Summary
Reproducibility in molecular and cellular studies is fundamental to scientific discovery. To establish the reproducibility of a well-defined long-term neuronal differentiation protocol, we repeated the cellular and molecular comparison of the same two iPSC lines across five distinct laboratories. Despite uncovering acceptable variability within individual laboratories, we detect poor cross-site reproducibility of the differential gene expression signature between these two lines. Factor analysis identifies the laboratory as the largest source of variation along with several variation-inflating confounders such as passaging effects and progenitor storage. Single-cell transcriptomics shows substantial cellular heterogeneity underlying inter-laboratory variability and being responsible for biases in differential gene expression inference. Factor analysis-based normalization of the combined dataset can remove the nuisance technical effects, enabling the execution of robust hypothesis-generating studies. Our study shows that multi-center collaborations can expose systematic biases and identify critical factors to be standardized when publishing novel protocols, contributing to increased cross-site reproducibility.
Keywords: induced pluripotent stem cell, reproducibility, cross-site experimental variation, cortical neurons, gene expression profile, proteomic profiles, single-cell sequencing, molecular profiling, stembancc, public-private partnership
Highlights
-
•
Cross-site reproducibility in iPSC-based molecular experiments is poor
-
•
Factor analysis-based normalization can be used to analyze nuisance variation
-
•
External validation of iPSC experimental molecular data is critical for reproducibility
-
•
Collaborative studies are needed to reveal systematic biases to improve reproducibility
In this article, Lakics and colleagues show that, while individual laboratories are able to identify consistent molecular and seemingly statistically robust differences between iPSC neuronal models, cross-site reproducibility is poor. Their findings support multi-center collaborations to expose systematic biases and identify critical factors to be standardized to improve reproducibility in iPSC-based molecular experiments.
Introduction
Reproducibility is a cornerstone of science. Yet, in recent years, a number of publications highlighted serious issues regarding this fundamental principle of scientific approach, to the extent that the expression “reproducibility crisis” was coined (Munafò et al., 2017, Baker, 2016). The more complex experimental procedures are, and the longer they are applied, the higher the possibility of introducing variability and noise during a research study. This is particularly critical for human induced pluripotent stem cells (iPSCs), which need to be differentiated using lengthy complex procedures in order to be used as novel in vitro models in basic science and drug discovery (Avior et al., 2016), but this increases the variability, such as well-to-well differences in cell density and cellular heterogeneity. Protocols for efficient generation of specific neuronal subtypes mimic human development both in the appearance of successive phenotypes, and also in duration, potentially taking more than 100 days in vitro (Shi et al., 2012a). Reproducibility is especially critical when comparing iPSC-derived cells from multiple donors to discover cellular disease phenotypes and their underlying pathways using unbiased omics experiments. While the reproducibility of transcriptomic (Li et al., 2014) and proteomic (Kim et al., 2007) approaches have been well established for simple cellular systems, no systematic studies have been performed to assess the cross-site reproducibility of these readouts after a long-term iPSC differentiation protocol, such as the derivation of human cortical neurons.
The goal of our study was to identify the extent of variability in an iPSC experiment conducted by multiple groups of the IMI StemBANCC (Innovative Medicines Initiative, Stem cells for biological assays of novel drugs and predictive toxicology) initiative, which aims to generate and interrogate a large collection of stem cell models for disease modeling and therapeutic research. For multi-site comparative studies, large public-private partnerships offer a unique framework due to the participation of both academic and industry organizations with strong scientific background in iPSC biology, representing a “best case scenario” to assess cross-site reproducibility.
Our cross-site analysis utilized a previously published neuronal differentiation protocol (Shi et al., 2012b). In this study we set out to assess the inter- and intra-laboratory reproducibility of transcriptomic and proteomic readouts using two iPSC lines and standard laboratory practices adhered to by all participating laboratories. The differentiation protocol nevertheless enables individual laboratories to apply their laboratory-specific approaches simulating the reproduction of a published method. The key questions in this study were firstly whether a laboratory would be able to separate the two iPSC lines at the molecular level, and secondly whether the identified molecular differences between the two lines were consistent between laboratories. Three academic and two industrial organizations participated in the study to simulate this real-life reproducibility scenario. In addition to bulk omics analyses and single-cell (SC) RNA sequencing (RNA-seq) to assess cellular heterogeneity, the reproducibility of a known cellular phenotype arising from a specific mutation in one of the iPSC lines has also been evaluated.
To our knowledge, this study represents the first comprehensive experiment to assess the intra- and inter-laboratory reproducibility of multiple readouts measured in an iPSC-derived in vitro model system containing differentiated human neurons. Despite acceptable intra-laboratory reproducibility of omics readouts and surprisingly good cross-site reproducibility of a previously identified cellular phenotype, omics datasets from different sites have large variation that masks specific differences, rendering it impossible to distinguish these two lines from each other in a combined dataset. SC analyses demonstrate that cell-type heterogeneity is an important confounder in these comparisons, with variation undermining the detection of differentially expressed (DE) genes, proteins, and pathways. However, we show that there are identifiable sources of variation that investigators can control and thereby increase biological signals in iPSC-based molecular studies. Besides strongly recommending to disclose these identified variation-inflating confounders in published iPSC differentiation protocols, our study also shows that collaborative approaches with larger sample numbers in cross-laboratory studies are valuable to detect and remove unwanted variation (Freytag et al., 2015).
Results
Experimental Design
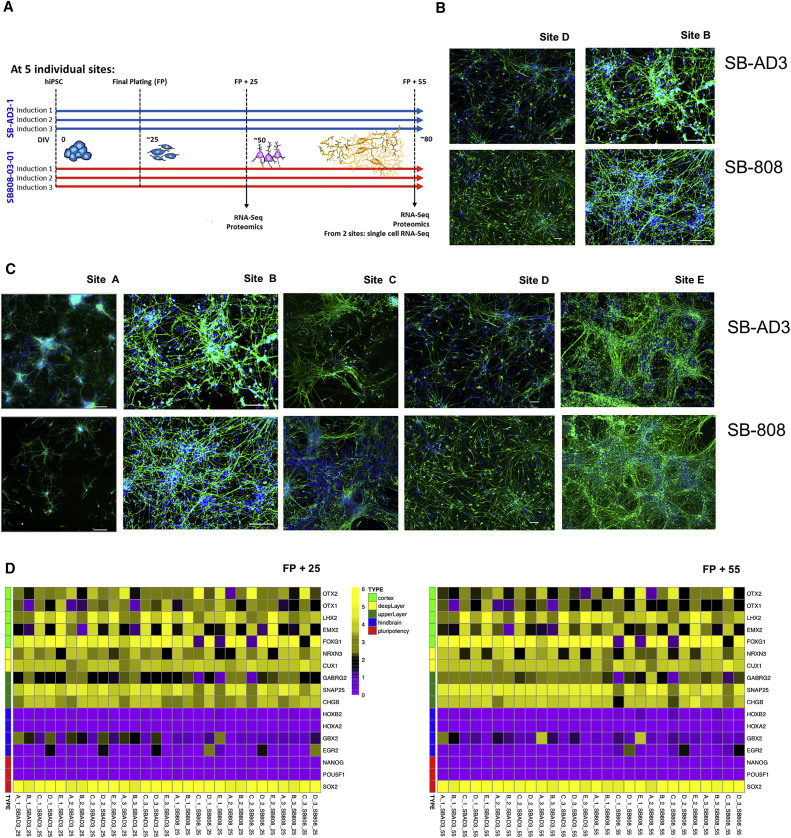
Five laboratories (referred as A, B, C, D, and E) received the same two fibroblast-derived human iPSC lines. One line was derived from a healthy control individual while the second one originated from a patient with familial Alzheimer’s disease carrying a presenilin 1 (PS1) mutation. Note that our study was not designed to examine the effects of this mutation per se but instead focusses on the reproducibility of the comparison of these two lines (see Discussion). All laboratories followed the same standard operating protocol (SOP) (see Supplemental Experimental Procedures) to differentiate the cultures into cortical projection neurons in three independent inductions (replicates) (Figure 1). Laboratory-specific variations and observations were recorded (see below and Table S1). Total RNA and cell lysates were collected at two time points during differentiation, specifically, after 25 and 55 days from the final plating (FP), respectively (representing ∼50 and ∼80 days in vitro differentiation from the iPSC state), and sent to central locations for RNA-seq and proteomic analyses (Figure 1A).
