Abstract
Eutrophication can play a significant role in seagrass decline and habitat loss. Microorganisms in seagrass sediments are essential to many important ecosystem processes, including nutrient cycling and seagrass ecosystem health. However, current knowledge of the bacterial communities, both beneficial and detrimental, within seagrass meadows in response to nutrient loading is limited. We studied the response of sediment bacterial and pathogen communities to nutrient enrichment on a tropical seagrass meadow in Xincun Bay, South China Sea. The bacterial taxonomic groups across all sites were dominated by the Gammaproteobacteria and Firmicutes. Sites nearest to the nutrient source and with the highest NH4 + and PO4 3− content had approximately double the relative abundance of putative denitrifiers Vibrionales, Alteromonadales, and Pseudomonadales. Additionally, the relative abundance of potential pathogen groups, especially Vibrio spp. and Pseudoalteromonas spp., was approximately 2‐fold greater at the sites with the highest nutrient loads compared to sites further from the source. These results suggest that proximity to sources of nutrient pollution increases the occurrence of potential bacterial pathogens that could affect fishes, invertebrates and humans. This study shows that nutrient enrichment does elicit shifts in bacterial community diversity and likely their function in local biogeochemical cycling and as a potential source of infectious diseases within seagrass meadows.
Keywords: bacteria, denitrification, eutrophication, putative pathogens, seagrass meadows
1. INTRODUCTION
Seagrass meadows are incredibly productive ecosystems (Hemminga & Duarte, 2000) and support a high diversity of microorganisms in their sediments (Bourque, Vega‐Thurber, & Fourqurean, 2015; García‐Martínez, López‐López, Calleja, Marbà, & Duarte, 2009), due to the release of root exudates (amino acids, sugars) and oxygen into the rhizosphere (Christiaen, McDonald, Cebrian, & Ortmann, 2013; Ingemann Jensen, Kühl, Glud, Jørgensen, & Priemé, 2005; Jensen, Kühl, & Priemé, 2007). Microbes play a fundamental role in several biogeochemical cycling processes, including the oxidation of organic carbon, nitrogen fixation, nitrification, denitrification, iron cycling, and sulfate reduction within the rhizosphere and surrounding sediments (Christiaen et al., 2013; Hemminga & Duarte, 2000; Marbà, Holmer, Gacia, & Barron, 2007). Most of these processes are primarily driven by bacterial communities (Jørgensen, 1982). For example, nitrogen fixation by cyanobacteria and sulfate‐reducing bacteria can significantly contribute to seagrass nutrient requirements (Hansen, Udy, Perry, Dennison, & Lomstein, 2000; Welsh, 2000). In addition, the presence of bacterial pathogens in coastal waters and sediments are important indicators of environmental health (Bally & Garrabou, 2007; Luna et al., 2010). In fact, seagrass meadows have been shown to be involved in reducing the abundance of bacterial pathogens linked to infections and diseases in humans and marine organisms (Lamb et al., 2017).
Unfortunately, seagrass beds have been severely degraded by anthropogenic disturbances and climate change, with rapid rate of decline worldwide (~7% per year; Waycott et al., 2009). In particular, seagrass beds have been adversely affected by eutrophication, and this pressure is predicted to increase over the coming decades due to increased coastal aquaculture, coastal development and concurrent runoff (Burkholder, Tomasko, & Touchette, 2007; Ralph, Tomasko, Moore, Seddon, & Macinnis‐Ng, 2006). In addition to directly affecting seagrass health through light reduction, ammonium toxicity and water‐column nitrate inhibition (Burkholder et al., 2007), eutrophication has been shown to affect sediment bacterial metabolism and function (Howard, Perez, Lopes, & Fourqurean, 2016; López, Duarte, Vallespinós, Romero, & Alcoverro, 1998). López et al. (1998) found that the addition of inorganic nitrogen to Posidonia oceanica sediments significantly increased ammonification rates and bacterial exoenzymatic activities, which resulted in enhanced bacterial decomposition of seagrass‐derived carbon. Additionally, the bacterial pathogen and disease occurrence have been consistently correlated with high nutrient loads in near‐shore environments (National Research Council, 2000; Vega Thurber et al., 2014; Zaneveld et al., 2016). However, studies that investigate the effect of nutrient loading on sediment bacterial community structure within seagrass meadows are otherwise rare (Guevara, Ikenaga, Dean, Pisani, & Boyer, 2014). Filling this gap could help us understand the factors driving nutrient cycling and the potential for disease outbreaks resulting from nutrient loading within seagrass meadows.
In this study, we used Next‐generation sequencing to answer the question: To what extent does nutrient enrichment affect the sediment bacterial community structure, including putative pathogens, in a seagrass meadow? We sampled sediments from mixed seagrass communities, with increasing distance from nutrient loads coming from fish cages in Xincun Bay, South China Sea (Liu et al., 2016; Zhang, Huang, & Jiang, 2014). Our previous research has indicated that high nutrient levels in Xincun Bay stimulate the seagrass, macroalgal and epiphytic biomass production, resulting in higher organic carbon availability for bacteria or contribution to sediment carbon stocks (Liu et al., 2016, 2017). We hypothesized that higher nutrient loads from the fish cages will alter the bacterial community structure and bacterial pathogens in seagrass sediments. The results of this study on how microbial community composition changes under eutrophication scenarios will help clarify the reciprocal relationships of microbes and the biogeochemical environment in degraded seagrass meadows, and increase our understanding of the microbial ecology in these ecologically and socioeconomically important ecosystems.
2. MATERIALS AND METHODS
2.1. Study area and sampling sites
The study was performed at Xincun Bay (18°24′34′′N–18°24′42′′N, 109°57′42′′E–109°57′58′′E), located in southeastern Hainan Island, South China Sea (Figure 1). In recent years, cage aquaculture, located near the entrance of the bay, has developed rapidly, and currently includes more than 450 floating cage units (Zhang et al., 2014). At the south‐eastern part of the fish farm, shallow‐water seagrass meadows dominated by Thalassia hemprichii (Ehrenb. ex Solms) Asch. and Enhalus acoroides (L.f.) Royle occupy an area of approximately 200 ha (Huang et al., 2006). Three transects (A, B, C) were selected according to the distance to fish farm in the seagrass bed and represented a nutrient load gradient (Liu et al., 2016). The first transect was 500 m from the fish farm, with the other transects ~800 m apart from each other. For each transect, independent samples were taken at sampling stations at 50, 400 and 750 m from the shore (stations 1, 2 and 3, respectively). E acoroides was not found at stations B1, C1 and C2, and T. hemprichii was not found at A2. Stations A1, A3, B2, B3, and C3 included both seagrass species (Figure 1). Each sample was annotated according to transect, station and seagrass species, for example, T. hemprichii in A1 can be represented as A1.T.
Figure 1.
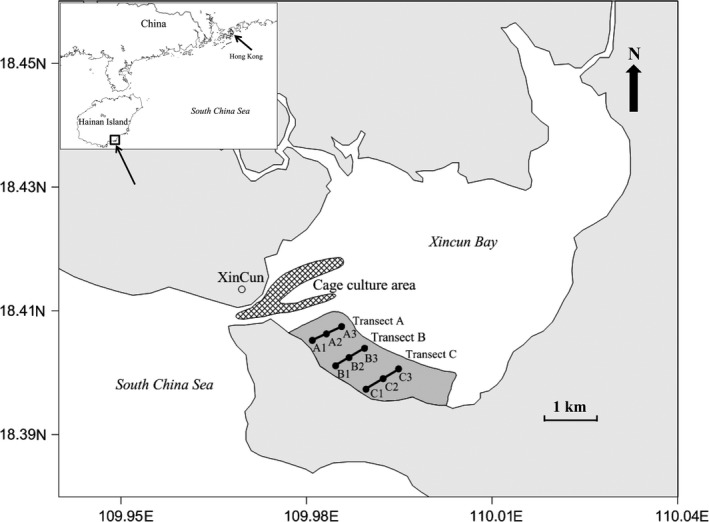
Sampling sites in Xincun Bay at Hainan Island in the South China Sea, which were divided by distance from the cage aquaculture (transects A, B and C) and distance from the shore (stations 1, 2 and 3)
2.2. Sample collection
During low tide (~ 10 cm depth), 1 L of seawater above the sediment was sampled for nutrient analysis. Additionally, a surface sediment sample (0–3 cm) was collected in both T. hemprichii and E. acoroides meadows at each site, using a sterile 10 cm diameter sampling core. Each sediment sample was divided into two subsamples, one subsample was frozen at −20°C for sediment organic carbon (SOC) and sediment total nitrogen (TN) analysis. The other subsample was preserved at −80°C until extraction of genomic DNA.
