Abstract
The sialylated glyconjugates (SGC) are found in abundance on the surface of brain cells, where they form a dense array of glycans mediating cell/cell and cell/protein recognition in numerous physiological and pathological processes. Metabolic genetic blocks in processing and catabolism of SGC result in development of severe storage disorders, dominated by CNS involvement including marked neuroinflammation and neurodegeneration, the pathophysiological mechanisms of which are still discussed. SGC patterns in the brain are cell and organelle-specific, dynamic and maintained by highly coordinated processes of their biosynthesis, trafficking, processing and catabolism. The changes in the composition of SGC during development and aging of the brain cannot be explained based solely on the regulation of the SGC-synthesizing enzymes, sialyltransferases, suggesting that neuraminidases (sialidases) hydrolysing the removal of terminal sialic acid residues also play an essential role. In the current review we summarize the roles of three mammalian neuraminidases: neuraminidase 1, neuraminidase 3 and neuraminidase 4 in processing brain SGC. Emerging data demonstrate that these enzymes with different, yet overlapping expression patterns, intracellular localization and substrate specificity play essential roles in the physiology of the CNS.
Keywords: Sialic acid, Polysialic acid, Lysosome, CNS, Ganglioside, Neuraminidase
Sialoglycoconjugates in mammalian brain
Sialoglycoconjugates (SGC) are biological macromolecules containing sialic acids (Sia) as part of their glycan chains usually at the terminal position. SGC are abundant on the surface as well as on the inner side of lysosomal and endosomal membranes in all mammalian cells, forming a dense array of complex sialylated glycans [1]. Due to the diverse physical and chemical properties of Sia (more than 50 types are described, distinguished by the presence of different side chains), SGC are ideally suited to serve as recognition markers for molecular and cellular interactions during cell signalling, proliferation, differentiation, interaction, migration, and adhesion (reviewed in [2–4]). The content of SGC strongly depends on the cell and tissue type, and changes during development [5].
In the central nervous system (CNS), two types of SGC, gangliosides and proteins containing polysialic acid (PSA) play determining roles. The gangliosides carry ~65% of all sialic acids in the mammalian brain and their levels in the brain are ~10-fold higher as compared to other organs. They are especially enriched in the membranes of neurons, usually as part of distinct lipid patches or rafts, and have several important functions essential for myelination, neuritogenesis, synaptic plasticity, and transmission of nervous impulses (reviewed in [6]). They associate laterally with membrane proteins, including receptors and ion channels, to modulate their activities. Ganglioside glycans extend into the extracellular space and interact with glycan-binding proteins to mediate cell-protein and cell-cell interactions. The brain ganglioside composition has a high complexity, but four related species: GM1a, GD1a, GD1b and GT1b dominate the adult mammalian brain. Each of them plays specific role in the brain function. For example, GM1a regulates Na+ channels, and in combination with GD1a, neuronal Ca2+ homeostasis and maintains neuronal viability, conductivity and excitability [7, 8]. Besides as described in details further down, GM1a plays an essential role in the multifascet mechanism for neurite outgrowth. By interaction with myelin-associated glycoprotein (MAG) GD1a and GT1b, mediate axon-myelin interactions and help axonal protection, regeneration and stabilization [9, 10]. In the developing brain dominant GD3 ganglioside plays a crucial role in maintaining the self-renewal capacity of neural stem cells, and neurogenesis by sustaining epidermal growth factor receptor (EGFR) signalling [11]. Not surprizingly, gene-targeted mice devoid of complex GM1a, GD1a, GD1b, GT1b showed weight loss, progressive motor and sensory dysfunction, and memory defects [12]. Similarly, mice with a deficiency in GD3 ganglioside and its derivatives had impaired brain neurogenesis in the hippocampus resulting from a progressive loss in neural stem cell population, and exhibited depression-like behavior [13].
4,8-Linked PSA chains are attached to a number of brain glycoproteins, the major beeng the Neural cell adhesion molecule (NCAM) and Synaptic cell adhesion molecule (SynCAM). On the neurons PSA play major roles by appropriately spacing the cells and shielding their surface receptors and ligands. Changes in PSA content have been implicated in the control of cell-cell and cell-matrix interactions, essential for appropriate brain development, adult brain plasticity, and nerve regeneration (reviewed in [6]). Dysregulation of PSA synthesis has been associated with multiple psychiatric disorders, including schizophrenia, bipolar disorder, and autism spectrum disorders [14, 15]. Besides NCAM and SynCAM, PSA have been identified on at least two brain microglia glycoproteins, Neurophilin 2 (NRP2) and E-selectin 1 (ESL-1). PSA are released upon activation of microglia and presumably play a role in negative feedback regulation reducing release of pro-inflammatory cytokines [16, 17].
In mammals, the synthesis of SGC is performed by a family of 20 sialyltransferases (ST) that catalyze the transfer of Sia from CMP-Sia to an acceptor carbohydrate [18]. Inactivating mutations in genes of the ST family have been linked to several human neurological disorders and behaviour abnormalities in the mouse strains, providing important clues about the function of SGC in the brain. In particular the deficiency of ST3GAL5 (GM3 synthase) causes Amish infantile epilepsy syndrome (OMIM 609056), an autosomal recessive syndrome associated with developmental stagnation and blindness [19]. Defects in the ST3GAL3 gene encoding β-galactoside-alpha-2,3-sialyltransferase-III cause autosomal recessive nonsyndromic mental retardation-12 (MRT12; OMIM 611090) [20] and early infantile epileptic encephalopathy-15 (EIEE15; OMIM 615006) [21]. In mouse models the loss of St8sia2 resulted in decreased social interaction associated with synaptic defects [22, 23] while genetic inactivation of St 3 gal4 caused a bleeding disorder associated with autosomal recessive thrombocytopenia due to abnormal sialylation of von Willebrand factor [24].
Neuraminidases 1, 3 and 4 are the main SGC processing enzymes in the brain
The enzymatic removal of Sia is mediated by neuraminidases (sialidases) that are present in the majority of vertebrates as well as in a variety of microorganisms. Four types of neuraminidases (NEU) designated as NEU1, NEU2, NEU3 and NEU4 are present in mammalians (reviewed in [25, 26]). These enzymes have different, yet overlapping substrate specificity, tissue expression and intracellular localization. NEU1 is active primarily against sialylated glycopeptides and oligosaccharides with negligible activity against gangliosides. NEU3 requires a hydrophobic aglycone [27], which makes it active mainly towards gangliosides [28] whereas NEU4 is active against all types of sialylated glycoconjugates including oligosaccharides, glycoproteins and gangliosides [29]. Neuraminidases also show different specificity at the level of glycan structures: all neuraminidases show high activity against α2,3-linked Sia [25], while α2,6-linked Sia are readily cleaved by NEU3 and NEU1 but not by NEU4. The presence of a branching 2-Fuc inhibits Neu4, but has almost no effect on NEU1 or NEU3. The nature of the sugar residue at the reducing end, either glucose (Glc) or N-acetyl-D-glucosamine (GlcNAc), only has a minor effect on all neuraminidases, whereas core structure, 1,3 or 1,4 bond between D-galactose (Gal) and GlcNAc, was found to be important for Neu4 strongly preferring the β3 (core 1) to the β4 (core 2) isomer [30]. Importantly, neuraminidases also cleave α2,8 linkages. NEU3 hydrolyses α2,8 and α2,3-Sia with the similar rate but NEU1 and NEU2 show preference for α2,3-Sia [25]. Both NEU1 and NEU4 are able to cleave 4,8-linked Sia polymers [31].
NEU1 is ubiquitously expressed with the highest levels in the kidney, pancreas, skeletal muscle, liver, lungs, placenta and brain [32]. In these tissues, NEU1 generally shows 10–20 times higher expression than NEU3 and NEU4, and ~103–102 higher expression than NEU2 [33]. NEU2 is generally expressed at lower levels with an exception for the placenta and testis [34]. NEU3 has the highest expression in the adrenal gland, skeletal muscle, heart, testis and thymus [35, 36], while NEU4 is highly expressed in the brain, skeletal muscle, heart, placenta and liver [29, 33, 37].
In the mouse brain, NEU1, NEU3 and NEU4 show similar expression, each enzyme contributing to ~30% of total acidic neuraminidase activity against the synthetic substrate, 2′-(4-methylumbelliferyl)-α-D-N-acetylneuraminic acid (4MU-NeuAc). In the brain tissues of Neu1, Neu3 or Neu4 knockout (KO) mice, the activity is reduced to 70% of that in the wild-type (WT) brain, while in the brain of Neu3/Neu4 double-knockout mice, the residual activity does not exceed 30% of the WT [27, 38]. Similar values were obtained by measuring neuraminidase activity in the presence of specific inhibitors of individual neuraminidase enzymes [39].
Importantly, while all 3 enzymes are expressed in the CNS neurons, they show unique expression patterns. Neu1 is expressed ubiquitously throughout the brain but more strongly in specific areas such as the CA3 region of the hippocampus and the cortex while Neu3 expression does not show a specific pattern [40]. In mice, NEU3 mRNA and protein levels in the hippocampus are high at early developmental stages (E15) and low in adulthood [41]. In contrast Neu4 expression in the mouse or rat brain is undetectable at the embryonic stage, but rapidly increases at 3–14 days after birth [42, 43] when the enzyme is expressed in neurons scattered throughout the brain with a greater density in the hippocampus and in the ventral cortex [40, 44]. The exact identity of these neurons still awaits investigation. Besides, a recent study suggested the existence of mechanisms for compensatory regulation between the Neu3 and Neu4 genes [38] since the levels of Neu3 and Neu4 mRNA were significantly increased in the brain tissues of Neu4 and Neu3 single KO mice, respectively [38].
