Abstract
Tumour-associated p53 missense mutants act as driver oncogenes impacting cancer progression, metastatic potential and drug resistance (Gain-of-Function, GOF)1. Mutant p53 protein stabilization is a prerequisite for GOF manifestation, however it does not represent an intrinsic property of p53 mutants, but rather requires secondary events2. Moreover, mutant p53 protein levels are often heterogeneous even within the same tumour, raising questions on the mechanisms that control local mutant p53 accumulation in some tumour cells but not in their neighbours 2,3. By investigating the cellular pathways that induce protection of mutant p53 from ubiquitin-mediated proteolysis, we found that HDAC6/Hsp90-dependent mutant p53 accumulation is sustained by RhoA geranylgeranylation downstream of the mevalonate (MVA) pathway, as well as by RhoA- and actin-dependent transduction of mechanical inputs, such as the stiffness of the extracellular environment. Our results provide evidence for an unpredicted layer of mutant p53 regulation that relies on metabolic and mechanical cues.
Mutation of the TP53 gene is the most frequent genetic lesion in human cancers1. Numerous studies have clearly established that missense p53 mutants are dependent on a transformed context for full activation of their malignant potential4. This includes oncogenic signalling in response of which mutant p53 proteins are post-translationally modified5, and tumour-specific mechanisms of protein hyper-stabilization6 that oppose the inherent instability of p53 mutants observed in normal tissues2. Remarkably, mutant p53 accumulation has been reported to be often spatially heterogeneous in individual tumours3,7, suggesting that its protein stability may be influenced by local environmental cues.
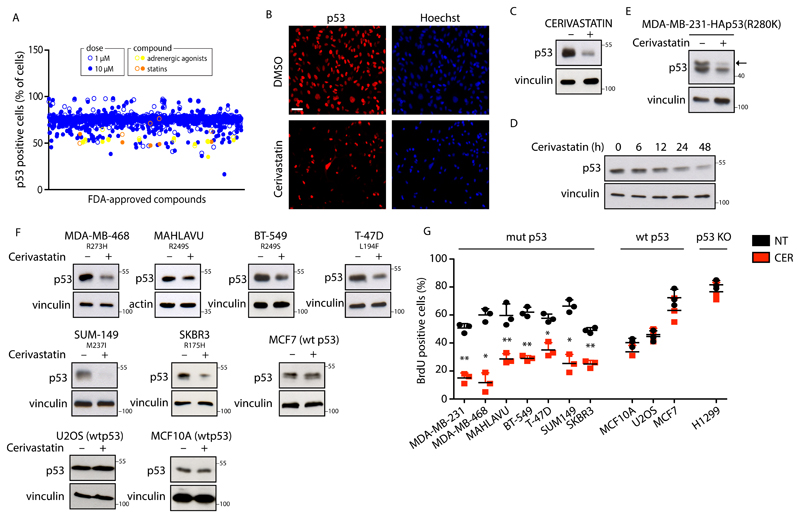
Aiming to identify cellular processes and biochemical pathways responsible for mutant p53 stabilization in cancer cells, we performed a high-content screening of FDA-approved drugs (Supplementary Figure 1A)8,9 in MDA-MB-231 breast cancer cells, in which we monitored the variations of mutant p53 levels after drug administration (Supplementary Table 1). Upon filtering results by reproducibility, toxicity, dose-dependence and manual inspection of images, the best hits associated with mutant p53 decrease were the cardiac glycoside Ouabain, the antipsychotics Spiperone and Thioridazine, the antiparasitic agent Ivermectin, as well as two entire classes of drugs, namely adrenergic agonists (e.g. Salmeterol) and mevalonate pathway inhibitors (statins) (Figure 1A-C, Supplementary Figure 1B, 1C, 5B and Supplementary Table 2). None of the compounds identified by our screening were able to reduce p53 mRNA levels, thus suggesting a post-transcriptional mechanism of action (Supplementary Figure 2A).
Figure 1. Statins reduce missense mutant p53 protein levels in cancer cells.
(A) Results of high-content screening. Mutant p53 positive cells were detected by immunofluorescence and quantified through automated image analysis. (B) Representative images of p53 immunofluorescence from the screening. MDA-MB-231 cells were treated with dimethylsulphoxide (DMSO) or with Cerivastatin 10 μM for 24h. Scale bar 15 μm. (C) MDA-MB-231 cells were treated with dimethylsulphoxide (DMSO) or with Cerivastatin (+) 10 μM for 48h. Representative western blots are shown. (D) MDA-MB-231 cells treated with Cerivastatin 10 μM for the indicated time points. Representative western blots are shown. (E) MDA-MB-231 cells expressing pcDNA3-HA-p53R280K vector treated with dimethylsulphoxide (DMSO) (-) or treated (+) with Cerivastatin 10 μM for 48h. Representative blots are shown. (F) The indicated cell lines were treated with dimethylsulphoxide (DMSO)(-) or with Cerivastatin 10 μM for 48h. Representative blots are shown. (G) Quantification of BrdU-positive cells. The indicated cell lines were treated with Cerivastatin (CER) 0.1 μM for 48h. Error bars represent mean ± s.d. from n=3 biologically independent experiments. **P < 0.01 *P < 0.05; P values (from left to right): 0.008; 0.016; 0.007; 0.008; 0.039; 0.017; 0.005; 0.266; 0.801; 0.221, 0.341. Two-tailed Student’s t-test is used. All experiments were repeated three times, apart from 1A, which was performed twice. Source data for panel 1G is available in Supplementary Table 6. Unprocessed scans of blots are shown in Supplementary Figure 6.
Confirming the reliability of our approach, adrenergic agonists (e.g. Isoprenaline) and Ouabain have been already identified as p53 destabilizing agents10,11. Thus, for further analysis we focused on statins, a class of drugs clinically used to lower cholesterol plasma levels in patients with cardiovascular disease. Statins act by inhibiting HMG-CoA reductase (HMGCR) which catalyses mevalonic acid (MVA) synthesis, i.e. the first rate-limiting step of the metabolic pathway leading to cholesterol biosynthesis, namely the mevalonate pathway12. Cerivastatin and Simvastatin behaved similarly in reducing mutant p53 levels (Figure 1C and Supplementary Figure 2B, 5B) and Cerivastatin induced mutant p53 cytoplasmic localization (Supplementary Figure 1D). Decrease of mutant p53 levels upon administration of Cerivastatin was time-dependent and maximal after 48 hours (Figure 1D and Supplementary Figure 5B). Cerivastatin also reduced the levels of exogenously expressed mutant p53 R280K in MDA-MB-231 cells (Figure 1E and Supplementary Figure 5B).
To assess whether the mevalonate pathway is a general regulator of mutant p53 levels, we analysed the effects of Cerivastatin on a panel of human tumour cell lines harbouring different p53 mutants or wild-type p53. As shown in Figure 1F, the treatment caused a substantial reduction of mutant p53 protein levels in all the tested cell lines (Figure 1F and Supplementary Figure 2C), while wild-type p53 levels were unaffected (Figure 1F and Supplementary Figure 1I and 5B). Consistently, statin treatment reduced the proliferation of cells expressing missense mutant p53, with no effect on cells expressing wild-type p53 or null for p53 (Figure 1G). Cerivastatin also induced mild cell death in MDA-MB-231 cells, however, inhibition of apoptosis by Z-VAD treatment did not prevent mutant p53 reduction, suggesting that mutant p53 levels were not altered as a consequence of cell death (Supplementary Figure 1E). In line with this result, induction of apoptosis in MDA-MB-231 cells by doxorubicin or 5-FU treatment did not reduce mutant p53 levels (Supplementary Figure 1F).
