Summary
X-linked hyper-IgM syndrome (XHIM) is a primary immunodeficiency due to mutations in CD40 ligand that affect immunoglobulin class switch recombination and somatic hypermutation. The disease is amenable to gene therapy using retroviral vectors, but dysregulated gene expression resulted in abnormal lymphoproliferation in mouse models, highlighting the need for alternative strategies. Here, we demonstrate the ability of both the TALEN and CRISPR/Cas9 platforms to efficiently drive integration of a normal copy of the CD40L cDNA delivered by Adeno-Associated Virus. Site-specific insertion of the donor sequence downstream of the endogenous CD40L promoter maintained physiologic expression of CD40L while overriding all reported downstream mutations. High levels of gene modification were achieved in primary human hematopoietic stem cells (HSC) as well as in cell lines and XHIM patient-derived T cells. Notably, gene corrected HSC engrafted in immunodeficient mice at clinically-relevant frequencies. These studies provide the foundation for a permanent curative therapy in XHIM.
Introduction
X-linked hyper-IgM syndrome (XHIM) is a primary immunodeficiency characterized by the absence of IgG, IgA, and IgE with normal to elevated IgM caused by defects in the CD40LG gene that encodes CD40 ligand (CD40L) expressed on the surface of activated T lymphocytes. CD40L binds to CD40 on B lymphocytes and is essential in the interaction between T and B cells that induces class switch recombination of the immunoglobulin heavy chain gene. XHIM affected individuals are profoundly susceptible to bacterial and opportunistic infections with a propensity for autoimmunity and malignancies.(Hayward et al., 1997; Levy et al., 1997)
Long-term prognosis for XHIM remains poor with a median survival time of approximately 25 years from diagnosis.(de la Morena et al., 2017) Immunologic reconstitution with allogeneic hematopoietic stem cell transplant (HSCT) is a definitive treatment but has been associated with a high incidence of acute graft-versus-host disease (GvHD), hepatic sinusoidal obstruction syndrome, exacerbation of infections, and death.(Gennery et al., 2014; Mitsui-Sekinaka et al., 2015) Despite the curative nature of HSCT, there is no overall difference in survival between patients treated with or without HSCT, although those who undergo transplantation report improved general well-being as measured by Karnofsky/Lansky scores.(de la Morena et al., 2017) Thus, there is a clear need for the development of alternative approaches to permanently and safely restore immune competence in XHIM. Gene therapy has been shown to be effective in other primary immune deficiencies (ADA SCID, X-linked SCID, chronic granulomatous disease, Wiskott-Aldrich syndrome), and novel platforms for gene editing including TALENs and CRISPR/Cas9 have shown tremendous promise.
In mouse models of XHIM, gene therapy through retroviral vectors corrects the immune defect but unregulated expression of the gene results in abnormal lymphoproliferation, reflecting the need for close control of CD40LG.(Brown et al., 1998; Sacco et al., 2000) In addition, viral vectors that can express the CD40L gene in a physiologically regulated manner have not been achieved in vivo.(Romero et al., 2011) Retroviral vectors also pose a significant risk of insertional oncogenesis, emphasizing the need for other therapeutic options.
Targeted gene correction and in-frame-targeted gene addition that utilizes the endogenous promoter represents a major advancement in gene therapy. The discovery of meganucleases in the mid-1990’s (e.g. Sce-I) marked the beginning of using endonucleases that could create double-strand breaks and promote homologous recombination by almost five orders of magnitude.(Epinat et al., 2003; Porteus and Baltimore, 2003) In the last decade, there has been a bloom in gene editing technologies with Zinc Finger Nucleases, Transcription Activator-Like Effector Nucleases (TALENs), and the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-associated protein 9 (Cas9). One such approach of gene editing was recently shown to be feasible in XHIM using TALENs to site-specifically insert a normal copy of the CD40L cDNA in human T cells.(Hubbard et al., 2016)
With its ease of use and nearly limitless DNA sequences that can be targeted, CRISPR/Cas9 is emerging as the preferred approach for gene editing.(Cong et al., 2013) The technology originated from a bacterial immune surveillance system and continues to evolve and transform the field of genome engineering. There have been a few studies demonstrating CRISPR/Cas9 to be effective in PID but none have been in XHIM. Here we demonstrate an approach using both the TALEN and CRISPR/Cas9 platforms to allow site-specific incorporation of a human codon-divergent CD40L cDNA at the 5’UTR of the gene in both primary patient T lymphocytes and human CD34+ HSC, bypassing all known downstream disease-causing mutations and maintaining expression of the therapeutic gene under control of the endogenous CD40L promotor. In contrast to previous work using TALENs in differentiated T cells(Hubbard et al., 2016), we focused on HSC-based genome-editing strategies which can provide permanent and robust immune reconstitution.
Results
Validation of TALEN-Mediated Gene Modification at CD40L in Cell Lines and Patient-Derived T Lymphocytes
For our first set of experiments, several TALEN pairs were designed to cleave within the 5’UTR of the CD40LG gene (Fig. S1A). In K562 cells, allelic disruption rates averaged 32 ± 3% as measured by Surveyor nuclease assay (CEL I) (Fig. S1B). When a donor template encoding a promoterless green fluorescent protein (GFP) reporter flanked by homology sequences that parallel the TALEN cut site was co-electroporated, ‘In-Out’ PCR demonstrated targeted GFP integrants (Fig. S1C-D). Introduction of TALEN expression plasmids and the GFP donor to Jurkat T cells, a CD40L-expressing T cell leukemia line, achieved up to 12% overall GFP expression, demonstrating permanent and stable gene integration (Fig. S1E). Incubation of treated cells with phytohemagglutinin (PHA) to stimulate lymphocyte proliferation and increase CD40L expression upregulated GFP expression in a dose dependent manner, suggesting that the GFP cassette was integrated under control of the endogenous CD40LG promoter (Fig S1F-G).
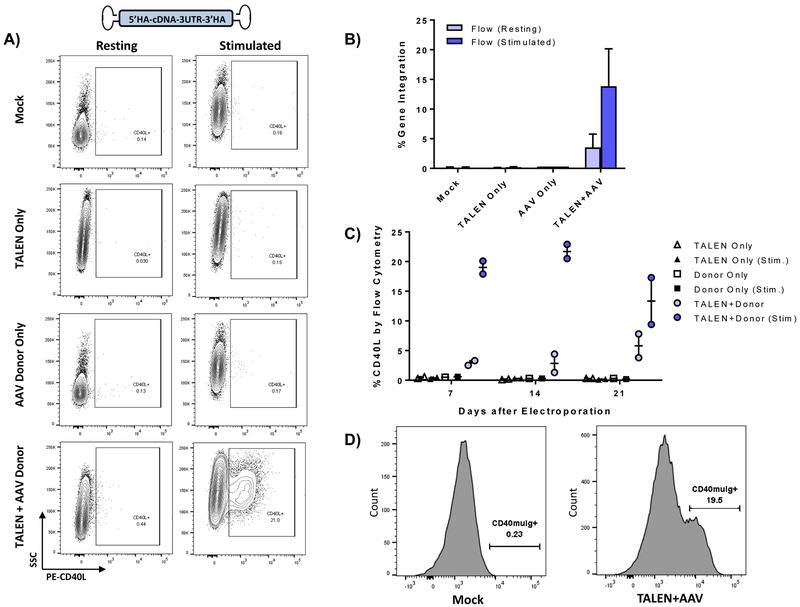
Following demonstration of targeted integration at CD40LG in cell lines, CD4+ T cells derived from XHIM patients were electroporated with TALEN mRNA and transduced with either an integrase-deficient lentivirus (IDLV) or adeno-associated virus serotype 6 (AAV6) vector containing a corrective, codon-divergent hCD40L cDNA cassette flanked by homology arms. As expected, despite high transduction of primary T cells by a GFP IDLV vector (Fig. S2A), XHIM T lymphocytes treated with both TALEN mRNA and the corrective cDNA IDLV (MOI 100) expressed only minimal (<1%) CD40L expression by flow cytometry (Fig. S2B). Exon-spanning PCR utilizing a reverse primer overlying two adjacent codons in the donor cDNA cassette demonstrated integration through gel electrophoresis, and targeted integrants were quantified at rates of <0.5% by digital droplet PCR (ddPCR) (Fig. S2C-D). In contrast, XHIM T cells transduced with an identical cDNA donor packaged as a recombinant AAV6 vector following TALEN mRNA electroporation expressed low levels of CD40L at baseline, with upregulation to >20% CD40L expression upon anti-hCD3/anti-hCD28 immune stimulation, comparable to CD40L expression in stimulated T cells from healthy donors (24.2 ± 4.2%) (Fig. 1A-B). Viability and fold expansion of treated T cells as measured by trypan blue was similar in control and treatment groups (Fig. S3A-B). Restoration of CD40L was dose dependent with increasing AAV6 MOI without significantly affecting viability and fold expansion. (Fig. S3C-E) Furthermore, corrected XHIM T cells demonstrated physiologic activation patterns to immune stimuation (Fig. 1C) and normal receptor-binding activity to recombinant chimeric CD40-muIg (Fig. 1D), a functional assessment of CD40L that identifies all patients with defective CD40L in the clinical setting.(Abraham and Aubert, 2016)
Figure 1. Targeted Integration in XHIM T cells of the CD40L cDNA donor delivered by an adeno associated virus (AAV6).
