Abstract
Background
Meningiomas are adult brain tumors originating in the meningeal coverings of the brain and spinal cord, with significant heritable basis. Genome-wide association studies (GWAS) have previously identified only a single risk locus for meningioma, at 10p12.31.
Methods
To identify a susceptibility locus for meningioma, we conducted a meta-analysis of 2 GWAS, imputed using a merged reference panel from the 1000 Genomes Project and UK10K data, with validation in 2 independent sample series totaling 2138 cases and 12081 controls.
Results
We identified a new susceptibility locus for meningioma at 11p15.5 (rs2686876, odds ratio = 1.44, P = 9.86 × 10–9). A number of genes localize to the region of linkage disequilibrium encompassing rs2686876, including RIC8A, which plays a central role in the development of neural crest-derived structures, such as the meninges.
Conclusions
This finding advances our understanding of the genetic basis of meningioma development and provides additional support for a polygenic model of meningioma.
Keywords: genome-wide association study, meningioma, polygenic, risk, single-nucleotide polymorphism
Importance of the study
Meningiomas are adult tumors arising in the meninges and account for around a third of all primary brain tumors. Evidence for common genetic variation contributing to meningioma predisposition has been provided by a GWAS, which identified a risk locus at chromosome 10p12.31. To gain further insight into the inherited susceptibility of meningioma, we performed a meta-analysis of 2 GWAS and 2 independent validation series comprising 2138 cases and 12081 controls, and report the identification of a new risk locus for meningioma at 11p15.5. A number of genes localize to this locus, including RIC8A, which plays a central role in the development of neural crest-derived structures, such as the meninges. This is only the second study, and the largest, to robustly associate common genetic variation as a risk factor for meningioma.
Meningiomas are adult tumors arising in the membranous layers surrounding the brain and spinal cord and account for around a third of all primary brain tumors.1–3 The incidence of meningioma is 2-fold higher in females than in males, and the disease is more common in individuals with African ancestry.1 Although mortality rates are relatively low, meningioma is associated with substantial morbidity.
Compared with malignant glial tumors, meningioma has been relatively understudied with regard to etiologic risk factors. Indeed, excluding exposure to ionizing radiation, no environmental factor has consistently been associated with tumor risk.2,3 Evidence for an inherited predisposition to meningioma is provided by the elevated risk seen in neurofibromatosis4 and Gorlin syndrome.5 While the risk of meningioma associated with these disorders is high, they are rare and collectively contribute little to the 3-fold increased risk of the tumor in the relatives of meningioma patients.6,7 Evidence for common genetic variation contributing to meningioma predisposition has been provided by a genome-wide association study (GWAS),8,9 which identified a risk locus at chromosome 10p12.31.10,11
To gain a further insight into inherited susceptibility to meningioma, we performed a meta-analysis of a previously published GWAS10 and a new unpublished GWAS, thereby providing increased study power to identify new risk loci and reduce the likelihood of false positives.12 Following replication genotyping in 2 additional independent series, we report the identification of a new risk locus for meningioma mapping to chromosome 11p15.5.
Methods
Ethics
Collection of patient samples and associated clinicopathological information in this study was completed with written informed consent and relevant ethical review board approval at the respective centers in accordance with the tenets of the Declaration of Helsinki. Specifically, these centers are for the German-GWAS: the ethics committees of the Medical Faculty of the University of Bonn and University Hospital Essen; USA-GWAS: the institutional review boards at Yale University School of Medicine, Brigham and Women’s Hospital, the University of California at San Francisco, The MD Anderson Cancer Center, Duke University School of Medicine, the Kaiser Foundation Research Institute, and the State of Connecticut Department of Public Health Human Investigation Committee; UK-replication: the South East Multicentre Research Ethics Committee and the Scottish Multicentre Research Ethics Committee; Danish-replication: the Danish ethical committee system, the Danish Data Protection Board, and the Danish Ministry of Justice.
Genome-Wide Association Studies
This meta-analysis was completed based on 2 GWAS datasets (Supplementary Table S1). The diagnosis of meningioma (ICD-10 D32/C70) was established in accordance with World Health Organization (WHO) guidelines.
The German-GWAS comprised 834 cases (250 male) and 2103 controls (1047 male). The German-GWAS case-control study has been described previously.10 Case subjects were patients who underwent surgery for meningioma at the University of Bonn Medical Center between 1996 and 2008. Control subjects were healthy individuals with no past history of malignancy from the Heinz Nixdorf Recall (HNR) study.13 DNA was extracted from samples using conventional methodologies and quantified using PicoGreen (Invitrogen). Genotyping of cases and controls was conducted using either Infinium HD Human660w-Quad or OmniExpress Beadchips according to the manufacturer’s protocols (Illumina).
