Abstract
Escherichia coli McrA (EcoKMcrA) acts as a methylcytosine and hydroxymethylcytosine dependent restriction endonuclease. We present a biochemical characterization of EcoKMcrA that includes the first demonstration of its endonuclease activity, small angle X-ray scattering (SAXS) data, and a crystal structure of the enzyme in the absence of DNA. Our data indicate that EcoKMcrA dimerizes via the anticipated C-terminal HNH domains, which together form a single DNA binding site. The N-terminal domains are not homologous to SRA domains, do not interact with each other, and have separate DNA binding sites. Electrophoretic mobility shift assay (EMSA) and footprinting experiments suggest that the N-terminal domains can sense the presence and sequence context of modified cytosines. Pyrrolocytosine fluorescence data indicate no base flipping. In vitro, EcoKMcrA DNA endonuclease activity requires Mn2+ ions, is not strictly methyl dependent, and is not observed when active site variants of the enzyme are used. In cells, EcoKMcrA specifically restricts DNA that is modified in the correct sequence context. This activity is impaired by mutations of the nuclease active site, unless the enzyme is highly overexpressed.
INTRODUCTION
Restriction enzymes (REases) provide a genetic barrier for incoming DNA. Typically, such systems rely on DNA methylation patterns to distinguish self from non-self. Most REases identify invading DNA by a lack of modifications, while leaving genomic DNA untouched. However, there is also a growing group of REases which do the exact opposite and recognize invading DNA by its modifications.
The Escherichia coli immigration control systems for modified DNA were initially identified genetically based on the susceptibility of bacterial strains to phages containing modified DNA, or to plasmids that were methylated in certain sequence contexts. Genetic studies identified modified cytosine restriction A and B (mcrA and mcrB) as barriers for DNA modified in CCGG (M.HpaII) and RGCGCY (M.HaeII) sequence contexts, respectively (1). The same genes had previously been named rglA and rglB, where rgl stands for ‘restricts glucose less phage’. The nomenclature was based on the observation that ablation of these genes affected propagation of glucosylation deficient phages with 5-hydroxymethylcytosine (5hmC) residues incorporated in the DNA, but not glucosylation proficient phages with DNA containing glucosyl-5hmC (2). Later, mutants in mrr, already known to affect restriction of methylated DNA, were also found to impair restriction of cytosine modified DNA in the mcrA-mcrB double negative background (3).
Since their genetics driven discovery, much information has accumulated on the biochemical properties of the modification dependent E. coli restriction endonucleases. Mrr is a remote homologue of the MspJI family of REases (4), which use an SRA domain for interaction with methylated DNA and a PD-(D/E)XK domain (the exact motif of this family is PD-QXK) for DNA cleavage (5–7). McrB consists of a AAA+ domain (8), and a structurally characterized specificity determining domain (McrB-N) (9) and assembles into a protein complex with the PD-(D/E)XK endonuclease McrC (10). The mechanistic basis for McrA modification dependence remains entirely unclear, making the enzyme an attractive target for structural studies.
McrA from E. coli K strains (EcoKMcrA) restricts modified DNA only when the methylation is in an appropriate sequence context. For example, it does not restrict DNA that has been methylated by Dcm in the C5mCWGG context, and therefore can co-exist with Dcm in E. coli. In contrast, the enzyme is very effective in restricting DNA that has been methylated by M.HpaII in the C5mCGG context. Until now, it has been impossible to reproduce DNA cleavage by EcoKMcrA in the test tube, even using plasmids that are restricted in cells (11). However, it has been shown that the enzyme exhibits methylation dependent DNA binding, which can be assayed biochemically. Electrophoretic mobility shift assays (EMSAs) suggest that the EcoKMcrA specificity is Y5mCGR (12), i.e. slightly broader than required to cleave M.HpaII methylated DNA (13). How the enzyme achieves its sequence and modification specificity is incompletely understood. EcoKMcrA has the highest affinity to fully (symmetrically) methylated DNA, but the enzyme also binds hemimethylated DNA more than 10-fold tighter than unmodified DNA. In ‘natural’ DNA substrates, the base opposite the 5mC is always a guanine. In synthetic substrates, replacement of the G base by A, C, T or U inhibits binding, and only inosine is tolerated (12).
EcoKMcrA, like many other REases, is a dimer according to size exclusion chromatography (11). Sequence analysis suggests the presence of an N-terminal domain that is not readily amenable to bioinformatics analysis and was implicated in the modification readout (14), and a C-terminal domain that can be confidently classified as an HNH nuclease (15). HNH domains are present in many restriction and homing endonucleases, as well as Cas9 proteins. They are built around a structural Zn2+ ion, and a second divalent catalytic metal cation. In EcoKMcrA, the Zn2+ ligands, the core ββα-Me motif with active site metal ligands and the nucleophile activating histidine are all predicted to be present. Therefore, the difficulties to demonstrate its activity in vitro have been surprising.
EcoKMcrA is not only of interest in its own right, but it can also be regarded as the prototype of a larger family of related enzymes. BLASTP searches readily identify proteins from eubacteria and archaea as similar to EcoKMcrA. However, in many cases, the similarity is clearly limited to the HNH domain. When only proteins with similarity to EcoKMcrA spanning the entire protein region are considered, the range of host organisms narrows down to the γ-proteobacteria, more specifically Enterobacteriaceae and a few additional species such as Aeromonas, Kushneria and Vibrio. Interestingly, the bacteria harboring clear EcoKMcrA orthologues are vertebrate commensals or pathogens. This finding supports the idea, originally based on the sequence specificity of the enzyme, that EcoKMcrA may also act as a barrier against the influx of CpG methylated DNA from the host (12).
Outside the γ-proteobacteria, ScoMcrA from the actinobacterial Streptomyces coelicolor has been described in some detail (16). The classification of ScoMcrA as an McrA protein is based on clear sequence similarity of the C-terminal nuclease domains. The N-terminal domains are rather different judging from amino acid sequence. ScoMcrA was originally discovered for its restricting activity on phosphorothioated DNA (16). The enzyme has type IV REase activity in the test tube, and has been shown to cleave 12–16 nucleotides away from a 5mC site and 16–28 nucleotides away from a site of phosphorothioation (16). It is still unknown whether the activity of ScoMcrA against phosphorothioated DNA is shared by the enzyme from E. coli.
Here, we present a biochemical characterization of EcoKMcrA that includes the first demonstration of its in vitro activity and the crystal structure of the enzyme in the absence of DNA together with small-angle X-ray scattering (SAXS) data obtained in solution. The data show that EcoKMcrA dimerizes via the HNH domains and has three separate DNA binding sites, one in the central channel formed by the nuclease domains, and two in the N-terminal domains. EMSA and DNase I footprinting data identify the N-terminal domains as the modification specific regions of EcoKMcrA.
MATERIALS AND METHODS
Cloning
The expression vectors for EcoKMcrA with N- and C-terminal His6-tags (MGHHHHHHEF-EcoKMcrA1-277 and EcoKMcrA1-277-GHHHHHHG) were made by ligating the ecokmcrA gene into the pET15bm and pLATE31 plasmids, respectively (ThermoFisher Scientific). The pLATE31 plasmid was also used for the expression construct of the EcoKMcrA N-terminal fragment (EcoKMcrA1-174-GHHHHHHG, henceforth dubbed ‘EcoKMcrA-N’). Mutations were introduced using the ‘quick change’ protocol (17). Sequencing confirmed the intended sequences for all EcoKMcrA variants except for the N-terminally histidine tagged protein, where the presence of a K196E mutation was revealed.
Overproduction
Cells were grown at 37°C to OD600 of 0.7, induced by the addition of IPTG (0.2 mM for EcoKMcrA-His6 and EcoKMcrA-N-His6, and 0.5 mM for His6-EcoKMcrA). Protein expression was carried out either overnight at 16°C (EcoKMcrA-His6 and EcoKMcrA-N-His6) or for 4 h at 22°C (His6-EcoKMcrA).
