Summary
Chimeric antigen receptor anti-CD19 (CAR19)-T cell immunotherapy-induced clinical remissions in CD19+ B cell lymphomas are often short lived. We tested whether CAR19-engineering of the CD1d-restricted invariant natural killer T (iNKT) cells would result in enhanced anti-lymphoma activity. CAR19-iNKT cells co-operatively activated by CD1d- and CAR19-CD19-dependent interactions are more effective than CAR19-T cells against CD1d-expressing lymphomas in vitro and in vivo. The swifter in vivo anti-lymphoma activity of CAR19-iNKT cells and their enhanced ability to eradicate brain lymphomas underpinned an improved tumor-free and overall survival. CD1D transcriptional de-repression by all-trans retinoic acid results in further enhanced cytotoxicity of CAR19-iNKT cells against CD19+ chronic lymphocytic leukemia cells. Thus, iNKT cells are a highly efficient platform for CAR-based immunotherapy of lymphomas and possibly other CD1d-expressing cancers.
Keywords: iNKT, CAR immunotherapy, B cell malignancies
Graphical Abstract

Highlights
-
•
Bespoke protocol for CAR19-iNKT cell transduction and clinical scale expansion
-
•
Higher CAR19-iNKT than CAR19-T cell expandability and killing of CD19+CD1d+ targets
-
•
CAR19-iNKT cell reactivity potentiation by αGalCer and ATRA
-
•
Prolonged survival and brain lymphoma eradication of CAR19-iNKT cell-treated mice
Rotolo et al. show that anti-CD19 chimeric antigen receptor (CAR19)-engineered CD1d-restricted invariant NKT cells (iNKT) are more effective than CAR19-T cells against CD1d-expressing lymphomas, including those in the brain. De-repression of CD1d expression further enhances the anti-tumor efficacy of CAR19-iNKT.
Significance
Anti-CD19 chimeric antigen receptor (CAR19)-T cell immunotherapy induces complete remissions in B cell lymphomas. However, more than half of patients relapse, highlighting the need for improving the efficacy of CAR-based immunotherapy. By exploiting the inherent biological properties of iNKT cells, we demonstrate that CAR19-iNKT cells exert a more powerful anti-tumor effect against B cell malignancy compared with CAR19-T cells, underpinned by dual targeting of CD19 and CD1d on target cells. This includes ability of CAR19-iNKT cells to eradicate brain tumors and to induce sustained second remissions. Anti-tumor efficacy can be further enhanced by transcriptional modulation of CD1D. Thus, iNKT cells are a versatile and effective platform for CAR immunotherapy of lymphoma and possibly of other cancers.
Introduction
Despite impressive early clinical efficacy, application of chimeric antigen receptor (CAR)-T cell immunotherapy for B cell malignancies is limited by disease relapse and tumor escape by downregulation of the commonly targeted CD19 antigen (Neelapu et al., 2017, Schuster et al., 2017). In addition, ability to induce sustained complete remissions with a single infusion of CAR-T cells and/or to exert a memory-like effect through long-term persistence, which would induce second remission of relapsing disease, are desirable but still not fully attained CAR-T cell attributes (Brudno and Kochenderfer, 2018). Recent efforts have focused on developing CAR targeting to tumor-associated antigens (Ruella et al., 2016) and on editing the endogenous T cell receptor (TCR) of the CAR-modified T cells (Eyquem et al., 2017, Qasim et al., 2017) to mitigate induction of acute graft-versus-host disease (aGVHD), allowing the use of unrelated donor-derived, allogeneic CAR-T cells. However, this entails additional genetic engineering that increases the complexity of CAR-T cell manufacturing.
Invariant natural killer T (iNKT) cells are rare but powerful immunoregulatory and effector T cells, playing a pivotal anti-tumor role (Bendelac et al., 2007, Exley et al., 2011, Salio et al., 2014). iNKT cells are restricted by CD1d, a non-polymorphic, glycolipid-presenting HLA I-like molecule expressed on B cells, antigen-presenting cells (APCs), and some epithelial tissues (Exley et al., 2000, Nickoloff et al., 1999). We and others have demonstrated the ability of donor iNKT cells to protect from experimental and clinical aGVHD in the context of allogeneic stem cell transplantation (Chaidos et al., 2012, Leveson-Gower et al., 2011, Rubio et al., 2016, Schneidawind et al., 2014). We also previously found co-expression of CD19 and CD1d in many subtypes of B cell lymphoma, including the incurable marginal zone lymphoma (MZL) and mantle cell lymphoma (MCL) (Kotsianidis et al., 2011), while in B cell chronic lymphocytic leukemia (CLL), the commonest hematologic malignancy, expression of CD1d is lower than in normal B cells or absent (Gorini et al., 2017, Kotsianidis et al., 2011).
We therefore hypothesized that equipping iNKT cells with CAR19 would potentially achieve dual targeting of CD1d and CD19, thus enhancing the overall anti-lymphoma effect. Given the protective impact of allogeneic iNKT cells against human aGVHD (Chaidos et al., 2012, Rubio et al., 2016), CAR-iNKT cells could be developed from healthy donors for “off-the-shelf” use. Previous work demonstrated the feasibility of CAR engineering of iNKT cells and their pre-clinical activity against neuroblastoma and CD1d−CD19+ B cell lymphoma lines but not patient-derived lymphoma cells (Heczey et al., 2014, Tian et al., 2016). However, in vivo anti-tumor response required repeated cell dosing and/or adjuvant IL-2 administration (Heczey et al., 2014, Tian et al., 2016). In addition, comparative analysis of CAR-T and -iNKT cells and exploration of the relative contributions of CD1d-versus CAR19-CD19-dependent interactions in CAR19-iNKT cell activation are lacking.
Results
Optimized Protocol for Generation of Poly-functional CAR-iNKT Cells
There is a dearth of information as to how best to CAR-engineer iNKT cells. To determine optimal conditions for efficient lentiviral CAR19 transduction and subsequent CAR19-iNKT cell expansion, we tested four different protocols using second- (19-28-z) or third-generation (19-28-OX40-z) CAR against CD19 (Figure S1A). In a stepwise approach (Figures S1B–S1E), conditions tested include transduction of sorted iNKT cells upfront versus post initial expansion in the presence of the iNKT cell agonist alpha-galactosylceramide (αGalCer); activation and expansion using anti-CD3/CD28-mediated stimulation versus CD1d-expressing APC plus αGalCer. Through paired comparisons, we first determined that upfront transduction of pre-selected and not of pre-expanded iNKT cells results in the highest transduction efficiency (protocol 3; Figures S1B–S1E) and next, use of IL-15 but not of IL-2 during the CD3/CD28-based activation phase and the first week post CAR19 transduction preserved viability of iNKT cells (Figures S1B–S1E).
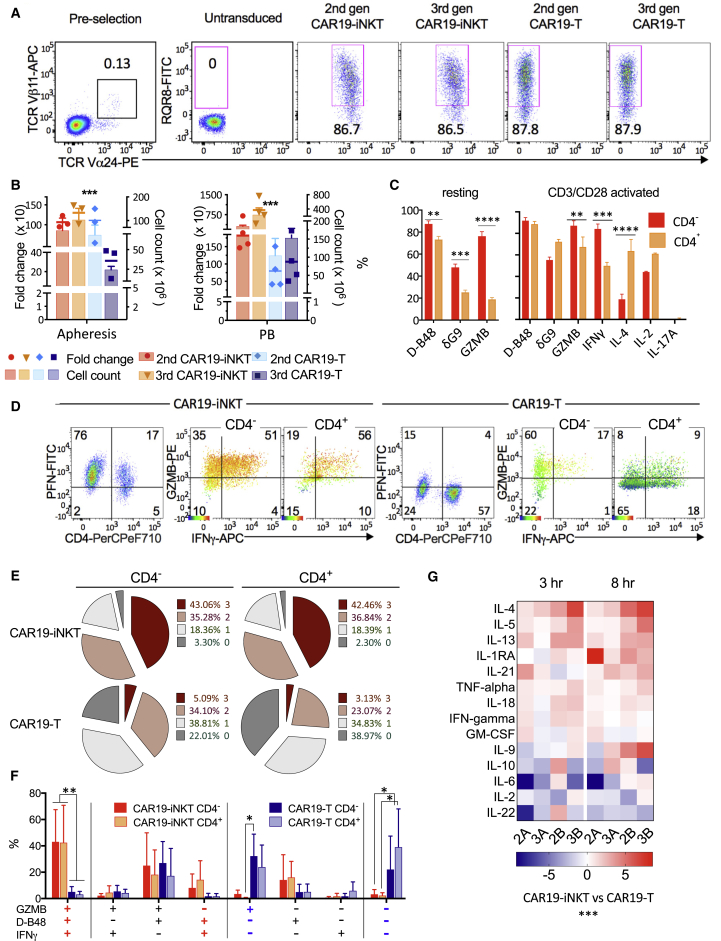
Overall, we found that the optimal approach (protocol 4), comprising upfront selection and lentiviral CAR19 transduction of CD3/CD28-activated iNKT cells in the presence of autologous APC and IL-15, consistently generates highly transduced and viable CAR-iNKT (and CAR-T) cells (Figures 1A and S1B–S1E) and, over a period of 3 weeks, it results in significantly higher expansion and absolute numbers of CAR19-iNKT than CAR19-T cells (Figure 1B). This protocol is efficient irrespective of the source of iNKT cells; i.e., fresh or frozen, normal donor, or patient-derived lymphocytes (Figure S1F). Importantly, it also ensures the preservation of the CD4– fraction of iNKT cells (Figure S1G), which, compared with their CD4+ counterparts, have a more polarized Th1 cytokine profile and express higher levels of cytotoxic granules (Gumperz et al., 2002). Indeed, we found that resting CD4– CAR19-iNKT cells express significantly higher levels of perforin and granzyme B and, upon activation, more granzyme B and interferon-γ (IFNγ) but less IL-4 than the CD4+ subset (Figures 1C and S1H). Compared with their CAR19-T counterparts, a higher proportion of CAR19-iNKT cells express IFNγ, perforin, and granzymes (Figure 1D) and a significantly higher proportion (40% versus <5%, p < 0.01) are tri-functional; i.e., co-express these three molecules (Figures 1D–1F). Of note also, while >20% of CAR19-T cells secreted none of the above three molecules, the corresponding proportion for CAR19-iNKT cells was <3%. Further, CAR19-iNKT cells secrete higher levels of Th1/2 cytokines than CAR19-T cells over an 8 hr period of activation (Figure 1G).
Figure 1.
Optimized Protocol for Generation of Poly-functional CAR19-iNKT Cells
(A) Flow cytometric identification of iNKT cells as TCRVα24+Vα11+ pre-selection and expression of second- and third-generation CAR19 in TCRVα24− T and TCRVα24+ iNKT cells as assessed by staining against the marker RQR8 3 days after lentiviral transduction.
(B) Expansion and absolute numbers of CAR19-T and CAR19-iNKT cells over 3 weeks using lymphapheresis (left) or peripheral blood (PB; right) (n = 3 and 4 respectively). p values are for CAR19-iNKT versus CAR19-T cells using Friedman test.
(C) Intracellular expression of cytokines in resting (n = 10) and anti-CD3/CD28-bead-activated (for 4 hr; n = 6) CD4− and CD4+ CAR19-iNKT cells. Flow cytometric analysis was performed as shown in (D). D-B48 and δG9 monoclonal antibodies identify total and granule-associated perforin respectively. GZMB, granzyme B.
(D) Representative example of flow cytometric intracellular analysis of shown cytokines in CD4− and CD4+ CAR19-T and CAR19-iNKT cells. In GZMB/IFNγ dot plots, intensity of perforin expression is projected as a heatmap according to the shown color scale. PFN, perforin.
(E) Proportions of cells co-expressing zero to three cytokines (mean of four independent experiments).
(F) Proportions of specific cytokines co-expressed by CD4− or CD4+ CAR19-T and CAR19-iNKT cells.
(G) Multiple cytokine secretion after 3 and 8 hr of activation of second- and third-generation (2 and 3) CAR19-T and CAR19-iNKT cells from two healthy donors (A and B). Heatmap shows normalized CAR19-iNKT/CAR19-T cell ratios.
Error bars represent SEM.
Asterisks indicate p values as follows: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. See also Figure S1.
Co-operative Activation of CAR19-iNKT Cells
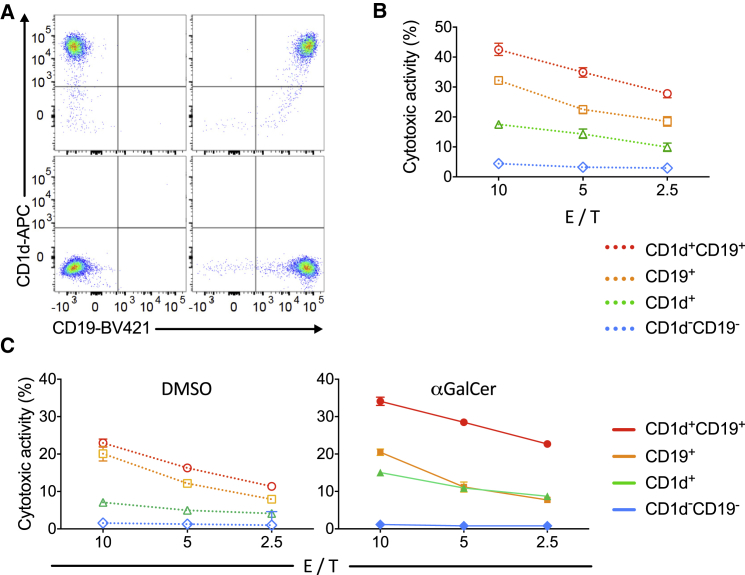
Next, we tested whether equipping iNKT cells with a CAR19 that powerfully activates T cells when it engages CD19 would affect the ability of iNKT cells to functionally interact with CD1d, the sole restricting element of the iTCR (Brossay et al., 1998, Exley et al., 1997, Nieda et al., 1999, Takahashi et al., 2000). For this purpose, we engineered the CD1d−CD19− K562 cells to express CD1d and CD19 singly or in combination (Figure 2A). We found that killing by CAR19-iNKT cells of CD1d+CD19−, CD1d−CD19+, and CD1d+CD19+ targets proceeded incrementally (Figure 2B). In the presence of αGalCer, a CD1d-presented glycolipid that selectively activates iNKT cells, we observed further enhancement of CAR19-iNKT cell cytotoxicity against CD1d+-only and CD1d+CD19+ targets but not against CD1d−CD19− and CD1d−CD19+ targets (Figure 2C). We conclude that the expression and engagement of CAR19 does not affect the ability of iNKT cells to be activated in the presence of CD1d, suggesting preservation of the ability of CAR19-iNKT cells to be activated through CD1d interaction. These findings also support the hypothesis that dual targeting of CD1d and CD19 results in co-operative killing of target cells and can be further enhanced by the use of αGalCer, thus further underscoring the importance of functional interaction of CAR19-iNKT cells with CD1d on target cells.
Figure 2.
Co-operative Activation of CAR19-iNKT Cells
(A) Dot plots showing expression of CD19 and CD1d in parental and derivative K562 cells after retroviral transduction of corresponding cDNAs.
(B) Cytotoxic activity of second-generation CAR19-iNKT cells against parental CD1d–19– K562 cells (WT) or K562 cells expressing CD1d and CD19 singly or in combination at the indicated E/T cell ratios (representative of three experiments).
