Abstract
Bacteria produce a variety of surface-exposed polysaccharides important for cell integrity, biofilm formation, and evasion of the host immune response. Synthesis of these polymers often involves the assembly of monomer oligosaccharide units on the lipid carrier undecaprenyl-phosphate at the inner face of the cytoplasmic membrane. For many polymers, including cell wall peptidoglycan, the lipid-linked precursors must be transported across the membrane by flippases to facilitate polymerization at the membrane surface. Flippase activity for this class of polysaccharides is most often attributed to MOP (Multidrug/Oligosaccharidyl-lipid/Polysaccharide) family proteins. Little is known about how this ubiquitous class of transporters identifies and translocates its cognate precursor over the many different types of lipid-linked oligosaccharides produced by a given bacterial cell. To investigate the specificity determinants of MOP proteins, we selected for variants of the WzxC flippase involved in Escherichia coli capsule (colanic acid) synthesis that can substitute for the essential MurJ MOP-family protein and promote transport of cell wall peptidoglycan precursors. Variants with substitutions predicted to destabilize the inward-open conformation of WzxC lost substrate specificity and supported both capsule and peptidoglycan synthesis. Our results thus suggest that specific substrate recognition by a MOP transporter normally destabilizes the inward-open state, promoting transition to the outward-open conformation and concomitant substrate translocation. Furthermore, the ability of WzxC variants to suppress MurJ inactivation provides strong support for the designation of MurJ as the flippase for peptidoglycan precursors, the identity of which has been controversial.
Keywords: peptidoglycan, cell wall, MurJ, flippase, MOP-family transporters
ABBREVIATED SUMMARY
From cell walls in bacteria to protein glycosylation in eukaryotes, surface exposed polysaccharides are built on polyprenol-phosphate lipid carriers. Monomer units are typically assembled at the cytoplasmic face of the membrane and require translocation to the cell surface for polymerization/assembly. MOP-family proteins are a major class of transporters associated with this flippase activity, and in this report we present evidence that transport proceeds via destabilization of the inward-open state of the transporter by specific substrate binding.
INTRODUCTION
Bacterial cells produce a variety of cell surface polysaccharides. Polymers like peptidoglycan (PG) and teichoic acids (TAs) are critical for cell integrity and shape maintenance, whereas capsular polysaccharides and O-antigens play central roles in virulence and the evasion of host defenses (Whitfield, 2006). Despite their vast structural diversity, the majority of surface polysaccharides are made by one of three types of synthesis and export mechanisms: synthase-dependent, ABC transporter-dependent, or Wzy-dependent pathways (Whitfield, 2006). Most complex polysaccharides with 3–6 sugars in their repeating unit are made by the widely-distributed Wzy-dependent pathway (Islam and Lam, 2014). Examples include O-antigens of gram-negative bacteria and many capsular polysaccharides. For these polymers, the monomeric oligosaccharide unit is synthesized at the inner face of the cytoplasmic membrane on the lipid carrier undecaprenyl phosphate (Und-P) (Islam and Lam, 2014; Ruiz, 2015). For polymerization, the oligosaccharide moiety must be transported across the membrane and exposed at the membrane surface by a class of transporters referred to as flippases (Islam et al., 2012; Ruiz, 2015). Notably, this overall lipid-linked synthesis strategy is near universal as it is also employed by eukaryotic cells to generate oligosaccharides for N-linked protein glycosylation (Helenius et al., 2002).
Lipid-linked sugar flippase activity for polysaccharide synthesis has principally been ascribed to one of two types of transporters: ABC (ATP-binding cassette) systems, and MOP (Multidrug/Oligosaccharidyl-lipid/Polysaccharide) family proteins (Ruiz, 2015). MOP-type flippases are typically associated with Wzy-dependent polysaccharide synthesis pathways, and several lines of evidence indicate that they have a strong preference for translocating the specific lipid-linked oligosaccharide produced by their cognate synthetic pathway (Hong and Reeves, 2014; Michael A Liu et al., 2015; Hong et al., 2017). How these transporters specifically recognize their substrates over the many different types of lipid-linked oligosaccharides produced in a bacterial cell is not currently known. The transport mechanism is also poorly understood, but the recently solved structures of the MOP-family protein MurJ from Thermosipho africanus and Escherichia coli has provided important clues (Kuk and Lee, 2017; Zheng et al., n.d.).
MurJ was identified several years ago as a protein essential for PG biogenesis. It was proposed to be the long-sought after flippase that translocates the final PG precursor lipid II (Ruiz, 2008; Sham et al., 2014), which consists of the disaccharide N-acetylmuramic acid (MurNAc)-β-1-4-N-acetylglucosamine (GlcNAc) with a pentapeptide attached to the MurNAc sugar via a lactyl group. Like other polysaccharide synthesis pathways, lipid II is assembled on the inner face of the cytoplasmic membrane and must be translocated before it can be polymerized and crosslinked to form the PG matrix that fortifies bacterial cells against osmotic rupture (Ruiz, 2015). Importantly, the functional assignment of MurJ as the PG lipid II flippase remains controversial in the field of PG biogenesis because in vitro assays have detected what appears to be lipid II flippase activity for the SEDS (shape, elongation, division, sporulation) family protein FtsW (Mohammadi et al., 2011; Ruiz, 2015).
