NADPH-dependent cinnamaldehyde and hexenal reductase affects the composition of green leaf volatiles and is involved in indirect plant defenses.
Abstract
Green leaf volatiles (GLVs), including six-carbon (C6) aldehydes, alcohols, and esters, are formed when plant tissues are damaged. GLVs play roles in direct plant defense at wound sites, indirect plant defense via the attraction of herbivore predators, and plant-plant communication. GLV components provoke distinctive responses in their target recipients; therefore, the control of GLV composition is important for plants to appropriately manage stress responses. The reduction of C6-aldehydes into C6-alcohols is a key step in the control of GLV composition and also is important to avoid a toxic buildup of C6-aldehydes. However, the molecular mechanisms behind C6-aldehyde reduction remain poorly understood. In this study, we purified an Arabidopsis (Arabidopsis thaliana) NADPH-dependent cinnamaldehyde and hexenal reductase encoded by At4g37980, named here CINNAMALDEHYDE AND HEXENAL REDUCTASE (CHR). CHR T-DNA knockout mutant plants displayed a normal growth phenotype; however, we observed significant suppression of C6-alcohol production following partial mechanical wounding or herbivore infestation. Our data also showed that the parasitic wasp Cotesia vestalis was more attracted to GLVs emitted from herbivore-infested wild-type plants compared with GLVs emitted from chr plants, which corresponded with reduced C6-alcohol levels in the mutant. Moreover, chr plants were more susceptible to exogenous high-dose exposure to (Z)-3-hexenal, as indicated by their markedly lowered photosystem II activity. Our study shows that reductases play significant roles in changing GLV composition and, thus, are important in avoiding toxicity from volatile carbonyls and in the attraction of herbivore predators.
Plants form and emit a wide variety of volatile organic compounds. Volatiles from flowers attract pollinators and increase the fitness of plants by promoting efficient reproduction, and those from fruits attract seed-dispersing animals and help plants to find new habitats (Dudareva et al., 2013). Vegetative organs, such as leaves, stems, and roots, also produce and emit volatiles, and this process generally is induced by various types of biotic and abiotic stresses as a defense response (Pierik et al., 2014).
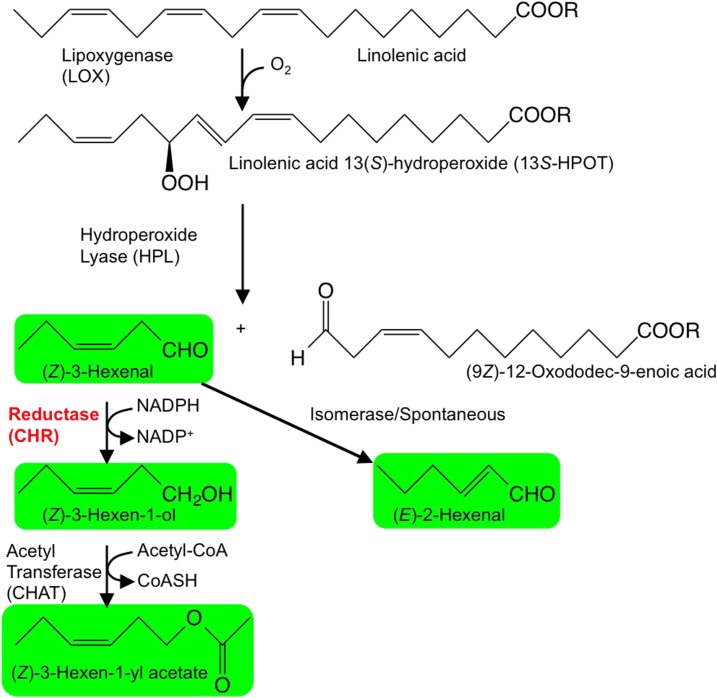
Green leaf volatiles (GLVs) are expressed ubiquitously with other groups of volatile compounds, such as terpenoids and amino acid derivatives. GLVs are derivatives of fatty acids and include six-carbon (C6) aldehydes, alcohols, and esters (Fig. 1; Matsui, 2006; Scala et al., 2013). In intact and healthy plant tissues, GLV levels generally are low, but when tissues suffer stresses associated with the disruption of cells, such as herbivore damage or attack by necrotrophic fungi, GLV-forming pathways are activated rapidly to yield large quantities of GLVs at damaged tissues (Matsui, 2006; Scala et al., 2013; Ameye et al., 2017). GLVs at damaged tissues participate in direct plant defense by preventing the invasion of harmful microbes (Shiojiri et al., 2006a; Kishimoto et al., 2008). GLVs also function as infochemicals that are an indirect plant defense via the attraction of natural predators of infesting herbivores (Kessler and Baldwin, 2001; Shiojiri et al., 2006a, 2006b; Joo et al., 2018). GLVs also have been associated with plant-plant communication (Sugimoto et al., 2014).
Figure 1.
The GLV synthetic pathway in plants. Linolenic acid or linolenoyl residues of lipids are oxygenated by lipoxygenase (LOX) to yield corresponding hydroperoxides. These are cleaved by hydroperoxide lyase (HPL) to form C6-aldehydes and 12-carbon ω-oxo carboxylic acids (or esters when lipids are the starting material). A portion of (Z)-3-hexenal is spontaneously or enzymatically isomerized to form (E)-2-hexenal, and a portion of (Z)-3-hexenal is reduced by reductases to (Z)-3-hexen-1-ol. Further acetylation leads to the formation of (Z)-3-hexen-1-yl acetate.
Because the chemical properties of GLV components are distinct, it is assumed that each plays a distinctive role. For example, C6-aldehydes were shown to be involved prominently in direct defense against the necrotrophic fungal pathogen Botrytis cinerea, whereas C6-alcohols and -esters were less involved (Kishimoto et al., 2008). The formyl groups in C6-aldehydes are potently reactive with nucleophiles, such as amines and alcohols, and contribute to the suppression of pathogen growth. When carbon-carbon double bonds are conjugated to the formyl group, as in (E)-2-hexenal, the α,β-unsaturated carbonyl moiety behaves as a more potent electrophile and is prone to reactions with various biological molecules, specifically reactions that eliminate their intrinsic functions (Farmer and Mueller, 2013). Even though (Z)-3-hexenal is not a reactive carbonyl species, because the two double bonds are interrupted with one methylene group, bis-allylic hydrogen is prone to removal, often resulting in oxygenation to yield 4-hydroxy-(E)-2-hexenal or 4-oxo-(E)-2-hexenal (Pospíšil and Yamamoto, 2017). These oxygenated hexenals are highly reactive carbonyl species, and (Z)-3-hexenal has been identified specifically as a potentially reactive compound (Matsui et al., 2012). C6-aldehydes were reported to be more effective bacteriostatic chemicals than C6-alcohols, and the incorporation of a double bond in either the C2 or C3 position enhances their activities (Nakamura and Hatanaka, 2002), albeit the formyl group is not always a prerequisite for antimicrobial activity (Prost et al., 2005).
GLVs are either attractive or repulsive to arthropods, depending on their composition and on the arthropod species (Wei and Kang, 2011; Scala et al., 2013). Females of the parasitic wasp Cotesia glomerata are attracted to (E)-2-hexenal and (Z)-3-hexen-1-yl acetate but not to (Z)-3-hexen-1-ol (Shiojiri et al., 2006b). Moreover, (Z)-3-hexen-1-yl acetate increased predation rates of a generalist natural predator toward eggs of the herbivore Manduca quinquemaculata on Nicotiana attenuata plants (Kessler and Baldwin, 2001). The composition of GLVs elicited by herbivores also differs between dawn and dusk, as indicated by Joo et al. (2018), who showed that GLV aldehydes and alcohols were less abundant in comparison with GLV esters at dawn whereas GLV ester levels were lower at dusk. Higher predation rates on Manduca sexta eggs by Geocoris spp. were observed with dawn GLV composition in nature, suggesting that the predator distinguished between GLV compositions (Joo et al., 2018).
Concerning plant-plant communication, (Z)-3-hexen-1-ol emitted from Spodoptera litura-infested tomato (Solanum lycopersicum) leaves was shown to be absorbed by neighboring tomato plants to form the defense compound (Z)-3-hexen-1-yl primeveroside (Sugimoto et al., 2014). It also was observed that the secretion of extrafloral nectar by lima beans (Phaseolus lunatus) to attract predatory and parasitoid insects (ants and wasps) was induced significantly after exposure of these plants to (Z)-3-hexen-1-yl acetate vapors (Kost and Heil, 2006). These accumulating lines of evidence indicate that the composition of GLVs is crucial for plants to aptly cope with enemies and foes and that the reduction of C6-aldehydes to C6-alcohol is a crucial step in the control of GLV composition.
LOX acts on linolenic/linoleic acids, either in their free forms or as acyl groups in glycerolipids, to form corresponding fatty acid/lipid hydroperoxides, which are cleaved subsequently by HPL to form C6-aldehydes such as n-hexanal or (Z)-3-hexenal (Fig. 1; Matsui, 2006; Scala et al., 2013; Mwenda and Matsui, 2014). A portion of (Z)-3-hexenal is isomerized to (E)-2-hexenal in some plant species (Kunishima et al., 2016; Spyropoulou et al., 2017), and a factor in the oral secretions of M. sexta catalyzes this isomerization in N. attenuata, resulting in a higher attraction of the predator Geocoris spp. (Allmann and Baldwin, 2010). Other portions of C6-aldehydes are reduced to corresponding C6-alcohols, and portions of C6-alcohols are converted further into corresponding acetates by acetyl-CoA transferase (D’Auria et al., 2007). It was assumed that NAD(H)-dependent alcohol dehydrogenase (EC 1.1.1.1) is involved in these reduction steps (Hatanaka, 1993), and this may be the case with completely disrupted plant tissues. Accordingly, in completely homogenized leaves of Arabidopsis (Arabidopsis thaliana) mutants that were deficient in alcohol dehydrogenase (AtADH1; At1g77120), n-hexan-1-ol and 3-hexen-1-ol were formed at 62% and 51% of the levels found in parent wild-type plants, respectively (Bate et al., 1998). In completely homogenized tomato tissues with increased and suppressed expression levels of SlADH2, which is the tomato homolog of AtADH1, n-hexan-1-ol and (Z)-3-hexen-1-ol levels correlated roughly with ADH activities (Speirs et al., 1998). In contrast, in partially wounded leaf tissues of Arabidopsis, (Z)-3-hexenal was formed at disrupted tissues and diffused into the neighboring intact tissues, where it was reduced to (Z)-3-hexen-1-ol in an NADPH-dependent manner (Matsui et al., 2012). Hence, an NADPH-dependent C6-aldehyde reductase is likely accountable for the reduction of C6-aldehydes to C6-alcohols. Because plant leaves are commonly damaged only in part, as following consumption by an herbivore, such an NADPH-dependent reductase may be ecophysiologically relevant, but one has not been identified yet.
