Significance
Despite the progress made by postmortem cytoarchitectonic analyses and animal electrophysiology in studying the structure and function of the hippocampal circuitry, complex anatomical challenges have prevented a detailed understanding of its substructural organization in living humans. By integrating high-resolution structural and resting-state functional neuroimaging, we demonstrate two main axes of substructural organization in the human hippocampus: one that respects its long axis and a second that follows patterns of hippocampal infolding and significantly correlates with an intracortical microstructure. Given the importance of the hippocampus for cognition, affect, and disease, our results provide an integrated hippocampal coordinate system that is relevant to cognitive neuroscience, clinical neuroimaging, and network neuroscience.
Keywords: hippocampus, connnectome, microstructure, MRI, neuroimaging
Abstract
The hippocampus plays key roles in cognition and affect and serves as a model system for structure/function studies in animals. So far, its complex anatomy has challenged investigations targeting its substructural organization in humans. State-of-the-art MRI offers the resolution and versatility to identify hippocampal subfields, assess its microstructure, and study topographical principles of its connectivity in vivo. We developed an approach to unfold the human hippocampus and examine spatial variations of intrinsic functional connectivity in a large cohort of healthy adults. In addition to mapping common and unique connections across subfields, we identified two main axes of subregional connectivity transitions. An anterior/posterior gradient followed long-axis landmarks and metaanalytical findings from task-based functional MRI, while a medial/lateral gradient followed hippocampal infolding and correlated with proxies of cortical myelin. Findings were consistent in an independent sample and highly stable across resting-state scans. Our results provide robust evidence for long-axis specialization in the resting human hippocampus and suggest an intriguing interplay between connectivity and microstructure.
The hippocampus is a complex structure located in the medial temporal lobe that has been implicated in a broad range of cognitive functions, notably declarative memory (1) and spatial navigation (2), but also emotional reactivity, emotional memory, and resilience (3, 4). Hippocampal pathology is linked to multiple brain disorders, including Alzheimer’s disease (5), drug-resistant epilepsy (6), posttraumatic stress disorder (7), as well as schizophrenia (8). Contemporary accounts of the hippocampus suggest that its wide-ranging role in cognition emerges from its status as a cortical hub (9, 10). The hippocampus forms abundant connections with regions of anterior and posterior cortex that allow it to coordinate widespread network activity. Despite its relevance for both theoretical and applied domains of neuroscience, the structural–functional principles that govern how the hippocampus is embedded within the larger cortical system remain to be established in humans.
Understanding the hippocampus in the broader cortical landscape is complicated by the microstructure of the region itself. A large body of postmortem anatomical evidence from humans and animals shows the hippocampus is composed of cytoarchitectonically different subfields, notably the subiculum, cornu ammonis (CA1–CA4), and dentate gyrus (DG). These subfields follow the hippocampal infolding in a medial/lateral fashion, and each subfield plays a specific role within the hippocampal circuitry suggested by largely distinct anatomical connections to other regions (11–14). In addition to the between-subfield differences, converging evidence from animal electrophysiology (15) and human neuroimaging suggests a functional distinction between anterior and posterior hippocampal segments (16), a pattern also referred to as “long-axis specialization.”
Recent advances in high-field magnetic resonance imaging (MRI) acquisition offer the possibility to delineate hippocampal subfields in vivo (17, 18) and to document its subregional anatomy in awake, healthy adults. Initiatives such as the Human Connectome Project (HCP) (19) have made it feasible to integrate markers of tissue microstructure, such as the ratio of T1- over T2-weighted images (T1w/T2w) (20), with macroscale connectivity information obtained from resting-state functional MRI (rs-fMRI) in large populations of participants. In the neocortex, studies integrating these complementary features have leveraged the potential of “multiscale” neuroimaging to characterize localized properties of individual regions with unprecedented detail (21) and to identify overarching topographic principles that govern the interplay between structure and function (22).
The current work used high-resolution structural and functional neuroimaging to identify microstructural features of hippocampal organization in healthy participants. Taking advantage of the openly accessible HCP dataset, we identified common and distinct patterns of intrinsic functional connectivity across different subfields. Using these data, we applied manifold learning techniques (23) to map local transitions in connectivity across thousands of subfield surface points. Our analysis revealed a principal gradient of connectivity that corresponded to established anatomical landmarks along the hippocampal long axis, and that was reproducible using metaanalytical coactivations derived from the task based functional MRI literature. Metaanalytic decoding supported an anterior-to-posterior functional gradient, emphasizing domains related to memory and emotional reactivity in anterior segments. A second gradient along a medial/lateral axis, followed hippocampal folding and strongly correlated with T1w/T2w, suggesting the possibility of interactions between tissue microstructure and macroscale function. Our results were reproducible across hemispheres, within subjects across different scanning sessions, and in an independent replication HCP subsample.