Purification
EcoKMcrA-His6 and EcoKMcrA-N-His6 variants were purified by chromatography through HisTrap HP chelating and HiTrap Heparin HP columns (GE Healthcare). For His6-EcoKMcrA purification, a gel filtration in 50 mM Tris–HCl pH 7.6, 200 mM NaCl, 1 mM EDTA, 1 mM DTT replaced the Heparin step. Proteins were concentrated by ultrafiltration, and stored in 20 mM Tris–HCl, 200 mM KCl, 1 mM DTT and 50% (v/v) glycerol, pH 8.0. Protein concentrations were determined from A280 measurements using the theoretical extinction coefficients calculated with the ProtParam tool available at http://web.expasy.org/protparam.
Oligonucleotides
Oligoduplex substrates used in this study are listed in Supplementary Table S1. Radioactive labeling was performed with [γ-33P]ATP (PerkinElmer) or [γ-32P]ATP (Hartmann Analytics) and T4 polynucleotide kinase (ThermoFisher Scientific). Oligoduplexes were assembled by annealing of the corresponding radiolabeled and unlabeled strands.
Electrophoretic mobility shift analysis
DNA binding of EcoKMcrA and EcoKMcrA-N was analyzed using 33P-labeled 12 or 30 bp oligoduplexes. DNA in the final concentration of 0.05–0.5 μM was obtained by mixing 2–4 nM of radiolabeled DNA with an appropriate amount of unlabeled oligoduplex. It was incubated with the protein (final concentrations varied from 0.02 to 2 μM) for 15 min in 20 μl of the binding buffer (40 mM Tris-acetate, pH 8.3 at 25°C, 0.1 mg/ml BSA) supplemented with 10% (v/v) glycerol and 1 mM EDTA. Free DNA and protein-DNA complexes were separated by electrophoresis through 8% w/v polyacrylamide gels (29:1 acrylamide/bisacrylamide, gel length:width:thickness 22:15:0.1 cm) in 40 mM Tris-acetate (pH 8.3) for 45 min at 5 V/cm. Low power consumption during electrophoresis runs (∼2 W or 110 V × ∼18 mA per electrophoretic unit containing two gels and 1 l of electrophoretic buffer) ensured that the gels remained at room temperature (∼22°C), well below the melting temperature of the oligoduplexes used in the assay (>40°C). Radiolabeled DNA and protein–DNA complexes were detected using the Cyclone phosphorimager and the OptiQuant software (Packard Instrument).
Oligonucleotide cleavage experiments
DNA cleavage experiments were performed at 37°C in the reaction buffer (50 mM KCl, 50 mM Tris–HCl, pH 7.8 at 37°C and 0.1 mg/ml BSA) supplemented with appropriate concentrations (0.01–10 mM) of MgCl2, MnCl2, CoCl2, CaCl2, NiCl2, CuSO4 or ZnCl2. In the oligoduplex cleavage assay, the reaction mixtures contained 0.5 μM EcoKMcrA (dimer) and 0.2 μM DNA. At timed intervals the reactions were stopped by addition of the denaturing loading dye solution (95% v/v formamide, 25 mM EDTA) and analyzed by denaturing PAGE.
Plasmid restriction assay
BL21(DE3) (McrA-) E. coli cells with no intrinsic antibiotic resistance were made competent. The cells were then transformed with pLATE31 plasmids (AmpR) containing wt EcoKMcrA, its H228A or H229A variants, its N-terminal fragment (residues 1–174) or REM14 (unrelated protein, UniProt ID: Q9FX77). The cells were made competent again using Amp selection (at all growing steps the media contained 1% glucose). The cells containing each of the pLATE31 plasmids were transformed with pACYC184 plasmids (CmR) containing either no methyltransferase (‘empty’) or M.HpaII. The pACYC184 plasmids were amplified in ER2267 (McrA-) E. coli strain and verified by sequencing. 30 ng (3 μl) was used for transformation of 0.1 ml competent cells. At the recovery step after heat-shock the cells were supplemented with 0.9 ml of LB medium containing 1% glucose. After 1 h of shaking cells were gently sedimented and resuspended in 0.6 ml of glucose-free medium. 0.1 ml of transformants were spread on 3 types of LB-agar plates with (i) Amp, Cm, 1% glucose; (ii) Amp, Cm; (iii) Amp, Cm and 0.2 mM IPTG.
DNase I footprinting
50 nM of 33P-labeled unmethylated and hemimethylated 50 bp DNA was preincubated at 22°C with 0.25–0.5 μM of wt EcoKMcrA homodimer or 0.5–1 μM EcoKMcrA-N monomer in 10 μl of the binding buffer. After 15 min, 2.5 μl of DNase I (ThermoFisher Scientific, 0.004 U/μl) in the binding buffer supplemented with 5 mM CaCl2 and 12.5 mM MgCl2 were subsequently added. After 2 min at 22°C the DNase I reactions were quenched by the addition of the denaturing loading dye solution (95% v/v formamide, 75 mM EDTA and bromophenol blue). Samples were separated on a high-resolution denaturing polyacrylamide gel (gel concentration 20% w/v, acrylamide:bis-acrylamide ratio 29:1, urea concentration 8 M, gel temperature 60°C, standard Tris–borate–EDTA buffer).
Small angle X-ray scattering (SAXS) experiments
The small angle X-ray scattering data for EcoKMcrA and EcoKMcrA-N were collected at the P12 EMBL beamline of the PETRA III ring of the DESY synchrotron in Hamburg, Germany (18), equipped with Pilatus 2M detector (Dectris). Detector to sample distance was 3.1 m. X-ray wavelength was 0.124 nm. The capillary temperature was set to 20°C, sample changer temperature was 10°C. 20 frames with exposure time 0.05 s were collected for each sample. Data were collected in the range from 0.076 to 4.665 nm−1. Key SAXS data parameters are summarized in Supplementary Table S2 and SAXS-based estimates of molecular mass are listed in Supplementary Table S3. All samples were transferred into the buffer containing 20 mM Tris–HCl pH 7.5, 200 mM KCl, 0.1 mM EDTA, 0.01% (w/v) sodium azide and 1 mM DTT using desalting columns NAP5 (GE Healthcare). Samples of 30 μl of EcoKMcrA-N as well as full length protein were mixed equimolarly with hemimethylated 12 bp DNA oligoduplex. EcoKMcrA and EcoKMcrA-N were step-wise concentrated by ultrafiltration and centrifuged 10–15 min before data collection. The ATSAS 2.7.1 (r6669) software suite (19) was used for data analysis. Indirect Fourier transform and parametrization of the data was performed using GNOM (Version 5.0) (20). DAMMIN (21) and DAMMIF (22) programs were used for ab initio shape determination. Ten runs of each program were averaged by DAMAVER package (23) and the most probable shape was determined. Models were overlaid with SUBCOMB (24). The comparison of the SAXS data with the crystal structure was performed with CRYSOL (25). The molecular masses were estimated using SAXS data measured in a relative scale by SAXSMoW (v2.1) implemented as server (http://saxs.ifsc.usp.br) (26). MW was also evaluated from Porod volumes calculated by DATPOROD and dummy atom modeling programs (DAMMIN and DAMMIF) and using the method of Rambo and Tainer (27) implemented in DATVC.
Crystallization and data collection
The EcoKMcrA K196E variant was crystallized in the absence of DNA by the vapor diffusion technique in sitting drops from 1:1 mixtures of the protein solution in 50 mM Tris–HCl, pH 7.6, 200 mM NaCl, 1 mM EDTA, 1 mM DTT with the reservoir buffer containing 0.1 M HEPES–CsOH, pH 7.3, 30% MPD. The diffraction data were collected at 1.22021 Å wavelength at the P14 beamline of the DESY synchrotron in Hamburg and at the 1.2833 and 1.2777 Å wavelengths at the MX 14.2 beamline of the BESSY synchrotron in Berlin. All data were processed with XDS (28). The data collected at the PETRA ring of DESY reached the highest 2.85 Å resolution.