Figure 1.
Experimental Outline of the Study
(A) iPSC lines from two genotypes were differentiated at five different sites with three individual inductions at each site. The given samples were taken at FP (final plating) + 25 days and FP + 55 days.
(B) Representative iPSC-induced cortical neurons at FP + 10 days in culture, immunolabeled with Tuj-1 (green) and DAPI (blue) derived from SBAD3 and AD SB808 cell lines. Neurons grown in two different laboratories are shown (sites D and B). Scale bars, 50 μm (site D), 100 μm (site B).
(C) Cortical neuronal inductions from CTR and PS1 cells 10–20 days after FP, showing presence of neuron-specific βIII-tubulin (green) for sites B, C, D, and E or MAP2 (green) for site A and nuclear marker DAPI (blue). Scale bars, 100 μm (site A); 100 μm (site B); 100 μm (site C); 50 μm (site D); 100 μm (site E).
(D) Heatmaps of gene expression (log10[fragments per kilobase of transcript per million mapped reads]) at the two time points (FP + 25 left and FP + 55 right) of cortical neuron markers, hindbrain markers, and pluripotency markers for 57 StemBANCC samples confirm the presence of expected neuronal markers and the absence of all but SOX2 non-neuronal markers.
Molecular Profiles Show Strong Similarity within Laboratories and Clearly Separate by Cell Line Genotype
To assess the reproducibility of transcriptomic readouts we first examined whether each laboratory was able to demonstrate a clear segregation between the two iPSC lines at a molecular level. Detection of differential molecular profiles between the two lines might be expected due to their differing genetic and epigenetic backgrounds. It is important for molecular studies of iPSC-based models that genotypic differences between lines are identifiable.
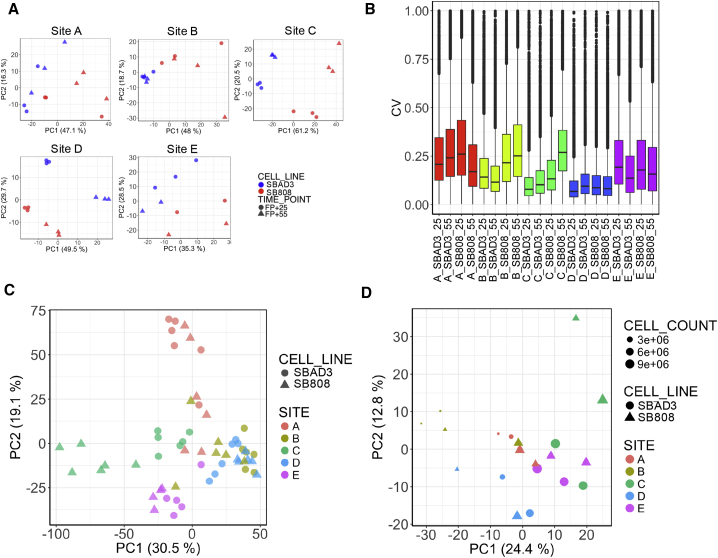
Applying RNA-seq, the expression of variable numbers of protein-coding genes across different samples were detected (with at least one count), with about 70% (13,373) of the 19,086 protein-coding genes expressed across all 57 samples. In further analyses we considered only this set of 13,373 commonly expressed genes. Principal-component analysis (PCA) on the transcriptomic profiles from individual sites illustrated clear separation between the samples from the two cell lines in all five laboratories at both early and late time points (Figure 2A), indicating that genetic background or genotype is a clear source of variation within laboratories. Each laboratory performed three independent cortical differentiations, and the consistency within each laboratory is evident by the greater similarity in gene expression profiles of the three replicates of the same genotype compared with the gene expression profiles between genotypes (Figure 2A). The Euclidean distances calculated between the gene expression profiles of each sample show that, within each laboratory, the expression profiles derived from replicates of the same line are significantly closer to each other than those between replicates of different lines for four out of five laboratories (Figure S1A).
Figure 2.
Detection of Laboratory as One Source of Unwanted Variability
(A) Individual laboratory experiments separate by cell line. PCA plots within laboratory (sites A–E) on 13,373 genes expressed across all samples (normalized gene counts were used) show clear separation between the samples from the two cell lines at both early and late time points.
(B) Degree of variability between replicates within the same laboratory. Boxplots showing the coefficients of variation calculated between gene expression values across replicates within each laboratory, cell line, and time point. Box-and-whisker graphs represent distributions, where the span of the box is the interquartile range (IQR) and includes the median (bold line). The ends of the upper and lower whiskers represent the data point with the maximum distance from the third and first quartiles, respectively, but no further than 1.5 × IQR. Data beyond the end of the whiskers are outliers.
(C) Samples cluster by laboratory in a combined PCA. First two principal components from a PCA on gene expression of 13,373 protein-coding genes that are expressed in all samples clearly cluster samples based on laboratory of origin.
(D) Laboratory and cell count are major confounders in protein-based PCA. First two principal components from a PCA on 1,034 proteins expressed across all samples; color coding according to laboratory; shapes correspond to cell line and sizes to averaged cell count.
See also Figure S1.
The power to identify DE genes is strongly dependent on the experimental variance. A measure of this variance, the coefficient of variation (CV) of the transcriptomic dataset varied between laboratories. While the CV showed no clear genotype or maturation time trends (Figure 2B), the differing CV for each laboratory resulted in a large difference in the number of DE genes between genotypes controlling for time point variation (Figure S1B). Unsurprisingly, the highest number of DE genes was found in laboratory D, which shows the lowest degree of dispersion between replicates.
Cross-Site Comparison of Molecular Profiles Show Poor Reproducibility
Having demonstrated that each laboratory exhibited a clear segregation based on gene expression profiles, we asked whether the molecular differences were consistent between laboratories. Despite the use of a detailed SOP, in a combined dataset containing data from all five partners, we found the laboratory was the dominant source of variation, masking any genotypic effects (Figure 2C). Importantly, only 15 DE genes are found in common between all laboratories indicating a remarkably low degree of cross-laboratory reproducibility (Figure S1B). The low number of overlapping genes may be a consequence of three laboratories (A, B, and E) detecting only a small number of DE genes. Certainly, sites C and D, which had the lowest CV and the highest number of DE genes, showed about ∼50% overlap. At the pathway level, the similarity in enriched gene ontology (GO) terms for these five lists of DE genes is highly variable with semantic similarity comparison values ranging from 0.36 to 0.64 (Figure S1B). In summary, despite extensive efforts to replicate the same experiment, we observe significant variation that would confound any inter-laboratory comparison.
