PORK1 is identified and characterized as a receptor-like kinase involved in tomato responses to the endogenous peptide systemin.
Abstract
Endogenous peptides regulate plant immunity and growth. Systemin, a peptide specific to the Solanaceae, is known for its functions in plant responses to insect herbivory and pathogen infections. Here, we describe the identification of the tomato (Solanum lycopersicum) PEPR1/2 ORTHOLOG RECEPTOR-LIKE KINASE1 (PORK1) as the TOMATO PROTEIN KINASE1b (TPK1b) interacting protein and demonstrate its biological functions in systemin signaling and tomato immune responses. Tomato PORK1 RNA interference (RNAi) plants with significantly reduced PORK1 expression showed increased susceptibility to tobacco hornworm (Manduca sexta), reduced seedling growth sensitivity to the systemin peptide, and compromised systemin-mediated resistance to Botrytis cinerea. Systemin-induced expression of Proteinase Inhibitor II (PI-II), a classical marker for systemin signaling, was abrogated in PORK1 RNAi plants. Similarly, in response to systemin and wounding, the expression of jasmonate pathway genes was attenuated in PORK1 RNAi plants. TPK1b, a key regulator of tomato defense against B. cinerea and M. sexta, was phosphorylated by PORK1. Interestingly, wounding- and systemin-induced phosphorylation of TPK1b was attenuated when PORK1 expression was suppressed. Our data suggest that resistance to B. cinerea and M. sexta is dependent on PORK1-mediated responses to systemin and subsequent phosphorylation of TPK1b. Altogether, PORK1 regulates tomato systemin, wounding, and immune responses.
INTRODUCTION
Pathogenic microbes and insect pests featuring diverse virulence strategies attack plants, threatening their growth and productivity. In response, plants have developed counteractive defense mechanisms that limit initial ingress or restrict the damage by microbial and pest intruders. Signals derived from pathogens or released from plant cells, owing to the different virulence activities of pathogens and insect pests are recognized by extra- or intracellular receptors to activate the plant defense system. Significant advances have been made toward understanding the mechanisms of recognition, signaling, and plant responses to pathogen-derived signals especially in model plant systems. Early stages of infection by necrotrophic fungi involve degradation of the cuticle and cell walls at the site of infection, which generate cell wall fragments and cutin monomers that are considered damage-associated molecular patterns, danger signals that trigger basal immune responses (Hahn et al., 1981; Schweizer et al., 1996; Kauss et al., 1999; D’Ovidio et al., 2004; Decreux and Messiaen, 2005). Endogenous peptides produced as a result of pathogen infection, mechanical wounding, or insect herbivory are also considered danger signals that activate plant immune responses. Arabidopsis thaliana Pep1, a 23-amino acid peptide, is derived from PROPEP1, which is a 92-amino acid Pep1 precursor. The PROPEP1 gene expression is induced by wounding, methyl jasmonate, and ethylene (Huffaker et al., 2006). The tomato (Solanum lycopersicum) systemin, an 18-amino acid peptide, was isolated from wounded tomato leaf extracts in a screen for compounds that induce protease inhibitor (PI) expression (Pearce et al., 1991). PIs are expressed in response to insect or pathogen attacks in wounded and systemic tissues. Tomato systemin is cleaved from the C-terminal region of a 200-amino acid Prosystemin protein (McGurl et al., 1992). A systemin binding protein of 50 kD was also isolated from plasma membranes of tomato leaves, which was suggested to be a systemin processing protease (Schaller and Ryan, 1994). The processing of Prosystemin to produce the biologically active peptide is mediated by the tomato aspartate-specific protease, which hydrolyzes Prosystemin at two aspartate residues flanking the systemin sequence (Beloshistov et al., 2018). Transgenic expression of Prosystemin results in constitutive accumulation of PI proteins and generates a mobile wound signal which was thought to be systemin (McGurl et al., 1994). Later, grafting experiments using tomato mutants defective in wound signaling, jasmonic acid (JA) biosynthesis, or JA responses suggested that JA or a derivative may serve as the systemic signal for wound signaling (Li et al., 2002).
Tomato systemin and Arabidopsis pep1 perception and signaling are functionally similar systems that activate plant immune responses against pathogens and insect pests. Arabidopsis pep1 activates expression of the plant defensin PDF1.2 and synthesis of reactive oxygen species, which mark activation of immune responses. Constitutive expression of PROPEP1, the precursor of pep1, causes constitutive expression of PDF1.2 and enhanced resistance to the necrotrophic oomycete Pythium irregulare in Arabidopsis (Huffaker et al., 2006; Huffaker and Ryan, 2007). In tomato, overexpression of Prosystemin confers resistance to fungal pathogens and insect herbivores (Coppola et al., 2015). By contrast, tomato antisense Prosystemin plants exhibited diminished systemic wound induction of proteinase inhibitors (McGurl et al., 1992), increased susceptibility to Botrytis cinerea infection, and feeding by Manduca sexta larvae (Orozco-Cardenas et al., 1993; El Oirdi et al., 2011). These observations indicate analogous signaling systems in different plant species with functions in plant defense.
In Arabidopsis, two closely related receptor like kinases (RLKs) PEP1 RECEPTOR1 (PEPR1) and PEPR2 recognize Arabidopsis peptides (Pep1–Pep6) (Yamaguchi et al., 2006, 2010). The PEPR1- and PEPR2-mediated peptide responses are required for resistance to pathogens and insect herbivory (Liu et al., 2013; Klauser et al., 2015). BRASSINOSTEROID INSENSITIVE1-ASSOCIATED KINASE1 (BAK1) serves as a coreceptor to multiple immune receptors including PEPR1 and PEPR2 (Postel et al., 2010; Schulze et al., 2010). Arabidopsis Pep1 induces PEPR1 heterodimerization with BAK1, which was demonstrated to be important for the activation of PEPR1 (Tang et al., 2015). Receptor-like cytoplasmic kinases are a subclass of RLKs that are genetic and biochemical components of immune response signaling acting early in the signaling step. BIK1 and BIK1-related kinases in Arabidopsis and other plant species integrate signals from various ligand-receptor recognition complexes and signal to downstream components, although the details of the link to the activation of defensive molecules remain relatively unclear (Zhang et al., 2010; Liu et al., 2013; Tang et al., 2017).
Tomato systemin peptide has been studied for nearly three decades. However, the identity of the systemin receptor remained unresolved until recently. Initially, by using a photoaffinity analog of systemin, a 160-kD cell surface receptor protein with features of the systemin receptor was isolated from membranes of Solanum peruvianum suspension cultured cells (Scheer and Ryan, 1999). Similarly, a biochemical approach was used to identify a specific, high-affinity binding site in membrane preparations of S. peruvianum (Meindl et al., 1998). Subsequently, it was determined that the SR160 protein is the tomato homolog of Arabidopsis BRASSINOSTEROID INSENSITIVE1 (SlBRI1), which bound to the radiolabeled photoaffinity systemin and was considered the systemin receptor (Scheer and Ryan, 2002). Subsequently, however, SlBRI1 was found to be dispensable for systemin-induced growth responses, ion flux, and gene expression (Holton et al., 2007; Lanfermeijer et al., 2008; Malinowski et al., 2009; Wang et al., 2018). More recently, a pair of tomato RLKs, Systemin Receptor 1 (SYR1) and SYR2, were implicated in systemin perception with SYR1 reported as a high-affinity bona fide systemin receptor (Wang et al., 2018). Previously, we demonstrated that TOMATO PROTEIN KINASE 1b (TPK1b) regulates plant responses to insects and necrotrophic fungi (Abuqamar et al., 2008).
In this report, we identified and characterized the TPK1b-interacting RLK, PEPR1/2 ORTHOLOG RECEPTOR-LIKE KINASE1 (PORK1) and demonstrate its functions in regulating responses to wounding or systemin. PORK1 is closely related to the newly described SYR1 and SYR2 proteins (Wang et al., 2018). Tomato PORK1 RNA interference (PORK1 RNAi) plants with diminished PORK1 expression display attenuated growth sensitivity to systemin, compromised systemin-mediated resistance to B. cinerea, as well as increased susceptibility to tobacco hornworm (M. sexta). Interestingly, silencing of PORK1 in plants that overexpress the systemin precursor Prosystemin abrogated resistance to B. cinerea. Mechanical wounding and systemin-induced expression of Proteinase Inhibitor II (PI-II) was severely attenuated in PORK1 RNAi plants. PI-II is a wound-induced PI that interferes with the digestive proteinases from insect and mammalian herbivores (Green and Ryan, 1972). Furthermore, silencing of PORK1 compromised wounding- and systemin-activated phosphorylation of TPK1b. Collectively, our data show that PORK1 is a key determinant of systemin responses in tomato, and TPK1b serves as a substrate in systemin-induced phosphorylation and systemin signaling that likely regulates defense responses to fungal pathogens and insects.
