Abstract
Innate immunity in animals including humans encompasses the complement system, which is considered an important host defense mechanism against Aspergillus fumigatus, one of the most ubiquitous opportunistic human fungal pathogens. Previously, it has been shown that the alkaline protease Alp1p secreted from A. fumigatus mycelia degrades the complement components C3, C4, and C5. However, it remains unclear how the fungal spores (i.e. conidia) defend themselves against the activities of the complement system immediately after inhalation into the lung. Here, we show that A. fumigatus conidia contain a metalloprotease Mep1p, which is released upon conidial contact with collagen and inactivates all three complement pathways. In particular, Mep1p efficiently inactivated the major complement components C3, C4, and C5 and their activation products (C3a, C4a, and C5a) as well as the pattern-recognition molecules MBL and ficolin-1, either by directly cleaving them or by cleaving them to a form that is further broken down by other proteases of the complement system. Moreover, incubation of Mep1p with human serum significantly inhibited the complement hemolytic activity and conidial opsonization by C3b and their subsequent phagocytosis by macrophages. Together, these results indicate that Mep1p associated with and released from A. fumigatus conidia likely facilitates early immune evasion by disarming the complement defense in the human host.
Keywords: complement system, complement, Aspergillus, infectious disease, inflammation, Aspergillus fumigatus, complement system, immune evasion, metalloprotease, phagocytosis
Introduction
The complement system is an important arm of the innate immunity that is triggered within minutes after the entry of foreign bodies. Because complement components are present in ample amount in the tissue fluids and blood, they can efficiently recognize and eliminate the invading microorganisms (1). The diverse strategies employed by the complement system for elimination of pathogens are as follows: (i) formation of a membrane attack complex (MAC)5 on the microbial membrane resulting in their lysis; (ii) promotion of microbial phagocytosis by opsonization and their uptake through complement receptors; (iii) recruitment of immune cells by anaphylatoxins; and (iv) enhancement of T and B cell responses (2–6).
In contrast, Aspergillus fumigatus, a saprophytic fungus but an opportunistic airborne pathogen, is known to cause a spectrum of diseases depending on the immune status of an individual. Hosts with immune hypersensitivity are predisposed to allergic aspergillosis and aspergilloma (7–9), whereas those with compromised immune status are susceptible to invasive aspergillosis (10). Immunocompetent individuals, however, are capable of efficiently eliminating the inhaled conidia by innate immune mechanisms (11–13). The last 2 decades have seen a significant increase in the invasive aspergillosis cases because of the widespread use of immunosuppressant drugs, particularly in hemato-oncology patients and transplant recipients (14–17). This has stimulated a consequential interest in understanding the virulence factors employed by the fungus to evade innate immune responses.
A. fumigatus conidia (asexual spores), and sometimes its hyphal fragments, are known to predominantly enter the lung-space through breathing air (18). Earlier studies have shown that both conidial as well as hyphal morphotypes of this pathogen are capable of activating the complement system (19). In addition, it has also been shown that the binding of activated complement component C3b per unit surface of conidia inversely correlates with its virulence. For example, A. fumigatus allows less C3b deposition on its conidia as compared with the nonpathogenic Aspergillus species (20). Furthermore, complement deficiency correlates with dissemination of fungal elements and enhanced susceptibility to fatal invasive systemic infection (21). However, because of the presence of a thick cell wall, it is less likely that the A. fumigatus conidia/hyphae are lysed by complement attack through MAC formation. Hence, clearance of A. fumigatus is expected to be due to complement-mediated opsonization that facilitates phagocytosis and/or release of anaphylatoxins that can enhance A. fumigatus killing by the innate immune cells (22–24).
A successful establishment of Aspergillus infection therefore would require efficient evasion of the complement system as both conidia and hyphae come in contact with complement in the lung. Consistent with this notion, this pathogen has developed a panoply of evasion strategies that include the following: avoiding recognition by complement (25, 26); acquisition of host complement regulators (e.g. factor H (FH), FH-like protein 1 (FHL-1), FH-related protein 1 (FHR-1), and C4b-binding protein (C4BP) (27, 28)); production of complement regulators/inhibitors (29); and degradation of complement components (30).
Secreted proteases from infectious microorganisms are known to be involved in the evasion of the complement system (31–33). One of the A. fumigatus alkaline proteases, Alp1p, secreted by the hyphal morphotype, is known to degrade the complement components (30); this protease effectively degrades all the major components of the complement system: C3, C4, and C5. It is believed that Alp1p plays an important role during cerebral aspergillosis (34). However, Alp1p is only secreted by A. fumigatus hyphal morphotype, and therefore, it is logical to posit that protease(s) other than Alp1p may subvert complement at the conidial stage, thus avoiding its efficient opsonization and engulfment by the phagocytes immediately after its inhalation. In this study, we looked for complement-degrading protease(s) associated with conidia. We observed that the culture supernatant collected upon conidial inoculation in the medium containing collagen or albumin (i.e. the components present in the lung environment) contains complement-degrading activity. The protease was identified as a metalloprotease, Mep1p, that showed proteolytic activity toward the major complement proteins C3, C4, and C5 as well as the pattern recognition molecules properdin, MBL, and ficolin-1, which are involved in the activation of complement pathways, leading to inhibition of activation of all the three complement pathways as well as complement-dependent phagocytosis. Together, our data indicate that degradation of complement proteins by Mep1p is one of the important mechanisms exploited by the A. fumigatus conidia in evading the early host defense posed by the complement system.
Results
A. fumigatus conidia-associated Mep1p degrades complement components
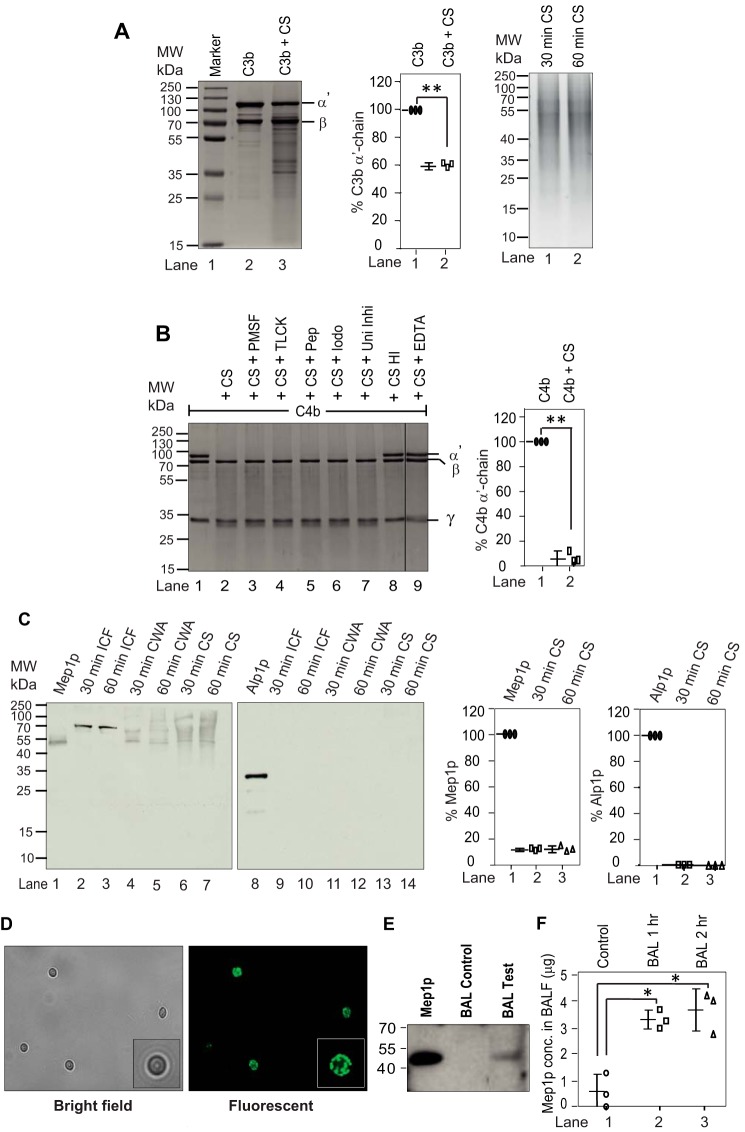
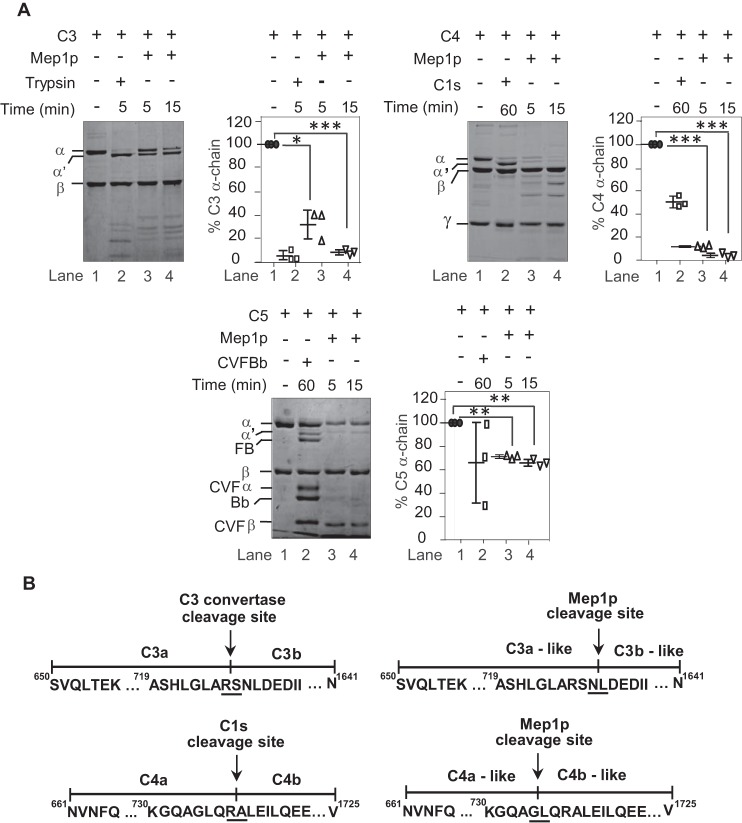
The A. fumigatus morphotype that enters the human lungs and first exposed to the alveolar environment is mainly A. fumigatus conidia. However, human bronchoalveolar lavage is reported to contain complement proteins (35), which prompted us to examine whether A. fumigatus conidia store protease(s) capable of subverting the complement system in the lungs. We thus cultured the wildtype (WT) conidia in the medium containing collagen, to mimic the lung environment, for less than 2 h and assessed the activity of the culture supernatant (CS) against human complement proteins C3b and C4b, which are expected to be generated as a result of complement activation induced by the conidia. The CS showed limited cleavage of C3b but efficiently cleaved C4b; the proteolytic activity was specifically directed against the α′-chain of C4b (Fig. 1, A and B). This indicated that a protease (or proteases) stored in the conidia, and released early on, is responsible for cleaving C4b. Next, to determine the class of conidial protease responsible for cleaving C4b, we inhibited the protease activity by adding various inhibitors. Intriguingly, the activity was inhibited only by EDTA apart from heat inactivation suggesting that C4b is cleaved by a metalloprotease (Fig. 1B).
Figure 1.
Cleavage of complement proteins by A. fumigatus culture supernatant and secretion of proteases from A. fumigatus conidia in collagen-containing medium. A, cleavage of C3b by A. fumigatus culture supernatant (CS). C3b (2 μg) was incubated for 60 min at 37 °C with the CS (1 μg of protein) obtained after 60 min of conidial inoculation in Tris buffer, pH 7.4 (total reaction volume of 30 μl). The reaction mixtures were then run on 10% SDS-PAGE, and the cleaved fragments of C3b were visualized by staining the gel with Coomassie Blue (left panel). The middle panel shows the % cleavage of C3b α′-chain by CS. Data represent mean ± S.D. of three experiments (**, p < 0.005). The right panel shows the CS collected at 30 and 60 min. B, cleavage of C4b by A. fumigatus CS and its inhibition by various protease inhibitors. C4b (2 μg) was incubated for 30 min at 37 °C with the CS (1 μg of protein) obtained after 60 min of conidial inoculation in Tris buffer, pH 7.4 (total reaction volume of 30 μl). The reaction mixtures were then run on 10% SDS-PAGE, and cleaved fragments of C4b were visualized by staining the gel with Coomassie Blue (left panel). The splice line between the lanes 8 and 9 indicates that the gel was spliced at this point. Inhibitors used are as follows: PMSF, phenylmethylsulfonyl fluoride (1 mm); TLCK, tosyl-l-lysyl-chloromethane hydrochloride (1 mm); Pep, pepstatin (11 μg/ml); Iodo, iodoacetamide (1 mm); Uni Inhi, universal inhibitor without EDTA (1×); EDTA, (10 mm). HI indicates heat-inactivated. CS effectively cleaved C4b, which could be inhibited by EDTA suggesting the involvement on metalloprotease in C4b cleavage. The right panel shows the % cleavage of C4b α′-chain by CS in the presence of the indicated inhibitor (**, p < 0.005). Data represent mean ± S.D. of three experiments. C, A. fumigatus conidia were cultured in collagen-containing medium for 30 min or 60 min. The CS was then collected, and the conidia were broken open to collect the intracellular fraction (ICF) and the cell wall autolysate (CWA). The intracellular fraction contained Mep1p in the pro-form (70 kDa), and the cell wall fraction and the culture supernatant contained Mep1p in the activated form (42 kDa); none of the fractions contained Alp1p (left panels). The right panels show % release of Mep1p and Alp1p compared with the control lane (1 μg of Mep1p or Alp1p). Data represent mean ± S.D. of three experiments. D, detection of Mep1p in the permeabilized conidia by immunofluorescence. E, Mep1p in BALF of mice after exposure to WT conidia for 2 h. Released Mep1p was detected by Western blot analysis using polyclonal anti-Mep1p antibody. F, Mep1p in BALF is also quantified by ELISA (mean ± S.D. of three experiments; values represent the Mep1p concentration in 5 ml of BALF collected from each mouse; *, p < 0.05).