Results
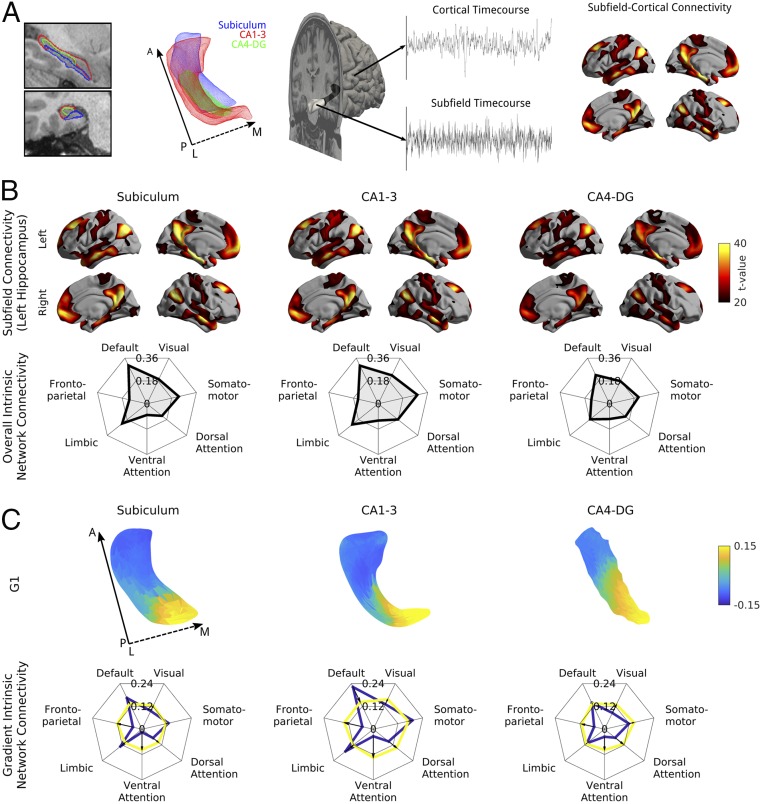
We studied the HCP S900 release, an open-access neuroimaging data aggregation and dissemination initiative (19). Using a recently developed technique, we automatically segmented hippocampal subfields (i.e., subiculum, CA1–3, and CA4–DG) from the HCPs high-resolution T1-weighted MRI data (17) and generated medial surfaces that run through the core of each subfield (24). These medial surfaces allowed for surface-wide sampling of T1w/T2w intensity (a marker of cortical microstructure) and rs-fMRI time series. We computed Pearson correlations between rs-fMRI time series of each hippocampal surface point and each neocortical region, as defined by a previous multimodal parcellation that integrated functional and anatomical markers (21). This generated hippocampal–neocortical connectomes, which describe the coupling of functional signals between all hippocampal subfield locations and all cortical regions (Fig. 1A). We selected one group of unrelated healthy young adults [n = 217 (122 women), mean ± SD age = 28.5 ± 3.7 y] as a discovery dataset and used another group [n = 134 (77 women), age = 28.7 ± 3.8 y] for validation. Details on the subject selection can be found in SI Appendix, Supplementary Materials and Methods and Fig. S1.
Fig. 1.
Analysis of functional connectome embedding of left hippocampal subfields. (A) Subfield-wide connectivity analysis. Segmentations and surfaces were automatically extracted for subiculum (blue), CA1–3 (red), and CA4–DG (green). Segmentations are shown in a T1w scan and as a mesh from a superior view with solid and dashed arrows denoting posterior (P) to anterior (A) and lateral (L) to medial (M) directions, respectively. rs-fMRI time series were extracted along the medial surfaces of each of these subfields and the neocortical surface. The mean time series of each subfield was computed and correlated with all cortical vertices resulting in a subfield-specific connectivity map. (B) Common and distinct functional connectivity of left hemispheric subfields (Left column, subiculum; Center column, CA1–3; Right column, CA4–DG) and neocortex as well as their connectivity to seven intrinsic networks, derived from a previous functional community detection (31). Surface-based findings were corrected for multiple comparisons and additionally thresholded at t > 20 to highlight only the most prominent connections. (C) The first principal component of intrinsic functional connectivity along hippocampal subfields (Left column, subiculum; Center column, CA1–3; Right column, CA4–DG) describes an anterior/posterior gradient. Based on a hippocampal–cortical connectivity matrix, we performed diffusion map embedding, an unsupervised manifold learning technique. The surfaces display the loadings of the first component, and the spider plots show connectivity patterns of the bottom (anterior, blue) and top (posterior, yellow) 25% of vertices to the seven intrinsic functional communities. Solid and dashed arrows denote posterior to anterior and lateral to medial direction, respectively.
The First Gradient of Functional Connectivity Describes the Hippocampal Long Axis.
In line with recent between-subfield functional connectivity analyses (25), there were differences in overall subfield connectivity profiles (SI Appendix, Fig. S2); however, all were closely integrated within default mode and temporolimbic networks (Fig. 1B). To study spatial transitions of hippocampal connectivity to neocortical regions across individual subfield surface points, we used a nonlinear diffusion embedding technique that was previously applied to map spatial gradients in neocortical connectivity (26). The first three gradients (G1–G3) were analyzed, as these explained 52% of variance and corresponded to the clearest elbow in the scree plot (SI Appendix, Fig. S3). G1 described an anterior/posterior axis across the hippocampus as a whole (Fig. 1C) and explained 31% of variance (SI Appendix, Fig. S3). Anterior regions had greater connectivity to default mode, limbic, and somatomotor areas, while posterior regions showed greater connectivity with posterior areas, especially visual and dorsal/ventral attention networks. To confirm that G1 related to long-axis specialization, we verified its correspondence with landmarks of anterior/posterior anatomy using manually drawn segmentations of hippocampal head, body, and tail from an independent dataset (27) (Fig. 2).