RESULTS
PORK1 Interacts with TPK1b
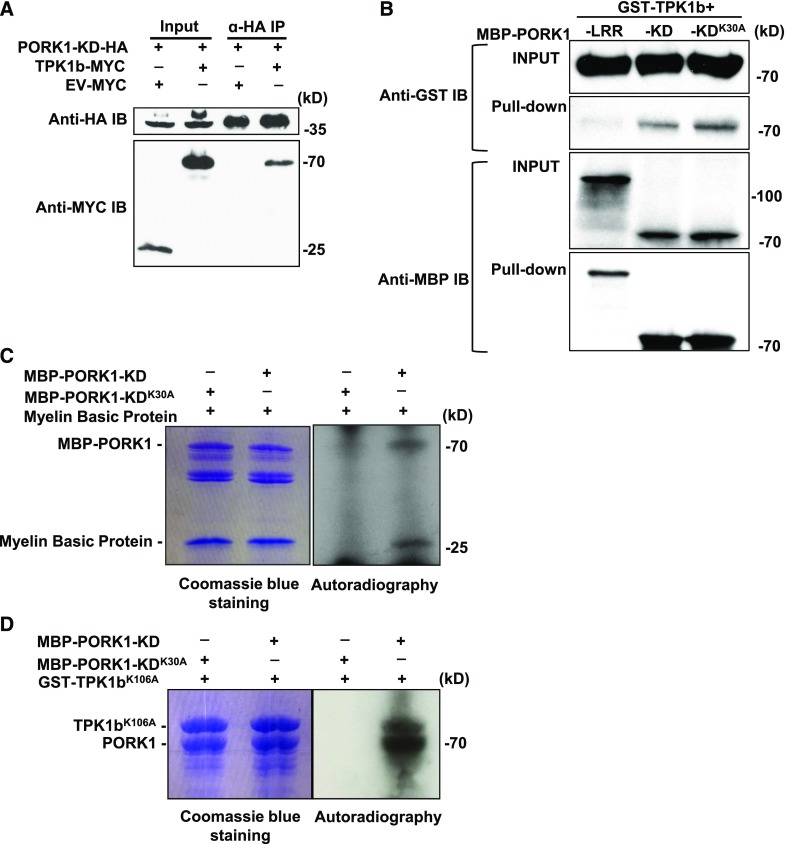
To dissect the molecular and biochemical mechanisms that underpin the functions of TPK1b, we sought to identify RLKs that function as its upstream regulators in tomato immune response signaling. The tomato RLK encoded by the Solyc03g123860 gene complexed with TPK1b in coimmunoprecipitation (co-IP) assays (Figure 1A; Supplemental Figure 1A). Phylogenetic analysis revealed that tomato Solyc03g123860 encodes a protein with higher sequence similarity to Arabidopsis PEPR1 and PEPR2 compared with other RLKs in the tomato genome (Supplemental Figures 2A to 2C); we thus designated it tomato PORK1. PORK1 harbors multiple extracellular LRRs, a short transmembrane, and an intracellular kinase domain, all features common to other RLKs (Supplemental Figure 2A). RLKs, SlBRI1, the receptor for brassinolide hormone (Lin et al., 2013), and tomato FLAGELLIN SENSING2 (SlFLS2), the receptor for the bacterial flagellin (Mueller et al., 2012), also interacted with TPK1b in co-IP assays (Supplemental Figures 1B and 1C). TPK1b interacted with kinase domains of all three proteins but not with the control co-IP when the empty vector expressing the MYC-epitope tag was coexpressed with PORK1 (Figure 1A; Supplemental Figures 1A to 1C). Since the biological functions of SlFLS2 and SlBRI1 have been reported previously (Robatzek et al., 2007; Bajwa et al., 2013), we chose to further study the function of PORK1 in tomato defense signaling. To validate direct interaction, we used TPK1b and PORK1 recombinant proteins for in vitro pull-down assays. The GST-tagged TPK1b (GST-TPK1b) was bound with MBP tagged PORK1 kinase domain (MBP-PORK1-KD), but not with the LRR domain of PORK1 protein (MBP-PORK1-LRR), which was used as a negative control (Figure 1B). The data demonstrate direct physical interaction between PORK1 and TPK1b proteins.
Figure 1.
PORK1 Interaction and Phosphorylation with TPK1b.
(A) TPK1b interacts with PORK1 kinase domain (KD). TPK1b-MYC and the kinase domain of PORK1 (PORK1-KD-HA) were coexpressed in N. benthamiana through Agrobacterium tumefaciens-mediated transient expression. PORK1-KD-HA was immunoprecipitated (IP) with anti-HA antibody coupled agarose beads along with TPK1b-MYC. The bound proteins were detected by immunoblot with the indicated antibodies.
(B) Direct interaction between PORK1 and TPK1b in in vitro binding assay. Equal amounts of recombinant proteins were mixed as shown in the input. After washing, GST-TPK1b was bound with MBP-PORK1-KD and MBP-PORK1-KDK30A, but not with the MBP-PORK1-LRR, which is used as a negative control.
(C) PORK1 is a functional kinase with autophosphorylation and transphosphorylation activities.
(D) PORK1 phosphorylates TPK1b in in vitro kinase assay.
In (C) and (D), MBP-PORK1-KD, MBP-PORK1-KDK30A, or GST-TPK1bK106A were expressed and purified from E. coli using amylose resin columns. MBP-PORK1-KD or MBP-PORK1-KDK30A was incubated with Myelin Basic Protein (C) or TPK1bK106A (D) in a kinase buffer containing [γ-32P]ATP. Phosphorylation was detected by autoradiography. Coomassie blue staining shows equal loading of protein samples. EV, empty vector. The experiments were repeated at least two times with similar results.
PORK1 Is Structurally and Phylogenetically Related to Arabidopsis PEPR1 and PEPR2 RLKs
Sequence comparison and phylogenetic analyses of PORK1 and other tomato RLKs was conducted to determine the relationship between PORK1 and other RLKs. Tomato PORK1 and tomato receptor for brassinosteroids, an RLK that was previously considered as the receptor for systemin (Scheer and Ryan, 2002), share lower sequence identity (data not shown). Consistent with this, BRI is an outlier in the phylogenetic analyses. By contrast, Arabidopsis PEPR1 and PORK1 (Solyc03g123860) share ∼50% identity, while other sequences in the tomato genome share no more than 37% identity to the Arabidopsis PEPR1. Likewise, PORK1 shares 48% identity to Arabidopsis PEPR2, while other sequences share no more than 36% identity. Tomato RLKs PORK1 and Arabidopsis PEPR1 and PEPR2 proteins share similar structures and conserved domains (Supplemental Figure 2C). Despite the evolutionarily conserved nature of RLKs, the sequence identity between PORK1 and other tomato proteins is relatively low (38% identity). Interestingly, there are 20 LRR-RLKs on tomato chromosome 3 located between positions 52.3 and 70.7 MB, linked to PORK1, but the functions of most of these RLKs are unknown. Tomato cold shock protein receptor (CORE), the receptor for the bacterial cold-shock proteins, and SYR1 (Solyc03g082470) and SYR2 (Solyc03g082450) (Wang et al., 2018) are among the cluster of 20 RLKs located in the same genomic region. Among these, SYR1 and SYR2 form a closely related clade to the tomato PORK1 and Arabidopsis PEPRs (Supplemental Figure 2B). There is a general structural similarity between this cluster of tomato RLKs although the specific sequence alignments reveal that PORK1 shares only 29 to 38% overall identity to these RLKs.
PORK1 Phosphorylates TPK1b
To determine PORK1 biochemical function, the recombinant PORK1 kinase domain (MBP-PORK1-KD) was used for kinase activity assays. The PORK1 kinase inactive recombinant protein carrying Ala (A) substitution at Lys (K) residue at position 30 (MBP-PORK1-KDK30A) was generated as a control. The MBP-PORK1-KD protein showed autophosphorylation activity that was visible as a band of 70 kD corresponding to the combined molecular mass of PORK1-KD and maltose binding protein, while MBP-PORK1-KDK30A substitution eliminated autophosphorylation and phosphorylation of Myelin Basic Protein, a commonly used commercial kinase substrate (Figure 1C), which demonstrates that PORK1 is a functional kinase. To determine whether TPK1b is a substrate for PORK1, kinase inactive TPK1bK106A protein was incubated with PORK1-KD or PORK1-KDK30A in an in vitro kinase assay. PORK1-KD, but not PORK1-KDK30A, phosphorylates TPK1bK106A (Figure 1D).
PORK1 Is Required for Systemin-Mediated Resistance to B. cinerea and M. sexta Feeding
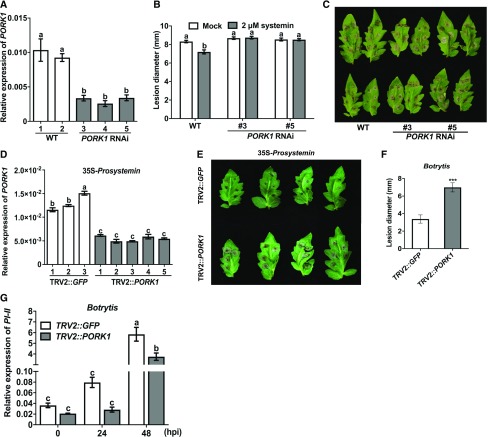
To study the biological function of PORK1, transgenic tomato RNAi plants that are reduced for PORK1 gene expression were generated using gene-specific fragments of the 5′ end of the PORK1 gene. Three independent homozygous transgenic PORK1 RNAi lines #3, #4, and #5 that showed substantial reduction of PORK1 gene expression were selected and used in subsequent studies (Figure 2A). The silencing specificity of PORK1 was confirmed by analyzing the expressions of two PORK1-related genes SYR1 and SYR2 (Solyc03g082470 and Solyc03g082450) in PORK1 RNAi plants. The RT-qPCR data revealed that there was no significant suppression of these genes (Supplemental Figures 3A and 3B). To elucidate the function of PORK1 in systemin-induced resistance to B. cinerea, leaves from tomato wild-type cultivar (Castlemart II, CMII) and PORK1 RNAi plants were treated with systemin or water (mock), and a day later, inoculated with B. cinerea. Mock-pretreated tomato leaves from wild type and two PORK1 RNAi lines exhibited comparable disease lesion size, whereas the wild-type leaves pretreated with systemin exhibited significantly improved resistance (Figures 2B and 2C). By contrast, PORK1 RNAi plants were significantly compromised in systemin-induced resistance to B. cinerea.
Figure 2.
PORK1 Mediates (Pro)systemin-Induced Resistance to B. cinerea.
(A) RT-qPCR analysis of PORK1 transcript levels in wild-type and PORK1 RNAi plants.
(B) and (C) B. cinerea disease lesion size (B) and disease symptoms in mock (water) or systemin-pretreated plants (C). Data represent mean ± se (n ≥ 90). Detached leaves were evenly sprayed with water (mock) or 2 µM systemin, in both cases containing 0.01% of Silwet L-77. Different letters indicate statistically significant differences. Multiple comparisons were calculated by two-way ANOVA followed by Bonferroni post-hoc test (P < 0.05; Supplemental File 1).
(D) VIGS of tomato PORK1 in the background of 35S-Prosystemin tomato plants.
(E) PORK1-silenced 35S-Prosystemin plants show enhanced disease symptoms. Pictures of disease symptoms are form 3 d after inoculation with B. cinerea. Disease assays were conducted on plants with reduced PORK1 expression shown under (D).
(F) Mean lesion diameter at 3 d after inoculation. Data represent means ± se (n ≥ 70). Asterisks indicate statistically significant differences (Student’s t test, P < 0.001; Supplemental File 1).
(G) PI-II expression in GFP- or PORK1-silenced 35S:Prosystemin plants. GFP (mock)- or PORK1-silenced plants were spray inoculated with B. cinerea spore suspension and RNA extracted from leaf tissues at different time points.
In (A), (D), and (G), gene expression was analyzed by RT-qPCR and relative expression levels were calculated by the comparative cycle threshold method using tomato Actin as a reference. Mean values represent ± sd (n = 9) from three independent biological replicates and three technical repeats. Different letters indicate statistically significant differences. Multiple comparisons were calculated by two-way ANOVA followed by Bonferroni post-hoc test (P < 0.05; Supplemental File 1). hpi, hours postinoculation. The presented data are representative of at least three independent experiments.
The systemin peptide which has Ala substitution at position 17 with Thr (systeminT17A) has been used as an inactive systemin in previous studies (Pearce et al., 1993; Wang et al., 2018). Consistent with this, systeminT17A failed to enhance plant resistance to B. cinerea (Supplemental Figure 4A). Arabidopsis Pep1, a peptide recognized by PEPR1/2 receptor in Arabidopsis, was unable to induce tomato resistance to B. cinerea. Conversely, systemin activated resistance in wild-type tomato plants but not in Arabidopsis (Supplemental Figures 4B and 4C). These data suggest that the recognition of these two peptides is specific to the corresponding species despite the overall sequence and structural similarity of PEPR1/2 and PORK1 proteins as well as the two peptides performing parallel functions in the two plant systems.