(C) Cytotoxic activity of second-generation CAR19-iNKT cells against parental CD19–CD1d– K562 cells or K562 cells expressing CD19 and CD1d singly or in combination with (right) and without (left) pre-pulsing of targets with 100 ng/mL αGalCer. E/T cell ratios are as shown (representative of two experiments).
Error bars represent SEM of triplicate assays.
Enhanced Short- and Long-Term Reactivity of CAR19-iNKT Cells against B Lineage Malignancies
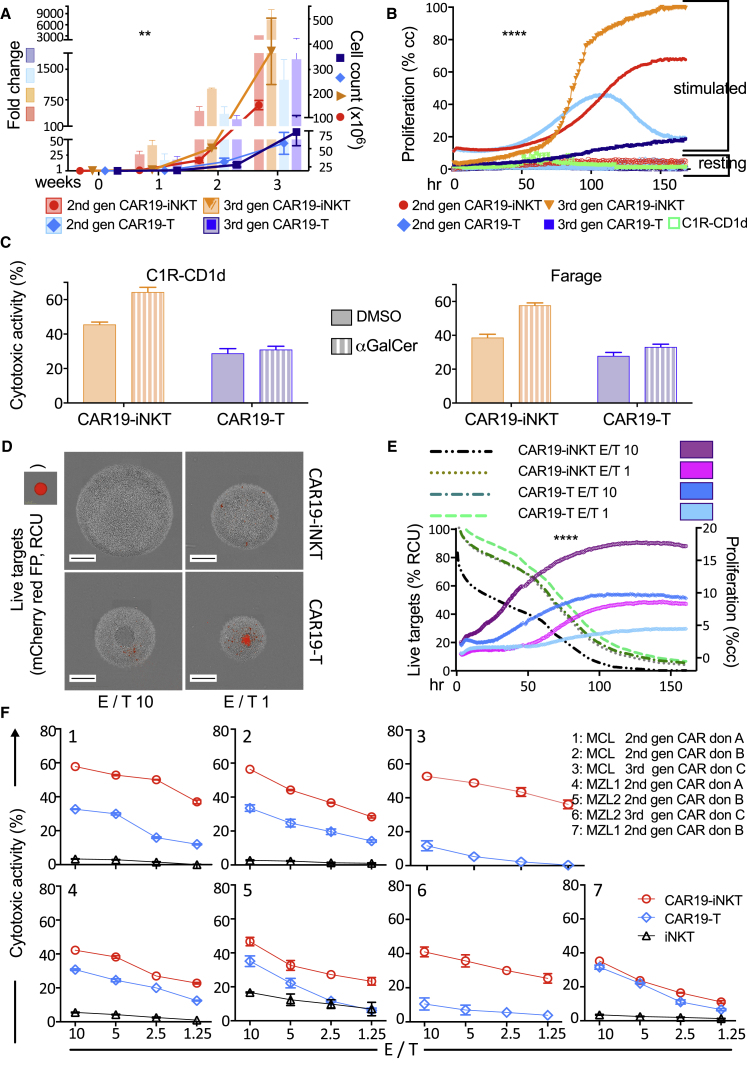
We next examined the short- and long-term in vitro reactivity of CAR19-iNKT cells in a B lineage cell context. First, we confirmed that CAR19-engineered iNKT cells are reactive against mature B lineage cell lines in a manner proportional to the expression levels of CD19 and CD1d (Figures S2A and S2B). We started dissecting the functional profile of CAR19-iNKT cells by comparing the proliferative potential of second- and third-generation CAR19-iNKT cells with their same-donor CAR19-T cell counterparts. Over a period of 3 weeks, we found a significantly higher expansion and higher absolute numbers of CAR19-iNKT cells, which were more striking for third-generation CAR (Figure 3A). This was corroborated in real-time imaging of second- and third-generation CAR19-iNKT versus CAR19-T cell proliferation in the presence of CD19+CD1d+ B cells over a period of 7 days (Figure 3B).
Figure 3.
Enhanced Short- and Long-Term Reactivity of CAR19-iNKT Cells against B Lineage Malignancies
(A) Second- and third-generation CAR19-T and CAR19-iNKT cell expansion (fold change) and absolute cell numbers (cell count) over a period of 3 weeks (n = 4). p value is for CAR19-iNKT versus CAR19-T cells using Friedman test. Error bars represent SEM.
(B) Proliferation analysis of second- and third-generation CAR19-T and CAR19-iNKT cells in the presence (stimulated) or not (resting) of irradiated CD1d+CD19+ (C1R-CD1d) cells over 7 days. p value is for CAR19-iNKT versus CAR19-T cells using Friedman test.
(C) Cytotoxicity of third-generation CAR19-T and -NKT cells against C1R-CD1d (representative of n = 3) and Farage lymphoma cell lines (representative of n = 2) pre-loaded or not with αGalCer. Error bars represent SEM of triplicate assays.
(D) IncuCyte images of representative wells showing the final effector (gray) and live target cells (red) after 7 days. Effectors were second-generation CAR19-T and CAR19-NKT cells. Targets were CD19+ ARH-77-CD1d cells expressing mCherry red fluorescent protein. Scale bar represents 400 μm.
(E) Seven-day trajectory of effector and target cell proliferation and elimination respectively as per (D). p value is for CAR19-iNKT versus CAR19-T cells using Friedman test.
(F) Cytotoxicity of second-generation CAR19-iNKT, CAR19-T, and of untransduced iNKT cells against lymphoma cells from one patient with mantle cell lymphoma (MCL; top) and two patients with marginal zone B lymphoma (MZL; bottom) using three different T/iNKT cell healthy donors (A, B, and C). Error bars represent SEM of triplicate assays.
∗∗p < 0.01; ∗∗∗∗p < 0.0001. Cc, cell confluency; FP, fluorescent protein; RCU, red calibrated units. See also Figures S2 and S3.
In cytotoxicity assays, we found higher reactivity of CAR19-iNKT cells than CAR19-T effectors against the CD19+CD1d+ double-positive C1R-CD1d cells and Farage lymphoma cells, an effect that was further enhanced by αGalCer (Figure 3C). These findings were extended in an assay in which proliferation of effector cells (i.e., CAR19-T or CAR19-iNKT cells) and survival of CD19+CD1d+ target cells were concomitantly imaged in real time over a period of 7 days (Figures 3D and 3E). Indeed, the cytotoxic and proliferative activity of CAR19-iNKT cells at an effector-to-target (E/T) ratio of 1:1 was similar to those of CAR19-T cells at an E/T ratio of 10:1, suggesting a functional equivalence of CAR19-iNKT:CAR19-T of 10:1.
To better reflect the clinical context, we set up cytotoxicity assays using as targets primary CD19+CD1d+ lymphoma cells from one patient with blastic variant of MCL and two patients with MZL, which co-express CD1d and CD19 as we previously reported (Kotsianidis et al., 2011) (Figure S3A). In six out of seven assays involving three healthy donors, CAR19-iNKT cells were more cytotoxic than CAR19-T cells (Figure 3F). Further characterization of lymphoma cell killing by assessment of cell size and 7-AAD retention (Figure S3B) confirmed superior killing by CAR19-iNKT cells of all patient lymphoma cells (Figures S3C and S3D). In these “same-tube” assays we also observed low to no killing of monocytes (Figure S3E), which express higher levels of CD1d than malignant B cells (Figure S3F) (Exley et al., 2000) but not CD19, suggesting a low “on-target, off-tumor” reactivity of CAR19-iNKT cells.
Transcriptional and Epigenetic Basis for Enhancing CAR-iNKT Cell Reactivity
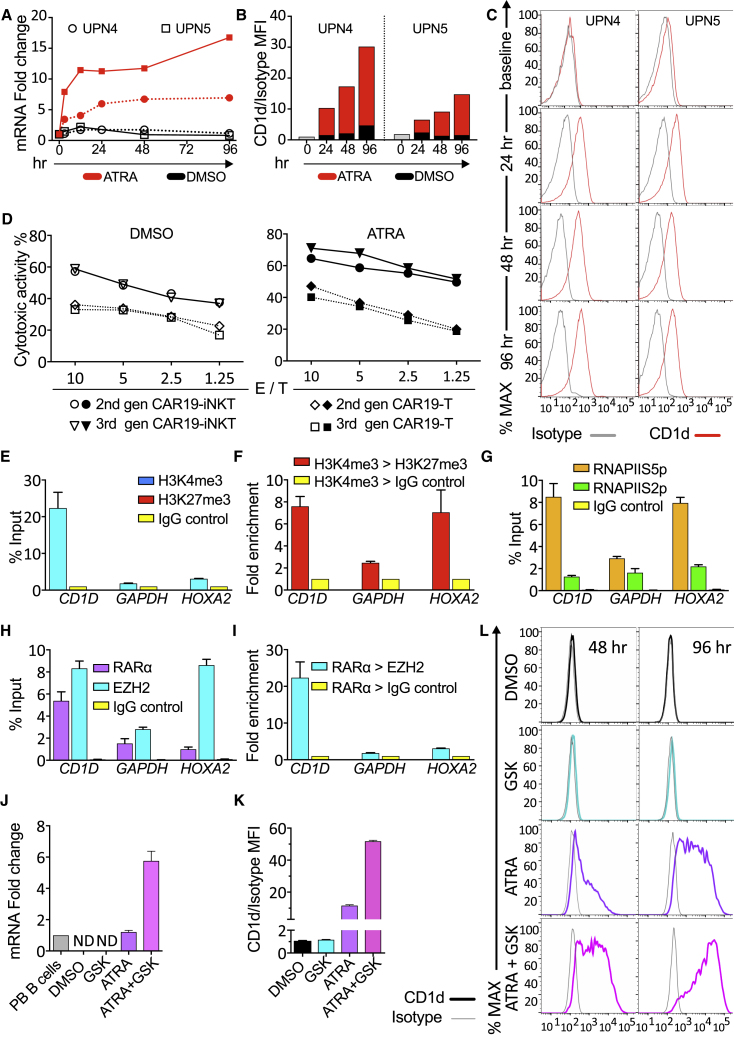
As previously reported, we found that CLL cells express low or no CD1d in comparison with normal B cells (Gorini et al., 2017, Kotsianidis et al., 2011) (Figure S4A). We found that CAR19-iNKT cells effectively killed CLL cells and their cytotoxicity was further enhanced in the presence of αGalCer, suggesting that dual targeting of CLL cells can be further enhanced by CD1d presentation of αGalCer despite the very low level of CD1d expression (Figure S4B). Previous work reported transcriptional regulation of CD1D expression in human B cells by the RARα ligand all-trans retinoic acid (ATRA) (Allan et al., 2011). Accordingly, we observed that CD1D mRNA and cell surface protein expression increased in a time-dependent manner after treatment with clinically relevant concentrations of ATRA (Figures 4A–4C and S4C–S4E) without affecting cell viability (Figure S4F). Moreover, αGalCer-pre-loaded CLL cells were more effectively killed by CAR19-iNKT cells than by CAR19-T cells (Figure 4D) and, upon ATRA pre-treatment of CLL cells (Figure S4G), the cytotoxic activity of CAR19-iNKT but not of CAR19-T cells increased further (Figure 4D).
Figure 4.
Transcriptional and Epigenetic Basis for Enhancing CAR19-iNKT Cell Reactivity
(A) CD1D mRNA quantification by qPCR in CLL cells from two patients upon ATRA treatment (10−6 M) for 0–96 hr.
(B and C) Bar charts (B) and flow cytometry histograms (C) showing CD1d expression on malignant B cells upon ATRA treatment and mean fluorescent intensity (MFI) analysis of CD1d expression in comparison with isotype control.
(D) Cytotoxicity of second- and third-generation CAR19-T and -NKT cells against αGalCer-pulsed CLL cells pre-treated with 0.1% DMSO control or 10−6 M ATRA. Error bars represent SEM of triplicate assays.
(E) ChIP-qPCR assay for H3K4me3 and H3K27me3 enrichment in the promoter of CD1D using IgG as control in U266 cells. GAPDH is an active gene control, while HOXA2 is a repressed gene control. ChIP data are shown as a percentage of the input chromatin.
(F) ChIP-re-ChIP qPCR assay showing fold enrichment of H3K27me3 or IgG control after immunoprecipitation (IP) against H3K4me3.
(G) ChIP-qPCR assay against RNA Pol II for Ser5 over Ser2 phosphorylated form at the promoter of the indicated genes.
(H) ChIP-qPCR assay against RARα, EZH2, and Ig control at the promoters of the genes shown.
(I) ChIP-re-ChIP qPCR assay showing enrichment of EZH2 or IgG control after IP against RARα in U266 cells for –(I) (n = 3).
(J) qPCR quantification of CD1D mRNA in U266 cells treated with 0.1% DMSO, 10−6 M GSK343, 10−6 M ATRA or 10−6 M GSK343 plus 10−6 M ATRA. Values are normalized to CD1D mRNA expression levels in normal peripheral PB B cells (n = 3). ND, not detectable.
(K and L) Relative MFI analysis (K) and histogram depiction (L) of CD1d expression in comparison with isotype control in U266 cells from the same experiment shown in (J).
Error bars represent SEM. See also Figure S4.
We dissected the epigenetic and transcriptional basis of CD1d regulation in the U266 cell line as a paradigm of a B lineage malignant cell with transcriptional repression of CD1D expression (Figure S4H). Using chromatin immunoprecipitation (ChIP) and re-ChIP assays we found enrichment of both H3K4me3-activating and H3K27me3-repressive histone marks at the CD1D promoter (Figures 4E and 4F). Notably, a similar bivalent histone state was observed in primary CLL cells (Figure S4I). In U266 cells we also demonstrated enrichment of the Ser5- but not Ser2-phosphorylated form of RNA PolII (Figure 4G), consistent with a bivalent, poised transcriptional state of CD1D. Importantly, histone bivalency at the CD1D promoter was associated with high levels of RARα binding as well as of EZH2, the Polycomb complex methyl-transferase responsible for H3K27me3 marks (Simon and Kingston, 2009) (Figure 4H), with direct interaction of EZH2 and RARα (Figure 4I), suggesting a co-operative transcriptional repressive function upon CD1D. In a pharmacological approach, although an EZH2 inhibitor had no discernible effect on transcription and surface expression of CD1d, it co-operatively enhanced the effect of ATRA on CD1d expression (Figures 4J–4L). These findings provide the mechanistic basis for developing CAR-iNKT cell immunotherapy in conjunction with transcriptional and epigenetic manipulation of CD1D.