A) Primary XHIM patient CD4+ T-cells were electroporated with TALEN mRNA and transduced with an AAV6 codon-divergent CD40L cDNA donor. Expression of CD40L was measured by flow cytometry in resting T cells and after stimulation with anti-hCD3/anti-hCD28 microbeads. B) Average gene modification rates as measured by flow cytometry with and without stimulation. Data are presented as mean ± SD. n=8–10 biological replicates, 2 XHIM donors. C) CD40L expression trends by flow cytometry in XHIM T cells electroporated with TALEN and AAV donor and re-stimulated over time in culture. D) CD40L function was assessed by binding to a fluorescent-labeled chimeric CD40-muIg and flow cytometry. Data in C were analyzed by Wilcoxon Rank-Sum Test. NS = not significant. See also Figure S3.
CRISPR-Cas9 Mediated Gene Correction at CD40LG in XHIM Patient-Derived T lymphocytes
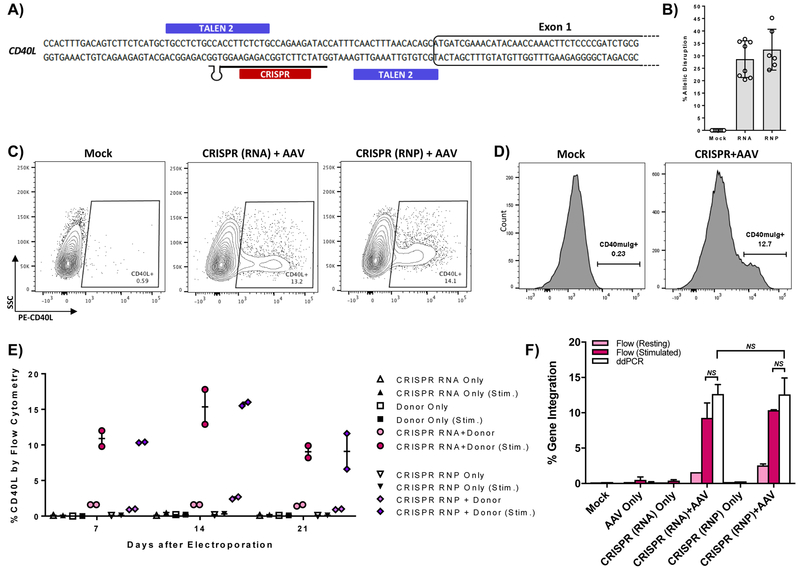
We next evaluated the efficiency of CRISPR mediated gene editing in XHIM primary T cells using a guide RNA (gRNA) also targeting the 5’ UTR of CD40LG (Fig. 2A). CRISPR reagents were delivered as gRNA pre-complexed to Cas9 protein as ribonucleoproteins (RNP) or as two RNA components (gRNA and Cas9 mRNA) to compare their efficacy in primary T cells. Allelic disruption rates in XHIM T cells averaged 30.6% and 33.7% when treated with gRNA and Cas9 mRNA or RNP, respectively. (Fig. 2B) AAV6 cDNA donor transduction following gRNA/Cas9 electroporation resulted in restoration of CD40L expression, with no differences using either form of Cas9 (Fig. 2C).
Figure 2. CRISPR/Cas9 mediated gene editing of the CD40L gene in XHIM T cells.
A) CRISPR gRNA binding site in the 5’UTR of the CD40L gene. B) Allelic disruption rates in XHIM CD4+ T cells using gRNA with Cas9 mRNA or complexed to recombinant Cas9 protein as RNP were measured by Surveyor nuclease assay (n=6–10 biological replicates in primary T cells from 4 different XHIM patients). C) Representative flow cytometry plots of gene-modified XHIM T cells after stimulation with anti-CD3/CD28 beads. D) CD40-muIg binding by CRISPR/Cas9 modified XHIM T cells to assess CD40L protein function. E) Expression of CD40L by flow cytometry in XHIM T cells treated with CRISPR/Cas9 RNA or RNP and AAV donor and re-stimulated over time in culture. F) Average gene integration rates in primary XHIM T cells measured by flow cytometry and ddPCR (n=2–4 biological replicates, 2 XHIM T cell donors). ddPCR was designed with a forward primer upstream of the 5’ homology arm, a reverse primer spanning exons 1 & 2, and a probe specific for the codon-optimized exon 1. Data in F were analyzed by Wilcoxon Rank-Sum Test. NS = not significant. See also Figure S3.
Modified T cells showed restored CD40L function as measured by receptor-binding to chimeric CD40-muIg by flow cytometry (Fig. 2D). Additionally, when cells were rested in culture and analyzed at one week intervals, CD40L expression consistently returned to baseline levels and responded physiologically to re-stimulation for weeks after treatment (Fig. 2E). In a separate set of experiments, cDNA integrants were directly quantified through ddPCR. The observed close correlation between gene modification rates determined by ddPCR and the levels of CD40L expression by flow cytometry indicated that ddPCR analysis could be used as a surrogate measurement for CD40L gene correction and expression in experiments utilizing healthy donor cells (Fig. 2F).
TALEN- and CRISPR/Cas9- Mediated Gene Correction of CD40LG in Mobilized Peripheral Blood CD34+ Cells
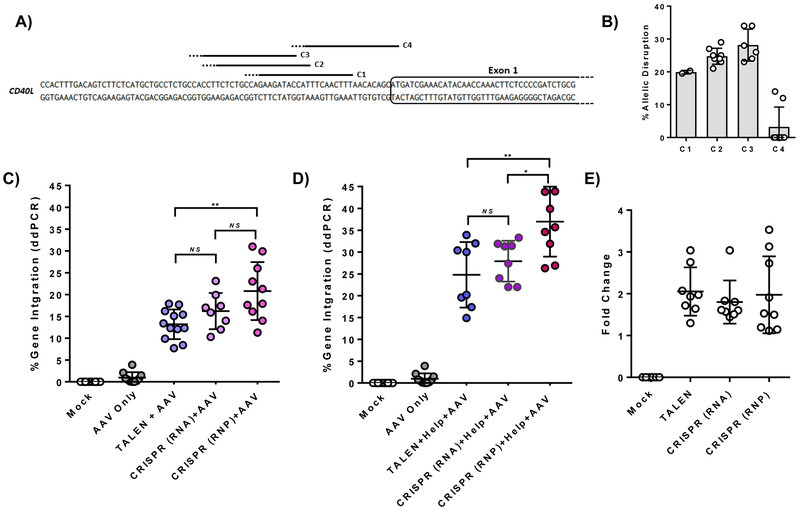
The potential for site-specific gene addition to be a curative therapy in XHIM through autologous transplantation was evaluated with both platforms using human granulocyte-colony stimulating factor (G-CSF) mobilized peripheral blood stem cells (PBSC). Electroporation of peripheral blood CD34+ cells with TALEN mRNA resulted in allelic disruption rates of 29.1 ± 7.8% (n=8 biological replicates, 4 PBSC donors). Multiple CRISPR gRNA targeting the 5’UTR were also evaluated in PBSC (Fig. 3A). Allelic disruption rates in HSC averaged 33% with CRISPR 3 (C3), which was used in all further CRISPR based experiments (Fig. 3B).
