Abstract
Background
Recent evidence supports that the gut microbiota may be involved in the pathophysiology of non-alcoholic fatty liver disease (NAFLD), and may also offer avenues for treatment or prevention.
Methods
We investigated the associations among gut microbiota, diet and hepatic fat fraction (HFF) in 107 adolescents. Magnetic Resonance Imaging (MRI) was used to assess HFF, and 16S rRNA gene sequencing was performed on collected fecal samples. Dietary intake was assessed using Food Frequency Questionnaires. We examined the association between gut microbiota alpha diversity and HFF, and assessed the predictive accuracy for HFF of 1) taxonomic composition, 2) dietary intake, 3) demographic and comorbid conditions, and 4) the combination of these.
Results
Lower alpha diversity was associated with higher HFF (β=−0.19, 95% CI −0.36, −0.02). Selected taxa explained 17.7% (95% CI: 16.0–19.4%) of the variation in HFF. The combination of 2 of these taxa, Bilophila and Paraprevotella, with dietary intake of monounsaturated fatty acids and BMI z-scores explained 32.0% (95% CI: 30.3–33.6%) of the variation in HFF.
Conclusions
The gut microbiota is associated with HFF in adolescents and may be useful to help identify youth who would be amenable to gut microbiota-based interventions.
Introduction
Non-alcoholic fatty liver disease (NAFLD) is a spectrum of conditions ranging from simple steatosis to progressive nonalcoholic steatohepatitis (NASH) and is the most common cause of chronic liver disease in the United States, as well as many other places in the world.(1) Prevalence of NAFLD has been increasing rapidly in recent years in conjunction with increases in obesity, which is a primary main risk factor for fatty liver, though it also occurs in lean individuals. (2) Current prevalence estimates range from 10–30% among U.S. adults and 3–10% in children.(2) Early interventions for NAFLD include dietary and lifestyle counseling, as well as vitamin supplementation.(3,4) In order to curb the long-term health effects of NAFLD, it is important to improve and expand the options for early interventions, and to gain a deeper understanding of the biological processes underlying the condition, which may also inform prevention efforts.
Both animal and observational studies in humans provide evidence that the pathogenesis of NAFLD may involve the gut microbiota through various mechanisms, such as effects on the metabolism of lipids and cholesterol, triglyceride storage, hepatic inflammation, and regulation of lipogenesis and gluconeogenesis.(5,6) Diet may also play a role in NAFLD, either directly or through gut microbiota-mediated effects. Total carbohydrates, specifically sugars and fructose, and total fat, particularly saturated fat, have been linked to increased risk for NAFLD; monounsaturated fats may be protective.(6,7) The fermentation of fiber by gut microbiota produces short chain fatty acids (SFCAs), which then affect energy metabolism, the immune system and adipose tissue expansion, and may play a role in gluconeogenesis in the liver.(6,8) Prebiotics or prebiotic foods, which are a type of fiber that are not digested in the small intestine but are fermented in the colon by the gut microbiota, have been suggested as a possible treatment for NAFLD, possibly in conjunction with probiotics.(9) A better understanding of the relationships between diet, the gut microbiota and NAFLD may help to inform or target such interventions.
In this study, we examined whether there is an association between gut microbiota and hepatic fat fraction (HFF), both alone and in conjunction with dietary information, in an adolescent population from the Exploring Perinatal Outcomes among Children (EPOCH) cohort. We assessed the accuracy of using the following information to predict HFF: 1) taxonomic composition, 2) dietary intake information, 3) demographic and comorbid conditions, and 4) the combination of all of the above.
Materials & Methods
Study Cohort
EPOCH is a historical prospective study of 604 mother / child pairs. Adolescents were identified through the Kaiser Permanente of Colorado Perinatal database based on exposure to gestational diabetes mellitus during singleton pregnancies. A research visit with data collection took place during 2012–2016, while the children were 12–19 years, and a fecal sample was requested from a randomly selected subsample of 240 participants. The subsample included 1 exposed participant for every 2 unexposed, matched on gender and race/ethnicity. Many participants chose to not provide a sample, or were unable to do so within the requested time frame. Thus, fecal samples were successfully collected from 120 participants. Of those, one sample failed to sequence, two had poor quality reads, four were missing dietary information, and six were missing the outcome measure of hepatic fat fraction, leaving a sample size of 107 for the primary analyses. Analyses involving waist circumference or insulin resistance included 106 participants with complete data. The study was approved both by the Colorado Multiple Institutional Review Board and Human Participant Protection Program. All participants provided written informed consent and youth provided written assent.
