Abstract
Biotechnological production of fuels, chemicals and proteins is dependent on efficient production systems, typically genetically engineered microorganisms. New genome editing methods are making it increasingly easy to introduce new genes and functionalities in a broad range of organisms. However, engineering of all these organisms is hampered by the lack of suitable gene expression tools. Here, we describe a synthetic expression system (SES) that is functional in a broad spectrum of fungal species without the need for host-dependent optimization. The SES consists of two expression cassettes, the first providing a weak, but constitutive level of a synthetic transcription factor (sTF), and the second enabling strong, at will tunable expression of the target gene via an sTF-dependent promoter. We validated the SES functionality in six yeast and two filamentous fungi species in which high (levels beyond organism-specific promoters) as well as adjustable expression levels of heterologous and native genes was demonstrated. The SES is an unprecedentedly broadly functional gene expression regulation method that enables significantly improved engineering of fungi. Importantly, the SES system makes it possible to take in use novel eukaryotic microbes for basic research and various biotechnological applications.
INTRODUCTION
The progress in DNA sequencing is rapidly uncovering the genetic richness of life. Parallel development of computational methods, novel algorithms, databases and increased computing power is now making it possible to extract meaningful information from the exponentially growing pool of sequence data. However, our capacity to read DNA sequences and to characterize novel microbial species is highly exceeding our possibilities in utilizing this information for basic research and biotechnology applications. The ability to design and write the code for biological systems can be harnessed to its full effect only if we have robust and predictable genetic tools that enable efficient engineering of biological systems to perform the designed tasks.
Nature contains a vast number of microbial species of which only a minute fraction is currently being used in biotechnological processes. Introducing novel production hosts with superior native characteristics (such as high resistance to extreme conditions, specific metabolic traits or efficient protein secretion) could offer interesting opportunities for the industry, and for detailed functional studies of the different microbes. To achieve this, essential tools would be robust and predictable gene expression systems. The lack of suitable expression systems, and slow progress in their development for each individual organism, are significantly hindering the use of novel hosts. The ability to engineer an increasing number of microbes would enable us to more efficiently study the richness of biological properties that have developed during evolution.
Wide applicability of an expression system across different organisms and production conditions requires minimal interference from endogenous gene expression regulation. There are several reports on engineering and characterization of gene expression systems that confer high level of functional independence over the native transcription regulation. This independence is typically achieved by the use of heterologous hybrid-transcription factors composed of independent DNA-binding and transcription activation domains of bacterial and viral origin, respectively (1–7). Most of these systems are regulated by an externally added compound, such as estradiol, testosterone or doxycycline (1,2,4,8,9), which due to cost, represents a potential hindrance for industrial feasibility. Although these systems are important tools for proof-of-concept studies, they have not been demonstrated to control gene expression in other species than those they were designed for. The species-specificity arises from the fact that the essential parts (such as artificial transcription factors) are expressed using native promoters.
Recently, we established an expression system for Saccharomyces cerevisiae (10) in which, in contrast to the preceding systems, the transcription factor was expressed using a core promoter (CP) instead of a full-length promoter. Similarly to several existing systems, our expression system made use of heterologous DNA-binding and transactivation domains in the designed (synthetic) transcription factor. Expression of the transcription factors constitutively, at low level, using the CP alone, enabled highly constitutive expression of target genes that contained in their promoters upstream binding sites (BS) for the transcription factor. By changing the number of transcription factor BS, and the CPs, a wide range of target gene expression levels was achieved in S. cerevisiae without the use of external inducer compounds.
Here, we describe a novel synthetic expression system (SES) that builds on the modular architecture of the previously established gene expression system for S. cerevisiae (10). The expression system is composed of a set of CPs and transcription factor modules originating from heterologous organisms. We demonstrate that the SES system is functional in a broad range of yeast and filamentous fungi species and that it allows adjustable and strong expression of target genes to higher levels than attainable with the strongest widely used promoters in the tested species. Importantly, in contrast to typical endogenous promoter-based systems, the system provides means for growth condition insensitive gene expression control. As a validation of its potential we show that the SES system enables engineering of fungal species that have not been previously genetically engineered (yeasts Candida apicola and Zygosaccharomyces lentus). The SES system represents a disruptive technology that now enables efficient utilization of nature’s microbial diversity in research and biotechnology applications.
MATERIALS AND METHODS
Strains and media
Parental strains which were used in this study are listed in Supplementary Table S1.
Bacterial cultivations were done in lysogeny broth (LB) with ampicillin (10 g/l tryptone (Becton Dickinson), 5 g/l yeast extract (Becton Dickinson), 0.17 M NaCl and 100 μg/ml ampicillin (Sigma-Aldrich)). Media which used in experiments with yeast and filamentous fungi: YPD (20 g/l bacto peptone (Becton Dickinson); 10 g/l yeast extract, 20 g/l D-glucose); SCD (6.7 g/l yeast nitrogen base (Becton Dickinson), 20 g/l D-glucose, 790 mg/l complete supplement mixture (Formedium)); SGM (20 g/l spent grain, 20 g/l spent grain extract, 60 g/l lactose, 37.8 mM (NH4)2SO4, 36.7 mM KH2PO4, 2.4 mM MgSO4, 4.1 mM CaCI2, 15.6 μM CoCI2, 18.0 μM FeSO4, 4.9 μM ZnSO4 and 9.5 μM MnSO4); Repressing medium with glucose used in bioreactors (10g/l glucose, 20 g/l yeast extract, 37.8 mM (NH4)2SO4, 36.7 mM KH2PO4, 2.4 mM MgSO4, 4.1 mM CaCI2, 15.6 μM CoCI2, 18.0 μM FeSO4, 4.9 μM ZnSO4 and 9.5 μM MnSO4); YPGel (20 g/l bacto peptone, 10 g/l yeast extract, 30 g/l gelatine). In case of agar plate cultivations, 20 g/l agar was added to liquid medium in addition to the components mentioned above. To produce Aspergillus niger and Trichoderma reesei conidia, potato dextrose agar plates were used—PDA (39 g/l potato dextrose agar (Becton Dickinson)). Cellulose containing plates were used as an inducing medium with T. reesei (20 g/l cellulose, 1.25 g/l yeast extract, 0.11 M KH2PO4, 37.8 mM (NH4)2SO4, 2.4 mM MgSO4, 4.1 mM CaCI2, 0.1% Triton X-100, 20 g/l agar, 15.6 μM CoCI2, 18.0 μM FeSO4, 4.9 μM ZnSO4 and 9.5 μM MnSO4).
Cloning
All the plasmids used in this study were cloned according to the manufacturer’s protocol using Gibson assembly (New England Biolabs), restriction enzyme-based techniques (Thermo Fisher Scientific), or by utilizing yeast homologous recombination (11,12). Kapa Hifi enzyme (Kapa Biosystems) was used for all polymerase chain reaction (PCR) reactions. Ligation and Gibson assembly mixes were transformed into Escherichia coli TOP10 by electroporation and the correct plasmids were identified by analytical digestions and sequencing. Sequences of the CPs, genes and other sequences are shown in Supplementary Tables S2–4. Synthetic DNA fragments and primers used in this work were obtained from Integrated DNA Technologies.
Core promoter screen
CPs of selected genes of S. cerevisiae, A. niger and T. reesei were ordered as synthetic DNA (Supplementary Table S2), and their functionality was tested in a S. cerevisiae–based screen. Candidate CPs were selected based on the following criteria; (i) they are part of a promoter of highly expressed gene, which was based on RNA sequencing (transcriptomics) data for A. niger (13) and T. reesei (14); (ii) they contain a TATA sequence (possible TATA-box) in their structure, upstream of the start codon, where the distance of the TATA sequence and the start codon (ATG) is no more than 180 bp and no less than 80 bp (in case of multiple sequences fulfilling the description, the TATA-box is defined as the TATA sequence with smallest distance from the start codon); (iii) the transcription start site is located downstream of the TATA sequence. The final selected CP sequence included a region spanning 50 bp upstream of the TATA-box to the start codon. Typical length of the CP varied approximately from 140 to 200 bp. Also 55 bp homology-regions matching the linearized target plasmid were introduced into CP synthetic DNA to allow plasmid assembly by in vivo homologous recombination in S. cerevisiae.
The gene blocks were co-transformed into S. cerevisiae with linearized centromeric plasmid which contained four BS for the LexA-based synthetic transcription factor (sTF) (forming a synthetic UAS for the tested CPs), mCherry reporter gene, and LEU2 selection marker (Figure 1B). The in vivo homologous recombination assembly created a LexA-sTF dependent synthetic promoter which controlled the expression of mCherry. Transformation was done into strains H3899 (WT) (Supplementary Table S1) and H4489. H4489 was constructed by transforming strain H3900 with NsiI linearized pHI3i-Tcp-sTF16 (10) to generate a strain expressing LexA-VP16 sTF constitutively from TDH3cp. Transformation was done into both strains to identify CP’s basal transcription level (H3899, no sTF expression) as well as its performance once activated with sTF (H4489) (Figure 1B).