Mep1p and Alp1p are the major endoproteases secreted at neutral pH by A. fumigatus when grown in the presence of protein as the nitrogen source (36). We thus next examined whether conidia release Mep1p after culture in liquid medium containing collagen. Western blot analysis revealed that Mep1p, but not Alp1p, was released by the conidia into the collagen medium early on (Fig. 1C; 30–120 min CS were tested, and they were positive for Mep1p but not for Alp1p; Western blotting for 30 and 60 min CS is presented). Mep1p was also released into the medium containing albumin but not into other classical Aspergillus culture media (Aspergillus minimal medium, BRIAN medium, or Sabouraud medium; data not shown), suggesting that the release of Mep1p is medium-specific. Moreover, permeabilization of conidial cell wall followed by probing with anti-Mep1p antibody revealed the presence of Mep1p, suggesting that Mep1p is indeed stored in the conidial cell wall (Fig. 1D). To ascertain whether Mep1p is also released in the lung during infection, we challenged mice with A. fumigatus conidia for 2 h and collected the bronchoalveolar lavage fluid (BALF). As shown in Fig. 1E, there was a band corresponding to Mep1p on Western blotting upon probing SDS-PAGE–separated BALF using polyclonal anti-Mep1p antibody, suggesting that Mep1p is also released into the lung environment. These results were also confirmed by ELISA (Fig. 1F).
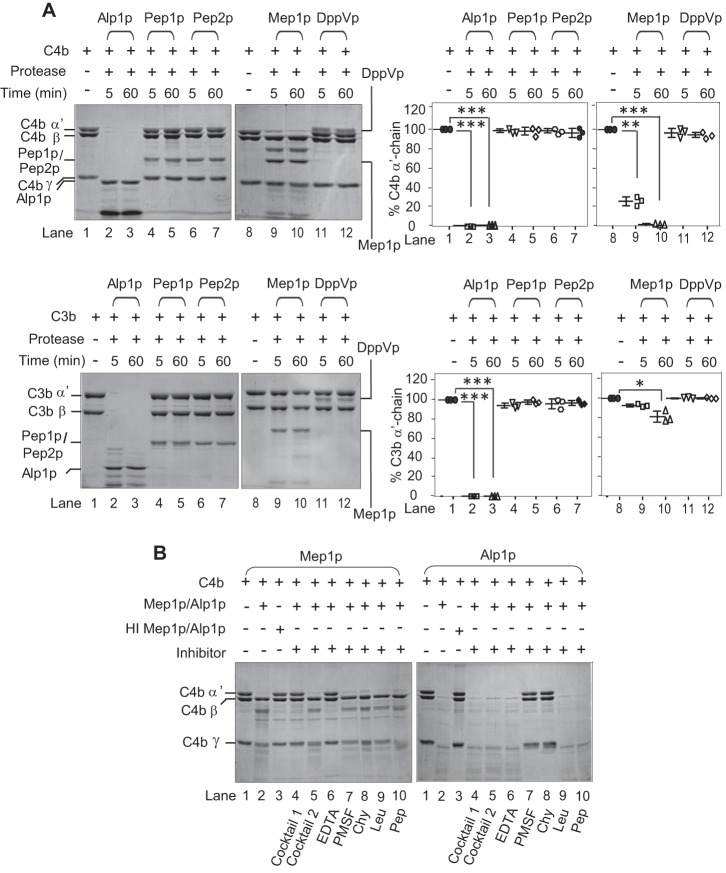
Consequently, to ascertain that Mep1p does have complement-degrading activity, we then expressed it using the Pichia expression system (Fig. S1). We also expressed Alp1p for comparison purposes and three other major A. fumigatus-secreted proteases (Pep1p, Pep2p, and DppVp; Fig. S1) (37). Examination of proteolytic activity of these recombinant proteases against C3b and C4b showed that, unlike Alp1p, Mep1p possesses proteolytic activity only toward C4b; the other three proteases did not show any proteolytic activity toward C3b or C4b at the reaction conditions tested (Fig. 2A). Furthermore, as expected, the C4b-degrading activity of Mep1p could be inhibited by EDTA (Fig. 2B and Fig. S2) but not by other protease inhibitors.
Figure 2.
Proteolytic activity of A. fumigatus proteases toward human complement proteins C4b and C3b. A, proteolytic activity of recombinant proteases (Alp1p, Pep1p, Pep2p, Mep1p, or DppVp) was observed by incubating 1 μg of each of the proteases with 3 μg of C4b (upper left panels) or C3b (lower left panels) for 5 or 60 min in Tris buffer at 37 °C. The cleavage products were visualized by running the samples on 10% SDS-PAGE under reducing conditions and staining them with Coomassie Blue. Among these proteases, only Alp1p and Mep1p showed proteolytic activity toward C4b. The right panels show the % cleavage of α′-chain of C3b and C4b (mean ± S.D. of three experiments; *, p < 0.05; **, p < 0.005; ***, p < 0.0005). B, inhibition of proteolytic activity of Mep1p by various classes of protease inhibitors. Mep1p (0.25 μg) was incubated with C4b (3 μg) in the presence of the indicated inhibitor in Tris buffer for 60 min at 37 °C, and the cleaved fragments were visualized by running the samples on 10% SDS-PAGE and staining with Coomassie Blue. Inhibitors: cocktail 1, complete mini mixture (1×) (Roche Applied Science); cocktail 2, complete mini, EDTA-free (1×) (Roche Applied Science); EDTA (10 mm); PMSF (1 mm); chymostatin (Chy) (100 μg/ml); leupeptin (Leu) (2.5 μg/ml); pepstatin (Pep) (11 μg/ml). The percentage of C4b cleaved was quantitated by densitometric analysis of the α′-chain band and presented as mean ± S.D. of three experiments in Fig. S2. The proteolytic activity of Mep1p for C4b was inhibited by EDTA, whereas that of Alp1p was inhibited by the serine protease inhibitors. Molecular weights: C4b α′-chain, 88,000; C4b β-chain, 75,000; C4b γ-chain, 33,000; C3b α′-chain, 105,000; C3b β-chain, 75,000.
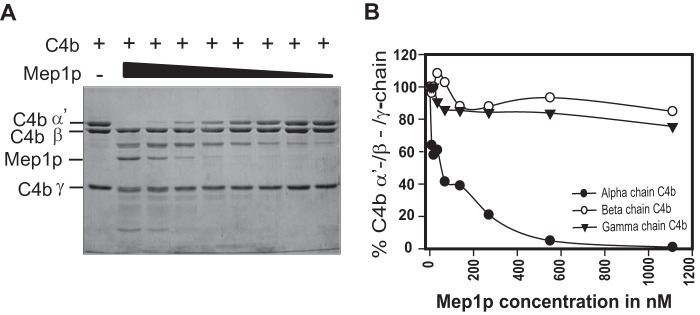
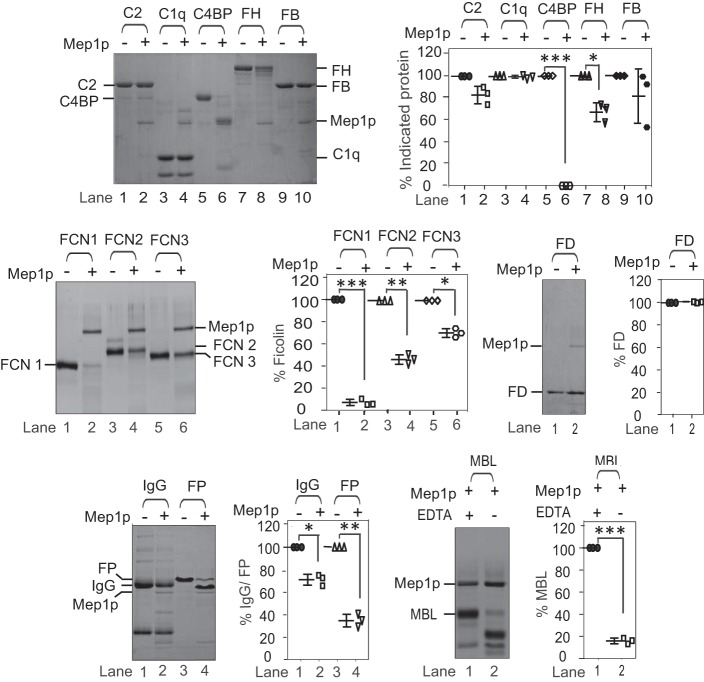
To determine the efficiency, different concentrations of Mep1p were incubated with C4b. The α′-chain of C4b was completely degraded by 580 nm Mep1p (Fig. 3, A and B). To examine whether the proteolytic activity of Mep1p is specific toward C4b, or is directed against other complement components as well, Mep1p was incubated with different complement components: C1q, C2, factor B, factor D, properdin, MBL, ficolins (-1, -2, and -3); complement regulators like C4BP and factor H (FH); and IgG. Mep1p efficiently cleaved properdin (FP), MBL, ficolin-1, and C4BP and showed limited activity toward ficolin-2, -3, IgG, and FH suggesting Mep1p targets multiple complement components (Fig. 4).
Figure 3.
Cleavage of C4b by various concentrations of Mep1p. A, human C4b (3 μg) was incubated with the graded concentrations (1100 to 9 nm) of Mep1p in Tris buffer for 60 min at 37 °C, and cleavage products were resolved on 10% SDS-PAGE under reducing conditions and stained with Coomassie Blue. B, graphical representation of the amount of various C4b chains remained (quantitated by densitometry) after incubation with graded concentrations of Mep1p. Data were normalized by considering the uncleaved protein chains as 100%. Molecular weights: C4b α′-chain, 88,000; C4b β-chain, 75,000; C4b γ-chain, 33,000.
Figure 4.
Proteolytic activity of Mep1p toward different complement proteins. The proteolytic activity of Mep1p was observed by incubating 3 μg of C2, C1q, C4b-binding protein (C4BP), factor H (FH), factor B (FB), IgG, properdin (FP), MBL, ficolin 1 (FCN1), ficolin 2 (FCN2), or ficolin 3 (FCN3), or 1 μg of factor D (FD) with 0.5 μg of Mep1p in 20 μl of Tris buffer for 60 min at 37 °C. The cleavage products were visualized by running the samples on 10% SDS-PAGE under reducing conditions and staining them with Coomassie Blue. The panels on the right side of each gel show % cleavage of the respective protein. Data represent mean ± S.D. of three experiments (*, p < 0.05; **, p < 0.005; ***, p < 0.0005). Mep1p showed strong proteolytic activity toward C4BP, MBL, FCN1, and FP and weak activity toward FH and IgG. Molecular weights: C2, 93,000; C1q, 25,000; C4BP (α-chain), 72,000; FH, 155,000; FB, 93,000; FCN1, 35,500; FCN2, 40,500; FCN3, 37,000; FD, 24,000; IgG H-chain, 50,000; IgG L-chain; 25,000; FP, 53,000; MBL, 33,500.
Mep1p cleaves C3, C4, and C5 into C3b-, C4b-,and C5b-like fragments, which are further inactivated either directly by Mep1p or indirectly by the physiological regulators
Earlier studies with Staphylococcus aureus (32), Tannerella forsythia (38) and snake venom (39) metalloproteases, and a serine protease from Neisseria meningitidis (40) have shown that they convert complement components C3, C4, or C5 into C3b-, C4b-, and C5b-like cleaved products. Hence, we proceeded to determine whether Mep1p also has the ability to cleave C3, C4, and C5. It is clear from Fig. 5A that incubation of Mep1p with these major complement components results in conversion of their α-chains into α′-like chains leading to generation of C3b-, C4b-, and C5b-like fragments. N-terminal sequence analyses of the Mep1p-generated α′-chains of C3b- and C4b-like fragments revealed that the Mep1p cleavage sites on these proteins are 1–3 residues away from the physiological convertase-cleaving sites; the α′-chain of the C5b-like fragment could not be sequenced (Fig. 5B).
