Abstract
The receptor cycle of type I peroxisomal matrix protein import is completed by ubiquitination of the membrane-bound peroxisome biogenesis factor 5 (Pex5p) and its subsequent export back to the cytosol. The receptor export is the only ATP-dependent step of the whole process and is facilitated by two members of the AAA family of proteins (ATPases associated with various cellular activities), namely Pex1p and Pex6p. To gain further insight into substrate recognition by the AAA complex, we generated an N-terminally linked ubiquitin–Pex5p fusion protein. This fusion protein displayed biological activity because it is able to functionally complement a PEX5-deletion in Saccharomyces cerevisiae. In vitro assays revealed its interaction at WT level with the native cargo protein Pcs60p and Pex14p, a constituent of the receptor docking complex. We also demonstrate in vitro deubiquitination by the deubiquitinating enzyme Ubp15p. In vitro pulldown assays and cross-linking studies demonstrate that Pex5p recognition by the AAA complex depends on the presence of the ubiquitin moiety and is mediated by Pex1p.
Keywords: peroxisome, protein targeting, protein translocation, protein sorting, ATPases associated with diverse cellular activities (AAA), ubiquitin
Introduction
Peroxisomes are ubiquitous cell organelles with a soluble matrix surrounded by a single lipid bilayer membrane. These organelles are involved in a broad range of metabolic processes, most notably displayed by a wide phenotypic range of peroxisomal disorders caused by peroxisome malfunction (1, 2). A special feature of peroxisomes, which clearly distinguishes them from other cellular organelles, is their ability to import folded, oligomerized and even co-factor bound proteins into the peroxisomal matrix (3, 4). To this end, matrix proteins are equipped with a targeting sequence, either a C-terminal PTS1 (peroxisomal targeting signal 1) or an N-terminal PTS2, which are recognized and bound in the cytosol by the import receptor Pex5p or Pex7p, respectively (5, 6). The receptors ferry the cargoes to the peroxisomal membrane, where they bind to a docking complex and become part of a highly dynamic and transient translocation pore (7, 8). How cargo translocation occurs mechanistically is still unknown, but available data are clear in that the cargo-free receptors are exported back to the cytosol in an ATP-dependent manner with ubiquitin serving as an export signal (9–11).
Among the different peroxisomal matrix protein import pathways, the so far best understood is the PTS1 pathway with Pex5p as the related import receptor. Pex5p comprises two separated and functionally distinct domains. The C-terminal domain of the receptor consists of an array of tetratricopeptide repeat domains and directly binds the PTS1 motif (12). The N-terminal domain is intrinsically disordered and capable of mediating all transport steps of the receptor cycle, including docking and pore formation (13–15). Within the N-terminal region, Pex5p is modified by the attachment of ubiquitin moieties. Two kinds of receptor ubiquitination are known, namely mono- and polyubiquitination (16). Pex5p polyubiquitination occurs on conserved lysine residues of Pex5p (Lys18/Lys24 in Saccharomyces cerevisiae) and leads to receptor degradation via the proteasome (17–20). In contrast, monoubiquitin is attached as an exception to the rule via a thioester bond to a conserved cysteine of Pex5p (Cys6 in S. cerevisiae) and enables the receptor recycling for further rounds of matrix protein import (21). In the cytosol, the monoubiquitin moiety is removed either in a nonenzymatic manner by a nucleophilic attack of GSH or enzyme-catalyzed by a ubiquitin hydrolase to allow another import cycle (22–24).
Studies on yeast and human fibroblasts illustrated that extraction of ubiquitinated Pex5p from the peroxisomal membrane is carried out by Pex1p and Pex6p (10, 25). Both proteins display similar architecture and belong to the family of ATPases associated with diverse cellular activities (AAAs).3 Studies on Pex1p and Pex6p from S. cerevisiae revealed that both peroxins form a heterohexameric complex (26, 27). The complex is present in the cytosol, as well as attached to the peroxisomal membrane. Specifically, the AAA complex is dynamically recruited to the peroxisomal membrane via a nucleotide-dependent interaction of Pex6p with the cytosolic domain of the tail-anchored membrane protein Pex15p or its orthologues in mammals (Pex26p) and plants (APM9) (28). Despite its crucial role for the dislocation, the functional relevance of the monoubiquitination of Pex5p, as well as the exact molecular mechanism of substrate recognition and extraction from the membrane, remains unclear.
Here we analyzed substrate recognition by the AAA complex by use of a linear N-terminal ubiquitin–Pex5p fusion protein. We demonstrate that this fusion functionally complements a PEX5-deletion in S. cerevisiae. Moreover, ubiquitin–Pex5p binds the cargo protein Pcs60p and Pex14p at the WT level. Based on in vitro pulldown assays and cross-linking studies, we conclude that the ubiquitination of Pex5p is a prerequisite for recognition by the AAA complex, with Pex1p representing the main binding partner.
Results
A main step of the peroxisomal import cycle is the release of the receptors from the peroxisomal membrane back to the cytosol. It was demonstrated for yeast and mammalian cells that the membrane release of the PTS1 receptor Pex5p is catalyzed by the AAA peroxins Pex1p and Pex6p (10, 25). Modification of the receptor with either mono- or polyubiquitin moieties on Pex5p turned out to be a prerequisite for this release from the membrane either for receptor recycling or for its degradation, respectively (16). It seems very likely that ubiquitin serves as an export signal, which primes the receptor molecule for the recognition by the AAA-type ATPase complex. To fulfill its function in Pex5p export, a direct or indirect interaction of the AAA complex with Pex5p and/or ubiquitin is required, the nature of which so far remains unknown.
To address this issue, we analyzed the ability of Pex1p and Pex6p for Pex5p binding by means of the yeast two-hybrid system. Gal4p fusions of the activation domain and the DNA-binding domain with various peroxins were co-expressed in different pairwise combinations in the S. cerevisiae host strain PJ69–4A (29), and interactions were monitored by histidine/adenine auxotrophy. In line with published results, Pex5p displayed clear interaction to the receptor docking constituent Pex14p as indicated by growth of the corresponding yeast reporter strain (Fig. 1 and Ref. 30). In addition, we confirmed Pex6p interaction with Pex1p as well as with the cytosolic part of its anchor protein Pex15p (Fig. 1 and Ref. 31). These results demonstrate that our Gal fusion proteins are expressed and properly folded to allow protein–protein interactions. The controls included show that expression of either of the fusion proteins alone did not support transcription activation of the reporter genes. However, when Gal fusions of the peroxisomal AAA proteins Pex1p or Pex6p were co-expressed with Pex5p, no growth of the reporter strain was observed in medium lacking histidine/adenine. The finding indicates an inability of Pex1p–Pex6p in binding of the PTS1 receptor under these conditions (Fig. 1).
Figure 1.
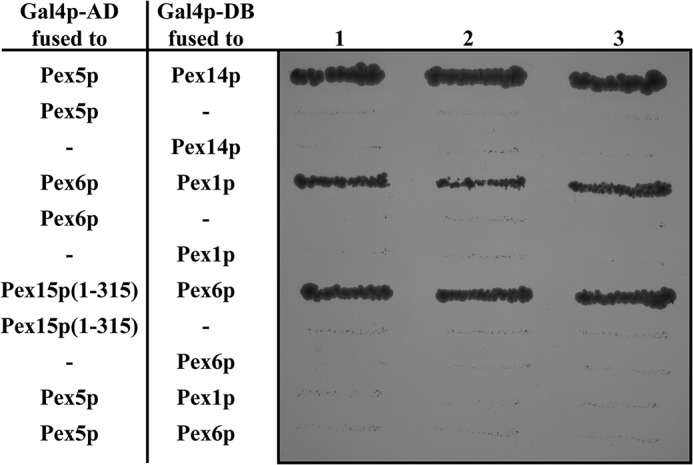
Yeast two-hybrid interaction assay of Pex5p and Pex1p–Pex6p. The S. cerevisiae strain PJ69-4A was transformed with proteins of interest fused to either the activation (AD) or binding domain (BD) of Gal4p as indicated. Interactions were monitored by histidine/adenine auxotrophy. Neither Pex1p nor Pex6p show an interaction with Pex5p under these conditions. The controls comprise the known interactions Pex5p–Pex14p, Pex1p–Pex6p, and Pex15p(1–315)-Pex6p, which confirm that all constructs are expressed and folded correctly.
In vitro binding of Pex6p to ubiquitin
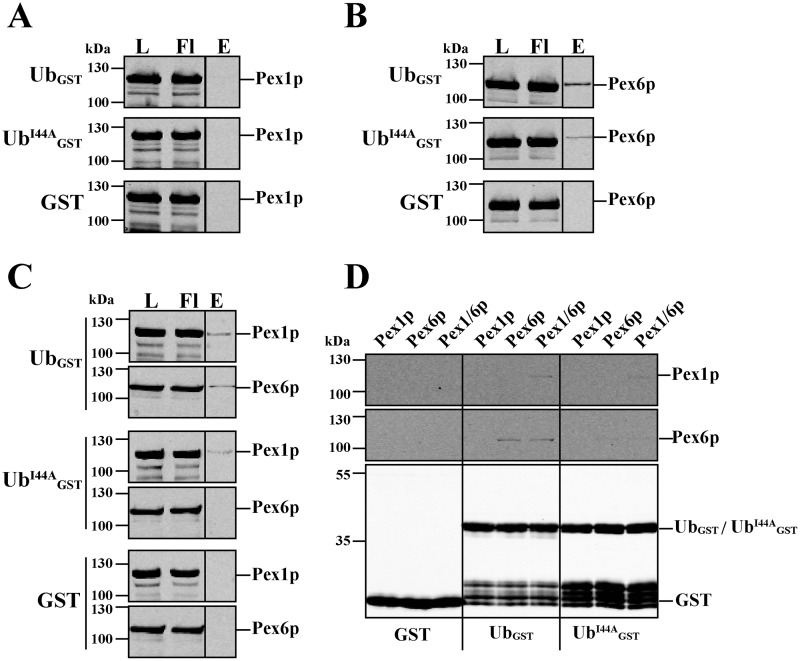
One possible reason for the observed lack of interaction between Pex5p and Pex1p–Pex6p might be that ubiquitin itself mediates the contact of the modified receptor to the peroxisomal AAA complex. To address this question, we also analyzed the interaction of recombinant ubiquitin and Pex1p–Pex6p by in vitro binding studies. Recombinant yeast Pex1p and Pex6p both fused to an N-terminal hexahistidyl tag (His6) and Pex1p in addition to a C-terminal GST tag were expressed separately in Escherichia coli and purified according to (32). To obtain the Pex1p–Pex6p complex, suspensions of Pex1p- and Pex6p-expressing cells were mixed prior to cell disruption, and complex isolation comprised successive affinity- and size-exclusion chromatography steps (32). It is important to note that during the isolation procedure, the GST tag was removed from Pex1p by thrombin cleavage with the GST tag remaining on the affinity column. Ubiquitin was expressed and purified as a GST fusion protein (UbGST). The fusion protein was eluted from the column by addition of GSH, which was subsequently dialyzed out of the obtained eluate fraction.
