Summary
Acinetobacter baumannii has emerged as a leading nosocomial pathogen, infecting a wide range of anatomic sites including the respiratory tract and the bloodstream. In addition to being multi‐drug resistant, little is known about the molecular basis of A. baumannii pathogenesis. To better understand A. baumannii virulence, a combination of a transposon‐sequencing (TraDIS) screen and the neutropenic mouse model of bacteremia was used to identify the full set of fitness genes required during bloodstream infection. The lytic transglycosylase MltB was identified as a critical fitness factor. MltB cleaves the MurNAc‐GlcNAc bond of peptidoglycan, which leads to cell wall remodeling. Here we show that MltB is part of a complex network connecting resistance to stresses, membrane homeostasis, biogenesis of pili and in vivo fitness. Indeed, inactivation of mltB not only impaired resistance to serum complement, cationic antimicrobial peptides and oxygen species, but also altered the cell envelope integrity, activated the envelope stress response, drastically reduced the number of pili at the cell surface and finally, significantly decreased colonization of both the bloodstream and the respiratory tract.
1. INTRODUCTION
Worldwide, 700,000 deaths are associated with multi‐drug resistant infections per year. If no new antimicrobials are developed, it is estimated that by 2050, the number of deaths associated with these infections will reach 10 million per year, which would exceed those due to cancer and diabetes combined (Shallcross et al., 2015; Willyard, 2017). Recently, the World Health Organization (WHO) reported a list of drug‐resistant bacteria that pose a great threat to human health and for which new antimicrobials are needed (Lawe‐Davies and Bennett, 2017). Accordingly, Acinetobacter baumannii is considered as the number one priority among these bacterial pathogens.
A. baumannii, a gram‐negative, encapsulated bacterium, has emerged as a leading nosocomial pathogen, particularly in intensive care units specializing in respiratory care, trauma and burns (Wong et al., 2017). This bacterium infects a wide range of anatomic sites including the respiratory tract, bloodstream, wounds, urinary tract and meninges (Wong et al., 2017). The high prevalence of infection in immunocompromised, catheterized patients or those suffering from chronic lung diseases is concerning as multi‐drug resistance leaves few, or in some cases, no antimicrobial treatment options (Geisinger and Isberg, 2017; Wong et al., 2017). On average, 62,200 and 1,000,000 bacterial infections per year are caused by A. baumannii in the United States and worldwide respectively (Spellberg and Rex, 2013). Also alarming is its mortality rate, which is about 50% and 36% for ventilator‐associated pneumonia and bloodstream infections respectively (Fagon et al., 1996; Garnacho et al., 2003; Seifert et al., 1995; Wisplinghoff et al., 2004).
Extensive work has been performed to understand the mechanisms mediating drug resistance in A. baumannii. However, its pathobiology is not well understood. Indeed, just a subset of virulence factors has been identified thus far. Capsule, lipooligosaccharide, metal acquisition systems (iron and zinc), secretion systems (Type I, II and VI) and outer membrane proteins (OmpA, Omp33‐66) are among the virulence factors that have been identified (Harding et al., 2017; Wong et al., 2017). To better understand the virulence of A. baumannii, we and other groups have performed transposon‐based screening in vivo (Gebhardt et al., 2015; Subashchandrabose et al., 2016; Wang et al., 2014). While shedding some light on the fitness factors required during infections, these studies have used either a hypovirulent strain or an invertebrate model of infection.
To identify the full set of fitness genes required during bloodstream infection, we performed Transposon‐Directed Insertion site Sequencing (TraDIS) using the virulent, multi‐drug resistant bloodstream isolate AB0057 (Hujer et al., 2006) and a murine model of bacteremia (Smith et al., 2010). The lytic transglycosylase MltB was among the top fitness factors identified in the screen. MltB, a member of the lytic transglycosylase (LT) family, cleaves the glycosidic bond between N‐acetylmuramic acid (MurNAc) and N‐acetylglucosamine (GlcNAc) residues of peptidoglycan (PG), concommittantly forming a 1,6‐anhydro bond in the MurNAc residue (Dik et al., 2017; Höltje et al., 1975; Scheurwater et al., 2008). These enzymes are involved in remodeling of the PG layer and releasing PG fragments (1,6‐anhydro‐muropeptides), and consequently, important for cell wall integrity (Dik et al., 2017). Recently, it was shown that LTs are important for pathogenesis in Neisseria gonorrhoeae, Brucella abortus and Edwardsiella tarda (Bao et al., 2017; Knilans et al., 2017; Liu et al., 2012; Ragland et al., 2017).
In the current study, we demonstrate that the lytic transglycosylase MltB is a critical fitness factor during bacteremia and pneumonia as it connects resistance to stresses, membrane homeostasis, biogenesis of pili and, ultimately, in vivo fitness.
2. RESULTS
2.1. Transposon insertion sequencing screen for in vivo fitness genes
To better understand the pathobiology of multi‐drug resistant A. baumannii (MDRAB) in a vertebrate model, TraDIS experiments were performed using the neutropenic murine model of bacteremia. Due to the limited genetic tools and markers available for use in the MDRAB AB0057 strain, we first engineered a kanamycin‐susceptible strain by creating an in‐frame, markerless, deletion‐mutant of the kanamycin resistance gene AB57_0288. When tested in the neutropenic murine model of bacteremia, this strain colonized the spleen and the liver as well as the parental strain AB0057 (Supporting Information Fig. S1). Since the AB57_0288 mutant strain is as virulent as the parental strain, and is susceptible to kanamycin, in this study, we considered the AB57_0288 mutant as the WT strain. Then, we used the EZ‐Tn5 transposome complex, along with the TypeOne™ Restriction Inhibitor (Epicentre), to generate a random transposon library of 49,628 mutants. The random distribution of the transposon across the chromosome was confirmed by sequencing 20 mutants (Supporting Information Table S1). In total, the library consists of 25,821 unique insertions and according to Goodman et al. (2011), an open reading frame was considered inactivated when at least three insertions were mapped into it.
To determine the full set of genes required during bloodstream infection, the transposon library was divided into five pools of 10,000 mutants and each pool was used to infect four mice each (20 mice in total). Twenty‐four hours post‐inoculation (hpi), the spleens and livers were collected, homogenized and samples from both organs were either plated for CFU enumeration (Supporting Information Fig. S2) or for genomic DNA isolation. Transposon‐gDNA junctions were amplified by PCR from the input and output pools and analyzed by Illumina sequencing to determine the relative abundance of each transposon mutant. Reads were mapped to the chromosome of strain AB0057 and a fitness index was calculated for each transposon mutant after passage into the bloodstream, as previously described by our group (Subashchandrabose et al., 2013; Subashchandrabose et al., 2016). By including the annotated pseudogenes, as well as the transposon insertions within 200 bp from the start codon of the gene, a total of 1,826 genes showed a fitness defect of at least two‐fold and a p value < 0.01 in the spleen (Supporting Information Table S2; Top 25 shown in Table 1). These broad criteria were chosen to identify any potential fitness factors, including the ones in which the transposon is inserted into the respective regulatory region. Although the number of candidate fitness factors seems high, Gebhardt et al. (2015), using the MDRAB AB5075 strain and the Galleria mellonella model of infection, found a comparable number of fitness factors having a fitness defect of at least 2.0 in their Data Set S1.
Table 1.
Top 25 candidate fitness factors.
| Locus tag | Function | Log2 Fitness defecta | P value |
|---|---|---|---|
| AB57_3427 | sulfurtransferase | –10.82 | 0.0096 |
| AB57_0688 | 2‐amino‐4‐hydroxy‐6‐hydroxymethyldihydropteridine pyrophosphokinase | –10.31 | 0.0078 |
| AB57_2595 | LuxR family transcriptional regulator | –10.18 | 0.0003 |
| AB57_3365 | hypothetical protein | –10.11 | 0.0034 |
| AB57_3288 | hypothetical protein | –9.66 | 0.0091 |
| AB57_0173 | hypothetical protein | –9.63 | 0.0052 |
| AB57_3551 | toluene tolerance protein | –9.60 | 0.0062 |
| AB57_3300 | hypothetical protein | –9.57 | 0.0001 |
| AB57_1860 | DNA‐binding protein HU‐beta | –9.52 | 0.0044 |
| AB57_1154 | hypothetical protein | –9.48 | 0.0075 |
| AB57_2881 | diadenosine tetraphosphatase | –9.47 | 0.0031 |
| AB57_3680 | acetyltransferase | –9.37 | 0.0062 |
| AB57_2749 | lytic transglycosylase | –9.36 | 0.0000 |
| AB57_2865 | NUDIX hydrolase | –9.24 | 0.0022 |
| AB57_0530 | 50S ribosomal protein L33 | –9.24 | 0.0001 |
| AB57_0531 | 50S ribosomal protein L28 | –9.24 | 0.0001 |
| AB57_0459 | hypothetical protein | –9.21 | 0.0052 |
| AB57_0336 | glutamyl‐Q tRNA(Asp) ligase | –9.17 | 0.0000 |
| AB57_3836 | phosphoserine phosphatase | –9.16 | 0.0035 |
| AB57_2526 | phosphoribosylamine–glycine ligase | –9.15 | 0.0001 |
| AB57_3700 | endonuclease | –9.15 | 0.0050 |
| AB57_RS18195 | hypothetical protein | –9.09 | 0.0094 |
| AB57_2745 | membrane protein | –9.08 | 0.0000 |
| AB57_0172 | PadR family transcriptional regulator | –9.08 | 0.0094 |
| AB57_0693 | hypothetical protein | –9.07 | 0.0038 |
Calculated fitness defect in the spleen at 24 hpi.
The most representative functional categories of the putative fitness factors identified in the spleen included amino acid transport and metabolism, transcription, general function, translation, and cell wall, membrane and envelope biogenesis (Fig. 1A). To confirm the TraDIS analysis, deletion mutants of six candidate fitness factors among these categories, and having a broad range of fitness defects (Table 2), were constructed and their in vivo fitness was determined using the neutropenic murine model of bloodstream infection. Mono‐infections were performed by injecting 107 CFU of either the WT or each mutant construct into the bloodstream of the neutropenic mouse via the tail vein. At 24 hpi, bacterial burden in the spleen and the liver was determined by CFU enumeration. By comparing the bacterial load between the WT strain and the mutant strains in these organs, five of six mutants showed a fitness defect, corresponding to a validation rate of ≥83% (Fig. 1B and C), which is typical of what we have previously observed in different Tn‐seq experiments in other species (Subashchandrabose et al., 2013; Bachman et al., 2015; Subashchandrabose et al., 2016; Anderson et al., 2017; Armbruster et al., 2017).
Figure 1.