Structure determination and refinement
The molecular replacement attempts proved unsuccessful and the structure was solved with the help of experimental phasing. The data collected at DESY and BESSY were not isomorphous enough to allow for a successful MAD approach. The SAD phasing based on the highest resolution data collected at 1.22021 Å and performed with the help of autoSHARP (29) which internally uses the SHELXC/D/E pipeline (30) proved most efficient. The procedure originally found two strong zinc peaks which were later complemented with additional two weaker metal peaks. The distances between the four metal sites pointed to the presence of a two-fold symmetry axis. Two-fold symmetry was expected based on the dimeric character of the EcoKMcrA enzyme but did not show up as a peak in the 180° self-rotation map. We determined the symmetry operator and averaged the preliminary density maps with the help of DM program (31). The resulting maps were input to the LJS routine of autoSHARP that alternatingly uses BUCCANEER (32) and PARROT (33) programs for model building and density modification. The resulting model indicated that only the C-terminal domain that anchors the metal ions followed the two fold symmetry. The model was manually improved with the N- and C-terminal domains of the better resolved protomer. The resulting structure was refined with BUSTER (Global Phasing Ltd, Cambridge, United Kingdom) and rebuilt with BUCCANEER (32). The alternating cycles of manual model improvement and further automatic rebuilding led to the assembly of the complete two protomers. The structure was refined with REFMAC (34) and COOT (35) programs. The final refinement statistics are presented in Supplementary Table S4. The atomic coordinates and the corresponding structure factors were deposited at PDB under the 6GHC accession code.
RESULTS
EcoKMcrA activity in vitro
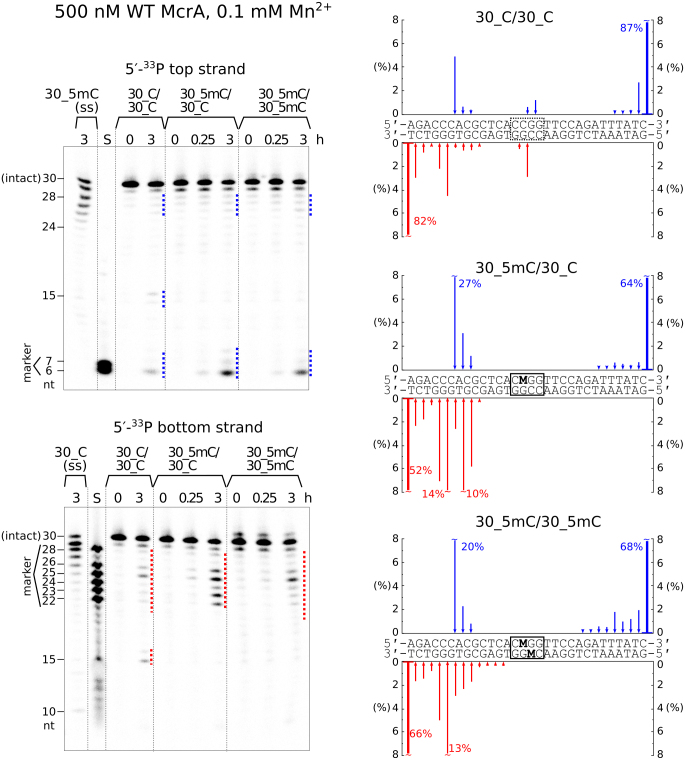
EcoKMcrA restricts M.HpaII methylated plasmids in cells, but so far all attempts to detect this activity in vitro have failed (11,12). We tested the catalytic activity of EcoKMcrA using 30 nucleotide long deoxyoligoduplex substrates with unmodified, hemi- or fully methylated C5mCGG sites in two different positions, with either top or bottom strand radiolabeled (Supplementary Table S1). The pilot cleavage reactions were performed with hemimethylated substrates in a buffer supplemented with various amounts of divalent metal ions (Supplementary Figure S1). Cleavage rates were highest for the Mn2+ ions in the concentration range of 0.01–0.2 mM. In the presence of Mn2+, it was possible to demonstrate the concerted cleavage of both DNA strands, most likely with a single nucleotide stagger (leading to 3′ overhangs). The extent of cleavage tended to be promoted by DNA methylation, but the effect was substrate dependent (Figure 1 and Supplementary Figure S2). The oligonucleotides were predominantly cleaved at multiple positions 5–15 nt upstream of the C5mCGG sequence. The unmethylated DNA was cut at identical positions, with additional cleavage observed at the unmodified CCGG sites (Figure 1 and Supplementary Figure S2). Wild type EcoKMcrA also displayed methylation independent exonuclease activity on single-stranded oligonucleotides (Figure 1).
Figure 1.
DNA cleavage by EcoKMcrA. The reactions were performed with 0.5 μM wild type EcoKMcrA (dimer) and 0.2 μM of unmodified, hemi- or fully methylated oligoduplex DNA or single stranded DNA, at 37°C in a buffer supplemented with 0.1 mM Mn2+. Top strand and bottom strand cleavage positions on the gels are marked by blue and red dotted lines, respectively. Gel lanes ‘S’ contained radiolabeled single-stranded oligonucleotides corresponding to the 5′-terminal fragments of the respective DNA strands (sizes in nucleotides are shown on the sides of the gels). The amounts of the bottom and top strand cleavage products after 3 hours of digestion are plotted as blue/red arrows along the oligoduplex sequences. The uncleaved oligonucleotides (the 0 hour lanes) contained a detectable fraction (∼10%) of shorter fragments. The reported cleavage data was corrected for the amount of these contaminants.
Based on a prediction of the EcoKMcrA active site (15), the H228A, H229A, H252A and H256A variants of the enzyme were expected to be catalytically impaired or inactive. Indeed, none of these mutants generated any cleavage products (Supplementary Figure S3), excluding a contaminating endonuclease as the source of the observed activity. Together, the data suggest that EcoKMcrA is catalytically active in vitro, but the activity requires high enzyme concentrations, and in such conditions the enzyme exhibits only a moderate preference for methylated DNA.
EcoKMcrA was active not only on short linear DNA oligoduplexes, but also on supercoiled plasmids and phage λ DNA (Supplementary Figure S4). Plasmid/phage DNA cleavage reactions required high (0.5 μM) enzyme concentrations, as the yield of cleavage products was much lower at 50 nM concentration. Under these conditions, there was surprisingly little effect of methylation in dcm (C5mCWGG) or HpaII (C5mCGG) context. Full digestion of the supercoiled unmethylated and methylated plasmids required 2 h incubation with 0.5 μM enzyme.
The low activity of EcoKMcrA and non-stringent preference for methylated DNA prompted us to test the in vitro activity of the enzyme using further substrates. The enzyme was active on PCR products created with a modified dNTP mix containing 5-hydroxymethylcytidine triphosphate, but it also cleaved the DNA of the Lambda and T4GT7 phages (the latter is a hydroxymethylation- and glucosylation-deficient variant of T4 phage) (Supplementary Figure S5A). However, control digestions using MspJI (5mCNNRN9/), HpaII (C/CGG) and MluCI (/AATT) endonucleases suggested that T4GT7 and Lambda contained, in addition to the predominant unmodified cytosine, also 5mC, resulting from the passage through the Dcm+ cells (Supplementary Figure S5B). DNA from T4 phage was resistant to digestion, demonstrating that EcoKMcrA does not cleave DNA with glucosylated 5hmC bases. The control digestions with H228A variant of the enzyme indicated only residual activity.
Next, we compared the cleavage of modified and unmodified substrates in isolation and in a competition assay. PCR DNAs containing either cytosine or 5hmC were tested separately and in a mixture additionally including 5mC modified XP12 phage DNA. In competition experiments as well as for isolated substrates, EcoKMcrA digested predominantly the modified DNA, whereas the unmodified DNA was barely affected. In this assay, the residual activity of the H228A variant was not observed. Controls with HpaII and MspJI exhibited the expected cleavage preferences (Supplementary Figure S6).
EcoKMcrA nuclease activity is required for plasmid and phage restriction
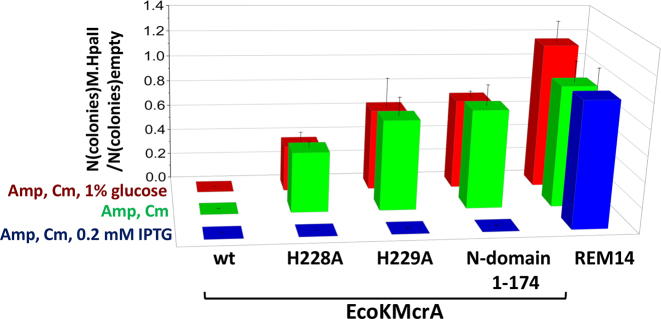
The weak in vitro activity of EcoKMcrA made us question whether the catalytic activity of EcoKMcrA was relevant for restriction in cells. In order to test this, we generated E. coli BL21(DE3) cells (McrA-) expressing wild type EcoKMcrA, its H228A, H229A and N-terminal variants (all catalytically impaired), or a control protein (REM14), from a plasmid maintained in the cells using ampicillin selection. We also prepared pACYC184 plasmid, carrying a chloramphenicol resistance (CmR) gene, without insert (‘empty’), or with an insert coding for M.HpaII. The CCGG sites in this vector were confirmed by HpaII digestion to be unmethylated and methylated, respectively. After transformation of the two plasmids into cells overexpressing EcoKMcrA or its variants, cells were subjected to double antibiotic selection, and survivor colonies were counted. For low or moderate protein overexpression (glucose repression or no induction), full restriction required both the M.HpaII methylation of the test plasmid, and the presence of an intact EcoKMcrA active site. High protein overexpression did not affect the colony formation only when the plasmid bearing a gene for an unrelated control protein was used (Figure 2 and Supplementary Figure S7).