In the cell, NEU3 is localized in the caveolae microdomains of plasma, and endosomal membranes, i.e. compartments associated with the endocytic/recycling pathway [45, 46]. The enzyme was also detected on the surface of exosomes [47]. NEU3 was extracted from the membranes by sodium carbonate suggesting that it is a peripheral membrane protein [47], however a recent report provided certain evidence that NEU3 is an S-acylated transmembrane protein with the C-terminus exposed to the cytosol and the rest of the protein to the extracellular space [48]. NEU1 is localized at the lysosomal and plasma membranes [49, 50]. Similarly to NEU3 the nature of its interaction with the membrane is the subject of debate. Lukong et al. have demonstrated that NEU1 is an integral membrane protein with a C-terminal transmembrane domain and a cytosolic lysosomal targeting signal [49]. They also proposed that in the lysosome, the transmembrane domain of NEU1 can be further cleaved, resulting in a soluble form of the enzyme that has been purified from several mammalian tissues (reviewed in [51]). In contrast, structural models of NEU1 (reviewed in [26]) do not predict the existence of trans-membrane segments, suggesting that NEU1 rather associates with the membrane by lipid anchors, or binds to a membrane protein. A recent report, however, proposed that NEU1 contains two novel potential transmembrane domains with one of them also involved in the dimerization of the enzyme [52]. The NEU4 gene is spliced into two forms differing in the first 12 N-terminal amino acids [33, 53]. The short isoform was found predominantly associated with ER membranes [33, 53], whereas the long form was reported to be present in lysosomes [29] and mitochondria [33, 54]. Because the substrate specificities of NEU1, NEU3 and NEU4 partially overlap, their distinct cellular and subcellular distribution may be the key to understanding the physiological functions of these enzymes. In contrast to other mammalian, bacterial or viral neuraminidases, NEU1 is activated by association with the lysosomal serine carboxypeptidase cathepsin A (protective protein, CTSA). CTSA, NEU1, and lysosomal β-galactosidase (GLB1) form a lysosomal multienzyme complex, where CTSA activates NEU1 and protects NEU1 and GLB1 against proteolytic degradation by lysosomal peptidases (reviewed in [51]).
NEU1: Lysosomal catabolism, missfolded proteins and neurodegenerative diseases
Genetic deficiency of NEU1 in humans results in a severe metabolic disease, sialidosis (OMIM #256550), caused by the lysosomal storage of sialylated glycoproteins and oligosaccharides (reviewed in [55]). In addition, the genetic deficiency of CTSA results in the secondary deficiencies of NEU1 and GLB1, and causes the lysosomal storage disorder (LSD) galactosialidosis (GS OMIM #256540) (reviewed in [56]). Both disorders clinically manifest with skeletal and gait abnormalities, progressive impaired vision, bilateral macular cherry-red spots, ataxia, seizures and myoclonus syndrome. Severe early-onset forms are also associated with a dysmorphic phenotype, dysostosis multiplex, mental retardation and hepatosplenomegaly [57, 58]. A recent MRI study involving 11 attenuated sialidosis patients described increased mean diffusivity throughout the brain together with compromised white matter tracts integrity, the most severely affected cortical region being the occipital lobe. They also detected decreased functional connectivity from the temporal and occipital lobes to the hippocampus and parahippocampus and diffused cortical atrophy with posterior focal lesions potentially related to cortical blindness due to an altered neural network and a compromised visual pathway in the patients [59]. However, due to the rarity of the disease (1:4,000,000 live births [60]) and unavailability of CNS materials from human patients, the majority of the insights into the pathological CNS changes in sialidosis and GS came from studying the mouse models of the diseases. KO mouse models of sialidosis (Neu1 KO) and galactosialidosis (Ctsa KO) develop phenotypes closely resembling severe systemic disease in human patients [57, 58]. Interestingly, the hypomorph Ctsa mouse with secondary 90% deficiency of NEU1 does not develop sialidosis suggesting that 10% of residual enzymatic activity is sufficient to support the required rate of lysosomal SGC catabolism [61]. The first reports on both sialidosis and GS mice described extensive brain pathology with lysosomal storage primarily observed in microglia and perivascular macrophages and to lesser extent in neurons. Neurodegeneration involving primarily Purkinje cells in the cerebellum was increased in GS as compared with sialidosis mice, which the authors attributed to the deficiency of Ctsa activity in the GS mouse [57, 58]. However, this hypothesis has not been further confirmed in the Ctsa knock-in model expressing an enzymatically mutant CTSA protein with the inactivating S190A mutation in the active site [61].
Further studies of neurodegeneration in the Neu1 KO mice revealed the presence of amyloid bodies primarily clustered in the CA3 region of the hippocampus and resembling Alzheimer β-amyloid plaques [62]. The majority of these bodies were also immunoreactive for ubiquitin and neurofilaments. The authors showed that the amyloid precursor protein is a substrate of NEU1 and is oversialylated and accumulated in the lysosomes in the brain tissues of Neu1 KO mice, perhaps due to a reduced catabolic rate. They further detected increased extracellular release of Aβ peptides by excessive lysosomal exocytosis, previously attributed to increased levels of the lysosomal membrane protein 1 (LAMP-1) showing elevated sialylation and a longer half-life in the lysosome in the absence of NEU1 [63]. Moreover, inactivation of the Neu1 gene in the transgenic 5XFAD mice overexpressing human mutant amyloid precursor protein (APP) augmented formation of the plaques whereas intracerebral injections of adeno-associated viruses (AAVs) expressing NEU1 and CTSA could slow down or revert the amyloidogenic process, suggesting that reduced NEU1 brain levels could represent a risk factor for developing Alzheimer disease in humans. It is important to note, however, that the formation of cerebral amyloid plaques, lipofuscine granules and pTau aggregates has been reported for several mouse models of LSDs including MPS IIIA, B and C, MPS I and others, and has been attributed to autophagy block and impaired catabolism of missfolded proteins (see for example [64] or [65]) (Fig. 1). Therefore, it remains to be investigated to what extent the Neu1 KO mice show augmented amyloidogenesis as compared with other neurological LSDs.
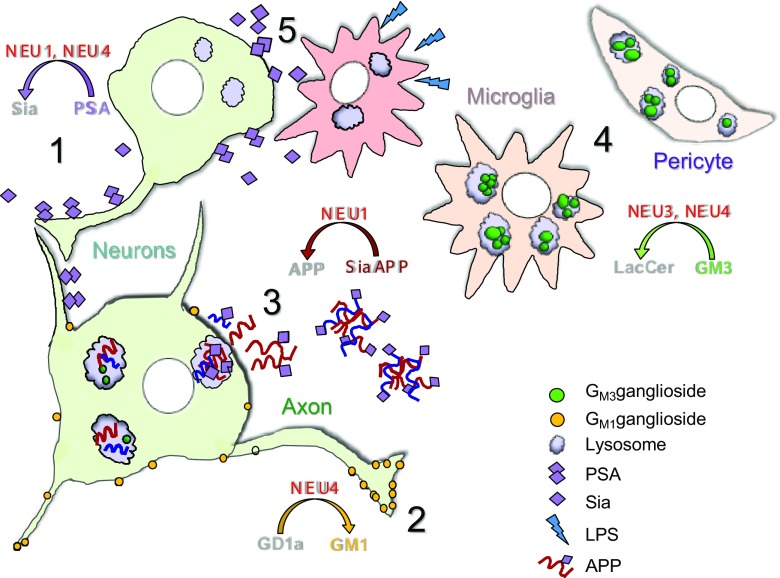
Fig. 1.
Proposed roles of neuraminidases in the CNS. 1. NEU1 and/or NEU4 reduce PSA content on neuronal surface during neuronal proliferation, differentiation, migration as well as during long-term potentiation of synapses. 2. NEU4 generates GM1 ganglioside from GD1a ganglioside at the tip of the axon and induces its growth. 3. NEU1 catalyzes lysosomal degradation of sialylated glycoproteins including APP. In sialidosis in the absence of Neu1, oversialylated APP is abnormally processed in lysosomes and the generated Aβ–peptide is released in the extracellular space. 4. NEU3 and NEU4 convert GM3 to LacCer in the lysosomes. In the Neu3/Neu4 double knockout mice, GM3 is stored in microglia, vascular pericytes and neurons, causing micro- and astrogliosis, neuroinflammation, and accumulation of lipofuscin bodies. 5. During neuroinflammation overexpressed NEU1 desialylates glycoproteins on cortical neurons
It also have been proposed that NEU1 is involved in the degradation of another missfolded pathogenic brain protein, prion (PrPSc). PrPSc is a conformationally altered, self-replicating state of the prion protein PrPC. Importantly, PrPSc is sialylated by sialyltransferases of the St6Gal family and this modification plays a major role in evading innate immunity and infecting a host [66]. Significantly higher amounts of the C1 fragment relative to full-length PrPC were detected in the brains of Neu1 KO mice as compared to WT, Neu3 or Neu4 KO mice, suggesting that catabolism of PrPC involves a desialylation of its proteolytic C1 fragments by NEU1 and consequent fast degradation of the desialylated products [67]. However, further studies are necessary to understand if NEU1 plays any role in predisposition to prion disease.