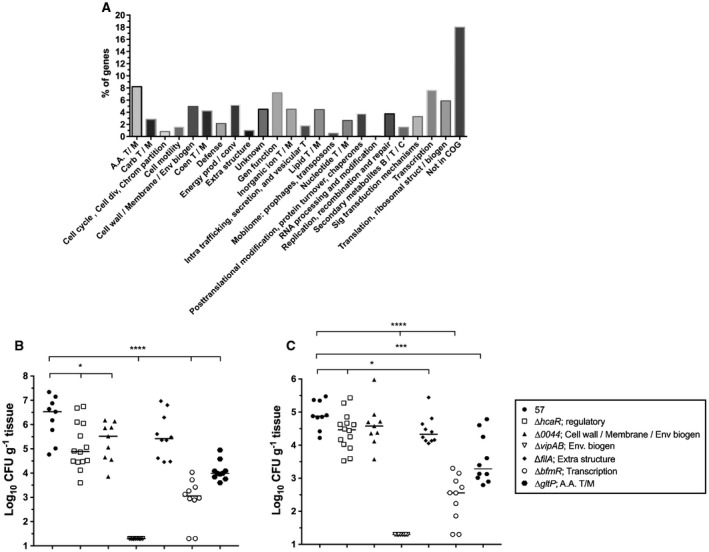
Validation of candidate AB0057 fitness factors during bloodstream infection. A. Classification of the candidate fitness factors required during bloodstream infection according to their Cluster of Orthologous Group (COG). B–C. Candidate fitness factors were confirmed using the neutropenic murine model of bloodstream infected. (B) Colonization of the spleen. (C) Colonization of the liver. CBA/J mice were infected with 107 CFU of either the WT (57) or the mutant strains by tail vein injection. At 24 hpi, mice were sacrificed, spleen and liver were harvested, and the bacterial burden was determined by CFU enumeration on LB agar. Bacterial numbers are presented as the log10 CFU g–1 of tissue. Each data point represents a sample from an individual mouse, and horizontal bars indicate the median values. Statistical significance was calculated by the Mann‐Whitney test (*p < 0.05; ***p < 0.0005; ****p < 0.0001). Abbreviation: A.A. T/M: Amino acids transport/metabolism, Carb T/M: Carbohydrates transport/metabolism, Cell div: Cell division, Chrom: Chromosome, Env biogen: Envelope biogenesis, Coen T/M: Coenzymes transport/metabolism, Prod/conv: production/conversion, Extra: extracellular, Gen: General, T/M: Transport/metabolism, Intra: Intracellular, T: Transport, B/T/C: Biosynthesis/transport/catabolism, Sig: Signal, Struct/biogen: Structure/biogenesis, 57: WT (AB0057Km).
Table 2.
Fitness factors validated in the neutropenic murine model of bacteremia.
| Locus tag | Genes | Function/COG category | TraDIS FDa in spleen | bFD in spleen | bFD in liver |
|---|---|---|---|---|---|
| AB57_0486 | hcaR | Transcriptional regulator/Transcription | 23 | 4 × 101 | 2.5 × 100 |
| AB57_0044 | AB57_0044 | Lytic transglycosylase/Cell Wall, membrane and env. biogen. | 352 | 1 × 101 | NS |
| AB57_0094‐95 | vipAB | Vi polysacharide biosynthesis proteins/Cell wall, membrane and env. biogen. | 436 | 105 | 3.6 × 103 |
| AB57_0739 | filA | Type III pili subunit/Extracellular structure | 177 | NSc | 3.3x100 |
| AB57_0796 | bfmR | Transcriptional regulator/Transcription | 221 | 3 × 103 | 2 × 102 |
| AB57_1698 | gltP | Proton/sodium‐glutamate symport protein/A.A. T/M | 50 | 3.5 × 102 | 3.7 × 101 |
Abbreviation: Env. biogen.: Envelope biogenesis, A.A. T/M: Amino acid transport and metabolism
FD; Fitness defect
Fitness defect was calculated by comparing the colonization burden between the WT and the mutant strain.
NS: not significant
2.2. The lytic transglycosylase gene mltB encodes a fitness factor
In addition to protecting cells from the environment, the cell envelope provides structural integrity to the cell and is associated with fitness in both in vitro and in vivo systems. To ensure its function, homeostasis of the cell envelope is tightly regulated. Due to its crucial importance, we were not surprised to observe 5% of the candidate fitness factors are involved in cell wall, membrane and envelope biogenesis. From this functional category, four genes belonging to the lytic transglycosylase family were identified as candidate fitness factors (Supporting Information Table S3), where the lytic transglycosylase mltB (AB57_2749) showed the greatest fitness defect in the spleen, 655‐fold, and overall, has the 13th greatest fitness defect among all candidate fitness factors (Table 1). An in silico analysis showed that MltB of A. baumannii strain AB0057 has 41% amino acid sequence identity and 56% amino acid similarity with the E. coli homolog MltB. Furthermore, it is also predicted to possess the characteristic SLT_2 and MltB superfamily domains (Supporting Information Table S3).
By cleaving the β‐1,4 glycosidic bond between the MurNAc and the GlcNAc residues of the PG, MltB is involved in the remodeling of PG and releasing of soluble fragments (Dik et al., 2017). Due to its role in maintaining the cell wall integrity, we sought to characterize the contribution of mltB in pathogenesis of A. baumannii. To confirm that mltB encodes a fitness factor in vivo, we constructed an in‐frame, markerless deletion mutant of mltB and tested whether its inactivation affects fitness in the neutropenic murine model of bloodstream infection. Mice were inoculated with 107 CFU of either the WT or the mltB mutant, and at 24 hpi, colonization of the spleen, liver and kidneys was determined by CFU enumeration. Compared to the WT strain, colonization of the spleen, liver and kidneys by the mltB mutant was decreased 185‐, 4‐ and 800‐fold respectively (Fig. 2A–C). Since mltB is the second gene of a two gene‐operon, downstream of mrdB, we complemented the mutation by cloning the entire operon with its native promoter into pABBR_Km and transformed the plasmid into the ∆mltB mutant. Complemention of the mutation in trans restored the WT fitness level in all organs (Fig. 2A–C) and confirmed that mltB encodes a crucial fitness factor in strain AB0057. Importantly, inactivation of mltB does not affect growth rate when cultured in vitro (LB and M9 supplemented with glucose and casamino acids) (Fig. 2D, E), demonstrating that attenuation of the mltB mutant in vivo was not simply due to a growth defect.
Figure 2.
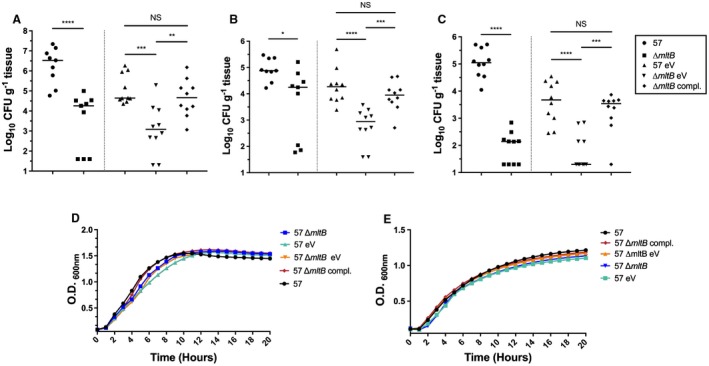
Colonization of the bloodstream by the mltB mutant. Colonization of the spleen (A), liver (B) and kidneys (C) was determined by infecting CBA/J mice with 107 CFU of either the WT strain (57) or its derivative strains. At 24 hpi, mice were sacrificed, organs were harvested, and the bacterial burden was determined by CFU enumeration on LB agar (57 and ∆mltB) and LB‐Km agar (57 eV, 57 ∆mltB eV and 57 ∆mltB compl.). Bacterial numbers are presented as the log10 CFU g–1 of tissue. Each data point represents a sample from an individual mouse, and horizontal bars indicate the median values. Statistical significance was calculated by the Mann‐Whitney test (*p < 0.05; **p < 0.01 ***p < 0.0005; ****p < 0.0001; NS, not significant). D and E. Growth of the WT (57) and its derivative strains. (D) LB. (E) M9 minimal medium supplemented with 0.4% Glucose and 0.2% casamino acids. Results from in vitro experiments are the mean values and standard deviations of three biological experiments. For ease of reading, standard deviations were removed from graphs D and E. Abbreviation: 57: WT (AB0057Km); eV: empty vector (pABBR_Km); compl.: complemented (pABBR_Km‐mrdB‐mltB).
2.3. MltB contributes to stress resistance
As MltB is involved in peptidoglycan turnover and integrity, we hypothesized that its fitness defect in the bloodstream is associated with increased susceptibility to stresses including bactericidal activity of serum, cationic antimicrobial peptides, oxidative and acid stresses, and osmotic shock. To test this hypothesis, we first sought to determine whether the ∆mltB mutant was more susceptible to the bactericidal activity of serum by incubating 107 CFU ml–1 of the WT, ∆mltB and complemented strains with 90% active human serum. The number of CFU was monitored every 60 min for a period of 3 hr. At 3 hpi, the number of CFU recovered from the mltB mutant was 10‐fold lower than the WT and the complemented strain (Fig. 3A; HS). To validate whether the increase in susceptibility of the ∆mltB mutant to human serum was mediated by its complement‐mediated bactericidal activity and not to a decreased fitness in serum itself, the strains were incubated in 90% heat‐inactivated human serum for 3 hr. At 3 hpi, no difference in viability was observed between the WT, the mltB mutant and the complemented strain (Fig. 3A; HI), confirming that the increased susceptibility of the mutant construct was due to the bactericidal activity of complement.
Figure 3.
