SUMMARY
Latrunculin A (LatA), a toxin from the red sea sponge Latrunculia magnifica, is the most widely used reagent to depolymerize actin filaments in experiments on live cells. LatA binds actin monomers and sequesters them from polymerization [1, 2]. Low concentrations of LatA result in rapid (tens of seconds) disassembly of actin filaments in animal [3] and yeast cells [2]. Depolymerization is usually assumed to result from sequestration of actin monomers. Our observations of single muscle actin filaments by TIRF microscopy showed that LatA bound ATP-actin monomers with a higher affinity (Kd = 0.1 μM) than ADP-Pi-actin (Kd = 0.4 μM) or ADP-actin (Kd = 4.7 μM). LatA also slowly severed filaments and increased the depolymerization rate at both ends of filaments freshly assembled from ATP-actin to the rates of ADP-actin. This rate plateaued at LatA concentrations >60 μM. LatA did not change the depolymerization rates of ADP- actin filaments or ADP-Pi-actin filaments generated with 160 mM phosphate in the buffer. LatA did not increase the rate of phosphate release from bulk samples of filaments assembled from ATP-actin. Thermodynamic analysis showed that LatA binds weakly to actin filaments with a Kd >100 μM. We propose that concentrations of LatA much lower than this Kd promote phosphate dissociation only from both ends of filaments, resulting in depolymerization limited by the rate of ADP-actin dissociation. Thus, one must consider both rapid actin depolymerization and severing in addition to sequestering actin monomers when interpreting the effects of LatA on cells.
eTOC blurb
Fujiwara et al. address how Latrunculin-A (LatA) depolymerizes actin filaments. Observations of single filaments revealed that LatA not only sequesters actin monomers but also promotes subunit dissociation from the ends of filaments assembled from ATP-actin monomers by rapid “aging” due to fast phosphate dissociation from terminal ADP-Pi-subunits.
RESULTS AND DISCUSSION
Control Experiments on Effects of Dye Labels on Actin Assembly and Disassembly
We investigated how LatA affects the elongation and depolymerization of muscle actin filaments by TIRF microscopy using actin labeled with Oregon Green conjugated to Cys374 (OG) [4] or Alex488 conjugated to a lysine (Alexa488) [6, 8, 33]. Labeling is known to influence actin polymerization [4, 9], so we carried out control experiments to understand more thoroughly how labeling might influence our experiments. We confirmed that both of these labels as well rhodamine on Cys374 (TMR) influence the rates of association of MgATP-actin monomers and of dissociation of subunits assembled from Mg-ATP-actin monomers depending on the fraction of labeled subunits in the monomer pool and filaments (Figures S1 and S2). Of the three labels, TMR had the strongest effect [9], since even 10% slowed elongation and increased the rate of depolymerization to >5 subunits/s. For example, 30% TMR-actin reduced the association rate constant by more than half, while 30% Alexa488- or OG-actin elongated at the rates measured by electron microscopy without labels [10].
Actin depolymerization was initiated by washing away free actin monomer with the polymerization buffer containing ATP. After washing away monomers from freshly polymerized ATP-actin filaments, the initial rate of depolymerization was slower with high molar fractions of actin labeled with Oregon green or TMR (red filled symbols in Figures S1G-S1I). ATP-actin filaments with low molar fractions of the three dyes depolymerized differently (Figure S2). Filaments of OG-Mg-ATP-actin depolymerized at a slow, constant rate over 400 s of observation (Figure S2B). The rate was similar to the dissociation rate constant of ATP-actin measured by extrapolation of plots of elongation rate vs. the concentration of OG-ATP-actin (Figure S1B) or unlabeled actin [10] to zero actin. Filaments assembled from Alexa488-Mg-ATP-actin depolymerized slowly at first, but the disassembly rate increased over time up to the rate of ADP-actin subunits (Figure S2A) as reported by Jégou and coworkers [8]. This progressive increase in the rate of depolymerization requires dissociation of phosphate, since phosphate in the buffer slows the depolymerization of both ends of Alexa488-Mg-ADP-actin filaments in a concentration-dependent fashion [6]. The slow, initial rates of depolymerization barbed ends of filaments assembled from Alexa488Mg-actin were in a range from −2.5 to −3.4 subunits/s over a range of fractions of labeled actin (Figures S1A and S1G), while the dissociation rate constant from linear fittings of positive elongation rates vs. the concentration of Alexa488-Mg-actin was −1.3 subunits/s (Figure S1A), similar to the rate constant for unlabeled Mg-ATP-actin (1.4 subunits/s) measured by electron microscopy [10]. This is consistent with ATP-actin and ADP-Pi-actin being the species dissociating during the initial slow phase of depolymerization of Alexa488Mg-ATP-actin filaments, followed by faster depolymerization as Pi is released rapidly from barbed-ends of Alexa488-Mg-ATP-actin filaments, as shown by Jégou and coworkers [8]. Filaments labeled with TMR depolymerized at a constant rate of ~−5 subunits/s immediately after washing away monomers (Figure S2C).
Thus, dye labels have different effects on the rate of depolymerization of filaments assembled from Mg-ATP-actin, likely due to their influence on phosphate dissociation. However, we do not know how the fraction of ADP-Pi-actin in unlabeled filaments influences the depolymerization rate. Therefore, we tested LatA on both OG- and Alexa488-actin and found similar results but used just one labeled actin for certain experiments.
LatA Binds Actin Monomers and Inhibits Actin Filament Elongation
We investigated how LatA binds ATP-, ADP-Pi- and ADP-actin monomers by using TIRF microscopy to measure how LatA inhibits filament elongation (Figure 1). We polymerized muscle-actin monomers in 10 mM imidazole buffer (pH 7) with 2 mM Mg2+, 50 mM KCl and 1 mM EGTA to sequester Ca2+. For experiments on Mg-ATP-actin we used 20% of OG-actin. For experiments with ADP-Pi- and ADP-actin and some experiments on Mg-ATP-actin, we use 20% Alexa488-actin, because of the low fluorescence of OG-ADP-actin. We used higher concentrations of ADP-actin to have useful numbers of filaments in an observation field. A low density of NEM-myosin tethered actin filaments to the slide. The filaments pivoted about these attachment points from which we measured contour lengths to both ends of each filament [4]. Barbed ends grew much faster than pointed ends, which we could not measure as accurately under our conditions (Figure 1A).
Figure 1. Effect of LatA on elongation of filaments by ATP-, ADP- and ADP-Pi-actin monomers.