The USA-GWAS comprised 772 cases (217 male) and 7720 controls (2966 male). Case patients eligible for the study included all persons diagnosed between 2006 and 2013 with a histologically confirmed intracranial meningioma among residents of the states of California, Connecticut, Massachusetts, North Carolina, and Texas. Case patients were diagnosed between the ages of 20 and 79 and were identified through the Rapid Care Ascertainment systems and state tumor registries at their respective study sites. Controls were obtained through random-digit dialing performed by an outside consulting firm (Kreider Research and Consulting) (n = 689) or are from The Resource for Genetic Epidemiology Research on Aging (GERA) cohort (n = 7031).14,15 Controls obtained through random-digit dialing were frequency matched with case patients by 5-year age interval, sex, and state of residence. Patients with a prior history of meningioma and/or a brain lesion of unknown pathology were not eligible for inclusion. The GERA cohort comprises 110266 adult members of the Kaiser Permanente Medical Care Plan, Northern California Region (KPNC). Participants were enrolled through participation in a mailed study conducted in 2007 of all KPNC adult members who had been members for more than 2 years. Respondents who completed consent forms were mailed saliva collection kits (Oragene). We sampled 7031 individuals from 56848 non-Hispanic white individuals whose data passed quality control for inclusion in the control group, to ensure 1:10 matching between cases and controls in the USA-GWAS, thereby optimizing study power, since there is little benefit of additional controls thereafter.16 Genotyping of cases and controls of all USA-GWAS subjects was completed using Affymetrix Axiom EUR arrays according to the manufacturer’s protocols.
Statistical Analysis
The quality control procedure described by Anderson et al17 was applied to each GWAS individually (Supplementary Table S1). To identify samples with discordant sex information, the mean homozygosity rate across X-chromosome markers was computed and samples were excluded if this rate contradicted the reported sex or was inconclusive (a rate between 0.2 and 0.8). We next excluded individuals if they exhibited an elevated genotype failure rate (>3%) or an outlying heterozygosity rate (±3 standard deviations from the mean). To identify duplicated or related individuals, the degree of shared ancestry between pairs of individuals was computed (using identity by descent, IBD). If a pair of individuals had an IBD score >0.185, then the individual with the lowest variant call rate was excluded. Individuals with a non-European ancestry were identified by merging data from 3 HapMap version II populations (CEU, JPT/CHB, and YRI) and conducting principal component analysis on the merged individuals. Individuals with a second principal component score less than 0.072 were excluded. Variants were excluded if they had a high missing data rate (>5%), if the genotyping call rates differed between the cases and the controls (P < 10−5 using Fisher’s exact test), if they had a minor allele frequency (MAF) <0.01, or if they deviated significantly from Hardy–Weinberg equilibrium (HWE, P < 10–5). Individuals were phased using SHAPEIT version 2.r837 software18 and a merged reference panel (EGAD00001000776, the European Genome-phenome Archive) containing data from the 1000 Genomes Project19 (Phase 3) and the UK10K.20 GWAS data were imputed to more than 10 million single nucleotide polymorphisms (SNPs) using IMPUTE version 2.3.021 and the same reference panel. Imputation was conducted separately for each of the studies. In each dataset, the data were pruned to the set of variants common to the cases and controls before imputation. Tests of association between the directly genotyped and imputed SNPs and meningioma were performed using logistic regression under an additive genetics model using SNPTEST version 2.5.2.22 Poorly imputed SNPs (information measure <0.8), SNPs with a low MAF (<0.005), and SNPs that deviated from HWE (P < 10−5) were excluded. To evaluate the possibility of differential genotyping of cases and controls and the adequacy of the case-control matching, quantile-quantile (Q-Q) plots of the test statistics were generated (Supplementary Figure 1). The computed inflation factor λ is based on the 90% least significant SNPs.23 In each study, the effects of population stratification were limited by including in the analysis the first 2 and 3 principal components for the German and USA series, respectively. Eigenvectors for each of the GWAS datasets were computed using EIGENSOFT version 4.2.24
Meta-analyses of the individual GWAS were completed using the β estimates and standard errors from each study and the fixed-effects inverse-variance method implemented in META version 1.7.25 Cochran’s Q-statistic and the I2 statistic were used to test for heterogeneity and estimate the proportion of the total variation that is due to heterogeneity.26 Meta-analysis was only completed for an SNP if it passed the quality thresholds in all considered GWAS. SNPTEST was used to perform conditional association analysis. SNP associations at P < 5 × 10−8 in the meta-analyses are considered genome-wide significant.27 Despite imposing a stringent significance threshold of P < 5 × 10−8 for declaring a GWAS association as being significant, it is possible that some such associations might still be false positives. To further assess the robustness of an association, Wakefield has proposed the application of an approximate Bayes factor to calculate the Bayes false discovery probability (BFDP).28 We estimated the BFDP based on a plausible odds ratio of 1.2 and a prior probability of 0.0001.29