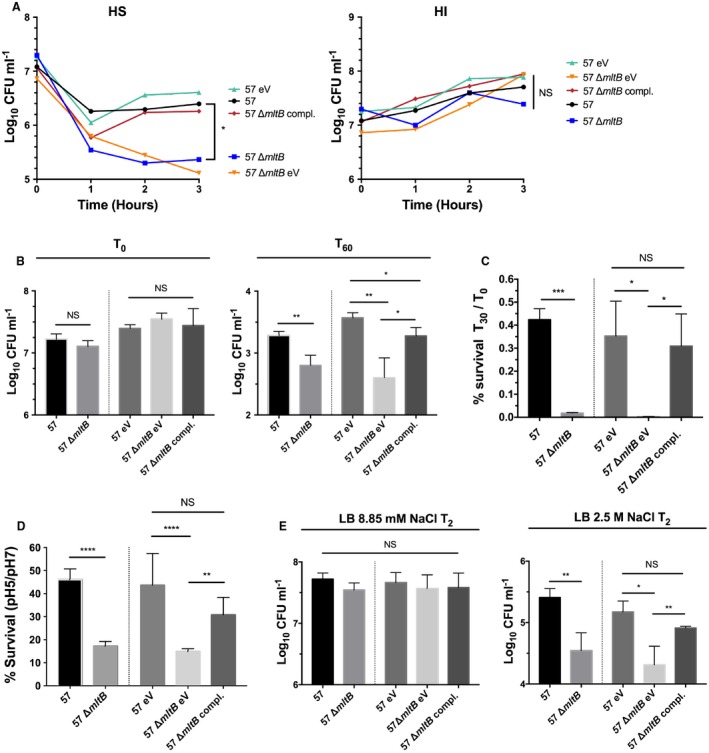
Resistance to stress by the mltB mutant. Resistance to stresses was determined by incubating 107 CFU ml–1 of the WT (57) and its derivative strains to different stressors. A. Survival in 90% human active serum (HS) and growth in 90% heat‐inactivated human serum (HI). The number of surviving CFUs was quantified by CFU enumeration on LB agar every hr. For ease of reading, standard deviations were removed. B. Survival in the presence of 1 µg ml‐1 of polymyxin B. The number of CFUs recovered at 60 min (T60) was determined by CFU enumeration on LB agar and compared to time 0 (T0). C. Survival to oxidative stress (2.5 mM H2O2). Percent survival in 2.5 mM H2O2 was determined by dividing the number of CFU recovered at 30 min (T30) post‐inoculation by the number of CFUs at time 0 (T0). D. Survival in an acidic environment. The percent survival in an acidic environment was determined by dividing the number of CFU recovered at 60 min (T60) post‐inoculation in LB‐pH5 by the number of CFUs recovered in LB‐pH7. E. Survival to osmotic shock. The number of CFUs recovered in LB containing 8.85 mM or 2.5 M NaCl at 2 hr (T2) post‐inoculation was determined by CFU enumeration on LB agar. Results are the mean values and standard deviations of three independent experiments. Statistical significance was calculated by the Student’s t‐test (*p < 0.05; **p < 0.01; ***p < 0.005; NS: Not significant). Abbreviation: 57: WT (AB0057Km), eV: empty vector (pABBR_Km), compl.: complemented (pABBR_Km‐mrdB‐mltB).
Capsule is among the factors contributing to pathogenesis and resistance to serum, notably by interfering with opsonophagocytosis and complement‐mediated killing (Merino et al., 1992; Whitfield, 2006; Bachman et al., 2015; Diao et al., 2017; Anderson et al., 2017). To test whether the increased human serum susceptibility of the ∆mltB mutant was due to a defect in capsule production, we used the Maneval’s stain coupled with light microscopy (Maneval, 1941) and the mucoviscosity assay (Bachman et al., 2015) to assess its production among the WT, ∆mltB and the complemented strains. Although slight differences were observed by microscopy between the strains, the ∆mltB mutant is 2.25‐times less mucoviscous than the WT strain and complementation of the mutation restored the mucoviscosity to the WT level (Supporting Information Fig. S3). The contribution of mltB in resistance to stresses was also validated in the MDRAB strain AB5075 and its isogenic mltB mutant. First, to confirm the role of mltB to the bactericidal activity of the human serum, we tested the survival rate to 90% active human serum as well as their growth in heat‐inactivated serum. At 3 hpi, the number of CFU recovered from the mltB mutant was 327‐times fewer than the AB5075 strain (Supporting Information Fig. S4A; HS). Susceptibility of the AB5075 ∆mltB strain to human serum was also due to the complement‐mediated bactericidal activity, as no difference in growth was observed between the mutant strain and AB5075 in heat‐inactivated human serum (Supporting Information Fig. S4A; HI). As for the AB0057 strain, the mltB mutant of strain AB5075 was 2.43‐less mucoviscous than the WT strain (Supporting Information Fig. S5).
To defend itself against infection, the host secretes antimicrobial peptides as part of its innate defense system. We tested whether the ∆mltB mutant was more susceptible to polymyxin B, a cationic antimicrobial peptide that disrupts membrane integrity. Therefore, 107 CFU ml–1 of the WT, ∆mltB and complemented strains were incubated with 1 µg ml‐1 of polymyxin B for 60 min. As expected, the mltB mutant was more susceptible than the WT strain as the number of CFU recovered from the mltB mutant was 3.1‐fold lower while complementation of the mutation restored the number of CFU to the WT level (Fig. 3B). Similarly, inactivation of mltB in strain AB5075 increased susceptibility to polymyxin B 3.2‐fold compared to the WT strain (Supporting Information Fig. S4B).
The host also protects itself against infection by the production of oxidative, acid and osmotic stresses. To address the contribution of mltB in response to these stresses, we challenged the WT, ∆mltB and complemented mutant with H2O2, HCl and high concentrations of NaCl. First, the AB0057 and the mltB mutant were incubated in the presence of 2.5 mM H2O2 for 30 min. Under this condition, the mltB mutant was 24.7‐times more sensitive to H2O2 compared to the WT strain, and complementation of mltB in trans restored the number of CFU of the mutant strain to the WT level (Fig. 3C). In agreement, the ∆mltB mutant of AB5075 was 17‐times more susceptible than the WT strain to H2O2 (Supporting Information Fig. S4C).
To test whether the mltB mutant is more susceptible to acid, the WT, ∆mltB and the complemented mutant were cultured in LB‐pH5 and survival in acidic environment was evaluated by comparing to their growth in LB‐pH7. At 1 hpi, the mltB mutant showed a 2.7‐fold lower survival rate at low pH than the WT strain. Indeed, the percent survival of the mltB mutant in LB‐pH5 was 17.2%, while the WT strain was 46.1%. In addition, survival of the complemented mutant at low pH was 30.8%, which partially restored survival of the mltB mutant to the WT level (Fig. 3D). In strain AB5075, the same trend was observed as the mltB mutant was two‐times more susceptible to low pH than the WT strain (Supporting Information Fig. S4D).
Finally, we tested whether the mutant strain was more susceptible to osmotic shock. To do so, the WT and the ∆mltB mutant were incubated for 2 hr in the presence of 2.5 M NaCl and the number of surviving cells was enumerated on LB agar. Under this condition, the mltB mutant was 7.3‐times more susceptible to high osmolarity than the WT strain cells (Fig. 3E). In addition to calculating the survival rate in high osmolarity, we wanted to determine the growth rate of the ∆mltB mutant in the presence of different concentration of NaCl. Strains were cultured in LB containing 100, 250, 500 and 750 mM NaCl and the growth rate was monitored by measurement of OD600 every 30 min. Although no growth defect was observed at 100 and 250 mM NaCl, the ∆mltB mutant was unable to grow at 500 and 750 mM NaCl (Supporting Information Fig. S6); complementation of the mutation restored the growth rate of the mltB mutant to the WT level (Fig. 3E and Supporting Information Fig. S6). Surprisingly, mltB did not seem to contribute to osmotic shock in the AB5075 strain as the mltB mutant was as resistant to high osmolarity as the WT strain (Supporting Information Fig. S4E and S7).
Taken together, these results demonstrated that the sensitivity of the mltB mutant to stresses may explain, at least in part, its fitness defect observed during bloodstream infection.
2.4. MltB contributes to the cell envelope homeostasis
Since the mltB mutant has a fitness defect in vivo and is more susceptible to stresses targeting the cell envelope, we hypothesized that the mltB mutant would have altered membrane integrity and consequently, be subject to an envelope stress response (ESR). To test this hypothesis, we first determined whether the membrane of the mltB mutant was more permeable than the WT strain by performing a propidium iodide assay. A mix of Syto 9 and propidium iodide dyes, where Syto 9 (green) stains the nucleic acid all cells and propidium iodide (red) only stains the nucleic acid of damaged (permeable) membrane cells, was used to score the % of permeable cells. By calculating the number of propidium iodide positive cells (red) over the total number of cells, we observed that the membrane of the mltB mutant is two‐times more permeable than the WT strain (Fig. 4A and B), and complementation restored the membrane permeability of the mutant strain to the WT level. To confirm that the increase in the number of permeable cells in the mltB mutant was due to an increased permeability of the membrane, and not to an increased cell death, the number of CFU following the staining was enumerated on LB agar plates. As shown in Fig. 4C, no difference was observed between strains, confirming the increased membrane permeability in the mltB mutant.
Figure 4.
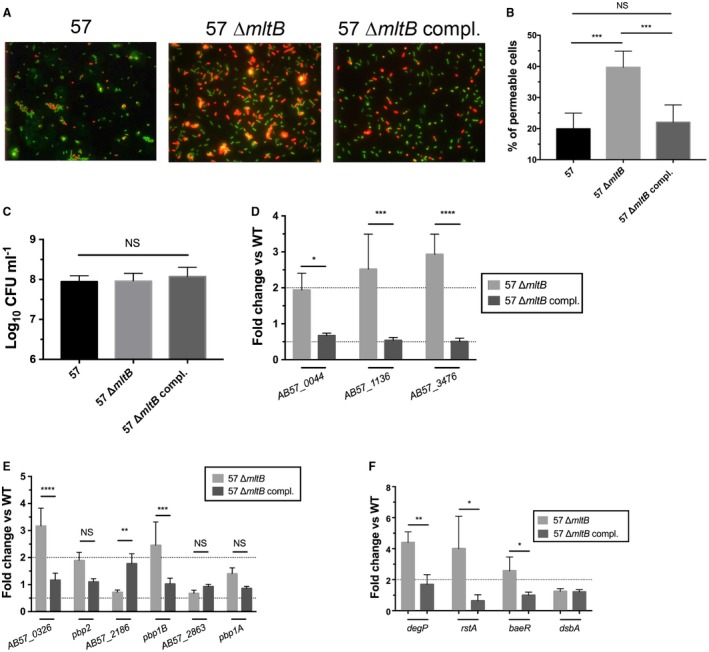
Membrane homeostasis is altered in the mltB mutant. A. Visualization of membrane permeability using fluorescent microscopy. The WT (57) and its derivative strains were stained with a mix of Syto 9 and propidium iodide dyes. Syto 9 dye (green) stains the nucleic acid of all bacteria while the propidium iodide (red) stains the nucleic acid of permeable cells. Images are representative of three independent experiments. B. Percentage of permeable cells was calculated by dividing the number of permeable cells (red) by the total number of bacteria. C. Cell viability from panel (A) and (B) was determined by CFU enumeration after the Syto 9 and propidium iodide staining. D–E–F. Gene expression between the WT, ∆mltB and the complemented strain. (D) Lytic transglycosylases. (E) Penicillin‐binding protein. (F) The envelope stress response (ESR). Gene expression was evaluated by qRT‐PCR and compared between the WT, ∆mltB and the complemented strain. The dashed line corresponds to the cutoff for a significant difference in expression. All results are the mean values and standard deviations of three independent experiments. Statistical significance was calculated by the Student’s t‐test (B and C) and by two‐way ANOVA with Sidak’s multiple comparisons test (D, E and F) (*p < 0.05; **p < 0.01; ***p < 0.0005; ****p < 0.0001; NS, not significant). Abbreviation: 57, WT (AB0057Km); eV: empty vector (pABBR_Km); compl., complemented (pABBR_Km‐mrdB‐mltB).
