Abstract
Background
Individuals with Crohn’s disease frequently require ileocecal resection (ICR), and inflammation often recurs in the neoterminal ileum following surgery. Fructooligosaccharide (FOS) is a fermentable prebiotic that stimulates the growth of bifidobacteria and may promote anti-inflammatory activity. The aim of this study was to determine if supplementation of a postICR diet with FOS in a mouse model would be effective in stimulating the growth of bifidobacteria and reducing systemic and local inflammation.
Methods
ICR was performed in IL10-/- mice (129S1/SvlmJ) with colitis. Following surgery, nonICR control and ICR mice were fed a chow diet ± 10% FOS for 28 days. Serum, colon, and terminal ileum (TI) were analyzed for cytokine expression by MesoScale discovery platform. DNA extracted from stool was analyzed using 16s rRNA sequencing and qPCR. Expression of occludin and ZO1 was assessed using qPCR. Short-chain fatty acid (SCFA) concentrations were assessed using gas chromatography.
Results
ICR led to increased systemic inflammation (P < 0.05) and a significant decline in fecal microbial diversity (P < 0.05). Mice on the FOS diet had a greater reduction in microbial diversity and also had worsened inflammation as evidenced by increased serum IL-6 (P < 0.05) and colonic IFNγ and TNFα (P < 0.05). Expression of occludin and ZO1 were significantly reduced in FOS-supplemented mice. There was a correlation between loss of diversity and the bifidogenic effectiveness of FOS (r = -0.61, P < 0.05).
Conclusions
FOS-supplementation of a postICR diet resulted in a decrease in fecal bacterial diversity, reduction in barrier function, and increased gut inflammation.
Keywords: experimental models, fructooligosaccharide, microbiome, surgery, Crohn’s disease
Mammals have evolved to house diverse microbial communities at many sites throughout the body that has resulted in an intimate relationship between microbes and their mammalian host.1 These relationships are at the forefront of intestinal health and disease.2 Abnormal microbial composition, or microbial dysbiosis, has been described in numerous autoimmune and chronic human diseases.2, 3 Crohn’s disease (CD), a subtype of inflammatory bowel disease (IBD), has been extensively studied in this regard, and although there appears to be a relationship between host inflammation and microbial dysbiosis, the extent, importance, and nature of that dysbiosis is unclear.3, 4 This relationship is further complicated by the fact that microbes can produce active metabolites that interact with the intestinal mucosa, or influence the activity of other microorganisms. One such interaction is cross-feeding, whereby the end product of a microbe’s fermentation process serves as a substrate for another microbe’s metabolic processes.5, 6 Intestinal microbial diversity decreases in the presence of active inflammatory processes, with certain bacterial communities, such as Proteobacteria, thriving under inflammatory conditions and other strict anaerobes, such as bifidobacteria, disappearing.7 A loss of these anaerobic species also removes many physiologically active metabolites that have anti-inflammatory and immune regulatory actions, thus potentially exacerbating inflammation.
Many patients with CD require an intestinal resection at some point, with the most common resection consisting of removal of the final portion of the small intestine and first portion of colon, called ileocecal resection (ICR).8 The postoperative CD gut offers an interesting opportunity to examine the relationship between microbes and inflammation. Surgery aims to remove all actively diseased tissue, thus providing a proverbial inflammatory clean slate. In addition, ICR also will remove a large proportion of bacteria at the diseased site, and presumably leaves the patient with a microbial clean slate. However, despite this, it has been shown that anywhere from 30–90% of patients suffer from endoscopic recurrence of disease at the site of their anastomoses within one year.9–11 It is possible that this recurrence is driven by a persistently dysbiotic microbiota left after surgery that is perpetuated and initiates disease recurrence. This concept is supported by the findings that probiotic use initiated immediately after surgery has a beneficial effect at early time points.12
The addition of prebiotics to the diet is a method of targeting specific microbes to increase their abundance. Fructooligosaccharide (FOS) is a prebiotic that can be utilized by bifidobacteria and has been shown to increase relative abudance of both Lactobacillus and Bifidobacterium in the gut.13, 14 Given the lack of bifidobacteria in the inflammatory disease state7 and the ability of bifidobacteria to produce and enable the production of putatively anti-inflammatory metabolites such as acetate and butyrate,5, 6 we hypothesized that in a murine model of postICR CD, the addition of FOS would increase the relative abundance of bifidobacteria and subsequently decrease local and systemic inflammation following surgery.
METHODS
Animal Model
The animal use protocols employed were approved by the animal care committee at the University of Alberta. IL-10 -/- (129 Sv/Ev) mice underwent ICR as previously described.15 The terminal ileum (TI) was transected proximal to the cecum and the descending colon was divided at the distal cecum. The anastomosis was created using 8-0 Prolene (Ethicon) sutures in a simple interrupted fashion and the abdominal wall was closed. Mice who had undergone ICR were maintained on liquid diet for 2 days preoperatively and 2 days postoperatively. Nonoperative control mice received 4 days of liquid diet but did not undergo surgery. Following the completion of the liquid diet, mice were started on either the control diet (Control group (n = 5) and ICR-C group (n = 5)) or the FOS-supplemented diet (FOS group (n = 6) and ICR-FOS group (n = 6)) (Supplementary Figure 1). Experimental diets contained a standard mouse chow, LabDiet 5001 (LabDiet, USA) (29% protein, 55% carbohydrates, and 13% fat; 3.8 kcal/g; PMI Nutrition International, Richmond, IN, USA) supplemented with the addition of a dietary fiber. Control diet contained additional cellulose to 10% by dry weight. FOS-supplemented diet contained 10% FOS by dry weight.
All mice were 12 weeks of age at the initiation of the experiment. Overall survival was 92% with 2 mice dying postoperatively before the introduction of either the control or experimental diet. Animals were weighed on days 0 (day of surgery), 14, 21, and 28 of the experiment. Food was weighed and replaced on days 0, 3, 7, 10, 14, 17, 21, 24, and 28. Stools were collected before the initiation of liquid diet, and on days 14 and 28. Animals were housed 2 per cage and littermates were randomized across treatment groups.