The PCA plot (Figure 2C) and the heatmap of Spearman's correlations (Figure S1C) revealed three potential outliers (SB808 line, laboratory C, here specific issues with detachment of cell monolayer were observed, see Table S1 on metadata). Nevertheless, in general, recorded variation in experimental procedures noted by individual laboratories did not explain the detected cross-laboratory sample variability (Figure S1C). The above observations suggest that much of the inter-laboratory variation arises from additional confounders that increase the within-laboratory variance.
To investigate whether cross-site variability in gene expression was also present at the proteomic level, lysates from replicate wells of the same 57 samples were pooled and analyzed (see Experimental Procedures). Similarly to the transcriptomic samples, the low number of overlapping proteins detected across all samples (only 10% of the 10,483 proteins observed in at least one sample) indicated that the abundance of various proteins is highly variable. For further analyses, we retained only those 1,037 proteins that were observed in all samples. As observed for the transcriptomics data, PCA and heatmap of Spearman's rank correlations of protein abundances did not show clustering of samples by genotype (Figures 2D and S1D). We noticed that, despite normalizing for total protein, the first principal component clearly captures a strong cell-number-related effect in addition to a laboratory-dependent effect. Taken together, the transcriptomic and proteomic profiles demonstrated a strong inter-laboratory variation that masks variation due to the genetic background of each iPSC line.
Factor Analysis Reveals the Transcriptional Axis of Maturation in iPSC-Derived Neurons and Confirms Robust Cortical Neuronal Differentiation
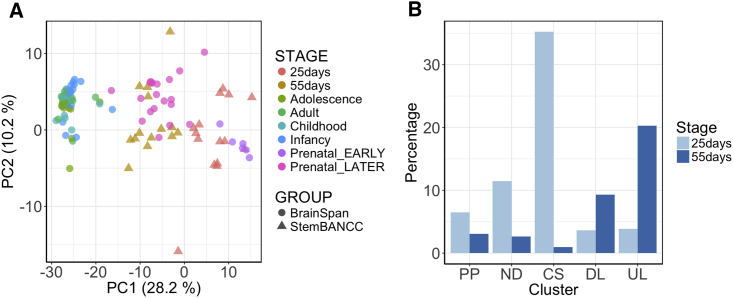
It is evident that cross-site comparisons can be significantly hampered by site-specific confounders, but collaborative studies that generate a large number of samples can have the power to identify nuisance technical effects. We applied a factor analysis-based method called remove unwanted variation (RUV) (Risso et al., 2014). This method can capture nuisance technical effects and RUV in the form of factors, while retaining variation associated with the biological covariate of interest. To demonstrate the utility of factor analysis in revealing biological signals, we first used the approach to determine the transcriptional determinants of in vitro neuronal maturation, exploiting the two time points, FP + 25 and FP + 55, in our samples. Consistent with the reported fetal nature of neurons derived from pluripotent stem cells (Handel et al., 2016), hierarchical clustering of the bulk transcriptomic profiles of 57 samples demonstrated their overall similarity to fetal postmortem brain samples from the BrainSpan Atlas of the Developing Human Brain (Sunkin et al., 2013) (Figure S2A). We performed normalization using RUV on samples from a single line (see Supplemental Experimental Procedures) to expose a clear time point variation that was not masked by any cell line-dependent effect on maturation.
PCA of the RUV-normalized gene expression profiles showed clustering of samples by time points (Figure 3A). BrainSpan samples projected onto the PCA coordinates of normalized iPSC neuron-maturity expression profiles recapitulated the direction of human neuronal maturation (Figure 3A) better than those projected onto PCA coordinates of non-normalized gene expression levels (Figure S2B). Accordingly, the post-RUV expression signature clearly separated the early and late stages of differentiation in BrainSpan fetal samples, and is in line with the observed direction of maturation in our samples. To confirm a neurodevelopmental role for genes whose expression varies in this component space, we selected those genes that maximally contributed in either direction to the identified transcriptional axis of maturation (principal component one) with a gene loading on this axis greater/less than ±0.01. To validate the biological role in neuronal maturation of the contributing genes, we used the CORTECON dataset (van de Leemput et al., 2014), which identified gene clusters representative of changes in temporal gene expression of in vitro cerebral cortex development from human embryonic stem cells. We observed that the genes characteristic to the less mature stage in STEMBANCC samples (with positive scores on PC1) were enriched in CORTECON gene cluster specific to the early developmental stages, namely the “cortical specification” cluster and that are active from days 10 to 20 after differentiation (van de Leemput et al., 2014). The set of genes representing the more mature stage (with negative scores on PC1) was instead significantly enriched in the “upper layer generation” cluster with an expression peak from day 60, as expected (Figure 3B). This analysis also confirmed that the laboratories were successful with their differentiation protocol in producing cells with cortical specification at the early stage and upper layer cortical neurons as expected with this protocol at the later stages.
Figure 3.
RUV-Corrected Gene Expression Reveals Differences of Maturation in Data
(A) Identification of a transcriptional axis of neuron maturation. BrainSpan samples are projected on the principal components calculated on the gene expression data in the present study, after RUV correction. It can be seen that the principal components of gene expression separate both STEMBANCC and BrainSpan samples based on developmental stages.
(B) Genes contributing to the identified transcriptional axis of neuron maturation are consistent with external data. The bar plot shows the percentages of time point-specific genes (selected based on gene loadings from PCA of the samples after RUV correction) falling into each CORTECON gene cluster representative of neuron developmental stages: 25-day-specific genes are enriched in pluripotency (PP), neuron development (ND), and cortical specification (CS) clusters; 55-day-specific genes are enriched in deep layer neuron generation (DL) and upper layer neuron generation (UL) clusters.
See also Figure S2.
Factor Analyses Reveal Genotype-Related Differential Molecular Expression
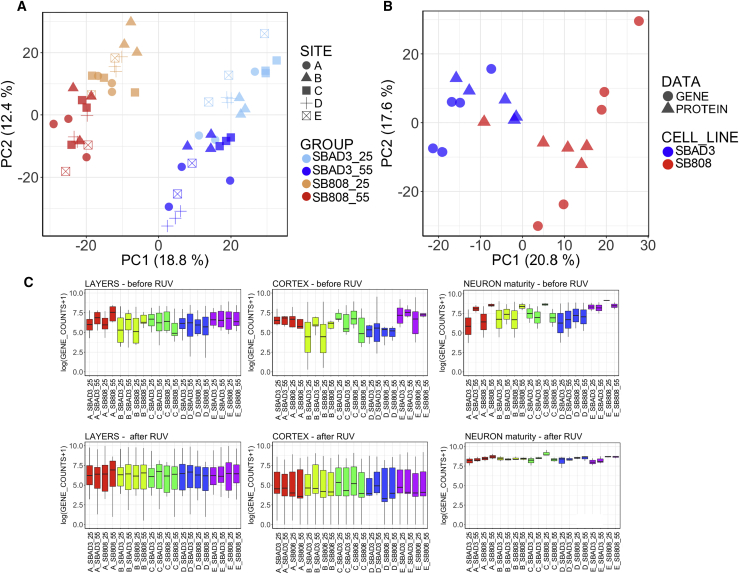
To examine the sources of experimental variation, we applied RUV across all samples, retaining both cell line and time point variations. After RUV correction of gene counts on the first five estimated factors of variation, samples in the combined dataset cluster clearly by cell line and by time point (Figure 4A). Consequently, the number of DE genes detected between the 2 iPSC lines across all samples combined increased (from 1,873 before RUV correction to 3,051 after RUV correction) and between time points (from 2,186 before RUV correction to 3,868 after RUV correction) across all samples (see Table S2 for a complete list of DE genes). Examining a set of neuron-specific stage markers that are expected to be expressed in the differentiating samples (see Supplemental Experimental Procedures), the large distributional differences that were evident between samples in the non-normalized data (Figure 4C, top) were reduced upon removal of the identified variance factors (Figure 4C, bottom).