Figure 2.
Plasmid restriction assay. Quantification of the restriction of empty and M.HpaII carrying plasmids by E. coli cells expressing either wild type EcoKMcrA, two different active site variants of the enzyme, its N-terminal fragment lacking the catalytic domain or an unrelated protein (REM14). The pACYC184 plasmid carrying a chloramphenicol resistance gene was assayed in BL21(DE3) (McrA-). The plasmid was either empty and thus without cytosine modifications or carried a gene for M.HpaII methyltransferase and thus was 5mC modified in the C5mCGG sequence context assumed to be among the EcoKMcrA targets.
The dependence of restriction on the catalytic activity of EcoKMcrA was further confirmed using a phage restriction assay with Lambda (partial dcm modification), T4gt (5hmC containing) and T4 (g5hmC containing) phages. T4 and Lambda phages were not significantly restricted for any tested phage titer. In contrast, T4gt phage was efficiently restricted for all tested titers by C2566 E. coli cells expressing wt EcoKMcrA. In contrast, the catalytically impaired variants (H228A and H229A), and the N-terminal fragment of the enzyme, only prevented plaque formation for low phage titer (Supplementary Figure S8).
EcoKMcrA structure
Crystallization of EcoKMcrA in the presence of dsDNA was not successful. However, we could grow crystals of the K196E variant in the absence of DNA. Crystals diffracted to 2.85 Å resolution at synchrotron beamlines (Supplementary Table S4). The anomalous signal resulting from the presence of Zn2+ ions was used for structure solution by experimental phasing at high energy remote wavelength (SAD approach).
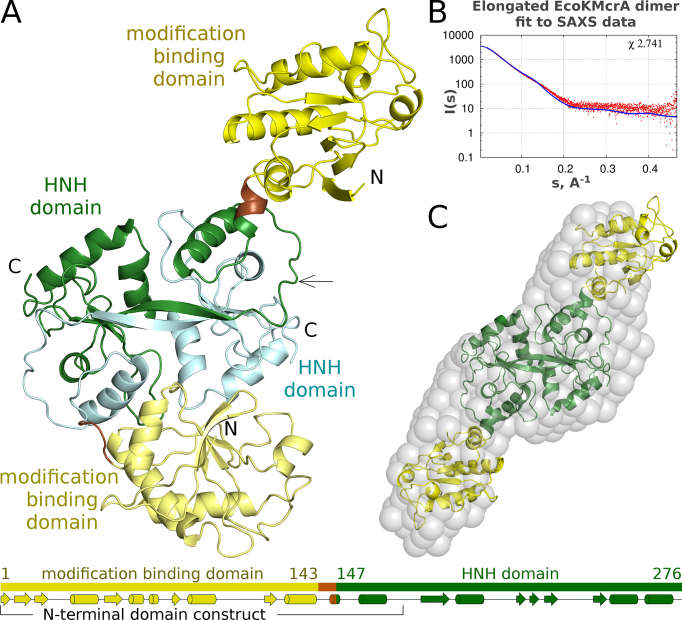
The EcoKMcrA structure confirms the expected two-domain architecture of the enzyme, with a previously uncharacterized N-terminal domain, and a C-terminal HNH domain predicted by prior bioinformatic work (Figure 3A). The nuclease domains clearly follow the two-fold symmetry, the N-terminal domains do not. This arrangement results in two distinct conformations of full length protomers, a compact and an extended one. We place the boundary between the N- and C-terminal domains and the linker at the switch points between the structurally similar and dissimilar regions. This choice assigns residues 147–276 to the HNH domain and residues 1–143 to the N-terminal domain. The drastically different relative orientations of the two domains in the protomers are therefore due to different conformations of a short linker region spanning only a few residues (144–146) and folding into a short helix in the extended form of the protomer (Supplementary Figure S9).
Figure 3.
Structure of EcoKMcrA. (A) Ribbon diagram of the EcoKMcrA dimer in the asymmetric unit of the crystals, colored according to domains. The domain organization of each protomer of is shown below. (B) Comparison of a symmetrized model of the EcoKMcrA dimer based on the more elongated protomer, with small-angle X-ray scattering (SAXS) data for the protein in the absence of DNA. (C) EcoKMcrA model ab initio calculated from the SAXS data (grey spheres) overlaid with the symmetrized dimer that was used for the calculation of the predicted small angle X-ray scattering data presented in panel B.
The overall structure of EcoKMcrA is in agreement with the previously reported dimeric form of the enzyme and suggests that the N-terminal domains on their own should be monomers. This was tested for the N-terminal EcoKMcrA fragment (residues 1–174) by analytical sizing chromatography in high salt conditions, to prevent unspecific protein interactions with the column material. The fragment boundaries were selected prior to the structure solution and thus the N-terminal domain includes a domain swapped fragment of the HNH-domain. As a control, we confirmed that full-length EcoKMcrA migrated as a dimer (apparent mass 54.4 kDa, compared to the expected mass for the dimer 64.5 kDa). In contrast, the N-terminal fragment eluted as a monomer (expected mass including His6-tag 20.6 kDa, apparent mass 20.1 kDa). Experiments in the presence of DNA could not be done meaningfully, because the required high ionic strength of the buffers (0.5 M) interfered with protein DNA binding, resulting in separate elution of the two components (Supplementary Figure S10).
In the crystal structure, the N-terminal domains pack very differently against the HNH-domain dimer. One domain interacts quite extensively, the other one makes contacts only to the crystallographic neighbors. According to DynDom (36), the relative domain orientations of the two protomers differ by 150° (Supplementary Figure S9). This suggested that the linkers are flexible and allow for multiple domain orientations. Alternatively, one of the observed conformations might result from the crystal packing effects.
To determine the solution conformations and to independently measure the molecular masses, we carried out small angle X-ray scattering (SAXS) experiments using full-length EcoKMcrA, and its N-terminal fragment. Data were collected both in the absence of DNA and in the presence of a 12-mer oligoduplex containing a single hemimethylated CCGG site (two oligoduplexes per EcoKMcrA dimer) (Supplementary Figure S11, Supplementary Table S2). We further calculated the ab initio shape of the enzyme in solution using SAXS data and the DAMMIF software (22) with no symmetry restraints. This shape was compared with the crystallized dimer, and the symmetrized dimers composed of the protomers in one conformation only. The symmetric, elongated dimer fitted best, judging from the real space fit and the χ parameters, which measure the agreement between calculated and observed scattering data in reciprocal space (Figure 3BC and Supplementary Figure S12). The SAXS masses for the full-length protein (between 58.7 and 77.6 kDa) agreed with the expected mass of the dimer even better than the gel filtration data. The SAXS masses for EcoKMcrA-N (between 16.8 and 23.7 kDa) confirmed the monomeric state of the N-terminal fragment in isolation (Supplementary Table S3).
EcoKMcrA N-terminal domain structure
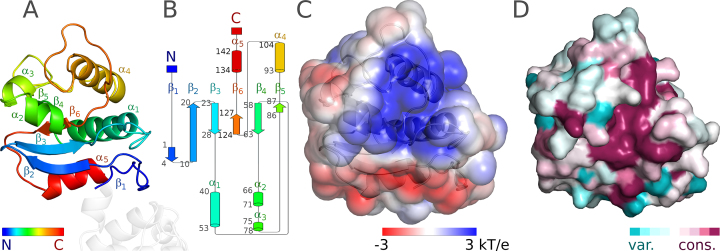
The N-terminal domain of EcoKMcrA is organized around the six-stranded antiparallel β-sheet. The edge strand β5 is much shorter than the strands in the center of the sheet, as expected, perhaps to avoid aggregation. The presence and length of strand β1 might result from the N-terminal His6 tag. Strands β1–β2, β2–β3, β4–β5 are connected by hairpins. Among these, the hairpin between strands β4–β5 is notable because it contains two short α-helices (α2 and α3), which pack approximately against the sheet. The α-helices of the β3–α1–β4 and β5–α4–β6 motifs, and another helix at the C-terminus of the domain (α5) also pack against the sheet, but on the other side (Figure 4AB).
