APE2 and ZDP have overlapping functions in base excision repair of DNA lesions produced during active DNA demethylation and by environmental stresses.
Abstract
Base excision repair (BER) is essential for active DNA demethylation and DNA damage repair in mammals and plants. Here, we provide genetic and biochemical evidence that APURINIC/APYRIMIDINIC ENDONUCLEASE2 (APE2) plays overlapping roles with ZINC FINGER DNA 3′-PHOSPHOESTERASE (ZDP) in active DNA demethylation and DNA damage repair in Arabidopsis thaliana. Simultaneous mutation of APE2 and ZDP causes DNA hypermethylation at more than 2000 loci, most of which are not hypermethylated in ape2 or zdp single mutants. The zdp and ape2 single mutants exhibit normal development, but the zdp ape2 double mutants display pleiotropic developmental defects and are supersensitive to the DNA alkylating reagent methyl methanesulfonate. The gradual accumulation of DNA lesions in the zdp ape2 seedlings is accompanied by constitutive activation of the DNA damage response and alteration of the cell cycle. Interestingly, knockout of the key DNA demethylase REPRESSOR OF SILENCING1 reduces the magnitude of DNA lesion accumulation and the DNA damage response in the zdp ape2 mutants, suggesting that a proportion of the DNA damage in the zdp ape2 mutants arises from incomplete active DNA demethylation. Lastly, we find that APE2 has 3′-phosphatase activity and strong 3′-5′ exonuclease activity in vitro. Together, our results suggest that APE2 and ZDP, two BER proteins, play overlapping roles in the maintenance of epigenome and genome stability in plants.
INTRODUCTION
Base excision repair (BER) is an essential mechanism that repairs damage to DNA bases, including alkylation, deamination, and oxidation (Krokan and Bjørås, 2013; Drohat and Coey, 2016). BER is also required for the repair of apurinic/apyrimidinic (AP) sites resulting from spontaneous depurination and processing of the 3′ blocking ends of DNA single-strand breaks (SSBs) (Krokan and Bjørås, 2013; Wallace, 2014; Spampinato, 2017). Furthermore, recent studies show that BER functions in active DNA demethylation downstream of DNA glycosylases in animals and plants (Zhu, 2009; Gong and Zhu, 2011; Wu and Zhang, 2014; Weber et al., 2016). At present, the BER machinery functioning in active DNA demethylation is not completely understood.
In plants, DNA glycosylases REPRESSOR OF SILENCING1 (ROS1), DEMETER-LIKE2 (DML2), DML3, and DEMETER (DME) directly excise 5-methylcytosine (5mC) using their glycosylase activity, initiating active DNA demethylation (Gong et al., 2002; Agius et al., 2006; Gehring et al., 2006; Morales-Ruiz et al., 2006; Ortega-Galisteo et al., 2008; Zhu, 2009). Because these enzymes also have AP lyase activity, the phosphodiester backbone can be cleaved via β-elimination reaction or successive β, δ-elimination reactions, producing a gap with a 3′-phosphor-α, β-unsaturated aldehyde (3′-PUA) or a 3′-phosphate (3′-P), respectively (Agius et al., 2006; Gehring et al., 2006). Either 3′-PUA or 3′-P must be processed to form a 3′-hydroxyl (3′-OH) terminus so that downstream DNA polymerases and ligases can act to fill in the gap. Among the three known AP endonuclease-like proteins in Arabidopsis thaliana, APE1L has robust 3′-phosphodiesterase activity and is the major enzyme responsible for the conversion of 3′-PUA to 3′-OH (Lee et al., 2014; Li et al., 2015). Zinc finger DNA 3′-PHOSPHATASE (ZDP) transforms 3′-P into 3′-OH (Martínez-Macías et al., 2012). APE1L-mediated 3′-PUA processing and ZDP-mediated 3′-P processing represent two different branches of the active DNA demethylation pathway downstream of ROS1 and its homologs, although APE1L also possesses weak 3′-phosphatase activity and may play an overlapping role with ZDP in the processing of 3′-P terminus (Li et al., 2015). Mutations in APE1L or ZDP cause DNA hypermethylation at ∼3500 and 1500 loci, respectively; both of these sets of loci show substantial overlap with the hyper-differentially methylated regions (hyper-DMRs) in the rdd triple mutant (ros1 dml2 dml3) (Li et al., 2015). Despite alterations in DNA methylation, ape1l and zdp single mutants do not exhibit an obvious developmental phenotype, like ros1 and rdd mutants. However, simultaneous mutation of APE1L and ZDP causes embryonic lethality associated with DNA hypermethylation of multiple imprinted genes in the endosperm (Lee et al., 2014; Li et al., 2015). Another Arabidopsis AP endonuclease-like protein, ARP, was also found to have 3′-phosphodiesterase and 3′ phosphatase activities. However, mutations in ARP alone did not result in extensive DNA hypermethylation under normal growth conditions (Lee et al., 2014; Li et al., 2015).
The Arabidopsis AP endonuclease-like protein APE2, like APE1L and ARP, harbors an endonuclease/exonuclease/phosphatase (EEP) domain that has highly sequence similarity to Escherichia coli exonuclease III (Mol et al., 1995), Xenopus laevis APE2 (Willis et al., 2013; Wallace et al., 2017), and human APE1 (Mol et al., 2000) and APE2 (Hadi and Wilson, 2000). Arabidopsis APE2 has an additional GRF-type zinc finger (Zf-GRF) domain that is not present in APE1L and ARP, but shows high sequence similarity to yeast APE2, X. laevis APE2, mouse APE2, and human APE2. Human APE2 preferentially repairs mismatched bases using its 3′-5′ exonuclease activity (Burkovics et al., 2006). X. laevis APE2 facilitates SSB end resection and recruitment of checkpoint proteins ATR and Chk1 to damaged sites (Willis et al., 2013; Wallace et al., 2017). In Arabidopsis, simultaneous mutation of APE1L and APE2 is embryonic lethal (Murphy et al., 2009), although mutation of either gene alone does not lead to defective growth or development. Furthermore, a recent study showed that APE2 possesses endonuclease activity (Li et al., 2015). Based on available data, we hypothesize that APE2 may function in the BER process to repair DNA damage arising during active DNA demethylation and stresses in plants.
Consequently, the main objectives of this work were (1) to examine the function of APE2 in BER during active DNA demethylation and DNA damage repair in response to the DNA alkylating reagent methyl methanesulfonate (MMS) and (2) to determine the mechanism underlying the function of APE2. Central to this study is the development and assessment of a series of single and double mutants for APE2 and ZDP. The double mutants of APE2 and ZDP exhibited extensive DNA hypermethylation and multiple developmental defects. Further analysis of the zdp ape2 double mutants yielded convincing evidence that APE2 is required for efficient BER of DNA lesions produced during active DNA demethylation and by environmental stresses, and its roles and 3′-phosphatase activity overlap with those of ZDP.
RESULTS
Locus-Specific Effects of ape2 and zdp ape2 on DNA Methylation
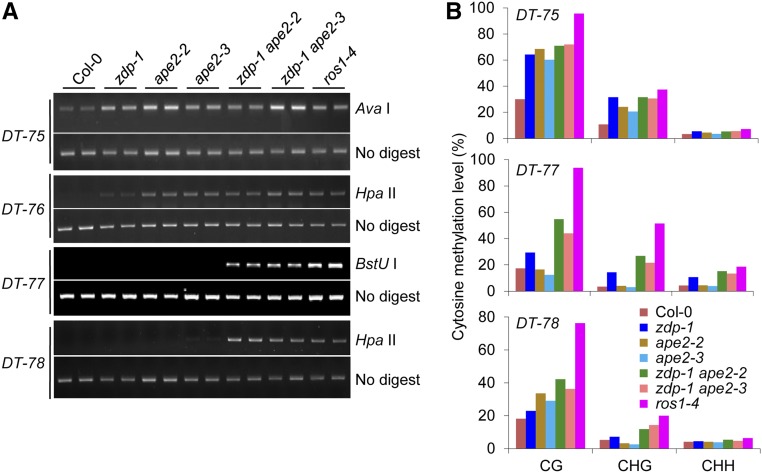
To investigate the possible role of APE2 in active DNA demethylation, two alleles of APE2 with T-DNA insertions in the Col background, ape2-2 and ape2-3, were obtained (Supplemental Figure 1A). To determine the combined effect of ZDP and APE2 mutations on active DNA demethylation, ape2-2 and ape2-3 were crossed with zdp-1 to create the zdp-1 ape2-2 and zdp-1 ape2-3 double mutants. Chop-PCR (PCR after digestion with DNA methylation-sensitive restriction enzymes) was employed to examine DNA methylation levels at representative endogenous demethylation target (DT) loci in the wild-type and mutant plants. More than 1000 DT loci were previously identified in increased DNA methylation1-1 (idm1-1), a mutant detective in ROS1 targeting (Qian et al., 2012). Chop-PCR marker loci DT-75 to DT-78, which reside in the 3′ regions of genes AT1G26380, AT1G26390, AT1G26400, and AT1G26410, respectively, were chosen for our analysis. The results revealed that ape2-2 and ape2-3 showed DNA hypermethylation at DT-75 and DT-76 (Figure 1A). However, at DT-77 and DT-78, DNA hypermethylation could only be detected in zdp-1 ape2-2 and zdp-1 ape2-3, but not in zdp-1, ape2-2, and ape2-3 (Figure 1A). Analysis of DNA methylation levels at these loci using bisulfite sequencing confirmed our chop-PCR results (Figure 1B). These results suggest that APE2 is required for active DNA demethylation. Intriguingly, the functions of APE2 and ZDP at some genomic loci are overlapping.
Figure 1.
Multiple Endogenous Loci Show DNA Hypermethylation upon APE2 Depletion or ZDP/APE2 Double Depletion.
(A) Analysis of DNA methylation levels at loci DT-75, DT-76, DT-77, and DT-78 by chop-PCR. AvaI, HpaII, and BstUI are DNA methylation-sensitive restriction enzymes. DNA hypermethylation results in increased levels of the PCR products. Undigested DNA was amplified as a control. DT-75, DT-76, DT-77, and DT-78 reside in 3′ regions of genes AT1G26380, AT1G26390, AT1G26400, and AT1G26410, respectively. Also see Supplemental Figure 2B for the precise genomic locations of these DT loci.
(B) Analysis of DNA methylation levels at loci DT-75, DT-77, and DT-78 by locus-specific bisulfite sequencing. The overall percent methylation of cytosine sites in different sequence contexts is presented.