2.3. Sample preparation and analysis
2.3.1. Seawater nutrients, SOC and sediment TN analysis
The seawater was filtered onto precombusted GF/F filters (Whatman, 450°C, 3 hr). The filtered seawater was analyzed for NO3 −–N, NO2 −–N, NH4 +–N, and PO4 3−, using an AQ‐2 Automated Discrete Analyzer (Seal Analytical Inc.). The sum of NO3 −–N, NO2 —N, and NH4 +–N represents the total concentration of dissolved inorganic nitrogen (DIN). The frozen (−20°C) sediment samples were freeze‐dried and composited by plot in order to make the samples homogeneous. The composite samples were sieved through a 500‐μm screen to remove coarse sediment and detrital materials. The samples were then ground and homogenized with a mortar and pestle. Sediment was acidified with 1 mol/L HCl overnight at room temperature to remove carbonate. Acidified sediments were washed with distilled water and dried at 40°C in an oven. SOC (acidified) and sediment TN (unacidified) was determined, using an elemental analyzer (Vario EL, Elemental Analyser systeme GmbH, Germany).
2.3.2. DNA extraction, PCR amplification and Illumina sequencing
DNA was extracted from 0.5 g of sediment (wet weight), using an EZNA® Soil DNA Kit (Omega Bio‐Tek Inc., Norcross, GA, USA), according to the manufacturer's instructions. The quality and quantity of the extracted DNA were verified by measuring the OD260/280 (≈1.8) with a Nanodrop 2000 spectrophotometer and agarose gel electrophoresis, respectively. Primer set 515F/806R was used to amplify the V4 region of 16S rRNA gene (Mckirdy et al., 2010). Sequencing was performed on an Illumina MiSeq platform at Novogene Genomics Technology Co. Ltd, Beijing, China.
2.3.3. Sequence analyses, OTU clustering and bioinformatics analysis
Sequences were analyzed, using the QIIME version 1.9.1 pipeline (Caporaso et al., 2010b). Raw sequences were demultiplexed and quality filtered, using the default parameters in QIIME. Sequences were then clustered into operational taxonomic units (OTUs), which was defined as >97% 16S rRNA gene sequence similarity, using UCLUST (Edgar, 2010) with the open reference clustering protocol. The resulting representative sequences set were aligned, using PyNAST (Caporaso et al., 2010a) and given a taxonomic classification, using RDP (Wang, Garrity, Tiedje, & Cole, 2007), retrained with the Greengenes version 13.5 (McDonald et al., 2012). The resulting OTU table was used to determine the Chao 1, Shannon, and rarefaction diversity indices, using QIIME. METAGENassist was used for Weighted Unifrac analysis (Arndt et al., 2012). In addition, we reviewed papers containing information pertinent to the putative pathogens (genus level) that can cause fish and invertebrate disease or death or cause human illness through the consumption of seafood or skin contact. A list of putative pathogen OTUs (total abundance of all samples >0.1) was compiled for further statistical analysis.
2.4. Statistical analysis
The raw data were log‐ or exponent‐transformed in order to fulfill the assumptions of homogeneity and normality in cases where these assumptions were not met. Since the effect of nutrient loading was the treatment we were primarily interested in testing, we ran a preliminary one‐way analysis of variance (ANOVA) to examine the effect of distance to shore on seawater nutrients (DIN, NO3 −–N, NO2 −–N, NH4 +–N and PO4 3−), SOC, sediment TN, and sediment C/N. Distance from the shore was not a significant treatment affecting the response variables (p > .05), and thus were pooled for further statistical tests. A one‐way ANOVA tested the effect of transect on seawater nutrients, while a two‐way ANOVA analyzed the effects of transect and seagrass species on the SOC, sediment TN and sediment C/N. Whenever significant differences were observed from an ANOVA, a Tukey's post hoc test was run to identify the significantly different components of dependent variables.
The weighted UNIFRAC resemblance matrix (Table S1) was used for the bacterial community analyses, while the filtered putative pathogen counts were used for pathogen community analyses. For both datasets, a preliminary one‐way permutational multivariate ANOVA (PERMANOVA) showed that distance to shore was not a significant treatment (Pseudo‐F = 0.8236, P‐perm = 0.569). A two‐way PERMANOVA was subsequently performed to determine the statistically significant differences among the transects and seagrass species. All main and interaction effects with α < 0.05 were considered statistically significant. A similarity percentage analysis (SIMPER) was used to identify the bacterial and pathogenic taxa driving the differences in treatments. The relationships between environmental parameters (seawater nutrients, SOC and sediment TN) and the entire bacterial community relative abundance data were evaluated by distance‐based redundancy analyses (db‐RDA) (Legendre & Anderson, 1999). The above‐mentioned statistical analyses were performed with PRIMER 6 & PERMANOVA+ (Clarke & Gorley, 2006) and IBM SPSS Statistics 19.0 software, respectively.
3. RESULTS
3.1. Variations of seawater nutrient and sediment parameters
The DIN and PO4 3− concentrations of the seawater ranged from 4.57 to 15.29 μmol·L−1 and 0.32 to 0.97 μmol·L−1, respectively (Table 1). The DIN species was dominated by NH4 +. Significantly higher DIN (F = 75.71, p = .000), NH4 + (F = 135.72, p = .000) and PO4 3− (F = 14.73, p = .005) concentrations were found in transect A compared to the other transects (Tukey's post hoc test). The SOC, sediment TN, and sediment C/N ranged from 0.09% to 0.73%, 0.015% to 0.053% and 7.40 to 16.07, respectively, with the highest SOC, TN, and C/N found at site A1.T (Table 1). Additionally, the sediment C/N ratios were not observed to be significantly different among transects (F = 2.08, p = .188); however, there was significantly higher SOC (F = 8.936, p = .009) and sediment TN (F = 16.53, p = .001) content observed in transect A compared to the other transects (Tukey's post hoc test). SOC, sediment TN, and sediment C/N were not affected by the seagrass species and the interactions of transect and seagrass species (p > .05).
Table 1.
Seawater nutrients and sediment elemental content among all the sampling stations
| Parameters | Stations | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A1 | A2 | A3 | B1 | B2 | B3 | C1 | C2 | C3 | ||
| DIN (μmol·L−1) | 12.86 | 15.29 | 12.71 | 5.62 | 5.16 | 6.41 | 4.87 | 5.51 | 4.57 | |
| NH4 + (μmol·L−1) | 10.68 | 12.46 | 11.38 | 4.07 | 3.75 | 4.88 | 3.56 | 3.86 | 3.09 | |
| NO3 − (μmol·L−1) | 1.94 | 2.67 | 1.12 | 1.35 | 1.22 | 1.48 | 1.14 | 1.48 | 1.29 | |
| NO2 − (μmol·L−1) | 0.24 | 0.16 | 0.21 | 0.19 | 0.19 | 0.05 | 0.16 | 0.17 | 0.19 | |
| PO4 3− (μmol·L−1) | 0.97 | 0.68 | 0.73 | 0.42 | 0.47 | 0.32 | 0.45 | 0.40 | 0.35 | |
| Thalassia hemprichii | SOC (%) | 0.73 | 0.33 | 0.16 | 0.14 | 0.17 | 0.13 | 0.09 | 0.14 | |
| Sediment TN (%) | 0.053 | 0.039 | 0.025 | 0.021 | 0.018 | 0.015 | 0.01 | 0.013 | ||
| Sediment C/N | 16.07 | 9.87 | 7.47 | 7.78 | 11.02 | 10.11 | 10.5 | 12.56 | ||
| Enhalus acoroides | SOC (%) | 0.26 | 0.22 | 0.32 | 0.1 | 0.16 | 0.15 | |||
| Sediment TN (%) | 0.041 | 0.029 | 0.035 | 0.015 | 0.025 | 0.021 | ||||
| Sediment C/N | 7.40 | 8.85 | 10.67 | 7.78 | 7.47 | 8.33 | ||||
3.2. Total bacterial community structure
Sequencing of the sediment microbial communities, using the 16S rRNA gene resulted in more than 388 k sequences and 2.5 k OTUs from all the sampling sites. On average, each sample had ~ 28,000 sequences and ~1,850 OTUs (Table S2). The α‐diversity indices (Chao1, Shannon, and rarefaction) indicated that there was lower diversity in transect A compared to the other transects (Table S2).