In support of a flippase function for MurJ, it was found to be required for lipid II translocation in Escherichia coli using an in vivo flippase assay whereas SEDS proteins were dispensable for this activity (Sham et al., 2014). The structure of MurJ from T. africanus also supports a transporter function (Kuk and Lee, 2017). In the crystals, the protein adopted an inward-open conformation with a solvent accessible cavity capable of accommodating the lipid II head group. Related structures of a different subfamily of MOP transporters involved in drug efflux had previously been solved in an outward-open conformation (He et al., 2010), and chemical probing of MurJ structure in vivo (Butler et al., 2013) indicates that MurJ can adopt such a conformation in cells. Evolutionary coupling (EC) analysis (Hopf et al., 2014) also shows that MurJ must adopt an additional conformation not accounted for in the inward-open crystal structure, since pairs of distant residues on the cytosolic face so strong co-evolution. An outward-open model of MurJ similarly leaves predicted interactions unsatisfied on the periplasmic face of the protein, indicating that at least two distinct conformations are subject to evolutionary selective pressure (Zheng et al., n.d.). Thus, it has been proposed that MurJ and other MOPS family proteins mediate transport/flipping via an alternating access model involving the interconversion between the inward- and outward-facing conformations, alternately allowing the lipid II headgroup to access the cytosolic and periplasmic faces of the membrane (Islam and Lam, 2014; Ruiz, 2015; Kuk and Lee, 2017; Zheng et al., n.d.).
To investigate the specificity determinants of MOP proteins, we selected for variants of the WzxC flippase involved in Escherichia coli capsule (colanic acid) synthesis (Stevenson et al., 1996) that gain translocation activity for peptidoglycan precursors and can substitute for the essential MurJ MOP-family protein in the cell wall synthesis pathway. Variants with amino acid changes predicted to destabilize the inward-open conformation of WzxC lost substrate specificity and supported both capsule and peptidoglycan synthesis. Our results thus suggest that specific substrate recognition by a MOP transporter normally functions to destabilize the inward-open state, promoting transition to the outward-open conformation and concomitant substrate translocation. Furthermore, the ability of WzxC variants to suppress MurJ inactivation provides strong support for the designation of MurJ as the flippase for peptidoglycan precursors.
RESULTS
Identification of WzxC variants that can substitute for MurJ
WzxC is a MOP-family transporter in E. coli required for the synthesis of the colanic acid capsule (Stevenson et al., 1996), the production of which is induced by activation of the Rcs envelope stress response (Majdalani and Gottesman, 2005). The colanic acid precursor is a hexasaccharide [L-fucose-(pyruyl-D-galactose-D-glucouronic acid-D-galactose)-O-acetyl-L-fucose-D-glucose] built on the Und-P lipid carrier. WzxC has been implicated in the transport (flipping) of this lipid-linked intermediate (Stevenson et al., 1996). Although WzxC is the closest relative of MurJ in E. coli (≈12% sequence identity), its substrate structure differs greatly from the lipid II PG precursor that MurJ has been implicated in flipping. It is therefore not surprising that WzxC fails to substitute for MurJ and promote growth when MurJ is depleted (Fig. 1, row 1). However, we thought it might be possible to identify altered WzxC proteins that gain the ability to flip the PG precursor and rescue a MurJ defect. We reasoned that the isolation and characterization of such variants would provide useful information about what determines substrate specificity in MOP-family flippases and potentially reveal new insights into the transport mechanism.
Figure 1. WzxC variants can substitute for MurJ.
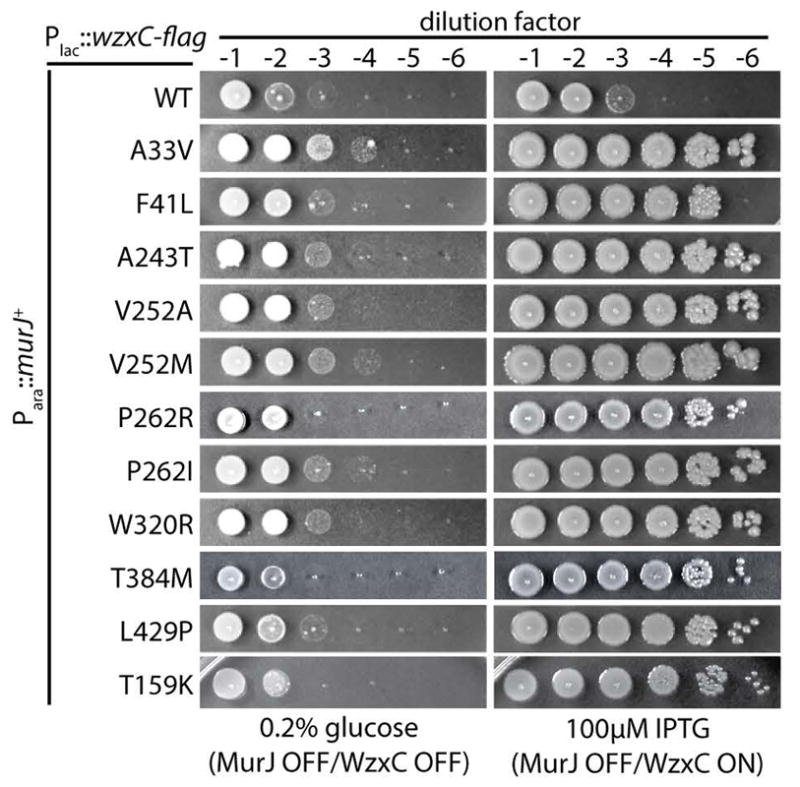
Cells of CS7 [Para::murJ] harboring plasmids encoding C-terminally FLAG-tagged WzxC (WT) or the indicated derivatives were grown in LB medium with arabinose overnight. Following normalization for culture density, serial dilutions (10−1 to 10−6) were prepared and 5 μl of each were spotted onto LB plates supplemented with either glucose or IPTG. Plates were photographed after incubation at 37°C for ≈16 hours. All of the strains grew similarly on plates supplemented with arabinose under this condition. An additional growth experiment with additional concentrations of IPTG are presented in SI Appendix, Fig. S2.