Tomato plants constitutively expressing Prosystemin (35S-Prosystemin) exhibit increased resistance to necrotrophic pathogens (Coppola et al., 2015). To determine whether PORK1 contributes to Prosystemin-mediated fungal resistance, PORK1 gene expression was silenced in 35S-Prosystemin plants using virus-induced gene silencing (VIGS). PORK1 gene expression in individual plants was examined by RT-qPCR to identify PORK1-silenced plants (Figure 2D). 35S-Prosystemin plants with reduced PORK1 expression were significantly compromised in resistance to B. cinerea (Figures 2E and 2F). Leaves from PORK1-silenced 35S-Prosystemin plants had disease lesions double in size relative to those observed in the mock-silenced (TRV2:GFP) plants. The increased B. cinerea susceptibility of PORK1-silenced 35S-Prosystemin plants was accompanied by compromised B. cinerea induction of PI-II gene expression (Figure 2G). Thus, PORK1 is required for systemin-mediated gene expression and resistance to B. cinerea.
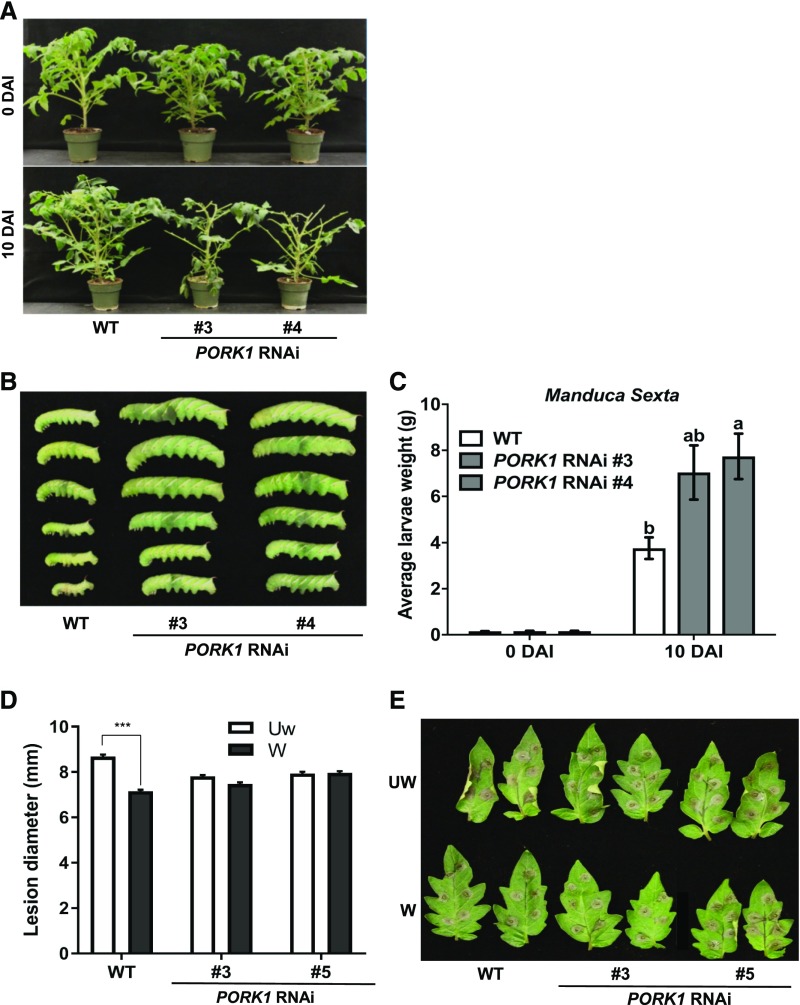
To determine whether PORK1 contributes to defense against insect attack, a feeding trial was conducted using tobacco hornworm (M. sexta) larvae. After a 10-d feeding trial, the size and average weights of larvae that fed on the PORK1 RNAi plants were significantly greater than those on wild-type plants (Figures 3B and 3C). In addition, the larvae consumed substantially more foliage of PORK1 RNAi than wild-type plants (Figure 3A). These results support the conclusion that PORK1 contributes to defense against larval feeding, similar to TPK1b function in resistance to M. sexta (Abuqamar et al., 2008).
Figure 3.
PORK1 RNAi Plants Show Reduced Resistance to Tobacco Hornworm Larvae.
(A) Wild-type and PORK1 RNAi plants at the beginning and end of tobacco hornworm (M. sexta) feeding trial.
(B) Size of larvae recovered 10 d after feeding trial.
(C) Average weights of larvae at the beginning and at 10 d after feeding trial. Data represent mean ± se (n = 10). Multiple comparisons were calculated by one-way ANOVA test (P < 0.05; Supplemental File 1). Different letters indicate statistically significant differences. DAI, days after inoculation.
(D) and (E) B. cinerea disease lesion size (D) and disease symptoms in wounded (W) and unwounded (UW) plants (E). Data represent means ± se (n ≥ 120). Asterisks indicate statistically significant differences (Student’s t test, P < 0.001). The experiments were repeated at least three times with similar results.
To determine whether the function of PORK1 is specific to insect herbivory or extends to responses to mechanical wounding, we studied wounding-mediated plant resistance. In Arabidopsis, wounding activates a strong resistance to B. cinerea (Chassot et al., 2008), and wounding and insect herbivory induce overlapping plant defense responses (Green and Ryan, 1972). When plants were wounded prior to B. cinerea inoculation, based on disease symptom and lesion size, PORK1-RNAi plants showed a reduced wounding-induced plant resistance relative to wild-type plants, which suggests that PORK1 plays an important role in that resistance (Figures 3D and 3E).
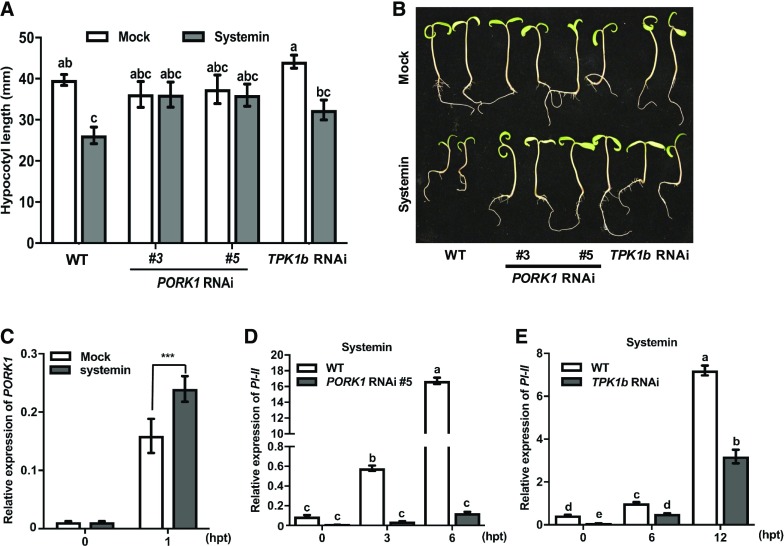
PORK1 Is Required for Tomato Seedling Growth Response to Systemin Peptide
To establish the functional connection between PORK1 and systemin, we examined the sensitivity of tomato seedling growth to systemin peptide. When tomato wild-type seedlings were grown on Murashige and Skoog (MS) medium containing synthetic systemin peptide, the hypocotyl lengths were reduced dramatically compared with seedlings grown on plain MS medium (Figures 4A and 4B). By contrast, the hypocotyl growth of PORK1 RNAi lines was less affected. TPK1b RNAi seedlings displayed intermediate insensitivity to systemin relative to PORK1 RNAi plants but were significantly more insensitive than the wild type. The data suggest PORK1 plays a greater role in regulating systemin responses while TPK1b functions in systemin signaling may also be shared by other receptor like cytoplasmic kinases. There were no significant differences in root length or seedling biomass between wild-type and PORK1 RNAi plants when grown on systemin (data not shown).
Figure 4.
PORK1 Contributes to Systemin-Mediated Seedling Growth Responses and Gene Expression.
(A) and (B) Tomato PORK1 RNAi lines display hypocotyl growth insensitivity (A) and loss of growth responses (B) to systemin. Seeds were geminated and grown on MS medium with or without 10 nM systemin. The hypocotyl lengths were measured at 12 d. Values represent mean ± se (n > 10) from each genotype. Multiple comparisons were calculated by two-way ANOVA followed by Bonferroni post-hoc test (P < 0.05). Different letters indicate statistically significant differences.
(C) to (E) The expression of PORK1 (C) and PI-II ([D] and [E]) genes in response to systemin. In (C), detached leaves of wild-type tomato plants were immersed in 50 nM systemin or water (mock). Values represent ± sd (n = 9) from three independent biological replicates and three technical replicates. Asterisks indicate significant difference (two-way ANOVA, P < 0.001). In (D) and (E), values represent mean ± sd (n = 9) from three independent biological replicates and three technical replicates. Quantification of each gene expression level was normalized with tomato Actin gene. The multiple comparisons were calculated by two-way ANOVA followed by Bonferroni post-hoc test (P < 0.05; Supplemental File 1). Different letters indicate statistically significant differences. hpt, hours post-treatment. At least two independent experiments were conducted showing similar results.
To establish the molecular link between systemin and PORK1, we examined systemin-induced PORK1 expression. Interestingly, PORK1 expression was induced significantly at 1 h after systemin treatment as compared with mock-treated plants (Figure 4C). In addition, systemin-induced expression of PI-II was severely compromised in PORK1 RNAi plants (Figure 4D; Supplemental Figure 5A). TPK1b RNAi plants also displayed significantly reduced PI-II gene expression in response to systemin (Figure 4E), but the level of reduction was not as dramatic as PORK1 RNAi plants, implying that PORK1 has a specific role in systemin-induced PI-II expression.