Figure 4.
Structure of the N-terminal domain of EcoKMcrA. (A) Ribbon diagram in rainbow coloring from N- to C-terminus (blue to red). (B) Topology diagram generated with Pro-origami (50). (C) Electrostatic potential at the solvent excluded surface, blue represents positively charged regions, red negatively charged regions. (D) ConSurf (37) JTT conservation scores were mapped to the solvent accessible surface. Coloring is from cyan (poorly conserved regions) to magenta (for highly conserved regions). Panels A, C and D show the domain in the same orientation.
Overall, the N-terminal domain of EcoKMcrA is almost charge neutral (18 arginines and lysines, versus 18 glutamates and aspartates, sequence based isoelectric point pI 6.9). However, when the charges or the electrostatic potential are mapped to the protein surface, a clear pattern emerges. The side of the protein distal to the linker is generally positively charged, whereas the side more proximal to the linker is negatively charged. The main regions of positive charge are the β5–α4–β6 motif (94–120) and the β4–β5 hairpin (70–81). Altogether, this patch can be described as a groove between helices α1 on one side and α2 and α4 on the other (wrapping from one face of the β-sheet to the other). We strongly suspect that this groove of EcoKMcrA is a DNA binding region (Figure 4C).
Analysis of the sequence conservation of full length EcoKMcrA homologues shows that only about half of the positively charged residues are conserved. We carried out an unbiased analysis of sequence conservation in the N-terminal domain with the help of the ConSurf server (37). We used default settings, i.e. we did not require the presence of an HNH domain accompanying the N-domain in the orthologues. Among the surface regions, the positively charged region was the most conserved (Figure 4D and Supplementary Figure S13).
Modification specificity of the EcoKMcrA N-terminal domain
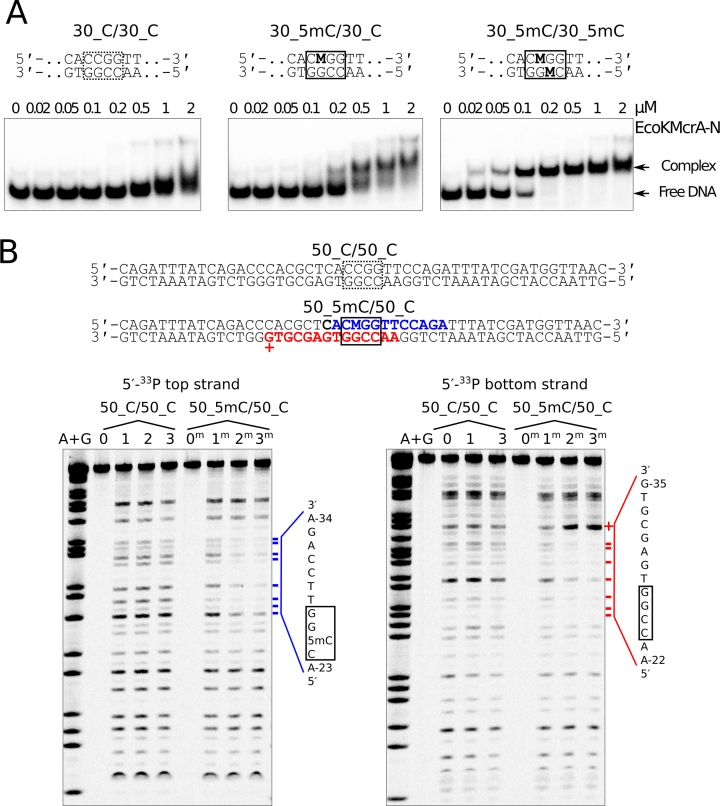
The domain architecture of EcoKMcrA is reminiscent of other methyl directed endonucleases, which are built as fusions of nuclease and modification binding domains. This makes the N-terminal domain of EcoKMcrA the prime candidate for the recognition of methylated DNA. We carried out electrophoretic mobility shift experiments using the N-terminal fragment of EcoKMcrA and the DNA that was previously used in the activity assay. The data indicated tighter binding of the methylated DNA compared to the unmodified DNA, confirming that EcoKMcrA-N is the methylation specific domain (Figure 5A). Methyl specific binding was also confirmed by the DNase I footprint of EcoKMcrA-N. The protection from DNase I cleavage was observed only for the methylated DNA (Figure 5B). The protected region around the 5mC was identical to the region covered by full-length EcoKMcrA (Supplementary Figure S14), but for the full-length enzyme, we additionally observed scattered footprint for unmethylated DNA. This suggests that EcoKMcrA-N has higher overall selectivity for the methylated site than the full-length enzyme. The methyl-independent DNA binding observed with EcoKMcrA is therefore likely due to the DNA interaction with the C-terminal HNH domains.
Figure 5.
DNA binding by the N-terminal fragment of EcoKMcrA (EcoKMcrA-N). (A) Electrophoretic mobility shift assay. DNA was unmodified, hemi- or fully methylated (central sequences shown on top; ‘M’ denotes 5mC). DNA concentration was 50 nM, concentrations of EcoKMcrA-N are indicated above gel lanes. Positions of free DNA and protein-DNA complexes are indicated. (B) DNase I footprint. Top: the region of the DNA protected from DNase I cleavage by EcoKMcrA-N binding is shown in bold font. The bottom strand G-35 position that becomes more susceptible to DNase I treatment upon EcoKMcrA-N binding is shown by ‘+’. Bottom: DNA protection by EcoKMcrA-N. Gel lanes ‘0’ contained untreated unmodified DNA, ‘1’ – the DNA treated with DNase I in the absence of EcoKMcrA-N, lanes ‘2’ and ‘3’ – the DNA treated with DNase I in the presence of 0.5 and 1 μM EcoKMcrA-N, respectively. Lanes ‘0m’, ‘1m’, ‘2m’ and ‘3m’ contained analogous samples prepared with hemimethylated DNA. Size markers (lanes ‘A+G’) were generated using a standard Maxam-Gilbert sequencing reaction. Positions of the DNA protected from DNase I cleavage upon EcoKMcrA-N binding are marked by blue (top strand) or red (bottom strand) dashes. The sequences of the protected regions are shown on the right-hand side of the gels.
No indication for modified base flipping by EcoKMcrA in fluorescence assay using pyrrolocytosine
In order to check whether nucleotide flipping played a role in the interactions of the EcoKMcrA N-terminal domain with DNA, we used a fluorescence based assay. The method relies on the properties of pyrrolocytosine (pC), which due to quenching exhibits about twice lower fluorescence in double stranded compared to single stranded DNA. Meaningful use of the pC analogue requires that the enzyme accepts it instead of the physiologically relevant cytosine bases. In the case of EcoKMcrA, a hemimodified 12-mer oligoduplex containing pC bound more tightly than unmodified control, but less tightly than the duplex containing 5mC (Supplementary Figure S15A). The dsDNA duplex containing pC had very similar properties in the absence of the protein, and in the presence of either full length EcoKMcrA or its N-terminal fragment (Supplementary Figure S15B). This negative result speaks against nucleotide flipping, but could also be due to quenching of the flipped pC fluorescence by aromatic amino acid residues of a binding pocket. According to the crystal structure, such a pocket is unlikely to be present in the N-terminal fragment of EcoKMcrA. Therefore, the data suggest that the enzyme recognizes modified cytosine bases in the context of dsDNA without nucleotide flipping.