Figure 4.
Impact of Unwanted Variance Removal by RUV Correction
(A) RUV separates sample gene expression profiles by cell line and time point. First two principal components from a PCA on gene expression over all samples after RUV correction.
(B) RUV separates sample gene expression and protein abundance profiles by cell line. First two principal components from a “second” PCA on both pooled gene and protein expression adjusted for PC1 of “first” PCA. This first PC1 captures the differences between protein and gene expression, therefore adjustment makes the two datasets more comparable. Gene expression values and protein abundances are RUV corrected separately on the two datasets for FP + 55 time point.
(C) RUV normalizes the expression of marker genes expected to be similarly expressed across all samples. Gene expression on log scale of gene markers of three neuron-specific stages before (top row) and after RUV correction (bottom row). Box-and-whisker graphs represent distributions, where the span of the box is the interquartile range (IQR) and includes the median (bold line). The ends of the upper and lower whiskers represent the data point with the maximum distance from the third and first quartiles, respectively, but no further than 1.5 × IQR. Data beyond the end of the whiskers are outliers.
Similar to the transcriptomic analysis, when RUV correction was applied to protein abundances (available for FP + 55 time point only), good separation between the two iPSC lines was observed. After this normalization, a combined PCA shows that both transcriptomic and proteomic samples cluster together by iPSC line, indicative of a correlation between the two data types (Figure 4B). This is further supported by the increase in the number of differential abundant (DA) proteins (0 before RUV correction, 205 after RUV correction) and consequently in the percentage of overlapping DE genes and DA proteins between laboratories after RUV correction (0% before RUV correction, 14% after RUV correction).
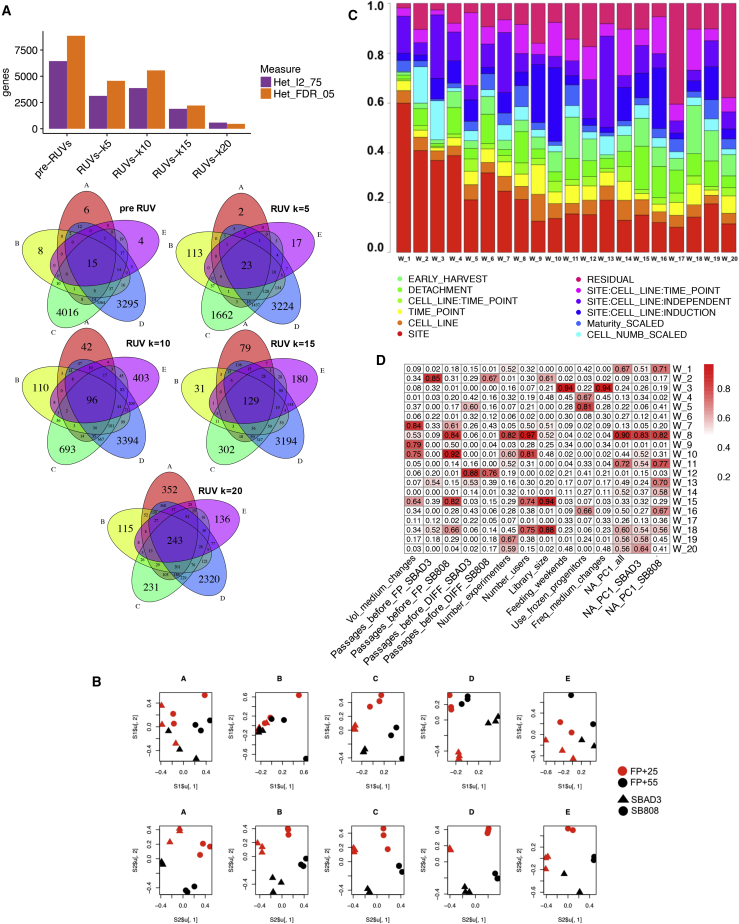
To study further the effect of RUV on the reproducibility across laboratories, we measured the extent of homogeneity between laboratories in evaluating the same biological effect (see Supplemental Experimental Procedures). The number of genes showing high heterogeneity across all samples (I2 ≥ 75%) decreased as a function of the number of RUV factors that are regressed out from the data (from 6,443 genes before RUV correction to 584 genes after RUV correction on 20 factors, Figure 5A, top). In addition, an increase in overlapping DE genes between cell lines across laboratories was also observed (from 15 before RUV to 243 after RUV correction on 20 factors, Figure 5A, middle; Table S3). The post-RUV PCA plots for each laboratory clearly reveal that the segregation by both time point and genotype is more evident than pre-RUV (Figure 5B). The I2 measure and variance analysis at the gene expression level before RUV and after RUV (Figures 5A, top and S3) confirm that the laboratory-dependent source of variation was properly removed from the data to expose the variation of interest. Thus, given sufficient power, technical variability, including hidden laboratory-dependent variation, can be corrected and enables detection of the biological signal.
Figure 5.
Analyses of Factors Explaining the Unwanted Variance and Laboratory Heterogeneity
(A) Increased reproducibility of gene expression difference between lines across laboratories after RUV correction. Number of genes showing high heterogeneity across sites before and after RUV based on 5% false discovery rate (FDR) threshold (Het_FDR_05) and on 75% I2 threshold (Het_I2_75) (Top). Overlap of DE genes between cell lines across sites before and after removal of 5, 10, 15, and 20 RUV factors (Venn diagrams, bottom).
(B) Separation between the lines in singular value decomposition plots helps explain the different number of DE genes between the two covariates of interest before and after RUV.
(C) “Laboratory” is a major confounder corrected by RUV. Each bar summarizes the proportions of variance captured by RUV factors (W_1 to W_20) and explained by known potential confounders.
(D) Laboratory variance is correlated to several experimental variations. The matrix shows the linear correlations between means of laboratory-specific RUV factors and known laboratory-specific potential confounders plus neuron-astrocyte axis scores (NA_PC1) described in Supplemental Experimental Procedures section and in Figure S4B.
See also Figures S3 and S4.
Identification of Experimental Variables Inflating Gene Expression Variance
We next examined the known and investigator-recorded covariates that correlated with the RUV factors. As expected, the variable “SITE” explained 60%, 40%, 38%, and 39% of the variance in factors 1, 2, 3, and 4, respectively (Figure 5C). The second most contributing source of variation captured by RUV was attributable to starting the experiment on different days for technical replicates (15%, 22%, 20%, and 38% of the variance in factor 1, 3, 7, and 13, respectively; Figure 5C). In general, the proportions of variance in RUV factors that could be explained by the remaining candidate confounders were moderate to low. Among these, 18% and 15% of variance in factors 2 and 3, respectively, were explained by differences in cell counts.