PORK1 Contributes to Systemin- and Wound-Induced Gene Expression
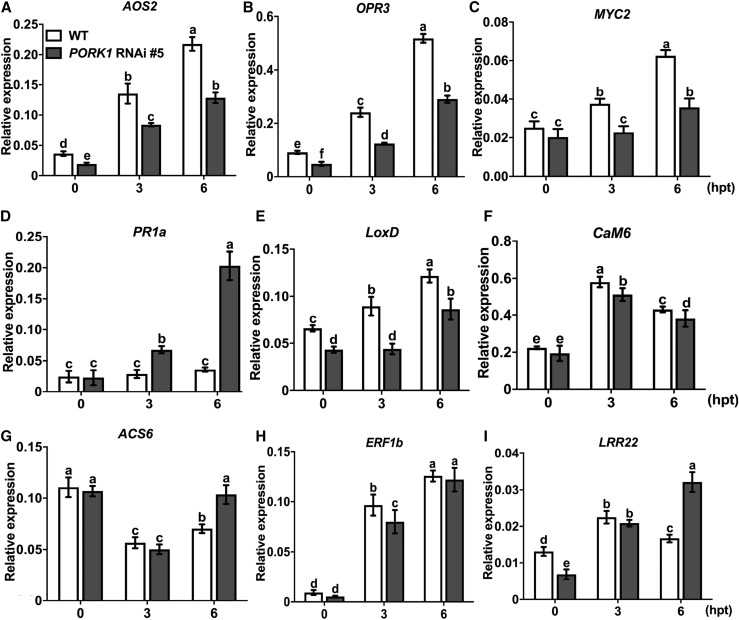
Previously, systemin has been shown to regulate defense responses through the octadecanoid for JA biosynthesis (Howe and Ryan, 1999; Li et al., 2003). The expression of components of this pathway, tomato AOS2, OPR3, and MYC2, was impaired in PORK1 RNAi plants (Figures 5A to 5C). By contrast, the PR1a gene which is widely used as a marker for activation of defense responses especially in relation to systemic acquired resistance, was highly induced in PORK1 RNAi plants in response to systemin (Figure 5D). There was a markedly contrasting expression between PR1 and PI-II in PORK1 RNAi plants, suggesting the antagonistic interactions between JA and salicylic acid (SA), which may occur downstream of PORK1 (Figures 4D and 5D). However, systemin-induced expression of NPR1, a major regulator of SA responses, was lower in nontreated plants but increased at 6 h after systemin treatment (Supplemental Figure 5B). Tomato COI1 (Li et al., 2004) expression was largely independent of PORK1, although 4 h after systemin treatment, the PORK1 RNAi plants showed significantly lower gene expression (Supplemental Figure 5C). Increased expressions of LIPOXYGENASE D (LoxD) and CALMODULIN (CaM) genes after systemin treatment have been shown previously (Heitz et al., 1997; Bergey and Ryan, 1999; Kandoth et al., 2007). Compared with wild-type plants, the expression of LoxD was reduced in PORK1 RNAi plants but that of CaM6 expression was induced by systemin treatment in wild-type plants comparable with PORK1 RNAi plants. The data suggest that PORK1-mediated plant defense is not mediated by the calmodulin signaling pathway (Figures 5E and 5F). Ethylene biosynthesis and responsive genes ACS6 and ERF1b did not show a consistent and significant difference in gene expression in response to systemin in wild-type and PORK1 RNAi plants, implying that PORK1 function may not affect ethylene regulated responses (Figures 5G and 5H).
Figure 5.
PORK1 Is Required for Systemin-Induced Gene Expression.
The expression of AOS2 (A), OPR3 (B), MYC2 (C), PR1a (D), LoxD (E), CaM6 (F) ACS6 (G), ERF1b (H), and LRR22 (I) genes in response to systemin in leaf tissues. Values represent mean ± sd (n = 9) from three independent biological replicates and three technical replicates. Expression is presented relative to expression level of each gene after normalization to the Actin gene. The multiple comparisons were calculated by two-way ANOVA followed by Bonferroni post-hoc test (P < 0.05; Supplemental File 1). Different letters indicate statistically significant differences. hpt, hours post-treatment. Data are representative of at least three independent experiments.
The tomato LRR22 gene has been defined as a marker for PAMP-triggered immunity studies in tomato and Nicotiana benthamiana (Nguyen et al., 2010; Taylor et al., 2012). In PORK1 RNAi plants, the basal expression of tomato LRR22 gene was lower than in the wild type but higher at 6 h after systemin treatment, suggesting that this particular gene may not correlate with systemin-triggered responses (Figure 5I). The accumulation of reactive oxygen species (ROS), which is commonly used as a readout for PAMP-triggered immunity, was not detected when tomato plants were treated with systemin (Supplemental Figure 6A). The tomato plants showed a strong ROS activity in response to flg22 peptide, which was used as an experimental control in our assays. These data suggest that ROS activity may not be a good marker for systemin responses in tomato. Activation of MAPK phosphorylation, downstream of different PAMP recognition, occurs in many different immune responses. The basal level of MPK3/MPK6 phosphorylation was slightly higher in the PORK1 RNAi plants. At 5 and 10 min after systemin treatment, there was not any significant difference in the MPK3/MPK6 phosphorylation between PORK1 RNAi and wild-type plants. Systemin-induced MPK3/MPK6 phosphorylation increased in both wild-type and PORK1 RNAi plants (Supplemental Figures 6B and 6C). PORK1 RNAi plants showed an increase in MPK3/MPK6 phosphorylation at 30 and 60 min after systemin treatment. Altogether, PORK1 is required for systemin-induced gene expression and in some instances PORK1 RNAi plants showed enhanced expression of genes at a later stage of systemin treatment, suggesting antagonism between JA and SA pathways following recognition of systemin.
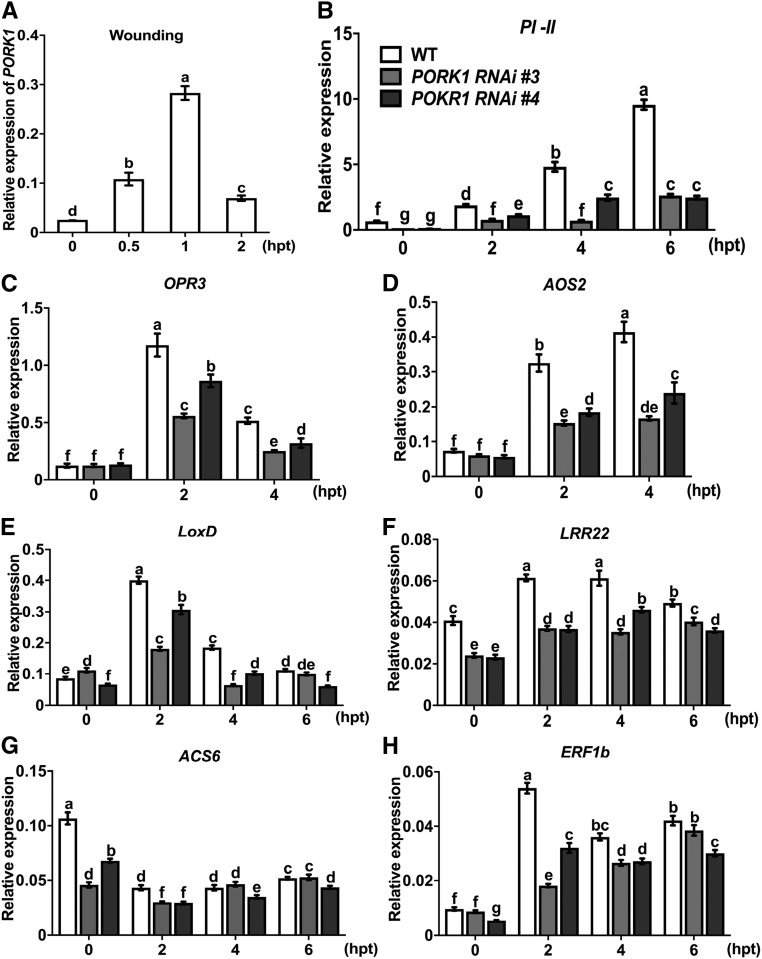
Because the effects of insect damage could be partially simulated by mechanical wounding (Green and Ryan, 1972), we examined wound-induced expression of PORK1 and other defense-related genes. PORK1 expression was significantly and rapidly induced by wounding, reaching peak expression level at 1 h after wounding and declining afterwards (Figure 6A). Wound-induced PI-II expression was compromised in PORK1 RNAi plants compared with wild-type plants (Figure 6B). Previous studies demonstrated that wound response signaling is largely dependent on accumulation of jasmonic family of compounds, including JA and methyl jasmonate (Sun et al., 2011). Expressions of both tomato AOS2 and OPR3, which encode key enzymes in the JA biosynthesis pathway (Wasternack et al., 2006), were lower in PORK1 RNAi plants but upregulated in wild-type plants in response to wounding (Figures 6C and 6D). The tomato LoxD, LRR22, and ERF1 genes, which were induced by wounding in tomato wild-type plants, showed reduced wound-induced expression in PORK1 RNAi plants, suggesting that PORK1 modulates expression of many defense related genes in response to wounding (Figures 6E to 6H).
Figure 6.
PORK1 Is Required for Wound-Induced Gene Expression.
(A) RT-qPCR showing the induction of PORK1 expression after mechanical wounding.
(B) to (H) The expression of PI-II (B), OPR3 (C), AOS2 (D), LoxD (E), LRR22 (F), ACS6 (G), and ERF1b (H) in responses to mechanical wounding was reduced in PORK1 RNAi plants. Values represent mean ± sd (n = 9) from three independent biological replicates and three technical repeats. Quantification of each gene expression level was normalized to the level of Actin expression. Different letters indicate significant differences. The multiple comparisons were calculated by two-way ANOVA followed by Bonferroni post-hoc test (P < 0.05; Supplemental File 1). hpt, hours post-treatment. Data are representative of at least three independent experiments.
The gene expression data revealed that PORK1 may play slightly different roles in systemin- and wound-induced gene expression. Overall, wounding-induced expression of a variety of genes clearly hinges upon functional PORK1. The role of PORK1 in systemin- and wound-induced gene expression is similar but not identical, suggesting some overlap and unique patterns. It is possible that other factors acting downstream of PORK1 may contribute to the slightly different regulation of systemin- or wounding-induced gene expression.
Wounding and Systemin Induce PORK1 and TPK1b Phosphorylation
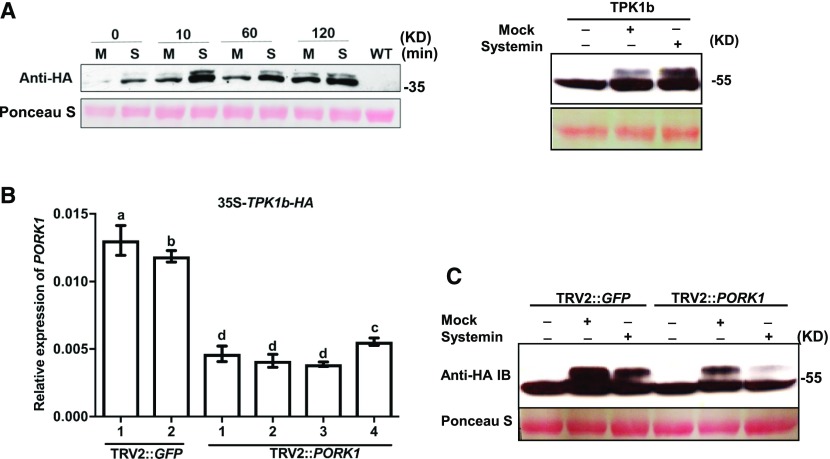
Experiments described in the preceding sections demonstrate that PORK1 is involved in tomato wound and systemin response signaling. To determine the biochemical relationship between PORK1 and TPK1b, we studied the regulation of PORK1 and TPK1b kinase activities. The TPK1b-HA and PORK1-KD-HA were transiently coexpressed in N. benthamiana, followed by mock (water) or systemin treatment. The amount of PORK1 was increased in response to systemin and reached its highest level 10 min after treatment, which also was accompanied by a slight mobility shift due to phosphorylation (Figure 7A). Immunoblot analyses using antiphosphoserine/threonine antibody confirmed the phosphorylation of PORK1 (Supplemental Figure 7A). Inclusion of protein phosphatase in the reaction led to reduction of PORK1-KD-HA phosphorylation (Supplemental Figure 7B). Interestingly, mock infiltration, which caused wounding of leaf tissue, also induced a slight increase in the phosphorylation of TPK1b-HA and PORK1-KD-HA (Figure 7A).
