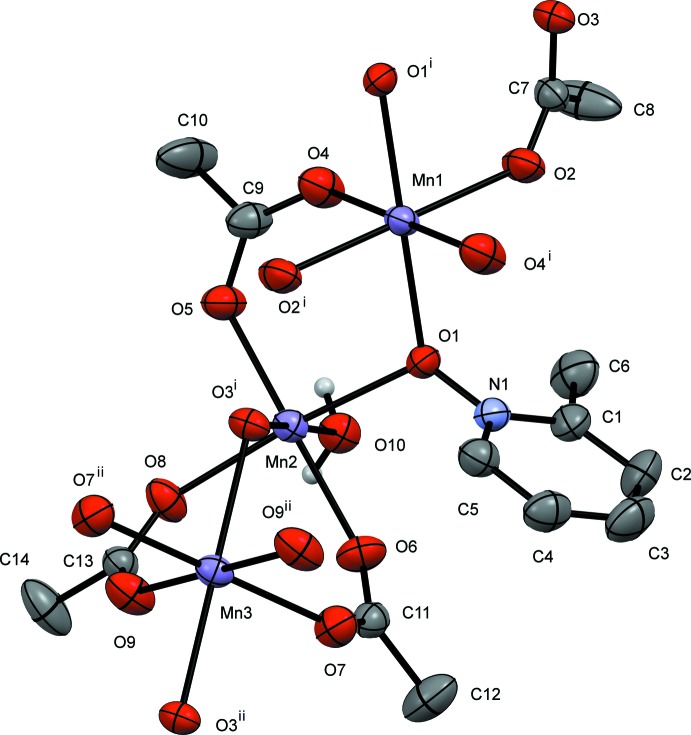
Figure 2.
A view of compound II, showing the atom labeling. Displacement ellipsoids are drawn at the 50% probability level, H atoms not involved in hydrogen bonding have been omitted for clarity. [Symmetry codes: (i) −x + 1, −y + 1, −z + 1; (ii) −x + 1, −y, −z + 2.]
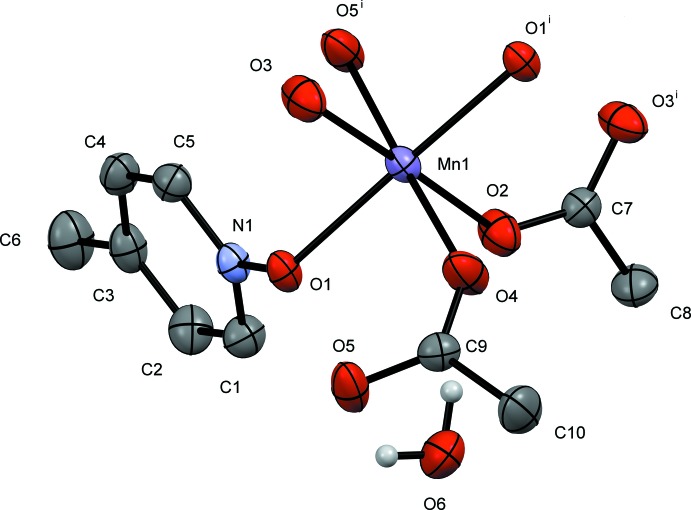
Figure 3.
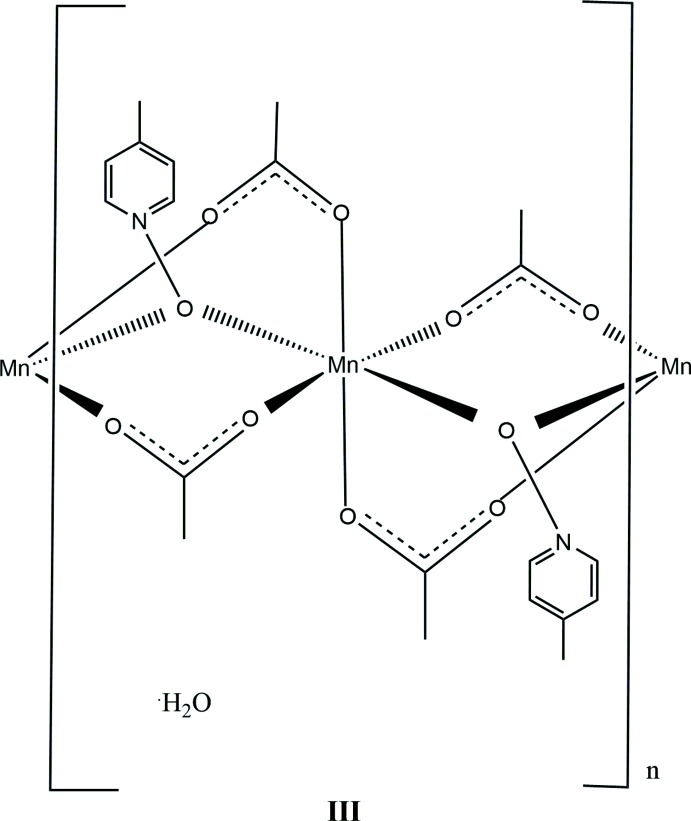
A view of compound III, showing the atom labeling. Displacement ellipsoids are drawn at the 50% probability level, H atoms not involved in hydrogen bonding have been omitted for clarity. [Symmetry code: (i) −x + 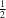 , y −
, y − 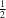 , −z +
, −z +  .]
.]
Structural commentary
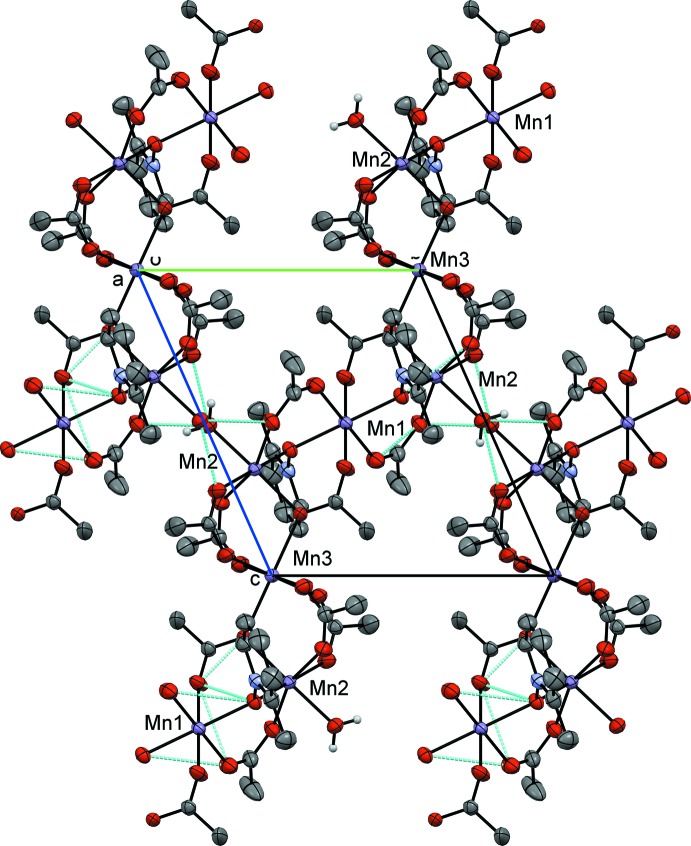
General structural details. The pyridine N-oxide (PNO) complex, compound I, is a repeating tetrameric coordination polymer that crystallizes in the monoclinic space group C2/c. The manganese atoms align as an Mn3, Mn2, Mn1, Mn2, chain. The structure can be formulated in the simplest empirical relationship as [Mn4(PNO)2(OAc)8(H2O)2]n. Examining the molecule across the AB vertex, Mn3 and Mn1 sit in a repeating line of Mn3, Mn1, Mn3 atoms. The Mn2 atoms all sit along a different line in this orientation. The atom-to-atom connectivity in the Mn3,Mn2,Mn1,Mn2 repeating unit can best be described as zigzag (Fig. 4 ▸). In this orientation, the pyridine rings also stack; however, they are not π stacked because of the separation distance caused by the methyl group of an acetate ligand in between each aromatic group. Interpolymeric chain hydrogen bonding is observed from the water molecule (O10) on Mn2 to an oxygen atom (O6) on an Mn2-bound acetate ligand (Table 1 ▸). The structure contains a six-coordinate metal center at each MnII atom with all six donor atoms being oxygen. Mn1 sits on an inversion center and is bound trans by two μ2-1,1-PNOs (to Mn2), trans by two μ2-1,3-acetates (to Mn2), and trans by two μ3-1,3,3-acetates (to both Mn2 and Mn3). Mn2 is also six-coordinate with a μ2-1,1-PNO (from Mn1), a μ2-1,3-acetate (from Mn1), and a μ3-1,3,3-acetate (from Mn1 and Mn3). Further, the octahedral environment is completed by a water of hydration, a μ2-1,1-acetate (to Mn3), and a μ2-1,3-acetate (to Mn3). Mn3 also sits on an inversion cente, showing an octahedral enviroment where all the six coordinated oxygen atoms belong to acetate ligands. The coordination sphere comprises two μ3-1,3,3-acetates (uniquely bound to Mn2 and Mn1), two μ2-1,1-acetates (to Mn2) and two μ2-1,3-acetates (to Mn2).
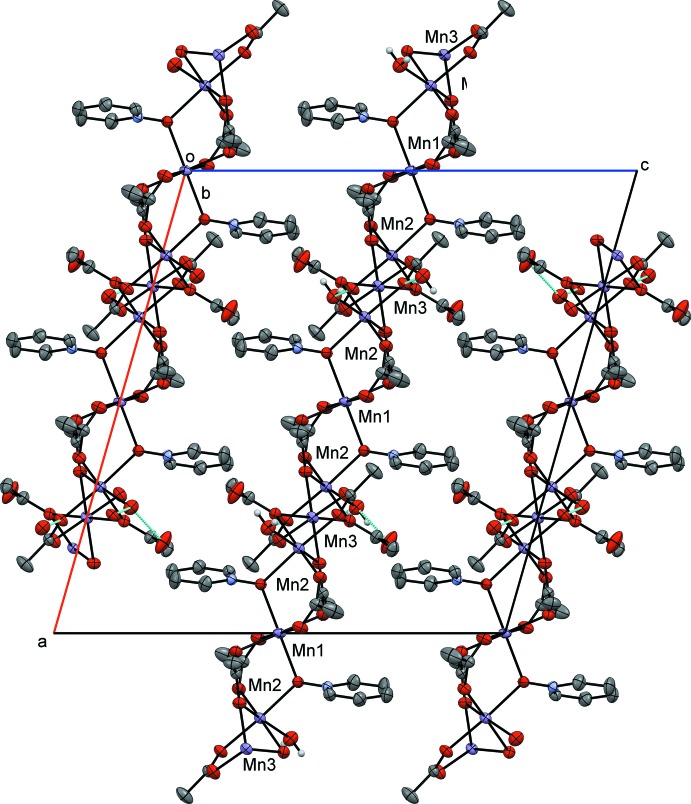
Figure 4.
Crystal packing diagram of compound I, viewed along the b axis. Displacement ellipsoids are drawn at the 50% probability level, H atoms not involved in hydrogen bonding have been omitted for clarity, hydrogen bonds are rendered in blue.