In this study, we purified and identified an NADPH-dependent reductase preferring (Z)-3-hexenal, which we named CINNAMALDEHYDE AND HEXENAL REDUCTASE (CHR). The involvement of CHR in changing the composition of GLVs was verified using a T-DNA knockout mutant disrupted in the corresponding CHR gene. Moreover, chr mutant lines were employed to assess the role of CHR in the indirect defense of Arabidopsis via GLV composition. CHR’s involvement in the detoxification of (Z)-3-hexenal also was examined.
RESULTS
Purification of Hexenal Reductase
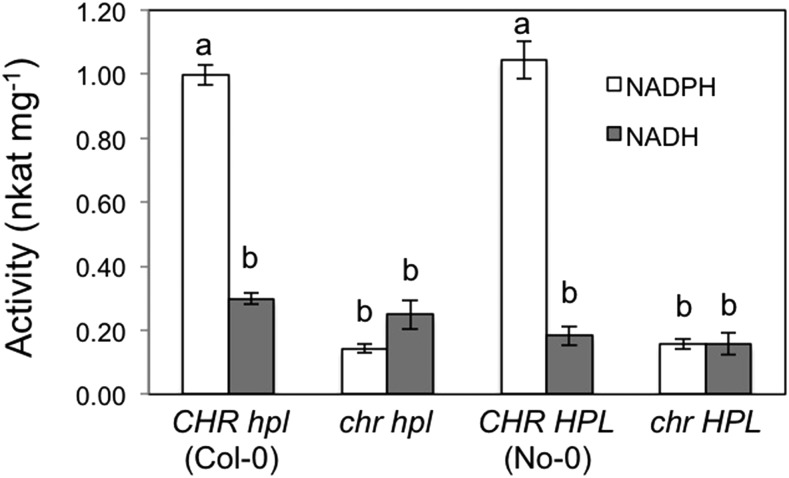
Following exposure to (Z)-3-hexenal vapors, intact Arabidopsis leaves absorb the compound into leaves, reduce it to (Z)-3-hexen-1-ol, and then reemit it from the leaves to the atmosphere (Matsui et al., 2012). Therefore, substantial enzyme activity for converting (Z)-3-hexenal to (Z)-3-hexen-1-ol already is present in Arabidopsis leaves under normal growth conditions, even without stress treatments and tissue disruptions to trigger GLV formation. When crude extracts from Arabidopsis leaves (No-0) were exposed to (Z)-3-hexenal, exogenous NADPH was consumed (see Fig. 4 below). Moreover, the rate of NADH consumption was less than 20% of that found with NADPH under the same reaction conditions. Taken with our previous results (Matsui et al., 2012), this result indicated that Arabidopsis leaves carry an NADPH-dependent (Z)-3-hexenal reductase.
Figure 4.
NADPH consumption depending on (Z)-3-hexenal in wild-type and mutant Arabidopsis leaves. Activities were determined in four biological replicates (n = 4) and are presented as averages ± se. Different letters indicate significant differences, as identified using two-way ANOVA and Tukey’s posthoc tests (P < 0.05).
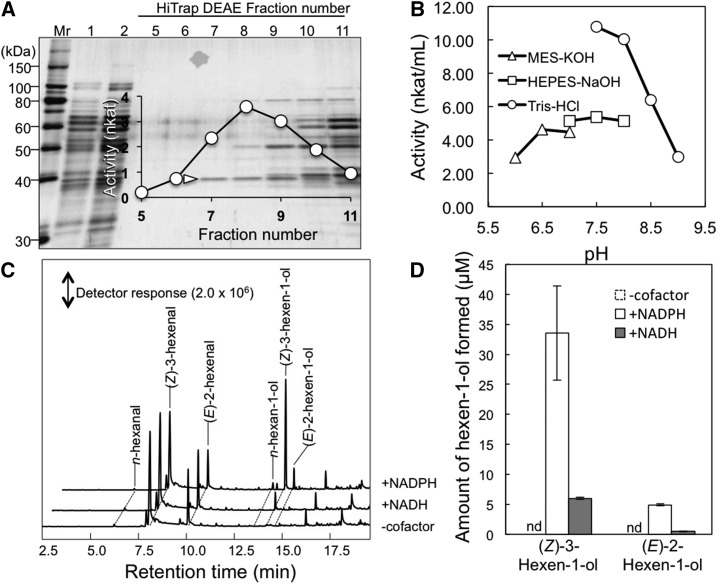
This reductase was purified from crude Arabidopsis leaf extracts using ammonium sulfate fractionation followed by three consecutive chromatography steps (Table 1). During these steps, reductase activity was identified as a single peak with a substantial yield, and SDS-PAGE analyses of fractions that were separated using a HiTrap DEAE column in the third chromatography step indicated that a protein of 38 kD correlated with the observed metabolic activity (Fig. 2A). In the presence of (Z)-3-hexenal, the purified enzyme consumed NADPH most actively at pH 7.5 (Fig. 2B). When a mixture of (Z)-3-hexenal and (E)-2-hexenal (154 and 46 µm, respectively) was used as a substrate in the absence of cofactor, the formation of the corresponding alcohol was hardly detected (Fig. 2C). The addition of NADH slightly facilitated the reduction of hexenals, whereas NADPH resulted in a more extensive formation of (Z)-3-hexen-1-ol and (E)-2-hexen-1-ol (Fig. 2D). However, n-hexanal formation through reduction of the carbon-carbon double bond in (Z)-3-hexenal and (E)-2-hexenal was barely increased, indicating that the enzyme has little double bond reductase activity.
Table 1. Summary of the (Z)-3-hexenal reductase purification process from Arabidopsis leaves.
| Purification Step | Total Protein | Total Activity | Specific Activity | Purification | Yield |
|---|---|---|---|---|---|
| mg | nkat | nkat mg−1 | -fold | % | |
| Crude extract | 1,707 | 1,062 | 1.66 | 1 | 100 |
| Ammonium sulfate | 1,634 | 592 | 26.0 | 0.58 | 55.7 |
| Butyl-Toyopearl | 83.7 | 90.0 | 10.8 | 1.73 | 8.47 |
| Hydroxyapatite | 0.28 | 21.2 | 6.62 | 119 | 1.99 |
| HiTrap DEAE | 0.031 | 3.94 | 7.88 | 209 | 0.37 |
Figure 2.
Purification and properties of hexenal reductase. A, Protein profile of the fractions collected using HiTrap DEAE chromatography. Equal volumes of fractions were loaded onto each lane, and proteins were stained with Coomassie Blue. (Z)-3-Hexenal reductase activities of each fraction (fractions 5–11) are shown. The 38-kD protein used in amino acid sequence analyses is indicated with a white arrowhead. Mr, Molecular mass marker. B, pH activity profile of the partially purified enzyme. C, Gas chromatography-mass spectrometry (GC-MS) chromatograms of products formed from (Z)-3-hexenal in the absence or presence of the cofactor NADH or NADPH. D, Quantities of (Z)-3-hexen-1-ol from C presented as averages ± se (n = 4). Because (Z)-3-hexen-1-ol was not formed in the absence of cofactor, the bars corresponding to activity are indicated by nd, for not detected.
Identification of a C6-Aldehyde Reductase Gene
Internal amino acid sequences from the purified protein showed complete identity with Arabidopsis CINNAMYL ALCOHOL DEHYDROGENASE7 (AtCAD7 [At4g37980]; Supplemental Fig. S1). The AtCAD7 gene also is known as ELICITOR-ACTIVATED GENE3 (Kiedrowski et al., 1992), and the protein comprises 357 amino acids and has a molecular mass of 38,245 D, close to that of the purified enzyme estimated in SDS-PAGE analyses (Fig. 2A). A MotifFinder search (http://www.genome.jp/tools/motif/) of the Pfam database showed that the protein contains ADH_N and ADH_zinc_N motifs. Cytosolic localization also was predicted based on the amino acid sequence using SUBA3 (http://suba.plantenergy.uwa.edu.au/).