Regulation of brain polysialic acids: NEU4 or NEU1?
PSA levels on the brain cells undergo rapid changes both during development and in the adulthood and neuraminidases together with metalloproteases have been proposed to play a role in this process. For example, rapid negative changes in neuraminidase activity in response to treatment with cortical steroids or chronic stress were detected in rat hippocampus and linked to increased PSA levels suggesting the existence of a pathway for regulation of synaptic plasticity by a brain neuraminidase(s) [68]. Takahashi [69] et al. demonstrated that the long form of murine NEU4 could hydrolyze oligoSia and PSA chains in vitro at a much higher rate than NEU1, NEU2 or NEU3, and could also remove PSA from NCAM when added to mouse brain tissue homogenates. Overexpression of NEU4 in ST8SiaIV-transfected neuroblastoma Neuro2a cells decreased PSA levels whereas silencing the endogenous Neu4 in mouse embryonic hippocampal primary neurons increased both the PSA levels on neurites and their outgrowth. In contrast, transfection of the neurons with a NEU4-encoding plasmid reduced both the number of neurites and their length, allowing the authors to suggest that the enzyme is involved in both PSA degradation and regulation of neuronal differentiation [69]. Indirectly this conclusion was also supported by the author’s previous work demonstrateing that the expression of NEU4 in the mouse brain hippocampus peaked at post-natal days 3–14 coinciding with a massive reduction of PSA in the brain [43].
It is important to note, however, that because of the overlapping specificity of neuraminidases (for example NEU1. NEU2 and NEU4 show a comparable capacity to degrade 2,8-linked Neu5Ac polymers [31]), the results obtained by silencing or overexpressing individual isoforms in cellulo cannot be considered as an ultimate proof of their physiological function. Moreover, further work suggested that NEU1 but not NEU4 catabolizes PSA and trims it off brain cell surface glycoproteins. In particular, NEU1 has been implicated in stripping PSA from the surface of hippocampal granule cells (GCs) and is essential for proper migration of these neurons into the innermost region of the granule cell layer (GCL) of the hippocampus [70]. NEU1 knockdown in newly generated GCs in vivo both increased PSA presence on these cells and attenuated proper termination of GC migration in the innermost GCL suggesting that NEU1-mediated PSA removal is essential for proper neuronal lamination [70]. Sumida et al. [71] reported that NEU1 also cleaves PSA on brain microglial cells. They demonstrated that NEU1 is rapidly released as a part of exovesicles by microglial cell line Ra2 upon an inflammatory stimulus. The secreted NEU1 clears PSA on the Ra2 surface and triggers the release of brain-derived neurotrophic factor retained by PSA molecules [71]. Most recently, Demina et al. (submitted) demonstrated that NEU1 is involved in the reduction of PSA on neuronal proteins following postnatal lipopolysaccharide (LPS) exposure in rat pups (a model of brain infections in preterm born children). Following intracranial injection of LPS, they detected a significant increase of the acidic neuraminidase activity and Neu1 mRNA in the upper cortical layers. This was accompanied by a long-term reduction of sialylation of N-glycans on brain glycoproteins and reduced PSA levels on the surface of neurons in the upper somatosensory cortex. The authors hypothesize that neonatal LPS exposure results in specific and sustained induction of NEU1, causing long-lasting negative changes in sialylation and polysialylation of glycoproteins on brain cells and potentially contributing to pathophysiological consequences of perinatal infectious exposure (Fig. 1).
Catabolism of brain gangliosides: NEU1, NEU3 or NEU4?
Metabolic genetic blocks in processing and catabolism of gangliosides result in the development of severe disorders, gangliosidoses, such as GM1 gangliosidosis, Tay-Sachs and Sandhoff diseases, which are all dominated by CNS involvement with marked neuroinflammation and neurodegeneration (reviewed in [72]). The ganglioside degradation pathway involves several desialylation steps (i.e. conversion of GD1a to GM1a, GM3 to lactosyl ceramide (LacCer) etc.) presumably catalyzed by the enzymes of the NEU family. However, no gangliosidoses caused by genetic deficiencies of human neuraminidases have been identified so far. Of all mammalian neuraminidases, NEU3 and NEU4 have the highest in vitro activity against gangliosides (reviewed in [25, 26, 73]) and are major candidates for being ganglioside catabolizing enzymes. In support of this, GM3 accumulation has been observed in a NEU3-silenced human glioblastoma cell line [74]. However, the Neu4 and Neu3 KO mouse models do not develop massive lysosomal storage of any ganglioside or severe clinical phenotypes resembling those of the Glb1 KO mouse (model of GM1 gangliosidosis) or β-hexosaminidase deficient Hexb KO mouse (model of Sandhoff disease) [40, 75]. The Neu3/Neu4 double-KO mice, however, store GM3 in the lysosomes of the brain’s phagocytic cells, microglia and pericytes, which gain a foamy appearance, accumulate multivesicular bodies similar to those present in GM1 and GM2 gangliosidoses, get activated, and secret inflammatory cytokines [38]. While brain phagocytic cells stored the majority of GM3 gangliosides, they are also accumulated in neurons, resulting in appearance of lysosomes containing whorls of membranes as well as enlarged lipofuscin bodies comprising multivesicular structures [38]. In vitro, neuraminidase activity against GM3 has been reduced to below detection level. However, GM3 increase in the brains of Neu3/Neu4 double-KO mice was relatively modest as compared with those caused by deficiencies of the essential ganglioside hydrolases such as β-galactosidase (GM1 gangliosidosis) or β-hexosaminidases A and B (GM2 gangliosidosis), suggesting the existence of alternative catabolic pathways.
One of such pathways exists in the mouse model of Tay-Sachs disease (Hexa−/− mouse), where the stored GM2 ganglioside is converted to GA2, then further hydrolyzed by β-hexosaminidase B to LacCer [76]. The pathway does not exist in humans, presumably because of lower levels of brain ganglioside neuraminidase activity. Three neuraminidases (NEU1, NEU3 and NEU4) have been proposed to be responsible for desialylation of GM2 in the mouse brain. First, Igdoura et al., showed a reduction of GM2 in the neuroglia cell line from a Tay-Sachs patient overexpressing human NEU1 [77]. They speculated that pharmacological induction or activation of this enzyme could be exploited for the treatment of Tay-Sachs disease [77]. Further studies, however, demonstrated that NEU1 has negligible activity against ganglioside substrates, in contrast with NEU3 and NEU4 [29]. In agreement with this, Hexa−/-CathAS190A-neo mice with secondary 90% deficiency of NEU1 in the tissues do not show an increase in brain GM2 levels as compared with Hexa−/− mice [78]. Li 2001 et al. tested if recombinant NEU2 and NEU3 could desialylate GM1 and GM2 gangliosides in vitro and found that the mouse NEU3 but not NEU2 was able to convert GM1 and GM2 into their respective asialo-derivatives in the presence of human or mouse GM2 activator protein [79]. Seyrantepe et al. further demonstrated that transfection with a NEU4-expressing plasmid can also rescue GM2 metabolism in the Tay-Sachs neuroglial cells [44]. However, double Neu4/Hexa KO mice showed only partially aggravated GM2 storage and clinical phenotype still milder than the Sandhoff mouse model [44]. These data suggested that NEU4 is not the only sialidase contributing to the metabolic bypass in Hexa−/− mice. The question of which neuraminidase catabolizes GM2 was answered when Pan et al. [38] demonstrated that NEU3 deficiency in Hexa−/− mice resulted in a vast (at least 5-fold) increase of stored brain GM2, suggesting that NEU3 is the enzyme responsible for the metabolic bypass. Independently, Seyrantepe et al. [80] conducted a thorough phenotypic characterization of double-knockout Hexa−/-Neu3−/− mice and determined that, in contrast with Hexa−/− mice that had a late onset at 10–11 months, the double-KO mice showed rapid deterioration with slow movement, ataxia and tremors and had a life span of only 1.5 to 4.5 months, similar to that of the Sandhoff Hexb−/− mice. The Hexa−/-Neu3−/− mice also had progressive neurodegeneration and astrogliosis mimicking the neuropathological and clinical abnormalities of the early-onset Tay-Sachs patients [80].
Potential role of the long NEU4 isoform (NEU4L) in the degradation of the b-series ganglioside GD3 is of special interest. GD3 is highly expressed in mouse and human embryonic neural stem cells but it becomes a minor component of the mature brain (reviewed in [6]). Hasegawa et al. published an observation that Neu4 RNA and protein levels are rapidly decreasing during the apoptosis of neuroblastoma cells induced by the overexpression of tyrosinase that generated toxic catechol metabolites [81]. This process was concomitant with the increase of the mitochondrial pool of GD3, presumably degraded by NEU4L. At the same time, the authors did not present evidence that the increase of GD3 was the consequence of the NEU4 decrease or if accumulation of mitochondrial GD3 was involved in the neuronal apoptosis induced by catechol metabolites.
Generation of GM1 ganglioside and induction of axonal growth: NEU3 or NEU4?
Ganglioside content and composition of the mammalian brain constantly change over an individual’s lifetime. Early in development (such as E14 in the rat), the mammalian brain is dominated by GM3 and GD3 gangliosides [82, 83]. By the time of birth, GM3 and GD3 become minor components in the brain, with GD1a and GT1b taking their place. Finally, in the mature brain, GM1a, GD1a, GD1b, and GT1b are present at comparable levels, together representing ~97% of gangliosides [84].