For in vitro binding, purified UbGST was preincubated either with Pex1p or Pex6p alone or with the assembled Pex1p–Pex6p complex. Thereafter the samples were loaded onto GSH–agarose, unbound proteins were removed, and bound proteins eluted by addition of GSH. It turned out that Pex1p was bound neither to UbGST nor GST alone, which served as control for the experiments (Fig. 2A). In contrast, Pex6p alone, as well as the Pex1p–Pex6p complex, displayed clear binding to the ubiquitin fusion protein, but not to the GST control (Fig. 2, B and C). From these results we conclude that ubiquitin is bound to the peroxisomal AAA complex via direct binding to Pex6p. It is known that a high number of proteins is capable of ubiquitin binding but with different binding motifs. However, the majority of these binding motifs interact with a hydrophobic region within ubiquitin with isoleucine on position 44 as central amino acid residue (33). To analyze whether this “hydrophobic patch” around isoleucine 44 might also play an important role in the binding to the Pex1p–Pex6p complex, mutant ubiquitin bearing an I44A substitution (UbI44A) was included in our experiments. Because Pex1p did not bind to WT ubiquitin, it was not surprising that it also did not bind to the mutant ubiquitin (Fig. 2A). Binding capacity of Pex6p and the Pex1p–Pex6p complex to UbI44A was significantly reduced when compared with WT ubiquitin (Fig. 2, B and C), supporting the idea of a commune ubiquitin-binding motif most likely within Pex6p.
Figure 2.
In vitro pulldown assay of ubiquitin and Pex1p–Pex6p. Recombinant GST and C-terminal GST fusion constructs of WT and mutant (I44A) ubiquitin were combined with purified recombinant HisPex1p (A), HisPex6p (B), or the assembled HisPex1p–HisPex6p complex (C) and loaded onto GSH–agarose. Bound proteins were eluted with buffer containing 50 mm reduced GSH. Equal volumes of load, flow through (Fl), and 10× concentrated eluate fractions were subjected to immunoblot analysis using antibodies against Pex1p, Pex6p, and GST. A comparison of all eluate fractions is shown in D. Pex6p alone and in combination with Pex1p, but not Pex1p alone, did bind ubiquitin. A maximum of 1–2% of loaded AAA proteins could be recovered in the eluate. The I44A mutation reduced binding to Pex6p.
Biological activity of the N-terminal Ub(1–6Δ)Pex5p
In vivo the PTS1 receptor Pex5p is modified by the attachment of a monoubiquitin moiety via a thioester bond at a conserved cysteine (Cys6 in S. cerevisiae) (17–20), which enables the receptor recycling for further rounds of matrix protein import (21). Because receptor ubiquitination is a prerequisite for Pex1p–Pex6p binding, we aimed to mimic and arrest this very dynamic and transient state of Pex5p for in vivo and in vitro analysis. To this end, we generated a construct in which the first six amino acid residues of Pex5p containing the conserved cysteine residue were replaced by ubiquitin (Fig. 3A, Ub(Δ1–6)Pex5p). Biological activity of the fusion protein was analyzed by testing its capability to complement the peroxisomal mutant phenotype of a PEX5-deficient yeast deletion strain. The pex5Δ strain displays a growth defect on oleic acid as the sole carbon source caused by an import defect of a group of peroxisomal matrix proteins (PTS1 and non-PTS proteins) (34). We transformed WT and pex5Δ cells with a plasmid expressing the fluorescence marker mCherry fused to type 1 targeting signal Ser-Lys-Leu (SKL). Fluorescence microscopy analysis of oleic acid-induced WT cells producing mCherry-SKL revealed the presence of distinct fluorescent spots, indicative for a peroxisomal localization of the synthetic peroxisomal matrix protein (Fig. 3B). Such spots were missing in pex5Δ cells, which display in contrast to WT an overall cytosolic labeling, demonstrating the specific import defect of this mutant (34). Although the import defect was functionally complemented by expression of WT Pex5p, Pex5p lacking the first six amino acid residues ((1–6Δ)Pex5p), including the conserved cysteine residue, was unable to restore the import defect of pex5Δ cells. Conversely, the fluorescence pattern of the mutant expressing the Ub–Pex5p fusion protein was indistinguishable from that of WT cells (Fig. 3B). Thus, the replacement of amino acids 1–6 of Pex5p by ubiquitin restored peroxisomal import of the pex5Δ mutant, demonstrating that the N-terminal tagging with ubiquitin does not interfere with the biological function of Pex5p. On the contrary, the presence of the ubiquitin moiety can account for the lack of the first six amino acids, especially the conserved cysteine, which otherwise is essential for Pex5p function in peroxisome biogenesis.
Figure 3.
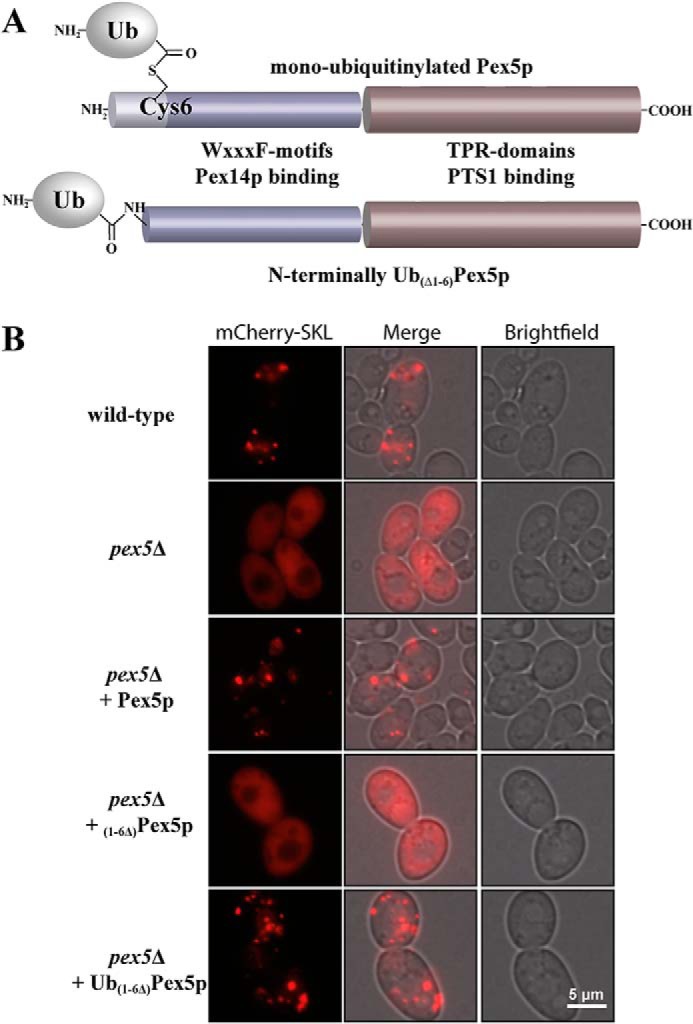
Functional analysis of N-terminally monoubiquitinated Pex5p. A, schematic comparison of the native and the artificial N-terminally monoubiquitinated Pex5p. Pex5p is monoubiquitinated at a conserved cysteine at position 6, resulting in a thioester-bonded ubiquitin. We genetically replaced the first six amino acids of Pex5p with a linear fusion to ubiquitin. As indicated, the N-terminal domain contains WXXXF motifs involved in Pex14p binding. The C-terminal region of Pex5p contains tetratricopeptide repeat domains that are responsible for PTS1 cargo recognition. B, the mutant pex5Δ strain of S. cerevisiae was transformed with indicated plasmid-encoded Pex5p variants and subjected to fluorescence microscopy. Nontransformed pex5Δ and WT strains served as controls. Peroxisomal matrix protein import was visualized by the plasmid-encoded reporter protein mCherry-SKL. In WT or complemented mutant cells, the peroxisomal localization of the marker protein appears as a typical punctate staining. Mutant pex5Δ cells and mutant cells expressing the nonfunctional N-terminally truncated Pex5p ((1–6Δ)Pex5p) are characterized by a peroxisomal import defect and mislocalization of the peroxisomal marker to the cytosol, which is indicated by the overall fluorescence. The fusion of ubiquitin to the N-terminally truncated Pex5p restored its ability to complement the pex5Δ mutant as indicated by the punctate fluorescence pattern, indicative of a functional peroxisomal protein import.
Ubiquitination does not alter Pex5p binding to cargo and Pex14p
In its function as PTS1 receptor, Pex5p binds peroxisomal matrix proteins harboring this type of the peroxisomal targeting signal. One representative of this group is Pcs60p, an oxalyl-CoA synthetase that belongs to the family of AMP-binding proteins (35, 36). The PTS1 of Pcs60p was demonstrated to be crucial for peroxisomal targeting (35). Moreover, a direct binding to the PTS1 receptor was previously reported (37). To investigate the influence of the linear fusion of ubiquitin to the N terminus of Pex5p on receptor cargo recognition, we tested whether presence of the ubiquitin moiety alters the in vitro binding of Ub–Pex5p to Pcs60p. To this end, Pex5p, Ub(Δ1–6)Pex5p, and Pcs60p were fused to an N-terminal GST tag. The genes coding for the different fusion proteins were expressed separately in E. coli, and soluble fractions of cells were loaded onto GSH–agarose. The GST–Pex5p variants were eluted with GSH and dialyzed to remove the GSH, whereas Pcs60p was removed by thrombin cleavage with the GST tag remaining on the agarose column. Equal portions of GST–Pex5p, GST–Ub(Δ1–6)Pex5p, or GST alone were combined with the purified Pcs60p and loaded onto GSH–agarose. After 1 h of incubation, the columns were washed, and elution of bound proteins was carried out by addition of reduced GSH. Samples of load, flow-through, and eluate fractions were analyzed by SDS–PAGE and Coomassie staining. The analysis revealed that no Pcs60p co-eluted together with the GST control. In contrast, efficient binding was observed when GST–Pex5p was bound to the agarose (Fig. 4A), which is in line with our previous reports of a direct Pex5p/Pcs60p interaction (37). Pcs60p was also co-eluted when GST–Ub(Δ1–6)Pex5p was bound to the column. The binding efficiency was similar to WT Pex5p (Fig. 4A). Thus, the fusion to ubiquitin does not alter cargo binding to Pex5p in vitro.