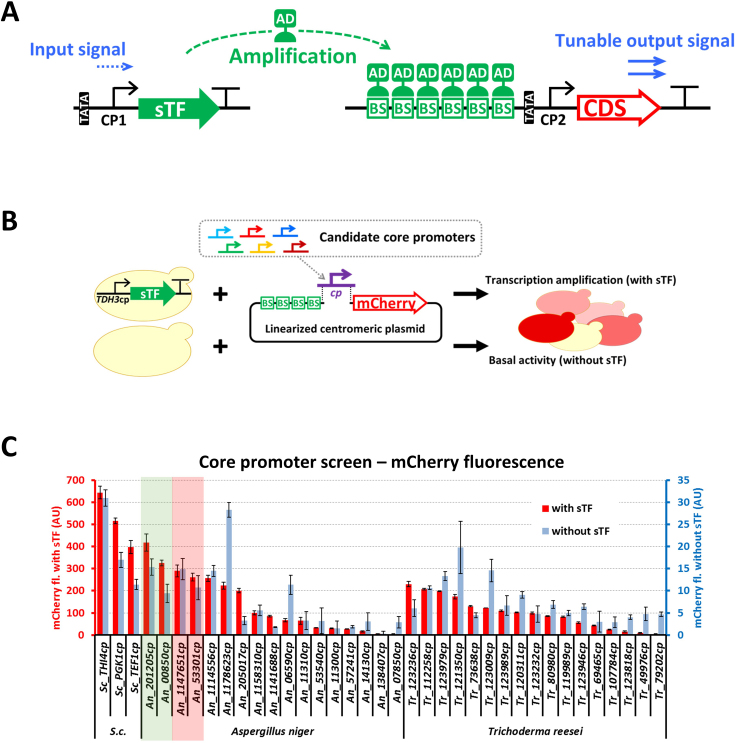
Figure 1.
The SES and the CP performance screen in Saccharomyces cerevisiae. (A) A schematic presentation of the SES concept. Expression of an sTF is achieved by a CP (CP1), which provides low and highly constitutive expression (input signal). The sTF recognizes BS in the upstream part of a target gene synthetic promoter, which can be designed to provide on-demand (amplification) output by varying the number of BSs and by the choice of a CP (CP2). (B) The setup for the CP screen in S. cerevisiae. Diverse CPs produced as synthetic DNA fragments were assembled in vivo (co-transformed) with a linearized centromeric plasmid in a S. cerevisiae strain expressing the LexA-VP16 sTF (and in parallel in an isogenic strain lacking the sTF). Two parameters were assessed, (i) ‘Transcription amplification’ which represents the capacity of the CP to trigger transcription initiation in the presence of the sTF, and (ii) ‘Basal activity’ which represents the ability of the CP to initiate transcription without sTF. (C) The mCherry fluorescence of the strains with the tested candidate CPs. The fluorescence of strains with the sTF (red bars) are plotted on the left y-axis, and the fluorescence of strains without the sTF (blue bars) are plotted on y-axis on the right. S. cerevisiae (S.c.) native CPs (THI4cp, PGK1cp and TEF1cp) were included as examples of highly functional species-specific CPs. The CPs from Aspergillus niger and Trichoderma reesei represent candidates from evolutionary distant organisms with a potential to retain their function in a broad spectrum of hosts. Coloured shading marks the four heterologous CPs (based on the transcription amplification function), which were selected to construct SES-A (green shading;) and SES-B (pink shading) expression systems. Values and error bars represent the mean and standard deviation from three biological replicates.
Pooled colonies were tested from each yeast transformation for mCherry fluorescence. This was done to allow a quick CP performance test without the need for tedious selection of colonies with sequence-confirmed assembled plasmids. To assess the relevance of the setup, for one transformation (with the 533cp), 40 random colonies were picked, plasmids isolated and sequenced and also fluorescence measurement performed. All plasmids contained the integrated CP; however in some (<10%) plasmids, the CP was incorrectly assembled. Fluorescence variation in colonies with correctly assembled plasmids was minimal, and colonies with incorrect plasmids showed no fluorescence. The average fluorescence of the tested individual colonies corresponded to the fluorescence of the pooled colonies (data not shown).
Fluorescence measurement was done as described in the section ‘Fluorescence measurement of yeast species with fluorometer’, except the pre-cultivations were conducted on SCD agar plates lacking histidine and leucine.
SES expression cassettes
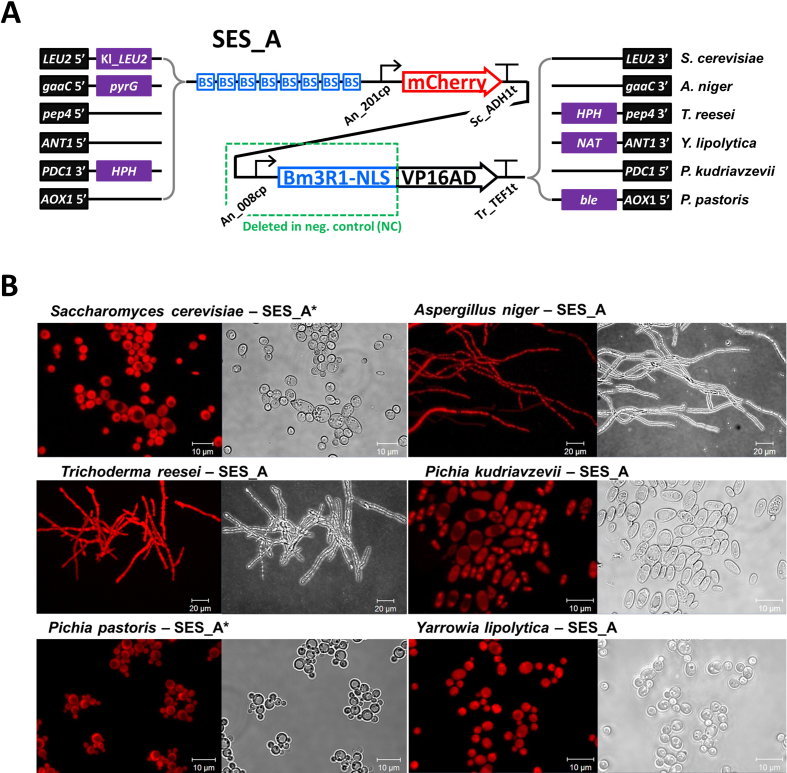
SES expression cassettes were constructed as single DNA molecules containing expression cassettes, both for mCherry and for sTF. The SES systems were cloned into species-specific integration plasmids, which contained a selectable marker gene and flanks for in-genome integration by homologous recombination (Supplementary Table S5). The mCherry expression cassettes contained a synthetic promoter which consisted of eight Bm3R1-specific sTF BS (Supplementary Table S4) and either An-201cp (SES-A) or An-114cp (SES-B), and the mCherry reporter gene (encoding red fluorescence protein), which was followed by the S. cerevisiae ADH1 terminator. The sTF expression cassette contained either An-008cp (SES-A) or An-533cp (SES-B), the sTF (BM3R1-NLS-VP16) encoding gene and the T. reesei TEF1 terminator. The corresponding negative control (NC) cassettes lacked the CP for the sTF, as indicated by a green dashed box in Figure 2A and Supplementary Figure S1A).
Figure 2.
Construction of SES and the assessment of function in a range of fungal hosts. (A) A schematic presentation of the SES system (SES-A), including positions of selection markers and genome-integration flanks for the listed species. The An_00850 CP (An_008cp) was used for the expression of the sTF (Bm3R1-NLS-VP16). The mCherry expression was controlled by the sTF via the sTF-specific BS positioned upstream of the An_201205 CP (An_201cp). The NC lack the region spanning the CP (An_008cp) and a large portion of the sTF. Each cassette was integrated in the specified genomic locus in single copy. (B) Fluorescent microscopy of the different microbial species harbouring the SES-A or SES-A* (asterisk indicates Saccharomyces cerevisiae codon optimized sTF). Uniform mCherry fluorescence signal was observed in all cells in the tested species. A corresponding bright field image is shown for each strain. The micrograph acquisition settings and the contrast were adjusted to maximize the visualization of the red fluorescence.
The SES-A expression cassettes with 0, 1, 2 and 4 sTF BS for A. niger and Pichia kudriavzevii were constructed analogously to the SES-A systems for these hosts, with the only difference being the number of sTF BS. In the case of P. kudriavzevii, also NC cassettes with 0, 1, 2 and 4 sTF BS were constructed on the basis the SES-A-NC.
The SES-B+NAT expression cassettes for C. apicola and Z. lentus were constructed by combining 5′- and 3′-genome integration homology-regions with the SES-B cassette and an expression cassette for nourseothricin selection marker (NAT). The NAT marker cassette contained two sTF BS, An-201cp, NAT ORF and Yarrowia lipolytica TEF1 terminator.
The SES-C expression cassette contained cbh1 gene (coding region identical to genomic sequence in T. reesei including naturally occurring introns). The mCherry–ScADH1 terminator part of the T. reesei SES-B construct was replaced by the cbh1–TrPDC1 terminator DNA fragment. The resulting SES-C system was subsequently cloned into a T. reesei plasmid with cbh1-locus targeting flanks (Supplementary Table S5).
Expression systems based on the established promoters
The sequences of promoters commonly used for (heterologous) gene expression in S. cerevisiae (PGK1 pr.), Y. lipolytica (TEF1 pr.), Pichia pastoris (GAP1 pr.) and A. niger (Aspergillu nidulans gpdA pr.) are listed in Supplementary Table S12. To assess their functionality in S. cerevisiae, these promoters were cloned into an integration plasmid forming four control constructs where the promoter was placed upstream of the mCherry-ScADH1 terminator fragment (Supplementary Figure S5A). In addition, Y. lipolytica, and A. niger specific constructs with the corresponding promoters were made where the species-specific promoter was placed upstream of the mCherry ScADH1 terminator fragment (Supplementary Figure S5A). All these constructs had identical selection markers and the genome integration DNA-flanks as the corresponding SES cassettes. In case of P. pastoris, the mCherry- ScADH1 terminator fragment was cloned downstream of the GAP1 promoter into the pGAPZ_A vector (Thermo Fisher Scientific) between the EcoRI and NotI sites.
Transformations
Transformations of SES expression cassettes and the cassettes with the established promoters were done by targeted, single copy integration into the host organism’s genome. The integration loci and length of the integration flanks are shown in Supplementary Table S5.