To select for altered substrate specificity variants of WzxC, the wzxC gene was mutagenized using error-prone PCR and cloned into a medium copy vector under control of the lactose promoter (Plac). The resulting plasmid library was transformed into a MurJ-depletion strain where native murJ was engineered to be under control of the arabinose promoter (Para). This promoter replacement renders the strain dependent on the presence of arabinose in the medium for growth. When the depletion strain harboring the wzxC plasmid library was plated on LB medium lacking arabinose but supplemented with isopropyl-β-D-thiogalactopyranoside (IPTG) to induce wzxC, surviving colonies arose at a low frequency (10−4). To distinguish between survivors with mutations allowing arabinose-independent expression of murJ from the desired wzxC mutants capable of substituting for murJ, plasmids were purified from the isolates and transformed back into the parental MurJ-depletion strain. The resulting transformants were then tested for growth on IPTG-containing medium with or without arabinose supplementation. Plasmids conferring arabinose-independent growth were then isolated and their wzxC insert was sequenced. Many of the primary isolates harbored wzxC clones with multiple mutations (Table S1). To identify the functionally relevant substitutions, we used site-directed mutagenesis to construct plasmids encoding C-terminal FLAG-tagged WzxC variants (WzxC-FLAG) with single amino acid changes corresponding to those identified in the original mutant isolates. The FLAG-tag did not appear to interfere with WzxC activity as the fusion was capable of supporting capsule production in a ΔwzxC strain (Fig. S1). Importantly, wild-type WzxC-FLAG also failed to promote growth upon MurJ depletion (Fig. 1, row 1). In total, eleven single amino acid substitutions in WzxC-FLAG representing changes throughout the length of the 492 amino acid protein were found to be sufficient for suppression of MurJ depletion (Fig. 1, rows 2–11). The wzxC alleles varied in the strength of the observed suppression phenotype, with most being able to promote growth of the MurJ depletion strain upon induction with 25 μM IPTG, but two requiring 75–100 μM IPTG to achieve full suppression (Fig. 1, rows 2–1, Fig. S2). Immunoblot analysis using anti-FLAG antibodies indicated that all of the WzxC-FLAG variants were produced at levels comparable to, or slightly lower than, the wild-type protein (Fig. S3). Thus, the WzxC variants do not gain the ability to substitute for MurJ simply due to their overproduction.
To determine if the altered WzxC proteins could fully substitute for MurJ, we assessed the ability of eight variants for their ability to support the growth of a murJ deletion. A ΔmurJ::KanR allele constructed in a background with a complementing murJ plasmid was used as a donor for P1 phage transduction of the deletion into MurJ+ strains harboring the wzxC plasmids. For all the mutants tested, transductants were successfully isolated on medium containing IPTG for wzxC induction, and deletion/replacement of the native murJ gene was confirmed in each case (Fig. S4). We chose two of these strains to further characterize their growth and morphology. Full induction of wzxC(V252M) or wzxC(A33V) from plasmid pDF2 or pDF15, respectively promoted a growth rate for the ΔmurJ strain that was comparable to that of wild-type (Fig. 2A). Additionally, the WzxC derivatives promoted a relatively normal rod-shaped morphology in the ΔmurJ background with a moderate cell division defect observed for cells producing WzxC(V252M) (Fig. 2B–D). We conclude that the WzxC variants identified in the selection are capable of overcoming the lethality of a murJ deletion and in at least one case can promote relatively normal cell growth and division. Therefore, we will henceforth refer to the identified variants as MJWzxC derivatives.
Figure 2. Growth and morphology of ΔmurJ cells producing WzxC variants.
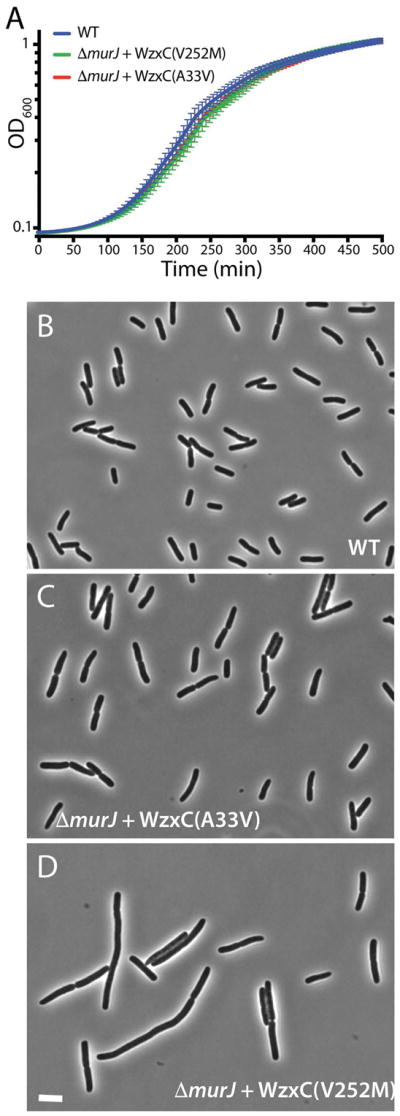
A. Cells of TB28 [WT], CS3/pDF2 [ΔmurJ/Plac::wzxC(V252M)-flag], and CS3/pDF3[ΔmurJ/Plac::wzxC(A33V)-flag] were grown in LB supplemented with 1 mM IPTG at 30°C in a 96-well plate. Growth was monitored by regular OD600 measurements in a plate reader. Plots are of the means of 10 technical replicates, and the error bars show the standard deviation. B–D. Cells of the same strains grown to an OD600 of 0.2–0.3 were imaged by phase-contrast microscopy. Bar equals 4 μm.