Though there was no significant effect on bacterial community structure associated with seagrass species (Pseudo‐ F = 2.1021, P‐perm = 0.115) or the transect x species interaction (Pseudo‐ F = 1.988, P‐perm = 0.129), the effect of transect location on the bacterial communities was marginally significant (Pseudo‐ F = 2.3613, P‐perm = 0.079). SIMPER analysis showed that the 10 most abundant orders accounted for more than 60% of the dissimilarity between transects and was dominated by the Gammaproteobacteria and Firmicutes (Figure 2 and Figure S1, Table S3). In transect A, the average relative abundance of Vibrionales, Alteromonadales, and Pseudomonadales, belonging to denitrifying bacteria groups, were up to twofold higher compared to the other two transects, while the Bacillales and Clostridiales were higher in transects B and C (Figure 2). Furthermore, the relative abundance of Clostridiales, Bacillales, and Exiguobacterales increased from transect A to transect C, while that of Pseudomonadales showed the opposite trend (Figure 2).
Figure 2.
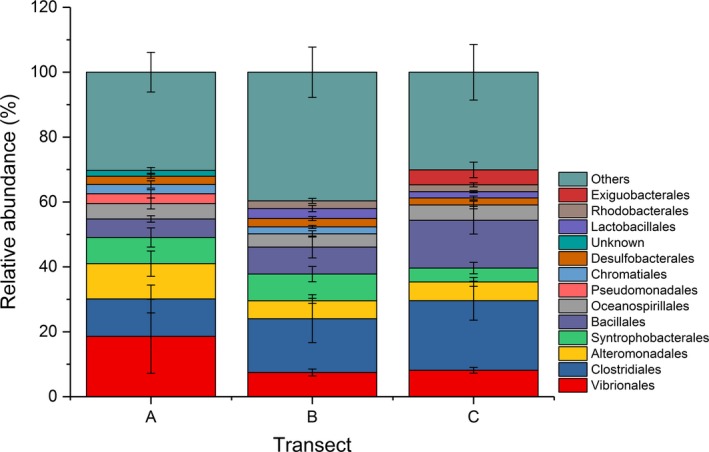
Microbial community composition of the top 10 most abundant orders averaged over each transect. Values show means and 1 standard error (n = 4–5). Relative abundances at each site are provided in Figure S1
The db‐RDA analyses of the 16S rRNA gene data explained 48.1% of the variation in the first two axes (Figure 3). The db‐RDA analyses confirmed the clear separation of transects according to above‐mentioned environmental factors, although much of this difference looks to be driven by one sample (A1.T). Axis 2 (db‐RDA2) separated the sites furthest from fish farms sites (including transect B and C) from the nearer sites (transect A) in Xincun Bay. The transect profiles reflected an apparent shift in SOC and nutrient concentrations between A and B/C (Figure 3). There were strong negative correlations between SOC and db‐RDA1 (r = −.755; Table 2), and strong positive correlations between NH4 + (r = .532), PO4 3− (r = .52) and db‐RDA2, respectively (Table 2).
Figure 3.
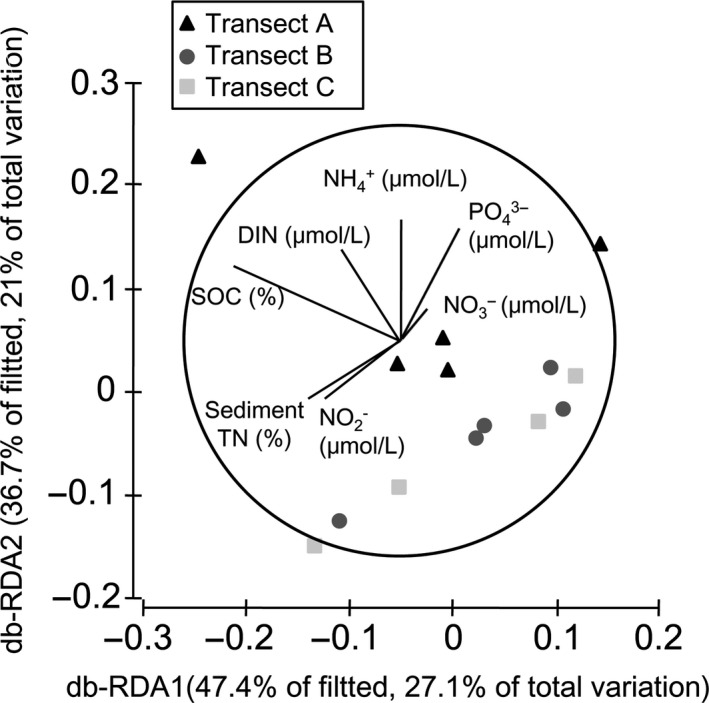
Distance‐based Redundancy Analysis (db‐RDA) ordination of microbial community data (Weighted UNIFRAC resemblance matrix calculated from relative abundance data) fitted to environmental variables. The plot represents a db‐RDA ordination based upon the Bray–Curtis distance of all the sampling sites. Correlations can be found in Table 2
Table 2.
Multiple partial correlations between Distance‐based Redundancy Analysis (db‐RDA) coordinate axes and environmental variables
| Variable | db‐RDA1 | db‐RDA2 |
|---|---|---|
| SOC (%) | −0.755 | 0.334 |
| Sediment TN (%) | −0.342 | −0.263 |
| DIN (μmol/L) | −0.264 | 0.418 |
| NH4 + (μmol/L) | 0.022 | 0.532 |
| NO3 − (μmol/L) | 0.112 | 0.148 |
| NO2 − (μmol/L) | −0.398 | −0.262 |
| PO4 3− (μmol/L) | 0.269 | 0.52 |
3.3. Putative pathogens
There were 18 potentially opportunistic, putative pathogens found in the seagrass sediments in this study, including those associated with human, fish, invertebrate, and mammal diseases (Table 3). The putative pathogens (at genus level) accounted for about 24.25% of total bacterial community among all the transects. In general, these taxa presented in higher relative abundances in transect A, particularly at the T. hemprichii stations in transect A, than other all other stations (Figure 4 and Figure S2). Although the PERMANOVA indicated that the overall bacterial community composition was not significantly different among the three transects, total pathogenic relative abundance in transect A was 1.8 times that of the other two transects. The average dissimilarities of transect A versus transect B and transect A versus transect C were twice as that of transect B versus transect C (Table S4). Abundance differences in Vibrio spp. and Pseudoalteromonas spp. contributed to more than 60% of the dissimilarities between transect A and the other transects (Table S4). Moreover, the relative abundances of Vibrio spp. and Pseudoalteromonas spp. were both more than twofold higher in transect A compared to transects B and C.
Table 3.
