Significance
Regulating gene expression is critical to the normal function of a biological cell, and misregulation of gene expression often leads to a diseased cell. In eukaryotic cells, DNA genetic information is wrapped around histone proteins into chromatin. Modification of histone proteins with the acetyl chemical group (i.e., acetylation) is an important mechanism for activating gene expression. Our biochemical and structural studies of the Gcn5 histone acetyltransferase and its partner protein Ada2 explain how Ada2 activates Gcn5’s enzymatic activity without directly contacting the histone substrate, as previously proposed. Instead, Ada2 indirectly influences binding of the enzyme cofactor acetyl-CoA, a mechanism not previously observed for histone modification enzymes.
Keywords: epigenetics, chromatin biology, histone modification, X-ray crystallography
Abstract
The Gcn5 histone acetyltransferase (HAT) subunit of the SAGA transcriptional coactivator complex catalyzes acetylation of histone H3 and H2B N-terminal tails, posttranslational modifications associated with gene activation. Binding of the SAGA subunit partner Ada2 to Gcn5 activates Gcn5’s intrinsically weak HAT activity on histone proteins, but the mechanism for this activation by the Ada2 SANT domain has remained elusive. We have employed Fab antibody fragments as crystallization chaperones to determine crystal structures of a yeast Ada2/Gcn5 complex. Our structural and biochemical results indicate that the Ada2 SANT domain does not activate Gcn5’s activity by directly affecting histone peptide binding as previously proposed. Instead, the Ada2 SANT domain enhances Gcn5 binding of the enzymatic cosubstrate acetyl-CoA. This finding suggests a mechanism for regulating chromatin modification enzyme activity: controlling binding of the modification cosubstrate instead of the histone substrate.
The acetylation of lysine residues in the histone protein component of chromatin is an important means of regulating gene expression (1–3). The acetyl mark recruits acetyl-lysine binding modules such as the bromodomain found in many gene-regulatory chromatin modification and remodeling enzymes (4, 5). Furthermore, acetylation can neutralize the positive charge of the lysine side chain, leading to destabilization of the nucleosome complex of histone proteins and DNA (2, 6, 7).
Gcn5 is one of the best characterized histone acetyltransferase (HAT) enzymes. Like other members of the Gcn5-related N-acetyltransferase family of HAT enzymes, Gcn5 contains a 160-residue catalytic domain necessary for histone acetylation (8). This catalytic domain transfers the acetyl group from acetyl-CoA to the recipient lysine residue on the histone peptide substrate (9). Kinetic studies of the Gcn5 HAT domain have determined that the catalytic mechanism employs an ordered Bi-Bi mechanism with acetyl-CoA binding first, followed by histone peptide binding, and that formation of this ternary complex is required for catalysis (10). Crystallographic and NMR studies of the Gcn5 HAT domain from yeast, human, and Tetrahymena provide structural descriptions for acetyl-CoA and histone peptide binding (11–16).
Gcn5 is found in the cell as part of multisubunit gene-regulatory complexes, including the SAGA and variant SAGA complexes (17–21). Whereas Gcn5 acetylates histone H3 weakly on its own and is not able to acetylate histones packaged into nucleosomes, the SAGA complex robustly acetylates nucleosomal histones (22). We and others have previously shown that Gcn5 forms a complex with SAGA subunits Ada2 and Ada3, and that this Ada2/Ada3/Gcn5 complex is necessary and sufficient for SAGA’s physiological HAT activity on nucleosomes (23, 24). We also showed that Ada2 activates Gcn5’s HAT activity on histone peptides, but Ada3 is required for nucleosomal HAT activity (24).
Deletion analysis of Gcn5 and Ada2 have defined regions required for their protein interaction and for HAT activity (23, 25–27). Approximately 20 residues of Gcn5 immediately following the HAT domain are necessary for Ada2 to bind to Gcn5, and consequently this region (Gcn5 residues 260–280) has been defined as the Ada2 interaction domain (26, 27). The SANT (Swi3, Ada2, N-Cor, and TFIIB) domain of Ada2 plays a critical role in Ada2’s interaction with Gcn5, as well as histone and nucleosomal HAT activity by the SAGA complex (27–29). It has been suggested that the SANT domain in Ada2 and other chromatin enzymes functions as a histone tail-binding module to present histone tails for efficient catalysis (28). The SANT domain’s structural similarity to the helix-turn-helix DNA-binding domain of Myb transcription factor also suggests the possibility that the Ada2 SANT domain could influence nucleosomal HAT activity by binding nucleosomal DNA (27).
The mechanistic basis for how the Ada2 SANT domain increases Gcn5’s catalytic activity has been hampered by the lack of structural information for how Ada2 interacts with Gcn5. We present here crystal structures of the Ada2/Gcn5 complex. The structures show how the SANT and ZZ domains of Ada2 interact with N- and C-terminal extensions to the Gcn5 HAT domain. The Ada2 SANT domain does not appear to act as a histone tail-binding module, as it is positioned away from the Gcn5 peptide-binding pocket. Instead, our studies indicate that the Ada2 SANT activates Gcn5’s HAT activity by enhancing binding of the acetyl-CoA cosubstrate.
Results
Crystallization and Structure Determination of Ada2/Gcn5 Complexes.
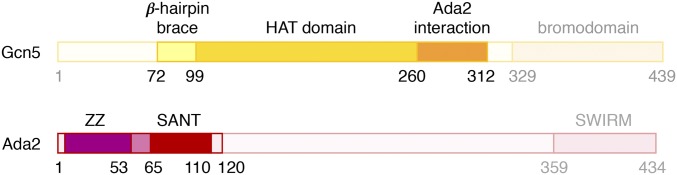
We had previously determined through deletion analysis that Gcn5(67-328) was sufficient to form a complex with Ada2(1-120) with robust HAT activity (30). This region of Gcn5 includes N- and C-terminal extensions to the HAT catalytic domain (residues 99–259) and the previously determined Ada2-interacting domain (residues 260–280; Fig. 1). Ada2(1-120) includes the SANT domain known to enhance Gcn5’s catalytic activity as well as the ZZ zinc-binding domain.
Fig. 1.
Structural and functional domains of Gcn5 and Ada2. The Gcn5 residues observed in Ada2/Gcn5 crystal form 1 (residues 72–312) are shown in saturated colors, and the residues outside this region are shown in faded colors. A similar convention is used to highlight the Ada2 residues observed in Ada2/Gcn5 crystal form 1 (residues 1–120).