Data collection
The primary outcome variable in this study was hepatic fat fraction (HFF) obtained by Magnetic Resonance Imaging (MRI). Hepatic imaging was performed using a modification of the Dixon method by Hussain involving multi-breath-hold double gradient echo sequences.(10,11) HFF was calculated from the mean pixel signal intensity data, for each flip angle acquisition. An HFF of 5% or greater is commonly used as an indicator for mild fatty liver.(12)
Data collection also involved completion of the Block Kid’s Food Questionnaire,(13) a semi-quantitative food frequency questionnaire (FFQ) developed for children aged 8 years and older, which assesses 85 food items consumed in the last week, frequency and average portion size. Height was measured by SECA stadiometer, and weight was measured using an electronic SECA scale, as described previously.(14) Age- and sex-specific BMI z-scores were calculated using CDC reference standards, (15) and weight groups were defined using percentiles of BMIfor- age: underweight=less than 5th percentile; normal weight=5th to 85th percentile; overweight=85th up to 95th percentile; obese=95th percentiles or above.(16) Waist circumference was measured according to the National Health and Nutrition Examination Survey protocol.(17) Blood samples were obtained at the EPOCH study visit after an overnight fast, and glucose, triglycerides, and alanine aminotransferase (ALT) were measured, as described previously.(14) HOMA-IR [homeostasis model of insulin resistance: fasting glucose (mmol/l) × fasting insulin (µU/ml)/22.5] was used as a marker of insulin resistance. Race / ethnicity was self-reported using 2000 U.S. census definitions and categorized as Hispanic (any race), non-Hispanic white, non- Hispanic African-American, and non-Hispanic other. Maternal level of education and total household income at the time of birth were self-reported during the office visit. Maternal diabetes status was physician-diagnosed using a standard two-step screening protocol (18) and ascertained from the Kaiser Permanente of Colorado Perinatal database, an electronic database linking the neonatal and perinatal medical record. The database was also used to determine delivery mode at birth.
Collection and processing of fecal samples
Before their study visit, a subsample (described above) of participants were asked for a microbiome stool sample. If they agreed to provide a sample, they were sent instructions as well as a Fisher Scientific BBL™ CultureSwab™ kit, a dual swab system for the sterile collection and transport of fecal microbiological samples. Participants were asked to take the sample as close as possible to the time of the in-person visit, and in all cases, it was the same day as the interview. The participants kept the sample at room temperature until the time of the interview, when the samples were frozen and stored at −80°F. DNA was extracted using the standard Power Soil Kit protocol (MoBio). Extracted DNA was PCR amplified with barcoded primers targeting the V4 region of 16S rRNA as detailed in Yatsunenko et al.(20) Control water samples that had undergone the same DNA extraction and PCR amplification procedures were also sequenced. Each PCR product was quantified using PicoGreen (Invitrogen), and equal amounts (ng) of DNA from each sample were pooled and cleaned using the UltraClean PCR Clean-Up Kit (MoBio). Sequences were generated on a MiSeq personal sequencer (Illumina, San Diego, CA).
Denoising and OTU picking
Reads were first quality filtered and trimmed to a uniform length based on average position of first low quality base pair among all samples. The R package DADA2(19) was then run on default parameters to denoise the data and find exact sequence abundances across samples. These sequences were then used as input for open reference Operational Taxonomic Unit (OTU) picking using QIIME 1.9 (20) with a 99% identity threshold to determine OTUs. OTUs are groups of organisms based on sequence similarity. Greengenes 13.8 was used as a reference database of near-full length sequences,(21) and unassigned sequences were clustered into de novo OTUs using UCLUSTref.(22) Analyses were standardized at the minimum sequence depth, 2537 sequences per sample, to avoid biases. The OTUs were summarized at the most specific known level of taxonomy for all analyses of taxonomic composition.
Statistical Methods
We dichotomized HFF using the same cutoff as typically used for adults to define mild steatosis: HFF≥5%(12) and compared demographic characteristics by NAFLD status using chi-squared tests for categorical variables and Wilcoxon rank-sum tests for continuous variables. Alpha diversity and UniFrac principal coordinates analyses (PCoA) were conducted using QIIME.(23) PCOA plots were colored by HFF and by weight group.
Alpha diversity
Alpha diversity measures the microbial diversity of each sample. There are many alpha diversity measures, and they differ in how they weight richness and evenness and whether they incorporate phylogenetic distance. We chose Shannon Diversity Index as our primary measure of alpha diversity because it gives equal weight to evenness and richness. We used linear regression models to examine the association between alpha diversity of gut microbiota and hepatic fat fraction (HFF). We used a square root transformation in order to normalize HFF to meet the assumptions of linear regressions. We controlled for sex, age, race/ethnicity, parental education, exposure to gestational diabetes in utero and delivery method at birth.