EcoKMcrA HNH domain fold and active site
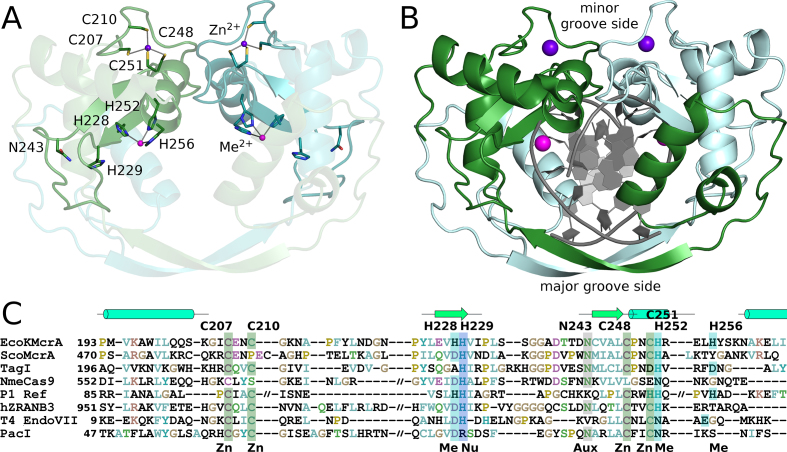
HNH endonucleases are built around a structural Zn2+ ion, and an active site divalent metal ion (Figure 6). The Zn2+ ion is typically coordinated by four cysteine residues in two CxxC motifs. Both CxxC motifs are present in the EcoKMcrA sequence, and in the structure indeed coordinate a Zn2+ ion, presumably taken from the expression cells and retained through the purification procedure. The HNH endonucleases are also known as ββα-Me endonucleases (where ββα denotes three consecutive secondary structure elements and Me stands for the active site metal ion). The ββα motif is canonically placed after the first CxxC motif, and overlaps with the second CxxC motif (Figure 6A).
Figure 6.
HNH domain dimer of EcoKMcrA. (A) Ribbon representation of the structure of the EcoKMcrA HNH domain. The structural Zn2+ ions, and the active site metal ions (Zn2+ in the crystal structure, and most likely Mn2+ in the physiological state), are shown together with their ligands. (B) Model of EcoKMcrA DNA complex, created based on the comparison with the Hpy99I restriction endonuclease. The blue and pink balls represent structural Zn2+ and active site metal ions, respectively. (C) Sequence alignment of the HNH core region of EcoKMcrA and related endonucleases: ScoMcrA (WP_011029780), TagI (WP_084609162), NmeCas9 (PDB: 5VGB), P1 phage Ref protein (PDB: 3PLW), human ZRANB3 endonuclease (PDB: 5MKW), T4 phage endonuclease VII (PDB: 1E7D) and PacI endonuclease (PDB: 3LDY). Zn – Zn2+ ligand, Me – active site metal ligand, Nu – nucleophile activator, Aux – auxiliary HNH residue.
All moniker residues of the HNH motif are present in EcoKMcrA in their standard places. The first histidine residue is located near the end of the first β-strand, and is generally thought to act as the activator for the water that is incorporated into the substrate. In EcoKMcrA, this residue is H229. The nucleophile activator histidine is typically preceded by a ligand for the active site metal ion. In the EcoKMcrA structure, the preceding residue is a histidine (H228), which is indeed a metal ion ligand and is required for activity (Supplementary Figure S3).
The asparagine residue of the HNH motif is typically located at the N-terminus of the second β-strand, and has a structural role. The residue in EcoKMcrA is N243, which donates hydrogen bonds from its side chain carboxamide group to the carbonyl oxygen atoms of I231 and A238, and accepts a hydrogen bond from the NH of I231. The interactions stabilize the extended loop between the two β-strands of the ββα-Me motif, and judging from the high conservation of this residue, seem important for the integrity of the motif.
The second histidine of the HNH motif is located in the ββα core α-helix and is typically a ligand for the active site metal ion. In EcoKMcrA, this residue is H252. A further metal ion ligand, H256, follows one helical turn downstream. Judging from the in vitro activity of the alanine variants, both histidine residues are required for activity (Supplementary Figure S3).
The active site metal ion in EcoKMcrA is thus coordinated by H228, H252 and H256. In the crystal structure, this metal ion is likely Zn2+ judging from the anomalous diffraction signal. According to the biochemical data, it is a Mn2+ ion in the productive EcoKMcrA complex. Commonly observed hexa-coordination of the Mn2+ ion provides more than sufficient ligand acceptor sites for interactions with non-bridging oxygen of the scissile phosphate and the leaving group oxygen that would be expected in a productive complex.
EcoKMcrA HNH domain dimerization mode
The HNH domains form a two-fold symmetric dimer, held together by an interface area of ∼3250 Å2 according to the PISA web server (38). At the center of the dimer, there is a very prominent channel, which could accommodate the bound DNA. Comparison of the HNH domains of EcoKMcrA and Hpy99I (39) or PacI (40) confirms the predicted DNA binding mode, and also defines major and minor groove sides of the DNA. In the ‘crossover’ region (residues 179–191), the polypeptide chain adopts an extended, β-strand conformation, which engages in main chain hydrogen bonding with its counterpart from the other protomer in a canonical antiparallel mode. The elNEMO server (41), which predicts normal modes of protein motion in solution based on molecular dynamics calculations, shows that the contact region between subunits on the major groove side is very flexible indeed. However, for EcoKMcrA to open up on this side to latch onto DNA, the entire ‘crossover’ interaction would have to be undone, and thus the crossover region is more likely to act as a hinge.
On the minor groove side of the modelled DNA, the protomers also interact, but the interface is quite different. The contacts involve a long loop preceding the ββα core motif (residues 215–224) and a fragment comprising the α-helix of the motif (residues 249–260). The helix is expected from the structures of other HNH endonucleases to wedge into the DNA minor groove. The two regions approach each other in such a manner that the region 249–260 of one protomer interfaces predominantly with the 215–224 region of the other protomer. Their interactions are mostly mediated by amino acid side chains, and contribute less than one third of the complete interface area. As they involve the catalytic core regions, these interactions are likely to fix the stagger for concerted cleavages of both DNA strands.
Two binding sites for the modified DNA in the EcoKMcrA dimer
The structural data suggest that EcoKMcrA should have one joint DNA binding site in the nuclease domain, and two separate modification dependent DNA binding domains. In order to verify this model, we first tested wild type EcoKMcrA for preferential binding of methylated over unmethylated DNA in an EMSA assay, using either 30mer or 12mer duplexes. As observed for the N-terminal domain alone, binding was promoted by DNA methylation. However, in agreement with the DNase I footprinting results, for 30 nt long substrates the dependence on methylation was more pronounced for EcoKMcrA-N than for the full length enzyme (Supplementary Figure S16A). We interpret these data as evidence that the nuclease domain contributes some unspecific affinity to DNA for the longer substrates, but not the short oligoduplexes that are largely covered by the N-terminal domain. EMSA experiments with full-length enzyme, but not EcoKMcrA-N, showed the presence of two bands of retarded electrophoretic mobility (Figure 5A and Supplementary Figure S16A), as reported earlier, and attributed to a monomer dimer equilibrium (12). In the light of the biochemical and structural data reported here, the two shifted bands more plausibly correspond to EcoKMcrA with either one or two bound oligoduplexes. To test this model, we carried out EMSA experiments with modified oligoduplexes of two different lengths mixed in various ratios (Supplementary Figure S16B). We observed the presence of three methyl-dependent complexes of distinct mobilities, which is consistent with the two independent binding sites for methylated DNA in the EcoKMcrA dimer. Next, we verified whether cleavage could be made more efficient when the nuclease domain was positioned by two closely spaced methylation sites. We have tested various combinations of hemi- and fully methylated substrates with two different distances between the modification sites. Consistent with the weak digestion of densely modified phage DNA, we observed at most a mild increase in cleavage rates with the new substrates (Supplementary Figure S17).
A synthetic cytosine modification-dependent restriction endonuclease
Wild-type EcoKMcrA has very moderate cleavage activity and nuclease specificity. We fused an attenuated variant of the 76 amino acid nuclease domain of the N.ϕGamma nickase of Bacillus anthracis (42) to wt EcoKMcrA, or its inactive H252A variant. The N.ϕGamma nickase is sequence specific and nicks the top strand of YC↓GGT (‘↓’ indicates top strand cleavage) or bottom strand of the ACC↑GR (‘↑’ indicates bottom strand cleavage) (42). The resulting enzyme nicked M.HpaII methylated, but not unmodified pBR322, at sites matching its nuclease target sequence (Supplementary Figure S18).