Figure 3. TALEN and CRISPR/Cas9 editing of the CD40L gene in primary human CD34+ peripheral blood stem cells (PBSC).
A) Binding sites and B) allelic disruption rates measured by Surveyor nuclease assay of four CRISPR gRNA (C1, C2, C3, and C4) targeting the CD40L 5’UTR in PBSC (n=8, 4 PBSC donor sources). Solid line indicates gRNA sequence; dotted line indicates PAM sequence. C) Gene integration rates in PBSC treated with TALEN mRNA vs. CRISPR gRNA/Cas9 mRNA vs. CRISPR RNP and a CD40L cDNA AAV6 donor (n=8–12, 4 PBSC donor sources). D) Gene integration rates of PBSC treated with the addition of adenoviral E4orf6/E1b55k H354 “helper” proteins co-expressed from mRNA. E) Fold change in gene integration rates with the addition of adenoviral helper proteins. Data in C and D were analyzed by Wilcoxon Rank-Sum Test. *p≤0.05, **p≤0.01, NS = not significant. See also Figure S4.
PBSC were analyzed three days after nuclease electroporation and cDNA AAV6 transduction. As shown in Figure 4C, modification rates in TALEN-treated cells averaged 13.2 ± 3.4% measured by ddPCR. Integration was dose-dependent with increasing AAV MOI up to 1e5, although there was decreased viability and expansion of cells at higher vector concentrations (Fig. S4A-E). Importantly, even higher rates of targeted gene insertion were observed in samples treated with CRISPR and the cDNA donor. Targeted integration averaged 16.2 ± 4.2% (gRNA, Cas9 mRNA) and 20.8 ± 6.6% (RNP) irrespective of PBSC donor source (Fig. 3C). Significantly, samples receiving CRISPR as RNP had improved viability and fold expansion when compared to those receiving Cas9 mRNA (Fig. S4F-G).
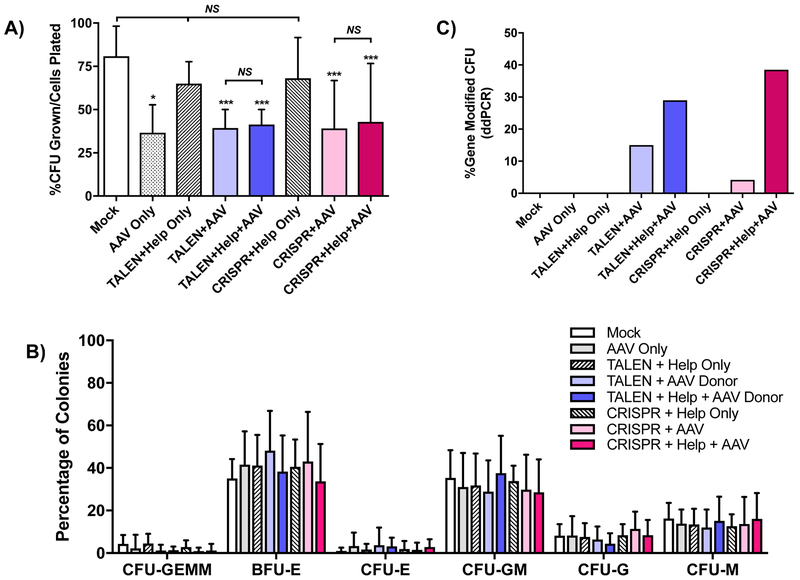
Figure 4. Colony-forming unit (CFU) assay of CD34+ PBSC after treatment with TALEN or CRISPR, AAV6 CD40L cDNA donor and adenoviral helper proteins.
A) PBSC treated with combinations of TALEN mRNA or CRISPR RNP, AAV6 CD40L cDNA donor and the adenoviral E4orf6/E1b55k helper proteins plated in methylcellulose CFU assay. Numbers of colonies were enumerated after 12–14 days and are represented as percent CFU over total cells plated. Data were analyzed by Wilcoxon Rank-Sum Test. Data are presented as mean ± SD. n=4 experiments, 3 PBSC donors. *p≤0.05, ***p≤0.001, NS = not significant. B) The percentages of the different colony types formed were enumerated. CFU-GEMM (CFU-granulocyte/erythroid/macrophage/megakaryocyte), BFU-E (burst-forming unit-erythroid), CFU-E (CFU-erythroid), CFU-GM (CFU-granulocyte/macrophage), CFU-G (CFU-granulocyte), CFU-GM (CFU-macrophage). C) The percentages of CFU gene-modified at the CD40LG 5’ UTR were determined by ddPCR analysis of genomic DNA from individual CFU.
Adenovirus helper proteins enhance CRISPR-mediated gene disruption and increase AAV transduction in T cells and cell lines.(Lentz and Samulski, 2015; Gwiazda et al., 2016) To evaluate the ability of these proteins to augment targeted gene integration at CD40LG in HSC, adenoviral serotype 5 helper proteins (E4Orf6 and E1B55K H354 mutant) were co-introduced as mRNA during electroporation with TALENs or CRISPRs. Figure 3 (D & E) shows that PBSC samples treated with helper protein achieved a 2-fold enhancement in gene modification (p<0.0001) regardless of nuclease platform. Addition of the adenoviral helper protein mRNA did not impair cell viability and fold expansion (Fig. S4D-G).
In Vitro Differentiation of Gene-Modified HSPC
To evaluate if gene-modified CD34+ PBSC retain the capacity to differentiate into multiple erythro-myeloid lineages, gene edited PBSC were cultured in methylcellulose medium containing cytokines that promote stem cell differentiation in the colony forming unit (CFU) progenitor cell assay. There was no difference in clonogenic potential between the nuclease platforms, and the addition of adenoviral helper proteins did not affect colony formation. However, samples transduced with AAV6, alone or with nuclease, yielded lower number of colonies compared to mock treated (Fig. 4A). All control, TALEN-, and CRISPR-Cas9 treatment arms produced similar frequencies of myeloid and erythroid clones (Fig. 4B). Gene integration analysis of individual clones demonstrated editing frequencies comparable to cells cultured in short term myeloid conditions, with similar increases in those treated with adenoviral helper proteins (Fig. 4C).
In Vivo Assessment of Hematopoietic Repopulating Potential in NSG Mice
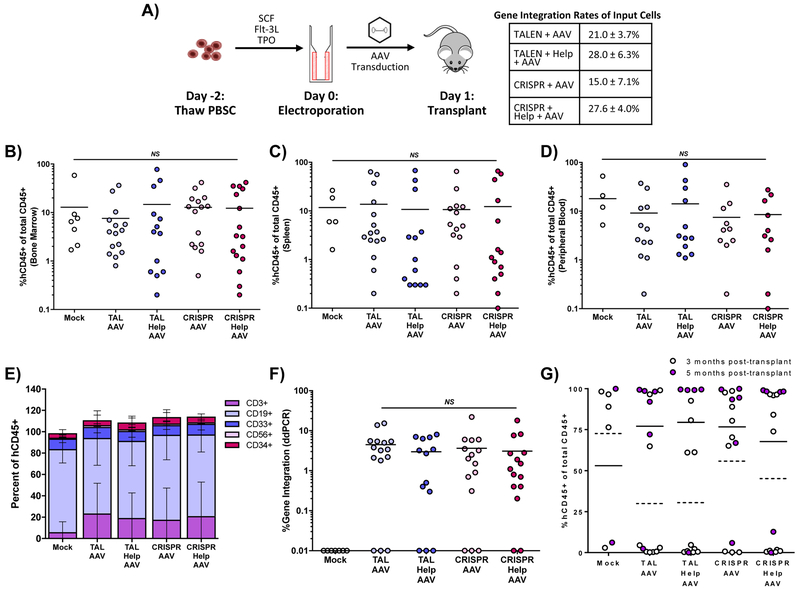
The repopulating capacity of gene-modified HSC was evaluated through transplantation of immunodeficient non-obese diabetic (NOD)-severe combined immunodeficiency (SCID) Il2rg−/− (NSG) mice. Edited PBSC were transplanted into sub-lethally irradiated NSG mice through intrahepatic injection between days of life 3 and 7 (Fig 5A). All transplanted mice displayed human engraftment at 12–20 weeks post-transplant in the bone marrow, spleen, and peripheral blood as measured by human CD45 of total CD45 (Fig. 5B-D). Enumeration of T cells (CD3), B cells (CD19), myeloid progenitors (CD33), NK cells (CD56), and hematopoietic progenitors (CD34) by flow cytometry showed normal lineage distribution in the bone marrow (Fig. 5E).