Taxonomic composition
In order to understand the interrelationships of the taxa, we used the graphical lasso technique as implemented in the R package qgraph, calculating the correlation using the Pearson’s correlation as input to the R function ccrepe, which was designed specifically for sparse compositional data such as these.(19) The minimum prevalence threshold for applying the ccrepe function was nonzero abundance in ≥10 samples, and we used this same cutoff for all analyses involving the gut microbiota taxa.
Assessment of relationship between taxonomic composition and cardiometabolic measures
We evaluated the association between the gut microbiota composition and HFF using a Microbiome Regression-Based Kernel Association Test (MiRKAT)(19) of the Bray-Curtis, weighted and unweighted UniFrac distance matrices. Each of these metrics compares the dissimilarity across samples in slightly different ways: Bray-Curtis compares presence / absence, weighted UniFrac compares both phylogeny and abundance, and unweighted UniFrac compares phylogeny alone. These models were run first unadjusted, and again controlling for sex, age, race/ethnicity, parental income, parental education, exposure to gestational diabetes in utero, and delivery mode at birth. We performed similar models with BMI z-score, waist circumference (N=106), and HOMA-IR (N=106) as the outcomes of interest.
Evaluation of Association Between HFF and Taxonomic Composition, Diet and Comorbidities
In order to gain understanding of the role of gut microbiota and diet in the generation of hepatic fat, we used random forests. We applied the R function VSURF (Variable Selection using Random Forests)(19) to select features that are highly associated with HFF from the following groups of predictors: 1) taxa meeting the minimum threshold of presence in ≥10 samples (N=76, pictured in Supplemental Figure S1); 2) dietary total daily kilocalories and the following macronutrients as percent of total intake: total fat, saturated fat, polyunsaturated fat, monounsaturated fat, total protein, total carbohydrates, sugars, soluble fiber, and insoluble fiber; 3) demographic and comorbid conditions: sex, age, race/ethnicity, parental income, parental education, exposure to gestational diabetes in utero, delivery mode at birth and current BMI z-score; and 4) the combination all of the these. The VSURF function is a multi-step algorithm that identifies the most important features for the prediction of HFF. We performed a sensitivity analysis for the random forest of dietary information, additionally including meat, fructose, vitamin D, vitamin A, and coffee / tea intake, which may be related to fatty liver.(6,7)
Whereas a linear regression would fit only a linear relationship between the predictors and the outcome, random forests allow for any type of relationship, including complex interactions. Random forests also do not provide regression coefficients, thus, we used various tools to understand the nature of the relationships between the predictors and the outcome as well as inter-relationships between the predictors, including partial plots and interaction plots. Partial plots show the adjusted relationship between the predictors and HFF, i.e. all the other selected features are held constant, as in multiple regression. We also used repeated cross-validation (3 folds, 100 repetitions) in order to evaluate the accuracy of the random forests. All analyses were performed using Qiime 1.9(20) and R v3.4.1.(19)
Results
Population Characteristics
The prevalence of NAFLD (defined as HFF≥5%) in this cohort was 7.5% (N=8; Table 1), which is similar to other estimates among youth in the United States, ranging between 3 and 10%.(2) Among those with NAFLD, the median hepatic fat fraction was 7.7% (IQR 6–8.5%; Range 5.2–11.1%); among those without NAFLD, it was 1.9% (IQR: 1.3–2.5; Range 0–4.7%). The overall prevalence of overweight was 15% and obesity was 20.6%. Individuals with NAFLD were all either overweight or obese. Compared to those without NAFLD, they had larger waist circumference, higher alanine aminotransferase (a measure of liver function), higher HOMA-IR and were more likely to be Hispanic. The parents of individuals with NAFLD tended to have a lower level of education, and the mothers had higher pre-pregnancy BMI. Other demographic, comorbidity and dietary information tended to be similar by NAFLD status (Table 1).
Table 1.