Replication Studies
Ten promising SNP associations from the meta-analysis of the 2 GWAS were taken forward for de novo replication (Supplementary Table S2). Promising associations were prespecified as loci with SNP association P-values <10−5, which also had support from additional correlated SNPs mapping to the same genetic region (ie, r2 > 0.5 and P < 10−3). The UK-replication series comprised 439 cases (ICD10 D32/C70) from the INTERPHONE study30 and 1865 population-based controls with no past history of any malignancy, ascertained through the National Study of Colorectal Cancer Genetics.31 The Danish-replication series comprised 115 cases (ICD-O 9530–9537) from the INTERPHONE study and 411 controls with no past history of cancer, ascertained through the Danish Central Population Registry. Replication genotyping of UK and Danish samples was performed using allele-specific PCR KASP chemistry (LGC). Primers are detailed in Supplementary Table S3. Thirty-four samples were excluded from the UK-replication series for having 3 or more failed calls. Call rates for each genotyped SNP were >98% in the remaining UK samples. Six samples were excluded from the Danish-replication series due to the failed call of the genotyped SNP.
Sequencing
To assess the fidelity of imputation of rs7124615, a subset of 126 cases and 56 controls from the German-GWAS series, selected to be enriched for the presumptive T allele, were sequenced using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems) in conjunction with ABI 3700xl semi-automated sequencers (Applied Biosystems). We did not detect the presence of the T allele in any of the samples. The SNP rs7124615 maps to a highly repetitive region, suggesting that this SNP may be incorrectly annotated to this region. Primer sequences are detailed in Supplementary Table S3.
Heritability Analysis
We used genome-wide complex trait analysis (GCTA) to estimate the heritability ascribed to the genotyped SNPs across all autosomes and each individual autosome.32 SNPs were excluded based on high missing rate (>5%), low MAF (<0.01), or evidence of deviation from HWE (P < 0.05). Individuals identified as being closely related were also excluded. Restricted maximum likelihood analysis was run using a genetic relationship matrix for each pair of samples. The lifetime risk of meningioma was used to transform the estimated heritability to the liability scale, as previously advocated when calculating the heritability of common lethal diseases such as cancer.33 The lifetime risk of brain and nervous system tumors is 0.62%,34 meningioma accounts for 36% of primary brain tumors,35 and we therefore estimated the lifetime risk of meningioma to be 0.224%. We followed the methodology of Yang et al36 to adjust for incomplete linkage disequilibrium between the genotyped and causal SNPs at a range of MAF thresholds between 0.1 and 0.5. Heritability was estimated for the German and USA series individually and a meta-analysis of the results was completed under a fixed-effects model. We additionally used the phenotype correlation–genotype correlation (PCGC) regression method to estimate the heritability ascribed to the genotyped SNPs across all autosomes,37 using the genetic relationship matrix and lifetime risk estimate that was used with GCTA. We adjusted for population structure when estimating heritability using the GCTA and PCGC regression approaches by including as covariates the first 2 and 3 principal components for the German and USA series, respectively. Estimates of individual variance in risk associated with meningioma risk SNPs was carried out using the method described in Pharoah et al.38
Expression Quantitative Trait Loci Analysis
Publicly available data from 47 tissues from the Genotype-Tissue Expression (GTEx) project39 v7 release were used to examine the relationship between SNP genotype and gene expression. We set a significance threshold for the expression quantitative trait loci (eQTL) analysis of P < 2.01 × 10−5, corresponding to a Bonferroni correction for 2491 tests (53 genes across 47 tissues).
Summary-Level Mendelian Randomization Analysis
To examine the relationship between meningioma risk loci and gene expression we performed a summary-level Mendelian randomization (SMR) analysis, as per Zhu et al.40 Briefly, GWAS summary statistics files were generated from the meta-analysis. Reference files were generated using data from the 1000 Genomes Project (Phase 3) and UK10K. As previously advocated, only probes with at least one eQTL P-value of <5.0 × 10−8 were considered for SMR analysis. We set a threshold for the SMR test of PSMR < 1.01 × 10−4, corresponding to a Bonferroni correction for 496 tests (496 probes with a top eQTL P < 5.0 × 10−8 across 47 tissues). For the HEIDI (heterogeneity in dependent instruments) test, P-values <0.05 were taken to indicate significant heterogeneity.