Figure 7.
Regulation of PORK1 and TPK1b Phosphorylation.
(A) Protein accumulation and phosphorylation of PORK1 (left panel) and TPK1b (right panel) are enhanced in response to wounding and systemin. PORK1-KD-HA and TPK1b-HA were infiltrated into N. benthamiana leaves. At 36 h, water (ddH2O) or 1 µM systemin was infiltrated, and tissue samples collected at different time points. Immunoblots were used to detect protein levels and mobility shifts of proteins. Ponceau S staining shows equal loading of total protein. M, mock; S, systemin. At least two independent experiments were repeated showing the similar results.
(B) RT-qPCR analysis of PORK1 transcript levels in GFP- or PORK1-silenced TPK1b-HA overexpressing plants. The expression of PORK1 was normalized to that of Actin. Values represent mean ± sd (n = 9) from three independent biological replicates and three technical replicates. Different letters indicate statistically significant differences. Multiple comparisons were calculated by one-way ANOVA followed by Bonferroni post-hoc test (P < 0.05; Supplemental File 1). Data are representative of two independent experiments.
(C) PORK1 is required for systemin-induced TPK1b phosphorylation. GFP- and PORK1-silenced TPK1b-overexpressing plants selected from (B) were infiltrated with water or systemin to induce TPK1b phosphorylation. TPK1b expression and phosphorylation were detected by immunoblotting. Mobility shifts of the proteins are owing to changes in phosphorylation. The experiment was repeated three times with similar results.
Since wounding and systemin could activate PORK1 and TPK1b phosphorylation in a similar manner and PORK1 directly phosphorylates TPK1b, we hypothesized that PORK1 is the upstream regulator of TPK1b in wounding- or systemin-induced signaling pathway. To test this hypothesis, PORK1 expression was silenced through VIGS in transgenic 35S-TPK1b-HA tomato plants. PORK1 expression in individual plants were examined by RT-qPCR (Figure 7B), and systemin was infiltrated into leaves of these PORK1 or mock-silenced (TRV2:GFP) transgenic plants. Systemin peptide and wounding (mock)-induced TPK1b-HA phosphorylation was reduced in PORK1-silenced plants compared with control plants (Figure 7C).
DISCUSSION
The systemin peptide, produced by species in the Solanaceae, has been known since 1991 for its role in regulating defense against insect herbivory (Pearce et al., 1991). Systemin was first isolated from wounded tomato leaves and regulates the synthesis of diverse antinutritional molecules, signaling proteins, and proteases contributing to defense (Bishop et al., 1981; McGurl et al., 1992; Ryan, 2000). In this report, we establish the biological and biochemical functions of PORK1, a TPK1b-interacting RLK, in wound/systemin signaling and systemin-mediated plant responses to fungal infection and insect herbivory. TPK1b and PORK1 perform overlapping functions in fungal and insect resistance, systemin-mediated gene expression, and transcriptional and posttranslational regulations by wounding and systemin. Phosphorylation of TPK1b in response to systemin or wounding was compromised when PORK1 was suppressed, implicating PORK1 as the upstream kinase for TPK1b. Importantly, suppression of PORK1 in the background of 35S:Prosystemin plants compromised systemin-mediated resistance to B. cinerea. Interestingly, mechanical wounding- and systemin-induced expressions of PI-II and JA-responsive genes were severely compromised in PORK1 and TPK1b RNAi plants. Furthermore, tomato seedling growth responses to systemin suggest PORK1 is required for growth responses to systemin. Previously, SlBRI1 was shown to bind to systemin (Scheer and Ryan, 2002). However, subsequent studies demonstrated that SlBRI1 is not the physiological systemin receptor, as it was found to be dispensable for systemin signaling (Holton et al., 2007; Malinowski et al., 2009; Wang et al., 2018).
Recently, tomato Solyc03g082470 (SYR1) and Solyc03g082450 (SYR2) encoding RLKs were demonstrated to regulate responses to systemin, and SYR1 was shown to display high-affinity binding to systemin (Wang et al., 2018). However, tomato PORK1 RNAi lines carrying functional SYR1 and SYR2 (Wang et al., 2018) show loss of responses to systemin, which suggests that PORK1 is critical for systemin-mediated plant responses. It is possible that PORK1 is involved in systemin-related functions by acting with other components including SYR1 and SYR2. Similar observations have been made previously for plant responses to the fungal PAMP chitin where multiple LysM RLKs with different affinities for chitin are involved (Petutschnig et al., 2010; Cao et al., 2014). This is further supported by the sequence relatedness of PORK1 and SYR proteins and their biological functions in systemin-regulated plant responses. The possibility of heterodimer or complex formation for systemin recognition among RLKs warrants further studies. Regardless, PORK1 is essential for systemin-induced downstream gene expression, seedling growth responses, insect herbivory, and Prosystemin-mediated resistance to B. cinerea infection, which reinforces its critical role as a component of systemin signaling.
PORK1 is a typical RLK, which often function in protein-protein and protein-ligand interactions, suggesting PORK1 may play roles in signal recognition and transduction (Sakamoto et al., 2012). Despite extensive studies on Arabidopsis RLKs as receptors for pathogen-derived signals and peptides, the function of most tomato RLKs and their downstream components are understudied. Recent reports describe the function of tomato RLKs FLS2, FLS3, and CORE, which detect the bacterial flagellin and bacterial cold-shock protein, respectively (Robatzek et al., 2007; Hind et al., 2016; Wang et al., 2016). The tomato LysM RLK mediates responses to Pseudomonas syringae, and the tomato I-3 gene, encoding an S-receptor-like kinase, confers resistance to Fusarium oxysporum f. sp lycopersici RLK (Zeng et al., 2012; Catanzariti et al., 2015). In addition, the tomato RLKs EIX1 and EIX2 serve as receptors for fungal xylanase (Ron and Avni, 2004; Bar et al., 2010). Tomato FLS2, BRI1, and PORK1 complex with TPK1b in co-IP assays. FLS2 has been studied in Arabidopsis and tomato (Chinchilla et al., 2007; Robatzek et al., 2007; Mueller et al., 2012), but the biological function of the interaction between TPK1b, FLS2, and BRI proteins is unclear. While TPK1b and PORK1 RNAi plants do not show any altered response to P. syringae and to brassinolide, our data clearly demonstrate that the two genes are required for plant responses to systemin, making the TPK1b and PORK1 interaction relevant for biological functions mediated by systemin.
Despite the divergence of the primary sequences of tomato systemin and Arabidopsis peptides, there appears to be a parallel system of peptide response signaling in the two plant systems that utilize the same overall mechanism and structurally related RLKs. Based on the phenotypic data, and loss of wound- and peptide-induced gene expression, PORK1 mediates responses to systemin similar to the pep1 responses in Arabidopsis mediated by PEPR1 and PEPR2. PORK1 shares 50% identity to Arabidopsis PEPR1 and 48% identity to PEPR2, while no other RLKs in the tomato genome matched that level of identity to Arabidopsis PEPRs. PEPR1 is the receptor for Pep1-6 and PEPR2 is a receptor for Pep1 and Pep2 (Yamaguchi et al., 2006), which are known to affect plant defense responses to fungal and bacterial pathogens. The Arabidopsis peptides and tomato systemin also trigger comparable responses in the two plant species. However, systemin induces responses only in tomato, similar to pep1, which activates responses in Arabidopsis but not tomato. Interestingly, PEPRs interact with BIK1 to mediate ethylene- and Pep1-induced plant immunity against B. cinerea (Liu et al., 2013), similar to TPK1b interactions with multiple tomato RLKs. Our data suggest that systemin activates PORK1-mediated TPK1b phosphorylation, which then may activate multiple intracellular targets. In Arabidopsis, BIK1 serves as a kinase substrate for multiple RLKs, including FLS2, EFR, and PEPR1/PEPR2 (Lu et al., 2010; Zhang et al., 2010; Liu et al., 2013). Similarly, TPK1b may complex with multiple RLKs, suggesting that it integrates responses from diverse recognition events.
Recent studies have identified peptide-receptor pairs involved in plant growth and immunity (Song et al., 2016). In the case of systemin, components of its signaling leading to the synthesis of proteinase inhibitors have been studied extensively. Tissue or cell damage caused by mechanical wounding, necrotrophic pathogen infection, or feeding by larvae of insect herbivores induces accumulation of Prosystemin. The proteolytic processing of Prosystemin generates systemin, which is then secreted to the extracellular space to be recognized by PRRs such as SYR1. Systemin activates phosphorylation of TPK1b by PORK1 and possibly other RLKs, SYR1 and SYR2 included, which likely provides the regulatory link to downstream signaling pathways, such as activation of the octadecanoid pathway for JA biosynthesis, defense gene expression, and the synthesis of protease inhibitors. This conclusion is consistent with PORK1-dependent expression of PI-II and of the gene for allene oxide synthase, which catalyzes the first step in the biosynthesis of JA from lipoxygenase-derived hydroperoxides of free fatty acids (Sivasankar et al., 2000). Jasmonates are key players in wounding- and systemin-induced expression of PI genes (Farmer et al., 1992; McGurl et al., 1992; Orozco-Cardenas et al., 1993). In addition, tomato mutants coi1, spr1, spr2, spr6, and def1, which are defective in JA biosynthesis and signaling, exhibited impaired wounding- and systemin-mediated defense gene expression and resistance to insect herbivores (Howe et al., 1996; Lee and Howe, 2003; Li et al., 2003; Schilmiller and Howe, 2005; Sun et al., 2011). Tomato MPK1 and MPK2 also contribute to systemin-mediated expression of PIs and resistance to feeding by tobacco hornworm larvae (Kandoth et al., 2007).