Demographic, comorbidity, and dietary information for adolescents in the EPOCH cohort, by NAFLD status (defined as HFF≥5%)
| Variable | Total (N=107) | NAFLD = 0 (N=99) |
NAFLD = 1 (N=8) |
P-Value |
|---|---|---|---|---|
|
| ||||
| N (%) or Median (IQR) | ||||
|
|
||||
| Male | 55 (51.4%) | 50 (50.5%) | 5 (62.5%) | 0.72 |
| Race | ||||
| Non-Hispanic White | 52 (48.6%) | 49 (49.5%) | 3 (37.5%) | 0.02 |
| Hispanic | 40 (37.4%) | 35 (35.4%) | 5 (62.5%) | |
| Non-Hispanic Black | 12 (11.2%) | 12 (12.1%) | 0 (0.0%) | |
| Non-Hispanic Other | 3 (2.8%) | 3 (3.0%) | 0 (0.0%) | |
| Parental Education Level | ||||
| High School | 15 (14.0%) | 11 (11.1%) | 4 (50.0%) | 0.001 |
| Some college/Associate degree | 33 (30.8%) | 31 (31.3%) | 2 (25.0%) | |
| Bachelor's degree | 31 (29.0%) | 29 (29.3%) | 2 (25.0%) | |
| Graduate degree | 28 (26.2%) | 28 (28.3%) | 0 (0.0%) | |
| Household income | ||||
| $16,000 through $34,999 | 4 (3.7%) | 4 (4.0%) | 0 (0.0%) | 0.02 |
| $35,000 through $74,999 | 40 (37.4%) | 34 (34.3%) | 6 (75.0%) | |
| $75,000 or more | 59 (55.1%) | 57 (57.6%) | 2 (25.0%) | |
| Don't know/Missing | 4 (3.7%) | 4 (4.0%) | 0 (0.0%) | |
| Delivery Mode | ||||
| C-section | 25 (23.4%) | 24 (24.2%) | 1 (12.5%) | 0.68 |
| Vaginal | 82 (76.6%) | 75 (75.8%) | 7 (87.5%) | |
| Exposure to DM in utero | 30 (28.0%) | 28 (28.3%) | 2 (25.0%) | >0.99 |
| Pre-pregnancy BMI | 26.1 (22.8–30.3) | 25.9 (22.5–30.3) | 30.9 (25.8–38.4) | 0.01 |
|
| ||||
| In person visit | ||||
|
| ||||
| Age (years) | 15.6 (15.1–16.5) | 15.7 (15.1–16.5) | 15.6 (15.6–16.4) | 0.16 |
| BMI | 22.5 (19.8–26.0) | 21.9 (19.5–24.9) | 32.2 (27.6–36.0) | <.001 |
| BMI-for-age Z | 0.5 (−0.3 to 1.5) | 0.4 (−0.3 to 1.2) | 2.0 (1.7–2.5) | <.001 |
| Weight group (based on BMI percentile) | ||||
| Underweight | 4 (3.7%) | 4 (4.0%) | 0 (0.0%) | <.001 |
| Normal weight | 65 (60.7%) | 65 (65.7%) | 0 (0.0%) | |
| Overweight | 16 (15.0%) | 14 (14.1%) | 2 (25.0%) | |
| Obese | 22 (20.6%) | 16 (16.2%) | 6 (75.0%) | |
| Waist circumference | 76.4 (70.6–86.3) | 75.4 (70.3–84.5) | 93.0 (92.3–106.4) | <.001 |
| Hepatic Fat Fraction | 2.0 (1.4–2.8) | 1.9 (1.3–2.5) | 7.7 (6.0–8.5) | <.001 |
| Alanine aminotransferase (U/L) | 25.0 (20.0–31.0) | 24.0 (19.5–30.0) | 31.0 (29.0–39.0) | 0.002 |
| HOMA-IR (Homeostasis model of insulin resistance) | 3.0 (2.3–4.4) | 2.8 (2.2–4.3) | 6.1 (4.6–8.4) | <.001 |
|
| ||||
| Diet | ||||
|
| ||||
| Kilocalories | 1547 (1217–2078) | 1567 (1217–2091) | 1497 (1085–1899) | 0.5 |
| % Carbohydrates | 48.6 (44.2–52.3) | 48.6 (43.7–52.3) | 48.1 (44.8–52.3) | 0.22 |
| % Added Sugars | 24.6 (20.5–28.2) | 24.5 (20.5–28.2) | 25.1 (19.8–28.4) | 0.43 |
| % Fiber | 3.0 (2.5–3.9) | 3.0 (2.5–3.9) | 2.6 (2.4–3.8) | 0.72 |