Table 1. Hydrogen-bond geometry (Å, °) for I .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O10—H10A⋯O9i | 0.85 (2) | 1.85 (2) | 2.652 (2) | 157 (3) |
| O10—H10B⋯O6ii | 0.83 (2) | 1.98 (2) | 2.786 (2) | 166 (3) |
Symmetry codes: (i) 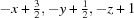 ; (ii)
; (ii) 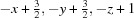 .
.
The 2-methylpyridine N-oxide (2MePNO) complex, compound II, is similar to I in that it is a repeating tetrameric coordination polymer. The polymer crystallizes in the triclinic system, space group P
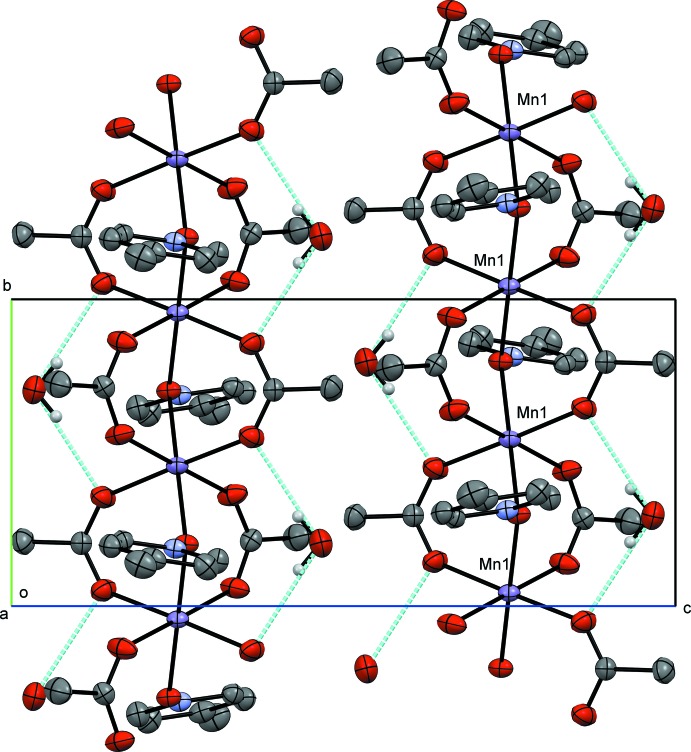
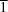 . The manganese atoms align as an Mn3, Mn2, Mn1, Mn2 chain similar to I with Mn1 and Mn3 sitting on inversion centers. Examining the molecule across the BC vertex, as in I, the Mn3 and Mn1 sit in a line whereas the Mn2 atoms all sit along a different line with respect to this orientation. The atom-to-atom connectivity in the Mn3, Mn2, Mn1, Mn2 repeating unit can best be described as zigzag (Fig. 5 ▸). In this orientation, the 2-methylpyridine ring planes are twisted by 85.31 (2)° with respect to the Mn1/O2/Mn2 plane with all the methyl groups pointing in two symmetry-related directions. As observed in I, interpolymeric chain hydrogen bonding is observed from the water molecule (O10) on Mn2 to an oxygen atom on the adjacent Mn2 on the next polymer. However, symmetry dictates that the hydrogen bonding is to oxygen atoms (O5 and O8) on two acetates bound to Mn2 (Table 2 ▸). The structure can be formulated with the same empirical stoichiometry as I, [Mn4(2MePNO)2(OAc)7(H2O)4]n. Compound II has one important variation from the PNO derivative outlined above. There is no evidence of the μ2-1,1-acetate bridge found above. While the singular μ3-1,3,3-acetate bridge is retained between Mn2 and Mn3, the μ2-1,1 has been replaced by a μ2-1,3 acetate bridge. This is likely because of the steric demands of the 2-methyl substituent.
. The manganese atoms align as an Mn3, Mn2, Mn1, Mn2 chain similar to I with Mn1 and Mn3 sitting on inversion centers. Examining the molecule across the BC vertex, as in I, the Mn3 and Mn1 sit in a line whereas the Mn2 atoms all sit along a different line with respect to this orientation. The atom-to-atom connectivity in the Mn3, Mn2, Mn1, Mn2 repeating unit can best be described as zigzag (Fig. 5 ▸). In this orientation, the 2-methylpyridine ring planes are twisted by 85.31 (2)° with respect to the Mn1/O2/Mn2 plane with all the methyl groups pointing in two symmetry-related directions. As observed in I, interpolymeric chain hydrogen bonding is observed from the water molecule (O10) on Mn2 to an oxygen atom on the adjacent Mn2 on the next polymer. However, symmetry dictates that the hydrogen bonding is to oxygen atoms (O5 and O8) on two acetates bound to Mn2 (Table 2 ▸). The structure can be formulated with the same empirical stoichiometry as I, [Mn4(2MePNO)2(OAc)7(H2O)4]n. Compound II has one important variation from the PNO derivative outlined above. There is no evidence of the μ2-1,1-acetate bridge found above. While the singular μ3-1,3,3-acetate bridge is retained between Mn2 and Mn3, the μ2-1,1 has been replaced by a μ2-1,3 acetate bridge. This is likely because of the steric demands of the 2-methyl substituent.
Figure 5.
Crystal packing diagram of compound II, viewed along the a axis. H atoms not involved in hydrogen bonding have been omitted for clarity, hydrogen bonds are rendered in blue.
Table 2. Hydrogen-bond geometry (Å, °) for II .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O10—H10D⋯O5i | 0.84 (2) | 2.04 (2) | 2.821 (3) | 155 (3) |
| O10—H10E⋯O8i | 0.85 (2) | 1.94 (2) | 2.727 (3) | 155 (3) |
Symmetry code: (i) 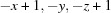 .
.
The 4-methylpyridine N-oxide (4MePNO) complex, compound III, is a repeating coordination polymer with one unique MnII ion that crystallizes in the monoclinic system, space group P21/n. In the coordination polymer, the manganese atoms are aligned along the b-axis direction. The structure can be formulated as [Mn(4MePNO)2(OAc)4(H2O)]n. The six-coordinate metal center is bridged by two oxygen atoms from μ2-1,1 4MePNO and four μ2-1,3 acetate bridges. The 4MePNO complex molecules alternate above and below the line formed by the manganese atoms. Unlike I and II, compound III only forms intramolecular hydrogen bonding in the polymeric chain (Table 3 ▸, Fig. 6 ▸). The water observed in the lattice forms a hydrogen bond at 2.21 (2) Å with the O2 atom belonging to one of the acetate μ2-1,3 acetate bridges.
Table 3. Hydrogen-bond geometry (Å, °) for III .
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O6—H6D⋯O3i | 0.84 (2) | 2.23 (2) | 3.052 (3) | 165 (4) |
| O6—H6E⋯O2 | 0.84 (2) | 2.21 (2) | 3.035 (3) | 170 (4) |
Symmetry code: (i) 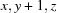 .
.
Figure 6.
Crystal packing diagram of compound III, viewed along the a axis. H atoms not involved in hydrogen bonding have been omitted for clarity, hydrogen bonds are rendered in blue.
Specific structural details. In I, the bond distances involving Mn1 lie between 2.1822 (15) Å (Mn1—O2) and 2.1207 (16) Å (Mn1—O4) whereas all bond angles are within 2.5° of 90°. These angles and distances are similar to those for other MnII acetate structures (see for example Dave et al., 1993 ▸ and Ciunik & Głowiak, 1980 ▸). The O1 atom of the PNO ligand bridges Mn1 at 2.168 (2) Å and Mn2 at 2.211 (2) Å which is unremarkable for compounds of MnII and pyridine N-oxide (Sniekers et al., 2017 ▸; Mondal et al., 2012 ▸). Mn2 shows a short bond distance to O5 (a μ2-1,3-acetate bridging from Mn1) of 2.1062 (15) Å and a long distance of 2.2671 (14) Å from O8, which is a μ2-1,1-acetate bridging to Mn3. The water molecule (O10), see Table 1 ▸, is found at 2.1506 (16) Å at a distance similar to that reported previously. (Mondal et al., 2012 ▸) and also hydrogen bonded to O9 (unbound acetate oxygen from μ2-1,1-acetate bridging across Mn2 and Mn3) at 2.652 (2) Å. The O3—Mn2—O10 bond angle is severely distorted from 180° to 162.68 (6)°. The other two trans bond angles around Mn2 are approximately 175°. The Mn3 bond distances span from 2.1338 (15) (Mn3—O7) to 2.2194 (14) Å (Mn3—O8). The O3—Mn3—O8 bond angle is somewhat constrained at 78.19 (5)° whereas the remaining angles are all nearly 90°.
The bond distances involving Mn1 in compound II lie between 2.129 (2) Å (Mn1—O4) and 2.2061 (19) Å (Mn1—O1) which are normal for MnII acetate compounds of this type (Dave et al., 1993 ▸ and Ciunik & Głowiak, 1980 ▸). The long bond distance is to the oxygen originating from the bridging 2MePNO and is 0.0398 Å longer than the Mn1—O1 PNO bond in I but similar to those reported previously (Sniekers et al., 2017 ▸; Mondal et al., 2012 ▸). The bond angles are within 5° of the expected 90° for octahedral systems with the most constrained angle being O1—Mn1—O2 [85.03 (7)°]. Mn2 has its shortest bond distance to O6 (a μ2-1,3-acetate bridging from Mn3) of 2.136 (2) Å, whereas its longest distance is 2.239 (2) Å to O10, the terminal water molecule. The μ2-1,1-2MePNO (O1) bond distance to Mn2 is also long [2.2300 (18) Å]. The μ3-1,3,3-acetate also links Mn2 and Mn3 via the O3 atom. The O5—Mn2—O10 bond angle is significantly distorted with a value of 81.63 (8)° as is the O6—Mn2—O10 bond angle of 78.61 (8)°. The μ3-1,3,3-acetate oxygen (O3) forms a long bond with Mn3 as well, observed at 2.3091 (18) Å. The other Mn3—O bond distances are unremarkable at approximately 2.15 Å. The bond angles around Mn3 are all within 4° of 90° in the six-coordinate Mn3 environment.
In compound III, the Mn1—O1 (4MePNO) bond length is the longest of those in this study, with the metal center at 2.203 (3) Å (Sniekers et al., 2017 ▸; Mondal et al., 2012 ▸) whereas the acetate Mn1—O bond distances range from 2.134 (3) Å to 2.179 (2) Å. The bond angles around the metal center are all within 5° of 90°, with the acetate O—Mn1—O angles being slightly larger, whereas the O(acetate)—Mn1—O(4MePNO) angles are slightly compressed. [For similar compounds, see for example Ciunik & Głowiak (1980 ▸) and Dave et al. (1993 ▸).] The water is in the lattice and forms a hydrogen bond at 2.21 (2) Å with an O2 atom belonging to one of the acetate μ2-1,3 acetate bridges.
Supramolecular features
The packing of I forms a polymeric structure bisecting the a axis and b axis. Because of the complexity of the structure, many of the details were outlined above. The structure is not linear but forms a zigzag chain in which the bridging acetates and N-oxide ligands connect the Mn ions. There is no evidence for π stacking but interpolymeric chain hydrogen bonding is present.
Compound II forms a similar polymeric structure to I, with the chain bisecting the unit cell at (½, 0, 1) and (½, 1, 0). The chain sets up in a similar fashion as I; however, μ2-1,3 and μ3-1,3,3 are the bridges observed while the μ2-1,1 bridge noted in I is absent.