Enhanced In Vivo Anti-tumor Activity of CAR19-iNKT Cells
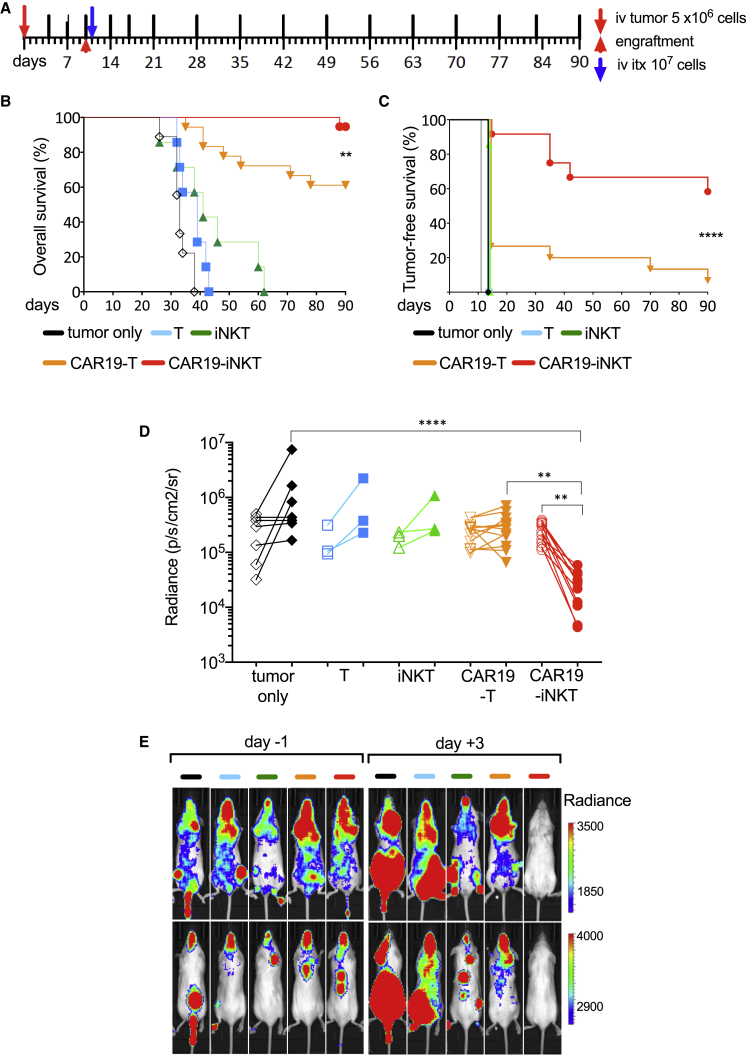
We compared second-generation CAR19-iNKT versus CAR19-T cells in a systemic in vivo model of CD1d+CD19+ B cell malignancy (C1R-CD1d cells; Figure 5A) based on a single infusion of immunotherapy in tumor-engrafted NSG mice and monitoring with serial bioluminescence imaging of the tumor burden. Animals treated with unmodified T or iNKT cells had poorer survival compared with mice receiving CAR-based immunotherapy (Figure 5B). However, compared with the CAR19-T cell-treated group, the CAR19-iNKT cell-treated group displayed a significantly improved overall (Figure 5B) and tumor-free survival (Figure 5C). This could be at least in part explained by a significantly swifter decline of tumor burden observed within 3 days following transfer of CAR19-iNKT cells (Figures 5D and 5E).
Figure 5.
Enhanced In Vivo Anti-tumor Activity of CAR19-iNKT Cells
(A) In vivo experiment layout. 5 × 106 Luciferase-expressing C1R-CD1d cells were intravenously (i.v.) transferred to NSG mice. Tumor growth was monitored by bioluminescence (BLI). After engraftment was confirmed by increasing photon activity in two consecutive BLI scans taken at least 72 hr apart, 107 effector cells (or same volume PBS control) were i.v. transferred followed by BLI monitoring of tumor burden at the schedule shown. Timings of BLI imaging are shown as top vertical lines.
(B and C) Overall (B) and tumor-free (C) survival of tumor-bearing mice untreated (n = 12) or treated with untransduced T cells (n = 7), untransduced iNKT cells (n = 7), second-generation CAR19-T cells (n = 19, of which four were excluded from tumor-free survival analysis. See note § in Figure 6C) or CAR19-iNKT cells (n = 19, of which seven were excluded from tumor-free survival analysis; see note § below). Data are from two independent experiments. Shown p values are for comparison of CAR19-T versus CAR19-iNKT cell-treated animals.
(D) Cumulative data as per (E) from one cohort of mice showing tumor burden as assessed by BLI radiance on days −1 and +3.
(E) Representative examples of ventral and dorsal BLI views of tumor burden on days −1 (pre-treatment) and +3 (post treatment) from the same cohort of mice shown in (D).
∗∗p < 0.01; ∗∗∗∗p < 0.0001. See also Figure S5.
Since human T cells may induce aGVHD in xenograft models (Alcantar-Orozco et al., 2013), we monitored body weight and clinical aGVHD score (Cooke et al., 1996). We found that body weight increased comparably in all experimental groups (Figure S5A), thus excluding the occurrence of clinically significant aGVHD, which is associated with >10% loss of weight (Cooke et al., 1996); in line with this, the aGVHD score, on a scale of 0–10, was 0 in all groups (Figure S5B).
CAR19-iNKT Cells Eradicate Intracranial and Relapsed Disease
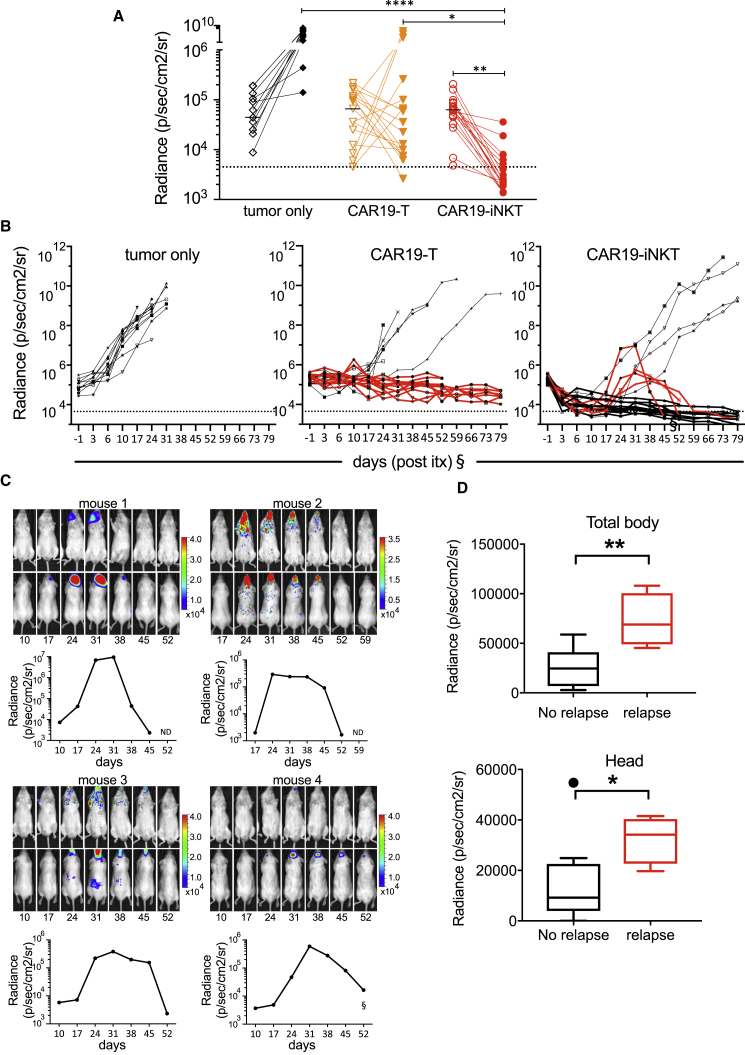
In the tumor model described herein, we observed enhanced bioluminescence over the head even at the time of engraftment, before commencement of immunotherapy. Correlation of bioluminescence with MRI, magnetic resonance spectroscopy, and histologic analysis, suggested the presence of intracranial meningeal, brain, and pituitary disease in most animals (Figures S5C–S5E and Table S1). While brain lymphoma persisted above the threshold of detection in all but one animal receiving CAR19-T cells as well as in all untreated animals, they were eliminated in 13/18 CAR19-iNKT cell-treated animals (Figure 6A). This would be consistent with CAR19-iNKT cells gaining entry into the brain and effectively controlling brain disease. In all tumor-only, T-, iNKT-, and CAR19-T cell-treated animals (n = 32; i.e., 11/11, 7/7, 7/7, and 7/19 animals respectively), systemic and brain lymphoma progressed without evidence of first or, in the case of one CAR19-T cell-treated animal, second remission. By contrast, in four CAR19-iNKT cell-treated mice, after initial clearance of systemic and brain lymphoma, relapse developed primarily in the brain at a later stage (Figures 6B and 6C) and in all four mice this relapsed disease eventually regressed, leading to long-term survival (Figures 6B and 6C), a finding consistent with persistence and ability for secondary anti-tumor responses by CAR19-iNKT cells. Importantly, in CAR19-T cell-treated animals with progressive brain disease that died during the course of the experiment, histological analysis revealed both CD3+ and CD19+ cell infiltration (Figure S5F), suggesting failure of CAR19-T cells to control brain lymphoma. The same analysis was performed in CAR19-iNKT cell-treated animals at the time of primary endpoint; i.e., at termination of the experiment on day 90. At that time, neither CD3+ or CD19+ cells were detected in the brains of the CAR19-iNKT cell-treated animals, including those four achieving second remission (Figure S5G), suggesting that they receded following eradication of relapsed disease.
Figure 6.
Eradication of Relapsed Lymphoma in CAR19-iNKT Cell-Treated Mice
(A) BLI photon activity in the head of control (n = 11), CAR19-T (n = 18), and CAR19-iNKT (n = 18) cell-treated animals at engraftment and at completion of the experiment. Detection threshold (dotted line) was set as the lowest BLI activity value recorded in the head at engraftment in the whole cohort of animals used.
(B) BLI activity recorded throughout the duration of the experiment in tumor-only control, CAR19-T, and CAR19-iNKT-treated groups. Second remission was achieved in four animals receiving CAR19-iNKT immunotherapy but in none within the other groups.
(C) Longitudinal BLI images and dynamics of radiance activity are shown for each of the four animals. Relapse occurred between 17 and 26 days following immunotherapy. In three mice, a second complete remission was documented by BLI. In the fourth mouse, a partial remission could be documented, due to restricted access to the IVIS instrument from day 70 (+59 post immunotherapy [itx]) (§). However, all mice survived until the end of the experiment with no clinical signs of tumor progression as assessed postmortem by fluorescence imaging, flow cytometry, or immunohistochemistry (not shown).
§In four and seven CAR19-T and CAR19-iNKT-treated animals, respectively, tumor burden could not be monitored by BLI from day 70 (+59 post itx) due to restricted access to the IVIS instrument. However, all mice were maintained in the study until achievement of survival endpoint or the end of the experiment.
(D) Total body and head BLI activity in relapsed and non-relapsed CAR19-iNKT cell-treated animals on day 3 post immunotherapy. Horizontal line in box-whisker plots shows median and upper and lower horizontal lines of box represent 75th and 25th percentile respectively, and whiskers represent 95th and fifth percentiles.
∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.0001. See also Figures S5 and S6.
Closer analysis of the four CAR19-iNKT cell-treated relapsed animals showed that both their total body and brain tumor burden 3 days following immunotherapy were significantly higher, by 2.8-fold and 3.7-fold, respectively, than in the non-relapsed CAR19-iNKT cell-treated animals, suggesting that failure to induce deep remission early post immunotherapy would result in overt relapse (Figure 6D).
As well as through dual CD19 and CD1d targeting, achievement of remission of primary and relapsed brain disease in CAR19-iNKT cell-treated animals might be related to enhanced potential for chemotaxis toward the tumor cells and to enter the brain. Previous work demonstrated that a much higher expression of the chemokine receptors CCR1, CCR2, CCR4, CCR5, CCR6, and CXCR3 by human iNKT cells than conventional effector or memory T cells underpins the ability of iNKT cells to migrate more effectively to peripheral, extra-lymphoid tissues (Kim et al., 2002). Further, human iNKT cells can infiltrate subcutaneous lymphoid tumors in NOD/SCID mice (Bagnara et al., 2009). Consistent with the chemokine receptor repertoire of human iNKT cells, transcriptome analysis of the CAR19-iNKT and CAR19-T cells generated from the healthy donor used in three out of the four animals with second brain lymphoma remission showed that CCR1, CCR2, CCR5, CCR6, CXCR3, and CCR3 are also expressed at higher levels in CAR19-iNKT than CAR19-T cells and only expression of CCR4 is modestly higher in CAR19-T cells (Figures S6A–S6C). Further, analysis and comparison of the mRNA chemokine receptor pattern of murine iNKT versus conventional T cells revealed a very similar pattern as in their human counterparts (Figures S6D and S6E), highlighting an evolutionary conserved potential of iNKT cells to migrate to peripheral tissues.
To gain some insight into the potential of the lymphoma cells used in our model to incite chemotaxis, we assessed chemokine expression by transcriptome analysis of the parental C1R-CD1d cells. We found that, with the exception of CCL2 (the CCR2 ligand), the lymphoma cells express the chemokines CLL3 (ligand for CCR1, CCR4, and CCR5), CCL5 (ligand for CCR1, CCR3, and CCR5), CCL20 (ligand for CCR6), CCL22 (ligand for CCR4), and CCL10 (ligand for CXCR3; Figures S6A and S6F). Of note, the CCR6-CCL20 axis is a critical determinant for T cell migration to the brain through the choroidal plexus (Sallusto et al., 2012).
Finally, entry of T cells into the brain through the blood-brain barrier (BBB) requires that they express integrin α4β1 (VLA-4). We found higher expression of the corresponding ITGA4 and ITGB1 genes by CAR19-iNKT than by CAR19-T cells (Figure S6G) (Sallusto et al., 2012).
Based on the above and published data, we postulate that the inherent chemotactic and migratory properties of CAR19-iNKT cells contribute to their enhanced in vivo anti-lymphoma activity and their potential to gain access to and eradicate primary or relapsed brain disease.
Discussion
Here, we exploit biological properties of iNKT cells to overcome shortcomings of conventional CAR-T cells and enhance the potential of CAR-based immunotherapy.
Our optimized protocol for CAR-iNKT cell production is unlike other CAR-T or CAR-iNKT cell generation protocols in that a “scale-down” approach, entailing CAR modification of a relatively small number of pre-selected iNKT cells (104–106 cells), precedes the scaling-up phase. The described approach is robust enough to deliver high-purity, clinical-scale cell product within 3 weeks from a variety of iNKT cell sources, and it requires a fraction of lentiviral supernatant used in conventional CAR transduction protocols; as such, it would be expected to significantly curtail cost of CAR-iNKT cell production. With an average expansion of >3,000-fold, starting with 2 × 105 purified iNKT cells will generate in excess of 5 × 108 CAR19-iNKT cells, thus covering cell doses of 1 × 106 to 5 × 106 cells/kg used in conventional CAR19-T cell immunotherapy protocols (Brudno and Kochenderfer, 2018). Whether a similar or even lower cell dosing range will be required for optimal CAR19-iNKT cell clinical immunotherapy will be determined in future clinical trials.
Upfront transduction of selected iNKT cells and the subsequent expansion of CAR19-iNKT cells are another two important features that distinguish our approach from other protocols of CAR-iNKT generation that entail CAR transduction of pre-expanded iNKT cells and use of IL-2 (Heczey et al., 2014, Tian et al., 2016). The use of IL-15 is likely to have significantly contributed to the survival and the robust, and sustained expansion of CAR19-iNKT cells. We found that, compared with IL-2, IL-15 maintains higher levels of iNKT cell viability during lentiviral transduction and during the first week post transduction, a critical requirement for the subsequent phase of expansion. Indeed, IL-15 has been shown to inhibit activation-induced cell death of T cells (Marks-Konczalik et al., 2000) and to drive iNKT cell terminal effector-like differentiation as well as expression of effector molecules such as granzymes and IFNγ by iNKT cells, while promoting cell survival through induction of BCL2 family proteins (Gordy et al., 2011).
We provide direct evidence that iNKT cells retain functionality of their endogenous iTCR-dependent activation in the presence of CAR19-mediated activation, in a glycolipid- as well as CD1d-dependent manner. The weaker iNKT cell-CD1d axis acts in a co-operative manner with the stronger CAR19-CD19 axis for iNKT cell activation. Downstream of these interactions, CAR19-iNKT cells with a more robust proliferative, cytokine secretion profile and enhanced cytotoxic activity outperform same-donor CAR19-T cells.