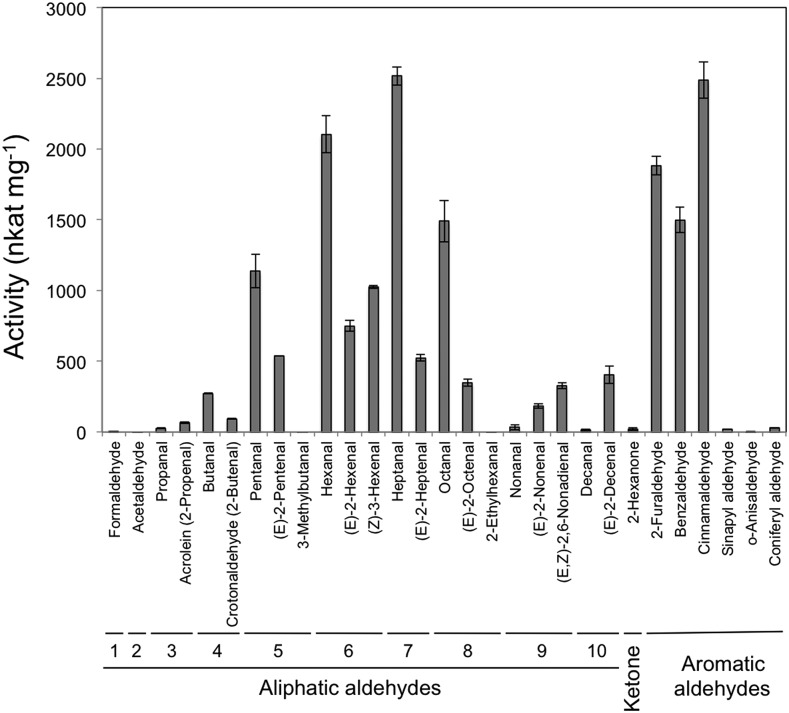
A recombinant protein encoded by the open reading frame of At4g37980 was expressed as a C-terminal His-tagged protein in Escherichia coli and then was purified (Supplemental Fig. S2). With straight-chain saturated aldehydes ranging from one to 10 carbons in length, the recombinant enzyme displayed highest activity with n-heptanal (Fig. 3), followed by n-hexanal, n-octanal, and then n-pentanal. The enzyme showed little activity toward formaldehyde and acetaldehyde. Among the unsaturated aldehydes examined in this study, (Z)-3-hexenal was the best substrate, followed by (E)-2-hexenal and (E)-2-pentenal, and the C6 ketone n-hexan-2-one was a poor substrate. The enzyme exhibited relatively high activities with aromatic aldehydes, such as 2-furaldehyde, benzaldehyde, and cinnamaldehyde, but little activity was observed with sinapyl aldehyde and coniferyl aldehyde, which are intermediates in the lignin biosynthesis pathway and are good substrates for AtCAD4 and AtCAD5 (Kim et al., 2004). Activity profiles of enzymes from Arabidopsis leaves with seven representative aldehydes were similar to those observed with the recombinant enzyme (Supplemental Fig. S3). The enzyme also obeyed Michaelis-Menten kinetics with (Z)-3-hexenal, n-heptanal, and cinnamaldehyde (Supplemental Fig. S4), and Km values were estimated to be 32.7, 84, and 8.46 µm, respectively (Table 2). Based on the properties of the enzyme encoded by At4g37980, we renamed the gene CHR.
Figure 3.
Substrate specificity of recombinant Arabidopsis CHR expressed in E. coli. Data are presented as averages ± se (n = 4).
Table 2. Km and Vmax values of recombinant Arabidopsis CHR with (Z)-3-hexenal, n-heptanal, and cinnamaldehyde.
| Substrate | Km | Vmax |
|---|---|---|
| µm | nkat mg−1 | |
| (Z)-3-Hexenal | 32.7 ± 30.90 | 347 ± 128.0 |
| n-Heptanal | 84.0 ± 34.51 | 286 ± 52.53 |
| Cinnamaldehyde | 8.46 ± 7.172 | 587 ± 103.0 |
Involvement of CHR in GLV Composition
To examine the in vivo functions of CHR, an Arabidopsis strain with a T-DNA insertion in the third exon of At4g37980 (Salk_001773) was prepared (Supplemental Fig. S5). T-DNA-disrupted strains with the Col-0 background grew normally and did not differ from the parent strain (Col-0; Supplemental Fig. S5C), but because Arabidopsis Col-0 has a 10-bp deletion in the HPL gene, its ability to form GLV aldehydes is impaired (Duan et al., 2005). Thus, we crossed the T-DNA-disrupted line (chr hpl) with Arabidopsis No-0 carrying active HPL (CHR HPL), and a strain that was homozygous for the At4g37980 T-DNA insertion and the functional HPL gene (chr HPL) was selected from the F2 generation for further analysis (Supplemental Fig. S5). In the presence of NADPH, enzymatic reduction of (Z)-3-hexenal to (Z)-3-hexen-1-ol in crude homogenates from chr hpl and chr HPL plants was suppressed to approximately 14% of that for Col-0 (CHR hpl) and No-0 (CHR HPL) plants (Fig. 4). Activities with NADH in extracts from chr hpl and chr HPL plants, however, were not significantly different from those for Col-0 (CHR hpl) and No-0 (CHR HPL) plants.
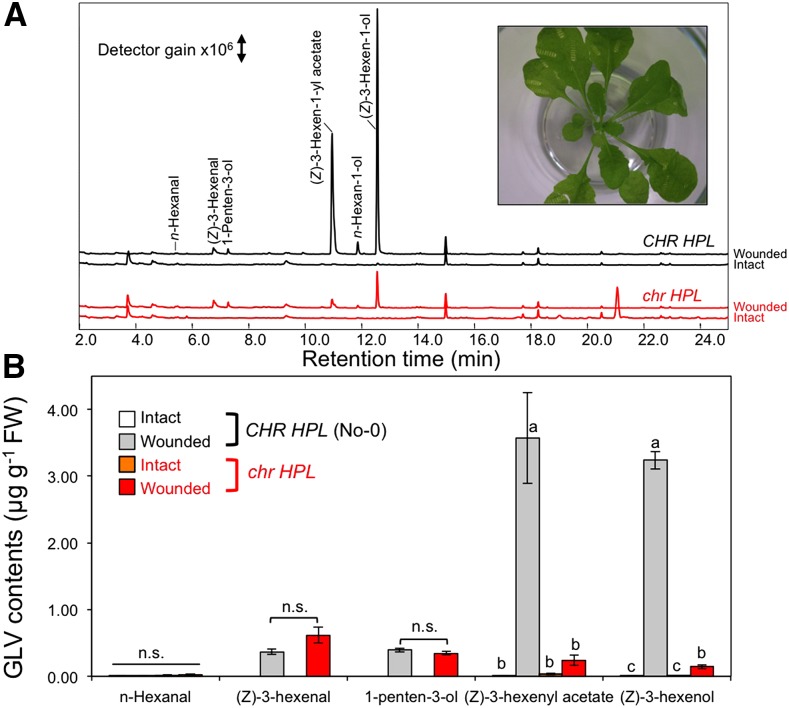
Intact leaves from No-0 (CHR HPL) or mutant (chr HPL) plants hardly emitted volatile compounds, but partial mechanical wounding promoted a GLV burst in these leaves (Fig. 5). The resulting n-hexanal and (Z)-3-hexenal quantities did not differ significantly between No-0 (CHR HPL) and the mutant (chr HPL), whereas the amounts of (Z)-3-hexen-1-yl acetate and (Z)-3-hexen-1-ol were significantly lower from the mutant (chr HPL) plants. Moreover, the amount of 1-penten-3-ol, which was formed depending on LOX but not on HPL (Mochizuki et al., 2016), was relatively unaffected by the presence of functional CHR. Because total amounts of GLVs differed between these two genotypes, we investigated whether their abilities to form C6-aldehydes through LOX and HPL also differ. After completely disrupting the leaves of No-0 (CHR HPL) and mutant (chr HPL) plants, the GLV pathway was mostly arrested after the HPL step that forms C6-aldehydes, and reduction reactions hardly proceeded, largely due to NADPH deficiencies (Matsui et al., 2012). Moreover, the formation of (Z)-3-hexenal, 1-penten-3-ol, (E)-2-hexenal, and (Z)-2-penten-1-ol was observed from both genotypes, and (Z)-3-hexen-1-ol, n-hexan-1-ol, and (Z)-3-hexen-1-yl acetate were detected at low levels in both No-0 (CHR HPL) and mutant (chr HPL) leaves (Supplemental Fig. S6). The quantities of (Z)-3-hexenal that were formed after the complete disruption of mutant leaves were slightly higher than those for No-0 leaves, suggesting that the ability to form (Z)-3-hexenal from lipids/fatty acids was not the prominent cause of lowered (Z)-3-hexen-1-ol and (Z)-3-hexen-1-yl acetate production after the partial mechanical wounding of chr HPL leaves.
Figure 5.
Formation and emission of volatiles from partially wounded Arabidopsis leaves. A, Representative chromatograms of volatiles from intact leaves (bottom chromatogram of each genotype) and partially wounded leaves (top chromatogram of each genotype). Wild-type (No-0, CHR HPL) and mutant (chr HPL) signals are shown in black and red, respectively. The inset shows a representative image of a mutant (chr hpl) plant after partial wounding. B, Quantities of GLVs from intact and partially wounded wild-type and mutant leaves. Data are averages ± se (n = 4). Different letters represent significant differences, as identified using two-way ANOVA and Tukey’s posthoc tests (P < 0.01); n.s., no significant difference. FW, Fresh weight.
CHR Is Involved in Indirect Defenses
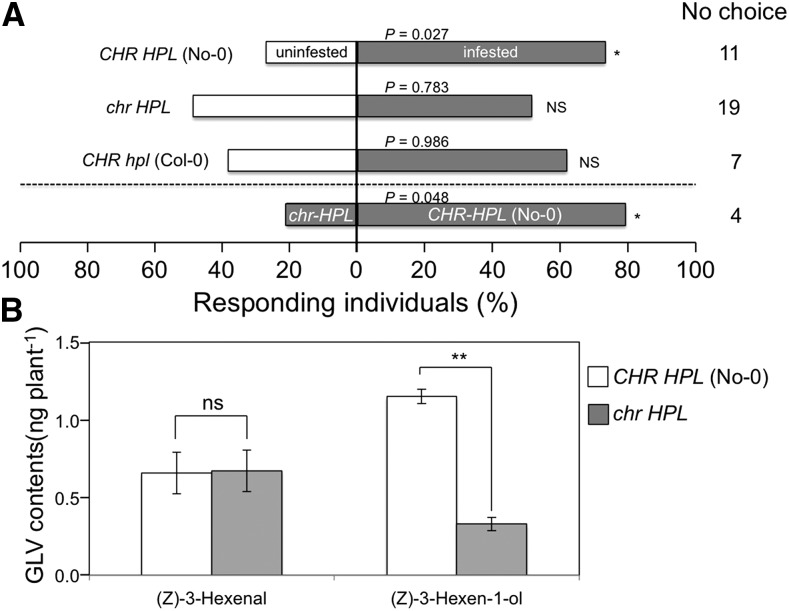
We observed flight responses of the parasitoid wasp Cotesia vestalis to the volatiles emitted from Arabidopsis leaves that had been infested for 24 h with larvae of the diamond back moth Plutella xylostella. In experiments with No-0 (CHR HPL), C. vestalis females preferred P. xylostella-damaged plants over intact plants, but they did not show any preference when Col-0 (CHR hpl) plants were used (Fig. 6A). No preference was observed with the mutant (chr HPL) either, and the numbers of wasps that landed on P. xylostella-infested plants and intact plants did not differ significantly. Examinations of wasp preferences also showed significant preferences for herbivore-infested No-0 (CHR HPL) plants over herbivore-infested mutant (chr HPL) plants. Validating experiments showed similar areas of consumption by P. xylostella on leaves of CHR HPL and chr HPL plants (Supplemental Fig. S7).