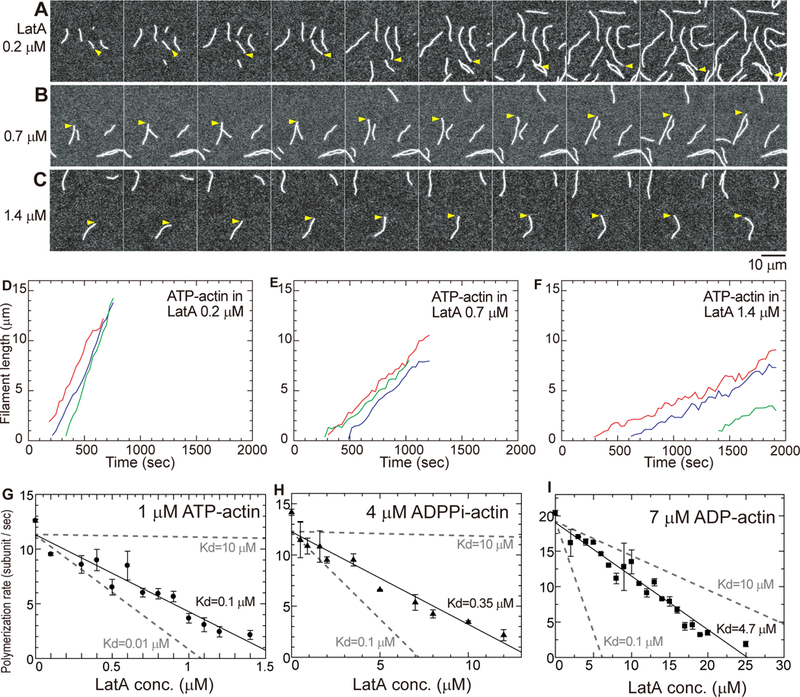
(A-C) Time series (time in upper left corner) of TIRF micrographs of 1 μM MgATP-actin (20% Oregon green labeled) polymerizing in the presence of a range of concentrations of LatA: (A) 0.2 μM; (B) 0.7 μM; and (C) 1.4 μM. Yellow arrowheads mark the barbed end of a typical filament in each sample. The polymerization buffer consisted of 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 10 mM imidazole, pH 7.0, 100 mM DTT, 0.2 mM ATP, 0.5% (wt/vol) methylcellulose (1500 cP). Scale bar is 10 μm. Time-lapse movies are published as supporting information on the journal web site (Video S1). The reactions were started at time 0 by mixing Mg-ATP-actin monomers (20% labeled with Oregon green) and LatA in polymerization buffer. (D-F) Time course of the elongation of the barbed-ends of 3 randomly-chosen (indicated with different colors) actin filaments by 1 μM Mg-ATP-actin monomers in (D) 0.2 μM LatA, (E) 0.7 μM LatA and (F) 1.4 μM LatA. (G-I) Dependence of mean elongation rates on the concentration of LatA. Polymerization rates were measured as the mode of the ensemble length changes of 6–13 typical actin filaments during 30 s intervals for ~800 s at each LatA concentration. The error bars are standard deviations (SD) of the average polymerization rates of individual filaments. Kds from experiments (black) were estimated from linear fitting (solid lines) as in Coué et al. [1]. The gray dashed lines are the theoretical linear fittings with Kds of 0.1 or 10 μM. (G) 1 μM Mg-ATP-actin; (H) 4 μM MgADP-Pi-actin (20% Alexa Fluor 488 labeled-ADP-actin with 160 mM sodium phosphate in polymerization buffer); and (I) 7 μM Mg-ADP-actin (20% Alexa Fluor 488 labeled with 160 mM sodium sulfate in polymerization buffer). See also Video S1.
LatA has two effects on polymerization of Mg-ATP-actin, both expected from the monomer sequestration mechanism [1]. First, LatA reduced the number of actin filaments, indicating that it slows spontaneous nucleation (Figures 1A–1C), which is very sensitive to the monomer concentration. Second, LatA reduced the rate of elongation of barbed-ends (Figures 1D, 1E and 1F) consistent with previous observations on bulk samples [5]. These two effects account for why the 1:1 complex of LatA with actin monomers does not polymerize [1, 5]. LatA also inhibited the elongation of filaments by ADP-Pi- or ADP-actin monomers (Figures 1H and 1I).
We estimated Kds for LatA binding to ATP-, ADP-Pi- or ADP-actin monomers from the dependence of the elongation rates at the barbed end on the concentrations of LatA (Figures 1G, 1H and 1I). The analysis assumes that free actin monomers associate with filament ends with the normal rate constants [6], while actin monomers bound to LatA do not contribute to elongation. Thus, elongation rates provide a direct measure of the free and sequestered actin monomer concentrations over a range of LatA concentrations.
The Kds for LatA binding were 0.1 μM to Mg-ATP-actin monomers, 0.4 μM to Mg-ADP-Pi-actin monomers and 4.7 μM to Mg-ADP-actin monomers. The affinity for Mg-ATP-actin is similar to previous measurements with less KCl in the buffer [1, 5]. Our experiments show for the first time that the bound nucleotide and phosphate strongly influence the affinity of actin monomers for LatA. The thermodynamics are clear. Hydrolysis of bound ATP reduces the affinity of actin monomers for LatA by 4-fold. Subsequent dissociation of the γ-phosphate from Mg-ADP-Pi-actin monomers reduces the affinity of Mg-ADP-actin monomers for LatA by 12-fold (0.4 to 4.7μM), so by detailed balance, bound LatA increases the affinity of Mg-ADP-actin monomers for phosphate by 12-fold (60 to 5.1 mM, left side of Figure 4 below). Knowledge of these reactions of nucleotides and phosphate with actin monomers is required to understand LatA binding to actin filaments, as explained below.
Figure 4. Thermodynamics and kinetics of LatA interactions with actin monomers, actin filament barbed ends and actin filament interior subunits with bound ATP, ADPPi or ADP.
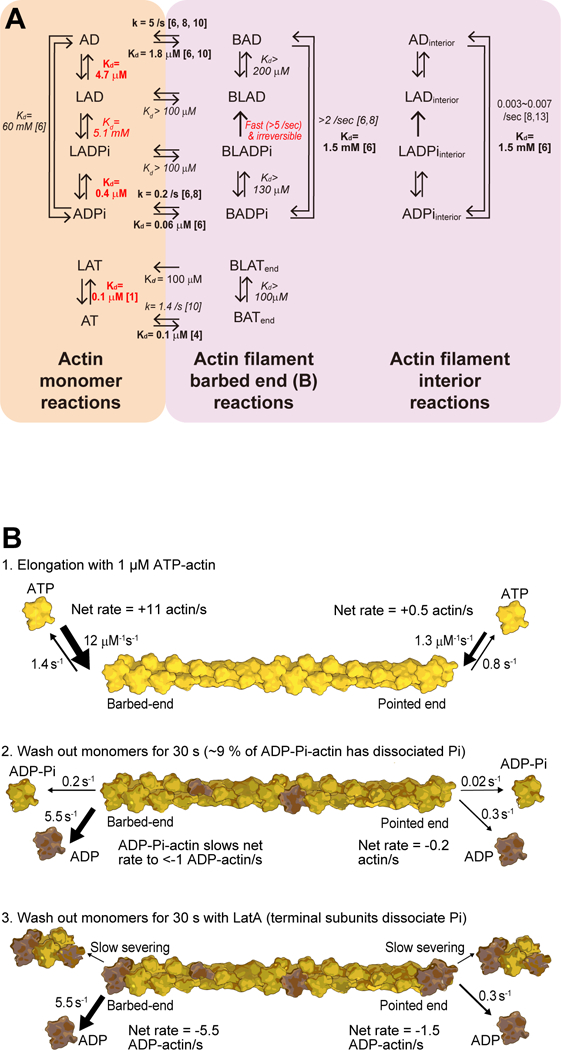
(A) Thermodynamic cycles. Abbreviations: A, actin molecule; L, Latrunculin A; T, ATP; D, ADP: Pi, phosphate; B, barbed end. Numbers next to arrows are rate constants (k) or equilibrium dissociation constants (Kd). (Left) Monomeric actin reactions. (Middle) Reactions on actin filament barbed ends. (Right) Reactions of interior subunits in actin filaments. Experimentally measured parameters are shown in bold type with reference numbers to previous work or in red for this study. Other parameters shown in italics were calculated using detailed balance. (B) Drawings of reactions that occur during (1) elongation by ATP-actin monomers, (2) after allowing ATP-actin filaments to age for 60 s and (3) after allowing ATP-actin filaments to age for 60 s in the presence of 60 μM LatA. Three shades of yellow show ATP-actin, ADP-Pi-actin and ADP-actin subunits.