In E. coli, it has been observed that LTs possess extensive functional redundancy (Heidrich et al., 2002; Lee et al., 2013). Since, the genome of AB0057 encodes four predicted lytic transglycosylases (LTs) (Supporting Information Table S3) (Hamidian et al., 2017), we sought to determine whether the other LTs were induced or repressed in the mltB mutant as a compensatory mechanism that might explain the increase membrane permeability of the mltB mutant. Expression of AB57_0044, AB57_1136 and AB57_3476 was quantified by qRT‐PCR and was compared between the WT, the ∆mltB mutant and the complemented strain. Genes AB57_1136 and AB57_3476 were induced 2.52‐ and 2.93‐fold respectively, in the ∆mltB mutant, and complementation of the mutation restored the WT expression level (Fig. 4D).
Penicillin‐binding proteins (PBP) are a major factor in cell wall biosynthesis. Indeed, the glycosyltransferase and transpeptidase domains of PBP catalyze the final steps of the growth of the PG layer thus conferring its 3D structure (Sung et al., 2009; Typas et al., 2011). Since PBPs are important for cell envelope integrity, and this integrity is altered in the ∆mltB mutant, we hypothesized that expression of PBP genes was affected in the mltB mutant. To test this hypothesis, expression of AB57_0326, pbp2, AB57_2186, pbp1B, AB57_2861 and pbp1A was determined by qRT‐PCR and was compared between the WT, the mltB mutant and the complemented strain. As expected, in the mltB mutant, genes AB57_0326 and pbp1B were induced 3.17‐ and 2.46‐fold respectively, and complementation of the mutation restored the WT expression level (Fig. 4E).
To determine whether inactivation of mltB affected the peptidoglycan structure, the muropeptides from the WT, ∆mltB and the complemented strains were prepared following growth in LB. To mimic growth in the bloodstream, strains were also grown in 50% heat‐inactivated human serum. The extracted muropeptides were then separated by high‐performance liquid chromatography to quantify any difference between strains (Glauner, 1988). As shown in Supporting Information Table S4, no major differences in the muropeptides composition were observed between the WT and the ∆mltB mutant in both conditions, suggesting that inactivation of mltB induces subtle changes in PG composition.
The ESR is a system that senses environmental changes and stresses; and prompts the cell to respond appropriately (Leblanc et al., 2011). Since, the ∆mltB mutant is more susceptible to stresses and shows altered membrane integrity, along with the induction of LTs and PBPs, we tested whether the ESR was induced in the mltB mutant. The ESR has been extensively studied in Enterobacteriaceae (Macritchie and Raivio, 2009; Cabeza et al., 2007; Guest and Raivio, 2016). However, this stress response is not well understood in A. baumannii. By screening homologues of the ESR members in A. baumannii, we quantified, by qRT‐PCR, expression of degP, rstA, baeR and dsbA in the mltB mutant. When compared to the WT strain, expression of degP, rstA and baeR was induced 4.4‐, 4.0‐ and 2.6‐fold in the mltB mutant, while expression of dsbA was not different from the WT strain (Fig. 4F). Complementation of the mltB deletion restored expression of degP, rstA and baeR to the WT level, which demonstrates that inactivation of mltB induced the ESR.
Taken together, these results demonstrate the contribution of mltB in maintenance of the cell envelope homeostasis in A. baumannii. In addition, these results may explain, at least in part, the increased sensitivity to stresses as well as the fitness defect observed in the neutropenic murine model of bacteremia.
2.5. MltB influences binding to abiotic surfaces and epithelial cells
Membrane homeostasis is important for proper assembly and function of membrane‐bound structures. Since this homeostasis is altered in the mltB mutant, we hypothesized that assembly of pili at the cell surface, for example, would be perturbed in the mltB mutant. As pili and adhesins are important factors contributing to binding to both abiotic and biotic surfaces, we first sought to determine, using the crystal violet binding assay, whether the mltB mutant was less able to bind to an abiotic surface and form a biofilm on polystyrene surface. When cultured at 30°C in LB for 24 hr under static conditions, the mltB mutant was 5.0‐times less able to form a biofilm than the WT and complemented strains (Fig. 5A). Similarly, inactivation of mltB in strain AB5075 reduced biofilm formation 2.7‐fold compared to the WT strain (Supporting Information Fig. S8A).
Figure 5.

Pili assembly in the mltB mutant of strain AB0057. A. Biofilm formation in LB at 30°C for 24 hr under static conditions. Crystal violet binding assay was used to monitor biofilm formation. B. Adherence to A549 lung epithelial cells. C. Transmission electron microscopy of the WT (57) and its derivative strains at magnification 20,000×. Images show a typical field of view. Arrows show location of the pili on cell surfaces. D. Colonization of the lungs by the WT (57) and its isogenic mltB mutant. CBA/J mice were infected by intranasal aspiration with 107 CFU of either the WT (57) or the mltB mutant. At 24 hpi, mice were sacrificed, lungs were harvested, and the bacterial burden was determined by CFU enumeration on LB agar. Bacterial numbers are presented as the log10 CFU g–1 of tissue. Each data point represents a sample from an individual mouse, and horizontal bars indicate the median values. Results from in vitro experiments are the mean values and standard deviations of three biological experiments. Statistical significance was calculated by the Student’s t‐test (A and C), and by the Mann–Whitney test (E) (*p < 0.05; **p < 0.01; ***p < 0.0001; NS, not significant). Abbreviations: 57, WT (AB0057Km); eV, empty vector (pABBR_Km); compl., complemented (pABBR_Km‐mrdB‐mltB).
We then wanted to determine whether inactivation of mltB influenced adhesion to host epithelial cells. To address this question, we performed adhesion assay on the human alveolar basal epithelial cells A549. Two hours after addition of bacteria to the tissue culture cells, the ∆mltB mutant adhered to epithelial cells in 3.7‐times fewer numbers than the WT strain, and complementation restored adhesion of the mutant strain to the WT level (Fig. 5B). This was also true for strain AB5075, as inactivation of mltB reduced adhesion to epithelial cells 3.5‐fold compared to the WT strain (Supporting Information Fig. S8B).
It was recently shown that, instead of adhering to the tissue culture cells, some A. baumannii strains preferentially bind to inert surfaces (Lazaro‐Diez et al., 2016). To confirm that the decreased adherence of the mltB mutant to A549 cells was actually associated to a defect in binding to the cell line, instead of to the polystyrene surface of the wells, the adherence assay was performed as described above with the exception that no cells were present in the wells. Two hours after addition of the bacteria to the wells, adhesion to the polystyrene surface was quantified, as above, and no significant difference was observed between the WT, mltB mutant and the complemented strain (Supporting Information Fig. S9A). Accordingly, no significant difference was observed between the WT strain of AB5075 and its mltB isogenic mutant (Supporting Information Fig. S9B). These results confirm that the adhesion defect of the mltB mutant is associated to the decrease ability to bind to A549 cells, and not to the wells of the microtiter plates used.
Since biofilm formation and adhesion to epithelial cells were both decreased in the mltB mutant, we hypothesized that mltB is important for assembly of pili at the cell surface. Accordingly, we hypothesized that inactivation of mltB reduces the presence of these structures at the cell surface. To test this hypothesis, transmission electron microscopy was performed on the WT, ∆mltB mutant, and complemented strains. The WT and the complemented strains presented long appendages, consistent with pili (Alvarez‐Fraga et al., 2016; Moon et al., 2017) at their cell surface. The numbers of pili‐like structures were drastically reduced in the ∆mltB mutant (Fig. 5C), confirming the role of mltB in assembly of pili at the cell surface.
2.6. MltB also significantly contributes to colonization of the respiratory tract
As the number of pili at the cell surface was drastically reduced in ∆mltB, which affected its biofilm formation and adhesion to alveolar epithelial cells, we wondered whether the mltB mutant had a fitness defect during pneumonia. Mouse pulmonary infection was induced by nasal aspiration with 107 CFU per mouse and at 24 hpi, the lungs were harvested, and the colonization burden was evaluated by CFU enumeration. As expected, the ∆mltB strain colonized the lungs with 192‐times fewer CFU than the WT strain (Fig. 5D). As for during bloodstream infection, these results confirm the role of mltB in colonization of the host.
Taken together, these results show that in addition to decrease resistance to stresses and to increase membrane permeability, inactivation of mltB also affects assembly of pili at the cell surfaces, which could be connected to inhibition of biofilm formation, adhesion to host epithelial cells and ultimately, may explain the fitness defects in the murine model of bloodstream and pneumonia infections.
3. DISCUSSION
With its high infection prevalence, mortality rate and resistance to multiple antibiotics, A. baumannii has emerged as a pathogen of concern that poses a serious threat to human health. Indeed, among the bacteriological pathogens for which new antimicrobials are urgently needed, A. baumannii is the number one priority according to the WHO (Lawe‐Davies and Bennett, 2017; Willyard, 2017). Despite having 79 complete genomes and 1795 contig sequences of multiple strains of A. baumannii, the identity of the genes essential for pathogenesis in mammalian hosts is not well known. Indeed, the mechanisms by which A. baumannii colonizes the host, avoids the immune system, and incites tissue damage are not yet well defined. By identifying and characterizing the fitness factors required for survival in vivo, we will be able to design strategies to combat its infections.
To unveil the fitness factors important for colonization of the host, we used a combination of transposon‐based screening (TraDIS) (Langridge et al., 2009) and the neutropenic murine model of bacteremia (Smith et al., 2010). Herein, we identified a total of 1,826 putative fitness factors (Supporting Information Table S2). Although this number seems high, Gebhardt et al. (2015), in their Data Set S1, found a similar number of fitness factors having a fitness defect of at least 2.0 in the MDRAB AB5075 strain using the G. mellonella model of infection. In addition to identifying known fitness factors, such as the Type I and II Secretion Systems (Johnson et al., 2015; Harding et al., 2016; Harding et al., 2017), iron and zinc acquisition systems (Gaddy et al., 2012; Hood et al., 2012; Mortensen et al., 2014; Subashchandrabose et al., 2016) and capsule synthesis (Russo et al., 2010; Gebhardt et al., 2015; Geisinger and Isberg, 2015), which we confirmed in our screen, we also identified novel fitness factors (Supporting Information Table S2), such as the lytic transglycosylase MltB.
By comparing the candidate fitness factors identified in our study with other transposon‐based screening, 109‐ and 46‐ genes were common to the ATCC17978 strain identified in the pneumonia (Wang et al., 2014) and bacteremia (Subashchandrabose et al., 2016) models of infection respectively (Supporting Information Table S5). The use of different strains (hypovirulent vs. virulent), model of infection (cyclophosphamide vs. RB6‐5C6 treated mice) and preparation of the inoculum prior to infection may be among the factors explaining the discrepancies between our current study and the one we previously published (Subashchandrabose et al., 2016). Interestingly, by comparing the putative fitness factors identified in MDRAB AB5075 strain from the G. mellonella larvae model of infection (Gebhardt et al., 2015), 628 genes were in common with ours (Supporting Information Table S5). The mltB gene is among them.