Figure 4.
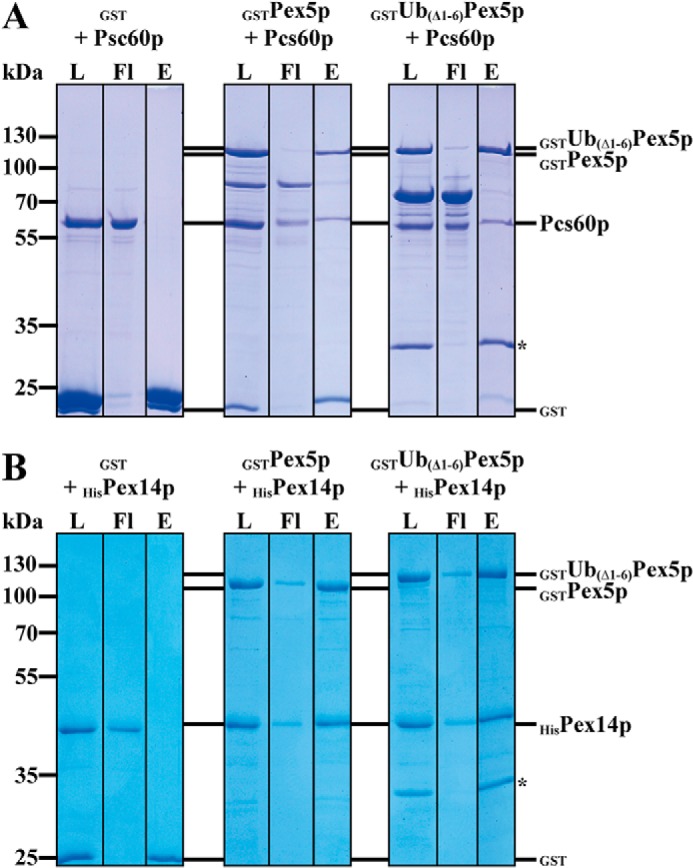
Analysis of cargo recognition and Pex14p interaction of N-terminally ubiquitinated Ub(1–6Δ)Pex5p. Purified GST and N-terminal GST fusion constructs of Pex5p and Ub(1–6Δ)Pex5p were combined with the PTS1 cargo protein Pcs60p (A) and the peroxisomal membrane docking protein HisPex14p (B) and loaded onto GSH–agarose. Bound proteins were eluted with buffer containing 50 mm reduced GSH. Equal volumes of load, flow through (Fl), and 3× concentrated eluate fractions were subjected to SDS–PAGE and Coomassie staining. The asterisk marks a degradation product of GSTUb(1–6Δ)Pex5p, which represents GSTUb. The N-terminally monoubiquitinated Pex5p binds its PTS1 cargo (Pcs60p) and the docking protein (Pex14p) with a similar efficiency as WT Pex5p. The amounts of recovered Pcs60p and HisPex14p in the eluates were 8–10 and 30%, respectively.
Once the receptor is cargo-loaded, the receptor–cargo complex binds to the peroxisomal membrane. Because of its direct interaction with Pex5p and the increase of binding when Pex5p is loaded with a PTS1-containing peptide, Pex14p has been proposed to serve as the docking site for the cytosolic receptor–cargo complex (30). Here we compared the Pex14p binding of the ubiquitin–Pex5p fusion and Pex5p. As described above, GST–Pex5p, GST–Ub(Δ1–6)Pex5p, and as control GST alone were expressed in E. coli and purified. We generated a Pex14p variant fused to an N-terminal His6 tag, which was separately expressed in E. coli and purified by affinity chromatography with nickel–nitrilotriacetic acid. As described above for Pcs60p, the different Pex5p variants were incubated with purified Pex14p, loaded on GSH—agarose, and eluted. As judged by Coomassie stain, Pex14p co-eluted with GST–Pex5p but also with Ub(Δ1–6)Pex5p in equal amounts, indicative of an efficient interaction (Fig. 4B). Signal-intensity measurements of the Pex5p and Pex14p protein bands of the Coomassie-stained gel indicated that similar amounts of Pex14p bound to the column, independent of whether Pex5p or Ub(Δ1–6)Pex5p served as bait (data not shown). Taken together, our results demonstrate that binding neither to Pcs60p nor to Pex14p is influenced by the exchange of the extreme N terminus of Pex5p by ubiquitin.
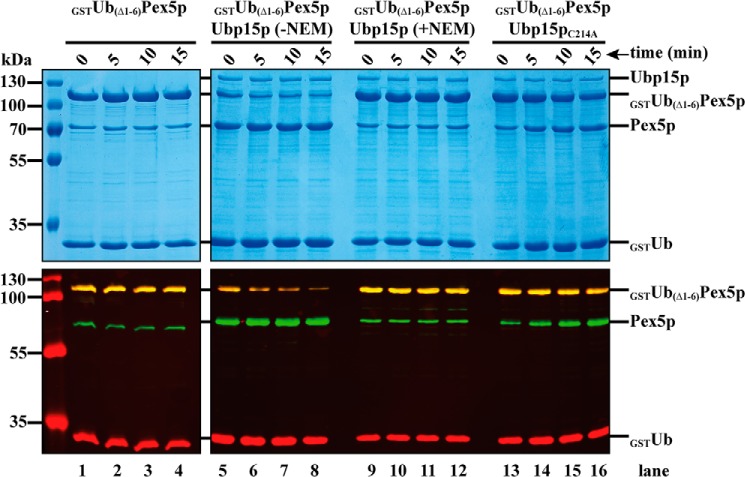
In vitro deubiquitination of the linear ubiquitin–Pex5p fusion by Ubp15p
Pex5p is mono- or polyubiquitinated in vivo, and the ubiquitin moiety has to be removed from Pex5p presumably during or after receptor export to allow another round of import. The cleavage of ubiquitin from a substrate protein is generally carried out by ubiquitin hydrolases also known as deubiquitinating enzymes (DUBs) (38). S. cerevisiae expresses more than 20 putative DUBs, which all except one are encoded by nonessential genes (39). Thus, individual deletions result in only subtle phenotypes, suggesting redundancy in their functions (40, 41). Among the yeast DUBs, Ubp15p was identified as a component of the AAA complex, and cells affected in Ubp15p display a stress-related partial protein import defect of PTS1 proteins (22). Based on in vitro cleavage assays, it was demonstrated that Ubp15p is capable of removing ubiquitin from Pex5p. Here we addressed the question of whether Ubp15p exhibits its deubiquitinating activity also on the linear ubiquitin Pex5p fusion protein (Ub(Δ1–6)Pex5p). To this end, we performed in vitro ubiquitin-cleavage assays. Heterologously expressed and isolated Ubp15p was incubated with GST-tagged Ub(Δ1–6)Pex5p, and the reaction was stopped by freezing in liquid nitrogen after zero (control) or later time points as indicated. Cleavage was monitored by Coomassie-stained SDS gels and immunoblot analysis. In the absence of Ubp15p, the Pex5p fusion remained mainly intact; only a slight cleavage of the joint region between Pex5p and the GST–Ub moiety occurred even at time point 0 (Fig. 5, lanes 1–4). In the presence of Ubp15p, a time-dependent decrease of the fusion protein was observed (Fig. 5, lanes 5–8). An increase of cleavage products can be seen already at 0 min, indicating that a large portion of the fusion protein was cleaved even before the temperature was shifted to 37 °C (Fig. 5, lanes 5–8). To exclude that this cleavage was based on proteases co-purified with Ubp15p or in a nonenzymatic manner by a nucleophilic attack of GSH as described in vivo for Pex5p from rat (23), we carried out the experiment after preincubation of Ubp15p with N-ethylmaleimide (NEM), which is known to inhibit deubiquitinating enzymes (42). Under these conditions, no additional cleavage of Pex5p was observed (Fig. 5, lanes 9–12). Sequence alignment of Ubp15p with other UBPs indicated that Cys214 of Ubp15p most likely represents an amino acid residue, which is crucial for the deubiquitinating activity (42). In fact, a Cys214 to Ala substitution introduced into the full-length Ubp15p (Ubp15pC214A) was shown to be enzymatically affected but not completely inactive (22). Accordingly, GST–Ub was slightly cleaved off from GST–Ub(Δ1–6)Pex5p, but most of the fusion protein remained stable when incubated with Ubp15pC214A (Fig. 5, lanes 13–16). Taken together, our data are clear in that like the natively ubiquitinated Pex5p, the linear Ub(Δ1–6)Pex5p fusion also displays a target for deubiquitination by Ubp15p.
Figure 5.
In vitro deubiquitination of N-terminally ubiquitinated Ub(1–6Δ)Pex5p. WT Ubp15p and the catalytically inactive mutant Ubp15pC214A were treated with or without NEM and incubated with purified GST-tagged Ub(1–6Δ)Pex5p. Samples taken at the indicated time points were subjected to SDS–PAGE and Coomassie staining (upper panels) or immunoblot analysis (lower panels) using antibodies against GST (red) and Pex5p (green). The increase of Pex5p and free GST-tagged ubiquitin (GSTUb) in the sample containing the catalytically active and non-NEM-treated Ubp15p shows that the N-terminally ubiquitinated Ub(1–6Δ)Pex5p is deubiquitinated by Ubp15p in vitro.