Until now, it has been assumed that the brain ganglioside composition is mainly regulated by changes in the rate of their biosynthesis in the Golgi, catalyzed by glycosyltransferases including sialyltransferases (reviewed in [6]). Genetic deficiencies of individual glycosyltransferases in human patients or in gene-targeted mouse models result in both changes in ganglioside profiles and severe neurological manifestations (reviewed in [6]). These mice lack the whole series of gangliosides: GD1b and GT1b in St8sia1 knockout (KO) mice, GM1, GD1a, GD1b, and GT1b in B4galnt1 KO mice or GD1a and GT1b in St 3 gal2/3-double KO mice [85, 86].
At the same time, multiple studies provide evidence that neuronal GM1 ganglioside is also generated by neuraminidases and that this process is essential for axonal growth and regeneration. During peripheral axon growth, GM1a binds to laminin-1 and induces its focal aggregation, in turn enhancing the relocation of tropomyosin receptor kinase A (TrkA) in lipid rafts and the subsequent activation of downstream signaling molecules [87]. Clustering of GM1 with laminin-1, along with laminin-1 self-assembly, also promotes relocation and enrichment of β1-integrin in lipid rafts and enhances combined laminin-integrin signaling to trigger neurite outgrowth [87]. Galectin-1 (Gal-1) was recently reported to cross-link GM1 ganglioside and α5 β1-integrin [88]. In normal cerebellar granule neurons and neuroblastoma NG108–15 cells, this led to a transient increase in intracellular Ca2+ by opening TRPC5 channels whereas cells from mice lacking major gangliosides, including GM1, showed retarded axonogenesis, underscoring the importance of GM1 for this process [88]. Conversion of GD1a to GM1 also reduces affinity for myelin-associated glycoprotein (MAG), identified as a ganglioside binding partner participating in in axon-myelin interactions [89]. Ganglioside-MAG interactions stabilize the axonal myelination, but they can also inhibit axon outgrowth and regeneration (reviewed in [6]). Moreover, in some types of nerve cells, MAG inhibition of axon outgrowth was reversed by cleaving the terminal sialic acids from GD1a and GT1b by exogenous sialidase (reviewed in [6]).
Several studies argued that NEU3 is the enzyme generating GM1 gangliosides at the growing axonal tip. First, Rodriguez et al. demonstrated that treatment of cultured differentiating hippocampal embryonic mouse neurons with pan-neuraminidase inhibitor, 2,3-dehydro-2-deoxy-N-acetylneuraminic acid (DANA) blocks axonal elongation, whereas transfection of the cells with NEU3 dramatically enhances axon growth and accelerates the polarization of cytoskeletal proteins [41]. Overexpression also resulted in the regrowth of the original axon but not of other neurites [41]. Proshin et al. reported that in human neuroblastoma NB-1 cells, expression of NEU3 was increased by dibutyryl cAMP which also induced neurite outgrowth [90]. Moreover, NB-1 cells transfected with a NEU3-expressing plasmid demonstrated more prominent outgrowth of neurites with axon-like characteristics, even in the absence of dibutyryl cAMP. Finally Da Silva et al. deciphered the mechanism by which NEU3 may induce axonal growth [91]. They demonstrated that NEU3 (referred to in the paper as plasma membrane ganglioside sialidase, PMGS) asymmetrically accumulates at the tip of one neurite of the unpolarized rat neuron, inducing actin instability. Suppression of NEU3 expression by siRNA and inhibition of neuraminidase activity by DANA blocked axonal generation, whereas neurons overexpressing a hemagglutinin (HA)-NEU3 fusion protein showed enhanced local TrkA activity, which further triggered phosphatidylinositol-3-kinase (PI3K)- and Rac1-dependent inhibition of RhoA signaling and actin depolymerization in the growing axon [91]. HA-NEU3 and TrkA could be co-immunoprecipitated from the neuronal membrane extracts, suggesting that they interact in the cell. The authors also claimed that pTrkA and HA-NEU3 interacted with GM1, because they could coprecipitate both proteins using cholera toxin B-modified beads. However, this experiment needs to be taken with a grain of salt since recent studies show that, besides GM1 ganglioside, cholera toxin also interacts with a number of gangliosides and even fucosylated glycoproteins [92].
Surprisingly, Valaperta et al. [93] reported that shRNA reduction of NEU3 in Neuro2a murine neuroblastoma cells did not inhibit axonal growth, but in contrast caused neurite elongation and appearance of the axonal marker protein Tau [93]. In NEU3 silenced cells, both GM1 and GD1a were decreased while GM2 increased, which was inconsistent with the hypothesis about the generation of GM1 from GD1a by NEU3 [93]. Most recently, Kappagantula et al. confirmed that NEU3 generates GM1 during the regeneneration of axons in the peripheral nerves but not of retinal axons in the CNS [94]. In peripheral axons of adult rats, NEU3 was activated and GM1/GD1a and GM1/GT1b ratios increased whereas no change in GM1/GD1a ratio or NEU3 expression was observed in retinal axons after axotomy, despite the presence of NEU3 in these cells. Finally, Woronowicz et al. proposed that TrkA is directly activated by a neuronal neuraminidase that cleaves a specific sialyl alpha-2,3-linked beta-galactosyl sugar residue on the receptor [95]. They reported that PC12 cells transfected with a TrkA-expressing plasmid as well as primary cortical neurons showed an increase of plasma neuraminidase activity in response to brain-derived neurotrophic factor (BDNF) [95]. Both the increase in neuraminidase activity and neurite outgrowth could be suppressed by an inhibitor of influenza virus neuraminidase, Tamiflu (Zanamavir), but not by DANA which, according to the authors, is specific towards NEU2 and NEU3 [95]. Other reports however have shown that DANA equally inhibits all 4 mamalian neuraminidases (IC50 between 10 and 50 μM), while Zanamavir shows the highest specificity towards NEU3 (IC50 4 μM), is less active against NEU2 and NEU4 (IC50 8 and 50 μM, respectively) and hardly shows any activity against NEU1 (IC50 > 500 μM) [27, 96–98]. Therefore, it is hard to conclude based on the reported data the activation of which NEU isoenzyme has been detected by the authors.
Importantly, as in the case of PSA removal described above, the main proof for the NEU3’s role in GM1 generation/axonal growth has been obtained in vitro either by treating embryonic neuronal cultures with unspecific neuraminidase inhibitor DANA or by overexpressing NEU3, which leaves open the question whether this also happens in the brain, where neuronal differentiation occurs in the presence of different cell types expressing different neuraminidases. This question has been partially addressed by the study of Pan [38] et al. in which they detected reduced GM1 in proximity to myelin layers in hippocampal and cortical neurons from both Neu4 KO and double Neu3/Neu4 KO mice. About 50% reduced GM1 and 30% increased GD1a levels were detected in total brains from both strains as compared with the WT levels. GM1a gangliosides were also reduced at the growing tip of the axons in cultured cortical neurons of the Neu3/Neu4 double-KO mice (Fig. 1). Besides, NEU3/NEU4 combined deficiency significantly decreased neuritogenesis in vitro and the density of MAP2-positive neurofilaments in vivo. The authors conclude that the simplest explanation for low neuronal levels of GM1 ganglioside is that it is produced from GD1a by the action of NEU4 while admitting that further analysis of the ganglioside metabolism is necessary to prove this point [38].
Rapid changes in cerebral neuraminidase activity associated with memory consolidation and seizures: NEU1?
Since the level of sialic acids on the cell surface as well as inside the cell changes during long-term potentiation (LTP), brain neuraminidase(s) were suggested to participate in neuronal plasticity. Initial evidences for this involvement were obtained by treating acute rodent hippocampal slices with pan-neuraminidase inhibitor DANA. In particular, Isaeva et al. demonstrated that DANA treatment increased the firing frequency and amplitude of spontaneous synchronous oscillations, the frequency of multiple unit activity and the tonic phase of seizure-like activity in the low-magnesium model of ictogenesis [99]. Pretreatment of hippocampal cultured brain slices with DANA also increased the density of simple and perforated synapses in the CA1 stratum radiatum region. Interestingly, high concentrations of calcium eliminated the effect of DANA on synaptic density, suggesting that synaptogenesis observed in response to neuraminidase treatment is an activity-dependent process [99].
More recently, the development of sensitive fluorogenic neuraminidase substrates and methods of live imaging allowed to investigate whether such processes occur in vivo. Minami et al. developed a highly sensitive histochemical imaging probe for sialidase activity, BTP3-Neu5Ac, which is cleaved by all 4 neuraminidases and demonstrated that, at neutral pH, the probe stained mossy fiber terminal fields in rat hippocampal acute slices [100]. The authors further showed that neuraminidase inhibitor DANA significantly impaired LTP at mossy fiber-CA3 pyramidal cell synapse. In rats, DANA injection into the hippocampal CA3 region of NEU4 knockdown rats prolonged the escape latency time in Morris water maze test suggesting that the regulation of sialylation by NEU4 is involved in hippocampal memory processing [100]. The same team further reported that neuraminidase activity at the CA3 stratum of rat acute hippocampal slices, visualized with BTP3-Neu5Ac, was rapidly increased in response to LTP-inducible high-frequency stimulation. This was accompanied by a rapid increase in free Sia levels, presumably shed from the neuronal surface [101]. Neuronal desialylation also occurred in live animals during hippocampus-dependent memory formation in a contextual fear-conditioning paradigm [101].