In MDA-MB-231 cells, Cerivastatin shortened the half-life of both endogenous and overexpressed mutant p53 in a proteasome-dependent manner (Figure 2A and Supplementary Figure 1G, 1H), and caused an increase of mutant p53 poly-ubiquitinated forms (Figure 2B). Based on this and other evidence 2,13 we hypothesized that statins could reactivate the ubiquitin-dependent degradation of mutant p53 in cancer cells. Indeed, administration of the MDM2 inhibitor Nutlin, as well as specific knockdown of MDM2 caused a significant rescue of mutant p53 stability in Cerivastatin-treated MDA-MB-231 cells (Figure 2A, 2C, 2D and Supplementary Figure 1J, 5B), thus suggesting that statin treatment impacts on mutant p53 levels via the MDM2 ubiquitin ligase. Of note, neither treatment was able to completely rescue mutant p53 levels, implying the existence of additional mechanisms mediating mutant p53 inhibition by statins.
Figure 2. Statins unleash MDM2-mediated degradation of mutant p53 by disrupting its interaction with Hsp90.
(A) Evaluation of mutant p53 half-life in MDA-MB-231 cells. Cells were pre-treated with Cerivastatin (1 μM), alone or with nutlin (10 μM) and after 24h cells were treated with cycloheximide (CHX) (50 μM) for the indicated times. MG132 (50 μM) was added to inhibit the proteasome. Representative western blots with the indicated antibodies are shown. The graph indicates normalized quantification of mutant p53 protein amounts. Each replicate from n=3 biologically independent experiments is shown. *P = 0.012. Two-tailed Student’s t-test. (B) Left: MDA-MB-231 cells were transfected with constructs expressing HA-ubiquitin and pcDNA3-p53R280K and then treated with Cerivastatin 1 μM for 48h. Right: mutant p53 was immunoprecipitated from lysates and anti-HA blot was performed to detect ubiquitinated forms of mutant p53. (C) MDA-MB-231 cells were treated with Nutlin-3 10 μM for 12h and then Cerivastatin 10 μM was added to the medium for additional 48 h. Representative blots are shown. (D) MDA-MB-231 cells were treated with Cerivastatin 10 μM for 48h after transfection with indicated siRNA for 24h. Representative blots are shown. (E) Schematic representation of the mechanism of mutant p53 stabilization by Hsp90 in cancer cells. (F) Mutant p53 was immunoprecipitated from lysates of MDA-MB-231 cells, untreated (-) or treated with Cerivastatin 1 or 10 μM for 24h. Co-immunoprecipitated Hsp90 and MDM2 were detected by Western blot. All experiments were repeated three independent times with similar results. Source data for panel 2A is avaibable in Supplementary Table 6. Unprocessed scans of blots are shown in Supplementary Figure 6.
In cancer cells, mutant p53 proteins are engaged in stable complexes with the Hsp90 chaperone, which is often upregulated during transformation14. This interaction results in marked reduction of mutant p53 ubiquitination by the E3 ligase MDM2 (Figure 2E)15,16. As shown in Figure 2F, Cerivastatin caused the dissociation of Hsp90 from mutant p53, while leaving unaffected its binding to MDM2 (Supplementary Figure 5B). These data suggest that inhibition of the mevalonate pathway in cancer cells restores the inherent instability of mutant p53 by functionally disrupting the molecular mechanism protecting mutant p53 from inhibition by MDM2.
It has been shown that HDAC6-mediated Hsp90 deacetylation fosters mutant p53 stabilization17,18. Of note, statins have been found to inhibit HDAC enzymatic activity in tumour cells19,20. We thus hypothesized that HMGCR inhibition could reduce HDAC6 activity, leading to Hsp90 acetylation (inactivation) and mutant p53 destabilization. As shown in Supplementary Figure 2D, cerivastatin treatment indeed inhibited HDAC6, leading to increased tubulin acetylation. Of note, cerivastatin treatment induced an increase of Hsp90 acetylation, that was comparable to that induced by Sulforaphane (SFN, a HDAC6 inhibitor17,21) (Supplementary Figure 2E). These results suggest that HMGCR inhibition reduces Hsp90 activity as a consequence of HDAC6 inhibition.
All enzymes belonging to the mevalonate pathway are under direct transcriptional control of SREBPs (sterol regulatory element-binding proteins) transcription factors (Figure 3A)22. Knockdown of either SREBP1 or SREBP2 in MDA-MB-231 cells coincided with a significant decrease of mutant p53 protein levels (Figure 3B and Supplementary Figure 5B). Conversely, activation of endogenous SREBP1/2 by culturing cells in lipoprotein-depleted medium (LDS), increased mutant p53 protein levels, and this effect was nullified by inhibiting the mevalonate pathway with statins (Figure 3C and Supplementary Figure 5B). Remarkably, mutant p53 protein levels were rescued in Cerivastatin-treated MDA-MB-231 cells by addition of MVA, consistent with mutant p53 levels being dependent on intracellular mevalonate levels (Figure 3D and Supplementary Figure 5B). These results demonstrate that activation of the SREBP/mevalonate pathway promotes the accumulation of mutant p53 protein in cancer cells.
Figure 3. SREBP/mevalonate pathway controls mutant p53 levels via GGPP.
(A) Schematic overview of the mevalonate pathway. Enzymes are shown in red and inhibitors in blue. (B) MDA-MB-231 cells transfected with siRNAs targeting either SREBP1 (BP1) or SREBP2 (BP2) or SREBP1/2 together (BP1/2) for 48h. Representative western blots with the indicated antibodies are shown. SCD-1, a SREBP1/2 target involved in lipid metabolism, was used as positive control. (C) MDA-MB-231 cells were grown in medium supplemented with 10% FBS or 2% Lipoprotein-depleted serum (LDS) with or without Cerivastatin (CER) 10 μM for 48h. Representative blots are shown. (D) MDA-MB-231 cells were treated with Cerivastatin 10 μM (CER) alone or with mevalonic acid (MVA) 0.5 mM for 48h. Representative blots are shown. (E) MDA-MB-231 cells were treated with different inhibitors: Cerivastatin (CER) 10 μM, Zoledronic Acid (ZA) 50 μM, geranylgeranyl transferase I inhibitor (GGTI-298) 10 μM, squalene synthase inhibitor (YM-53601) 20 μM, farnesyl transferase inhibitor (FTI-277) 20 μM, for 48h. Representative blots are shown. (F) MDA-MB-231 cells were treated with GGTI-298 (10 μM) for the indicated times. Representative blots are shown. (G) MDA-MB-231 cells were treated with Cerivastatin 10 μM (CER) either alone or in combination with geranygeranyl pyrophosphate 20 μM (CER+GGPP) for 48h. Representative blots are shown. (H) Colony formation assay. The indicated cell lines were treated with Cerivastatin (CER) 0.1 μM either alone or in combination with geranygeranyl pyrophosphate 20 μM (CER + GGPP) for 6 days. (I) Mutant p53 was immunoprecipitated from lysates of MDA-MB-231 cells either untreated (-) or treated (+) with GGTI-298 10 μM for 24h. Co-immunoprecipitated Hsp90 was detected by western blot. All experiments were repeated three independent times with similar results. Unprocessed scans of blots are shown in Supplementary Figure 6.