Crystallization trials of Ada2(1-120)/Gcn5(67-328) produced single 200 × 200 × 200-µm crystals, but these crystals diffracted X-rays only to 8 Å (30). Postcrystallization dehydration beneficial for other macromolecular crystals did not improve the diffraction. Crystals with similar morphology and diffraction properties were also grown by using Ada2(1-120)/Gcn5(67-317) (31), truncated slightly on the Gcn5 C-terminal end. However, efforts to grow crystals with improved diffraction properties by (i) manipulating the precise N or C termini of yeast Ada2 and Gcn5, (ii) employing other species of Ada2 and Gcn5, (iii) engineering surface entropy mutations, or (iv) employing fusion proteins with rigid linkers were unsuccessful (30, 31).
Abs, especially mAbs, have been employed as crystallization chaperones to grow crystals of intractable macromolecules such as membrane proteins (32–34). Instead of preparing monoclonal antibodies, we used phage display to screen in vitro for synthetic Abs (sABs), which are functionally and structurally similar to Fabs (35). Biochemical screening against the Ada2(1-120)/Gcn5(67-317) complex produced 17 sABs, 12 of which bound to the Ada2(1-120)/Gcn5(67-317) complex in size-exclusion chromatography binding experiments. Ada2(1-120)/Gcn5(67-317)/sAB complexes corresponding to 9 of the 12 sABs were purified by size exclusion and subjected to crystallization experiments. Three such complexes produced crystals, two of which (referred to as Ada2/Gcn5 crystal forms 1 and 2) diffracted X-rays to 2.7 Å and 2.0 Å, respectively.
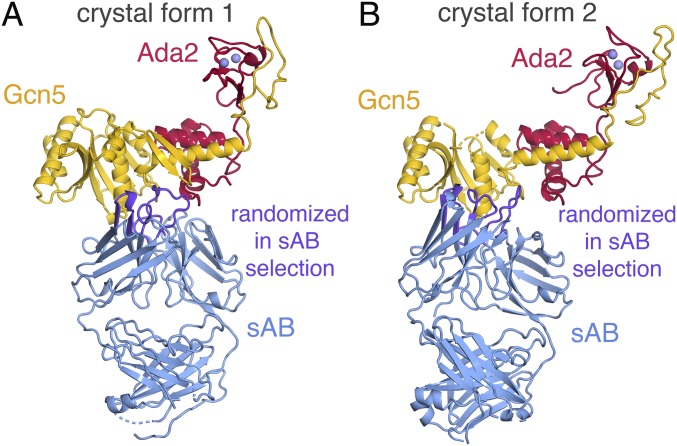
We determined the crystal structures of Ada2/Gcn5 crystal forms 1 and 2 by molecular replacement, using as search models the NMR structure of a SANT domain [Protein Data Bank (PDB) ID code 2ELK], the crystal structure of yeast Gcn5 HAT domain (PDB ID code 1YGH) (11), and the crystal structure of an sAB molecule used to crystallize a protein targeting complex (PDB ID code 4XTR) (36). The crystallographic asymmetric unit for the Ada2/Gcn5 crystal form 1 contains one copy each of Ada2, Gcn5, and the sAB (Fig. 2A), whereas the asymmetric unit for Ada2/Gcn5 crystal form 2 contains two copies of each (SI Appendix, Fig. S1). As expected, the CDRs of the sABs form the antigen (Ada2/Gcn5) binding site (Fig. 2). Interestingly, the two different sABs in crystal forms 1 and 2 interact with similar regions of the Ada2/Gcn5 complex, with the sAB in crystal form 1 making more extensive interactions with Ada2 and Gcn5, whereas the sAB in crystal form 2 contacts Gcn5 predominantly (Fig. 2). The sABs are responsible for the critical crystal contacts for crystal forms 1 and 2 (SI Appendix, Fig. S2), highlighting the integral role of the sABs in producing these crystals.
Fig. 2.
Ada2/Gcn5 crystal forms 1 and 2 structures. (A) Crystal form 1 with Ada2, Gcn5, the sAB, and zinc ions shown in red, yellow, blue, and lavender, respectively. The sAB regions randomized in the selection process are shown in purple. Internal disordered regions of 10 residues or less are shown in dashes. (B) Crystal form 2 with the same coloring conventions as for A.
Interactions Between Ada2 and Gcn5.
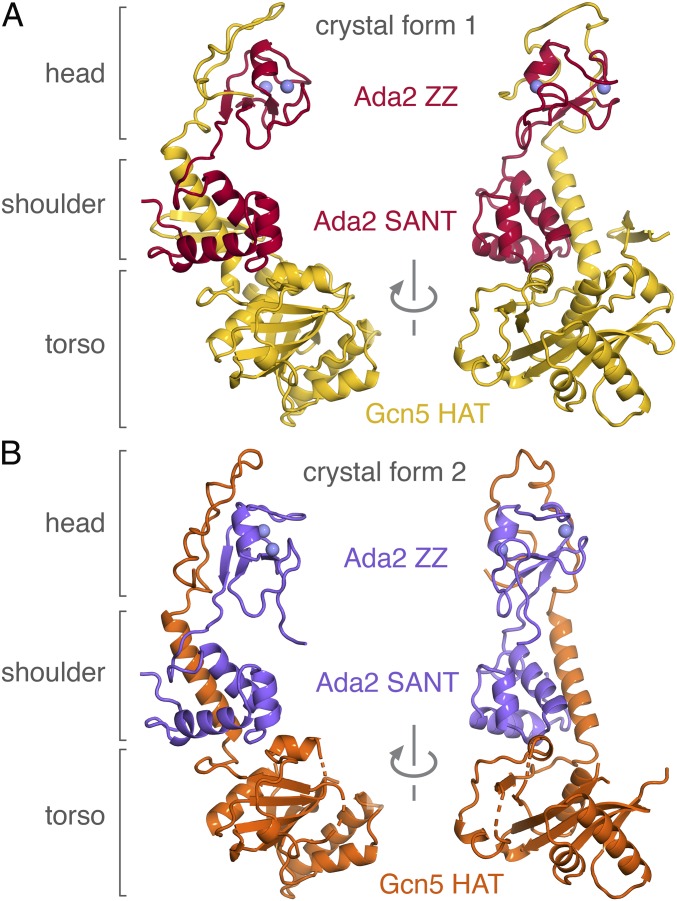
The crystal structures of yeast Ada2/Gcn5 crystal forms 1 and 2 show an elongated three-lobed complex with dimensions 80 × 50 × 40 Å (Fig. 3). The three lobes correspond to the torso, shoulders, and head of the complex. The complex’s Gcn5 HAT domain (Gcn5 residues 99–259), which constitutes the torso lobe, is very similar in structure to the yeast Gcn5 HAT domain on its own (rmsd = 0.51 Å for all atoms), with only localized differences in the flexible histone peptide binding loop, the CoA binding loop, and the N-terminal region (SI Appendix, Fig. S3A). The N- and C-terminal extensions to the Gcn5 HAT domain interact with the Ada2 ZZ and SANT domains predominantly through hydrophobic contacts to form the shoulder and head lobes. The shoulder lobe is composed of an α-helix corresponding to Gcn5 residues 260–280 (immediately C-terminal to the HAT domain) flanked by the Ada2 SANT domain on one side and a β-hairpin from the Gcn5 N-terminal extension to the HAT domain (“β-hairpin brace,” residues 72–98) on the other side. This Gcn5 α-helix corresponds to the same region identified as the Ada2 interaction region in biochemical deletion experiments (26). The head lobe is comprised of the Gcn5 extended chain residues 283–312, which cradle the Ada2 ZZ domain (the two zinc atoms in the ZZ domain can be thought of as eyes in the head region; Fig. 3). The extensive and multivalent interaction between Ada2 and Gcn5 provides a structural explanation for why partial deletion of the Ada2 ZZ domain or the SANT domain did not prevent formation of the Ada2/Gcn5 complex or the SAGA complex (27, 28).