Histological Analyses
Sections of perianastomotic ileum and colon were taken at day 28 and fixed in 10% buffered formalin, embedded in paraffin, and cut to 5 µm. These sections were stained with either Masson trichome or hematoxylin/eosin (H&E). Histologic injury was scored by a pathologist blinded to the treatment groups using a validated 10 point scale that includes enterocyte injury, epithelial hyperplasia, lamina propria lymphocytes, and lamina propria neutrophils. Collagen deposition was scored from 0–2 using the Masson trichome stains as previously described.16
Gut Microbial Composition
Genomic DNA was extracted from stool samples using FastDNA Spin Kit for Feces (MP Biomedicals, Lachine, QC, Canada) and quantified using PicoGreen DNA quantification kit (Invitrogen, Carlsbad, CA, USA). Sequencing was performed using the MiSeq Illumina platform. Genomic DNA was subjected to a NaCl and ethanol precipitation procedure to remove contaminants that interfere with PCR. The protocol involved the addition of 5M NaCl to a total 5% of the DNA sample and precipitation of the DNA with 1 sample volume of ice-cold anhydrous ethanol. The samples were left at -20C for 30 minutes then centrifuged at 10,000g for 15 minutes. The liquid was discarded and 1 volume of ice-cold 75% ethanol was added to the pellet as a wash. The samples were centrifuged, decanted, and left to dry at room temperature for 30 minutes. The DNA pellet was solubilized in EB buffer (Qiagen, USA). Microbial composition was assessed using Illumina’s established 16S rRNA amplicon sequencing method and the MiSeq sequencing platform. No deviations from the manufacturer’s protocol were used. Briefly, a segment of the V3 and V4 region of the 16S gene was amplified with gene specific primers (aligning to 341bp and 805bp in the gene) that also include an adapter sequence overhang: Bact_16s_ILL1_341mF 5-TCG TCG GCA GCG TCA GAT GTG TAT AAG AGA CAG CCT ACG GGN GGC WGC AG-3, Bact_16s_ILL1_805mR 5- GTC TCG TGG GCT CGG AGA TGT GTA TAA GAG ACA GGA CTA CHV GGG TAT CTA ATC C-3. This PCR reaction was cycled 25 times and the resulting reaction was purified using bead-based clean-up followed by an 8 cycle PCR reaction using Illumina’s proprietary bar-coding primers that also align to the adapter sequence. After a second clean-up, the bar-coded libraries were diluted, denatured, pooled, and run using a V3 300bp reagent cartridge on the MiSeq system.
Bacterial composition was estimated from the data using Quantitative Insights into Microbial Ecology (QIIME 1.9.1) pipelines.17 In brief, QIIME was used to analyze for phylogenetic and operational taxonomic unit (OTU). First of all, it was used to de-multiplex the barcoded reads and perform chimera filtering. Filtered sequence reads were grouped into OTUs at a sequence similarity level of 97%, which approximates species-level phylotypes. Taxonomy of the OTUs was assigned and sequences were aligned with RDP classifier and Pynast. To evaluate the alpha diversities of each microbiota community, we calculated the Shannon diversity metric. A Linear Discriminant Analysis effect size tool, which employs a Kruskal-Wallis sum-rank test (P < 0.05), a Wilcoxon rank-sum test (P < 0.05), and subsequent linear discriminant analysis was used to identify and estimate the effect size of each differentially abundant population.
16s rRNA qPCR
Genomic DNA was extracted from stools using a FastDNA Spin Kit (MP Biomedicals). The manufacturer’s protocol was followed, and the resultant DNA was quantified using a PicoGreen assay (Invitrogen). The samples were then diluted to 50ng/ml and re-quantified with PicoGreen. Reactions containing 6µL of H20, 10µL of Fast SYBR Green Master Mix, and 1µL of 10uM forward and reverse primer were added to 2µL of target DNA for quantitative PCR (qPCR) in MicroAmp 96 well optical plates. PCR conditions in the 7900HT instrument were 5 minutes at 50 degrees C, 5 minutes at 95 degrees C, 15 seconds at 95 degrees C, and 1 minute at 60 degrees C, with a melting curve step progressing between 60 to 95 degrees C in 12 minutes. DNA copy number was determined by comparison to standard curves constructed from purified PCR product quantified using PicoGreen. Gene copy per nanogram of genomic DNA was determined in relation to the starting concentration of genomic DNA in each reaction.
RNA Extraction and qPCR
Total RNA was isolated and extracted from mouse TI tissue. Tissues were snap frozen, then homogenized and extracted in Trizol according to the manufacture’s protocol (Invitrogen Corporation, Carlsbad, CA, USA). An RNeasy mini kit (Qiagen, Inc., Mississauga, ON, USA) was used for purification, and purity was assessed using a Nanodrop 1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Complementary DNA was synthesized by the the High Capacity cDNA Reverse Transcription Kit (Invitrogen). Quantitative real-time polymerase chain reaction (qPCR) was performed using Taqman Gene Expression Assays (Applied Biosystems, Foster City, CA, USA) for 18s rRNA, Occludin and ZO1. Each sample was run in triplicate using the 7900HT Fast Real-Time PCR System and Sequence Detection System v2.4 software (Applied Biosystem) and the relative quantification was calculated using the ΔΔCt method and compared to expression of 18S rRNA.
Assessment of Serum Lipopolysaccharide (LPS)
A HEK-blue cell system (InvivoGen) was used to analyze the serum concentration of LPS. The cells were cultured in a 96-well plate and 5µL of serum diluated 1:10 was incubated with the cells for 6 hours. LPS activation of the system catalyzes the detection medium to turn blue. Activation was quantified by measuring absorption at 650 nm for quantification.
Measurement of Cytokines
Segments of perianastomotic colon and ileum were snap frozen at collection. They were then homogenized in 7.5X tissue weight in PBS and 0.05% Tween 20. They were centrifuged at 10,000 rpm for 10 minutes and the resultant supernatant was assessed for cytokine expression. Cytokine values are corrected for tissue weight. INF-γ, IL-1β, IL-2, IL-4, IL-5, IL-6, KC/GRO, IL12p70, and TNF-α were assessed using the Meso Scale discovery system (Meso Scale Diagnostics, Gaithersburg, MD, USA) as per the manufactures protocol.