As SITE was the strongest cause of variability, we attempted to correlate SITE-specific variation in RUV factors to particular experimental effects by fitting linear models regressing site-specific RUV means outputted from the variance component analysis on each site-specific metadata variable in turn. Several covariates, namely iPSC passage number before differentiation and the number of passages before FP, media volume changes, feeding at weekends, and use of frozen neural progenitor cells, were highly correlated with several factors (Figure 5D).
We also examined the variation underlying expression at the gene-specific level by fitting a regression model (MCMCglmm) (Hadfield, 2010) between gene counts and the known covariates. The analysis enabled the identification of genes that may underlie the covariate. The top 100 genes related to “DETACHMENT” were enriched in GO terms related to regulation of cell cycle, apoptosis, and DNA metabolism, while those similarly associated with “CELL_COUNT” are enriched in cellular respiration and lipid metabolism pathways, and those genes related to “TIME_POINT” were involved mainly in neuron differentiation processes (complete lists of GO terms in Table S4).
Cell Type Heterogeneity Is a Major Source of Inter-laboratory Variation
Cellular heterogeneity can be a major confounder in tissue and cell culture comparison (Sandor et al., 2017), and could represent an important source of inflated variance within and between laboratories. To investigate variation in the cellular composition of our iPSC-derived cell populations, we generated the individual transcriptional profiles of 1,440 fluorescence-activated cell sorting (FACS)-sorted iPSC-derived cortical neurons produced by two of five participant laboratories (sites D and E for each of the two cells lines at the FP + 55 time point; see Experimental Procedures). After discarding low-quality cell libraries (see Supplemental Experimental Procedures), 771 SC transcriptomes were available for subsequent analysis.
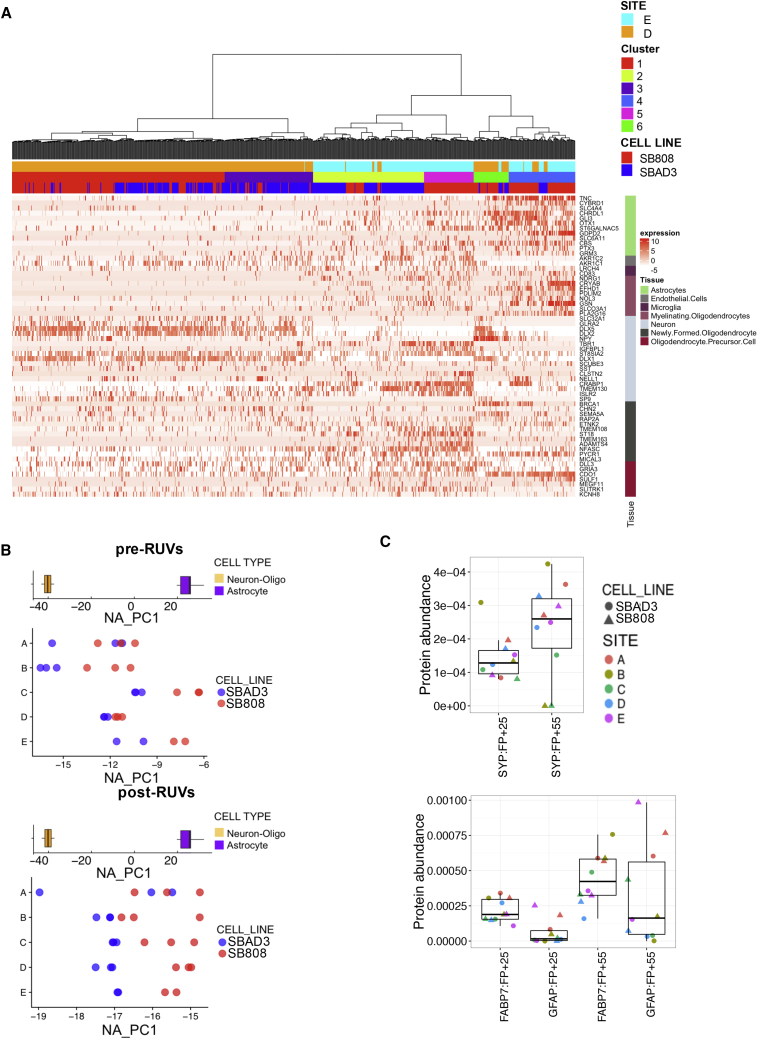
Using unsupervised hierarchical clustering on the expression profiles we identified four and five subpopulations of cells within the SB808 and SBAD3 cell populations, respectively (see Supplemental Experimental Procedures, Figures 6A and S5A). The cortical differentiation protocol we used has been extensively validated and efficiently produces high yields of cortical excitatory neurons as well as astrocytes. We assessed the presence of neuron-, glial-, and other cell-type-specific markers within each subpopulation (see Supplemental Experimental Procedures). For each line, we found that the largest cell subpopulation was uniquely and significantly enriched in neuron-specific markers (Figures 6A and S5A). The second largest subpopulation was also enriched for a distinct set of neuron-specific markers (cluster 2 for SB808 and SBAD3) (Figures 6A and S5A). Other subpopulations were enriched in astrocyte markers (e.g., clusters 3 and 4 for SB808 and cluster 4 for SBAD3) (Figure S5).
Figure 6.
Distinct Cellular Populations Are Identified within the iPSC-Derived Neuronal Populations That Potentially Drive Differences in the Bulk Transcriptomic Comparisons
(A) Cellular heterogeneity in individual cells across cell lines and laboratories. The heatmap of single-cell transcription data reveals six distinct cellular populations in SB808 and SBAD3 lines in two laboratories according to their expression of a set of cell identity marker genes (see Supplemental Experimental Procedures).
(B) A neuron-astrocyte axis of gene expression variation illustrates cell type is a major contributor to cell line variation across all laboratories. The projection of before (top) and after (bottom) RUV correction bulk transcriptomic expression patterns (FP + 55) onto neuronal-glia gene expression identity axis (top) shows that glia-neuronal identity contributes to the expression variation (bottom).
(C) Increase of neuron- and astrocyte-specific protein abundances at later time points in all laboratories. Protein abundance of Human Protein Atlas neuron-specific (top) and glia-specific (bottom) proteins. Protein abundances of a neuron marker at different time points (top). A significant increase of SYP is observed from FP + 25 to FP + 55 time point (p < 0.05). Protein abundances of two astrocyte markers at different time points (bottom). A significant increase of FABP7 and GFAP is observed from FP + 25 to FP + 55 time points (p < 0.05). GFAP specifically shows an increase in all SB808 lines compared with SBAD3 lines within each laboratory. Box-and-whisker graphs represent distributions, where the span of the box is the interquartile range (IQR) and includes the median (bold line). The ends of the upper and lower whiskers represent the data point with the maximum distance from the third and first quartiles, respectively, but no further than 1.5 × IQR. Data beyond the end of the whiskers are outliers.
See also Figure S5.
While Shi et al. (2012b), who described the protocol, observed astrocytes forming after day 45, we found here (1) that glial cells represented a large proportion (20.8%), (2) that the fraction of glial cells varied from site to site (15% site D versus 21% site E), and (3) that the fraction of glial cells was higher in the SB808 line than in the SBAD3 line (29% SB808 versus 12% SBAD3).