Despite promising pre-clinical activity, the expectation that third-generation CAR-T cells would lead to enhanced anti-tumor effect has not materialized in pre-clinical or clinical studies so far (Till et al., 2012, Zhao et al., 2015). Our data shows that the proliferative potential of third-generation CAR19-iNKT cells is considerably better than that of CAR19-T cells, suggesting that iNKT cells might provide an optimal platform for clinical development of third-generation CARs. In particular, given the previously reported requirement for the co-stimulatory OX40L-OX40 axis for optimal iNKT cell-mediated anti-tumor responses (Zaini et al., 2007, Zhou, 2007), the inclusion of OX40 in a third-generation CAR configuration might be advantageous.
In line with the dual targeting hypothesis, enhanced cytotoxic effect of CAR-iNKT cells against B cell lines extends to include primary CD1d+CD19+ MZL and MCL cells. Therefore, enhanced anti-tumor reactivity of CAR-iNKT cells requires interaction of CAR19-iNKT cells with CD1d on target cells. While it is possible that this interaction also involves the iTCR, definitive delineation of its direct role in the activation of CAR19-iNKT cells via interaction with CD1d would require deletion of the iTCR.
In previous work involving NSG mice with humanized hematopoiesis, including myelopoiesis, anti-GD2 CAR-T but not CAR-iNKT cells incited aGVHD (Heczey et al., 2014). Since xenogeneic aGVHD requires presence of human myeloid as well as of T cells (Schroeder and DiPersio, 2011), it is likely that lack of clinical aGVHD in our model was due to transfer to NSG mice solely of highly purified CAR19-modified or unmodified T or iNKT cells. Nevertheless, given the protective effect of allogeneic iNKT cells, and in particular of the CD4− fraction, against aGVHD (Chaidos et al., 2012, Rubio et al., 2016), effective CAR19-iNKT cell immunotherapy can in principle be delivered using iNKT cells sourced from healthy donors. In this context, the consistent preservation of the CD4– fraction of iNKT cells in our manufacturing protocol, but not in others (Heczey et al., 2014, Tian et al., 2016), is another important salient feature. Further, in contrast to current efforts to delete the TCR of conventional CAR-T cells to abrogate TCR-major histocompatibility complex interactions and thus reduce the risk of aGVHD imparted by allogeneic CAR-T cells (Eyquem et al., 2017, Qasim et al., 2017), CAR19-iNKT cells impart protection from aGVHD with their iTCR intact.
CAR19-T cell immunotherapy has been considerably less successful in CLL (Mato et al., 2017) as compared with other B lineage malignancies. Our work provides the cellular and molecular rationale to employ CAR19-iNKT cells as a potentially more effective immunotherapeutic approach for CLL. By dissecting the epigenetic landscape that restricts transcription of CD1D in CLL cells, namely transcriptional repression via a co-operative effect of RARα and Polycomb complex, we define a clinically applicable pharmacological approach that enhances the in vitro effect of CAR19-based immunotherapy against CLL cells. Further enhancement of the anti-lymphoma effect could be imparted by the use of αGalCer, to which CAR19-iNKT cells remain selectively responsive, resulting in increased cytolysis of CLL cells.
The potential toxicity of CAR-iNKT cell immunotherapy, especially the “off-tumor, on-target” targeting of CD1d-expressing cells, will eventually be determined in clinical trials. However, we found that CAR19-iNKT cell cytotoxicity against monocytes, the highest CD1d-expressing blood cells, is very low and comparable with that of CAR-T cells. Although the mechanism of differential CD1d-dependent reactivity of CAR19-iNKT cells against monocytes and lymphoma B cells remains to be determined, it may reflect the differential endogenous glycolipid repertoire presented by CD1d in each cell type (Metelitsa et al., 2001). Previous work showed that normal CD14+ monocytes within peripheral blood mononuclear cells (PBMCs) do not activate autologous iNKT cells (Bosma et al., 2012), and human primary B cells incite considerably lower iNKT cell reactivity than autologous lymphoma B cells (Webb et al., 2016), suggesting that CD1d+ primary non-malignant cells are less sensitive than malignant cells to iNKT cell reactivity.
Since normal B cells express both CD1d and CD19, B cell aplasia, as is the case with CAR19-T cells, will also be expected to develop with CAR19-iNKT cell immunotherapy. Of note, adoptive transfer to patients with cancer of in vitro expanded, autologous iNKT cells resulted in no more than grade 2 toxicity (Exley et al., 2017).
In concordance with the in vitro findings, our in vivo model of CD1d+CD19+ B lineage malignancy clearly demonstrated the enhanced anti-tumor effect of CAR19-iNKT over CAR-T cells in terms of both tumor-free and overall survival. This effect is associated with a significantly more robust anti-tumor effect in the first few days post immunotherapy, perhaps a reflection of the physiological role of iNKT cells as the immune cells that modulate an emerging immune response well before conventional T cells (Brigl et al., 2003). It is likely that the combination of swift responses against two tumor-associated targets (i.e., CD19 and CD1d) will induce deeper responses, will limit immune escape associated with loss of the CAR target, and will eventually curtail disease relapse. Highlighting further the importance of early deep remissions, CAR19-iNKT cell-treated animals with the highest tumor burden early following immunotherapy subsequently relapsed, although eventually a second remission was seen in all four animals. This is consistent with the observation that sustained long-term remissions in patients with acute lymphoblastic leukemia (ALL) treated with CAR19-T cell immunotherapy are primarily achieved in those with early complete remissions (Brudno and Kochenderfer, 2018, Park et al., 2018).
In previous work (Heczey et al., 2014, Tian et al., 2016), anti-tumor activity of CAR-iNKT cells required either repeated cell dosing (neuroblastoma) or adjuvant administration of IL-2 (CD19+CD1d– lymphoma), implying a reduced in vivo fitness of CAR-iNKT cells. This contrasts with our findings that a single dose of CAR19-iNKT is sufficient for a drastic anti-lymphoma effect. While the reasons for these differences are not clear, they might be related to differences in the manufacturing process discussed above.
While CAR19-T cells failed to control brain disease, which in most cases was the cause of death, CAR19-iNKT cells almost uniformly eradicated established tumors, implying ability to access the brain. Based on the analysis of the expressed chemokine and corresponding chemokine receptor repertoire in tumor and effectors cells and on published observations (Kim et al., 2002), we postulate that CAR19-iNKT cells gain access to the brain through the BBB and the choroidal plexus and migrate toward the tumor cells. Consistent with this, previous work demonstrated the ability of human iNKT cells to infiltrate subcutaneous lymphoid tumors in NOD/SCID mice (Bagnara et al., 2009).
While a significantly higher disease burden, including in the brain, 3 days after immunotherapy might have underpinned disease relapse in 4 CAR19-iNKT cell-treated animals, the mechanism of subsequent complete regression of relapsed brain disease remains to be defined and it will be an important focus for further investigation. Previous work demonstrated that, determined by the CDR3β region of the Vβ11 chain of the iTCR, human iNKT cells differ widely in terms of iTCR binding affinity to CD1d and its ability to incite proliferation, IFNγ secretion, and cytotoxic activity against CD1d+ targets (Matulis et al., 2010). We speculate that such highly active and fitter CAR19-iNKT cells were at low frequency at the beginning of the experiment and, by selectively surviving, were more effective against the relapsing primary tumor. In contrast to CAR19-T cells, which surrounded and failed to control progressing brain lymphoma, we did not observe CAR19-iNKT cell infiltration of the brain tissue; it is possible that such infiltration receded by the time histological analysis was performed several weeks after clearance of relapsed disease.
Overall, these findings provide the impetus for further development of iNKT cell-based immunotherapy for other types of brain cancers as well as brain lymphoma.
Our in vitro work using CD19+CD1d− targets suggests that CAR19-iNKT would also be effective against CD19+CD1d− B cell malignancies, thus extending the application of CAR-iNKT cell technology to the entire spectrum of B lineage tumors. At the progenitor end of the spectrum, this, as well as common ALL, could also include the adverse prognosis MLL-rearranged CD19+CD1d+ ALL (Fais et al., 2005), while, at the mature end of the spectrum, multiple myeloma (MM), a cancer of plasma cells that we previously showed to express CD1d (Spanoudakis et al., 2009), could be targeted by CAR-iNKT cells. In the case of MM, anti-BCMA CAR immunotherapy has shown promising early clinical efficacy (Rotolo et al., 2016) and thus we envisage that CAR-BCMA-iNKT cells would further improve the prospects of effective immunotherapy against this incurable blood cancer.
In summary, we provide the mechanistic cellular and molecular rationale for developing iNKT cells as a more effective and versatile platform than conventional T cells for CAR-based immunotherapy against CD1d+ B lineage malignancies. Since allogeneic iNKT cells do not incite aGVHD, CAR-iNKT cell immunotherapy would also be suitable for off-the-shelf universal use.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| goat anti-mouse FITC-F(ab’)2 fragment, polyclonal | Invitrogen | Cat# A24519; RRID: AB_2535988, Lot# 46-101-040115 |
| mouse anti-human CD34-APC, QBend10 | Abcam | Cat# FAB7227A; RRID: AB_10972777, Lot# ACOE01 |
| mouse anti-human CD34-FITC, QBend10 | Abcam | Cat# ab78165; RRID: AB_1566006, Lot# GR214126-11 |
| mouse anti-human CD3-PerCP-Cy5.5, OKT3 | eBioscience | Cat# 45-0037-42; RRID: AB_1566006, Lot# E11758-1634 |
| mouse anti-human CD8-Pe-Cy7, RPA-T8 | eBioscience | Cat# 25-0088-42; RRID: AB_1659702, Lot# E10669-1635 |
| mouse anti-human CD4-eFluor450, OKT4 | eBioscience | Cat# 48-0048-42; RRID: AB_2016674, Lot# E10910-1633 |
| mouse anti-human TCRVα24-PE, C15 | Beckman Coulter | Cat# IM2283; RRID: AB_131321, Lot# 33 |
| mouse anti-human TCRVβ11-APC, C21 | Beckman Coulter | Cat# A66905, Lot# 15 |
| mouse anti-human TCRVα24Jα18-FITC, 6B11 | BD Pharmingen™ | Cat# 558371; RRID: AB_397078, Lot# 73592 |
| mouse anti-human CD19-BV421, HIB19 | Biolegend | Cat# 302234; RRID: AB_302234, Lot# B218631 |
| mouse anti-human CD1d-APC, 51.1 | Biolegend | Cat# 350307; RRID: AB_10642029, Lot# B187498 |
| mouse anti-human CD1d-APC, 42.1 | BD Pharmingen™ | Cat# 563505, Lot# 6119717 |
| mouse anti-human CD5-PerCP-eFluor710, UCHT2 | eBioscience | Cat# 46-0059-42; RRID: AB_1834407, Lot# E10854-1633 |
| mouse anti-human CD3-APC-eFluor780, OKT3 | eBioscience | Cat# 47-0038-42; RRID: AB_1272042, Lot# E08437-1636 |
| mouse anti-human CD3-eFluor450, UCHT1 | eBioscience | Cat# 48-0037-42; RRID: AB_1272055, Lot# E08482-1633 |
| mouse anti-human CD4-PerCP-eFluor710, SK3 | eBioscience | Cat# 46-0047-42; RRID: AB_1834401, Lot# E10835-1634 |
| mouse anti-human CD8a-BUV395, RPA-T8 | BD Horizon™ | Cat# 563795; RRID: AB_2722501, Lot# 6084982 |
| mouse anti-human Perforin-FITC, delta-G9 | eBioscience | Cat# 11-9994-41; RRID: AB_1944476, Lot# E11376-1630 |
| mouse anti-human Perforin-FITC, B-D48 | 2BScientific | Cat# 854.952.010; RRID: AB_2169096, Lot# F131108 |
| mouse anti-human GranzymeB-PE, GB11 | eBioscience | Cat# 12-8899-41; RRID: AB_1659718, Lot# E10678-1639 |
| mouse anti-human IFNgamma-APC, 4S.B3 | eBioscience | Cat# 17-7319-41; RRID: AB_10852838, Lot# E14061-104 |
| rat anti-human IL-2-PE, MQ1-17H12 | eBioscience | Cat# 12-7029-42; RRID: AB_2572651, Lot# E17170-102 |
| mouse anti-human IL-4-PeCy7, 8D4-8 | eBioscience | Cat# 25-7049-41; RRID: AB_1659722, Lot# E10585-1632 |
| mouse anti-human IL17A APC-eFluor780 eBio64, DEC17 | eBioscience | Cat# 47-7179-42; RRID: AB_11043559, Lot# E15331-105 |
| mouse anti-human CD3-Pe/Cy7, OKT3 | Biolegend | Cat# 317333; RRID: AB_317333, Lot# B224617 |
| mouse anti-human CD56-Pe/Cy7, 5.1H11 | Biolegend | Cat# 362509; RRID: AB_2563926, Lot# B230713 |
| mouse anti-human CD11b-Pe/Cy7, ICRF44 | Biolegend | Cat# 301322; RRID: AB_830644, Lot# B166859 |
| mouse anti-human CD14-Pe/Cy7, HCD14 | Biolegend | Cat# 325617; RRID: AB_830690, Lot# B161190 |
| mouse anti-human CD16-Pe/Cy7, B73.1 | Biolegend | Cat# 360707; RRID: AB_2562950, Lot# B203769 |
| mouse IgG2bk-APC isotype control, MPC11 | Biolegend | Cat# 400322; RRID: AB_326500, Lot# B202281 |
| mouse IgG1k-APC isotype control, MOPC-21 | BD Pharmingen™ | Cat# 550854; RRID: AB_398467, Lot# 7061976 |
| mouse IgG1-FITC isotype control, 11711 | R&D Systems | Cat# IC002F; RRID: AB_357241, Lot# LGY0914051 |