Measurements of Short-Chain Fatty Acids (SCFA)
The concentration of SCFA in feces were determined using gas chromatography: 0.2g of feces was homogenized in 800µL of 0.1N hydrochloric acid then 200µl of 25% phosphoric acid was added. This was centrifiuged at 3000g for 10 minutes and the supernatant was added to internal standard solution (150 mg of 4-methyl-valeric acid, S381810, Sigma-Aldrich) and 5% phosphoric acid in a glass chromatography tube, mixed well, and kept at room temperature for 30 minutes. The supernatant was analyzed for SCFA (ie, acetic, propionic, butyric, and isobutyric) using a Varian model 3400 Gas Chromatograph (Varian, Walnut Creek, CA) with a Stabilwax-DA column (30-m × 0.25-mm i.d.; Restek, Bellefonte, PA). A flame-ionization detector was used with an injector temperature of 170°C and a detector temperature of 190°C.
Statistical Analysis
All data are presented as the mean ± the standard error of the mean. Statistical analysis was performed using STATA v13.1. Student’s t test was used when comparing 2 groups. Two-way analysis of variance (ANOVA) was used to compare the significance when the model consisted of both diet and ICR. Principal component analysis (PCA) plots of bacterial populations and heatmaps of cytokine expression were created using Metaboanalyst 3.0 after logarithmic transformation of the data.18
RESULTS
Effect of Surgery
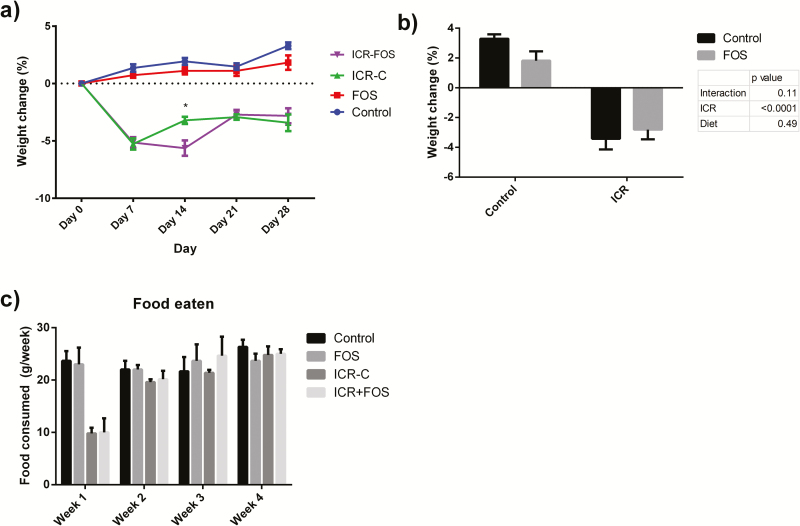
The surgical insult associated with ICR led to a mean weight loss of 5.2 ± 0.3% in all surgical mice at one week (Fig. 1A). On day 28, at the time of killing, mice who had undergone ICR had a significantly lower weight than at day 0 (P < 0.01) (Fig. 1A) and also as compared to their nonoperative counterparts (P < 0.01) (Fig. 1B). There was a delay in the restoration of weight in ICR-FOS mice compared to ICR-C mice (P = 0.01) (Fig. 1A). All ICR mice consumed significantly less food during the first week (P < 0.01), but food consumption was similar across all groups in subsequent weeks (Fig. 1C).
FIGURE 1.
FOS-supplementation was associated with delayed recovery following ICR. (A) Mean percent weight change ± SEM from baseline at 0, 7, 14, 21, and 28 days. Mice experienced weight loss following ICR. ICR-C mice experienced recovery more quickly than ICR-FOS mice, with significantly different weights on day 14 (represented by *, Student’s t-test, P = 0.01). (B) ICR mice lost significantly more weight over the course of the experiment than their nonsurgical counterparts (Two-way ANOVA, Ϯ represents P < 0.0001). (C,) Following ICR, mice consumed less food in the first week (Student’s t-test compared to control group. Ϯ represents P < 0.0001). Food consumption was similar in weeks 2 through 4.
Control: n = 5; Control-FOS: n = 6; ICR-C: n = 5; ICR-FOS: n = 6
Effect of Surgery and FOS on Inflammation and Fibrosis
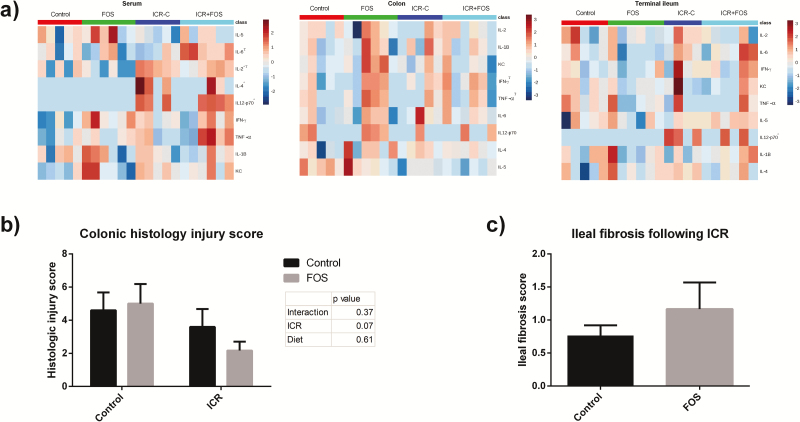
PostICR mice demonstrated increased levels of serum inflammatory cytokines, evidenced by increased IL-2, IL-12, and IL-4 (P < 0.05) (Fig. 2A). Serum IL-6 was significantly elevated in ICR-FOS mice compared to ICR-C mice (P = 0.02) (Fig. 2A). Although FOS-supplementation increased colonic IFNγ (P = 0.05) and TNFα (P = 0.04) (Fig. 2A), there were no significant differences in histological scores in the colon between the groups (Fig. 2B). ICR-FOS mice demonstrated a nonsignificant increase in fibrosis scores compared to ICR-C mice (1.17 versus 0.6 P = 0.28)(Fig. 2C). ICR was associated with elevated IL-12 in the terminal ileum (Fig. 2A).
FIGURE 2.