In Arabidopsis, APE2 has two isoforms that are produced by alternative splicing (Supplemental Figure 1A). The longer isoform of APE2 harbors an EEP domain at the N terminus and a Zf-GRF domain at the C terminus. The shorter isoform of APE2 has an intact Zf-GRF domain, but lacks one-third of the coding sequence of the EEP domain, which contains amino acids essential for the enzymatic activities of APE2 (Supplemental Figure 1B) (Lee et al., 2014). RT-PCR analysis revealed that both isoforms of APE2 were disrupted in ape2-2, while only the longer isoform was disrupted in ape2-3 (Supplemental Figure 1C).
Genome-Wide Effects of ape2 and zdp ape2 on DNA Methylation
To investigate the global impact of depletion of APE2 and ZDP on DNA methylation, we next performed whole-genome bisulfite sequencing (WGBS) using DNA extracted from the ape2-2, zdp-1, zdp-1 ape2-2, and ros1-4 mutants. Consistent with previous findings (Qian et al., 2012; Li et al., 2015), zdp-1 had ∼1400 hypermethylated loci, while ros1-4 had more than 4000 hypermethylated loci (Supplemental Figure 2A; listed in Supplemental Data Set 1). The ape2-2 mutant had DNA hypermethylation and DNA hypomethylation at 918 and 673 loci, respectively. The number of hypermethylated loci was increased to 2166, but the number of hypomethylated loci was decreased to 405 by the addition of the zdp-1 mutation (Supplemental Figure 2A; listed in Supplemental Data Set 1). All of the chop-PCR marker loci (DT-75, DT-76, DT-77, and DT-78) were detected to be hypermethylated in zdp-1 ape2-2 and ros1-4 by WGBS (Supplemental Figure 2B), validating the accuracy of our WGBS method. In this study, we focused on DNA hypermethylation caused by ape2-2 and zdp-1ape2-2.
To determine whether APE2 depletion affects DNA demethylation in specific genomic regions, we divided genomic regions into genic regions, intergenic regions, transposable elements (TEs) outside of genic regions, and TEs overlapping or within genic regions. Whereas genic regions account for more than 60% of the hyper-DMRs in ape2-2 and zdp-1 ape2-2, <40% of the hyper-DMRs in ros1-4 are distributed in genic regions (Supplemental Figure 2C). These results indicate that APE2 depletion preferentially affects DNA demethylation at genic regions.
Transcript levels of ROS1 and DML2 were not affected by the APE2 and ZDP mutations that we examined, excluding the possibility that these APE2 and ZDP mutations indirectly caused changes in DNA methylation levels (Supplemental Figure 3). Transcript levels of several key genes in the RNA-directed DNA methylation (RdDM) pathway also remained unaltered in all of the mutants produced for this study (Supplemental Figure 3). Unexpectedly, transcript levels of multiple genes responsible for maintaining DNA methylation, including METHYLTRANSFERASE1 (MET1), CHROMOMETHYLASE3 (CMT3), VARIANT IN METHYLATION1 (VIM1), and VIM2 (Law and Jacobsen, 2010), were significantly elevated in the zdp ape2 double mutants (Supplemental Figure 3), indicating enhanced maintenance of DNA methylation in the zdp ape2 double mutants.
APE2 and ZDP Play Overlapping Roles in Active DNA Demethylation
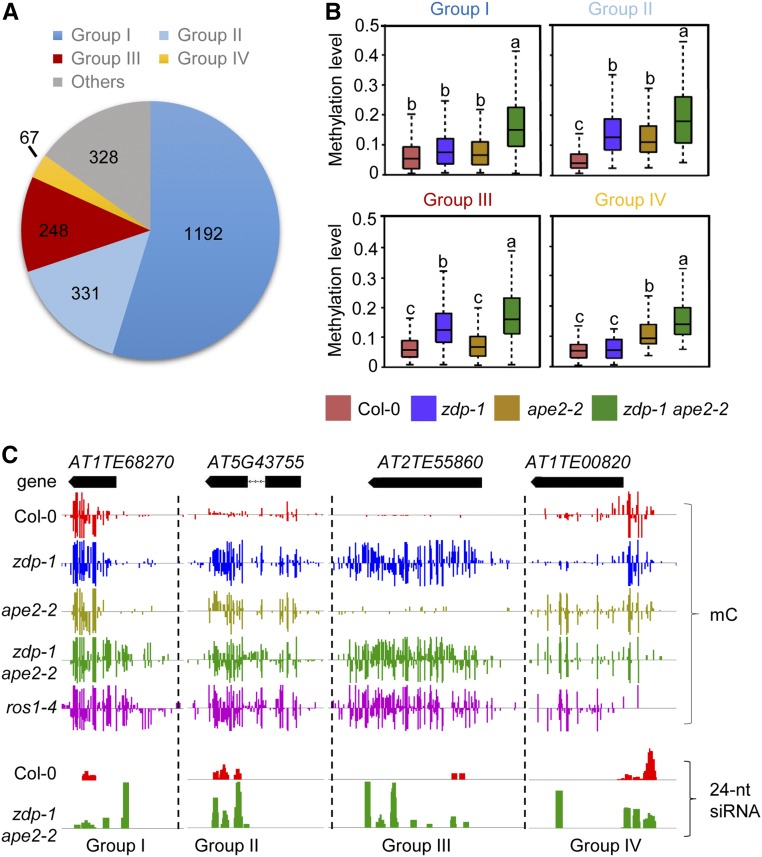
To analyze the extent of functional overlapping between APE2 and ZDP, the hypermethylated loci in zdp-1 ape2-2 were classified into four groups according to their dependence on APE2 and ZDP for DNA demethylation (Figure 2A). Group I loci are not affected by mutation of APE2 or ZDP alone, but these loci are hypermethylated in the zdp ape2 double mutants, indicating that APE2 and ZDP play overlapping functions at these loci. Group II loci are hypermethylated in both single mutants, but their DNA methylation levels are further increased in the double mutant, indicating that the activities of ZDP and APE2 at these loci are additive. Group III and IV include loci that are hypermethylated in one single mutant, but not in the other, indicating that either ZDP or APE2 participates in DNA demethylation at these loci (Figure 2B and 2C). All of the categorized loci are annotated in the hyper-DMR list for the zdp-1ape2-2 mutant in Supplemental Data Set 1. Group II loci, exemplified by AT5G43755 (Figure 2C), account for ∼15% of the hypermethylated loci in zdp-1 ape2-2. Chop-PCR marker loci DT-75, DT-76, and DT-78 were categorized into group II. Approximately 11% of the loci belonged to group III (exemplified by AT2TE55860), whereas ∼3% of the loci belonged to group IV (exemplified by AT1TE00820). Strikingly, group I, which includes loci AT1TE68270 (Figure 2C), DT-77, and DT-79, is the largest group and accounts for more than 50% of the hypermethylated loci in zdp-1 ape2-2, suggesting a high level of functional overlapping between APE2 and ZDP (Figures 2B and 2C). The distribution of TEs, genes, and intergenic regions in each group of hyper-DMRs is similar to that in total hyper-DMRs of zdp-1 ape2-2 except that TEs overlapping or within genic regions are underrepresented in group III of hyper-DMRs (Supplemental Figure 2C).
Figure 2.
APE2 and ZDP Play Overlapping Roles in Active DNA Demethylation.
(A) Pie chart showing proportions of different groups of hyper-DMRs in zdp-1 ape2-2. Group I: Loci not affected by the single APE2 or ZDP mutation, but hypermethylated upon double mutations of APE2 and ZDP. Group II: Loci hypermethylated in both of the two single mutants. DNA methylation levels further increase in the double mutant. Groups III and IV: Loci hypermethylated in one single mutant, but not in the other.
(B) Box plots showing the DNA methylation levels of the four groups of loci as categorized in (A). Dark horizontal line, median; edges of boxes, 25th (bottom) and 75th (top) percentiles; whiskers, minimum and maximum percentage of DNA methylation. Significant differences between two groups are marked with different letters (P < 10−15, Mann-Whitney U test).
(C) Snapshots in the Integrated Genome Browser showing the levels of DNA methylation and 24-nucleotide siRNAs at the indicated representative loci.
APE2 and ZDP Act Downstream of ROS1
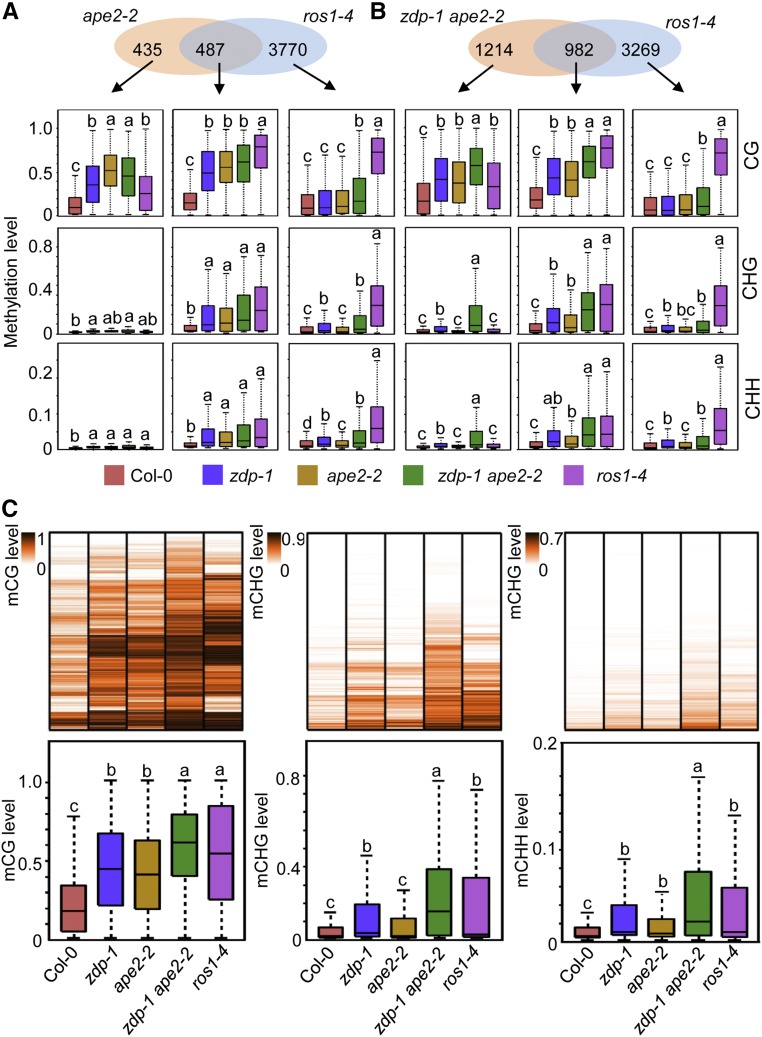
To confirm whether APE2, like ZDP, functions in ROS1-mediated active DNA demethylation, we compared the hyper-DMRs in ape2-2 and zdp-1 ape2-2 to those in ros1-4. Approximately 52.7% and 44.7% of the hyper-DMRs in ape2-2 and zdp-1 ape2-2, respectively, overlapped with the hyper-DMRs in ros1-4 (Supplemental Figure 2A); these findings indicated significant enrichment (P = 5.38 × 10−98 for ape2-2; P = 1.36 × 10−86 for zdp-1 ape2-2) compared with that expected by random chance for a data set of this size. At the overlapping hyper-DMRs, the levels of DNA methylation in all sequence contexts were elevated in all examined mutants (Figures 3A and 3B). At the ape2-2-specific and zdp-1 ape2-2-specific hyper-DMRs, CG methylation levels in all mutants were apparently elevated, although the levels of DNA methylation in the CHG and CHH contexts were largely unaltered or slightly elevated (Figures 3A and 3B). The similar elevation of CG methylation observed in ape2-2, zdp-1 ape2-2, and ros1-4 suggests that the actual overlaps between the sets of hyper-DMRs in ape2-2 and zdp-1 ape2-2 and hyper-DMRs in ros1-4 are likely larger than those reported here. Due to stringent analysis parameters, however, many of the hyper-DMRs in ape2-2 and zdp-1 ape2-2 were not counted to overlap with those in ros1-4. We further examined the methylation levels of different mutants in regions that were found to be hypermethylated in zdp-1 ape2-2 (Figure 3C). The overall CG, CHG, and CHH DNA methylation levels of these regions in ros1-4 were significantly higher than those in the wild-type plants. DNA methylation patterns in ape2-2 and/or zdp-1 ape2-2 resembles those in ros1-4 not only at the whole-genome level, but also at specific loci. As shown in Figure 2C, DNA methylation at AT1TE68270 spread to neighboring regions in zdp-1 ape2-2. Such spreading of DNA methylation is typically observed in the ros1 mutants. Moreover, the levels of DNA methylation at AT5G43755 and AT2TE55860 in zdp-1 ape2-2 were nearly identical to those in ros1-4.
