Abstract
The ability to process molecules available in the environment into useable building blocks characterizes catabolism in contemporary cells and was probably critical for the initiation of life. Here we show that a catabolic process in collectively autocatalytic sets of RNAs allows diversified substrates to be assimilated. We modify fragments of the Azoarcus group I intron and find that the system is able to restore the original native fragments by a multi-step reaction pathway. This allows in turn the formation of catalysts by an anabolic process, eventually leading to the accumulation of ribozymes. These results demonstrate that rudimentary self-reproducing RNA systems based on recombination possess an inherent capacity to assimilate an expanded repertoire of chemical resources and suggest that coupled catabolism and anabolism could have arisen at a very early stage in primordial living systems.
INTRODUCTION
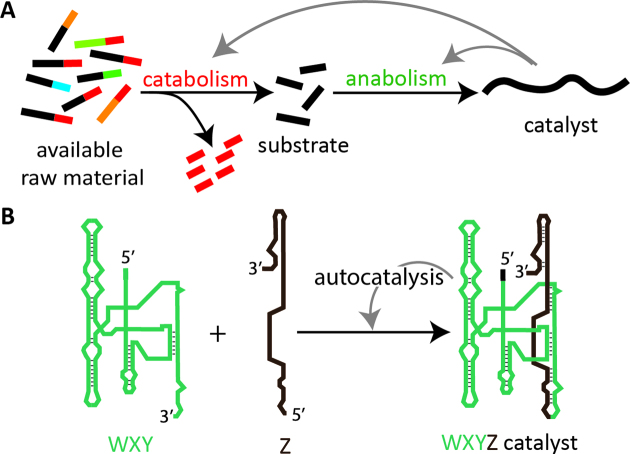
Collective autocatalytic sets (CASs) (1–3), where an ensemble of molecules can reproduce each other, have been envisaged as a possible scenario for the origin of life (4–10). A fundamental feature of such sets is their ability to self-sustain using substrates available in the environment (the food set) (6,10,11), a property which has reached a very high level of complexity and diversity in contemporary metabolisms. In the context of origin of life and the RNA world, where directly useable substrates were limited (12–17), it would have been advantageous for a self-reproducing system to thrive on a broad range of resources by pre-processing them (Figure 1A).
Figure 1.
Coupled catabolism and anabolism in self-reproducing systems. (A) Schematic showing how catabolism in a self-reproducing system can process unusable raw material into usable substrates. (B) Autocatalytic synthesis of covalent RNA catalyst (WXYZ) from inactive RNA substrates (WXY and Z) using an anabolic autocatalytic process in the Azoarcus ribozyme system.
In this regard, RNAs derived from the group I intron (18) of the Azoarcus bacterium (19) are an attractive model, as they can self-reproduce and recycle RNA materials by recombination reactions (17). They are 200 nt long RNA recombinases (WXYZ, Figure 1B) which can catalyze their own assembly from the fragments WXY + Z via trans-esterification in an autocatalytic process (20). For the assembly, they exploit Watson–Crick interactions between the 3 nt at both extremities of WXY fragments (the internal guide sequence ‘IGS’ and the target sequence ‘tag’ at the 5’ and 3’ ends, respectively). The IGS and tag of the fragments can be engineered to form autocatalytic recombination networks (21). However, the autocatalytic character of this system has so far only been demonstrated in a purely anabolic manner, relying on designed substrates obtained by fragmentation of the ribozyme (22). In a more realistic prebiotic setting, the initial reaction mixtures would likely consist of a much broader range of molecules. This could cause self-reproduction of CASs to be inhibited or stopped for several reasons: (i) the available molecules cannot be used as substrates by the catalysts; (ii) available molecules can be used as substrates, but lead to futile products that are not capable of catalysis; (iii) the available molecules are assembled to form novel catalysts, but these catalysts do not allow formation of an autocatalytic cycle.
Here, we mimic a prebiotic environment containing substrates that cannot be used directly to form autocatalytic ribozymes by fueling the Azoarcus ribozyme system with only modified RNA substrates. We observe that unmodified fragments are, nevertheless, reformed, leading to the production of wild-type catalysts (WXYZ). Kinetic and biochemical analyses show that the transformation of the raw material is catalyzed by the reaction products via a multi-step reaction pathway involving a series of specific but unexpected binding interactions between the RNA substrates and the ribozyme. The combination of catabolic and anabolic steps enables collective autocatalysis. We furthermore show that CASs comprising multiple species are maintained in similar conditions. These results highlight a form of rudimentary catabolism, where the catalysts transform the available resources into building blocks that drive their own formation.