| mouse IgG1k-PeCy7 isotype control, P3.6.2.8.1 | eBioscience | Cat# 25-4714-41; RRID: AB_1548707, Lot# E10143-1634 |
| mouse IgG1k-APCeFluor780 isotype control, P3.6.2.8.1 | eBioscience | Cat# 47-4714-80; RRID: AB_1271993, Lot# E08469-1636 |
| mouse IgG1-Pe isotype control, X40 | BD Biosciences | Cat# 345816, Lot# 3352966 |
| rat IgG2ak-Pe isotype control, eBR2a | eBioscience | Cat# 12-4321-41; RRID: AB_1518774, Lot# E10138-1631 |
| rabbit anti-histone H3 tri methyl K4 antibody, polyclonal | Abcam | Cat# ab8580; RRID: AB_306649, Lot# GR3175719-1 |
| mouse anti-histone H3 tri methyl K27, mAbcam 6002 | Abcam | Cat# ab6002; RRID: AB_305237, Lot# GR130002-6 |
| rabbit anti-RNA polymerase II CTD repeat YSPTSPS phospho S2, polyclonal | Abcam | Cat# ab5131; RRID: AB_449369, Lot# GR126665-3 |
| rabbit anti-RNA polymerase II CTD repeat YSPTSPS phosphor S5, polyclonal | Abcam | Cat# ab5095; RRID: AB_304749, Lot# GR132981-2 |
| rabbit anti-EZH2, polyclonal | Abcam | Cat# ab195409, Lot# GR3100231-14 |
| mouse anti-retinoic acid receptor alpha, H1920 | Abcam | Cat# ab41934; RRID: AB_777683, Lot# GR298513-4 |
| rabbit IgG, polyclonal | Millipore | Cat# PP64; RRID: AB_97852, Lot# 2146049 |
| mouse IgG, polyclonal | Millipore | Cat# 12-371B; RRID: AB_2617156, Lot# 2138001 |
| mouse anti–human CD19, BT51E | Novocastra | Cat# PA0843 |
| rabbit anti–human CD19, polyclonal | Abcam | Cat# ab99965; RRID: AB_10672632 |
| mouse anti–human CD3, PS1 | Novocastra | Cat# RTU-CD3-PS1; RRID: AB_563542 |
| rabbit anti–human CD3, SP7 | Abcam | Cat# ab16669; RRID: AB_443425 |
| Bacterial and Virus Strains | ||
| Turbo Competent E.coli, F′ ´proA+B+ lacIq Δ lacZ M15/ fhuA2 Δ(lac-proAB) glnV gal R(zgb- 210::Tn10)TetS endA1 thi-1 Δ(hsdS-mcrB)5 | New England Biolabs | Cat# C2984I |
| All lenti- and retro-viruses were VSV-G pseudotyped | This paper | N/A |
| Biological Samples | ||
| Healthy donors and B lymphoproliferative disorders (MCL, MZL, CLL) patients’ peripheral blood/apheresis | Hammersmith Hospital | N/A |
| Xenografts tissues | This paper | N/A |
| Chemicals, Peptides, and Recombinant Protein | ||
| KRN7000 (α-galactosylceramide) | BioVision | Cat# 2152 250 |
| All-trans retinoic acid (ATRA) | Sigma-Aldrich | Cat# 2625 |
| GSK343 | Sigma-Aldrich | Cat# SML0766 |
| Dimethyl sulfoxide (CryoSure-DMSO) | WAK-Chemie Medical GmbH | Cat# WAK-DMSO-10 |
| Polybrene | Sigma-Aldrich | Cat# H9268 |
| Poly-L-ornithine | Sigma-Aldrich | Cat# P4957 |
| Dynabeads™ Human T-Activator CD3/CD28 for T Cell Expansion and Activation | Gibco™ | Cat# 11131D |
| Human IL-2 IS, premium grade | Miltenyi Biotec | Cat# 130-097-745 |
| Human IL-15, premium grade | Miltenyi Biotec | Cat# 130-095-765 |
| Cell Stimulation Cocktail (500X) | eBioscience | Cat# 00-4970-93 |
| Monensin Solution (1000x) 1 mL | eBioscience | Cat# 00-4505-51 |
| Brefeldin A Solution (1000X) 1 mL | eBioscience | Cat# 00-4506-51 |
| CellTrace™ Violet | Invitrogen™ | Cat# C34571 |
| 7-AAD | Invitrogen™ | Cat# 00-6993-50 |
| 4',6-Diamidino-2-Phenylindole, Dilactate (DAPI) | Invitrogen™ | Cat# D3571 |
| Isoflurane | Zoetis UK | Vm 42058/4195 |
| D-luciferin | Goldbio | Cat# LUCK-1G |
| Formalin solution, neutral buffered, 10% | Sigma-Aldrich | Cat# HT501128 |
| Acetic acid, zinc chloride and formaldehyde solution (AZF) | Genta Medical | N/A |
| Xylenes | Sigma-Aldrich | Cat# 214736 |
| Ethanol | Sigma-Aldrich | Cat# E7023 |
| Haematoxylin | Sigma-Aldrich | Cat# H3136 |
| Eosin | Sigma-Aldrich | Cat# 230251 |
| Hydrochloric Acid | Sigma-Aldrich | Cat# 1.09057 |
| Sodium citrate tribasic dihydrate | Sigma-Aldrich | Cat# C8532 |
| Tween-20 | Sigma-Aldrich | Cat# P1379 |
| Triton-X100 | Sigma-Aldrich | Cat# X100 |
| Saponin | Sigma-Aldrich | Cat# 47036 |
| Normal goat serum | Abcam | Cat# ab7481 |
| Gadovist (Gadobutrol) | Bayer | PL 00010/0535 |
| Critical Commercial Assays | ||
| ProcartaPlex HumanCytokine Panel 1B (25 plex) | Invitrogen™ | Cat# EPX250-12166-901 |
| Taqman Gene Expression Master Mix and Assays | Applied Biosystems | Cat# 4369016 |
| Deposited Data | ||
| CAR-iNKT/CAR-T cell/C1R cell line RNA-seq data | This paper | EGAS00001003176 |
| Experimental Models: Cell Lines | ||
| Lenti-X-293T | Clontech | Cat# 632180 |
| K562 | ATCC | Cat# ATCC CCL-243 |
| ARH-77 | DSMZ | Cat# ACC 512 |
| KMS-12-BM | DSMZ | Cat# ACC 551 |
| NCI-H929 | DSMZ | Cat# ACC 163 |
| U266 | DSMZ | Cat# ACC 9 |
| C1R and C1R-CD1d | Prof Vincenzo Cerundolo, University of Oxford, Oxford, UK | N/A |
| Farage | Prof Ronald Gartenhaus, University of Maryland School of Medicine, Baltimore, USA | N/A |
| Experimental Models: Organisms/Strains | ||
| NOD/SCID/IL-2Rγ-null NSG | Charles River | N/A |
| Oligonucleotides | ||
| 19-CD28ζ CAR forward: 5’-CCCAGCACCTCCCGTGGCCGGCCCGTCA GTCTTCTGGGTCCTGGTGGTGG |
This paper | N/A |
| 19-CD28ζ CAR reverse: 5’-CGATAAGCTTGATAT CAAGCTTGCATGCCTGCAGGTCATCTGGGTG |
This paper | N/A |
| 19-CD28OX40ζ CAR forward: 5’-CCCAGCACCTCCCGTGGCCGGCCC GTCAGTCTTTTGGGTGCTGGTGGTG |
This paper | N/A |
| 19-CD28OX40ζ CAR reverse: 5’-CGATAAGCTTGATA TCAAGCTTGCATGCCTGCAGGTTAGCGAGGAGGC |
This paper | N/A |
| CD1D Hs00939888_m1 | Applied Biosystems | Cat# 4331182 |
| ACTB Hs99999903_m1 | Applied Biosystems | Cat# 4331182 |
| GAPDH Hs03929097_g1 | Applied Biosystems | Cat# 4331182 |
| U266 ChIP, CD1D forward: 5’-CCCTGAGAAAGTGACCTTGG |
This paper | N/A |
| U266 ChIP, CD1D reverse: 5’-TGGCTGTTAGCTTTCAGTTCC |
This paper | N/A |
| U266 ChIP, GAPDH forward: 5’-CCGGGAGAAGCTGAGTCATG |
This paper | N/A |
| U266 ChIP, GAPDH reverse: 5-TTTGCGGTGGAAATGTCCTT |
This paper | N/A |
| U266 ChIP, HOXA2 forward: 5’-AGGAAAGATTTTGGTTGGGAAG |
This paper | N/A |
| U266 ChIP, HOXA2 reverse: 5’-AAAAAGAGGGAAAGGGACAGAC |
This paper | N/A |
| CLL ChIP, CD1d distal promoter forward: 5'-TGGACGTCCGAGAGGTAAGAG |
This paper | N/A |
| CLL ChIP, CD1d distal promoter reverse: 5'-CACAGTAACCTGGAGATCCACTA |
This paper | N/A |
| CLL ChIP, CD1d proximal promoter forward: 5'-AATGATGCTGGGGTGTGAGG |
This paper | N/A |
| CLL ChIP, CD1d proximal promoter reverse: 5'-GCACGGCCTGCAAGATTATG |
This paper | N/A |
| CLL ChIP, CD1d exon 2 forward: 5'-CTCCAGATCTCGTCCTTCGC |
This paper | N/A |
| CLL ChIP, CD1d exon 2 reverse: 5'-CTGGGACCAAGGCTTCAGAG |
This paper | N/A |
| Recombinant DNA | ||
| 19-IgGFc-CD28ζ CAR | Martin Pule | N/A |
| 19-IgGFc-CD28OX40ζ CAR | Martin Pule | N/A |
| 19-CD28ζ CAR | This paper | N/A |
| 19-CD28OX40ζ CAR | This paper | N/A |
| SFG-dCD19 | Martin Pule | N/A |
| pHRSIN-CD1d | Vincenzo Cerundolo | N/A |
| SFG-CD1d | This paper | N/A |
| SFG-CD1d.2A.dCD19 | This paper | N/A |
| SFG-ffLuciferase.tdRFP | Whilding et al., 2017 | N/A |
| Software and Algorithms | ||
| FlowJo v10 | www.flowjo.com | http://docs.flowjo.com/ |
| IncuCyte™ software | EssenBio/Sartorius | https://www.essenbioscience.com/en/resources/incucyte-zoom-resources-support/ |
| Living Image software | PerkinElmer | http://www.perkinelmer.com/lab-products-and-services/resources/in-vivo-imaging-software-downloads.html |
| Paravision 6.01 | Bruker, BioSpin | https://www.bruker.com/service/support-upgrades/software-downloads/mri.html |
| OsiriX | Pixmeo SARL | http://www.osirix-viewer.com/download_form/download_form.php |
| jMRUI | MRUI Consortium | http://www.jmrui.eu/license-and-download/ |
| GraphPad Prism 7 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| FastQC (version 0.64) | Babraham Bioinformatics https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ | https://toolshed.g2.bx.psu.edu/repository/display_tool?repository_id=ca249a25748b71a3&render_repository_actions_for=tool_shed&tool_config=%2Fsrv%2Ftoolshed%2Fmain%2Fvar%2Fdata%2Frepos%2F000%2Frepo_829%2FrgFastQC.xml&changeset_revision=28d39af2dd06 |
| bcl2fastq2 Conversion Software v2.18 | http://emea.support.illumina.com/sequencing/sequencing_software/bcl2fastq-conversion-software/downloads.html# | https://support.illumina.com/sequencing/sequencing_software/bcl2fastq-conversion-software/downloads.html |
| STAR (version 2.5.3a) | (Dobin et al., 2013) | https://github.com/alexdobin/STAR |
| R package Rsubread (version 1.24.2) | (Liao et al., 2013) | https://bioconductor.org/packages/release/bioc/html/Rsubread.html |
| R package edgeR v3.20.9 | (Robinson et al., 2010) | https://bioconductor.org/packages/release/bioc/html/edgeR.html |
| TopHat 2.0.14 | (Kim et al., 2013) | http://ccb.jhu.edu/software/tophat/downloads/ |
| Cufflinks 2.2.1 | (Trapnell et al., 2010) | http://cole-trapnell-lab.github.io/cufflinks/releases/v2.2.1/ |
| Other | ||
| Anti-iNKT, microbeads, human | Miltenyi Biotec | Cat# 130-094-842 |
| CD34 MicroBead Kit UltraPure, human | Miltenyi Biotec | Cat# 130-100-453 |
| NucleoSpin RNA kit | Macherey Nagel | Cat# 740955 |
| RevertAid first strand cDNA synthesis kit | Thermo Fisher Scientific | Cat# K1621 |
| Qubit RNA HS Assay kit | Life Technologies | Cat# Q32852 |
| RNA 6000 pico kit | Agilent | Cat# 5 067-1513 |
| NEBNext® Ultra™ II Directional RNA Library Prep Kit for Illumina with rRNA depletion | New England Biolabs | Cat# E7760S/L |
| NEBNext poly(A) mRNA Magnetic Isolation Module | New England Biolabs | Cat# E7490S/L |
| NEBNext Ultra RNA Library Prep kit for Illumina | New England Biolabs | Cat# E7530S/L |
| Qubit dsDNA High Sensitivity Assay kit | Life Technologies | Cat# Q32851 |
| Bioanalyser High Sensitivity DNA kit | Agilent | Cat# 5067-4626 |
| NEBNext® Library Quant Kit for Illumina | New England Biolabs | Cat# E7630S/L |
Contact for Reagent and Resource Sharing
Further information and requests for reagents may be directed to, and will be fulfilled by the corresponding author Anastasios Karadimitris (a.karadimitris@imperial.ac.uk).
Experimental Model and Subject Details
Primary Cells
Healthy volunteer peripheral blood (PB) and lymphapheresis samples as well as PB samples from mantle cell lymphoma (MCL), marginal zone lymphoma (MZL) and chronic lymphocytic leukemia (CLL) patients were obtained after written informed consent and research ethics committee approval (Research Ethics Committee reference: 11/H0308/9). Peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation and were used as a source of either CD3+ lymphoid cells for CAR engineering or CD19+ tumor cell targets for functional assays. In order to generate CAR iNKT cells, TCRVα24Jα18+ lymphocytes were immunomagnetically purified from PBMC and apheresis mononuclear cells using anti-human iNKT cell microbeads (Miltenyi Biotec). Primary cells were maintained in RPMI1640, 10% FBS, 2% glutamine, 1% Penicillin-Streptomycin, 1% sodium pyruvate; 1% essential and non-essential aminoacids, 10 mM Hepes buffer (Sigma-Aldrich) and 5.5x10-5 M beta-mercaptoethanol (Gibco®, Life Technologies). For CD3+ cells, human IL2 and/or IL-15 (premium grade, Miltenyi Biotec) were added at 100-200 IU/ml unless otherwise stated.
Systemic Xenograft Tumor Model
Six-week old NOD/SCID/IL-2Rγ-null (NSG) sex-matched mice were handled in accordance with the 1986 Animal Scientific Procedures Act and under a United Kingdom Government Home Office–approved project licence 70/8586. The animals were housed at the Hammersmith Central Biomedical Services (CBS) facility, Imperial College London. On day 0 all animals received 5 × 106 luciferase-expressing C1R-CD1d cells by tail vein (iv) injection, followed by bioluminescence imaging (BLI) monitoring twice a week. Upon confirmation of engraftment, defined on the basis of increased photon signal in two consecutive scans performed 72 hr apart, on day 11 the mice were randomized to no treatment or immunotherapy with either T, iNKT, 2nd generation CAR19-T or 2nd generation CAR19-iNKT cells generated from the same donor. Thereafter, BLI was performed twice a week until day 21 and weekly until the end of experiment on day 90. Primary endpoints were overall survival and tumor-free survival. Secondary endpoint was brain tumor-free survival. All mice were sacrificed according to protocol when either experimental or humane endpoints were reached.
Cell Lines
The adherent Human Embryonic Kidney 293 (Lenti-X-293T) cells were purchased from Clontech and used for lentivirus production. Lenti-X-293T cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) (Sigma-Aldrich), supplemented with 10% Fetal Bovine Serum (FBS) (Sigma-Aldrich), 2 mM L-Glutamine and 10 ml/L Penicillin-Streptomycin (Stem Cell Technologies). The K562 cells were obtained from ATCC, while the ARH-77, KMS-12-BM, NCI-H929 and U266 cell lines were purchased from DSMZ. C1R and C1R-CD1d cell lines were provided by Prof Cerundolo (Medical Research Council (MRC) Human Immunology Unit, Weatherall Institute of Molecular Medicine, University of Oxford, Oxford). The Farage cell line was kindly donated by Prof Ronald Gartenhaus (University of Maryland School of Medicine, Baltimore, MD USA). All suspension cell lines were grown in RPMI-1640 medium (Sigma-Aldrich) supplemented with 10-20% Fetal Bovine Serum (FBS) (Sigma-Aldrich), 2 mM L-Glutamine and 10 ml/L Penicillin-Streptomycin (Stem Cell Technologies).