Figure 5.
Proteolytic activity of Mep1p toward C3, C4, and C5. A, cleavage of C3, C4, and C5 by Mep1p. Three micrograms of purified C3, C4, or C5 were incubated with 0.5 μg of Mep1p in Tris buffer for the indicated time period at 37 °C and analyzed on SDS-PAGE. The reactions were run on 7.5, 9, and 6% SDS-PAGE for C3, C4, and C5, respectively, under reducing conditions. The controls were formed by incubating C3 with trypsin or C4 with C1s or C5 with CVF Bb, i.e. C5 convertase. Mep1p converts C3, C4, and C5 into C3b-/C3a-like, C4b-/C4a-like, and C5b-/C5a-like fragments. The panels on the right side of each gel show % cleavage of α-chains of the respective protein. Data represent mean ± S.D. of three experiments (*, p < 0.05; **, p < 0.005; ***, p < 0.0005). B, identification of the Mep1p cleavage site on C3 and C4 by N-terminal sequencing of the cleaved α′-chains. The α′-chains of C3b-like and C4b-like obtained after the Mep1p cleavage as described in A were subjected to N-terminal sequencing. The panels show the cleavage site for the respective convertases and Mep1p on C3 (upper sequences) and C4 (lower sequences). Mep1p cleaved C3 and C4 close to their physiological cleavage site by the respective convertases. Molecular weights: C3 α-chain, 110,000; C3 β-chain, 75,000; C4 α-chain, 97,000; C4 β-chain, 75,000; C4 γ-chain, 33,000; C5 α-chain, 115,000; C5 β-chain, 75,000; FB, 93,000; Bb, 60,000; CVF α-chain, 68,000; CVF β-chain, 48,000.
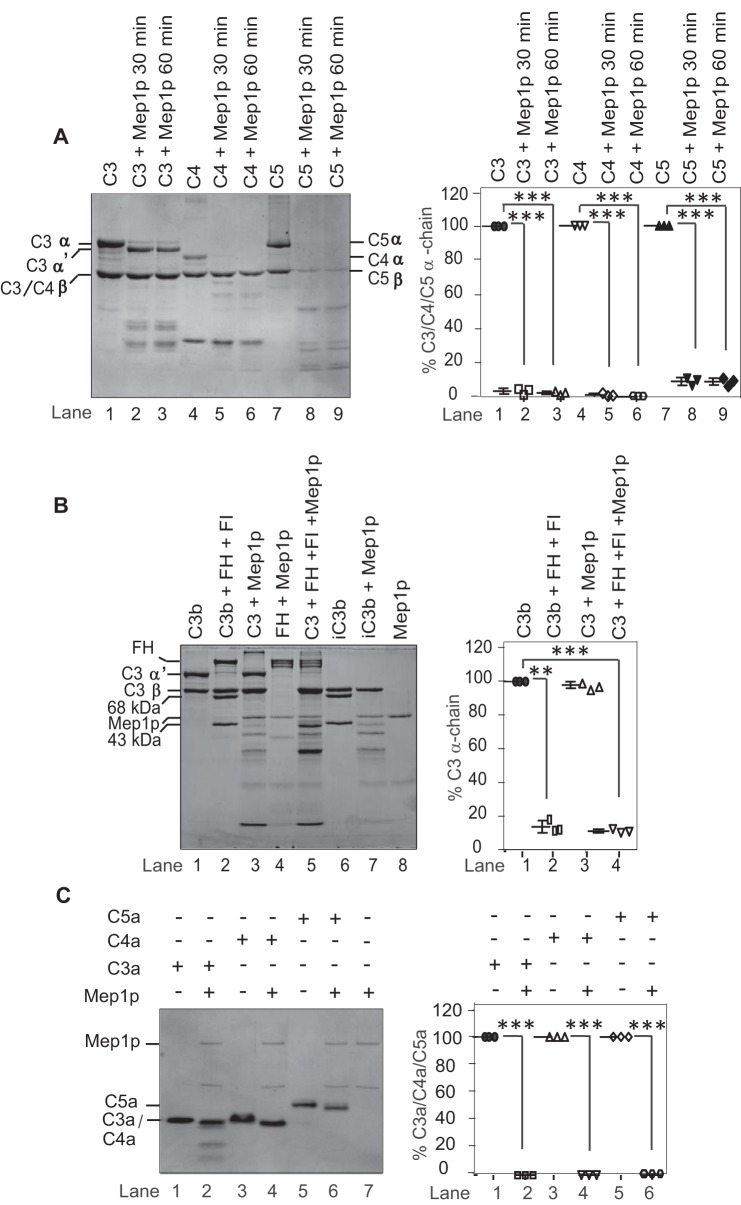
Because cleavage of C3, C4, and C5 would result in better clearance of conidia (41, 42), we asked whether the generated C3b-, C4b-, and C5b-like fragments are stable or are further cleaved and inactivated by Mep1p. Mep1p efficiently cleaved the generated C4b-like and C5b-like fragments; α′-chains of C4b- and C5b-like fragments were completely degraded upon prolonged incubation with Mep1p. However, Mep1p was unable to cleave the α′-chain of the C3b-like fragment (Fig. 6A). Nevertheless, such generation of a C3b-like fragment would occur in the fluid phase away from the conidial surface, and hence we tested whether the Mep1p-generated C3b-like fragment is cleaved by the physiological complement regulators FH and factor I (FI). But, of note, we did observe that Mep1p is also capable of cleaving FH (Fig. 4), which raised the following question: does cleaved FH possess the cofactor activity? Our results demonstrated that the Mep1p-cleaved FH does retain this activity. In this experiment, Mep1p was first incubated with FH, and then C3 and FI were added to the same reaction mixture. During incubation, Mep1p cleaved C3 into a C3b-like fragment, which was further degraded indicating that even though FH is cleaved by Mep1p it retains the cofactor activity (Fig. 6B). FH supports FI-mediated cleavage of C3b into inactivated C3b or iC3b, which possesses cleaved α′-chain (α′-chain is cleaved into N-terminal α-68-kDa and C-terminal α-43-kDa fragments; Fig. 6B, lane 6). Because we did not observe cleaved α-chain fragment (α-68-kDa and C-terminal α-43-kDa fragments) in the presence of Mep1p (Fig. 6B, lane 5), we examined whether these fragments are unstable in the presence of Mep1p. Incubation of purified iC3b with Mep1p resulted in cleavage of both these fragments (Fig. 6B, lane 7), suggesting that iC3b is further cleaved by Mep1p to a C3d-like fragment.
Figure 6.
Inactivation of C3, C4, C5, and the anaphylatoxins by Mep1p and demonstration that Mep1p-cleaved factor H retains its cofactor activity. A, 3 μg of purified native C3, C4, or C5 were incubated with Mep1p (0.5 μg) in Tris buffer for 30 or 60 min at 37 °C. The cleaved products were analyzed by 10% SDS-PAGE under reducing conditions, and the gel was stained with Coomassie Blue. For both time points, the α-chain of C3 was only partially cleaved by Mep1p, whereas the α-chains of C4 and C5 were completely cleaved by Mep1p. The β-chain of C5 was also partially cleaved by Mep1p. The right panel shows the % cleavage of C3, C4, and C5 α-chains. Data represent mean ± S.D. of three experiments (***, p < 0.0005). B, Mep1p-cleaved factor H retains its cofactor activity. Lane 1, C3b (3 μg) alone; lane 2, C3b (3 μg) was degraded when incubated with factor H (FH) (1 μg) and factor I (FI) (150 ng); lane 3, C3 (3 μg) was converted to its C3b-like fragment when incubated with Mep1p (0.5 μg); lane 4, FH (1 μg) was cleaved by Mep1p (0.5 μg). Lane 5, in the presence of Mep1p (0.5 μg) and FI (0.15 μg), both C3 (5 μg) and FH (1 μg) were cleaved suggesting Mep1p cleaved FH retains the cofactor activity; lane 6, iC3b (3 μg) alone; lane 7, iC3b (3 μg) was further cleaved to C3d-like fragment when incubated with Mep1p (0.5 μg); lane 8, Mep1p (0.5 μg) alone. The right panel shows the % C3 α-chain cleavage (mean ± S.D. of three experiments; **, p < 0.005; ***, p < 0.0005). C, cleavage of C3a, C4a, and C5a by Mep1p. One microgram of purified C3a, C4a, or C5a was incubated with 0.5 μg of Mep1p in Tris buffer for 60 min at 37 °C and analyzed on a 16% Tricine gel under reducing condition. The right panel shows the percent cleavage of C3a, C4a, and C5a by Mep1p (mean ± S.D. of three experiments; ***, p < 0.0005). Mep1p cleaved all the anaphylatoxins. Molecular weights: C3 α-chain, 110,000; C3 β-chain, 75,000; C4 α-chain, 97,000; C4 β-chain, 75,000; C4 γ-chain, 33,000; C5 α-chain, 115,000; C5 β-chain, 75,000; C3b α′-chain, 105,000; C3b β-chain, 75,000; FH, 155,000; C3a, 9,083; C4a, 8,757; C5a glycosylated, 10,400.
Physiological activation of C3, C4, and C5 into C3b, C4b, and C5b results in generation of anaphylatoxins C3a, C4a, and C5a. We therefore also examined whether Mep1p inactivates C3a, C4a, and C5a. Incubation of C3a, C4a, and C5a with Mep1p resulted in the cleavage of these anaphylatoxins into smaller fragments (Fig. 6C). Following N-terminal sequencing and high-resolution Orbitrap MS analysis of the Mep1p-cleaved C3a, C4a, and C5a, it was observed that all three anaphylatoxins were cleaved at the C terminus, which removed the C-terminal arginine, suggesting that Mep1p inactivates the anaphylatoxins (Table 1 and Fig. S3). Moreover, C3a and C4a were also trimmed at their respective N termini (Table 1).
Table 1.
Mep1p cleavage site on C3a, C4a, and C5a as determined by N-terminal sequencing and mass spectrometry
| Peptide | Sequence | N-terminal sequence | Molecular mass |
|
|---|---|---|---|---|
| Expecteda | Observedb | |||
| C3a | 650SVQLTEKRMDKVGKYPKELRKCCEDGMRENPMRFSCQRRTRFISLGEACKKVFLDCCNYITELRRQHARASHLGLAR726 | SVQLT | 9094.65 | 9082.5102 |
| Mep1p-cleaved C3ac | LTEKRMDKVGKYPKELRKCCEDGMRENPMRFSCQRRTRFISLGEACKKVFLDCCNYITELRRQHARASH | LTEKR | 8258.0335 | |
| C4a | 661NVNFQKAINEKLGQYASPTAKRCCQDGVTRLPMMRSCEQRAARVQQPDCREPFLSCCQFAESLRKKSRDKGQAGLQR737 | NVNFQ | 8764.07 | 8752.2559 |
| Mep1p-cleaved C4ac | FQKAINEKLGQYASPTAKRCCQDGVTRLPMMRSCEQRAARVQQPDCREPFLSCCQFAESLRKKSRDKGQAG | FQKAI | 8355.0326 | |
| C5a | 660TLQKKIEEIAAKYKHSVVKKCCYDGACVNNDETCEQRAARISLGPRCIKAFTECCVVASQLRANISHKDMQLGR733 | TLQKK | 8273.63 | 10585.8926 |
| Mep1p-cleaved C5ac | TLQKKIEEIAAKYKHSVVKKCCYDGACVNNDETCEQRAARISLGPRCIKAFTECCVVASQLRANISHKD | TLQKK | 10000.5840 | |
a Data are based on the primary sequence.
b Data are based on mass spectrometry.
c The Mep1p-cleaved C3a, C4a, and C5a obtained as described in Fig. 6C were subjected to N-terminal sequencing and mass spectrometry analysis by Orbitrap. Mep1p cleaved and inactivated all the anaphylatoxins by removing the C-terminal arginine.
Mep1p inhibits all three major pathways of the complement system
The results presented above using purified complement components suggest that Mep1p targets the early complement components like properdin, MBL, ficolins, C3, and C4 as well as one of the terminal components C5. It therefore suggested that such Mep1p-mediated inactivation of complement would result in inhibition of all three major pathways of complement activation. Hence, we next determined whether Mep1p is capable of inhibiting complement pathways when whole human serum is used as a source of complement. For this, we employed hemolytic assays as well as the Wieslab complement screen ELISA.