Figure 5. In vivo assay of CD34+ PBSC in NSG mice after treatment with TALEN or CRISPR, AAV6 CD40L cDNA donor and adenoviral helper proteins.
A) Schematic of NSG mouse transplants and gene integration of input cells as measured by ddPCR. Engraftment of gene modified human PBSC in B) bone marrow, C) spleen, and D) peripheral blood of transplanted mice, determined as the percentage of human CD45+ cells of all human and murine CD45+ cells. E) Lineage distribution of human cells engrafted in NSG mice in the bone marrow 12 weeks post-transplant using fluorescent-labeled antibodies to human T cells (CD3), human B cells (CD19), human myeloid cells (CD33), human NK cells (CD56), and human progenitors (CD34). F) Gene editing determined by ddPCR in bone marrow 12–20 weeks after transplant. G) Thymic engraftment analyzed at 3 and 5 months post-transplant. Data were analyzed by Wilcoxon Rank-Sum Test. **p≤0.01, NS = not significant. See also Figure S5.
Gene modification rates in samples of the bulk transplanted PBSC cultured in vitro averaged 21%, 28%, 15%, and 28% in the TALEN/AAV donor, TALEN/helper/AAV donor, RNP/AAV donor, and RNP/helper/AAV donor samples, respectively (Fig. 5A). Gene integration was detectable in the bone marrow 12–20 weeks after transplant in 80% of mice. Of these mice, integration rates ranged from 0.3% to 22% with a mean of 4.4% across all treatment groups (Fig. 5F). While adenoviral helper protein augmented gene integration rates in vitro in short term cultures, this effect was not observed long term in NSG mice.
Thymic reconstitution was detectable in 60% of mice, with frequency of engraftment trending higher in those analyzed at 5 months compared to 3 months post-transplant (Fig. 5G). Physiologic ratios of CD4+ and CD8+ T cells were found in all thymi with T cell reconstitution (Fig. S5A). In addition, T cell receptor V beta repertoire was quantified by flow cytometry in one mouse from each arm and found to be equivalent amongst all treatment groups compared to mice transplanted with unmodified PBSC (Fig. S5B).
Assessment of Nuclease Specificity and Characterization of HDR-Mediated Junctions
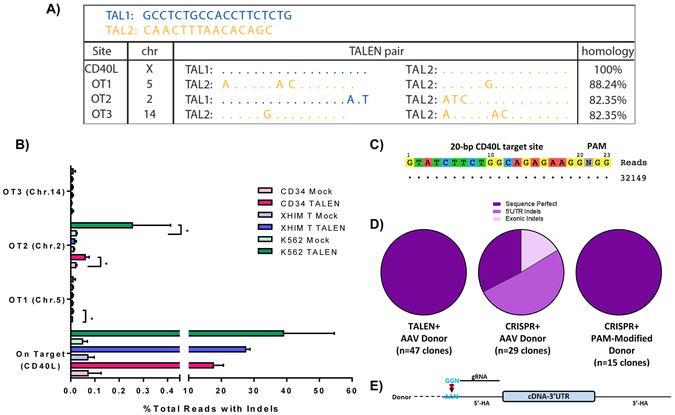
The specificities of both TALENs and CRISPR gRNA were assessed using in silico prediction algorithms as well as unbiased genome-wide assays. In silico predicted TALEN off-target sites were identified using the PROGNOS (Predicted Report Of Genome-wide Nuclease Off-target Sites) tool(Fine et al., 2014) and evaluated in K562 cells treated with TALEN expression plasmids. By CEL I, allelic disruption was only detected on-target, with no differences in banding patterns between mock and TALEN treated samples for each of the predicted five off-target sites (Fig. S6A). In silico predicted off-target sites for CRISPR 3 were identified using the CRISPR Design Tool (crispr.mit.edu/)(Hsu et al., 2013), and interrogation of these predicted off-target sites by Surveyor nuclease assay demonstrated no detectable gene disruption at the top three sites listed (Fig. S6C).
Genome-wide profiling of off-target activity was also performed in TALEN and CRISPR treated K562 cells based on the propensity for exogenous DNA sequences to be trapped at sites of DSB, whether on- or off-target. For TALENs, a non-homologous IDLV was introduced to TALEN-treated cells in a method previously described.(Gabriel et al., 2011) Clustered integration site analysis of captured IDLV revealed three off-target loci in chromosomes 5 (OT1), 2 (OT2), and 14 (OT3) at areas with relatively high levels of homology to the CD40L target (Fig. 6A). While no allelic disruption was detected by CEL I at these sites in K562 cells (Fig. S6B), high-throughput sequencing of off-target sites in PBSC, XHIM CD4+ T cells, and K562 cells treated with TALEN mRNA (PBSC, XHIM T cell) or expression plasmids (K562) demonstrated statistically significant gene disruption at OT1 (chromosome 5) in PBSC and OT2 (chromosome 2) in PBSC and K562 cells compared to mock (Fig. 6B). Off-target cleavage activities of CRISPR 3 were assessed similarly using capture of a short double-stranded oligodeoxynucleotide at DSBs through GUIDE-seq (Genome-Wide, Unbiased Identification of DSBs Enabled by Sequencing)(Tsai et al., 2015) with no off-target sites identified (Fig. 6C).
Figure 6. Off-Target Analysis and Characterization of HDR-Mediated Junctions.
A) Off-target sites as identified by GFP-IDLV trapping in TALEN-treated K562 cells. Allowing for homodimerization of TALEN arms, degree of homology to the on-target binding site is illustrated. B) Targeted re-sequencing of on-target and off-target sites by high-throughput sequencing in PBSC, XHIM CD4+ T cells, and K562 cells treated with TALEN mRNA (PBSC, XHIM T cell) or expression plasmids (K562). Data were analyzed by Wilcoxon Rank-Sum Test. *p≤0.05. C) Off target analysis of CRISPR 3 by GUIDE-seq in K562 cells treated with CRISPR RNP and double-stranded oligodeoxynucleotides. D) Characterization of HDR-mediated junctions between the endogenous CD40L genomic locus and the 5’ end of the integrated donor cassette in samples treated with either TALEN or CRISPR and the codon-optimized donor. Samples treated with CRISPRs required a donor containing a PAM site modification depicted in E) to prevent re-binding of the CRISPR gRNA causing unwanted repeat cleavage after cDNA integration. See also Figure S6.