A list of putative pathogens of human, fishes, and invertebrates identified in this study
| Taxon | Infectious organisms | References |
|---|---|---|
| Arcobacter spp. | Human | Collado, Inza, Guarro, and Figueras (2008) |
| Bacillus spp. | Fishes and invertebrates | Webster (2007) and Austin, Austin, Austin, and Austin (2012) |
| Chryseobacterium spp. | Fishes | Austin et al. (2012) |
| Clostridium spp. | Human | Gorbach and Thadepalli (1975) |
| Corynebacterium spp. | Human | Roux et al. (2004) |
| Edwardsiella spp. | Human and Fishes | Bullock and Herman (1985) and Obasohan, Agbonlahor, and Obano (2010) |
| Flavobacterium spp. | Fishes | Farkas (1985) |
| Francisella spp. | Fishes | Mauel, Soto, Moralis, and Hawke (2007) |
| Halomonas spp. | Human | Stevens, Hamilton, Johnson, Kim, and Lee (2009) |
| Mycobacterium spp. | Human and Fishes | Primm, Lucero, and Falkinham (2004) and Watral and Kent (2007) |
| Mycoplasma spp. | Human and invertebrates | Paillard, Le Roux, and Borrego (2004) and Waites, Katz, and Schelonka (2005) |
| Pseudoalteromonas spp. | Invertebrates | Chistoserdov, Gubbala, Smolowitz, Mirasol, and Hsu (2005) |
| Pseudomonas spp. | Human and invertebrates | Gilardi (1972) and Webster (2007) |
| Psychrobacter spp. | Human | Bowman (2006) |
| Shewanella spp. | Human and invertebrates | Li et al. (2010) and Janda (2014) |
| Streptococcus spp. | Fishes | Baeck, Kim, Gomez, and Park (2006) |
| Tenacibaculum spp. | Fishes | Austin et al. (2012) |
| Vibrio spp. | Human, fishes and invertebrates | Colwell and Grimes (1984), Janda, Powers, Bryant, and Abbott (1988) and Vaseeharan and Ramasamy (2003) |
Figure 4.
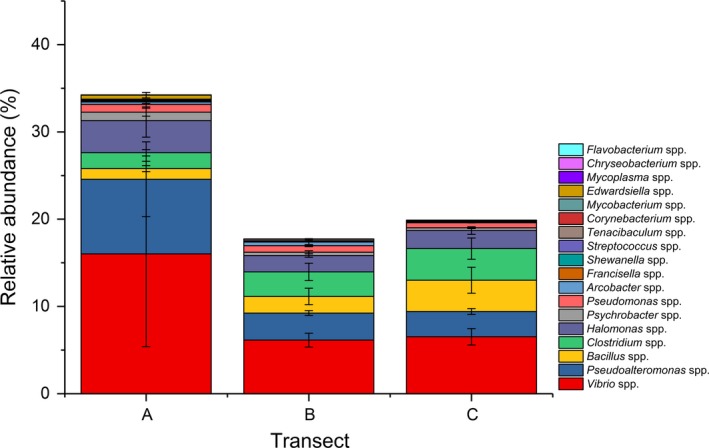
Presence of putative pathogens at the genus level averaged over the three transects. Values show means and 1 standard error (n = 4–5). Relative abundances at each site are provided in Figure S2
4. DISCUSSION
The objective of this work was to study the effects of aquaculture‐associated nutrient loading on the bacterial community structure in seagrass sediments, including the presence of putative pathogens. Overall the nutrient concentrations of this study were generally higher than other nutrient‐impacted seagrass beds (Apostolaki, Holmer, Marbà, & Karakassis, 2010; Guevara et al., 2014), which can mainly be attributed to the large amount of floating fish cage units in a nearly closed‐system Xincun Bay. On a local scale, the nutrient concentrations in the seawater were highest in the areas closest to the fish farming area, which was similar to that observed in previous studies carried out in Xincun Bay (Zhang et al., 2014). Liu et al. (2016) revealed that the SOC source in seagrass meadows of Xincun Bay was of marine autochthonous origin, with more algal organic carbon contribution in the high nutrient areas. This indicated that the aquaculture‐induced nutrient loading enhanced more labile organic carbon inputs, and the import of aquaculture‐sourced particulate organic matter to seagrass meadows was negligible. Therefore, the results of this study provided evidence that eutrophic inputs, including labile organic carbon inputs, could be shifting the sediment bacterial community toward functional groups that can utilize the excess nutrients. The elevated denitrifying communities and putative pathogens in the high nutrient‐loaded area (transect A) coupled with the reduced seawater nutrient load and shift in microbial communities in transects B and C also suggest that the seagrass plants themselves and their associated microbial communities could be playing a role in minimizing the influence and spread of nutrients and pathogens to nearby areas, potentially through metabolic and biogeochemical cycling and filtration.
4.1. Characteristics of bacterial and pathogen community structure
In this study, high bacterial community diversity (Shannon index of this study: 3.72–8.72; other studies: 3.71–4.00; Ikenaga, Guevara, Dean, Pisani, & Boyer, 2010; Guevara et al., 2014) could contribute to the maintenance, function and stability of the environment, and thus improve the resilience of ecosystems suffering from human disturbance (Parnell, Crowl, Weimer, & Pfrender, 2009). Consistent with a previous study in Xincun Bay (Jiang et al., 2015), the Gammaproteobacteria and Firmicutes were the dominant bacterial taxonomic groups in the surface sediments. Fish farming typically generates a large amount of feces that are abundant in Firmicutes (Wu et al., 2010), which explains the relative high abundance of Firmicutes in this study. However, this study showed some dissimilar results compared with other regional seagrass meadows’ studies that showed sediments primarily dominated by Deltaproteobacteria and Bacteroidetes (García‐Martínez et al., 2009; Guevara et al., 2014; Ikenaga et al., 2010). Most Deltaproteobacteria are sulfate reducers and occur in anaerobic conditions (de Moraes, Franco, Pellizari, & Sumida, 2014; López‐García et al., 2003). Bacteroidetes were associated with substrates rich in organic carbon (Fierer, Bradford, & Jackson, 2007). The sandy substrate and the low sediment organic matter in the seagrass beds of Xincun Bay (Liu et al., 2016, 2017) likely promote deeper oxygen penetration, which could reduce the microbially mediated sulfate reduction metabolic pathways in sediments (Bourque et al., 2015). This possibly explains the relatively low abundances of the Deltaproteobacteria and Bacteroidetes in this study.
Along similar trends, the pathogenic groups were mainly assigned to Gammaproteobacteria and Firmicutes. Our sequences aligned with 18 of 42 potentially pathogenic genera described in Lamb et al. (2017). Lamb et al. (2017) reported that the relative abundance of potentially pathogenic genera was less than 1% in seawater in seagrass meadows at Spermonde Archipelago, Indonesia. This concentration was much lower than the sediment pathogenic relative abundance (24%) in this study. It has been previously shown that pathogen abundance can be up to 100‐fold greater in sediment when compared with the water column (Ghaderpour et al., 2014; Perkins et al., 2014), and this may indicate that the sediment in seagrass meadows may be a sink for pathogens in the water column that move across a seagrass canopy. However, more work needs to be done to link the high relative abundances of putative pathogenic genera to the actual pathogenic species or strain as well as to actual cell counts in order to fully understand the risks of pathogenic accumulation to human and ecosystem health.
4.2. Responses of bacterial and pathogenic community structure to nutrient load
We found preliminary evidence that sediment bacterial community structure could be influenced by the proximity to point source nutrients loads. The db‐RDA2 axis indicated high nutrient load sampling stations were separated from the other sampling stations, although the PERMANOVA results indicated no statistical difference. These disparate results were likely due to the variability in seagrass meadow diversity as well as low replication sampling within a transect. However, despite the low statistical power, the db‐RDA analysis indicated that NH4 + and PO4 3− contents were important drivers of the microbial community structure between transect A and transects B and C, that is, proximity to nutrient source. It was previously shown that microbial communities of salt marsh sediments can also be stabilized in response to nutrient loading (Bowen et al., 2011; Kearns et al., 2016). In this study, it is possible that NH4 + and PO4 3−, typically the limiting factors for heterotrophic bacteria production (Kirchman, 1994; Thingstad, Zweifel, & Rassoulzadegan, 1998; Wheeler & Kirchman, 1986), are directly synthesized into bacterial biomass and promoting bacterial growth. Furthermore, higher eutrophic conditions can also lead to higher labile organic carbon inputs, which was apparent in transect A (Liu et al., 2016). Elevated nutrient load in combination with labile organic carbon availability favors the fast‐growing r‐strategist microbes (Fernandes, Kirchman, Michotey, Bonin, & LokaBharathi, 2014; Pinhassi & Berman, 2003; Trevathan‐Tackett et al., 2017) and is likely linked to the higher relative abundances of Gammaproteobacteria, like Vibrionales, Alteromonadales, and Pseudomonadales, in transect A.