Fig. 3.
Comparison of Ada2/Gcn5 crystal forms 1 and 2. (A) Crystal form 1 with Gcn5 shown in yellow and Ada2 in red. (B) Crystal form 2 with Gcn5 in orange and Ada2 in purple.
Conformational Flexibility Within Ada2/Gcn5 Complex.
Although the overall structure of the Ada2/Gcn5 complex is similar in crystal forms 1 and 2, there are notable conformational differences away from the Gcn5 HAT domain in the two crystal forms. The torso lobes (Gcn5 HAT domain) and the shoulder lobes (which contain Ada2 SANT domain) of crystal forms 1 and 2 align very well (rmsd = 0.44 Å for all atoms), but the head lobes (which contain the Ada2 ZZ domain) are rotated by 16° in the two crystal forms (SI Appendix, Fig. S4A). The head lobes of crystal forms 1 and 2 themselves are very similar (rmsd = 0.47 Å for all atoms), and the differences between crystal forms 1 and 2 are mostly isolated to rigid body rotation of the head lobes (SI Appendix, Fig. S4B). Crystal form 1’s head lobe is rotated toward the torso lobe compared with crystal form 2’s head lobe. The shoulder lobe is rigidly positioned next to the torso lobe because of the tight packing of the Ada2 SANT domain in the shoulder lobe against the Gcn5 HAT domain that constitutes the torso lobe. In contrast, there are relatively few interactions binding the head lobe to the shoulder domain. These observations suggest that the head lobe of the Ada2/Gcn5 complex is flexibly linked to the shoulder and torso lobes in solution. The different Ada2/Gcn5 complex conformations were stabilized in crystal forms 1 and 2 by the sAB molecules, which mediate the critical crystal contacts. We speculate that this conformational flexibility within the Ada2/Gcn5 complex contributed to the relatively poor internal order of our original Ada2/Gcn5 crystals grown in the absence of the sAB crystallization chaperones.
Mechanism of HAT Activation by Ada2.
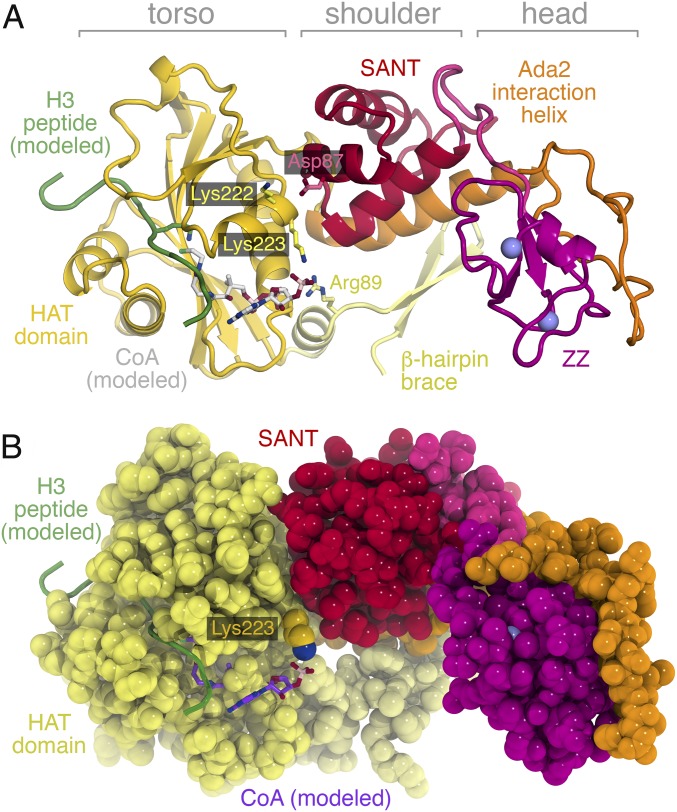
The Ada2/Gcn5/sAB complex crystals were grown in the absence of substrate histone peptides or CoA. However, given the strong structural similarity between the yeast and Tetrahymena Gcn5 HAT domains and the absence of significant conformational changes between the HAT domain in Gcn5 vs. Ada2/Gcn5, we could model by simple superposition the substrate peptide and CoA binding based on the Tetrahymena crystal structures containing an 11- or 19-residue H3 peptide and CoA (13, 15) (Fig. 4). The Gcn5 HAT domain binds the histone peptide substrate in a pocket that is positioned away from the Ada2 SANT domain. If the Ada2 SANT domain and the Gcn5 Ada2 interaction helix make up the shoulder of the Ada2/Gcn5 complex, the histone peptide substrate-binding pocket is located in the lower torso of the complex. No direct contacts between the modeled 19-residue H3 peptide and the Ada2 SANT or ZZ domains are possible. [The closest contact between the modeled H3 peptide and the Ada2 SANT domain is 16.8 Å. This and other analysis (detailed later) was made with the use of Ada2/Gcn5 crystal form 1 because of fewer unstructured loops in this crystal form despite the lower resolution, but the structural conclusions are consistent with both crystal forms.] It is therefore difficult to account for Ada2’s activation of Gcn5’s HAT activity, especially given that we did not observe significant conformational changes within the Gcn5 HAT domain in the Ada2/Gcn5 complex.
Fig. 4.