In addition to supplying de novo biosynthesis of cholesterol, the mevalonate pathway is essential for production of other key metabolites, such as farnesyl pyrophosphate and geranylgeranyl pyrophosphate (Figure 3A)23. Interestingly, only the geranylgeranyl transferase 1 (GGTase1) inhibitor GGTI-298 and the farnesyl di-phosphate synthase (FDPS) inhibitor zoledronic acid (ZA) were able to phenocopy the effect of statins on mutant p53, whereas the inhibition of squalene synthase (FDFT) by YM-53601 and farnesyl transferase (FNTA) by FTI-277 were ineffective (Figures 3E and 3F and Supplementary Figure 5B). GGTI-298 treatment induced MDM2-dependent mutant p53 ubiquitination (Supplementary Figure 2F), due to inhibition of HDAC6, consequent increase of Hsp90 acetylation and dissociation of mutant p53 (Figure 3I and Supplementary Figure 2D-E and 5B). The effects of GGTI-298 were partially rescued by either proteasome or MDM2 inhibition (Supplementary Figure 2G-H). Geranylgeranyl pyrophosphate (GGPP) addition to statin-treated cells, instead, rescued both mutant p53 levels (Figures 3G and Supplementary Figure 5B) and activation (as measured by the expression of the “ten genes” signature5, Supplementary Figure 4A) as well as cell proliferation (Figure 3H and Supplementary Figure 4B). These data indicate that protein geranylgeranylation is required for mutant p53 accumulation in cancer cells.
The main biological role of geranylgeranylation is anchoring proteins to cellular membranes24. Rho-GTPases represent major GGTase1 targets and statins have been found to inhibit the enzymatic activity of RhoA8. Interestingly, RhoA has been recently identified as a key mediator of the anti-tumour activity of statins, and a crucial target of mutant p53 in controlling cellular metabolism8,12,25,26. Therefore, we asked whether RhoA might control mutant p53 levels downstream of MVA. Strikingly, inhibition of RhoA, by means of either short interfering RNA or treatment with its specific inhibitor C3 toxin, was able to reduce mutant p53 protein levels in different cancer cell lines (Figure 4A, 4B and Supplementary Figures 2I-J, 5B). As expected, treatment of MDA-MB-231 cells with C3 shortened mutant p53 half-life (Supplementary Figure 2K). Conversely, over-expression of a constitutively active form of RhoA (RhoA-G14V) in MDA-MB-231 cells induced mutant p53 accumulation and activation (Figure 4C and Supplementary Figure 2L, 4C and 5B) that were completely prevented by treatment with GGTase1 inhibitor (Figure 4C and Supplementary Figure 5B). Importantly, in cells expressing a statin-sensitive but GGTI-insensitive GFP–RhoA mutant (RhoA-G14V-F)8,27, GGTase1 inhibition was less efficient in reducing mutant p53 levels and cell proliferation (Figure 4D and Supplementary Figure 2M-N, 5B), thus proving that RhoA prenylation is required for mutant p53 accumulation. Finally, knock-down of RhoA expression reduced the binding of Hsp90 to mutant p53 (Figure 4F and Supplementary Figure 5B). Altogether, these results implicate the activation of RhoA in the regulation of mutant p53 protein levels.
Figure 4. RhoA geranylgeranylation controls mutant p53 levels downstream of mevalonate pathway.
(A) The indicated cell lines were transfected with two independent siRNAs targeting RhoA for 72h. Representative blots are shown. (B) MDA-MB-231 cells were treated with C3 toxin (100 ng/ml) for 48 h. Representative blots are shown. (C) MDA-MB-231 cells stably expressing control vector (CTL) or the active form of RhoA (RhoA-G14V) were treated with increasing amount of GGTI-298 (0; 10μM; 20μM) for 48h. Representative blots are shown. (D) Fluorescence microscopy analysis of MDA-MB-231 cells stably expressing the construct coding for a mutant RhoA-G14V bearing a farnesylation consensus sequence (GFP-RhoA-G14V-F) either left untreated (-) or treated with Cerivastatin 1 μM or with GGTI-298 10μM for 48h. Scale bar 15 μm. (E) MDA-MB-231 cells stably expressing control vector (CTL) or GFP-RhoA-G14V-F were left untreated (-) or treated with GGTI-298 10 μM or ZA 50 μM for 48 h. Representative blots are shown. (F) Mutant p53 was immunoprecipitated from lysates of MDA-MB-231 cells transfected with control (CTL) or two independent RhoA siRNAs (siR#1 and siR#2) for 72h. Co-immunoprecipitated Hsp90 was detected by western blot. All experiments were repeated three independent times with similar results. Unprocessed scans of blots are shown in Supplementary Figure 6.
RhoA is a master regulator of actin cytoskeleton rearrangements and its activity is fundamental for transducing mechanical inputs generated by the extracellular/intracellular environment28. Thus, we asked whether acto-myosin dynamics control mutant p53 levels downstream of Mevalonate/RhoA signalling. First, we analysed the impact of the mevalonate pathway on actomyosin dynamics by treating cells with Cerivastatin, and monitoring phalloidin and pMLC2(Ser19) staining, as readouts of F-actin polymerization and of RhoA-dependent contractile myosin, respectively. Strikingly, inhibition of mevalonate pathway by statin or ZA caused a dramatic reduction in both F-actin polymerization and MLC2 phosphorylation, as also observed upon Latrunculin A treatment, suggesting that the mevalonate pathway is functionally required for RhoA-dependent mechanotransduction in vitro (Figure 5A and Supplementary Figure 3A-B). A direct demonstration of actin cytoskeleton de-structuring has been provided by the significant reduction in cell stiffness observed in ZA and statin treated cells (p<0.001), and measured by means of atomic force microscopy (AFM) (Figure 5B). Similar effects have been observed in blebbistatin or Rho-Kinase (ROCK) inhibitor treated cells (Figure 5B). Interestingly, geranylgeranyl pyrophosphate was able to reverse statin effects increasing cellular stiffness to levels comparable to those observed in untreated cells (Figure 5B). To test whether disruption of actin cytoskeleton impacts on mutant p53 levels, we administrated Latrunculin A to MDA-MB-231 cells and stained cells for F-actin and p53. As shown in Figure 5C, Latrunculin A elicited attenuation of F-actin stress fibers and concomitant reduction of mutant p53 levels (Figure 5C and Supplementary Figure 3E). Similarly, actin contractility inhibition or Rho-Kinase inhibition reduced mutant p53 levels (Figure 5B and Supplementary Figure 3C-D). Of note, in statin-treated cells the effect of mevalonate addition on mutant p53 levels was nullified by Latrunculin A (Supplementary Figure 3F). These results suggest that RhoA-dependent acto-myosin dynamics impact mutant p53 levels in tumour cells.
Figure 5. Mechanical cues control mutant p53 levels and activity via RhoA/actin cytoskeleton.
(A) MDA-MB-231 cells treated with Latrunculin A 0.5 μM (LAT.A) or Cerivastatin 10μM for 48h. Scale bar 15 μm. (B) Cell stiffness 48h after treatment with indicated compounds. Data for NT come from n=291 cells (pooled across 5 independent experiments), for ZOL, CER and CER-GGPP from n=171, n=156 and n=163 cells, respectively (pooled across 3 independent experiments each), while for BLEB and Y from n=114 and n=110 cells, respectively (pooled across 2 independent experiments each). Box plots range from 25th to 75th percentiles, bold lines inside the box represent the median. P values obtained by two-sample Kolmogorov-Smirnov test based on probability distribution. **P<0.01.