Herein, we determined the contribution of the lytic transglycosylase mltB to in vivo fitness of A. baumannii AB0057. First, we confirmed that mltB encodes an important fitness factor since its inactivation highly compromises colonization of the bloodstream (Fig. 2A–C) and the respiratory tract (Fig. 5D), but does not affect in vitro growth rate (Fig. 2D, E). Second, we showed that mltB is important for resistance to stresses associated to bloodstream infection (Fig. 3 and Supporting Information Fig. S6). Third, we demonstrated that mltB contributes to cell envelope integrity and to a lesser extent, capsule production (Fig. 4 and Supporting Information Fig. S3). Finally, we presented evidences that mltB influences assembly of pili at the cell surface, affects biofilm formation, as well as adherence to human alveolar basal epithelial cells (Fig. 5). The phenotypes described above were also validated in the MDRAB strain AB5075 (Supporting Information Fig. S4, S5 and S8).
The contribution of LTs to pathogenesis was recently observed in N. gonorrhoeae, B. abortus and E. tarda (Liu et al., 2012; Bao et al., 2017; Knilans et al., 2017; Ragland et al., 2017). However, the molecular mechanisms connecting LTs to pathogenesis are not well defined, especially in A. baumannii. Here, we showed that inactivation of mltB impaired resistance to stresses associated with bloodstream infection, such as the bactericidal activity of serum, cationic antimicrobial peptide, oxidative and acid stresses as well as to osmotic shock (Fig. 3 and Supporting Information Fig. S4 and S6). Since the host elicits similar stresses to combat infection, the increased susceptibility of the mltB mutant to these stresses may help explain, at least in part, its decreased virulence in the murine model of bloodstream and pulmonary infections. Our results are also in agreement with what was observed in N. gonorrhoeae and E. tarda. Indeed, in N. gonorrhoeae, a lgtA‐lgtD double mutant is more susceptible to lysozyme and neutrophil elastase (Ragland et al., 2017), while in E. tarda, inactivation of mltA reduces survival in minimal medium, as well as increasing susceptibility to high osmolarity (Liu et al., 2012).
The cell envelope protects the cells against environmental insults, such as the immune system, temperature, pH, osmolarity, toxic compounds and antibiotics (Guest and Raivio, 2016). To adapt to these assaults, bacteria have evolved several ESRs to sense these attacks, monitor defects or damages and to restore the cell envelope homeostasis (Grabowicz and Silhavy, 2017). Accordingly, along with the increased susceptibility to stresses, the mltB mutant presents signs of alteration of its cell envelope integrity. Indeed, inactivation of mltB increases membrane permeability and induces the ESR (Fig. 4A, B and F). Our data are also in agreement with what was observed in N. gonorrhoeae and Pseudomonas aeruginosa in regard to membrane permeability (Lamers et al., 2015; Ragland et al., 2017). However, the mechanisms by which inactivation of LTs leads to increased membrane permeability are unclear.
To determine whether inactivation of mltB affects the composition of the PG, and explains the alteration of the cell envelope integrity of the mutant strain, a complete analysis of the PG composition between the WT, ∆mltB and the complemented strains was performed (Supporting Information Table S4). Not surprisingly, no obvious difference between the WT and the mltB mutant were observed. Accordingly, other groups observed that inactivation of LTs or PBPs may not dramatically affect the composition of the PG (Jorgenson et al., 2014; Boll et al., 2016; Kohler et al., 2007). It is possible that inactivation of mltB induces subtle changes in the peptidoglycan structure that we could not detect. Since it is proposed that LTs are functionally equivalent (Koraimann, 2003, Lee et al., 2013; Scheurwater et al., 2008; Wu et al., 2016; Dik et al., 2017), it is possible that a compensatory mechanism is activated in the mltB mutant to overcome the loss of mltB and may explain the absence of difference in the PG composition between the WT and the mltB mutant. Indeed, expression of two LTs (AB57_1136 and AB57_3476) were induced in the mltB mutant (Fig. 4D).
In addition, we also observed that two PBP (AB57_0326 and pbp1B) and the ESR were induced in the mltB mutant as well (Fig. 4E, F). In E. coli, it was observed that LTs, PBPs and the ESR are interconnected. Indeed, activation of the ESR induces expression of the LT gene slt (Bernal‐Cabas et al., 2015), while inactivation of PBPs activates the ESR (Bernal‐Cabas et al., 2015). It is proposed these three systems are part of a complex regulatory network involved in maintaining the cell envelope integrity. Our results strongly support this hypothesis since inactivation of mltB induces expression of two LTs, two PBPs and activates the ESR. Thus, the molecular mechanisms connecting mltB, LTs, PBPs and the ESR, as well as their contribution to the phenotypes observed in the ∆mltB mutant remain to be determined.
In addition to being involved in membrane homeostasis, it is postulated that peptidoglycan degrading enzymes, such as LTs and endopeptidases, act as bacterial ‘space‐making’ autolysins (Scheurwater et al., 2008; Stohl et al., 2013). Accordingly, it was observed they are required for assembly of flagella and/or pili and in Caulobacter crescentus, Neisseria gonorrhoeae, and Rhodobacter sphaeroides (Viollier and Shapiro, 2003; Stohl et al., 2013; Herlihey et al., 2016), the type III secretion system in Xanthomonas campestris pv. Vesicatoria (Hausner et al., 2017) and the type VI secretion system in E. coli and A. baumannii (Weber et al., 2016; Santin and Cascales, 2017). Given these observations, it was not surprising to note that the ∆mltB mutant was devoid of pili at its cell surface (Fig. 5C). Lack of pili at the cell surface of the mltB mutant appears to be associated to its reduction in biofilm formation as well as adherence to human alveolar basal epithelial cells A549 (Fig. 5A, B), and may explain, at least in part, its decreased in vivo fitness. In addition, in N. gonorrhoeae, it was observed that inactivation of the dd‐carboxypeptidase and endopeptidase NGO1686 gene, encoding a peptidoglycan degrading enzyme, increased susceptibility to H2O2, which is associated with the lack of piliation of the mutant strain (Stohl et al., 2012; Stohl et al., 2013). In our study, it may be possible that in the ∆mltB mutant, the increased sensitivity of the mltB mutant to H2O2 is due to the dramatic decrease of pili at its cell surface. The contribution of these pili to adherence, resistance to H2O2 as well as in pathogenesis remains to be determined.
In summary, in this study we have demonstrated that mltB encodes an important fitness factor during bloodstream and pulmonary infections. In addition, we have shown that mltB is part of a complex network connecting membrane homeostasis, resistance to stresses, assembly of pili and consequently, pathogenesis. Furthermore, since its crucial importance in the physiology of not only A. baumannii, but also other pathogens, MltB or other LTs, could be considered a prime target for the development of therapeutics agents to manage or prevent infections.
4. EXPERIMENTAL PROCEDURES
4.1. Bacterial strains, plasmids and growth media
Strains and plasmids used in this study are listed in Supporting Information Table S6. Bacteria were cultured in Lysogeny Broth (LB) at 37°C. Bacteria were also cultured in M9 minimal medium supplemented with 0.4% glucose and 0.2% casamino acids. Antibiotics and reagents were added as required at the following concentrations: kanamycin, 50 µg ml–1; ampicillin, 100 µg ml–1; zeocin, 10 µg ml–1 (E. coli) and 200 µg ml–1 (A. baumannii), amikacin, 10 µg ml–1; diaminopimelic acid (DAP), 50 µg ml–1 and sucrose, 10% wt/vol.
4.2. Construction of non‐polar mutants, transposon library and complemented strain
Primers used in this study are listed in Supporting Information Table S7. Non‐polar mutants were generated using homologous recombination (Aranda et al., 2010) and allelic exchange (Donnenberg and Kaper, 1991). Homologous recombination was used to delete the AB57_0288 gene (conferring kanamycin resistance). Briefly, the AB57_0288 gene, flanked by ~1 Kb, was PCR‐amplified and cloned into the plasmid pSU2719. Then, a recombineering approach (Yu et al., 2000) was used to replace the AB57_0288 gene with the sh_ble cassette flanked with the FRT sites from plasmid pKD_zeo. Then, the mutated fragment was PCR‐amplified, with the 1 Kb flanking region, and electroporated into AB0057 as described by Aranda et al. (2010). Following confirmation of the homologous recombination, the sh_ble cassette was excised using a Km‐modified version of pAT03 (Tucker et al., 2014), pAT03_Km. The AB0057 ∆AB57_0288 strain was considered as the WT strain in this study and named AB0057Km (WT).
Random Tn5 transposon insertion mutants were generated in A. baumannii strain AB0057Km (WT). Briefly, EZ‐Tn5 transposome (Epicentre) complexes were electroporated into AB0057Km (WT) according to Jacobs et al. (2014). To increase transformation efficiency, the TypeOne™ Restriction Inhibitor (Epicentre) was added to the electroporation mixture. Based on the size of the genome (4.05 Mb), 34,000 transposon insertion mutants were required to achieve 99.99 % genome saturation coverage confidence (Zilsel et al., 1992). In total, 49,628 transposon mutants were generated and archived in pools of 5,000 mutants.
Generation of in‐frame markerless mutants was achieved by allelic exchange using a modified version of pCVD442 plasmid (Donnenberg and Kaper, 1991). Briefly, the 5ʹ end of the gene to be deleted possessed at least 1 Kb including the initiation codon and a 6‐nt restriction site, while the 3ʹ region consisted of at least 1 Kb including the last 7 codons and a 6‐nt restriction site. The 5ʹ and 3ʹ regions were cloned into pCVD442_MCS_Amk, which results in the in‐frame deletion of the internal region of the gene of interest. The construct was transformed into the donor strain MGN617, and was transferred to the AB0057Km (WT) strain by conjugation. Transconjugants were selected on LB agar containing amikacin. Individual colonies were cultured 2 hr in LB broth, diluted and spread on LB agar plates containing 10% (wt/vol) sucrose to select the second recombination event. Sucrose‐resistant and amikacin‐sensitive isolates were screened by PCR to confirm deletion of the gene of interest.
Complementation of the mltB deletion was achieved by cloning the mrdB‐mltB operon, including 183 nt upstream of mrdB, in the pABBR_Km plasmid.
4.3. Mouse infection experiments
All procedures involving the use of mice were performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals (8th edition) and were approved by the University Committee on Use and Care of Animals at the University of Michigan (PRO00007111).