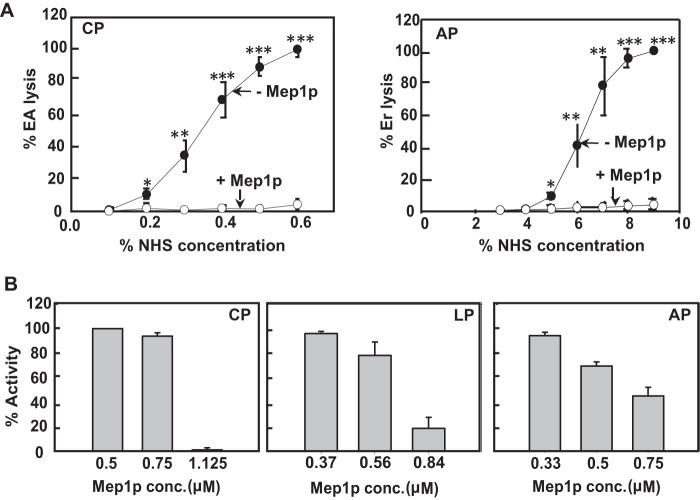
To measure the effect of Mep1p on complement using hemolytic assays, Mep1p was first incubated with 10% normal human serum (NHS) in the presence of Ca2+, and then the hemolytic activity of the treated sera was examined for the classical and alternative pathways under appropriate conditions. Mep1p showed significant inhibitory effects on both classical as well as alternative pathway-mediated lysis of erythrocytes. The addition of 0.6 μm Mep1p to NHS completely blocked the activation of both the pathways (Fig. 7A), indicating that it is a potent inactivator of the classical as well as alternative complement pathways.
Figure 7.
Mep1p inhibits activation of all the three major complement pathways. A, examination of the effect of Mep1p on the classical and alternative pathways by employing hemolytic assays. Normal human serum (NHS; 10%) was incubated with Mep1p (1.5 μg) in appropriate buffer conditions (see “Experimental procedures”) for 60 min at 37 °C and then the graded concentration of this mixture was further incubated with the sensitized sheep erythrocytes (EA) for CP activation or rabbit erythrocytes (Er) for the AP activation. The percentage of lysis of erythrocytes indicate the percentage activity of CP (left panel) and AP (right panel). Results are expressed as mean ± S.D. of three individual experiments. Mep1p inhibited both CP and AP. (*, p < 0.05; **, p < 0.005; ***, p < 0.001). B, examination of the effect of Mep1p on the classical, alternative, and lectin pathways by employing ELISA assays. NHS (∼30%) was incubated with the indicated concentration of Mep1p for 60 min at 37 °C, and then the effect on the activation of various pathways was measured by the Wieslab ELISA kit. The IC50 of Mep1p for the inhibition of CP, AP, and LP were 0.9, 0.7, and 0.7 μm, respectively. Mep1p effectively inhibited all the three complement pathways.
To further validate these results and obtain a quantitative estimate of the inhibitory effect of Mep1p on the individual complement pathways, we used the commercially available ELISA-based functional Wieslab assay kit. Mep1p showed inhibition of the classical, alternative, and lectin pathways with IC50 values of about 0.9, 0.7, and 0.7 μm, respectively (Fig. 7B). Thus, the order of inhibition was lectin pathway = alternative pathway > classical pathway.
Mep1p inhibits C3b deposition on conidia and consequently their phagocytosis
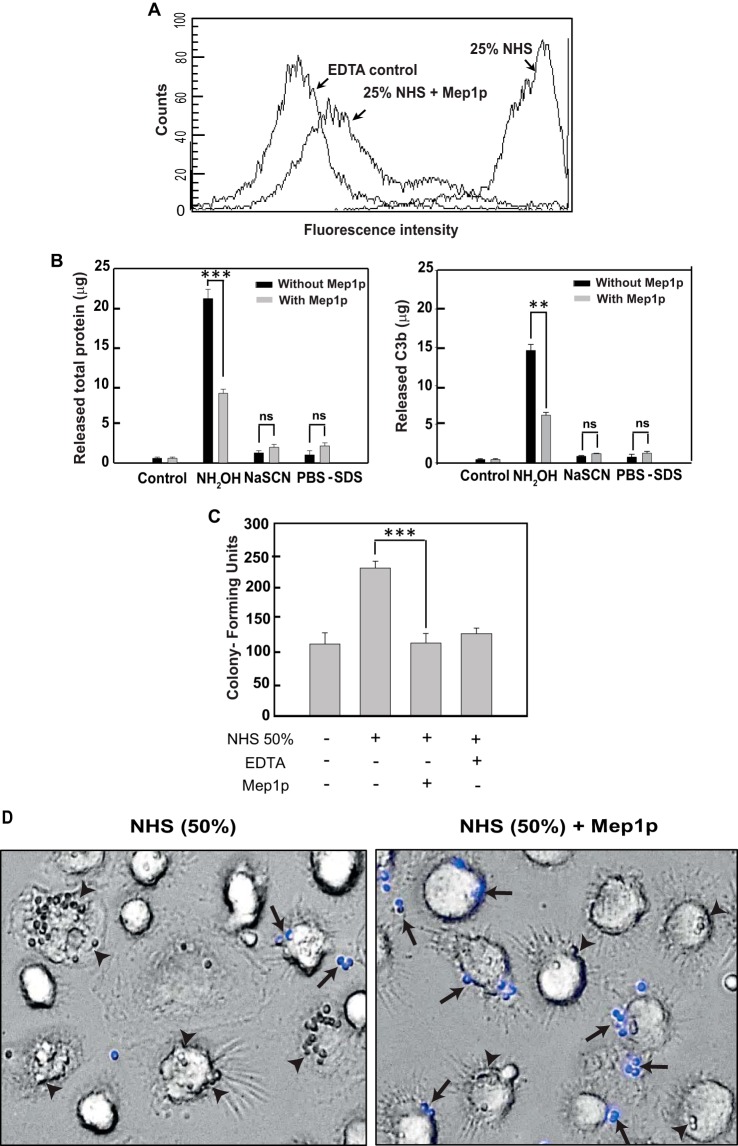
Phagocytosis is an essential step in clearing inhaled A. fumigatus conidia and is suggested to be dependent on the conidial opsonization by complement components that are recognized by the complement receptors on the phagocytes (43). As Mep1p showed inactivation of different complement pathways, we also investigated whether it inhibits C3b deposition on the conidial surface when serum is used as a source of complement. Herein, heat-inactivated dormant conidia were incubated with NHS or NHS pretreated with Mep1p, and the amount of C3b deposited on the conidial surface was detected by FITC-labeled anti-C3 antibody using flow cytometry. Incubation of conidia with NHS resulted in considerable deposition of C3b molecules on the conidial surface, and this deposition was inhibited by EDTA, which blocks complement activation. Importantly, pretreatment of NHS with Mep1p resulted in significant reduction in C3b deposition on conidia (Fig. 8A), suggesting that Mep1p indeed depletes C3 from the serum.
Figure 8.
Mep1p inhibits C3b deposition on conidia and their phagocytosis. A, inhibition of C3b deposition by Mep1p. NHS (50%) was incubated with or without Mep1p (3 μg) in GVB2+ buffer for 60 min at 37 °C, and then this reaction mixture was further incubated with heat-inactivated conidia (1 × 106). Control sample contained 20 mm EDTA. Bound C3b was detected by FACS using FITC-conjugated F(ab′)2 anti-C3 goat IgG. Mep1p considerably inhibited C3b deposition on conidia. B, C3b are bound to the conidial surface by ester linkage. Opsonized conidia were treated with NH2OH, NaSCN, or PBS/SDS, and the eluted proteins were estimated by Bradford assay (left panel) and ELISA (right panel; for details see the “Experimental procedures”) (**, p < 0.01; ***, p < 0.001). ns, not significant. In the case of opsonized conidia in the presence of Mep1p, the treatment of NH2OH released a significantly less amount of C3b. Mep1p inhibited covalent attachment of C3b on the surface of conidia. C and D, inhibition of phagocytosis by Mep1p. Conidia (1 × 106) were opsonized by incubating with NHS (50%; 200 μl) for 60 min at 37 °C in the presence or absence of Mep1p (1 μg/200 μl 50% NHS), and the treated conidia were incubated with macrophages. Opsonizing conidia with NHS in the presence of Mep1p significantly decreased the conidial uptake by human monocyte-derived macrophages by at least 60% (***, p < 0.001). D, arrowheads indicate phagocytosed and arrows indicate adherent/nonphagocytosed conidia. The adherent/nonphagocytosed conidia were detected by calcofluor white staining. Mep1p treatment reduced the number of phagocytosed conidia.
To determine whether the deposited C3b molecules are linked to the conidial surface via covalent linkage, we treated opsonized conidia with NH2OH, NaSCN, or PBS/SDS and estimated the released protein by Bradford assay and ELISA using anti-C3b antibody. The majority of protein bound to conidia was released by NH2OH, but not NaSCN or PBS/SDS (Fig. 8B), and this NH2OH-releasable protein fraction was anti-C3b antibody-positive on ELISA, indicating that C3b molecules are indeed attached to the conidial surface by the ester linkage. However, preincubation of NHS with Mep1p followed by conidial opsonization resulted in a significant decrease in the NH2OH-releasable C3b from the opsonized conidial surface (p < 0.01; Fig. 8B).
Furthermore, to study the effect of Mep1p on complement-dependent phagocytosis, conidia were first opsonized with NHS in the presence or absence of Mep1p followed by feeding them to human monocyte-derived macrophages. There was a significant decrease in the phagocytosis upon opsonizing conidia with NHS in the presence of Mep1p, indicating that the complement-depleting role of Mep1p adversely affects conidial opsonization and phagocytosis (Fig. 8, C and D).
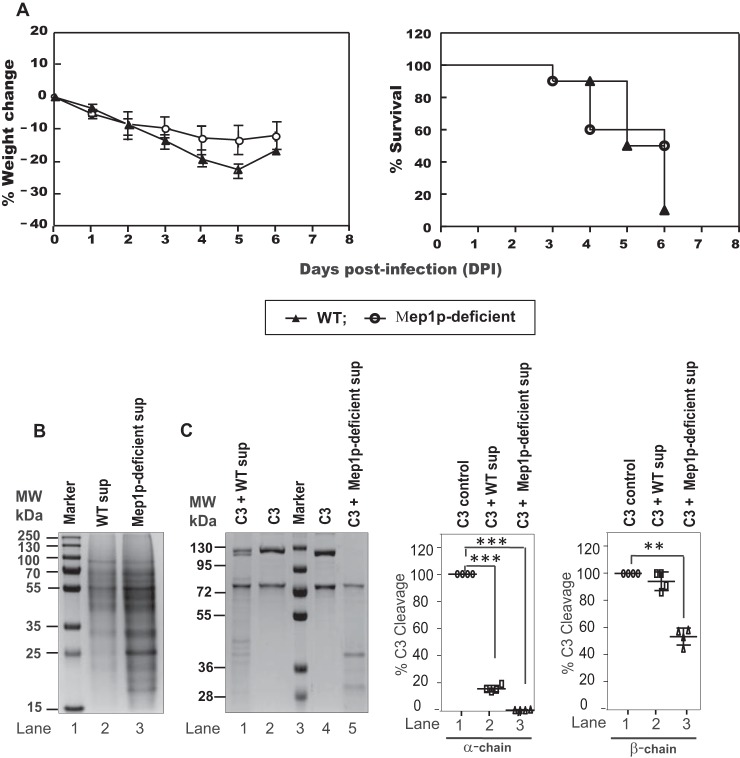
In vivo study using a murine model
An in vivo experiment was performed using a cyclophosphamide-immunosuppressed mouse model. However, there was no significant difference between the body weight and survival curves of the mice intranasally challenged with the WT and Mep1p-deficient conidia (Fig. 9A). Nevertheless, when SDS-PAGE was performed with the Mep1p-deficient conidial collagen culture supernatant, the protein profile was different from that of the WT collagen culture supernatant (Fig. 9B). Moreover, unlike the WT conidial collagen culture supernatant that could not degrade C3, at least not beyond the C3b stage, the mutant conidial collagen culture supernatant produced substantial degradation of C3 (both α- and β-chains) (Fig. 9C). This could be because of the stress due to MEP1 deletion altering the released protein composition, which is still capable of degrading the complement proteins but in a different manner, and hence obscuring the effect of MEP1 deletion in the in vivo mouse model.
Figure 9.
Virulence of Mep1p-deficient and WT strain in cyclophosphamide-treated murine infection model. A, weight loss and survival of cyclophosphamide-immunosuppressed mice challenged with WT or Mep1p-deficient conidia (10 mice per group and two replications; totally 20 mice per group). Mice with weight loss of more than 30% of its original weight were sacrificed and considered dead for calculations of mortality. Although the survival was higher in the mouse group challenged by Mep1p-deficient (50%) compared with the mouse group challenged by WT (10%) conidia, the difference was not statistically significant (p = 0.28; log-rank test). B, SDS-PAGE showing secreted protein profiles of WT and Mep1p-deficient mutant in the collagen culture supernatant. The gel was stained using Coomassie Blue. C, SDS-PAGE showing degradation of C3 α- as well as β-chain by WT and Mep1p-deficient conidial collagen culture supernatant. In this experiment, C3 was incubated with culture supernatant for 1 h at 37 °C. The right panels show the percent cleavage of C3 α- and β-chains by WT and Mep1p-deficient culture sup (mean ± S.D. of three experiments; **, p < 0.005; ***, p < 0.0005).