(C) MDA-MB-231 cells treated with Latrunculin A 0.5 μM (LAT.A) for 48h. Scale bar 15 μm. (D) Lysates of MDA-MB-231-derived xenograft tumours from saline- or zoledronic acid (ZA)-treated mice. (E) Upper: schematic overview of the experiment. Lower: mammary epithelial cells were plated on soft or stiff fibronectin-coated hydrogels for 3 days with or without Cerivastatin 10μM. (F) The indicated cell lines were either grown on plastic, or plated on soft fibronectin-coated hydrogels for 3 days. Cells grown on plastic were also treated with Latrunculin A 0.5 μM (LAT.A) or Cerivastatin 10μM for the last 48h. (G) SUM149 cells stably expressing control vector (CTL) or the active form of RhoA (G14V) were plated on plastic or on fibronectin-coated soft hydrogels for 3 days. (H) Mutant p53 was immunoprecipitated from lysates of MDA-MB-231 cells grown on plastic, or plated on soft hydrogels for 3 days. (I) MDA-MB-231 cells were plated on soft or stiff hydrogels for 3 days with or without Sulforaphane (SFN) 20μM. (J) Average gene expression values of “mutant p53 signature” genes in missense mutant p53 breast cancer samples, classified according to the “stiffness” signature. (K) Average gene expression values of “YAP/TAZ signature” genes in missense mutant p53 breast cancer samples, classified according to the “stiffness” signature. Data are shown as individual samples (n = 117 independent breast cancer patients; dots) and the mean ± s.e.m. (standard error of the mean; back lines). P-value <0.0001 in a two-tailed unpaired t-test. Experiments in 1A, 1E, 1F, 1G, 1H and 1I were repeated three independent times with similar results. The experiment in 5D was performed once. Unprocessed scans of blots are shown in Supplementary Figure 6.
We then evaluated the role of the mevalonate pathway in tumour mechanosignalling and the effects of this on mutant p53 accumulation in vivo. To this aim, we used archival tumour tissues from nude mice orthotopically injected with MDA-MB-231 cells and treated with ZA. In these mice, zoledronic acid caused a significant reduction of tumour growth8. Tumour tissues were then analysed for markers of mechanosignalling and for mutant p53 levels. Of note, tumour samples from mice receiving zoledronic acid showed a robust reduction of mechanosignalling, as indicated by decreased phosphorylation of focal adhesion kinase (pFAK residue Tyr397) and of myosin light chain 2 (pMLC2 residue Ser19) (Figure 5D). Accordingly, ZA treatment significantly decreased mutant p53 levels (Figure 5D). Interestingly, AFM analysis of tumour tissues showed that ZA treatment also reduced the stiffness of the cancer cells and of the extracellular matrix within the tumour (Supplementary Figure 3O).
Aberrant tissue tensional homeostasis is a feature of several epithelial cancers29. In this context, RhoA becomes activated by tissue rigidity and is part of an integrated mechanoregulatory circuit linking extracellular matrix stiffness to cytoskeletal tension (Supplementary Figure 3G-H)29. Based on these premises, we postulated that matrix stiffness might act as an environmental input promoting mutant p53 stabilization through Mevalonate/RhoA-mediated mechanosignalling. To verify this hypothesis, we employed a pre-neoplastic experimental system, i.e. mammary epithelial cells (MECs) isolated from 8-week old p53R172H/R172H knock-in mice. These cells were grown either on a soft fibronectin-coated hydrogel matching the compliance of normal mammary tissue (Young’s modulus: 0.5 kPa)29,30, or on a stiff fibronectin-coated hydrogel (Young’s modulus: 50 kPa) (Figure 5E and Supplementary Figure 5B). As expected, mutant p53 levels were almost undetectable in MECs plated on soft matrix, confirming that the protein is intrinsically unstable in non-transformed tissues (Figure 5E). Strikingly, however, mutant p53 accumulated at high levels when MECs were grown on a hyperstiff matrix, and inhibition of the mevalonate pathway by Cerivastatin treatment prevented its accumulation (Figure 5E). Cancer cells grown on soft fibronectin-coated hydrogels also showed significantly lower mutant p53 protein levels and activation (measured by the “ten genes” signature5) (Figure 5F and Supplementary Figure 3H and 3N) as wells as reduced levels of the enzymes involved in protein geranylgeranylation (namely Ggps1 and GGTase-1) and RhoA activation (Supplementary Figure 3G-I). Of note, inhibition of actin polymerization by Latrunculin A and treatment with Cerivastatin both reduced mutant p53 levels and activation with similar efficiency (Figure 5F and Supplementary Figure 5B). Overexpression of activated RhoA (RhoA-G14V) in cells grown on soft matrix efficiently promoted stabilization of mutant p53 to levels observed in cells grown on plastic (Figure 5G and Supplementary Figure 5B). Notably, in cells grown on soft hydrogels, Hsp90 is acetylated due to HDAC6 inhibition, and consequently less efficient in binding mutant p53 (Figure 5H, Supplementary Figure 2D-E and 5B). Finally, while growing cells on stiff matrix induced mutant p53 accumulation, the concomitant inhibition of HDAC6 prevented this effect (Figure 5I and Supplementary Figure 3J, 3K and 5B), thus demonstrating the functional requirement of HDAC6 for mutant p53 stabilization downstream of RhoA/mechanical cues. These results demonstrate that mutant p53 proteins are not intrinsically stable in tumour cells, rather their accumulation is strongly influenced by the Mevalonate/RhoA axis and by mechanical cues such as the stiffness of the extracellular environment.
We then decided to verify whether increased mechanosignalling correlates with mutant p53 activation in human tumours. To this aim, we performed and validated gene expression analysis on MDA-MB-231 cells grown on either soft or stiff surface and generated a “stiffness” signature, composed of genes induced in stiff as compared to soft matrix (Supplementary Figure 4D and Supplementary Table 5). Next, we interrogated a dataset of 117 missense mutant p53 human breast cancers and classified tumours as having high or low expression of this “stiffness” signature. When we monitored the activation of mutant p53 in these tumours, by analysing expression of a published “mutant p53 signature”31, we found that tumours classified as “highly stiff” were also characterized by strong activation of mutant p53 (Figure 5J). As positive control, YAP/TAZ signature32 was also upregulated in the same specimens32–34 indicating a parallel between activation of a widely validated mechanosensitive cascade and of mutant p53 in primary human tumours (Figure 5K and Supplementary Figure 5A). These observations confirm that mutant p53 is more active in cells with activated mechanosignalling. Of note, YAP/TAZ were not involved in mutant p53 stabilization, as YAP or YAP/TAZ knockdown had no effects on mutant p53 levels in stiff matrix or in cells overexpressing the active form of RhoA (Supplementary Figure 3L-M).
In cancer cells, the mevalonate pathway can be activated by a direct mutant p53-SREBP interaction, which sustains SREBP transcriptional activity35. Combined with our finding that SREBP activation impacts on mutant p53 levels, this evidence supports the concept that in cancer cells mutant p53 can give rise to a positive feed-back loop in which, by forcing mevalonate pathway activation and GGPP biosynthesis, mutant p53 can sustain its own stabilization. RhoA geranylgeranylation turns out to be an essential process in these events. This interplay could possibly require other proteins, such as HDAC636,17,37. The Rho pathway may work in concert with other, possibly redundant or tumour-specific mechanisms to stabilize mutant p53. For example, activation of the Hsp40/DNAJ chaperone by the mevalonate pathway has been recently found to control mutant p53 stability38 through the ubiquitin ligase CHIP. Although the involvement of cell mechanics was not investigated in that study, our work nonetheless defines a different pathway, involving Hsp90 and MDM2.
More importantly, we demonstrate that pharmacologic inhibition of the Mevalonate/RhoA axis (e.g. by statins or zoledronic acid or GGTI) can interfere with the transduction of mechanical inputs in vitro and in vivo and thereby prevent the stabilization of mutant p53 and of other oncogenes such as YAP/TAZ8, ultimately restraining diverse malignant cancer phenotypes.
Mutant p53 accumulation is often spatially heterogeneous within primary tumours, with mutant p53 over-expressing tumour foci associated with fibrous stroma3. Our work shows that the extracellular microenvironment controls mutant p53 stabilization and activation. Based on this, we speculate that the heterogeneity of mutant p53 expression observed within individual tumours might be explained by different mechanical niches existing within the diseased tissues30. The presence of fibrotic “stiff” lesions, along with influencing treatment efficacy39, is associated with poor prognosis40, enhanced growth and survival signalling, as well as with invasive and pro-metastatic features29. Our results implicate that conditions that increase ECM stiffening (fibrosis) and cell contractility in mutant p53 tumours, could induce stabilization of mutant p53 and activation of its oncogenic properties, ultimately facilitating malignant progression.