Data Availability
Genotype data from GERA are available from dbGaP (accession phs000674.v2.p2). The 1000 Genomes Project and UK10K imputation panel data are available from the European Genome-phenome Archive (accession EGAD00001000776). Remaining data are available from the authors upon request.
Results
Association Analysis
We analyzed GWAS SNP data passing quality control for 1606 cases and 9823 controls of European ancestry from 2 studies: a previously reported GWAS of 834 cases and 2103 controls (German-GWAS)10 and a new GWAS of 772 cases and 7720 controls (USA-GWAS) from Yale University, Brigham and Women’s Hospital, The MD Anderson Cancer Center, Duke University School of Medicine, and the University of California San Francisco (Supplementary Tables S1 and S4). To increase genomic resolution, we used data from the 1000 Genomes Project and UK10K to impute >9 million SNPs. Q-Q plots for SNPs with a MAF >1% post imputation did not show evidence of substantive overdispersion (λ between 0.99 and 1.04; Supplementary Figure S1). We computed joint odds ratios and 95% CIs under a fixed-effects model for each SNP and associated per allele principal component corrected P-values for all cases versus controls from the 2 series (Fig. 1, Supplementary Figure 2).
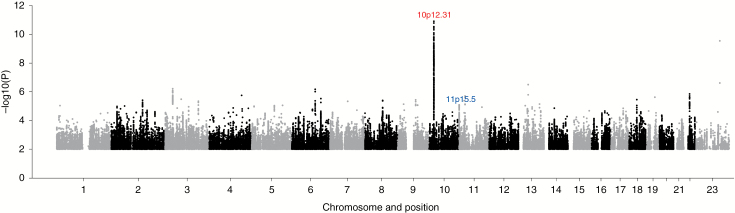
Fig. 1.
Manhattan plot of association P-values. Shown are the genome-wide P-values (2-sided) of >9 million successfully imputed SNPs in 1606 cases and 9823 controls. Text labeled in red is about previously identified risk loci and text labeled in blue is about newly identified risk loci. Imputation of rs7124615 was not supported by sequencing and this SNP is therefore not represented.
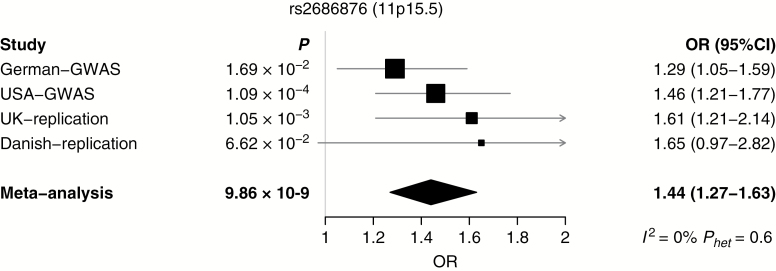
The strongest association was provided by SNP rs530000334 (P = 1.41 × 10−11), which maps to the previously identified risk locus at 10p12.31 (Fig. 1). Excluding the poorly imputed SNP rs7124615 at 11p15.5, no other association was genome-wide significant. We sought independent validation of promising associations (ie, P < 10−5) at 10 loci where support was provided by SNPs in linkage disequilibrium (r2 > 0.5 and P < 10−3) by genotyping additional case-control series from the UK and Denmark (Supplementary Table S2). In a combined analysis of the GWAS and replication datasets for these select SNPs, the only genome-wide association was shown by rs2686876, also at 11p15.5 (P = 9.86 × 10−9; Table 1, Fig. 2, Supplementary Table S2). The BFDP for this association was 1.8%, thereby supporting the robustness of the association. At both 11p15.5 and 10p12.31, a conditional analysis of SNP genotypes provided no evidence for additional independent signals at either risk locus.
Table 1.
Summary results for the SNP from the newly reported locus associated with meningioma risk
| SNP | Locus | Position (bp) * | Risk Allele | Study | Case RAF | Control RAF | OR (95% CI) | P-value |
|---|---|---|---|---|---|---|---|---|
| rs2686876 | 11p15.5 | 258909 | T | German-GWAS | 0.927 | 0.902 | 1.29 (1.05–1.59) | 1.69 × 10–2 |
| USA-GWAS | 0.938 | 0.910 | 1.46 (1.21–1.77) | 1.09 × 10–4 | ||||
| GWAS phase meta-analysis | 1.38 (1.20–1.59) | 8.19 × 10 –6 | ||||||
| UK-replication | 0.955 | 0.921 | 1.61 (1.21–2.14) | 1.05 × 10–3 | ||||
| Danish-replication | 0.943 | 0.905 | 1.65 (0.97–2.82) | 6.62 × 10–2 | ||||
| Replication phase meta-analysis | 1.62 (1.26–2.08) | 1.73 × 10 –4 | ||||||
| Combined GWAS/replication phase meta-analysis | 1.44 (1.27–1.63) | 9.86 × 10 –9 |
RAF, risk allele frequency; OR, odds ratio; CI, confidence interval. *Position is based on NCBI build 37.