Ribbon and space-filling representations of the Ada2/Gcn5 complex with modeled H3 peptide substrate (green) and CoA (white or purple) cosubstrate from PDB ID code 1PU9. (A) Cartoon representation of the Ada2/Gcn5 complex showing Gcn5 Lys223 and Arg89 side chains interacting with the modeled CoA cosubstrate. The Ada2 SANT and ZZ domains are positioned too far away to directly interact with the H3 peptide substrate or the CoA cosubstrate. Ada2 Asp87 and Gcn5 Lys222 side chains are also shown. (B) Space-filling representation of the Ada2/Gcn5 complex highlights the wall created by the Ada2 SANT domain to constrain the Lys223 side chain and promote interaction with the CoA cosubstrate.
Histone acetylation by Gcn5 requires two substrates: the histone peptide and acetyl-CoA. Gcn5 forms a ternary complex with its two substrates before catalysis, with acetyl-CoA binding first, followed by the histone peptide (10). Like the modeled histone peptide, the modeled CoA does not directly contact Ada2 in our structure: the closest contact is between the 3′ phosphate group of CoA and the SANT domain at approximately 6 Å. Although the electron density for the Gcn5 Lys223 side chain is weak (SI Appendix, Figs. S5 and S6), this modeled CoA 3′ phosphate group appears to hydrogen-bond with the terminal amino group of Gcn5 Lys223 from the HAT domain and to make additional electrostatic interactions with the guanidinium group of Gcn5 Arg89 from the β-hairpin brace N-terminal extension to the HAT domain (Fig. 4).
The Gcn5 Lys223 side chain that contacts the modeled CoA 3′ phosphate group lies on the same surface that interacts with the Ada2 SANT domain. In fact, the Ada2 SANT domain forms a wall that constrains the position of the Gcn5 Lys223 side chain (Fig. 4B). The main chain oxygen of Leu80 in the Ada2 SANT domain makes very close contact with the Ca atom of Gcn5 Lys223 (3.1 Å), whereas the main chain oxygen of Thr79 is near (4.4 Å) the apparent position of the epsilon carbon of the Lys223 side chain. It is worth noting that only main-chain atoms of Ada2 contact Gcn5 Lys223 and that no amino acid side chain from the Ada2 SANT domain appears to interact with Gcn5 Lys223. These structural observations suggest that Ada2 activates Gcn5’s HAT activity because the Ada2 SANT domain constrains the Gcn5 Lys223 side chain to adopt a conformation appropriate for interacting with CoA through the CoA phosphate group.
To test this hypothesis, we prepared point mutations in Gcn5 and Ada2 and examined the HAT activity against an H3 peptide (Table 1). Compared with Gcn5 alone, the Ada2/Gcn5 complex had more than 100-fold higher catalytic efficiency (kcat/Km) in 10 µM acetyl-CoA, resulting from a 26-fold increase in the catalytic rate constant and a fourfold decrease in the Michaelis constant (SI Appendix, Fig. S7). Removing the Gcn5 Lys223 side chain dramatically reduced the catalytic efficiency of the Ada2/Gcn5 complex (∼18-fold reduced vs. WT complex) by affecting the kcat and the Km. In fact, the Km of the Ada2/Gcn5(K223A) mutation was greater than that of Gcn5 on its own. Mutation of Arg89 had a smaller effect: the kcat/Km of the Ada2/Gcn5(R89A) complex was only 2.7-fold lower than that of the WT Ada2/Gcn5 complex. Mutating Ada2 D87A had a statistically insignificant effect, consistent with the incidental contact between Ada2 Asp87 and Gcn5 Lys222, compared with the much more extensive interactions between the Ada2 SANT domain and the Gcn5 HAT domain and the Gcn5 Ada2 interaction helix (Fig. 4). Removing both K223 and R89 side chains in the Ada2/Gcn5 complex eliminated the Ada2-mediated enhancement of Gcn5’s HAT activity, with a similar kcat/Km to that of Gcn5 alone. At much higher concentrations of acetyl-CoA [10× higher than the Kd (as detailed later) or 600 µM when the Kd could not be determined], only Ada2/Gcn5 complexes with mutations in K223 or R89 showed significantly increased kcat/Km, which was attributable mostly to decreases in Km (Table 1 and SI Appendix, Fig. S8). The partial compensation of increased acetyl-CoA concentrations for the removal of K223 and R89 is consistent with the proposed roles of these side chains in recruiting acetyl-CoA. We also find that the HAT activity of the Ada2/Gcn5 complex was not altered significantly when assayed in the presence of the sAB used to crystallize the Ada2/Gcn5 complex in crystal form 1, indicating that the antibody does not affect the catalytic activity of Ada2/Gcn5. This agrees with our structural finding that the sAB binds Ada2 and Gcn5 away from the H3 peptide and acetyl-CoA binding sites in Gcn5.
Table 1.
HAT activity and AcCoA isothermal calorimetry binding data for Gcn5(67-317) and Ada2(1-120)/Gcn5(67-317) variants
| Samples | Catalysis in 10 μM AcCoA | Catalysis at higher AcCoA concentrations | AcCoA binding | ||||||||
| Ada2 | Gcn5 | sAB | [AcCoA], µM | kcat, s−1 | Km, µM | kcat/KM, M−1⋅s−1 | [AcCoA], µM | kcat, s−1 | Km, µM | kcat/KM, M−1⋅s−1 | AcCoA Kd, µM |
| − | + | − | 10 | 0.048 ± 0.003 | 749 ± 112 | 65 | 100 | 0.102 ± 0.014 | 1,348 ± 400 | 75 | 7.8 ± 3 |
| − | K223A | − | — | — | — | — | 600 | 0.047 ± 0.003 | 802 ± 123 | 59 | ND |
| + | + | − | 10 | 1.28 ± 0.12 | 189 ± 62 | 6,790 | 24 | 1.27 ± 0.076 | 187 ± 38 | 6,780 | 2.3 ± 0.2 |
| + | K223A | − | 10 | 0.414 ± 0.029 | 1110 ± 175 | 372 | 600 | 0.430 ± 0.048 | 291 ± 103 | 1,480 | NA |
| + | R89A | − | 10 | 0.917 ± 0.085 | 364 ± 97 | 2,520 | 400 | 0.724 ± 0.078 | 187 ± 78 | 3,870 | 36.2 ± 5.5 |
| + | R89A,K223A | − | 10 | 0.096 ± 0.006 | 1930 ± 212 | 50 | 600 | 0.209 ± 0.023 | 771 ± 182 | 271 | NA |
| D87A | + | − | 10 | 1.06 ± 0.11 | 191 ± 76 | 5,570 | 30 | 1.15 ± 0.14 | 213 ± 97 | 5,420 | 2.7 ± 0.6 |
| + | + | + | 10 | 1.34 ± 0.096 | 156 ± 41 | 8,570 | 30 | 1.51 ± 0.16 | 156 ± 59 | 9,670 | ND |
Columns 4–7 show catalysis results in 10 μM AcCoA, columns 8–11 show catalysis results in near-saturating conditions of AcCoA, and column 12 shows AcCoA binding data. conc, concentration; NA, not applicable (because no binding detected); ND, not determined.