Methods
Cell lines
MDA-MB-231 (p53 R280K), MDA-MB-468 (p53 R273H), SUM149 (p53 M237I) and BT-549 (p53 R249S), SK-BR-3 (p53 R175H), T47D (p53 L194F), Hs-578-T (p53 V157F) are human breast cancer cell lines. Mahlavu (p53 R249S) are human hepatocellular carcinoma cells. U118MG (p53 R213Q) are human glioblastoma cells. U2OS, osteosarcoma cell line, and MCF-7, human adenocarcinoma cell line, express wild-type p53, while H1299, a non-small cell lung cancer cell line, are p53 null.
MDA-MB-231, MDA-MB-468, BT-549, SKBR-3, U2OS, U118MG and T47D cells were cultured in DMEM (LONZA) supplemented with 10% FBS (Fetal Bovine Serum) and with 1% antibiotics (penicillin 100U/mL and streptomycin 10 μg/mL). SUM149 cells were cultured in DMEM/F12 (LONZA) (1:1) supplemented with 10% FBS (Fetal Bovine Serum) and with 1% antibiotics. Hs-578-T were cultured in DMEM (Thermo Fisher) supplemented with 10% FBS, glutamine and antibiotics and 10 μg/ml insulin (Sigma). Mahlavu cells were cultured in EMEM (Sigma) supplemented with 10% FBS (Fetal Bovine Serum), with 1% antibiotics (penicillin 100 U/mL and streptomycin 10 μg/mL), 1% MEM NEAA (Minimum essential medium non-essential amino acids) and 1% Glutamax. H1299 cells were cultured in RPMI medium RPMI 1640 with 10% FBS and 1% antibiotics. MCF7 cells were cultured in EMEM (Sigma) supplemented with 10% FBS (Fetal Bovine Serum), with 1% antibiotics (penicillin 100 U/mL and streptomycin 10 μg/mL) and 1% MEM NEAA (Minimum essential medium non-essential amino acids.
MDA-MB-231 cells, stable expressing GFP-RhoA-G14V and the mutant GFP-RhoA-G14V-F, were maintained in DMEM (LONZA) supplemented with 10% FBS (Fetal Bovine Serum) and with 1% antibiotics (penicillin 100 U/mL and streptomycin 10 μg/mL) and with addition of selection antibiotics.
MEFs infected with pLPC-RASV12 were maintained as previously described5.
Mammary epithelial cells (MECs) were isolated from p53R172H/R172H knock-in mice as previously described9,41 and seeded on top of 50 kPa or 0.5 kPa Easy Coat hydrogels (Cell guidance system) and harvested after three days. Hydrogels for the experiments in Figures 5 were as in Aragona et al., 201133.
Reagents and plasmids
The library of FDA-approved drugs (Screen-Well FDA-Approved Drug Library, 640 chemical compounds dissolved at 10 mM in dimethylsulphoxide) was obtained from Enzo Life Sciences.
The following compounds were purchased from Sigma Aldrich: Cerivastatin (SML0005), Ouabain (O3125), Spiperone (S7395), Ivermectin (I888), Salmeterol (S5068), Simvastatin (S6196), GGTI-298 (G5169), Geranylgeranyl Pyrophosphate (#G6025), Zoledronic Acid (SML0223), Mevalonic acid (41288), Cycloheximide (C7698 and C1988), Blebbistatin (B0560), Y-27632 dihydrochloride (Y0503). Latrunculin-A (sc-202691) was purchased from Santa Cruz Biotechnology. The following compounds were purchased from Cayman: YM-53601 (18113), FTI-277 (F9803), Nutlin-3 (10004372) and SAHA (10009929). MG132 (474790) was purchased from Calbiochem. Thioridazine hydrochloride (1306110) was purchased from Tocris. C3-RhoA inhibitor I (CT04) was purchased from Cytoskeleton. Lipoprotein Depleted Serum (LDS) (880100-2) was purchased from Biocompare (DBA). Z-Vad FMK is from ENZO Life Science (ALX-260-020-M001). 5-Fluorouracile (5FU) is from Teva. Doxorubicin hydrochloride is from Sigma (D1515). Picro Sirus Red Stain Kit (Connective Stain) (ab 150681) was purchased from Abcam.
pEGFP–RhoA-G14V was a gift from C. Schneider (Laboratorio Nazionale CIB, Italy). The retroviral constructs (pLPC) coding for GFP–RhoA-G14V (CLVL) and GFP– RhoAV14-F (CVLS) were generated by PCR mutagenesis from pEGFP–RhoA-G14V8.
pcDNA3-p53R280K was previously described5.
High Content Screening
For the screening experiments, MDA-MB-231 cells (3.0×103 per well) were seeded on black clear-bottom 384-well plates (PerkinElmer). Twenty-four hours later, the FDA-approved drugs were transferred robotically from library stock plates (0.1 mM and 1 mM in DMSO) to the plates containing the cells; controls were added to columns 1, 2, 23 and 24 of each plate. Cells were fixed at 48h after plating, i.e. 24h after addition of drugs, and processed immediately for immunofluorescence. Briefly, cells were fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.5% Triton X-100 in phosphate buffered saline (PBS) solution for 10 min, followed by 30 min blocking in 3% FBS. Cells were then incubated with a mouse antibody against mutant p53 (DO-1, Santa Cruz Biotechnology) diluted in blocking solution for 1 h. Cells were further washed with PBS and incubated for 1h with a secondary antibody conjugated to Alexa Fluor-594 (Life Technologies), and stained with Hoechst 33342 (Life Technologies).
Image acquisition was performed using an ImageXpress Micro automated high-content screening fluorescence microscope (Molecular Devices) at a 10× magnification; a total of 9 images were acquired per wavelength, well and replicate, corresponding to ca. 4,500 cells analysed per experimental condition and replicate. Image analysis to identify cells presenting mutant p53 signal was performed using the ‘Multi-Wavelength Translocation’ application module implemented in MetaXpress software (Molecular Devices).
Screening was performed in duplicate, at two drug concentrations (1 μM and 10 μM); final concentration of DMSO in the culture medium was 1% (v/v) for all experimental conditions. The screening was performed at the ICGEB High-Throughput Screening Facility (http://www.icgeb.org/high-throughput-screening.html).
Transfections
siRNA transfections were performed with Lipofectamine RNAi-MAX (Life technologies) in antibiotics-free medium according to manufacturer instructions. Sequences of siRNAs are reported in Supplementary Table 3. Negative control siRNA was: AllStars negative control siRNA Qiagen 1027281. In Supplementary Figure 3J, siHDAC6 was transfected two times to improve the efficiency of HDAC6 protein reduction.
DNA transfections were performed in MDA-MB 231 cell lines with Lipofectamine 2000 (Invitrogen) in antibiotic-free medium according to the manufactured instructions.
For retrovirus production, low-confluence HEK-293GP packaging cells were transfected with appropriate vectors by calcium phosphate, in combination with pMD2ENV coding for envelope proteins. After 48–72 h the virus-containing medium was filtered and added to target cells. Cells were selected with puromycin (0.5 μg mL-1).
Quantitative Real-Time PCR
Cells were harvested in Qiazol lysis reagent (Qiagen) for total RNA extraction, and contaminant DNA was removed by DNase treatment. qRT-PCR analyses were carried out on retrotranscribed cDNAs with Quantitect reverse transcription kit (Qiagen) and analyzed with Biorad CFX Manager software. Experiments were performed at least three times, with duplicate replicates. The quantification is based on the 2-ΔΔCt method using the housekeeping gene histone 3 (H3) as normalizer. PCR oligonucleotide sequences (F= Forward, R= Reverse) are reported in Supplementary Table 4. To monitor activation of mutant p53 we measured mRNA expression of the “ten genes” signature as previously described5.