Fig. 2.
Forest plot of effect size and direction for the SNP from the newly reported locus associated with meningioma risk.
Most meningiomas (>80%) are WHO grade I tumors, with the remainder grade II (atypical, 15%) and grade III (anaplastic) meningioma41; males are more likely than females to have atypical or aggressive lesions. We assessed the relationship between 11p15.5 genotype and WHO grade, sex, and age at diagnosis by case-only analysis. WHO grade was not available for all USA-GWAS, UK-replication, and Danish-replication cases and therefore the WHO grade case-only analysis was restricted to the German-GWAS cases. Case-only analyses of sex and age at diagnosis were conducted in all series. These analyses provided no evidence for association between rs2686876 and WHO grade, sex, or age at diagnosis, consistent with a generic effect of genotype on meningioma risk (Supplementary Table S5).
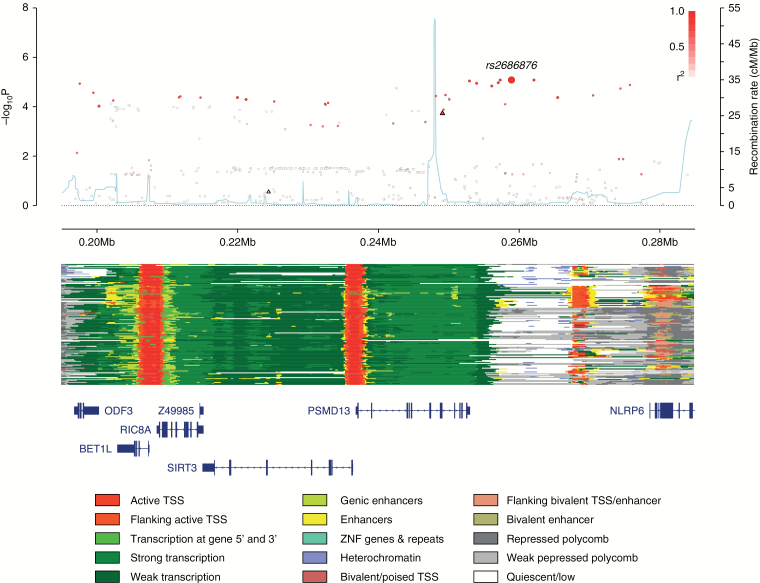
A number of genes localize to the region of linkage disequilibrium encompassing rs2686876 (Fig. 3). They include RIC8A, a homolog of C. elegans Ric8/synembryn that encodes a highly conserved G protein regulator. Intriguingly RIC8A plays a central role in the development of neural crest-derived structures including the meninges.42 To gain insight into the biological basis underlying the 11p15.5 association, we first evaluated each of the risk SNPs as well as the correlated variants (r2 > 0.8) using the online resources HaploReg v4,43 RegulomeDB,44 and SeattleSeq45 for evidence of functional effects (Supplementary Table S6). These data revealed active chromatin states overlapping SNPs correlated with rs2686876.
Fig. 3.
Regional plot of the 11p15.5 association. Plot (drawn using visPig48) shows association results of both genotyped (triangles) and imputed (circles) SNPs in the GWAS samples and recombination rates; −log10P-values (y-axis) of the SNPs are shown according to their chromosomal positions (x-axis). The sentinel SNP is shown as a large circle and is labeled by its rsID. The color intensity of each symbol reflects the extent of linkage disequilibrium with the top genotyped SNP, white (r2 = 0) through to dark red (r2 = 1.0). Genetic recombination rates, estimated using 1000 Genomes Project samples,19 are shown with a light blue line. Physical positions are based on National Center for Biotechnology Information (NCBI) build 37 of the human genome. Also shown are the chromatin-state segmentation tracks for 127 cell types and tissues from ENCODE and the Roadmap Epigenomics Consortium,49 generated using ChromHMM50 and the Wash U Epigenome Browser,51 and the positions of genes and transcripts mapping to the region of association. ANO9 is located 128 kb centromeric of the plotted region.