Interestingly, rapid changes in neuraminidase activity (decrease in cerebral cortex and cerebellum and increase in brainstem, hypothalamus and hippocampus) were also reported for the brains of WT kindled epileptic and non-epileptic seizing rats [102]. The authors speculate that these changes in activity may reflect functional and anatomical plastic changes associated with seizures [102]. Although providing a potentially interesting observation, the study does not allow to conclude which neuraminidases were responsible for the above changes.
Conclusion
Multiple data endorse the important biological roles played by mammalian neuraminidases in the development and functioning of the CNS providing a rationale for identifying the main biological targets of neuraminidases in the brain. The availability of mouse models and recent development of high-throughput methods for analysis of sialoglycoconjugates through their metabolic labeling, advanced mass-spectrocopy and live imaging deliver a unique opportunity to compare the sialome (total complements of sialylated glycoproteins) of cells, tissues or organs from WT and KO mice. These animal models also provide a key tool for functional studies to link biochemical deficits of neuraminidases to biological phenotypes, which can only be defined by using the living organism. In addition, recent advances in the development of selective inhibitors of human neuraminidases [39, 97] provide researchers with pharmacological methods to target these enzymes in vitro and in vivo.
NEU1 deficiency causes severe systemic disorders, sialidosis and GS associated with pathological changes in the CNS. Although monogenic disorders caused by deficiencies of other neuraminidases are not described, it is possible that changes in sialylation can cause effects strong enough to influence predisposition to common neurological diseases (such as Alzheimer, Parkinson or ASD) but not strong enough to cause a Mendelian disease, therefore contributing to “hidden heritability” in human populations. Further studies are necessary to test this hypothesis by linking changes in neuraminidase activity and cell sialylation to pathological phenotypes.
Acknowledgements
This work was supported in part by the operating grants 156345 and 148863 from the Canadian Institutes of Health Research (http://www.cihr-irsc.gc.ca/) and G2 from the Canadian Glycomic Network to A.V.P. The authors thank Rachel Héon-Roberts and Annie Nguyen for critical reading of the manuscript.
Conflicts of interest
The authors declare that they have no conflicts of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
References
- 1.Cohen M, Varki A. The sialome--far more than the sum of its parts. OMICS. 2010;14:455–464. doi: 10.1089/omi.2009.0148. [DOI] [PubMed] [Google Scholar]
- 2.Kelm S, Schauer R. Sialic acids in molecular and cellular interactions. Int. Rev. Cytol. 1997;175:137–240. doi: 10.1016/S0074-7696(08)62127-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lehmann F, Tiralongo E, Tiralongo J. Sialic acid-specific lectins: occurrence, specificity and function. Cell. Mol. Life Sci. 2006;63:1331–1354. doi: 10.1007/s00018-005-5589-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allende ML, Proia RL. Lubricating cell signaling pathways with gangliosides. Curr. Opin. Struct. Biol. 2002;12:587–592. doi: 10.1016/S0959-440X(02)00376-7. [DOI] [PubMed] [Google Scholar]
- 5.Jones CJ, Aplin JD, Mulholland J, Glasser SR. Patterns of sialylation in differentiating rat decidual cells as revealed by lectin histochemistry. J. Reprod. Fertil. 1993;99:635–645. doi: 10.1530/jrf.0.0990635. [DOI] [PubMed] [Google Scholar]
- 6.Schnaar RL, Gerardy-Schahn R, Hildebrandt H. Sialic acids in the brain: gangliosides and polysialic acid in nervous system development, stability, disease, and regeneration. Physiol. Rev. 2014;94:461–518. doi: 10.1152/physrev.00033.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nowycky MC, Wu G, Ledeen RW. Glycobiology of ion transport in the nervous system. Adv Neurobiol. 2014;9:321–342. doi: 10.1007/978-1-4939-1154-7_15. [DOI] [PubMed] [Google Scholar]
- 8.Ledeen RW, Wu G. Ganglioside function in calcium homeostasis and signaling. Neurochem. Res. 2002;27:637–647. doi: 10.1023/A:1020224016830. [DOI] [PubMed] [Google Scholar]
- 9.Yang LJ, Zeller CB, Shaper NL, Kiso M, Hasegawa A, Shapiro RE, Schnaar RL. Gangliosides are neuronal ligands for myelin-associated glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 1996;93:814–818. doi: 10.1073/pnas.93.2.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sheikh KA, Sun J, Liu Y, Kawai H, Crawford TO, Proia RL, Griffin JW, Schnaar RL. Mice lacking complex gangliosides develop Wallerian degeneration and myelination defects. Proc. Natl. Acad. Sci. U. S. A. 1999;96:7532–7537. doi: 10.1073/pnas.96.13.7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J, Yu RK. Interaction of ganglioside GD3 with an EGF receptor sustains the self-renewal ability of mouse neural stem cells in vitro. Proc. Natl. Acad. Sci. U. S. A. 2013;110:19137–19142. doi: 10.1073/pnas.1307224110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tajima O, Egashira N, Ohmi Y, Fukue Y, Mishima K, Iwasaki K, Fujiwara M, Inokuchi J, Sugiura Y, Furukawa K, Furukawa K. Reduced motor and sensory functions and emotional response in GM3-only mice: emergence from early stage of life and exacerbation with aging. Behav. Brain Res. 2009;198:74–82. doi: 10.1016/j.bbr.2008.10.024. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Cheng A, Wakade C, Yu RK. Ganglioside GD3 is required for neurogenesis and long-term maintenance of neural stem cells in the postnatal mouse brain. J. Neurosci. 2014;34:13790–13800. doi: 10.1523/JNEUROSCI.2275-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brennaman LH, Maness PF. Developmental regulation of GABAergic interneuron branching and synaptic development in the prefrontal cortex by soluble neural cell adhesion molecule. Mol. Cell. Neurosci. 2008;37:781–793. doi: 10.1016/j.mcn.2008.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vawter MP. Dysregulation of the neural cell adhesion molecule and neuropsychiatric disorders. Eur. J. Pharmacol. 2000;405:385–395. doi: 10.1016/S0014-2999(00)00568-9. [DOI] [PubMed] [Google Scholar]
- 16.Werneburg S, Muhlenhoff M, Stangel M, Hildebrandt H. Polysialic acid on SynCAM 1 in NG2 cells and on neuropilin-2 in microglia is confined to intracellular pools that are rapidly depleted upon stimulation. Glia. 2015;63:1240–1255. doi: 10.1002/glia.22815. [DOI] [PubMed] [Google Scholar]
- 17.Werneburg S, Buettner FF, Erben L, Mathews M, Neumann H, Muhlenhoff M, Hildebrandt H. Polysialylation and lipopolysaccharide-induced shedding of E-selectin ligand-1 and neuropilin-2 by microglia and THP-1 macrophages. Glia. 2016;64:1314–1330. doi: 10.1002/glia.23004. [DOI] [PubMed] [Google Scholar]
- 18.Takashima S. Characterization of mouse sialyltransferase genes: their evolution and diversity. Biosci. Biotechnol. Biochem. 2008;72:1155–1167. doi: 10.1271/bbb.80025. [DOI] [PubMed] [Google Scholar]
- 19.Simpson MA, Cross H, Proukakis C, Priestman DA, Neville DC, Reinkensmeier G, Wang H, Wiznitzer M, Gurtz K, Verganelaki A, Pryde A, Patton MA, Dwek RA, Butters TD, Platt FM, Crosby AH. Infantile-onset symptomatic epilepsy syndrome caused by a homozygous loss-of-function mutation of GM3 synthase. Nat. Genet. 2004;36:1225–1229. doi: 10.1038/ng1460. [DOI] [PubMed] [Google Scholar]
- 20.Hu H, Eggers K, Chen W, Garshasbi M, Motazacker MM, Wrogemann K, Kahrizi K, Tzschach A, Hosseini M, Bahman I, Hucho T, Muhlenhoff M, Gerardy-Schahn R, Najmabadi H, Ropers HH, Kuss AW. ST3GAL3 mutations impair the development of higher cognitive functions. Am. J. Hum. Genet. 2011;89:407–414. doi: 10.1016/j.ajhg.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edvardson S, Baumann AM, Muhlenhoff M, Stephan O, Kuss AW, Shaag A, He L, Zenvirt S, Tanzi R, Gerardy-Schahn R, Elpeleg O. West syndrome caused by ST3Gal-III deficiency. Epilepsia. 2013;54:e24–e27. doi: 10.1111/epi.12050. [DOI] [PubMed] [Google Scholar]