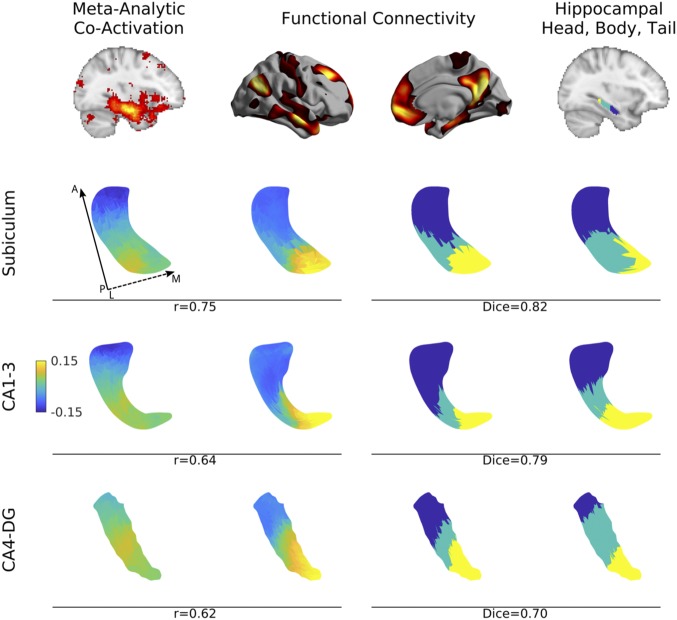
Fig. 2.
Long-axis specialization of the left subiculum (second row), CA1–3 (third row), and CA4–DG (fourth row) across different modalities. The principal gradient of both metaanalytic task-fMRI coactivation (first column) and rs-fMRI (second column) ran in anterior/posterior direction. K-means clusters (k = 3) of the rs-fMRI connectivity derived gradient (third column) overlapped strongly with manual segmentations of hippocampal head, body, and tail based on a previous atlas (27) (fourth column). Correlation coefficient values denote the association between metaanalytic coactivation and functional connectivity within each subfield. Dice indices denote geometric overlaps between k-means clusters of functional connectivity and the hippocampal head, body, and tail. Hippocampal surfaces are shown from a superior view. Solid and dashed arrows denote posterior to anterior and lateral to medial direction, respectively. For findings in the right hemisphere, see SI Appendix, Fig. S4.
Applying k-means (k = 3) clustering on G1 achieved the highest overlap with the anatomy-based tripartite subdivision of the hippocampus (Dice = 0.78 over all subfields, Fig. 2), while overlaps for G2 (Dice = 0.46) and G3 (Dice = 0.42) were numerically smaller. Findings were similar in the right hippocampus (SI Appendix, Fig. S4). To further verify the correspondence between G1 and long-axis specialization, we performed automated coactivation analysis of task-based fMRI literature (28). Specifically, we applied diffusion embedding to metaanalytical coactivation matrices based on 11,406 studies, with identical parameters as for rs-fMRI gradient mapping. This analysis also revealed a principal gradient in long-axis direction, which correlated strongly vertex-wise with the G1 derived from rs-fMRI (left, r = 0.66; right, r = 0.63; Fig. 2) but less so with the corresponding G2 (left, r = 0.28; right, r = 0.27) and G3 (left, r = 0.42; right, r = 0.36) as assessed by Steiger’s test (all differences significant at P < 0.01) (29).
To assess cognitive differences across the principal gradient, we performed an automated reverse inference of the top and bottom 33% of the gradient using Neurosynth (28). Overall, functional terms correlated more strongly with the anterior hippocampus than with its posterior counterpart (SI Appendix, Fig. S5). Specifically, the posterior hippocampus has somewhat weaker associations with memory-related terms, and far weaker associations with emotion-related terms. This corresponds with results of anterior/posterior functional connectivity to intrinsic networks (Fig. 1C), which indicated less distinctive temporolimbic and default mode connectivity of the posterior hippocampus.
The Second Principal Gradient Reflects Internal Hippocampal Microstructure.
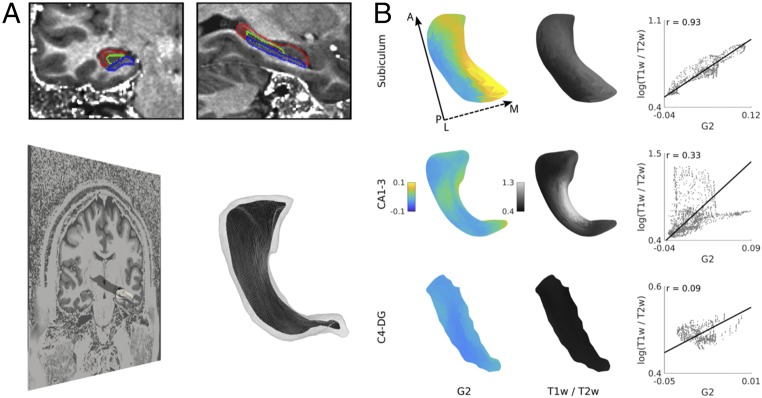
G2 explained 12% of variance (SI Appendix, Fig. S3) and described a medial/lateral gradient that largely followed hippocampal infolding, with highest loadings in subiculum, lowest in CA4–DG, and intermediary loadings in CA1–3 and correlated strongest with T1w/T2w intensity (Fig. 3, Left and see SI Appendix, Fig. S6, Right). Despite high significance of the overall model testing for an association between G2 and T1w/T2w intensity (P < 0.001), structure/function correlations differed across subfields, with G2 correlating highest to T1w/T2w in subiculum (left, r = 0.93; right, r = 0.87), followed by CA1–3 (r = 0.33; r = 0.29) and CA4–DG (r = 0.09; r = 0.45); with differences in CA4–DG possibly relating to low G2 variance causing unstable correlation estimates (SI Appendix, Figs. S3 and S6). Across subfields, correlations between G2 and T1w/T2w (left, r = 0.46; right, r = 0.45) were higher than those between the other gradients and T1w/T2w, as assessed by Steiger’s test (29) (G1 left, r = 0.01; G1 right, r = 0.23; G3 left, r = 0.19; G3 right, r = 0.14; all differences significant at P < 0.01). To address specificity to cortical microstructure, we repeated our analyses after correcting for local columnar volume (an index of local gray matter morphology), partial volume effects for cerebrospinal fluid (derived from a Gaussian mixture tissue classification) (30), and estimated rs-fMRI temporal signal-to-noise ratio (31). Notably, correlations between G2 and T1w/T2w remained significant after these corrections. We also correlated T1w/T2w of our HCP subjects to hippocampal quantitative T1 relaxation maps (qT1) from an independent sample of healthy adults from a previous study (32). Despite some variability across subfields, mean hippocampal T1w/T2w correlated significantly to qT1 (CA1–3: left/right, r = −0.85/−0.82; subiculum, r = −0.55/0.38; CA4–DG, r = −0.17/−0.47; see SI Appendix, Fig. S7).