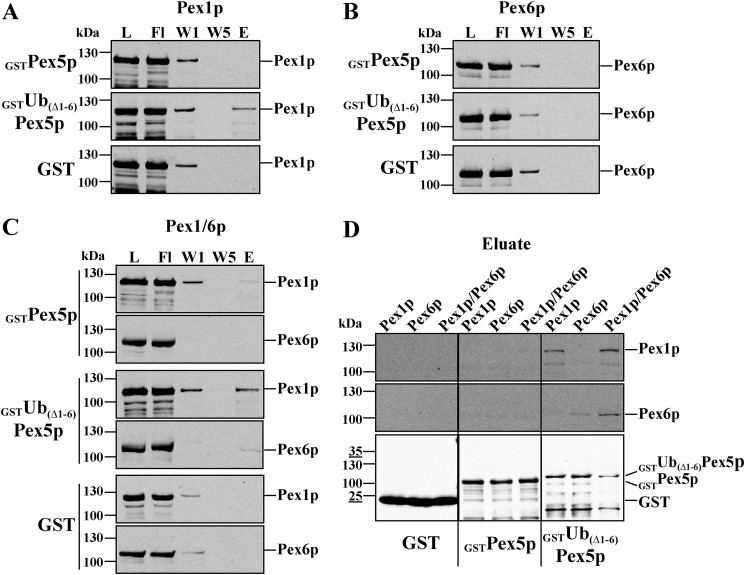
Ubiquitin-dependent binding of Pex5p to the Pex1p–Pex6p complex
Our studies show that the Pex1p–Pex6p complex can bind ubiquitin (Fig. 2). However, the natural substrate for the Pex1p–Pex6p-dependent release is monoubiquitinated Pex5p, which in our study is mimicked by the linear Ub(Δ1–6)Pex5p fusion. Therefore, in the following we tested the binding of Ub(Δ1–6)Pex5p to Pex1p and/or Pex6p by in vitro pulldown assays. To this end, heterologously expressed and purified GST fusions of Pex5p or Ub(Δ1–6)Pex5p, as well as GST alone (control), were incubated with isolated Pex1p, Pex6p, or the Pex1p–Pex6p complex. The samples were loaded onto GSH–agarose columns, and after extensive washing bound proteins were eluted with GSH. Neither Pex1p, Pex6p, nor the Pex1p–Pex6p complex co-eluted with GST as bait (Fig. 6). In contrast, the Pex1p–Pex6p complex co-eluted together with GST–Ub(Δ1–6)Pex5p (Fig. 6C), indicating that Ub(Δ1–6)Pex5p can physically interact with the Pex1p–Pex6p complex. Because the nonubiquitinated Pex5p did not interact with the AAA complex, the data also show that the interaction depends on the presence of ubiquitin. Also Pex1p alone did bind Ub(Δ1–6)Pex5p (Fig. 6A). A small amount of Ub(Δ1–6)Pex5p seems to associate with Pex6p; this, however, is only seen upon longer exposure (Fig. 6D). In contrast to Pex1p, which did bind to Ub(Δ1–6)Pex5p with nearly the same efficiency as the Pex1p–Pex6p complex, binding of Pex6p alone was much less efficient (Fig. 6D). These findings indicate that both Pex1p and Pex6p are able to interact with Ub(Δ1–6)Pex5p, but the data also suggest that Pex1p might be the primary binding partner for Ub(Δ1–6)Pex5p.
Figure 6.
Ubiquitin-dependent in vitro interaction of Pex5p with Pex1p and Pex6p. Purified GST and N-terminal GST fusion constructs of Pex5p and Ub(1–6Δ)Pex5p were combined with HisPex1p (A), HisPex6p (B), or the assembled HisPex1p–HisPex6p complex (C) and loaded onto GSH–agarose. Bound proteins were eluted with buffer containing 50 mm reduced GSH. Equal volumes of each relevant step were subjected to immunoblot analysis using antibodies against Pex1p, Pex6p, and GST. For comparison, the immunoblot analysis of higher amounts of eluates is shown in D. Interaction of Pex1p and Pex6p with the receptor Pex5p strongly depends on the presence of ubiquitin fused to Pex5p. A maximum of 2.5% of loaded AAA protein was recovered in the eluates. L, load; Fl, flow through; W1, wash 1; W5, wash 5; E, 10× concentrated eluate.
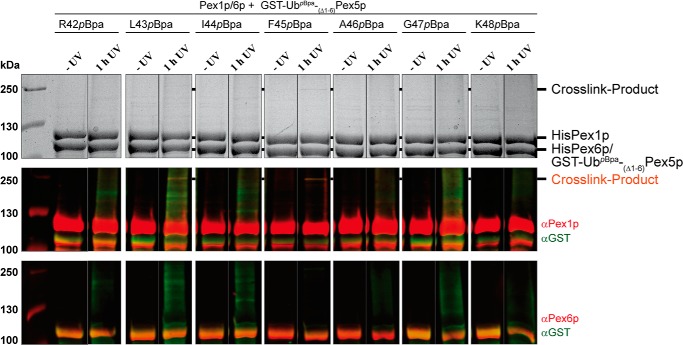
Site-specific in vitro cross-linking of the Pex1p–Pex6p–Ub(Δ1–6)Pex5p complex
To gain deeper insight into the recruitment of the ubiquitinated PTS1 receptor to the Pex1p–Pex6p complex, we aimed to stabilize the complex by use of site-specific in vitro photocross-linking (43). Our initial experiments demonstrate that the interaction of ubiquitin with the Pex1p–Pex6p complex is weakened when the isoleucine at position 44 of the ubiquitin is replaced by alanine (Fig. 2). This result indicates that ubiquitin binds to the AAA complex via a hydrophobic patch with Ile44 as central amino acid residue. Such an interaction is typical for binding of ubiquitin to many ubiquitin-binding proteins (33). For our cross-linking experiments, we genetically incorporated the photocross-linking amino acid para-benzoyl-phenylalanine (pBpa) into the corresponding ubiquitin part of recombinant Ub(Δ1–6)Pex5p. The pBpa is encoded by the amber codon TAG, and its incorporation into the proteins takes place during translation, using an orthogonal tRNA/aminoacyl-tRNA synthetase pair. The pBpa-containing proteins efficiently form stable complexes with their partners upon UV irradiation at 365-nm wavelength (43, 44). In our case, the base triplets encoding amino acid residues 42–48 of ubiquitin were individually changed to amber codons. Expression of the Ub(Δ1–6)Pex5p variants was carried out in the presence of an evolved Methanococcus jannashii tRNA/aminoacyl-tRNA synthetase pair, 0.02% arabinose and 1 mm pBpa in E. coli strain BL21 (DE3). Proteins were expressed as GST fusions, and their isolation was carried out by affinity chromatography on GSH–agarose with GSH elution. SDS–PAGE analysis showed no difference between the purified amber mutant Ub(Δ1–6)Pex5p and the WT protein (data not shown).
Isolated Ub(Δ1–6)Pex5p variants were incubated with the purified Pex1p–Pex6p complex for 1 h in the presence and absence of UV light. Subsequently, samples were subjected to SDS–PAGE followed by either Coomassie staining or immunodetection. In the Coomassie-stained gel, a very thin band of a high molecular weight protein was visible for the F43pBpa variant and the F45pBpa variant (Fig. 7). This band was only visible when the sample was treated with UV light, indicative of a specific cross-link product. Immunoblot analysis revealed that the band represents a cross-linking product of Pex1p and the linear ubiquitin–Pex5p fusion (Fig. 7). No cross-linking product was found with Pex6p. Thus, we conclude that Pex1p directly binds the ubiquitin part of the fusion protein Ub(Δ1–6)Pex5p. Our results show that the Pex1p–Pex6p complex can bind Ub(Δ1–6)Pex5p via its ubiquitin moiety and that this interaction is mediated by a physical binding of ubiquitin to Pex1p.
Figure 7.
Site-specific photocross-linking of monoubiquitinated Pex5p and Pex1p. Purified GST-tagged Ub(1–6Δ)Pex5p constructs harboring different ubiquitin amino acid substitutions by the cross-linker pBpa were combined with purified HisPex1p–HisPex6p complex and irradiated with UV light (365 nm). Samples taken before and after UV treatment were subjected to SDS–PAGE and Coomassie staining (top panels) or immunoblot analysis with specific antibodies against GST (middle and bottom panels, green), Pex1p (middle panels, red), and Pex6p (bottom panels, red). All depicted lanes in the top, middle, and bottom panels, respectively, belong to the same gel/blot. The Leu43 and the Phe45 substitutions resulted in a cross-link product of Pex1p and GST–Ub(1–6Δ)Pex5p that appeared as a very thin band stained by Coomassie and was detected with GST and Pex1p antibodies, but not with Pex6p antibodies.
Discussion
In this study we report on the importance of Pex5p ubiquitination for its association with the AAA proteins Pex1p–Pex6p. We identify Pex1p as the main binding factor and demonstrate that ubiquitination does not affect Pex5p binding to its cargo or to the docking complex at the peroxisomal membrane.
A crucial step in the peroxisomal import cycle for PTS1 proteins is the release of the unloaded PTS1 receptor Pex5p from the peroxisomal membrane back to the cytosol to allow another round of import. This export step 1) is responsible for the ATP dependence of the overall process, 2) depends on monoubiquitination of the receptor, and 3) is performed by a complex of the peroxisomal AAAs Pex1p and Pex6p (21, 45). In contrast to conventional targets for ubiquitination, Pex5p is special, formed by ubiquitination of a lysine residue, a thioester bond is formed between ubiquitin and a conserved cysteine of Pex5p (21, 46, 47). This conserved cysteine residue proved to be essential for the biological function of Pex5p (49–51). In line with this finding, a Pex5p truncation of the first six amino acid residues, including the conserved cysteine, is incapable of restoring the PTS1 import deficiency of a pex5Δ strain (Fig. 3).
In addition to lysine or cysteine ubiquitination, ubiquitin can also be conjugated to the free α-amino group of the first residue of a target protein (52). The biological role of such an N-terminal ubiquitination remains elusive because of a low number of examples. It is speculated that it may serve as a target for further polyubiquitination, which is a well-known degradation signal recognized by the proteasome (52, 53). With respect to Pex5p, it turned out that not the type of target residue but its position is of major relevance for the functionality of the receptor. For instance, Pex5p function was preserved upon replacement of the conserved cysteine by lysine (49, 51, 54). To mimic the monoubiquitinated version of the PTS1 receptor, we used here the biological template of a linear N-terminal ubiquitin fusion and replaced the extreme N terminus of Pex5p by ubiquitin. Expression of the linear ubiquitin–Pex5p fusion in pex5Δ restored the peroxisomal import defect, despite a reduced steady-state level of about 50% compared with the plasmid-encoded Pex5p WT (data not shown), indicating that the linear fusion of the receptor with ubiquitin fulfills all requirements for proper matrix protein import (Fig. 3). The result demonstrates that the N-terminal ubiquitin tagging can functionally replace the cysteine-dependent monoubiquitination.