In general, the Gammaproteobacteria represent abundant denitrifying communities in marine sediments (Bhatt, Zhao, Monteil‐Rivera, & Hawari, 2005). Here, the results indicate that the carbon‐limited conditions of this study area, indicated by low sediment C/N ratios and SOC content (this study: average value was 0.22%; global data: average value was 1.8%; Kennedy et al., 2010) and high proportion of microbial biomass carbon in SOC (previous study in Xincun Bay: average value was 15.74%; Liu et al., 2016), would favor denitrification processes (Burgin & Hamilton, 2007). The process of nitrate being converted to N2 would be enhanced in high nutrient areas due to the potential high denitrification activity. This may be benefiting nearby areas by reducing nitrate loads in areas closest to the fish farming area. Conversely, at the relative low nutrient concentration areas, there were increases in Firmicutes, including members of the Bacillales, Clostridiales, and Exiguobacterales. Liu et al. (2016, 2017) have found that bacterial biomass and labile organic carbon decreased with decreasing nutrient concentrations in Xincun Bay. The Firmicutes is relative stable group of bacteria under various substrate availability conditions (Kampmann et al., 2012), therefore the increase in their relative abundance at the lower nutrient transects might be due to the relative shifts in other bacterial groups.
The pathogenic population was about twofold greater in high nutrient load areas than that of the relatively lower nutrient areas. This was consistent with previous theoretical and empirical studies that nutrient enrichment often enhances pathogen abundance due to high resource availability, including abundant labile carbon substrates (Johnson et al., 2010; Lafferty & Holt, 2003; Liu et al., 2016; McKenzie & Townsend, 2007). Previous work on the causative agent of cholera (Vibrio cholerae) provided a good example of how a pathogen can be affected by nutrient load and related marine plankton bloom (McKenzie & Townsend, 2007; Vezzulli, Pruzzo, Huq, & Colwell, 2010). Secondly, high nutrient loads have led to a decline in seagrass biomass and cover in this study area (Liu et al., 2016), which could reduce their ‘pathogen filtering’ abilities within the seagrass meadows (Lamb et al., 2017). Thirdly, the elevated nutrient load could also increase pathogen fitness and virulence or change host susceptibility (Johnson et al., 2010; McKenzie & Townsend, 2007). In this study, Vibrio spp. and Pseudoalteromonas spp. were mainly responsible for the differences in pathogen abundances under different nutrient conditions. The nutrient load induced by fish farms could heighten the prevalence risk of infectious diseases in natural fishery resources as well as aquaculture organisms in Xincun Bay (Iwamoto, Ayers, Mahon, & Swerdlow, 2010; Johnson et al., 2010; Oetama et al., 2016). Additionally, there are more than 12 species of Vibrio known as human pathogens (Blazer, 1988), resulting in disease as a consequence of toxin intake (Martinez‐Urtaza et al., 2012; Oetama et al., 2016). Furthermore, the sediment pathogens can be grazed associated with sediment organic matter at the bottom, by meiofauna, worms, prawns, cockles, and demersal fishes, sequentially may move up the food chain and reach humans (Ghaderpour et al., 2014).
5. CONCLUSION
We found that higher nutrient load into a seagrass meadow elevated the relative abundance of denitrifying communities as well as potential pathogen groups. Changes of sediment bacteria and pathogen relative abundance not only should be attributed to nutrient enrichment, but also associated labile organic carbon. On one hand, these findings imply that high nutrient enrichment enhanced the potential denitrification activity, yet present a risk of infectious diseases in nature. What is unknown, however, is how the bacterial and pathogenic activities change in response to nutrient loading. As a matter for future research, we recommend meta‐transcriptomic and metabolomics analyses of seagrass sediment microbial communities to evaluate functional gene expression associated with nutrient cycling and virulence products among different nutrient load levels.
CONFLICT OF INTEREST
None declared.
Supporting information
ACKNOWLEDGMENTS
This research was supported by the National Basic Research Program of China (2015CB452905, 2015CB452902), the National Natural Science Foundation of China (nos. 41730529), and the National Specialized Project of Science and Technology (2015FY110600).
Liu S, Jiang Z, Deng Y, et al. Effects of nutrient loading on sediment bacterial and pathogen communities within seagrass meadows. MicrobiologyOpen. 2018;7:e600 10.1002/mbo3.600
REFERENCES
- Apostolaki, E. , Holmer, M. , Marbà, N. , & Karakassis, I. (2010). Metabolic imbalance in coastal vegetated (Posidonia oceanica) and unvegetated benthic Ecosystems. Ecosystems, 13, 459–471. 10.1007/s10021-010-9330-9 [DOI] [Google Scholar]
- Arndt, D. , Xia, J. , Liu, Y. , Zhou, Y. , Guo, A. C. , Cruz, J. A. , Sinelnikov, I. , Budwill, K. , Nesbø, C. L. , Wishart, D. S. . (2012). METAGENassist: A comprehensive web server for comparative metagenomics. Nucleic Acids Research, 40, W88–W99. 10.1093/nar/gks497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin, B. , Austin, D.A. , Austin, B. , & Austin, D.A. (2012). Bacterial fish pathogens Heidelberg, Germany: Springer. [Google Scholar]
- Baeck, G. W. , Kim, J. H. , Gomez, D. K. , & Park, S. C. (2006). Isolation and characterization of Streptococcus sp. from diseased flounder (Paralichthys olivaceus) in Jeju Island. Journal of Veterinary Science, 7, 53–58. 10.4142/jvs.2006.7.1.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bally, M. , & Garrabou, J. (2007). Thermodependent bacterial pathogens and mass mortalities in temperate benthic communities: A new case of emerging disease linked to climate change. Global Change Biology, 13, 2078–2088. 10.1111/j.1365-2486.2007.01423.x [DOI] [Google Scholar]
- Bhatt, M. , Zhao, J.‐S. , Monteil‐Rivera, F. , & Hawari, J. (2005). Biodegradation of cyclic nitramines by tropical marine sediment bacteria. The Journal of Industrial Microbiology and Biotechnology, 32, 261–267. 10.1007/s10295-005-0239-9 [DOI] [PubMed] [Google Scholar]
- Blazer, V. S. (1988). Bacterial fish pathogens. Environmental Biology of Fishes, 21, 77–79. 10.1007/BF02984445 [DOI] [Google Scholar]
- Bourque, A. S. , Vega‐Thurber, R. , & Fourqurean, J. W. (2015). Microbial community structure and dynamics in restored subtropical seagrass sediments. Aquatic Microbial Ecology, 74, 43–57. 10.3354/ame01725 [DOI] [Google Scholar]
- Bowen, J. L. , Ward, B. B. , Morrison, H. G. , Hobbie, J. E. , Valiela, I. , Deegan, L. A. , & Sogin, M. L. (2011). Microbial community composition in sediments resists perturbation by nutrient enrichment. ISME Journal, 5, 1540–1548. 10.1038/ismej.2011.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman, J. P . (2006). The genus Psychrobacter In Balows A., Trüper H. G., Dworkin M., Harder W. & Schleifer K. H. (Eds.), The prokaryotes (pp. 920–930). New York, NY, USA: Springer; 10.1007/0-387-30746-X [DOI] [Google Scholar]
- Bullock, G. , & Herman, R. L . (1985). Edwardsiella infections of fishes. U.S. Fish and Wildlife Service, Fish disease leaflet 71, Kearneysville, West Virginia.