The SES and the control-promoter cassettes (for selected hosts), including the selection marker and the integration flanks, were released from plasmids by NotI restriction enzyme digestion prior to transformation of S. cerevisiae, P. kudriavzevii, Y. lipolytica and A. niger. MssI enzyme digestion was used prior to transformation of T. reesei, C. apicola and Z. lentus. The plasmids for P. pastoris were only linearized by the digestion of MssI (SES systems), or AvrII (GAP1 promoter control).
The yeast integrative plasmids were transformed into yeast parental strains (Supplementary Table S1) with the standard lithium acetate protocol (15), except of C. apicola. The method for T. reesei transformation was described previously (16).
Aspergillus niger transformations were done using a protoplast method. A. niger conidia (∼2 × 108) were cultivated overnight at +28°C (250 rpm) in 250 ml YPGel. Mycelium was vacuum filtrated and washed with water and cold KMC solution (1 M KCl, 25 mM CaCl2, 10 mM Tris–HCl, pH 5.8). Protoplasts were prepared by digesting the cell wall of mycelium by incubation for 3 h at +30°C (60 rpm) with 20 ml of Caylase C4 (Cayla) solution (1 mg enzyme/1 ml KMC). Protoplasts were cooled down on ice and isolated by vacuum filtration through a sterilized fabric. Protoplasts were washed (4 min, 1500 g, +2°C) with 20 ml of cold KMC followed by a wash with 20 ml of cold STC solution (1.33 M D-sorbitol, 10 mM Tris–HCl, 50 mM CaCl2, pH 8.0). Protoplasts were suspended into 200 μl of STC solution. One hundred μl of protoplast suspension was mixed with 3.5 μg of donor DNA (NotI digested plasmid) and 30 μl of CRISPR-solution (1 μM Cas9 protein, 1 μM synthetic crRNA (an equimolar mix of three different targeting RNAs, Supplementary Table S6), and 1 μM tracrRNA (all IDT)). A total of 100 μl of transformation solution (25% PEG 6000, 50 mM CaCl2, 10 mM Tris–HCl, pH 7.5) was added followed by a 20-min incubation on ice. A total of 2 ml of transformation solution was added and the mixture was incubated for 5 min at RT. A total of 4 ml of STC was added followed by 7 ml of molten (50°C) top agar (200 g/l D-sorbitol, 6.7 g/l of yeast nitrogen base, complete supplement mixture without uracil (Formedium) and 20 g/l D-glucose). The mixture was poured on a selection plate lacking uracil (identical composition as the top agar). Cultivation was done at +28°C for 5 days.
Candida apicolatransformation was done by the same method as the A. niger transformation, with following modifications: The protoplasts were prepared from cells cultivated overnight at +30°C (250 rpm) in 50 ml of YPD. The washed cells were incubated in 20 ml of Lysing Enzymes from Trichoderma harzianum (Sigma-Aldrich) solution (1 mg enzyme/1 ml KMC) for 40 min at +30°C (60 rpm). Single targeting RNA was used (Supplementary Table S6). The selection agent in the top agar as well as in the agar plates was 200 μg/ml nourseothricin. Cultivation was done at +30°C for 4 days.
SES- and control-promoter strain genome-integration analysis
Single colonies from transformation plates were isolated and regrown on appropriate selection plates. The clones were analysed to select the ones with correct integration at the intended locus and a single genomic copy of the integrated SES cassettes or the control cassettes with established promoters. Genomic DNA was extracted from the yeasts clones with phenol chloroform extraction method, and from the T. reesei and A. niger clones by using the Phire Plant Direct PCR kit (Thermo Fisher Scientific) according to manufacturer’s protocol. Phenol chloroform method was conducted by disrupting cells using Precellys24 homogenizer (Bertin Instruments). A total of 600 μl of glass beads, 600 μl 1 × TE (pH 7.5) and 600 μl of phenol–chloroform–isoamyl–alcohol solution (50% phenol, 48% chloroform, 2% isoamylalcohol) were added and shaken vigorously. After centrifugation the supernatant was diluted 100× in water to obtain the template for the subsequent copy number and integration site PCR analysis. Copy number analysis was conducted with quantitative reverse transcriptase-PCR (qRT-PCR) Lightcycler 480II (Roche) using LightCycler® 480 SYBR Green I Master (Roche) according to manufacturer’s instructions. The qPCR signal of the SES target (mCherry and Bm3R1) was normalized to a reference gene of the host (RT-PCR primers listed in Supplementary Table S7). The correct single-integrations clones conferred value ∼1 (in case of diploid P. kudriavzevii value ∼0.5). The correctness of the integration loci was confirmed by PCR of the genomic DNA, where the amplified DNA region spanned the integrated construct and the genomic DNA outside of the integration flanks.
Fluorescence measurement of yeast species with fluorometer
Yeast cells (S. cerevisiae, Y. lipolytica, P. kudriavzevii, P. pastoris and C. apicola) were cultivated at +30°C on YPD plates for 24 h before starting the experiment, Z. lentus was cultivated at +24°C on YPD plates for 48 h. A total of 4 ml of SCD medium in 24-well plate was inoculated to initial optical density of 0.2 (OD600). Three parallel replicates were cultivated for each strain. Cells were cultivated for 18 h (+30°C/+24°C in case of Z. lentus, 800 rpm) followed by centrifugation and resuspension in 200 μl of water. A total of 200 μl of the suspension was transferred to Black Cliniplate (Thermo Fisher Scientific) and mCherry fluorescence was measured with Varioskan (Thermo Electron Corporation). The excitation and the emission wavelengths were 587/610 nm, measurement time was 500 ms using a 12 nm excitation bandwidth. Cell density (OD600) measurement was done for normalizing mCherry fluorescence measurement results. For this, cells were diluted 100× in water, and then OD600 was measured with Varioskan (photometric measurement mode, wavelength = 600 nm, bandwidth = 5 nm, measurement time = 100 ms) using a transparent microtiter plate (Nunc 96F, Thermo Fisher Scientific). Normalization was done by dividing the mCherry measurement value with OD600 result.
Fluorescence measurement of T. reesei with fluorometer
Trichoderma reesei conidia were used to inoculate 50 ml YPGel pre-cultivations. Cultivations were carried out at +28°C (200 rpm) for 24 h. Mycelium was centrifuged, supernatant removed and pellet resuspended in water. The suspensions were used to inoculate four ml SCD cultivations (three replicates each) in a 24-well plate to an initial density of 0.5 (OD600). Cells were cultivated 18 h at + 28°C (800 rpm) followed by centrifugation and resuspension in 600 μl of water. A total of 200 μl of the suspension was transferred to Black Cliniplate (Thermo Fisher Scientific) and mCherry fluorescence was measured with Varioskan in the same way as described above in the section ‘Fluorescence measurement of yeast species with fluorometer’. OD600 was measured by preparing 10× dilutions from the cell suspension. OD600 values were used for normalizing the fluorescence results to cell density.
Flow cytometry
Yeast cultivations were conducted in a same way as described above in ‘Fluorescence measurement of yeast species with fluorometer’. Cells were diluted into 1× phosphate-buffered saline (PBS) solution and measurements were done with FACSAria III (BD). A total of 10 000 events were recorded and results were normalized by dividing mCherry fluorescence values by cell size (forward scatter, FSC-A). The green (561 nm) laser and 610/20 nm filter were used for the mCherry fluorescence measurements, the voltage settings used in flow cytometry analysis are shown in Supplementary Table S8. In the case of A. niger and T. reesei strains, the flow cytometry was done using conidia. Strains were first sporulated on PDA plates (7 days), conidia were collected and diluted in 1× PBS prior to the flow cytometry analysis. Minor gating was performed on data to exclude obvious errors, such as dust particles, cell clusters and conidia aggregates. Flow cytometry results were analysed with IBM SPSS statistics software.
Sample collection for transcription analysis
Samples were collected after 18 h of cultivation, washed with cold water before freezing the pellets in liquid nitrogen and stored at −80°C. Transcription analysis was performed using two biological replicates. From each biological replicate, two technical replicates were conducted.
Yeast cells (S. cerevisiae, Y. lipolytica, P. kudriavzevii and P. pastoris) were cultivated for transcription analysis as described in chapter ‘Fluorescence measurement of yeast species with fluorometer’.
Trichoderma reesei mycelia were cultivated for transcription analysis as described in section ‘Fluorescence measurement of T. reesei with fluorometer’. For transcription analysis of bioreactor cultures, samples were collected at the specified time points from the bioreactor. The mycelium was isolated by vacuum filtration and quenched in liquid nitrogen.
Aspergillus niger conidia were used to inoculate 50 ml of YPGel for pre-cultivations. Mycelia were pre-grown in 250-ml Erlenmeyer flasks for 24 h at 28°C, 200 rpm and harvested by vacuum filtration. Collected mycelia were resuspended in 50 ml of SCD and divided in 4 ml parallel cultivations in 24-well plates, and cultivated for 18 h at 28°C, 800 rpm. Samples for transcriptional analysis were collected by vacuum filtration and quenched in liquid nitrogen.
RNA extraction and cDNA synthesis
RNeasy Mini Kit (Qiagen) was used for total RNA extraction from each tested organism. Approximately 100 μg (wet weight) of yeast cells or the mycelium samples were disrupted using Precellys24 homogenizer (Bertin Instruments). A total of 600 μl of glass beads (acid-washed; 425–600 μm; Sigma-Aldrich) and 600 μl of RLT buffer were added to the cells on ice, and shaken (instrument settings: 6500 − 2 × 30 s). After centrifugation of the samples, 350 μl of the supernatant was subjected to the RNeasy protocol 1c for yeast. The RNA concentration and purity was determined by Nanodrop 2000 instrument (Thermo Fisher Scientific), and the integrity of RNA assessed by Agilent 2100 Bioanalyzer. A total of 1 μg of the total RNA was treated with DNase (DNase I RNase-free, Thermo Fisher Scientific) to remove residual genomic DNA. Transcription First Strand cDNA Synthesis Kit (Roche) was used for the cDNA synthesis according to manufacturer’s protocol with anchored-oligo(dT)18 primer.