The monoubiquitination primes the PTS1 receptor for its export back to the cytosol, which is an essential step in the import of matrix proteins (55, 56). However, it is still an open question whether this is the only function of the monoubiquitination in the import process. According to the idea of the export-driven import model, the ubiquitin-mediated and ATP-dependent export of the receptor might be functionally interconnected to protein translocation across the peroxisomal membrane (57). This idea is contradicted by data from mammalian cells, suggesting that unloading of the receptor; thus release of the PTS1 cargo protein occurs prior to and thus independently of monoubiquitination of Pex5p (58–60). However, the time point when the native receptor is modified during the receptor cycle and import process is still not clearly defined. In principle, Pex5p monoubiquitination could coincide in time with any of the steps of the receptor cycle prior to its export back to the cytosol, an idea that was also proposed by Williams et al. (21). We demonstrate that binding of Pex5p to its cargo and to Pex14p is not affected by its ubiquitination (Fig. 4), indicating that Pex5p monoubiquitination alone seems not to play a crucial role in cargo binding in the cytosol, docking of the receptor–cargo complex to the peroxisomal membrane, or release of the cargo during the import process.
To explore the possibility that the ubiquitin moiety provides a protein interface, which triggers association with the receptor export machinery, we focused on Pex1p and Pex6p as potential binding partners of the ubiquitinated receptor. These peroxins belong to the family of AAA proteins (61–63), typically involved in processes such as protein unfolding and degradation or disassembly of protein complexes (64). They were shown to form a heterohexameric complex, organizing as a trimer of dimers (26, 27, 65). Pex1p and Pex6p have been implicated in the recycling of Pex5p, because they were found to act in the terminal steps of matrix protein import in the yeast Pichia pastoris (66) and ATP was reported to be indispensable for receptor export (10). Work with human cells as well as S. cerevisiae showed that indeed Pex1p and Pex6p are necessary for Pex5p export (10, 25). However, our knowledge about the mechanism of action is still scarce. Here we show that the Pex1p–Pex6p complex directly interacts with monoubiquitinated Pex5p in vitro. In line with recently published results, we found no direct interaction with unmodified Pex5p (67). This is in contrast to mammalian Pex1p, which exists as a homo-oligomer in the cytosol and a hetero-oligomer with Pex6p on peroxisome membranes (68). A surface plasmon resonance–based assay demonstrated that the mammalian Pex1p homo-oligomer binds directly to nonubiquitinated Pex5p, although at a low affinity (69). The study, however, did not investigate the contribution of ubiquitin to the interaction. In our study, the interaction of Pex5p to the AAA complex, in particular Pex1p, strongly depends on the presence of the ubiquitin moiety fused to the N terminus of Pex5p (Fig. 6). Furthermore, the hydrophobic patch around isoleucine 44 of ubiquitin seems to be involved in the interaction. The mutation of this amino acid to alanine in UbGST resulted in a decreased binding to the AAA proteins (Fig. 2), and site-specific photocross-linking was only successful when the cross-linker was inserted next to the essential Ile44 residue (Fig. 7). The involvement of the hydrophobic patch of ubiquitin for recognition is typical for many ubiquitin-binding proteins (33, 70). The N-terminal domains of both Pex1p and Pex6p have been shown to comprise two double-ψ-β-barrel domains (65), which were previously identified as ubiquitin-binding domains in the related AAA protein p97 and its adapter protein Ufd1 (71).
Interestingly, our data suggest that Pex1p and Pex6p have distinct binding capabilities for ubiquitin with respect to the surrounding protein context. Although Pex6p was able to bind GST-tagged ubiquitin, Pex1p showed a higher specificity for the Ub–Pex5p fusion construct. The N-terminal domains of Pex1p are flexibly located above the double-ring structure of the AAA complex (26, 65), which makes them a likely candidate for the initial contact with the ubiquitinated receptor. Pex1p might therefore be able to specifically recognize the ubiquitin modification at the conserved cysteine of Pex5p. Pex6p, however, seems to have a more general affinity toward ubiquitin. It could be imagined that after initial binding by Pex1p, the ubiquitin moiety is handed over to Pex6p to position it for deubiquitination by Ubp15p, which is associated with the D1 domain of Pex6p (22), whereas Pex5p is directed to the pore of the AAA complex for further processing. Alternatively, the N-terminal domains of Pex6p might provide an additional quality control factor to efficiently capture polyubiquitinated Pex5p species.
Materials and methods
Strains, plasmids, and primers
The strains, plasmids, and sequences of oligonucleotides used are listed in Tables 1–3. The plasmids pIG26–pIG32 for expression of the cross-linker constructs of GSTUbpBpa-(1–6Δ)Pex5p were generated by QuikChange site-directed mutagenesis according to the manufacturer's instructions (Stratagene) using selected primer pairs and the template pIG24.
Table 1.
S. cerevisiae strains used in this study
Table 2.
Plasmids used in this study
| Plasmid | Description | Source or reference | Oligonucleotides |
|---|---|---|---|
| pPC86–PEX5 | GalAD–PEX5 | Ref. 81 | |
| pPC86–PEX6 | GalAD–PEX6 | Ref. 74 | |
| pPC86–PEX15(1–315) | GalAD–PEX15 (1–315) | Ref. 82 | |
| pPC97–PEX14 | GalBD–PEX14 | Ref. 30 | |
| pBM5 | GalBD–PEX1 | Ref. 74 | |
| pBM21 | GalBD–PEX6 | Ref. 31 | |
| pGEX-4T3 | GST | Invitrogen | |
| pET–Ub–V–GST | UBIQUITIN–GST | Ref. 83 | |
| pET–UbI44A–V–GST | UBIQUITIN(I44A)–GST | Ref. 83 | |
| pRSFDuet–His6Pex1 | HIS6–PEX1 | Ref. 84 | |
| pRSFDuet–HisPex1cGST | HIS6–PEX1–GST | Ref. 84 | |
| pRSFDuet–HisPex6 | HIS6–PEX6 | Ref. 84 | |
| pGEX4T2–GST–Pex5 | GST–PEX5 | Ref. 81 | |
| pIG24 | Ub(Δ1–5)PEX5 | This study | RE3176/3178 |
| pNH01 | GST-PCS60 | Ref. 37 | |
| pET9d–His6–PEX14 | HisPex14p | Ref. 48 | |
| pGEX4T2–GST–Ub(R42amber)–Pex5 | GST–Ub (R42pBpa)– (Δ1–6)Pex5p | This study | RE3589/3890 |
| pGEX4T2–GST–Ub(L43amber)–Pex5 | GST–Ub (L43pBpa)– (Δ1–6)Pex5p | This study | RE3591/3592 |
| pGEX4T2–GST–Ub(I44amber)–Pex5 | GST–Ub (I44pBpa)- (Δ1–6)Pex5p | This study | RE3587/3588 |
| pGEX4T2–GST–Ub(F45amber)–Pex5 | GST–Ub (F45pBpa)– (Δ1–6)Pex5p | This study | RE3593/3594 |
| pGEX4T2–GST–Ub(A46amber)–Pex5 | GST–Ub (A46pBpa)– (Δ1–6)Pex5p | This study | RE3595/3596 |
| pGEX4T2-GST-Ub(G47amber)-Pex5 | GST–Ub (G47pBpa)– (Δ1–6)Pex5p | This study | RE3597/3598 |
| pGEX4T2–GST–Ub(K48amber)–Pex5 | GST–Ub (K48pBpa)– (Δ1–6)Pex5p | This study | RE3599/3600 |
| pHP17 | Yeast expression vector, PEX5 | Ref. 20 | |
| pFM03 | Yeast expression vector, Ub (Δ1–6)PEX5 | This study | RE3176/3178 |
| pFM01 | Yeast expression vector, (Δ1–6)PEX5 | This study | RE3013/3014 |
| pGEX–4T–2–UBP15 | GST–UBP15 | Ref. 22 | |
| pGEX–4T–2–UBP15(C214A) | GST–UBP15 (C214A) | Ref. 22 | |
| pEVOL–RS | t-RNA and t-RNA–synthetase | P. Schultz, Scripps Research Institute, La Jolla, CA |
Table 3.
Oligonucleotides used in this study
| Designation | Sequence (5′ → 3′) |
|---|---|
| RE3013 | CCGGTCGACATGCAGATCTTCGTCAAGACGTTAACCGGT |
| RE3014 | GTAGACCTTACATCTTGTCTTAAGACTAAGAGGTGGTGCGTCGACGCC |
| RE3176 | GGGGGATCCATGGACGTAGGAAGTTGCTCAG |
| RE3178 | GCGGCCGCTCAAAACGAAAATTCTCCTTTAA |
| RE3587 | GCATTCCACCTGATCAACAAAGATTGTAGTTTGCCGGTAAGCAGC |
| RE3588 | GCTGCTTACCGGCAAACTACAATCTTTGTTGATCAGGTGGAATGC |
| RE3589 | GGAAGGCATTCCACCTGATCAACAATAGTTGATCTTTGCCGGTAAG |
| RE3590 | CTTACCGGCAAAGATCAACTATTGTTGATCAGGTGGAATGCCTTCC |
| RE3591 | CACCTGATCAACAAAGATAGATCTTTGCCGGTAAGCA |
| RE3592 | TGCTTACCGGCAAAGATCTATCTTTGTTGATCAGGTG |
| RE3593 | CCACCTGATCAACAAAGATTGATCTAGGCCGGTAAGCAGC |
| RE3594 | GCTGCTTACCGGCCTAGATCAATCTTTGTTGATCAGGTGG |
| RE3595 | TTCCACCTGATCAACAAAGATTGATCTTTTAGGGTAAGCAGCTCGAG |
| RE3596 | CTCGAGCTGCTTACCCTAAAAGATCAATCTTTGTTGATCAGGTGGAA |
| RE3597 | CAACAAAGATTGATCTTTGCCTAGAAGCAGCTCGAGGACGGTAGA |
| RE3598 | TCTACCGTCCTCGAGCTGCTTCTAGGCAAAGATCAATCTTTGTTG |
| RE3599 | ACAAAGATTGATCTTTGCCGGTTAGCAGCTCGAGGA |
| RE3600 | TCCTCGAGCTGCTAACCGGCAAAGATCAATCTTTGT |
Yeast expression of Pex5p was performed using the plasmid pHP17 (45). For yeast expression of (1–6Δ)Pex5p, truncated Pex5p was amplified from pHP17 using primer pair RE3177/RE3178 and introduced behind the Pex5p promoter in a pRS416 plasmid using BamHI and NotI restriction sites, resulting in the plasmid pFM01. For yeast expression of Ub–(1–6Δ)Pex5p, synthetic ubiquitin was first amplified from YEp96 (72) with the primer pair RE3013/RE3014 and subsequently cloned into the plasmid pHP17 using a SalI restriction site. Truncated (1–6Δ)Pex5p was amplified from pHP17 with primer pair RE3176/RE3178 and introduced into the ubiquitin construct, resulting in the plasmid pFM03.