Our data implicate PORK1 and TPK1b as early components of the systemin response pathway. Interestingly, most components of this pathway affect responses to fungal infection and insect pests, consistent with the phenotypes of PORK1 RNAi and TPK1b RNAi plants that are susceptible to feeding by tobacco hornworm larvae and are impaired in B. cinerea resistance. PORK1 and TPK1b regulate the systemin-mediated gene expression that underpins the defense functions of systemin. Analysis of the genome-wide regulatory effect of PORK1 during systemin responses and its similarity and differences with fungal-induced gene expression will help dissect what downstream components of PORK1 are important. In addition, the regulatory output from PORK1- and TPK1b-mediated phosphorylation events leading to transcriptional regulation of target genes as well as the functional and regulatory relationships with the tomato SYR1 and SYR2 proteins, and the exact role of PORK1 at the early stages of systemin responses require further study. The functional and biochemical relationship between tomato PROPEP6 (Lori et al., 2015) and PORK1 also needs further analyses.
METHODS
Plant Growth
Tomato plants (Solanum lycopersicum) cultivar Castlemart II were grown in plastic pots containing compost soil (Sun Gro Metro Mix 510) in a growth chamber with a photoperiod extended to 16 h under fluorescent lights (160 W mol−1 m−2 s−1) at a temperature of 20 to 24°C. The 35S-Prosystemin transgenic line was obtained from Gregg Howe (Michigan State University). 35S-TPK1b-HA and PORK1 RNAi transgenic plants were generated at the Boyce Thompson Institute for Plant Research transformation facility. The homozygous lines were identified by selecting on MS medium containing 75 µg/mL of kanamycin, and silencing of PORK1 expression in RNAi transgenic plants was confirmed with quantitative RT-PCR. All tomato plants were fertilized (Miracle-Gro Tomato Food) alternated with tap water.
Seedling Growth Sensitivity Assay
Tomato seeds were surface sterilized with 70% ethanol for 5 s and 10 min in sterilization solution containing 35% commercial bleach solution (5.25% [w/v] sodium hypochlorite) and 0.1% Tween 20 (Bio-Rad). The seeds were put in Murashige and Skoog Basal Salt Mixture (Phyto Technology Laboratories) medium containing 0.8% agarose with or without 10 nM systemin (GenScript). The germinated seedlings were grown for 10 to 12 d before measurements were taken on seedling growth.
Fungal Culture and Disease Assay
The Botrytis cinerea strain B05-10 was cultured on V8 agar medium (36% V8 juice, 0.1% CaCO3, and 2% Bacto-agar) and incubated at room temperature for 8 to 12 d. Conidia were suspended in 1% Sabouraud Maltose Broth buffer (Becton, Dickinson and Company). B. cinerea disease assay was performed by drop inoculation of 5 μL of 1 × 105 spores/mL spore suspension on detached leaves. Disease lesions were measured at 3 d after inoculation. For the systemin-induced resistance assay, detached leaves were sprayed with water or 2 µM systemin supplemented with 0.01% Silwet L-77 and incubated overnight in a container with high relative humidity at which point they were inoculated with B. cinerea as described above.
Insect Feeding Assay, Wounding, and Systemin Treatment
Eggs of tobacco hornworm (Manduca sexta) were hatched at 24°C in a growth chamber for 5 d on an artificial diet (M. sexta diet; Southland Products). One or two larvae were weighed and then placed on individual plants, which were kept in an observation cage (BioQuip Products) and allowed to feed up to 10 d. The larvae were weighed again at the end of assay. For the wound response assays, 5-week-old tomato plants were mechanically wounded as described previously (Abuqamar et al., 2008). Dented forceps were used to press across the middle veins three times at different positions. For systemin-induced gene expression, tomato leaves were cut at petioles and dipped into 10 nM systemin solution. For systemin-induced phosphorylation of TPK1b and PORK1-KD, 1 µM systemin was syringe-infiltrated into Nicotiana benthamiana leaves expressing PORK1-KD-HA or TPK1b-HA protein for 1 h before the leave tissues are homogenized for protein expression.
VIGS
Silencing of PORK1 was conducted using Tobacco Rattle Virus (TRV)-based vector system as described (Liu et al., 2002). To generate pTRV2-PORK1 construct, 550 bp from 5′ end of cDNA was cloned into pYL156 (Wu et al., 2011). pTRV2-GFP and pTRV2-PORK1 and pTRV1 constructs were transformed into Agrobacterium tumefaciens strain GV3101, which was infiltrated into the cotyledons of tomato seedlings. Four weeks after infiltration, leaves from each plant were collected for examining PORK1 gene expression. Plants with silenced PORK1 expression were selected for further experiments.
Co-IP and Immunoblot Analysis
Co-IP assay was conducted on protein extracts from N. benthamiana leaves transiently expressing the various constructs. Leaves were frozen in liquid nitrogen and homogenized in cold extraction buffer containing 20 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM NaF, 10 mM β-glycerophosphate, 0.1% Tween 20, protease inhibitor cocktail (Sigma-Aldrich), 1 mM PMSF, 1 mM DTT, 1 mM Na3VO4, and 0.1% Triton X-100. Tissue lysate was collected and spun down several times by centrifugation at 13,500g to get rid of insoluble components. The supernatant was incubated in anti-HA conjugated agarose beads (Sigma-Aldrich) at 4°C overnight. The beads were washed with extraction buffer three times, boiled for 10 min with 6× SDS loading buffer, and spun down. The proteins in the supernatant were separated in SDS-PAGE and visualized by immunoblot using the anti-HA tag (ab9110; Abcam) and the anti-Myc tag (ab9106; Abcam) polyclonal antibodies.
In Vitro Pull-Down Assay
The production of recombinant proteins is described below. The full-length recombinant protein TPK1b tagged with GST (GST-TPK1b) was prewashed with amylose beads and then mixed with MBP-tagged PORK1 recombinant protein from LRR domain (LRR), kinase domain (KD), or kinase domain with point mutation (KDK30A). The mixture was incubated in binding buffer (10 mM HEPES, pH 7.5, 200 mM NaCl, 1 mM EDTA, 10% glycerol, and 0.5% Triton X-100) under gentle shaking at 4°C 1 h with maltose beads. After that, beads were washed with binding buffer four times and resuspended into binding buffer with 6× SDS loading buffer for immunoblot analysis by anti-GST antibody (New England Biolabs) and anti-MBP antibody (Abcam).
Purification of Recombinant Proteins and in Vitro Kinase Assay
For purification of recombinant PORK1-KD, PORK1-KDK30A, and TPK1bK106A, the cDNA was cloned with the restriction sites BamHI, PstI, XmnI, and XbaI into maltose binding protein expression vector pMAL-c2X (New England Biolabs) and transformed into Escherichia coli BL21 cells. The expression and purification were conducted as described (Riggs, 2000). For PORK1-KD protein, BL21 cells containing PORK1-KD construct were cultured at 37°C until the concentration of cells reached A600 = 0.5. IPTG was added to a final concentration of 1 mM to induce the expression of PORK1-KD at room temperature. Cells were centrifuged down and resuspended in column buffer containing 20 mM Tris-HCl (pH 7.4), 200 mM NaCl, and 1 mM EDTA. Cells were disrupted by sonication and spun down by centrifugation. The proteins in the supernatant were loaded in an affinity column containing amylose resin, which binds to maltose binding protein fused recombinant protein. The column was washed with column buffer and proteins were eluted from the column through elution buffer. The concentration of the recombinant proteins was determined by Quick Start Bradford Protein Assay (Bio-Rad). In vitro kinase assay was performed as described (Abuqamar et al., 2008) by incubating 5 µg of active kinase PORK1-KD, or PORK1-KDK30A and 5 µg substrate TPK1bK106A, or the artificial kinase substrate MBP in a 20-μL reaction buffer containing 20 mM Tris-HCl, pH 7.5, 10 mM MnCl2, 1 mM DTT, 1 mM EGTA, 100 µM ATP, and 5 µCi [γ-32P]ATP at 25°C for 25 min. The reaction was stopped by adding 6× SDS loading buffer. The phosphorylation of MBP or TPK1bK106A was detected by autoradiography after separation on 12% or 8% SDS-PAGE, respectively.
Protein Phosphorylation
To study phosphorylation levels, PORK1-KD-HA was expressed in N. benthamiana leaves through agroinfiltration. Protein was extracted at 36 h after infiltration and separated on SDS-PAGE gel and the PORK1-KD-HA protein, and its phosphorylation were detected by probing anti-HA antibody (Sigma-Aldrich) and antiphosphoserine/threonine (ECM Biosciences), respectively. To confirm phosphorylation events, PORK1-KD-HA was transiently expressed in N. benthamiana and total protein was extracted and treated with calf intestinal alkaline phosphatase (New England Biolabs). After incubation for 1 h at 37°C, 6× SDS loading buffer was added and boiled for further immunoblot analysis by anti-HA antibody (Sigma-Aldrich).
Vector Construction
To make the pTRV2-PORK1 construct, 550 bp from 5′ end of cDNA was amplified by PCR using pTRV2-PORK1/LP (5′-TGCTCTAGAACTTCTGATGGCACCGCTTTA-3′, the XbaI site is underlined) and pTRV2-PORK1/RP (5′-CCGCTCGAGTTTCCTATGGAAGAGGGAATT-3′, the XhoI site is underlined). After restriction digestion, PCR products were cloned into pYL156 vector.
Generation of PORK1 RNAi construct was similar to procedures described previously (Abuqamar et al., 2008) with some modifications. The 500 bp from 5′ end of cDNA was amplified by PCR from tomato cDNA using PORK1 RNAi/LP (5′-GCACTAGTCCATGGCCTCCATTCCTTCTC-3′, SpeI and NcoI sites are underlined) and PORK1 RNAi/RP (5′-ATGGATCCGGCGCGCCAAGTTCCCCATTGAAA-3′, BamHI and AscI sites are underlined). A two-step cloning procedure was used to clone the inverted repeats into the binary vector pGSA1165. In the first step, PCR products and vector were cleaved by AscI and NcoI and ligated by T4 DNA Ligase (New England Biolabs). For the second step, PCR products and the plasmid generated from the first step were cleaved using BamHI and SpeI and ligated again. The second insert is placed in an inverted orientation with respect to the first one, resulting in an inverted repeat separated by a GUS fragment.
To generate 35S-TPK1b-HA overexpression construct for transient expression in N. benthamiana, TPK1b full-length cDNA was amplified from tomato cDNA by PCR with TPK1b-HA/LP (5′-TCCCCGCGGTATGGGGATATGTTTGAGTGCT-3′, SacII site is underlined) and TPK1b-HA/RP (5′-TGCTCTAGATTTAGCGTAAGGGGGAGAAGC-3′, XbaI site is underlined). After restriction digestion, PCR products were cloned into pCAMBIA99-1 vector (Abuqamar et al., 2008). Using similar methods, we also generated 35S-PORK1-HA, 35S-PORK1-KD-HA, 35S-SlBRI1-KD-HA, and 35S-SlFLS2-KD-HA. The primers used for construction of these vectors are listed in Supplemental Table 2.