MJWzxC variants can support PG lipid II flipping in vivo
The ability of the MJWzxC variants to suppress the essentiality of MurJ suggests that they have gained the ability to transport the lipid II precursor for PG biogenesis. To test this possibility, we took advantage of an in vivo assay for lipid II flipping (Sham et al., 2014). To detect lipid II transport, cells were radiolabeled with the PG precursor [3H]-meso-diaminopimelic acid (mDAP) and treated with Colicin M (ColM). This toxin invades the periplasm and cleaves flipped lipid II, generating a soluble pyrophospho-disaccharide pentapeptide that is subsequently converted to disaccharide tetrapeptide by periplasmic carboxypeptidases. When MurJ is functional, ColM cleavage of flipped lipid II generates a new soluble radiolabeled product and destroys the labeled lipid fraction (Ghachi et al., 2006; Sham et al., 2014) (Fig. 3). The MurJ variant, MurJ(A29C), is sensitive to the Cys-modifying reagent (2-sulfonatoethyl)methanethiosulfanate (MTSES). When radiolabeled cells relying on this MurJ derivative for lipid II translocation are treated with MTSES, the lipid fraction is protected from ColM cleavage and the soluble ColM product is not observed (Sham et al., 2014) (Fig. 3), indicating that flipping is blocked. A plasmid producing WzxC(WT) did not alleviate this block (Fig. 3). However, production of WzxC(V252M) restored lipid II cleavage by ColM in MurJ-inactivated cells and promoted the accumulation of the soluble ColM product (Fig. 3). We therefore conclude that the MJWzxC derivatives have gained the ability to facilitate lipid II transport.
Figure 3. Support of PG lipid-II flipping in vivo by a WzxC variant.
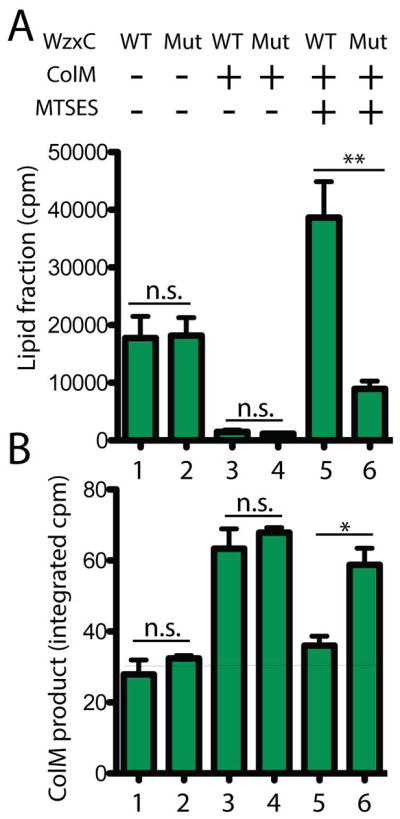
Cells of CAM290 [murJ(A29C)] harboring plasmid pCS124 [Plac::wzxC(WT)-flag] or pDF2 [Plac::wzxC(V252M)-flag] were grown in labeling medium to an OD600 of 0.2. [3H]-mDAP was then added to radiolabel PG precursors. After a 15 min labeling period, MTSES and ColM were added to block MurJ(A29C) activity and cleave flipped lipid II, respectively. Just prior to cell lysis, cells were collected by centrifugation and fractionated to measure radioactivity in the PG lipid precursor pool (A) and soluble ColM cleavage product (B). WT and Mut denote WzxC(WT) and WzxC(V252M), respectively. The means and the standard error of means (SEMs) from three experiments are shown. P-values were calculated with two-tailed unpaired Student’s t-test. *, p < 0.05; **, p < 0.01; n.s., not significant. cpm = counts per minute.
Most MJWzxC derivatives have lost substrate specificity
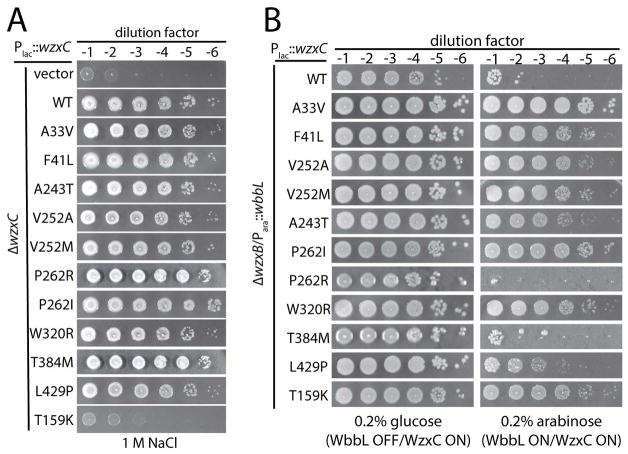
We next investigated whether the MJWzxC variants that support lipid II translocation retain the ability to promote colanic acid capsule synthesis. Production of the capsule is induced by the Rcs stress response system when cells are grown in high-salt medium (Sledjeski and Gottesman, 1996). Cells unable to make capsule grew poorly on LB medium containing 1.0 M NaCl (Fig. 4A, row 1, Fig. S1). This growth defect was observed for mutants blocked at the first step in the pathway (ΔwcaJ) or at the WzxC step. Thus, the phenotype does not require the build up of lipid-linked colanic acid precursors that would likely reduce the pool of lipid carrier available to other pathways like PG synthesis (Jorgenson and Young, 2016). Production of wild-type WzxC-FLAG as well as most of the MJWzxC-FLAG derivatives restored the growth of ΔwzxC cells on LB with 1.0M NaCl (Fig. 4A, rows 2–13). The exception was WzxC(T159K), which suppressed MurJ depletion, but not the ΔwzxC phenotype. Thus, the majority of the MJWzxC proteins appear to retain WzxC function.
Figure 4. Transport of colanic acid and O-antigen precursors by MJWzxC variants.