Quantitative RT-PCR
The RT-PCR reactions were prepared in white LightCycler 480 96-well plates (Roche) by combining 2.5 μl of 100× diluted cDNA samples, 2.5 μl of primer mixes (8 μM each) and 15 μl of 1.5× diluted LightCycler 480 SYBR Green I Master (Roche). The reactions were performed using the Lightcycler 480II instrument (Roche), and the analysis was done with the accompanied software: (i) Advanced Relative Quantification Tool (high confidence program) for calculating fold-expression ratios toward the selected normalization gene signal, (ii) Melting Curve Genotyping Analysis to validate the identity of the PCR products. The tested genes were translation elongation factor one alpha from each selected organism, mCherry and Bm3R1 (sTF). Their expression levels were normalized to ubiquitin-protein ligase encoding gene (UBC6 in S. cerevisiae and its closest homologs in other species). The use of the UBC6 as the transcription normalization control was motivated by the extraordinary stability of its expression in various conditions reported in S. cereviase (17) and also seen in T. reesei (14). In the case of T. reesei, also expression of cbh1 and actin-encoding genes were tested. The primers used in transcription analyses, the target genes including references to genome databases for each organism and the length of each RT-PCR amplicon are shown in Supplementary Table S9.
Comparison of the SES system with selected established promoters
The strains of yeast species (S. cerevisiae, Y. lipolytica and P. pastoris) carrying the SES-A/SES-A* system and the strains with expression systems based on the established promoters were compared by fluorometry. The strains were pre-cultivated overnight on selection plates at +30°C. A total of 4 ml of SCD medium (or SCD + 100 μg/ml zeocin in the case of P. pastoris strains) was inoculated to OD600 of 0.2 in 24-well plates. Cultivations were maintained 18 h at +30°C (800 rpm). The cells were pelleted by centrifugation, resuspended into 200 μl of water and fluorescence was measured as described in the section ‘Fluorescence measurement of yeast species with fluorometer’.
In case of A. niger, transcription analysis was used for the comparison of the SES-A system with the expression system based on the A. nidulans gpdA promoter. A total of 50 ml of YPDGel medium was inoculated with ∼2.5×107 conidia and cultivated for 24 h at +28°C (250 rpm). The mycelium was collected and washed with cold water prior to freezing with liquid nitrogen. RNA extraction and cDNA synthesis was done as described in the sections ‘RNA extraction and cDNA synthesis’ and ‘Quantitative RT-PCR’.
Fluorescence microscopy
Localization of mCherry was visualized in all organisms using a Zeiss LSM 710 laser scanning confocal microscope (Carl Zeiss, Oberkochen) with a 63× water immersion objective (excitation at 543 nm and detection at 586–670 nm). Yeast cells were prepared for microscopy similarly as in ‘Fluorescence measurement of yeast species with fluorometer’ experiments. Filamentous fungi mycelia were prepared by inoculating and cultivating conidia in 50 ml YPGel (28°C, 200 rpm, 2 days). Samples showing A. niger and T. reesei during conidiogenesis (sporulation) were collected from PDA or cellulose-containing agar plates.
CBHI protein production
The use of the SES system for protein production was demonstrated by producing CBHI protein in T. reesei and the performance of the expression system was compared to production of native CBHI in the parental strain. The strain carrying SES system and the parental strain (M2068) were cultivated in 1 l bioreactors either in cellulase-inducing medium (SGM) as batch cultivations, or in repressing medium (containing glucose) at 28°C as fed-batch cultivations. The pH (pH 5.5) and the glucose feed were controlled via the control paradigm DELTABAS, as described previously (18). The cultivations were carried out for 5 days and the samples from days 1 (18 h), 3 (66 h), and 5 (117 h) were analysed for the total protein and CBHI production into the culture media (secreted protein) and for the transcription of the selected genes.
For the total protein analysis, 1.5 μl of each culture supernatant was mixed with 15 μl of 1× sodium dodecyl sulphate (SDS) loading buffer (100 ml/l glycerol, 25 ml/l β-mercaptoethanol, 0.5 g/l OrangeG dye (Sigma-Aldrich), 10 g/l SDS and 31.2 mM Tris–HCl, pH 6.8), boiled and loaded on a 4–20% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) gradient gel (BioRad) together with dilutions of purified CBHI protein as a standard. The gel was stained with colloidal coomassie stain (PageBlue Protein Staining Solution; Thermo Fisher Scientific) according to manufacture’s protocol. The visualization of the stained gel was performed on the Odyssey CLx Imaging System instrument (LI-COR Biosciences). Total protein concentration in the culture supernatants were calculated based on the CBHI standard with the Image Studio™ Software (LI-COR Biosciences).
For the western blot analysis, 0.075 μl of each culture supernatant was mixed with 15 μl of 1× SDS loading buffer, boiled and loaded on a 4–20% SDS-PAGE gradient gel (Criterion TGX, BioRad) together with dilutions of purified CBHI protein. The gel was transferred onto a nitrocellulose membrane (Trans-Blot Turbo, BioRad), and the membrane was blocked with Odyssey blocking solution (LI-COR Biosciences). The CBHI protein was detected with the mouse anti-CBHI primary antibody (mab261, 1:5000 dilution in TBST), and goat anti-mouse-IRDye-680RD secondary antibody (LI-COR Biosciences; 1:10000 dilution in TBST). The composition of TBST solution was 10 mM Tris–HCl, 4.4 g/l NaCl, 0.5 ml/l Tween-20 (Fluka), pH 8.0. The visualization of the signal was performed on the Odyssey CLx Imaging System instrument (LI-COR Biosciences). The CBHI concentration in the culture supernatants was calculated based on the CBHI standard with the Image Studio™ Software (LI-COR Biosciences).
Partial genome sequence analysis of Zygosaccharomyces lentus
The ACT1 gene was amplified from purified genomic DNA of Z. lentus with primers (Zb_ACT1_F: ATGGAGAAAA TCTGGCACCA CACCTTCTAC, and Zb_ACT1_R: TTAGAAACAC TTGTGGTGGA CGATGGATGG) designed based on the related sequenced species Zygosaccharomyces bailii (http://fungi.ensembl.org/Zygosaccharomyces_bailii_isa1307/Info/Index). The PCR product was sequenced (Supplementary Table S10), and the qPCR primers designed based on the sequenced DNA. To obtain suitable sequence for the genome integration, the Z. lentus genomic DNA was digested with MunI restriction enzyme, the DNA fragments ligated into EcoRI-digested plasmid pRSET_A (Invitrogen) and several plasmids containing inserts longer than 1000 bp were sequenced. One of the fragments contained a genomic region encompassing YIP3 CDS with up- and down-stream regions (Supplementary Table S11), which was used for targeting the SES cassette into the genome of Z. lentus.
Analysis of SES system’s genetic stability
The long-term expression stability of the SES system was tested by conducting extended, regularly diluted cultivations. First, yeast strains carrying the SES-A (Y. lipolytica and P. kudriavzevii) or SES-A* (S. cerevisiae and P. pastoris) cassettes were pre-cultivated on selection plates overnight at +30°C. These cultivations were used to inoculate 25 ml of YPD (YPD + 100 μg/ml zeocin in case of P. pastoris) medium to initial OD600 of 0.2. The cultivations were continued for 24 h at +30°C (250 rpm), after which 150 μl of the cultures were used to inoculate new 25 ml of YPD (or YPD + 100 μg/ml zeocin in case of P. pastoris). The re-inoculation was repeated every 24 h for 7 days (Supplementary Figure S8A). The fluorescence was measured at days 1, 3, 5 and 7. Samples for the measurements were prepared by collecting 4 ml of cultures, pelleting the cells by centrifugation and resuspending into 400 μl of water. The fluorescence measurement was done as described in the section ‘Fluorescence measurement of yeast species with fluorometer’. Three biological replicates (independent serial cultivations) were performed for each strain.
Analogous tests for the SES system stability were performed in T. reesei and A. niger strains, which were first pre-cultivated in 50 ml YPGel medium (inoculated by conidia/spores) for 3 days at + 28°C (250 rpm). A total of 2.5 ml of the pre-cultures were used for inoculation of 50 ml YPDGel medium (YPGel with 20 g/l D-glucose). Cultivations were continued for 24 h at + 28°C (250 rpm), after which 2.5 ml of the cultures were used to inoculate another 50 ml YPGel medium. This was repeated every 24 hours for seven days. The samples were collected at 1, 3, 5 and 7 days. For T. reesei, 4 ml culture samples were collected by pelleting the mycelia by centrifugation, and resuspension into 2 ml of water for the fluorescence measurement. The fluorescence was measured as described in the section ‘Fluorescence measurement of yeast species with fluorometer’. For A. niger, transcription analysis was performed because the mycelium morphology does not allow analysis by fluorometry. The mycelium was collected and washed with cold water prior freezing with liquid nitrogen. The A. niger transcription samples were processed as described in the sections ‘RNA extraction and cDNA synthesis’ and ‘Quantitative RT-PCR’. Two biological replicates (independent serial cultivations) were performed for each strain.