Figure 6.
Role of CHR in C. vestalis flight responses and in volatile emission following P. xylostella infestation. A, Flight responses of C. vestalis to P. xylostella-infested (gray) and uninfested (white) Arabidopsis CHR HPL (No-0), chr HPL, and CHR hpl (Col-0) plants (40 individuals; above the dashed line) and to P. xylostella-infested chr HPL and CHR HPL (No-0) plants (60 individuals; below the dashed line). *, 0.05 > Ptotal > 0.01 (replicated G test); NS, no significant difference. C. vestalis that did not choose either plant (no-choice subjects) were not included in the statistical analysis. B, (Z)-3-Hexenal and (Z)-3-hexen-1-ol emissions from wild-type (No-0, CHR HPL) and mutant (chr HPL) plants after a 24-h infestation with P. xylostella larvae. The quantities of volatiles in headspaces of glass vials containing three plants were collected after a 24-h infestation period using solid-phase microextraction (SPME) fibers for 30 min. Data are presented as averages ± se (n = 3). **, P < 0.01, as identified using Student’s t test; ns, no significant difference. Quantities of (Z)-3-hexen-1-yl acetate were below the detection limit (less than 0.8 ng in the pot).
The quantities of volatiles emitted at 24 h after the onset of P. xylostella infestation of mutant (chr HPL) and No-0 (CHR HPL) plants were quite low, and the volatiles (Z)-3-hexenal and (Z)-3-hexen-1-ol were detected under the experimental conditions employed here with the selected ion monitoring mode of GC-MS. In these analyses, (Z)-3-hexen-1-yl acetate contents were below the detection limit (less than 0.8 ng in a glass volatile collection jar). The quantities of (Z)-3-hexenal emitted from the two genotypes at 24 h after the onset of P. xylostella infestation also were almost the same, whereas those of (Z)-3-hexen-1-ol were significantly lower from infested mutant (chr HPL) plants than from infested No-0 (CHR HPL) plants (Fig. 6B).
Involvement of CHR in the Detoxification of Hexenal
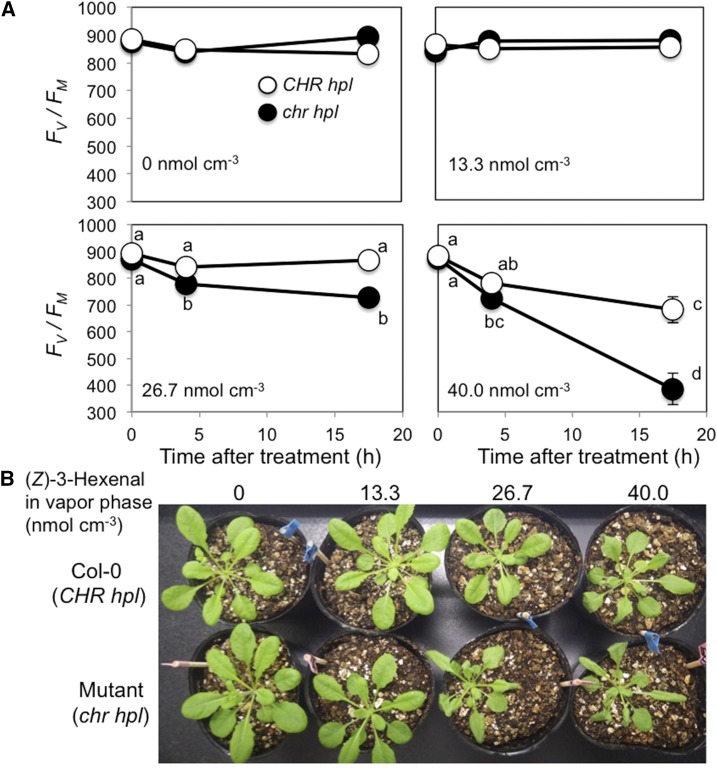
The chlorophyll fluorescence parameter Fv/Fm (for maximum photochemical efficiency of PSII in the dark-adapted state) was used in a bioassay as an indicator of cell deterioration. When Col-0 (CHR hpl) plants were exposed to (Z)-3-hexenal vapor at 13.3 or 26.7 nmol cm−3, Fv/Fm remained relatively unchanged. Fv/Fm decreased significantly when Col-0 plants were exposed to 40 nmol cm−3 (Z)-3-hexenal (Fig. 7A), as we observed previously (Matsui et al., 2012). Mutant plants (chr hpl) treated with 13.3 nmol cm−3 (Z)-3-hexenal maintained normal Fv/Fm values; however, Fv/Fm decreased significantly after 4 h of treatment with 26.7 nmol cm−3 (Z)-3-hexenal. Following treatment with 40 nmol cm−3 (Z)-3-hexenal, Fv/Fm of the mutant plants decreased more quickly than that of Col-0 (CHR hpl), and after 17.5 h, Fv/Fm of the mutant plants (chr hpl) was significantly lower than that of Col-0 (CHR hpl). Both Col-0 (CHR hpl) and mutant (chr hpl) plants showed no visible symptoms after treatment with 13.3 nmol cm−3 (Z)-3-hexenal (Fig. 7B). However, treatment with 26.7 nmol cm−3 (Z)-3-hexenal caused the leaves of mutant plants to wilt slightly and curl at their margins, and 40 nmol cm−3 (Z)-3-hexenal treatment caused obvious damage in both genotypes, with more serious effects seen in the mutant plants.
Figure 7.
Changes in PSII activity in Arabidopsis wild-type (Col-0, CHR hpl) and mutant (chr hpl) leaves during exposure to (Z)-3-hexenal. A, Aboveground parts of Arabidopsis wild-type (white circles) and mutant (black circles) plants were exposed to 0, 13.3, 26.7, or 40 nmol cm−3 (Z)-3-hexenal vapor in closed glass jars, and PSII activity was estimated using the chlorophyll fluorescence parameter Fv/Fm. Data are averages ± se (n = 8). Differences were identified using two-way ANOVA and Tukey’s posthoc test. Different letters indicate significant differences (P < 0.05). B, Representative images of Col-0 (CHR hpl) and mutant (chr hpl) plants at 17.5 h after the onset of exposure to 0, 13.3, 26.7, or 40 nmol cm−3 (Z)-3-hexenal vapor.
DISCUSSION
In this study, we confirmed that At4g37980, which had been designated previously as CAD7, encodes an NADPH-dependent aldehyde reductase that prefers aliphatic aldehydes of five to eight carbons in length and simple aromatic aldehydes as substrates. Crude extracts from leaves of the At4g37980 T-DNA knockout mutant had significantly lower enzyme activities to oxidize NADPH and reduce (Z)-3-hexenal. Furthermore, knockout mutant plants with active HPL formed significantly lower concentrations of (Z)-3-hexen-1-ol and (Z)-3-hexen-1-yl acetate after the partial mechanical wounding of leaves. (Z)-3-Hexen-1-ol formation after herbivore infestation also was suppressed in knockout mutant plants with active HPL. These results indicate that the enzyme encoded by At4g37980 converts (Z)-3-hexenal to (Z)-3-hexen-1-ol during GLV bursts that are elicited by the partial wounding of Arabidopsis leaves. Accordingly, we renamed the gene CHR.
CHR was designated originally as a member (AtCAD7) of the CAD gene family, which comprises nine putative AtCAD genes (AtCAD1–AtCAD9) according to a previous classification (Sibout et al., 2003; Kim et al., 2004). Kim et al. (2004) showed that recombinant CHR (AtCAD7) activities against intermediates of the lignin biosynthetic pathway, such as p-coumaryl aldehyde, caffeyl aldehyde, coniferyl aldehyde, 5-hydroxyconiferyl aldehyde, or sinapyl aldehyde, were considerably lower than those of recombinant AtCAD4 or AtCAD5 enzymes. AtCAD4 and AtCAD5 were later confirmed to be CADs of the lignin biosynthesis pathway in floral stems of Arabidopsis, as indicated by the lignin biosynthetic activities in corresponding knockout mutant lines (Sibout et al., 2005). In agreement, Klason lignin contents in T-DNA knockout mutants of CHR (AtCAD7) were similar to those in corresponding wild-type Arabidopsis (Eudes et al., 2006). Taken with these observations, this study indicates that CHR has limited involvement in lignin biosynthesis; rather, its primary role is to convert (Z)-3-hexenal to (Z)-3-hexen-1-ol during GLV bursts.