RNA Extraction and Analysis of Gene Expression
Total RNA was isolated with Trizol reagent from tomato or Arabidopsis leaf tissues according to the manufacturer’s instructions (Invitrogen). After extraction, RNA is treated with DNase I (New England Biolabs). RNA concentration was measured using Nanodrop 2000c (Thermo Scientific). The first-strand cDNA was synthesized from 2 µg of total RNA using AMV reverse transcriptase (New England Biolabs). RT-qPCR analysis was performed on Mx3000P real-time PCR detection system (Stratagene) using iTaq Universal SYBR Green Supermix (Bio-Rad). Tomato Actin was used as an endogenous reference gene for normalization. Three technical replicates of the qRT-qPCR assay were used for each sample with a minimum of three biological replicates. Each biological replicate was sampled from three individual plants. Each experiment was repeated three times. Expression levels were calculated by the comparative cycle threshold (Ct) method. Primers used for RT-qPCR are listed in Supplemental Table 1.
ROS Assay and Immunoblot of MPK3/MPK6 Phosphorylation
For ROS assays, the protocol was modified from luminol-based assay in Arabidopsis thaliana (Bisceglia et al., 2015). Briefly, four leaf disks were collected from each tomato plant and three plants from each line were used for each experiment. Leaf disks were washed with water three times every 30 min and kept overnight in the dark at room temperature. On the second day, leaf disks were soaked into solution containing luminol, horseradish peroxidase, and 1 µM systemin or flg22, and luminescence detection was conducted for at least 40 min in Infinity M200 Pro plate reader (Tecan). For MPK3/MPK6 phosphorylation, tomato leaves were treated with 1 µM systemin and sampled at 0, 5, 10, 30, and 60 min after treatment. Total protein was extracted with the same method described above. The phosphorylated MPK3 and MPK6 were detected by anti-P44/P42 antibody (Cell Signaling Technology).
Phylogenetic Analysis
Arabidopsis PEPR1 and PEPR2 amino acid sequences were used for building the phylogenetic tree by the software Molecular Evolutionary Genetics Analysis version 7.0 (MEGA7) (Kumar et al., 2016). Sequences were aligned using ClustalW (Thompson et al., 1994) using default gap opening penalty and gap extension penalty for pairwise alignment and multiple alignment. The aligned sequences (Supplemental Data Set 1) were used to construct a rooted neighbor-joining phylogenetic tree (Saitou and Nei, 1987). SlBRI1 served as outlier that is divergent from other PEPR1 and PEPR2 related proteins.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL databases under the following accession numbers: TPK1b, EU555286; PORK1, XM_004235463; SlFLS2, XM_004233044; SlBRI1, NM_001309251; AOS2, NM_001287778; ERF1b, AY192367; LoxD, U37840; SlLRR22, XM_004245095; PI-II, K03291; AtPEPR1, NM_105966; AtPEPR2, NM_101638; ACS6, NM_001247235; OPR3, NM_001246944; PR1a, NM_001247199; CaM6, NM_001247857; and MYC2, NM_001324483.
Supplemental Data
Supplemental Figure 1. Tomato PORK1, BRI1, and FLS2 interact with TPK1b.
Supplemental Figure 2. Tomato PORK1 is closely related to Arabidopsis PEPR1 and PEPR2.
Supplemental Figure 3. Expression of the Systemin receptors 1 and 2 is not affected in PORK1 RNAi plants.
Supplemental Figure 4. Tomato systemin and Arabidopsis Pep1 peptides activate plant responses only in their respective host plants.
Supplemental Figure 5. PI-II, NPR1, and COI1 gene expression in response to systemin in PORK1 RNAi plants.
Supplemental Figure 6. Accumulation of reactive oxygen species and MPK3/MPK6 phosphorylation in response to systemin.
Supplemental Figure 7. PORK1 phosphorylation detected by mobility shift and antiphosphorylation-specific antibody is sensitive to CIP phosphatase.
Supplemental Table 1. Primers used in this article.
Supplemental Table 2. Systemin sequence synthesized and used in this article.
Supplemental Data Set 1. Text file of the alignment used for the phylogenetic analysis in Supplemental Figure 2B.
Supplemental File 1. ANOVA tables.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
This research was funded by grants from the National Science Foundation (IOS-1456594) and the Next-Generation BioGreen 21 Program, Rural Development Administration, Republic of Korea (SSAC, Project PJ01137902), and by U.S.-Israel Binational Agricultural Research and Development Fund Grant IS-4937-16. We thank Gregg Howe (Michigan State University) for the Prosystemin overexpression plants.
AUTHOR CONTRIBUTIONS
S.X. conducted most of the experiments and wrote the article. C.-J.L. conducted the phosphorylation and interactions assays and contributed to the writing of the manuscript. N.J. conducted systemin mediated fungal resistance and seedling sensitivity assays. S.L. contributed to the gene expression analysis, writing of the article, and designing the experiments. I.K. and M.G. designed and advised the insect feeding trials. D.-J.Y. and S.Y.L. were involved with the project ideas and participated in writing the article. T.M. designed the research, analyzed the data, wrote the article, and directed the project.
References
- Abuqamar S., Chai M.F., Luo H., Song F., Mengiste T. (2008). Tomato protein kinase 1b mediates signaling of plant responses to necrotrophic fungi and insect herbivory. Plant Cell 20: 1964–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajwa V.S., Wang X., Blackburn R.K., Goshe M.B., Mitra S.K., Williams E.L., Bishop G.J., Krasnyanski S., Allen G., Huber S.C., Clouse S.D. (2013). Identification and functional analysis of tomato BRI1 and BAK1 receptor kinase phosphorylation sites. Plant Physiol. 163: 30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar M., Sharfman M., Ron M., Avni A. (2010). BAK1 is required for the attenuation of ethylene-inducing xylanase (Eix)-induced defense responses by the decoy receptor LeEix1. Plant J. 63: 791–800. [DOI] [PubMed] [Google Scholar]
- Beloshistov R.E., et al. (2018). Phytaspase-mediated precursor processing and maturation of the wound hormone systemin. New Phytol. 218: 1167–1178. [DOI] [PubMed] [Google Scholar]
- Bergey D.R., Ryan C.A. (1999). Wound- and systemin-inducible calmodulin gene expression in tomato leaves. Plant Mol. Biol. 40: 815–823. [DOI] [PubMed] [Google Scholar]
- Bisceglia B.G., Gravino M., Savatin D.V. (2015). Luminol-based assay for detection of immunity elicitor-induced hydrogen peroxide production in Arabidopsis thaliana leaves. Bioprotocols 5: e1685. [Google Scholar]
- Bishop P.D., Makus D.J., Pearce G., Ryan C.A. (1981). Proteinase inhibitor-inducing factor activity in tomato leaves resides in oligosaccharides enzymically released from cell walls. Proc. Natl. Acad. Sci. USA 78: 3536–3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Liang Y., Tanaka K., Nguyen C.T., Jedrzejczak R.P., Joachimiak A., Stacey G. (2014). The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. eLife 3: 03766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzariti A.M., Lim G.T.T., Jones D.A. (2015). The tomato I-3 gene: a novel gene for resistance to Fusarium wilt disease. New Phytol. 207: 106–118. [DOI] [PubMed] [Google Scholar]
- Chassot C., Buchala A., Schoonbeek H.J., Métraux J.P., Lamotte O. (2008). Wounding of Arabidopsis leaves causes a powerful but transient protection against Botrytis infection. Plant J. 55: 555–567. [DOI] [PubMed] [Google Scholar]
- Chinchilla D., Boller T., Robatzek S. (2007). Flagellin signalling in plant immunity. Adv. Exp. Med. Biol. 598: 358–371. [DOI] [PubMed] [Google Scholar]
- Coppola M., Corrado G., Coppola V., Cascone P., Martinelli R., Digilio M.C., Pennacchio F., Rao R. (2015). Prosystemin overexpression in tomato enhances resistance to different biotic stresses by activating genes of multiple signaling pathways. Plant Mol. Biol. Report. 33: 1270–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ovidio R., Mattei B., Roberti S., Bellincampi D. (2004). Polygalacturonases, polygalacturonase-inhibiting proteins and pectic oligomers in plant-pathogen interactions. Biochim. Biophys. Acta 1696: 237–244. [DOI] [PubMed] [Google Scholar]
- Decreux A., Messiaen J. (2005). Wall-associated kinase WAK1 interacts with cell wall pectins in a calcium-induced conformation. Plant Cell Physiol. 46: 268–278. [DOI] [PubMed] [Google Scholar]
- El Oirdi M., El Rahman T.A., Rigano L., El Hadrami A., Rodriguez M.C., Daayf F., Vojnov A., Bouarab K. (2011). Botrytis cinerea manipulates the antagonistic effects between immune pathways to promote disease development in tomato. Plant Cell 23: 2405–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer E.E., Johnson R.R., Ryan C.A. (1992). Regulation of expression of proteinase inhibitor genes by methyl jasmonate and jasmonic acid. Plant Physiol. 98: 995–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green T.R., Ryan C.A. (1972). Wound-induced proteinase inhibitor in plant leaves: a possible defense mechanism against insects. Science 175: 776–777. [DOI] [PubMed] [Google Scholar]
- Hahn M.G., Darvill A.G., Albersheim P. (1981). Host-pathogen interactions: XIX. The endogenous elicitor, a fragment of a plant cell wall polysaccharide that elicits phytoalexin accumulation in soybeans. Plant Physiol. 68: 1161–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz T., Bergey D.R., Ryan C.A. (1997). A gene encoding a chloroplast-targeted lipoxygenase in tomato leaves is transiently induced by wounding, systemin, and methyl jasmonate. Plant Physiol. 114: 1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hind S.R., et al. (2016). Tomato receptor FLAGELLIN-SENSING 3 binds flgII-28 and activates the plant immune system. Nat. Plants 2: 16128. [DOI] [PubMed] [Google Scholar]
- Holton N., Caño-Delgado A., Harrison K., Montoya T., Chory J., Bishop G.J. (2007). Tomato BRASSINOSTEROID INSENSITIVE1 is required for systemin-induced root elongation in Solanum pimpinellifolium but is not essential for wound signaling. Plant Cell 19: 1709–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe G.A., Ryan C.A. (1999). Suppressors of systemin signaling identify genes in the tomato wound response pathway. Genetics 153: 1411–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe G.A., Lightner J., Browse J., Ryan C.A. (1996). An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell 8: 2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A., Ryan C.A. (2007). Endogenous peptide defense signals in Arabidopsis differentially amplify signaling for the innate immune response. Proc. Natl. Acad. Sci. USA 104: 10732–10736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A., Pearce G., Ryan C.A. (2006). An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. USA 103: 10098–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth P.K., Ranf S., Pancholi S.S., Jayanty S., Walla M.D., Miller W., Howe G.A., Lincoln D.E., Stratmann J.W. (2007). Tomato MAPKs LeMPK1, LeMPK2, and LeMPK3 function in the systemin-mediated defense response against herbivorous insects. Proc. Natl. Acad. Sci. USA 104: 12205–12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauss H., Fauth M., Merten A., Jeblick W. (1999). Cucumber hypocotyls respond to cutin monomers via both an inducible and a constitutive H2O2-generating system. Plant Physiol. 120: 1175–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klauser D., Desurmont G.A., Glauser G., Vallat A., Flury P., Boller T., Turlings T.C., Bartels S. (2015). The Arabidopsis Pep-PEPR system is induced by herbivore feeding and contributes to JA-mediated plant defence against herbivory. J. Exp. Bot. 66: 5327–5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33: 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfermeijer F.C., Staal M., Malinowski R., Stratmann J.W., Elzenga J.T.M. (2008). Micro-electrode flux estimation confirms that the Solanum pimpinellifolium cu3 mutant still responds to systemin. Plant Physiol. 146: 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G.I., Howe G.A. (2003). The tomato mutant spr1 is defective in systemin perception and the production of a systemic wound signal for defense gene expression. Plant J. 33: 567–576. [DOI] [PubMed] [Google Scholar]