All cell lines were tested for mycoplasma contamination using the MycoAlert Mycoplasma Detection Kit (Lonza). The K562, C1R and ARH-77 cell lines were transduced to express the human CD19 and/or CD1d. For the purposes of in vitro functional assays, ARH-77 cells were also modified with the pHR-SIN plasmid described above to co-express the mCherry red fluorescent protein together with CD1d+. The C1R-CD1d cell line was modified with the luciferase-tdRFP plasmid for in vitro and in vivo monitoring by fluorescence and bioluminescence imaging (BLI) respectively.
Method Details
Vectors and Constructs
To generate CD19-specific CAR iNKT and T cells, a 2nd generation 19-IgGFc-CD28ζ and 3rd generation 19-IgGFc-CD28OX40ζ CAR constructs with FMC63-derived scFv (Almasbak et al., 2015) were kindly donated by Dr Martin Pule, University College London. To protect CAR-expressing cells from in vivo opsonisation (Almasbak et al., 2015), the original IgGFc-derived spacers were modified by removing the CH2 and CH3 extracellular domains and the resulting new constructs were re-cloned into pSew lentiviral vector via overlapping PCR using Gibson assay (New England Biolabs). The primer pairs used were: 2nd generation CAR 5’- CCCAGCACCTCCCGTGGC
CGGCCCGTCAGTCTTCTGGGTCCTGGTGGTGG and 5’- CGATAAGCTTG
ATATCAAGCTTGCATGCCTGCAGGTCATCTGGGTG; 3rd generation CAR
5’- CCCAGCACCTCCCGTGGCCGGCCCGTCAGTCTTTTGGGTGCTGGTG
GTG and 5’- CGATAAGCTTGATATCAAGCTTGCATGCCTGCAGGTTAGCG
AGGAGGC. The modular structure is provided in Figure S1A. The RQR8 marker/suicide (Philip et al., 2014) gene was maintained upstream of the CARs with an intervening FMD-2A peptide to allow early detection of CAR-transduced cells as previously described (Philip et al., 2014). To generate CD19 and CD1d single- or double-expressing -cells, two sequences encoding for the human CD19 and CD1d, obtained from Dr Martin Pule and Prof Vincenzo Cerundolo, University of Oxford, respectively, were cloned singly or together with an interposed FMD2A fragment into a retroviral SFG vector. For the purposes of real-time in vitro monitoring of CAR cell cytotoxic activity, mCherry-labeled CD1d+ B cell targets were generated by using a lentiviral pHR-SIN plasmid encoding for a hCD1d-mCherry fusion protein (Prof Vincenzo Cerundolo). To detect tumor cells and monitor tumor growth in vivo, firefly luciferase was co-expressed with tandem dimer Tomato red fluorescent protein (tdRFP) in a single SFG vector as previously described (Whilding et al., 2017).
Pharmacologic Agents
α-galactosylceramide (KRN7000, Cambridge Bioscience), all-trans retinoic acid (ATRA, Sigma-Aldrich) and the EZH2 inhibitor GSK343 (Sigma-Aldrich) were purchased in lyophilized form. Stock solutions were prepared in 100% dimethyl sulfoxide (DMSO) at 1 mg/ml (0.001 M), 3 mg/ml (0.01 M), 15 mg/ml (0.03 M) respectively. The DMSO solution of α-galactosylceramide (αGalCer) was completely dissolved by heating at 80° for 1 hr, aliquoted and stored at -20°C until use. Prior to use, a working solution was prepared by heating for another 60 seconds at 80°C, followed by dilution in PBS at 100 μg/ml (1000x). ATRA and GSK343 were used to assess the transcriptional regulation of CD1d in primary CLL cells and the U266 cell line. The ATRA solution in DMSO was freshly prepared before each experiment protected from light and diluted in PBS to 1 mM (1000X) for immediate use. The GSK343 stock solution was stored at -20°C and diluted in PBS to 1 mM (1000x) prior to use. CLL cells were treated with 10-6 M ATRA. The U266 cells were harvested during their exponential growth and treated with either 10-6 M ATRA or 10-6M GSK343 or a combination of both. 0.01% DMSO was used as control. In all cases, the cells were incubated for up to 96 hr before proceeding to RT-PCR, flow cytometry and ChIP/re-ChIP assays at the indicated time points.
Retroviral and Lentiviral Vector Constructs, Viral Production and Transduction
VSV-G pseudotyped retroviruses and lentiviruses were generated by transfection of 80% confluent Lenti-X-293T cells with the transfer, packaging and envelope plasmids using the CaCl2 method (Kutner et al., 2009). pCMV-Gag-Pol and pVSV-G were used for retrovirus, or pRsv-REV, pMDlg-pRRE and pMD2G were used for lentivirus respectively. Virus supernatant was harvested at 48 and 72 hr post transfection, centrifuged and filtered through a 0.45 μm cellulose acetate filter, concentrated by ultracentrifugation at 23,000 × g 4°C for 120 min and re-suspended with 1 × RPMI-1640 medium (Sigma-Aldrich). Cell lines were transduced with retrovirus in the presence of 8 μg/ml polybrene (Sigma-Aldrich). Two days later, transduction efficiency was determined by flow cytometry as percentage of CD19+ and/or CD1d+ cells or tdRFP+ cells. Where required, positive cells were sorted by immuno-magnetic selection or fluorescence-activated cell sorting (FACS) and further expanded in RPMI-1640 supplemented with 10% Fetal Bovine Serum (FBS, Gibco) and 1% Penicillin-Streptomycin (Pen/Strep, Stem Cell Technologies) (standard culture medium). To generate CAR-engineered T cells, mononuclear cells were activated with Dynabeads Human T-Activator CD3/CD28 (Gibco™) at 1:1 beads-to-cell ratio in complete cell medium prepared as described above, supplemented with IL-15 (Miltenyi Biotec). Activated T were transduced and expanded as reported below for iNKT cells.
Generation of CAR19-iNKT Cells as per Protocol 4
TCRVα24Jα18+ lymphocytes were immunomagnetically sorted from PBMC or apheresis mononuclear cells using anti-human iNKT cell microbeads (Miltenyi Biotec). Purified iNKT cells were seeded in round-bottom 96-wells at 1:1 ratio with irradiated (3500 rad) autologous mononuclear cells (iAPC) and activated with Dynabeads Human T-Activator CD3/CD28 (Gibco™) at 1:1 beads-to-cell ratio in T cell medium at a density of 1-5 x 104 cells per ml. IL-15 (Miltenyi Biotec) at 30 IU/ml and 150 IU/ml was added at the time of seeding and 12 hr later respectively. Within 24-48 hr, activated iNKT cells were transduced with concentrated CAR lentivirus using an MOI of 2-5 in the presence of 4 μg/ml polybrene, with spinoculation for 90 min at 1000G. After 8-12 hr, cells were resuspended in fresh medium supplemented with 150 IU/ml of IL-15 and let rest for 2 to 4 days before assessment for viability and CAR expression. Transduction efficiency was determined by flow cytometry as percentage of RQR8+ cells relative to untransduced controls as previously described (Philip et al., 2014). Where required, CAR+ cells were immunomagnetically sorted by using anti-CD34-microbeads (Miltenyi), re-plated as above and re-stimulated with 1:1 irradiated C1R-CD1d cells, IL-15 (30 IU/ml) and αGalCer (100 ng/ml), with additional 150 IU/ml of IL-15 12-24 hr later. After 7 days cells were counted, assessed for purity by flow cytometry using a F(ab')2-Goat anti-Mouse antibody (Invitrogen) and switched to a low IL-15 (down to 20 IU/ml) medium for additional 7 days towards resting phase prior to functional studies. Alternatively, CAR19-iNKT cells were propagated with IL-15 (100 IU/ml) every other day for up to 4-6 days, harvested during exponential growth phase, cryopreserved in 10% DMSO and stored in liquid nitrogen until use.
Antibodies and Intracellular Staining
CAR+ cells were identified by using the mouse anti-human APC-CD34 (QBend10, R&D Systems) or FITC-CD34 (QBend10, Abcam) monoclonal antibody (MoAb) against the RQR8 marker or the goat anti-mouse FITC-F(ab’)2 fragment (Invitrogen™) against the CAR hinge. For T cell phenotyping, the following antibodies were used: mouse anti-human PerCP-Cy5.5-CD3 (OKT3, eBioscience), Pe-Cy7-CD8 (RPA-T8, eBioscience) and eFluor450-CD4 (OKT4, eBioscience). iNKT cells were defined as TCRVα24+, TCRVβ11+, TCRVα24Jα18+ T cells using mouse anti-human PE-TCRVα24 (C15, Beckman Coulter), APC-TCRVβ11 (C21, Beckman Coulter), or FITC-TCRVα24Jα18+ (6B11, BD) pairwise combinations. Either a minimum of 0.5 x106 CD3+ cells or 200 iNKT cells were recorded to facilitate accurate calculation of the total, CD4+ and CD4− iNKT cell frequencies. The B cell panel included the mouse anti-human BV421-CD19 (HIB19, BD Biosciences), APC-CD1d (51.1, Biolegend), PerCP-eFluor710-CD5 (UCHT2, eBioscience), APC-eFluor780-CD3 (OKT3, eBioscience). 4',6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI) staining was used to exclude dead cells. For intracellular cytokine profiling, T cells were fixed and permeabilized using BD Cytofix/Cytoperm Plus kit as per the recommendation of the manufacturer, followed by staining with mouse anti-human eFluor450-CD3 (UCHT1, eBioscience), PerCP-eFluor710-CD4 (SK3, eBioscience), BUV395-CD8a (RPA-T8, eBioscience), FITC-Perforin (δ-G9, eBioscience and B-D48, 2BScientific), PE-GranzymeB (GB11, eBioscience), APC-IFNγ (4S.B3, eBioscience), PE-IL-2 (MQ1-17H12, eBioscience), PeCy7-IL-4 (8D4-8, eBioscience) and APC-eFluor780-IL17A (eBio64DEC17, eBioscience). Cytokine profile was assessed in resting cells and upon stimulation for 4 hr with Dynabeads (1:1) or PMA/ionomycin (eBioscience) or C1R CD1d cells (1:1) in the presence of Monensin and Brefeldin (eBioscience).
Multiplex Cytokine Quantification Assays
CAR-modified cells were stimulated for 3 and 8 hr with Dynabeads at 1:1 ratio. Supernatants were collected and analyzed with a Human ProcartaPlex immunoassay (Invitrogen) using the Luminex assay according to the manufacturer’s instructions.
Proliferation Assays
Up to 104 irradiated C1R-CD1d cells were plated in poly-L-ornithine-coated, flat-bottom 96-well microplates. Where required, αGalCer or vehicle were added for 4 hr before the addition of up to 104 CAR-modified cells in RPMI 1640 medium supplemented with 10% FBS, 1% Pen/Strep and 20 IU ml−1 IL-15. The cell plate was maintained at 37°C and 5% CO2 into the IncuCyte ZOOM® instrument (Sartorius) for up to one week without any further manipulation. CAR cell proliferation was determined by the IncuCyte™ software, with a 24 hr scanning scheduled for every 1 hr with 10x or 4x objectives and using the confluence algorithm according to the manufacturer’s instructions (EssenBio protocol 8000-0331-A00).
In Vitro Real-Time Monitoring of CAR Cell Cytotoxic Activity
CD1d-mCherry-transduced ARH-77 cells were seeded in a 96-well round-bottom ultra-low attachment (ULA) microplate (Corning) with standard culture medium at a density of 500 cells/well and let settle for 1 hr. Effector cells were then re-suspended in standard medium with 20 IU ml−1 IL-15 and added at the indicated ratio in triplicates. Wells with targets alone and effectors were included as controls. The microplate was maintained at 37°C and 5%CO2 into the IncuCyte ZOOM® instrument for up to one week without any further manipulation and scanned using a phase and a red channel every 1 hr with 10x objective during the first 48 hr and 4x objective from day 3 until the end of the experiment. Targets cells were monitored as red fluorescent objects and quantified with IncuCyte™ software by using red mean image fluorescence (MIF) and red fluorescence area (μm2/image) according to the manufacturer’s instructions (EssenBio protocol 8000-0330-B00). CAR cell proliferation was determined as described in the paragraph ‘Long-term proliferation assay’.
Cytotoxicity Assays
CellTrace™ Violet (Invitrogen)-labeled targets were incubated at the indicated ratios with effector cells for 3 hours. Where required, targets were labeled with an antibody mix containing PeCy7-conjugated mouse anti-human CD3 (OKT3, Biolegend), CD56 (5.1H11, Biolegend), CD11b (ICRF44, Biolegend), CD14 (HCD14, Biolegend) and CD16 (B73.1, Biolegend) mAbs to allow discrimination between CD19+ and CD19- mononuclear cells. As controls, targets and effectors alone were simultaneously incubated to determine spontaneous cell death. Where indicated, targets were pre-incubated with αGalCer or vehicle at 37°C for 4 hr before addition of the effector cells. Cells were then harvested and 7-AAD was added prior to flow cytometric analysis on BD Fortessa Flow Cytometer, using BD FACSDiva software version 6.0. Specific cytotoxic activity was determined as ((% sample (7-AAD+, Violet+) − % spontaneous (7-AAD+, Violet+)) / (100 - %spontaneous (7-AAD+, Violet+)) x 100. All assays were run in duplicates or triplicates. Data analysis was performed using FlowJo 10.2.
Gene Expression Analysis
Total RNA from primary CLL cells and U266 cell line was extracted using the NucleoSpin RNA kit (Macherey Nagel), followed by cDNA synthesis with RevertAid first strand cDNA synthesis kit (Thermo Fisher Scientific), as per the manufacturer’s instructions. For gene expression quantification, RQ-PCR of template cDNA was performed in triplicate on StepOnePlus™ Real-Time PCR System using Taqman Gene Expression Master Mix and Assays (Applied Biosystems). CD1d transcript levels were determined relative to the reference genes ACTB and GAPDH, using the ΔΔCT method. Taqman probes were CD1D Hs00939888_m1, ACTB Hs99999903_m1 and GAPDH Hs03929097_g1.
Chromatin Immunoprecipitation Assays
Chromatin immunoprecipitation (ChIP) combined with real-time quantitative polymerase chain reaction (ChIP-RQ-PCR) was performed for anti-H3K4me3, anti-H3K27me3, anti-RNA polymerase II CTD phospho Ser2, anti-RNA polymerase II CTD phospho Ser5, anti-EZH2, anti-RARα and control IgG as previously described (Caputo et al., 2013). For Re-ChIP assays, the wash and chromatin elution steps of the first IP were performed with protease inhibitor-containing buffers and the first elution was performed by incubating the magnetic beads in 10 mM DTT/TE for 30 min at 37°C. The eluate of the first IP was diluted at least 20X in ChIP dilution buffer, followed by a second IP according to the same protocol as above. Expression of immunoprecipitated DNA was calculated, either relative to input DNA or DNA immunoprecipitated by control IgG antibody, using the ΔΔCT method.