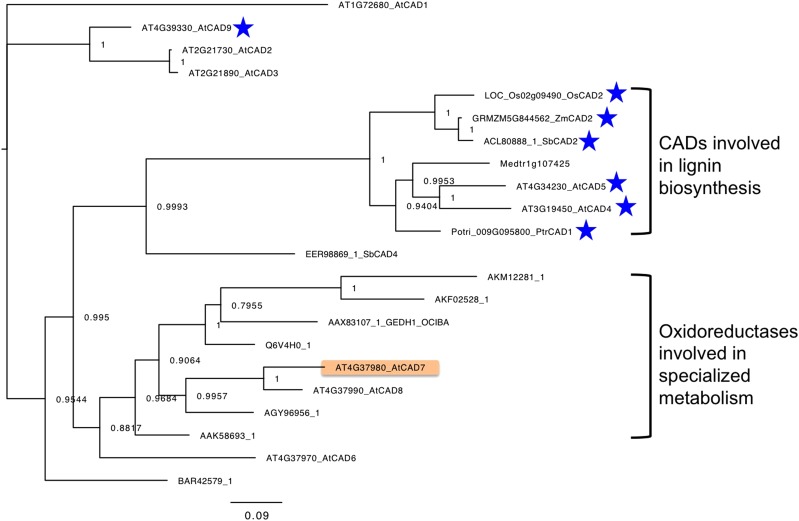
BLASTP searches were performed against National Center for Biotechnology Information Protein Reference Sequences, and those with BLAST scores higher than 250 and with biochemically confirmed functions were chosen. A phylogenetic tree was then constructed with these sequences and those of AtCAD1 to AtCAD9 (Fig. 8). We focused on two clades of the tree, and the first of these, the CAD clade, comprises Oryza sativa OsCAD2 (Zhang et al., 2006), Populus tremula × Populus alba PtrCAD1 (Van Acker et al., 2017), Arabidopsis AtCAD4 and AtCAD5 (Sibout et al., 2005; Anderson et al., 2015), Zea mays ZmCAD2 (Chen et al., 2012), and Sorghum bicolor SbCAD2 (Saballos et al., 2009; Sattler et al., 2009). The in vivo functions of all of these as CADs of the lignin synthesis pathway have been confirmed in expression studies of respective mutant and wild-type genes. Arabidopsis CHR was located in another clade with oxidoreductases of terpenoid biosynthesis, including Catharanthus roseus tetrahydroalstonin synthase (Stavrinides et al., 2015), C. roseus tabersonin 3-reductase (Qu et al., 2015), C. roseus 8-hydroxygeraniol dehydrogenase (Krithika et al., 2015), Ocimum basilicum geraniol dehydrogenase (Iijima et al., 2006), and Zingiber officinale geraniol dehydrogenase (Iijima et al., 2014), as proteins with biochemically confirmed functions. Populus tremuloides sinapyl alcohol dehydrogenase (Bomati and Noel, 2005) and AtCAD6 and AtCAD8 (Kim et al., 2004), which were thought to be involved in lignin biosynthesis, also were located within the clade; however, their in vivo functions have not been completely confirmed. For example, AtCAD8 knockout mutants showed no differences in lignin contents compared with those in the corresponding wild-type plants, even though recombinant AtCAD8 had slight activities against intermediates of the lignin biosynthetic pathway (Eudes et al., 2006). Accordingly, we propose that the genes in the CAD clade are true CADs and are involved in monolignol synthesis, whereas those encoding the ORSM clade are less involved in monoliginol synthesis but take primary roles as oxidoreductases that contribute to the biosynthesis of specialized metabolites. In further studies, TBLASTN searches were performed using CHR as a query with the whole-genome sequences of six representative plant species (Populus trichocarpa, Glycine max, tomato, Z. mays, O. sativa, and Marchantia polymorpha). Subsequent phylogenetic tree analysis of genes with BLAST scores higher than 250 also gave a tree with two clades, comprising ORSM family and true CAD family members (Supplemental Fig. S8). These observations imply that ancestral genes for oxidoreductases diversified functionally into specialized metabolism and lignin biosynthesis, warranting detailed studies of in vitro and in vivo functions of the present genes.
Figure 8.
Phylogenetic analysis of CADs and oxidoreductases involved in specialized metabolism (ORSMs). Proteins are listed with BLASTP analyses that were performed using Arabidopsis CHR as a query. Proteins with BLAST scores higher than 250 were chosen, and those studied at least at the protein level were selected to construct the tree (Supplemental Table S2). In vivo functions of the CADs in lignin synthesis have been characterized in mutant expression studies for the genes indicated by blue stars. The scale bar represents 0.09 amino acid substitutions per site.
Formerly, NADH-dependent ADH (EC 1.1.1.1) was thought to be involved in the reduction of (Z)-3-hexenal to (Z)-3-hexen-1-ol in the GLV biosynthetic pathway (Bicsak et al., 1982; Lai et al., 1982; Dolferus and Jacobs, 1984). Accordingly, several studies examined the effects of mutations or genetic manipulation of these ADH genes on volatiles. Overexpression or antisense suppression of SlADH2 in tomato led to higher or lower amounts of C6-alcohols, respectively, formed after complete tissue disruption (Speirs et al., 1998). Given that ADH from tomato fruits reduced C6-aldehydes in vitro (Bicsak et al., 1982) and that SlADH2 was highly expressed in fruits (Van Der Straeten et al., 1991), SlADH2 likely is involved in the reduction of C6-aldehydes, at least in completely disrupted tissue. GLVs that formed from completely disrupted leaves of Arabidopsis adh1 mutants showed reduced amounts (about 50% of that in the parental line) of C6-alcohols, and quantities of (E)-2-hexenal were approximately 10-fold higher than those detected in wild-type plants (Bate et al., 1998). Overexpression of VvADH2 in grapevine (Vitis vinifera) also resulted in increased 2-hexenal and hexan-1-ol contents of leaves that were disrupted through freeze/thaw treatment and subsequent buffer extraction, whereas antisense suppression did not affect the concentrations of these metabolites (Tesniere et al., 2006). These results suggest that NADH-dependent ADH is involved at least partly in the reduction of C6-aldehydes to C6-alcohols in totally disrupted plant tissues. As we reported previously, C6-aldehydes are formed mostly in disrupted tissues after the partial mechanical wounding of leaves and diffuse into neighboring intact tissues, where reduction proceeds to form C6-alcohols in an NADPH-dependent manner (Matsui et al., 2012). This study indicates that CHR is involved largely in the reduction of C6-aldehydes to C6-alcohols in partially wounded Arabidopsis leaf tissue. Accordingly, the reduction of C6-aldehydes into C6-alcohols in totally and partially disrupted Arabidopsis tissue is performed by different enzymes, such as NADH-dependent ADHs and NADPH-dependent CHR, respectively. Further studies are expected to extend these findings into other plant species.
GLVs are employed by some herbivores to find food and by some predators and parasitoids to find their prey (Matsui, 2006; Scala et al., 2013). Arthropod behaviors vary depending on each component of GLVs. For example, C. glomerata is a parasitoid of Pieris rapae larvae and was attracted by (Z)-3-hexen-1-yl acetate and (E)-2-hexenal but comparatively not by (Z)-3-hexen-1-ol (Shiojiri et al., 2006b). Higher (E)/(Z) ratios of GLVs formed by M. sexta-infested N. attenuata resulted in greater egg predation by Geocoris spp. (Allmann and Baldwin, 2010). (E)-2-Hexenal was more attractive to C. vestalis, a parasitoid of P. xylostella larvae, and (Z)-3-hexen-1-ol was favored under certain conditions (Yang et al., 2016). In this context, it is assumed that the adjustment of GLV composition is beneficial for plants, allowing the management of both beneficial and harmful arthropods. We demonstrated that CHR in Arabidopsis plants is involved in the adjustment of GLV composition and showed that (Z)-3-hexen-1-ol production in response to infestations of P. xylostella larvae was lower in knockout mutant (chr HPL) plants than in wild-type (CHR HPL) plants. The volatiles emitted from the P. xylostella-infested No-0 (CHR HPL) plants also were sufficient to attract C. vestalis. However, C. vestalis failed to distinguish between mutant (chr HPL) Arabidopsis plants that were infested with P. xylostella larvae and intact mutant plants. C. vestalis also failed to distinguish between infested and intact plants that lacked GLV formation (Col-0; CHR hpl). These observations indicate that (Z)-3-hexen-1-ol is required for C. vestalis to find its prey on Arabidopsis plants, suggesting that CHR plays a role in this Arabidopsis plant-herbivore-carnivore system.
We reported previously that exogenous (Z)-3-hexanal vapor was harmful to Arabidopsis plants (Matsui et al., 2012), likely because the aldehyde moiety is potently reactive and is readily oxidized to 4-hydroxy/oxo-(E)-2-hexenal, which is highly reactive with biological molecules (Farmer and Mueller, 2013). Here, we show that the exposure of mutant (chr hpl) Arabidopsis plants to (Z)-3-hexenal vapor led to significant malfunctions of photosynthesis, likely due to their inability to reduce (Z)-3-hexenal. Particularly high concentrations of (Z)-3-hexenal were used in the vapor phase in this study, specifically 26.7 and 40 nmol cm−3, at which a difference in susceptibility between the mutant (chr hpl) and Col-0 (CHR hpl) plants was observed. However, the local concentration of (Z)-3-hexenal in the liquid phase of disrupted tissues can reach up to approximately 1.5 µmol cm−3 (Matsui et al., 2012); therefore, intact leaf tissues adjacent to disrupted tissues should cope with high concentrations of (Z)-3-hexenal. Thus, the ability of CHR to reduce (Z)-3-hexenal is one of the systems required to avoid the toxicity of potentially harmful GLV-aldehydes, at least under the assay conditions employed in this study. Nevertheless, the ecophysiological significance of (Z)-3-hexenal-detoxification by CHR should be confirmed by further studies. These may include the direct determination of (Z)-3-hexenal concentration in intact leaf tissues adjacent to tissues disrupted by partial wounding, direct determination of the amount of (Z)-3-hexenal that is taken into intact Arabidopsis leaf tissues from the vapor phase, or investigation of the precise mechanism through which (Z)-3-hexenal has harmful effects on leaf tissues.
MATERIALS AND METHODS
Plant Materials
Arabidopsis (Arabidopsis thaliana) ecotypes Col-0 (CHR hpl) and No-0 (CHR HPL), as well as a T-DNA tagged mutant (chr hpl, background Col-0; Salk_001773), were obtained from the Arabidopsis Biological Resource Center. Because wild-type Col-0 has a deletion in its HPL gene and lacks the ability to form GLVs (Duan et al., 2005), a chr hpl mutant with the Col-0 background (Salk_001773) was crossed with No-0, and homozygotes with the T-DNA-disrupted CHR gene (chr mutant) and the active HPL gene (chr HPL) were isolated from the F2 population using genomic PCR analyses with the primers listed in Supplemental Table S1. Plants were germinated in soil (Tanemaki Baido; Takii Seeds) in plastic pots (6 cm i.d.), incubated at 4°C in the dark for 2 to 3 d, and then grown in a chamber at 22°C under fluorescent light (60 µmol m−2 s−1) with a 14-h-light/10-h-dark photoperiod.