Antibodies
The antibodies used for western blot and immunofluorescence were: anti-p53 (1:1000; DO-1, Santa Cruz Biotechnology), anti-Actin (1:5000; C11, Sigma), anti-GAPDH (1:5000; MAB374, Millipore), anti-SREBP1 (2A4) (1:500; sc13551, Santa Cruz Biotechnology), anti-SREBP2 (1:500; 557037, BD Bioscience), anti-SCD-1 (1:1000; ab19862, Abcam), Anti-Vinculin (1:5000; V4505 Sigma), anti-Hsp90 (1:1000; sc13119, Santa Cruz Biotechnology), anti-HDAC6 (H-300) (1:1000; sc-11420, Santa Cruz Biotecnology), Acetylated-Lysine Antibody (1:1000; 9441, Cell Signaling), anti-α-Tubulin (1:5000, T5168, Sigma), anti-acetylated-tubulin (1:1000; T6793, Sigma), anti GFP (1:1000; home-made), anti-MDM2 (SMP14; SC-965, Santa Cruz Biotechnology), anti-MLC2 (1:1000; 3672S, Cell Signaling), anti-pMLC2 (phospho Ser19) (1:1000; 3675S; Cell signalling), anti-FAK (C-20) (1:1000; sc-558, Santa Cruz Biotechnology), anti-pFAK (phospho Y397) (1:1000; ab81298, Abcam), anti-YAP (1:1000; sc-15407; Santa Cruz Biotechnology), anti-TAZ (1:1000; HPA007415, Sigma), Anti-PSMA2 (1:1000; sc-54671; Santa Cruz). Phalloidin is A12379 (Alexa Fluor), Anti-BrdU antibody (RPN202) is GE Healthcare. anti- cleaved PARP p85 fragment pAb is from Promega (G7341).
Immunofluorescence and Western Blot
Immunofluorescence staining was performed as previously described8. Briefly, cells were fixed in 4% paraformaldehyde for 10min, washed in PBS, permeabilized with Triton 0.1% for 10 min and blocked in PBS FBS 3% for 30 min. Antigen recognition was done by incubating primary antibody for 1 h at 37°C and with Goat anti-mouse Alexa Fluor 568 (Life Technologies) as secondary antibody for 30 min at 37°C. Nuclei were counterstained with Hoechst 33342 (Life Technologies).
Western blot analysis was performed as previously described8. The protein stability determination was performed as previously described5. Immunoblots were quantified using the ImageJ program, which measures the integrated density of bands corrected for background. In Supplementary Figure 6 is shown the average ratio density from at least 3 experiments ± s.d.. Two-tailed Student’s t-tests, was performed to determine statistical significance.
Co-immunoprecipitation
Co-IP experiments with endogenous proteins were performed using Co-IP buffer (NaCl 120mM, Tris-HCl pH8 20mM, EDTA 1mM, NP40 0,5%) with protease inhibitors. Samples were cleared by centrifugation for 30 min at 13,000g at 4 °C and incubated for 2h at 4 °C with anti-p53 antibody. After 1 h incubation with protein G-Sepharose (GE Healthcare), immunoprecipitates were washed three times in Co-IP buffer, resuspended in sample buffer, and analyzed by immunoblotting.
For ubiquitination assays, cells were lysed in 2% SDS, 150 mM NaCl, 10 mM Tris-HCl, pH 8.0, 1 mM PMSF, 5 mM NaF, 1mM Na3VO4, 0,5% (v/v) sodium deoxycholate with protease inhibitor cocktail (Sigma-Aldrich) and Ubiquitin Aldeyde 50 ng/mL. Cell lysates were diluted in IP buffer: 10 mM Tris-HCl, pH8.0, 150 mM NaCl, 2 mM EDTA, 1% Triton. The anti-p53 antibody (DO-1; Santa Cruz) was covalently bound to protein G Sepharose (Amersham Biosciences, GE Healthcare, Munich, Germany) using 5 mg/mL dimethylpimelimidate (Pierce Biosciences, Thermo Fisher Scientific, Bonn, Germany).
Isolation of GTP-loaded RhoA GTPase
The GTP-loaded form of RhoA was pulled down with GST–RHOTEKIN beads (Cytoskeleton), according to the manufacturer’s instructions.
Colony-formation assay
5000 cells were plated on 6 cm plates. The day after the medium was supplemented with drugs as indicated in the figures. After 6 days, cells were fixed with 4% paraformaldehyde (PFA) and stained for 30 min with Giemsa (FLuka) diluted solution 1:5 in water. Plates washed with water and dried were scanned. Colonies quantification was performed using Image J.
Brdu incorporation assay
Cells (3×104) were plated in 24-well plates. The day after the medium was supplemented with DMSO or Cerivastatin as indicated in figures. After 48 h from the treatment, the DNA precursor bromodeoxyuridine (BrdU) (20μM) was added to the medium for 2-12h before fixation. Briefly, cells were fixed in 4% paraformaldehyde for 10 min, washed in PBS, permeabilized with Triton 0.1% for 10 min and washed 3 times with NaOH 50mM solution and washed in PBS. Primary anti BrdU antibody solution (1:2 dilution), to detect bromodeoxyuridine (BrdU) incorporated, was used for 2 h at 37°C and Goat anti-mouse Alexa Fluor 568 (Life Technologies) as secondary antibody for 1h a 37°C. Nuclei were counterstained with Hoechst 33342 (Life Technologies).
Mice and animal care
For in vivo studies, one million of MDA-MB-231 cells were resuspended in 100 μL of DMEM, injected into the mammary fat of previously anesthetized 7 weeks old SCID female mice (1–3% isoflurane, Merial Italia S.p.A, Italy) as previously described8. At day 12 after cell injection, mice were subjected to intravenous injection of zoledronic acid ([1-hydroxy-2- (1H-imidazoledronic acid-1-yl) ethylidene] (200 μg/Kg body weight), every 4 days until the end of the experiment (day 40). At day 40 the animals were sacrificed and the primary tumours were extracted and directly frozen in liquid nitrogen. Tissues were lysed for immunoblot analysis or sectioned at 5 μm, fixed, and stained either with hematoxylin and eosin (H&E) for histological analysis or with Picro Sirius and hematoxylin to perform AFM analysis. Four tumors per group were used. The mice were used and housed in a specific pathogen-free (SPF) animal facility. Procedures involving animals and their care were performed in conformity with institutional guidelines (D.L. 116/92 and subsequent complementing circulars) and all experimental protocols were approved by the ethical Committee of the University of Padua (CEASA). the study is compliant with all relevant ethical regulations regarding animal research.
Microarray analysis for stiffness signature
For microarrays of genes regulated by matrix stiffness uptake, MDA-MB-231 cells were plated on soft fibronectin-coated hydrogels, as compared to the same cells grown on fibronectin-coated rigid surfaces (i.e. plastic). For each experimental condition, four biological replicates were prepared and processed in parallel. Total RNA was extracted using TriPure (Roche). RNA quality and purity were assessed on the Agilent Bioanalyzer 2100 (Agilent Technologies, Waldbronn, Germany); RNA concentration was determined using the NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies Inc.). Labeling and hybridization were performed according to Affymetrix One Cycle Target Labeling protocol on HG-U133 Plus 2.0 arrays (Affymetrix).