We further assessed the role of Gcn5(K223A) by examining its effect in the Gcn5 protein or in the Ada2/Gcn5 complex. We find that Gcn5(K223A) has a smaller effect, particularly on the Km for histone H3 in saturating or near-saturating concentrations of acetyl-CoA. Compared with Gcn5 alone, the Km of histone H3 for Ada2/Gcn5 is increased 7.2 fold, whereas the equivalent Km for Ada2/Gcn5(K223A) is increased only 2.8 fold (Table 1). This is consistent with a Gcn5 K223-dependent role for Ada2 in Ada2/Gcn5’s HAT activity.
The HAT activity results suggest that the Gcn5 Lys223 side chain plays a major role by interacting with acetyl-CoA. To investigate this directly, we measured acetyl-CoA binding by isothermal calorimetry (Table 1 and SI Appendix, Fig. S9). We find that Gcn5 binds acetyl-CoA with a Kd of 7.8 µM, similar to the 8.5-µM Kd measured by equilibrium dialysis for the yeast Gcn5 catalytic domain (37). The Ada2/Gcn5 complex binds acetyl-CoA with a Kd of 2.3 µM, or 3.4-fold more tightly. Strikingly, the Ada2/Gcn5(K223A) and the Ada2/Gcn5(R89A,K223A) complexes fail to bind acetyl-CoA, whereas the Ada2/Gcn5(R89A) complex binds 15.7× less tightly than the WT Ada2/Gcn5 complex. The Ada2(D87A)/Gcn5 complex binds acetyl-CoA with a similar affinity as the WT complex, consistent with the insignificant effect of this mutation on the HAT activity. These acetyl-CoA binding studies indicate that yeast Gcn5 Lys223 plays a critical role in binding acetyl-CoA and that Ada2 enhances acetyl-CoA binding by Gcn5 even though Ada2 does not appear to interact directly with acetyl-CoA.
We also used calorimetry to investigate whether Ada2 affects binding of Gcn5’s binding of its H3 peptide substrate. We find that Gcn5 and Ada2/Gcn5 bind the H3 peptide with similar affinities, suggesting that Ada2 does not activate Gcn5 by promoting histone peptide binding (SI Appendix, Fig. S10 and Table S2). Similar results were obtained in the presence of CoA.
Discussion
The crystal structures of the Ada2/Gcn5 complex provide insights into how the Gcn5 catalytic HAT domain, together with N- and C-terminal extensions, binds to the Ada2 ZZ and SANT domains. Extensive contacts mediate the interactions between the two proteins, suggesting that they cooperatively assemble to form their stable structure, as it is likely that many of the structural elements in the complex would be unfolded on their own. For example, the β-hairpin brace and Ada2 interaction helix of Gcn5 are stabilized by their intramolecular interactions and the intermolecular interaction of Gcn5’s Ada2 interaction helix with the Ada2 SANT domain. Gcn5 residues 283–312 form an extended chain platform to bind to the Ada2 ZZ domain, and are likely disordered in the absence of Ada2.
The structures suggest a mechanism for how Ada2 increases Gcn5’s HAT activity. Several studies have documented that the Ada2 SANT domain is required for the activation of Gcn5’s HAT activity in the Ada2/Gcn5 subcomplex or the full SAGA complex (27–29). A kinetic analysis of the Ada2/Gcn5 complex showed that deleting the Ada2 SANT domain reduced H3 peptide substrate binding and catalysis, but apparently not acetyl-CoA binding (28). This finding led to the hypothesis that the Ada2 SANT domain functions as a histone tail-binding/presentation module. Our studies suggest a different mechanism. We find that the Ada2 SANT is far removed from the Gcn5 peptide binding pocket and also too far to make direct contact with CoA. However, it does contact Gcn5 Lys223, which itself appears to directly interact with CoA through the CoA 3′ phosphate group. The SANT domain creates a physical barrier that constrains the conformational flexibility of the Lys223 side chain and could help direct the side chain for productive interaction with the 3′ phosphate group of acetyl-CoA. As binding of acetyl-CoA by Gcn5 is the necessary first step in the catalytic mechanism before binding a histone peptide (10), enhancing acetyl-CoA binding will increase Gcn5’s HAT activity. The Ada2 SANT domain thus functions to complete the CoA binding pocket on Gcn5. The Gcn5 N-terminal extension of an α-helix plus the β-hairpin brace also contributes toward this acetyl-CoA binding pocket, with Arg89 residue apparently interacting with the same 3′ phosphate group of acetyl-CoA (Fig. 4). In hindsight, it is not surprising that Ada2 can influence Gcn5’s HAT activity by modulating interactions with acetyl-CoA, as acetyl-CoA is a cosubstrate in the histone acetylation reaction.
Our results define what has been a nebulous role of Lys223 in Gcn5’s HAT activity. Partially pure recombinant Gcn5(K223A) displayed reduced HAT activity in vitro and a more modest effect of the mutant on transcriptional activity in yeast (38). It should be noted that the steady-state transcriptional assay used in this study masks SAGA complex’s now recognized role in generating newly synthesized mRNA transcripts (39). A triple Gcn5(F221A,K222A,K223A) mutant was found to be strongly defective for growth and in vivo transcriptional activity (40), but a specific effect of K223A cannot be determined from this result because subsequent crystal structures show that F221 is part of the hydrophobic core of the Gcn5 HAT domain. Another study found that Gcn5 (K223A,T227A,K228A,E229A) and Gcn5(K223A,T227A,K242A) surface mutations severely affected HAT activity in vitro and displayed partial growth defects (27). In crystal structures of Tetrahymena Gcn5 HAT domain bound to H3 peptide CoA, the equivalent lysine points its side chain away from the CoA phosphate group (13). Interestingly, the only structure of the yeast Gcn5 HAT domain crystallized without CoA or substrate peptide shows Lys223 in a similar conformation to what we observed in Ada2/Gcn5 (11).