Figure 3.
APE2 Functions Downstream of ROS1 in Active DNA Demethylation.
(A) and (B) Venn diagrams showing the number of hyper-DMRs that overlap between ape2-2 and ros1-4 (A) and between zdp-1 ape2-2 and ros1-4 (B). Box plots show the methylation levels of each class of hyper-DMRs in the indicated genotypes. Significant differences between two genotypes are marked with different letters (P < 10−15, Mann-Whitney U test).
(C) DNA methylation analysis in different mutant plants. Upper panel: Heat map showing CG, CHG, and CHH methylation levels at hyper-DMRs caused by the zdp-1 ape2-2 mutation in the indicated genotypes. Lower panel: Box plots showing CG, CHG, and CHH methylation levels at hyper-DMRs caused by the zdp-1 ape2-2 mutation in the indicated genotypes. Significant differences between two genotypes are marked with different letters (P < 10−15, Mann-Whitney U test).
DNA hypermethylation is usually associated with accumulation of 24-nucleotide small interfering RNAs (siRNAs), which guide methylation of homologous DNA loci during RdDM (Law and Jacobsen, 2010). In the ros1 mutants, 24-nucleotide siRNAs accumulated at ROS1 target loci (Tang et al., 2016). We next compared the levels of 24-nucleotide siRNAs in the wild-type and zdp-1 ape2-2 plants by performing small RNA sequencing. The overall levels of 24-nucleotide siRNAs in the wild-type and zdp-1 ape2-2 plants were comparable (Supplemental Figure 4A). The level of 24-nucleotide siRNAs at zdp-1 ape2-2-caused hypo-DMRs dropped in zdp-1 ape2-2 (Supplemental Figure 4B). However, the level of 24-nucleotide siRNAs at zdp-1 ape2-2-caused hyper-DMRs, as exemplified at the chop-PCR marker loci (Supplemental Figure 2B), was significantly higher in zdp-1 ape2-2 (Supplemental Figure 4B). Specifically, about a quarter of the CG hyper-DMRs and half of CHG and CHH hyper-DMRs in zdp-1 ape2-2 have 24-nucleotide siRNA reads (Supplemental Figure 4C). These results suggest that these hypermethylated loci are RdDM targets. Together, our results suggest that APE2, either alone or working together with ZDP, functions in ROS1-mediated active DNA demethylation.
The zdp ape2 Double Mutants Display Pleiotropic Developmental Defects
Despite alterations in DNA methylation, the zdp-1, ape2-2, and ape2-3 single mutants manifest no obvious developmental phenotype under normal growth conditions. However, the zdp-1 ape2-2 and zdp-1 ape2-3 double mutants display pleiotropic phenotypes, including retarded shoot and root growth, and narrow and slightly serrated leaves at the adult stage (Figure 4; Supplemental Figure 5). Approximately 5% of the double mutants were found to have flattened stems (Supplemental Figures 5B and 5C), suggesting the occurrence of stochastic incidents during development. The pleiotropic phenotypes observed in the double mutants were fully rescued by transgenic expression of ZDP or APE2 (Figure 4), suggesting that dysfunction of ZDP and APE2 is responsible for the defects. Because only simultaneous depletion of ZDP and APE2, but not depletion of either protein, results in developmental defects, we conclude that ZDP and APE2 play overlapping roles in plant development.
Figure 4.
The zdp ape2 Double Mutants Show Pleiotropic Developmental Defects.
(A) Root growth retardation of the zdp ape2 double mutants. ZDP com and APE2 com represent proZDP:ZDP-3Flag zdp-1 ape2-2 and proAPE2:APE2-3Myc zdp-1 ape2-2, respectively.
(B) Phenotypes of 30-d-old seedlings of each genotype grown on soil.
DNA Lesions Accumulate in the zdp ape2 Double Mutants
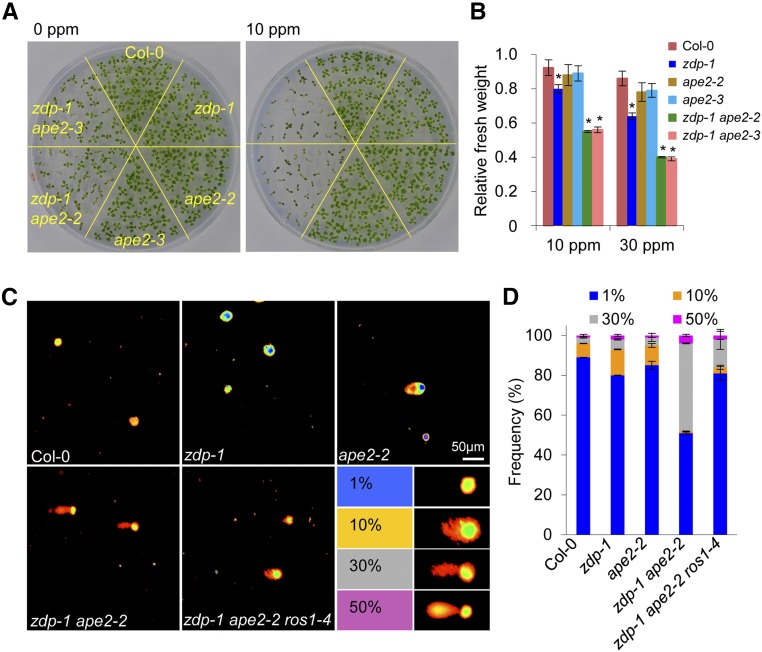
The developmental defects of the zdp ape2 double mutants were reminiscent of defects observed in many other DNA repair deficient mutants, indicating impairment of DNA repair in the zdp ape2 double mutants. To test this, we examined the sensitivity of the single and double mutants to MMS. The sensitivity of the ape2 single mutants to MMS was not different from that of the wild-type plants, and the zdp-1 single mutants showed slightly reduced survival after MMS treatment (Figures 5A and 5B), in agreement with previous findings (Martínez-Macías et al., 2012). In contrast, the zdp ape2 double mutants were highly sensitive to MMS treatment, even at a concentration as low as 10 ppm (Figure 5A). Measurement of plant growth by measuring the fresh weight of plants also revealed that the zdp ape2 double mutants were much more sensitive to MMS in comparison with the Col-0 and single mutant plants (Figure 5B).
Figure 5.
The zdp ape2 Double Mutants Are Sensitive to DNA Damage.
(A) MMS-sensitive phenotype of seedlings of the indicated genotypes. Photographs of 14-d-old seedlings grown on MS medium supplemented with 0 or 10 ppm of MMS were taken.
(B) Relative fresh weights of seedlings of the indicated genotypes. The ratios of fresh weight grown on MS medium with 10 or 30 ppm MMS to that grown on MS medium without MMS were calculated. The error bars represent the mean ± sd of three biological replicates from independent pools of seedlings; asterisks indicate significant differences between Col-0 and the indicated mutant (P < 0.05, two-tailed Student's t test).
(C) Representative comet images of nuclei from 22-d-old leaves of Col-0, zdp-1, ape2-2, zdp-1 ape2-2, and zdp-1 ape2-2 ros1-4. DNA damage was expressed as the ratio of the signal intensity of a comet tail versus that of the entire nucleus.
(D) Stacked bar histogram showing the distribution of different grades of DNA damage in Col-0, zdp-1 ape2-2, and zdp-1 ape2-2 ros1-4 nuclei. The error bars represent the mean ± sd based on at least 200 comets from three independent gels.
To directly measure DNA strand breaks in the zdp ape2 double mutants, we performed comet assays using the alkaline unwinding/neutral electrophoresis (A/N) protocol, which measures DNA SSBs as well as double-strand breaks (DSBs) (Figures 5C and 5D). Comet signals were divided into four grades based on the ratio of signal intensity from a comet tail versus that from the entire nucleus (Luo et al., 2012). In Col-0, zdp-1, and ape2-2, cells with <1% and 10% DNA damage signal accounted for more than 90% of all cells. In zdp-1 ape2-2, the numbers of cells with <1% and 10% DNA damage were greatly reduced, but the numbers of cells with 30% and more than 50% DNA damage signal were greatly increased (Figures 5C and 5D).