(A) Cells of CS38 [ΔwzxC] harboring an empty vector (vector) or vectors encoding the indicated FLAG-tagged WzxC variant were grown and plated on LB medium with 1M NaCl as described in Figure 1. Complementation of the ΔwzxC phenotype results in the formation of mucoid colonies on the high-salt medium. (B) Cells of CS39/pCS160 [ΔwzxB/Para::wbbL] harboring the same WzxC-encoding plasmids were grown and diluted as descried in Figure 1 followed by plating on LB medium with 100 μM IPTG (to induce WzxC production) plus either glucose or arabinose (to repress or induce O-antigen production, respectively) as indicated.
To further investigate the range of potential substrates capable of being utilized by the MJWzxC variants, we tested their ability to participate in O-antigen synthesis. These polysaccharides decorate the lipopolysaccharides (LPS) that form the outer leaflet of the outer membrane in gram-negative bacteria (Raetz et al., 2007). E. coli K-12 strains, including the MG1655 derivatives used here, do not make O-antigen polymers. This defect is due to an insertion element that disrupts wbbL, which encodes the enzyme catalyzing the committed step for the synthesis of the O-16 antigen with the repeating unit D-galactose-D-glucose-L-rhamnose-(D-glucose)-D-N-acetylglucosamine (D Liu and Reeves, 1994). The flippase for this pathway is thought to be WzxB (Marolda, 2004). Inactivation of wzxB is not lethal in WbbL− strains. However, ectopic expression of wbbL in ΔwzxB cells is lethal, presumably due to the accumulation of lipid-linked O-16 precursors and the sequestration of Und-P lipid carrier from the PG synthesis pathway (Marolda et al., 2006; Jorgenson and Young, 2016). Unlike wild-type WzxC-FLAG, co-production of many of the MJWzxC-FLAG variants wth WbbL rescued the WbbL-induced lethality of ΔwzxB cells and partially restored O-antigen production (Fig. 4B and S5). However, two variants, WzxC(P262R) and WzxC(T384M) that complemented the MurJ-depletion and ΔwzxC phenotypes failed to substitute for WzxB. On the other hand, WzxC(T159K), which failed to complement ΔwzxC rescued both the ΔwzxB and MurJ-depletion phenotypes. From these results, we conclude that the MJWzxC variants have largely lost substrate specificity, allowing them to function in a variety of polysaccharide synthesis pathways.
Substitutions in the MJWzxC variants are predicted to destabilize the inward-open conformation
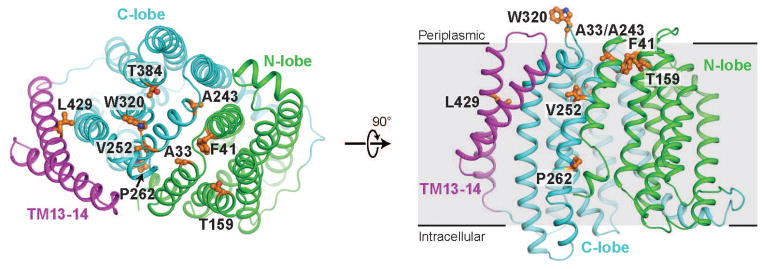
In order to better understand the molecular basis for the effects of the MJWzxC mutations, we constructed a homology model of WzxC using the crystal structure of E. coli MurJ (Zheng et al., n.d.) as a template (Fig. 5 and S5). We expected that most of the specificity altering changes in WzxC would occur in the aqueous cavity of the transporter where substrate is predicted to bind. However, when mapped onto the model WzxC structure, the majority of residues altered in the MJWzxC variants clustered at or near the periplasmic face of the protein. Many of these substitutions occur in residues that mediate inter-domain contact between the N-lobe and C-lobe on the periplasmic side (A33, A243, F41 and W320). For example, the pseudo-symmetry related pair of residues A33 and A243 are in small, sterically restricted spaces (Fig. S5A). Mutation of these to Val or Thr is incompatible with the inward-open homology model due to steric clash with neighboring residues, and so is expected to destabilize the inward-open conformation. Similarly, the buried N-lobe residue F41 engages in extensive hydrophobic contacts with L248, F316, and other residues in the C-lobe, so its mutagenic substitution with Leu may destabilize the inward-open conformation (Fig. S5A). Likewise, W320 sits near the interface between the two lobes, and the nonconservative substitution with Arg likely weakens interdomain interactions on the periplasmic side (Fig. S5A). It is noteworthy that most of these substitutions are modest. They may simply raise the energy of the inward-open state, altering the equilibrium between inward-open and outward-open states without causing a complete loss of function.
Figure 5. Structural analysis of specificity-broadening mutations in WzxC.
A homology model of WzxC is shown, with the sites of mutations highlighted in orange sticks. At left, the protein is viewed from the periplasmic side, and at right is viewed parallel to the membrane plane. With only a few exceptions, the mutations cluster at the periplasmic face of the protein near the interface between the N- and C-terminal lobes of WzxC.
Other substitutions are not located at the interdomain interface but rather are found in or near the central cavity of the enzyme. For instance, P262 and V252 sit in the lateral gate between TM1 and TM8 (Fig. S5B). Both residues may be directly involved in substrate binding or play a critical role in conformation transition. Their alteration may expand the range of substrates accepted by transporter by affecting substrate binding affinity. Notably, L429 is located neither in the central cavity nor in the extracellular gate, but rather is found in TM13 (Fig. S5C). Its mutation to proline is incompatible with α-helical geometry, and must force a distortion in the helix. The connection between this effect and broadened substrate specificity is unclear, but attests to a functionally important role for TMs 13 and 14, which are absent in most MOP family flippases.