ICR and FOS-supplementation led to increased systemic and enteric cytokine expression. (A) Heatmap of relative cytokine expression in the serum, terminal ileum, and colon after logarithmic transformation. ICR mice and FOS-supplemented mice expressed significantly higher levels of IL-2, IL-12, and IL-4 (P < 0.05). (Two-way ANOVA, * represents a change induced by ICR and Τ represents a change induced by FOS, P≤0.05).(B) Histologic injury scores given as a combined score for enterocyte injury, epithelial hyperplasia, and lymphocyte and neutrophil infiltration into the lamina propria. Two-way ANOVA revealed no effect of FOS or ICR on histologic injury score as a whole or in individual components of the score. (C) Fibrosis score assessing collagen deposition in the TI following ICR. (P = 0.28)
Control: n = 5; Control-FOS: n = 6; ICR-C: n = 5; ICR-FOS: n = 6
Barrier Function
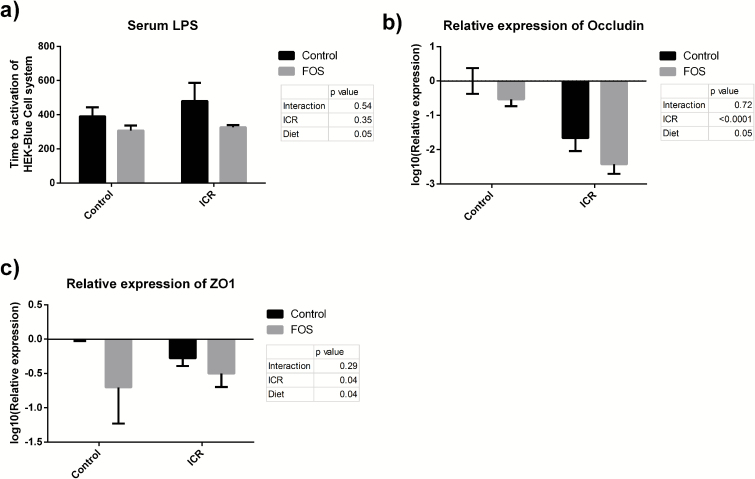
FOS-supplementation led to increased levels of serum LPS compared to control diet (P = 0.05) (Fig. 3A), suggesting a loss of barrier function. A loss of barrier function was also supported by the findings of a decreased expression in RNA of the tight junction proteins, occludin, and ZO1 secondary to FOS-supplementation (P = 0.05 and P = 0.04, respectively) and ICR (P < 0.01 and P = 0.04, respectively) (Figs. 3B and C).
FIGURE 3.
FOS-supplementation worsened barrier function. (A) Time to activation of a HEK-Blue cell system sensitive to LPS by serum. A lower time to activation of this cell system is indicative of higher levels of serum LPS. Presented as mean time to activation ± SEM. FOS-supplementation was significantly associated with increased LPS (P = 0.05) (Two-way ANOVA). (B) Relative mRNA expression of occludin following logarithmic transformation in the TI of mice presented as mean expression ± SEM. Both FOS-supplementation (P = 0.05) and ICR (P < 0.01) were associated with decreased expression. (C) Relative mRNA expression of ZO1 in the TI of mice presented as mean expression ± SEM. Both FOS-supplementation (P = 0.04) and ICR (P = 0.04) were associated with decreased expression.
Control: n = 5; Control-FOS: n = 6; ICR-C: n = 5; ICR-FOS: n = 6
Microbial Composition
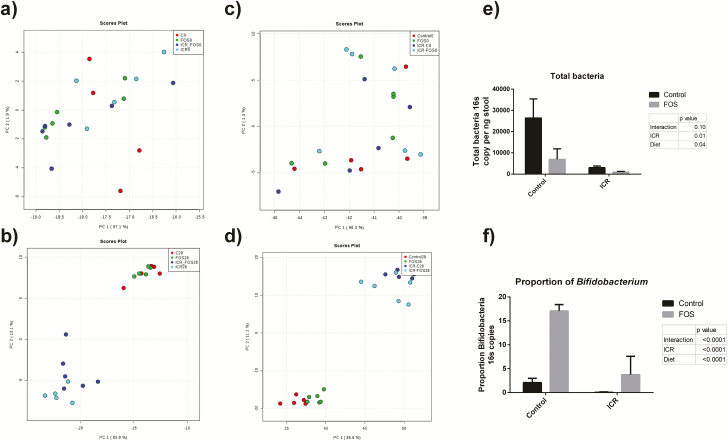
The baseline microbiota was analyzed before the initiation of the liquid diet and was similar in all mice (Figs. 4A and C; Supplementary Figs. 2–6). The qPCR analysis revealed a large drop in the total number of fecal bacteria following surgery (P < 0.01) (Fig. 4E). FOS-supplementation was associated with a further decrease in levels of fecal bacteria (P = 0.04). As expected, although there was decreased bifidobacteria postICR in all surgical mice, FOS-supplementation did result in an increase in bifidobacteria in both controls and ICR mice, albeit to a much decreased level in the postICR mice (P < 0.0001) (Fig.4F).
FIGURE 4.
ICR causes significant shift in bacterial populations, and a reduction in total fecal bacteria and bifidobacteria. a) PCoA plots of the Bray-Curtis index showing clustering at the phyla (a) and family (c) level at baseline prior to the initiation of liquid diet and surgery. At day 28, the ICR groups clustered separately from the control groups at the phyla (b) and family (d) levels. (e) Total fecal bacteria per ng of stool in feces at day 28 of all groups. ICR and FOS-supplementation were associated with decreased fecal bacteria (P < 0.01, P = 0.04). (f) Relative proportions of bifidobacteria in feces at day 28 of all groups. ICR decreased the proportion of bifidobacteria (P < 0.01), while FOS supplementation increased the proportion of bifidobacteria (P < 0.01).