The DNA Damage Response Is Constitutively Activated in the zdp ape2 Mutants
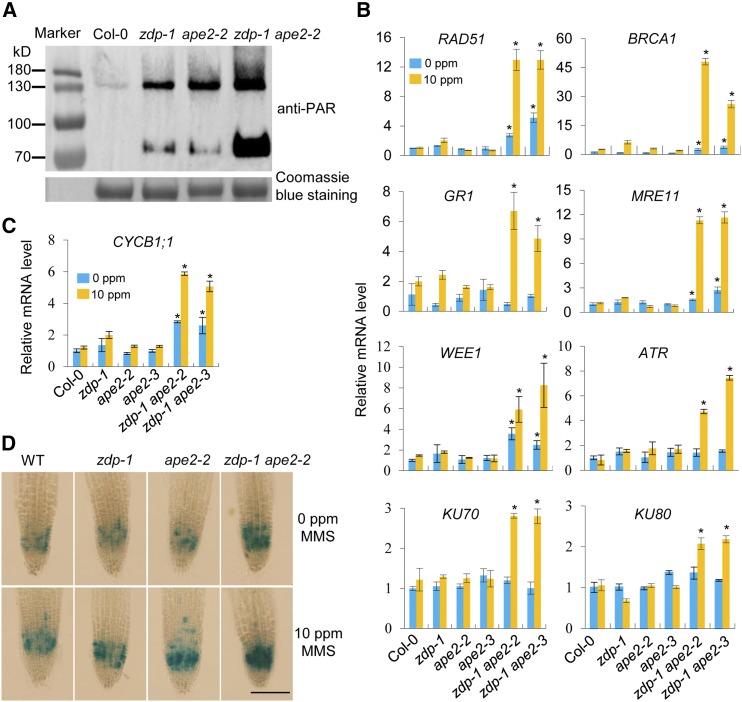
Protein poly(ADP-ribosyl)ation, catalyzed by poly(ADP-ribose) polymerases, is an important posttranslational modification involved in early responses to base excision repair, SSB repair, and DSB repair (Liu et al., 2017). To test for activation of the DNA damage response (DDR), we detected the levels of poly(ADP-ribosyl)ation (PAR) in Col-0, zdp-1, ape2-2, and zdp-1 ape2-2 using an anti-PAR antibody (Figure 6A). The levels of PARylation are mildly increased in zdp-1 and ape2-2 and greatly increased in zdp-1 ape2-2, compared with Col-0. To further determine whether accumulation of DNA lesions in the zdp ape2 mutants induces expression of genes involved in DDR, we detected the expression levels of selected genes by performing RT-qPCR. Our results revealed that, in comparison with those of the wild-type plants, the expression levels of RAD51, BRCA1, MRE11, and WEE1 in the zdp/ape2 mutants, but not in the ape2 and zdp-1 single mutants, were significantly higher under normal growth conditions (Figure 6B). Moreover, the expression levels of these genes in the zdp/ape2 mutants were increased dramatically upon MMS treatment (Figure 6B). GR1, ATR, KU70, and KU80 were not activated in any of the mutants under normal growth conditions. However, expression of these genes was induced greatly by MMS treatment in the zdp ape2 mutants (Figure 6B). These results suggest that some DDR genes are constitutively activated in the zdp ape2 mutants.
Figure 6.
The zdp ape2 Double Mutants Show Constitutive Activation of DDR.
(A) Detection of the levels of poly(ADP-ribosyl)ation in the indicated genotypes using an anti-PAR antibody. Equivalent loading of each lane was verified using Coomassie blue stain.
(B) Relative expression levels of genes involved in DDR in the indicated genotypes under normal and MMS-treated conditions as determined by RT-qPCR. The error bars represent the mean ± sd of three biological replicates from independent pools of seedlings. Asterisks indicate significant differences between Col-0 and the indicated mutants (P < 0.05, two-tail Student’s t test).
(C) Relative expression levels of a cell cycle marker gene CYCB1;1 in the indicated genotypes under normal and MMS-treated conditions as determined by RT-qPCR. The error bars represent the mean ± sd of three biological replicates from independent pools of seedlings. Asterisks indicate significant differences between Col-0 and the indicated mutants (P < 0.05, two-tail Student’s t test).
(D) GUS staining of root tips from 7-d-old seedlings of the pCYCB1;1:Dbox-GUS reporter line in wild-type, zdp-1, ape2-2, and zdp-1 ape2-2 backgrounds. Bar = 100 μm.
To investigate whether activation of the DDR in the zdp ape2 mutants arrests the cell cycle, we detected the expression level of the cyclin CYCB1;1 gene, which is specifically activated at the G2-to-M phase transition (Doerner et al., 1996; Shaul et al., 1996; Criqui et al., 2001), as well as other cell cycle related genes, including CDKB1;1, CYCA1;1, CYCA2;4, and CYCB1;2 (Vandepoele et al., 2002). In comparison with the other groups, expression of CYCB1;1 was significantly higher in the zdp ape2 double mutants; moreover, its expression level in the zdp ape2 double mutants was further increased by exposure to MMS (Figure 6C). CDKB1;1, CYCA1;1, CYCA2;4, and CYCB1;2 were only activated in the double mutants after MMS treatment (Supplemental Figure 6). We also introduced a CYCB1;1-GUS expression system with the gene encoding GUS driven by the CYCB1;1 promoter (Colón-Carmona et al., 1999) into the wild-type, zdp-1, ape2-2, and zdp-1 ape2-2 plants. In comparison with the other groups, the GUS signal was remarkably more intense in the zdp-1 ape2-2 plants; this signal was also strengthened by MMS treatment (Figure 6D). Collectively, these results suggest that the zdp ape2 double mutants showed DNA damage-triggered cell cycle arrest.
Constitutive DNA Damage in the zdp ape2 Double Mutants Is Partially a Result of Defective DNA Demethylation
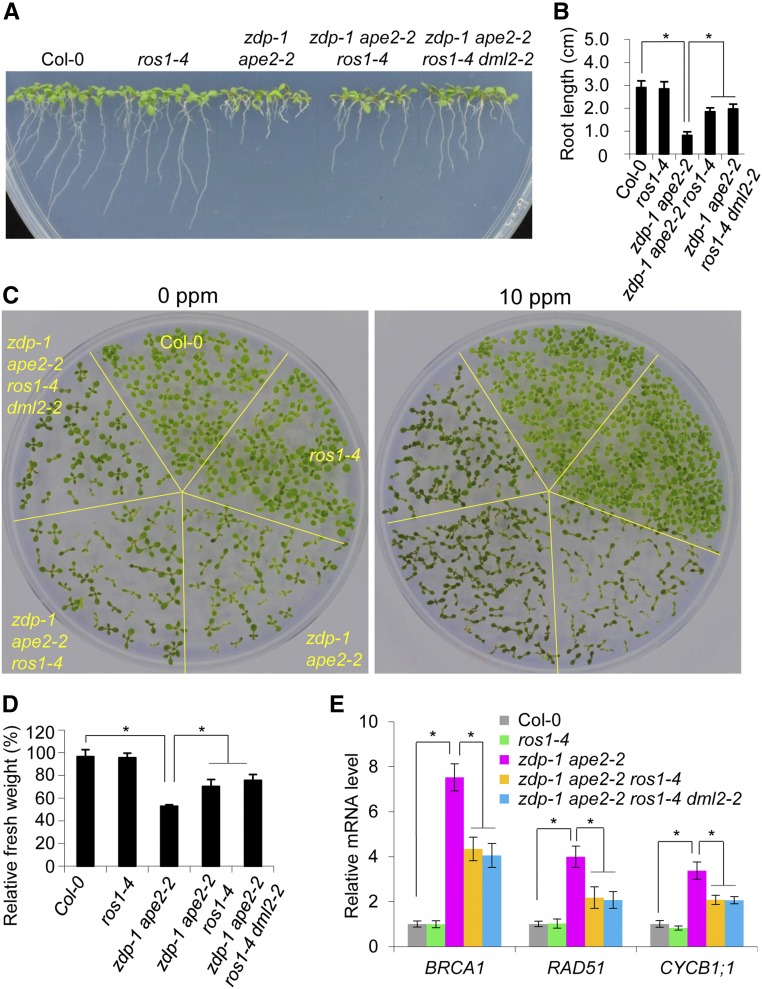
Because ROS1 may excise 5mC even in the absence of downstream enzymes, the products of 5mC excision by ROS1, when kept unprocessed and unrepaired, could form an important source of DNA damage (Drohat and Coey, 2016). To test this possibility, we introduced the ros1-4 mutation into the zdp-1 ape2-2 mutant. Dysfunction of ROS1 partially rescued the root growth retardation and MMS-sensitive phenotypes of zdp-1 ape2-2 (Figures 7A to 7D) and partially recovered the number of cells with minimal DNA damage in comet assays (Figure 5D). Furthermore, our RT-qPCR results revealed that activation of homologous recombination-related genes BRCA1 and RAD51 and cyclin gene CYCB1;1 was significantly compromised by the introduction of the ros1-4 mutation (Figure 7E). Our results strongly suggest that DNA damage in the zdp ape2 double mutants arises partially from defective processing of ROS1 products. The addition of the dml2-2 mutation to the zdp-1 ape2-2 ros1-4 mutant plants did not strengthen the rescue effect (Figure 7), suggesting that ROS1 plays a dominant role in producing DNA lesions in the zdp ape2 double mutants.
Figure 7.
ROS1 Mutation Partially Rescues Constitutive Activation of the DNA Damage Response in the zdp-1 ape2-2 Plants.
(A) Root growth retardation in the indicated genotypes.
(B) Quantification of the root length of seedlings of each genotype. The root lengths of ∼30 seedlings (8 d old) of each genotype were measured. The error bars represent the mean ± se of three biological replicates from independent pools of seedlings. Asterisks indicate significant differences between the indicated genotypes (P < 0.05, two-tail Student’s t test).
(C) MMS-sensitive phenotype of seedlings of the indicated genotypes. Photographs were taken of seedlings (14 d old) grown on MS medium supplemented with 0 or 10 ppm of MMS.
(D) Relative fresh weights of seedlings of the indicated genotypes. The ratios of fresh weight grown on MS medium with 10 ppm MMS to that grown on MS medium without MMS were calculated. The error bars represent the mean ± sd of three biological replicates from independent pools of seedlings. Asterisks indicate significant differences between the indicated genotypes (P < 0.05, two-tailed Student’s t test).
(E) Relative expression levels of BRCA1, RAD51, and CYCB1;1 in the indicated genotypes as determined by RT-qPCR. The error bars represent the mean ± sd of three biological replicates from independent pools of seedlings. Asterisks indicate significant differences between the indicated genotypes (P < 0.05, two-tailed Student’s t test).