MATERIALS AND METHODS
Materials
All chemicals were purchased from Sigma-Aldrich (unless specified otherwise). 4-(2-Hydroxyethyl)-1-piperazinepropanesulfonic acid, EPPS was purchased from Alfa Aesar (Product no.: J60511, CAS no.: 16052-06-5). For all the reactions water was used from ThermoFisher Scientific (UltraPureTM DNAse/RNase free, Product no.: 10977035) or from the MilliQ water purifier system (Millipore). RNA concentrations were measured on a NanoDrop-1000 UV-spectrophotometer (Peqlab). Denaturing polyacrylamide gels were prepared using gel stock solution from Roth and run in 1× TBE (Tris-Borate ethylenediaminetetraacetic acid (EDTA), prepared from 10× TBE from Roth). All analysis were performed using 12% denaturing polyacrylamide gels containing 8.3 M urea and run for at least 2–3 h at constant power of 24 W. Gels were stained with 1× GelRedTM (Biotium). Gel analysis and calculation of conversions were carried out with ImageJ software (https://imagej.nih.gov/ij/). All DNA oligonucleotides were obtained from IDT DNA technologies (https://eu.idtdna.com) and are described in Supplementary Table S1.
RNA preparation
dsDNA templates for in vitro transcription reactions were produced using standard PCR reactions. For PCR, ∼25 pg of plasmid bearing dsWXYZ sequence was mixed with 1× PCR buffer (ThermoFisher Scientific), 0.5 μM of each forward and reverse primer (Supplementary Table S1), 0.2 mM of each dNTP, 0.02 U/μl Hot Start Phusion polymerase (ThermoFisher Scientific, Product no.: F-549L) and thermocycled as follows: step 1: 98°C/30 s, step 2: 98°C/10 s, step 3: 57°C/30 s, step 4: 72°C/30 s with 24 additional cycles from step 2 to 4 and final extension at 72°C/5 min. The purity of the dsDNA was checked on a 2% agarose gel (stained with GelRed™, run under standard electrophoresis conditions; 1× Tris-acetate-EDTA (TAE), 110 V, 40 min). After PCR, amplified dsDNA templates were isopropanol precipitated, pellets were washed with 70% ethanol, dissolved in water and used directly for in vitro transcription. In vitro transcription reactions were performed at 100 μl scale with dsDNA mixed with 1× transcription buffer (ThermoFisher Scientific), 16 mM additional MgCl2, 4 mM of each NTP and 10 U/μl of T7 RNA polymerase (ThermoFisher Scientific, Product no.: EP0111) and incubated at 37°C for 4 h. The reaction was stopped by addition of gel loading buffer (70% formamide containing 0.1% of each xylene cyanol and bromophenol blue) and purified on 12% denaturing polyacrylamide gels using standard electrophoresis conditions (1× TBE buffer, run at 24 W for 2–3 h). Transcript bands were excised and eluted in 0.3 M Na-Acetate pH 5.5 overnight at room temperature. The eluted solution was isopropanol precipitated, washed with 70% Ethanol, dissolved in water and concentrations were measured.
Autocatalytic trans-esterification reaction
For the autocatalytic trans-esterification reactions RNA substrates were mixed in water to give a final concentration of 0.5 μM for each RNA. For the cooperative network formation each of the three WXY substrates were mixed to give a final concentration of 0.5 μM for each of the three RNAs (1.5 μM total) and the other substrate (Z or Z-mod) was added to a final concentration of 1.5 μM (to give 1:1 stoichiometry of WXY with Z or Z-mod). To fold the RNA, the mixture was heated at 80°C for 3 min and gradually cooled down to 20°C (at a rate of 0.1°C/s). Then 30 mM of EPPS (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) buffer pH 7.4 and 100 mM MgCl2 was added and the reaction was incubated at 48°C. For gel-based kinetic analysis, at each time point 2 μl of reaction was taken out and mixed with gel loading buffer containing 70% formamide, 0.01% of each xylene cyanol and bromophenol blue, and ∼2 equivalents of EDTA (ethylenediaminetetraacetic acid) and analyzed on denaturing polyacrylamide gels.