We explored whether there were any associations between rs2686876 genotype and the transcript levels of genes within 1 Mb using eQTL data on 47 tissues generated by the GTEx project39 (Supplementary Table S7). After accounting for multiple testing (53 genes across 47 tissues; P < 2.01 × 10−5), significant eQTL for ANO9 were observed in brain caudate basal ganglia (P = 8.30 × 10−7) and brain putamen basal ganglia (P = 2.58 × 10−6), for BET1L in esophagus mucosa (P = 9.03 × 10−6) and for PSMD13 in brain anterior cingulate cortex (P = 1.36 × 10−5). ANO9 upregulation has been observed in colorectal cancer46 and has been associated with poor prognosis in pancreatic cancer.47 The rs2686876 meningioma risk allele was, however, conversely associated with lower ANO9 expression at the 2 eQTLs. While the risk allele of rs2686876 is associated with higher RIC8A expression at nominal significance levels (P < 0.05) in 15 of the 47 tissues, the associations were not significant after correction for multiple testing.
We used SMR analysis to test for a concordance between signals from GWAS and cis eQTL for genes within 1 Mb of the sentinel and correlated SNPs (r2 > 0.8) at the 11p15.5 locus and derived bXY statistics, which estimate the effect of gene expression on meningioma risk (Supplementary Table S8). After accounting for multiple testing, the SMR analysis failed to provide overwhelming evidence to implicate a specific gene.
Discussion
We have provided the first evidence that implicates variation at 11p15.5 as a determinant of meningioma risk. To our knowledge this is only the second study, and the largest, to robustly associate common genetic variation as a risk factor for meningioma.
Although functional studies will be required, dysregulation of RIC8A provides an attractive basis of the 11p15.5 association a priori. RIC8A has an essential role in the development of the mammalian central nervous system, maintaining the integrity of pial basement membrane and modulating cell division.42 Intriguingly, conditional Ric8a knockout mice have been reported to exhibit defects in meningeal layer formation.42
Thus far, variation at only 2 loci has been robustly shown to affect meningioma risk.10 To estimate the potential heritability of meningioma attributable to all common variation, we applied GCTA32 and PCGC regression37 to the GWAS datasets (Supplementary Table S9). Combining data from the 2 GWAS indicates that the heritability associated with common variation is 27.9% (±4.4%).
The identification of risk variants at 11p15.5 provides further evidence for common genetic variation influencing meningioma risk and suggests the involvement of specific genes in tumor development. Since variation at 10p12.31 and 11p15.5 account for only ~4% of the familial risk of meningioma (Supplementary Table S10), it is likely that further risk variants for meningioma will be identified through additional and larger GWAS.
Funding
This work was supported by Cancer Research UK (C1298/A8362 supported by the Bobby Moore Fund), which provided principal funding for the study in the UK. The UK and Danish INTERPHONE studies were supported by the European Union Fifth Framework Program “Quality of Life and Management of Living Resources” (contract number QLK4-CT-1999-01563) and the Union for International Cancer Control (UICC). The UICC received funds for this purpose from the Mobile Manufacturers’ Forum and Global System for Mobile Communications Association. Provision of funds via the UICC was governed by agreements that guaranteed INTERPHONE’s complete scientific independence. These agreements are publicly available at http://www.iarc.fr. The UK centers were also supported by the Mobile Telecommunications and Health Research Programme, and the Northern UK center was supported by the Health and Safety Executive, Department of Health and Safety Executive and the UK Network Operators (O2, Orange, T-Mobile, Vodafone, and “3”).
In Germany, funding was provided to M.S. and J.S. by the Deutsche Forschungsgemeinschaft (Si 552, Schr 285), the Deutsche Krebshilfe (70-2385-Wi2, 70-3163-Wi3, 10-6262), and BONFOR.
The German-GWAS made use of genotyping data from the population based HNR study. The HNR study is supported by the Heinz Nixdorf Foundation (Germany). Additionally, the study is funded by the German Ministry of Education and Science and the German Research Council (Project SI 236/8-1, SI236/9-1, ER 155/6-1). The genotyping of the HNR subjects was financed by the German Centre for Neurodegenerative Disorders, Bonn.
In the United States, funding was provided by the Brain Science Foundation, the Meningioma Mommas, and NIH R01 grants CA109468, CA109461, CA109745, CA108473, CA109475, CA052689 and CA151933, and P50CA097257.
Supplementary Material
Acknowledgments
We are grateful to all patients and individuals for their participation and we would also like to thank the clinicians and other hospital staff, cancer registries, and study staff in respective centers who contributed to sample and data collection. We are indebted to R. Mahlberg (Bonn), who provided technical support.
Conflict of interest statement. None declared.