Discussion
A. fumigatus is known to secrete proteases that include serine proteases, metalloproteases, aspartic proteases, and dipeptidyl-peptidases, and the abundance of each protease varies depending on the surrounding medium. The primary function of A. fumigatus secreted proteases is to degrade organic matter from the environment or the host tissue either to obtain nutrients or for invasion (14, 44). In this study, we show that Mep1p, a metalloprotease released from A. fumigatus conidia, is capable of cleaving several complement molecules, thus inactivating the complement system, an integral part of the host innate immunity. We demonstrate that Mep1p directly cleaves MBL, ficolins, properdin, and C4 and C5 molecules. It also inactivates C3, the central component of the complement system, nevertheless, indirectly. It first generates the C3b-like fragment, but in fluid phase, which is further degraded by the host regulators FH and FI. Additionally, we show that Mep1p is proficient in inactivating the anaphylatoxins/chemoattractants C3a, C4a, and C5a, which are generated during the activation of C3, C4, and C5, respectively. Together, this study demonstrates that Mep1p inhibits activation of all three complement pathways as well as effector functions of the complement system by cleaving and inactivating the complement components.
A. fumigatus was shown to down-regulate complement activation on its surface by recruiting FH and C4BP, the host complement regulators, and by secreting Alp1p, an alkaline protease that degrades host complement components present in the surrounding milieu (27, 28, 30). The possible primary function of Alp1p is degradation of the host structural proteins collagen/elastin, facilitating fungal invasion and acquisition of nutrients (45). In support of this role, Alp1p was found to be secreted into the in vitro culture medium containing collagen or albumin. Of note, Alp1p was found to be secreted by the mycelial morphotype. However, it was shown that Mep1p is secreted in a substantial amount from ALP1 deletion mutant mycelia, which is sufficient for proteolytic degradation of collagen (36). Moreover, antibodies against Mep1p were detected in the sera from patients suffering from aspergilloma as compared with healthy individuals, suggesting that Mep1p is potentially secreted during infection (36). Interestingly, we observed that Mep1p is released into in vitro culture medium, specifically containing collagen or albumin, during the early growth phase (i.e. within 30–60 min of conidial contact with the culture medium mimicking the lung environment). These data along with immunolabeling assay that showed positive fluorescence for dormant conidial cell wall with anti-Mep1p antibody suggest that Mep1p is associated with A. fumigatus conidia and is likely to play a specific role in conidial survival.
We show that Mep1p blocks the alternative complement pathway (AP) by two different ways: 1) by generating C3b-like fragment in the fluid phase, facilitating its degradation by FH and FI; and 2) by cleaving properdin, which is a positive regulator of the AP C3-convertase (C3bBb) capable of stabilizing AP C3-convertase (46). Igs also contribute to AP activation by binding to the pathogen surface and facilitating the recruitment and activation of C3 (47, 48); however, Mep1p showed limited IgG-cleavage activity. Furthermore, we show that Mep1p also restricts the lectin pathway (LP) activation by two different ways: by targeting MBL, which initiates the pathway, and by degrading the α-chain of C4. Both the AP and LP are activated immediately within minutes after sensing the pathogens and do not require the presence of antibodies. A recent report has shown that in the absence of antibodies, MBL is the key initiator of complement on the resting conidia, whereas AP amplifies the complement activation initiated by MBL (49). Thus, the release of Mep1p from conidia is expected to protect this morphotype from these complement pathways. We observed that in addition to AP and LP, Mep1p also targets the CP by degrading C4. Earlier studies have shown that in normal human serum, initiation of complement activation on conidia primarily comes from the classical pathway (22, 49). Thus, Mep1p is expected to subvert the complement system even in immunocompetent individuals.
The mechanism of Mep1p in degrading the major complement components C3, C4, and C5 was found to be divergent from that of Alp1p. There was a complete degradation of C3, C4, and C5 by Alp1p, whereas Mep1p initially cleaved C3, C4, and C5 close to physiological convertase cleavage sites and thus generated C3a-/C3b-like, C4a-/C4b-like, and C5a-/C5b-like fragments. It further degraded the C4b-like and C5b-like fragments, dampening the activation of complement pathways directly, particularly the CP and LP, as well as the terminal pathway by inhibiting the MAC formation. The cleavage of C3, C4, and/or C5 into C3b-, C4b-, and C5b-like forms is also known to be mediated by metalloproteases of multiple pathogens such as S. aureus (aureolysin (32)), T. forsythia (karilysin (50)), T. forsythia (mirolysin (31)), Pseudomonas aeruginosa (elastase (51)), Leptospira strains (thermolysin (52)), as well as snake venom (39). However, the exact site of cleavage is not known in most cases except aurolysin, which cleaves C3 at the identical site as Mep1p. Apparently, these proteases seemed to convert C3, C4, and C5 into their active forms, but the conversion occurs in the fluid phase, facilitating their further degradation by host factors, as only pathogen-bound forms of these active components are likely to be protected (53).
Mep1p could also cleave and inactivate C3a, C4a, and C5a released during activation of C3, C4, and C5, respectively. This concurred with an earlier report that showed that gelatinase E, a metalloprotease from Enterococcus faecalis, could degrade C3a and dampen the chemoattractant response. Unlike gelatinase E, aureolysin from S. aureus was unable to cleave C3a directly; however, in the presence of human serum and with the help of host components aureolysin could inactivate C3a (32). Thus, direct inactivation of C3a/C5a by Mep1p may restrict migration of neutrophils and macrophages, leading to inefficient phagocytosis, clearance, and adaptive immune responses, hence creating favorable conditions for the germination of conidia.
A central question is whether Mep1p functions as a virulence factor of A. fumigatus? Earlier studies have demonstrated that there was no difference in the pathogenicity of the WT strain and the MEP1-ALP1 double-deficient mutant in the immunocompromised murine model (54). We also observed similar results; Mep1p-deficient A. fumigatus showed virulence similar to that of the WT strain in the immunosuppressed murine model. To determine whether Mep1p deletion induces a stress that alters the secretome, we examined the protein profiles of collagen culture supernatants of the WT and Mep1p-deficient A. fumigatus mutant by SDS-PAGE. Indeed, the protein profile of the mutant conidial collagen culture supernatant was more abundant and slightly different from that of the WT collagen culture supernatant. Moreover, unlike the WT conidial collagen culture supernatant that converted C3 α-chain into α′-form, the MEP1-deletion mutant conidial collagen culture supernatant completely degraded the C3 α-chain. Thus, A. fumigatus seems to have built protease redundancy to take care of the adverse changes in the environmental conditions.
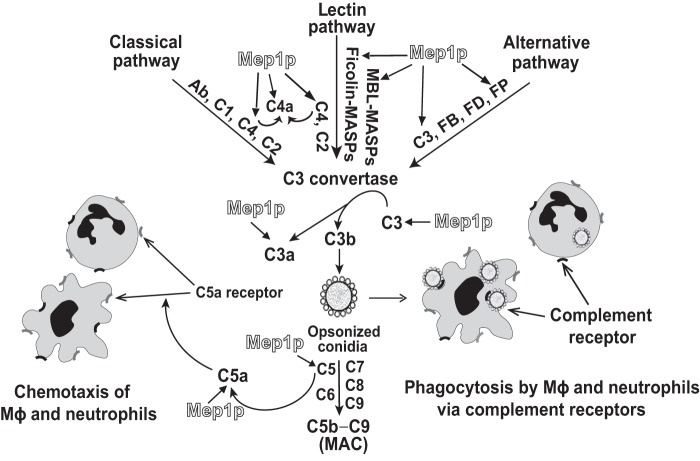
In summary, our study suggests that complement proteins, also found in the alveolar environment, facilitate A. fumigatus conidial opsonization and thereby their phagocytosis, as unopsonized conidial phagocytosis was significantly lower compared with the opsonized conidia. At the same time, A. fumigatus conidium is endowed with a metalloprotease, Mep1p, which is efficient in inhibiting all three major complement activation pathways, and as a consequence, complement-mediated opsonization and phagocytosis of conidia and generation of C5a (Fig. 10), a potent chemoattractant for neutrophils and macrophages (41). Furthermore, consistent with the earlier observations (55, 56), we show that Mep1p is secreted into the medium containing albumin/collagen, the constituents of the lung matrix (57–59). We thus propose that the release of Mep1p from conidia in the lung environment is likely to subvert the complement system and consequently inhibit the conidial clearance by phagocytes (Fig. 10). Our study thus adds to the understanding of early evasion mechanisms exploited by A. fumigatus conidia against host complement defense.
Figure 10.
Proposed model for Mep1p-mediated inactivation of complement by A. fumigatus. Mep1p released from WT conidia (Fig. 1) is efficient in cleaving pattern recognition molecules such as properdin, MBL, and ficolin-1 (Fig. 4), and major complement proteins C3, C4, and C5 (Figs. 5 and 6), resulting in reduced opsonization of conidia (Fig. 8) as well as the MAC formation on their surface. Inhibition of opsonization by Mep1p results in decreased phagocytosis of conidia by macrophages (Mϕ) (Fig. 8) and is also expected to result in decreased phagocytosis by neutrophils. Mep1p also inactivates C5a (Fig. 6 and Table 1), which is a potent chemoattractant for macrophages and neutrophils and thus is expected to reduce chemotaxis of these cells. Together the data presented here indicate that Mep1p-mediated complement subversion would result in decreased clearance of conidia, which would facilitate their colonization and invasion.
Experimental procedures
A. fumigatus strains, culture media, growth conditions, and cell fractionation
A. fumigatus clinical isolate CBS 144-89 and G10 (a nitrate reductase mutant of CBS 144-89) were used as the WT strains; MEP1 deletion was performed on the G10 background (36, 60, 61). These fungal strains were maintained on 2% malt agar slants at ambient temperature. Liquid culture media used were Aspergillus minimal, BRIAN, Sabouraud, collagen (0.1% insoluble collagen in water), and BSA (0.1% in water). To obtain culture supernatant, 10-day-old conidia (1 × 108) were harvested from the malt-agar slants using Tween/water (0.05% Tween 80), inoculated into 5 ml of culture medium, and incubated at 37 °C in a shaken condition for different time intervals. Culture in the collagen medium was passed through 0.45-μm mesh-size filters; thus, the collagen medium filtrate as well as the cultures in other media were subjected to centrifugation (4500 rpm, 10 min) to obtain the supernatant. Pelleted conidia from the above cultures were washed and broken using 0.5-mm glass beads in a FastPrep (MP Biomedicals); the contents were centrifuged (4000 rpm, 10 min), and the supernatant (intracellular fraction) was collected. The pellet (cell wall) was incubated with sodium acetate buffer (20 mm, pH 5.5) to obtain cell wall autolysate.
Serum, antibodies, proteins, and buffers
To obtain normal human serum (NHS), blood was collected from a healthy donor, allowed to clot at 37 °C for 30 min, centrifuged, and stored in aliquots at −80 °C until use. Polyclonal rabbit antibodies against Alp1p and Mep1p were raised as described earlier (54, 62). FITC-conjugated goat anti-C3 IgG F(ab′)2 antibody was purchased from MP Biomedicals (Santa Ana, CA). Complement components C5 and C1q were purchased from Calbiochem, and C2, C4, C4b, C4BP, and factor D were purchased from Complement Technology, Inc. (Tyler, TX). Human C3 (63), C3b (64), factor B (65), factor H (63), and CVF (66) were purified as described. Cyclophosphamide was purchased from Sigma. Buffers used were as follows: Tris buffer (50 mm Tris, 100 mm NaCl, and 2 mm MgCl2, pH 7.4); HEPES buffer (20 mm HEPES, 140 mm NaCl, 5 mm CaCl2, and 2.5 mm MgCl2, pH 7.4); acetate buffer (20 mm acetate, pH 4.5); veronal-buffered saline (VBS) (5 mm barbital, 145 mm NaCl, and 0.02% sodium azide, pH 7.4); VBS2+ (VBS with 0.5 mm MgCl2, and 0.15 mm CaCl2); gelatin veronal buffer (GVB) (VBS with 0.1% gelatin); GVB2+ (GVB with 0.5 mm MgCl2 and 0.15 mm CaCl2); GVB/EDTA (GVB with 10 mm EDTA); Mg-EGTA (0.1 m MgCl2 and 0.1 m EGTA); PBS/Tween (PBS-T) (10 mm sodium phosphate, 145 mm NaCl, and 0.05% (v/v) Tween 20, pH 7.4); and TBS/Tween (TBS-T) (20 mm Tris-HCl, 150 mm NaCl, and 0.05% (v/v) Tween 20, pH 7.5).