DISCUSSION
In this report, we isolated MJWzxC variants that have lost substrate specificity and gain the ability to transport the PG precursor lipid II and O-antigen precursors in addition to its native substrate for colanic acid synthesis. The location of the amino acid changes in WzxC resulting in this phenotype was surprising. Rather than altering the predicted substrate-binding region of the modeled structure, the changes largely map to portions of the protein located near the periplasmic face of the membrane. Based on the MurJ structure, this region of the protein is predicted to form contacts that stabilize the inward-open conformation of the transporter. Because many of the amino acid substitutions in the MJWzxC variants involve a change in side chain size or charge, we infer that they exert their effect on the transporter through the destabilization of the inward-open conformation. Thus, the genetic results support a model in which the stability of the inward-open conformation plays a key role in determining the substrate specificity of the transporter (Fig. 6).
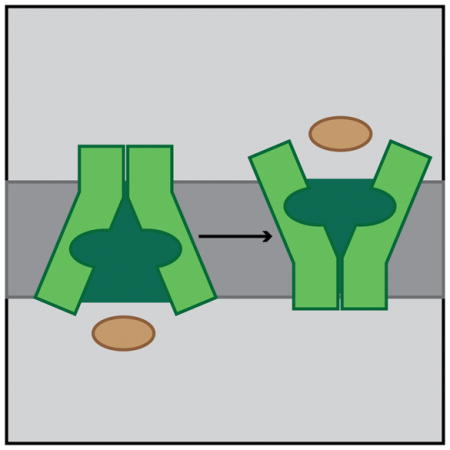
Figure 6. Model for substrate induced conformational changes in MOP family flippases.
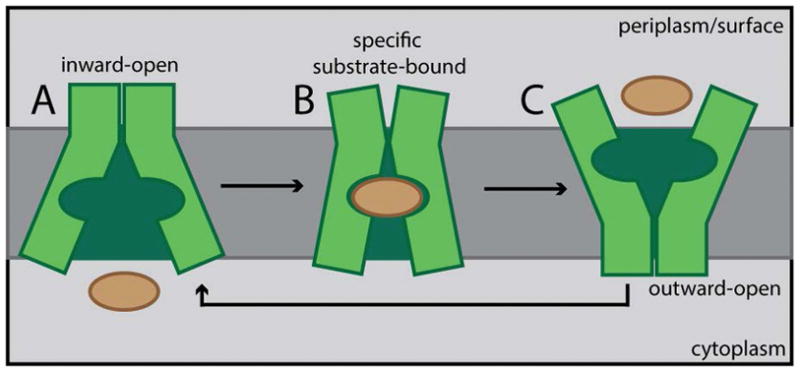
Shown is a schematic summarizing our model for substrate transport by MOP family flippases. Structural studies suggest that the inward-open conformation is the most stable state of the transporter. Based on our genetic results, we propose that specific substrate binding is required to break contacts at the outer face of the transporter to destabilize the inward-open conformation. Once these contacts are broken, a transition to the outward-open conformation can occur to allow for substrate release on the opposite face of the membrane. For simplicity, the lipid anchor of the substrate is not drawn.
We propose that for wild-type WzxC, a specific interaction between the native substrate and the hydrophillic core of the transporter is needed to break contacts at the periplasmic face of the membrane involved in stabilizing the inward-open conformation. Such a change would then facilitate the transition to the outward-open conformation and the release of substrate on the outside face of the membrane. Once substrate is released, the protein would then be free to transition back to the more stable inward-open conformation and begin another round of transport. Due to the changes in the MJWzxC variants, we envision that the protein can more readily interconvert between the two main conformations without the need for specific substrate binding. Thus, the transport of non-native precursors would be facilitated by the altered flippase. The main limitation in this case is likely to be the ability of the precursor sugar moiety to fit within the hydrophillic core of the altered transporter. WzxC was therefore an especially fortuitous choice for this specificity study. Its native substrate is relatively large such that the hydrophilic core of the MJWzxC variants is likely capable of accommodating and flipping a wider range of substrates than might be possible with other transporters.
The phenotype of a previously isolated mutant in a different flippase suggests that other transporters may function similarly to WzxC. TacF is a MOP family protein implicated in the transport of teichoic acid (TA) precursors of Streptococcus pneumoniae (Damjanovic et al., 2007). The TAs in this organisms are normally decorated with choline (Massidda et al., 2013). A TacF variant was identified that suppressed the choline-dependent growth phenotype of S. pneumoniae, presumably by allowing the transport of TA precursors lacking choline (Damjanovic et al., 2007). The change in this variant that alters the substrate choline requirement is located in a loop of TacF predicted to be at the outer surface of the membrane (Damjanovic et al., 2007). Similar to WzxC, this area is exactly where contacts that stabilize the inward-open conformation are likely to be made. Thus, the use of a specific substrate binding event to destabilize the inward-open state and promote a conformational transition may be a general component of the transport mechanism of MOP family flippases. Substrate-induced conformational changes have also been implicated in the transport mechanism of the (NSS) family of transporters (Billesbølle et al., 2015), suggesting that they may be involved in many different types of membrane transport processes.
In addition to a better understanding of the transport mechanism of MOP family flippases, the activities of the MJWzxC variants also provide insight into the process of PG biogenesis. Although flippase activity has yet to be demonstrated for MurJ in vitro, the finding that a protein implicated in flipping colanic acid precursors can substitute for MurJ in PG biogenesis makes it hard to argue that MurJ is anything other than the lipid II flippase. Furthermore, the ability of MJWzxC variants as well as other heterologous or promiscuous flippase proteins to maintain growth and viability upon MurJ inactivation and promote a relatively normal cell morphology suggests that lipid II transport does not need to occur in the context of specific multi-protein complexes with other PG biogenesis factors. Such complexes may be formed to render the process more efficient, especially during division where some problems were observed in ΔmurJ cells producing WzxC(V252M). However, specific interactions involving MurJ do not appear to be necessary for the construction of the PG layer.
In conclusion, our results highlight the utility of unbiased genetic selections to study the function of MOP family flippases. Further structural analysis of these transporters using the substitutions identified in the MJWzxC variants should facilitate the capture of additional conformations of these proteins and provide further insight into their transport mechanism.