Protein purification
Protein expression and purification of Pex1p, Pex6p, or the Pex1p–Pex6p complex was performed according to Ref. 32. Expression of recombinant proteins was induced with 0.4 mm isopropyl β-d-thiogalactopyranoside with expression conditions of 4 h at 30 °C for GSTPex5p, GSTPcs60p, His6Pex14p, UbGST, and UbI44AGST, as well as 20 h at 20 °C for GSTUb(Δ1–6)-Pex5p, GSTUbp15p, and GSTUbp15pC214A. For expression of cross-linking constructs of GSTUb–(1–6Δ)Pex5p, the medium additionally contained 0.02% arabinose and 1 mm pBpa.
The cells were harvested after expression and suspended in buffer I (50 mm Tris, 100 mm NaCl, 1 mm DTT, pH 7.4) (GSTPex5p, GSTUb–(1–6Δ)Pex5p, GSTUb–(1–6Δ)Pex5p cross-linkers, GSTUbp15p, GSTUbp15pC214A), buffer II (50 mm Tris, 150 mm NaCl, 1 mm DTT, pH 7.4) (GST, GSTPcs60p, UbGST, UbI44AGST), or buffer III (20 mm Tris, 500 mm NaCl, 5 mm imidazole, pH 7.9) (His6Pex14p). All buffers for cell lysis contained a selection of protease inhibitors (1 mm phenylmethylsulfonyl fluoride, 8 mm antipain, 0.3 mm aprotinin, 1 mm bestatin, 10 mm chymostatin, 5 mm leupeptin, 15 mm pepstatin) and 25 μg/ml DNase I.
The cells were broken by sonication, and the homogenate was centrifuged at 14,000 rpm for 1 h (rotor SS-34; Thermo Scientific). Supernatants containing soluble GST fusion proteins were loaded onto a GSH–agarose 4B matrix (Macherey-Nagel, Düren, Germany). After incubation for 1 h, unbound protein was washed off with 20-fold column volume of buffer. The proteins were eluted either with buffer containing 50 mm reduced GSH (GSTPex5p, GSTUb–(1–6Δ)Pex5p, and GSTUb–(1–6Δ)Pex5p cross-linkers, GST, UbGST, UbI44AGST) or after incubation with thrombin overnight to retain the GST tag on the column (GSTUbp15p, GSTUbp15pC214A, GSTPcs60p). The homogenate containing His6Pex14p was loaded onto a HisTrap HP column (GE Healthcare) using an ÄKTA Prime system (GE Healthcare). After washing with buffer III containing 60 mm imidazole, the protein was eluted with a continuous imidazole gradient of up to 1 m imidazole.
In vitro deubiquitination
Purified GST-tagged Ub–Pex5p fusion protein was incubated with or without Ubp15p for 15 min at 37 °C. Samples taken every 5 min were analyzed via SDS–PAGE and subsequent Coomassie staining or immunoblotting. For control purposes, Ubp15p activity was inhibited by preincubation with NEM or by point mutation C214A.
In vitro binding assay
For binding assays purified GST-tagged bait proteins were premixed with other proteins and incubated with 50 μl of GSH–agarose 4B (Macherey-Nagel) for 1 h. Estimated amounts of 50 μg (Figs. 2 and 6), 30 μg (Fig. 4A), and 15 μg (Fig. 4B) of each protein were used in these assays. Unbound protein was washed off with 10-fold column volume of buffer, and bound components were eluted with buffer containing 100 mm reduced GSH. For assays containing Pex1p and Pex6p, the bait proteins were dialyzed against AAA buffer before the experiment.
In vivo complementation
The WT S. cerevisiae strain UTL-7A and the corresponding pex5Δ strain were transformed with a plasmid containing mCherry with C-terminally fused PTS1. Additionally, the pex5Δ strain was transformed with an empty pRS416 plasmid, as well as plasmid-encoded WT Pex5p, (Δ1–6)Pex5p, or Ub–(Δ1–6)Pex5p. The cells were precultured in YNBG medium containing 0.3% glucose. Peroxisome proliferation was induced in YNBGO medium containing 0.1% glucose and 0.1% oleic acid.
Antibodies and immunoblotting
Immunoblot analysis was performed according to Harlow and Lane (73) with polyclonal rabbit antibodies raised against Pex1p (74), Pex6p (31), Pex5p (30), or monoclonal anti-GST (Signal). Primary antibodies were detected with an IRDye 800CW goat anti-rabbit IgG secondary antibody (LI-COR Bioscience, Bad Homburg, Germany) followed by detection using an IR imaging system (LI-COR Bioscience). Semiquantitative analyses of immunoblot signals were obtained using the IR Imaging System Application software version 3.0 (LI-COR Bioscience).
Fluorescence microscopy
Wide-field fluorescence microscopy imaging was performed on a Zeiss Axioskop50 fluorescence microscope (Zeiss). Images were taken with a Princeton Instruments 1300Y digital camera. The mCherry fluorescence was visualized with a 546/12-nm band-pass excitation filter, a 560-nm dichromatic mirror, and a 575–640-nm band-pass emission filter.
Miscellaneous
Semiquantitative analyses of immunoblot signals were obtained using the IR Imaging System Application software version 3.0 (LI-COR Bioscience). Images were processed with Photoshop CS5 (Adobe) and arranged in figures using Illustrator CS5 (Adobe). The applied two-hybrid assay was based on the described method by Fields and Sternglanz (75). Co-transformation of two-hybrid vectors into the strain PJ69-4A was performed according to Gietz and Woods (76). Transformed yeast cells were plated onto SD synthetic medium without tryptophan and leucine. β-Galactosidase filter assays were performed as described elsewhere (77). Plasmid expressing mCherry-SKL was kindly provided by B. Warscheid (Freiburg, Germany). All experiments shown here were performed at least two times with reproducible results.
Author contributions
R. E. and W. G. conceptualization; R. E. and W. G. supervision; R. E. funding acquisition; R. E., D. P. S., I. G., and W. G. validation; R. E., D. P. S., I. G., and W. G. investigation; R. E., D. P. S., I. G., and W. G. visualization; R. E., D. P. S., and I. G. methodology; R. E., D. P. S., and W. G. writing-original draft; D. P. S. and I. G. data curation; D. P. S. and I. G. formal analysis.
This work was supported by Deutsche Forschungsgemeinschaft Grant FOR1905 (to R. E.). The authors declare that they have no conflicts of interest with the contents of this article.
- AAA
- ATPases associated with diverse cellular activities
- PTS
- peroxisomal targeting signal
- DUB
- deubiquitinating enzyme
- NEM
- N-ethylmaleimide
- pBpa
- para-benzoyl-phenylalanine.
References
- 1. Waterham H. R., Ferdinandusse S., and Wanders R. J. (2016) Human disorders of peroxisome metabolism and biogenesis. Biochim. Biophys. Acta 1863, 922–933 10.1016/j.bbamcr.2015.11.015 [DOI] [PubMed] [Google Scholar]
- 2. Braverman N. E., Raymond G. V., Rizzo W. B., Moser A. B., Wilkinson M. E., Stone E. M., Steinberg S. J., Wangler M. F., Rush E. T., Hacia J. G., and Bose M. (2016) Peroxisome biogenesis disorders in the Zellweger spectrum: an overview of current diagnosis, clinical manifestations, and treatment guidelines. Mol. Genet. Metab. 117, 313–321 10.1016/j.ymgme.2015.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Glover J. R., Andrews D. W., and Rachubinski R. A. (1994) Saccharomyces cerevisiae peroxisomal thiolase is imported as a dimer. Proc. Natl. Acad. Sci. U.S.A. 91, 10541–10545 10.1073/pnas.91.22.10541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McNew J. A., and Goodman J. M. (1994) An oligomeric protein is imported into peroxisomes in vivo. J. Cell Biol. 127, 1245–1257 10.1083/jcb.127.5.1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Girzalsky W., Saffian D., and Erdmann R. (2010) Peroxisomal protein translocation. Biochim. Biophys. Acta 1803, 724–731 10.1016/j.bbamcr.2010.01.002 [DOI] [PubMed] [Google Scholar]
- 6. Brown L. A., and Baker A. (2003) Peroxisome biogenesis and the role of protein import. J. Cell. Mol. Med. 7, 388–400 10.1111/j.1582-4934.2003.tb00241.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Meinecke M., Cizmowski C., Schliebs W., Krüger V., Beck S., Wagner R., and Erdmann R. (2010) The peroxisomal importomer constitutes a large and highly dynamic pore. Nat. Cell Biol. 12, 273–277 10.1038/ncb2027 [DOI] [PubMed] [Google Scholar]
- 8. Montilla-Martinez M., Beck S., Klümper J., Meinecke M., Schliebs W., Wagner R., and Erdmann R. (2015) Distinct pores for peroxisomal import of PTS1 and PTS2 proteins. Cell Rep. 13, 2126–2134 10.1016/j.celrep.2015.11.016 [DOI] [PubMed] [Google Scholar]
- 9. Platta H. W., Debelyy M. O., El Magraoui F., and Erdmann R. (2008) The AAA peroxins Pex1p and Pex6p function as dislocases for the ubiquitinated peroxisomal import receptor Pex5p. Biochem. Soc. Trans. 36, 99–104 10.1042/BST0360099 [DOI] [PubMed] [Google Scholar]
- 10. Miyata N., and Fujiki Y. (2005) Shuttling mechanism of peroxisome targeting signal type 1 receptor Pex5: ATP-independent import and ATP-dependent export. Mol. Cell. Biol. 25, 10822–10832 10.1128/MCB.25.24.10822-10832.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schwerter D. P., Grimm I., Platta H. W., and Erdmann R. (2017) ATP-driven processes of peroxisomal matrix protein import. Biol. Chem. 398, 607–624 [DOI] [PubMed] [Google Scholar]
- 12. Stanley W. A., and Wilmanns M. (2006) Dynamic architecture of the peroxisomal import receptor Pex5p. Biochim. Biophys. Acta 1763, 1592–1598 10.1016/j.bbamcr.2006.10.015 [DOI] [PubMed] [Google Scholar]
- 13. Carvalho A. F., Costa-Rodrigues J., Correia I., Costa Pessoa J., Faria T. Q., Martins C. L., Fransen M., Sá-Miranda C., and Azevedo J. E. (2006) The N-terminal half of the peroxisomal cycling receptor Pex5p is a natively unfolded domain. J. Mol. Biol. 356, 864–875 10.1016/j.jmb.2005.12.002 [DOI] [PubMed] [Google Scholar]