RESULTS
Identification of universal core promoters
We recently showed that in S. cerevisiae the TDH3 CP (CP) alone can be used to drive the expression of a synthetic transcription factor (sTF) at a low and constitutive level (10). The low sTF level was sufficient to strongly activate a downstream target gene, controlled by a synthetic sTF-dependent promoter. The low transcription activity of the CP (initial input signal) was amplified by binding of the sTF to the synthetic promoter, leading to a significant expression level of the target gene (output signal). The output expression level could be adjusted from low to very high by modulating the number of sTF BS and the choice of the CP region in the synthetic promoter (Figure 1A) (10). However, this particular expression system (containing S. cerevisiae CPs) had limited or no functionality in other eukaryotic microbes. We hypothesized that by using CP(s) with broad inter-species functionality, it might be possible to construct a generic expression system that can be used for gene expression control in a broad range of eukaryotic organisms.
To identify such CPs we carried out a screen in S. cerevisiae (Figure 1B). CPs selected for the test (∼200 bp region of a promoter immediately upstream of the start codon) originated from filamentous fungi, A. niger and T. reesei, and they were chosen from genes that displayed high transcription levels under various growth conditions (13,14). We reasoned that due to the highly conserved core transcription machinery among eukaryotes (19,20), such CPs could have sufficient ability to initiate transcription in a broad spectrum of species. We were primarily interested in CPs that can form a strong sTF-dependent synthetic promoter, thus we used in the screen a S. cerevisiae strain constitutively expressing a LexA-VP16 sTF (Supplementary Table S3). This strain was transformed with a mix of linearized centromeric plasmid and a library of candidate CPs. After in vivo assembly of the centromeric plasmid with the candidate CP, the expression system was formed, where the tested CP became part of the sTF-dependent synthetic promoter driving the expression of a reporter gene, mCherry (Figure 1B). The level of reporter fluorescence in the resulting strains was used for quantitative assessment of the CP’s functionality (Figure 1C, plotted on the left y-axis). In addition, another S. cerevisiae strain, which lacked the sTF, was used in a parallel screen to assess the basal expression level of the tested CPs in the absence of the sTF (Figure 1C, plotted on the right y-axis). The absence of the sTF resulted in substantially lower expression of the mCherry reporter gene. Three S. cerevisiae native CPs (THI4cp, PGK1cp and TEF1cp) were included in the screen as examples of highly active native CPs. The screen identified several heterologous CPs, which were highly functional in S. cerevisiae. The CP activity with upstream bound sTF was used as the main selection criterion, and the CPs showing similar activity levels as the native yeast CPs were selected for development of the SES (Figure 1C, green and pink shading).
SES enables high level gene expression in diverse eukaryotic microbes
The CPs (An_201205cp ≈ An_201cp and An_00850cp ≈ An_008cp) that displayed highest sTF-associated activity among the non-yeast CPs were used to construct a SES, SES-A (Figure 2A). The system was designed to contain two expression cassettes: (i) the reporter cassette with mCherry gene under the control of a synthetic promoter consisting of eight sTF-BS upstream of the An_201cp; and (ii) the sTF cassette with the An_008cp controlling the sTF expression. The sTF was composed of the (A. niger codon optimized) Bm3R1 DNA binding protein (21), SV40 nuclear localization signal, and the VP16 activation domain (7). The Bm3R1 was chosen based on literature (22,23) and our previous experiments with diverse bacterial DNA-binding proteins in S. cerevisiae (24) and A. niger (our unpublished observation), where the Bm3R1-based sTF showed highest capacity to activate expression. In addition to the SES-A system, an analogous SES, SES-B, was constructed which contained another pair of well-performing CPs identified in the screen (Supplementary Figure S1).
The SES-A system with species-specific selection marker genes and genome-integration DNA-flanks (Figure 2A), was integrated in single-copy into the genomes of the yeasts S. cerevisiae, Pichia (Komagataella) pastoris, P. kudriavzevii and Y. lipolytica, and in the filamentous fungi A. niger and T. reesei. These organisms form a group of evolutionarily distant fungal hosts that are industrially relevant for chemical and fuel production (S. cerevisiae, Y. lipolytica, A. niger), for protein production (P. pastoris and T. reesei), or represent an attractive multi-tolerant host with limited tools for gene expression engineering (P. kudriavzevii (25)). The resulting strains were analysed qualitatively by confocal fluorescence microscopy for production of mCherry (Figure 2B). The analysis revealed strong and uniform fluorescence throughout the cell population in liquid cultures in all tested hosts. In addition, a highly constitutive expression pattern was observed also during formation of conidia (sporulation) of filamentous fungi on solid media (Supplementary Figure S2).
To quantitate the performance of the SES system, we analysed transcription levels of the mCherry, the sTF and the TEF gene (encoding translation elongation factor 1 alpha; TEF1 in yeasts, tef1a in T. reesei, and tefA in A. niger), which is one of the most highly expressed genes in eukaryotes (26). In all hosts, mCherry transcription levels were similar or higher than the level of TEF (Figure 3A). In T. reesei, the transcript level of mCherry was in fact more than 60-fold higher than the level of tef1a. The NC version of the SES system (SES-NC), where no sTF encoding DNA was present, resulted in negligible expression of mCherry in all hosts (Figure 3A), except of P. kudriavzevii, where moderate levels of mCherry transcription was detected in the strain carrying the SES-NC (∼10% compared to the corresponding complete SES system). This suggests that in P. kudriavzevii, the Bm3R1-BS may be recognised by an unidentified, endogenous transcription factor(s). Indeed, when the number of BS was decreased or no BS were used in the NC-SES system, the mCherry expression diminished (Supplementary Figure S3).
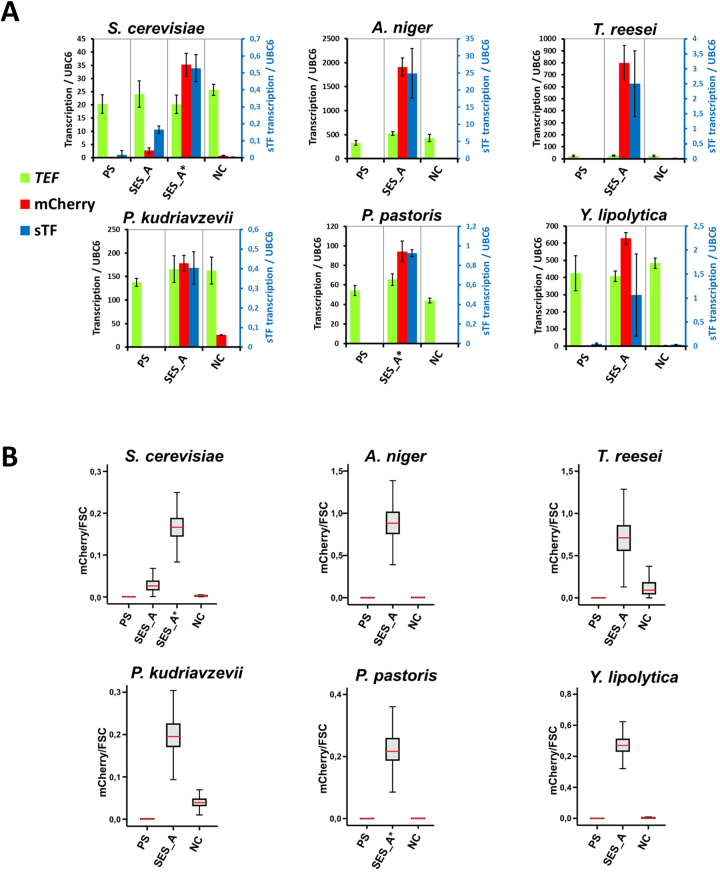
Figure 3.
Quantification of the SES performance in fungal hosts. (A) Transcription analysis of the strains. The mCherry transcript levels were compared to the transcript levels of endogenous TEF genes in each host. Transcript levels of the sTF are shown on the secondary y-axis. PS denotes the parental strains (without SES). In Saccharomyces cerevisiae, in addition to the SES-A, also a modified version was tested having the S. cerevisiae-codon-optimized sTF gene, which is indicated by an asterisk (SES-A*). In Pichia pastoris, only the SES-A* version was used. The transcription of the UBC6 gene homologs (Supplementary Table S9) in each species were used for normalization. Values and error bars represent the mean and standard deviation from two biological (four technical) replicates. (B) Analysis of mCherry expression by flow-cytometry. Flow-cytometry was performed on cells (yeast species) or conidia (filamentous fungi). The box plots show the fluorescence intensity (mCherry) normalized by the particle (cell/conidia) size (FSC—forward scatter) for ∼10 000 cells/conidia from each strain. The horizontal red line (inside the grey box) represents the median value, the grey box represents the interquartile range (IQ range), the bottom line of grey box represents the 25% percentile value, the top line of grey box represents the 75% percentile value. The whiskers in box plot together with the IQ range, represent about 99% of all measured instances (cells/conidia) (for numerical values see Supplementary Figure S4A). Alternative fluorescence analysis of the strains was performed by quantitative fluorometry (Supplementary Figure S4B and C).
In S. cerevisiae, the use of the SES system resulted in relatively low level of the mCherry transcription. This was most likely caused by suboptimal translation efficiency of the sTF mRNA (A. niger codon optimized) in this host with an AT-rich genome. In support of this, the S. cerevisiae codon optimised sTF resulted in high expression of mCherry (Figure 3A, SES-A*). In P. pastoris, another organism with an AT-rich genome, only S. cerevisiae codon optimised sTF was used in the SES-A system (Figure 3A, SES-A*).