All data analyses were performed in R (version 3.2.4) using Bioconductor libraries (BioC 3.2) and R statistical packages. Probe level signals were converted to expression values using robust multi-array average procedure RMA (Irizarry et al, 2003) of Bioconductor affy package. Differentially expressed genes were identified using Significance Analysis of Microarray algorithm coded in the samr R package (Tusher et al, 2001). In SAM, we estimated the percentage of false-positive predictions (i.e., false discovery rate, FDR) with 100 permutations. To identify genes associated with stiffness in cells of mammary origin (stiffness signature), we compared the expression levels of MDA-MB-231 cells grown on stiff (Young’s modulus: 50 kPa) and soft hydrogels (Young’s modulus: 0.5 kPa) and selected those probe sets with an FDR ≤ 1% and a fold change ≥ 1.5. This selection resulted in 220 probe sets induced in stiff matrix (Supplementary Table 5).
Atomic Force Microscopy analysis
Atomic force microscopy (AFM) was used to investigate cell mechanical properties. In particular, elastic assessment of stiffness was done taking advantage of the force spectroscopy capabilities of a Smena AFM (NT-MDT Co., Moscow, Russia) mounted on an inverted fluorescence microscope (Nikon Eclipse Ti-U). In brief, force spectroscopy measures the deflection of the AFM cantilever when it is pushed against a surface. Deflection data were subsequently converted in a force vs. indentation curve based on cantilever spring constant and displacement knowledge [J.L.Alonso, W.H.Goldmann, Feeling the forces: atomic force microscopy in cell biology, Life Sci. 72 (2003) 2553e2560]. Compliance of the material under the tip was determined in this work fitting the data with a Hertzian model of surface indentation [I.N. Sneddon, The relation between load and penetration in the axisymmetric Boussinesq problem for a punch of arbitrary profile, Int. J. Eng. Sci.3 (1965) 47e57.]. Single-cell stiffness experiments were performed measuring AFM indentation positioning the tip apex on the central part of the cell, roughly corresponding to nucleus position, by means of optical visualization. Cells were measured in 1× PBS buffer at room temperature. For each sample 60 randomly choose cells were measured and analyzed. Tissue section’s stiffness was studied evaluating the stiffness of the cells inside the tissue§, marking their nuclei via Hematoxylin staining, and ECM stiffness, pointing it out by the collagen and amyloid specific dye Picro Sirius Red Stain. AFM force curves were obtained on each sample (4 control, not treated, samples and 4 zoledronate treated ones) randomly acquiring about 60 curves on blue, Hematoxylin positive, areas and about 60 curves on red, Picro Sirius Red positive, areas (see Fig. S3). A total number of 210 and 245 stiffness values were obtained for controls and treated cells, respectively. 240 and 239 curves were instead acquired for control and treated ECMs. During AFM characterization sections were maintained immersed in 1× PBS buffer at room temperature. Cantilever used was a tip-less probe characterized by a spring constant of about 0.03 nN/nm (HQ:CSC38 cantilevers from MikroMasch Co. – Tallinn, Estonia) at the end of which a 18 µm in diameter silica bead (Thermo Fisher Scientific, CA, USA) was glued using UV curable glue (Norland Products Inc., NJ, USA). Force spectroscopy measurements were performed at constant speed (2.5 µm/s) and triggered to a maximum force applied to the sample of 5 nN. Elastic modulus values (E), in kPa, were determined by fitting obtained force-displacement curves with an Hertzian model for the tip used taking advantage of the NOVA (NT-MDT Co., Moscow, Russia) control and analysis software.
Statistics and data processing were performed using Igor Pro software (www.wavematrics.com) and R statistical computing software (www.R-project.org). Significance of data differences was established as equality of probability distributions via the Kolmogorov–Smirnov test.
Breast cancer gene expression data
We downloaded the METABRIC collection, comprising gene expression data and clinical annotations for 997 breast cancer samples, from the European Genome-Phenome Archive (EGA, http://www.ebi.ac.uk/ega/) under accession number EGAD00010000210 (2012). Original Illumina probe identifiers have been mapped to Entrez gene IDs using the Bioconductor illuminaHumanv3.db annotation package for Illumina HT-12 v3 arrays obtaining log2 intensity values for a total of 19,761 genes. The TP53 status of 117 samples annotated as “missense” mutant p53 was derived from Silwal-Pandit et al. (2014)42.
Average signature expression and signature scores
Average signature expression of the mutant-p53 signature31 and of the YAP/TAZ activity-signature32 has been calculated as the standardized average expression of all signature genes in sample subgroups. Samples have been classified as “stiffness high” or “stiffness low” summarizing the standardized expression levels of the “stiffness” signature genes into a combined score with zero mean. The values shown in graphs are thus adimensional.
Statistics and reproducibility
The experiment for Figure 1A was repeated two times. The experiment in Figure 5D has been completed once. All other experiments are representative of at least 3 independent repeats. Error bars represent mean ± s.d. from n=3 biological replicates. P values were determined using two-tailed Student’s t-test as noted in the figure legends.
Supplementary Material
Acknowledgement
We thank A. Testa for discussions and proofreading the manuscript. We acknowledge G. Pastore for technical support. We thank Stefano Giulitti for preparation of hydrogels. We acknowledge support by the Italian Health Ministry (RF-2011-02346976 to GDS and GR-2011-02348707 to DS), the Italian University and Research Ministry (PRIN-2015-8KZKE3), Cariplo Foundation (Grant n. 2014-0812), and Beneficentia-Stiftung to GDS. This work was supported by grants from the Associazione Italiana per la Ricerca sul Cancro (AIRC) and AIRC Special Program Molecular Clinical Oncology ‘5 per mille’ (Grant n. 10016) to G.D.S., S.B., A.R., S.P. and AIRC IG (Grant n. 17659) to GDS. This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No 670126-DENOVOSTEM) and an AIRC PI-Grant and by Epigenetics Flagship project CNR-Miur grants to S.P. M.M. is supported by the FIRB RBAP11Z4Z9 project from the Italian Ministry of Education and the FCT Investigator Programme IF/00694/2013 from the Portuguese Foundation for Science and Technology (FCT), Portugal. R.B. is a fellow of the Fondazione Italiana per la Ricerca sul Cancro (FIRC).
Footnotes
Data availability.