Authorship
R.S.H. and E.B.C. developed the project and provided overall project management; A.J.C., S.E.D., and R.S.H. drafted the manuscript. A.J.C. and S.E.D. performed bioinformatic and statistical analyses; P.B. performed project management and supervised genotyping; A.H. performed genotyping. Within the United States, E.B.C. obtained funding and managed the overall project; E.B.C., J.L.W., M.W., M.B., and J.M.S. oversaw institutional review board approval, patient recruitment and sample acquisition; L.C. supervised data collection and project management; L.L. and H.M.H. oversaw specimen storage and DNA isolation; J.L.W. and H.M.H. performed all genotyping; I.S. and K.M.W. performed bioinformatic and statistical analyses. Within Germany, M.S., J.S. and A.S. obtained funding, and were responsible for patient recruitment and sample acquisition, P.H. and M.M.N. oversaw DNA isolation and genotyping of the HNR controls and obtained funding for this, K-H.J. provided samples. Within Denmark, S.B.L. and C.J. conducted subject recruitment and sample collection. All authors contributed to the final manuscript.
References
- 1. Kohler BA, Ward E, McCarthy BJ, et al. Annual report to the nation on the status of cancer, 1975–2007, featuring tumors of the brain and other nervous system. J Natl Cancer Inst. 2011;103(9):714–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Braganza MZ, Kitahara CM, Berrington de González A, Inskip PD, Johnson KJ, Rajaraman P. Ionizing radiation and the risk of brain and central nervous system tumors: a systematic review. Neuro Oncol. 2012;14(11):1316–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wiemels J, Wrensch M, Claus EB. Epidemiology and etiology of meningioma. J Neurooncol. 2010;99(3):307–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Evans DG. Neurofibromatosis type 2 (NF2): a clinical and molecular review. Orphanet J Rare Dis. 2009;4:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lo Muzio L. Nevoid basal cell carcinoma syndrome (Gorlin syndrome). Orphanet J Rare Dis. 2008;3:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cahill KS, Claus EB. Treatment and survival of patients with nonmalignant intracranial meningioma: results from the surveillance, epidemiology, and end results program of the National Cancer Institute. Clinical article. J Neurosurg. 2011;115(2):259–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Claus EB, Calvocoressi L, Bondy ML, Schildkraut JM, Wiemels JL, Wrensch M. Family and personal medical history and risk of meningioma. J Neurosurg. 2011;115(6):1072–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bush WS, Moore JH. Chapter 11: Genome-wide association studies. PLoS Comput Biol. 2012;8(12):e1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sud A, Kinnersley B, Houlston RS. Genome-wide association studies of cancer: current insights and future perspectives. Nat Rev Cancer. 2017;17(11):692–704. [DOI] [PubMed] [Google Scholar]
- 10. Dobbins SE, Broderick P, Melin B, et al. Common variation at 10p12.31 near MLLT10 influences meningioma risk. Nat Genet. 2011;43(9):825–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Egan KM, Baskin R, Nabors LB, et al. Brain tumor risk according to germ-line variation in the MLLT10 locus. Eur J Hum Genet. 2015;23(1):132–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chapman K, Ferreira T, Morris A, Asimit J, Zeggini E. Defining the power limits of genome-wide association scan meta-analyses. Genet Epidemiol. 2011;35(8):781–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schmermund A, Möhlenkamp S, Stang A, et al. Assessment of clinically silent atherosclerotic disease and established and novel risk factors for predicting myocardial infarction and cardiac death in healthy middle-aged subjects: rationale and design of the Heinz Nixdorf RECALL Study. Risk factors, evaluation of coronary calcium and lifestyle. Am Heart J. 2002;144(2):212–218. [DOI] [PubMed] [Google Scholar]
- 14. Banda Y, Kvale MN, Hoffmann TJ, et al. Characterizing race/ethnicity and genetic ancestry for 100,000 subjects in the genetic epidemiology research on adult health and aging (GERA) cohort. Genetics. 2015;200(4):1285–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kvale MN, Hesselson S, Hoffmann TJ, et al. Genotyping informatics and quality control for 100,000 subjects in the genetic epidemiology research on adult health and aging (GERA) cohort. Genetics. 2015;200(4):1051–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mukherjee S, Simon J, Bayuga S, et al. Including additional controls from public databases improves the power of a genome-wide association study. Hum Hered. 2011;72(1):21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nat Protoc. 2010;5(9):1564–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2011;9(2):179–181. [DOI] [PubMed] [Google Scholar]
- 19. Auton A, Brooks LD, Durbin RM, et al. ; 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang J, Howie B, McCarthy S, et al. Improved imputation of low-frequency and rare variants using the UK10K haplotype reference panel. Nat Commun. 2015;6:8111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39(7):906–913. [DOI] [PubMed] [Google Scholar]