- 22.Calandreau L, Marquez C, Bisaz R, Fantin M, Sandi C. Differential impact of polysialyltransferase ST8SiaII and ST8SiaIV knockout on social interaction and aggression. Genes Brain Behav. 2010;9:958–967. doi: 10.1111/j.1601-183X.2010.00635.x. [DOI] [PubMed] [Google Scholar]
- 23.Angata K, Long JM, Bukalo O, Lee W, Dityatev A, Wynshaw-Boris A, Schachner M, Fukuda M, Marth JD. Sialyltransferase ST8Sia-II assembles a subset of polysialic acid that directs hippocampal axonal targeting and promotes fear behavior. J. Biol. Chem. 2004;279:32603–32613. doi: 10.1074/jbc.M403429200. [DOI] [PubMed] [Google Scholar]
- 24.Ellies LG, Ditto D, Levy GG, Wahrenbrock M, Ginsburg D, Varki A, Le DT, Marth JD. Sialyltransferase ST3Gal-IV operates as a dominant modifier of hemostasis by concealing asialoglycoprotein receptor ligands. Proc. Natl. Acad. Sci. U. S. A. 2002;99:10042–10047. doi: 10.1073/pnas.142005099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miyagi T, Yamaguchi K. Mammalian sialidases: physiological and pathological roles in cellular functions. Glycobiology. 2012;22:880–896. doi: 10.1093/glycob/cws057. [DOI] [PubMed] [Google Scholar]
- 26.Monti E, Miyagi T. Structure and function of mammalian Sialidases. Top. Curr. Chem. 2015;366:183–208. doi: 10.1007/128_2012_328. [DOI] [PubMed] [Google Scholar]
- 27.Zou Y, Albohy A, Sandbhor M, Cairo CW. Inhibition of human neuraminidase 3 (NEU3) by C9-triazole derivatives of 2,3-didehydro-N-acetyl-neuraminic acid. Bioorg. Med. Chem. Lett. 2010;20:7529–7533. doi: 10.1016/j.bmcl.2010.09.111. [DOI] [PubMed] [Google Scholar]
- 28.Hasegawa T, Yamaguchi K, Wada T, Takeda A, Itoyama Y, Miyagi T. Molecular cloning of mouse ganglioside sialidase and its increased expression in Neuro2a cell differentiation. J. Biol. Chem. 2000;275:8007–8015. doi: 10.1074/jbc.275.11.8007. [DOI] [PubMed] [Google Scholar]
- 29.Seyrantepe V, Landry K, Trudel S, Hassan JA, Morales CR, Pshezhetsky AV. Neu4, a novel human lysosomal lumen sialidase, confers normal phenotype to sialidosis and galactosialidosis cells. J. Biol. Chem. 2004;279:37021–37029. doi: 10.1074/jbc.M404531200. [DOI] [PubMed] [Google Scholar]
- 30.Smutova V, Albohy A, Pan X, Korchagina E, Miyagi T, Bovin N, Cairo CW, Pshezhetsky AV. Structural basis for substrate specificity of mammalian neuraminidases. PLoS One. 2014;9:e106320. doi: 10.1371/journal.pone.0106320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies LR, Pearce OM, Tessier MB, Assar S, Smutova V, Pajunen M, Sumida M, Sato C, Kitajima K, Finne J, Gagneux P, Pshezhetsky A, Woods R, Varki A. Metabolism of vertebrate amino sugars with N-glycolyl groups: resistance of alpha2-8-linked N-glycolylneuraminic acid to enzymatic cleavage. J. Biol. Chem. 2012;287:28917–28931. doi: 10.1074/jbc.M112.365056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonten E, van der Spoel A, Fornerod M, Grosveld G, d'Azzo A. Characterization of human lysosomal neuraminidase defines the molecular basis of the metabolic storage disorder sialidosis. Genes Dev. 1996;10:3156–3169. doi: 10.1101/gad.10.24.3156. [DOI] [PubMed] [Google Scholar]
- 33.Yamaguchi K, Hata K, Koseki K, Shiozaki K, Akita H, Wada T, Moriya S, Miyagi T. Evidence for mitochondrial localization of a novel human sialidase (NEU4) Biochem. J. 2005;390:85–93. doi: 10.1042/BJ20050017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koseki K, Wada T, Hosono M, Hata K, Yamaguchi K, Nitta K, Miyagi T. Human cytosolic sialidase NEU2-low general tissue expression but involvement in PC-3 prostate cancer cell survival. Biochem. Biophys. Res. Commun. 2012;428:142–149. doi: 10.1016/j.bbrc.2012.10.028. [DOI] [PubMed] [Google Scholar]
- 35.Wada T, Yoshikawa Y, Tokuyama S, Kuwabara M, Akita H, Miyagi T. Cloning, expression, and chromosomal mapping of a human ganglioside sialidase. Biochem. Biophys. Res. Commun. 1999;261:21–27. doi: 10.1006/bbrc.1999.0973. [DOI] [PubMed] [Google Scholar]
- 36.Monti E, Bassi MT, Papini N, Riboni M, Manzoni M, Venerando B, Croci G, Preti A, Ballabio A, Tettamanti G, Borsani G. Identification and expression of NEU3, a novel human sialidase associated to the plasma membrane. Biochem. J. 2000;349:343–351. doi: 10.1042/bj3490343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Comelli EM, Amado M, Lustig SR, Paulson JC. Identification and expression of Neu4, a novel murine sialidase. Gene. 2003;321:155–161. doi: 10.1016/j.gene.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 38.Pan X, De Aragao CBP, Velasco-Martin JP, Priestman DA, Wu HY, Takahashi K, Yamaguchi K, Sturiale L, Garozzo D, Platt FM, Lamarche-Vane N, Morales CR, Miyagi T, Pshezhetsky AV. Neuraminidases 3 and 4 regulate neuronal function by catabolizing brain gangliosides. FASEB J. 2017;31:3467–3483. doi: 10.1096/fj.201601299R. [DOI] [PubMed] [Google Scholar]
- 39.Guo T, Datwyler P, Demina E, Richards MR, Ge P, Zou C, Zheng R, Fougerat A, Pshezhetsky AV, Ernst B, Cairo CW. Selective inhibitors of human neuraminidase 3. J. Med. Chem. 2018;61:1990–2008. doi: 10.1021/acs.jmedchem.7b01574. [DOI] [PubMed] [Google Scholar]
- 40.Seyrantepe V, Canuel M, Carpentier S, Landry K, Durand S, Liang F, Zeng J, Caqueret A, Gravel RA, Marchesini S, Zwingmann C, Michaud J, Morales CR, Levade T, Pshezhetsky AV. Mice deficient in Neu4 sialidase exhibit abnormal ganglioside catabolism and lysosomal storage. Hum. Mol. Genet. 2008;17:1556–1568. doi: 10.1093/hmg/ddn043. [DOI] [PubMed] [Google Scholar]
- 41.Rodriguez JA, Piddini E, Hasegawa T, Miyagi T, Dotti CG. Plasma membrane ganglioside sialidase regulates axonal growth and regeneration in hippocampal neurons in culture. J. Neurosci. 2001;21:8387–8395. doi: 10.1523/JNEUROSCI.21-21-08387.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hasegawa T, Feijoo Carnero C, Wada T, Itoyama Y, Miyagi T. Differential expression of three sialidase genes in rat development. Biochem. Biophys. Res. Commun. 2001;280:726–732. doi: 10.1006/bbrc.2000.4186. [DOI] [PubMed] [Google Scholar]
- 43.Shiozaki K, Koseki K, Yamaguchi K, Shiozaki M, Narimatsu H, Miyagi T. Developmental change of sialidase neu4 expression in murine brain and its involvement in the regulation of neuronal cell differentiation. J. Biol. Chem. 2009;284:21157–21164. doi: 10.1074/jbc.M109.012708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Seyrantepe, V., Lema, P., Caqueret, A., Dridi, L., Bel Hadj, S., Carpentier, S., Boucher, F., Levade, T., Carmant, L., Gravel, R. A., Hamel, E., Vachon, P., Di Cristo, G., Michaud, J. L., Morales, C. R., and Pshezhetsky, A. V. (2010) Mice doubly-deficient in lysosomal hexosaminidase A and neuraminidase 4 show epileptic crises and rapid neuronal loss. PLoS Genet 6 [DOI] [PMC free article] [PubMed]
- 45.Wang Y, Yamaguchi K, Wada T, Hata K, Zhao X, Fujimoto T, Miyagi T. A close association of the ganglioside-specific sialidase Neu3 with caveolin in membrane microdomains. J. Biol. Chem. 2002;277:26252–26259. doi: 10.1074/jbc.M110515200. [DOI] [PubMed] [Google Scholar]
- 46.Zanchetti G, Colombi P, Manzoni M, Anastasia L, Caimi L, Borsani G, Venerando B, Tettamanti G, Preti A, Monti E, Bresciani R. Sialidase NEU3 is a peripheral membrane protein localized on the cell surface and in endosomal structures. Biochem. J. 2007;408:211–219. doi: 10.1042/BJ20070503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paolini L, Orizio F, Busatto S, Radeghieri A, Bresciani R, Bergese P, Monti E. Exosomes secreted by HeLa cells shuttle on their surface the plasma membrane-associated Sialidase NEU3. Biochemistry. 2017;56:6401–6408. doi: 10.1021/acs.biochem.7b00665. [DOI] [PubMed] [Google Scholar]
- 48.Rodriguez-Walker M, Daniotti JL. Human Sialidase Neu3 is S-Acylated and behaves like an integral membrane protein. Sci. Rep. 2017;7:4167. doi: 10.1038/s41598-017-04488-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lukong KE, Seyrantepe V, Landry K, Trudel S, Ahmad A, Gahl WA, Lefrancois S, Morales CR, Pshezhetsky AV. Intracellular distribution of lysosomal sialidase is controlled by the internalization signal in its cytoplasmic tail. J. Biol. Chem. 2001;276:46172–46181. doi: 10.1074/jbc.M104547200. [DOI] [PubMed] [Google Scholar]