Microarray data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under the accession code GSE93529. Gene expression data and clinical annotations for 997 breast cancer samples in the METABRIC collection were downloaded from European Genome-Phenome Archive (EGA),http://www.ebi.ac.uk/ega/) under accession number EGAD00010000210. Source data for Figures 1G, 2A, S1J, S2I, S2K, S3I, S4A-D, 5B are provided in Supplementary Table 6. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Author contributions
E.I., G.S., K.L., R.B., A.Z. and L.A. performed the experiments. A.R. performed mice experiments. M.M. performed the high-content screening. S.B. performed bioinformatic analysis. D.S. and L.U.S. performed AFM experiments. G.S., E.I. and G.D.S designed experiments. G.S., F.M., S.P. and G.D.S. wrote the manuscript.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Mantovani F, Walerych D, Sal G Del. Targeting mutant p53 in cancer: a long road to precision therapy. FEBS J. 2016 doi: 10.1111/febs.13948. [DOI] [PubMed] [Google Scholar]
- 2.Terzian T, et al. The inherent instability of mutant p53 is alleviated by Mdm2 or p16 INK4a loss. Genes Dev. 2008;22:1337–1344. doi: 10.1101/gad.1662908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koga T, et al. Heterogeneous distribution of P53 immunoreactivity in human lung adenocarcinoma correlates with MDM2 protein expression, rather than with P53 gene mutation. Int J Cancer. 2001;95:232–239. doi: 10.1002/1097-0215(20010720)95:4<232::aid-ijc1040>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 4.Freed-Pastor WA, Prives C. Mutant p53: One name, many proteins. Genes Dev. 2012;26:1268–1286. doi: 10.1101/gad.190678.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Girardini JE, et al. A Pin1/Mutant p53 Axis Promotes Aggressiveness in Breast Cancer. Cancer Cell. 2011;20:79–91. doi: 10.1016/j.ccr.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Ashcroft M, Vousden KH. Regulation of p53 stability. Oncogene. 1999;18:7637–7643. doi: 10.1038/sj.onc.1203012. [DOI] [PubMed] [Google Scholar]
- 7.Bouchalova P, et al. Mutant p53 accumulation in human breast cancer is not an intrinsic property or dependent on structural or functional disruption but is regulated by exogenous stress and receptor status. J Pathol. 2014;233:238–246. doi: 10.1002/path.4356. [DOI] [PubMed] [Google Scholar]
- 8.Sorrentino G, et al. Metabolic control of YAP and TAZ by the mevalonate pathway. Nat Cell Biol. 2014;16:357–366. doi: 10.1038/ncb2936. [DOI] [PubMed] [Google Scholar]
- 9.Sorrentino G, et al. Glucocorticoid receptor signalling activates YAP in breast cancer. Nat Commun. 2017;8 doi: 10.1038/ncomms14073. 14073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, et al. Cardiac glycosides inhibit p53 synthesis by a mechanism relieved by Src or MAPK inhibition. Cancer Res. 2009;69:6556–6564. doi: 10.1158/0008-5472.CAN-09-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hara MR, et al. A stress response pathway regulates DNA damage through β(2)-adrenoreceptors and β-arrestin-1. Supplemental data. Nature. 2011;53:1–7. doi: 10.1038/nature10368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mullen PJ, Yu R, Longo J, Archer MC, Penn LZ. The interplay between cell signalling and the mevalonate pathway in cancer. Nat Rev Cancer. 2016 doi: 10.1038/nrc.2016.76. [DOI] [PubMed] [Google Scholar]
- 13.Lukashchuk N, Vousden KH. Ubiquitination and degradation of mutant p53. Mol Cell Biol. 2007;27:8284–95. doi: 10.1128/MCB.00050-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malkin D. Li-fraumeni syndrome. Genes Cancer. 2011;2:475–84. doi: 10.1177/1947601911413466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esser C, Scheffner M, Höhfeld J. The chaperone-associated ubiquitin ligase CHIP is able to target p53 for proteasomal degradation. J Biol Chem. 2005;280:27443–27448. doi: 10.1074/jbc.M501574200. [DOI] [PubMed] [Google Scholar]
- 16.Nagata Y, et al. The stabilization mechanism of mutant-type p53 by impaired ubiquitination: the loss of wild-type p53 function and the hsp90 association. Oncogene. 1999;18:6037–49. doi: 10.1038/sj.onc.1202978. [DOI] [PubMed] [Google Scholar]
- 17.Li D, Marchenko ND, Moll UM. SAHA shows preferential cytotoxicity in mutant p53 cancer cells by destabilizing mutant p53 through inhibition of the HDAC6-Hsp90 chaperone axis. Cell Death Differ. 2011;18:1904–1913. doi: 10.1038/cdd.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kovacs JJ, et al. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell. 2005;18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 19.Lin YC, et al. Statins increase p21 through inhibition of histone deacetylase activity and release of promoter-associated HDAC1/2. Cancer Res. 2008;68:2375–2383. doi: 10.1158/0008-5472.CAN-07-5807. [DOI] [PubMed] [Google Scholar]
- 20.Feig JE, et al. Statins promote the regression of atherosclerosis via activation of the CCR7-dependent emigration pathway in macrophages. PLoS One. 2011;6 doi: 10.1371/journal.pone.0028534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clarke JD, Hsu A, Yu Z, Dashwood RH, Ho E. Differential effects of sulforaphane on histone deacetylases, cell cycle arrest and apoptosis in normal prostate cells versus hyperplastic and cancerous prostate cells. Mol Nutr Food Res. 2011;55:999–1009. doi: 10.1002/mnfr.201000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown MS, Goldstein JL. The SREBP pathway: Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 23.Repko EM, Maltese WA. Post-transitional isoprenylation of cellular proteins is altered in response to mevalonate availability. Journal of Biological Chemistry. 1989;264:9945–9952. [PubMed] [Google Scholar]
- 24.Zhang FL, Casey PJ. Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 25.Wang Z, et al. Interplay of mevalonate and Hippo pathways regulates RHAMM transcription via YAP to modulate breast cancer cell motility. Proc Natl Acad Sci U S A. 2014;111:E89–98. doi: 10.1073/pnas.1319190110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang C, et al. Tumour-associated mutant p53 drives the Warburg effect. Nat Commun. 2013;4:1–15. doi: 10.1038/ncomms3935. 2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mi W, et al. Geranylgeranylation signals to the Hippo pathway for breast cancer cell proliferation and migration. Oncogene. 2014:1–12. doi: 10.1038/onc.2014.251. [DOI] [PubMed] [Google Scholar]
- 28.Ingber DE. Cellular mechanotransduction: putting all the pieces together again. FASEB J. 2006;20:27. doi: 10.1096/fj.05-5424rev. [DOI] [PubMed] [Google Scholar]
- 29.Butcher DT, Alliston T, Weaver VM. A tense situation: forcing tumour progression. Nat Rev Cancer. 2009;9:108–22. doi: 10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janmey Pa, Miller RT. Mechanisms of mechanical signaling in development and disease. J Cell Sci. 2011;124:9–18. doi: 10.1242/jcs.071001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller LD, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. PNAS. 2005;102:13550–5. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zanconato F, et al. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol. 2015;17:1218–1227. doi: 10.1038/ncb3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Aragona M, et al. A Mechanical Checkpoint Controls Multicellular Growth through YAP/TAZ Regulation by Actin-Processing Factors. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 34.Zanconato F, Cordenonsi M, P S. YAP/TAZ at the Roots of Cancer. Cancer Cell. 2016 doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Freed-Pastor WA, et al. Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell. 2012;148:244–258. doi: 10.1016/j.cell.2011.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Destaing O, et al. A novel Rho-mDia2-HDAC6 pathway controls podosome patterning through microtubule acetylation in osteoclasts. J Cell Sci. 2005;118:2901–2911. doi: 10.1242/jcs.02425. [DOI] [PubMed] [Google Scholar]
- 37.Boyault C, Sadoul K, Pabion M, Khochbin S. HDAC6, at the crossroads between cytoskeleton and cell signaling by acetylation and ubiquitination. Oncogene. 2007;26:5468–76. doi: 10.1038/sj.onc.1210614. [DOI] [PubMed] [Google Scholar]
- 38.Parrales A, et al. DNAJA1 controls the fate of misfolded mutant p53 through the mevalonate pathway. Nat Cell Biol. 2016;18:1233–1243. doi: 10.1038/ncb3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Netti PA, Berk DA, Swartz MA, Grodzinsky AJ, Jain RK. Role of extracellular matrix assembly in interstitial transport in solid tumours. Cancer Res. 2000;60:2497–503. [PubMed] [Google Scholar]
- 40.Colpaert CG, et al. The presence of a fibrotic focus in invasive breast carcinoma correlates with the expression of carbonic anhydrase IX and is a marker of hypoxia and poor prognosis. Breast Cancer Res Treat. 2003;81:137–147. doi: 10.1023/A:1025702330207. [DOI] [PubMed] [Google Scholar]
- 41.Rustighi A, et al. Prolyl-isomerase Pin1 controls normal and cancer stem cells of the breast. EMBO Mol Med. 2014;6:99–119. doi: 10.1002/emmm.201302909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silwal-Pandit L, et al. TP53 mutation spectrum in breast cancer is subtype specific and has distinct prognostic relevance. Clin Cancer Res. 2014;20:3569–3580. doi: 10.1158/1078-0432.CCR-13-2943. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.