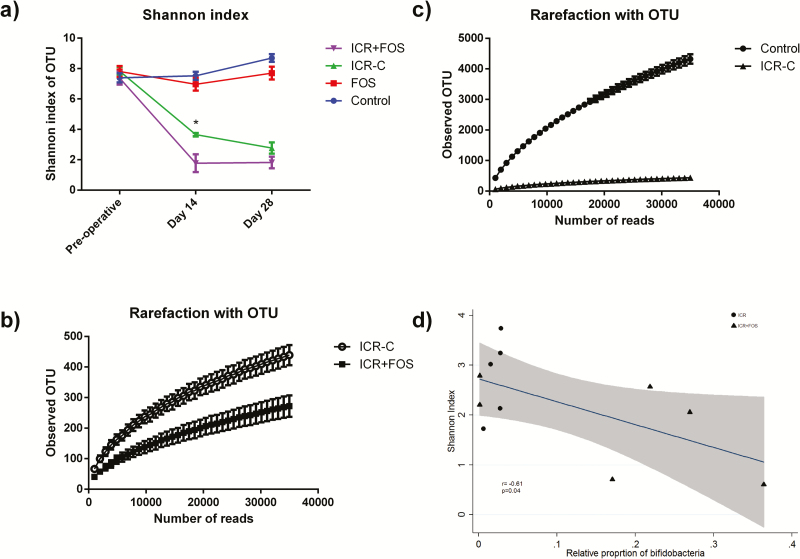
ICR was responsible for a precipitous decrease in bacterial diversity as measured by the Shannon index (Control diet: 7.39 ± 0.30 v ICR-C: 2.77 ± 0.37 P = 0.01) and in richness as evidenced by the rarefaction curves (Figs. 5A and B). The decreased diversity was exacerbated by the addition of a FOS-supplemented diet at 14 days (ICR-C: 3.66 ± 0.11 v. ICR-FOS 1.77 ± 0.59 P = 0.02) (Fig. 5A). Fourteen days following ICR, the Firmicutes phylum dominated (Supplementary Fig. 5; Table 1), and the dominant family in both ICR-C and ICR-FOS groups was Lactobacillaceae (Supplementary Fig. 6; Table 1). However, Lactobacillaceae accounted for nearly all reads in the ICR group supplemented with FOS (90.6 ± 4.9%), whereas a more modest level was seen in the ICR-C group (52.6 ± 4.7%). The increase in Lactobacillaceae was at the expense of Enterobacteriaceae and Enterococcaceae that were relatively more abundant in ICR-C mice at 14 days (Table 1, Supplementary Fig. 4). At 28 days following ICR, the relationship between FOS-supplementation and decreased diversity persisted (ICR-C: 2.77 ± 0.37 v. ICR-FOS: 1.82 ± 0.38 P = 0.11), although interestingly by this point ICR-FOS mice showed an elevation in Bacteroidetes that was not seen in the ICR-C mice (Table 1). ICR-FOS mice also saw an increase in Proteobacteria that was not seen in ICR mice. There was a correlation between the loss of diversity and bifidogenic effectiveness of FOS as measured by qPCR (r = -0.61; P = 0.047) (Fig. 5D).
FIGURE 5.
ICR leads to a decrease in bacterial diversity and richness that is exacerbated by FOS-supplementation. (A) Mean Shannon diversity index value ± SEM of each treatment group through the course of the experiment. At day 14 ICR-C mice possessed greater diversity than ICR-FOS (represented by *, Student t test, P = 0.02). (B) Rarefaction curve representing species richness. Data points represent mean OTU observed at a certain number of reads ± SEM. ICR lessened species richness. (C) Rarefaction curve representing species richness. Data points represent mean OTU observed at a certain number of reads ± SEM. FOS-supplementation postICR further decreased species richness. D) Plot of Shannon index value compared to the proportion of bifidobacteria in the stool at day 28. As bifidogenesis increased, diversity decreased. (r = -0.61, P = 0.05)
Table 1:
Effect of FOS-Supplementation PostICR on Bacterial Populations at 14 and 28 Days
| Relative abundance (%) | Day 14 | LDA Effect size | Day 28 | LDA Effect size | ||
|---|---|---|---|---|---|---|
| ICR-C % (SEM) |
ICR-FOS (SEM) |
ICR-C (SEM) |
ICR-FOS (SEM) |
|||
| Phylum | ||||||
| Firmicutes | 99.10 (0.09) |
99.96 (0.01) |
3.82 | 99.8 (<0.1) |
89.9 (4.5) |
4.81 |
| Bacteroidetes | 0.03 (<0.01) |
0.02 (0.01) |
NS | 0.02 (<0.01) |
3.51 (3.14) |
4.33 |
| Proteobacteria | 0.01 (0.0) |
0.64 (0.34) |
NS | 0.03 (0.02) |
4.62 (9.51) |
NS |
| Class | ||||||
| Bacilli | 99.35 (0.59) |
99.88 (0.03%) |
3.87 | 99.54 (0.22) |
81.29 (5.91) |
NS |
| Gammaproteobacteria | 0.64 (0.15) |
<0.01 (<0.01) |
3.88 | 0.02 (0.01) |
4.50 (3.81) |
NS |
| Bacteroidia | 0.03 (<0.01) |
0.02 (0.01) |
NS | 0.02 (0.02) |
3.51 (4.50) |
4.54 |
| Clostridia | 0.06 (0.05) |
0.69 (1.31) |
NS | 0.28 (0.35) |
9.01 (13.95) |
NS |
| Family | ||||||
| Lactobacillaceae | 52.58 (4.71) |
90.61 (4.97) |
5.27 | 80.00 (7.21) |
78.41 (6.32) |
NS |
| Enterococcaceae | 32.8 (7.26) |
2.31 (1.37) |
5.20 | 8.77 (3.79) |
1.46 (0.51) |
NS |
| Enterobacteriaceae | 0.63 (0.15) |
<0.01 (<0.01) |
3.94 | 0.02 (0.01) |
4.50 (3.81) |
NS |
| Clostridiaceae | 0.01 (0.02) |
<0.01 (<0.01) |
NS | 0.01 (0.01) |
9.23 (14.5) |
NS |
| Bacteroidaceae | <0.01 (<0.01) |
<0.01 (<0.01) |
NS | <0.01 (<0.01) |
3.45 (3.12) |
4.55 |
| Lachnospiraceae | 0.77 (1.52) |
0.03 (0.05) |
NS | 0.27 (0.39) |
0.08 (0.1) |
NS |
| Genus | ||||||
| Lactobacillus | 52.58 (4.70) |
90.61 (4.97) |
5.49 | 80.00 (7.21) |
78.41 (6.32) |
NS |
| Clostridium | <0.01 (<0.01) |
<0.01 (<0.01) |
NS | <0.01 (<0.01) |
9.37 (14.7) |
NS |
| Enterococcus | 25.79 (6.13) |
0.71 (0.41) |
5.36 | 5.05 (2.75) |
0.66 (0.31) |
4.36 |
| Citrobacter | <0.01 (<0.01) |
<0.01 (<0.01) |
NS | <0.01 (<0.01) |
4.38 (3.72) |
4.44 |
| Staphylococcus | 0.02 (0.01) |
0.42 (0.37) |
NS | <0.01 (<0.01) |
0.01 (0.01) |
4.22 |
Significant values as calculated using the Linear Discriminant Analysis effect size tool, which employs a Kruskal-Wallis sum-rank test (P < 0.05), a Wilcoxon rank-sum test (P < 0.05), and subsequent linear discriminant analysis to identify and estimate the effect size of each differentially abundant population.