MATERIALS AND METHODS
Media, Bacterial Strains and Plasmids
Strains used in this study are listed in SI Appendix, Table S2. Unless otherwise specified, E. coli cells were grown in lysogeny broth (LB) under aeration at 37°C. Where indicated, arabinose and glucose are added to a final concentrations of 0.2% (w/v). The antibiotics ampicillin, chloramphenicol, and kanamycin were used at a final concentration of 25 μg/ml. Spectinomycin was added to a final concentration of 40 μg/ml. Plasmids and oligonucleotides used in this study are listed in SI Appendix, Table S3 and S4, respectively.
Selection of WzxC variants that can suppress MurJ essentiality
Strain CS7 [Para::murJ] was transformed with a mutagenized wzxC plasmid library (Plac::wzxC) (see SI appendix for details). Transformants were scraped from the agar surface into 5ml of LB medium and the resulting cell suspension was serially diluted and plated on LB agar supplemented with chloramphenicol, 0.2% glucose, and IPTG. Isolates that required induction of the wzxC plasmid with IPTG for growth in the absence of murJ expression (0.2% glucose) were selected for sequencing.
Growth measurements and microscopy
Overnight cultures of TB28 [WT], CS3/pDF2 [ΔmurJ/Plac::wzxC(V252M)-flag], and CS3/pDF3[ΔmurJ/Plac::wzxC(A33V)-flag] were grown in LB supplemented with 200μM IPTG and, for all strains but TB28, with 25 μg/ml chloramphenicol. The cells were then washed once with fresh LB, subcultured 1:500 in LB with 1mM IPTG, and grown at 30°C for 3h to an OD600 of 0.2–0.3. For microscopy, the cells were then fixed with 2.5% formaldehyde and 0.04% glutaraldehyde in 60mM sodium phosphate buffer, pH 7.4, for 20 min at RT and washed 1× with PBS (137mM NaCl, 2.7mM KCl, 1mM CaCl2, 0.5mM MgCl2, 10mM Na2HPO4, 1.8mM KH2PO4, pH 7.4) before imaging on 2% agarose pads, using Nikon TE2000 inverted microscope outfitted with a CFI Plan Apo DM 100× 1.4NA phase contrast objective and an Andor Zyla 4.2 PLUS sCMOS camera. For the growth curves, the cells were diluted into LB containing 1 mM IPTG to a starting OD600 of 0.005, and 200μL aliquots were distributed into a 96-well plate. Cells were grown in a Tecan Infinite 200 Pro plate reader at 30°C with constant shaking, and OD600 readings were taken every 5min.
Detection of lipid II flippase activity using Colicin M
Lipid II translocation across the inner membrane was monitored using the previously described colicin M assay (Sham et al., 2014). Cells of CAM290/pCS124 [murJ(A29C)/Plac::wzxC] and CAM290/pDF2 [murJ(A29C)/Plac::wzxC(V252M)] were grown in LB medium with chloramphenicol overnight. Cultures were diluted 100 fold in 40ml of the labeling medium with 100 μM IPTG (M9 supplemented with 0.1% (w/v) casamino acids, 0.2% (w/v) maltose, 0.1mg/ml of lysine, threonine and methionine) and grown at 37°C with aeration. When the culture OD600 reached 0.2, 15μl of 1.5 μCi/μl of 3H-mDAP (ARC) was added to 10ml of the culture and incubated for 15 minutes at 37°C. When indicated, colicin M and MTSES were added to a final concentration of 500 ng/ml and 0.4 mM, respectively. The cultures were then incubated for 10 minutes and chilled immediately on ice. Cells were collected by centrifugation at 8,000 × g for 2 minutes at 4°C and resuspended in 1ml of preheated water. Samples were then boiled for 30 minutes and processed to measure the soluble colicin M product and PG lipid precursors as described previously (Sham et al., 2014).
Homology model construction
A homology model of E. coli WzxC was constructed in MODELLER (Webb and Sali, 2002) using a multi-template modeling protocol with the crystal structures of MurJ from E. coli (Zheng et al., n.d.) and T. africanus (Kuk and Lee, 2017) (PDB ID: 5T77) serving as templates.
Supplementary Material
Acknowledgments
The authors would like to thank all members of the Bernhardt, Rudner, and Kruse labs for helpful advice and discussions. This work was supported by the National Institutes of Health (R01AI083365 and AI099144 to TGB, and CETR U19 AI109764 to TGB and ACK).