Sanger sequencing
To confirm the identity of the different products formed with different substrate combinations (Figure 2; c2, c3, c4) the sequences were analyzed by Sanger sequencing. For each reaction, the expected size product band was excised from polyacrylamide gel, RNA were eluted in 0.3 M sodium-acetate pH 5.5 overnight and isopropanol precipitated. The pellets were washed three-times with 70% ethanol and re-suspended in 30 μl water. A total of 5 μl was used for reverse-transcription (RT) using 2.5 μM of primer complementary to the 3’ end of the Z fragment (primer 11, Supplementary Table S1). For the sequencing of WXY-mod, RNA was additionally tailed with poly-guanosine (23) by Escherichia coli polyA polymerase (New England Biolabs, Product no.: M0276L) using 2 mM guanosine triphosphate (GTP) and the standard protocol. For the RT of poly-guanosine tailed samples, primer 24 was used and for PCR amplification primer 25 and primer 15 were used as forward and reverse primers, respectively. The RT reaction (20 μl total volume) was performed in 1× first-strand buffer (250 mM Tris–HCl (pH 8.3), 375 mM KCl, 15 mM MgCl2), 0.2 mM dNTPs, 5 mM DTT and 10 U/μl of enzyme (SuperScript® III Reverse Transcriptase; ThermoFisher Scientific; Product no.: 18080093). The reaction was incubated at 55°C for 1 h and samples were then purified using 1.2 equivalent of AMPure XP magnetic beads (Beckman Coulter, Product no.: A63881) following the manufacturer’s protocol. A total of 10 μl of purified RT products were then PCR amplified using the same protocol as for in vitro transcription using primer 1 as the forward primer and primer 11 as the reverse primer (Supplementary Table S1). Polymerase chain reaction (PCR) products were purified using AMPure XP magnetic beads. Approximately 20 ng of purified PCR products were ligated into pJET1.2/blunt vector using CloneJET PCR Cloning Kit (ThermoFisher Scientific; Product no.: K1232). A total of 5 μl of ligation mix was mixed with 50 μl of MegaX DH10B™ T1R Electrocomp™ Cells (ThermoFisher Scientific; Product No.: C640003), kept for 2 min on ice, then at 42°C for 1 min 30 s and then put back on ice. A total of 1 ml of LB medium was added and cells were grown for 1 h at 37°C before plating on a LB agar plate supplemented with 100 μg/ml ampicillin and plates were incubated at 37°C overnight. The positive colonies were checked by colony PCR: picked colonies were added into 3 μl water from which half was used for colony PCR and other half to start a 5 ml overnight culture at 37°C in LB medium for positive colonies. Plasmids were extracted from overnight cultures using NucleoSpin® Plasmid kit (Macherey-Nagel; Product no.: 740588.250) and sequenced using the GATC ‘Light-run’ Sanger sequencing service.
Figure 2.
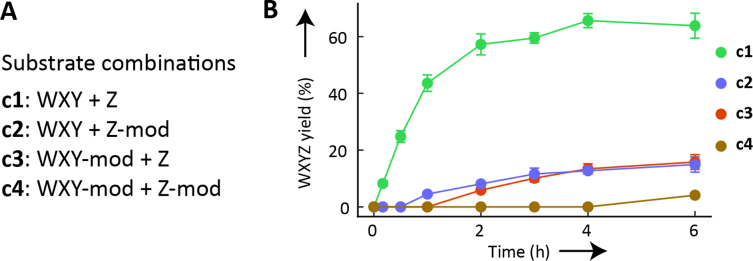
Synthesis of WXYZ catalyst with modified RNA substrates. (A) Different substrate combinations used to study the catabolic properties of the Azoarcus ribozyme system. (B) Kinetics of the synthesis of WXYZ using all four substrate combinations (c1 to c4). The reported yield is the substrate to product conversion in percentage. Error bars represent ±1 standard deviation (triplicates).
Kinetic modeling
The kinetic model used here was developed using a similar approach as described previously (22). From a small set of assumptions (covalent bond formation is reversible and catalysts can be covalent ribozymes or non-convalent complexes between the two substrate fragments), we derived a set of 14 chemical reactions (Supplementary Table S2) for the complete model. We also constructed reduced models based on additional assumptions and computed a set of model selection criteria (Supplementary Table S3) to investigate whether the number of rate constants could be reduced. Our approach is described in detail in the Supplementary Text. For all models, the reaction rates were fitted using Scipy (24) nonlinear least-squares problem solving routine with ‘trf’ method (Trust Region Reflective algorithm) and ‘cauchy’ loss function (results are similar with ‘linear’ loss function). Reactions and fitted rates are summarized in Supplementary Table S2.
Network genotyping
In order to obtain the genotypic distribution, the reaction products WXYZ ribozymes were sequenced by Illumina high-throughput sequencing. RNA samples after 6 h of reaction were reverse transcribed using a specific primer (Primer 13, see Supplementary Table S1) and PCR amplified following the protocol above (see Sanger sequencing section). PCR was performed in two steps to sequentially add the Illumina sequencing adaptors. For the first step, we used primer 14 and primer 15 and amplified the samples for total of 18 cycles. After the first step, samples were purified using AMPure XP magnetic beads (Beckman Coulter, Product no.: A63881) and subjected to an additional 10 cycles of amplification with primer 16 and primer 17. After this, the final amplicons were purified with AMPure XP magnetic beads (Beckman Coulter, Product no.: A63881), quantified using Quibit (Qubit® 3.0 Fluorometer) and subjected to 2 × 150 paired-end nanoMiseq (Institut Curie High Throughput Sequencing platform, Paris). The relative number of Unique Molecular Identifiers (UMIs) (25) for each genotype is de-convoluted using a custom software pipeline developed in house.