NS- not significant
Fecal Short-Chain Fatty Acids
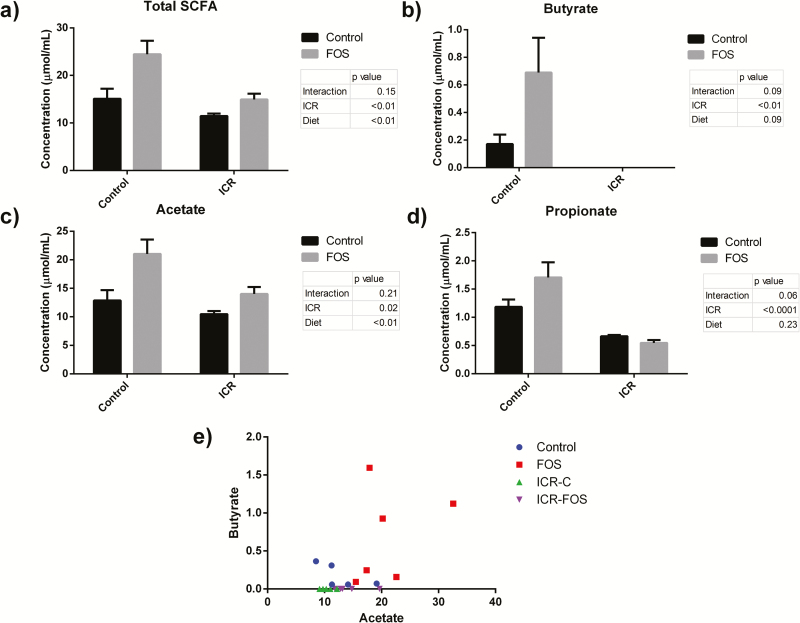
The concentration of short-chain fatty acids (SCFA) was measured in the stool at day 28. As expected, FOS-supplementation led to an increase in the total concentration of fecal SCFA (P < 0.01) and acetate (P < 0.01) (Fig. 6). ICR depleted fecal acetate, propionate, and total SCFA, and completly eliminated fecal butyrate (Fig. 6). Acetate is an end product of FOS metabolism by bifidobacteria, and accordingly there was a correlation in ICR mice between the proportion of fecal bifidobacteria and the level of acetate in the stool (r = 0.76, P < 0.01).
FIGURE 6.
Effect of FOS-supplementation on fecal SCFA levels. (A) Total mean fecal SCFA concentration ± SEM at day 28. ICR was associated with a depletion of SCFA (P < 0.01), whereas FOS-supplementation increased total fecal SCFA (P < 0.01).((B) Mean fecal butyrate concentration ± SEM at day 28. ICR mice had a complete absence of stool butyrate. FOS-supplementation in control mice was associated with a trend toward increased fecal butyrate (P = 0.09). (C) Mean fecal acetate concentration ± SEM at day 28. ICR was associated with a depletion of acetate (P = 0.02), while FOS-supplementation was associated with increased fecal acetate (P < 0.01). (D) Mean fecal propionate concentration ± SEM at day 28. ICR was associated with a depletion of propionate (P < 0.001). (E) Fecal butyrate was seen following FOS-supplementation in nonICR mice, but no butyrate was detected in ICR mice, regardless of FOS-supplementation.
DISCUSSION
In this study, we demonstrate that FOS-supplementation of a postICR diet results in a bloom of Lactobacillaceae and subsequent decrease in fecal bacterial diversity along with an impairment in barrier function and increased inflammation in IL-10-/- mice.
Recurrence of CD following ICR has been a challenging problem. Recently, there has been a linkage identified between certain bacterial populations and the loss of microbial diversity seen in CD disease recurrence.19, 20 A study by Fedorak et al demonstrated the potential of microbiota manipulations, in the form of the probiotic combination VSL#3, in altering the course of postoperative CD recurrence.12 Our model showing massive gut microbial changes induced by surgery is consistent with the previous literature.21, 22 This consisted of a decrease in total bacterial density following ICR, and a stark decrease in both bacterial diversity and richness. This was associated with the loss of specific bacterial groups, including bifidobacteria following ICR. These microbial changes were associated with the loss of beneficial microbial metabolites, especially butyrate, which was not present in the feces of postICR mice. These changes were accompanied by an apparent worsening of barrier function. Ultimtely ICR was associated with increased expression of systemic and enteric proinflammatory cytokine expression.
In previous studies, FOS-supplementation has been shown to stimulate the growth of bifidobacteria, increase the production of butyrate, and decrease colonic inflammation.23, 24 Bifidobacteria also have been shown to decrease the secretion of inflammatory cytokines by intestinal epithelial cells and by immune cells.25–28 Based on these findings, we attempted to improve the delterious course of postICR inflammation in a murine model of postoperative CD recurrence by inducing the growth of protective microbes through supplementation of the diet with FOS. In terms of bifidogenesis, FOS was successful; the relative amount of bifidobacteria was higher in mice whose diets were supplemented with the prebiotic. It has been suggested that the beneficial effect of bifidobacteria may be mediated by the increased production of SCFA.29 However, although FOS-supplementation was successful in increasing fecal SCFA concentrations, particularly in nonsurgical mice, there was no increase in butyrate production in ICR mice. Indeed, despite a small increase in acetate induced by FOS feeding in the ICR mice, the overall condition of ICR-FOS mice was worsened compared to ICR-C mice, as evidenced by elevated colonic, ileal, and systemic inflammatory cytokines and a delay in the restoration of weight following surgery.