LatA Accelerates Depolymerization of ATP-actin Filaments
We used TIRF microscopy to compare the effects of LatA on depolymerization of filaments assembled from muscle Mg-ATP-actin labeled with Oregon Green on Cys374 or Alexa488 on a lysine residue (Figure 2 and S3). Mg-ATP-actin is the physiologically relevant species, since most of the unpolymerized actin has bound ATP [7]. Each experiment began by observing the elongation of the filaments to establish their polarity. Then we washed free actin monomers out of the chamber with polymerization buffer alone or buffer containing a range of concentrations of LatA at the times indicated in Figures 2A, 2B, and 2C. The initial rate of depolymerization of Alexa488-actin filaments was faster (−1.7 subunits/s) than OG-actin filaments (−0.3 subunits/s) (Figure S2).
Figure 2. Effect of LatA on the dissociation of subunits from the ends of filaments assembled from Mg-ATP-actin.
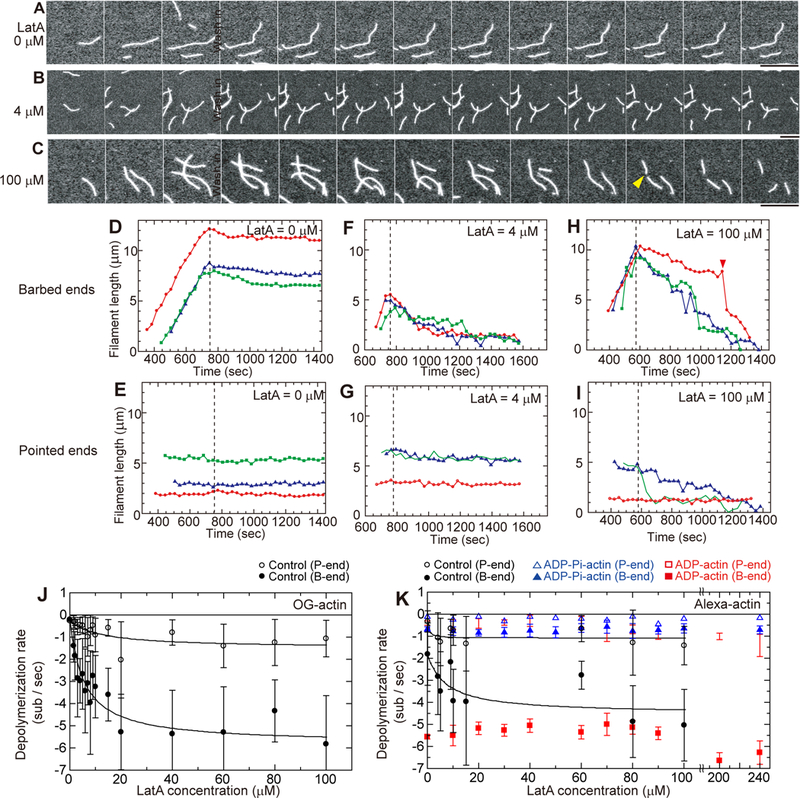
(A-C) Time series (time on each panel) of TIRF micrographs during assembly and disassembly of actin filaments. At time 0, 1 μM Mg-ATPactin (20% Oregon green labeled) was mixed with polymerization buffer, loaded into the flow cell and observed to identify the rapidly growing barbed ends and the slowly growing pointed ends. At 760 s, free actin was washed from the chamber with polymerization buffer containing (A) no LatA, (B) 4 μM LatA or (C) 100 μM LatA. The yellow arrowhead in C marks a severing event at 1250 s. Scale bars = 10 μm. Movies are published as supporting information on the journal web site (Videos S2 and S3). (D-I) Time courses of length changes at the ends of samples of 3 typical actin filaments. Actin monomers were washed out at the times indicated by the vertical dashed lines and replaced by polymerization buffer with a range of LatA concentrations. (D, F, H) Barbed ends. (E, G, I) Pointed ends. (D and E) no LatA, (F and G) 4 μM LatA and (H and I) 100 μM LatA. (J and K) Dependence of the rates of dissociation of actin from barbed ends (filled circles) and pointed ends (open circles) on the concentration of LatA in polymerization buffer. Mean initial rates were determined from linear fits of lengths vs. time of each filament during the first 90 s of depolymerization. Error bars are standard deviations of the means of the initial rates. (J) Depolymerization rates of freshly polymerized Mg-ATP-actin with 20% Mg-ATP-OG-actin in polymerization buffer. (K) Depolymerization rates of filaments assembled from 20% Alexa488-actin monomers with three different bound nucleotides: (black symbols) Mg-ATP-actin filaments; (red symbols) Mg-ADP-actin filaments; and (blue symbols) Mg-ADP-Pi-actin filaments (ADPactin with 160 mM sodium phosphate in polymerization buffer). See also STAR Methods, Figures S1, S2, S3, Video S2 and S3.
LatA increased the initial rate of depolymerization of the barbed ends of filaments assembled from either Mg-ATP-OG-actin and Mg-ATP-Alexa488-actin with similar concentration-dependence and plateaus at LatA concentrations >60 μM (Figure 2J) at rates of ~−5.5 subunits/s (Figures 2J and 2K). This maximum depolymerization rate with LatA is the same as the rate constant for dissociation of ADP-actin, the maximum known intrinsic depolymerization rate. The LatA concentration-dependence for depolymerizing barbed ends of ATP-actin filaments gave apparent Kds of 6.0 μM OG-actin filaments and 7.3 μM for Alexa488-actin filaments (Figure 2J), similar to the affinity of LatA for Mg-ADP-actin monomers. This behavior establishes that LatA binds directly to actin filaments, although it does not identify the location along the filaments.
The effects of LatA on the depolymerization of pointed ends of ATP-actin filaments were harder to measure owing to the low rates, but the effects were similar to barbed ends (Figures 2E, 2G, 2I, 2J and 2K). LatA increased the rate of depolymerization of pointed ends of both Alexa488-actin filaments and OG-actin filaments paragraph from about −0.2 to −1.4 subunits/s with apparent Kds between 5 and 10 μM.
Effects of Bound Nucleotide on Actin Depolymerization in the Presence of LatA
We used Mg-ADP-Alexa488-actin to test the effects of LatA on depolymerization of ADP- and ADP-Pi-actin, because the fluorescence of ADP-OG-actin filaments is low. We generated ADP-Pi-actin with 160 mM phosphate in the buffer, a saturating concentration at both ends in our previous work [6].