- 50.Vinogradova MV, Michaud L, Mezentsev AV, Lukong KE, El-Alfy M, Morales CR, Potier M, Pshezhetsky AV. Molecular mechanism of lysosomal sialidase deficiency in galactosialidosis involves its rapid degradation. Biochem. J. 1998;330(Pt 2):641–650. doi: 10.1042/bj3300641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pshezhetsky AV, Ashmarina M. Lysosomal multienzyme complex: biochemistry, genetics, and molecular pathophysiology. Prog. Nucleic Acid Res. Mol. Biol. 2001;69:81–114. doi: 10.1016/S0079-6603(01)69045-7. [DOI] [PubMed] [Google Scholar]
- 52.Maurice P, Baud S, Bocharova OV, Bocharov EV, Kuznetsov AS, Kawecki C, Bocquet O, Romier B, Gorisse L, Ghirardi M, Duca L, Blaise S, Martiny L, Dauchez M, Efremov RG, Debelle L. New insights into molecular Organization of Human Neuraminidase-1: Transmembrane topology and dimerization ability. Sci. Rep. 2016;6:38363. doi: 10.1038/srep38363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Monti E, Bassi MT, Bresciani R, Civini S, Croci GL, Papini N, Riboni M, Zanchetti G, Ballabio A, Preti A, Tettamanti G, Venerando B, Borsani G. Molecular cloning and characterization of NEU4, the fourth member of the human sialidase gene family. Genomics. 2004;83:445–453. doi: 10.1016/j.ygeno.2003.08.019. [DOI] [PubMed] [Google Scholar]
- 54.Bigi A, Morosi L, Pozzi C, Forcella M, Tettamanti G, Venerando B, Monti E, Fusi P. Human sialidase NEU4 long and short are extrinsic proteins bound to outer mitochondrial membrane and the endoplasmic reticulum, respectively. Glycobiology. 2010;20:148–157. doi: 10.1093/glycob/cwp156. [DOI] [PubMed] [Google Scholar]
- 55.Thomas, G. H. (2001) Disorders of glycoprotein degradation: α-mannosidosis, β-mannosidosis, fucosidosis, and sialidosis. In The Metabolic and Molecular Bases of Inherited Disease (Scriver CR, B. A., sly WS, Valle D. Ed.), McGraw-hill professional, New York. Pp
- 56.d'Azzo A, Andria G, Strisciuglio G, Galjaard H. Galactosialidosis. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic and Molecular Bases of Inherited Disease. 8th International student. New York: McGraw-Hill Professional; 2001. pp. 3811–3826. [Google Scholar]
- 57.Zhou XY, Morreau H, Rottier R, Davis D, Bonten E, Gillemans N, Wenger D, Grosveld FG, Doherty P, Suzuki K, Grosveld GC, d'Azzo A. Mouse model for the lysosomal disorder galactosialidosis and correction of the phenotype with overexpressing erythroid precursor cells. Genes Dev. 1995;9:2623–2634. doi: 10.1101/gad.9.21.2623. [DOI] [PubMed] [Google Scholar]
- 58.de Geest N, Bonten E, Mann L, de Sousa-Hitzler J, Hahn C, d'Azzo A. Systemic and neurologic abnormalities distinguish the lysosomal disorders sialidosis and galactosialidosis in mice. Hum. Mol. Genet. 2002;11:1455–1464. doi: 10.1093/hmg/11.12.1455. [DOI] [PubMed] [Google Scholar]
- 59.Lu CS, Ng SH, Lai SC, Kao LY, Liu L, Lin WY, Wu YM, Chen YL, Wang JJ. Cortical damage in the posterior visual pathway in patients with sialidosis type 1. Brain Imaging Behav. 2017;11:214–223. doi: 10.1007/s11682-016-9517-6. [DOI] [PubMed] [Google Scholar]
- 60.Meikle PJ, Hopwood JJ, Clague AE, Carey WF. Prevalence of lysosomal storage disorders. JAMA. 1999;281:249–254. doi: 10.1001/jama.281.3.249. [DOI] [PubMed] [Google Scholar]
- 61.Seyrantepe V, Hinek A, Peng J, Fedjaev M, Ernest S, Kadota Y, Canuel M, Itoh K, Morales CR, Lavoie J, Tremblay J, Pshezhetsky AV. Enzymatic activity of lysosomal carboxypeptidase (cathepsin) a is required for proper elastic fiber formation and inactivation of endothelin-1. Circulation. 2008;117:1973–1981. doi: 10.1161/CIRCULATIONAHA.107.733212. [DOI] [PubMed] [Google Scholar]
- 62.Annunziata I, Patterson A, Helton D, Hu H, Moshiach S, Gomero E, Nixon R, d'Azzo A. Lysosomal NEU1 deficiency affects amyloid precursor protein levels and amyloid-beta secretion via deregulated lysosomal exocytosis. Nat. Commun. 2013;4:2734. doi: 10.1038/ncomms3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yogalingam G, Bonten EJ, van de Vlekkert D, Hu H, Moshiach S, Connell SA, d'Azzo A. Neuraminidase 1 is a negative regulator of lysosomal exocytosis. Dev. Cell. 2008;15:74–86. doi: 10.1016/j.devcel.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martins C, Hulkova H, Dridi L, Dormoy-Raclet V, Grigoryeva L, Choi Y, Langford-Smith A, Wilkinson FL, Ohmi K, DiCristo G, Hamel E, Ausseil J, Cheillan D, Moreau A, Svobodova E, Hajkova Z, Tesarova M, Hansikova H, Bigger BW, Hrebicek M, Pshezhetsky AV. Neuroinflammation, mitochondrial defects and neurodegeneration in mucopolysaccharidosis III type C mouse model. Brain. 2015;138:336–355. doi: 10.1093/brain/awu355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ohmi K, Kudo LC, Ryazantsev S, Zhao HZ, Karsten SL, Neufeld EF. Sanfilippo syndrome type B, a lysosomal storage disease, is also a tauopathy. Proc. Natl. Acad. Sci. U. S. A. 2009;106:8332–8337. doi: 10.1073/pnas.0903223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Katorcha E, Baskakov IV. Analyses of N-linked glycans of PrP(Sc) revealed predominantly 2,6-linked sialic acid residues. FEBS J. 2017;284:3727–3738. doi: 10.1111/febs.14268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Katorcha E, Klimova N, Makarava N, Savtchenko R, Pan X, Annunziata I, Takahashi K, Miyagi T, Pshezhetsky AV, d'Azzo A, Baskakov IV. Loss of cellular Sialidases does not affect the Sialylation status of the prion protein but increases the amounts of its Proteolytic fragment C1. PLoS One. 2015;10:e0143218. doi: 10.1371/journal.pone.0143218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wielgat P, Walesiuk A, Braszko JJ. Effects of chronic stress and corticosterone on sialidase activity in the rat hippocampus. Behav. Brain Res. 2011;222:363–367. doi: 10.1016/j.bbr.2011.03.070. [DOI] [PubMed] [Google Scholar]
- 69.Takahashi K, Mitoma J, Hosono M, Shiozaki K, Sato C, Yamaguchi K, Kitajima K, Higashi H, Nitta K, Shima H, Miyagi T. Sialidase NEU4 hydrolyzes polysialic acids of neural cell adhesion molecules and negatively regulates neurite formation by hippocampal neurons. J. Biol. Chem. 2012;287:14816–14826. doi: 10.1074/jbc.M111.324186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sajo M, Sugiyama H, Yamamoto H, Tanii T, Matsuki N, Ikegaya Y, Koyama R. Neuraminidase-dependent degradation of Polysialic acid is required for the lamination of newly generated neurons. PLoS One. 2016;11:e0146398. doi: 10.1371/journal.pone.0146398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sumida M, Hane M, Yabe U, Shimoda Y, Pearce OM, Kiso M, Miyagi T, Sawada M, Varki A, Kitajima K, Sato C. Rapid trimming of cell surface Polysialic acid (PolySia) by exovesicular Sialidase triggers release of preexisting surface Neurotrophin. J. Biol. Chem. 2015;290:13202–13214. doi: 10.1074/jbc.M115.638759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sandhoff K, Harzer K. Gangliosides and gangliosidoses: principles of molecular and metabolic pathogenesis. J. Neurosci. 2013;33:10195–10208. doi: 10.1523/JNEUROSCI.0822-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pshezhetsky AV, Ashmarina LI. Desialylation of surface receptors as a new dimension in cell signaling. Biochemistry (Mosc) 2013;78:736–745. doi: 10.1134/S0006297913070067. [DOI] [PubMed] [Google Scholar]
- 74.Takahashi K, Proshin S, Yamaguchi K, Yamashita Y, Katakura R, Yamamoto K, Shima H, Hosono M, Miyagi T. Sialidase NEU3 defines invasive potential of human glioblastoma cells by regulating calpain-mediated proteolysis of focal adhesion proteins. Biochim. Biophys. Acta. 2017;1861:2778–2788. doi: 10.1016/j.bbagen.2017.07.023. [DOI] [PubMed] [Google Scholar]