The unintened consequence of heightened inflammation following FOS-supplementation postICR may be due to the effect of the fiber on overall microbial diversity. FOS-supplementation exacerbated the loss of diversity and richness induced by surgery, and in fact, the effectiveness of FOS-supplementation in increasing bifidobacteria postICR actually correlated with a decrease in bacterial diversity. Lactobacillaceae appeared to benefit most from FOS-supplementation following ICR. Both of the selective pressures applied during this study, namely ICR and FOS-supplemenation, proved to enhance the fitness of Lactobacillaceae and led to this family becoming overwhelmingly dominant following ICR in FOS-supplemented mice. A loss of microbial diversity in the gut has been associated with many health conditions, including recurrent C. difficle infection,30 psoriatic arthritis,31 and CD.32 Adequate diversity levels are considered necessary for the function of an ecosystem. The importance of diversity is almost certainly multifactorial, although one mechanism may be the production of specific metabolites that are only possible through cross-feeding. Cross-feeding is the process where the breakdown products of polysaccharides from primary degraders are used as secondary substrates by different microbes.5 A prominent example of this phenomenon is the increase of butyrate production by the Faecalibacterium genera following acetate production by bifidobacteria.6 The reliance of cross-feeding on microbial diversity may explain the pattern of fecal butyrate seen in this experiement. Those mice that did not undergo sugery were able to preserve a high-level of diversity, and consequently had a signifigant degree of cross-feeding resulting in butyrate production. The loss of diversity following surgery crippled the ability of the microbiome to engage in metabolic cross-feeding. Therefore, despite FOS stimulating increased fecal acetate, the overall intestinal ecosystem was not conducive to butyrate formation.
The route of FOS-administration in this project was through solid diet, as opposed to liquid oral gavage. A previous study using a twice daily gavage of FOS demonstrated a beneficial effect of FOS in reducing the severity of DSS colitis23; however, another study using a food delivery system for FOS, also in the DSS model, did not show a benefit.33 Whether these differing outcomes are due to the delivery method or alternatively a reflection of the timing of adminstration or the different models of colitis remains to be determined. However, the effect of any type of fiber supplementation would be affected by the baseline composition of microbes, and whether or not the microbes that are able to utilize the particular substrate are present or absent. Thus, in our ICR model, as opposed to a prevention model using DSS to induce colitis, the severe loss of microbial diversity that occurred following surgery would have effectively reduced the number of microbial species capable of utilizing the FOS.
The systemic and local inflammation seen following ICR and FOS-supplementation may have been partially due to impaired barrier function. This effect has been previously demonstrated following FOS-supplementation.33, 34 In this previous work FOS-supplementation decreased the expression of 2 tight junction proteins, occludin and ZO1, and led to an increase level of serum LPS suggesting a loss of barrier function. Moreau et al33 also showed FOS-supplementation to be ineffective at maintaining gut barrier function in a DSS model. Interestingly, ICR-mice on the FOS supplementation also had an increase in Proteobacteria by day 28, suggesting that FOS induced a more favourable oxygenated lumenal environment for the growth of these potentially inflammatory organisms.
The immediate postoperative period in CD has often been viewed as a favorable time in which to initiate therapies, as it is seen as a clean slate, devoid of disease pathology. Although that may be true in terms of pathologic inflammation, we demonstrate here that in terms of microbial pertubations this is not the case. In reality, the postoperative period represents a time of significant dysbiosis, and treatments initiated in this time frame must take that into account. In this case providing a competitive advantage to those bacteria that use FOS as a substrate, specifically Lactobacillaceae, diminished microbial diveristy and subsequently resulted in worsened barrier function, increased levels of Citrobacteria, and increased inflammation. Future attempts at manipulation of bacterial populations in the profoundly dysbiotic gut must address community diversity as well as propagation of specific beneficial species.
SUPPLEMENTARY DATA
Supplementary data are available at Inflammatory Bowel Diseases online.
ACKNOWLEDGEMENTS
Author contributions: Karen L. Madsen, Troy Perry, Richard Fedorak, Bryan Dicken, and Michael Laffin designed the study; Michael Laffin, Naomi Hotte, and Heekuk Park conducted the research; Karen Madsen, Michael Laffin,and Heekuk Park carried out the data analysis; Aducio Thiesen performed the histological assessment; Michael Laffin, Heekuk Park, and Karen Madsen wrote the manuscript; All authors read, edited, and approved the final manuscript. The authors wish to thank The Applied Genomics Center at the University of Alberta for sequencing.
Glossary
Abbreviations:
- CD
Crohn's disease
- FOS
Fructooligosaccharide
- ICR
Ileocecal resection
- SCFA
Short chain fatty acids
- TI
Terminal Ileum.
Conflict of Interest: The authors have no conflicts to declare.
Support: Funding by the Canadian Institutes for Health Research (Grant number: 93675).