Fig. 3.
Association between left hippocampal second gradient and T1w/T2w intensities. (A) To assess the association between functional gradients and hippocampal microstructure, we mapped hippocampal segmentations to T1w/T2w images and extracted T1w/T2w intensities at each vertex of the hippocampal medial surfaces. (B) Systematic correlation analyses indicated highest correlations between surface-sampled T1w/T2w and the second gradient, which runs along the hippocampal infolding. Solid and dashed arrows denote posterior to anterior and lateral to medial direction, respectively. For findings in the right hemisphere, see SI Appendix, Fig. S6.
The third principal component, G3, explained a further 10% in connectome variance (SI Appendix, Fig. S3), but it did not follow as clear a pattern as G1 and G2, likely representing a mixture of both. Indeed, it described both anterior/posterior (specifically in subiculum and CA4–DG) and medial/lateral gradients (in CA1–3 and anterior subiculum).
Reliability and Reproducibility.
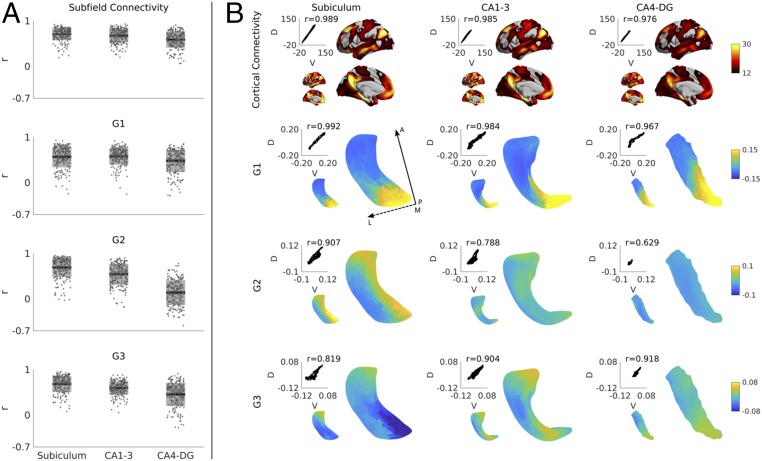
Overall subfield-to-cortex connectivity and within-subfield gradient findings were consistent across the four different scans in the HCP dataset, demonstrating excellent test/retest stability (Fig. 4A). Highest stability was found in subiculum, intermediary in CA1–3, and lowest in CA4–DG. When directly comparing discovery and validation cohorts (Fig. 4B), we obtained markedly similar results likely due to the large sample size and long scanning time allowing group-level averages to approach the population average. Again, findings were most consistent for subiculum (r > 0.82), followed by CA1–3 (r > 0.79), and finally CA4–DG (r > 0.63). Notably, we repeated gradient clustering and T1w/T2w correlation analyses and found comparable results. Specifically, the three-cluster subdivision of G1 overlapped most strongly with hippocampal head, body, and tail (Dice = 0.73), and G2 correlated most strongly and significantly with T1w/T2w (P < 0.001).
Fig. 4.
(A) Test/retest stability of left hippocampal connectivity and gradients and (B) reproducibility in an independent HCP subsample. Dark gray zones in the boxplots denote 95% confidence intervals; light gray zones denote one SD. Small and large surfaces represent data of the discovery (D) and validation (V) cohorts, respectively. Scatterplots show the vertex-wise correspondence between groups, with Pearson correlation values denoted above the scatterplot. Solid and dashed arrows denote posterior to anterior and medial to lateral direction, respectively.
Although our main findings were based on hippocampal–cortical connectomes, we could reconstruct virtually identical gradients when also incorporating subcortical regions (brainstem, amygdala, caudate nucleus, nucleus accumbens, putamen, pallidum, and thalamus) into the target regions. In fact, both left and right hemispheric G1–G3 based on whole-brain target regions showed correlations >0.85 with the original hippocampal–cortical gradients, indicating stability (SI Appendix, Fig. S8).