Compound III forms a polymeric chain which is observed in the b-axis direction. Each manganese(II) atom is bridged by a single 4MePNO and two μ2-1,3 acetate ions. The 4MePNO bridging ligands are alternating up and down in the a-axis direction. There is no evidence for π stacking due to the long distance found in the structure with the aromatic rings at separations of 7.334 (6) Å.
Database survey
A search in the Cambridge Structural Database (CSD Version 5.39, November 2017 update; Groom et al., 2016 ▸) for aromatic N-oxides and acetate ligands bound to manganese returned 36 entries. Seven of the entries contain derivatives of picolinic N-oxides, thirteen involve derivatives of dipyridal N-oxide and fifteen include di- or tri-acetate ligands. Similar N-oxides with simple benzoate in the list include pyridine N-oxide (YIYRAA; Sarma et al., 2008 ▸), and the p-nitrobenzoate with PNO (TIXKER01 and TIXKER; Sarma et al., 2008 ▸). Another report by Sarma and co-workers includes the p-hydroxy, o-nitro and p-chlorobenzoate derivatives with PNO (POYRAX; Sarma et al., 2009 ▸).
Synthesis and crystallization
The manganese(II) coordination polymers were all synthesized by a similar method. 0.245 g (1.00 mmol) manganese(II) acetate tetrahydrate (MnAc2·4H2O, FW 245 g mol−1) was dissolved in a minimal amount (20 mL) of methanol. 2 molar equivalents of the appropriate N-oxide (0.191 g pyridine N-oxide, PNO; 0.220 g 2-methylpyrdine N-oxide, 2MePNO; 0.220 g 4-methylpyridine N-oxide, 4MePNO) were similarly dissolved in 10 mL of methanol. The N-oxide alcoholic solution was added in one portion to the MnII one. The combined reaction mixture was stirred for 30 minutes, filtered and the filtrate was allowed to evaporate by slow diffusion. X-ray quality crystals were obtained by precipitation from the mother liquor. A final wash with a minimal amount of methanol was performed to assist with removal of excess N-oxide.
Compound I. Yield 0.0642 g, 27.0 (%), decomposition/melting temperature = 433–437 K (turns to a brown liquid). Selected IR bands (ATR, FT–IR, KBr composite, cm−1): 3356 (2, br), 3118 (w), 1558 (s), 1495 (m), 1477 (m), 1418 (m), 1424 (m), 1216 (m), 1025 (w), 835 (m), 783 (m), 653 (w).
Compound II. Yield 0.0484 g, 20.4 (%), decomposition/melting temperature. 417–423 K (turns to a brown liquid). Selected IR bands (ATR, FT–IR, KBr composite, cm−1): 3348 (m, br), 1558 (s), 1495 (m), 1417 (s), 1209 (m), 846 (m), 783 (s), 655 (s).
Compound III. Yield 0.0892 g, 29.7 (%), decomposition/melting temperature. 405–411 K (turns to a black liquid). Selected IR bands (ATR, FT–IR, KBr composite, cm−1): 3412 (m, br), 1652 (w), 1574 (s), 1491 (s), 1424 (m), 1213 (s), 833 (m), 763 (s), 668 (s).
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 4 ▸. All carbon-bound H atoms were positioned geometrically and refined as riding, with C—H = 0.95 or 0.98 Å and U iso(H) = 1.2U eq(C) or U iso(H) = 1.5U eq(C) for C(H) and CH3 groups, respectively. In order to ensure chemically meaningful O—H distances for the bound water molecules in the compounds, the oxygen-to-hydrogen distances were restrained to a target value of 0.84 (2) Å (using a DFIX command in SHELXL2017; Sheldrick, 2015b ▸) and U iso(H) = 1.5U eq(O).
Table 4. Experimental details.
| I | II | III | |
|---|---|---|---|
| Crystal data | |||
| Chemical formula | [Mn4(C2H3O2)8(C5H5NO)2(H2O)2] | [Mn4(C2H3O2)8(C6H7NO)2(H2O)2] | [Mn(C2H3O2)2(C6H7NO)]·H2O |
| M r | 918.34 | 946.40 | 300.17 |
| Crystal system, space group | Monoclinic, C2/c | Triclinic, P
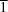
|
Monoclinic, P21/n |
| Temperature (K) | 173 | 173 | 173 |
| a, b, c (Å) | 19.936 (7), 10.603 (4), 18.692 (7) | 9.7704 (3), 10.5882 (7), 11.4720 (2) | 11.100 (3), 7.334 (3), 15.9808 (4) |
| α, β, γ (°) | 90, 105.925 (4), 90 | 65.76 (2), 83.84 (2), 65.512 (15) | 90, 96.500 (11), 90 |
| V (Å3) | 3800 (2) | 982.0 (2) | 1292.6 (6) |
| Z | 4 | 1 | 4 |
| Radiation type | Mo Kα | Mo Kα | Mo Kα |
| μ (mm−1) | 1.38 | 1.34 | 1.04 |
| Crystal size (mm) | 0.4 × 0.4 × 0.2 | 0.3 × 0.3 × 0.2 | 0.2 × 0.05 × 0.05 |
| Data collection | |||
| Diffractometer | Rigaku XtaLAB mini | Rigaku XtaLAB mini | Rigaku XtaLAB mini |
| Absorption correction | Multi-scan (REQAB; Rigaku, 1998 ▸) | Multi-scan (REQAB; Rigaku, 1998 ▸) | Multi-scan (REQAB; Rigaku, 1998 ▸) |
| T min, T max | 0.885, 1.00 | 0.842, 1.00 | 0.850, 1.00 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 19620, 4332, 3855 | 10542, 4503, 3551 | 13236, 2957, 2224 |
| R int | 0.046 | 0.058 | 0.074 |
| (sin θ/λ)max (Å−1) | 0.649 | 0.650 | 0.651 |
| Refinement | |||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.032, 0.083, 1.08 | 0.041, 0.101, 1.06 | 0.045, 0.105, 1.07 |
| No. of reflections | 4332 | 4503 | 2957 |
| No. of parameters | 249 | 259 | 173 |
| No. of restraints | 2 | 2 | 2 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.31, −0.36 | 0.62, −0.46 | 0.35, −0.40 |
Supplementary Material
Crystal structure: contains datablock(s) I, II, III. DOI: 10.1107/S205698901801232X/zl2728sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698901801232X/zl2728Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S205698901801232X/zl2728IIsup3.hkl
Structure factors: contains datablock(s) III. DOI: 10.1107/S205698901801232X/zl2728IIIsup4.hkl
Additional supporting information: crystallographic information; 3D view; checkCIF report
supplementary crystallographic information
catena-Poly[manganese(II)-µ3-acetato-di-µ2-acetato-[aquamanganese(II)]-µ2-acetato-µ-(pyridine N-oxide)-manganese(II)-µ3-acetato-µ2-acetato-µ-(pyridine N-oxide)-[aquamanganese(II)]-di-µ2-acetato] (I) . Crystal data
| [Mn4(C2H3O2)8(C5H5NO)2(H2O)2] | F(000) = 1872 |
| Mr = 918.34 | Dx = 1.605 Mg m−3 |
| Monoclinic, C2/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 19.936 (7) Å | Cell parameters from 5012 reflections |
| b = 10.603 (4) Å | θ = 2.1–27.5° |
| c = 18.692 (7) Å | µ = 1.38 mm−1 |
| β = 105.925 (4)° | T = 173 K |
| V = 3800 (2) Å3 | Prism, colorless |
| Z = 4 | 0.4 × 0.4 × 0.2 mm |
catena-Poly[manganese(II)-µ3-acetato-di-µ2-acetato-[aquamanganese(II)]-µ2-acetato-µ-(pyridine N-oxide)-manganese(II)-µ3-acetato-µ2-acetato-µ-(pyridine N-oxide)-[aquamanganese(II)]-di-µ2-acetato] (I) . Data collection
| Rigaku XtaLAB mini diffractometer | 4332 independent reflections |
| Radiation source: Sealed Tube | 3855 reflections with I > 2σ(I) |
| Graphite Monochromator monochromator | Rint = 0.046 |
| Detector resolution: 13.6612 pixels mm-1 | θmax = 27.5°, θmin = 2.1° |
| profile data from ω–scans | h = −25→25 |
| Absorption correction: multi-scan (REQAB; Rigaku, 1998) | k = −13→13 |
| Tmin = 0.885, Tmax = 1.00 | l = −24→24 |
| 19620 measured reflections |
catena-Poly[manganese(II)-µ3-acetato-di-µ2-acetato-[aquamanganese(II)]-µ2-acetato-µ-(pyridine N-oxide)-manganese(II)-µ3-acetato-µ2-acetato-µ-(pyridine N-oxide)-[aquamanganese(II)]-di-µ2-acetato] (I) . Refinement
| Refinement on F2 | Hydrogen site location: mixed |