Furthermore, the fidelity of editing the CD40LG locus was characterized by Sanger sequencing of the junctions between the homologous template and the endogenous DNA. Using a forward primer upstream of the 5’ homology arm and a reverse primer spanning the codon-optimized donor, all TALEN treated samples that were transduced with the AAV donor were found to be sequence-perfect (Fig. 6D). CRISPR treated cells were also sequence perfect when their corresponding donors contained PAM modifications to prevent re-cleavage after donor integration which has been observed by other groups (Fig. 6D-E).(Satomura et al., 2017)
Discussion
Major advancements in the field of genome-editing over the last decade have allowed targeted gene therapy to become a realistic potential therapeutic option for monogenic diseases of the blood and immune systems. This is particularly applicable to defects in genes that require close regulation, and from a clinical standpoint, offers definitive treatment for individuals who are too chronically ill to undergo myeloablative doses of bone marrow conditioning and immune suppression or do not have an HLA-matched donor. The ability to integrate the corrective cDNA cassette at the 5’UTR of the gene and retain its normal chromosomal context in human HSC represents a potential curative therapeutic option for those affected by XHIM. Our results show that both TALEN and CRISPR/Cas9 platforms can achieve targeted insertion of a normal human CD40L cDNA at the CD40LG gene locus in CD34+ hematopoietic stem/progenitor cells (HSPC) in addition to differentiated human T lymphocytes. Notably, gene corrected HSPC engrafted in immunodeficient mice at clinically-relevant frequencies. Taken together these studies provide the foundation for a permanent curative therapy in XHIM.
Nuclease reagents and corrective cDNA donors were initially tested in primary XHIM patient-derived T lymphocytes. The codon optimized/divergent cDNA donor was assembled to contain 3’ UTR of the CD40L gene, as stability of the mRNA transcript is dependent on binding of a polypyrimidine tract binding protein (PTB) to the 3’UTR.(Anderson, 2010; Saifuddin, Yang and Covey, 2013) Additionally, PTB binding to the 3’UTR plays an important role in activated T cell viability and proliferation(Covey, Porta and Nicodemos, 2015), which must be maintained for long-lasting immune correction in vivo.(Vavassori and Covey, 2009) While both TALEN and CRISPR/Cas9 platforms can achieve good rates of gene editing in vitro, there have been to date no studies to deem one superior over the other. For this reason, both technologies were applied to primary T cells and HSC in short-term cultures followed by assessment of HSC long-term in vivo in NSG mice. We were able to recapitulate the results of prior work using TALENs and established that CRISPR/Cas9 could also attain high rates of targeted gene integration and restore physiologically-regulated CD40L expression and function in CD40L-deficient T cells.
In CD34+ HSC, even higher rates of targeted gene integration were consistently achieved than could be attained in primary T cells across multiple G-CSF mobilized PBSC donors. Gene correction was consistently higher in CRISPR compared to TALEN treated samples. Cas9 delivered as either mRNA or as recombinant Cas9 protein directly complexed to the gRNA as RNP were equally efficient, although there was improved viability and fold expansion of cells for RNP-treated cells. Neither of the nuclease systems in themselves had significant effects on the clonogenic potential of HSC when measured by CFU assay, although there was some toxicity due to AAV6 donor transduction that was not affected by the type of nuclease used.
Addition of adenoviral helper protein mRNA with the AAV CD40L cDNA donor consistently doubled cDNA donor integration rates in CD34+ PBSC assayed in vitro regardless of nuclease platform without changes to short-term viability, fold expansion, and clonogenic potential by CFU assay. However, despite significantly augmented rates of gene integration in HSC in vitro, this benefit was not maintained long term in vivo in murine studies. In experiments sorting out cells at different stages of differentiation, it appeared this result was not attributable to a lack of gene editing in the immunophenotypic long-term (LT) HSC population, with rates of targeted integration in the LT-HSC similar to those in the progenitor cell population (data not shown). Instead, long-term engraftment in NSG mice may arise from a more quiescent fraction of stem cells that are more resistant to homologous recombination and therefore unlikely to be affected by adenoviral helper proteins.
There were several advantages to using the CRISPR/Cas9 platform at the CD40LG locus. Across multiple experiments and PBSC donors sources, CRISPRs achieved higher on-target activity than TALENs targeting the 5’UTR of the CD40LG gene. Additionally, the gRNA with the highest on target activity also possessed higher specificity than TALENs, which had detectable levels of off-target activity at a relatively homologous site in the human genome. While there is currently no gold standard for detection of unwanted nuclease activity in clinical trials, we utilized cell-based assays such as IDLV capture and GUIDE-seq, considered to be more physiologically relevant than purely in vitro assays. Additionally, high throughput sequencing results of TALEN OT1 and OT2 identified by IDLV capture demonstrated statistically significant off-target allelic disruption in PBSC and K562 cells treated with TALENs. The higher level of off-target activity in K562 cells was likely due to a more prolonged presence of plasmid DNA compared to mRNA in cells, although use of TALEN mRNA did not prevent off-target activity in primary cells. Efforts to mitigate unintended nuclease activity in TALEN-treated cells were unsuccessful. Obligate heterodimeric forms of FokI had minimal on-target activity for eight TALEN pairs tested, and only those utilizing the wild type FokI nuclease domain retained sufficient activity to allow meaningful rates of gene integration.
In the in vivo studies, G-CSF mobilized PBSC that were gene-modified with either platform engrafted at levels equivalent to mock treated cells in bone marrow, spleen, and peripheral blood of NSG mice analyzed at 3 months and 5 months post-transplantation. More than two-thirds of mice had detectable gene editing in the bone marrow occurring over a large range and lower than observed in in vitro manipulated cells. Reductions in gene editing rates from in vitro to in vivo studies have been reported by other groups using similar gene editing reagents in hematopoietic stem cells.(Genovese et al., 2014; Hoban et al., 2015; De Ravin et al., 2016; Schiroli et al., 2017) Selection methods using reporter genes may be able to overcome the problem(Dever et al., 2016), but this method has not been shown to be scalable to human trials of gene editing due a significant loss of HSC numbers. However, despite the reduced editing rates in vivo, it is likely that in XHIM a proportionally small number of gene-corrected T cells will be sufficient to allow enough class-switching to significantly ameliorate or cure the disease. Only low levels of normal CD40L expression are needed to result in correction of the immune deficiency based on murine models.(Brown et al., 1998; Sacco et al., 2000) In humans, female carriers who have X-inactivation in hematopoietic lineages that is skewed towards expression of the defective CD40LG allele are asymptomatic at even very low frequencies (~10%) of peripheral blood mononuclear cells expressing CD40L.(Hollenbaugh et al., 1994) In one report on allogeneic transplantation for primary immunodeficiency, one of the patients with XHIM survived despite having only 4% CD40L expression in the periphery.(Petrovic et al., 2009)
While this work is limited to a single primary immunodeficiency, successful translation to the clinic will have significant implications for all other monogenic diseases of the hematopoietic system. In all, the results reported here set the foundation for developing a curative therapy for XHIM through site-specific gene editing. Although murine models may not fully recapitulate autologous transplantation in humans, the rates of targeted gene insertion are nonetheless likely to result in clinical improvement in those with XHIM. As we and others in the field continue to focus on the technical and biological challenges of increasing gene integration in long-term repopulating HSC, it is expected that improved outcomes will ultimately lead to clinical trials of gene editing for XHIM.