Discussion
For decades, the hippocampus has been considered a model system in neuroscience for understanding how local structure and circuit properties interact to produce cognition. Despite the progress made by cytoarchitectonic studies and animal electrophysiology, its complex anatomy has challenged targeted neuroimaging investigations of its substructural organization in living humans. Harnessing recently developed hippocampal segmentation techniques on high-resolution MRI data, we identified topographic and anatomical principles that govern how different hippocampal subfields are embedded in macroscale functional networks. Specifically, we utilized connectome compression techniques to map axes of spatial variations in connectivity at a scale below that of individual subfields and performed systematic multimodal correlations that detailed their association with tissue microstructure and segmental hippocampal anatomy. We identified two principal gradients of functional connectivity that ran in anterior/posterior and medial/lateral direction, respectively. A series of validation experiments confirmed that the principal gradient corresponded to long-axis anatomical subdivisions as well as to metaanalytical gradients, while the second correlated highly with surface-based markers of intracortical microstructure. These associations were consistent in both left and right hippocampus, and occurred above and beyond differences in the exact connectivity profiles of particular subfields. Collectively, our findings provide evidence for two main axes of substructural organization in the human hippocampus and suggest a close association between macroscale connectome integration, hippocampal long-axis organization, and allocortical microstructure.
Increased availability of high-resolution MRI has highlighted the possibility for segmenting subfields in vivo using both manual and automatic techniques (17, 33, 34), permitting detailed investigations of hippocampal organization. Specifically, submillimeter resolution provided by 3T and beyond can resolve strata rich in white matter as well as the hippocampal sulcus, landmarks reliably separating CA subfields and subiculum from DG. Based on subfields that were automatically segmented in the HCP dataset (17, 18), we employed advanced postprocessing techniques to generate medial surface representations running through the core of each (24). This approach allowed for a spatially specific multiparameter sampling throughout the entire hippocampus, while retaining its interlocked anatomical organization. By unfolding the hippocampus, we could systematically assess connectivity for each subfield surface point and thus move beyond paradigms that assessed whole-hippocampal (35) or whole-subfield connectivity (25). The dense and continuous representation of hippocampal subfield surface connectivity offered a substrate for diffusion map embedding, a nonlinear dimension reduction algorithm operating on connectomes that was recently applied to describe the topography of neocortical rs-fMRI connections (23). In the hippocampus, this data-driven approach revealed a primary axis of connectivity variation along the anterior/posterior axis in all subfields, converging with a large body of animal work showing gradual changes in anatomical connectivity, gene expression, and electrophysiological properties along the hippocampal long axis (15). Coregistering MRI landmarks (i.e., hippocampal head, body, and tail) from an independent dataset (27), we demonstrated a robust correspondence to macroscopic segments of long-axis anatomy. Furthermore, gradient mapping based on Neurosynth-derived metaanalytical coactivations from previously published fMRI studies (28) recovered a similar principal gradient, supporting the well-established correspondence of task-based and task-free networks (36, 37) and anchoring our rs-fMRI findings to task data supporting long-axis specialization in humans (38). Notably, Neurosynth-based reverse inference suggested gradual shifts in both memory-related terms as well as terms relating to emotional reactivity between anterior and posterior segments, providing a cognitive and affective basis for the observed functional gradients. While our results provide a first outlook at how connectivity, microstructure, and functional substrates of cognitive processes may covary along hippocampal subfields, future studies that combine task-free and task-based neuroimaging paradigms tailored to mesiotemporal lobe functions in an adequately powered sample will provide additional opportunities to validate and expand our approach.
Since classic cytoarchitectonic studies, neuroanatomists have emphasized the interplay between local tissue properties and macroscopic connectivity of individual brain regions in understanding their underlying functional roles. In fact, numerous studies have integrated both features for areal boundary characterization (21, 39, 40). In the hippocampus, animal studies have shown that cytoarchitectonically defined subfields express selective connectivity fingerprints to other nodes within the hippocampus and beyond (12). Subiculum has long been recognized as one of the main interfaces of the hippocampus, with bidirectional interconnections to entorhinal and other cortical areas (41, 42). The DG, on the other hand, is thought to act mainly as an input structure, which subsequently relays neural information into a CA3–CA1 pathway, possibly instantiating computations related to pattern separation and completion (14). Different hippocampal subfields have unique developmental trajectories, specifically with respect to internal cortical myelination. DG and subiculum have a more protracted developmental time course, while CA subfields undergo a more rapid myeloarchitectural maturation (43, 44). The diverging developmental patterns of individual subfields may interact with the processes that produce long-axis structural/functional specialization, as can be seen in age-related changes in both children and adults in functional activation patterns (45) and structural markers (46). To bridge local microstructural properties of the hippocampus with macroscale connectivity information in vivo, we related rs-fMRI connectome gradients to surface-sampled T1w/T2w image intensity, a proxy for intracortical myelin (20). Although the exact contribution of cortical myelin to T1w/T2w is not fully understood, hippocampal T1w/T2w values sampled in the current work resembled gradual changes in R1, the inverse of quantitative T1 relaxation times (R1 = 1/qT1), which we recently mapped in a different sample of healthy controls and patients with drug-resistant epilepsy (32). Importantly, the topography of T1w/T2w findings resembled the second connectome gradient, describing mainly a medial/lateral pattern. Notably, this association remained significant after extensive corrections for potential confounds, including morphology, partial volume effects, and signal-to-noise ratio, indicating a specific structure/function link between G2 and markers of intracortical microstructure. Although there is currently no established methodology to accurately and reliably map intrahippocampal fibers on HCP-style in vivo MRI data, the differentiation across subfields inherent to G2 may also reflect the course of internal hippocampal circuits. In particular, the perforant pathway is believed to run from entorhinal regions toward the subicular complex, which it may perforate to reach CA and subsequently DG subfields. A recent study has leveraged polarized light imaging to trace the course of the perforant pathway in human and nonhuman primate ex vivo data (47) and there has been work based on postmortem diffusion MRI data (48). While there are also previous applications based on in vivo MRI (47, 49), these studies have relied on specialized diffusion MRI sequences targeting the medial temporal lobe. Future work could combine such targeted imaging with rs-fMRI to assess the relationship between fine-grained intrahippocampal circuity and functional gradients obtained from resting-state connectivity.