| % Insoluble Fiber | 1.9 (1.5–2.5) | 2.0 (1.5–2.5) | 1.6 (1.5–2.4) | 0.68 |
| % Soluble Fiber | 1.1 (0.9–1.4) | 1.1 (0.9–1.4) | 1.0 (0.9–1.3) | 0.54 |
| % Fat | 38.0 (34.4–41.8) | 38.0 (34.4–41.8) | 37.1 (34.0–40.1) | 0.16 |
| % SFA | 13.5 (11.9–15.3) | 13.4 (11.9–15.0) | 14.2 (12.4–15.4) | 0.71 |
| % PUFA | 5.7 (4.8–6.7) | 5.8 (4.8–6.8) | 5.0 (4.5–5.7) | 0.10 |
| % MUFA | 15.4 (13.7–16.8) | 15.5 (13.7–16.8) | 14.7 (13.6–15.8) | 0.15 |
| % Protein | 34.2 (31.3–38.3) | 34.2 (31.2–38.2) | 35.9 (32.7–40.3) | 0.53 |
|
| ||||
| Alpha Diversity | ||||
|
| ||||
| Shannon Diversity Index | 4.8 (4.4–5.2) | 4.8 (4.4–5.2) | 4.5 (4.2–5.0) | 0.32 |
| PD whole tree | 10.8 (9.0–12.2) | 10.9 (9.0–12.2) | 9.2 (8.9–12.1) | 0.6 |
| Observed species | 67.0 (55.0–78.0) | 67.0 (56.0–78.0) | 59.5 (52.0–78.5) | 0.63 |
Alpha Diversity
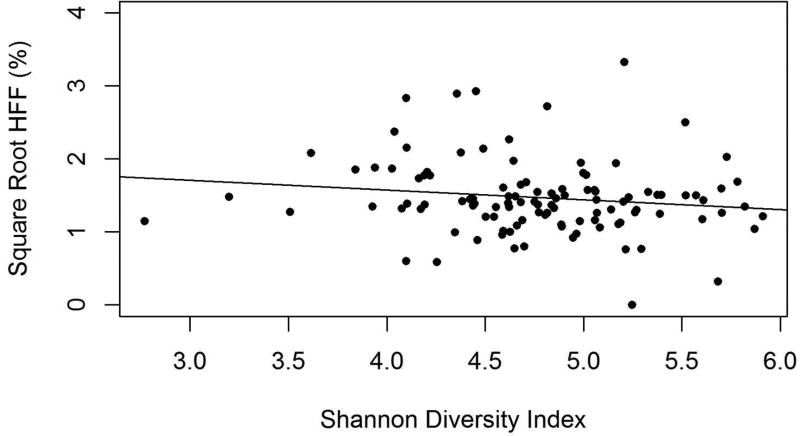
Alpha diversity measures (Shannon Diversity Index, Phylogenetic Distance and Observed Species) did not differ significantly by dichotomous NAFLD status (Table 1). However, we also used regressions to evaluate the relationship between one of these alpha diversity measures, Shannon Diversity, which takes into account both richness and evenness, and the continuous measure of HFF. We found that in unadjusted models, there was a trend towards lower Shannon Diversity with increasing levels of liver fat (β=−0.15, 95% CI −0.33, 0.02; p-value=0.07; Figure 1). The effect became statistically significant when controlling for sex, age, race / ethnicity, parental education, exposure to diabetes in utero and delivery method at birth (β=−0.20, 95% CI −0.37, −0.03; p-value=0.03).
Figure 1.
The relationship between hepatic fat fraction (HFF) with square root transformation and Shannon Diversity Index of gut microbiota of adolescents in the EPOCH cohort. Shannon diversity is significantly lower with higher HFF, when controlling for race/ethnicity, sex, age, parental education, exposure to diabetes in utero and delivery method at birth (β=−0.20, 95% CI −0.37, −0.03; p-value=0.03).
Taxonomic Composition
The fecal microbiota across our samples was dominated by the phyla Firmicutes and Bacteroidetes, particularly the genus of Bacteroides, those from the families of Lachnospiraceae and Ruminococcaceae, as is typically observed in Western populations (Supplemental Figure S2).(24) In order to examine relationships between bacterial taxa, we formed a network using the Graphical LASSO technique(25) (Supplemental Figure S1). This network showed a large cluster of co-occurring taxa that included representatives of a wide array of phyla, as well as many smaller clusters, although the majority of taxa were not in clusters.