| Least-squares matrix: full | H atoms treated by a mixture of independent and constrained refinement |
| R[F2 > 2σ(F2)] = 0.032 | w = 1/[σ2(Fo2) + (0.0348P)2 + 3.1638P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.083 | (Δ/σ)max = 0.001 |
| S = 1.08 | Δρmax = 0.31 e Å−3 |
| 4332 reflections | Δρmin = −0.36 e Å−3 |
| 249 parameters | Extinction correction: SHELXL-2018/1 (Sheldrick 2015), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 2 restraints | Extinction coefficient: 0.00109 (13) |
| Primary atom site location: dual |
catena-Poly[manganese(II)-µ3-acetato-di-µ2-acetato-[aquamanganese(II)]-µ2-acetato-µ-(pyridine N-oxide)-manganese(II)-µ3-acetato-µ2-acetato-µ-(pyridine N-oxide)-[aquamanganese(II)]-di-µ2-acetato] (I) . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
catena-Poly[manganese(II)-µ3-acetato-di-µ2-acetato-[aquamanganese(II)]-µ2-acetato-µ-(pyridine N-oxide)-manganese(II)-µ3-acetato-µ2-acetato-µ-(pyridine N-oxide)-[aquamanganese(II)]-di-µ2-acetato] (I) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Mn1 | 0.500000 | 0.500000 | 0.500000 | 0.02291 (11) | |
| O1 | 0.60564 (7) | 0.50006 (13) | 0.57232 (8) | 0.0254 (3) | |
| C1 | 0.60680 (13) | 0.5206 (2) | 0.69567 (13) | 0.0368 (5) | |
| H1 | 0.594602 | 0.605051 | 0.687011 | 0.044* | |
| N1 | 0.61602 (8) | 0.44789 (16) | 0.64043 (9) | 0.0226 (3) | |
| Mn2 | 0.68359 (2) | 0.53844 (3) | 0.51070 (2) | 0.01989 (10) | |
| C2 | 0.61555 (15) | 0.4693 (3) | 0.76534 (13) | 0.0455 (6) | |
| H2 | 0.609182 | 0.519044 | 0.803946 | 0.055* | |
| O2 | 0.45997 (7) | 0.59052 (14) | 0.58476 (8) | 0.0290 (3) | |
| O3 | 0.34900 (7) | 0.64803 (13) | 0.54422 (8) | 0.0251 (3) | |
| C3 | 0.63378 (13) | 0.3441 (2) | 0.77773 (13) | 0.0398 (6) | |
| H3 | 0.640102 | 0.308856 | 0.824684 | 0.048* | |
| Mn3 | 0.750000 | 0.250000 | 0.500000 | 0.02146 (11) | |
| C4 | 0.64253 (12) | 0.2717 (2) | 0.71959 (13) | 0.0355 (5) | |
| H4 | 0.654963 | 0.187201 | 0.727123 | 0.043* | |
| O4 | 0.51209 (8) | 0.68135 (14) | 0.45732 (9) | 0.0319 (3) | |
| O5 | 0.62046 (8) | 0.67838 (14) | 0.44529 (8) | 0.0287 (3) | |
| C5 | 0.63271 (11) | 0.3254 (2) | 0.65014 (12) | 0.0280 (4) | |
| H5 | 0.637647 | 0.276949 | 0.610427 | 0.034* | |
| C6 | 0.41374 (10) | 0.67368 (19) | 0.57075 (11) | 0.0241 (4) | |
| O6 | 0.75532 (7) | 0.56777 (13) | 0.44447 (8) | 0.0269 (3) | |
| C7 | 0.43584 (13) | 0.8095 (2) | 0.58470 (15) | 0.0402 (6) | |
| H7A | 0.459117 | 0.836106 | 0.548603 | 0.060* | |
| H7B | 0.467028 | 0.817737 | 0.633739 | 0.060* | |
| H7C | 0.395451 | 0.861292 | 0.580629 | 0.060* | |
| O7 | 0.79968 (8) | 0.37538 (13) | 0.44121 (8) | 0.0299 (3) | |
| O8 | 0.74244 (7) | 0.10035 (13) | 0.41514 (7) | 0.0239 (3) | |
| C8 | 0.56142 (11) | 0.72708 (19) | 0.43642 (12) | 0.0268 (4) | |
| C9 | 0.54817 (15) | 0.8513 (3) | 0.39541 (19) | 0.0567 (8) | |
| H9A | 0.520255 | 0.837263 | 0.345358 | 0.085* | |
| H9C | 0.591852 | 0.888472 | 0.394560 | 0.085* | |
| H9B | 0.523979 | 0.907121 | 0.420264 | 0.085* | |
| O9 | 0.69630 (12) | −0.01467 (16) | 0.31454 (10) | 0.0552 (6) | |
| C10 | 0.79817 (10) | 0.49193 (19) | 0.42916 (11) | 0.0235 (4) | |
| O10 | 0.72800 (9) | 0.68131 (14) | 0.59148 (9) | 0.0324 (3) | |
| H10A | 0.7598 (12) | 0.645 (3) | 0.6246 (13) | 0.049* | |
| H10B | 0.7398 (15) | 0.7520 (19) | 0.5805 (16) | 0.049* | |
| C11 | 0.85076 (14) | 0.5440 (2) | 0.39252 (16) | 0.0435 (6) | |
| H11A | 0.833091 | 0.536035 | 0.339502 | 0.065* | |
| H11B | 0.893627 | 0.497746 | 0.409250 | 0.065* | |
| H11C | 0.859103 | 0.631314 | 0.405432 | 0.065* | |
| C12 | 0.70807 (11) | 0.0878 (2) | 0.34617 (11) | 0.0286 (4) | |
| C13 | 0.68183 (13) | 0.2058 (2) | 0.30226 (12) | 0.0362 (5) | |
| H13A | 0.691076 | 0.277449 | 0.334898 | 0.054* | |
| H13B | 0.632510 | 0.198775 | 0.279886 | 0.054* | |
| H13C | 0.705221 | 0.216242 | 0.264085 | 0.054* |
catena-Poly[manganese(II)-µ3-acetato-di-µ2-acetato-[aquamanganese(II)]-µ2-acetato-µ-(pyridine N-oxide)-manganese(II)-µ3-acetato-µ2-acetato-µ-(pyridine N-oxide)-[aquamanganese(II)]-di-µ2-acetato] (I) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Mn1 | 0.0195 (2) | 0.0200 (2) | 0.0295 (2) | 0.00260 (15) | 0.00725 (17) | 0.00445 (17) |
| O1 | 0.0230 (7) | 0.0277 (8) | 0.0258 (7) | 0.0009 (6) | 0.0073 (5) | 0.0085 (6) |
| C1 | 0.0477 (14) | 0.0283 (12) | 0.0336 (12) | 0.0043 (10) | 0.0098 (10) | −0.0067 (9) |
| N1 | 0.0228 (8) | 0.0231 (8) | 0.0224 (8) | 0.0014 (6) | 0.0069 (6) | 0.0022 (7) |
| Mn2 | 0.02113 (16) | 0.01489 (16) | 0.02513 (17) | 0.00127 (10) | 0.00883 (12) | 0.00102 (11) |
| C2 | 0.0627 (17) | 0.0480 (15) | 0.0268 (12) | 0.0038 (13) | 0.0141 (11) | −0.0095 (11) |
| O2 | 0.0275 (8) | 0.0293 (8) | 0.0305 (8) | 0.0055 (6) | 0.0082 (6) | 0.0002 (6) |
| O3 | 0.0239 (7) | 0.0178 (7) | 0.0331 (8) | 0.0015 (5) | 0.0070 (6) | −0.0026 (6) |
| C3 | 0.0437 (14) | 0.0487 (15) | 0.0263 (11) | 0.0020 (11) | 0.0087 (10) | 0.0076 (10) |
| Mn3 | 0.0249 (2) | 0.0140 (2) | 0.0275 (2) | 0.00299 (15) | 0.01058 (17) | 0.00005 (16) |
| C4 | 0.0398 (13) | 0.0320 (12) | 0.0347 (12) | 0.0047 (10) | 0.0102 (10) | 0.0081 (10) |
| O4 | 0.0289 (8) | 0.0233 (7) | 0.0466 (9) | 0.0034 (6) | 0.0155 (7) | 0.0078 (7) |
| O5 | 0.0283 (8) | 0.0244 (7) | 0.0350 (8) | 0.0064 (6) | 0.0113 (6) | 0.0075 (6) |
| C5 | 0.0318 (11) | 0.0255 (10) | 0.0288 (10) | 0.0044 (8) | 0.0117 (8) | 0.0026 (8) |
| C6 | 0.0266 (10) | 0.0215 (10) | 0.0257 (10) | −0.0009 (8) | 0.0095 (8) | −0.0011 (8) |
| O6 | 0.0292 (8) | 0.0204 (7) | 0.0361 (8) | 0.0031 (6) | 0.0173 (6) | 0.0035 (6) |
| C7 | 0.0323 (12) | 0.0270 (12) | 0.0611 (16) | −0.0051 (9) | 0.0124 (11) | −0.0075 (11) |
| O7 | 0.0394 (9) | 0.0180 (7) | 0.0390 (8) | 0.0044 (6) | 0.0218 (7) | 0.0033 (6) |
| O8 | 0.0282 (7) | 0.0188 (7) | 0.0248 (7) | 0.0045 (5) | 0.0072 (6) | 0.0000 (5) |
| C8 | 0.0285 (11) | 0.0204 (10) | 0.0325 (11) | 0.0026 (8) | 0.0101 (8) | 0.0039 (8) |
| C9 | 0.0467 (16) | 0.0354 (14) | 0.096 (2) | 0.0137 (12) | 0.0331 (15) | 0.0358 (15) |
| O9 | 0.0897 (16) | 0.0254 (9) | 0.0329 (9) | 0.0061 (9) | −0.0127 (9) | −0.0053 (7) |
| C10 | 0.0256 (10) | 0.0221 (10) | 0.0251 (10) | 0.0022 (8) | 0.0107 (8) | 0.0017 (8) |
| O10 | 0.0397 (9) | 0.0176 (7) | 0.0364 (9) | −0.0023 (6) | 0.0045 (7) | −0.0003 (6) |
| C11 | 0.0506 (15) | 0.0286 (12) | 0.0652 (17) | 0.0046 (10) | 0.0394 (13) | 0.0088 (11) |
| C12 | 0.0334 (11) | 0.0239 (10) | 0.0262 (10) | 0.0025 (8) | 0.0042 (8) | −0.0014 (8) |
| C13 | 0.0454 (13) | 0.0314 (12) | 0.0294 (11) | 0.0092 (10) | 0.0062 (10) | 0.0042 (9) |
catena-Poly[manganese(II)-µ3-acetato-di-µ2-acetato-[aquamanganese(II)]-µ2-acetato-µ-(pyridine N-oxide)-manganese(II)-µ3-acetato-µ2-acetato-µ-(pyridine N-oxide)-[aquamanganese(II)]-di-µ2-acetato] (I) . Geometric parameters (Å, º)
| Mn1—O1i | 2.1680 (15) | C4—H4 | 0.9300 |
| Mn1—O1 | 2.1681 (15) | C4—C5 | 1.381 (3) |
| Mn1—O2i | 2.1822 (15) | O4—C8 | 1.251 (3) |
| Mn1—O2 | 2.1822 (15) | O5—C8 | 1.254 (2) |
| Mn1—O4i | 2.1207 (16) | C5—H5 | 0.9300 |
| Mn1—O4 | 2.1207 (16) | C6—C7 | 1.508 (3) |
| O1—N1 | 1.351 (2) | O6—C10 | 1.262 (2) |
| O1—Mn2 | 2.2113 (15) | C7—H7A | 0.9600 |