APE2 Has 3′-Phosphatase Activity and Strong 3′-5′ Exonuclease Activity
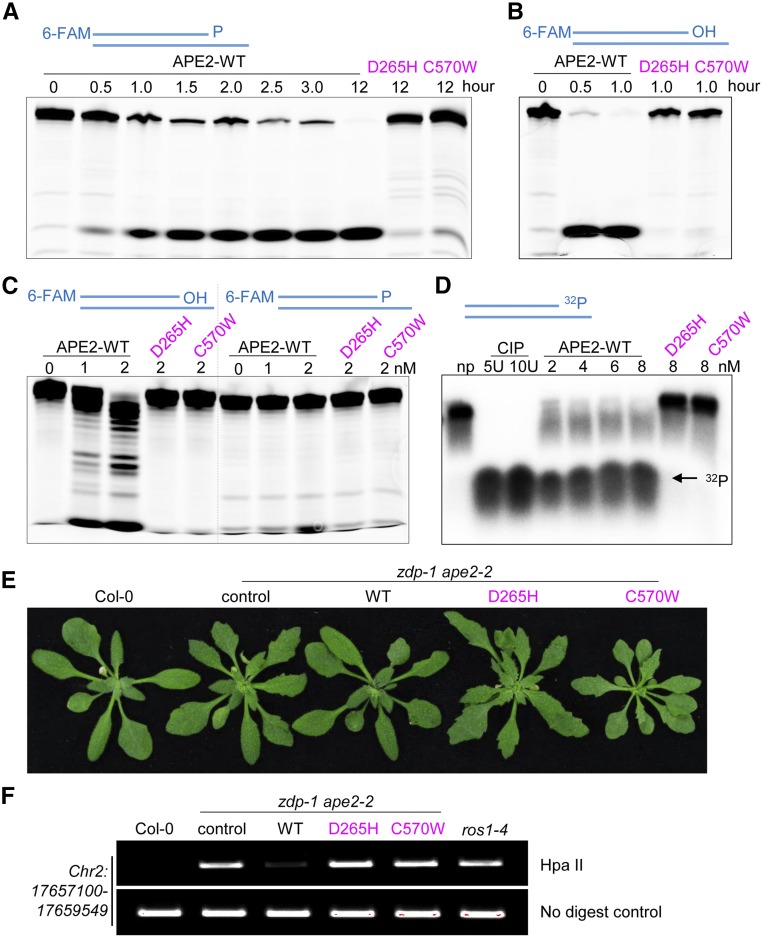
To investigate the molecular basis of the functional overlapping between APE2 and ZDP, we sought to examine whether APE2, like ZDP, has 3′-phosphatase activity. 6-FAM-labeled DNA substrate with a 3′-P terminus was incubated with purified APE2. APE2 was able to process the DNA substrate, as revealed by reduction of the DNA substrate (Figure 8A). However, a product with a 3′-OH terminus was not observed. To exclude the possibility that a low resolution of DNA molecules hindered detection of 3′-OH-terminated intermediate product, DNA substrate with a 3′-P terminus was incubated with calf-intestinal alkaline phosphatase (CIP). The formation of 3′-OH-terminated product was readily detected, indicating adequate gel resolution (Supplemental Figure 7D). When using APE2, however, such product could not be detected, even though we observed loss of 3′-P-terminated DNA substrate (Supplemental Figure 7D). Because human APE2 has strong 3′-5′ exonuclease activity (Burkovics et al., 2006), we speculated that the strand with the 3′-OH terminus was quickly degraded by the exonuclease activity of APE2. Consistent with this idea, under the same reaction conditions, DNA substrate with a 3′-OH terminus was degraded by APE2 within 30 min (Figure 8B). Our data favor a scenario where APE2 processes 3′-P into 3′-OH, after which its exonuclease activity resects DNA. A DNA ladder representing the intermediate products was detected when DNA substrate with a 3′-OH terminus was incubated with a lower concentration of APE2. However, at such a low concentration, APE2 was unable to process 3′-P-terminated DNA substrate (Figure 8C). These results suggest that APE2 has stronger 3′-5′ exonuclease activity, which hinders direct detection of its 3′-phosphatase activity using this method.
Figure 8.
APE2 Has Weak DNA 3′ Phosphatase Activity and Strong DNA 3′-5′ Exonuclease Activity in Vitro.
(A) APE2 activity against DNA substrate with a 3′-P terminus. The wild-type or mutant form of APE2 proteins (10 nM) was incubated with 3′-P terminated DNA substrate (10 nM) for the indicated times.
(B) APE2 activity against DNA substrate with a 3′-OH terminus. Reactions were performed as described in (A) with the exception that the 3′-P in the DNA substrate was replaced by a 3′-OH.
(C) Stronger exonuclease activities of APE2. APE2 proteins at low concentrations (1 or 2 nM) were incubated with 3′-P terminated or 3′-OH terminated DNA substrate (10 nM) for 30 min.
(D) Release of free phosphate residue from 3′-32P-labeled DNA substrate by APE2. APE2 proteins at different concentrations were incubated with 3′-32P-labeled DNA substrate (0.05 pM) for 3 h. CIP (5 and 10 units) was used as positive controls; np, no protein control.
(E) Phenotypes of 30-d-old zdp-1ape2-2 seedlings complemented by wild-type or mutant forms of APE2.
(F) Analysis of the DNA methylation level at the Chr2:17657100-17659549 locus by chop-PCR. HapII is a methylation-sensitive restriction enzyme. DNA hypermethylation results in an increased level of the PCR product. Undigested DNA was amplified as a control.
To provide direct evidence for 3′-phosphatase activity of APE2, we then turned to use DNA substrate labeled with radioactive 32P at its 3′ end and measured the release of free phosphate residues by APE2. Incubation of 3′-32P-labeled DNA substrate with APE2 for 3 h, like incubation with CIP, resulted in nearly complete release of free phosphate residues (Figure 8D), demonstrating that APE2 has 3′-phosphatase activity.
To determine whether conserved residues in the EEP domain and Zf-GRF domain are required for the enzymatic activities of APE2, we introduced two amino acid changes (D265H and C570W) using site-directed mutagenesis (Supplemental Figures 7A to 7C). Both of the amino acid changes abolished the enzymatic activities of APE2 (Figures 8A to 8D). Furthermore, wild-type APE2 complemented the DNA hypermethylation and developmental phenotypes of the zdp-1 ape2-2 plants, but both of the mutant forms of APE2 failed to do so (Figures 8E and 8F), although the mutant forms of APE2 were expressed at the same level as wild-type APE2 (Supplemental Figure 7E). These results suggest that the EEP domain and Zf-GRF domain are essential for the enzymatic activities of APE2 and for the involvement of APE2 in active DNA demethylation and plant development.
DISCUSSION
Active DNA demethylation in animals and plants entails the BER pathway. Previous studies have demonstrated that active DNA demethylation in plants comprises two functional branches downstream of DNA glycosylases ROS1, DML2, DML3, and DME. One branch, catalyzed by DNA phosphatase ZDP, converts ROS1/DME β/δ-elimination product 3′-P to 3′-OH (Martínez-Macías et al., 2012), whereas another branch, catalyzed by AP endonuclease APE1L, converts ROS1/DME β-elimination product 3′-PUA to 3′-OH (Lee et al., 2014; Li et al., 2015). Our study shows that the AP endonuclease APE2 also functions in active DNA demethylation. The roles of APE2 and ZDP in active DNA demethylation are highly overlapping. In addition to DNA hypermethylation, the zdp ape2 double mutants show constitutive activation of the DDR. The roles of APE2 and ZDP in DNA damage repair are overlapping as well. APE2 has 3′-phosphatase activity, which explains the functional overlapping between ZDP and APE2 in active DNA demethylation and DNA repair. Previously only the AP endonuclease activity of APE2, which is required for repair of AP sites, has been found. Our findings expand our knowledge of APE2’s activities and highlight APE2 as a component of active DNA demethylation in plants.
APE2 and ZDP Play Overlapping Roles in Active DNA Demethylation
APE2 depletion alone led to DNA hypermethylation at 918 loci. The zdp ape2 double mutants showed DNA hypermethylation at 2166 loci. Surprisingly, more than half of the hypermethylated loci in the zdp-1 ape2-2 plants had normal DNA methylation levels in either the zdp-1 or ape2-2 plants, but became hypermethylated when both APE2 and ZDP were depleted (Figure 2). These findings suggest that APE2 and ZDP play overlapping roles in the maintenance of nuclear epigenome stability.
APE1L was previously found to have 3′ phosphatase activity (Li et al., 2015); therefore, processing of 3′-P to 3′-OH at some ROS1/DME target loci may also require APE1L. However, because APE1L and APE2 have different spatiotemporal expression patterns (Lee et al., 2014), APE1L and APE2 likely control the processing of 3′-P to 3′-OH at different loci and at different times, even though they both have overlapping roles with ZDP. Differential expression and use of AP endonucleases in different cells have been previously reported. In mouse germinal center B cells, APE1 is downregulated, but APE2 is upregulated to promote error-prone repair during somatic hypermutation of antibody variable region genes (Stavnezer et al., 2014).
The addition of the ros1-4 mutation partially rescued the constitutive DNA damage phenotype of the zdp ape2 plants, suggesting that, in the absence of ZDP and APE2, ROS1 still executes its function and excises 5mC. This raises the question of why, if ROS1 removes 5mC, the zdp ape2 mutants still display a DNA hypermethylation phenotype. It has been reported that human APE1 has a higher affinity to AP sites compared with 8-oxoguanine DNA glycosylase 1 (OGG1). The association of APE1 with the AP site displaces OGG1 from its substrate and thus accelerates turnover of OGG1 (Hill et al., 2001; Sidorenko et al., 2007). We hypothesize that APE2 in Arabidopsis, like human APE1, may play a role in displacing ROS1 and increasing the turnover and catalytic efficiency of ROS1. In the absence of APE2, release of ROS1 from its substrate is affected, and turnover rate of ROS1 is decreased, which could lead to DNA hypermethylation. Moreover, the activity of DME, and possibly also ROS1, on one DNA strand was reported to be inhibited by abasic sites on the other DNA strand (Gehring et al., 2006). In the absence of APE2 and/or other enzymes, some of the abasic sites remain unrepaired. Thus, the activity of ROS1 against 5mC on the opposite DNA strand is inhibited. This could also explain why DNA hypermethylation was detected in the ape2 and zdp ape2 mutants.
Notably, 44.7% hyper-DMRs in zdp-1 ape2-2 were found to be ROS1 targets and the zdp-1 ape2-2 double mutant appears to have more hyper-DMRs located within genes than ros1-4 (Supplemental Figure 2). Because ZDP and APE2 could also function downstream of other DNA demethylases such as DME, DML2, and DML3, which have different expression patterns and target preference. Thus, it is possible that DMRs identified in zdp-1 ape2-2 do not completely overlap those in ros1-4 and DMR distribution of zdp-1 ape2-2 is not identical to that of ros1-4. On the other hand, elevated expression of MET1, CMT3, VIM1, and VIM2 was detected in zdp-1 ape2-2 and zdp-1 ape2-3. For hyper-DMRs that are uniquely identified in the double mutants but not ROS1 targets, DNA hypermethylation could be caused by elevated expression of MET1, CMT3, VIM1, and VIM2.