ChIP Primers
For the experiments with U266 cells, ChIP primers were designed to analyze the upstream regulatory element of the CD1D gene, approximately 1.5kb from the ATG translational start site, consistent with the reported location of the RARE (Chen and Ross, 2007). As controls, the upstream regulatory regions of HOXA2, a putative target of polycomb mediated repression, known to be marked by bivalent histone modifications, and GAPDH, as transcriptionally active housekeeping gene, were also evaluated. The primer sequences pairs used were: CD1D 5-CCCTGAGAAAGTGACCTTGG and 5’-TGGCTGTTAGCTTTCAGTTCC, GAPDH 5’-CCGGGAGAAGCTGAGTCATG and 5-TTTGCGGTGGAAATGTCCTT, HOXA2 5’-AGGAAAGATTTTGGTTGGGAAG and 5’-AAAAAGAGGGAAAGGGACAGAC. For the experiments with primary CLL cells, 3 primer sets were designed to analyze 2 regions upstream the ATG start codon at -3047 (distal, DP) and -1240 (proximal, PP) and 1 region within exon 2 at +382 (I2P). The corresponding sequences were: DP 5'-TGGACGTCCGAGAGGTAAGAG and 5'-CACAGTAACCTGGAGATCCACTA, PP 5'-AATGATGCTGGGGTGTGAGG and 5'-GCACGGCCTGCAAGATTATG, I2P 5'-CTCCAGATCTCGTCCTTCGC 5'-CTGGGACCAAGGCTTCAGAG.
Systemic Xenograft Tumor Model
On day 0, six-week old NOD/SCID/IL-2Rγ-null (NSG) sex-matched mice received 5 × 106 luciferase-expressing C1R-CD1d cells by tail vein (iv) injection, followed by bioluminescence imaging (BLI) monitoring twice a week. Upon confirmation of engraftment, defined on the basis of increased photon signal in two consecutive scans performed 72 hr apart, on day 11 the mice were randomized to no treatment or immunotherapy with either T, iNKT, 2nd generation CAR19-T or 2nd generation CAR19-iNKT cells generated from the same donor. Thereafter, BLI was performed twice a week until day 21 and weekly until the end of experiment on day 90. Primary endpoints were overall survival and tumor-free survival. Secondary endpoints were acute Graft-versus-Host disease (aGvHD) and brain tumor-free survival. aGvHD was assessed at least twice a week by means of a five-parameters scoring system incorporating non-tumor related weight loss greater than 10%, hunched posture, reduced activity, raffled fur and scaling skin, as previously described (Cooke et al., 1996). Specifically, each criterion was graded from 0 to 2 according to severity and finally a clinical aGvHD score (min 0 = no GvHD, max 10 = severe aGvHD) was obtained. All mice were sacrificed according to protocol when either experimental or humane endpoints were reached.
Bioluminescence Imaging (BLI)
Bioluminescence images were collected on an IVIS Lumina XR III Imaging System using Living Image software (PerkinElmer). Before imaging, mice were anesthetized and maintained under inhalational anesthesia via a nose cone with 2% isoflurane (Zoetis UK)/medical oxygen. All mice received a single intraperitoneal (IP) injection of 150 mg/Kg D-luciferin (Goldbio) in PBS 10 min before scanning. Up to three mice were imaged simultaneously in a 12.5 cm field of view (FOV) with minimum target count of 30,000 and exposure times ranging from 0.5 to 2 min at medium binning, with additional images acquired at low binning levels to maximize sensitivity and spatial resolution where required. Both ventral and dorsal scans were acquired for each mouse. The dorsal and ventral signals were quantitated separately through region of interest (ROI) analysis using Living Image software and expressed in radiance (unit of photons/s/cm2/sr) as a total signal summation normalized to the ROI area. Where required, normalized background signal from similarly sized ROIs was subtracted.
Magnetic Resonance Imaging (MRI) and MR Spectroscopy (MRS)
Brain tumors were assessed and monitored with MRI and MRS in 12 animals. All MRI scans were performed on a pre-clinical 9.4 T scanner (94/20 USR Bruker BioSpec; Bruker Biospin, Ettlingen, Germany) housed at the Biological Imaging Centre, Imperial College London. Mice were anesthetized as described above and positioned prone in a dedicated mouse bed provided with a circulating warm water heat mat to control body temperature. Respiration and body temperature were continuously monitored (1030-MR, SA Instruments, Stony Brook, NY, USA) and the amount of isoflurane and heat delivered were adjusted through the MRI scans to maintain the respiratory rate within the range of 35-45 breaths per min and the body temperature at 36.5°C. Brain images were acquired with Paravision 6.01 (Bruker, BioSpin) using an 86 mm inner diameter volume transmit quadrature coil combined with an actively decoupled mouse brain array receiver. The imaging datasets consisted of T1 weighted FLASH and T2 weighted RARE sequences in sagittal, axial and coronal orientation obtained within 10 min and 25 min respectively after iv injection of Gadovist (Gadobutrol, Bayer). The contrast agent was diluted in 0.9% saline and used at a concentration of 0.3 mmol/kg in all but 1 mouse. For T1 FLASH images the following settings were applied: T1 sagittal: TR/TE = 250/2.6 ms; FOV = (18 x 14) mm2, in plane spatial resolution (58 x 56) μm2, slice thickness 500 μm, 10 μm slice gap, 20 slices, scan time 6 min 30 s; T1 axial: TR/TE=320/2.6 ms; FOV = (16 x 14) mm2, in plane spatial resolution (62 x 61) μm2, slice thickness 500 μm, 10 μm slice gap, 30 slices, scan time 6 min 30 s, T1 coronal: TR/TE = 200/3 ms, FOV = (14 x x16) μm2, in plane resolution (34 x 62) μm2, 500 μm slice thickness, slice gap 50 μm, 10 slices. Scan time 3 min 50 s. T2 RARE images were generated with: T2 sagittal: TR/TE = 3000/40 ms, FOV = (18 x 14) mm2, in plane spatial resolution (70 x 55) μm2, slice thickness 500 μm, 10 μm slice gap, 20 slices, scan time 5 min. T2 axial: TR/TE = 2500/45 ms, FOV = (14 x 14) μm2, in plane resolution (55 x 55) μm2, 700 μm slice thickness, slice gap 50 μm, 12 slices. Scan time 4 min. T2 coronal: TR/TE = 3600/40 ms, FOV = (18 x 14) mm2, in plane spatial resolution (70 x 55) μm2, slice thickness 500 μm, 10 μm slice gap, 30 slices, scan time 6 min. All images were analyzed using OsiriX software. Image quality and contrast enhancement were determined based on assessment of contrast-to-noise ratio between the areas of clinical relevance (regions of interests (ROI) with higher signal intensity, SA) and same-size, reference areas with no evidence gadolinium uptake (ROI with lower signal intensity, SB), scaled to image noise, according to the following formula: CNR = | SA – SB | / σo, where σo is the standard deviation of the pure image noise. For MRS, the voxel was positioned within the pituitary gland avoiding inclusion of surrounding tissue. Field-map based shimming (up to 4th order) was performed prior to the MRS acquisition to optimize the main field homogeneity in the voxel of interest. Single voxel spectra (SVS) were acquired at both long and short echo times (LTE and STE respectively), with: LTE PRESS: TR/TE = 2500/100 ms, voxel size (2 x 1.2 x 1.35) mm3, total scan time 13 min 20 s; STE STEAM: 2500/3 ms, voxel size (2 x 1.2 x 1.35) mm3, total scan time 13 min 20 s. Relative quantification of Creatine/NAA, Choline/NAA ratios was computed from the LTE spectra. The spectra were pre-processed (phased, apodized) and quantified afterwards using AMARES (jMRUI software).
Histologic Analysis
Mouse tissues were fixed in 10% Neutral Buffered Formalin for up to 36hr. Animal heads underwent additional treatment with aceto-zinc formalin for 12 hr and Gooding and Stewart’s decalcification fluid for 8.5 hr prior to further processing. All samples were then processed on a Tissue Tek Sakura VIP 5. Blocks were embedded in paraffin wax and sectioned on Polysine coated slides at 1 μm (organs) or 2 μm (heads). Tissue sections were stained either with Haematoxylin and Eosin using a Leica ST5020 autostainer or anti-human CD19 and CD3 primary antibodies using a Leica DAB polymer staining kit on a Leica Bond III staining platform. Stained sections were viewed and images acquired using an EVOS XL Imaging System (ThermoFisher Scientific).
RNA-sequencing and Analysis
Total RNA was isolated from resting CAR19-iNKT and -T cells by using NucleoSpin® RNA (Macherey-Nagel) omitting carrier RNA. Treatment with DNAse I was performed to remove all traces of DNA. Total RNA concentration was measured using Qubit® RNA HS Assay Kit on a Qubit® 2.0 Fluorometer (Thermo Fisher Scientific), whereas RNA integrity was assessed on an Agilent Bioanalyzer RNA 6000 Pico Chip. Next, RNA sequencing libraries were prepared using NEBNext® Ultra™ II Directional RNA Library Prep Kit for Illumina with rRNA depletion (NEB #E7760S/L), according to the manufacturer`s protocol, applying the algorithm for intact RNA (RIN >7). Sequencing was performed on an Illumina HiSeq4000 sequencer to obtain 75bp paired-end reads. Base calling and demultiplexing and conversion to fastq file were performed using the ‘Real Time Analysis’ (RTA) and bcl2fastq2 v2.18 illumina software respectively. Reads were subsequently mapped onto the Genome Reference Consortium Human genome build 38 (GRCh38) using STAR (version 2.5.3a)(Dobin et al., 2013) with a threshold of 10,000 alignments per read (command: --alignTranscriptsPerReadNmax 100000), allowing generation of chimeric outputs for downstream analysis. Processing of large reads files was completed by further increasing the STAR limits (command: --limitGenomeGenerateRAM 20000000000). Raw counts were obtained by using the ‘Rsubread’ v1.24.2 Bio-conductor package (Liao et al., 2013). Normalisation analysis was carried out using the ‘edgeR’ v3.20.9 Bio-conductor package (Robinson et al., 2010).
For RNA-seq of C1R-CD1d parental cells, the same procedures were followed, except for the fact that libraries were prepared .using the NEBNext poly(A) mRNA Magnetic Isolation Module and the NEBNext Ultra RNA Library Prep kit for Illumina (New Engand Biolabs) and sequenced on an Illumina HiSeq 2500 platform to obtain paired-end 100bp reads. Reads obtained from parental C1R-CD1d RNA-seq experiments were aligned using TopHat (v2.0.14) (Kim et al., 2013) against the Hg19 human reference genome using the default settings. ‘Cuffdiff’ part of the Cufflinks package (Trapnell et al., 2010) was used for data normalization.
Quantification and Statistical Analysis
Sample Size
In vitro assays were planned to achieve at least 3 biological replicates per set of experiments. For in vivo experiments, sample size was determined to show a 30% survival improvement in CAR19-iNKT vs CAR19-T -treated animals at 5% significance and 80% power.
Replicates
The in vitro data were reproduced in technical duplicates or triplicates using samples from different donors and lymphoma patients as a source of CAR-engineered and target cells respectively. For in vivo studies, to show enhanced anti-lymphoma activity of CAR19-iNKT compared to CAR19-T cells, after completing two pilot experiments, two repeats were performed, obtaining similar results.
In Vivo Randomization
At the beginning of the in vivo experiment a numeric ID was randomly assigned to each mouse. At the time engraftment, all animals were assessed in a consecutive order according to their ID, from the smaller to the larger. Same sex mice with comparable weight were assigned to one of the 5 treatment arms only based on the bioluminescent signal, aiming to ensure an equal distribution of tumor burden across the groups.
Statistical Analysis
Statistical analysis was performed on GraphPad Prism 7 software. For comparisons between two groups, the Wilcoxon (paired) or Mann–Whitney U (unpaired) test were used, with correction for multiple t tests according to the two-stage step-up method of Benjamini, Krieger and Yekutieli. For comparison between more than two groups, either one of the following tests were performed depending on the number of variables: non-parametric Friedman (paired) or Kruskal-Wallis (unpaired) test with post-hoc Dunn’s test (one variable, non-parametric for one-way ANOVA) or two-way ANOVA adjusted by Tukey (more than one variables). Survival was calculated using the Kaplan-Meier method, with log rank analysis for comparing survival between groups. All experimental data are presented as mean ± standard error mean (SEM) unless otherwise stated. All p values given are two-tailed values. A p value below 0.05 was considered significant.
Data and Software Availability
The RNA-seq data generated in this study will be available from the European Genome-Phenome Archive (EGAS00001003176).
Acknowledgments
The authors acknowledge funding support from Bloodwise, the Imperial National Institute for Research Biomedical Research Center (BRC), and Imperial Confidence in Concept.
We acknowledge the expertise of the LMS-NIHR BRC flow cytometry facility, the Imperial College London Multiple Sclerosis and Parkinson's Tissue Bank, and the Imperial BRC Genomics facility. We also thank Dr Martin Pule for providing the original CAR19 constructs and the Feldhahn and Di Giovanni laboratories for their technical support during the histological studies.
Author Contributions
Conceptualization: A.K. and A.R.; Methodology: A.K., A.R., and J.M.; Investigation: A.R., V.S.C., M.H., N.B., O.D., M.S.C., K.G., K.P., and S.I.; Formal Analysis: C.K., N.B., X.X., and D.S.P.; Writing – Original Draft: A.K. and A.R.; Writing – Review and Editing: all authors; Funding Acquisition: A.K. and A.R.; Resources: K.N.; Supervision: A.K. and J.M.
Declaration of Interests
J.M. is chief scientific officer of Leucid Bio, which is a spinout company focused on development of cellular therapeutic agents.
A.K. and A.R. are inventors on a patent application related to this work, filed by Imperial College London.
All other authors declare no competing interests.
Published: October 8, 2018
Footnotes
Supplemental Information includes six figures and one table and can be found with this article online at https://doi.org/10.1016/j.ccell.2018.08.017.