| C1—H1 | 0.9300 | C7—H7B | 0.9600 |
| C1—N1 | 1.341 (3) | C7—H7C | 0.9600 |
| C1—C2 | 1.377 (3) | O7—C10 | 1.255 (2) |
| N1—C5 | 1.341 (3) | O8—C12 | 1.290 (2) |
| Mn2—O3i | 2.2404 (15) | C8—C9 | 1.510 (3) |
| Mn2—O5 | 2.1062 (15) | C9—H9A | 0.9600 |
| Mn2—O6 | 2.1551 (15) | C9—H9C | 0.9600 |
| Mn2—O8ii | 2.2671 (14) | C9—H9B | 0.9600 |
| Mn2—O10 | 2.1506 (16) | O9—C12 | 1.228 (3) |
| C2—H2 | 0.9300 | C10—C11 | 1.506 (3) |
| C2—C3 | 1.380 (4) | O10—H10A | 0.848 (17) |
| O2—C6 | 1.250 (2) | O10—H10B | 0.829 (17) |
| O3—Mn3iii | 2.2028 (15) | C11—H11A | 0.9600 |
| O3—C6 | 1.278 (2) | C11—H11B | 0.9600 |
| C3—H3 | 0.9300 | C11—H11C | 0.9600 |
| C3—C4 | 1.380 (3) | C12—C13 | 1.509 (3) |
| Mn3—O7ii | 2.1338 (15) | C13—H13A | 0.9600 |
| Mn3—O7 | 2.1338 (15) | C13—H13B | 0.9600 |
| Mn3—O8 | 2.2194 (14) | C13—H13C | 0.9600 |
| Mn3—O8ii | 2.2194 (14) | ||
| O1i—Mn1—O1 | 180.00 (7) | O7—Mn3—O7ii | 180.0 |
| O1i—Mn1—O2i | 91.91 (6) | O7—Mn3—O8 | 91.58 (6) |
| O1—Mn1—O2i | 88.09 (6) | O7—Mn3—O8ii | 88.43 (6) |
| O1i—Mn1—O2 | 88.09 (6) | O7ii—Mn3—O8ii | 91.57 (6) |
| O1—Mn1—O2 | 91.91 (6) | O7ii—Mn3—O8 | 88.43 (6) |
| O2—Mn1—O2i | 180.0 | O8—Mn3—O8ii | 180.00 (5) |
| O4i—Mn1—O1i | 92.47 (6) | C3—C4—H4 | 120.2 |
| O4—Mn1—O1 | 92.47 (6) | C3—C4—C5 | 119.7 (2) |
| O4i—Mn1—O1 | 87.53 (6) | C5—C4—H4 | 120.2 |
| O4—Mn1—O1i | 87.53 (6) | C8—O4—Mn1 | 130.47 (13) |
| O4i—Mn1—O2 | 91.36 (6) | C8—O5—Mn2 | 139.17 (14) |
| O4—Mn1—O2 | 88.64 (6) | N1—C5—C4 | 119.5 (2) |
| O4—Mn1—O2i | 91.36 (6) | N1—C5—H5 | 120.2 |
| O4i—Mn1—O2i | 88.64 (6) | C4—C5—H5 | 120.2 |
| O4—Mn1—O4i | 180.0 | O2—C6—O3 | 122.61 (19) |
| Mn1—O1—Mn2 | 112.12 (6) | O2—C6—C7 | 118.29 (19) |
| N1—O1—Mn1 | 117.31 (11) | O3—C6—C7 | 119.09 (18) |
| N1—O1—Mn2 | 128.21 (11) | C10—O6—Mn2 | 129.45 (13) |
| N1—C1—H1 | 120.3 | C6—C7—H7A | 109.5 |
| N1—C1—C2 | 119.5 (2) | C6—C7—H7B | 109.5 |
| C2—C1—H1 | 120.3 | C6—C7—H7C | 109.5 |
| C1—N1—O1 | 118.17 (18) | H7A—C7—H7B | 109.5 |
| C5—N1—O1 | 119.51 (16) | H7A—C7—H7C | 109.5 |
| C5—N1—C1 | 122.28 (19) | H7B—C7—H7C | 109.5 |
| O1—Mn2—O3i | 85.37 (6) | C10—O7—Mn3 | 135.59 (13) |
| O1—Mn2—O8ii | 89.66 (5) | Mn3—O8—Mn2ii | 97.00 (6) |
| O3i—Mn2—O8ii | 76.44 (5) | C12—O8—Mn2ii | 128.45 (13) |
| O5—Mn2—O1 | 92.21 (6) | C12—O8—Mn3 | 134.54 (13) |
| O5—Mn2—O3i | 107.68 (6) | O4—C8—O5 | 126.06 (19) |
| O5—Mn2—O6 | 87.13 (6) | O4—C8—C9 | 116.96 (19) |
| O5—Mn2—O8ii | 175.59 (6) | O5—C8—C9 | 116.97 (19) |
| O5—Mn2—O10 | 88.66 (6) | C8—C9—H9A | 109.5 |
| O6—Mn2—O1 | 176.02 (6) | C8—C9—H9C | 109.5 |
| O6—Mn2—O3i | 91.08 (6) | C8—C9—H9B | 109.5 |
| O6—Mn2—O8ii | 91.27 (6) | H9A—C9—H9C | 109.5 |
| O10—Mn2—O1 | 88.66 (6) | H9A—C9—H9B | 109.5 |
| O10—Mn2—O3i | 162.77 (6) | H9C—C9—H9B | 109.5 |
| O10—Mn2—O6 | 95.25 (6) | O6—C10—C11 | 117.96 (18) |
| O10—Mn2—O8ii | 87.39 (6) | O7—C10—O6 | 124.82 (19) |
| C1—C2—H2 | 120.1 | O7—C10—C11 | 117.21 (18) |
| C1—C2—C3 | 119.9 (2) | Mn2—O10—H10A | 106 (2) |
| C3—C2—H2 | 120.1 | Mn2—O10—H10B | 124 (2) |
| C6—O2—Mn1 | 123.62 (13) | H10A—O10—H10B | 113 (3) |
| Mn3iii—O3—Mn2i | 98.27 (6) | C10—C11—H11A | 109.5 |
| C6—O3—Mn2i | 120.04 (12) | C10—C11—H11B | 109.5 |
| C6—O3—Mn3iii | 138.28 (13) | C10—C11—H11C | 109.5 |
| C2—C3—H3 | 120.4 | H11A—C11—H11B | 109.5 |
| C2—C3—C4 | 119.2 (2) | H11A—C11—H11C | 109.5 |
| C4—C3—H3 | 120.4 | H11B—C11—H11C | 109.5 |
| O3i—Mn3—O3iv | 180.00 (7) | O8—C12—C13 | 117.86 (18) |
| O3i—Mn3—O8ii | 78.19 (5) | O9—C12—O8 | 123.5 (2) |
| O3iv—Mn3—O8ii | 101.81 (5) | O9—C12—C13 | 118.66 (19) |
| O3iv—Mn3—O8 | 78.19 (5) | C12—C13—H13A | 109.5 |
| O3i—Mn3—O8 | 101.81 (5) | C12—C13—H13B | 109.5 |
| O7—Mn3—O3i | 89.75 (6) | C12—C13—H13C | 109.5 |
| O7ii—Mn3—O3iv | 89.75 (6) | H13A—C13—H13B | 109.5 |
| O7ii—Mn3—O3i | 90.25 (6) | H13A—C13—H13C | 109.5 |
| O7—Mn3—O3iv | 90.25 (6) | H13B—C13—H13C | 109.5 |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) −x+3/2, −y+1/2, −z+1; (iii) x−1/2, y+1/2, z; (iv) x+1/2, y−1/2, z.
catena-Poly[manganese(II)-µ3-acetato-di-µ2-acetato-[aquamanganese(II)]-µ2-acetato-µ-(pyridine N-oxide)-manganese(II)-µ3-acetato-µ2-acetato-µ-(pyridine N-oxide)-[aquamanganese(II)]-di-µ2-acetato] (I) . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O10—H10A···O9ii | 0.85 (2) | 1.85 (2) | 2.652 (2) | 157 (3) |
| O10—H10B···O6v | 0.83 (2) | 1.98 (2) | 2.786 (2) | 166 (3) |
Symmetry codes: (ii) −x+3/2, −y+1/2, −z+1; (v) −x+3/2, −y+3/2, −z+1.
catena-Poly[[manganese(II)]-µ3-acetato-µ2-acetato-µ-(2-methylpyridine N-oxide)-[aquamanganese(II)]-di-µ2-acetato-manganese(II)-di-µ2-acetato-µ3-acetato-[aquamanganese(II)]-µ2-acetato-µ-(2-methylpyridine N-oxide)] (II) . Crystal data
| [Mn4(C2H3O2)8(C6H7NO)2(H2O)2] | Z = 1 |
| Mr = 946.4 | F(000) = 484 |
| Triclinic, P1 | Dx = 1.600 Mg m−3 |
| a = 9.7704 (3) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 10.5882 (7) Å | Cell parameters from 2512 reflections |
| c = 11.4720 (2) Å | θ = 2.0–27.5° |
| α = 65.76 (2)° | µ = 1.34 mm−1 |
| β = 83.84 (2)° | T = 173 K |
| γ = 65.512 (15)° | Prism, colorless |
| V = 982.0 (2) Å3 | 0.3 × 0.3 × 0.2 mm |
catena-Poly[[manganese(II)]-µ3-acetato-µ2-acetato-µ-(2-methylpyridine N-oxide)-[aquamanganese(II)]-di-µ2-acetato-manganese(II)-di-µ2-acetato-µ3-acetato-[aquamanganese(II)]-µ2-acetato-µ-(2-methylpyridine N-oxide)] (II) . Data collection
| Rigaku XtaLAB mini diffractometer | 4503 independent reflections |
| Radiation source: Sealed Tube | 3551 reflections with I > 2σ(I) |
| Graphite Monochromator monochromator | Rint = 0.058 |
| Detector resolution: 13.6612 pixels mm-1 | θmax = 27.5°, θmin = 2.3° |
| profile data from ω–scans | h = −12→12 |
| Absorption correction: multi-scan (REQAB; Rigaku, 1998) | k = −13→13 |
| Tmin = 0.842, Tmax = 1.00 | l = −14→14 |
| 10542 measured reflections |
catena-Poly[[manganese(II)]-µ3-acetato-µ2-acetato-µ-(2-methylpyridine N-oxide)-[aquamanganese(II)]-di-µ2-acetato-manganese(II)-di-µ2-acetato-µ3-acetato-[aquamanganese(II)]-µ2-acetato-µ-(2-methylpyridine N-oxide)] (II) . Refinement
| Refinement on F2 | Hydrogen site location: mixed |
| Least-squares matrix: full | H atoms treated by a mixture of independent and constrained refinement |
| R[F2 > 2σ(F2)] = 0.041 | w = 1/[σ2(Fo2) + (0.0271P)2 + 0.5206P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.101 | (Δ/σ)max < 0.001 |
| S = 1.06 | Δρmax = 0.62 e Å−3 |
| 4503 reflections | Δρmin = −0.46 e Å−3 |
| 259 parameters | Extinction correction: SHELXL-2018/1 (Sheldrick 2015), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 2 restraints | Extinction coefficient: 0.0045 (10) |
| Primary atom site location: dual |
catena-Poly[[manganese(II)]-µ3-acetato-µ2-acetato-µ-(2-methylpyridine N-oxide)-[aquamanganese(II)]-di-µ2-acetato-manganese(II)-di-µ2-acetato-µ3-acetato-[aquamanganese(II)]-µ2-acetato-µ-(2-methylpyridine N-oxide)] (II) . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
catena-Poly[[manganese(II)]-µ3-acetato-µ2-acetato-µ-(2-methylpyridine N-oxide)-[aquamanganese(II)]-di-µ2-acetato-manganese(II)-di-µ2-acetato-µ3-acetato-[aquamanganese(II)]-µ2-acetato-µ-(2-methylpyridine N-oxide)] (II) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Mn1 | 0.500000 | 0.500000 | 0.500000 | 0.02633 (15) | |