Cellular Subpopulations Can Show Opposing Differential Gene Expression and Introduce Considerable Bias in Comparative Studies
The SC analysis revealed cell culture subpopulations of differing proportions between two sites (Figure S5A) that could affect the differential gene expression analysis. Interestingly we found that subpopulations 2, 3, and 4 also expressed a small number of genes representing oligodendrocyte or microglia markers (Figure 6A). The expression of genetic markers of other cell types not intended to be induced by our differentiation may represent either a genuine developmental feature of these cells or an artifact of in vitro differentiation, where the epigenetic silencing of other cell type-specific genes is not fully effective––in either case the heterogeneity could significantly bias differential gene expression between lines and between sites.
After discounting technical artifacts (e.g., plate effects), we found that the DE genes and pathways varied significantly between iPSC-derived subpopulations. Most notably we observed that gene expression differences between the two lines were negatively correlated between subpopulations 1 and 6, and thus directly obscure the detection of DE genes (Figure S5D). To evaluate how the results of DE analyses can be biased by the observed cell heterogeneity, we randomly sampled populations of 100 cells from each line and observed the stability of DE genes and pathways. Of 192 originally observed DE genes, only 10 genes (5%) that were associated with a very low p value (<10−20) were consistently reported with a false-negative rate <5% (Figure S5B). This clearly demonstrated that cell heterogeneity can yield a major bias in the comparison of gene expression profiles between iPSC-derived cells and that only the most significant DE genes are detectable through the heterogeneity. Indeed, upregulated and downregulated DE genes for each laboratory in the bulk transcriptomic study were significantly overrepresented among specific subpopulations of the SC study (upregulated in subpopulation numbers 1, 2, and 3, and downregulated in subpopulation numbers 1, 4, and 6, respectively, hypergeometric test; Tables S2 and S3; Figure S5C).
Cellular Composition Varies Both by Laboratory and Cell Line upon Differentiation
Given the results obtained at the SC level, we asked whether the observed heterogeneity in cell identity (neuronal and non-neuronal populations) could explain the variance in the bulk transcriptomic data due to the test site. For this, we used available RNA-seq data from purified human brain cell types to identify gene expression variation associated with cell type and extended the list of marker genes employed in the SC analyses (see Experimental Procedures). Examining the variation between genotypes across all sites, we found that the SBAD3 upregulated genes were enriched in neuronal markers (p = 2.6 × 10−18, after RUV correction), while the SB808 upregulated genes were enriched in non-neuronal markers (p = 3.2 × 10−11 for astrocytes after RUV correction). Furthermore, a clear separation by genotype on principal components reflecting the above-mentioned similarity of SBAD3 lines to neurons and of SB808 lines to non-neuronal cells was evident when the samples were projected on the PCA coordinates of the human brain cell types both before and after RUV correction (Figure 6B). However, before RUV correction, there is significant systematic variation evident in the neuronal/non-neuronal composition in the lines cultured by different laboratories following the same protocol. Indeed, the neuronal/non-neuronal composition of the lines cultured by each laboratory is well-correlated with the “laboratory” contribution to RUV factors 1, 8, and several other factors (Figure 5D). Thus, the predisposition of each line toward generating cell populations with distinct proportions of neurons and non-neuronal cells is an important driver of gene expression differences between the two cell lines at the bulk transcriptome level. This is also reflected at the protein level, whereby glial marker proteins (FABP7 and GFAP) showed increased abundance at the FP + 55 time point and that GFAP is more abundant in SB808 samples than in SBAD3 samples for all laboratories (Figure 6C). Interestingly, variation in neuronal/non-neuronal composition of the two lines did not increase from the early to the later time point when we explained RUV factors through known covariates (Figure 5C) and compared Euclidean distances between time points (Figure S1A).
Discussion
In this study we examined the reproducibility of a long-term neuronal differentiation protocol undertaken in five laboratories. Further, we also intended to identify “hidden” factors important to a robust method and quantify the extent to which they contribute to experimental variation in molecular data. We therefore focused on repeated differentiations of two lines with different genetic backgrounds to assess whether it was possible to consistently distinguish two iPSC lines after neuronal differentiation, using molecular readouts. In our multi-center experiment, we deliberately chose not to investigate multiple disease and control donor lines in order to focus on reproducibility rather than identifying novel disease phenotypes. With a detailed and shared protocol applied across all partner laboratories, we controlled variability in the differentiation process to the extent it is usually disclosed in published protocols, while expecting variation between the cell lines due to their differing genotypes (the sum of all other potential genetic/epigenetic differences between the two lines), as well as originating from specific laboratory practices not harmonized across test sites.
Our approach found (1) that genotype-driven gene expression variation is detectable by a laboratory in particular when within-laboratory consistency is high; (2) that genotypic effects are masked in aggregated molecular data from multiple laboratories due to site-specific confounders; (3) that cell-type compositional heterogeneity varied both by laboratory and by cell line, and contributed significantly to the masking of genotypic effects in multi-site studies; (4) that prolonged cell culture after FP did not significantly increase inter-laboratory variance and that much of the cell-type compositional heterogeneity was likely determined during neural patterning and shortly after FP; and (5) that normalization methods were able to remove nuisance site-specific effects to reveal biological signals including genotypic effects. Our study therefore underscores the importance of recognizing, recording and reporting experimental variables, and, where possible, using appropriate statistical methods to remove unwanted variability, in order to generate more reproducible molecular studies based on differential gene and protein expression phenotypes.
The application of in vitro human disease models using iPSC lines is a potentially transformative approach for understanding disease mechanisms, novel target discovery, and personalized medicine. Unsurprisingly, the majority of efforts have been on monogenic forms of disease where there are strong genotype-phenotype relationships due to large effect sizes of the gene mutation. The expectation for such disorders is that, at a cellular level, highly penetrant mutations would cause easily detectable in vitro molecular and cellular phenotypes. Although not the focus of our study, when we examined the biochemical phenotype in line SB808, which carried a familial Alzheimer’s disease mutation in the PS1 gene, we did indeed detect a highly robust change in specific β-amyloid peptide ratios (Szaruga et al., 2015) when compared with the iPSC line without the PS1 mutation (Figure S6). This strong cross-site reproducibility has likely been observed because (1) the altered production of β-amyloid peptides is proximal to the PS1 mutation, and (2) is due to the robustness of the ratio-based readout.
The molecular analysis by contrast showed very little overlap between sites despite the detailed, shared protocol, and attempts to minimize technical variability, including processing and analyzing omics samples at the same laboratories, enabling us to focus on the differentiation process-related confounders. Within a laboratory, there was much less variance, and gene expression profiles of the two lines clearly segregated even before RUV analysis. Two sites, C and D, showing low levels of dispersion between replicates, produced a large number of genes that were significantly differentially expressed. When considering all 5 sites, only 15 genes were consistently different between the two cell lines prior to normalization, compared with over 200 genes mutually detected after factor analysis-based removal of the unwanted variation. The pre-RUV overlap of the DE genes of sites C and D was quite high, but much of this overlap may have been artifactual, since for site C the number of DE genes fell from 7,524 to 3,354 after removing 5 factors, and to 1,480 genes after removing 20 factors. This suggests that a laboratory could generate “private” gene expression lists with high confidence based upon highly significant p values as in our study, but, unless sources of variance are explored, it is difficult to know whether such DE lists are biologically relevant. This is important because molecular studies by individual laboratories are often used to generate hypotheses for further investigations, and therefore our study raises significant concerns that many of the detected DE genes can be an artifactual.