APE2 and ZDP Play Overlapping Roles in DNA Repair
The zdp-1 mutant is slightly sensitive to MMS. However, the zdp ape2 double mutants were found to be hypersensitive to MMS. The zdp ape2 double mutants grown under normal conditions showed accumulation of DNA strand breaks, activated expression of homologous recombination-related genes, and arrested cell cycle at the G2/M transition. Exposure of the zdp ape2 double mutants to a low concentration of MMS heightened the DDR (Figures 5 and 6). These results indicate that ZDP and APE2, two BER proteins, play overlapping roles in the maintenance of nuclear genome stability. Similar constitutive activation of the DDR was found in plants with mutations in proteins involved in DNA repair, including ASYMMETRIC LEAVES1/2 ENHANCER7 (Luo et al., 2012), the largest subunit of replication factor C (RFC1) (Liu et al., 2010b), POLα/ICU2, a catalytic subunit of DNA polymerase α (Liu et al., 2010a), POLD2, the second small subunit of DNA polymerase δ (Zhang et al., 2016b), ABO4/POL2a/TIL1, a catalytic subunit of DNA polymerase ε (Yin et al., 2009), and FLAP ENDONUCLEASE1 (Zhang et al., 2016a). The strong similarity in the effects of mutating these DNA repair proteins and those produced by mutating ZDP and APE2 suggest that these proteins may function in the same pathway. Future studies should determine which DNA polymerase acts downstream of ZDP and APE2 to facilitate active DNA demethylation and DNA repair.
Enzymatic Activities of APE2
Despite the presence of the shorter isoform of APE2, DNA hypermethylation was detected in ape2-3 and ape2-2 and ape2-3 cause identical DNA demethylation defects at the DT loci we examined (Figure 1; Supplemental Figure 1C). Furthermore, zdp-1 ape2-2 and zdp-1 ape2-3 have identical developmental phenotypes and molecular features. These results suggest that the longer isoform of APE2 may play a critical role in active DNA demethylation and plant development. We used this isoform for our experimental detection of enzymatic activities. We found that the longer isoform of APE2 has 3′-phosphatase activity (Figures 8A to 8D). Even with the longer isoform of APE2, previous efforts to detect its enzymatic activities were unsuccessful (Li et al., 2015), probably because APE2 has strict substrate specificity and reaction condition or the in vitro APE2 activity on some types of DNA substrates are too weak to be detected. The DNA clamp PCNA stimulates the 3′-phosphodiesterase and 3′-5′ exonuclease activities of human APE2 (Tsuchimoto et al., 2001; Burkovics et al., 2009). It is possible that APE2 activity in Arabidopsis can also be enhanced by PCNAs or other proteins. Identification of the binding partners of APE2 and the regulatory mechanisms governing its enzymatic activities could facilitate detection of its enzymatic activities on a variety of DNA substrates, including ROS1/DME products. The enzymatic activities of APE2 were abolished by mutation in the EEP catalytic core or mutation in the Zf-GRF domain responsible for DNA binding (Figure 8), suggesting that interaction with DNA and the catalytic domain are both essential for the enzymatic activities of APE2. A recent study revealed that mutation in the Zf-GRF domain also influences the binding of X. laevis APE2 to DNA and its 3′-5′ exonuclease activity (Wallace et al., 2017), indicating that the importance of the Zf-GRF domain to APE2 could be highly conserved among different species.
One may ask why both ZDP and APE2 participate in active DNA demethylation and DNA damage repair at some loci, given that ZDP alone has robust 3′ phosphatase activity. In addition to its 3′ phosphatase activity, our results reveal that APE2 has strong 3′-5′ exonuclease activity. Such activity could be useful for efficient removal of a stretch of DNA with multiple 5mC or damaged bases and the ensuing long-patch BER. Therefore, we propose that APE2 is particularly important for active DNA demethylation and DNA damage repair through long-patch DNA synthesis. Mutations in both ZDP and APE2 might block both single-nucleotide insertion and long-patch DNA synthesis at a particular locus (Córdoba-Cañero et al., 2009), thus leading to DNA hypermethylation and deficiencies in DNA damage repair. However, unlike simultaneous mutation of APE1L and ZDP, simultaneous mutation of APE2 and ZDP does not affect plant fertility, suggesting that APE2 and ZDP are dispensable for gametogenesis and seed development.
Possible Overlapping Roles of APE2 and PNKP in Active DNA Demethylation and DNA Repair in Animals
In contrast to direct removal of 5mC by DNA glycosylases in plants, active DNA demethylation in mammalian cells begins with deamination of 5mC to thymine (Cortellino et al., 2011) or oxidation of 5mC to 5-hydroxymethylcytosine (5 hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) (Tahiliani et al., 2009; He et al., 2011; Ito et al., 2011). Thymine, 5fC, or 5caC can be removed by monofunctional DNA glycosylase TDG to produce an AP site (Cortellino et al., 2011; He et al., 2011; Maiti and Drohat, 2011; Weber et al., 2016). A recent study revealed that DNA glycosylases NEIL1 and NEIL2 cooperate with TDG during 5fC and 5caC removal (Schomacher et al., 2016); they displace TDG and accelerate TDG turnover. As NEIL1 and NEIL2 have lyase activity, they cleave the DNA backbone (Schomacher et al., 2016). When a gap with a 3′-P terminus is generated, an enzyme with 3′ phosphatase activity is required for 3′-P processing. However, it remains unknown whether the individual mutation of APE2 or PNKP (the human homolog of the ZDP gene) (Jilani et al., 1999) or the simultaneous mutations of APE2 and PNKP cause DNA demethylation abnormalities in human cells. Human APE2 and X. laevis APE2 are implicated in BER (Burkovics et al., 2006) and SSB repair (Willis et al., 2013; Wallace et al., 2017). PNKP participates in BER (Wiederhold et al., 2004), SSB repair (Whitehouse et al., 2001), and DSB repair (Chappell et al., 2002). It also remains unknown whether APE2 and PNKP collaborate in the repair of DNA damage. Further investigations are warranted to examine the possibility that APE2 and PNKP also play overlapping roles in animals.
METHODS
Plant Materials, Generation of Transgenic Plants, and Growth Conditions
All Arabidopsis thaliana lines used in this study were of the Col-0 ecotype. The T-DNA insertion mutant lines Sail_201_C04 (ape2-2) and Salk_120629re-seq (ape2-3) were obtained from the ABRC. The zdp-1 (SAIL_60_C08) and ros1-4 (Salk_045303) mutants were previously described (Martínez-Macías et al., 2012). For complementation of the mutants, ZDP and APE2 genomic DNA fragments with promoter regions of ∼2 kb were amplified from wild-type Col-0 genomic DNA by PCR and cloned into the pCAMBIA1305 vector. The primers used are listed in Supplemental Data Set 2. Mutations were introduced into the proAPE2:APE2-3Myc construct through site-directed mutagenesis with the QuikChange II XL site-directed mutagenesis kit (Agilent Technologies). Agrobacterium tumefaciens strain GV3101 carrying a ZDP or APE2 construct was used to transform the mutant plants via the standard floral dipping method (Clough and Bent, 1998). Primary transformants were selected on 0.5× Murashige and Skoog (MS) plates containing 25 mg/L hygromycin. Homozygous transgenic lines were used for phenotypic analysis. The seedlings were grown on 0.5× MS solid medium and later on soil in a 16-h-light (Philips; TLD, 36 W/865)/8-h-dark cycle at 23°C. For DNA damage sensitivity assays, seeds from different genotypes were sown on 0.5× MS medium containing different concentrations of MMS.
Chop-PCR
Genomic DNA was extracted from 14-d-old seedlings using the DNeasy plant mini kit (Qiagen), digested with a methylation-sensitive restriction enzyme (AvaI, HpaII, or BstUI), and subjected to PCR with ExTaq (Takara). Hypermethylated DNA is resistant to digestion by AvaI, HpaII, and BstUI, and could be amplified to yield a band. Undigested DNA was amplified as a control. The primers used for chop-PCR are listed in Supplemental Data Set 2.
Locus-Specific Bisulfite Sequencing
Genomic DNA was extracted from 14-d-old seedlings using the DNeasy plant mini kit and treated with sodium bisulfite using the BisulFlash DNA modification kit (Epigentek) according to the manufacturer’s instructions. Bisulfite-treated DNA was used for PCR using ExTaq (Takara). The PCR products were subcloned into the T-easy vector (Promega). At least 20 independent clones of each sample were sequenced. DNA methylation levels were measured using Kismeth software (http://katahdin.mssm.edu/kismeth). The primers used for locus-specific bisulfite sequencing are listed in Supplemental Data Set 2.
WGBS and Data Analysis
Genomic DNA (∼5 μg) was extracted from 14-d-old seedlings using the DNeasy plant mini kit and sent to BGI for bisulfite treatment, library preparation, and sequencing. The quality-ensured libraries were sequenced on an Illumina HiSeq 4000 instrument to produce 100-bp paired-end reads. Low-quality paired reads, including reads with more than 10% N bases, reads with more than 50% bases with sequencing quality value <5, and reads with residual length <37 bases after trimming of the adaptor sequences, were removed. Clean reads were mapped to the TAIR 10 genome using BSMAP (allowing two mismatches). DMRs were identified according to the methods of Zhang et al. (2013) with some modifications. In brief, only cytosines with a depth of at least four in all libraries were retained for further analysis. Comparison of DNA methylation levels for 50-bp sliding windows of 200 bp between the wild-type and mutant plants was performed using Fisher’s exact test with a P value cutoff of 0.05. To control for false discovery rates, the raw P values were adjusted by the Benjamini-Hochberg procedure. Windows with ≥7 differentially methylated cytosines (defined as C with P < 0.01 in Fisher’s exact test) and ≥2-fold change in DNA methylation levels were defined as DMRs. DMRs in close vicinity (<100 bp) were merged together.
Real-Time RT-PCR/RT-PCR
Total RNA was extracted from 14-d-old seedlings using the RNeasy plant mini kit (Qiagen), and contaminating DNA was removed with the RNase-free DNase set (Qiagen). About 1 µg of total RNA was used for first-strand cDNA synthesis with the SuperScript III first-strand synthesis system (Invitrogen). The cDNA reaction mixture was diluted five times. For real-time PCR, 1 μL of diluted cDNA was used as the template in a PCR reaction with iQ SYBR Green Supermix (Bio-Rad). The expression levels of selected genes were normalized to that of TUBULIN4. For regular PCR, 1 μL of diluted cDNA was used as the template in a PCR reaction with ExTaq (Takara). The primers used for PCR are listed in Supplemental Data Set 2.