- Burgin, A. J. , & Hamilton, S. K. (2007). Have we overemphasized the role of denitrification in aquatic ecosystems? A review of nitrate removal pathways. Frontiers in Ecology and the Environment, 5, 89–96. 10.1890/1540-9295(2007)5[89:HWOTRO]2.0.CO;2 [DOI] [Google Scholar]
- Burkholder, J. M. , Tomasko, D. A. , & Touchette, B. W. (2007). Seagrasses and eutrophication. Journal of Experimental Marine Biology and Ecology, 350, 46–72. 10.1016/j.jembe.2007.06.024 [DOI] [Google Scholar]
- Caporaso, J. G. , Bittinger, K. , Bushman, F. D. , DeSantis, T. Z. , Andersen, G. L. , & Knight, R. (2010a). PyNAST: A flexible tool for aligning sequences to a template alignment. Bioinformatics, 26, 266–267. 10.1093/bioinformatics/btp636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso, J. G. , Kuczynski, J. , Stombaugh, J. , Bittinger, K. , Bushman, F. D. , Costello, E. K. , Fierer, N. , Peña, A. G. , Goodrich, J. K. , Gordon, J. I. , Huttley, G. A. . (2010b). QIIME allows analysis of high‐throughput community sequencing data. Nature Methods, 7, 335–336. 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdov, A. Y. , Gubbala, S. L. , Smolowitz, R. , Mirasol, F. , & Hsu, A. (2005). A microbiological assessment of epizootic shell disease in the American lobster indicates its strictly dermal etiology Lobster shell disease workshop (pp. 12–20). Boston, MA: University of Massachusetts. [Google Scholar]
- Christiaen, B. , McDonald, A. , Cebrian, J. , & Ortmann, A. C. (2013). Response of the microbial community to environmental change during seagrass transplantation. Aquatic Botany, 109, 31–38. 10.1016/j.aquabot.2013.03.008 [DOI] [Google Scholar]
- Clarke, K. , & Gorley, R . (2006). User manual/tutorial. Plymouth: PRIMER‐E Ltd, 93. [Google Scholar]
- Collado, L. , Inza, I. , Guarro, J. , & Figueras, M. J. (2008). Presence of Arcobacter spp. in environmental waters correlates with high levels of fecal pollution. Environmental Microbiology, 10, 1635–1640. 10.1111/j.1462-2920.2007.01555.x [DOI] [PubMed] [Google Scholar]
- Colwell, R. , & Grimes, D. (1984). Vibrio diseases of marine fish populations. Helgoländer Meeresunters, 37, 265–287. 10.1007/BF01989311 [DOI] [Google Scholar]
- de Moraes, P. C. , Franco, D. C. , Pellizari, V. H. , & Sumida, P. Y. G. (2014). Effect of plankton‐derived organic matter on the microbial community of coastal marine sediments. Journal of Experimental Marine Biology and Ecology, 461, 257–266. 10.1016/j.jembe.2014.08.017 [DOI] [Google Scholar]
- Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 26, 2460–2461. 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Farkas, J. (1985). Filamentous Flavobacterium sp. isolated from fish with gill diseases in cold water. Aquaculture, 44, 1–10. 10.1016/0044-8486(85)90037-7 [DOI] [Google Scholar]
- Fernandes, S. O. , Kirchman, D. L. , Michotey, V. D. , Bonin, P. C. , & LokaBharathi, P. (2014). Bacterial diversity in relatively pristine and anthropogenically‐influenced mangrove ecosystems (Goa, India). The Brazilian Journal of Microbiology, 45, 1161–1171. 10.1590/S1517-83822014000400006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer, N. , Bradford, M. A. , & Jackson, R. B. (2007). Toward an ecological classification of soil bacteria. Ecology, 88, 1354–1364. 10.1890/05-1839 [DOI] [PubMed] [Google Scholar]
- García‐Martínez, M. , López‐López, A. , Calleja, M. L. , Marbà, N. , & Duarte, C. M. (2009). Bacterial community dynamics in a seagrass (Posidonia oceanica) meadow sediment. Estuaries and Coasts, 32, 276–286. 10.1007/s12237-008-9115-y [DOI] [Google Scholar]
- Ghaderpour, A. , Nasori, K. N. M. , Chew, L. L. , Chong, V. C. , Thong, K. L. , & Chai, L. C. (2014). Detection of multiple potentially pathogenic bacteria in Matang mangrove estuaries, Malaysia. Marine Pollution Bulletin, 83, 324–330. 10.1016/j.marpolbul.2014.04.029 [DOI] [PubMed] [Google Scholar]
- Gilardi, G. (1972). Infrequently encountered Pseudomonas species causing infection in humans. Annals of Internal Medicine, 77, 211–215. 10.7326/0003-4819-77-2-211 [DOI] [PubMed] [Google Scholar]
- Gorbach, S. L. , & Thadepalli, H. (1975). Isolation of Clostridium in human infections: Evaluation of 114 cases. Journal of Infectious Diseases, 131, S81–S85. 10.1093/infdis/131.Supplement.S81 [DOI] [PubMed] [Google Scholar]
- Guevara, R. , Ikenaga, M. , Dean, A. L. , Pisani, C. , & Boyer, J. N. (2014). Changes in sediment bacterial community in response to long‐term nutrient enrichment in a subtropical seagrass‐dominated estuary. Microbial Ecology, 68, 427–440. 10.1007/s00248-014-0418-1 [DOI] [PubMed] [Google Scholar]
- Hansen, J. W. , Udy, J. W. , Perry, C. J. , Dennison, W. C. , & Lomstein, B. A. (2000). Effect of the seagrass Zostera capricorni on sediment microbial processes. Marine Ecology Progress Series, 199, 83–96. 10.3354/meps199083 [DOI] [Google Scholar]
- Hemminga, M. A. , & Duarte, C. M. (2000). Seagrass ecology Cambridge, UK: Cambridge University Press. [Google Scholar]
- Howard, J. L. , Perez, A. , Lopes, C. C. , & Fourqurean, J. W. (2016). Fertilization changes seagrass community structure but not blue carbon storage: Results from a 30‐year field experiment. Estuaries and Coasts, 39, 1422–1434. 10.1007/s12237-016-0085-1 [DOI] [Google Scholar]
- Huang, X. P. , Huang, L. M. , Li, Y. H. , Xu, Z. Z. , Fong, C. W. , Huang, D. J. , Han, Q. , Huang, H. , Tan, Y. , Liu, S. . (2006). Main seagrass beds and threats to their habitats in the coastal sea of South China. Chinese Science Bulletin, 51, 136–142. 10.1007/s11434-006-9136-5 [DOI] [Google Scholar]
- Ikenaga, M. , Guevara, R. , Dean, A. L. , Pisani, C. , & Boyer, J. N. (2010). Changes in community structure of sediment bacteria along the Florida coastal everglades marsh–mangrove–seagrass salinity gradient. Microbial Ecology, 59, 284–295. 10.1007/s00248-009-9572-2 [DOI] [PubMed] [Google Scholar]
- Ingemann Jensen, S. , Kühl, M. , Glud, R. N. , Jørgensen, L. B. , & Priemé, A. (2005). Oxic microzones and radial oxygen loss from roots of Zostera marina . Marine Ecology Progress Series, 293, 49–58. 10.3354/meps293049 [DOI] [Google Scholar]
- Iwamoto, M. , Ayers, T. , Mahon, B. E. , & Swerdlow, D. L. (2010). Epidemiology of seafood‐associated infections in the United States. Clinical Microbiology Reviews, 23, 399–411. 10.1128/CMR.00059-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janda, J. M. (2014). Shewanella: A marine pathogen as an emerging cause of human disease. Clinical Microbiology Newsletter, 36, 25–29. 10.1016/j.clinmicnews.2014.01.006 [DOI] [Google Scholar]
- Janda, J. , Powers, C. , Bryant, R. , & Abbott, S. (1988). Current perspectives on the epidemiology and pathogenesis of clinically significant Vibrio spp. Clinical Microbiology Reviews, 1, 245–267. 10.1128/CMR.1.3.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, S. I. , Kühl, M. , & Priemé, A. (2007). Different bacterial communities associated with the roots and bulk sediment of the seagrass Zostera marina . FEMS Microbiology Ecology, 62, 108–117. 10.1111/j.1574-6941.2007.00373.x [DOI] [PubMed] [Google Scholar]