Expression and purification of A. fumigatus proteases
A. fumigatus complementary DNAs (cDNAs) encoding the proteins of interest were obtained by polymerase chain reaction (PCR) using DNA prepared from 106 clones of a λgt11 cDNA library previously constructed (60). Primers were derived from genomic DNA sequences of the genes. Two hundred nanograms of the target DNA, 10 μl of each sense and antisense oligonucleotides at a concentration of 42 mmol/liter, and 8 μl of deoxynucleotide mix (containing 10 mmol/liter of each dNTP) were dissolved in 100 μl of PCR buffer (10 mmol/liter Tris-HCl, pH 8.3, 50 mmol/liter KCl, and 1.5 mmol/liter MgCl2). To each reaction, 2.5 units of AmpliTAQ DNA polymerase (PerkinElmer Life Sciences) were added. The reaction mixtures were incubated 5 min at 94 °C, subjected to 25 cycles of 0.5 min at 94 °C, 0.5 min at 55 °C, 0.5 min at 72 °C, and finally incubated 10 min at 72 °C. Expression plasmids (pHIL-S1) were constructed by cloning the cDNA PCR products in Pichia pastoris expression vectors. The PCR products were purified using PCR purification kit (Roche Diagnostics, Germany) and digested by restriction enzymes for which a site was previously designed at the 5′ end of the primers. P. pastoris transformation, selection of transformants, and production of recombinant enzymes in methanol medium were performed as described previously (67, 68). All His6-tagged proteins bound to a Probond column (Invitrogen); after washing the column with a 20 mmol/liter phosphate buffer, pH 6.0, containing 0.5 mol/NaCl, proteins were eluted from the Ni2+ column with 50 mmol/liter histidine. Identity of purified proteases was verified by sequencing them by MS using AB-Sciex 4800 MALDI-TOF/TOF analyzer. Purity of the proteases was examined by densitometry of the Coomassie Blue-stained SDS-PAGE (Fig. S1). The purity of all the proteins exceeded 92%, except Mep1p, which was ∼80%; the ∼18-kDa band in Mep1p is the auto-proteolysis degradation fragment of the protease as determined by sequencing of the fragment by Orbitrap MS.
Complement degradation assay
To assess complement-degrading activity, culture supernatant (the volume corresponding to 1 μg of proteins) was incubated with 2 μg of C3b/C4b for 30 or 60 min at 37 °C in a total volume of 30 μl. The reaction was then stopped by adding SDS-PAGE sample buffer containing β-mercaptoethanol, incubated at 95 °C for 5 min, and subjected to 10% SDS-PAGE. To examine the proteolytic activity of various purified A. fumigatus proteases, 1 μg of the protease (Alp1p, Mep1p, Pep1p, Pep2p, or DPPV) was incubated with 3 μg of the complement protein for 5 or 60 min at 37 °C in 20 μl of Tris buffer. The reaction then was stopped by adding SDS-PAGE sample buffer containing DTT and resolved on 10% SDS-PAGE. To examine the inhibition of proteases with various inhibitors, the above reaction was performed upon preincubating proteases with various inhibitors: leupeptin (2.5 μg/ml), chymostatin (100 μg/ml), pepstatin (11 μg/ml), phenylmethylsulfonyl fluoride (1 mm), EDTA (10 mm), complete mini mixture (1×) (Roche Applied Science); complete mini-mixture, EDTA-free (1×) (Roche Applied Science). Heat-inactivated (95 °C for 10 min) proteases were included as negative controls. The percent cleavage of proteins was quantitated by densitometric analysis and presented as mean ± S.D. of three experiments. Data were normalized considering the uncleaved protein as 100%.
C3, C4, and C5 cleavage assay
To study the cleavage of C3, C4, and C5 by Mep1p, 500 ng of Mep1p was incubated with 3 μg of native C3, C4, or C5 in 20 μl of Tris buffer at 37 °C for the indicated time points. The reaction was stopped by adding SDS-PAGE sample buffer containing DTT, and samples were resolved in 10% SDS-PAGE. For N-terminal sequencing of the cleaved fragments, samples run on SDS-PAGE were transferred onto polyvinylidene difluoride membranes (ProBlott, ABI) and sequenced. Positive controls for cleavage of C3, C4, and C5 to C3b, C4b, and C5b, respectively, were formed as described below. For C3, 3 μg of C3 was incubated with 3 ng of trypsin in 20 μl of 80 mm ammonium bicarbonate buffer, pH 8, for 5 min at 37 °C. For C4, 3 μg of C4 was incubated with 1 μg of activated C1s in 20 μl of VBS2+ for 60 min at 37 °C. For C5, 3 μg of C5 was incubated with the C5 convertase (formed by incubating 3 μg of CVF with 2.5 μg of factor B, 80 ng of factor D in a total volume of 20 μl for 60 min at 37 °C) in 20 μl of VBS for 60 min at 37 °C. The cleavage of the complement components was analyzed using 10% SDS-PAGE under reducing conditions. The gel was stained with Coomassie Blue. The percentage of cleavage was quantitated by densitometric analysis and presented as mean ± S.D. of three experiments. Data were normalized considering the uncleaved protein as 100%.
Hemolytic assays and ELISA
Hemolytic assays were performed as described (64). In brief, to determine the effect of Mep1p on the classical pathway, 10% NHS in GVB2+ was pretreated with 0.6 μm Mep1p for 60 min at 37 °C. Thereafter, graded concentrations of this reaction were mixed with 5 μl of antibody-coated sheep erythrocytes (1 × 109/ml) and incubated for 60 min at 37 °C after adjusting the volume to 250 μl. The reactions were stopped by keeping the samples on ice and centrifuged. The percentage of lysis was determined by measuring the absorbance of the supernatant at 405 nm. To determine the effect on the alternative pathway, 10% NHS in GVB was preincubated with 0.6 μm Mep1p in the presence of 1.5 mm CaCl2 for 60 min at 37 °C. The reaction was then stopped by adding 1.5 mm EGTA. Various concentrations of this reaction mix were then added to 10 μl of rabbit erythrocytes (1 × 109/ml in GVB) in 100 μl of GVB containing 5 mm each of MgCl2 and EGTA and incubated for 20 min at 37 °C. The reaction was stopped by adding 200 μl of GVBE and centrifuged. The absorbance of the supernatant was read at 405 nm to determine the percentage of lysis.
To determine the effect of Mep1p on various pathways using the commercially available Wieslab complement system screen kit (Euro-Diagnostica, Malmo, Sweden), 4 μl of NHS was preincubated at 37 °C for 60 min with graded concentrations of Mep1p in the presence of 1.5 mm CaCl2 in a total volume of 13 μl of reaction mixture in GVB. These reaction mixtures were then diluted as per the manufacturer's instructions and added to wells precoated with IgM, mannan, or LPS to determine the effect on classical, lectin, or alternative pathways. The ELISA plate was then developed following the manufacturer's instructions.
C3b deposition on dormant conidia
Flow cytometry
In this assay, 50% NHS in GVB2+ was preincubated with 1.5 μm Mep1p for 60 min at 37 °C in a 50 μl of reaction mixture. Thereafter, 25 μl of Mep1p-pretreated NHS was mixed with 1 × 106 heat-inactivated conidia in 50 μl in GVB2+ and incubated at 37 °C for 60 min. The conidia were washed three times with GVBE containing 0.05% Tween 20, and the C3b deposited was detected by FITC-conjugated F(ab′)2 anti-C3 goat IgG (dilution 1:1000) on FACSCalibur (BD Biosciences). Data were analyzed using CellQuestPro software (BD Biosciences).
Biochemical assay
Conidia (5 × 108) harvested from malt-agar slants after 10 days of growth were washed twice with 0.05% Tween/water and five times with phosphate-buffered saline/SDS (PBS/SDS (0.1%)) solution before C3b opsonization by incubation with 50 μl of 40% normal human serum (NHS) in HEPES buffer, pH 7.4, for 30 min at 37 °C, and in the absence or presence of recombinant Mep1p of concentration (1 μg/50 μl), with intermittent mixing every 5 min. Afterward, the conidia were washed for five times with PBS/SDS and incubated with 1 m hydroxylamine (NH2OH; 1 h at 37 °C) in 0.2 m sodium bicarbonate (NaHCO3, pH 10.0), 3.5 m sodium thiocyanate (NaSCN, pH 7.0), or PBS/SDS (each volume, 200 μl). The contents were centrifuged, and the supernatants were collected for protein quantification by the Bradford assay, and C3b was quantified by ELISA. Briefly, a standard curve for C3b was obtained by coating fold-dilutions of C3b followed by adding anti-C3b antibody conjugated to horseradish peroxidase (HRP), and then coated C3b was estimated using ortho-phenylenediamine (OPD) as HRP substrate. Furthermore, C3b content in the NH2OH, NaSCN, and PBS/SDS released material was estimated similarly by coating them on an ELISA plate, using HRP-conjugated anti-C3b antibody and OPD. Supernatant obtained from unopsonized conidia incubated with NH2OH-NaHCO3 reagent was treated as the control.
Conidial phagocytosis in the absence or presence of Mep1p
Conidial opsonization
Conidia (1 × 106) were opsonized with 12.5% NHS in HEPES buffer, in the absence or presence of Mep1p (1 μg) at 37 °C for 20 min, with intermittent mixing every 3 min. Conidia were also treated with heat-inactivated NHS or with NHS in the presence of EDTA (20 mm). As a negative control, conidia were incubated with HEPES buffer alone. Following that, the reaction was arrested by adding EDTA; the contents were centrifuged; and conidia were separated, washed twice with HEPES buffer, resuspended in 50 μl of HEPES buffer, and then added to macrophage culture.
Phagocytosis
Human peripheral blood mononuclear cells (PBMCs) were utilized to generate monocyte-derived macrophages. These were isolated from the whole-blood samples collected from anonymous healthy donors after written consent were obtained from Hôpital Saint-Louis (Paris, France), through the Etablissement Français du Sang (Paris, France). The use of this material was approved by the ethics committees of Institut Pasteur and the Etablissement Français du Sang (convention 12/EFS/023). To generate monocyte-derived macrophages, human PBMCs were isolated from blood, and 2 × 106 PBMCs were seeded in 12-well culture plates and incubated in RPMI medium overnight at 37 °C in a CO2 incubator. The medium was aspirated, and adherent monocytes were washed twice with medium. The differentiation of monocytes into macrophages was facilitated by culturing with GM-CSF (10 ng/ml) in complete RPMI (RPMI, heat-inactivated fetal calf serum, antibiotics, and HEPES) for 6 days at 37 °C in a CO2 incubator. The medium was aspirated, and the monocyte-derived macrophages were washed with medium. Opsonized conidia resuspended in HEPES buffer were then added to the macrophages (1 × 106 conidia per well) and incubated in a CO2 incubator for 60 min at 37 °C. Thereafter, medium was aspirated, and macrophages were washed twice with RPMI and lysed upon the addition of 100 μl of 1% Triton X-100 for 30 min at 4 °C. The lysate, which contained the phagocytosed conidia, was collected, and the volume was made up to 1 ml with water. After appropriate dilution, 25 μl of lysate was spread over malt agar. The agar plates were incubated at 37 °C for 24 h followed by counting the colony-forming units. Simultaneously, in a duplicate experiment, culture plate wells were observed under microscope (bright-field microscopy) every minute for 60 min. At 60 min, the nonphagocytosed conidia were observed by labeling with calcofluor white.
C3a, C4a, and C5a cleavage by Mep1p
For SDS-PAGE analysis, 1 μg each of C3a, C4a, or C5a was incubated with Mep1p (0.5 μg) in Tris buffer for 15 min at 37 °C in a total volume of 20 μl. The reaction was then stopped by adding the sample buffer, and samples were loaded onto 16% Tricine gel. The cleavage of the anaphylatoxin was analyzed by staining the gel with Coomassie Blue. The uncleaved and cleaved anaphylatoxins were also subjected to N-terminal sequencing as described above for C3, C4, and C5. For determination of the intact molecular mass by MS, 3 μg of C3a, C4a, or C5a was incubated with Mep1p (0.5 μg) in Tris buffer for 60 min at 37 °C in a total reaction volume of 10 μl. The samples were then diluted in acetonitrile/H2O solution (1:1) so that their final concentration was 20 ng/μl. The mass analysis was performed as described below.
Intact mass analysis of uncleaved and Mep1p-cleaved C3a, C4a, and C5a using LC-Q-Exactive Plus mass spectrometer
Molecular mass measurement of uncleaved and Mep1p-cleaved C3a, C4a, and C5a was achieved using the Q-Exactive Plus MS coupled to Dionex Vanquish UHPLC system (ThermoFisher Scientific). Briefly, 5 μl of the sample was injected into UHPLC system equipped with a C18 reverse phase column (100 × 2.1 mm, 1.9 μm). Reverse phase separation of the protein sample was attained with solvent A (0.1% formic acid in 100% LC-MS grade water) and solvent B (0.1% formic acid in 100% LC-MS grade acetonitrile) using a 15-min gradient (5–70% B for 11 min followed by 5% B for 4 min) at a flow rate of 0.3 ml/min. The reverse-phase eluent was nebulized into the MS through the HESI-Source (Heated Electrospray Ionization). The MS acquisition parameters were as follows: the instrument was operated in the positive mode with an electrospray voltage of 4.2 kV, capillary temperature 275 °C, source temperature 200 °C, sheath gas 25, auxiliary gas 10, resolution 70,000, IT 100 ms, AGC 1.00E6, 10-μm scans, and m/z range of 500–2000. The acquisition parameters were fed into the instrument using the Tune Plus software version 2.8, and sample acquisitions were attained using the Xcalibur software version 4.0 (ThermoFisher Scientific). Acquired spectra were deconvoluted, and the intact mass analysis was attained using the BioPharma Finder software version 2.0 (ThermoFisher Scientific).