Small RNA Sequencing and Data Analysis
Total RNA was isolated from 14-d-old seedlings using TRIzol reagent. The libraries for small RNA sequencing were constructed according to the manufacturer’s instructions (NEBNext small RNA library prep set for Illumina). The libraries were cleaned using Beckman AMpure XP Beads and sequenced on an Illumina SE50 instrument. Raw paired-end reads were trimmed and quality controlled by SeqPrep (https://github.com/jstjohn/SeqPrep) and Sickle (https://github.com/najoshi/sickle) with default parameters. Clean reads of 18 to 32 nucleotides were mapped to the TAIR 10 genome by TopHat (http://tophat.cbcb.umd.edu/), after which perfectly matched reads were counted. Data analysis was performed according to the methods of Zhang et al. (2013). Briefly, 24-nucleotide small RNA reads were normalized to reads per 10 million based on the total number of clean reads mapped to the genome. To visualize their global pattern, normalized 24-nucleotide small RNA reads in nonoverlapping 500-nucleotide windows were plotted across five chromosomes.
Histochemical Analysis of Cell Cycle Arrest
The CYCB1;1:GUS reporter line (Colón-Carmona et al., 1999) was introduced into the zdp-1, ape2-2, and zdp-1 ape2-2 mutant backgrounds by crossing, and homozygous plants were used for GUS activity analysis. GUS staining was performed as described previously (Zhang and Li, 2013).
Comet Assay
The comet assay was performed according to a previously described method (Wang and Liu, 2006) using a CometAssay kit from Trevigen. Briefly, 22-d-old leaves were chopped by a razor in a buffer containing 1× PBS and 20 mM EDTA at 4°C. The samples were filtered through a 60-mm nylon mesh. Ten microliters of the filterable debris was mixed with 100 mL of low-melting-point agarose (1% concentration and prewarmed at 37°C) and added onto Trevigen-precoated slides. After solidification, the slides were incubated in lysis solution (2.5 M NaCl, 100 mM EDTA, pH 10.0, 10 mM Tris, 1% sodium lauryl sarcosinate, and 1% Triton X-100) for 1 h at 4°C and then incubated in alkaline solution (0.3 N NaOH and 5 mM EDTA) for 40 min to allow for unwinding of the DNA. After neutralization with 1× TBE three times for 5 min each time, the slides were run at 1 V/cm for 10 min in 1× TBE buffer and then incubated in 70% ethanol for 5 min. After air-drying, the slides were stained with SYBR green nucleic acid stain and then photographed by epifluorescence microscopy. Comet Score software (http://www.autocomet.com) was used to evaluate the levels of DNA damage.
Protein Expression and Purification
The full-length APE2 cDNA was cloned into the pET28a vector to generate the construct pET28a-APE2 that expresses His-APE2 fusion protein. A point mutation in the EEP domain (D265H) or Zf-GER domain (C570W) was introduced into the pET28a-APE2 construct through site-directed mutagenesis with the QuikChange II XL site-directed mutagenesis kit (Agilent Technologies). The constructs were transformed into Escherichia coli BL21 (DE3). Protein expression was induced with 0.1 mM IPTG for 16 h at 16°C. Wild-type and mutant APE2 proteins were purified according to QIAexpressionist, a handbook for high-level expression and purification of 6xHis-tagged proteins (Qiagen).
Enzymatic Assays
For detection of APE2’s capability to process 3′-P-terminated DNA substrate, a 25-nucleotide primer containing 6-FAM at its 5′ terminus and a phosphate group at its 3′ terminus was annealed to a 30-nucleotide oligomer template to generate a DNA substrate. The DNA substrate with 3′-blocking phosphate (10 nM) was incubated with commercial CIP (NEB) or purified His-APE2 proteins in 1× CutSmart buffer containing 50 mM potassium acetate (pH 7.9), 20 mM Tris-acetate, 10 mM magnesium acetate, and 100 μg/mL BSA (NEB) at 22°C for the indicated time. For detection of the 3′-5′ exonuclease activity of APE2, a DNA substrate with a 3′-OH terminus was generated by annealing a 25-nucleotide primer containing 6-FAM at its 5′ terminus and a hydroxyl group at its 3′ terminus to a 30-nucleotide oligomer template. Incubation of the 3′-OH-terminated DNA substrate with purified APE2 proteins was performed in the same buffer and under the same conditions as described above. Reactions were stopped by the addition of 2× TBE-urea sample buffer (Invitrogen). The reaction products were denatured for 10 min at 95°C, cooled for 2 min at 4°C, and resolved on a 20% TBE-urea gel. The gel was imaged using a Typhoon 9500 imager (GE Healthcare). The structures and sequences of the substrates used in this study are listed in Supplemental Table 1.
For detection of APE2’s capability to release free phosphate residue, DNA substrate with 3′-32P terminus was generated according to a previously described method (Akishev et al., 2016). Briefly, a 25-nucleotide primer was 3′-end labeled using 3′-α[32P]-dATP (cordycepin 5′-triphosphate; Perkin-Elmer) to generate a 26-nucleotide up-strand primer, which was then treated with recombinant human tyrosyl-DNA phosphodiesterase 1 (Tdp1). Tdp1 removed 3′-terminal cordycepin nucleoside, generating an oligonucleotide primer with a phosphate residue at its 3′ end (Lebedeva et al., 2013). The resulting 25-nucleotide 3′-32P fragment was annealed to the 30-nucleotide complementary down-strand primer and then used as substrate to measure 3′-phosphatase activity of APE2. The oligonucleotide sequences of these primers are the same as those of 6-FAM labeled primers.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: ZDP (AT3G14890), APE2 (AT4G36050), ROS1 (AT2G36490), DML2 (AT3G10010), RDR2 (AT4G11130), DCL3 (AT3G43920), DRM2 (AT5G14620), NRPD1 (AT1G63020), NRPE1 (AT2G40030), AGO4 (AT2G27040), MET1 (At5G49160), CMT3 (At1G69770), VIM1 (At1G57820), VIM2 (At1G66050), VIM3 (At5G39550), RAD51 (AT5G20850), BRCA1 (AT4G21070), GR1 (AT3G52115), MRE11 (AT5G54260), KU70 (AT1G16970), KU80 (AT1G48050), CYCB1;1 (AT4G37490), CYCA1;1 (AT1G44110), CYCA2;4 (AT1G80370), DT-75 (AT1G26380), DT-76 (AT1G26390), DT-77 (AT1G26400), and DT-78 (AT1G26410). The WGBS data were deposited in NCBI under SRP119887, and the small RNA-seq data were deposited in NCBI under SRP119987).
Supplemental Data
Supplemental Figure 1. Characterization of APE2 T-DNA insertion mutants.
Supplemental Figure 2. DNA methylome analysis of the wild-type, zdp-1, ape2-2, zdp-1 ape2-2, and ros1-4 mutant plants.
Supplemental Figure 3. Gene expression levels in the ape2 and zdp ape2 mutants.
Supplemental Figure 4. Profiling of 24-nucleotide siRNA levels in the zdp-1 ape2-2 plants.
Supplemental Figure 5. Pleiotropic developmental defects of the zdp ape2 double mutants.
Supplemental Figure 6. Gene expression levels in the ape2 and zdp ape2 mutants.
Supplemental Figure 7. Sequence alignments for the EEP and Zf-GRF domains of APE2 proteins, purification of APE2 proteins, and detection of APE2 proteins from transgenic plants.
Supplemental Table 1. Sequences of DNA oligonucleotides used for the generation of APE2 substrates.
Supplemental Data Set 1. List of hyper-DMRs and hypo-DMRs in the ape2-2, zdp-1, zdp-1 ape2-2, and ros1-4 plants.
Supplemental Data Set 2. Primers used in this study.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank Murat Saparbaev and Shitang Huang for their assistance in the DNA phosphatase assay using 3′-α[32P]-dATP, Peter Doerner for providing reporter lines, and Daowen Wang for suggestions on this project. This work was funded by grants from the Ministry of Science and Technology of China (Grant 2016YFA0500800) and the National Natural Science Foundation of China (Grants 31522005 and 31571326) to W.Q.