The critical role of yeast Gcn5 Lys223 in binding acetyl-CoA is likely to be preserved in Gcn5 orthologs, as this lysine residue is strictly conserved from yeast to plants to mammals (SI Appendix, Fig. S11). In contrast, the minor role of yeast Gcn5 Arg89 in binding acetyl-CoA among Gcn5 orthologs is more difficult to predict. Although the sequence alignment in SI Appendix, Fig. S11 suggests that Gcn5 Arg89 is not conserved among species, an arginine amino acid two residues before this position in many species could possibly function to bind acetyl-CoA.
The Ada2/Gcn5 complexes in the two different crystal forms highlights a general conformational flexibility, in particular that of the head lobe containing the Ada2 ZZ domain, which is tethered to the Ada2 SANT domain-containing shoulder lobe via relatively mobile connections. The functional significance of this conformational flexibility within the Ada2/Gcn5 complex is uncertain. This is because Ada2/Gcn5 is only part of the Ada3/Ada2/Gcn5 HAT module, itself part of the full SAGA complex. It is possible that the conformational flexibility between the head and shoulder lobes is an artifact of the isolated and truncated Ada2/Gcn5 subcomplex investigated here and that binding of Ada3 or remaining regions of Ada2 or Gcn5 will fix the orientation between the head and shoulder domains. It is also possible that the conformational flexibility between the head and shoulder domains could play a functional role within the SAGA complex for nucleosomal acetylation.
Our determination of the crystal structure of the Ada2/Gcn5 complex relied on the use of synthetic antibodies as crystallization chaperones, an approach that is gaining increasing attention because of its demonstrated successes in crystallizing recalcitrant protein systems. In this regard, an important recent advance has been the use of phage display to generate sABs. It has proven to produce a much larger variety of binders, much faster and cheaper than traditional monoclonal approaches. Importantly, the methodology allows for selection conditions that can be designed to produce sABs with desired characteristics; for instance, sABs that bind to a particular surface epitope or conformational state of the target protein. Notably, the Ada2/Gcn5 biopanning conditions were highly biased toward selecting sABs that recognized the assembled complex, not the individual protein components. This stabilizes the macromolecular assemblage during crystallization into the form that is most biologically relevant. We speculate that our crystals of the Ada2/Gcn5 grown in the absence of sABs were limited to 8-Å diffraction because of the flexible link between the head and shoulder lobes observed in the two crystal forms. Examination of the crystal packing in Ada2/Gcn5 crystal forms 1 and 2 shows how the respective synthetic antibodies build the crystal through antibody/antibody interactions (SI Appendix, Fig. S2), and the resulting packing stabilizes the otherwise flexible Ada2/Gcn5 complex.
In summary, our structural and biochemical studies reveal mechanistic insights for how an accessory or partner protein can influence the enzymatic activity of a HAT: by affecting binding of the acetyl-CoA substrate instead of the histone substrate. We suspect that similar mechanisms will be employed by other histone modification enzymes.
Materials and Methods
Protein Expression and Purification.
Ada2 residues 1–120 [N-terminally tagged with hexahistidine and maltose binding protein (MBP)] and Gcn5 residues 67–317 were coexpressed by using the pST44 polycistronic expression vector (41, 42). Site-directed mutations were introduced by using QuikChange-based procedures (43) employing Q5 DNA polymerase (New England Biolabs). Protein expression in BL21(DE3)pLysS host strains were induced with 0.2 mM IPTG at 18 °C.
Ada2/Gcn5 complexes were purified from cell lysates by cobalt metal affinity chromatography (Talon resin; Clontech), followed by TEV protease cleavage to remove the HISMBP tag and SourceS cation-exchange chromatography (GE Healthcare). As Gcn5(67-317) proved difficult to purify on its own, we first purified the Ada2/Gcn5 complex and then used mildly denaturing conditions (3 M urea) to dissociate the complex and to isolate Gcn5(67-317) by SourceS chromatography. The SourceS Gcn5(67-317) pool was then further purified by Superdex 200 Increase 10/300 size-exclusion chromatography (GE Healthcare).
sAB Selection, Characterization, Expression, and Purification.
The generation and screening of complex-specific sABs has been described previously (44). Two N-terminally Avi-His-TEV-tag modified Ada2/Gcn5 complexes were prepared whereby the affinity tag was placed on Ada2 or Gcn5. Both proteins were expressed, biotinylated in vivo, purified, and tested for biotinylation efficiency. Three rounds of phage display selection were performed by using 100 nM (round 1), 50 nM (round 2), and 10 nM (round 3) biotinylated protein targets and each time eluted with TEV protease (Thermo). After successful selection, the specificity of candidate sABs was assayed against both Avi-tagged complexes by using a single point competitive ELISA (44). Clones that exhibited strong and specific binding to both antigens were sequenced. We obtained more than 30 sequence-unique sABs, and 17 were nominated for further biochemical characterization. All sABs were characterized for their ability to stabilize Ada2/Gcn5 complex by gel filtration in 20 mM Hepes, 100 mM NaCl, 10 mM β-mercaptoethanol, pH 7.5, buffer on a Superdex 200 Increase 10/300 column (GE Healthcare). Only sABs that formed the most homogenous complex were systematically tested in crystallization trials.
sAB clones were expressed and purified by using dual-column chromatography using Protein A affinity (HiTrap MabSelect SuRe, 5 mL), immediately followed by ion exchange (Resource S, 1 mL) as previously described (44). Samples were dialyzed against 20 mM Hepes, 100 mM NaCl, pH 7.5, concentrated to more than 10 mg/mL and frozen in 100-μL aliquots.
yGcn5/yAda2/sAB Crystallization and Structure Determination.
Gcn5/Ada2 complex was incubated with 1.2 molar excess of sAB and purified by Superdex 200 Increase 10/300 size-exclusion chromatography in 20 mM Hepes, pH 7.5, 100 mM NaCl, 10 mm 2-mercaptoethanol. Pooled fractions were concentrated to approximately 10 mg/mL and used for microbatch under oil crystallization trials (45). Crystal form 1 containing 65H10_1 synthetic antibodies was grown in 10 mM Tris⋅Cl, pH 8.5, 200 mM Li2SO4, and 25% wt/vol PEG3350, whereas crystal form 2 containing 63E9_24 synthetic antibodies was grown in 100 mM succinic acid, pH 7.0, and 15% wt/vol PEG3350.
Before flash-cooling in liquid nitrogen, crystal form 1 crystals were soaked in the crystallization solution supplemented with 20% vol/vol glycerol in 5% glycerol steps, whereas crystal form 2 crystals were soaked in their crystallization solution supplemented with 25% wt/vol PEG3350 in 5% PEG3350 steps.