Addressing this concern, our work found that, despite the numerous sources of potential confounders, it is possible to detect consistently replicated signals if there is a sufficient number of samples to power an appropriate statistical approach and due consideration is given to complexity of iPSC-differentiated cell cultures. The presence of multiple cellular subpopulations differing between two labs was confirmed by the SC transcriptome study. Strikingly, we observed that differential gene expression patterns between the two cell lines in one cellular subpopulations can have the opposite pattern in another subpopulation. We found that, in a simulated heterogeneous bulk transcriptome based on our SC data, only the most significant and strongest gene expression (p < 10−20) differences between the two cell lines were detectable. The differing propensity in cellular fates of the two iPSC lines produced by our standardized differentiation protocols was evident in the bulk transcriptome data both before and after removing unwanted variation, demonstrating systematic variation in culture cellular composition associated with both genotype and laboratory. More rigorous quality control of cellular composition upon differentiation at a series of intermediary time points may help improve the consistency of results between laboratories. However, we also found that different subpopulations within a culture can be characterized by aberrant expression of cell identity markers from cell types that are not present in the culture such as microglia or oligodendrocytes. Immunohistochemistry or functional studies such as calcium imaging or electrophysiology may not reveal these cell subpopulations. These cells may therefore represent a potentially important cause of variance that will be hidden to quality control measures unless these include SC profiling. Altogether our study shows that cellular heterogeneity can introduce significant bias in differential gene expression experiments and likely represents the major contributor to inflating within-laboratory variance and to inter-laboratory variability.
One of the important aspects of our study was to identify those factors which are correlated to the RUV factors, potentially explaining the increased cross-site variability. Not surprisingly, our computational analysis indicated that SITE (i.e., laboratory) is the most influential source of variation (explaining between 40% and 60% of the variance in the first 4 RUV factors), followed by the practice of starting the differentiation of progenitors on different days as opposed to plating on the same day. To further identify the sources of inter-laboratory variability, we correlated site-specific variation in RUV factors to experimental practices known to be different for the various test sites. This analysis allowed us to pinpoint experimental variables which were highly correlated to several RUV factors, and potentially hampered cross-site reproducibility. Among these were a number of factors, some of which are often not disclosed in published differentiation protocols, such as iPSC passage number before differentiation, the number of passages before FP, media volume changes, feeding at weekends, and use of frozen neural progenitor cells. Many of these factors likely alter the epigenetic and cellular programs that determine progenitor cell fate choices, including neuronal-glial balance to thereby contribute to the heterogeneity and variance. Based on our study we strongly suggest that these should be a standard part of every published differentiation protocol to increase the chance of robust reproducibility of iPSC-based studies.
Reproducibility in biomedical science is a major cause of concern and has impacted the pharmaceutical industry, where study reproducibility is a pre-requisite for target discovery, assay development, and a successful drug discovery program. The potentially underlying causes for the lack of reproducibility have been extensively scrutinized and attributed to factors such as poor study design and inappropriate statistical methods, as well as the culture of grant funding and publication biases. Moreover, as our paper illustrates, an individual laboratory conducting hypotheses generating molecular studies, without external reference, or further validation studies cannot know whether their significant differential gene findings are due to a systematic bias in their laboratory or arising from the biological condition under study. It is therefore critical that any potential hypotheses are validated including through the use of literature evidence and increasingly available complementary datasets such as human brain tissue and animal model studies. Collaborative approaches, especially large public-private partnerships involving multiple test centers, if carefully designed, offer a powerful solution to performing studies which yield reproducible mechanistic insights. These multi-center experiments also reveal important learnings for the individual laboratories by identifying experimental practices to be disclosed when publishing iPSC differentiation protocols to increase their reproducibility.
In our paper, we have shown that, while cellular heterogeneity of the iPSC cultures differentiated at various laboratories arising from site-specific practices as well as other cryptic factors can mask almost all biological effects, these confounders can be identified and overcome. The computational biology approaches employed here revealed and removed the site-specific biases, enabled access to the underlying biology, and identified publication best practices.
Experimental Procedures
See further details in the Supplemental Experimental Procedures.
Generation and Maintenance of STEMBANCC iPSC Lines
The human iPSC lines SBAD3-1 and SB808-03-04 (the latter carried the Alzheimer's disease-related PS1 intron 4 mutation) were derived from human skin biopsy fibroblasts following signed informed consent, with approval from the UK NHS Research Ethics Committee (REC: 13/SC/0179) and were derived as part of the IMI-EU sponsored StemBANCC consortium. iPSC generation was performed using the CytoTune-iPS 2.0 Sendai Reprogramming Kit (A16517) from Thermo Fisher Scientific (Waltham, MA).
Bulk Transcriptomic Experiment
For transcriptomic analyses, 12 samples were generated in each laboratory: 3 replicates of the SBAD3 cell line and 3 replicates of the SB808 cell line at each of the 2 time points (Figure 1A). Two samples were excluded from analysis due to problems during the differentiation process and another one because of a contamination issue during RNA-seq, leading to a total of 57 samples being available for transcriptomic analysis.
SC RNA-Seq Experiment
SC Isolation
SC suspensions were generated using Accutase dissociation followed by SC filtration of iPSC-derived cortical neurons. The success of the suspension was manually confirmed on a hemocytometer. The SC suspension was sorted into a 96-well PCR plate containing a lysis mix. Sorting gates were set to include only live (DAPI-negative) single cells. Stream alignment and sort efficiency was checked using Accudrop beads (Becton Dickinson).
SC RNA-Seq Library Preparation and Sequencing
Single iPSC-derived cortical neurons were isolated by FACS onto 96-well plates in 2 μL lysis buffer (Trombetta et al., 2014). Each plate included 4 bulks, each obtained by extracting total RNA from 4,000 cells using RNeasy Micro Kit (QIAGEN) and using 1/14th of the extracted RNA solution. Libraries were prepared following the Smart-Seq2 protocol described by Picelli et al. (2013) and Trombetta et al. (2014). Each sample was spiked with the equivalent of 1 μL of 1:10,000,000 dilution of the ERCC RNA Spike-In Mix 1 (Thermo Fisher Scientific). Libraries were pooled in 288- or 384-plexes and each pool sequenced on 1 lane of HiSeq 4000 at 75 bp paired end.
Proteomics
Proteomic Sample Processing, Measurement, and Data Analysis
Cells from three wells were detached in ice-cold PBS, pooled, and snap frozen for proteomics analysis. Prior to digestion, cell pellets were dissolved in lysis buffer, and obtained lysates were pooled from triplicate wells, replicated to increase the number of detectable proteins leading to a total of 20 samples for subsequent analysis, and spun at 1,000 × g. Samples were subject to in-solution proteolytic tryptic digestion and analyzed using 2D-LC-MS. Proteins were identified using Waters ProteinLynx Global server v.3.0.1 and Progenesis Bioinformatic software (non-linear dynamics) as described previously (Heywood et al., 2015).