- 23. Clayton DG, Walker NM, Smyth DJ, et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nat Genet. 2005;37(11):1243–1246. [DOI] [PubMed] [Google Scholar]
- 24. Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. [DOI] [PubMed] [Google Scholar]
- 25. Liu JZ, Tozzi F, Waterworth DM, et al. ; Wellcome Trust Case Control Consortium Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42(5):436–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558. [DOI] [PubMed] [Google Scholar]
- 27. Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32(4):381–385. [DOI] [PubMed] [Google Scholar]
- 28. Wakefield J. A Bayesian measure of the probability of false discovery in genetic epidemiology studies. Am J Hum Genet. 2007;81(2):208–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Phelan CM, Kuchenbaecker KB, Tyrer JP, et al. ; AOCS study group; EMBRACE Study; GEMO Study Collaborators; HEBON Study; KConFab Investigators; OPAL study group Identification of 12 new susceptibility loci for different histotypes of epithelial ovarian cancer. Nat Genet. 2017;49(5):680–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cardis E, Richardson L, Deltour I, et al. The INTERPHONE study: design, epidemiological methods, and description of the study population. Eur J Epidemiol. 2007;22(9):647–664. [DOI] [PubMed] [Google Scholar]
- 31. Penegar S, Wood W, Lubbe S, et al. National study of colorectal cancer genetics. Br J Cancer. 2007;97(9):1305–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee SH, Harold D, Nyholt DR, et al. ; ANZGene Consortium; International Endogene Consortium; Genetic and Environmental Risk for Alzheimer’s disease Consortium Estimation and partitioning of polygenic variation captured by common SNPs for Alzheimer’s disease, multiple sclerosis and endometriosis. Hum Mol Genet. 2013;22(4):832–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hankey BF, Ries LA, Edwards BK. The surveillance, epidemiology, and end results program: a national resource. Cancer Epidemiol Biomarkers Prev. 1999;8(12):1117–1121. [PubMed] [Google Scholar]
- 35. Ostrom QT, Gittleman H, Farah P, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro Oncol. 2013;15(Suppl 2):ii1–ii56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yang J, Benyamin B, McEvoy BP, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42(7):565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Golan D, Lander ES, Rosset S. Measuring missing heritability: inferring the contribution of common variants. Proc Natl Acad Sci U S A. 2014;111(49):E5272–E5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pharoah PD, Antoniou AC, Easton DF, Ponder BA. Polygenes, risk prediction, and targeted prevention of breast cancer. N Engl J Med. 2008;358(26):2796–2803. [DOI] [PubMed] [Google Scholar]
- 39. Consortium GT. Human genomics. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhu Z, Zhang F, Hu H, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48(5):481–487. [DOI] [PubMed] [Google Scholar]
- 41. Mawrin C, Perry A. Pathological classification and molecular genetics of meningiomas. J Neurooncol. 2010;99(3):379–391. [DOI] [PubMed] [Google Scholar]
- 42. Kask K, Ruisu K, Tikker L, et al. Deletion of RIC8A in neural precursor cells leads to altered neurogenesis and neonatal lethality of mouse. Dev Neurobiol. 2015;75(9):984–1002. [DOI] [PubMed] [Google Scholar]
- 43. Ward LD, Kellis M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016;44(D1):D877–D881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22(9):1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ng SB, Turner EH, Robertson PD, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461(7261):272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li C, Cai S, Wang X, Jiang Z. Identification and characterization of ANO9 in stage II and III colorectal carcinoma. Oncotarget. 2015;6(30):29324–29334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jun I, Park HS, Piao H, et al. ANO9/TMEM16J promotes tumourigenesis via EGFR and is a novel therapeutic target for pancreatic cancer. Br J Cancer. 2017;117(12):1798–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Scales M, Jäger R, Migliorini G, Houlston RS, Henrion MY. visPIG—a web tool for producing multi-region, multi-track, multi-scale plots of genetic data. PLoS One. 2014;9(9):e107497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kundaje A, Meuleman W, Ernst J, et al. ; Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518(7539):317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9(3):215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhou X, Maricque B, Xie M, et al. The human epigenome browser at Washington University. Nat Methods. 2011;8(12):989–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genotype data from GERA are available from dbGaP (accession phs000674.v2.p2). The 1000 Genomes Project and UK10K imputation panel data are available from the European Genome-phenome Archive (accession EGAD00001000776). Remaining data are available from the authors upon request.