References
- Billesbølle CB, Krüger MB, Shi L, Quick M, Li Z, Stolzenberg S, et al. Substrate-induced Unlocking of the Inner Gate Determines the Catalytic Efficiency of a Neurotransmitter:Sodium Symporter. J Biol Chem. 2015;290:26725–26738. doi: 10.1074/jbc.M115.677658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler EK, Davis RM, Bari V, Nicholson PA, Ruiz N. Structure-function analysis of MurJ reveals a solvent-exposed cavity containing residues essential for peptidoglycan biogenesis in Escherichia coli. J Bacteriol. 2013;195:4639–4649. doi: 10.1128/JB.00731-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damjanovic M, Kharat AS, Eberhardt A, Tomasz A, Vollmer W. The Essential tacF Gene Is Responsible for the Choline-Dependent Growth Phenotype of Streptococcus pneumoniae. J Bacteriol. 2007;189:7105–7111. doi: 10.1128/JB.00681-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Ghachi M, Bouhss A, Barreteau H, Touzé T, Auger G, Blanot D, Mengin-Lecreulx D. Colicin M exerts its bacteriolytic effect via enzymatic degradation of undecaprenyl phosphate-linked peptidoglycan precursors. J Biol Chem. 2006;281:22761–22772. doi: 10.1074/jbc.M602834200. [DOI] [PubMed] [Google Scholar]
- He X, Szewczyk P, Karyakin A, Evin M, Hong WX, Zhang Q, Chang G. Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature. 2010;467:991–994. doi: 10.1038/nature09408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius J, Ng DTW, Marolda CL, Walter P, Valvano MA, Aebi M. Translocation of lipid-linked oligosaccharides across the ER membrane requires Rft1 protein. Nature. 2002;415:447–450. doi: 10.1038/415447a. [DOI] [PubMed] [Google Scholar]
- Hong Y, Reeves PR. Diversity of o-antigen repeat unit structures can account for the substantial sequence variation of wzx translocases. J Bacteriol. 2014;196:1713–1722. doi: 10.1128/JB.01323-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Liu MA, Reeves PR. Progress in Our Understanding of Wzx Flippase for Translocation of Bacterial Membrane Lipid-Linked Oligosaccharide. J Bacteriol. 2017;200:e00154–17. doi: 10.1128/JB.00154-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopf TA, Schärfe CPI, Rodrigues JPGLM, Green AG, Kohlbacher O, Sander C, et al. Sequence co-evolution gives 3D contacts and structures of protein complexes. elife. 2014;3:65. doi: 10.7554/eLife.03430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam ST, Lam JS. Synthesis of bacterial polysaccharides via the Wzx/Wzy-dependent pathway. Can J Microbiol. 2014;60:697–716. doi: 10.1139/cjm-2014-0595. [DOI] [PubMed] [Google Scholar]
- Islam ST, Fieldhouse RJ, Anderson EM, Taylor VL, Keates RAB, Ford RC, Lam JS. A cationic lumen in the Wzx flippase mediates anionic O-antigen subunit translocation in Pseudomonas aeruginosa PAO1. Molecular Microbiology. 2012;84:1165–1176. doi: 10.1111/j.1365-2958.2012.08084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgenson MA, Young KD. Interrupting Biosynthesis of O Antigen or the Lipopolysaccharide Core Produces Morphological Defects in Escherichia coli by Sequestering Undecaprenyl Phosphate. J Bacteriol. 2016;198:3070–3079. doi: 10.1128/JB.00550-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuk ACY, Lee SY. Crystal structure of the MOP flippase MurJ in an inward-facing conformation. Nat Struct Mol Biol. 2017;24:171–176. doi: 10.1038/nsmb.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Reeves PR. Escherichia coli K12 regains its O antigen. Microbiology. 1994;140:49–57. doi: 10.1099/13500872-140-1-49. [DOI] [PubMed] [Google Scholar]
- Liu MA, Stent TL, Hong Y, Reeves PR. Inefficient translocation of a truncated O unit by a Salmonella Wzx affects both O-antigen production and cell growth. FEMS Microbiol Lett. 2015;362:725. doi: 10.1093/femsle/fnv053. [DOI] [PubMed] [Google Scholar]
- Majdalani N, Gottesman S. The Rcs phosphorelay: a complex signal transduction system. Annu Rev Microbiol. 2005;59:379–405. doi: 10.1146/annurev.micro.59.050405.101230. [DOI] [PubMed] [Google Scholar]
- Marolda CL. Wzx proteins involved in biosynthesis of O antigen function in association with the first sugar of the O-specific lipopolysaccharide subunit. Microbiology. 2004;150:4095–4105. doi: 10.1099/mic.0.27456-0. [DOI] [PubMed] [Google Scholar]
- Marolda CL, Tatar LD, Alaimo C, Aebi M, Valvano MA. Interplay of the Wzx translocase and the corresponding polymerase and chain length regulator proteins in the translocation and periplasmic assembly of lipopolysaccharide O antigen. J Bacteriol. 2006;188:5124–5135. doi: 10.1128/JB.00461-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massidda O, Nováková L, Vollmer W. From models to pathogens: how much have we learned about Streptococcus pneumoniae cell division? Environ Microbiol. 2013;15:3133–3157. doi: 10.1111/1462-2920.12189. [DOI] [PubMed] [Google Scholar]
- Mohammadi T, van Dam V, Sijbrandi R, Vernet T, Zapun A, Bouhss A, et al. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J. 2011;30:1425–1432. doi: 10.1038/emboj.2011.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz CRH, Reynolds CM, Trent MS, Bishop RE. Lipid A modification systems in gram-negative bacteria. Annu Rev Biochem. 2007;76:295–329. doi: 10.1146/annurev.biochem.76.010307.145803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz N. Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc Natl Acad Sci USA. 2008;105:15553–15557. doi: 10.1073/pnas.0808352105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz N. Lipid Flippases for Bacterial Peptidoglycan Biosynthesis. Lipid Insights. 2015;8s1 doi: 10.4137/LPI.S31783. LPI.S31783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham LT, Butler EK, Lebar MD, Kahne D, Bernhardt TG, Ruiz N. Bacterial cell wall. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science. 2014;345:220–222. doi: 10.1126/science.1254522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledjeski DD, Gottesman S. Osmotic shock induction of capsule synthesis in Escherichia coli K-12. J Bacteriol. 1996;178:1204–1206. doi: 10.1128/jb.178.4.1204-1206.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson G, Andrianopoulos K, Hobbs M, Reeves PR. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. J Bacteriol. 1996;178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B, Sali A. Comparative Protein Structure Modeling Using MODELLER. Curr Protoc Bioinformatics. 2002;235:5.6.1–5.6.32. doi: 10.1002/0471250953.bi0506s47. [DOI] [PubMed] [Google Scholar]
- Whitfield C. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem. 2006;75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- Zheng S, Sham L-T, Brock K, Marks DS, Bernhardt TG, Kruse AC. Structure and function of the lipid II flippase MurJ from Escherichia coli. Proc Natl Acad Sci USA. doi: 10.1073/pnas.1802192115. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.