RESULTS
Effect of substrate modification on the formation of Azoarcus ribozymes
To investigate the impact of substrate modifications on the Azoarcus system, we modified the native substrates WXY and Z into WXY-mod and Z-mod, respectively, by appending a foreign sequence at the 3’ end (mod): a poly-adenosine stretch preceded by ‘agugcc’ (Supplementary Figure S1). These modifications are relevant in the context of prebiotic Earth where any random ligation, recombination or chemical addition could result in the appendage of a stretch of oligonucleotides. Homopolymers are plausible candidates for inert appendages as they cannot form stable folded structures, and spontaneous synthesis of long nucleic acids has so far been shown to be biased toward such homopolymers (26,27). We analyzed the effect of different combinations (Figure 2A) of these modified RNA substrates on the kinetics of self-assembly reactions (Figure 2B). Remarkably, the modifications do not block the reaction completely, as we observe the appearance of measurable amounts of WXYZ catalyst in each case (Figure 2B and Supplementary Figure S2). However, the modification drastically slows down the reaction (Figure 2B; compare c1 with c2, c3 or c4). When only one substrate is modified (c2 and c3), WXYZ synthesis starts rather slowly and reaches ∼15% of initial material after 6 h of reaction as opposed to ∼60% with non-modified substrates (c1). When both substrates are modified the WXYZ production drops to only ∼4% after 6 h (c4).
Additionally, we investigated other modifications of the substrate by changing the tail from poly-adenosine to poly-guanosine or poly-uridine and by changing the sequence preceding the tail (Supplementary Figure S3). In all these cases, we observed the formation of WXYZ catalysts.
These results highlight the inherent capacity of the Azoarcus ribozyme system to overcome the modification burden by somehow processing the available raw material. The simplest scenario to purge the modification would involve direct cleavage, as for example proposed in an earlier theoretical study (28), of mod part from the substrates WXY-mod and Z-mod. However, we do not observe direct cleavage of mod from Z-mod, even in the presence of WXYZ (Supplementary Figure S4). Instead mod is transferred from Z-mod to WXY to generate WXY-mod (Supplementary Figure S5). Conversely, transfer of mod from WXY-mod to Z is not detected (Supplementary Figure S6). Ultimately, the modification is slowly cleaved from WXY-mod, catalyzed by WXYZ, generating unmodified WXY (Supplementary Figure S7). Note that while the experiments were routinely performed at 100 mM MgCl2, we observed that these reaction steps lead to measurable amounts of products even down to 10 mM MgCl2 (Supplementary Figure S8).
Multi-step reaction pathway and kinetic modeling
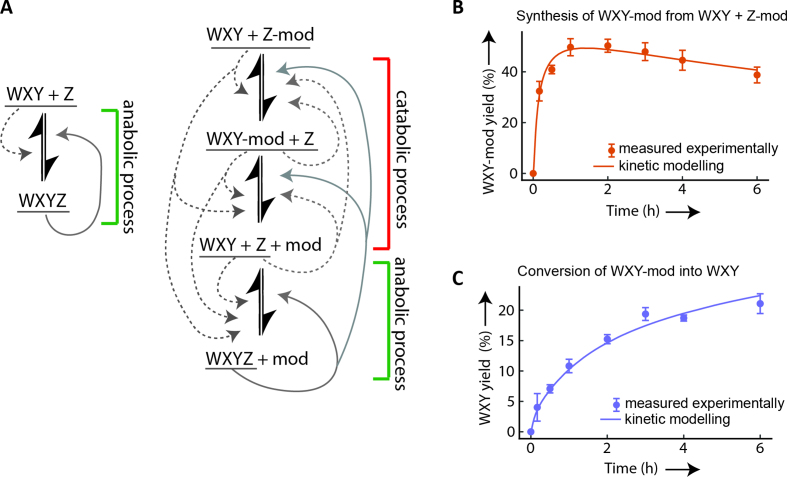
These observations suggest a reaction path (Figure 3A) where substrate composition c2 is converted into c3 by transfer of mod from Z-mod to WXY, followed by conversion into the canonical fragments (substrate combination c1), by cleavage of mod from WXY-mod which then react to generate WXYZ via the autocatalytic anabolic reaction described by Hayden et al. (22). The kinetic traces of the proposed intermediates WXY-mod and WXY are consistent with this reaction pathway: we observe the formation of WXY-mod, which peaks at almost 50% conversion within an hour, followed by a slow decrease (Figure 3B). Starting from combination c3 leads to conversion from WXY-mod to WXY which happens on slower timescales, with <10% of WXY-mod being converted to WXY within an hour (Figure 3C).