- 14. Gouveia A. M., Guimarães C. P., Oliveira M. E., Reguenga C., Sá-Miranda C., and Azevedo J. E. (2003) Characterization of the peroxisomal cycling receptor Pex5p import pathway. Adv. Exp. Med. Biol. 544, 219–220 10.1007/978-1-4419-9072-3_26 [DOI] [PubMed] [Google Scholar]
- 15. Schäfer A., Kerssen D., Veenhuis M., Kunau W. H., and Schliebs W. (2004) Functional similarity between the peroxisomal PTS2 receptor binding protein Pex18p and the N-terminal half of the PTS1 receptor Pex5p. Mol. Cell. Biol. 24, 8895–8906 10.1128/MCB.24.20.8895-8906.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Platta H. W., Brinkmeier R., Reidick C., Galiani S., Clausen M. P., and Eggeling C. (2016) Regulation of peroxisomal matrix protein import by ubiquitination. Biochim. Biophys. Acta 1863, 838–849 10.1016/j.bbamcr.2015.09.010 [DOI] [PubMed] [Google Scholar]
- 17. Kiel J. A., Emmrich K., Meyer H. E., and Kunau W. H. (2005) Ubiquitination of the PTS1 receptor, Pex5p, suggests the presence of a quality control mechanism during peroxisomal matrix protein import. J. Biol. Chem. 280, 1921–1930 10.1074/jbc.M403632200 [DOI] [PubMed] [Google Scholar]
- 18. Kragt A., Voorn-Brouwer T., van den Berg M., and Distel B. (2005) The Saccharomyces cerevisiae peroxisomal import receptor Pex5p is monoubiquitinated in wild type cells. J. Biol. Chem. 280, 7867–7874 10.1074/jbc.M413553200 [DOI] [PubMed] [Google Scholar]
- 19. Léon S., and Subramani S. (2007) A conserved cysteine residue of Pichia pastoris Pex20p is essential for its recycling from the peroxisome to the cytosol. J. Biol. Chem. 282, 7424–7430 10.1074/jbc.M611627200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Platta H. W., Girzalsky W., and Erdmann R. (2004) Ubiquitination of the peroxisomal import receptor Pex5p. Biochem. J. 384, 37–45 10.1042/BJ20040572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Williams C., van den Berg M., Sprenger R. R., and Distel B. (2007) A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J. Biol. Chem. 282, 22534–22543 10.1074/jbc.M702038200 [DOI] [PubMed] [Google Scholar]
- 22. Debelyy M. O., Platta H. W., Saffian D., Hensel A., Thoms S., Meyer H. E., Warscheid B., Girzalsky W., and Erdmann R. (2011) Ubp15p, an ubiquitin hydrolase associated with the peroxisomal export machinery. J. Biol. Chem. 286, 28223–28234 10.1074/jbc.M111.238600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grou C. P., Carvalho A. F., Pinto M. P., Huybrechts S. J., Sá-Miranda C., Fransen M., and Azevedo J. E. (2009) Properties of the ubiquitin–Pex5p thiol ester conjugate. J. Biol. Chem. 284, 10504–10513 10.1074/jbc.M808978200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Grou C. P., Francisco T., Rodrigues T. A., Freitas M. O., Pinto M. P., Carvalho A. F., Domingues P., Wood S. A., Rodríguez-Borges J. E., Sá-Miranda C., Fransen M., and Azevedo J. E. (2012) Identification of ubiquitin-specific protease 9X (USP9X) as a deubiquitinase acting on ubiquitin–peroxin 5 (PEX5) thioester conjugate. J. Biol. Chem. 287, 12815–12827 10.1074/jbc.M112.340158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Platta H. W., Grunau S., Rosenkranz K., Girzalsky W., and Erdmann R. (2005) Functional role of the AAA peroxins in dislocation of the cycling PTS1 receptor back to the cytosol. Nat. Cell Biol. 7, 817–822 10.1038/ncb1281 [DOI] [PubMed] [Google Scholar]
- 26. Ciniawsky S., Grimm I., Saffian D., Girzalsky W., Erdmann R., and Wendler P. (2015) Molecular snapshots of the Pex1/6 AAA+ complex in action. Nat. Commun. 6, 7331 10.1038/ncomms8331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gardner B. M., Chowdhury S., Lander G. C., and Martin A. (2015) The Pex1/Pex6 complex is a heterohexameric AAA+ motor with alternating and highly coordinated subunits. J. Mol. Biol. 427, 1375–1388 10.1016/j.jmb.2015.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grimm I., Erdmann R., and Girzalsky W. (2016) Role of AAA+-proteins in peroxisome biogenesis and function. Biochim. Biophys. Acta 1863, 828–837 10.1016/j.bbamcr.2015.10.001 [DOI] [PubMed] [Google Scholar]
- 29. James G. L., Goldstein J. L., Pathak R. K., Anderson R. G., and Brown M. S. (1994) PxF, a prenylated protein of peroxisomes. J. Biol. Chem. 269, 14182–14190 [PubMed] [Google Scholar]
- 30. Albertini M., Rehling P., Erdmann R., Girzalsky W., Kiel J. A., Veenhuis M., and Kunau W.-H. (1997) Pex14p, a peroxisomal membrane protein binding both receptors of the two PTS-dependent import pathways. Cell 89, 83–92 10.1016/S0092-8674(00)80185-3 [DOI] [PubMed] [Google Scholar]
- 31. Birschmann I., Stroobants A. K., van Den Berg M., Schäfer A., Rosenkranz K., Kunau W. H., and Tabak H. F. (2003) Pex15p of Saccharomyces cerevisiae provides a molecular basis for recruitment of the AAA peroxin Pex6p to peroxisomal membranes. Mol. Biol. Cell 14, 2226–2236 10.1091/mbc.e02-11-0752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Grimm I., Saffian D., Girzalsky W., and Erdmann R. (2016) Nucleotide-dependent assembly of the peroxisomal receptor export complex. Sci. Rep. 6, 19838 10.1038/srep19838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hurley J. H., Lee S., and Prag G. (2006) Ubiquitin-binding domains. Biochem. J. 399, 361–372 10.1042/BJ20061138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Van der Leij I., Franse M. M., Elgersma Y., Distel B., and Tabak H. F. (1993) PAS10 is a tetratricopeptide-repeat protein that is essential for the import of most matrix proteins into peroxisomes of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 90, 11782–11786 10.1073/pnas.90.24.11782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Blobel F., and Erdmann R. (1996) Identification of a yeast peroxisomal member of the family of AMP-binding proteins. Eur. J. Biochem. 240, 468–476 10.1111/j.1432-1033.1996.0468h.x [DOI] [PubMed] [Google Scholar]
- 36. Foster J., and Nakata P. A. (2014) An oxalyl-CoA synthetase is important for oxalate metabolism in Saccharomyces cerevisiae. FEBS Lett. 588, 160–166 10.1016/j.febslet.2013.11.026 [DOI] [PubMed] [Google Scholar]
- 37. Hagen S., Drepper F., Fischer S., Fodor K., Passon D., Platta H. W., Zenn M., Schliebs W., Girzalsky W., Wilmanns M., Warscheid B., and Erdmann R. (2015) Structural insights into cargo recognition by the yeast PTS1 receptor. J. Biol. Chem. 290, 26610–26626 10.1074/jbc.M115.657973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mevissen T. E. T., and Komander D. (2017) Mechanisms of deubiquitinase specificity and regulation. Annu. Rev. Biochem. 86, 159–192 10.1146/annurev-biochem-061516-044916 [DOI] [PubMed] [Google Scholar]
- 39. Huseinovic A., van Dijk M., Vermeulen N. P. E., van Leeuwen F., Kooter J. M., and Vos J. C. (2018) Drug toxicity profiling of a Saccharomyces cerevisiae deubiquitinase deletion panel shows that acetaminophen mimics tyrosine. Toxicol. in Vitro 47, 259–268 10.1016/j.tiv.2017.12.007 [DOI] [PubMed] [Google Scholar]
- 40. Amerik A. Y., Li S. J., and Hochstrasser M. (2000) Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol. Chem. 381, 981–992 [DOI] [PubMed] [Google Scholar]
- 41. Kouranti I., McLean J. R., Feoktistova A., Liang P., Johnson A. E., Roberts-Galbraith R. H., and Gould K. L. (2010) A global census of fission yeast deubiquitinating enzyme localization and interaction networks reveals distinct compartmentalization profiles and overlapping functions in endocytosis and polarity. PLoS Biol. 8, e1000471 10.1371/journal.pbio.1000471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hochstrasser M. (1996) Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30 [DOI] [PubMed] [Google Scholar]
- 43. Wang L., Brock A., Herberich B., and Schultz P. G. (2001) Expanding the genetic code of Escherichia coli. Science. 292, 498–500 10.1126/science.1060077 [DOI] [PubMed] [Google Scholar]
- 44. Chin J. W., Martin A. B., King D. S., Wang L., and Schultz P. G. (2002) Addition of a photocrosslinking amino acid to the genetic code of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 99, 11020–11024 10.1073/pnas.172226299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Platta H. W., El Magraoui F., Schlee D., Grunau S., Girzalsky W., and Erdmann R. (2007) Ubiquitination of the peroxisomal import receptor Pex5p is required for its recycling. J. Cell Biol. 177, 197–204 10.1083/jcb.200611012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Carvalho A. F., Pinto M. P., Grou C. P., Alencastre I. S., Fransen M., Sá-Miranda C., and Azevedo J. E. (2007) Ubiquitination of mammalian Pex5p, the peroxisomal import receptor. J. Biol. Chem. 282, 31267–31272 10.1074/jbc.M706325200 [DOI] [PubMed] [Google Scholar]
- 47. Okumoto K., Misono S., Miyata N., Matsumoto Y., Mukai S., and Fujiki Y. (2011) Cysteine ubiquitination of PTS1 receptor Pex5p regulates Pex5p recycling. Traffic 12, 1067–1083 10.1111/j.1600-0854.2011.01217.x [DOI] [PubMed] [Google Scholar]