X-ray diffraction data were collected at Advanced Photon Source NE-CAT beamline 24-ID-C (0.9792 Å; 100K). The diffraction data were processed by using XDS (46) and SCALA (47) software packages. Molecular replacement using PHASER (48) employed rigid body search models from the HAT domain of yGcn5 (PDB ID code 1YGH), a SANT domain homolog (PDB ID code 2ELK), and the sAB from the Get3–sAB complex (PDB ID code 4XTR). Structure refinement was performed with the programs REFMAC (49) and PHENIX (50) together with manual model building in COOT (51). Crystallographic data statistics are summarized in SI Appendix, Table S1. All molecular graphics were prepared with PyMOL (52).
HAT Assay.
HAT activity was measured by using a pyruvate dehydrogenase enzyme-coupled assay (53) with substrate H3 peptide residues 1–20 (ARTKQTARKSTGGKAPRKQL). Each reaction contained 1× TBA (50 mM Tris, 50 mM Bis⋅Tris, 100 mM sodium acetate, pH 7.5), 5 mM MgCl2, 1 mM DTT, 0.2 mM thiamine pyrophosphate, 0.2 mM NAD+, 2.5 mM pyruvate, 0.04 U pyruvate dehydrogenase, and 0.25 μM Ada2/Gcn5 or Gcn5 enzyme. Specified concentrations of acetyl-CoA were included, with the highest concentration of 600 µM chosen because we observed inhibition at greater than this concentration. The reaction mixture was incubated at room temperature for 5 min before the reaction was initiated by adding the enzyme. The reaction was monitored in real time at 340 nm in a Varian Cary 50 Bio spectrophotometer.
Isothermal Titration Calorimetry.
Calorimetric experiments were conducted at 25 °C with a MicroCal auto-iTC200 instrument (Malvern). The protein samples were dialyzed against the buffer containing 20 mM Hepes (pH 7.5), 100 mM NaCl, and 2 mM β-mercaptoethanol. The exothermic heat of the reaction for acetyl-CoA binding was measured by 25 sequential 1.5-µL injections of 100 µM acetyl-CoA into 200 µL of 10 µM Gcn5 or Ada2/Gcn5 complex, spaced at intervals of 180 s. To further saturate Ada2/Gcn5(R89A), 200 µM acetyl-CoA was titrated into 10 µM of this mutant complex. Histone H3 binding was measured with similar injections of 800 µM H3(1-20) peptide into 200 µL of 80 µM Gcn5 or Ada2/Gcn5 complex with or without 80 µM CoA present. The data were analyzed by using the Origin 7 software package (Microcal).
Supplementary Material
Acknowledgments
We are very grateful to the staff of APS NE-CAT beamlines 24-ID-C and 24-ID-E for their help during synchrotron data collection, J. Fecko and N. Yennawar (Penn State Huck Institute X-ray core facility) for assistance with isothermal calorimetry and crystallographic refinement, and members of the laboratory of S.T., Phil Bevilacqua, and the Penn State Center for Eukaryotic Gene Regulation for discussions. This work was supported by National Institutes of Health National Institute of General Medical Sciences Grants R01GM088236 (to S.T.), R01GM111651 (to S.T.), U01GM094588 (to A.A.K.), R01GM072688 (to A.A.K.), and U54HG006436 (to A.A.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID codes 6CW2 and 6CW3).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1805343115/-/DCSupplemental.
References
- 1.Verdin E, Ott M. 50 years of protein acetylation: From gene regulation to epigenetics, metabolism and beyond. Nat Rev Mol Cell Biol. 2015;16:258–264. doi: 10.1038/nrm3931. [DOI] [PubMed] [Google Scholar]
- 2.Suganuma T, Workman JL. Signals and combinatorial functions of histone modifications. Annu Rev Biochem. 2011;80:473–499. doi: 10.1146/annurev-biochem-061809-175347. [DOI] [PubMed] [Google Scholar]
- 3.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benton CB, Fiskus W, Bhalla KN. Targeting histone acetylation: Readers and writers in leukemia and cancer. Cancer J. 2017;23:286–291. doi: 10.1097/PPO.0000000000000284. [DOI] [PubMed] [Google Scholar]
- 5.Marmorstein R, Zhou M-M. Writers and readers of histone acetylation: Structure, mechanism, and inhibition. Cold Spring Harb Perspect Biol. 2014;6:a018762. doi: 10.1101/cshperspect.a018762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi JK, Howe LJ. Histone acetylation: Truth of consequences? Biochem Cell Biol. 2009;87:139–150. doi: 10.1139/O08-112. [DOI] [PubMed] [Google Scholar]
- 7.Galvani A, Thiriet C. Nucleosome dancing at the tempo of histone tail acetylation. Genes (Basel) 2015;6:607–621. doi: 10.3390/genes6030607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan H, Marmorstein R. Histone acetyltransferases: Rising ancient counterparts to protein kinases. Biopolymers. 2013;99:98–111. doi: 10.1002/bip.22128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tanner KG, et al. Catalytic mechanism and function of invariant glutamic acid 173 from the histone acetyltransferase GCN5 transcriptional coactivator. J BiolChem. 1999;274:18157–18160. doi: 10.1074/jbc.274.26.18157. [DOI] [PubMed] [Google Scholar]
- 10.Tanner KG, Langer MR, Kim Y, Denu JM. Kinetic mechanism of the histone acetyltransferase GCN5 from yeast. J Biol Chem. 2000;275:22048–22055. doi: 10.1074/jbc.M002893200. [DOI] [PubMed] [Google Scholar]
- 11.Trievel RC, et al. Crystal structure and mechanism of histone acetylation of the yeast GCN5 transcriptional coactivator. Proc Natl Acad Sci USA. 1999;96:8931–8936. doi: 10.1073/pnas.96.16.8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin Y, Fletcher CM, Zhou J, Allis CD, Wagner G. Solution structure of the catalytic domain of GCN5 histone acetyltransferase bound to coenzyme A. Nature. 1999;400:86–89. doi: 10.1038/21922. [DOI] [PubMed] [Google Scholar]
- 13.Rojas JR, et al. Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature. 1999;401:93–98. doi: 10.1038/43487. [DOI] [PubMed] [Google Scholar]
- 14.Schuetz A, et al. Crystal structure of a binary complex between human GCN5 histone acetyltransferase domain and acetyl coenzyme A. Proteins. 2007;68:403–407. doi: 10.1002/prot.21407. [DOI] [PubMed] [Google Scholar]
- 15.Clements A, et al. Structural basis for histone and phosphohistone binding by the GCN5 histone acetyltransferase. Mol Cell. 2003;12:461–473. doi: 10.1016/s1097-2765(03)00288-0. [DOI] [PubMed] [Google Scholar]