Virulence of Mep1p-deficient mutant in murine model of invasive pulmonary aspergillosis
Animal experiments performed in this study were approved by the ethical committee for animal experimentation Comité d'Éthique en Experimentation Animale (CETEA Project license number 2013-0020). A total of 40 male 8-week-old BALB/c mice with ∼25 g original weight (Janvier, France) were randomly divided into two even groups. The mice were immunosuppressed by intraperitoneal injections of 200 mg/kg cyclophosphamide (Sigma) on day −4 and day −1. Prior to infection on day 0, conidial suspensions of WT strain and Mep1p-deficient mutant strain were prepared fresh in PBS supplemented with 0.1% Tween 20. Each mouse was anesthetized by an intramuscular injection of a volume of 150 μl containing 10 mg/ml ketamine and 10 mg/ml xylazine. The anesthetized mice were then intranasally inoculated with a 25-μl volume containing 5 × 105 WT conidia (control group) or the Mep1p-deficient mutant conidia (test group). The weight and survival of the mice were monitored daily.
In vivo secretion of Mep1p from conidia in the lungs of murine model
A total of six male 8-week-old BALB/c mice were used in this assay (three mice per set, two biological replicates). Mice of the test group and the control group were intranasally inoculated with A. fumigatus conidial suspension of WT strain (2.5 × 108) or PBS, respectively. The BALF was collected using sterile PBS from each mouse 2 h post-challenge (total BALF volume was 5 ml). The BALF was centrifuged to remove the debris and subjected to albumin depletion (Thermo Scientific albumin depletion kit). Bradford assay was performed to determine the protein concentration of the BALF. To examine the presence of Mep1p in the BALF, each BALF sample (2.5 μg of protein) was mixed with the sample buffer, incubated at 95 °C for 5 min, and resolved in 10% SDS-PAGE. The separated proteins were transferred to a nitrocellulose membrane, and the presence of Mep1p was probed by polyclonal anti-Mep1p antibody. In parallel, BALF samples from both control and conidia inoculated mice were coated on the ELISA plates, followed by addition of rabbit anti-Mep1p antibody (1:2000 diluted in PBS/BSA) and incubation at room temperature for 1 h, and then addition of secondary anti-rabbit IgG (whole molecules; Sigma) conjugated to HRP and again incubation at room temperature for 1 h. Between each step, ELISA plate wells were washed three times with PBS containing 0.5% Tween 20. Then, using OPD as the HRP-substrate, bound peroxidase activity was measured, and the reaction was arrested by adding 4% H2SO4. These peroxidase activities were converted into metalloprotease released by performing ELISA upon coating fold-dilutions of rMep1p instead of BALF and thus obtaining a standard curve.
Statistical analysis
Data are presented as mean ± S.D., and statistical evaluation was performed using Student's t test (SigmaStat, Systat Software, Inc., San Jose, CA).
Author contributions
R. S., S. S. W. W., S. R., V. A., and A. S. conceptualization; J. K. P., J.-P. L., V. A., and A. S. resources; R. S., S. S. W. W., S. R., R. B., T. M., V. A., and A. S. formal analysis; A. S. and V. A. supervision; A. S., T. M., and V. A. funding acquisition; R. S., S. S. W. W., and S. R. validation; R. S., S. S. W. W., S. R., R. B., O. I.-G., M. M., K.-H. G., and J. K. P. investigation; R. S., S. S. W. W., and S. R. methodology; R. S. and S. S. W. W. writing-original draft; A. S. and V. A. project administration; R. S., S. S. W. W., S. R., J. K. P., T. M., V. A., and A. S. writing-review and editing.
Supplementary Material
Acknowledgments
We thank Venkatesh Naik (Proteomic Facility, National Centre for Cell Science, Pune, India) for sequencing purified A. fumigatus proteases, Hemendra Singh Panwar for help in preparing figures, and A. Walimbe for statistical analysis. We acknowledge the Department of Biotechnology, Government of India, Grant BT/PR10855/BRB/10/1330/2014 for funding Orbitrap mass spectrometer to the National Centre for Cell Science.
This work was supported in part by a bilateral COMASPIN grant from the Department of Science and Technology (DST) India and l'Agence Nationale de la Recherché (ANR) France (to A. S., T. M., and V. A.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3.
- MAC
- membrane attack complex
- CP
- classical pathway
- LP
- lectin pathway
- AP
- alternative pathway
- Mep1
- metalloprotease 1
- Alp1
- alkaline protease 1
- DPPV
- dipeptidyl peptidase V
- Pep1
- aspartic protease 1
- Pep2
- aspartic protease 2
- PMSF
- phenylmethylsulfonyl fluoride
- CS
- culture supernatant
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- FH
- factor H
- FI
- factor I
- FP
- properdin
- C4BP
- C4b-binding protein
- VBS
- veronal-buffered saline
- NHS
- normal human serum
- MBL
- mannose-binding lectin
- GVB
- gelatin veronal buffer
- OPD
- ortho-phenylenediamine
- PBMC
- peripheral blood mononuclear cell
- BALF
- bronchoalveolar lavage fluid
- HRP
- horseradish peroxidase
- CVF
- cobra venom factor.
References
- 1. Walport M. J. (2001) Complement. First of two parts. N. Engl. J. Med. 344, 1058–1066 10.1056/NEJM200104053441406 [DOI] [PubMed] [Google Scholar]
- 2. Carroll M. C., and Isenman D. E. (2012) Regulation of humoral immunity by complement. Immunity 37, 199–207 10.1016/j.immuni.2012.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Heeger P. S., and Kemper C. (2012) Novel roles of complement in T effector cell regulation. Immunobiology 217, 216–224 10.1016/j.imbio.2011.06.004 [DOI] [PubMed] [Google Scholar]
- 4. Dunkelberger J. R., and Song W. C. (2010) Role and mechanism of action of complement in regulating T cell immunity. Mol. Immunol. 47, 2176–2186 10.1016/j.molimm.2010.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lyubchenko T., dal Porto J., Cambier J. C., and Holers V. M. (2005) Coligation of the B cell receptor with complement receptor type 2 (CR2/CD21) using its natural ligand C3dg: activation without engagement of an inhibitory signaling pathway. J. Immunol. 174, 3264–3272 10.4049/jimmunol.174.6.3264 [DOI] [PubMed] [Google Scholar]
- 6. van den Elsen J. M., and Isenman D. E. (2011) A crystal structure of the complex between human complement receptor 2 and its ligand C3d. Science 332, 608–611 10.1126/science.1201954 [DOI] [PubMed] [Google Scholar]
- 7. Agarwal R., Chakrabarti A., Shah A., Gupta D., Meis J. F., Guleria R., Moss R., Denning D. W., and ABPA complicating asthma ISHAM working group. (2013) Allergic bronchopulmonary aspergillosis: review of literature and proposal of new diagnostic and classification criteria. Clin. Exp. Allergy 43, 850–873 10.1111/cea.12141 [DOI] [PubMed] [Google Scholar]
- 8. Lazovic B., Stajic Z., and Putnikovic B. (2012) Pulmonary aspergilloma. Med. Arch. 66, 420–422 10.5455/medarh.2012.66.420-422 [DOI] [PubMed] [Google Scholar]
- 9. Warris A. (2014) The biology of pulmonary Aspergillus infections. J. Infect. 69, Suppl. 1, S36–S41 10.1016/j.jinf.2014.07.011 [DOI] [PubMed] [Google Scholar]
- 10. Cadena J., Thompson G. R. 3rd., and Patterson T. F. (2016) Invasive aspergillosis: current strategies for diagnosis and management. Infect. Dis. Clin. North Am. 30, 125–142 10.1016/j.idc.2015.10.015 [DOI] [PubMed] [Google Scholar]
- 11. Gazendam R. P., van de, Geer A., Roos D., van den Berg T. K., and Kuijpers T. W. (2016) How neutrophils kill fungi. Immunol. Rev. 273, 299–311 10.1111/imr.12454 [DOI] [PubMed] [Google Scholar]
- 12. Philippe B., Ibrahim-Granet O., Prévost M. C., Gougerot-Pocidalo M. A., Sanchez Perez M. A., Van der Meeren A., and Latgé J. P. (2003) Killing of Aspergillus fumigatus by alveolar macrophages is mediated by reactive oxidant intermediates. Infect. Immun. 71, 3034–3042 10.1128/IAI.71.6.3034-3042.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Askew D. S., Kontoyiannis D. P., and Clemons K. V. (2014) Advances against aspergillosis: biology, host response, diagnosis and treatment. Mycopathologia 178, 321–324 10.1007/s11046-014-9819-4 [DOI] [PubMed] [Google Scholar]
- 14. Abad A., Fernández-Molina J. V., Bikandi J., Ramírez A., Margareto J., Sendino J., Hernando F. L., Pontón J., Garaizar J., and Rementeria A. (2010) What makes Aspergillus fumigatus a successful pathogen? Genes and molecules involved in invasive aspergillosis. Rev. Iberoam. Micol. 27, 155–182 10.1016/j.riam.2010.10.003 [DOI] [PubMed] [Google Scholar]
- 15. Al-Bader N., and Sheppard D. C. (2016) Aspergillosis and stem cell transplantation: an overview of experimental pathogenesis studies. Virulence 7, 950–966 10.1080/21505594.2016.1231278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Escobar N., Ordonez S. R., Wösten H. A., Haas P. J., de Cock H., and Haagsman H. P. (2016) Hide, keep quiet, and keep low: properties that make Aspergillus fumigatus a successful lung pathogen. Front. Microbiol. 7, 438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Paulussen C., Hallsworth J. E., Álvarez-Pérez S., Nierman W. C., Hamill P. G., Blain D., Rediers H., and Lievens B. (2017) Ecology of aspergillosis: insights into the pathogenic potency of Aspergillus fumigatus and some other Aspergillus species. Microb. Biotechnol. 10, 296–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. O'Gorman C. M., Fuller H., and Dyer P. S. (2009) Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature 457, 471–474 10.1038/nature07528 [DOI] [PubMed] [Google Scholar]
- 19. Kozel T. R., Wilson M. A., Farrell T. P., and Levitz S. M. (1989) Activation of C3 and binding to Aspergillus fumigatus conidia and hyphae. Infect. Immun. 57, 3412–3417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Henwick S., Hetherington S. V., and Patrick C. C. (1993) Complement binding to Aspergillus conidia correlates with pathogenicity. J. Lab. Clin. Med. 122, 27–35 [PubMed] [Google Scholar]
- 21. Hector R. F., Yee E., and Collins M. S. (1990) Use of DBA/2N mice in models of systemic candidiasis and pulmonary and systemic aspergillosis. Infect. Immun. 58, 1476–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Braem S. G., Rooijakkers S. H., van Kessel K. P., de Cock H., Wösten H. A., van Strijp J. A., and Haas P. J. (2015) Effective neutrophil phagocytosis of Aspergillus fumigatus is mediated by classical pathway complement activation. J. Innate Immun. 7, 364–374 10.1159/000369493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roy R. M., Paes H. C., Nanjappa S. G., Sorkness R., Gasper D., Sterkel A., Wüthrich M., and Klein B. S. (2013) Complement component 3C3 and C3a receptor are required in chitin-dependent allergic sensitization to Aspergillus fumigatus but dispensable in chitin-induced innate allergic inflammation. MBio. 4, e00162–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rambach G., Hagleitner M., Mohsenipour I., Lass-Flörl C., Maier H., Würzner R., Dierich M. P., and Speth C. (2005) Antifungal activity of the local complement system in cerebral aspergillosis. Microbes Infect. 7, 1285–1295 10.1016/j.micinf.2005.04.014 [DOI] [PubMed] [Google Scholar]
- 25. Tsai H. F., Chang Y. C., Washburn R. G., Wheeler M. H., and Kwon-Chung K. J. (1998) The developmentally regulated alb1 gene of Aspergillus fumigatus: its role in modulation of conidial morphology and virulence. J. Bacteriol. 180, 3031–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tsai H. F., Washburn R. G., Chang Y. C., and Kwon-Chung K. J. (1997) Aspergillus fumigatus arp1 modulates conidial pigmentation and complement deposition. Mol. Microbiol. 26, 175–183 10.1046/j.1365-2958.1997.5681921.x [DOI] [PubMed] [Google Scholar]