- 48. Kerssen D., Hambruch E., Klaas W., Platta H. W., de Kruijff B., Erdmann R., Kunau W. H., and Schliebs W. (2006) Membrane association of the cycling peroxisome import receptor Pex5p. J. Biol. Chem. 281, 27003–27015 10.1074/jbc.M509257200 [DOI] [PubMed] [Google Scholar]
- 49. Schwartzkopff B., Platta H. W., Hasan S., Girzalsky W., and Erdmann R. (2015) Cystein-specific ubiquitination protects the peroxisomal import receptor Pex5p against proteasomal degradation. Biosci. Rep. 35, e00215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Carvalho A. F., Grou C. P., Pinto M. P., Alencastre I. S., Costa-Rodrigues J., Fransen M., Sá-Miranda C., and Azevedo J. E. (2007) Functional characterization of two missense mutations in Pex5p-C11S and N526K. Biochim. Biophys. Acta 1773, 1141–1148 10.1016/j.bbamcr.2007.04.011 [DOI] [PubMed] [Google Scholar]
- 51. Ma C., Hagstrom D., Polley S. G., and Subramani S. (2013) Redox-regulated cargo binding and release by the peroxisomal targeting signal receptor, Pex5. J. Biol. Chem. 288, 27220–27231 10.1074/jbc.M113.492694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Varland S., Osberg C., and Arnesen T. (2015) N-terminal modifications of cellular proteins: the enzymes involved, their substrate specificities and biological effects. Proteomics 15, 2385–2401 10.1002/pmic.201400619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hochstrasser M. (1996) Protein degradation or regulation: Ub the judge. Cell 84, 813–815 10.1016/S0092-8674(00)81058-2 [DOI] [PubMed] [Google Scholar]
- 54. Nordgren M., Francisco T., Lismont C., Hennebel L., Brees C., Wang B., Van Veldhoven P. P., Azevedo J. E., and Fransen M. (2015) Export-deficient monoubiquitinated PEX5 triggers peroxisome removal in SV40 large T antigen-transformed mouse embryonic fibroblasts. Autophagy 11, 1326–1340 10.1080/15548627.2015.1061846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Francisco T., Rodrigues T. A., Dias A. F., Barros-Barbosa A., Bicho D., and Azevedo J. E. (2017) Protein transport into peroxisomes: knowns and unknowns. BioEssays 39, 201700047. [DOI] [PubMed] [Google Scholar]
- 56. Platta H. W., Hagen S., and Erdmann R. (2014) The peroxisomal exportomer. In Molecular Machines Involved in Peroxisome Biogenesis and Maintenance (Brocard C., and Hartig A., eds) pp. 347–370, Springer-Verlag, New York Inc., New York [Google Scholar]
- 57. Schliebs W., Girzalsky W., and Erdmann R. (2010) Peroxisomal protein import and ERAD: variations on a common theme. Nat. Rev. Mol. Cell Biol. 11, 885–890 10.1038/nrm3008 [DOI] [PubMed] [Google Scholar]
- 58. Francisco T., Rodrigues T. A., Freitas M. O., Grou C. P., Carvalho A. F., Sá-Miranda C., Pinto M. P., and Azevedo J. E. (2013) A cargo-centered perspective on the PEX5 receptor-mediated peroxisomal protein import pathway. J. Biol. Chem. 288, 29151–29159 10.1074/jbc.M113.487140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Alencastre I. S., Rodrigues T. A., Grou C. P., Fransen M., Sá-Miranda C., and Azevedo J. E. (2009) Mapping the cargo protein membrane translocation step into the PEX5 cycling pathway. J. Biol. Chem. 284, 27243–27251 10.1074/jbc.M109.032565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rodrigues T. A., Alencastre I. S., Francisco T., Brites P., Fransen M., Grou C. P., and Azevedo J. E. (2014) A PEX7-centered perspective on the peroxisomal targeting signal type 2-mediated protein import pathway. Mol. Cell. Biol. 34, 2917–2928 10.1128/MCB.01727-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Erdmann R., Wiebel F. F., Flessau A., Rytka J., Beyer A., Fröhlich K. U., and Kunau W.-H. (1991) PAS1, a yeast gene required for peroxisome biogenesis, encodes a member of a novel family of putative ATPases. Cell 64, 499–510 10.1016/0092-8674(91)90234-P [DOI] [PubMed] [Google Scholar]
- 62. Voorn-Brouwer T., van der Leij I., Hemrika W., Distel B., and Tabak H. F. (1993) Sequence of the PAS8 gene, the product of which is essential for biogenesis of peroxisomes in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1216, 325–328 10.1016/0167-4781(93)90166-B [DOI] [PubMed] [Google Scholar]
- 63. Beyer A. (1997) Sequence analysis of the AAA protein family. Protein Sci. 6, 2043–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hanson P. I., and Whiteheart S. W. (2005) AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6, 519–529 10.1038/nrm1684 [DOI] [PubMed] [Google Scholar]
- 65. Blok N. B., Tan D., Wang R. Y., Penczek P. A., Baker D., DiMaio F., Rapoport T. A., and Walz T. (2015) Unique double-ring structure of the peroxisomal Pex1/Pex6 ATPase complex revealed by cryo-electron microscopy. Proc. Natl. Acad. Sci. U.S.A. 112, E4017–E4025 10.1073/pnas.1500257112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Collins C. S., Kalish J. E., Morrell J. C., McCaffery J. M., and Gould S. J. (2000) The peroxisome biogenesis factors Pex4p, Pex22p, Pex1p, and Pex6p act in the terminal steps of peroxisomal matrix protein import. Mol. Cell. Biol. 20, 7516–7526 10.1128/MCB.20.20.7516-7526.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gardner B. M., Castanzo D. T., Chowdhury S., Stjepanovic G., Stefely M. S., Hurley J. H., Lander G. C., and Martin A. (2018) The peroxisomal AAA-ATPase Pex1/Pex6 unfolds substrates by processive threading. Nat. Commun. 9, 135 10.1038/s41467-017-02474-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Tamura S., Yasutake S., Matsumoto N., and Fujiki Y. (2006) Dynamic and functional assembly of the AAA peroxins, Pex1p and Pex6p, and their membrane receptor Pex26p. J. Biol. Chem. 281, 27693–27704 10.1074/jbc.M605159200 [DOI] [PubMed] [Google Scholar]
- 69. Tamura S., Matsumoto N., Takeba R., and Fujiki Y. (2014) AAA peroxins and their recruiter Pex26p modulate the interactions of peroxins involved in peroxisomal protein import. J. Biol. Chem. 289, 24336–24346 10.1074/jbc.M114.588038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sloper-Mould K. E., Jemc J. C., Pickart C. M., and Hicke L.. Distinct functional surface regions on ubiquitin. J. Biol. Chem. 276, 30483–30489 [DOI] [PubMed] [Google Scholar]
- 71. Park S., Isaacson R., Kim H. T., Silver P. A., and Wagner G. (2005) Ufd1 exhibits the AAA-ATPase fold with two distinct ubiquitin interaction sites. Structure 13, 995–1005 10.1016/j.str.2005.04.013 [DOI] [PubMed] [Google Scholar]
- 72. Ellison M. J., and Hochstrasser M. (1991) Epitope-tagged ubiquitin. A new probe for analyzing ubiquitin function. J. Biol. Chem. 266, 21150–21157 [PubMed] [Google Scholar]
- 73. Harlow E., and Lane D. (1988) Antibodies: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 74. Birschmann I., Rosenkranz K., Erdmann R., and Kunau W.-H. (2005) Structural and functional analysis of the interaction of the AAA–peroxins Pex1p and Pex6p. FEBS J. 272, 47–58 [DOI] [PubMed] [Google Scholar]
- 75. Fields S., and Sternglanz R. (1994) The two-hybrid system: an assay for protein–protein interactions. Trends Genet. 10, 286–292 10.1016/0168-9525(90)90012-U [DOI] [PubMed] [Google Scholar]
- 76. Gietz R. D., and Woods R. A. (1994) High efficiency transformation in Yeast. In Molecular Genetics of Yeast: Practical Approaches (Johnston J. A., ed) pp. 121–134, Oxford University Press, Oxford, UK [Google Scholar]
- 77. Rehling P., Marzioch M., Niesen F., Wittke E., Veenhuis M., and Kunau W.-H. (1996b) The import receptor for the peroxisomal targeting signal 2 (PTS2) in Saccharomyces cerevisiae is encoded by the PAS7 gene. EMBO J. 15, 2901–2913 [PMC free article] [PubMed] [Google Scholar]
- 78. Erdmann R., Veenhuis M., Mertens D., and Kunau W.-H. (1989) Isolation of peroxisome-deficient mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 86, 5419–5423 10.1073/pnas.86.14.5419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Girzalsky W., Rehling P., Stein K., Kipper J., Blank L., Kunau W. H., and Erdmann R. (1999) Involvement of Pex13p in Pex14p localization and peroxisomal targeting signal 2-dependent protein import into peroxisomes. J. Cell Biol. 144, 1151–1162 10.1083/jcb.144.6.1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. James P., Halladay J., and Craig E. A. (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144, 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Erdmann R., and Blobel G. (1996) Identification of Pex13p a peroxisomal membrane receptor for the PTS1 recognition factor. J. Cell Biol. 135, 111–121 10.1083/jcb.135.1.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Rosenkranz K., Birschmann I., Grunau S., Girzalsky W., Kunau W.-H., and Erdmann R. (2006) Functional association of the AAA-complex and the peroxisomal importomer. FEBS J. 273, 3804–3815 10.1111/j.1742-4658.2006.05388.x [DOI] [PubMed] [Google Scholar]
- 83. Meyer H. H., Wang Y., and Warren G. (2002) Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. EMBO J. 21, 5645–6552 10.1093/emboj/cdf579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Saffian D., Grimm I., Girzalsky W., and Erdmann R. (2012) ATP-dependent assembly of the heteromeric Pex1p-Pex6p-complex of the peroxisomal matrix protein import machinery. J. Struct. Biol. 179, 126–132 10.1016/j.jsb.2012.06.002 [DOI] [PubMed] [Google Scholar]