Mice were anesthetized with a weight‐appropriate dose (0.1 ml for a mouse weighing 20 gm) of ketamine/xylazine (80–120 mg kg–1 ketamine and 5–10 mg kg–1 xylazine) by IP injection (model of pneumonia). Mice were euthanized by inhalant anesthetic overdose followed by vital organ removal. All infections performed in this study were mono‐infection. Neutrophils were depleted by intraperitoneal injection of 500 µg of rat anti‐mouse monoclonal antibody (MAb) RB6‐8C5 (RB6) (BioXCell) 24 hpi (Conlan and North, 1994; van Faassen et al., 2007).
For the murine model of bacteremia, infections were performed as described previously (Smith et al., 2010), in which female CBA/J mice aged from 6‐ to 8‐week‐old were inoculated via tail vein injection with 107 CFU. At 24 hpi, mice were euthanized and the spleen, liver and kidneys were aseptically removed, homogenized, diluted and plated on LB‐agar plates to determine the colonization level in these organs.
For the murine model of pneumonia, infections were performed as described elsewhere (Jacobs et al., 2010) with slight modifications. Briefly, female CBA/J mice aged from 6‐ to 8‐week‐old were anesthetized with ketamine/xylazine and pneumonia was induced by intranasal inoculation of 107 CFU suspended in a volume of 20 µl of PBS (10 µl per nostril). At 24 hpi, mice were euthanized and lungs were aseptically removed, homogenized, diluted and plated onto LB agar plates to determine the bacterial burden.
4.4. In vivo screen for A. baumannii fitness factors
Mice were inoculated with transposon library pools as described above. Preparation of the input (inoculum) and output (24 hpi) pools were prepared as described by Anderson et al. (2017). Pools of 10,000 mutants (5 pools total; 50,000 mutants) were used to infect four mice each (20 mice total). Two aliquots of 1 ml of each inoculum suspension (input) were collected by centrifugation and stored at −80°C for subsequent isolation of genomic DNA.
4.5. Illumina sequencing
Illumina sequencing was performed as described by Subaschandrabose et al. (Subashchandrabose et al., 2013; Subashchandrabose et al., 2016). Briefly, genomic DNA from the input (5 pools of 2 inocula each) and the output (infected spleens, 5 pools of 4 mice each) was isolated by phenol/chloroform/isoamyl alcohol extraction. Genomic DNA (5 µg) was sheared to yield fragments of ≈300 bp (Covaris). Illumina TruSeq adapters were ligated to DNA fragments. Transposon‐gDNA junctions were enriched by PCR using the Tn‐specific primer and the TruSeq Indexed adapter_barcode primers (Supporting Information Table S7). Twenty‐five ng of the TruSeq libraries were used as template for 28 cycles of amplification. Amplicons were further processed for Illumina sequencing according to manufacturer’s recommendations and sequenced, using the Tn‐specific primer, on an Illumina HiSeq 2000 sequencer using the 50‐nucleotide single‐end read cycle. Libraries from input and output samples were sequenced on the same lane, in triplicate. Libraries preparation and sequencing were performed at the University of Michigan DNA core facility.
4.6. Mapping of transposon insertion sites
Reads from the input and output libraries starting with AGACAG, corresponding to the end of the transposon, and having more than 15 bp, were aligned to the genome of A. baumannii AB0057 (NCBI accession no. NC_011586.1) using the short‐read aligner BOWTIE (Langmead et al., 2009). One nucleotide mismatch was allowed during mapping to the chromosome. Fitness factors were identified by comparing the number of reads that map to a given chromosomal location in the input and output libraries based on the statistical cutoff of fold‐change >2.0 and adjusted p < 0.01.
4.7. Resistance of A. baumannii to human serum, polymyxin B, acid, oxidative stress and osmotic shock
Growth of A. baumannii in human serum was performed as previously described (Lamarche et al., 2005; Crepin et al., 2012). Briefly, bacteria were cultured overnight in LB broth at 37°C. Bacterial cultures were resuspended 1:100 in fresh medium and grown to mid‐log growth phase (OD600 = 0.6). Bacteria were washed with PBS and 107 CFU ml–1 were incubated either with 90% heat‐inactivated or 90% normal human serum (Innovative Research). Suspensions were incubated at 37°C and viable cell counts were determined at 0, 1, 2 and 3 hr post‐incubation on LB agar plates.
Resistance to polymyxin B was assessed as described by Crepin et al. (2012) with slight modifications. Briefly, bacteria were cultured as described above and 107 CFU ml‐1 were incubated with 1 µg ml‐1 of polymixin B for 60 min. The number of bacteria that survived the treatment was determined by CFU enumeration on LB agar.
Resistance to acid was performed as described by Lamarche et al. (2005). Bacteria were cultured to mid‐log phase of growth as described above and 107 CFU ml‐1 were resuspended in either LB (LB‐pH7) or 100 mM MES‐buffered LB (LB‐pH5). Percent survival at 1 hpi was calculated by dividing the number of CFU recovered from LB‐pH5 by the number of CFU recovered from LB‐pH7.
Resistance to oxidative stress was assessed by culturing bacteria as described above and by mixing 107 CFU ml–1 to LB supplemented with 2.5 mM H2O2. At 30 min post‐inoculation, viable cell counts were determined and percent survival was calculated by dividing the number of CFU recovered by the number of CFU at time 0.
Resistance to osmotic shock was measured as described previously (Lamers et al., 2015). Bacteria were cultured as described above and 107 CFU ml‐1l were mixed with either LB containing either 8.55 mM (low salt) or 2.5 mM (high salt) NaCl. The number of cells that survive the treatment was determined by CFU enumeration onto LB agar plate.
The growth of A. baumannii in the presence of defined concentrations of NaCl (0, 100, 250, 500 and 750 mM) was measured in LB medium. Strains were cultured overnight in LB without NaCl, washed once in PBS and the OD600 was adjusted to 0.1 in medium with the corresponding NaCl concentration. Growth was measured by OD600 determination every 30 min with a BioScreen C Analyzer at 37°C with continuous shaking.
4.8. Quantitative RT‐PCR
Strains were cultured as described above and RNA was extracted using TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer’s recommendations. RNA samples were submitted to a rigorous DNase treatment using Turbo DNA‐free (Ambion) to remove any DNA contamination. The iScript cDNA synthesis kit and the SsoFast Evagreen Supermix kit (Bio‐Rad) were used for qRT‐PCR analysis according to the manufacturer’s instructions. The gyrB gene was used as a housekeeping control (Anderson et al., 2017). Gene expression was calculated using the 2−ΔΔ CT method (Livak and Schmittgen, 2001). Genes with a fold‐change above or below the defined threshold of two were considered as differentially expressed. Primers used for qRT‐PCR analysis are listed in Supporting Information Table S7.
4.9. Measurement of membrane permeability using Syto 9 and propidium iodide dyes
Bacteria cultured to mid‐log phase of growth were exposed to BacLight viability dyes propidium iodide and Syto 9 (Thermo Fisher Scientific). Three fields per slide were captured per biological replicate and ~300 cells per biological replicate were counted. Percent of A. baumannii bacteria positive for propidium iodide staining, an indication of a permeable membrane, was calculated by dividing the propidium iodide‐positive bacteria by the total number of bacteria. Images were captured with a Zeiss Axioplan 2 epifluorescence microscope equipped with a 100× Plan‐Neofluor objective with a numerical aperture of 1.3. Images were analyzed and processed with FIJI (Schindelin et al., 2012).
4.10. Biofilm formation
Biofilm formation was measured as previously described (Subashchandrabose et al., 2013) with slight modifications. Briefly, bacteria were cultured in LB overnight, washed twice in PBS and normalized to an OD600 of 0.05 in 1 ml of fresh LB. Cultures were incubated in polystyrene culture tubes at 30°C for 24 hr under static conditions. Supernatants were aspirated and tubes were washed three times with water and stained with 1.5 ml of 1.0% crystal violet solution for 10 min. Biofilm‐bound crystal violet was dissolved in 2 ml of 33% acetic acid and absorbance was measured at 540 nm.
4.11. Adhesion assay
The adenocarcinomic human alveolar basal epithelial cells A549 (American Type Culture Collection ATCC® CCL‐185™) were cultured to confluence in 24‐well plates in Kaighn’s Modification of Ham’s F‐12 Medium (ATCC® 30‐2004™) supplemented with 10% heat‐inactivated fetal bovine serum (FBS) at 37°C and 5% CO2. Bacterial strains were cultured overnight in LB, washed twice with PBS and adjusted to 107 CFU ml–1 in Kaighn’s Modification of Ham’s F‐12 Medium supplemented with 10% heat‐inactivated FBS. The mixture was added to each well containing 106 A549 cells (MOI of 10). Bacterium‐host cell contact was enhanced by a 5‐min centrifugation at 600 × g. At 2 hr post‐incubation, cells were washed 3 times with DPBS (removing the non‐adherent bacteria), lysed with 0.25 % Triton X‐100 for 5 min and then, serially diluted for CFU enumeration. No difference in survival rate between strains were observed at 5 min post‐incubation with Triton X‐100 (data not shown). Quantification of cell‐associated bacteria was performed as previously described (Crepin et al., 2012).
4.12. Transmission electron microscopy
Transmission electron microscopy was performed as described previously (Subashchandrabose et al., 2013) with slight modifications. Briefly, bacterial strains were cultured overnight in LB, washed twice in PBS and adjusted in PBS to an OD600 of 1.0. Ten µl were pipetted onto Formvar/Carbon 300 Mesh Copper Grids (Ted Pella). Bacteria were allowed to adhere to the grids for 5 min, then excess liquid medium was wicked off with filter paper. Grids were washed once with 10 µl of deionized water, then stained for 5 min with 10 µl of 1% phosphotungstic acid (pH 6.8). Excess stain was removed; grids were washed with 10 µl of deionized water and dried. Grids were visualized using a JEOL JSM 1400 plus transmission electron microscope at Microscopy & Image Analysis Laboratory of the University of Michigan.
4.13. Peptidoglycan analysis
The PG was extracted and analyzed according to B. Glauner (1988). Briefly, A. baumannii strains were grown in LB or 50% heat‐inactivated human serum (Innovative Research) to ~108 CFU ml–1. Cells were then collected by centrifugation, resuspended in 6 ml ice‐cold water and lysed by drop wise addition to 6 ml boiling 8% SDS. The PG was purified and digested with the muramidase cellosyl (Hoechst, Frankfurtam Main, Germany) to release the muropeptides, which were reduced by sodium borohydride, and separated on a Prontosil 120‐3‐6C18 AQ reversed phase column (Bischoff, Leonberg, Germany). The eluted muropeptides were detected by their absorbance at 205 nm.
4.14. Statistical analyses
All data were analyzed using the GraphPad Prism 7 software program. A Mann–Whitney test was used to determine statistical significance for mono‐infection experiments. All other statistical analyses were determined by the Student’s t‐test and either one‐ or two‐way analysis of variance (ANOVA) with Tukey’s or Sidak’s multiple comparison test.