Figure 3.
Multi-step reaction pathway to catabolize the starting material. (A) Diagrammatic representation of the catabolic steps involved in the synthesis of substrates WXY and Z from the modified RNAs (WXY-mod and Z-mod) to produce WXYZ. Dotted and continuous gray arrow show feed-back by non-covalent and covalent catalyst, respectively. (B) Kinetic analysis of the formation of WXY-mod with substrate combination c2. (C) Kinetic analysis of the formation of WXY from WXY-mod with substrate combination c3. For both graphs (B) and (C), the circles represent experimental data obtained by polyacrylamide gel electrophoresis and the lines represents the data obtained from kinetic modeling (Supplementary Text). The reported yield is the substrate to product conversion in percentage. Error bars represent ±1 standard deviation (triplicates).
We then built a model to test whether the kinetics of species for different substrate combinations could be described by a single kinetic model of the entire reaction scheme. The model is based on the recombination mechanisms reported earlier for Azoarcus ribozymes (22) (Supplementary Table S2): non-covalent complexes of the substrates can catalyze recombination reactions at a slow rate (dashed gray arrows, Figure 3A) and covalent ribozymes at a higher rate (solid gray arrows, Figure 3A). We obtained a single set of parameters from the devised kinetic model by fitting to the experimental data (Figure 3B and C; Supplementary Figure S9). In particular, the model reproduces well the critical steps of the pathway: the rapid formation of WXY-mod and its slow consumption, as well as WXYZ formation. We further investigated the validity of the model by reducing the number of parameters and by computing different model selection criteria (Supplementary Figure S10 and Table S3).
Mechanisms of WXY-mod formation
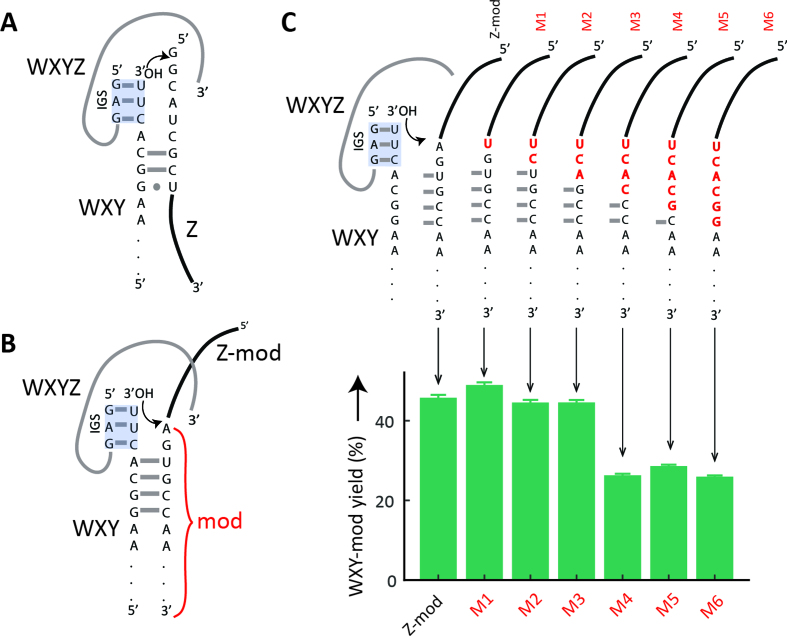
Based on the previously characterized reaction mechanism (20,29) and the product sequences (Supplementary Figure S2), we hypothesized that the transfer of the modification from Z-mod to WXY is mediated by Watson–Crick base-pairing of Z-mod to the 3’ end of WXY (Figure 4A). Whereas normally the 5’ end of the unmodified substrate Z base-pairs with the 3’ end of WXY, here, the region of Z-mod immediately 5’ to the poly-adenosine sequence binds to the 3’ end of WXY via four Watson–Crick base-pairs (Figure 4B). We probed the contribution of these nucleotides in the formation of WXY-mod by sequentially substituting them to disrupt the proposed Watson–Crick base-pairing and analyzed the amount of WXY-mod product formed (Figure 4C). Changing nucleotides that are not proposed to base-pair (mutants M1 and M2) as well as that proposed to form the first (A-U) base-pair (mutant M3) has no significant effect on WXY-mod formation. Additionally disrupting the second (G-C) base-pair (mutant M4) reduces the formation of WXY-mod by almost half (only ∼25% of WXY converted to WXY-mod in an hour). However, mutating all the other proposed base-pairing residues (mutants M5 and M6) does not reduce WXY-mod formation further, indicating that other interactions must be involved and could be important in the reaction mechanism.