LatA had minimal effects on the depolymerization of either end of filaments of MgADP-Alexa488-actin or Mg-ADP-Pi-Alexa488-actin (Figure 2K). Barbed ends of Mg-ADP-Alexa488-actin filaments shortened at −5.5 subunits/s at LatA concentrations from 0 to 100 μM. The rate was slightly faster at very high concentrations of LatA, but the difference was not statistically significant. Pointed ends of ADP-Alexa488-actin filaments shortened at ~−0.5 subunit/s at LatA concentrations from 0 to 50 μM and slightly faster at very high concentrations of LatA. Barbed ends of Mg-ADP-Pi-Alexa488-actin filaments shortened at ~1 subunit/s at LatA concentrations ranging from 0 to 100 μM (Figure 2K). Pointed ends of filaments of ADP-Pi-Alexa488-actin shortened very slowly at all LatA concentrations tested from 0 to 240 μM. Thus, a high concentration (160 mM) of phosphate slows the depolymerization of ADP-Alexa488-actin filaments even in the presence of LatA.
These experiments show that dissociation of the γ-phosphate is required for LatA to increase the rate of subunit dissociation from barbed ends to the rate of ADP-actin dissociation. In other words, LatA appears to accelerate aging of ATP-actin filaments.
LatA Severs Actin Filaments
In high concentrations of LatA (>40 μM) large segments of filaments (i.e. over 1.5 μm long) occasionally disappeared from the ends of depolymerizing filaments between the frames of our movies (Figures 2C and 2H, arrowheads, Video S3). We interpret these abrupt changes in length as severing events. Severing of short segments near the ends of filaments would be hard to distinguish from depolymerization of subunits from the end. If these small severing events dominated during depolymerization, the rate of depolymerization would increase without saturation at the concentrations of LatA tested owing to the low affinity of LatA for the middle of filaments. However, the actin depolymerization rate plateaued at low LatA concentrations (Figures 2J and 2K), indicating that small severing events do not affect the rates of depolymerization. In plots of the distributions of length changes in 30 s intervals, most events fall within a Gaussian distribution centered on the average rate of depolymerization. Severing events appear as large negative length changes outside Gaussian distributions of length changes in high LatA concentrations (Figure 3C, arrows). These severing events are rare (Figure 3D) with actin filaments alone (Figure 3A) or at LatA concentrations <40 μM (Figure 3B), but the frequency of severing increased with the concentration of LatA (Figure 3D). In the presence of 100 μM LatA severing accounted for 15% of the total length changes. Severing also contributed to the overall rate of actin depolymerization by increasing the number of ends. Severing is a second line of evidence that LatA binds directly to actin filaments.
Figure 3. Effects of LatA on actin filament severing and influence of actin filament capping and Pi release on depolymerization.
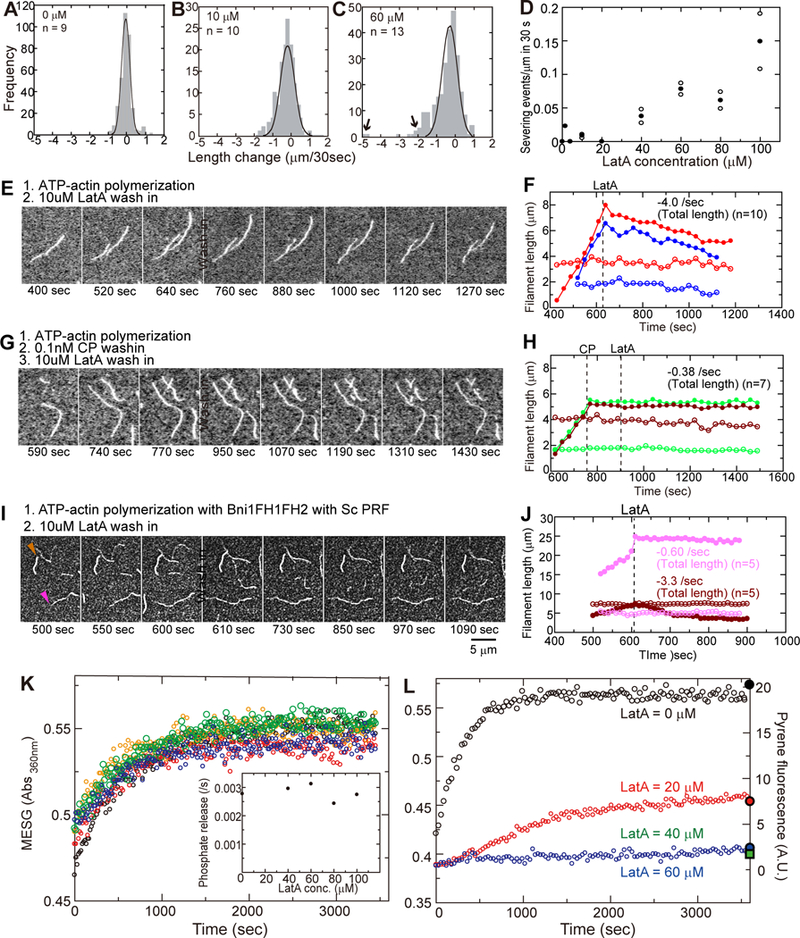
(A-C) Histograms of the frequencies of length changes at barbed ends during 30 s intervals with a range of concentrations of LatA. Large negative length changes outside the Gaussian distributions were due to severing (C, arrows). (D) Dependence of the severing rate on the concentration of LatA. () Severing rates in two or three independent experiments at each concentration of LatA with 7–20 actin filaments in the observation fields. (●) Average severing rates. (E, G, I) Time series (time at the bottom of each panel) of TIRF micrographs during assembly and disassembly of actin filaments. At time zero 1 μM Mg-ATP-actin (20% Oregon green labeled) was mixed with the polymerization buffer, loaded into the flow cell and observed to identify rapidly growing barbed ends and slowly growing pointed ends. At the times indicated by “wash in” the solution in the chamber was changed. (F, H, J) Time courses of filament lengths from a fiducial mark to barbed (filled) and pointed-ends (open) of 2 typical actin filaments (color coordinated). (E, F) Polymerization of 1 μM Mg-ATP-actin (20% Oregon green labeled) alone. At the indicated times, free actin was washed from the chamber with polymerization buffer containing 10 μM LatA. (G, H) At the left dashed line in (H) free actin was washed from the chamber with polymerization buffer and replaced with 0.1 μM capping protein (CP) in polymerization buffer. After incubating for 2 min, 10 μM LatA was loaded in the chamber at the right dashed line. (I, J) A mixture of 0.5 μM Mg-ATP-actin (20% Oregon green labeled), 5 nM formin (Bni1FH1FH2) and 1 μM Sc profilin in polymerization buffer was loaded into the observation chamber. At the time indicated by the dashed line in (J), free actin monomers, formins and profilin were washed from the chamber with polymerization buffer containing 10 μM LatA. Formin-bound filaments (barbed end marked with a pink arrowhead) are dimmer than filaments with free barbed ends (barbed end marked with a brown arrowhead). Colors in (J) are coordinated with (I). (K) Time course of the release of phosphate from filaments rapidly polymerized from unlabeled 40 μM Mg-ATP-actin in 2 min and then diluted at time zero to 20 μM into polymerization buffer with a range of concentrations of LatA: 0 (black); 40 μM (red); 60 μM (orange); 80 μM (green); and 100 μM (blue). (Inset) Observed rate constants for the release of phosphate vs. LatA concentration. (L) Time course of the release of phosphate during the polymerization of 20 μM Mg-ATPactin followed by the absorbance of MESG at 360 nm. Colors indicate the micromolar concentrations of LatA: 0 (black); 20 (red); 40 (green); and 60 (blue). The filled symbols on the right Y-axis are the fluorescence of pyrenyl-actin (5% label) measured after 1 h of polymerization. See also STAR Methods.