Author Contributions
V.V. led on the data analysis and data integration and interpretation. C.W. led the overall informatics team. J.S. and V.L. developed the study SOP. A.H., S.E.N., E.W., S.C., A.V., C.C., K.C., C.P., P.B., J.S., W.H., R.D.F., K.J., C.M., A.A., and F.W. performed the experimental work. M.L. and L.A. generated the iPSC lines. A.H., S.H., and S.E.N. performed the SC experimental work. A.B. performed the mass spectrometry proteomics. E.S. and J.N. performed the RNA sequencing. M.A., E.S., and J.N. performed the SC RNA sequencing. C.S. performed the SC analyses. J.S.R. contributed to the omics analysis. G.N. contributed to the statistical analyses and modeling. P.G., P.R., J.A., I.R., P.J.P., G.C.T., M.G., F.J.L., C.J.A., K.M., R.B., C.W., M.Z.C., and V.L. supervised the research teams, contributed to the experimental design and data interpretation. All authors were involved in manuscript writing and editing. C.W., M.Z.C., and V.L. developed the study concept and experimental design and coordinated the analysis.
Acknowledgments
The research leading to these results has received support from the Innovative Medicines Initiative Joint Undertaking under grant agreement no. 115439, resources of which are composed of financial contribution from the European Union's Seventh Framework Program (FP7/2007-2013) and EFPIA companies' in kind contribution. A.H., S.C., and M.Z.C. were also funded by the NIHR (Oxford BRC). K.M. and A.B. were also supported by the NIHR GOSH BRC. The views expressed are those of the author(s) and not necessarily of those of the NHS, the NIHR or the Department of Health. We also thank Dr. Quin Wills and Dr. Davis McCarthy for their advice regarding the processing of our single transcriptomic dataset and David Lakics for creating the illustrations in Figure 1A. Several of the authors (R.D.F., G.C.T., P.R., J.R., K.J., V.L., A.V., C.C., K.C., C.P., I.R., P.J.P., and M.G.) are employed by the pharmaceutical industry, and therefore, as they are affiliated with a commercial entity, declare competing financial interests. J.S. is a shareholder in Talisman Therapeutics. F.J.L. is a shareholder in Talisman Therapeutics and Gen2 Neuroscience. Honoraria or research funding has been received for M.Z.C. from Orion, Daiichi Sankyo, TEVA, and Novartis; P.G. from BioMarin, Actelion, Dipharma, and SOBI; K.M. from Genzyme, Actelion, and BioMarin.
Published: September 20, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, and six tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2018.08.013.
Contributor Information
Caleb Webber, Email: caleb.webber@dpag.ox.ac.uk.
M. Zameel Cader, Email: zameel.cader@ndcn.ox.ac.uk.
Viktor Lakics, Email: viktor.lakics@abbvie.com.
Accession Numbers
The accession number for the transcriptomic data reported in this paper is GEO: GSE118735.
Supplemental Information
References
- Avior Y., Sagi I., Benvenisty N. Pluripotent stem cells in disease modelling and drug discovery. Nat. Rev. Mol. Cell Biol. 2016;17:170–182. doi: 10.1038/nrm.2015.27. [DOI] [PubMed] [Google Scholar]
- Baker M. Is there a reproducibility crisis? A nature survey lifts the lid on how researchers view the 'crisis’ rocking science and what they think will help. Nature. 2016;533:452. [Google Scholar]
- Freytag S., Gagnon-Bartsch J., Speed T.P., Bahlo M. Systematic noise degrades gene co-expression signals but can be corrected. BMC Bioinformatics. 2015;16:309. doi: 10.1186/s12859-015-0745-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield J.D. MCMC methods for multi-response generalized linear mixed models: the MCMCglmm R package. J. Stat. Softw. 2010;33 [Google Scholar]
- Handel A.E., Chintawar S., Lalic T., Whiteley E., Vowles J., Giustacchini A., Argoud K., Sopp P., Nakanishi M., Bowden R. Assessing similarity to primary tissue and cortical layer identity in induced pluripotent stem cell-derived cortical neurons through single-cell transcriptomics. Hum. Mol. Genet. 2016;25:989–1000. doi: 10.1093/hmg/ddv637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heywood W.E., Galimberti D., Bliss E., Sirka E., Paterson R.W., Magdalinou N.K., Carecchio M., Reid E., Heslegrave A., Fenoglio C. Identification of novel CSF biomarkers for neurodegeneration and their validation by a high-throughput multiplexed targeted proteomic assay. Mol. Neurodegener. 2015;10:64. doi: 10.1186/s13024-015-0059-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.J., Zhan P., Feild B., Ruben S.M., He T. Reproducibility assessment of relative quantitation strategies for LC-MS based proteomics. Anal. Chem. 2007;79:5651–5658. doi: 10.1021/ac070200u. [DOI] [PubMed] [Google Scholar]
- Li S., Łabaj P.P., Zumbo P., Sykacek P., Shi W., Shi L., Phan J., Wu P.-Y., Wang M., Wang C. Detecting and correcting systematic variation in large-scale RNA sequencing data. Nat. Biotechnol. 2014;32:888–895. doi: 10.1038/nbt.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munafò R., Nosek B., Bishop D., Button K., Chambers C., Percie du Sert N., Simonsohn U., Wagenmakers E., Ware J., Ioannidis J. A manifesto for reproducible science. Nat. Hum. Behav. 2017;1:0021. doi: 10.1038/s41562-016-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S., Björklund Å.K., Faridani O.R., Sagasser S., Winberg G., Sandberg R. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods. 2013;10:1096–1098. doi: 10.1038/nmeth.2639. [DOI] [PubMed] [Google Scholar]
- Risso D., Ngai J., Speed T.P., Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 2014;32:896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandor C., Robertson P., Lang C., Heger A., Booth H., Vowles J., Witty L., Bowden R., Hu M., Cowley S.A. Transcriptomic profiling of purified patient-derived dopamine neurons identifies convergent perturbations and therapeutics for Parkinson’s disease. Hum. Mol. Genet. 2017;26:552–556. doi: 10.1093/hmg/ddw412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Kirwan P., Smith J., Robinson H.P.C., Livesey F.J. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nat. Neurosci. 2012;15:477–486. doi: 10.1038/nn.3041. S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Kirwan P., Livesey F.J. Directed differentiation of human pluripotent stem cells to cerebral cortex neurons and neural networks. Nat. Protoc. 2012;7:1836–1846. doi: 10.1038/nprot.2012.116. [DOI] [PubMed] [Google Scholar]
- Sunkin S.M., Ng L., Lau C., Dolbeare T., Gilbert T.L., Thompson C.L., Hawrylycz M., Dang C. Allen Brain Atlas: an integrated spatio-temporal portal for exploring the central nervous system. Nucleic Acids Res. 2013;41:D996–D1008. doi: 10.1093/nar/gks1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szaruga M., Veugelen S., Benurwar M., Lismont S., Sepulveda-Falla D., Lleo A., Ryan N.S., Lashley T., Fox N.C., Murayama S. Qualitative changes in human γ-secretase underlie familial Alzheimer’s disease. J. Exp. Med. 2015;212:2003–2013. doi: 10.1084/jem.20150892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta J.J., Gennert D., Lu D., Satija R., Shalek A.K., Regev A. Preparation of single-cell RNA-seq libraries for next generation sequencing. Curr. Protoc. Mol. Biol. 2014;107:4.22.1–4.22.17. doi: 10.1002/0471142727.mb0422s107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Leemput J., Boles N.C., Kiehl T.R., Corneo B., Lederman P., Menon V., Lee C., Martinez R.A., Levi B.P., Thompson C.L. CORTECON: a temporal transcriptome analysis of in vitro human cerebral cortex development from human embryonic stem cells. Neuron. 2014;83:51–68. doi: 10.1016/j.neuron.2014.05.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.