Figure 4.
Proposed binding of Z and Z-mod and effect of mutations in Z-mod. (A) Diagrammatic representation of the binding of Z to WXY in the case of non-modified substrates (20,29). (B) Proposed binding of Z-mod to WXY. (C) Sequence of mutated variants (M1-6, substitutions in red) of Z-mod and the yield of WXY-mod after 1 h reaction between WXY and non-mutated or mutated variants of Z-mod obtained by polyacrylamide gel electrophoresis. The reported yield is the substrate to product conversion in percentage. Error bars represent ±1 standard deviation (triplicates).
Cooperative network is not hampered by modified substrates
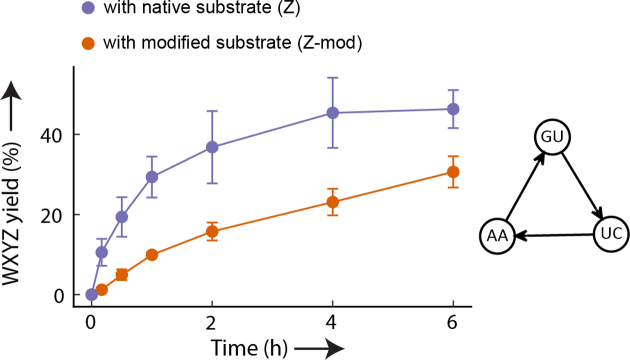
Azoarcus ribozymes are known to form multi-species autocatalytic networks where the growth of a species is dictated by how well its formation is catalyzed by other species in the network (21). These networks are formed by base-pairing interactions between the IGS (GMG) and the tag (CNU) of different fragments, where M and N are variable nucleotides (A, C, U or G) in IGS-tag combination, denoted as MN (20,21). For example, IGS ‘A’ (GAG) from WXYZ recognizes tag ‘U’ (CUU) in WXY to catalyze its assembly with Z. Appropriate combinations of such interactions allows the formation of a closed catalytic cycle as shown in Figure 5, right panel. However, it is unclear whether such networks can function when fueled with modified substrates. We thus constructed a three-membered network (30,31) with either modified (Z-mod) or canonical (Z) substrate and analyzed the synthesis of the RNA catalyst (WXYZ) (Figure 5). Even when only modified substrates are used, the Azoarcus ribozyme network is still functional and the yield of WXYZ is only slightly lower than with unmodified substrates (∼45% after 6 h with Z compared to ∼30% with Z-mod). This shows that the catabolic steps do not interfere with the collective dynamics. High-throughput sequencing confirmed that not only the growth (amount of WXYZ formed), but also the relative distribution of the members of the network are maintained (Supplementary Figure S11). In particular, we found the same ordering of the members as reported in an earlier study with the same network of non-modified fragments (31), where the ‘UC’ ribozyme is at higher concentration than the other two ribozymes (‘AA’ and ‘GU’).
Figure 5.
Formation of cooperative RNA network using modified substrates. Reactions were started by adding WXY fragments with different IGS-tag sequences (MN = AA, GU and UC, right panel) as used in previous studies (30,31). The reaction is either provided with native (Z, purple) or modified substrate (Z-mod, orange). Kinetic analysis was performed by analysing the formation of WXYZ at different time points using polyacrylamide gel electrophoresis. The reported yield is the substrate to product conversion in percentage. Error bars represent ±1 standard deviation (triplicates).
DISCUSSION
The work presented here demonstrates how catabolic steps in collectively autocatalytic sets can expand the range of usable substrates, which would have been highly advantageous in a heterogeneous prebiotic milieu. The assimilation of modified substrates here is enabled by the recombination activity of Azoarcus ribozymes, and involves a multi-step reaction pathway, which contrasts with other experimental autocatalytic systems (32–34). We have furthermore shown that a cooperative network made of several species and fueled with modified substrates is almost as efficient at synthesizing catalysts as with unmodified substrates and maintains the relative distribution of the members. Although RNA fragments used here are longer than what is usually imagined in prebiotic scenarios, it should be noted that Azoarcus ribozymes can be formed from much shorter RNA fragments (<50 nt) (21,35).
Similar catabolic processes may be transposed to smaller molecules CAS (e.g. the formose reaction, where sugars are formed from formaldehyde (36) as soon as the catalysts formed by the autocatalytic reactions are able to reshuffle chemicals. However it remains difficult to elaborate such scenarios at present, given the scarcity of experimental models.