LatA does not increase the rate of depolymerization from capped barbed ends
Capping actin filament barbed ends with either mouse capping protein or S. cerevisiae formin Bni1(FH1FH2)p prevented LatA from rapid depolymerizing filaments freshly polymerized from ATP-OG-actin monomers (Figures 3E–3J). LatA at 10 μM increased the rate of depolymerization of barbed-ends of control filaments without capping protein (Figures 3E and 3F) but not when the barbed ends were blocked with capping protein (Figures 3G and 3H). Similarly, after polymerizing Mg-ATP-OG-actin in the presence of 5 nM formin and 1 μM profilin, addition of LatA did not stimulate depolymerization of barbed ends associated with the formin (pink arrowhead in Figures 3I and 3J). Owing to the lower affinity of profilin for OG-labeled actin monomers than unlabeled monomers, the actin filaments associated with a formin were dimmer than filaments without a formin [11]. The bright actin filaments with free barbed ends served as an internal control in these experiments and depolymerized rapidly in 10 μM LatA (brown arrowhead). In the presence of 10 μM Lat A the total lengths of capped actin filaments shortened at about the rate of pointed ends: 0.4 subunits/s with capping protein and 0.6 subunits/s with formin. We obtained similar results with S. pombe formin Cdc12p(FH1FH2)p.
These experiments establish that LatA does not dissociate capping protein or formin FH2 domains from barbed ends. Thus, the main effect of LatA on actin depolymerization is dissociating subunits from free barbed ends, rather severing off small segments of filaments. However, these experiments do not distinguish whether rapid depolymerization of both ends comes from effects of LatA on the ends of filaments or along their whole lengths. Therefore, we tested if LatA influences the dissociation of phosphate from ADP-Pi-actin filaments.
LatA does not Promote Phosphate Release from the Bulk of ADP-Pi-actin Filaments
To explore how LatA depolymerizes actin filaments assembled from Mg-ATP-actin, we tested the effect of LatA on the time course of phosphate (Pi) release from the polymerized ADP-Pi-actin intermediate with a spectroscopic assay. We used a high concentration (40 μM) of ATP-actin monomers to assure rapid polymerization in 2 min during which virtually all of the subunits hydrolyzed the bound ATP. Owing to slow dissociation of the γ-phosphate [12], most subunits would have bound ADP-Pi. Then we monitored Pi release from the ADP-Piactin filaments with a 2-amino-6-mercapto-7-methyl purine riboside-phosphorylase (MESG) assay [12, 13]. Without LatA freshly polymerized ATP-actin released Pi over ~1000 s similar to previous measurements [12, 13]. No concentration of LatA up to 100 μM increased the rate of Pi-release from filaments freshly polymerized from ATP-actin (Figure 3K, inset). Thus, LatA does not promote phosphate dissociation from the bulk of the polymerized ADP-Pi-actin subunits.
On the other hand, high concentrations of LatA completely inhibited the spontaneous polymerization of ATP-actin monomers and phosphate release (Figure 3L) owing to monomer sequestration [1, 5]. With 20 μM LatA some actin monomers were free to polymerize slowly and released a small amount of phosphate.
LatA preferentially binds monomeric, rather than filamentous actin
Our experiments show that LatA not only sequesters actin monomers but also increases the rate of actin depolymerization. LatA binds in the nucleotide-binding cleft of the actin monomer without changing its conformation [14]. Extensive interactions of LatA with the sides of the nucleotide-binding cleft [2, 14] likely restrict the scissor-like motion that flattens actin subunits as required for incorporation into filaments [15, 16]. Locking the actin molecule in the monomeric conformation explains both sequestration and inhibition of the dissociation of a bound nucleotide from actin monomers with or without profilin [5]. The complex of LatA with ATP-actin monomers [14] resembles ATP-actin more than ADP-actin [17], so LatA does not move the “sensor” residues.
The incompatibility of LatA binding and assembly also explains how LatA severs actin filaments. Interactions of LatA with the sides of the nucleotide cleft will likely favor the monomeric conformation even in a filament, so LatA bound to a single subunit will destabilize the filament locally and lead to severing. The rare severing events require high concentrations of LatA owing to the very low affinity of LatA for polymerized actin. The interior subunits in actin filaments are not only flattened but the four surrounding subunits are likely to limit opening of the cleft to bind LatA. This unfavorable reaction of LatA with filaments is similar to the exceedingly low rates of nucleotide exchange in filaments [18]. Thus, severing by LatA caused by a subunit conformational change differs from severing proteins such as gelsolin and the WH2 domain-containing protein Cordon-Bleu, which sterically disrupt interactions between actin subunits [19–21].
Reciprocally, the thermodynamic cycles in Figure 4 illustrate that the flattened conformation of polymerized actin is very unfavorable for LatA binding. We know the affinity of LatA for ATP-, ADP-Pi- and ADP-actin monomers and the affinities of each of these nucleotide states for the ends of actin filaments (Figure 4 shows only barbed ends). We observe that none of the monomeric actin species (ATP-, ADP-Pi- or ADP-) with bound LatA elongates actin filaments, so the affinities of the ends of the filaments for actin monomers with bound LatA must be very low. Of course, LatA-actin complexes must interact transiently with the filament ends, but then dissociate rapidly, reflecting their low affinities. Thus, LatA-actin complexes do not contribute to or interfere with elongation by free actin monomers. Assuming that the Kds for the association of the complex of LatA:actin monomers are >100 μM, then by detailed balance the affinities of the barbed ends for LatA must also be very weak with Kds >100 μM.
Fast Depolymerization of ATP-actin Filaments Suggests LatA Rapidly Dissociates Phosphate from the Ends of Filaments
Our observations suggest that LatA increases dissociation of subunits from the ends of filaments assembled from ATP-actin by promoting dissociation of the γ-phosphate from terminal subunits with bound ADP and Pi followed by dissociation at the rate characteristic of ADP-actin. First, LatA increased the rate of subunit dissociation from both ends of freshly polymerized ATP-actin filaments up to the rates of ADP-actin subunits, regardless of whether the filaments were assembled from slowly depolymerizing OG-ATP-actin or rapidly depolymerizing Alexa488-ATP-actin. Second, LatA had no effect on dissociation of subunits from either end of filaments of ADP-actin or ADP-Pi-actin (in the presence of 160 mM phosphate) verifying that dissociation of the γ-phosphate is part of the mechanism.
However, the thermodynamics explained above raise questions about this hypothesis. First, the dependence of the depolymerization reactions on the concentration of LatA suggested a Kd of <10 μM (Figures 2J and 2K), while the thermodynamic cycles in Figure 4 indicate that the Kds for LatA binding to barbed ends are >200 μM for ADP-actin filaments and >130 μM for ADP-Pi-actin filaments.