The HCP initiative offers an open repository to study connectomic principles in healthy young adults using state-of-the-art neuroimaging data, with a sample size large enough for both discovery and validation. Our test/retest stability analysis within subjects and reanalysis of an independent HCP subsample provided an optimistic outlook on robustness of our results. Several other open access initiatives with a similar emphasis on high quality imaging and anatomically meaningful processing are emerging. Moving forward, these will help to address generalizability and to assess alterations of the observed interplay between hippocampal microstructure and macroscale features during neurodevelopment, aging, and brain disorders.
Materials and Methods
A detailed description of the subjects inclusions, image processing, and analysis methodology can be found in SI Appendix, Supplemental Materials and Methods. In brief, we selected healthy adults from the HCP S900 release for whom all four rs-fMRI and structural scans were available. We selected two cohorts without family relationships, both within and between cohorts, and acceptable image quality: discovery [n = 217 (122 women), mean ± SD age = 28.5 ± 3.7 y] and validation [n = 134 (77 women), age = 28.7 ± 3.8 y].
All MRI data used in this study were publicly available and anonymized. Participant recruitment procedures and informed consent forms, including consent to share deidentified data, were previously approved by the Washington University Institutional Review Board as part of the HCP.
Based on high-resolution T1-weighted images, we segmented CA1–3, CA4–DG, and subiculum using a patch-based algorithm in every subject (17). The algorithm employs a population-based patch normalization relative to a template library, which offers good time and space complexity. Notably, by operating on T1-weighted images only, the currently preferred anatomical contrast of many big data MRI initiatives, it avoids reliance on T2-weighted MRI data, a modality that may be prone to motion and flow artifacts, and that may be susceptible to intensity changes due to pathological changes in the hippocampal formation. In previous validations, this algorithm has shown high segmentation accuracy of hippocampal subfields (17). We then generated surfaces running through each subfield’s core (24), which allowed for the sampling of rs-fMRI time series and for hippocampal unfolding. We also sampled cortical time series using the surfaces provided by HCP and subcortical time series using segmentations from FSL FIRST (50). We correlated hippocampal and cortical time series, and used Fisher z transformations to render correlation coefficients more normally distributed. Subfield connectivity in Fig. 1B was mapped using linear and mixed-effects models in SurfStat [www.math.mcgill.ca/keith/surfstat/ (51)]. Diffusion embedding (ref. 26; Matlab code: https://github.com/MICA-MNI/micaopen/) identified principal gradients in rs-fMRI connectivity along subfield surfaces, with the anterior/posterior gradient shown in Fig. 1C and the medial/lateral gradient shown in Fig. 3B. We repeated diffusion embedding based on metaanalytical coactivation maps derived from Neurosynth in Fig. 2 (28).
To assess the relation between functional organization, hippocampal anatomy, and microstructure, we related rs-fMRI gradients to manual segmentations of hippocampal head, body, and tail in Fig. 2 (27) and to surface-sampled T1w/T2w intensity in Fig. 3B, a proxy for myelin content (20) (see also comparison between HCP-derived T1w/T2w intensities and quantitative T1 relaxation times from ref. 27) (SI Appendix, Fig. S7). Findings were consistent in the left and right hippocampus (SI Appendix, Figs. S2–S6, for right hemisphere findings). We demonstrated test/retest stability in all individuals from the discovery cohort in Fig. 4A, by correlating connectivity and gradients maps between two scans within each subject to the other two. Furthermore, we assessed reproducibility, by correlating subfield connectivity and gradient maps between the discovery and validation dataset in Fig. 4B.
Supplementary Material
Acknowledgments
R.V.d.W. was supported by the McGill Faculty of Medicine; S.L. was supported by a Jeanne Timmins Costello Fellowship; S.-J.H. was supported by the Canadian League Against Epilepsy; N.B. and A.B. were supported by Canadian Institutes of Health Research (CIHR) and received salary support from the Fonds de la Recherche du Quebec–Santé (FRQS). B.C.B. acknowledges research support from the National Science and Engineering Research Council of Canada (NSERC Grant Discovery 1304413), the Canadian Institutes of Health Research (CIHR Grant FDN-154298), SickKids Foundation (NI17-039), and Azrieli Center for Autism Research of the Montreal Neurological Institute, as well as salary support from FRQS (Chercheur Boursier). J.S. was supported by the European Research Council (Grant WANDERINGMINDS - ERC646927). Data were provided, in part, by the Human Connectome Project, Washington University, the University of Minnesota, and Oxford University Consortium (Principal Investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: This manuscript is based on open-access data from the Human Connectome Project, accessible via https://db.humanconnectome.org/app/template/Login.vm;jsessionid=BB92BF16664CB083801A9DD5140BF26B.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803667115/-/DCSupplemental.