Experimental Procedures
TALEN and CRISPR Construction
TALENs targeted to the CD40LG 5’ untranslated region were designed with the online TAL Effector Nucleotide Targeter 2.0. program(Cermak et al., 2011; Doyle et al., 2012) and assembled using the Golden Gate TALEN kit (Addgene; Cambridge, MA) and Golden Gate TALEN assembly protocol.(Cermak et al., 2011) TALENs were subsequently cloned into a pCAG mammalian expression vector. The T7 promoter was cloned into the vector preceding the TALEN sequence using a gene block containing the T7 promoter with the In-Fusion HD Cloning Kit (ClonTech Laboratories; Mountain View, CA). CRISPR single guide RNAs (gRNA) were designed using the online design tool created by the Zhang Lab. Paired oligonucleotides for gRNAs were synthesized (Integrated DNA Technologies; San Diego, CA), annealed, and cloned into the pX330-U6-Chimeric_BB-CBh-hSpCas9 expression vector (Addgene) as previously described(Hsu et al., 2013) to express both the sgRNA and the Streptococcus pyogenes Cas9. For use as an in vitro mRNA transcription template, hSpCas9 from the pX330 plasmid was cloned into an in-house-modified pGEM-5Zf(+) plasmid (Promega; Madison, WI) that includes optimized 5’ and 3’ UTR sequences and a modified 3’ UTR that encodes a run of 120 adenine nucleotides followed by an SpeI restriction site for linearization.(Warren et al., 2010)
In Vitro Transcription of TALEN mRNA, CRISPR gRNA, and Cas9 mRNA
TALEN expression plasmids were linearized with AgeI, purified with the PureLink PCR Purification Kit (Life Technologies; Waltham, MA), and used as the template for in vitro transcription using the mMESSAGE mMACHINE T7 Ultra Transcription Kit (ThermoFischer Scientific; Waltham, MA). The T7-Cas9 plasmid was linearized with SpeI and in vitro transcribed as above. gRNA were in vitro transcribed as single guide RNA (sgRNA) from a T7 promoter from either the pDR274 plasmid linearized with DraI using the MEGAshortscript T7 Transcription Kit (ThermoFisher Scientific) or from a PCR-generated DNA template using the HiScribe T7 Quick High Yield RNA Synthesis Kit (New England Biolabs; Ipswich, MA).(DeWitt and Wong, 2015)
Adenovirus 5 Helper Proteins
Adenovirus 5 helper proteins E4Orf6 and E1B55K H354(Yew, Kao and Berk, 1990) were ordered as commercially generated gblocks (Integrated DNA Technologies) and cloned into the modified pGEM-5Zf(+) plasmid downstream of the T7 promoter by Gibson Assembly (New England BioLabs). Expression plasmids were linearized with SpeI and used as templates for in vitro transcription as described above.
Donor Template Construction
The human codon-optimized CD40LG cDNA donor template was commercially synthesized (Integrated DNA Technologies) and contains all five exons (785bp) along with the 3’ UTR (944bp) of the human CD40LG gene followed by the polyA signal (24bp). The cDNA donor sequence is flanked by a 5’ homology arm that begins at the TALEN and CRISPR/Cas9 cut site and extends 162bp upstream and a 3’ homology arm that extends 405bp downstream of the TALEN cut site. The 5’ and 3’ homology arms were amplified from human cord blood HSC-derived genomic DNA using primers 5’-ctttacgtaacgtttttgctgg-3’ with 5’-agaagtttggttgtatgtttcgatcataaatggtatcttctggcagaga-3’ and 5’-ccagaagataccatttcaactttaac-3’ with 5’-gggcagagccaagatttga-3’ respectively. Splice overlap PCR was used to stitch together the 5’ homology arm and CD40LG cDNA with primers 5’-gccgccagtgtgctggaattcctttacgtaacgtttttgctgg-3’ and 5’-cagcctgcaaggtgacactgttcagagtttgagtaagccaaagg-3’. The 3’UTR and 3’ homology arm fragment was incorporated similarly using primers 5’-acagtgtcaccttgcaggc-3’ and 5’-gcagaattcgcccttgggcagagccaagatttga-3’. The two PCR products were ligated and cloned into the PCR2.1 TOPO backbone using the In-Fusion HD Cloning Kit (Clontech). The entire CD40LG cDNA donor with flanking homology arms donor sequence was cloned into a previously established lentiviral backbone (Dull 1998) in reverse orientation to avoid splicing during packaging of the virus. Lentiviral vectors were packaged as integrase-defective lentiviral vectors using the p8.2(Int-) packaging plasmid and concentrated; titers were determined as described.(Cooper et al., 2011) The CD40LG cDNA donor with flanking homology arms was also cloned into an AAV vector plasmid (pX601, Addgene) and produced as an AAV serotype 6 (Virovek, Hayward CA).
Surveyor Nuclease Assay
Surveyor Nuclease Assay was used to determine TALEN induced site-specific allelic disruption at the 5’UTR of the CD40LG gene. A 400–500 base pair region surrounding the TALEN and CRISPR binding sites of the CD40LG gene 5’ UTR was PCR amplified from 200 ng of genomic DNA from treated cells with primers 5’-gcaacgattgtgcgctctta-3’ and 5’-acacagcaaaaagtgctgacc-3’ using AccuPrime Taq Hi-Fi (ThermoFisher Scientific). Denaturation, reannealing, CEL I digestion, and electrophoretic and densitometry analysis were completed as described.(Joglekar et al., 2013)
Electroporation
K562 cells were cultured in RPMI 1640 (Mediatech; Tewksbury, MA) with 10% fetal bovine serum (Omega Scientific; Tarzana, CA) and 1x glutamine/penicillin/streptomycin (Gemini Bio Products; Sacramento, CA), electroporated with the Lonza 4D-Nucleofector System (program FF-120) in SF Cell Line solution at a cell density of 2×105 cells per 20uL reaction with 500–750 ng of either TALEN or CRISPR expression plasmids. XHIM patient derived peripheral blood mononuclear cells were obtained under a UCLA IRB-approved protocol (UCLA IRB #10–0011399) and cultured in RPMI with 20% fetal bovine serum, 1x glutamine/penicillin/streptomycin (Gemini Bio Products), 10 ng/mL hIL-2 (R&D Systems; Minneapolis, MN) and Dynabeads Human T-Activator CD3/CD28 (ThermoFischer Scientific) at a 3:1 bead:cell ratio to expand T lymphocytes for 5–7 days. One day prior to electroporation, CD4+ T lymphocytes were enriched from bulk CD3+ T cells by depletion of CD8+ lymphocytes using CD8 microbeads (Miltenyi Biotec 130–045-021; Bergisch Gladbach, Germany) passed through LD columns (Miltenyi Biotec). CD4+ T lymphocytes were electroporated using the ThermoFischer Neon Transfection System (1400V, 10ms, 3 pulses) in Opti-MEM at a cell density of 4.5×107 cells/mL with either 1.5ug of each TALEN arm as in vitro transcribed mRNA or 2.5 ug of in vitro transcribed CRISPR sgRNA and 2.5ug Cas9 mRNA or 5 ug of CRISPR sgRNA complexed to 50 pmol of Cas9 protein. Immediately post-electroporation, cells were transduced with codon-optimized cDNA IDLV or AAV6 vector. G-CSF mobilized peripheral blood CD34+ cells (HemaCare; Van Nuys, CA) were pre-stimulated for 48 hours in X-VIVO15 medium containing penicillin, streptomycin, glutamine, 50 ng/mL rhuSCF, 50 ng/mL rhuFlt-3 ligand, and 50 ng/mL rhuTPO (Peprotech; Rocky Hill, NJ). The CD34+ cells were then electroporated with the Harvard Apparatus BTX ECM 830 Square Wave Electroporator (255V, 5msec, 1 pulse) in BTXpress buffer at a cell density of 2×106 cells/mL. TALEN treated samples received 4 ug of each TALEN arm as in vitro transcribed mRNA. CRISPR samples received 5 ug gRNA and 5 ug Cas9 mRNA or 9 ug gRNA conjugated to 200 pmol Cas9 protein. CRISPR RNP samples receiving adenoviral helper protein mRNA were treated with half the amount of gRNA and Cas9 protein. Cells were transduced with donor AAV6 or IDLV after 10 minutes of rest at room temperature in X-VIVO15, penicillin, streptomycin, glutamine, and cytokines for 24 hours, at which point they were counted for viability and fold expansion and studied in vitro and in vivo.
Colony Forming Unit (CFU) Assay
CFU assays were performed using Methocult H4435 Enriched methylcellulose (StemCell Technologies, Vancouver, Canada. Cat. # 04445) according to manufacturer’s instructions with minor modifications. Briefly, 25 and 100 CD34+ PBSC were plated in duplicates into 35 mm gridded cell culture dishes. After 14 days of culture at 5% CO2, 37°C and humidified atmosphere, the different types of hematopoietic colonies were identified and counted. CFU were then plucked for genomic DNA isolation (NucleoSpin Tissue XS, Clontech Laboratories Inc., Mountain View, CA).