Analysis of the reactions in actin depolymerization experiments suggests a possible mechanism to reconcile these differences. We propose that LatA binding to subunits at or near the ends of the filaments results in rapid dissociation of the γ-phosphate from terminal ADP-Pi-actin subunits (the reaction BLADPi → BLAD in Figure 4). Given the very low affinity of LatA for ADP-actin filaments (>200 μM), LatA then dissociates rapidly generating an ADP-actin subunit on the end of the filament (BAD), which dissociates from the end at its normal rate (BAD → AD). LatA does not increase the rate of γ-phosphate dissociation from interior subunits in actin filaments, either because of the slower binding of LatA to interior subunits or because of the very slow rate of γ-phosphate dissociation from interior subunits.
A local effect of LatA on the ends of filaments is consistent with our previous work [6] and that of Jégou et al. [8] showing that phosphate dissociates much faster from terminal subunits (>2/s) than from subunits in the middle of filaments (0.003/s) [12]. Loose binding of the γ-phosphate to terminal subunits of actin filaments allows LatA binding to accelerate γphosphate dissociation. Thus, an unknown, small number of ADP-Pi-actin subunits at the ends of freshly polymerized filaments must differ in conformation from the bulk of the ADPPi-actin subunits in the middle of filaments.
Note that LatA interacts differently with monomeric and filamentous actin in two ways. First, despite its high affinity for ADP-Pi-actin monomers (Kd of 0.4 μM), LatA does not affect depolymerization of ADP-Pi-actin filaments when the buffer contains a saturating concentration of phosphate. Second, whereas LatA promotes phosphate binding to ADP-actin monomers, it speeds phosphate dissociation from ADP-Pi-actin subunits at the ends of filaments.
Thus, the structures of actin monomers and subunits inside and at the ends of filaments must all differ as observed in low-resolution reconstructions of pointed ends from electron micrographs [22] and our experiments on profilin and phosphate binding to actin monomers and filament barbed ends [6, 15]. Oda et al. [15] also suggested that the conformation of terminal subunits might differ from the bulk of the filament.
Mechanism of Fast Depolymerization of ATP-actin Filaments by LatA
Given the very low affinity of LatA for filaments, how can low concentrations of LatA promote phosphate release and depolymerization? One factor is that dissociation of phosphate is essentially irreversible in these experiments, since the buffer contains little phosphate. If phosphate release after LatA binding (BLADPi → BLAD) is faster than the rate of LatA binding to the end of the filament (BADPi ⇆ BLADPi), then the effect of LatA on the rate of conversion of ADP-Pi-actin to ADP-actin saturates when the rate of phosphate dissociation exceeds the rate of ADP-actin dissociation from the end of the filament (5.4 subunits/s at the barbed end and 0.2 subunits/s at the pointed end). Our experiment on the concentration dependence of LatA depolymerization involves 4 sequential reactions (BADPi ⇆ BLADPi → BLAD ⇆ BAD ⇆ AD), with phosphate release (BLADPi → BLAD) being irreversible. Therefore, the experiment measures the rate of LatA binding required to drive two first order reactions that are ultimately limited by the rate of ADP-actin dissociation from the end of the filament. Phosphate dissociation from terminal subunits limits disassembly, because a low fraction of ADP-Pi-actin subunits slows depolymerization disproportionately [6].
Similarities and differences in the mechanisms of LatA and profilin
The effects of LatA on filament dynamics are similar in two ways to those observed for profilin, a protein that inhibits some interactions of actin monomers with filaments and, at high concentrations, depolymerizes actin filaments. First, both LatA and profilin stimulate phosphate release from barbed ends [8]. Second, both LatA and profilin bind monomers more tightly than barbed ends. However, LatA prefers both ATP-actin monomers and ATP-actin barbed ends, while profilin binds most tightly to ATP-actin monomers and ADP-barbed ends [8, 25]. This difference suggests that that the profilin-binding sites of terminal ADP-actin subunits resemble those of actin monomers more closely than the sites on ATP-bound subunits.
In spite of these similarities, LatA and profilin promote filament depolymerization by different mechanisms, likely owing to their different binding sites. Whereas LatA promotes the dissociation of subunits from barbed ends at the typical ADP-actin rate, depolymerization in the presence of profilin can dramatically exceed this rate. This may be due to profilin binding to the terminal subunit causing a conformational change in actin that compromises its association with the neighboring subunits.
LatA Promotes Filament Disassembly in Cells by Accelerated Aging
In addition to confirming that LatA depletes the pool of actin monomers, slows all assembly reactions and thus stimulates net depolymerization, we have added two mechanisms by which LatA disassembles actin filaments in cells. First, promoting phosphate release from the ends of filaments accelerates their aging and increases the rates of subunit dissociation from both ends of filaments. We simulated the effects of LatA on depolymerization using equations and rates of phosphate release from Jegou et al., 2011 [8]. The rate of depolymerization increases by 60% if LatA increases phosphate release to 5/s or more than doubles when phosphate is released at 20/s. Second, severing can increase the numbers of filament ends for depolymerization.
Enhancing depolymerization from both ends accelerates disassembly of any uncapped filaments. One concern about this hypothesis is that many cellular actin filaments appear to be capped by capping protein [26]. Formin FH2 domains also prevent rapid depolymerization by LatA. On the other hand, regulatory proteins such as CARMIL may attenuate capping and generate free barbed ends [27]. Furthermore, spectroscopic observations on fish keratocyte lamellipodia reveled an abundance of short, diffusing actin filaments, many of which appear to be uncapped [28]. Experiments with cofilin mutants in fission yeast also suggest short filaments are a major intermediate in recycling actin subunits [29]. Short filaments are targets for LatA, and, given the high concentration of ends, may help explain how modest increases in the rate of subunit dissociation by LatA might rapidly depolymerize actin filaments in cells.
Given the low affinity of LatA for the bulk of filaments, severing by LatA plays a minor role with purified actin, but in cells side-binding proteins might influence severing by LatA. Monomer sequestration, rapid depolymerization and severing should be considered when interpreting the effects of LatA on the time course of actin filament depolymerization in cells [2, 30].
STAR METHODS
Detailed methods are provided in the online version of this paper and include the following:
Experimental model and subject details
Proteins and protein labeling
Method details
TIRF assay to measure actin filament lengths and depolymerization rates
Quantification and statistical analysis
Estimation of the affinity of LatA for actin monomers
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ikuko Fujiwara (fujiwara.ikuko@nitech.ac.jp).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Proteins and protein labeling
Actin was prepared from an acetone powder of rabbit skeletal muscle by one cycle of polymerization and depolymerization [31]. It was gel filtered on a column of Sephacryl S300 and stored in buffer G (2 mM Tris, pH 8.0, 0.2 mM ATP, 0.1 mM CaCl2, 0.5 mM dithiothreitol (DTT)) at 4°C. The cation bound to monomeric actin was converted from Ca2+ to Mg2+ right before each sample was prepared for observation. Monomeric actin in buffer G was mixed with 10x ME buffer (final concentrations: 1 mM EGTA, 0.1 mM MgCl2) and incubated on ice for 2 min [32]. Mg-ADP actin monomers were prepared by treating MgATP actin with soluble hexokinase and glucose [10] and used within 7 h. We labeled actin on cysteine 374 with Oregon Green 488 succinimidyl ester (Molecular Probes, ThermoFisher) as described [4] for use in experiments with ATP-actin. For experiments with ADP- and ADPPi-actin monomers, we labeled ATP-actin on a lysine with Alexa Fluor 488 succinimidyl ester (Molecular Probes, ThermoFisher) as described [33] before converting to ADP-actin, because the fluorescence emission is stronger.