References
- 1.Penfield W, Milner B. Memory deficit produced by bilateral lesions in the hippocampal zone. AMA Arch Neurol Psychiatry. 1958;79:475–497. doi: 10.1001/archneurpsyc.1958.02340050003001. [DOI] [PubMed] [Google Scholar]
- 2.Burgess N, Maguire EA, O’Keefe J. The human hippocampus and spatial and episodic memory. Neuron. 2002;35:625–641. doi: 10.1016/s0896-6273(02)00830-9. [DOI] [PubMed] [Google Scholar]
- 3.Phelps EA. Human emotion and memory: Interactions of the amygdala and hippocampal complex. Curr Opin Neurobiol. 2004;14:198–202. doi: 10.1016/j.conb.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 4.Franklin TB, Saab BJ, Mansuy IM. Neural mechanisms of stress resilience and vulnerability. Neuron. 2012;75:747–761. doi: 10.1016/j.neuron.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 5.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 6.Mouritze Dam A. Hippocampal neuron loss in epilepsy and after experimental seizures. Acta Neurol Scand. 1982;66:601–642. doi: 10.1111/j.1600-0404.1982.tb04528.x. [DOI] [PubMed] [Google Scholar]
- 7.Karl A, et al. A meta-analysis of structural brain abnormalities in PTSD. Neurosci Biobehav Rev. 2006;30:1004–1031. doi: 10.1016/j.neubiorev.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Altshuler LL, et al. An MRI study of temporal lobe structures in men with bipolar disorder or schizophrenia. Biol Psychiatry. 2000;48:147–162. doi: 10.1016/s0006-3223(00)00836-2. [DOI] [PubMed] [Google Scholar]
- 9.Moscovitch M, Cabeza R, Winocur G, Nadel L. Episodic memory and beyond: The hippocampus and neocortex in transformation. Annu Rev Psychol. 2016;67:105–134. doi: 10.1146/annurev-psych-113011-143733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Squire LR, Stark CE, Clark RE. The medial temporal lobe. Annu Rev Neurosci. 2004;27:279–306. doi: 10.1146/annurev.neuro.27.070203.144130. [DOI] [PubMed] [Google Scholar]
- 11.Hodgetts CJ, et al. Ultra-high-field fMRI reveals a role for the subiculum in scene perceptual discrimination. J Neurosci. 2017;37:3150–3159. doi: 10.1523/JNEUROSCI.3225-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Strien NM, Cappaert NL, Witter MP. The anatomy of memory: An interactive overview of the parahippocampal-hippocampal network. Nat Rev Neurosci. 2009;10:272–282. doi: 10.1038/nrn2614. [DOI] [PubMed] [Google Scholar]
- 13.Berron D, et al. Strong evidence for pattern separation in human dentate gyrus. J Neurosci. 2016;36:7569–7579. doi: 10.1523/JNEUROSCI.0518-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neunuebel JP, Knierim JJ. CA3 retrieves coherent representations from degraded input: Direct evidence for CA3 pattern completion and dentate gyrus pattern separation. Neuron. 2014;81:416–427. doi: 10.1016/j.neuron.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strange BA, Witter MP, Lein ES, Moser EI. Functional organization of the hippocampal longitudinal axis. Nat Rev Neurosci. 2014;15:655–669. doi: 10.1038/nrn3785. [DOI] [PubMed] [Google Scholar]
- 16.Poppenk J, Evensmoen HR, Moscovitch M, Nadel L. Long-axis specialization of the human hippocampus. Trends Cogn Sci. 2013;17:230–240. doi: 10.1016/j.tics.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 17.Caldairou B, et al. 2016. A surface patch-based segmentation method for hippocampal subfields. Medical Image Computing and Computer-Assisted Intervention, Lecture Notes in Computer Science (Springer, New York), pp 379–387.
- 18.Kulaga-Yoskovitz J, et al. Multi-contrast submillimetric 3 Tesla hippocampal subfield segmentation protocol and dataset. Sci Data. 2015;2:150059. doi: 10.1038/sdata.2015.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Essen DC, et al. WU-Minn HCP Consortium The WU-Minn Human Connectome Project: An overview. Neuroimage. 2013;80:62–79. doi: 10.1016/j.neuroimage.2013.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glasser MF, Van Essen DC. Mapping human cortical areas in vivo based on myelin content as revealed by T1- and T2-weighted MRI. J Neurosci. 2011;31:11597–11616. doi: 10.1523/JNEUROSCI.2180-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glasser MF, et al. A multi-modal parcellation of human cerebral cortex. Nature. 2016;536:171–178. doi: 10.1038/nature18933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huntenburg JM, et al. A systematic relationship between functional connectivity and intracortical myelin in the human cerebral cortex. Cereb Cortex. 2017;27:981–997. doi: 10.1093/cercor/bhx030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Margulies DS, et al. Situating the default-mode network along a principal gradient of macroscale cortical organization. Proc Natl Acad Sci USA. 2016;113:12574–12579. doi: 10.1073/pnas.1608282113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim H, et al. Multivariate hippocampal subfield analysis of local MRI intensity and volume: Application to temporal lobe epilepsy. Med Image Comput Comput Assist Interv. 2014;17:170–178. doi: 10.1007/978-3-319-10470-6_22. [DOI] [PubMed] [Google Scholar]
- 25.de Flores R, et al. Intrinsic connectivity of hippocampal subfields in normal elderly and mild cognitive impairment patients. Hum Brain Mapp. 2017;38:4922–4932. doi: 10.1002/hbm.23704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coifman RR, Lafon S. Diffusion maps. Appl Comput Harmon Anal. 2006;21:5–30. [Google Scholar]