Assessment of relationship between taxonomic composition and cardiometabolic measures
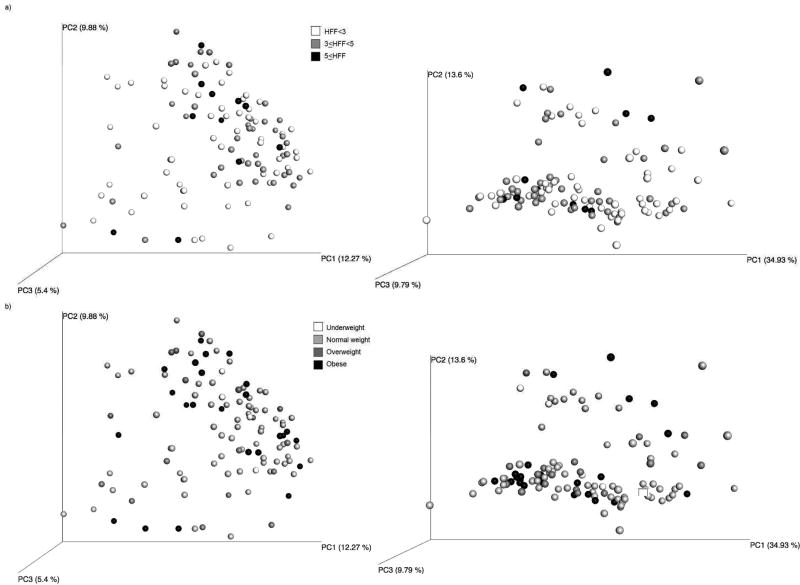
UniFrac-based PCoA plots of the gut microbiota samples of adolescents are shown in Figure 2, colored by (a) amount of HFF and (b) by weight group. Statistical models showed that qualitative differences in taxonomic phylogeny (unweighted UniFrac) were significantly associated with HFF (unadjusted p=0.01; adjusted p=0.02; Table 2), while quantitative differences in phylogenetic abundance (weighted UniFrac) were significantly associated with BMI z-score (unadjusted p=0.04; adjusted p=0.07). Presence / absence of taxa (Bray-Curtis) was associated with waist circumference in adjusted models (unadjusted p=0.08; adjusted p=0.02). None of the taxonomic composition measures examined were associated with HOMA-IR.
Figure 2.
Principal Coordinate Analysis plots of unweighted (left) and weighted (right) UniFrac distance by a) amount of hepatic fat fraction, and b) weight group. Statistical models showed a significant relationship between unweighted UniFrac distance and HFF (p=0.01), as well as between weighted UniFrac distance and BMI z-score (p=0.04).
Table 2.
Results of statistical models to assess the association between measures of taxonomic composition (Unweighted UniFrac, Weighted UniFrac and Bray-Curtis distance metrics) and metabolic measures, including HFF, BMI z-score, waist circumference, and HOMA-IR (homeostasis model of insulin resistance). Adjusted models controlled for the following covariates: sex, race/ethnicity, age, parental education, delivery method at birth and in utero exposure to diabetes.
| Unadjusted | Adjusted | |||||||
|---|---|---|---|---|---|---|---|---|
|
|
||||||||
| Outcome | Unweighted UniFrac |
Weighted UniFrac |
Bray-Curtis | Omnibus | Unweighted UniFrac |
Weighted UniFrac |
Bray-Curtis | Omnibus |
| HFF | 0.014 | 0.152 | 0.173 | 0.016 | 0.015 | 0.118 | 0.308 | 0.033 |
| BMI z-score | 0.563 | 0.041 | 0.257 | 0.079 | 0.453 | 0.068 | 0.253 | 0.158 |
| Waist circumference (N=106) | 0.252 | 0.763 | 0.081 | 0.157 | 0.209 | 0.772 | 0.017 | 0.047 |
| HOMA-IR (N=106) | 0.212 | 0.816 | 0.864 | 0.389 | 0.279 | 0.923 | 0.933 | 0.493 |
Evaluation of Association Between HFF and Taxonomic Composition, Diet and Comorbidities
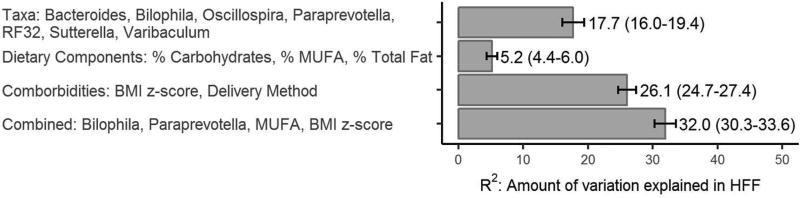
In order to identify which features were most associated with hepatic fat, we used a random forest feature selection process, the results of which are shown in Figure 3. The selected subset of gut microbiota most predictive of HFF included 7 taxa (R2: 17.7; 95% CI: 16.0, 19.4): Bilophila, Paraprevotella, Varibaculum, Sutterella, Oscillospira, Order RF32 with unclassified genus, and Bacteroides. Varibaculum was highly correlated many other taxa (as shown by the connected nodes in Supplemental Figure S1); Bacteroides showed a weak negative correlation with Prevotella copri; and Oscillospira and RF32 were positively correlated with each other.
Figure 3.