Supplemental Information
References
- Alcantar-Orozco E.M., Gornall H., Baldan V., Hawkins R.E., Gilham D.E. Potential limitations of the NSG humanized mouse as a model system to optimize engineered human T cell therapy for cancer. Hum. Gene Ther. Methods. 2013;24:310–320. doi: 10.1089/hgtb.2013.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan L.L., Stax A.M., Zheng D.J., Chung B.K., Kozak F.K., Tan R., van den Elzen P. CD1d and CD1c expression in human B cells is regulated by activation and retinoic acid receptor signaling. J. Immunol. 2011;186:5261–5272. doi: 10.4049/jimmunol.1003615. [DOI] [PubMed] [Google Scholar]
- Almasbak H., Walseng E., Kristian A., Myhre M.R., Suso E.M., Munthe L.A., Andersen J.T., Wang M.Y., Kvalheim G., Gaudernack G., Kyte J.A. Inclusion of an IgG1-Fc spacer abrogates efficacy of CD19 CAR T cells in a xenograft mouse model. Gene Ther. 2015;22:391–403. doi: 10.1038/gt.2015.4. [DOI] [PubMed] [Google Scholar]
- Bagnara D., Ibatici A., Corselli M., Sessarego N., Tenca C., De Santanna A., Mazzarello A., Daga A., Corvo R., De Rossi G. Adoptive immunotherapy mediated by ex vivo expanded natural killer T cells against CD1d-expressing lymphoid neoplasms. Haematologica. 2009;94:967–974. doi: 10.3324/haematol.2008.001339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendelac A., Savage P.B., Teyton L. The biology of NKT cells. Annu. Rev. Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- Bosma A., Abdel-Gadir A., Isenberg D.A., Jury E.C., Mauri C. Lipid-antigen presentation by CD1d(+) B cells is essential for the maintenance of invariant natural killer T cells. Immunity. 2012;36:477–490. doi: 10.1016/j.immuni.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigl M., Bry L., Kent S.C., Gumperz J.E., Brenner M.B. Mechanism of CD1d-restricted natural killer T cell activation during microbial infection. Nat. Immunol. 2003;4:1230–1237. doi: 10.1038/ni1002. [DOI] [PubMed] [Google Scholar]
- Brossay L., Chioda M., Burdin N., Koezuka Y., Casorati G., Dellabona P., Kronenberg M. CD1d-mediated recognition of an alpha-galactosylceramide by natural killer T cells is highly conserved through mammalian evolution. J. Exp. Med. 1998;188:1521–1528. doi: 10.1084/jem.188.8.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudno J.N., Kochenderfer J.N. Chimeric antigen receptor T-cell therapies for lymphoma. Nat. Rev. Clin. Oncol. 2018;15:31–46. doi: 10.1038/nrclinonc.2017.128. [DOI] [PubMed] [Google Scholar]
- Caputo V.S., Costa J.R., Makarona K., Georgiou E., Layton D.M., Roberts I., Karadimitris A. Mechanism of Polycomb recruitment to CpG islands revealed by inherited disease-associated mutation. Hum. Mol. Genet. 2013;22:3187–3194. doi: 10.1093/hmg/ddt171. [DOI] [PubMed] [Google Scholar]
- Chaidos A., Patterson S., Szydlo R., Chaudhry M.S., Dazzi F., Kanfer E., McDonald D., Marin D., Milojkovic D., Pavlu J. Graft invariant natural killer T-cell dose predicts risk of acute graft-versus-host disease in allogeneic hematopoietic stem cell transplantation. Blood. 2012;119:5030–5036. doi: 10.1182/blood-2011-11-389304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Ross A.C. Retinoic acid regulates CD1d gene expression at the transcriptional level in human and rodent monocytic cells. Exp. Biol. Med. (Maywood) 2007;232:488–494. [PMC free article] [PubMed] [Google Scholar]
- Cooke K.R., Kobzik L., Martin T.R., Brewer J., Delmonte J., Jr., Crawford J.M., Ferrara J.L. An experimental model of idiopathic pneumonia syndrome after bone marrow transplantation: I. The roles of minor H antigens and endotoxin. Blood. 1996;88:3230–3239. [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exley M., Garcia J., Balk S.P., Porcelli S. Requirements for CD1d recognition by human invariant Valpha24+ CD4-CD8- T cells. J. Exp. Med. 1997;186:109–120. doi: 10.1084/jem.186.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exley M., Garcia J., Wilson S.B., Spada F., Gerdes D., Tahir S.M., Patton K.T., Blumberg R.S., Porcelli S., Chott A., Balk S.P. CD1d structure and regulation on human thymocytes, peripheral blood T cells, B cells and monocytes. Immunology. 2000;100:37–47. doi: 10.1046/j.1365-2567.2000.00001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exley M.A., Friedlander P., Alatrakchi N., Vriend L., Yue S., Sasada T., Zeng W., Mizukami Y., Clark J., Nemer D. Adoptive transfer of invariant NKT cells as immunotherapy for advanced melanoma: a phase I clinical trial. Clin. Cancer Res. 2017;23:3510–3519. doi: 10.1158/1078-0432.CCR-16-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Exley M.A., Lynch L., Varghese B., Nowak M., Alatrakchi N., Balk S.P. Developing understanding of the roles of CD1d-restricted T cell subsets in cancer: reversing tumor-induced defects. Clin. Immunol. 2011;140:184–195. doi: 10.1016/j.clim.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyquem J., Mansilla-Soto J., Giavridis T., van der Stegen S.J., Hamieh M., Cunanan K.M., Odak A., Gonen M., Sadelain M. Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature. 2017;543:113–117. doi: 10.1038/nature21405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fais F., Tenca C., Cimino G., Coletti V., Zanardi S., Bagnara D., Saverino D., Zarcone D., De Rossi G., Ciccone E., Grossi C.E. CD1d expression on B-precursor acute lymphoblastic leukemia subsets with poor prognosis. Leukemia. 2005;19:551–556. doi: 10.1038/sj.leu.2403671. [DOI] [PubMed] [Google Scholar]
- Gordy L.E., Bezbradica J.S., Flyak A.I., Spencer C.T., Dunkle A., Sun J., Stanic A.K., Boothby M.R., He Y.W., Zhao Z. IL-15 regulates homeostasis and terminal maturation of NKT cells. J. Immunol. 2011;187:6335–6345. doi: 10.4049/jimmunol.1003965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorini F., Azzimonti L., Delfanti G., Scarfo L., Scielzo C., Bertilaccio M.T., Ranghetti P., Gulino A., Doglioni C., Di Napoli A. Invariant NKT cells contribute to chronic lymphocytic leukemia surveillance and prognosis. Blood. 2017;129:3440–3451. doi: 10.1182/blood-2016-11-751065. [DOI] [PubMed] [Google Scholar]
- Gumperz J.E., Miyake S., Yamamura T., Brenner M.B. Functionally distinct subsets of CD1d-restricted natural killer T cells revealed by CD1d tetramer staining. J. Exp. Med. 2002;195:625–636. doi: 10.1084/jem.20011786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heczey A., Liu D., Tian G., Courtney A.N., Wei J., Marinova E., Gao X., Guo L., Yvon E., Hicks J. Invariant NKT cells with chimeric antigen receptor provide a novel platform for safe and effective cancer immunotherapy. Blood. 2014;124:2824–2833. doi: 10.1182/blood-2013-11-541235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C.H., Butcher E.C., Johnston B. Distinct subsets of human Valpha24-invariant NKT cells: cytokine responses and chemokine receptor expression. Trends Immunol. 2002;23:516–519. doi: 10.1016/s1471-4906(02)02323-2. [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotsianidis I., Nakou E., Spanoudakis E., Bouchliou I., Moustakidis E., Miltiades P., Vadikolia C.M., Szydlo R., Karadimitris A., Tsatalas C. The diagnostic value of CD1d expression in a large cohort of patients with B-cell chronic lymphoproliferative disorders. Am. J. Clin. Pathol. 2011;136:400–408. doi: 10.1309/AJCP2F2DOXOTXHZA. [DOI] [PubMed] [Google Scholar]
- Kutner R.H., Zhang X.Y., Reiser J. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat. Protoc. 2009;4:495–505. doi: 10.1038/nprot.2009.22. [DOI] [PubMed] [Google Scholar]
- Leveson-Gower D.B., Olson J.A., Sega E.I., Luong R.H., Baker J., Zeiser R., Negrin R.S. Low doses of natural killer T cells provide protection from acute graft-versus-host disease via an IL-4-dependent mechanism. Blood. 2011;117:3220–3229. doi: 10.1182/blood-2010-08-303008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013;41:e108. doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks-Konczalik J., Dubois S., Losi J.M., Sabzevari H., Yamada N., Feigenbaum L., Waldmann T.A., Tagaya Y. IL-2-induced activation-induced cell death is inhibited in IL-15 transgenic mice. Proc. Natl. Acad. Sci. USA. 2000;97:11445–11450. doi: 10.1073/pnas.200363097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mato A.R., Thompson M.C., Nabhan C., Svoboda J., Schuster S.J. Chimeric antigen receptor T-cell therapy for chronic lymphocytic leukemia: a narrative review. Clin. Lymphoma Myeloma Leuk. 2017;17:852–856. doi: 10.1016/j.clml.2017.07.007. [DOI] [PubMed] [Google Scholar]
- Matulis G., Sanderson J.P., Lissin N.M., Asparuhova M.B., Bommineni G.R., Schumperli D., Schmidt R.R., Villiger P.M., Jakobsen B.K., Gadola S.D. Innate-like control of human iNKT cell autoreactivity via the hypervariable CDR3beta loop. PLoS Biol. 2010;8:e1000402. doi: 10.1371/journal.pbio.1000402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metelitsa L.S., Naidenko O.V., Kant A., Wu H.W., Loza M.J., Perussia B., Kronenberg M., Seeger R.C. Human NKT cells mediate antitumor cytotoxicity directly by recognizing target cell CD1d with bound ligand or indirectly by producing IL-2 to activate NK cells. J. Immunol. 2001;167:3114–3122. doi: 10.4049/jimmunol.167.6.3114. [DOI] [PubMed] [Google Scholar]
- Neelapu S.S., Locke F.L., Bartlett N.L., Lekakis L.J., Miklos D.B., Jacobson C.A., Braunschweig I., Oluwole O.O., Siddiqi T., Lin Y. Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N. Engl. J. Med. 2017;377:2531–2544. doi: 10.1056/NEJMoa1707447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickoloff B.J., Wrone-Smith T., Bonish B., Porcelli S.A. Response of murine and normal human skin to injection of allogeneic blood-derived psoriatic immunocytes: detection of T cells expressing receptors typically present on natural killer cells, including CD94, CD158, and CD161. Arch. Dermatol. 1999;135:546–552. doi: 10.1001/archderm.135.5.546. [DOI] [PubMed] [Google Scholar]
- Nieda M., Nicol A., Koezuka Y., Kikuchi A., Takahashi T., Nakamura H., Furukawa H., Yabe T., Ishikawa Y., Tadokoro K. Activation of human Valpha24 NKT cells by alpha-glycosylceramide in a CD1d-restricted and Valpha24 TCR-mediated manner. Hum. Immunol. 1999;60:10–19. doi: 10.1016/s0198-8859(98)00100-1. [DOI] [PubMed] [Google Scholar]
- Park J.H., Riviere I., Gonen M., Wang X., Senechal B., Curran K.J., Sauter C., Wang Y., Santomasso B., Mead E. Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. N. Engl. J. Med. 2018;378:449–459. doi: 10.1056/NEJMoa1709919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip B., Kokalaki E., Mekkaoui L., Thomas S., Straathof K., Flutter B., Marin V., Marafioti T., Chakraverty R., Linch D. A highly compact epitope-based marker/suicide gene for easier and safer T-cell therapy. Blood. 2014;124:1277–1287. doi: 10.1182/blood-2014-01-545020. [DOI] [PubMed] [Google Scholar]
- Qasim W., Zhan H., Samarasinghe S., Adams S., Amrolia P., Stafford S., Butler K., Rivat C., Wright G., Somana K. Molecular remission of infant B-ALL after infusion of universal TALEN gene-edited CAR T cells. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aaj2013. [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotolo A., Caputo V., Karadimitris A. The prospects and promise of chimeric antigen receptor immunotherapy in multiple myeloma. Br. J. Haematol. 2016;173:350–364. doi: 10.1111/bjh.13976. [DOI] [PubMed] [Google Scholar]
- Rubio M.T., Bouillie M., Bouazza N., Coman T., Trebeden-Negre H., Gomez A., Suarez F., Sibon D., Brignier A., Paubelle E. Pre-transplant donor CD4- invariant NKT cell expansion capacity predicts the occurrence of acute graft-versus-host disease. Leukemia. 2016;31:903–912. doi: 10.1038/leu.2016.281. [DOI] [PubMed] [Google Scholar]
- Ruella M., Barrett D.M., Kenderian S.S., Shestova O., Hofmann T.J., Perazzelli J., Klichinsky M., Aikawa V., Nazimuddin F., Kozlowski M. Dual CD19 and CD123 targeting prevents antigen-loss relapses after CD19-directed immunotherapies. J. Clin. Invest. 2016;126:3814–3826. doi: 10.1172/JCI87366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salio M., Silk J.D., Jones E.Y., Cerundolo V. Biology of CD1- and MR1-restricted T cells. Annu. Rev. Immunol. 2014;32:323–366. doi: 10.1146/annurev-immunol-032713-120243. [DOI] [PubMed] [Google Scholar]
- Sallusto F., Impellizzieri D., Basso C., Laroni A., Uccelli A., Lanzavecchia A., Engelhardt B. T-cell trafficking in the central nervous system. Immunol. Rev. 2012;248:216–227. doi: 10.1111/j.1600-065X.2012.01140.x. [DOI] [PubMed] [Google Scholar]
- Schneidawind D., Pierini A., Alvarez M., Pan Y., Baker J., Buechele C., Luong R.H., Meyer E.H., Negrin R.S. CD4+ invariant natural killer T cells protect from murine GVHD lethality through expansion of donor CD4+CD25+FoxP3+ regulatory T cells. Blood. 2014;124:3320–3328. doi: 10.1182/blood-2014-05-576017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder M.A., DiPersio J.F. Mouse models of graft-versus-host disease: advances and limitations. Dis. Model Mech. 2011;4:318–333. doi: 10.1242/dmm.006668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster S.J., Svoboda J., Chong E.A., Nasta S.D., Mato A.R., Anak O., Brogdon J.L., Pruteanu-Malinici I., Bhoj V., Landsburg D. Chimeric antigen receptor T cells in refractory B-cell lymphomas. N. Engl. J. Med. 2017;377:2545–2554. doi: 10.1056/NEJMoa1708566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J.A., Kingston R.E. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat. Rev. Mol. Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Spanoudakis E., Hu M., Naresh K., Terpos E., Melo V., Reid A., Kotsianidis I., Abdalla S., Rahemtulla A., Karadimitris A. Regulation of multiple myeloma survival and progression by CD1d. Blood. 2009;113:2498–2507. doi: 10.1182/blood-2008-06-161281. [DOI] [PubMed] [Google Scholar]
- Takahashi T., Nieda M., Koezuka Y., Nicol A., Porcelli S.A., Ishikawa Y., Tadokoro K., Hirai H., Juji T. Analysis of human V alpha 24+ CD4+ NKT cells activated by alpha-glycosylceramide-pulsed monocyte-derived dendritic cells. J. Immunol. 2000;164:4458–4464. doi: 10.4049/jimmunol.164.9.4458. [DOI] [PubMed] [Google Scholar]
- Tian G., Courtney A.N., Jena B., Heczey A., Liu D., Marinova E., Guo L., Xu X., Torikai H., Mo Q. CD62L+ NKT cells have prolonged persistence and antitumor activity in vivo. J. Clin. Invest. 2016;126:2341–2355. doi: 10.1172/JCI83476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Till B.G., Jensen M.C., Wang J., Qian X., Gopal A.K., Maloney D.G., Lindgren C.G., Lin Y., Pagel J.M., Budde L.E. CD20-specific adoptive immunotherapy for lymphoma using a chimeric antigen receptor with both CD28 and 4-1BB domains: pilot clinical trial results. Blood. 2012;119:3940–3950. doi: 10.1182/blood-2011-10-387969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb T.J., Carey G.B., East J.E., Sun W., Bollino D.R., Kimball A.S., Brutkiewicz R.R. Alterations in cellular metabolism modulate CD1d-mediated NKT-cell responses. Pathog. Dis. 2016;74 doi: 10.1093/femspd/ftw055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whilding L.M., Parente-Pereira A.C., Zabinski T., Davies D.M., Petrovic R.M.G., Kao Y.V., Saxena S.A., Romain A., Costa-Guerra J.A., Violette S. Targeting of Aberrant alphavbeta6 integrin expression in solid tumors using chimeric antigen receptor-engineered T cells. Mol. Ther. 2017;25:2427. doi: 10.1016/j.ymthe.2017.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaini J., Andarini S., Tahara M., Saijo Y., Ishii N., Kawakami K., Taniguchi M., Sugamura K., Nukiwa T., Kikuchi T. OX40 ligand expressed by DCs costimulates NKT and CD4+ Th cell antitumor immunity in mice. J. Clin. Invest. 2007;117:3330–3338. doi: 10.1172/JCI32693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Condomines M., van der Stegen S.J.C., Perna F., Kloss C.C., Gunset G., Plotkin J., Sadelain M. Structural design of engineered costimulation determines tumor rejection kinetics and persistence of CAR T cells. Cancer Cell. 2015;28:415–428. doi: 10.1016/j.ccell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D. OX40 signaling directly triggers the antitumor effects of NKT cells. J. Clin. Invest. 2007;117:3169–3172. doi: 10.1172/JCI33976. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.