- Jiang, Y.‐F. , Ling, J. , Wang, Y.‐S. , Chen, B. , Zhang, Y.‐Y. , & Dong, J.‐D. (2015). Cultivation‐dependent analysis of the microbial diversity associated with the seagrass meadows in Xincun Bay, South China Sea. Ecotoxicology, 24, 1540–1547. 10.1007/s10646-015-1519-4 [DOI] [PubMed] [Google Scholar]
- Johnson, P. T. , Townsend, A. R. , Cleveland, C. C. , Glibert, P. M. , Howarth, R. W. , McKenzie, V. J. , Rejmankova, E. , Ward, M. H. . (2010). Linking environmental nutrient enrichment and disease emergence in humans and wildlife. Ecological Applications, 20, 16–29. 10.1890/08-0633.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen, B. B. (1982). Mineralization of organic matter in the sea bed—the role of sulphate reduction. Nature, 296, 643–645. 10.1038/296643a0 [DOI] [Google Scholar]
- Kampmann, K. , Ratering, S. , Kramer, I. , Schmidt, M. , Zerr, W. , & Schnell, S. (2012). Unexpected stability of Bacteroidetes and Firmicutes communities in laboratory biogas reactors fed with different defined substrates. Applied and Environmental Microbiology, 78, 2106–2119. 10.1128/AEM.06394-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns, P. J. , Angell, J. H. , Howard, E. M. , Deegan, L. A. , Stanley, R. H. , & Bowen, J. L. (2016). Nutrient enrichment induces dormancy and decreases diversity of active bacteria in salt marsh sediments. Nature Communications, 10.1038/ncomms12881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy, H. , Beggins, J. , Duarte, C. M. , Fourqurean, J. W. , Holmer, M. , Marbà, N. , & Middelburg, J. J. (2010). Seagrass sediments as a global carbon sink: Isotopic constraints. Global Biogeochemical Cycles, 24, 1–8 10.1029/2010GB003848 [DOI] [Google Scholar]
- Kirchman, D. L. (1994). The uptake of inorganic nutrients by heterotrophic bacteria. Microbial Ecology, 28, 255–271. 10.1007/BF00166816 [DOI] [PubMed] [Google Scholar]
- Lafferty, K. D. , & Holt, R. D. (2003). How should environmental stress affect the population dynamics of disease? Ecology Letters, 6, 654–664. 10.1046/j.1461-0248.2003.00480.x [DOI] [Google Scholar]
- Lamb, J. B. , van de Water, J. A. , Bourne, D. G. , Altier, C. , Hein, M. Y. , Fiorenza, E. A. , Abu, N. , Jompa, J. , Harvell, C. D. . (2017). Seagrass ecosystems reduce exposure to bacterial pathogens of humans, fishes, and invertebrates. Science, 355, 731–733. 10.1126/science.aal1956 [DOI] [PubMed] [Google Scholar]
- Legendre, P. , & Anderson, M. J. (1999). Distance‐based redundancy analysis: Testing multispecies responses in multifactorial ecological experiments. Ecological Monographs, 69, 1–24. 10.1890/0012-9615(1999)069[0001:DBRATM]2.0.CO;2 [DOI] [Google Scholar]
- Li, H. , Qiao, G. , Gu, J.‐Q. , Zhou, W. , Li, Q. , Woo, S.‐H. , Xu, D. H. , Park, S. I. . (2010). Phenotypic and genetic characterization of bacteria isolated from diseased cultured sea cucumber Apostichopus japonicus in northeastern China. Diseases of Aquatic Organisms, 91, 223–235. 10.3354/dao02254 [DOI] [PubMed] [Google Scholar]
- Liu, S. , Jiang, Z. , Wu, Y. , Zhang, J. , Arbi, I. , Ye, F. , Huang, X. , Macreadie, P. I. . (2017). Effects of nutrient load on microbial activities within a seagrass‐dominated ecosystem: Implications of changes in seagrass blue carbon. Marine Pollution Bulletin, 117, 214–221. 10.1016/j.marpolbul.2017.01.056 [DOI] [PubMed] [Google Scholar]
- Liu, S. , Jiang, Z. , Zhang, J. , Wu, Y. , Lian, Z. , & Huang, X. (2016). Effect of nutrient enrichment on the source and composition of sediment organic carbon in tropical seagrass beds in the South China Sea. Marine Pollution Bulletin, 110, 274–280. 10.1016/j.marpolbul.2016.06.054 [DOI] [PubMed] [Google Scholar]
- López, N. I. , Duarte, C. M. , Vallespinós, F. , Romero, J. , & Alcoverro, T. (1998). The effect of nutrient additions on bacterial activity in seagrass (Posidonia oceanica) sediments. Journal of Experimental Marine Biology and Ecology, 224, 155–166. 10.1016/S0022-0981(97)00189-5 [DOI] [Google Scholar]
- López‐García, P. , Duperron, S. , Philippot, P. , Foriel, J. , Susini, J. , & Moreira, D. (2003). Bacterial diversity in hydrothermal sediment and epsilonproteobacterial dominance in experimental microcolonizers at the Mid‐Atlantic Ridge. Environmental Microbiology, 5, 961–976. 10.1046/j.1462-2920.2003.00495.x [DOI] [PubMed] [Google Scholar]
- Luna, G. , Vignaroli, C. , Rinaldi, C. , Pusceddu, A. , Nicoletti, L. , Gabellini, M. , Danovaro, R. , Biavasco, F. . (2010). Extraintestinal Escherichia coli carrying virulence genes in coastal marine sediments. Applied and Environmental Microbiology, 76, 5659–5668. 10.1128/AEM.03138-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbà, N. , Holmer, M. , Gacia, E. , & Barron, C . (2007). Seagrass beds and coastal biogeochemistry In Larkum A. W. D., Orth R. J. & Duarte C. M. (Eds.), Seagrasses: Biology, ecology and conservation (pp. 135–157). Dordrecht, Netherlands: Springer. [Google Scholar]
- Martinez‐Urtaza, J. , Blanco‐Abad, V. , Rodriguez‐Castro, A. , Ansede‐Bermejo, J. , Miranda, A. , & Rodriguez‐Alvarez, M. X. (2012). Ecological determinants of the occurrence and dynamics of Vibrio parahaemolyticus in offshore areas. ISME Journal, 6, 994–1006. 10.1038/ismej.2011.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauel, M. , Soto, E. , Moralis, J. , & Hawke, J. (2007). A piscirickettsiosis‐like syndrome in cultured Nile tilapia in Latin America with Francisella spp. as the pathogenic agent. Journal of Aquatic Animal Health, 19, 27–34. 10.1577/H06-025.1 [DOI] [PubMed] [Google Scholar]
- McDonald, D. , Price, M. N. , Goodrich, J. , Nawrocki, E. P. , DeSantis, T. Z. , Probst, A. , Andersen, G. L. , Knight, R. , Hugenholtz, P. . (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME Journal, 6, 610–618. 10.1038/ismej.2011.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie, V. J. , & Townsend, A. R. (2007). Parasitic and infectious disease responses to changing global nutrient cycles. EcoHealth, 4, 384–396. 10.1007/s10393-007-0131-3 [DOI] [Google Scholar]
- Mckirdy, D. M. , Thorpe, C. S. , Haynes, D. E. , Grice, K. , Krull, E. S. , Halverson, G. P. , & Webster, L. J. (2010). The biogeochemical evolution of the Coorong during the mid‐ to late Holocene: An elemental, isotopic and biomarker perspective. Organic Geochemistry, 41, 96–110. 10.1016/j.orggeochem.2009.07.010 [DOI] [Google Scholar]
- National Research Council (2000). Clean coastal waters—Understanding and reducing the problems from nutrient pollution: Washington (p. 450). DC: National Academy of Sciences Press. [Google Scholar]
- Obasohan, E. , Agbonlahor, D. , & Obano, E. (2010). Water pollution: A review of microbial quality and health concerns of water, sediment and fish in the aquatic ecosystem. African Journal of Biotechnology, 9, 423–427. [Google Scholar]