Enzyme Assays
Hexenal reductase activity was monitored according to the oxidation of NADPH by continuously following decreases in absorption at 340 nm using a photometer (UV-160A; Shimadzu) at 25°C. The oxidation of NADH also was monitored using absorption at 340 nm. (Z)-3-Hexenal was dispersed in 1% (w/v) Tween 20 to a concentration of 40 mm using a tip-type ultrasonic disruptor (UD-211; Tomy Seiko). Suspensions were kept under nitrogen gas at −20°C until use. In typical assays, 10-µL aliquots of 40 mm (Z)-3-hexenal in 1% (w/v) Tween 20 were mixed with the enzyme in 50 mm Tris-HCl (pH 8; 1 mL total). Slight changes in background absorption at 340 nm likely reflected the instability of the aqueous emulsion and were recorded for 2 to 4 min prior to starting the reaction with the addition of 10-µL aliquots of 10 mm NADPH (Oriental Yeast). The aldehydes used for determinations of substrate specificity (Fig. 2) are listed in Supplemental Figure S9, and all except sinapaldehyde and coniferaldehyde were suspended in 1% (w/v) Tween 20 as 10 mm stock solutions. Sinapaldehyde and coniferaldehyde were dissolved in dimethyl sulfoxide to prepare 20 mm stock solutions. At 1% (v/v), dimethyl sulfoxide showed little effect on CHR activity with (Z)-3-hexenal. To estimate Km and Vmax values, S-V plots were constructed and analyzed using Hyper 32 (version 1.0.0).
Enzyme activities also were evaluated using GC-MS analyses of the products. Briefly, enzymes were placed in 22-mL glass vials (Perkin Elmer) with 10-µL aliquots of 20 mm (Z)-3-hexenal (suspended in 1% [w/v] Tween 20) and 20-µL aliquots of 10 mm NADPH in buffer containing 50 mm Tris-HCl (pH 8; total volume, 1 mL). Vials were then closed tightly with rubber septa, and mixtures were incubated at 28°C for 5 min. The reactions were terminated by adding 1 mL of saturated CaCl2 solution. Vials were then sealed tightly with a butyl stopper and a crimp-top seal (National Scientific), and SPME fibers (50/30-μm DVB/Carboxen/PDMS; Supelco) were exposed to headspaces of vials for 30 min at 60°C. Fibers were then inserted into the port of a GC-MS instrument (QP-5050; Shimadzu) equipped with a 0.25-mm × 30-m Stabiliwax column (film thickness, 0.25 µm; Restek). The column temperature was programmed as follows: 40°C for 1 min, increasing by 15°C min−1 to 180°C, and then 180°C for 1 min. The carrier gas (He) was delivered at a flow rate of 1 mL min−1. The glass insert was an SPME Sleeve (Supelco), and splitless injections were performed with a sampling time of 1 min. To remove all compounds from the matrix, the fiber was held in the injection port for 10 min. Injector and interface temperatures were 200°C and 230°C, respectively. The mass detector was operated in electron-impact mode with an ionization energy of 70 eV. Compounds were identified using retention indexes and MS profiles of corresponding authentic specimens, and quantitative analyses were performed with standard curves that were generated for each compound using aqueous suspensions containing Tween 20. Standard solutions were mixed at various ratios and then mixed with 1-mL aliquots of saturated CaCl2 solution in a glass vial. Volatiles were analyzed using SPME-GC-MS as described above, and calibration curves were constructed for each compound.
Purification of Hexenal Reductase
Leaves (350 g fresh weight) were harvested from Arabidopsis (No-0) plants that were grown for 40 to 60 d, frozen in liquid nitrogen, and then homogenized in 0.5 L of 0.1 m Tris-HCl (pH 8) containing 4 mm DTT and 1 mm phenylmethane sulfonyl fluoride using a juicer-mixer (TM837; TESCOM). Debris was removed by squeezing juices through two layers of bleached cotton cloth, and the remaining juice was centrifuged at 10,000 rpm (R12-2 rotor; Hitachi Koki) at 4°C for 20 min to afford crude extract as the supernatant. The crude extract was then fractionated in ammonium sulfate (40%–60% saturation) and dissolved in 30% saturated ammonium sulfate in buffer A containing 10 mm Tris-HCl (pH 8.0) and 4 mm DTT.
After removing insoluble materials by centrifuging at 10,000 rpm (T15A36 rotor; Hitachi Koki) at 4°C for 10 min, supernatants were applied to a Toyopearl Butyl 650M column (2.8 × 18 cm; Tosoh) equilibrated with 30% saturated ammonium sulfate in buffer A. The column was then washed with 0.2 L of equilibration buffer, and the enzyme was eluted using a linear gradient of 30% to 0% saturated ammonium sulfated in buffer A (total of 0.3 L). Fractions with substantial activity were combined, desalted using a PD-10 column (GE Healthcare Life Sciences), equilibrated with buffer A, and then applied to Bio-Scale Mini CHT 40-µm Cartridges (5 mL; Bio-Rad Laboratories) equilibrated with 10 mm potassium phosphate buffer (pH 8) containing 4 mm DTT. After washing the columns with 30 mL of the same buffer, enzymes were eluted with a linear gradient of 10 to 400 mm potassium phosphate buffer (pH 8) supplemented with 4 mm DTT.
Fractions with substantial activity were combined, concentrated using an Amicon Ultra-15 (Merck), and then desalted in a PD-10 column with buffer A. Active fractions were applied to HiTrap DEAE Sepharose Fast-Flow resin (1 mL; GE Healthcare Life Sciences) equilibrated with buffer A. After washing with 10 mL of buffer A, the enzyme was eluted with a linear gradient of 0 to 0.5 m NaCl in buffer A. Hexenal reductase activity was determined in every fraction, and SDS-PAGE analyses revealed a protein band that correlated well with enzyme activity (Fig. 1).
After purification, the protein band of 38 kD excised from the SDS-PAGE gel was digested in gel using trypsin. Trypsin-digested peptides were then analyzed using HPLC-MS/MS, and the protein was identified. LC-MS/MS analyses were performed using a linear ion trap time-of-flight mass spectrometer (NanoFrontier eLD; Hitachi High-Technologies) coupled to a nanoflow HPLC instrument (NanoFrontier nLC; Hitachi High-Technologies). Trypsin-digested peptides were then separated using a MonoCap C18 Fast-Flow column (0.05 × 150 mm; GL Science) and eluted with a linear gradient of 2% to 40% (v/v) solvent B over 60 min at a flow rate of 200 nL min−1. Solvent A was 2% (v/v) acetonitrile and 0.1% (v/v) formic acid, and solvent B was 98% (v/v) acetonitrile with 0.1% (v/v) formic acid. The eluent was ionized using a nano-electrospray ionization source equipped with an uncoated SilicaTip (New Objective) and was analyzed using a linear ion trap time-of-flight mass spectrometer. Mass spectra were obtained in positive ion mode at a scan mass-to-charge ratio (m/z) of 100 to 2,000. MS/MS spectra were generated by collision-induced dissociation in the linear ion trap. To identify the protein, MS and MS/MS data were converted to an MGF file using NanoFrontier eLD Data Processing software (Hitachi High-Technologies) and then were analyzed using the MASCOT MS/MS Ions Search with the following parameters: database, NCBInr; enzyme, trypsin; missed cleavages, three; taxonomy, all entries; fixed modifications, carbamidomethyl (C); variable modifications, oxidation (HW) and oxidation (M); peptide tolerance, 0.2 D; MS/MS tolerance, 0.2 D; peptide charge, 1+, 2+, and 3+; and instrument, ESI-TRAP.
cDNA Cloning
Total RNA was extracted from mature Arabidopsis (Col-0) leaves using FavorPrep Plant Total RNA Purification Mini Kits (Favorgen Biotech). DNA was degraded using DNA-free Kits (Ambion, Thermo Fisher Scientific), and cDNA was synthesized using SuperScript VILO cDNA Synthesis Kits (Invitrogen). Subsequently, CHR cDNA was PCR amplified using the primers 5′-acatATGGGAAAGGTTCTTGAGAAG-3′ and 5′-gaagcttAGGAGTTGGCTTCATCGTGT-3′, which were designed according to the sequence of the gene (At4g37980). The resulting PCR products were cloned into pGEM T-Easy vectors (Promega) for sequencing. The PCR product was then subcloned into the NdeI-HindIII site of the pET24a vector (Merck), and the resulting plasmid was transfected into Escherichia coli Rosetta2 cells (Merck). Cells were grown in Luria broth supplemented with kanamycin (50 µg mL−1) and chloramphenicol (30 µg mL−1) at 37°C until the optical density at 600 nm reached 0.6 to 0.8. After chilling cultures on ice for 15 min, isopropyl β-d-1-thiogalactosylpyranoside was added to a concentration of 0.5 mm, and cells were then cultured at 16°C for 16 h.
Cells were recovered by centrifugation at 5,000g for 5 min at 4°C and resuspended in 25 mm Tris-HCl (pH 7.5) containing 0.1% (v/v) 2-mercaptoethanol, 0.45 m KCl, 0.1 mm phenylmethane sulfonyl fluoride, and 50 µg mL−1 lysozyme. After incubating on ice for 15 min with intermittent swirling, suspended cells were disrupted with a tip-type ultrasonic disruptor (UD-211; Tomy Seiko). Lysates were then cleared with centrifugation at 10,000g for 10 min at 4°C and applied directly to a His-Accept (Nacalai Tesque) column preequilibrated with 25 mm Tris-HCl (pH 7.5) containing 0.1% (v/v) 2-mercaptoethanol and 0.45 m KCl. After washing the column in the same buffer, the recombinant Arabidopsis CHR enzyme was eluted with the same buffer supplemented with 50 mm imidazole.