Author contributions
S.C. and H.L.T.M. designed the experiments. S.C., E.N.O., K.P., S.N.S and S.D.H. performed the experiments. S.C., E.N.O. and H.L.T.M. analyzed the data. H.L.T.M. and W.V. contributed funding and resources. S.C. and H.L.T.M. wrote the manuscript. All authors reviewed, edited and approved the manuscript.
Supporting information
Acknowledgements
We acknowledge Weisheng Wu and Lili Zhao (University of Michigan) for bioinformatic expertise; Sargurunathan Subaschandrabose (Wake Forest University), Mark T. Anderson, Christopher J. Alteri and Valerie Forsyth (University of Michigan) for their technical expertise. We thank Maria Sandkvist (University of Michigan) for her critical reading of the manuscript. We would also like to thank Bryan W. Davies (University of Texas, Austin), Charles M. Dozois (INRS – Institut Armand‐Frappier), Colin Manoil (University of Washington) and Michele S. Swanson (University of Michigan) for sharing strains, plasmids and instruments.
Funding Information
This work was supported by the Natural Sciences and Engineering Research Council of Canada PostDoctoral Fellowship (S.C), the Public Health Service Grant AI107184 from the National Institutes of Health, the University of Michigan Medical School Host Microbiome Initiative (H.L.T.M.) and the Wellcome Trust grant 101824/Z/13/Z (W.V.). The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript. The authors declare that they have no conflicts of interest with the contents of this article.
REFERENCES
- Alvarez‐Fraga, L. , Perez, A. , Rumbo‐Feal, S. , Merino, M. , Vallejo, J. A. , Ohneck, E. J. , et al. (2016) Analysis of the role of the LH92_11085 gene of a biofilm hyper‐producing Acinetobacter baumannii strain on biofilm formation and attachment to eukaryotic cells. Virulence 7: 443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, M. T. , Mitchell, L. A. , and Mobley, H. L. T. (2017) Cysteine biosynthesis controls Serratia marcescens phospholipase activity. J Bacteriol 199: e00159–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, M. T. , Mitchell, L. A. , Zhao, L. , and Mobley, H. L. T. (2017) Capsule production and glucose metabolism dictate fitness during Serratia marcescens bacteremia. MBio 8: e00740–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda, J. , Poza, M. , Pardo, B. G. , Rumbo, S. , Rumbo, C. , Parreira, J. R. , et al. (2010) A rapid and simple method for constructing stable mutants of Acinetobacter baumannii . BMC Microbiol 10: 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster, C. E. , Forsyth‐DeOrnellas, V. , Johnson, A. O. , Smith, S. N. , Zhao, L. , Wu, W. , et al. (2017) Genome‐wide transposon mutagenesis of Proteus mirabilis: Essential genes, fitness factors for catheter‐associated urinary tract infection, and the impact of polymicrobial infection on fitness requirements. PLoS Pathog 13: e1006434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachman, M. A. , Breen, P. , Deornellas, V. , Mu, Q. , Zhao, L. , Wu, W. , et al. (2015) Genome‐wide identification of Klebsiella pneumoniae fitness genes during lung infection. MBio 6: e00775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao, Y. , Tian, M. , Li, P. , Liu, J. , Ding, C. , and Yu, S. (2017) Characterization of Brucella abortus mutant strain ∆22915, a potential vaccine candidate. Vet Res 48: 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal‐Cabas, M. , Ayala, J. A. , and Raivio, T. L. (2015) The Cpx envelope stress response modifies peptidoglycan cross‐linking via the L, D‐transpeptidase LdtD and the novel protein YgaU. J Bacteriol 197: 603–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boll, J. M. , Crofts, A. A. , Peters, K. , Cattoir, V. , Vollmer, W. , Davies, B. W. , et al. (2016) A penicillin‐binding protein inhibits selection of colistin‐resistant, lipooligosaccharide‐deficient Acinetobacter baumannii . Proc Natl Acad Sci USA 113: E6228–E6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabeza, M. L. , Aguirre, A. , Soncini, F. C. , and Vescovi, E. G. (2007) Induction of RpoS degradation by the two‐component system regulator RstA in Salmonella enterica . J Bacteriol 189: 7335–7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlan, J. W. , and North, R. J. (1994) Neutrophils are essential for early anti‐Listeria defense in the liver, but not in the spleen or peritoneal cavity, as revealed by a granulocyte‐depleting monoclonal antibody. J Exp Med 179: 259–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crepin, S. , Houle, S. , Charbonneau, M. E. , Mourez, M. , Harel, J. , and Dozois, C. M. (2012) Decreased expression of type 1 fimbriae by a pst mutant of uropathogenic Escherichia coli reduces urinary tract infection. Infect Immun 80: 2802–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao, J. , Bouwman, C. , Yan, D. , Kang, J. , Katakam, A. K. , Liu, P. , et al. (2017) Peptidoglycan association of murein lipoprotein is required for KpsD‐dependent group 2 capsular polysaccharide expression and serum resistance in a uropathogenic Escherichia coli isolate. MBio 8: e00603–e00617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dik, D. A. , Marous, D. R. , Fisher, J. F. , and Mobashery, S. (2017) Lytic transglycosylases: concinnity in concision of the bacterial cell wall. Crit Rev Biochem Mol Biol 52: 503–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnenberg, M. S. , and Kaper, J. B. (1991) Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive‐selection suicide vector. Infect Immun 59: 4310–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagon, J. Y. , Chastre, J. , Domart, Y. , Trouillet, J. L. , and Gibert, C. (1996) Mortality due to ventilator‐associated pneumonia or colonization with Pseudomonas or Acinetobacter species: assessment by quantitative culture of samples obtained by a protected specimen brush. Clin Infect Dis 23: 538–542. [DOI] [PubMed] [Google Scholar]
- Gaddy, J. A. , Arivett, B. A. , McConnell, M. J. , Lopez‐Rojas, R. , Pachon, J. , and Actis, L. A. (2012) Role of acinetobactin‐mediated iron acquisition functions in the interaction of Acinetobacter baumannii strain ATCC 19606T with human lung epithelial cells, Galleria mellonella caterpillars, and mice. Infect Immun 80: 1015–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnacho, J. , Sole‐Violan, J. , Sa‐Borges, M. , Diaz, E. , and Rello, J. (2003) Clinical impact of pneumonia caused by Acinetobacter baumannii in intubated patients: a matched cohort study. Crit Care Med 31: 2478–2482. [DOI] [PubMed] [Google Scholar]
- Gebhardt, M. J. , Gallagher, L. A. , Jacobson, R. K. , Usacheva, E. A. , Peterson, L. R. , Zurawski, D. V. , et al. (2015) Joint transcriptional control of virulence and resistance to antibiotic and environmental stress in Acinetobacter baumannii. MBio 6: e01660– e01615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisinger, E. , and Isberg, R. R. (2015) Antibiotic modulation of capsular exopolysaccharide and virulence in Acinetobacter baumannii . PLoS Pathog 11: e1004691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisinger, E. , and Isberg, R. R. (2017) Interplay between antibiotic resistance and virulence during disease promoted by multidrug‐resistant bacteria. J Infect Dis 215: S9–S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glauner, B. (1988) Separation and quantification of muropeptides with high‐performance liquid chromatography. Anal Biochem 172: 451–464. [DOI] [PubMed] [Google Scholar]
- Goodman, A. L. , Wu, M. , and Gordon, J. I. (2011) Identifying microbial fitness determinants by insertion sequencing using genome‐wide transposon mutant libraries. Nat Protoc 6: 1969–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabowicz, M. , and Silhavy, T. J. (2017) Envelope stress responses: An interconnected safety net. Trends Biochem Sci 42: 232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guest, R. L. , and Raivio, T. L. (2016) Role of the Gram‐negative envelope stress response in the presence of antimicrobial agents. Trends Microbiol 24: 377–390. [DOI] [PubMed] [Google Scholar]
- Hamidian, M. , Venepally, P. , Hall, R. M. , and Adams, M. D. (2017) Corrected genome sequence of Acinetobacter baumannii strain AB0057, an antibiotic‐resistant isolate from lineage 1 of global clone 1. Genome Announc 5: e00836–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding, C. M. , Kinsella, R. L. , Palmer, L. D. , Skaar, E. P. , and Feldman, M. F. (2016) Medically relevant Acinetobacter species require a type II secretion system and specific membrane‐associated chaperones for the export of multiple substrates and full virulence. PLoS Pathog 12: e1005391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding, C. M. , Pulido, M. R. , Di Venanzio, G. , Kinsella, R. L. , Webb, A. I. , Scott, N. E. , et al. (2017) Pathogenic Acinetobacter species have a functional type I secretion system and contact‐dependent inhibition systems. J Biol Chem 292: 9075–9087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausner, J. , Hartmann, N. , Jordan, M. , and Buttner, D. (2017) The predicted lytic transglycosylase HpaH from Xanthomonas campestris pv. vesicatoria associates with the type III secretion system and promotes effector protein translocation. Infect Immun 85: e00788–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidrich, C. , Ursinus, A. , Berger, J. , Schwarz, H. , and Höltje, J. V. (2002) Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli . J Bacteriol 184: 6093–6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlihey, F. A. , Osorio‐Valeriano, M. , Dreyfus, G. , and Clarke, A. J. (2016) Modulation of the lytic activity of the dedicated autolysin for flagellum formation SltF by flagellar rod proteins FlgB and FlgF. J Bacteriol 198: 1847–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höltje, J. V. , Mirelman, D. , Sharon, N. , and Schwarz, U. (1975) Novel type of murein transglycosylase in Escherichia coli . J Bacteriol 124: 1067–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood, M. I. , Mortensen, B. L. , Moore, J. L. , Zhang, Y. , Kehl‐Fie, T. E. , Sugitani, N. , et al. (2012) Identification of an Acinetobacter baumannii zinc acquisition system that facilitates resistance to calprotectin‐mediated zinc sequestration. PLoS Pathog 8: e1003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hujer, K. M. , Hujer, A. M. , Hulten, E. A. , Bajaksouzian, S. , Adams, J. M. , Donskey, C. J. , et al. (2006) Analysis of antibiotic resistance genes in multidrug‐resistant Acinetobacter sp isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob Agents Chemother 50: 4114–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, A. C. , Hood, I. , Boyd, K. L. , Olson, P. D. , Morrison, J. M. , Carson, S. , et al. (2010) Inactivation of phospholipase D diminishes Acinetobacter baumannii pathogenesis. Infect Immun 78: 1952–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, A. C. , Thompson, M. G. , Gebhardt, M. , Corey, B. W. , Yildirim, S. , Shuman, H. A. , et al. (2014) Genetic manipulation of Acinetobacter baumannii . Curr Protoc Microbiol 35: 6G 2 1–11. [DOI] [PubMed] [Google Scholar]