In vivo studies
NOD/SCID/γ-chainnull (NSG) mice (NOD.Cg.Prkdcscid Il2rgtm1Wjl/SzJ; stock no. 005557; Jackson Laboratory, Bar Harbor, ME) were housed in accordance with an approved protocol by the UCLA Research Safety & Animal Welfare Administration (ARC #2008–167-23P). At all times animals were handled in laminar flow hoods and housed in a pathogen-free colony in a biocontainment vivarium. Newborn pups at 3–7 days of life of both genders were injected with 1 × 106 cells/pup via intrahepatic injection of unmodified or gene-modified human CD34-positive cells prepared as described above, one day after conditioning with 125cGy of sub-lethal body irradiation from a 137Ce source with attenuator, and allowed to engraft over 12–20 weeks as described.(De Oliveira et al., 2013) Human CD34-positive cells were incubated with 1.5 ug OKT3 (eBioscience, San Diego, CA) for approximately 1 hour before transplant to ensure depletion of any contaminating T cells. At the end of each experiment, mice were sacrificed by CO2 inhalation followed by cardiac puncture, and bone marrow, thymus, liver, and spleen were harvested for assays.
Statistics
Descriptive Statistics such as number of observations, mean and standard deviation were reported and presented graphically for quantitative measurements. Kruskal-Wallis test was used to assess overall difference among experimental groups, followed by post-hoc pairwise comparisons with Dwass, Steel, Critchlow-Fligner method(Dwass, 1960; Steel, 1960; Critchlow and Fligner, 1991) to adjust for multiple comparisons. For other pre-specified two-group comparisons, unpaired t test or Wilcoxon rank sum test were used if normality assumption was violated for the data. For all statistical investigations, tests for significance were two-tailed. A p-value of less than the 0.05 significance level was considered to be statistically significant. All statistical analyses were carried out using statistical software SAS version 9.4 (SAS Institute Inc. 2013).
Study Approval
XHIM patient derived peripheral blood mononuclear cells were obtained under a UCLA IRB-approved protocol (UCLA IRB #10–0011399). Written informed consent was received from participants prior to inclusion in the protocol. NOD/SCID/γ-chainnull (NSG) mice (NOD.Cg.Prkdcscid Il2rgtm1Wjl/SzJ were housed in accordance with an approved protocol by the UCLA Research Safety & Animal Welfare Administration (ARC #2008–167-23P).
Supplementary Material
Acknowledgements
These studies were supported by research grants from the California Institute for Regenerative Medicine (Quest Discovery Award, DISC2–10124 to C.Y.K.), Clinical Immunology Society/Baxalta Junior Faculty Award (C.Y.K.), UCLA Children’s Discovery and Innovation Institute Today’s and Tomorrow’s Children Fund Award (C.Y.K), Primary Immune Deficiency Treatment Consortium, a part of the NIH Rare Disease Clinical Research Network (2U54AI082973–06 to C.Y.K.), UCLA K12 Child Health Research Center Development Award (K12HD34610–19, K12HD34610–20 to C.Y.K.), the Hyper-IgM Foundation Research Award (C.Y.K.), and the Eli & Edythe Broad Center of Regenerative Medicine and Stem Cell Research (D.B.K.). M.D.H. was supported by the Interdisciplinary Training in Virology and Gene Therapy award (5 T32 AI060567) and the Whitcome Predoctoral Training Program, the UCLA Molecular Biology Institute. A.R.C. was supported by the Ruth L. Kirschstein National Research Service Award (GM007185). The authors thank Devin Brown for his technical assistance with the murine work. The Flow Cytometry Core and DNA Sequencing Core of the Eli & Edythe Broad Center of Regenerative Medicine & Stem Cell Research provided essential support. The authors thank Dr. Maria Garcia-Lloret for her thoughtful review and editing of this manuscript.
Footnotes
Declaration of Interests
The authors declare no competing interests.
References
- Abraham RS and Aubert G (2016). Flow Cytometry, a Versatile Tool for Diagnosis and Monitoring of Primary Immunodeficiencies. Clinical and Vaccine Immunology 23, pp. 254–271. doi: 10.1128/CVI.00001-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P (2010). Post-transcriptional regulons coordinate the initiation and resolution of inflammation. Nature Reviews Immunology 10, pp. 24–35. doi: 10.1038/nri2685. [DOI] [PubMed] [Google Scholar]
- Brown MP, Topham DJ, Sangster MY, Zhao J, Flynn KJ, Surman SL, Woodland DL, Doherty PC, Farr AG, Pattengale, et al. (1998). Thymic lymphoproliferative disease after successful correction of CD40 ligand deficiency by gene transfer in mice. Nature Medicine 4, pp. 1253–1260. doi: 10.1038/3233. [DOI] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ and Voytas DF (2011). Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Research 39, pp. 7879. doi: 10.1093/nar/gkr739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini, et al. (2013). Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339, pp. 819–823. doi: 10.1126/science.1229223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper AR, Patel S, Senadheera S, Plath K, Kohn DB and Hollis RP (2011). Highly efficient large-scale lentiviral vector concentration by tandem tangential flow filtration. Journal of Virological Methods 177, pp. 1–9. doi: 10.1016/j.jviromet.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covey L, Porta J.La and Nicodemos RM (2015). The RNA-binding protein, polypyrimidine tract-binding protein, affects multiple events in CD4 T cell activation through distinct processes. The Journal of Immunology 194, pp. 18. [Google Scholar]
- Critchlow DE and Fligner MA (1991). On distribution-free multiple comparisons in the one-way analysis of variance. Communications in Statistics - Theory and Methods 20, pp. 127–139. doi: 10.1080/03610929108830487. [DOI] [Google Scholar]
- Dever DP, Bak RO, Reinisch A, Camarena J, Washington G, Nicolas CE, Pavel-Dinu M, Saxena N, Wilkens AB, Mantri S, et al. (2016). CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature 539, pp. 384–389. doi: 10.1038/nature20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt MA and Wong J (2015). Cas9 RNP nucleofection for cell lines using Lonza 4D Nucleofector. Protocols.io. Available at: doi: 10.17504/protocols.io.dm649d. [DOI] [Google Scholar]
- Doyle EL, Booher NJ, Standage DS, Voytas DF, Brendel VP, Vandyk JK and Bogdanove AJ (2012). TAL Effector-Nucleotide Targeter (TALE-NT) 2.0: Tools for TAL effector design and target prediction. Nucleic Acids Research 40, pp. 117–122. doi: 10.1093/nar/gks608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwass M (1960). Some k-Sample Rank-Order Tests. In Contributions to Probability and Statistics, Olkin I, ed. (Stanford: Stanford University Press; ), pp.198–202. [Google Scholar]
- Epinat JC, Amould S, Chames P, Rochaix P, Desfontaines D, Puzin C, Patin A, Zanghellini A, Pâques F and Lacroix E (2003). A novel engineered meganuclease induces homologous recombination in yeast and mammalian cells. Nucleic Acids Research 31, pp. 2952–2962. doi: 10.1093/nar/gkg375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fine EJ, Cradick TJ, Zhao CL, Lin Y and Bao G (2014). An online bioinformatics tool predicts zinc finger and TALE nuclease off-target cleavage. Nucleic Acids Research 42, pp. 1–13. doi: 10.1093/nar/gkt1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel R, Lombardo A, Arens A, Miller J, Genovese P, Kaeppel C, Nowrouzi A, Bartholomae CC, Wang J, Friedman G, et al. (2011). An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nature Biotechnology 29, pp. 816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- Gennery AR, Khawaja K, Veys P, Bredius RGM, Notarangelo LD, Mazzolari E, Fischer A, Landais P, Cavazzana-Calvo M, Friedrich W, et al. (2014). Treatment of CD40 ligand deficiency by hematopoietic stem cell transplantation : a survey of the European experience, 1993–2002. Blood 103, pp. 1152–1157. doi: 10.1182/blood-2003-06-2014.A. [DOI] [PubMed] [Google Scholar]