- Li C., Liu G., Xu C., Lee G.I., Bauer P., Ling H.Q., Ganal M.W., Howe G.A. (2003). The tomato suppressor of prosystemin-mediated responses2 gene encodes a fatty acid desaturase required for the biosynthesis of jasmonic acid and the production of a systemic wound signal for defense gene expression. Plant Cell 15: 1646–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Li C., Lee G.I., Howe G.A. (2002). Distinct roles for jasmonate synthesis and action in the systemic wound response of tomato. Proc. Natl. Acad. Sci. USA 99: 6416–6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhao Y., McCaig B.C., Wingerd B.A., Wang J., Whalon M.E., Pichersky E., Howe G.A. (2004). The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16: 126–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Lu D., Gao X., Jiang S., Ma X., Wang Z., Mengiste T., He P., Shan L. (2013). Inverse modulation of plant immune and brassinosteroid signaling pathways by the receptor-like cytoplasmic kinase BIK1. Proc. Natl. Acad. Sci. USA 110: 12114–12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Dinesh-Kumar S.P. (2002). Virus-induced gene silencing in tomato. Plant J. 31: 777–786. [DOI] [PubMed] [Google Scholar]
- Liu Z., Wu Y., Yang F., Zhang Y., Chen S., Xie Q., Tian X., Zhou J.M. (2013). BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc. Natl. Acad. Sci. USA 110: 6205–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lori M., van Verk M.C., Hander T., Schatowitz H., Klauser D., Flury P., Gehring C.A., Boller T., Bartels S. (2015). Evolutionary divergence of the plant elicitor peptides (Peps) and their receptors: interfamily incompatibility of perception but compatibility of downstream signalling. J. Exp. Bot. 66: 5315–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D., Wu S., Gao X., Zhang Y., Shan L., He P. (2010). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl. Acad. Sci. USA 107: 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinowski R., Higgins R., Luo Y., Piper L., Nazir A., Bajwa V.S., Clouse S.D., Thompson P.R., Stratmann J.W. (2009). The tomato brassinosteroid receptor BRI1 increases binding of systemin to tobacco plasma membranes, but is not involved in systemin signaling. Plant Mol. Biol. 70: 603–616. [DOI] [PubMed] [Google Scholar]
- McGurl B., Pearce G., Orozco-Cardenas M., Ryan C.A. (1992). Structure, expression, and antisense inhibition of the systemin precursor gene. Science 255: 1570–1573. [DOI] [PubMed] [Google Scholar]
- McGurl B., Orozco-Cardenas M., Pearce G., Ryan C.A. (1994). Overexpression of the prosystemin gene in transgenic tomato plants generates a systemic signal that constitutively induces proteinase inhibitor synthesis. Proc. Natl. Acad. Sci. USA 91: 9799–9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meindl T., Boller T., Felix G. (1998). The plant wound hormone systemin binds with the N-terminal part to its receptor but needs the C-terminal part to activate it. Plant Cell 10: 1561–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller K., Bittel P., Chinchilla D., Jehle A.K., Albert M., Boller T., Felix G. (2012). Chimeric FLS2 receptors reveal the basis for differential flagellin perception in Arabidopsis and tomato. Plant Cell 24: 2213–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H.P., Chakravarthy S., Velásquez A.C., McLane H.L., Zeng L., Nakayashiki H., Park D.H., Collmer A., Martin G.B. (2010). Methods to study PAMP-triggered immunity using tomato and Nicotiana benthamiana. Mol. Plant Microbe Interact. 23: 991–999. [DOI] [PubMed] [Google Scholar]
- Orozco-Cardenas M., McGurl B., Ryan C.A. (1993). Expression of an antisense prosystemin gene in tomato plants reduces resistance toward Manduca sexta larvae. Proc. Natl. Acad. Sci. USA 90: 8273–8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G., Strydom D., Johnson S., Ryan C.A. (1991). A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 253: 895–897. [DOI] [PubMed] [Google Scholar]
- Pearce G., Johnson S., Ryan C.A. (1993). Structure-activity of deleted and substituted systemin, an 18-amino acid polypeptide inducer of plant defensive genes. J. Biol. Chem. 268: 212–216. [PubMed] [Google Scholar]
- Petutschnig E.K., Jones A.M.E., Serazetdinova L., Lipka U., Lipka V. (2010). The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J. Biol. Chem. 285: 28902–28911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postel S., Küfner I., Beuter C., Mazzotta S., Schwedt A., Borlotti A., Halter T., Kemmerling B., Nürnberger T. (2010). The multifunctional leucine-rich repeat receptor kinase BAK1 is implicated in Arabidopsis development and immunity. Eur. J. Cell Biol. 89: 169–174. [DOI] [PubMed] [Google Scholar]
- Riggs P. (2000). Expression and purification of recombinant proteins by fusion to maltose-binding protein. Mol. Biotechnol. 15: 51–63. [DOI] [PubMed] [Google Scholar]
- Robatzek S., Bittel P., Chinchilla D., Köchner P., Felix G., Shiu S.H., Boller T. (2007). Molecular identification and characterization of the tomato flagellin receptor LeFLS2, an orthologue of Arabidopsis FLS2 exhibiting characteristically different perception specificities. Plant Mol. Biol. 64: 539–547. [DOI] [PubMed] [Google Scholar]
- Ron M., Avni A. (2004). The receptor for the fungal elicitor ethylene-inducing xylanase is a member of a resistance-like gene family in tomato. Plant Cell 16: 1604–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan C.A. (2000). The systemin signaling pathway: differential activation of plant defensive genes. Biochim. Biophys. Acta 1477: 112–121. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Sakamoto T., Deguchi M., Brustolini O.J.B., Santos A.A., Silva F.F., Fontes E.P.B. (2012). The tomato RLK superfamily: phylogeny and functional predictions about the role of the LRRII-RLK subfamily in antiviral defense. BMC Plant Biol. 12: 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller A., Ryan C.A. (1994). Identification of a 50-kDa systemin-binding protein in tomato plasma membranes having Kex2p-like properties. Proc. Natl. Acad. Sci. USA 91: 11802–11806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer J.M., Ryan C.A. (1999). A 160-kD systemin receptor on the surface of Lycopersicon peruvianum suspension-cultured cells. Plant Cell 11: 1525–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer J.M., Ryan C.A. Jr (2002). The systemin receptor SR160 from Lycopersicon peruvianum is a member of the LRR receptor kinase family. Proc. Natl. Acad. Sci. USA 99: 9585–9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller A.L., Howe G.A. (2005). Systemic signaling in the wound response. Curr. Opin. Plant Biol. 8: 369–377. [DOI] [PubMed] [Google Scholar]
- Schulze B., Mentzel T., Jehle A.K., Mueller K., Beeler S., Boller T., Felix G., Chinchilla D. (2010). Rapid heteromerization and phosphorylation of ligand-activated plant transmembrane receptors and their associated kinase BAK1. J. Biol. Chem. 285: 9444–9451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer P., Felix G., Buchala A., Muller C., Metraux J.P. (1996). Perception of free cutin monomers by plant cells. Plant J. 10: 331–341. [Google Scholar]
- Sivasankar S., Sheldrick B., Rothstein S.J. (2000). Expression of allene oxide synthase determines defense gene activation in tomato. Plant Physiol. 122: 1335–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., et al. (2016). Signature motif-guided identification of receptors for peptide hormones essential for root meristem growth. Cell Res. 26: 674–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.Q., Jiang H.L., Li C.Y. (2011). Systemin/Jasmonate-mediated systemic defense signaling in tomato. Mol. Plant 4: 607–615. [DOI] [PubMed] [Google Scholar]
- Tang D., Wang G., Zhou J.M. (2017). Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell 29: 618–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J., Han Z., Sun Y., Zhang H., Gong X., Chai J. (2015). Structural basis for recognition of an endogenous peptide by the plant receptor kinase PEPR1. Cell Res. 25: 110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor K.W., Kim J.G., Su X.B., Aakre C.D., Roden J.A., Adams C.M., Mudgett M.B. (2012). Tomato TFT1 is required for PAMP-triggered immunity and mutations that prevent T3S effector XopN from binding to TFT1 attenuate Xanthomonas virulence. PLoS Pathog. 8: e1002768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins D.G., Gibson T.J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Albert M., Einig E., Fürst U., Krust D., Felix G. (2016). The pattern-recognition receptor CORE of Solanaceae detects bacterial cold-shock protein. Nat. Plants 2: 16185. [DOI] [PubMed] [Google Scholar]
- Wang L., Einig E., Almeida-Trapp M., Albert M., Fliegmann J., Mithöfer A., Kalbacher H., Felix G. (2018). The systemin receptor SYR1 enhances resistance of tomato against herbivorous insects. Nat. Plants 4: 152–156. [DOI] [PubMed] [Google Scholar]
- Wasternack C., Stenzel I., Hause B., Hause G., Kutter C., Maucher H., Neumerkel J., Feussner I., Miersch O. (2006). The wound response in tomato—role of jasmonic acid. J. Plant Physiol. 163: 297–306. [DOI] [PubMed] [Google Scholar]
- Wu C., Jia L., Goggin F. (2011). The reliability of virus-induced gene silencing experiments using tobacco rattle virus in tomato is influenced by the size of the vector control. Mol. Plant Pathol. 12: 299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Pearce G., Ryan C.A. (2006). The cell surface leucine-rich repeat receptor for AtPep1, an endogenous peptide elicitor in Arabidopsis, is functional in transgenic tobacco cells. Proc. Natl. Acad. Sci. USA 103: 10104–10109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Huffaker A., Bryan A.C., Tax F.E., Ryan C.A. (2010). PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell 22: 508–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng L., Velásquez A.C., Munkvold K.R., Zhang J., Martin G.B. (2012). A tomato LysM receptor-like kinase promotes immunity and its kinase activity is inhibited by AvrPtoB. Plant J. 69: 92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., et al. (2010). Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe 7: 290–301. [DOI] [PubMed] [Google Scholar]