To assess the mCherry protein accumulation and expression level distribution in the tested species, fluorescence signal was analysed by flow-cytometry in yeast cells and in conidia of the filamentous fungi (Figure 3B). In addition, quantitative fluorometry of the yeast cells and the T. reesei mycelia was performed as an alternative method for the fluorescence assessment (Supplementary Figure S4). Consistent with the transcription analysis, flow-cytometry revealed high level of red fluorescence in the hosts and demonstrated homogeneous mCherry fluorescence in the analysed populations. In S. cerevisiae, a substantial increase of mCherry production after codon optimization of the sTF was confirmed (Figure 3B, SES-A versus SES-A*). However, additional codon-optimization of the mCherry gene (high-GC content variant used in all SES versions) did not increase the fluorescence level any further (data not shown). In the T. reesei conidia, some level of fluorescence was observed with NC version of the SES-A system, whereas negligible expression was detected in mycelia both by qPCR (Figure 3A) and by fluorometry (Supplementary Figure S4). This observation indicates the existence of an unidentified mechanism that is responsible for low activation of the SES system during T. reesei conidiogenesis (sporulation).
The alternative expression system (SES-B) that was based on different CPs (An_1147651cp ≈ An_114cp and An_53301cp ≈ An_533cp) revealed similar performance as the SES-A system for all analysed parameters (Supplementary Figures S1 and 4). Similar performance of both A and B versions of the SES system suggest that the original screen for CPs carried out in a heterologous organisms S. cerevisiae provided a robust platform for CP functionality.
The function of the SES system was also tested by direct comparison with established expression systems, i.e. native or commonly used promoters. Single copy genome-integrated expression cassettes, containing the mCherry reporter gene under the control of typically used fungal promoters (Supplementary Figure S5A), were compared to the SES-A system in the selected hosts. The S. cerevisiae PGK1 promoter (27), Y. lipolytica TEF1 promoter (28), P. pastoris GAP1 promoter (29) and A. nidulans gpdA (30), were tested in S. cerevisiae, as well as individually in the natural hosts (Supplementary Figure S5B and C). The fluorometry analysis of the yeast strains, and the transcription analysis of the A. niger strains, revealed that the SES system considerably outperformed the established expression systems in each species. In addition, it was demonstrated in the S. cerevisiae example (Supplementary Figure S5B) that the species-specific promoters cannot be used for efficient gene expression in other hosts.
Cross-species expression level tuning by SES
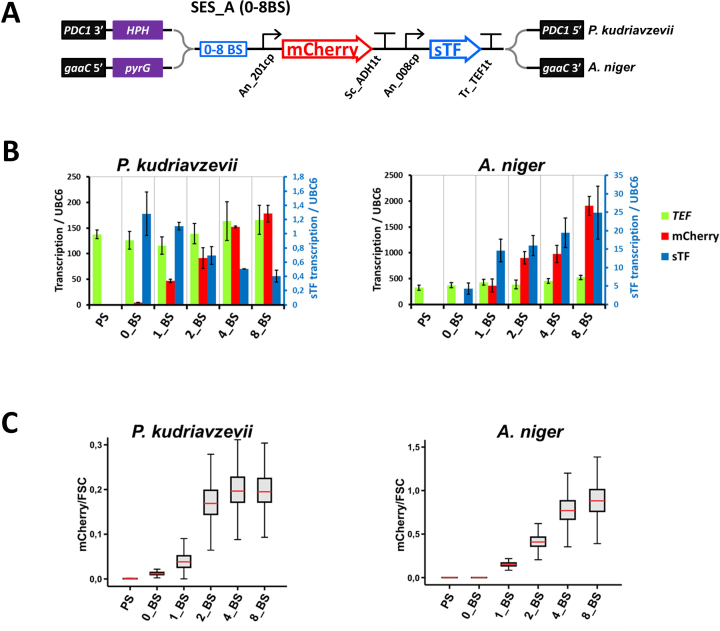
Components of biological systems (enzymes in metabolic pathways, subunits in protein complexes, etc.) need to be expressed at specific, often very dissimilar, levels to ensure optimal performance. For this purpose, numerous expression systems have been developed (31) that provide adjustable expression levels of target genes. The expression levels can be controlled by employing differently strong native promoters or their modified versions (8,30). In heterologous expression systems, inducing agents regulating the transcription levels (1,3,8,9,32), or selected CPs, orthogonal transcription factors and their BS (1,3,6,10,33,34) have also been successfully utilized. However, all of these systems are host-specific. To test whether the SES system enables expression level adjustment in diverse organisms, we used two distantly related species, P. kudriavzevii and A. niger. The SES-A system was modified to contain zero, one, two, four or eight sTF-BS upstream of the mCherry gene (Figure 4A), and it was integrated in single copy into genomes of these hosts. Depending on the number of sTF BS, the mCherry mRNA levels in P. kudriavzevii varied from negligible (0 sTF BS) to levels comparable to TEF1 (eight sTF BS) with about 40-fold expression range (Figure 4B). Notably, P. kudriavzevii is a diploid yeast where both alleles contribute to the transcription level of TEF1, as opposed to mCherry expression from a single SES-A cassette. In A. niger, expression levels varied from negligible to levels that exceeded 4-fold tefA expression with over a 9000-fold expression range (Figure 4B). Fluorescence flow cytometry analysis of P. kudriavzevii cells and A. niger conidia confirmed the adjustability of mCherry expression levels in these strains (Figure 4C).
Figure 4.
Expression level adjustability tests in Pichia kudriavzevii and Aspergillus niger. (A) A schematic presentation of the SES-A cassettes with zero to eight sTF-BS (0–8BS), including positions of selection markers and genome-integration flanks for P. kudriavzevii and A. niger. (B) Transcription analysis of the P. kudriavzevii and A. niger strains with the SES-A (0–8BS) systems. The mCherry, the TEF and the sTF transcript levels were monitored, and normalized to the transcript signal of the UBC6 homologs as in the Figure 3B. PS denotes parental strain (without SES). Values and error bars represent the mean and standard deviation from two biological (four technical) replicates. (C) Analysis of the mCherry fluorescence by flow-cytometry measurements of P. kudriavzevii cells and A. niger conidia. The box plots show the same features as in Figure 3B. The analysis of the mCherry expression in P. kudriavzevii was also performed by quantitative fluorometry (Supplementary Figure S2D).
SES enables improved protein production in T. reesei
To assess the utility of the SES system in an industrially relevant application, we employed T. reesei, which has the natural ability to secrete large amounts of proteins (35) and is commonly used for industrial production of enzymes (36). Typically, heterologous protein production is achieved by using the strong inducible cbh1 promoter. The media inducing this promoter leads to simultaneous co-production of a high number of native cellulolytic enzymes (36). This results in increased downstream purification costs for the protein(s) of interest. Therefore, an expression system that enables high production levels in conditions where the broad induction of cellulolytic enzymes does not occur would be highly beneficial.
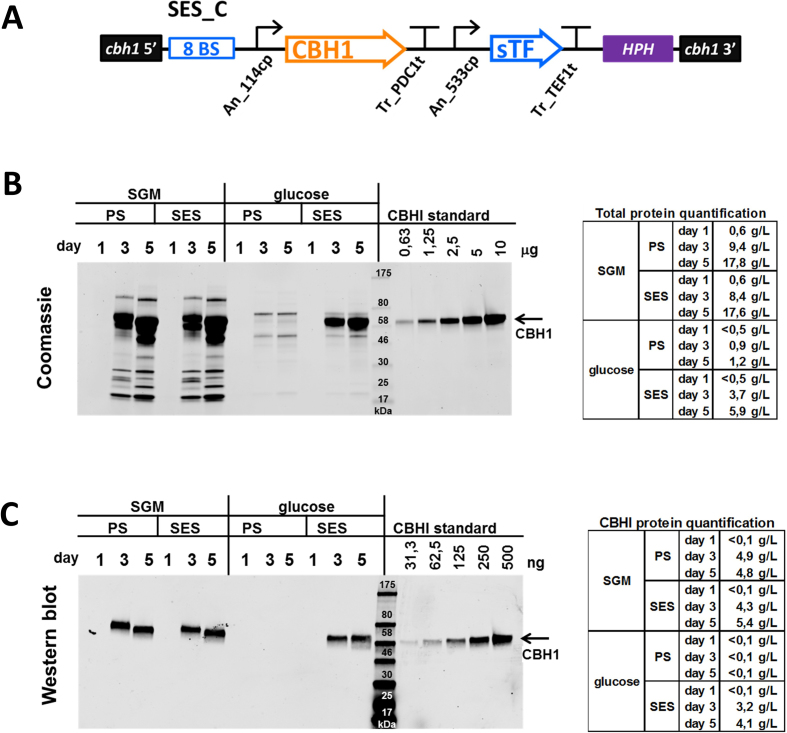
We modified the SES expression cassette by replacing the mCherry gene with a gene encoding T. reesei native cellobiohydrolase CBHI enzyme (Figure 5A). A T. reesei strain with two genome-integrated copies of the CBHI-SES cassette (SES-C) was tested in bioreactor cultivations. The SES-modified strain and the parental strain were cultivated either in a cellulase-inducing medium containing spent grain and lactose (SGM) commonly used for cbh1-promoter-driven production of proteins, or in a glucose-containing medium where the expression of large majority of native secreted enzymes (e.g. cellulases) is repressed. Both strains secreted highly similar protein mixtures (17.8 g/l and 17.6 g/l total protein on day 5, respectively) into the culture medium in the cellulase-inducing conditions (Figure 5B). However, when the strains were grown in the presence of glucose, only a small amount of protein (1.2 g/l, on day 5) was secreted by the parental strain, whereas the SES strain secreted 5.9 g/l of protein, which consisted mainly of CBHI (∼70% in contrast to ∼30% in SGM). In addition, the amount of the CBHI protein produced by the SES strain on glucose was similar to the amount produced by the parental strain in inducing conditions (4.1 g/l and 4.8 g/l, respectively) (Figure 5C).
Figure 5.