- 16.Clements A, et al. Crystal structure of the histone acetyltransferase domain of the human PCAF transcriptional regulator bound to coenzyme A. EMBO J. 1999;18:3521–3532. doi: 10.1093/emboj/18.13.3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L, Dent SYR. Functions of SAGA in development and disease. Epigenomics. 2014;6:329–339. doi: 10.2217/epi.14.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samara NL, Wolberger C. A new chapter in the transcription SAGA. Curr Opin Struct Biol. 2011;21:767–774. doi: 10.1016/j.sbi.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baker SP, Grant PA. The SAGA continues: Expanding the cellular role of a transcriptional co-activator complex. Oncogene. 2007;26:5329–5340. doi: 10.1038/sj.onc.1210603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spedale G, Timmers HTM, Pijnappel WWMP. ATAC-king the complexity of SAGA during evolution. Genes Dev. 2012;26:527–541. doi: 10.1101/gad.184705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koutelou E, Hirsch CL, Dent SYR. Multiple faces of the SAGA complex. Curr Opin Cell Biol. 2010;22:374–382. doi: 10.1016/j.ceb.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grant PA, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: Characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 23.Candau R, Berger SL. Structural and functional analysis of yeast putative adaptors. Evidence for an adaptor complex in vivo. J Biol Chem. 1996;271:5237–5245. doi: 10.1074/jbc.271.9.5237. [DOI] [PubMed] [Google Scholar]
- 24.Balasubramanian R, Pray-Grant MG, Selleck W, Grant PA, Tan S. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J Biol Chem. 2002;277:7989–7995. doi: 10.1074/jbc.M110849200. [DOI] [PubMed] [Google Scholar]
- 25.Candau R, et al. Identification of human proteins functionally conserved with the yeast putative adaptors ADA2 and GCN5. Mol Cell Biol. 1996;16:593–602. doi: 10.1128/mcb.16.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Candau R, Zhou JX, Allis CD, Berger SL. Histone acetyltransferase activity and interaction with ADA2 are critical for GCN5 function in vivo. EMBO J. 1997;16:555–565. doi: 10.1093/emboj/16.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sterner DE, Wang X, Bloom MH, Simon GM, Berger SL. The SANT domain of Ada2 is required for normal acetylation of histones by the yeast SAGA complex. J Biol Chem. 2002;277:8178–8186. doi: 10.1074/jbc.M108601200. [DOI] [PubMed] [Google Scholar]
- 28.Boyer LA, et al. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol Cell. 2002;10:935–942. doi: 10.1016/s1097-2765(02)00634-2. [DOI] [PubMed] [Google Scholar]
- 29.Barbaric S, Reinke H, Hörz W. Multiple mechanistically distinct functions of SAGA at the PHO5 promoter. Mol Cell Biol. 2003;23:3468–3476. doi: 10.1128/MCB.23.10.3468-3476.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kramer RM. 2002. Characterization and crystallization of the yADA2-yGCN5 complex. MS thesis (The Pennsylvania State University, University Park, PA)
- 31.Barrios AF. 2005. Characterization and crystallization of HAT subcomplexes. MS thesis (The Pennsylvania State University, University Park, PA)
- 32.Griffin L, Lawson A. Antibody fragments as tools in crystallography. Clin Exp Immunol. 2011;165:285–291. doi: 10.1111/j.1365-2249.2011.04427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lieberman RL, Culver JA, Entzminger KC, Pai JC, Maynard JA. Crystallization chaperone strategies for membrane proteins. Methods. 2011;55:293–302. doi: 10.1016/j.ymeth.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bukowska MA, Grütter MG. New concepts and aids to facilitate crystallization. Curr Opin Struct Biol. 2013;23:409–416. doi: 10.1016/j.sbi.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 35.Paduch M, et al. Generating conformation-specific synthetic antibodies to trap proteins in selected functional states. Methods. 2013;60:3–14. doi: 10.1016/j.ymeth.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mateja A, et al. Protein targeting. Structure of the Get3 targeting factor in complex with its membrane protein cargo. Science. 2015;347:1152–1155. doi: 10.1126/science.1261671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langer MR, Fry CJ, Peterson CL, Denu JM. Modulating acetyl-CoA binding in the GCN5 family of histone acetyltransferases. J Biol Chem. 2002;277:27337–27344. doi: 10.1074/jbc.M203251200. [DOI] [PubMed] [Google Scholar]
- 38.Kuo MH, Zhou J, Jambeck P, Churchill ME, Allis CD. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 1998;12:627–639. doi: 10.1101/gad.12.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baptista T, et al. SAGA is a general cofactor for RNA polymerase II transcription. Mol Cell. 2017;68:130–143.e5. doi: 10.1016/j.molcel.2017.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang L, Liu L, Berger SL. Critical residues for histone acetylation by Gcn5, functioning in Ada and SAGA complexes, are also required for transcriptional function in vivo. Genes Dev. 1998;12:640–653. doi: 10.1101/gad.12.5.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan S. A modular polycistronic expression system for overexpressing protein complexes in Escherichia coli. Protein Expr Purif. 2001;21:224–234. doi: 10.1006/prep.2000.1363. [DOI] [PubMed] [Google Scholar]
- 42.Tan S, Kern RC, Selleck W. The pST44 polycistronic expression system for producing protein complexes in Escherichia coli. Protein Expr Purif. 2005;40:385–395. doi: 10.1016/j.pep.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 43.Xia Y, Chu W, Qi Q, Xun L. New insights into the QuikChange™ process guide the use of Phusion DNA polymerase for site-directed mutagenesis. Nucleic Acids Res. 2015;43:e12. doi: 10.1093/nar/gku1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paduch M, Kossiakoff AA. Generating conformation and complex-specific synthetic antibodies. Methods Mol Biol. 2017;1575:93–119. doi: 10.1007/978-1-4939-6857-2_6. [DOI] [PubMed] [Google Scholar]
- 45.D’Arcy A, Sweeney AM, Haber A. Practical aspects of using the microbatch method in screening conditions for protein crystallization. Methods. 2004;34:323–328. doi: 10.1016/j.ymeth.2004.03.023. [DOI] [PubMed] [Google Scholar]
- 46.Kabsch W. Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr. 2010;66:133–144. doi: 10.1107/S0907444909047374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Evans P. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 48.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 50.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 52.Schrödinger, LLC 2015. The PyMOL Molecular Graphics System, Version 1.8.
- 53.Kim Y, Tanner KG, Denu JM. A continuous, nonradioactive assay for histone acetyltransferases. Anal Biochem. 2000;280:308–314. doi: 10.1006/abio.2000.4546. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.