AUTHOR CONTRIBUTIONS
J.L., W.L., and W.Q. designed the research. J.L., W.L., and Y.L. performed the experiments. J.L. and W.Q. analyzed the data. J.L., Y.L., and W.Q. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Agius F., Kapoor A., Zhu J.K. (2006). Role of the Arabidopsis DNA glycosylase/lyase ROS1 in active DNA demethylation. Proc. Natl. Acad. Sci. USA 103: 11796–11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akishev Z., Taipakova S., Joldybayeva B., Zutterling C., Smekenov I., Ishchenko A.A., Zharkov D.O., Bissenbaev A.K., Saparbaev M. (2016). The major Arabidopsis thaliana apurinic/apyrimidinic endonuclease, ARP is involved in the plant nucleotide incision repair pathway. DNA Repair (Amst.) 48: 30–42. [DOI] [PubMed] [Google Scholar]
- Burkovics P., Szukacsov V., Unk I., Haracska L. (2006). Human Ape2 protein has a 3′-5′ exonuclease activity that acts preferentially on mismatched base pairs. Nucleic Acids Res. 34: 2508–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkovics P., Hajdú I., Szukacsov V., Unk I., Haracska L. (2009). Role of PCNA-dependent stimulation of 3′-phosphodiesterase and 3′-5′ exonuclease activities of human Ape2 in repair of oxidative DNA damage. Nucleic Acids Res. 37: 4247–4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell C., Hanakahi L.A., Karimi-Busheri F., Weinfeld M., West S.C. (2002). Involvement of human polynucleotide kinase in double-strand break repair by non-homologous end joining. EMBO J. 21: 2827–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Colón-Carmona A., You R., Haimovitch-Gal T., Doerner P. (1999). Technical advance: spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20: 503–508. [DOI] [PubMed] [Google Scholar]
- Córdoba-Cañero D., Morales-Ruiz T., Roldán-Arjona T., Ariza R.R. (2009). Single-nucleotide and long-patch base excision repair of DNA damage in plants. Plant J. 60: 716–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortellino S., et al. (2011). Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146: 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criqui M.C., Weingartner M., Capron A., Parmentier Y., Shen W.H., Heberle-Bors E., Bögre L., Genschik P. (2001). Sub-cellular localisation of GFP-tagged tobacco mitotic cyclins during the cell cycle and after spindle checkpoint activation. Plant J. 28: 569–581. [DOI] [PubMed] [Google Scholar]
- Doerner P., Jørgensen J.E., You R., Steppuhn J., Lamb C. (1996). Control of root growth and development by cyclin expression. Nature 380: 520–523. [DOI] [PubMed] [Google Scholar]
- Drohat A.C., Coey C.T. (2016). Role of base excision “Repair” enzymes in erasing epigenetic marks from DNA. Chem. Rev. 116: 12711–12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M., Huh J.H., Hsieh T.F., Penterman J., Choi Y., Harada J.J., Goldberg R.B., Fischer R.L. (2006). DEMETER DNA glycosylase establishes MEDEA polycomb gene self-imprinting by allele-specific demethylation. Cell 124: 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z., Zhu J.K. (2011). Active DNA demethylation by oxidation and repair. Cell Res. 21: 1649–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z., Morales-Ruiz T., Ariza R.R., Roldán-Arjona T., David L., Zhu J.K. (2002). ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111: 803–814. [DOI] [PubMed] [Google Scholar]
- Hadi M.Z., Wilson D.M. III (2000). Second human protein with homology to the Escherichia coli abasic endonuclease exonuclease III. Environ. Mol. Mutagen. 36: 312–324. [PubMed] [Google Scholar]
- He Y.F., et al. (2011). Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 333: 1303–1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill J.W., Hazra T.K., Izumi T., Mitra S. (2001). Stimulation of human 8-oxoguanine-DNA glycosylase by AP-endonuclease: potential coordination of the initial steps in base excision repair. Nucleic Acids Res. 29: 430–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S., Shen L., Dai Q., Wu S.C., Collins L.B., Swenberg J.A., He C., Zhang Y. (2011). Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 333: 1300–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jilani A., Ramotar D., Slack C., Ong C., Yang X.M., Scherer S.W., Lasko D.D. (1999). Molecular cloning of the human gene, PNKP, encoding a polynucleotide kinase 3′-phosphatase and evidence for its role in repair of DNA strand breaks caused by oxidative damage. J. Biol. Chem. 274: 24176–24186. [DOI] [PubMed] [Google Scholar]
- Krokan H.E., Bjørås M. (2013). Base excision repair. Cold Spring Harb. Perspect. Biol. 5: a012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J.A., Jacobsen S.E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11: 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedeva N.A., Rechkunova N.I., Ishchenko A.A., Saparbaev M., Lavrik O.I. (2013). The mechanism of human tyrosyl-DNA phosphodiesterase 1 in the cleavage of AP site and its synthetic analogs. DNA Repair (Amst.) 12: 1037–1042. [DOI] [PubMed] [Google Scholar]
- Lee J., Jang H., Shin H., Choi W.L., Mok Y.G., Huh J.H. (2014). AP endonucleases process 5-methylcytosine excision intermediates during active DNA demethylation in Arabidopsis. Nucleic Acids Res. 42: 11408–11418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Córdoba-Cañero D., Qian W., Zhu X., Tang K., Zhang H., Ariza R.R., Roldán-Arjona T., Zhu J.K. (2015). An AP endonuclease functions in active DNA demethylation and gene imprinting in Arabidopsis [corrected]. PLoS Genet. 11: e1004905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Vyas A., Kassab M.A., Singh A.K., Yu X. (2017). The role of poly ADP-ribosylation in the first wave of DNA damage response. Nucleic Acids Res. 45: 8129–8141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Ren X., Yin H., Wang Y., Xia R., Wang Y., Gong Z. (2010a). Mutation in the catalytic subunit of DNA polymerase alpha influences transcriptional gene silencing and homologous recombination in Arabidopsis. Plant J. 61: 36–45. [DOI] [PubMed] [Google Scholar]
- Liu Q., Wang J., Miki D., Xia R., Yu W., He J., Zheng Z., Zhu J.K., Gong Z. (2010b). DNA replication factor C1 mediates genomic stability and transcriptional gene silencing in Arabidopsis. Plant Cell 22: 2336–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D., Bernard D.G., Balk J., Hai H., Cui X. (2012). The DUF59 family gene AE7 acts in the cytosolic iron-sulfur cluster assembly pathway to maintain nuclear genome integrity in Arabidopsis. Plant Cell 24: 4135–4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti A., Drohat A.C. (2011). Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: potential implications for active demethylation of CpG sites. J. Biol. Chem. 286: 35334–35338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Macías M.I., Qian W., Miki D., Pontes O., Liu Y., Tang K., Liu R., Morales-Ruiz T., Ariza R.R., Roldán-Arjona T., Zhu J.K. (2012). A DNA 3′ phosphatase functions in active DNA demethylation in Arabidopsis. Mol. Cell 45: 357–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol C.D., Kuo C.F., Thayer M.M., Cunningham R.P., Tainer J.A. (1995). Structure and function of the multifunctional DNA-repair enzyme exonuclease III. Nature 374: 381–386. [DOI] [PubMed] [Google Scholar]
- Mol C.D., Izumi T., Mitra S., Tainer J.A. (2000). DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected]. Nature 403: 451–456. [DOI] [PubMed] [Google Scholar]
- Morales-Ruiz T., Ortega-Galisteo A.P., Ponferrada-Marín M.I., Martínez-Macías M.I., Ariza R.R., Roldán-Arjona T. (2006). DEMETER and REPRESSOR OF SILENCING 1 encode 5-methylcytosine DNA glycosylases. Proc. Natl. Acad. Sci. USA 103: 6853–6858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy T.M., Belmonte M., Shu S., Britt A.B., Hatteroth J. (2009). Requirement for abasic endonuclease gene homologues in Arabidopsis seed development. PLoS One 4: e4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Galisteo A.P., Morales-Ruiz T., Ariza R.R., Roldán-Arjona T. (2008). Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol. Biol. 67: 671–681. [DOI] [PubMed] [Google Scholar]
- Qian W., et al. (2012). A histone acetyltransferase regulates active DNA demethylation in Arabidopsis. Science 336: 1445–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schomacher L., Han D., Musheev M.U., Arab K., Kienhöfer S., von Seggern A., Niehrs C. (2016). Neil DNA glycosylases promote substrate turnover by Tdg during DNA demethylation. Nat. Struct. Mol. Biol. 23: 116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul O., Mironov V., Burssens S., Van Montagu M., Inze D. (1996). Two Arabidopsis cyclin promoters mediate distinctive transcriptional oscillation in synchronized tobacco BY-2 cells. Proc. Natl. Acad. Sci. USA 93: 4868–4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorenko V.S., Nevinsky G.A., Zharkov D.O. (2007). Mechanism of interaction between human 8-oxoguanine-DNA glycosylase and AP endonuclease. DNA Repair (Amst.) 6: 317–328. [DOI] [PubMed] [Google Scholar]
- Spampinato C.P. (2017). Protecting DNA from errors and damage: an overview of DNA repair mechanisms in plants compared to mammals. Cell. Mol. Life Sci. 74: 1693–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavnezer J., Linehan E.K., Thompson M.R., Habboub G., Ucher A.J., Kadungure T., Tsuchimoto D., Nakabeppu Y., Schrader C.E. (2014). Differential expression of APE1 and APE2 in germinal centers promotes error-prone repair and A:T mutations during somatic hypermutation. Proc. Natl. Acad. Sci. USA 111: 9217–9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., Rao A. (2009). Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324: 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang K., Lang Z., Zhang H., Zhu J.K. (2016). The DNA demethylase ROS1 targets genomic regions with distinct chromatin modifications. Nat. Plants 2: 16169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimoto D., Sakai Y., Sakumi K., Nishioka K., Sasaki M., Fujiwara T., Nakabeppu Y. (2001). Human APE2 protein is mostly localized in the nuclei and to some extent in the mitochondria, while nuclear APE2 is partly associated with proliferating cell nuclear antigen. Nucleic Acids Res. 29: 2349–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandepoele K., Raes J., De Veylder L., Rouzé P., Rombauts S., Inzé D. (2002). Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14: 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace S.S. (2014). Base excision repair: a critical player in many games. DNA Repair (Amst.) 19: 14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace B.D., Berman Z., Mueller G.A., Lin Y., Chang T., Andres S.N., Wojtaszek J.L., DeRose E.F., Appel C.D., London R.E., Yan S., Williams R.S. (2017). APE2 Zf-GRF facilitates 3′-5′ resection of DNA damage following oxidative stress. Proc. Natl. Acad. Sci. USA 114: 304–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Liu Z. (2006). Arabidopsis ribonucleotide reductases are critical for cell cycle progression, DNA damage repair, and plant development. Plant Cell 18: 350–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A.R., Krawczyk C., Robertson A.B., Kuśnierczyk A., Vågbø C.B., Schuermann D., Klungland A., Schär P. (2016). Biochemical reconstitution of TET1-TDG-BER-dependent active DNA demethylation reveals a highly coordinated mechanism. Nat. Commun. 7: 10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse C.J., Taylor R.M., Thistlethwaite A., Zhang H., Karimi-Busheri F., Lasko D.D., Weinfeld M., Caldecott K.W. (2001). XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell 104: 107–117. [DOI] [PubMed] [Google Scholar]
- Wiederhold L., Leppard J.B., Kedar P., Karimi-Busheri F., Rasouli-Nia A., Weinfeld M., Tomkinson A.E., Izumi T., Prasad R., Wilson S.H., Mitra S., Hazra T.K. (2004). AP endonuclease-independent DNA base excision repair in human cells. Mol. Cell 15: 209–220. [DOI] [PubMed] [Google Scholar]
- Willis J., Patel Y., Lentz B.L., Yan S. (2013). APE2 is required for ATR-Chk1 checkpoint activation in response to oxidative stress. Proc. Natl. Acad. Sci. USA 110: 10592–10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Zhang Y. (2014). Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 156: 45–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Zhang X., Liu J., Wang Y., He J., Yang T., Hong X., Yang Q., Gong Z. (2009). Epigenetic regulation, somatic homologous recombination, and abscisic acid signaling are influenced by DNA polymerase epsilon mutation in Arabidopsis. Plant Cell 21: 386–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., et al. (2013). DTF1 is a core component of RNA-directed DNA methylation and may assist in the recruitment of Pol IV. Proc. Natl. Acad. Sci. USA 110: 8290–8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Li L. (2013). SQUAMOSA promoter binding protein-like7 regulated microRNA408 is required for vegetative development in Arabidopsis. Plant J. 74: 98–109. [DOI] [PubMed] [Google Scholar]
- Zhang J., Xie S., Zhu J.K., Gong Z. (2016a). Requirement for flap endonuclease 1 (FEN1) to maintain genomic stability and transcriptional gene silencing in Arabidopsis. Plant J. 87: 629–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Xie S., Cheng J., Lai J., Zhu J.K., Gong Z. (2016b). The second subunit of DNA polymerase delta is required for genomic stability and epigenetic regulation. Plant Physiol. 171: 1192–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J.K. (2009). Active DNA demethylation mediated by DNA glycosylases. Annu. Rev. Genet. 43: 143–166. [DOI] [PMC free article] [PubMed] [Google Scholar]