All polymerization and depolymerization assays were carried out at room temperature in polymerization buffer consisting of 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 10 mM imidazole, pH 7.0, 100 mM DTT, 0.2 mM ATP or ADP, 0.5 % (wt/vol) methylcellulose (1500 cP from Sigma-Aldrich). For experiments with Mg-ADP-Pi-actin monomers and filaments we added 160 mM sodium phosphate (pH 7.0) to the buffer. Control samples for these experiments had 160 mM sodium sulfate in the buffer with of ADP-actin monomers and filaments. Latrunculin-A (Sigma-Aldrich) was dissolved in dimethyl sulfoxide at a concentration of 10–20 mM and diluted into polymerization buffer.
METHOD DETAILS
TIRF assay to measure actin filament lengths and depolymerization rates
Observation chambers were made by assembling a coverslip (24 × 50-mm, No. 1) and a flint glass slide (25 × 75 × 1-mm). Glasses were cleaned by sonicating for 1 to 3 h of single cycle of alkali and acid wash and rinsed thoroughly in double distilled (dd) water. Clean slides were stored in absolute ethanol and were used within two weeks [4, 6]. A cleaned coverslip and flint glass slide were placed perpendicular and attached by placing 2 parafilm strips of about 5 mm width in between. The parafilm strips were compressed by hand pressure on the top of the slide to seal the sides of the chamber. The chamber was flamed briefly and cooled to stick the protruding parafilm strips to the open coverslip surface. Chambers were pretreated with 20 μL of 50 nM N-ethylmaleimide (NEM)-inactivated skeletal muscle myosin [4] at room temperature. Unbound NEM-myosin was washed with KMEI buffer (50 mM KCl, 1 mM EGTA, 1 mM MgCl2; 10 mM imidazole, pH 7.0). Actin in KMEI buffer was immediately added into the chamber to initiate polymerization. Once a useful density of filaments had grown on the slide, depolymerization was initiated by washing the chamber with polymerization buffer without actin monomers and with a range of concentrations of LatA. Samples were kept in the dark except when images were taken at 30 s intervals. For capping assays, the chamber was washed with polymerization buffer with 0.1 μM mouse capping protein [4] followed by introduction of 10 μM LatA in polymerization buffer. For formin experiments filaments were polymerized with 5 nM of a fragment of S. cerevisiae (Sc) formin Bni1p consisting of its FH1 and FH2 domains (Bni1(FH1FH2)p) and 1 μM Sc profilin [24], followed by washing with polymerization buffer with 10 μM LatA.
By tracing length of individual actin filaments using ImageJ software (NIH), we measured rates of depolymerization from plots of filament length vs. time (Figures 2 and S1) [4] or by recording the length change during each 30 s interval between images and plotting these changes in histograms (Figures 3A–3C) [6]. We fit these histograms with Gaussian distributions to determine the average rates of elongation (Figure 1) and subunit dissociation (Figures 3A–3C) and to identify severing events, which were outside the Gaussian distributions (Figure 3C).
QUANTIFICATION AND STATISTICAL ANALYSIS
Estimation of the affinity of LatA for actin monomers
Assuming the complex of LatA with monomeric actin cannot participate in elongation [1], actin elongation depends on the concentration of free actin monomers. Thus we measured the concentrations of free actin monomers under each condition from rates of actin elongation and the rate constants for each actin species.
Aa is the concentration of free actin, V is the elongation rate and k+ and k- are the association and dissociation rate constants for free ATP-, ADP- and ADP-Pi-actin monomers [6]. We estimated the dissociation equilibrium constants from the measured concentrations of free actin monomers and the complex of LatA with actin monomers, using the equation with Kaleida graph (Synergy Software):
Supplementary Material
Time lapse movies (810 s total) of the elongation of 1 μM Mg-ATP-actin filaments (20% Oregon green label) in the presence of 0.2 (top) and 1.4 μM LatA (bottom), related to Figure 1.
Time lapse movie (1350 s total) of the elongation and depolymerization of actin filaments (20% Oregon green label), related to Figure 2. The movie begins with filaments elongating in 1 μM Mg-ATP-actin in polymerization buffer, viewed in a flow chamber coated with 50 nM NEM-myosin. At the point indicated by “Buff wash in”-free actin monomers were washed out of the chamber with polymerization buffer without LatA.
Time lapse movie (1830 s total) of elongation by 1 μM Mg-ATP-actin (20% Oregon green label) followed by washing out the actin monomer with 100 μM LatA in polymerization buffer, viewed in a flow chamber coated with 50 nM NEM-myosin, related to Figure 2. At the point indicated by “LatA washin”-free actin monomers were washed out of the chamber with polymerization buffer with 100 μM LatA.
Highlights (up to 4).