| C1 | 0.9223 (3) | 0.1967 (3) | 0.5954 (3) | 0.0385 (7) | |
| O1 | 0.66323 (19) | 0.2610 (2) | 0.58767 (17) | 0.0294 (4) | |
| N1 | 0.7920 (3) | 0.2213 (2) | 0.6544 (2) | 0.0306 (5) | |
| C2 | 1.0513 (4) | 0.1617 (4) | 0.6641 (4) | 0.0526 (9) | |
| H2 | 1.143547 | 0.147895 | 0.624004 | 0.063* | |
| O2 | 0.6422 (2) | 0.5705 (2) | 0.35280 (18) | 0.0405 (5) | |
| Mn2 | 0.54423 (4) | 0.10659 (4) | 0.65107 (4) | 0.02574 (12) | |
| C3 | 1.0472 (4) | 0.1465 (4) | 0.7900 (4) | 0.0559 (9) | |
| H3 | 1.135892 | 0.121710 | 0.836731 | 0.067* | |
| O3 | 0.5590 (2) | 0.80401 (19) | 0.20131 (16) | 0.0292 (4) | |
| Mn3 | 0.500000 | 0.000000 | 1.000000 | 0.02716 (15) | |
| C4 | 0.9121 (4) | 0.1680 (4) | 0.8465 (3) | 0.0446 (8) | |
| H4 | 0.907481 | 0.155806 | 0.933430 | 0.053* | |
| O4 | 0.3972 (3) | 0.4521 (2) | 0.37791 (19) | 0.0434 (5) | |
| O5 | 0.3654 (2) | 0.2403 (2) | 0.49693 (19) | 0.0389 (5) | |
| C5 | 0.7842 (4) | 0.2072 (3) | 0.7767 (3) | 0.0375 (7) | |
| H5 | 0.690486 | 0.224241 | 0.814678 | 0.045* | |
| C6 | 0.9159 (4) | 0.2095 (4) | 0.4616 (3) | 0.0518 (9) | |
| H6A | 0.866513 | 0.147157 | 0.458109 | 0.078* | |
| H6B | 1.018458 | 0.174165 | 0.434390 | 0.078* | |
| H6C | 0.858321 | 0.315728 | 0.404389 | 0.078* | |
| O6 | 0.7494 (2) | −0.0675 (2) | 0.76008 (18) | 0.0411 (5) | |
| C7 | 0.6334 (3) | 0.6596 (3) | 0.2404 (2) | 0.0292 (6) | |
| O7 | 0.7340 (2) | −0.0951 (2) | 0.96318 (18) | 0.0391 (5) | |
| O8 | 0.4469 (2) | −0.0610 (2) | 0.72192 (18) | 0.0381 (5) | |
| C8 | 0.7129 (5) | 0.5940 (4) | 0.1464 (3) | 0.0695 (13) | |
| H8A | 0.651688 | 0.650930 | 0.064547 | 0.104* | |
| H8C | 0.728199 | 0.487456 | 0.179635 | 0.104* | |
| H8B | 0.810839 | 0.600750 | 0.133554 | 0.104* | |
| O9 | 0.4385 (3) | −0.1325 (2) | 0.93300 (18) | 0.0417 (5) | |
| C9 | 0.3347 (4) | 0.3642 (3) | 0.4000 (3) | 0.0388 (7) | |
| C10 | 0.2131 (5) | 0.4060 (4) | 0.3031 (4) | 0.0752 (13) | |
| H10A | 0.227713 | 0.316316 | 0.288714 | 0.113* | |
| H10B | 0.218744 | 0.485502 | 0.222086 | 0.113* | |
| H10C | 0.113896 | 0.443323 | 0.335646 | 0.113* | |
| O10 | 0.6635 (2) | 0.0116 (2) | 0.50856 (19) | 0.0347 (5) | |
| H10D | 0.669 (4) | −0.076 (2) | 0.529 (3) | 0.052* | |
| H10E | 0.615 (4) | 0.054 (4) | 0.436 (2) | 0.052* | |
| C11 | 0.8010 (3) | −0.1286 (3) | 0.8738 (3) | 0.0324 (6) | |
| C12 | 0.9586 (4) | −0.2532 (5) | 0.9039 (3) | 0.0574 (10) | |
| H12B | 1.028728 | −0.212929 | 0.850872 | 0.086* | |
| H12C | 0.959934 | −0.336348 | 0.885672 | 0.086* | |
| H12A | 0.989418 | −0.291080 | 0.994697 | 0.086* | |
| C13 | 0.4250 (3) | −0.1437 (3) | 0.8312 (3) | 0.0299 (6) | |
| C14 | 0.3769 (5) | −0.2656 (4) | 0.8398 (3) | 0.0583 (10) | |
| H14A | 0.291205 | −0.220931 | 0.777399 | 0.087* | |
| H14C | 0.347239 | −0.310285 | 0.926417 | 0.087* | |
| H14B | 0.461132 | −0.344649 | 0.821308 | 0.087* |
catena-Poly[[manganese(II)]-µ3-acetato-µ2-acetato-µ-(2-methylpyridine N-oxide)-[aquamanganese(II)]-di-µ2-acetato-manganese(II)-di-µ2-acetato-µ3-acetato-[aquamanganese(II)]-µ2-acetato-µ-(2-methylpyridine N-oxide)] (II) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Mn1 | 0.0309 (3) | 0.0219 (3) | 0.0235 (3) | −0.0121 (2) | −0.0010 (2) | −0.0047 (2) |
| C1 | 0.0297 (15) | 0.0323 (15) | 0.0463 (17) | −0.0089 (13) | 0.0010 (13) | −0.0126 (14) |
| O1 | 0.0254 (9) | 0.0257 (9) | 0.0335 (10) | −0.0095 (8) | −0.0038 (8) | −0.0082 (8) |
| N1 | 0.0301 (12) | 0.0259 (11) | 0.0320 (12) | −0.0113 (10) | −0.0046 (10) | −0.0066 (10) |
| C2 | 0.0262 (15) | 0.058 (2) | 0.064 (2) | −0.0109 (15) | −0.0015 (15) | −0.0215 (19) |
| O2 | 0.0388 (11) | 0.0326 (11) | 0.0319 (11) | −0.0104 (9) | 0.0061 (9) | −0.0015 (9) |
| Mn2 | 0.0309 (2) | 0.0255 (2) | 0.0225 (2) | −0.01314 (18) | 0.00076 (16) | −0.00934 (17) |
| C3 | 0.0438 (19) | 0.054 (2) | 0.065 (2) | −0.0127 (17) | −0.0211 (17) | −0.0211 (19) |
| O3 | 0.0372 (10) | 0.0228 (9) | 0.0237 (9) | −0.0106 (8) | 0.0045 (8) | −0.0081 (8) |
| Mn3 | 0.0340 (3) | 0.0254 (3) | 0.0215 (3) | −0.0123 (2) | 0.0022 (2) | −0.0089 (2) |
| C4 | 0.0488 (19) | 0.0428 (18) | 0.0398 (17) | −0.0160 (16) | −0.0093 (15) | −0.0144 (15) |
| O4 | 0.0600 (14) | 0.0399 (12) | 0.0350 (11) | −0.0272 (11) | −0.0071 (10) | −0.0100 (10) |
| O5 | 0.0449 (12) | 0.0278 (10) | 0.0385 (11) | −0.0133 (9) | −0.0110 (9) | −0.0065 (9) |
| C5 | 0.0430 (17) | 0.0346 (16) | 0.0351 (15) | −0.0177 (14) | 0.0004 (13) | −0.0118 (13) |
| C6 | 0.0410 (18) | 0.060 (2) | 0.0445 (19) | −0.0117 (17) | 0.0085 (15) | −0.0222 (18) |
| O6 | 0.0430 (12) | 0.0371 (11) | 0.0324 (11) | −0.0075 (10) | −0.0079 (9) | −0.0105 (9) |
| C7 | 0.0300 (14) | 0.0264 (13) | 0.0290 (14) | −0.0124 (11) | 0.0060 (11) | −0.0092 (11) |
| O7 | 0.0366 (11) | 0.0430 (12) | 0.0334 (11) | −0.0120 (10) | 0.0040 (9) | −0.0162 (10) |
| O8 | 0.0544 (13) | 0.0426 (12) | 0.0298 (10) | −0.0322 (11) | 0.0046 (9) | −0.0139 (9) |
| C8 | 0.109 (3) | 0.0298 (17) | 0.041 (2) | −0.006 (2) | 0.019 (2) | −0.0141 (16) |
| O9 | 0.0666 (15) | 0.0410 (12) | 0.0278 (10) | −0.0306 (11) | 0.0047 (10) | −0.0149 (10) |
| C9 | 0.0446 (17) | 0.0289 (15) | 0.0403 (16) | −0.0114 (13) | −0.0100 (14) | −0.0121 (13) |
| C10 | 0.090 (3) | 0.047 (2) | 0.077 (3) | −0.030 (2) | −0.052 (2) | 0.001 (2) |
| O10 | 0.0417 (11) | 0.0367 (11) | 0.0298 (10) | −0.0172 (10) | 0.0012 (9) | −0.0158 (10) |
| C11 | 0.0308 (14) | 0.0283 (14) | 0.0371 (15) | −0.0113 (12) | −0.0021 (12) | −0.0120 (13) |
| C12 | 0.0366 (18) | 0.066 (2) | 0.051 (2) | 0.0004 (17) | −0.0074 (16) | −0.0263 (19) |
| C13 | 0.0335 (14) | 0.0277 (14) | 0.0294 (14) | −0.0152 (12) | 0.0017 (11) | −0.0094 (12) |
| C14 | 0.096 (3) | 0.062 (2) | 0.048 (2) | −0.061 (2) | 0.019 (2) | −0.0251 (18) |
catena-Poly[[manganese(II)]-µ3-acetato-µ2-acetato-µ-(2-methylpyridine N-oxide)-[aquamanganese(II)]-di-µ2-acetato-manganese(II)-di-µ2-acetato-µ3-acetato-[aquamanganese(II)]-µ2-acetato-µ-(2-methylpyridine N-oxide)] (II) . Geometric parameters (Å, º)
| Mn1—O1 | 2.2061 (19) | C4—C5 | 1.374 (4) |
| Mn1—O1i | 2.2061 (19) | O4—C9 | 1.244 (4) |
| Mn1—O2i | 2.159 (2) | O5—C9 | 1.263 (3) |
| Mn1—O2 | 2.159 (2) | C5—H5 | 0.9500 |
| Mn1—O4 | 2.129 (2) | C6—H6A | 0.9800 |
| Mn1—O4i | 2.129 (2) | C6—H6B | 0.9800 |
| C1—N1 | 1.354 (4) | C6—H6C | 0.9800 |
| C1—C2 | 1.389 (4) | O6—C11 | 1.246 (3) |
| C1—C6 | 1.491 (4) | C7—C8 | 1.495 (4) |
| O1—N1 | 1.359 (3) | O7—C11 | 1.253 (3) |
| O1—Mn2 | 2.2300 (18) | O8—C13 | 1.258 (3) |
| N1—C5 | 1.346 (4) | C8—H8A | 0.9800 |
| C2—H2 | 0.9500 | C8—H8C | 0.9800 |
| C2—C3 | 1.385 (5) | C8—H8B | 0.9800 |
| O2—C7 | 1.232 (3) | O9—C13 | 1.247 (3) |
| Mn2—O3i | 2.2224 (18) | C9—C10 | 1.513 (4) |
| Mn2—O5 | 2.177 (2) | C10—H10A | 0.9800 |
| Mn2—O6 | 2.136 (2) | C10—H10B | 0.9800 |
| Mn2—O8 | 2.182 (2) | C10—H10C | 0.9800 |
| Mn2—O10 | 2.239 (2) | O10—H10D | 0.838 (18) |
| C3—H3 | 0.9500 | O10—H10E | 0.846 (18) |
| C3—C4 | 1.379 (5) | C11—C12 | 1.513 (4) |
| O3—Mn3ii | 2.3091 (18) | C12—H12B | 0.9800 |
| O3—C7 | 1.286 (3) | C12—H12C | 0.9800 |