This figure shows the results of feature selection procedures that choose the most important features for the prediction of hepatic fat fraction (HFF) in adolescents in the EPOCH cohort. Four groups of variables were explored: gut microbiota taxa, dietary components, comorbidities & demographic variables, and the combination of all these. For each group of variables, we indicate the selected features and amount of variation in HFF that is explained (R2 and the 95% confidence interval, CI).
The selected dietary components explained substantially less of the variation in HFF compared with the taxa (R2: 5.2%, 95% CI: 4.4, 6.0) and included dietary percentage of monounsaturated fats, carbohydrates and total fats. The sensitivity analysis including additional dietary components performed even worse, and none of the additional dietary components were among the selected features.
The selected demographic and comorbid features included current BMI z-score and delivery mode at birth; these features explained more of the variation in HFF than the taxa or dietary components alone (R2: 26.1%, 95% CI: 24.7, 27.4). The random forests using the combination of all of these groups of features had the best accuracy measures, and included BMI z-score, percent monounsaturated fats, Bilophila and Paraprevotella (R2: 32.0%, 95% CI: 30.3, 33.6).
The most important features identified by random forests may be positively or negatively related to the outcome, or they may be related through complex interactions with each other. Thus, we used various plotting methods as detailed in the methods section in order to interpret the results of the random forests (Supplemental Figures S3–4; summarized in Table 3).
Table 3.
This table summarizes the general relationships between hepatic fat fraction (HFF) and the features selected in the random forests for the prediction of HFF. Plots are used to understand the direction of the relationships between the predictors and the outcome, as well as the inter-relationships between the predictors, since random forests do not provide regression coefficients; the partial and interaction plots showing these relationships in detail are in the supplemental figures. Since random forests allow for complex interactions between the predictors, this table notes when there is no evidence of strong 2-way interactions and notes the interacting features when there is evidence of interactions. In order to fully understand the nature of these interactions, please see the supplemental figures.
| Group | Selected Features | Adjusted association with HFF |
Interactions? |
|---|---|---|---|
| Taxa | g_Varibaculum | Negative | No |
| o_RF32 | Positive | No | |
| g_Paraprevotella | Positive | No | |
| g_Sutterella | Positive | Bilophila | |
| g_Bilophila | Positive | Bacteroides, Oscillospira, | |
| g_Oscillospira | Negative | Bilophila | |
| g_Bacteroides | U-shaped: High HFF with low and high abundance, low HFF with moderate | Bilophila | |
|
| |||
| Diet | % Carbohydrates | Negative | % MUFA |
| % Total fat | Positive | % MUFA | |
| % MUFA | Negative | Carbohydrates, | |
|
| |||
| Comorbidities & Demographics | BMI z-score | Positive | Delivery mode |
| Delivery mode | Higher HFF with vaginal birth | BMI | |
|
| |||
| Combined Features | g_Bilophila | Positive | Paraprevotella |
| g_Paraprevotella | Positive | Bilophila, % MUFA | |
| % MUFA | Negative | Paraprevotella | |
| BMI z-score | Positive | ||
Most of the taxa highlighted by the random forests correlated positively with HFF, including Bilophila, Paraprevotella, Suturella, and RF32. Bacteroides showed a U-shaped pattern with hepatic fraction over levels of abundance; both low and high abundance corresponded to higher hepatic fat, while moderate levels corresponded with low hepatic fat. Oscillospira and Varibaculum were protective. Dietary components were not strongly associated with HFF, and they were highly correlated with each other, which makes it difficult to separate their effects. The adjusted relationships showed protective effects of monounsaturated fats and carbohydrates, and positive correlation between percent total fat and HFF. As would be expected, BMI z-scores positively correlated with HFF.
When comparing the groups of predictors of HFF, gut microbiota taxa show important value in terms of the prediction of HFF (Figure 3). Dietary components showed the weakest association with HFF. The most accurate predictions of HFF included BMI z-scores, as might be expected since obesity is a major risk factor for NAFLD.
Discussion
In this observational study of adolescents, our results support the hypothesis that there is an association between gut microbiota and hepatic fat fraction. We found associations between lower alpha diversity, taxonomic phylogeny, and specific gut microbiota taxa with HFF. We did not find a strong association between the dietary components examined and HFF, or strong interactions between gut microbiota taxa and dietary components. In this cohort, HFF was associated with qualitative differences in taxonomic composition (unweighted UniFrac), while BMI z-score was associated with quantitative differences in phylogenetic abundance (weighted UniFrac). Understanding this shift in the types of microorganisms present among individuals with more hepatic fat may shed light on the role of the gut microbiota in NAFLD.