- 75.Yamaguchi K, Shiozaki K, Moriya S, Koseki K, Wada T, Tateno H, Sato I, Asano M, Iwakura Y, Miyagi T. Reduced susceptibility to colitis-associated colon carcinogenesis in mice lacking plasma membrane-associated sialidase. PLoS One. 2012;7:e41132. doi: 10.1371/journal.pone.0041132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gravel R, Kaback MM, Proia RL, Sandhoff K, Suzuki K. The GM2 gangliosidoses. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The Metabolic and Molecular Bases of Inherited Diseases. 8. New York: McGraw-Hill; 2001. pp. 3827–3876. [Google Scholar]
- 77.Igdoura SA, Mertineit C, Trasler JM, Gravel RA. Sialidase-mediated depletion of GM2 ganglioside in Tay-Sachs neuroglia cells. Hum. Mol. Genet. 1999;8:1111–1116. doi: 10.1093/hmg/8.6.1111. [DOI] [PubMed] [Google Scholar]
- 78.Timur ZK, Akyildiz Demir S, Marsching C, Sandhoff R, Seyrantepe V. Neuraminidase-1 contributes significantly to the degradation of neuronal B-series gangliosides but not to the bypass of the catabolic block in Tay-Sachs mouse models. Mol Genet Metab Rep. 2015;4:72–82. doi: 10.1016/j.ymgmr.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Li SC, Li YT, Moriya S, Miyagi T. Degradation of G(M1) and G(M2) by mammalian sialidases. Biochem. J. 2001;360:233–237. doi: 10.1042/bj3600233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Seyrantepe V, Demir SA, Timur ZK, Von Gerichten J, Marsching C, Erdemli E, Oztas E, Takahashi K, Yamaguchi K, Ates N, Donmez Demir B, Dalkara T, Erich K, Hopf C, Sandhoff R, Miyagi T. Murine Sialidase Neu3 facilitates GM2 degradation and bypass in mouse model of Tay-Sachs disease. Exp. Neurol. 2018;299:26–41. doi: 10.1016/j.expneurol.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 81.Hasegawa T, Sugeno N, Takeda A, Matsuzaki-Kobayashi M, Kikuchi A, Furukawa K, Miyagi T, Itoyama Y. Role of Neu4L sialidase and its substrate ganglioside GD3 in neuronal apoptosis induced by catechol metabolites. FEBS Lett. 2007;581:406–412. doi: 10.1016/j.febslet.2006.12.046. [DOI] [PubMed] [Google Scholar]
- 82.Yu RK, Macala LJ, Taki T, Weinfield HM, Yu FS. Developmental changes in ganglioside composition and synthesis in embryonic rat brain. J. Neurochem. 1988;50:1825–1829. doi: 10.1111/j.1471-4159.1988.tb02484.x. [DOI] [PubMed] [Google Scholar]
- 83.Yu RK, Tsai YT, Ariga T. Functional roles of gangliosides in neurodevelopment: an overview of recent advances. Neurochem. Res. 2012;37:1230–1244. doi: 10.1007/s11064-012-0744-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tettamanti G, Bonali F, Marchesini S, Zambotti V. A new procedure for the extraction, purification and fractionation of brain gangliosides. Biochim. Biophys. Acta. 1973;296:160–170. doi: 10.1016/0005-2760(73)90055-6. [DOI] [PubMed] [Google Scholar]
- 85.Kawai H, Allende ML, Wada R, Kono M, Sango K, Deng C, Miyakawa T, Crawley JN, Werth N, Bierfreund U, Sandhoff K, Proia RL. Mice expressing only monosialoganglioside GM3 exhibit lethal audiogenic seizures. J. Biol. Chem. 2001;276:6885–6888. doi: 10.1074/jbc.C000847200. [DOI] [PubMed] [Google Scholar]
- 86.Sturgill ER, Aoki K, Lopez PH, Colacurcio D, Vajn K, Lorenzini I, Majic S, Yang WH, Heffer M, Tiemeyer M, Marth JD, Schnaar RL. Biosynthesis of the major brain gangliosides GD1a and GT1b. Glycobiology. 2012;22:1289–1301. doi: 10.1093/glycob/cws103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ichikawa N, Iwabuchi K, Kurihara H, Ishii K, Kobayashi T, Sasaki T, Hattori N, Mizuno Y, Hozumi K, Yamada Y, Arikawa-Hirasawa E. Binding of laminin-1 to monosialoganglioside GM1 in lipid rafts is crucial for neurite outgrowth. J. Cell Sci. 2009;122:289–299. doi: 10.1242/jcs.030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wu G, Lu ZH, Andre S, Gabius HJ, Ledeen RW. Functional interplay between ganglioside GM1 and cross-linking galectin-1 induces axon-like neuritogenesis via integrin-based signaling and TRPC5-dependent Ca(2)(+) influx. J. Neurochem. 2016;136:550–563. doi: 10.1111/jnc.13418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Collins BE, Kiso M, Hasegawa A, Tropak MB, Roder JC, Crocker PR, Schnaar RL. Binding specificities of the sialoadhesin family of I-type lectins. Sialic acid linkage and substructure requirements for binding of myelin-associated glycoprotein, Schwann cell myelin protein, and sialoadhesin. J. Biol. Chem. 1997;272:16889–16895. doi: 10.1074/jbc.272.27.16889. [DOI] [PubMed] [Google Scholar]
- 90.Proshin S, Yamaguchi K, Wada T, Miyagi T. Modulation of neuritogenesis by ganglioside-specific sialidase (Neu 3) in human neuroblastoma NB-1 cells. Neurochem. Res. 2002;27:841–846. doi: 10.1023/A:1020269326825. [DOI] [PubMed] [Google Scholar]
- 91.Da Silva JS, Hasegawa T, Miyagi T, Dotti CG, Abad-Rodriguez J. Asymmetric membrane ganglioside sialidase activity specifies axonal fate. Nat. Neurosci. 2005;8:606–615. doi: 10.1038/nn1442. [DOI] [PubMed] [Google Scholar]
- 92.Wands AM, Cervin J, Huang H, Zhang Y, Youn G, Brautigam CA, Matson Dzebo M, Bjorklund P, Wallenius V, Bright DK, Bennett CS, Wittung-Stafshede P, Sampson NS, Yrlid U, Kohler JJ. Fucosylated molecules competitively interfere with cholera toxin binding to host cells. ACS Infect Dis. 2018;4:758–770. doi: 10.1021/acsinfecdis.7b00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Valaperta R, Valsecchi M, Rocchetta F, Aureli M, Prioni S, Prinetti A, Chigorno V, Sonnino S. Induction of axonal differentiation by silencing plasma membrane-associated sialidase Neu3 in neuroblastoma cells. J. Neurochem. 2007;100:708–719. doi: 10.1111/j.1471-4159.2006.04279.x. [DOI] [PubMed] [Google Scholar]
- 94.Kappagantula S, Andrews MR, Cheah M, Abad-Rodriguez J, Dotti CG, Fawcett JW. Neu3 sialidase-mediated ganglioside conversion is necessary for axon regeneration and is blocked in CNS axons. J. Neurosci. 2014;34:2477–2492. doi: 10.1523/JNEUROSCI.4432-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Woronowicz A, Amith SR, De Vusser K, Laroy W, Contreras R, Basta S, Szewczuk MR. Dependence of neurotrophic factor activation of Trk tyrosine kinase receptors on cellular sialidase. Glycobiology. 2007;17:10–24. doi: 10.1093/glycob/cwl049. [DOI] [PubMed] [Google Scholar]
- 96.Zhang Y, Albohy A, Zou Y, Smutova V, Pshezhetsky AV, Cairo CW. Identification of selective inhibitors for human neuraminidase isoenzymes using C4,C7-modified 2-deoxy-2,3-didehydro-N-acetylneuraminic acid (DANA) analogues. J. Med. Chem. 2013;56:2948–2958. doi: 10.1021/jm301892f. [DOI] [PubMed] [Google Scholar]
- 97.Albohy A, Zhang Y, Smutova V, Pshezhetsky AV, Cairo CW. Identification of selective Nanomolar inhibitors of the human neuraminidase, NEU4. AACS Med. Chem. Lett. 2013;4:532–537. doi: 10.1021/ml400080t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Albohy A, Mohan S, Zheng RB, Pinto BM, Cairo CW. Inhibitor selectivity of a new class of oseltamivir analogs against viral neuraminidase over human neuraminidase enzymes. Bioorg. Med. Chem. 2011;19:2817–2822. doi: 10.1016/j.bmc.2011.03.039. [DOI] [PubMed] [Google Scholar]
- 99.Isaeva E, Lushnikova I, Savrasova A, Skibo G, Holmes GL, Isaev D. Blockade of endogenous neuraminidase leads to an increase of neuronal excitability and activity-dependent synaptogenesis in the rat hippocampus. Eur. J. Neurosci. 2010;32:1889–1896. doi: 10.1111/j.1460-9568.2010.07468.x. [DOI] [PubMed] [Google Scholar]
- 100.Minami A, Saito M, Mamada S, Ieno D, Hikita T, Takahashi T, Otsubo T, Ikeda K, Suzuki T. Role of Sialidase in long-term potentiation at mossy Fiber-CA3 synapses and Hippocampus-dependent spatial memory. PLoS One. 2016;11:e0165257. doi: 10.1371/journal.pone.0165257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Minami A, Meguro Y, Ishibashi S, Ishii A, Shiratori M, Sai S, Horii Y, Shimizu H, Fukumoto H, Shimba S, Taguchi R, Takahashi T, Otsubo T, Ikeda K, Suzuki T. Rapid regulation of sialidase activity in response to neural activity and sialic acid removal during memory processing in rat hippocampus. J. Biol. Chem. 2017;292:5645–5654. doi: 10.1074/jbc.M116.764357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Boyzo A, Ayala J, Gutierrez R, Hernandez RJ. Neuraminidase activity in different regions of the seizing epileptic and non-epileptic brain. Brain Res. 2003;964:211–217. doi: 10.1016/S0006-8993(02)03985-9. [DOI] [PubMed] [Google Scholar]