DISCUSSION
The N-terminal domain of EcoKMcrA binds to DNA in a methylation dependent way. EMSA assays indicate that the affinity is highest for fully-methylated DNA, lower for hemi-methylated DNA, and lowest for unmethylated DNA. The structure of the N-terminal domain of EcoKMcrA is surprisingly dissimilar from other modification specific DNA binding domains. In particular, the domain is not structurally similar to SRA domains and unrelated to the McrB-N domain responsible for the modification dependence of McrBC (9). The DALI server (43) suggested marginal similarity of the EcoKMcrA N-terminal domain to I-DreI, a dimeric meganuclease composed of homing endonucleases I-DmoI and I-CreI (44). PDBeFOLD (45) picked up fold similarities to MotA, a transcription factor from T4 phage (46) (Supplementary Figure S19). As T4 phage can infect E. coli, a common evolutionary origin of MotA and EcoKMcrA is plausible, in spite of the very low sequence similarity and different specificity (MotA binds to g5hmC-containing T4 phage DNA, EcoKMcrA restricts 5mC and 5hmC, but not g5hmC-containing DNA). Fold comparisons show that the connectivity of the β-sheet and two α-helices is largely preserved in the three proteins. The major difference is an insertion of residues 64–123 in EcoKMcrA that covers structural elements α2-4 and β5 (Figure 4 and Supplementary Figure S20).
The DNA binding mode of the N-terminal fragment of EcoKMcrA is still unknown. The pyrrolocytosine assay does not provide evidence for nucleotide flipping, but alone is not conclusive, because the fluorescence of the probe could be similarly quenched in a protein pocket as in the context of double stranded DNA. A structural model for DNA binding can be tentatively deduced from similarity to I-DmoI and MotA. The inferred binding mode places the DNA on the positively charged patch of the surface of the EcoKMcrA N-terminal domain, in the region of highest amino acid conservation. It implies that, as in the templates, the protein faces the major groove of the DNA, where –near the outer edges– methyl or hydroxymethyl groups of modified cytosine bases would be located. The model predicts a footprint of about one full DNA turn, in agreement with biochemical observations (Figure 5). The elongated symmetrized EcoKMcrA dimer with DNA bound to the conserved patch of the N-terminal domain surface agrees equally well with the experimental scattering for the EcoKMcrA-DNA complex, as the model for the elongated EcoKMcrA alone with its corresponding SAXS data (Supplementary Figure S20).
The C-terminal domain of EcoKMcrA is responsible for the nuclease activity. The domain has a typical HNH fold, with the expected active site residues, which are conserved in the EcoKMcrA family. It is therefore surprising that demonstration of in vitro endonucleolytic activity has been so difficult. In our hands, EcoKMcrA has weak, Mn2+ ion dependent, endonuclease activity on double- and single-stranded DNA. The specific requirement for Mn2+ ions was unexpected, because many other HNH endonucleases have less stringent preference for the divalent metal cations in their active sites. Mn2+ ions may be available physiologically, thanks to mechanisms for active uptake, presumably from the host (47). EcoKMcrA generated nicks in both strands of the oligoduplex. The nicking of two DNA strands is consistent with concerted action resulting in double strand breaks (DSB). The most prominent nicks indicate 1 nucleotide 3′ cleavage stagger (Figure 1).
The overall organization of EcoKMcrA with N-terminal modification specific, and C-terminal nuclease domain is reminiscent of the two-domain organization of the SRA-HNH (48), PD-(D/E)XK-SRA (PvuRts1I) (49) and SRA-PD-(D/E)XK (MspJI) (6) modification specific restriction endonucleases. In all these enzymes, the largely unspecific nuclease domains mediate dimerization, and expose a single joint DNA binding site, whereas the modification specific domains expose separate binding sites for modified DNA. Our EcoKMcrA crystal structure indicates that the nuclease domains of the enzyme indeed form a tight dimer, the conformation of which is compatible with concerted cleavage of two DNA strands. The results of the SAXS experiments confirm that EcoKMcrA is dimeric also in solution. Finally, the biochemical data indicate that each N-terminal domain binds double stranded DNA in modification dependent manner. However, the in vitro cleavage assay does not show a clear increase of the enzyme activity in the presence of two closely spaced modification sites.
Surprisingly, the EcoKMcrA in vitro activity is weak, and like the activity of the SRA-HNH endonuclease Sco5333 (ScoA3IV) (48), only partially methylation dependent. The activity of EcoKMcrA on unmethylated DNA and the lack of dependence on two modification sites are probably a consequence of high protein concentrations required to achieve any DNA cleavage at all. Why such high protein concentrations and long incubation times are required remains puzzling, especially in the light of sequence and structural data that point to the conservation of the active site residues. The low in vitro activity of EcoKMcrA also has to be reconciled with the efficacy of the enzyme as a restricting factor for phages or plasmids with cytosine modifications. It is possible that an optimal substrate for in vitro experiments has not been identified. Such a substrate might only be created in cells, for example as a side product of DNA replication or transcription. Alternatively, EcoKMcrA could require an activator or an effector nuclease, from the pool of host proteins. Our finding that the nuclease activity of EcoKMcrA is required for efficient phage or plasmid restriction makes it unlikely that the activity of the enzyme depends on the recruitment of another nuclease. The other possible explanations for the efficacy of EcoKMcrA as a restricting factor remain to be explored.
DATA AVAILABILITY
The atomic coordinates and corresponding structure factors were deposited at PDB under the 6GHC accession code.
Supplementary Material
ACKNOWLEDGEMENTS
The SAXS data were collected at the P12 beamline operated by EMBL Hamburg at the PETRA III storage ring (DESY, Hamburg, Germany). Crystal diffraction data were collected at the P14 beamline of the PETRA III storage ring (DESY, Hamburg, Germany), and at MX14.2 beamline of the BESSY storage ring (Berlin, Germany). We would like to thank Dr A. Kikhney for the assistance with SAXS data collection, and Drs Uwe Mueller, Manfred Weiss, and Gleb Bourenkov for assistance with crystallographic data collection. We also thank Geoff Wilson and Peter Weigele for providing phage DNA, and Kiersten Flodman for help with enzyme purification.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Science and Higher Education [0295/B/PO1/2008/34 to M.B., N N301 425038 to H.C.]; Polish National Science Centre (NCN) [UMO-2011/02/A/NZ1/00052, UMO-2014/13/B/NZ1/03991, UMO-2014/14/M/NZ5/00558 to M.B.]; Research Council of Lithuania [MIP-027/2012 to G.S.]; Part of this work was performed using Centre for Preclinical Research and Technology (CePT) infrastructure [European Union POIG.02.02.00-14-024/08-00 project]. Funding for open access charge: NCN [UMO-2014/13/B/NZ1/03991].
Conflict of interest statement. Dr Shuang-yong Xu is an employee of New England Biolabs.