In addition to the basic ingredients of fragment recycling and autocatalysis, the recovery of the unmodified catalytic sequences indicates an inherent form of selection at the molecular level. This selection may be related to the differential protection of functional and parasitic folds from recombination, as envisaged earlier (4). This idea is reminiscent of the concepts of dynamical combinatorial chemistry (37), where self-assembly is replaced by covalent recombination, and differential product stability may arise from thermodynamic as well as from kinetic factors. Interestingly, these ingredients are applied here to an autocatalytic system and suggest a mechanism to limit the extinction of early metabolic cycles by side reactions highlighted by Orgel (38).
Coupled catabolism and anabolism is a universal feature of contemporary living systems: diverse complex molecules are broken down into simpler building blocks that are used to construct new biomolecules. Although the conditions and mechanisms observed in this study differ from those observed in contemporary cellular life, it is striking to witness that coupled catabolism and anabolism could have arisen in a system with a much lower level of complexity, at an early stage in the RNA world.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Estelle Mendes, Nympha Elisa Sia and Bilal Mazhar for their technical assistance in the realization of the experiments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Union Seventh Framework Program (FP7/2007-2013) [294332 (EvoEvo)]; PSL Research University (OCAV project); Ecole Polytechnique for PhD fellowship (AMX) to S. Arsène; Ecole Doctorale FdV (Programme Bettencourt). Funding for open access charge: European Union Seventh Framework Program (FP7/2007–2013) [294332 (EvoEvo)].
Conflict of interest statement. None declared.
REFERENCES
- 1. Eigen M. Selforganization of matter and the evolution of biological macromolecules. Naturwissenschaften. 1971; 58:465–523. [DOI] [PubMed] [Google Scholar]
- 2. Eigen M., Schuster P.. The hypercycle. A principle of natural self-organization. Part A: emergence of the hypercycle. Naturwissenschaften. 1977; 64:541–565. [DOI] [PubMed] [Google Scholar]
- 3. Kauffman S.A. Autocatalytic sets of proteins. J. Theor. Biol. 1986; 119:1–24. [DOI] [PubMed] [Google Scholar]
- 4. Higgs P.G., Lehman N.. The RNA World: molecular cooperation at the origins of life. Nat. Rev. Genet. 2015; 16:7–17. [DOI] [PubMed] [Google Scholar]
- 5. Hordijk W., Steel M.. Chasing the tail: the emergence of autocatalytic networks. Biosystems. 2017; 152:1–10. [DOI] [PubMed] [Google Scholar]
- 6. Hordijk W., Steel M., Kauffman S.A.. The structure of autocatalytic sets: evolvability, enablement, and emergence. Acta. Biotheor. 2012; 60:379–392. [DOI] [PubMed] [Google Scholar]
- 7. Jain S., Krishna S.. Autocatalytic sets and the growth of complexity in an evolutionary model. Phys. Rev. Lett. 1998; 81:5684–5687. [Google Scholar]
- 8. Jain S., Krishna S.. A model for the emergence of cooperation, interdependence, and structure in evolving networks. Proc. Natl. Acad. Sci. U.S.A. 2001; 98:543–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lee D.H., Severin K., Ghadiri M.R.. Autocatalytic networks: the transition from molecular self-replication to molecular ecosystems. Curr. Opin. Chem. Biol. 1997; 1:491–496. [DOI] [PubMed] [Google Scholar]
- 10. Vasas V., Fernando C., Santos M., Kauffman S.A., Szathmáry E.. Evolution before genes. Biol. Direct. 2012; 7:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hordijk W., Steel M.. Detecting autocatalytic, self-sustaining sets in chemical reaction systems. J. Theor. Biol. 2004; 227:451–461. [DOI] [PubMed] [Google Scholar]
- 12. Chen X., Li N., Ellington A.D.. Ribozyme catalysis of metabolism in the RNA world. Chem. Biodivers. 2007; 4:633–655. [DOI] [PubMed] [Google Scholar]
- 13. Copley S.D., Smith E., Morowitz H.J.. The origin of the RNA world: co-evolution of genes and metabolism. Bioorg. Chem. 2007; 35:430–443. [DOI] [PubMed] [Google Scholar]
- 14. King G.A. Recycling, reproduction, and life's origins. Biosystems. 1982; 15:89–97. [DOI] [PubMed] [Google Scholar]
- 15. Orgel L.E. Prebiotic chemistry and the origin of the RNA world. Crit. Rev. Biochem. Mol. Biol. 2004; 39:99–123. [DOI] [PubMed] [Google Scholar]