Volatiles from Leaves
Aboveground parts of 37-d-old Arabidopsis plants with fully developed leaves were cut off with a razor blade, weighed, and then carefully placed in glass jars (200 mL, 63 mm i.d. × 70 mm). Cut surfaces were immersed in distilled water in microtube caps to avoid desiccation. The leaves were left intact or were partially (12.5% of total leaf area) wounded using forceps without injuring the midvein. The jar was then closed tightly, and an SPME fiber (50/30-µm DVB/Carboxen/PDMS; Supelco) was exposed to the headspace of the jar for 30 min at 25°C. Volatiles collected on the fiber were analyzed using GC-MS under the conditions used in the enzyme assays. Molecular ion chromatograms with m/z of 56 (for n-hexanal and n-hexan-1-ol), 69 [for (Z)-3-hexenal], and 82 [for (Z)-3-hexen-1-yl acetate, (Z)-3-hexen-1-ol, and (E)-2-hexen-1-ol] were used for quantification. Calibration curves were constructed from GC-MS analyses under the same conditions by placing known amounts of authentic compounds (suspended in 0.1% [w/v] Tween 20) onto the tips of cotton swabs in the glass jar. To analyze volatiles formed in thoroughly disrupted leaf tissues, leaves (about 60 mg fresh weight) were homogenized in 1-mL aliquots of 50 mm MES-KOH (pH 5.5) for 1 min using a mortar and pestle. To determine volatiles in intact leaves, buffer was replaced with solution containing 1 g mL−1 CaCl2 to arrest enzyme activities. Homogenates then were transferred into tubes by rinsing the mortar with 0.5-mL aliquots of the same buffer twice. Volatile compounds in homogenates were extracted with 2 mL of methyl tert-butyl ether containing 1 µg mL−1 nonanyl acetate. GC-MS analyses then were performed as described above.
To collect volatiles, three pots with CHR HPL (No-0) and chr HPL plants that had been infested for 24 h with Plutella xylostella larvae were placed in a glass jar (8.5-cm diameter × 11 cm) with a tight cover, and SPME fibers were exposed to headspaces for 30 min at 25°C. Volatiles then were analyzed using GC-MS under the conditions described above, but in the selected ion monitoring mode with m/z 69.1 [C6H10O-CHO]+ and 82.1 [C6H12O-H2O/C8H14O2-C2H4O2]+.
Flight Responses of Cotesia vestalis
P. xylostella larvae were collected from crucifer crops in a field in Kyoto City, Japan, and were reared in a climate-controlled room (25°C ± 2°C and 50%–70% relative humidity) with a 16-h photoperiod on leaves of Komatsuna (Brassica rapa var perviridis). C. vestalis from parasitized host larvae were collected from a field in Kyoto, Japan. Adult parasitoids were fed honey and maintained in plastic cages (25 × 35 × 30 cm) in a climate-controlled room (25°C ± 2°C and 50%–70% relative humidity) with a 16-h photoperiod for 3 d to ensure mating. Females were then transferred individually to glass tubes of 20 mm diameter and 130 mm length containing honey and were kept in a climate-controlled room (18°C ± 2°C and 50%–70% relative humidity) under continuous darkness. Females were never kept for more than 10 d. At least 1 h before the start of each experiment, oviposition-inexperienced female wasps were transferred to another climate-controlled room (25°C ± 2°C and 50°C–70% relative humidity) and exposed to continuous light.
Two groups of three pots containing 4- to 5-week-old nonflowering Arabidopsis plants with fully developed leaves were positioned about 25 cm apart in a cage (25 × 35 × 30 cm) with three windows covered by a nylon gauze and a door for introducing plants and wasps. To determine the effects of P. xylostella infestation on CHR HPL (No-0), CHR hpl (Col-0), and chr hpl Arabidopsis plants, five second-instar larvae of P. xylostella were placed onto one group (three pots) of plants. After 24 h, larvae and feces were removed and the cleaned plants were used as infested plants. The other group of plants was left for 24 h without larvae. Ten wasps then were released halfway between the two groups of plants, and the first landing by each wasp on a plant was recorded as its choice of the two groups. Upon landing on a plant, wasps were removed immediately from the cage using an insect aspirator. If the wasp did not land on any of the plants within 30 min, it was recorded as no-choice. The trial was repeated four times, and plants were replaced for each trial. Preferences of female C. vestalis for volatiles emitted from CHR HPL (No-0) and chr HPL that had been infested with P. xylostella for 24 h also were assessed.
Susceptibility to (Z)-3-Hexenal Vapor
Maximum quantum yields of PSII were estimated from chlorophyll fluorescence measurements using a pulse amplitude modulated (PAM) fluorometer (Mini-PAM photosynthesis yield analyzer; Walz). PSII yields were calculated as Fv/Fm (Maxwell and Johnson, 2000). The saturation pulse duration was 0.8 s and was applied with an intensity of approximately 8,300 mmol m−2 s−1. Four plants (Col-0 and chr hpl, 39 d old) were placed inside a glass cylinder (2,800 cm3), and 112-µL aliquots of CH2Cl2 containing 0.33, 0.67, or 1 m (Z)-3-hexenal were applied to cotton swabs that were placed 5 cm above the plant canopies. The flask then was sealed tightly with a glass plate and was incubated at 22°C in the light (60 µmol m−2 s−1). For each plant, fluorescence measurements were conducted at the midpoint of four fully matured leaves immediately before treatment or at 4 or 17.5 h after the onset of treatment. The plants were dark acclimated for at least 15 min before pulse amplitude modulation analysis.
Phylogenetic Analysis
Amino acid sequences of selected proteins were aligned initially using the multiple sequence alignment program MUSCLE (Edgar, 2004) at EMBL-EBI. The resulting alignments were inspected and edited manually to remove gaps and unconserved regions using the alignment editing program AliView 1.21 (Larsson, 2014). Phylogenetic trees were constructed using the phylogeny analysis program MrBayes 3.2.6 (Huelsenbeck and Ronquist, 2001) under the WAG model.
Accession Number
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number NP_195511.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Amino acid sequences as determined using MASCOT.
Supplemental Figure S2. SDS-PAGE analysis of recombinant Arabidopsis CHR protein with a His tag following isolation from E. coli.
Supplemental Figure S3. Substrate specificity of CHR purified from Arabidopsis leaves.
Supplemental Figure S4. S-V plots of recombinant Arabidopsis CHR with (Z)-3-hexenal, n-heptanal, and cinnamaldehyde.
Supplemental Figure S5. CHR and HPL loci and characterization of wild-type and mutant plants.
Supplemental Figure S6. Formation of GLVs after complete disruption of Arabidopsis leaves.
Supplemental Figure S7. Consumption of leaves by P. xylostella larvae.
Supplemental Figure S8. Phylogenetic tree of Arabidopsis CHR and related genes.
Supplemental Figure S9. Carbonyls used as substrates for enzyme assays.
Supplemental Table S1. Primer sequences used for genotyping.
Supplemental Table S2. List of genes used for constructing the phylogenetic tree in Figure 8.
Dive Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
In this research, we used the supercomputer of ACCMS, Kyoto University, for phylogenetic analyses. We thank Brian St. Aubin at Michigan State University for English proofreading.
Footnotes
This work was supported partly by a grant from the Japan Society for the Promotion of Science (KAKENHI 16H03283; to K.M.).
Articles can be viewed without a subscription.
References
- Allmann S, Baldwin IT (2010) Insects betray themselves in nature to predators by rapid isomerization of green leaf volatiles. Science 329: 1075–1078 [DOI] [PubMed] [Google Scholar]
- Ameye M, Allmann S, Verwaeren J, Smagghe G, Haesaert G, Schuurink RC, Audenaert K (2017) Green leaf volatile production by plants: a meta-analysis. New Phytol (in press) 10.1111/nph.14671 [DOI] [PubMed] [Google Scholar]
- Anderson NA, Tobimatsu Y, Ciesielski PN, Ximenes E, Ralph J, Donohoe BS, Ladisch M, Chapple C (2015) Manipulation of guaiacyl and syringyl monomer biosynthesis in an Arabidopsis cinnamyl alcohol dehydrogenase mutant results in atypical lignin biosynthesis and modified cell wall structure. Plant Cell 27: 2195–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bate NJ, Riley JCM, Thompson JE, Rothstein SJ (1998) Quantitative and qualitative differences in C6-volatile production from the lipoxygenase pathway in an alcohol dehydrogenase mutant of Arabidopsis thaliana. Physiol Plant 104: 97–104 [Google Scholar]
- Bicsak TA, Kann LR, Reiter A, Chase T Jr (1982) Tomato alcohol dehydrogenase: purification and substrate specificity. Arch Biochem Biophys 216: 605–615 [DOI] [PubMed] [Google Scholar]
- Bomati EK, Noel JP (2005) Structural and kinetic basis for substrate selectivity in Populus tremuloides sinapyl alcohol dehydrogenase. Plant Cell 17: 1598–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, VanOpdorp N, Fitzl D, Tewari J, Friedemann P, Greene T, Thompson S, Kumpatla S, Zheng P (2012) Transposon insertion in a cinnamyl alcohol dehydrogenase gene is responsible for a brown midrib1 mutation in maize. Plant Mol Biol 80: 289–297 [DOI] [PubMed] [Google Scholar]
- D’Auria JC, Pichersky E, Schaub A, Hansel A, Gershenzon J (2007) Characterization of a BAHD acyltransferase responsible for producing the green leaf volatile (Z)-3-hexen-1-yl acetate in Arabidopsis thaliana. Plant J 49: 194–207 [DOI] [PubMed] [Google Scholar]
- Dolferus R, Jacobs M (1984) Polymorphism of alcohol dehydrogenase in Arabidopsis thaliana (L.) Heynh.: genetical and biochemical characterization. Biochem Genet 22: 817–838 [DOI] [PubMed] [Google Scholar]
- Duan H, Huang MY, Palacio K, Schuler MA (2005) Variations in CYP74B2 (hydroperoxide lyase) gene expression differentially affect hexenal signaling in the Columbia and Landsberg erecta ecotypes of Arabidopsis. Plant Physiol 139: 1529–1544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva N, Klempien A, Muhlemann JK, Kaplan I (2013) Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol 198: 16–32 [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eudes A, Pollet B, Sibout R, Do CT, Séguin A, Lapierre C, Jouanin L (2006) Evidence for a role of AtCAD 1 in lignification of elongating stems of Arabidopsis thaliana. Planta 225: 23–39 [DOI] [PubMed] [Google Scholar]