- 27.Bernasconi N, et al. Mesial temporal damage in temporal lobe epilepsy: A volumetric MRI study of the hippocampus, amygdala and parahippocampal region. Brain. 2003;126:462–469. doi: 10.1093/brain/awg034. [DOI] [PubMed] [Google Scholar]
- 28.Yarkoni T, Poldrack RA, Nichols TE, Van Essen DC, Wager TD. Large-scale automated synthesis of human functional neuroimaging data. Nat Methods. 2011;8:665–670. doi: 10.1038/nmeth.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steiger JH. Tests for comparing elements of a correlation matrix. Psychol Bull. 1980;87:245–251. [Google Scholar]
- 30.Ashburner J, Friston KJ. Unified segmentation. Neuroimage. 2005;26:839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 31.Yeo BTT, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106:1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernhardt BC, et al. Preferential susceptibility of limbic cortices to microstructural damage in temporal lobe epilepsy: A quantitative T1 mapping study. Neuroimage. 2017 doi: 10.1016/j.neuroimage.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 33.Pipitone J, et al. Alzheimer’s Disease Neuroimaging Initiative Multi-atlas segmentation of the whole hippocampus and subfields using multiple automatically generated templates. Neuroimage. 2014;101:494–512. doi: 10.1016/j.neuroimage.2014.04.054. [DOI] [PubMed] [Google Scholar]
- 34.Yushkevich PA, et al. Automated volumetry and regional thickness analysis of hippocampal subfields and medial temporal cortical structures in mild cognitive impairment. Hum Brain Mapp. 2015;36:258–287. doi: 10.1002/hbm.22627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andrews-Hanna JR, Reidler JS, Sepulcre J, Poulin R, Buckner RL. Functional-anatomic fractionation of the brain’s default network. Neuron. 2010;65:550–562. doi: 10.1016/j.neuron.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith SM, et al. Correspondence of the brain’s functional architecture during activation and rest. Proc Natl Acad Sci USA. 2009;106:13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- 38.Strange BA, Fletcher PC, Henson RN, Friston KJ, Dolan RJ. Segregating the functions of human hippocampus. Proc Natl Acad Sci USA. 1999;96:4034–4039. doi: 10.1073/pnas.96.7.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carey D, Krishnan S, Callaghan MF, Sereno MI, Dick F. Functional and quantitative MRI mapping of somatomotor representations of human supralaryngeal vocal tract. Cereb Cortex. 2017;27:265–278. doi: 10.1093/cercor/bhw393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuehn E, et al. Body topography parcellates human sensory and motor cortex. Cereb Cortex. 2017;27:3790–3805. doi: 10.1093/cercor/bhx026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Mara S. The subiculum: What it does, what it might do, and what neuroanatomy has yet to tell us. J Anat. 2005;207:271–282. doi: 10.1111/j.1469-7580.2005.00446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Witter MP, Groenewegen HJ. The subiculum: Cytoarchitectonically a simple structure, but hodologically complex. Prog Brain Res. 1990;83:47–58. doi: 10.1016/s0079-6123(08)61240-6. [DOI] [PubMed] [Google Scholar]
- 43.Benes FM, Turtle M, Khan Y, Farol P. Myelination of a key relay zone in the hippocampal formation occurs in the human brain during childhood, adolescence, and adulthood. Arch Gen Psychiatry. 1994;51:477–484. doi: 10.1001/archpsyc.1994.03950060041004. [DOI] [PubMed] [Google Scholar]
- 44.Abrahám H, et al. Myelination in the human hippocampal formation from midgestation to adulthood. Int J Dev Neurosci. 2010;28:401–410. doi: 10.1016/j.ijdevneu.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 45.Langnes E, et al. 2018. Development and decline of the hippocampal long-axis specialization and differentiation during encoding and retrieval of episodic memories. bioRxiv:10.1101/323097. Preprint, posted May 15, 2018.
- 46.DeMaster D, Pathman T, Lee JK, Ghetti S. Structural development of the hippocampus and episodic memory: Developmental differences along the anterior/posterior axis. Cereb Cortex. 2014;24:3036–3045. doi: 10.1093/cercor/bht160. [DOI] [PubMed] [Google Scholar]
- 47.Zeineh MM, et al. Direct visualization and mapping of the spatial course of fiber tracts at microscopic resolution in the human hippocampus. Cereb Cortex. 2017;27:1779–1794. doi: 10.1093/cercor/bhw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Augustinack JC, et al. Direct visualization of the perforant pathway in the human brain with ex vivo diffusion tensor imaging. Front Hum Neurosci. 2010;4:42. doi: 10.3389/fnhum.2010.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yassa MA, Muftuler LT, Stark CE. Ultrahigh-resolution microstructural diffusion tensor imaging reveals perforant path degradation in aged humans in vivo. Proc Natl Acad Sci USA. 2010;107:12687–12691. doi: 10.1073/pnas.1002113107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Patenaude B, Smith SM, Kennedy DN, Jenkinson M. A Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage. 2011;56:907–922. doi: 10.1016/j.neuroimage.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Worsley KJ, et al. SurfStat: A Matlab toolbox for the statistical analysis of univariate and multivariate surface and volumetric data using linear mixed effects models and random field theory. Neuroimage. 2009;47(Suppl 1):S102. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.