- Oetama, V. S. P. , Hennersdorf, P. , Abdul‐Aziz, M. A. , Mrotzek, G. , Haryanti, H. , & Saluz, H. P. (2016). Microbiome analysis and detection of pathogenic bacteria of Penaeus monodon from Jakarta Bay and Bali. Marine Pollution Bulletin, 110, 718–725. 10.1016/j.marpolbul.2016.03.043 [DOI] [PubMed] [Google Scholar]
- Paillard, C. , Le Roux, F. , & Borrego, J. J. (2004). Bacterial disease in marine bivalves, a review of recent studies: Trends and evolution. Aquatic Living Resources, 17, 477–498. 10.1051/alr:2004054 [DOI] [Google Scholar]
- Parnell, J. J. , Crowl, T. A. , Weimer, B. C. , & Pfrender, M. E. (2009). Biodiversity in microbial communities: System scale patterns and mechanisms. Molecular Ecology, 18, 1455–1462. 10.1111/j.1365-294X.2009.04128.x [DOI] [PubMed] [Google Scholar]
- Perkins, T. L. , Clements, K. , Baas, J. H. , Jago, C. F. , Jones, D. L. , Malham, S. K. , & McDonald, J. E. (2014). Sediment composition influences spatial variation in the abundance of human pathogen indicator bacteria within an estuarine environment. PLoS ONE, 9:e112951 10.1371/journal.pone.0112951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinhassi, J. , & Berman, T. (2003). Differential growth response of colony‐forming α‐and γ‐proteobacteria in dilution culture and nutrient addition experiments from Lake Kinneret (Israel), the Eastern Mediterranean Sea, and the Gulf of Eilat. Applied and Environmental Microbiology, 69, 199–211. 10.1128/AEM.69.1.199-211.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primm, T. P. , Lucero, C. A. , & Falkinham, J. O. (2004). Health impacts of environmental mycobacteria. Clinical Microbiology Reviews, 17, 98–106. 10.1128/CMR.17.1.98-106.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph, P. J. , Tomasko, D. , Moore, K. , Seddon, S. , & Macinnis‐Ng, C. M . (2006). Human impacts on seagrasses: Eutrophication, sedimentation, and contamination In Larkum A. W. D., Orth R. J. & Duarte C. M. (Eds.), Seagrasses: Biology, ecology and conservation (pp. 567–593). Dordrecht, Netherlands: Springer. [Google Scholar]
- Roux, V. , Drancourt, M. , Stein, A. , Riegel, P. , Raoult, D. , & La Scola, B. (2004). Corynebacterium species isolated from bone and joint infections identified by 16S rRNA gene sequence analysis. Journal of Clinical Microbiology, 42, 2231–2233. 10.1128/JCM.42.5.2231-2233.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, D. A. , Hamilton, J. R. , Johnson, N. , Kim, K. K. , & Lee, J.‐S. (2009). Halomonas, a newly recognized human pathogen causing infections and contamination in a dialysis center: Three new species. Medicine, 88, 244–249. 10.1097/MD.0b013e3181aede29 [DOI] [PubMed] [Google Scholar]
- Thingstad, T. F. , Zweifel, U. L. , & Rassoulzadegan, F. (1998). P limitation of heterotrophic bacteria and phytoplankton in the northwest Mediterranean. Limnology and Oceanography, 43, 88–94. 10.4319/lo.1998.43.1.0088 [DOI] [Google Scholar]
- Trevathan‐Tackett, S. M. , Seymour, J. R. , Nielsen, D. A. , Macreadie, P. I. , Jeffries, T. C. , Sanderman, J. , Baldock, J. , Howes, J. M. , Steven, A. D. , Ralph, P. (2017). Sediment anoxia limits microbial‐driven seagrass carbon remineralization under warming conditions. FEMS Microbiology Ecology, 93: 6 10.1093/femsec/fix033 [DOI] [PubMed] [Google Scholar]
- Vaseeharan, B. , & Ramasamy, P. (2003). Control of pathogenic Vibrio spp. by Bacillus subtilis BT23, a possible probiotic treatment for black tiger shrimp Penaeus monodon . Letters in Applied Microbiology, 36, 83–87. 10.1046/j.1472-765X.2003.01255.x [DOI] [PubMed] [Google Scholar]
- Vega Thurber, R. L. , Burkepile, D. E. , Fuchs, C. , Shantz, A. A. , McMinds, R. , & Zaneveld, J. R. (2014). Chronic nutrient enrichment increases prevalence and severity of coral disease and bleaching. Global Change Biology, 20, 544–554. 10.1111/gcb.12450 [DOI] [PubMed] [Google Scholar]
- Vezzulli, L. , Pruzzo, C. , Huq, A. , & Colwell, R. R. (2010). Environmental reservoirs of Vibrio cholerae and their role in cholera. Environmental Microbiology Reports, 2, 27–33. 10.1111/j.1758-2229.2009.00128.x [DOI] [PubMed] [Google Scholar]
- Waites, K. B. , Katz, B. , & Schelonka, R. L. (2005). Mycoplasmas and ureaplasmas as neonatal pathogens. Clinical Microbiology Reviews, 18, 757–789. 10.1128/CMR.18.4.757-789.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q. , Garrity, G. M. , Tiedje, J. M. , & Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology, 73, 5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watral, V. , & Kent, M. L. (2007). Pathogenesis of Mycobacterium spp. in zebrafish (Danio rerio) from research facilities. Comparative Biochemistry and Physiology Part C: Toxicology & Pharmacology, 145, 55–60. [DOI] [PubMed] [Google Scholar]
- Waycott, M. , Duarte, C. M. , Carruthers, T. J. , Orth, R. J. , Dennison, W. C. , Olyarnik, S. , Calladine, A. , Fourqurean, J. W. , Heck, K. L. , Hughes, A. R. , Kendrick, G. A. (2009). Accelerating loss of seagrasses across the globe threatens coastal ecosystems. Proceedings of the National Academy of Sciences of the United States of America, 106, 12377–12381. 10.1073/pnas.0905620106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster, N. S. (2007). Sponge disease: A global threat? Environmental Microbiology, 9, 1363–1375. 10.1111/j.1462-2920.2007.01303.x [DOI] [PubMed] [Google Scholar]
- Welsh, D. T. (2000). Nitrogen fixation in seagrass meadows: Regulation, plant–bacteria interactions and significance to primary productivity. Ecology Letters, 3, 58–71. 10.1046/j.1461-0248.2000.00111.x [DOI] [Google Scholar]
- Wheeler, P. A. , & Kirchman, D. L. (1986). Utilization of inorganic and organic nitrogen by bacteria in marine systems. Limnology and Oceanography, 31, 998–1009. 10.4319/lo.1986.31.5.0998 [DOI] [Google Scholar]
- Wu, C. H. , Sercu, B. , Van De Werfhorst, L. C. , Wong, J. , DeSantis, T. Z. , Brodie, E. L. , Hazen, T. C. , Holden, P. A. , Andersen, G. L. (2010). Characterization of coastal urban watershed bacterial communities leads to alternative community‐based indicators. PLoS ONE, 5:e11285 10.1371/journal.pone.0011285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaneveld, J. R. , Burkepile, D. E. , Shantz, A. A. , Pritchard, C. E. , McMinds, R. , Payet, J. P. , Welsh, R. , Correa, A. M. , Lemoine, N. P. , Rosales, S. , Fuchs, C. (2016). Overfishing and nutrient pollution interact with temperature to disrupt coral reefs down to microbial scales. Nature Communications, 7:11833 10.1038/ncomms11833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J. P. , Huang, X. P. , & Jiang, Z. J. (2014). Physiological responses of the seagrass Thalassia hemprichii (Ehrenb.) Aschers as indicators of nutrient loading. Marine Pollution Bulletin, 83, 508–515. 10.1016/j.marpolbul.2013.12.056 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