- Johnson, T. L. , Waack, U. , Smith, S. , Mobley, H. , and Sandkvist, M. (2015) Acinetobacter baumannii is dependent on the type II secretion system and its substrate LipA for lipid utilization and in vivo fitness. J Bacteriol 198: 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgenson, M. A. , Chen, Y. , Yahashiri, A. , Popham, D. L. , and Weiss, D. S. (2014) The bacterial septal ring protein RlpA is a lytic transglycosylase that contributes to rod shape and daughter cell separation in Pseudomonas aeruginosa . Mol Microbiol 93: 113–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knilans, K. J. , Hackett, K. T. , Anderson, J. E. , Weng, C. , Dillard, J. P. , and Duncan, J. A. (2017) Neisseria gonorrhoeae lytic transglycosylases LtgA and LtgD reduce host innate immune signaling through TLR2 and NOD2. ACS Infect Dis 3: 624–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler, P. L. , Hamilton, H. L. , Cloud‐Hansen, K. , and Dillard, J. P. (2007) AtlA functions as a peptidoglycan lytic transglycosylase in the Neisseria gonorrhoeae type IV secretion system. J Bacteriol 189: 5421–5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koraimann, G. (2003) Lytic transglycosylases in macromolecular transport systems of Gram‐negative bacteria. Cell Mol Life Sci 60: 2371–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamarche, M. G. , Dozois, C. M. , Daigle, F. , Caza, M. , Curtiss, R. 3rd , Dubreuil, J. D. , et al. (2005) Inactivation of the pst system reduces the virulence of an avian pathogenic Escherichia coli O78 strain. Infect Immun 73: 4138–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamers, R. P. , Nguyen, U. T. , Nguyen, Y. , Buensuceso, R. N. , and Burrows, L. L. (2015) Loss of membrane‐bound lytic transglycosylases increases outer membrane permeability and beta‐lactam sensitivity in Pseudomonas aeruginosa . Microbiologyopen 4: 879–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead, B. , Trapnell, C. , Pop, M. , and Salzberg, S. L. (2009) Ultrafast and memory‐efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langridge, G. C. , Phan, M. D. , Turner, D. J. , Perkins, T. T. , Parts, L. , Haase, J. , et al. (2009) Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res 19: 2308–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawe‐Davies, O. , and Bennett, S. (2017) WHO publishes list of bacteria for which new antibiotics are urgently needed. URL www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-needed/en
- Lazaro‐Diez, M. , Navascues‐Lejarza, T. , Remuzgo‐Martinez, S. , Navas, J. , Icardo, J. M. , Acosta, F. , et al. (2016) Acinetobacter baumannii and A. pittii clinical isolates lack adherence and cytotoxicity to lung epithelial cells in vitro. Microbes Infect 18: 559–564. [DOI] [PubMed] [Google Scholar]
- Leblanc, S. K. , Oates, C. W. , and Raivio, T. L. (2011) Characterization of the induction and cellular role of the BaeSR two‐component envelope stress response of Escherichia coli . J Bacteriol 193: 3367–3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M. , Hesek, D. , Llarrull, L. I. , Lastochkin, E. , Pi, H. , Boggess, B. , et al. (2013) Reactions of all Escherichia coli lytic transglycosylases with bacterial cell wall. J Am Chem Soc 135: 3311–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, W. , Dong, N. , and Zhang, X. H. (2012) Overexpression of mltA in Edwardsiella tarda reduces resistance to antibiotics and enhances lethality in zebra fish. J Appl Microbiol 112: 1075–1085. [DOI] [PubMed] [Google Scholar]
- Livak, K. J. , and Schmittgen, T. D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Macritchie, D. M. , and Raivio, T. L. (2009) Envelope Stress Responses . EcoSal Plus 3. [DOI] [PubMed] [Google Scholar]
- Maneval, W. E. (1941) Staining bacteria and yeasts with acid dyes. Stain Technol 16: 13–19. [Google Scholar]
- Merino, S. , Camprubi, S. , Alberti, S. , Benedi, V. J. , and Tomas, J. M. (1992) Mechanisms of Klebsiella pneumoniae resistance to complement‐mediated killing. Infect Immun 60: 2529–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon, K. H. , Weber, B. S. , and Feldman, M. F. (2017) Subinhibitory concentrations of trimethoprim and sulfamethoxazole prevent biofilm formation by Acinetobacter baumannii through inhibition of Csu pili expression. Antimicrob Agents Chemother 61: e00778–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen, B. L. , Rathi, S. , Chazin, W. J. , and Skaar, E. P. (2014) Acinetobacter baumannii response to host‐mediated zinc limitation requires the transcriptional regulator Zur. J Bacteriol 196: 2616–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragland, S. A. , Schaub, R. E. , Hackett, K. T. , Dillard, J. P. , and Criss, A. K. (2017) Two lytic transglycosylases in Neisseria gonorrhoeae impart resistance to killing by lysozyme and human neutrophils. Cell Microbiol 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo, T. A. , Luke, N. R. , Beanan, J. M. , Olson, R. , Sauberan, S. L. , MacDonald, U. , et al. (2010) The K1 capsular polysaccharide of Acinetobacter baumannii strain 307–0294 is a major virulence factor. Infect Immun 78: 3993–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santin, Y. G. , and Cascales, E. (2017) Domestication of a housekeeping transglycosylase for assembly of a Type VI secretion system. EMBO Rep 18: 138–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheurwater, E. , Reid, C. W. , and Clarke, A. J. (2008) Lytic transglycosylases: bacterial space‐making autolysins. Int J Biochem Cell Biol 40: 586–591. [DOI] [PubMed] [Google Scholar]
- Schindelin, J. , Arganda‐Carreras, I. , Frise, E. , Kaynig, V. , Longair, M. , Pietzsch, T. , et al. (2012) Fiji: an open‐source platform for biological‐image analysis. Nat Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert, H. , Strate, A. , and Pulverer, G. (1995) Nosocomial bacteremia due to Acinetobacter baumannii. Clinical features, epidemiology, and predictors of mortality. Medicine 74: 340–349. [DOI] [PubMed] [Google Scholar]
- Shallcross, L. J. , Howard, S. J. , Fowler, T. , and Davies, S. C. (2015) Tackling the threat of antimicrobial resistance: from policy to sustainable action. Philos Trans R Soc Lond B Biol Sci 370: 20140082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, S. N. , Hagan, E. C. , Lane, M. C. , and Mobley, H. L. (2010) Dissemination and systemic colonization of uropathogenic Escherichia coli in a murine model of bacteremia. MBio 1: e00262–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellberg, B. , and Rex, J. H. (2013) The value of single‐pathogen antibacterial agents. Nat Rev Drug Discov 12: 963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohl, E. A. , Chan, Y. A. , Hackett, K. T. , Kohler, P. L. , Dillard, J. P. , and Seifert, H. S. (2012) Neisseria gonorrhoeae virulence factor NG1686 is a bifunctional M23B family metallopeptidase that influences resistance to hydrogen peroxide and colony morphology. J Biol Chem 287: 11222–11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohl, E. A. , Dale, E. M. , Criss, A. K. , and Seifert, H. S. (2013) Neisseria gonorrhoeae metalloprotease NGO1686 is required for full piliation, and piliation is required for resistance to H2O2‐ and neutrophil‐mediated killing. MBio 4: e00399–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subashchandrabose, S. , Smith, S. , DeOrnellas, V. , Crepin, S. , Kole, M. , Zahdeh, C. , et al. (2016) Acinetobacter baumannii genes required for bacterial survival during bloodstream infection . mSphere 1: e00013–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subashchandrabose, S. , Smith, S. N. , Spurbeck, R. R. , Kole, M. M. , and Mobley, H. L. (2013) Genome‐wide detection of fitness genes in uropathogenic Escherichia coli during systemic infection. PLoS Pathog 9: e1003788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, M. T. , Lai, Y. T. , Huang, C. Y. , Chou, L. Y. , Shih, H. W. , Cheng, W. C. , et al. (2009) Crystal structure of the membrane‐bound bifunctional transglycosylase PBP1b from Escherichia coli . Proc Natl Acad Sci USA 106: 8824–8829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker, A. T. , Nowicki, E. M. , Boll, J. M. , Knauf, G. A. , Burdis, N. C. , Trent, M. S. , et al. (2014) Defining gene‐phenotype relationships in Acinetobacter baumannii through one‐step chromosomal gene inactivation. MBio 5: e01313–01314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas, A. , Banzhaf, M. , Gross, C. A. , and Vollmer, W. (2011) From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol 10: 123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Faassen, H. , KuoLee, R. , Harris, G. , Zhao, X. , Conlan, J. W. , and Chen, W. (2007) Neutrophils play an important role in host resistance to respiratory infection with Acinetobacter baumannii in mice. Infect Immun 75: 5597–5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier, P. H. , and Shapiro, L. (2003) A lytic transglycosylase homologue, PleA, is required for the assembly of pili and the flagellum at the Caulobacter crescentus cell pole. Mol Microbiol 49: 331–345. [DOI] [PubMed] [Google Scholar]
- Wang, N. , Ozer, E. A. , Mandel, M. J. , and Hauser, A. R. (2014) Genome‐wide identification of Acinetobacter baumannii genes necessary for persistence in the lung. MBio 5: e01163–01114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber, B. S. , Hennon, S. W. , Wright, M. S. , Scott, N. E. , de Berardinis, V. , Foster, L. J. , et al. (2016) Genetic dissection of the type VI secretion system in Acinetobacter and identification of a novel peptidoglycan hydrolase, TagX, required for its biogenesis. MBio 7: e01253–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield, C. (2006) Biosynthesis and assembly of capsular polysaccharides in Escherichia coli . Annu Rev Biochem 75: 39–68. [DOI] [PubMed] [Google Scholar]
- Willyard, C. (2017) The drug‐resistant bacteria that pose the greatest health threats. Nature 543: 15. [DOI] [PubMed] [Google Scholar]
- Wisplinghoff, H. , Bischoff, T. , Tallent, S. M. , Seifert, H. , Wenzel, R. P. , and Edmond, M. B. (2004) Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis 39: 309–317. [DOI] [PubMed] [Google Scholar]
- Wong, D. , Nielsen, T. B. , Bonomo, R. A. , Pantapalangkoor, P. , Luna, B. , and Spellberg, B. (2017) Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin Microbiol Rev 30: 409–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, C. J. , Huang, Y. W. , Lin, Y. T. , and Yang, T. C. (2016) Inactivation of lytic transglycosylases increases susceptibility to aminoglycosides and macrolides by altering the outer membrane permeability of Stenotrophomonas maltophilia . Antimicrob Agents Chemother 60: 3236–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, D. , Ellis, H. M. , Lee, E. C. , Jenkins, N. A. , Copeland, N. G. , and Court, D. L. (2000) An efficient recombination system for chromosome engineering in Escherichia coli . Proc Natl Acad Sci USA 97: 5978–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilsel, J. , Ma, P. H. , and Beatty, J. T. (1992) Derivation of a mathematical expression useful for the construction of complete genomic libraries. Gene 120: 89–92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