- 27. Behnsen J., Hartmann A., Schmaler J., Gehrke A., Brakhage A. A., and Zipfel P. F. (2008) The opportunistic human pathogenic fungus Aspergillus fumigatus evades the host complement system. Infect. Immun. 76, 820–827 10.1128/IAI.01037-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vogl G., Lesiak I., Jensen D. B., Perkhofer S., Eck R., Speth C., Lass-Flörl C., Zipfel P. F., Blom A. M., Dierich M. P., and Würzner R. (2008) Immune evasion by acquisition of complement inhibitors: the mould Aspergillus binds both factor H and C4b-binding protein. Mol. Immunol. 45, 1485–1493 10.1016/j.molimm.2007.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Washburn R. G., Hammer C. H., and Bennett J. E. (1986) Inhibition of complement by culture supernatants of Aspergillus fumigatus. J. Infect. Dis. 154, 944–951 10.1093/infdis/154.6.944 [DOI] [PubMed] [Google Scholar]
- 30. Behnsen J., Lessing F., Schindler S., Wartenberg D., Jacobsen I. D., Thoen M., Zipfel P. F., and Brakhage A. A. (2010) Secreted Aspergillus fumigatus protease Alp1 degrades human complement proteins C3, C4, and C5. Infect. Immun. 78, 3585–3594 10.1128/IAI.01353-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jusko M., Potempa J., Mizgalska D., Bielecka E., Ksiazek M., Riesbeck K., Garred P., Eick S., and Blom A. M. (2015) A metalloproteinase mirolysin of Tannerella forsythia inhibits all pathways of the complement system. J. Immunol. 195, 2231–2240 10.4049/jimmunol.1402892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Laarman A. J., Ruyken M., Malone C. L., van Strijp J. A., Horswill A. R., and Rooijakkers S. H. (2011) Staphylococcus aureus metalloprotease aureolysin cleaves complement C3 to mediate immune evasion. J. Immunol. 186, 6445–6453 10.4049/jimmunol.1002948 [DOI] [PubMed] [Google Scholar]
- 33. Svoboda E., Schneider A. E., Sándor N., Lermann U., Staib P., Kremlitzka M., Bajtay Z., Barz D., Erdei A., and Józsi M. (2015) Secreted aspartic protease 2 of Candida albicans inactivates factor H and the macrophage factor H-receptors CR3 (CD11b/CD18) and CR4 (CD11c/CD18). Immunol. Lett. 168, 13–21 10.1016/j.imlet.2015.08.009 [DOI] [PubMed] [Google Scholar]
- 34. Rambach G., Dum D., Mohsenipour I., Hagleitner M., Würzner R., Lass-Flörl C., and Speth C. (2010) Secretion of a fungal protease represents a complement evasion mechanism in cerebral aspergillosis. Mol. Immunol. 47, 1438–1449 10.1016/j.molimm.2010.02.010 [DOI] [PubMed] [Google Scholar]
- 35. Bolger M. S., Ross D. S., Jiang H., Frank M. M., Ghio A. J., Schwartz D. A., and Wright J. R. (2007) Complement levels and activity in the normal and LPS-injured lung. Am. J. Physiol. Lung Cell. Mol. Physiol. 292, L748–L759 10.1152/ajplung.00127.2006 [DOI] [PubMed] [Google Scholar]
- 36. Monod M., Paris S., Sanglard D., Jaton-Ogay K., Bille J., and Latgé J. P. (1993) Isolation and characterization of a secreted metalloprotease of Aspergillus fumigatus. Infect. Immun. 61, 4099–4104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Monod M., Capoccia S., Léchenne B., Zaugg C., Holdom M., and Jousson O. (2002) Secreted proteases from pathogenic fungi. Int. J. Med. Microbiol. 292, 405–419 10.1078/1438-4221-00223 [DOI] [PubMed] [Google Scholar]
- 38. Koneru L., Ksiazek M., Waligorska I., Straczek A., Lukasik M., Madej M., Thogersen I. B., Enghild J. J., and Potempa J. (2016) Mirolysin, a LysargiNase from Tannerella forsythia, proteolytically inactivates the human cathelicidin, LL-37. Biol. Chem. 2016 /j/bchm.ahead-of-print/hsz-2016–0267/hsz-2016–0267.xml [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pidde-Queiroz G., Magnoli F. C., Portaro F. C., Serrano S. M., Lopes A. S., Paes Leme A. F., van den Berg C. W., and Tambourgi D. V. (2013) P-I snake venom metalloproteinase is able to activate the complement system by direct cleavage of central components of the cascade. PLoS Negl. Trop. Dis. 7, e2519 10.1371/journal.pntd.0002519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Del Tordello E., Vacca I., Ram S., Rappuoli R., and Serruto D. (2014) Neisseria meningitidis NalP cleaves human complement C3, facilitating degradation of C3b and survival in human serum. Proc. Natl. Acad. Sci. U.S.A. 111, 427–432 10.1073/pnas.1321556111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wetsel R. A., Kildsgaard J., and Haviland D. L. (2000) in Therapeutic Interventions in the Complement System (Lambris J. D., and Holers V. M., eds), pp. 113–153, Humana Press Inc., Totowa, NJ [Google Scholar]
- 42. Heinekamp T., Schmidt H., Lapp K., Pähtz V., Shopova I., Köster-Eiserfunke N., Krüger T., Kniemeyer O., and Brakhage A. A. (2015) Interference of Aspergillus fumigatus with the immune response. Semin. Immunopathol. 37, 141–152 10.1007/s00281-014-0465-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gresnigt M. S., Becker K. L., Smeekens S. P., Jacobs C. W., Joosten L. A., van der Meer J. W., Netea M. G., and van de Veerdonk F. L. (2013) Aspergillus fumigatus-induced IL-22 is not restricted to a specific Th cell subset and is dependent on complement receptor 3. J. Immunol. 190, 5629–5639 10.4049/jimmunol.1202601 [DOI] [PubMed] [Google Scholar]
- 44. Askew D. S. (2008) Aspergillus fumigatus: virulence genes in a street-smart mold. Curr. Opin. Microbiol. 11, 331–337 10.1016/j.mib.2008.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moutaouakil M., Monod M., Prévost M. C., Bouchara J. P., Paris S., and Latgé J. P. (1993) Identification of the 33-kDa alkaline protease of Aspergillus fumigatus in vitro and in vivo. J. Med. Microbiol. 39, 393–399 10.1099/00222615-39-5-393 [DOI] [PubMed] [Google Scholar]
- 46. Fearon D. T., and Austen K. F. (1975) Properdin: binding to C3b and stabilization of the C3b-dependent C3 convertase. J. Exp. Med. 142, 856–863 10.1084/jem.142.4.856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sahu A., and Pangburn M. K. (1994) Covalent attachment of human complement C3 to IgG: identification of the amino acid residue involved in ester linkage formation. J. Biol. Chem. 269, 28997–29002 [PubMed] [Google Scholar]
- 48. Rattan A., Pawar S. D., Nawadkar R., Kulkarni N., Lal G., Mullick J., and Sahu A. (2017) Synergy between the classical and alternative pathways of complement is essential for conferring effective protection against the pandemic influenza A(H1N1) 2009 virus infection. PLoS Pathog. 13, e1006248 10.1371/journal.ppat.1006248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rosbjerg A., Genster N., Pilely K., Skjoedt M. O., Stahl G. L., and Garred P. (2016) complementary roles of the classical and lectin complement pathways in the defense against Aspergillus fumigatus. Front. Immunol. 7, 473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jusko M., Potempa J., Karim A. Y., Ksiazek M., Riesbeck K., Garred P., Eick S., and Blom A. M. (2012) A metalloproteinase karilysin present in the majority of Tannerella forsythia isolates inhibits all pathways of the complement system. J. Immunol. 188, 2338–2349 10.4049/jimmunol.1101240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schmidtchen A., Holst E., Tapper H., and Björck L. (2003) Elastase-producing Pseudomonas aeruginosa degrade plasma proteins and extracellular products of human skin and fibroblasts, and inhibit fibroblast growth. Microb. Pathog. 34, 47–55 10.1016/S0882-4010(02)00197-3 [DOI] [PubMed] [Google Scholar]
- 52. Fraga T. R., Courrol Ddos S., Castiblanco-Valencia M. M., Hirata I. Y., Vasconcellos S. A., Juliano L., Barbosa A. S., and Isaac L. (2014) Immune evasion by pathogenic Leptospira strains: the secretion of proteases that directly cleave complement proteins. J. Infect. Dis. 209, 876–886 10.1093/infdis/jit569 [DOI] [PubMed] [Google Scholar]
- 53. Pangburn M. K., Ferreira V. P., and Cortes C. (2008) Discrimination between host and pathogens by the complement system. Vaccine 26, Suppl. 8, I15–I21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jaton-Ogay K., Paris S., Huerre M., Quadroni M., Falchetto R., Togni G., Latgé J. P., and Monod M. (1994) Cloning and disruption of the gene encoding an extracellular metalloprotease of Aspergillus fumigatus. Mol. Microbiol. 14, 917–928 10.1111/j.1365-2958.1994.tb01327.x [DOI] [PubMed] [Google Scholar]
- 55. Sharon H., Hagag S., and Osherov N. (2009) Transcription factor PrtT controls expression of multiple secreted proteases in the human pathogenic mold Aspergillus fumigatus. Infect. Immun. 77, 4051–4060 10.1128/IAI.00426-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hartmann T., Cairns T. C., Olbermann P., Morschhäuser J., Bignell E. M., and Krappmann S. (2011) Oligopeptide transport and regulation of extracellular proteolysis are required for growth of Aspergillus fumigatus on complex substrates but not for virulence. Mol. Microbiol. 82, 917–935 10.1111/j.1365-2958.2011.07868.x [DOI] [PubMed] [Google Scholar]
- 57. Fernández D., Russi S., Vendrell J., Monod M., and Pallarès I. (2013) A functional and structural study of the major metalloprotease secreted by the pathogenic fungus Aspergillus fumigatus. Acta Crystallogr. D Biol. Crystallogr. 69, 1946–1957 10.1107/S0907444913017642 [DOI] [PubMed] [Google Scholar]
- 58. Markaryan A., Morozova I., Yu H., and Kolattukudy P. E. (1994) Purification and characterization of an elastinolytic metalloprotease from Aspergillus fumigatus and immunoelectron microscopic evidence of secretion of this enzyme by the fungus invading the murine lung. Infect. Immun. 62, 2149–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sirakova T. D., Markaryan A., and Kolattukudy P. E. (1994) Molecular cloning and sequencing of the cDNA and gene for a novel elastinolytic metalloproteinase from Aspergillus fumigatus and its expression in Escherichia coli. Infect. Immun. 62, 4208–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Monod M., Paris S., Sarfati J., Jaton-Ogay K., Ave P., and Latgé J. P. (1993) Virulence of alkaline protease-deficient mutants of Aspergillus fumigatus. FEMS Microbiol. Lett. 106, 39–46 10.1111/j.1574-6968.1993.tb05932.x [DOI] [PubMed] [Google Scholar]
- 61. Thau N., Monod M., Crestani B., Rolland C., Tronchin G., Latgé J. P., and Paris S. (1994) Rodletless mutants of Aspergillus fumigatus. Infect. Immun. 62, 4380–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Monod M., Togni G., Rahalison L., and Frenk E. (1991) Isolation and characterisation of an extracellular alkaline protease of Aspergillus fumigatus. J. Med. Microbiol. 35, 23–28 10.1099/00222615-35-1-23 [DOI] [PubMed] [Google Scholar]
- 63. Mullick J., Bernet J., Panse Y., Hallihosur S., Singh A. K., and Sahu A. (2005) Identification of complement regulatory domains in vaccinia virus complement control protein. J. Virol. 79, 12382–12393 10.1128/JVI.79.19.12382-12393.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sahu A., Isaacs S. N., Soulika A. M., and Lambris J. D. (1998) Interaction of vaccinia virus complement control protein with human complement proteins: factor I-mediated degradation of C3b to iC3b1 inactivates the alternative complement pathway. J. Immunol. 160, 5596–5604 [PubMed] [Google Scholar]
- 65. Singh A. K., Mullick J., Bernet J., and Sahu A. (2006) Functional characterization of the complement control protein homolog of herpesvirus saimiri: ARG-118 is critical for factor I cofactor activities. J. Biol. Chem. 281, 23119–23128 10.1074/jbc.M603085200 [DOI] [PubMed] [Google Scholar]
- 66. Rawal N., and Pangburn M. K. (2000) Functional role of the noncatalytic subunit of complement C5 convertase. J. Immunol. 164, 1379–1385 10.4049/jimmunol.164.3.1379 [DOI] [PubMed] [Google Scholar]
- 67. Beggah S., Léchenne B., Reichard U., Foundling S., and Monod M. (2000) Intra- and intermolecular events direct the propeptide-mediated maturation of the Candida albicans secreted aspartic proteinase Sap1p. Microbiology 146, 2765–2773 10.1099/00221287-146-11-2765 . [DOI] [PubMed] [Google Scholar]
- 68. Borg-von Zepelin M., Beggah S., Boggian K., Sanglard D., and Monod M. (1998) The expression of the secreted aspartyl proteinases Sap4 to Sap6 from Candida albicans in murine macrophages. Mol. Microbiol. 28, 543–554 10.1046/j.1365-2958.1998.00815.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.