REFERENCES
- 1. Raleigh E.A., Wilson G.. Escherichia coli K-12 restricts DNA containing 5-methylcytosine. Proc. Natl. Acad. Sci. U.S.A. 1986; 83:9070–9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Revel H.R. Restriction of nonglucosylated T-even bacteriophage: properties of permissive mutants of Escherichia coli B and K12. Virology. 1967; 31:688–701. [DOI] [PubMed] [Google Scholar]
- 3. Kelleher J.E., Raleigh E.A.. A novel activity in Escherichia coli K-12 that directs restriction of DNA modified at CG dinucleotides. J. Bacteriol. 1991; 173:5220–5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zheng Y., Cohen-Karni D., Xu D., Chin H.G., Wilson G., Pradhan S., Roberts R.J.. A unique family of Mrr-like modification-dependent restriction endonucleases. Nucleic Acids Res. 2010; 38:5527–5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cohen-Karni D., Xu D., Apone L., Fomenkov A., Sun Z., Davis P.J., Kinney S.R., Yamada-Mabuchi M., Xu S.Y., Davis T. et al. . The MspJI family of modification-dependent restriction endonucleases for epigenetic studies. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:11040–11045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Horton J.R., Mabuchi M.Y., Cohen-Karni D., Zhang X., Griggs R.M., Samaranayake M., Roberts R.J., Zheng Y., Cheng X.. Structure and cleavage activity of the tetrameric MspJI DNA modification-dependent restriction endonuclease. Nucleic Acids Res. 2012; 40:9763–9773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Horton J.R., Wang H., Mabuchi M.Y., Zhang X., Roberts R.J., Zheng Y., Wilson G.G., Cheng X.. Modification-dependent restriction endonuclease, MspJI, flips 5-methylcytosine out of the DNA helix. Nucleic Acids Res. 2014; 42:12092–12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Neuwald A.F., Aravind L., Spouge J.L., Koonin E.V.. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999; 9:27–43. [PubMed] [Google Scholar]
- 9. Sukackaite R., Grazulis S., Tamulaitis G., Siksnys V.. The recognition domain of the methyl-specific endonuclease McrBC flips out 5-methylcytosine. Nucleic Acids Res. 2012; 40:7552–7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bujnicki J.M., Rychlewski L.. Grouping together highly diverged PD-(D/E)XK nucleases and identification of novel superfamily members using structure-guided alignment of sequence profiles. J. Mol. Microbiol. Biotechnol. 2001; 3:69–72. [PubMed] [Google Scholar]
- 11. Mulligan E.A., Dunn J.J.. Cloning, purification and initial characterization of E. coli McrA, a putative 5-methylcytosine-specific nuclease. Protein Expr. Purif. 2008; 62:98–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mulligan E.A., Hatchwell E., McCorkle S.R., Dunn J.J.. Differential binding of Escherichia coli McrA protein to DNA sequences that contain the dinucleotide m5CpG. Nucleic Acids Res. 2010; 38:1997–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Card C.O., Wilson G.G., Weule K., Hasapes J., Kiss A., Roberts R.J.. Cloning and characterization of the HpaII methylase gene. Nucleic Acids Res. 1990; 18:1377–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Anton B.P., Raleigh E.A.. Transposon-mediated linker insertion scanning mutagenesis of the Escherichia coli McrA endonuclease. J. Bacteriol. 2004; 186:5699–5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bujnicki J.M., Radlinska M., Rychlewski L.. Atomic model of the 5-methylcytosine-specific restriction enzyme McrA reveals an atypical zinc finger and structural similarity to betabetaalphaMe endonucleases. Mol. Microbiol. 2000; 37:1280–1281. [DOI] [PubMed] [Google Scholar]
- 16. Liu G., Ou H.Y., Wang T., Li L., Tan H., Zhou X., Rajakumar K., Deng Z., He X.. Cleavage of phosphorothioated DNA and methylated DNA by the type IV restriction endonuclease ScoMcrA. PLos Genet. 2010; 6:e1001253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zheng L., Baumann U., Reymond J.L.. An efficient one-step site-directed and site-saturation mutagenesis protocol. Nucleic Acids Res. 2004; 32:e115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Blanchet C.E., Spilotros A., Schwemmer F., Graewert M.A., Kikhney A., Jeffries C.M., Franke D., Mark D., Zengerle R., Cipriani F. et al. . Versatile sample environments and automation for biological solution X-ray scattering experiments at the P12 beamline (PETRA III, DESY). J. Appl. Crystallogr. 2015; 48:431–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Franke D., Petoukhov M.V., Konarev P.V., Panjkovich A., Tuukkanen A., Mertens H.D.T., Kikhney A.G., Hajizadeh N.R., Franklin J.M., Jeffries C.M. et al. . ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J. Appl. Crystallogr. 2017; 50:1212–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Svergun D.I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992; 25:495–503. [Google Scholar]
- 21. Svergun D.I. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999; 76:2879–2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Franke D., Svergun D.I.. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2009; 42:342–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Volkov V.V., Svergun D.I.. Uniqueness of ab-initio shape determination in small-angle scattering. J. Appl. Cryst. 2003; 36:860–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kozin M., Svergun D.I.. Automated matching/ of high- and low-resolution structural models. J. Appl. Cryst. 2001; 34:33–41. [Google Scholar]
- 25. Svergun D.I., Barberato C., Koch M.H.J.. CRYSOL - a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Cryst. 1995; 28:768–773. [Google Scholar]
- 26. Fischer H., Oliveira Neto M., Napolitano H.B., Polikarpov I., Craievich A.F.. Determination of the molecular weight of proteins in solution from a single small-angle X-ray scattering measurement on a relative scale. J. Appl. Cryst. 2010; 43:101–109. [Google Scholar]
- 27. Rambo R.P., Tainer J.A.. Accurate assessment of mass, models and resolution by small-angle scattering. Nature. 2013; 496:477–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kabsch W. Xds. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vonrhein C., Blanc E., Roversi P., Bricogne G.. Automated structure solution with autoSHARP. Methods Mol. Biol. 2007; 364:215–230. [DOI] [PubMed] [Google Scholar]
- 30. Sheldrick G.M. A short history of SHELX. Acta Crystallogr. A. 2008; 64:112–122. [DOI] [PubMed] [Google Scholar]
- 31. Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A. et al. . Overview of the CCP4 suite and current developments. Acta Crystallogr. D. Biol. Crystallogr. 2011; 67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D. Biol. Crystallogr. 2006; 62:1002–1011. [DOI] [PubMed] [Google Scholar]
- 33. Cowtan K. Recent developments in classical density modification. Acta Crystallogr. D. Biol. Crystallogr. 2010; 66:470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Murshudov G.N., Skubak P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., Winn M.D., Long F., Vagin A.A.. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D. Biol. Crystallogr. 2011; 67:355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 36. Taylor D., Cawley G., Hayward S.. Quantitative method for the assignment of hinge and shear mechanism in protein domain movements. Bioinformatics. 2014; 30:3189–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ashkenazy H., Erez E., Martz E., Pupko T., Ben-Tal N.. ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010; 38:W529–W533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Krissinel E., Henrick K.. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007; 372:774–797. [DOI] [PubMed] [Google Scholar]
- 39. Sokolowska M., Czapinska H., Bochtler M.. Crystal structure of the beta beta alpha-Me type II restriction endonuclease Hpy99I with target DNA. Nucleic Acids Res. 2009; 37:3799–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shen B.W., Heiter D.F., Chan S.H., Wang H., Xu S.Y., Morgan R.D., Wilson G.G., Stoddard B.L.. Unusual target site disruption by the rare-cutting HNH restriction endonuclease PacI. Structure. 2010; 18:734–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Suhre K., Sanejouand Y.H.. ElNemo: a normal mode web server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res. 2004; 32:W610–W614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gutjahr A., Xu S.Y.. Engineering nicking enzymes that preferentially nick 5-methylcytosine-modified DNA. Nucleic Acids Res. 2014; 42:e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Holm L., Laakso L.M.. Dali server update. Nucleic Acids Res. 2016; 44:W351–W355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chevalier B.S., Kortemme T., Chadsey M.S., Baker D., Monnat R.J., Stoddard B.L.. Design, activity, and structure of a highly specific artificial endonuclease. Mol. Cell. 2002; 10:895–905. [DOI] [PubMed] [Google Scholar]
- 45. Krissinel E., Henrick K.. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D. Biol. Crystallogr. 2004; 60:2256–2268. [DOI] [PubMed] [Google Scholar]
- 46. Cuypers M.G., Robertson R.M., Knipling L., Waddell M.B., Moon K., Hinton D.M., White S.W.. The phage T4 MotA transcription factor contains a novel DNA binding motif that specifically recognizes modified DNA. Nucleic Acids Res. 2018; 46:5308–5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Diaz-Ochoa V.E., Jellbauer S., Klaus S., Raffatellu M.. Transition metal ions at the crossroads of mucosal immunity and microbial pathogenesis. Front. Cell Infect. Microbiol. 2014; 4:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Han T., Yamada-Mabuchi M., Zhao G., Li L., Liu G., Ou H.Y., Deng Z., Zheng Y., He X.. Recognition and cleavage of 5-methylcytosine DNA by bacterial SRA-HNH proteins. Nucleic Acids Res. 2015; 43:1147–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kazrani A.A., Kowalska M., Czapinska H., Bochtler M.. Crystal structure of the 5hmC specific endonuclease PvuRts1I. Nucleic Acids Res. 2014; 42:5929–5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stivala A., Wybrow M., Wirth A., Whisstock J.C., Stuckey P.J.. Automatic generation of protein structure cartoons with Pro-origami. Bioinformatics. 2011; 27:3315–3316. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and corresponding structure factors were deposited at PDB under the 6GHC accession code.