SES-based production of CBHI protein in Trichoderma reesei. (A) A schematic presentation of the SES-C cassette, including position of the HPH selection marker and cbh1-locus genome-integration flanks, used for production of CBHI enzyme in T. reesei. (B) The analysis of total protein produced (secreted) in bioreactor cultivations of T. reesei. The parental (PS) as well as the SES strain were cultivated in cellulase-inducing medium (containing lactose and spent grain, SGM), and repressing medium (containing glucose). Culture media samples were analysed for total produced protein by SDS-PAGE gel and Coomassie staining (left panel). The total protein concentrations were calculated based on the purified CBHI protein standard (right panel). (C) The analysis of the CBHI protein produced (secreted) in bioreactor cultivations of T. reesei. The samples of B) were analysed by a western blot (left panel) with a specific anti-CBHI (mab261) antibody. The CBHI concentrations were calculated based on the purified CBHI protein standard (right panel).
Transcription analysis of the bioreactor samples (Supplementary Figure S6) revealed principal differences in CBHI production in the parental and the SES strains. In the parental strain, cbh1 transcription was strongly induced, albeit decreasing at latter stages of the culture in the SGM medium, as is typically observed (14). However, in the SES strain the cbh1 transcription displayed a much more stable profile in both media.
SES functionality in novel organisms
Numerous microbes with highly promising properties are currently underutilized both in research and in biotechnological applications. This is mainly due to lack of tools for their genetic engineering, and related to this, the need for tedious development of such tools for each organism individually. We set out to test the usability of the SES system in engineering of two such organisms, the highly osmotolerant yeasts species C. apicola (37) and Z. lentus (38) that, to our knowledge, have not been genetically engineered previously. A genome sequence draft of C. apicola has been published (37); however, Z. lentus genome has not been sequenced. To circumvent this deficit, we performed partial genome sequence analysis of Z. lentus to obtain a suitable non-essential locus for SES cassette integration. To enable selection of transformants in these novel hosts the SES system was modified to allow selection marker gene (NAT) expression control by the sTF (Supplementary Figure S7A). The SES cassette integration in the genomes of these species resulted in strains, which were not only able to grow in the presence of nourseothricin, but showed strong and uniform mCherry fluorescence as monitored by fluorescent microscopy (Supplementary Figure S7B), and fluorometry analysis (Supplementary Figure S7C). This demonstrates the potential of the SES system beyond established hosts, and opens up possibilities to introduce new organisms for a broad range of applications.
DISCUSSION
Introducing novel production hosts to industrial processes is currently limited by the need to develop an expression system for each host individually. In addition, the host-specific DNA parts are not orthogonal to the native regulation and may require specific cultivation conditions for optimal function. For example, the use of native DNA parts may prohibit the use of the cheapest available media or may require supplementation of various inducer compounds. Therefore, an expression system that would circumvent these issues, and yet allow efficient expression control in any host is in great demand.
This work provides a solution by establishing a robust and transferable SES that is functional in a remarkably broad range of eukaryotic microbes. The SES system enables expression of target gene(s) at similar or higher levels than very strong endogenous promoters or conventional gene expression tools. In addition, the SES can be used for on-demand modulation of target gene(s) expression levels, hence offering novel possibilities in orthogonal control for metabolic engineering, protein production, or other bioengineering purposes. Without host-specific optimization, we demonstrate SES utility in six yeasts and two filamentous fungi covering several industrially relevant fungal genera. Thus, SES displays unprecedented functionality in gene expression regulation across a wide range of eukaryotic hosts.
The functionality in diverse hosts arises from the key elements of SES, the selected CPs. We identified the highly functional universal CPs for SES by screening CPs from filamentous fungi (A. niger and T. reesei) in S. cerevisiae (Figure 1B and C) and found candidates, which proved to be highly functional as universal CPs. The candidate CPs were chosen based on the presence of a TATA-box in their structure, and on the relative level of gene expression, where these CPs are part of the promoter. Indeed, most eukaryotic promoters do not contain a canonical TATA-box (39–41) and it is likely that many of the so called ‘TATA-box-less’ CPs might also be universally functional. There were two features of the screened CPs essential for their use in the SES system: (i) ability to provide high transcription level of the target gene, when incorporated in the sTF-dependent promoter, and (ii) sufficient basal transcription activity, when used as a sole control element for the sTF expression. These two characteristics seemed to be positively correlated for majority of the tested CPs (Figure 1C).
We demonstrated that the SES system can provide superior performance in providing constitutive and high expression levels in diverse hosts. In fact, the expression levels achieved by the SES were considerably higher than expression levels of many established expression systems (commonly used native promoters) in the tested fungal hosts (Supplementary Figure S5). In addition, in A. niger and T. reesei, the SES enabled expression in conidia, hyphae and other tissues (A. niger conidiophore) (Supplementary Figure S1) indicating unprecedented stability of the expression outputs. This is in marked contrast e.g. to the activity of the native T. reesei cbh1 promoter that is commonly used for industrial production of heterologous proteins in T. reesei. The expression of mCherry under the cbh1 promoter resulted in a strong fluorescence signal only in T. reesei mycelia but not in conidia in the cbh1-inducing conditions (Supplementary Figure S1).
In many cases, protein purification costs may comprise up to 90% of the manufacturing costs. Thus, simplification of down-stream processing, for instance—by enhancing the purity of the product, could result in substantial savings. We used the SES system in the filamentous fungus T. reesei for production of a widely used industrial enzyme, cellobiohydrolase I (CBHI) and demonstrated high-level production (secretion into culture medium) of this protein in the presence of glucose (Figure 5). With this strategy we were able to produce significantly purer protein at levels comparable to the levels achieved by the well-established cbh1 promoter. This indicates that SES can enable high production levels for desired enzymes, or even tailored enzyme mixtures, without undesirable background activities in T. reesei. It should be emphasized that the SES system was not optimized for T. reesei, and the target gene transcription levels obtained were significantly lower than the maximal transcription levels with the fully induced cbh1 promoter (Supplementary Figure S4). The constitutive nature of the SES-controlled expression, however, provided means for a high final production titer of the CBHI protein. It is possible that other CPs, optimized for use in T. reesei would allow a stronger transcription output and improved production.
In contrast to protein production, synthesis of biochemicals in microbial fermentations involves enzymatic pathways to convert a substrate to a target compound. Efficient conversion requires the individual enzymes of a pathway to be present in optimal proportions to ensure maximal flux through the pathway. Highly dissimilar expression levels of the pathway enzymes may be needed, especially if some heterologous enzymes confer poor performance in the given host (42). Our results provide strong evidence that SES allows expression level tuning in diverse hosts, as demonstrated in P. kudriavzevii and A. niger (Figure 4B and C) with identical expression cassettes. In addition, the finely graded expression pattern was retained in A. niger in mycelium grown in liquid culture (Figure 4B) as well as in conidia formed on solid media (Figure 4C). This implies that the SES system allows, in multiple hosts, fine-tuned metabolic pathway optimization that is essential to reach industrially feasible production levels.
The genetic stability of an expression system is a key feature for any successful application. To reduce the probability for recombination between repetitive sTF-binding-sites, the SES system was designed to contain variable DNA-linkers between the individual BS. In addition, the sTF-BS can also be modified by introducing mutations, which can increase the variability, but do not negatively affect the sTF binding. We tested the stability of the SES performance in extended cultivations, and found no signs of expression level decline during repeated serial cultures over one week (Supplementary Figure S8).
Future development of industrial biotechnology will need introduction of novel production hosts. The need for new organisms is motivated by numerous factors, including use of novel substrates or production conditions (often harsh or toxic biomass hydrosylates/sidestream fractions), need for specific metabolic or physiological features, IPR issues, etc. However, introducing new hosts with superior native characteristics into a bioprocess can be a costly endeavour, especially if the development and establishment of expression tools has to be undertaken to genetically engineer the novel organism. We demonstrate with two little studied yeast species, C. apicola and Z. lentus, that SES provides an important, enabling tool for the establishment of new industrial hosts. In order to achieve selection of transformed colonies, and without prior knowledge about native or functional promoters in these species, we extended the SES system with a selectable marker gene to create a fully autonomous, universal expression system (Supplementary Figure S5A). By obtaining highly fluorescent colonies, we were able to demonstrate that the SES system can be used for high-level gene expression control also in these previously unexploited hosts.
In conclusion, the SES system is ready to be applied for studies investigating biological phenomena in a broad range of eukaryotic microorganisms that have previously been lacking gene expression tools. At the same time, SES can be used to optimize industrial production hosts, and increase the number of engineerable organisms that exhibit advantageous properties for biotechnological applications.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Mikko Arvas for valuable help in analysing the filamentous fungal transcriptomics data for the CP selection, and Toni Paasikallio, Merja Helanterä and Oriane Vedrines for excellent technical assistance. We thank Paula Jouhten and Robert Pylkkänen for critical comments on the manuscript.
Author Contributions’: D.M., A.R., C.P.L. and J.J. conceived the study and analysed the results; A.R. and D.M. conducted and performed most of the experiments, C.P.L. conducted the bioreactor cultivations; J.K. performed the transcription analysis in A. niger; A.K. performed the transcription analysis in T. reesei; L.R. performed the fluorescence microscopy; O.K. performed part of the yeast experiments; M.V. constructed SES-CBHI-producing T. reesei strain; J.J., D.M., A.R., C.P.L. and M.P. wrote the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Finnish Funding Agency for Innovation (TEKES) (Living factories project) [562/31/2014]; Academy of Finland [260957, 272550].
Conflict of interest statement. SES has been granted a patent (Finnish patent (FI127283B) 15 March 2018) and is the subject of patent applications (PCT/FI2017/050114 on 21 February 2017 and 015234 US on 18 December 2017) by D.M., A.R., J.J., C.P.L. and J.K.