LatA has a higher affinity for actin monomers than filamentous actin subunits
LatA sequesters actin monomers and also severs and depolymerizes ATP-actin filaments
LatA rapidly “ages” ADP-Pi-actin filaments by dissociating phosphate from their ends
Dyes conjugated to actin affect depolymerization of Mg-ATP-actin filaments
ACKNOWLEDGEMENTS
Research reported in this publication was supported by National Institute of General Medical Sciences of the National Institutes of Health under award number R01GM026338 to TDP, R01GM122787 to NC and JSPS KAKENHI Grant Number 15K21106 to IF. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors thank Dimitrios Vavylonis, Shuichi Takeda and Yuichiro Maéda for their advice on the interpretation of the experiments, and Jeffrey Kuhn for generously supplying a length analysis software.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES LIST
- 1.Coué M, Brenner SL, Spector I, and Korn ED (1987). Inhibition of actin polymerization by latrunculin A. FEBS Lett. 213, 316–318. [DOI] [PubMed] [Google Scholar]
- 2.Ayscough KR, Stryker J, Pokala N, Sanders M, Crews P, and Drubin DG (1997). High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J Cell Biol 137, 399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spector I, Shochet NR, Kashman Y, and Groweiss A (1983). Latrunculins: novel marine toxins that disrupt microfilament organization in cultured cells. Science 219, 493–495. [DOI] [PubMed] [Google Scholar]
- 4.Kuhn JR, and Pollard TD (2005). Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys. J. 88, 1387–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yarmola EG, Somasundaram T, Boring TA, Spector I, and Bubb MR (2000). Actin-latrunculin A structure and function. Differential modulation of actin-binding protein function by latrunculin A. J Biol Chem 275, 28120–28127. [DOI] [PubMed] [Google Scholar]
- 6.Fujiwara I, Vavylonis D, and Pollard TD (2007). Polymerization kinetics of ADP- and ADP-Pi-actin determined by fluorescence microscopy. Proc Natl Acad Sci U S A 104, 8827–8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenblatt J, Peluso P, and Mitchison TJ (1995). The bulk of unpolymerized actin in Xenopus egg extracts is ATP-bound. Mol. Biol. Cell 6, 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jégou A, Niedermayer T, Orbán J, Didry D, Lipowsky R, Carlier MF, and Romet-Lemonne G (2011). Individual actin filaments in a microfluidic flow reveal the mechanism of ATP hydrolysis and give insight into the properties of profilin. PLoS Biol 9, e1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amann KJ, and Pollard TD (2001). Direct real-time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection fluorescence microscopy. Proc Natl Acad Sci U S A 98, 15009–15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pollard TD (1986). Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J Cell Biol 103, 2747–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kovar DR, Harris ES, Mahaffy R, Higgs HN, and Pollard TD (2006). Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell 124, 423–435. [DOI] [PubMed] [Google Scholar]
- 12.Melki R, Fievez S, and Carlier M-F (1996). Continuous Monitoring of Pi release Following Nucleotide Hydrolysis in Actin or Tubulin Assembly Using 2-Amino6mercapto-7-methylpurine Ribonucleoside and Purine-Nucleoside Phosphorylase as an Enzyme-linked Assay. Biochemistry 35, 12038–12045. [DOI] [PubMed] [Google Scholar]
- 13.Blanchoin L, and Pollard TD (1999). Mechanism of interaction of Acanthamoeba actophorin (ADF/Cofilin) with actin filaments. J. Biol. Chem. 274, 15538–15546. [DOI] [PubMed] [Google Scholar]
- 14.Morton WM, Ayscough KR, and McLaughlin PJ (2000). Latrunculin alters the actin-monomer subunit interface to prevent polymerization. Nat Cell Biol 2, 376–378. [DOI] [PubMed] [Google Scholar]
- 15.Oda T, Iwasa M, Aihara T, Maeda Y, and Narita A (2009). Nature of the globular to fibrous actin transition. Nature 457, 441–445. [DOI] [PubMed] [Google Scholar]
- 16.Galkin VE, Orlova A, Vos MR, Schröder GF, and Egelman EH (2015). Near-atomic resolution for one state of F-actin. Structure 23, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graceffa P, and Dominguez R (2003). Crystal structure of monomeric actin in the ATP state. Structural basis of nucleotide-dependent actin dynamics. J. Biol. Chem. 278, 34172–34180. [DOI] [PubMed] [Google Scholar]
- 18.Pollard TD, Goldberg I, and Schwarz WH (1992). Nucleotide exchange, structure, and mechanical properties of filaments assembled from ATP-actin and ADP-actin. J. Biol. Chem. 267, 20339–20345. [PubMed] [Google Scholar]
- 19.Nag S, Larsson M, Robinson RC, and Burtnick LD (2013). Gelsolin: the tail of a molecular gymnast. Cytoskeleton (Hoboken) 70, 360–384. [DOI] [PubMed] [Google Scholar]
- 20.Jiao Y, Walker M, Trinick J, Pernier J, Montaville P, and Carlier MF (2014). Mutagenetic and electron microscopy analysis of actin filament severing by CordonBleu, a WH2 domain protein. Cytoskeleton (Hoboken) 71, 170–183. [DOI] [PubMed] [Google Scholar]
- 21.Husson C, Renault L, Didry D, Pantaloni D, and Carlier MF (2011). CordonBleu uses WH2 domains as multifunctional dynamizers of actin filament assembly. Mol Cell 43, 464–477. [DOI] [PubMed] [Google Scholar]
- 22.Narita A, Oda T, and Maeda Y (2011). Structural basis for the slow dynamics of the actin filament pointed end. EMBO J. 30, 1230–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goddette DW, and Frieden C (1986). Actin polymerization - the mechanism of action of cytochalasin-D. J. Biol. Chem. 261, 5974–5980. [PubMed] [Google Scholar]
- 24.Sampath P, and Pollard TD (1991). Effects of cytochalasin, phalloidin, and pH on the elongation of actin filaments. Biochemistry 30, 1973–1980. [DOI] [PubMed] [Google Scholar]
- 25.Courtemanche N, and Pollard TD (2013). Interaction of profilin with the barbed end of actin filaments. Biochemistry 52, 6456–6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hug C, Jay PY, Reddy I, McNally JG, Bridgman PC, Elson EL, and Cooper JA (1995). Capping protein levels influence actin assembly and cell motility in dictyostelium. Cell 81, 591–600. [DOI] [PubMed] [Google Scholar]
- 27.Fujiwara I, Remmert K, Piszczek G, and Hammer JA (2014). Capping protein regulatory cycle driven by CARMIL and V-1 may promote actin network assembly at protruding edges. Proc Natl Acad Sci U S A 111, E1970–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raz-Ben Aroush D, Ofer N, Abu-Shah E, Allard J, Krichevsky O, Mogilner A, and Keren K (2017). Actin Turnover in Lamellipodial Fragments. Curr Biol 27, 2963–2973.e2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Q, and Pollard TD (2013). Actin filament severing by cofilin dismantles actin patches and produces mother filaments for new patches. Curr Biol 23, 11541162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yonetani A, Lustig RJ, Moseley JB, Takeda T, Goode BL, and Chang F (2008). Regulation and targeting of the fission yeast formin cdc12p in cytokinesis. Mol. Biol. Cell 19, 2208–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spudich JA, and Watt S (1971). The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246, 4866–4871. [PubMed] [Google Scholar]
- 32.Yasuda R, Miyata H, and Kinosita K (1996). Direct measurement of the torsional rigidity of single actin filaments. J Mol Biol 263, 227–236. [DOI] [PubMed] [Google Scholar]
- 33.Mahaffy RE, and Pollard TD (2006). Kinetics of the formation and dissociation of actin filament branches mediated by Arp2/3 complex. Biophys. J. 91, 3519–3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuhn JR, and Pollard TD (2007). Single molecule kinetic analysis of actin filament capping. Polyphosphoinositides do not dissociate capping proteins. J Biol Chem. 282, 28014–28024. [DOI] [PubMed] [Google Scholar]
- 35.Courtemanche N, and Pollard TD (2012). Determinants of Formin Homology 1 (FH1) domain function in actin filament elongation by formins. J Biol Chem. 287, 7812–7820. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Time lapse movies (810 s total) of the elongation of 1 μM Mg-ATP-actin filaments (20% Oregon green label) in the presence of 0.2 (top) and 1.4 μM LatA (bottom), related to Figure 1.
Time lapse movie (1350 s total) of the elongation and depolymerization of actin filaments (20% Oregon green label), related to Figure 2. The movie begins with filaments elongating in 1 μM Mg-ATP-actin in polymerization buffer, viewed in a flow chamber coated with 50 nM NEM-myosin. At the point indicated by “Buff wash in”-free actin monomers were washed out of the chamber with polymerization buffer without LatA.
Time lapse movie (1830 s total) of elongation by 1 μM Mg-ATP-actin (20% Oregon green label) followed by washing out the actin monomer with 100 μM LatA in polymerization buffer, viewed in a flow chamber coated with 50 nM NEM-myosin, related to Figure 2. At the point indicated by “LatA washin”-free actin monomers were washed out of the chamber with polymerization buffer with 100 μM LatA.