| Mn3—O7iii | 2.157 (2) | C12—H12A | 0.9800 |
| Mn3—O7 | 2.157 (2) | C13—C14 | 1.511 (4) |
| Mn3—O9 | 2.1453 (19) | C14—H14A | 0.9800 |
| Mn3—O9iii | 2.1452 (19) | C14—H14C | 0.9800 |
| C4—H4 | 0.9500 | C14—H14B | 0.9800 |
| O1i—Mn1—O1 | 180.0 | O9—Mn3—O7 | 93.91 (8) |
| O2i—Mn1—O1 | 85.03 (7) | O9iii—Mn3—O7iii | 93.91 (8) |
| O2—Mn1—O1i | 85.03 (7) | O9iii—Mn3—O9 | 180.0 |
| O2—Mn1—O1 | 94.97 (7) | C3—C4—H4 | 120.1 |
| O2i—Mn1—O1i | 94.97 (7) | C5—C4—C3 | 119.8 (3) |
| O2—Mn1—O2i | 180.0 | C5—C4—H4 | 120.1 |
| O4i—Mn1—O1i | 91.37 (8) | C9—O4—Mn1 | 132.59 (19) |
| O4—Mn1—O1 | 91.37 (8) | C9—O5—Mn2 | 133.4 (2) |
| O4—Mn1—O1i | 88.63 (8) | N1—C5—C4 | 119.8 (3) |
| O4i—Mn1—O1 | 88.63 (8) | N1—C5—H5 | 120.1 |
| O4i—Mn1—O2i | 91.45 (8) | C4—C5—H5 | 120.1 |
| O4i—Mn1—O2 | 88.55 (8) | C1—C6—H6A | 109.5 |
| O4—Mn1—O2 | 91.45 (8) | C1—C6—H6B | 109.5 |
| O4—Mn1—O2i | 88.55 (8) | C1—C6—H6C | 109.5 |
| O4—Mn1—O4i | 180.0 | H6A—C6—H6B | 109.5 |
| N1—C1—C2 | 117.6 (3) | H6A—C6—H6C | 109.5 |
| N1—C1—C6 | 117.2 (3) | H6B—C6—H6C | 109.5 |
| C2—C1—C6 | 125.2 (3) | C11—O6—Mn2 | 136.82 (19) |
| Mn1—O1—Mn2 | 110.48 (7) | O2—C7—O3 | 123.2 (2) |
| N1—O1—Mn1 | 120.93 (14) | O2—C7—C8 | 117.5 (2) |
| N1—O1—Mn2 | 119.74 (14) | O3—C7—C8 | 119.4 (2) |
| C1—N1—O1 | 118.8 (2) | C11—O7—Mn3 | 134.16 (18) |
| C5—N1—C1 | 122.8 (3) | C13—O8—Mn2 | 134.65 (18) |
| C5—N1—O1 | 118.4 (2) | C7—C8—H8A | 109.5 |
| C1—C2—H2 | 119.5 | C7—C8—H8C | 109.5 |
| C3—C2—C1 | 121.0 (3) | C7—C8—H8B | 109.5 |
| C3—C2—H2 | 119.5 | H8A—C8—H8C | 109.5 |
| C7—O2—Mn1 | 140.53 (19) | H8A—C8—H8B | 109.5 |
| O1—Mn2—O10 | 88.73 (7) | H8C—C8—H8B | 109.5 |
| O3i—Mn2—O1 | 89.31 (7) | C13—O9—Mn3 | 139.90 (18) |
| O3i—Mn2—O10 | 176.15 (7) | O4—C9—O5 | 125.4 (3) |
| O5—Mn2—O1 | 97.41 (7) | O4—C9—C10 | 118.2 (3) |
| O5—Mn2—O3i | 101.91 (7) | O5—C9—C10 | 116.4 (3) |
| O5—Mn2—O8 | 87.26 (8) | C9—C10—H10A | 109.5 |
| O5—Mn2—O10 | 81.63 (8) | C9—C10—H10B | 109.5 |
| O6—Mn2—O1 | 86.79 (8) | C9—C10—H10C | 109.5 |
| O6—Mn2—O3i | 97.97 (7) | H10A—C10—H10B | 109.5 |
| O6—Mn2—O5 | 159.71 (8) | H10A—C10—H10C | 109.5 |
| O6—Mn2—O8 | 88.10 (8) | H10B—C10—H10C | 109.5 |
| O6—Mn2—O10 | 78.61 (8) | Mn2—O10—H10D | 112 (2) |
| O8—Mn2—O1 | 174.87 (7) | Mn2—O10—H10E | 115 (2) |
| O8—Mn2—O3i | 91.81 (7) | H10D—O10—H10E | 99 (3) |
| O8—Mn2—O10 | 89.86 (8) | O6—C11—O7 | 125.7 (3) |
| C2—C3—H3 | 120.6 | O6—C11—C12 | 116.1 (3) |
| C4—C3—C2 | 118.8 (3) | O7—C11—C12 | 118.3 (3) |
| C4—C3—H3 | 120.6 | C11—C12—H12B | 109.5 |
| Mn2i—O3—Mn3ii | 110.16 (7) | C11—C12—H12C | 109.5 |
| C7—O3—Mn2i | 117.09 (16) | C11—C12—H12A | 109.5 |
| C7—O3—Mn3ii | 132.72 (17) | H12B—C12—H12C | 109.5 |
| O3iv—Mn3—O3i | 180.0 | H12B—C12—H12A | 109.5 |
| O7iii—Mn3—O3i | 88.03 (7) | H12C—C12—H12A | 109.5 |
| O7—Mn3—O3iv | 88.03 (7) | O8—C13—C14 | 117.4 (2) |
| O7—Mn3—O3i | 91.97 (7) | O9—C13—O8 | 125.2 (3) |
| O7iii—Mn3—O3iv | 91.97 (7) | O9—C13—C14 | 117.3 (2) |
| O7—Mn3—O7iii | 180.0 | C13—C14—H14A | 109.5 |
| O9iii—Mn3—O3i | 88.85 (7) | C13—C14—H14C | 109.5 |
| O9iii—Mn3—O3iv | 91.15 (7) | C13—C14—H14B | 109.5 |
| O9—Mn3—O3iv | 88.85 (7) | H14A—C14—H14C | 109.5 |
| O9—Mn3—O3i | 91.15 (7) | H14A—C14—H14B | 109.5 |
| O9iii—Mn3—O7 | 86.09 (8) | H14C—C14—H14B | 109.5 |
| O9—Mn3—O7iii | 86.09 (8) |
Symmetry codes: (i) −x+1, −y+1, −z+1; (ii) x, y+1, z−1; (iii) −x+1, −y, −z+2; (iv) x, y−1, z+1.
catena-Poly[[manganese(II)]-µ3-acetato-µ2-acetato-µ-(2-methylpyridine N-oxide)-[aquamanganese(II)]-di-µ2-acetato-manganese(II)-di-µ2-acetato-µ3-acetato-[aquamanganese(II)]-µ2-acetato-µ-(2-methylpyridine N-oxide)] (II) . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O10—H10D···O5v | 0.84 (2) | 2.04 (2) | 2.821 (3) | 155 (3) |
| O10—H10E···O8v | 0.85 (2) | 1.94 (2) | 2.727 (3) | 155 (3) |
Symmetry code: (v) −x+1, −y, −z+1.
catena-Poly[[manganese(II)-di-µ2-acetato-µ-(4-methylpyridine N-oxide)] monohydrate] (III) . Crystal data
| [Mn(C2H3O2)2(C6H7NO)]·H2O | F(000) = 620 |
| Mr = 300.17 | Dx = 1.542 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| a = 11.100 (3) Å | Cell parameters from 2982 reflections |
| b = 7.334 (3) Å | θ = 2.1–27.5° |
| c = 15.9808 (4) Å | µ = 1.04 mm−1 |
| β = 96.500 (11)° | T = 173 K |
| V = 1292.6 (6) Å3 | Prism, colorless |
| Z = 4 | 0.2 × 0.05 × 0.05 mm |
catena-Poly[[manganese(II)-di-µ2-acetato-µ-(4-methylpyridine N-oxide)] monohydrate] (III) . Data collection
| Rigaku XtaLAB mini diffractometer | 2224 reflections with I > 2σ(I) |
| Detector resolution: 13.6612 pixels mm-1 | Rint = 0.074 |
| profile data from ω–scans | θmax = 27.6°, θmin = 2.1° |
| Absorption correction: multi-scan (REQAB; Rigaku, 1998) | h = −14→14 |
| Tmin = 0.850, Tmax = 1.00 | k = −9→9 |
| 13236 measured reflections | l = −20→20 |
| 2957 independent reflections |
catena-Poly[[manganese(II)-di-µ2-acetato-µ-(4-methylpyridine N-oxide)] monohydrate] (III) . Refinement
| Refinement on F2 | Hydrogen site location: mixed |
| Least-squares matrix: full | H atoms treated by a mixture of independent and constrained refinement |
| R[F2 > 2σ(F2)] = 0.045 | w = 1/[σ2(Fo2) + (0.0269P)2 + 1.0998P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.105 | (Δ/σ)max = 0.001 |
| S = 1.07 | Δρmax = 0.35 e Å−3 |
| 2957 reflections | Δρmin = −0.40 e Å−3 |
| 173 parameters | Extinction correction: SHELXL-2018/1 (Sheldrick 2015), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 2 restraints | Extinction coefficient: 0.0042 (9) |
catena-Poly[[manganese(II)-di-µ2-acetato-µ-(4-methylpyridine N-oxide)] monohydrate] (III) . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
catena-Poly[[manganese(II)-di-µ2-acetato-µ-(4-methylpyridine N-oxide)] monohydrate] (III) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| Mn1 | 0.25529 (4) | 0.54319 (6) | 0.75049 (3) | 0.02337 (15) | |
| O1 | 0.35635 (15) | 0.7980 (2) | 0.73689 (12) | 0.0243 (4) | |
| C1 | 0.5439 (2) | 0.8534 (4) | 0.69051 (19) | 0.0316 (7) | |
| H1 | 0.505841 | 0.869094 | 0.636076 | 0.038* | |
| N1 | 0.47864 (19) | 0.8127 (3) | 0.75356 (14) | 0.0235 (5) | |
| C2 | 0.6685 (3) | 0.8721 (4) | 0.7071 (2) | 0.0354 (7) | |
| H2 | 0.713428 | 0.902878 | 0.663478 | 0.042* | |
| O2 | 0.33022 (19) | 0.4486 (3) | 0.63859 (13) | 0.0372 (5) | |
| O3 | 0.3188 (2) | 0.1450 (3) | 0.63793 (13) | 0.0389 (5) | |
| C3 | 0.7278 (2) | 0.8459 (4) | 0.7874 (2) | 0.0323 (7) | |
| C4 | 0.6557 (3) | 0.8040 (4) | 0.85053 (19) | 0.0321 (7) | |
| H4 | 0.691779 | 0.786075 | 0.905330 | 0.039* | |
| O4 | 0.10483 (18) | 0.6385 (3) | 0.66703 (14) | 0.0402 (6) | |
| O5 | 0.09920 (19) | 0.9411 (3) | 0.66333 (14) | 0.0385 (5) | |
| C5 | 0.5315 (3) | 0.7885 (4) | 0.83299 (18) | 0.0284 (6) | |
| H5 | 0.484307 | 0.761524 | 0.875895 | 0.034* | |
| C6 | 0.8641 (3) | 0.8641 (5) | 0.8053 (2) | 0.0455 (8) | |
| H6A | 0.884813 | 0.903431 | 0.862398 | 0.068* | |
| H6B | 0.892537 | 0.952060 | 0.767630 | 0.068* | |
| H6C | 0.901330 | 0.748229 | 0.797218 | 0.068* | |
| C7 | 0.3391 (2) | 0.2954 (4) | 0.60495 (17) | 0.0259 (6) | |
| C8 | 0.3785 (3) | 0.2898 (5) | 0.51770 (19) | 0.0379 (7) | |