REFERENCES
- 1. McIsaac R.S., Gibney P.A., Chandran S.S., Benjamin K.R., Botstein D.. Synthetic biology tools for programming gene expression without nutritional perturbations in Saccharomyces cerevisiae. Nucleic Acids Res. 2014; 42:e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McIsaac R.S., Oakes B.L., Wang X., Dummit K.A., Botstein D., Noyes M.B.. Synthetic gene expression perturbation systems with rapid, tunable, single-gene specificity in yeast. Nucleic Acids Res. 2013; 41:e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ottoz D.S., Rudolf F., Stelling J.. Inducible, tightly regulated and growth condition-independent transcription factor in Saccharomyces cerevisiae. Nucleic Acids Res. 2014; 42:e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liang J., Ning J.C., Zhao H.. Coordinated induction of multi-gene pathways in Saccharomyces cerevisiae. Nucleic Acids Res. 2013; 41:e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mazumder M., McMillen D.R.. Design and characterization of a dual-mode promoter with activation and repression capability for tuning gene expression in yeast. Nucleic Acids Res. 2014; 42:9514–9522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ito Y., Yamanishi M., Ikeuchi A., Matsuyama T.. A highly tunable system for the simultaneous expression of multiple enzymes in Saccharomyces cerevisiae. ACS Synth. Biol. 2015; 4:12–16. [DOI] [PubMed] [Google Scholar]
- 7. Sadowski I., Ma J., Triezenberg S., Ptashne M.. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988; 335:563–564. [DOI] [PubMed] [Google Scholar]
- 8. Blount B.A., Weenink T., Vasylechko S., Ellis T.. Rational diversification of a promoter providing fine-tuned expression and orthogonal regulation for synthetic biology. PLoS One. 2012; 7:e33279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Belli G., Gari E., Piedrafita L., Aldea M., Herrero E.. An activator/repressor dual system allows tight tetracycline-regulated gene expression in budding yeast. Nucleic Acids Res. 1998; 26:942–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rantasalo A., Czeizler E., Virtanen R., Rousu J., Lahdesmaki H., Penttila M., Jantti J., Mojzita D.. Synthetic transcription amplifier system for orthogonal control of gene expression in Saccharomyces cerevisiae. PLoS One. 2016; 11:e0148320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oldenburg K.R., Vo K.T., Michaelis S., Paddon C.. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 1997; 25:451–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Raymond C.K., Pownder T.A., Sexson S.L.. General method for plasmid construction using homologous recombination. Biotechniques. 1999; 26:134–141. [DOI] [PubMed] [Google Scholar]
- 13. Kuivanen J., Wang Y.J., Richard P.. Engineering Aspergillus niger for galactaric acid production: elimination of galactaric acid catabolism by using RNA sequencing and CRISPR/Cas9. Microb. Cell Fact. 2016; 15:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pakula T.M., Nygren H., Barth D., Heinonen M., Castillo S., Penttila M., Arvas M.. Genome wide analysis of protein production load in Trichoderma reesei. Biotechnol. Biofuels. 2016; 9:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gietz R.D., Woods R.A.. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002; 350:87–96. [DOI] [PubMed] [Google Scholar]
- 16. Penttila M., Nevalainen H., Ratto M., Salminen E., Knowles J.. A versatile transformation system for the cellulolytic filamentous fungus Trichoderma reesei. Gene. 1987; 61:155–164. [DOI] [PubMed] [Google Scholar]
- 17. Teste M.A., Duquenne M., Francois J.M., Parrou J.L.. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol. Biol. 2009; 10:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bailey M.J., Tahtiharju J.. Efficient cellulase production by Trichoderma reesei in continuous cultivation on lactose medium with a computer-controlled feeding strategy. Appl. Microbiol. Biotechnol. 2003; 62:156–162. [DOI] [PubMed] [Google Scholar]
- 19. Albright S.R., Tjian R.. TAFs revisited: more data reveal new twists and confirm old ideas. Gene. 2000; 242:1–13. [DOI] [PubMed] [Google Scholar]
- 20. Burley S.K., Roeder R.G.. Biochemistry and structural biology of transcription factor IID (TFIID). Annu. Rev. Biochem. 1996; 65:769–799. [DOI] [PubMed] [Google Scholar]
- 21. Palmer C.N., Axen E., Hughes V., Wolf C.R.. The repressor protein, Bm3R1, mediates an adaptive response to toxic fatty acids in Bacillus megaterium. J. Biol. Chem. 1998; 273:18109–18116. [DOI] [PubMed] [Google Scholar]
- 22. Stanton B.C., Nielsen A.A., Tamsir A., Clancy K., Peterson T., Voigt C.A.. Genomic mining of prokaryotic repressors for orthogonal logic gates. Nat. Chem. Biol. 2014; 10:99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stanton B.C., Siciliano V., Ghodasara A., Wroblewska L., Clancy K., Trefzer A.C., Chesnut J.D., Weiss R., Voigt C.A.. Systematic transfer of prokaryotic sensors and circuits to mammalian cells. ACS Synth. Biol. 2014; 3:880–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rantasalo A., Kuivanen J., Penttila M., Jantti J., Mojzita D.. Synthetic toolkit for complex genetic circuit engineering in Saccharomyces cerevisiae. ACS Synth. Biol. 2018; 7:1573–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matsushika A., Negi K., Suzuki T., Goshima T., Hoshino T.. Identification and characterization of a novel Issatchenkia orientalis GPI-Anchored protein, IoGas1, required for resistance to low pH and salt stress. PLoS One. 2016; 11:e0161888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sasikumar A.N., Perez W.B., Kinzy T.G.. The many roles of the eukaryotic elongation factor 1 complex. Wiley Interdiscip. Rev. RNA. 2012; 3:543–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Partow S., Siewers V., Bjorn S., Nielsen J., Maury J.. Characterization of different promoters for designing a new expression vector in Saccharomyces cerevisiae. Yeast. 2010; 27:955–964. [DOI] [PubMed] [Google Scholar]
- 28. Muller S., Sandal T., Kamp-Hansen P., Dalboge H.. Comparison of expression systems in the yeasts Saccharomyces cerevisiae, Hansenula polymorpha, Klyveromyces lactis, Schizosaccharomyces pombe and Yarrowia lipolytica. Cloning of two novel promoters from Yarrowia lipolytica. Yeast. 1998; 14:1267–1283. [DOI] [PubMed] [Google Scholar]
- 29. Zhang A.L., Luo J.X., Zhang T.Y., Pan Y.W., Tan Y.H., Fu C.Y., Tu F.Z.. Recent advances on the GAP promoter derived expression system of Pichia pastoris. Mol. Biol. Rep. 2009; 36:1611–1619. [DOI] [PubMed] [Google Scholar]
- 30. Blumhoff M., Steiger M.G., Marx H., Mattanovich D., Sauer M.. Six novel constitutive promoters for metabolic engineering of Aspergillus niger. Appl. Microbiol. Biotechnol. 2012; 97:259–267. [DOI] [PubMed] [Google Scholar]
- 31. Leavitt J.M., Alper H.S.. Advances and current limitations in transcript-level control of gene expression. Curr. Opin. Biotechnol. 2015; 34:98–104. [DOI] [PubMed] [Google Scholar]
- 32. Meyer V., Wanka F., van Gent J., Arentshorst M., van den Hondel C.A., Ram A.F.. Fungal gene expression on demand: an inducible, tunable, and metabolism-independent expression system for Aspergillus niger. Appl. Environ. Microbiol. 2011; 77:2975–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leavitt J.M., Tong A., Tong J., Pattie J., Alper H.S.. Coordinated transcription factor and promoter engineering to establish strong expression elements in Saccharomyces cerevisiae. Biotechnol. J. 2016; 11:866–876. [DOI] [PubMed] [Google Scholar]
- 34. Redden H., Alper H.S.. The development and characterization of synthetic minimal yeast promoters. Nat. Commun. 2015; 6:7810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cherry J.R., Fidantsef A.L.. Directed evolution of industrial enzymes: an update. Curr. Opin. Biotechnol. 2003; 14:438–443. [DOI] [PubMed] [Google Scholar]
- 36. Saloheimo M., Pakula T.M.. The cargo and the transport system: secreted proteins and protein secretion in Trichoderma reesei (Hypocrea jecorina). Microbiology. 2012; 158:46–57. [DOI] [PubMed] [Google Scholar]
- 37. Vega-Alvarado L., Gomez-Angulo J., Escalante-Garcia Z., Grande R., Gschaedler-Mathis A., Amaya-Delgado L., Sanchez-Flores A., Arrizon J.. High-qualitydraft genome sequence of Candida apicola NRRL Y-50540. Genome Announc. 2015; 3:e00437-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Steels H., James S.A., Roberts I.N., Stratford M.. Zygosaccharomyces lentus: a significant new osmophilic, preservative-resistant spoilage yeast, capable of growth at low temperature. J. Appl. Microbiol. 1999; 87:520–527. [DOI] [PubMed] [Google Scholar]
- 39. Basehoar A.D., Zanton S.J., Pugh B.F.. Identification and distinct regulation of yeast TATA box-containing genes. Cell. 2004; 116:699–709. [DOI] [PubMed] [Google Scholar]
- 40. Yang C., Bolotin E., Jiang T., Sladek F.M., Martinez E.. Prevalence of the initiator over the TATA box in human and yeast genes and identification of DNA motifs enriched in human TATA-less core promoters. Gene. 2007; 389:52–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zuo Y.C., Li Q.Z.. Identification of TATA and TATA-less promoters in plant genomes by integrating diversity measure, GC-Skew and DNA geometric flexibility. Genomics. 2011; 97:112–120. [DOI] [PubMed] [Google Scholar]
- 42. Du J., Yuan Y., Si T., Lian J., Zhao H.. Customized optimization of metabolic pathways by combinatorial transcriptional engineering. Nucleic Acids Res. 2012; 40:e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.