- Farmer EE, Mueller MJ (2013) ROS-mediated lipid peroxidation and RES-activated signaling. Annu Rev Plant Biol 64: 429–450 [DOI] [PubMed] [Google Scholar]
- Hatanaka A. (1993) The biogeneration of green odour by green leaves. Phytochemistry 34: 1201–1218 [Google Scholar]
- Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755 [DOI] [PubMed] [Google Scholar]
- Iijima Y, Wang G, Fridman E, Pichersky E (2006) Analysis of the enzymatic formation of citral in the glands of sweet basil. Arch Biochem Biophys 448: 141–149 [DOI] [PubMed] [Google Scholar]
- Iijima Y, Koeduka T, Suzuki H, Kubota K (2014) Biosynthesis of geranial, a potent aroma compound in ginger rhizome (Zingiber officinale): molecular cloning and characterization of geraniol dehydrogenase. Plant Biotechnol 31: 525–534 [Google Scholar]
- Joo Y, Schuman MC, Goldberg JK, Kim SG, Yon F, Brütting C, Baldwin IT (2018) Herbivore-induced volatile blends with both “fast” and “slow” components provide robust indirect defence in nature. Funct Ecol 32: 136–149 [Google Scholar]
- Kessler A, Baldwin IT (2001) Defensive function of herbivore-induced plant volatile emissions in nature. Science 291: 2141–2144 [DOI] [PubMed] [Google Scholar]
- Kiedrowski S, Kawalleck P, Hahlbrock K, Somssich IE, Dangl JL (1992) Rapid activation of a novel plant defense gene is strictly dependent on the Arabidopsis RPM1 disease resistance locus. EMBO J 11: 4677–4684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, Kim MR, Bedgar DL, Moinuddin SGA, Cardenas CL, Davin LB, Kang C, Lewis NG (2004) Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. Proc Natl Acad Sci USA 101: 1455–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishimoto K, Matsui K, Ozawa R, Takabayashi J (2008) Direct fungicidal activities of C6-aldehydes are important constituents for defense responses in Arabidopsis against Botrytis cinerea. Phytochemistry 69: 2127–2132 [DOI] [PubMed] [Google Scholar]
- Kost C, Heil M (2006) Herbivore-induced plant volatiles induce an indirect defence in neighbouring plants. J Ecol 94: 619–628 [Google Scholar]
- Krithika R, Srivastava PL, Rani B, Kolet SP, Chopade M, Soniya M, Thulasiram HV (2015) Characterization of 10-hydroxygeraniol dehydrogenase from Catharanthus roseus reveals cascaded enzymatic activity in iridoid biosynthesis. Sci Rep 5: 8258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunishima M, Yamauchi Y, Mizutani M, Kuse M, Takikawa H, Sugimoto Y (2016) Identification of (Z)-3:(E)-2-hexenal isomerases essential to the production of the leaf aldehyde in plants. J Biol Chem 291: 14023–14033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai YK, Chandlee JM, Scandalios JG (1982) Purification and characterization of three non-allelic alcohol dehydrogenase isozymes in maize. Biochim Biophys Acta 706: 9–18 [DOI] [PubMed] [Google Scholar]
- Larsson A. (2014) AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30: 3276–3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui K. (2006) Green leaf volatiles: hydroperoxide lyase pathway of oxylipin metabolism. Curr Opin Plant Biol 9: 274–280 [DOI] [PubMed] [Google Scholar]
- Matsui K, Sugimoto K, Mano J, Ozawa R, Takabayashi J (2012) Differential metabolisms of green leaf volatiles in injured and intact parts of a wounded leaf meet distinct ecophysiological requirements. PLoS ONE 7: e36433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell K, Johnson GN (2000) Chlorophyll fluorescence: a practical guide. J Exp Bot 51: 659–668 [DOI] [PubMed] [Google Scholar]
- Mochizuki S, Sugimoto K, Koeduka T, Matsui K (2016) Arabidopsis lipoxygenase 2 is essential for formation of green leaf volatiles and five-carbon volatiles. FEBS Lett 590: 1017–1027 [DOI] [PubMed] [Google Scholar]
- Mwenda CM, Matsui K (2014) The importance of lipoxygenase control in the production of green leaf volatiles by lipase-dependent and independent pathway. Plant Biotechnol 31: 445–452 [Google Scholar]
- Nakamura S, Hatanaka A (2002) Green-leaf-derived C6-aroma compounds with potent antibacterial action that act on both Gram-negative and Gram-positive bacteria. J Agric Food Chem 50: 7639–7644 [DOI] [PubMed] [Google Scholar]
- Pierik R, Ballaré CL, Dicke M (2014) Ecology of plant volatiles: taking a plant community perspective. Plant Cell Environ 37: 1845–1853 [DOI] [PubMed] [Google Scholar]
- Pospíšil P, Yamamoto Y (2017) Damage to photosystem II by lipid peroxidation products. Biochim Biophys Acta 1861: 457–466 [DOI] [PubMed] [Google Scholar]
- Prost I, Dhondt S, Rothe G, Vicente J, Rodriguez MJ, Kift N, Carbonne F, Griffiths G, Esquerré-Tugayé MT, Rosahl S, et al. (2005) Evaluation of the antimicrobial activities of plant oxylipins supports their involvement in defense against pathogens. Plant Physiol 139: 1902–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Y, Easson MLAE, Froese J, Simionescu R, Hudlicky T, De Luca V (2015) Completion of the seven-step pathway from tabersonine to the anticancer drug precursor vindoline and its assembly in yeast. Proc Natl Acad Sci USA 112: 6224–6229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saballos A, Ejeta G, Sanchez E, Kang C, Vermerris W (2009) A genomewide analysis of the cinnamyl alcohol dehydrogenase family in sorghum [Sorghum bicolor (L.) Moench] identifies SbCAD2 as the brown midrib6 gene. Genetics 181: 783–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sattler SE, Saathoff AJ, Haas EJ, Palmer NA, Funnell-Harris DL, Sarath G, Pedersen JF (2009) A nonsense mutation in a cinnamyl alcohol dehydrogenase gene is responsible for the sorghum brown midrib6 phenotype. Plant Physiol 150: 584–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scala A, Allmann S, Mirabella R, Haring MA, Schuurink RC (2013) Green leaf volatiles: a plant’s multifunctional weapon against herbivores and pathogens. Int J Mol Sci 14: 17781–17811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiojiri K, Kishimoto K, Ozawa R, Kugimiya S, Urashimo S, Arimura G, Horiuchi J, Nishioka T, Matsui K, Takabayashi J (2006a) Changing green leaf volatile biosynthesis in plants: an approach for improving plant resistance against both herbivores and pathogens. Proc Natl Acad Sci USA 103: 16672–16676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiojiri K, Ozawa R, Matsui K, Kishimoto K, Kugimiya S, Takabayashi J (2006b) Role of the lipoxygenase/lyase pathway of host-food plants in the host searching behavior of two parasitoid species, Cotesia glomerata and Cotesia plutellae. J Chem Ecol 32: 969–979 [DOI] [PubMed] [Google Scholar]
- Sibout R, Eudes A, Pollet B, Goujon T, Mila I, Granier F, Séguin A, Lapierre C, Jouanin L (2003) Expression pattern of two paralogs encoding cinnamyl alcohol dehydrogenases in Arabidopsis: isolation and characterization of the corresponding mutants. Plant Physiol 132: 848–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibout R, Eudes A, Mouille G, Pollet B, Lapierre C, Jouanin L, Séguin A (2005) CINNAMYL ALCOHOL DEHYDROGENASE-C and -D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. Plant Cell 17: 2059–2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speirs J, Lee E, Holt K, Yong-Duk K, Steele SN, Loveys B, Schuch W (1998) Genetic manipulation of alcohol dehydrogenase levels in ripening tomato fruit affects the balance of some flavor aldehydes and alcohols. Plant Physiol 117: 1047–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spyropoulou EA, Dekker HL, Steemers L, van Maarseveen JH, de Koster CG, Haring MA, Schuurink RC, Allmann S (2017) Identification and characterization of (3Z):(2E)-hexenal isomerases from cucumber. Front Plant Sci 8: 1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavrinides A, Tatsis EC, Foureau E, Caputi L, Kellner F, Courdavault V, O’Connor SE (2015) Unlocking the diversity of alkaloids in Catharanthus roseus: nuclear localization suggests metabolic channeling in secondary metabolism. Chem Biol 22: 336–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Matsui K, Iijima Y, Akakabe Y, Muramoto S, Ozawa R, Uefune M, Sasaki R, Alamgir KM, Akitake S, et al. (2014) Intake and transformation to a glycoside of (Z)-3-hexenol from infested neighbors reveals a mode of plant odor reception and defense. Proc Natl Acad Sci USA 111: 7144–7149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesniere C, Torregrosa L, Pradal M, Souquet JM, Gilles C, Dos Santos K, Chatelet P, Gunata Z (2006) Effects of genetic manipulation of alcohol dehydrogenase levels on the response to stress and the synthesis of secondary metabolites in grapevine leaves. J Exp Bot 57: 91–99 [DOI] [PubMed] [Google Scholar]
- Van Acker R, Déjardin A, Desmet S, Hoengenaert L, Vanholme R, Morreel K, Laurans F, Kim H, Santoro N, Foster C, et al. (2017) Different routes for conifer- and sinapaldehyde and higher saccharification upon deficiency in the dehydrogenase CAD1. Plant Physiol 175: 1018–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Straeten D, Pousada RAR, Gielen J, Van Montagu M (1991) Tomato alcohol dehydrogenase: expression during fruit ripening and under hypoxic conditions. FEBS Lett 295: 39–42 [DOI] [PubMed] [Google Scholar]
- Wei J, Kang L (2011) Roles of (Z)-3-hexenol in plant-insect interactions. Plant Signal Behav 6: 369–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Zhang YN, Gurr GM, Vasseur L, You MS (2016) Electroantennogram and behavioral responses of Cotesia plutellae to plant volatiles. Insect Sci 23: 245–252 [DOI] [PubMed] [Google Scholar]
- Zhang K, Qian Q, Huang Z, Wang Y, Li M, Hong L, Zeng D, Gu M, Chu C, Cheng Z (2006) GOLD HULL AND INTERNODE2 encodes a primarily multifunctional cinnamyl-alcohol dehydrogenase in rice. Plant Physiol 140: 972–983 [DOI] [PMC free article] [PubMed] [Google Scholar]