| H8A | 0.351789 | 0.398675 | 0.487760 | 0.057* | |
| H8B | 0.343341 | 0.185322 | 0.488106 | 0.057* | |
| H8C | 0.465247 | 0.282015 | 0.521701 | 0.057* | |
| C9 | 0.0622 (2) | 0.7859 (4) | 0.63902 (18) | 0.0271 (6) | |
| C10 | −0.0435 (3) | 0.7767 (5) | 0.5705 (2) | 0.0461 (9) | |
| H10A | −0.083947 | 0.892610 | 0.565920 | 0.069* | |
| H10B | −0.099334 | 0.684427 | 0.584238 | 0.069* | |
| H10C | −0.014279 | 0.747388 | 0.517831 | 0.069* | |
| O6 | 0.3091 (2) | 0.7951 (4) | 0.53409 (15) | 0.0440 (6) | |
| H6D | 0.301 (4) | 0.882 (4) | 0.567 (2) | 0.066* | |
| H6E | 0.313 (4) | 0.708 (4) | 0.568 (2) | 0.066* |
catena-Poly[[manganese(II)-di-µ2-acetato-µ-(4-methylpyridine N-oxide)] monohydrate] (III) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Mn1 | 0.0222 (2) | 0.0195 (2) | 0.0284 (2) | −0.00149 (16) | 0.00291 (17) | −0.00006 (16) |
| O1 | 0.0159 (9) | 0.0210 (10) | 0.0355 (11) | −0.0003 (7) | 0.0009 (8) | −0.0011 (8) |
| C1 | 0.0253 (14) | 0.0377 (18) | 0.0317 (16) | −0.0018 (13) | 0.0033 (12) | 0.0015 (13) |
| N1 | 0.0167 (10) | 0.0223 (12) | 0.0311 (12) | −0.0014 (9) | 0.0014 (9) | −0.0015 (10) |
| C2 | 0.0279 (15) | 0.0411 (19) | 0.0388 (17) | −0.0032 (14) | 0.0105 (13) | −0.0054 (15) |
| O2 | 0.0438 (13) | 0.0300 (12) | 0.0403 (13) | −0.0022 (10) | 0.0155 (10) | −0.0046 (9) |
| O3 | 0.0518 (13) | 0.0287 (12) | 0.0396 (12) | 0.0006 (10) | 0.0192 (11) | 0.0044 (10) |
| C3 | 0.0227 (14) | 0.0291 (17) | 0.0445 (18) | 0.0001 (12) | 0.0006 (13) | −0.0065 (13) |
| C4 | 0.0298 (15) | 0.0320 (17) | 0.0327 (16) | 0.0002 (12) | −0.0047 (13) | −0.0015 (13) |
| O4 | 0.0329 (11) | 0.0293 (12) | 0.0547 (14) | −0.0023 (9) | −0.0114 (10) | 0.0081 (10) |
| O5 | 0.0343 (12) | 0.0250 (12) | 0.0529 (14) | 0.0027 (9) | −0.0093 (10) | −0.0067 (10) |
| C5 | 0.0283 (15) | 0.0279 (16) | 0.0289 (15) | 0.0011 (12) | 0.0031 (12) | −0.0002 (12) |
| C6 | 0.0270 (16) | 0.047 (2) | 0.061 (2) | −0.0010 (15) | 0.0000 (15) | −0.0082 (18) |
| C7 | 0.0215 (13) | 0.0306 (16) | 0.0259 (14) | 0.0000 (11) | 0.0036 (11) | −0.0001 (12) |
| C8 | 0.0426 (18) | 0.0417 (19) | 0.0307 (16) | −0.0055 (14) | 0.0097 (14) | 0.0014 (14) |
| C9 | 0.0221 (14) | 0.0310 (16) | 0.0284 (15) | 0.0022 (12) | 0.0033 (12) | 0.0028 (12) |
| C10 | 0.043 (2) | 0.043 (2) | 0.047 (2) | −0.0048 (16) | −0.0164 (17) | 0.0023 (16) |
| O6 | 0.0536 (15) | 0.0473 (16) | 0.0306 (12) | −0.0011 (13) | 0.0023 (11) | −0.0039 (10) |
catena-Poly[[manganese(II)-di-µ2-acetato-µ-(4-methylpyridine N-oxide)] monohydrate] (III) . Geometric parameters (Å, º)
| Mn1—O1i | 2.206 (2) | C4—C5 | 1.380 (4) |
| Mn1—O1 | 2.203 (2) | O4—C9 | 1.243 (3) |
| Mn1—O2 | 2.170 (2) | O5—C9 | 1.257 (3) |
| Mn1—O3ii | 2.179 (2) | C5—H5 | 0.9300 |
| Mn1—O4 | 2.134 (2) | C6—H6A | 0.9600 |
| Mn1—O5i | 2.136 (2) | C6—H6B | 0.9600 |
| O1—N1 | 1.358 (3) | C6—H6C | 0.9600 |
| C1—H1 | 0.9300 | C7—C8 | 1.508 (4) |
| C1—N1 | 1.339 (4) | C8—H8A | 0.9600 |
| C1—C2 | 1.386 (4) | C8—H8B | 0.9600 |
| N1—C5 | 1.348 (4) | C8—H8C | 0.9600 |
| C2—H2 | 0.9300 | C9—C10 | 1.513 (4) |
| C2—C3 | 1.387 (4) | C10—H10A | 0.9600 |
| O2—C7 | 1.254 (3) | C10—H10B | 0.9600 |
| O3—C7 | 1.254 (3) | C10—H10C | 0.9600 |
| C3—C4 | 1.391 (4) | O6—H6D | 0.841 (19) |
| C3—C6 | 1.514 (4) | O6—H6E | 0.839 (18) |
| C4—H4 | 0.9300 | ||
| O1—Mn1—O1i | 176.46 (6) | C5—C4—C3 | 120.9 (3) |
| O2—Mn1—O1i | 94.96 (8) | C5—C4—H4 | 119.5 |
| O2—Mn1—O1 | 86.72 (8) | C9—O4—Mn1 | 138.5 (2) |
| O2—Mn1—O3ii | 178.58 (8) | C9—O5—Mn1ii | 135.55 (19) |
| O3ii—Mn1—O1i | 86.37 (8) | N1—C5—C4 | 119.9 (3) |
| O3ii—Mn1—O1 | 91.92 (8) | N1—C5—H5 | 120.0 |
| O4—Mn1—O1 | 91.80 (8) | C4—C5—H5 | 120.0 |
| O4—Mn1—O1i | 85.20 (8) | C3—C6—H6A | 109.5 |
| O4—Mn1—O2 | 86.30 (9) | C3—C6—H6B | 109.5 |
| O4—Mn1—O3ii | 93.32 (9) | C3—C6—H6C | 109.5 |
| O4—Mn1—O5i | 177.62 (9) | H6A—C6—H6B | 109.5 |
| O5i—Mn1—O1 | 90.27 (8) | H6A—C6—H6C | 109.5 |
| O5i—Mn1—O1i | 92.69 (8) | H6B—C6—H6C | 109.5 |
| O5i—Mn1—O2 | 94.99 (9) | O2—C7—C8 | 117.7 (3) |
| O5i—Mn1—O3ii | 85.44 (9) | O3—C7—O2 | 125.5 (3) |
| Mn1—O1—Mn1ii | 112.64 (8) | O3—C7—C8 | 116.7 (3) |
| N1—O1—Mn1 | 123.84 (15) | C7—C8—H8A | 109.5 |
| N1—O1—Mn1ii | 118.48 (15) | C7—C8—H8B | 109.5 |
| N1—C1—H1 | 120.3 | C7—C8—H8C | 109.5 |
| N1—C1—C2 | 119.4 (3) | H8A—C8—H8B | 109.5 |
| C2—C1—H1 | 120.3 | H8A—C8—H8C | 109.5 |
| C1—N1—O1 | 118.9 (2) | H8B—C8—H8C | 109.5 |
| C1—N1—C5 | 121.5 (2) | O4—C9—O5 | 125.3 (3) |
| C5—N1—O1 | 119.5 (2) | O4—C9—C10 | 117.0 (3) |
| C1—C2—H2 | 119.3 | O5—C9—C10 | 117.6 (3) |
| C1—C2—C3 | 121.5 (3) | C9—C10—H10A | 109.5 |
| C3—C2—H2 | 119.3 | C9—C10—H10B | 109.5 |
| C7—O2—Mn1 | 134.09 (19) | C9—C10—H10C | 109.5 |
| C7—O3—Mn1i | 138.4 (2) | H10A—C10—H10B | 109.5 |
| C2—C3—C4 | 116.8 (3) | H10A—C10—H10C | 109.5 |
| C2—C3—C6 | 121.5 (3) | H10B—C10—H10C | 109.5 |
| C4—C3—C6 | 121.8 (3) | H6D—O6—H6E | 100 (4) |
| C3—C4—H4 | 119.5 |
Symmetry codes: (i) −x+1/2, y−1/2, −z+3/2; (ii) −x+1/2, y+1/2, −z+3/2.
catena-Poly[[manganese(II)-di-µ2-acetato-µ-(4-methylpyridine N-oxide)] monohydrate] (III) . Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O6—H6D···O3iii | 0.84 (2) | 2.23 (2) | 3.052 (3) | 165 (4) |
| O6—H6E···O2 | 0.84 (2) | 2.21 (2) | 3.035 (3) | 170 (4) |
Symmetry code: (iii) x, y+1, z.
Funding Statement
This work was funded by Armstrong State University grant .
References
- Ciunik, Z. & Głowiak, T. (1980). Acta Cryst. B36, 2029–2033.
- Dave, B. C., Czernuszewicz, R. S., Bond, M. R. & Carrano, C. J. (1993). Inorg. Chem. 32, 3593–3594.
- Dolomanov, O. V., Bourhis, L. J., Gildea, R. J., Howard, J. A. K. & Puschmann, H. (2009). J. Appl. Cryst. 42, 339–341.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Kang, L., Lynch, G., Lynch, W. & Padgett, C. (2017). Acta Cryst. E73, 1434–1438. [DOI] [PMC free article] [PubMed]
- Liu, J., Chen, L., Cui, H., Zhang, Y., Zhang, L. & Su, C.-Y. (2014). Chem. Soc. Rev. 43, 6011–6061. [DOI] [PubMed]
- Mondal, S., Guha, A., Suresh, E., Jana, A. D. & Banerjee, A. (2012). J. Mol. Struct. 1029, 169–174.
- Ren, X.-H., Wang, P., Cheng, J.-Y. & Dong, Y.-B. (2018). J. Mol. Struct. 1161, 145–151.
- Rigaku (1998). REQAB. Rigaku Corporation, Tokyo, Japan.
- Rigaku (2009). CrystalClear. Rigaku Corporation, Tokyo, Japan.
- Sarma, R. & Baruah, J. B. (2011). Solid State Sci. 13, 1692–1700.
- Sarma, R., Karmakar, A. & Baruah, J. B. (2008). Inorg. Chim. Acta, 361, 2081–2086.
- Sarma, R., Perumal, A. & Baruah, J. B. (2009). J. Coord. Chem. 62, 1513–1524.
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Sniekers, J., Malaquias, J. C., Van Meervelt, L., Fransaer, J. & Binnemans, K. (2017). Dalton Trans. 46, 2497–2509. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, II, III. DOI: 10.1107/S205698901801232X/zl2728sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698901801232X/zl2728Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S205698901801232X/zl2728IIsup3.hkl
Structure factors: contains datablock(s) III. DOI: 10.1107/S205698901801232X/zl2728IIIsup4.hkl
Additional supporting information: crystallographic information; 3D view; checkCIF report