A portion of this work has been presented at Digestive Disease Week 2016, San Diego, USA and Canadian Digestive Disease Week 2017, Banff, Canada
REFERENCES
- 1. Ley RE, Lozupone CA, Hamady M et al. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol. 2008;6:776–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bindels LB, Delzenne NM, Cani PD, Walter J. Towards a more comprehensive concept for prebiotics. Nat Rev Gastroenterol Hepatol. 2015;12:303–310. [DOI] [PubMed] [Google Scholar]
- 3. Packey CD, Sartor RB. Commensal bacteria, traditional and opportunistic pathogens, dysbiosis and bacterial killing in inflammatory bowel diseases. Curr Opin Infect Dis. 2009;22:292–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gevers D, Kugathasan S, Denson LA et al. The treatment-naive microbiome in new-onset crohn’s disease. Cell Host Microbe. 2014;15:382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. De Vuyst L, Leroy F. Cross-feeding between bifidobacteria and butyrate-producing colon bacteria explains bifdobacterial competitiveness, butyrate production, and gas production. Int J Food Microbiol. 2011;149:73–80. [DOI] [PubMed] [Google Scholar]
- 6. Rios-Covian D, Gueimonde M, Duncan SH et al. Enhanced butyrate formation by cross-feeding between Faecalibacterium prausnitzii and Bifidobacterium adolescentis. FEMS Microbiol Lett. 2015;362 [DOI] [PubMed] [Google Scholar]
- 7. Sha S, Xu B, Wang X et al. The biodiversity and composition of the dominant fecal microbiota in patients with inflammatory bowel disease. Diagn Microbiol Infect Dis. 2013;75:245–251. [DOI] [PubMed] [Google Scholar]
- 8. Peyrin-Biroulet L, Loftus EV Jr, Colombel JF, Sandborn WJ. The natural history of adult crohn’s disease in population-based cohorts. Am J Gastroenterol. 2010;105:289–297. [DOI] [PubMed] [Google Scholar]
- 9. Rutgeerts P. Strategies in the prevention of post-operative recurrence in crohn’s disease. Best Pract Res Clin Gastroenterol. 2003;17:63–73. [DOI] [PubMed] [Google Scholar]
- 10. Lee YW, Lee KM, Chung WC et al. Clinical and endoscopic recurrence after surgical resection in patients with crohn’s disease. Intest Res. 2014;12:117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Onali S, Calabrese E, Petruzziello C et al. Post-operative recurrence of crohn’s disease: A prospective study at 5 years. Dig Liver Dis. 2016;48:489–494. [DOI] [PubMed] [Google Scholar]
- 12. Fedorak RN, Feagan BG, Hotte N et al. The probiotic VSL#3 has anti-inflammatory effects and could reduce endoscopic recurrence after surgery for crohn’s disease. Clin Gastroenterol Hepatol. 2015;13:928–35.e2. [DOI] [PubMed] [Google Scholar]
- 13. Fishbein L, Kaplan M, Gough M. Fructooligosaccharides: a review. Vet Hum Toxicol. 1988;30:104–107. [PubMed] [Google Scholar]
- 14. Kaplan H, Hutkins RW. Fermentation of fructooligosaccharides by lactic acid bacteria and bifidobacteria. Appl Environ Microbiol. 2000;66:2682–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Perry T, Borowiec A, Dicken B et al. Murine ileocolic bowel resection with primary anastomosis. J Vis Exp. 2014:e52106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Borowiec AM, Sydora BC, Doyle J et al. Small bowel fibrosis and systemic inflammatory response after ileocolonic anastomosis in IL-10 null mice. J Surg Res. 2012;178:147–154. [DOI] [PubMed] [Google Scholar]
- 17. Caporaso JG, Kuczynski J, Stombaugh J et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xia J, Sinelnikov IV, Han B, Wishart DS. Metaboanalyst 3.0–making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251–W257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mondot S, Lepage P, Seksik P et al. ; GETAID. Structural robustness of the gut mucosal microbiota is associated with crohn’s disease remission after surgery. Gut. 2016;65:954–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. De Cruz P, Kang S, Wagner J et al. Association between specific mucosa-associated microbiota in crohn’s disease at the time of resection and subsequent disease recurrence: a pilot study. J Gastroenterol Hepatol. 2015;30:268–278. [DOI] [PubMed] [Google Scholar]
- 21. Perry T, Jovel J, Patterson J et al. Fecal microbial transplant after ileocolic resection reduces ileitis but restores colitis in IL-10-/- mice. Inflamm Bowel Dis. 2015;21:1479–1490. [DOI] [PubMed] [Google Scholar]
- 22. Devine AA, Gonzalez A, Speck KE et al. Impact of ileocecal resection and concomitant antibiotics on the microbiome of the murine jejunum and colon. Plos One. 2013;8:e73140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Winkler J, Butler R, Symonds E. Fructo-oligosaccharide reduces inflammation in a dextran sodium sulphate mouse model of colitis. Dig Dis Sci. 2007;52:52–58. [DOI] [PubMed] [Google Scholar]
- 24. Koleva PT, Valcheva RS, Sun X et al. Inulin and fructo-oligosaccharides have divergent effects on colitis and commensal microbiota in HLA-B27 transgenic rats. Br J Nutr. 2012;108:1633–1643. [DOI] [PubMed] [Google Scholar]
- 25. Heuvelin E, Lebreton C, Grangette C et al. Mechanisms involved in alleviation of intestinal inflammation by bifidobacterium breve soluble factors. Plos One. 2009;4:e5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Riedel CU, Foata F, Philippe D et al. Anti-inflammatory effects of bifidobacteria by inhibition of LPS-induced NF-kappab activation. World J Gastroenterol. 2006;12:3729–3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Madsen K, Cornish A, Soper P et al. Probiotic bacteria enhance murine and human intestinal epithelial barrier function. Gastroenterology. 2001;121:580–591. [DOI] [PubMed] [Google Scholar]
- 28. López P, Gueimonde M, Margolles A et al. Distinct bifidobacterium strains drive different immune responses in vitro. Int J Food Microbiol. 2010;138:157–165. [DOI] [PubMed] [Google Scholar]
- 29. Fukuda S, Toh H, Hase K et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature. 2011;469:543–547. [DOI] [PubMed] [Google Scholar]
- 30. Chang JY, Antonopoulos DA, Kalra A et al. Decreased diversity of the fecal microbiome in recurrent clostridium difficile-associated diarrhea. J Infect Dis. 2008;197:435–438. [DOI] [PubMed] [Google Scholar]
- 31. Scher JU, Ubeda C, Artacho A et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol. 2015;67:128–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Manichanh C, Rigottier-Gois L, Bonnaud E et al. Reduced diversity of faecal microbiota in crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moreau NM, Martin LJ, Toquet CS et al. Restoration of the integrity of rat caeco-colonic mucosa by resistant starch, but not by fructo-oligosaccharides, in dextran sulfate sodium-induced experimental colitis. Br J Nutr. 2003;90:75–85. [DOI] [PubMed] [Google Scholar]
- 34. Ten Bruggencate SJ, Bovee-Oudenhoven IM, Lettink-Wissink ML et al. Dietary fructooligosaccharides increase intestinal permeability in rats. J Nutr. 2005;135:837–842. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.