- 16. Robertson M.P., Joyce G.F.. The origins of the RNA world. Cold Spring Harb. Perspect. Biol. 2012; 4:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vaidya N., Walker S.I., Lehman N.. Recycling of informational units leads to selection of replicators in a prebiotic soup. Chem. Biol. 2013; 20:241–252. [DOI] [PubMed] [Google Scholar]
- 18. Cech T.R. Self-splicing of group I introns. Annu. Rev. Biochem. 1990; 59:543–568. [DOI] [PubMed] [Google Scholar]
- 19. Reinhold-Hurek B., Shub D.A.. Self-splicing introns in tRNA genes of widely divergent bacteria. Nature. 1992; 357:173–176. [DOI] [PubMed] [Google Scholar]
- 20. Draper W.E., Hayden E.J., Lehman N.. Mechanisms of covalent self-assembly of the Azoarcus ribozyme from four fragment oligonucleotides. Nucleic Acids Res. 2008; 36:520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vaidya N., Manapat M.L., Chen I.A., Xulvi-Brunet R., Hayden E.J., Lehman N.. Spontaneous network formation among cooperative RNA replicators. Nature. 2012; 491:72–77. [DOI] [PubMed] [Google Scholar]
- 22. Hayden E.J., von Kiedrowski G., Lehman N.. Systems chemistry on ribozyme self-construction: evidence for anabolic autocatalysis in a recombination network. Angew. Chem. Int. Ed. Engl. 2008; 47:8424–8428. [DOI] [PubMed] [Google Scholar]
- 23. Yehudai-Resheff S., Schuster G.. Characterization of the E. coli poly(A) polymerase: nucleotide specificity, RNA-binding affinities and RNA structure dependence. Nucleic Acids Res. 2000; 28:1139–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oliphant T.E. Python for Scientific Computing. Comput. Sci. Eng. 2001; 9:10–20. [Google Scholar]
- 25. Kivioja T., Vaharautio A., Karlsson K., Bonke M., Enge M., Linnarsson S., Taipale J.. Counting absolute numbers of molecules using unique molecular identifiers. Nat. Methods. 2011; 9:72–74. [DOI] [PubMed] [Google Scholar]
- 26. Huang W., Ferris J.P.. Synthesis of 35–40 mers of RNA oligomers from unblocked monomers. A simple approach to the RNA world. Chem. Commun. 2003; 0:1458–1459. [DOI] [PubMed] [Google Scholar]
- 27. Huang W., Ferris J.P.. One-step, regioselective synthesis of up to 50-mers of RNA oligomers by montmorillonite catalysis. J. Am. Chem. Soc. 2006; 128:8914–8919. [DOI] [PubMed] [Google Scholar]
- 28. Hordijk W., Steel M.. Autocatalytic sets in polymer networks with variable catalysis distributions. J. Math. Chem. 2106; 54:1997–2021. [Google Scholar]
- 29. Adams P.L., Stahley M.R., Kosek A.B., Wang J., Strobel S.A.. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004; 430:45–50. [DOI] [PubMed] [Google Scholar]
- 30. Yeates J.A.M., Hilbe C., Zwick M., Nowak M.A., Lehman N.. Dynamics of prebiotic RNA reproduction illuminated by chemical game theory. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:5030–5035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yeates J.A.M., Nghe P., Lehman N.. Topological and thermodynamic factors that influence the evolution of small networks of catalytic RNA species. RNA. 2017; 23:1088–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ashkenasy G., Jagasia R., Yadav M., Ghadiri M.R.. Design of a directed molecular network. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:10872–10877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kim D.E., Joyce G.F.. Cross-catalytic replication of an RNA ligase ribozyme. Chem. Biol. 2004; 11:1505–1512. [DOI] [PubMed] [Google Scholar]
- 34. Sievers D., von Kiedrowski G.. Self-replication of complementary nucleotide-based oligomers. Nature. 1994; 369:221–224. [DOI] [PubMed] [Google Scholar]
- 35. Jayathilaka T.S., Lehman N.. Spontaneous covalent self-assembly of the Azoarcus ribozyme from five fragments. Chembiochem. 2018; 19:217–220. [DOI] [PubMed] [Google Scholar]
- 36. Butlerov A.M. Einiges über die chemische Structur der Körper. Zeitschrift für Chem. 1861; 4:549–560. [Google Scholar]
- 37. Corbett P.T., Leclaire J., Vial L., West K.R., Wietor J.L., Sanders J.K., Otto S.. Dynamic combinatorial chemistry. Chem. Rev. 2006; 106:3652–3711. [DOI] [PubMed] [Google Scholar]
- 38. Orgel L.E. The implausibility of metabolic cycles on the prebiotic Earth. PLoS Biol. 2008; 6:e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.