- Genovese P, Schiroli G, Escobar G, Di Tomaso T, Firrito C, Calabria A, Moi D, Mazzieri R, Bonini C, Holmes MC, et al. (2014). Targeted genome editing in human repopulating haematopoietic stem cells. Nature 510, pp. 235–240. doi: 10.1038/nature13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwiazda KS, Grier AE, Sahni J, Burleigh SM, Martin U, Yang JG, Popp NA, Krutein MC, Khan IF, Jacoby K, et al. (2016). High Efficiency CRISPR/Cas9-mediated Gene Editing in Primary Human T-cells Using Mutant Adenoviral E4orf6/E1b55k “Helper” Proteins. Molecular Therapy 24, pp. 1–11. doi: 10.1038/mt.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward AR, Levy J, Facchetti F, Notarangelo L, Ochs HD, Etzioni A, Bonnefoy JY, Cosyns M and Weinberg A (1997). Cholangiopathy and tumors of the pancreas, liver, and biliary tree in boys with X-linked immunodeficiency with hyper-IgM. Journal of Immunology 158, pp. 977–983. Available at: http://www.ncbi.nlm.nih.gov/pubmed/8993019. [PubMed] [Google Scholar]
- Hoban MD, Cost GJ, Mendel MC, Romero Z, Kaufman ML, Joglekar AV, Ho M, Lumaquin D, Gray D, Lill GR, et al. (2015). Correction of the sickle cell disease mutation in human hematopoietic stem/progenitor cells. Blood 125, pp. 2597–2604. doi: 10.1182/blood-2014-12-615948.The. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbaugh D, Wu LH, Ochs HD, Nonoyama S, Grosmaire LS, Ledbetter JA, Noelle RJ, Hill H and Aruffo A (1994). The random inactivation of the X chromosome carrying the defective gene responsible for X-linked hyper IgM syndrome (X-HIM) in female carriers of HIGM1. Journal of Clinical Investigation 94, pp. 616–622. doi: 10.1172/JCI117377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nature Biotechnology 31, pp. 827–32. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard N, Hagin D, Sommer K, Song Y, Khan I, Clough C, Ochs HD, Rawlings DJ, Scharenberg AM and Torgerson TR (2016). Targeted gene editing restores regulated CD40L expression and function in X-HIGM T cells. Blood 127, pp. 2513–2522. doi: 10.1182/blood-2015-11-683235. [DOI] [PubMed] [Google Scholar]
- Joglekar AV, Hollis RP, Kuftinec G, Senadheera S, Chan R and Kohn DB (2013). Integrase-defective lentiviral vectors as a delivery platform for targeted modification of adenosine deaminase locus. Molecular Therapy 21, pp. 1705–1717. doi: 10.1038/mt.2013.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Morena MT, Leonard D, Torgerson TR, Cabral-Marques O, Slatter M, Aghamohammadi A, Chandra S, Murguia-Favela L, Bonilla FA, Kanariou M, et al. (2017). Long-term outcomes of 176 patients with X-linked hyper-IgM syndrome treated with or without hematopoietic cell transplantation. Journal of Allergy and Clinical Immunology 139, pp. 1282–1292. doi: 10.1016/j.jaci.2016.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentz TB and Samulski RJ (2015). Insight into the mechanism of inhibition of adeno-associated virus by the mre11/rad50/nbs1 complex. Journal of virology 89, pp. 181–94. doi: 10.1128/JVI.01990-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy J, Espanol-boren T, Thomas C, Fiscbe A, Bordigoni P, Resnick I, Fasth A, Aid B, Sanders EAM, Tabone M, et al. (1997). Clinical spectrum of X-linked hyper-IgM syndrome. The Journal of Pediatrics 131, pp. 47–54. doi: 10.1016//s0022-3476(98)70048-4. [DOI] [PubMed] [Google Scholar]
- Mitsui-Sekinaka K, Imai K, Sato H, Tomizawa D, Kajiwara M, Nagasawa M, Morio T and Nonoyama S (2015). Clinical features and hematopoietic stem cell transplantations for CD40 ligand deficiency in Japan. Journal of Allergy and Clinical Immunology 136, pp. 1018–1024. doi: 10.1016/j.jaci.2015.02.020. [DOI] [PubMed] [Google Scholar]
- De Oliveira SN, Ryan C, Giannoni F, Hardee CL, Tremcinska I, Katebian B, Wherley J, Sahaghian A, Tu A, Grogan T, et al. (2013). Modification of Hematopoietic Stem/Progenitor Cells with CD19-Specific Chimeric Antigen Receptors as a Novel Approach for Cancer Immunotherapy. Human Gene Therapy 24, pp. 824–839. doi: 10.1089/hum.2012.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic A, Dorsey M, Miotke J, Shepherd C and Day N (2009). Hematopoietic stem cell transplantation for pediatric patients with primary immunodeficiency diseases at All Children’s Hospital/University of South Florida. Immunologic Research 44, pp. 169–178. doi: 10.1007/s12026-009-8111-z. [DOI] [PubMed] [Google Scholar]
- Porteus MH and Baltimore D (2003). Gene Targeting in Human Cells. Science 300, p. 75390. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- De Ravin SS, Reik A, Liu PQ, Li L, Wu X, Su L, Raley C, Theobald N, Choi U, Song AH, et al. (2016). Targeted gene addition in human CD34+ hematopoietic cells for correction of X-linked chronic granulomatous disease. Nature Biotechnology 34, pp. 1–8. doi: 10.1038/nbt.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero Z, Torres S, Cobo M, Muñoz P, Unciti JD, Martín F and Molina IJ (2011). A tissue-specific, activation-inducible, lentiviral vector regulated by human CD40L proximal promoter sequences. Gene therapy 18, pp. 364–371. doi: 10.1038/gt.2010.144. [DOI] [PubMed] [Google Scholar]
- Sacco MG, Ungari M, Cato EM, Villa A, Strina D, Notarangelo LD, Jonkers J, Zecca L, Facchetti F and Vezzoni P (2000). Lymphoid abnormalities in CD40 ligand transgenic mice suggest the need for tight regulation in gene therapy approaches to hyper immunoglobulin M (IgM) syndrome. Cancer Gene Ther 7, pp. 1299–1306. doi: 10.1038/sj.cgt.0234 [DOI] [PubMed] [Google Scholar]
- Saifuddin A, Yang A and Covey L (2013). Phosphorylation and the regulation of CD40L mRNA stability by polypyrimidine tract binding protein (P1394). The Journal of Immunology 190, pp. 14. [Google Scholar]
- Satomura A, Nishioka R, Mori H, Sato K, Kuroda K and Ueda M (2017). Precise genome-wide base editing by the CRISPR Nickase system in yeast. Scientific Reports 7, pp. 1–10. doi: 10.1038/s41598-017-02013-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiroli G, Ferrari S, Conway A, Jacob A, Capo V, Albano L, Plati T, Castiello MC, Sanvito F, Gennery AR, et al. (2017). Preclinical modeling highlights the therapeutic potential of hematopoietic stem cell gene editing for correction of SCID-X1. Science Translational Medicine 9. doi: 10.1126/scitranslmed.aan0820. [DOI] [PubMed] [Google Scholar]
- Steel RG (1960). A Rank Sum Test for Comparing All Pairs of Treatments. Technometrics 2, pp. 197–207. [Google Scholar]
- Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, et al. (2015). GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nature Biotechnology 33, pp. 187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavassori S and Covey LR (2009). Post-transcriptional regulation in lymphocytes: the case of CD154. RNA biology 6, pp. 259–65. doi: 10.4161/rna.6.3.8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, et al. (2010). Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 7, pp. 618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yew PR, Kao CC and Berk AJ (1990). Dissection of functional domains in the adenovirus 2 early 1B 55K polypeptide by suppressor-linker insertional mutagenesis. Virology 179, pp. 795–805. doi: 10.1016/0042-6822(90)90147-J. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.