We observed lower adjusted alpha diversity with higher hepatic fat. Prior studies of NAFLD and NASH in pediatric populations have shown similar trends of lower alpha diversity with these conditions relative to healthy controls, and lower alpha diversity has also been associated with obesity. (26–28)
Some of the taxa associated with fatty liver in this study are highly associated with bile acids. The gut microbiota play an important role in modulating bile acid homeostasis, and bile acids likewise play an important, but not fully understood, role in NAFLD.(5,29) Bilophila was positively correlated with HFF in our study and thrives in the presence of bile, specifically taurine conjugated bile.(30) Interestingly, the bile acid pool shifts towards taurine conjugation in response to a diet high in taurine, which is predominantly in animal products.(31) One species of Bilophila in particular, B. wadworthia, has been consistently seen across studies as enriched in response to Western diets or those high in fat,(32,33) and is also linked to Th1-mediated intestinal inflammation;(32) it is thought that its by-products of hydrogen sulfide and secondary bile acids may degrade the gut mucosal barrier.(32) Oscillospira and Bacteroides are also associated with diets high in animal products,(34,35) and are likewise highly bile tolerant. However, these microbes showed different patterns in their relationships with HFF in our study.
Oscillospira was negatively related to HFF. This is not surprising since Oscillospira is generally associated with leanness and health,(36) and has previously been seen to be reduced with NAFLD and NASH in other pediatric populations.(26,27) However, its functions in the gut are not well understood.(36) Interestingly, the positive association between HFF and Bilophila was only observed at low levels of Oscillospira (Supplemental Figure S4). These microbes were not mutually exclusive; some individuals did have high levels of both Oscillospira and Bilophila. Since this is a cross-sectional epidemiologic study, we cannot offer conclusions about the biological underpinnings of this relationship, but it is possible that if Bilophila contributes toward fatty liver, Oscillospira counteracts its effects in some way.
High levels of HFF were seen at both very low and very high abundance of Bacteroides, whereas moderate HFF corresponded with moderate abundance of Bacteroides. Bacteroides is very common in the human gut, and has been associated with a “Western Diet”(37) but certain species of Bacteroides have also been negatively associated with obesity.(38)
Dietary components were not highly predictive of hepatic fat without other information. Monounsaturated fat, total fat and carbohydrates were highly correlated with each other; thus, it is difficult to disentangle their effects. The protective effect of monounsaturated fat was the most consistent association, and it agrees with prior related research.(7) Given that many of the taxa that were associated with HFF in our results are also associated with diets high in animal products, it is somewhat surprising that dietary components were not more predictive of fatty liver. Total fat was selected as important, which might correlate with a diet high in animal products, but other indicators of a diet high in animal products, for example protein, saturated fats, or meat were not associated with HFF. Accurately capturing diet has known challenges, and likely does not fully capture all of dietary components that may be on importance for the gut microbiota, such as prebiotic food intake. (39) Since prebiotics have been proposed as a potential avenue for treatment or prophylaxis of NAFLD,(9) we expected to see either importance of fiber (prebiotics are a specific type of fiber), or evidence of protective associations for HFF with taxa that have previously shown to bloom in response to prebiotics, such as Faecalibacteria, Eubacteria, or Akkermansia.(40) This was not the case, but it may reflect that there is diversity across individuals in the taxa present in “healthy” taxonomic composition.
This study has some important limitations. Since it is cross-sectional, we cannot draw any conclusions about the direction of associations between gut microbiota and liver fat. The highlighted taxa may contribute towards fatty liver or may be the result of obesity and fatty liver. We identified numerous taxa that may prove to be important in the pathophysiology of NAFLD, but larger longitudinal studies would be necessary to further understand and confirm our findings. This is an ethnically diverse cohort, but there was not enough sample size within each racial / ethnic group to examine specific patterns in the association between gut microbiota and HFF. One methodological limitation is that we did not explicitly separate the data into a training set and test set for the random forests due to a relatively small number of individuals with NAFLD. However, we were able to estimate the error rates of the random forests using 3-fold repeated cross-validation, which repeatedly separates two-thirds of the data into a training set, using the remaining third as a test set.
There are many strengths of this study as well. We have a measure of hepatic fat from MRI scans on over 100 adolescents, dietary information from a questionnaire designed specifically for children, and a good distribution of weight groups across values of hepatic fat. Due to the young age of the participants in this study, the confounding by alcohol intake and prescription medications is likely much less than in older populations. We used machine learning methods, which are particularly suited to the analysis of complex gut microbiota data.
Our results show associations of the microbiota diversity and composition with fatty liver in adolescents. The taxa highlighted in our results support the notion that gut microbiota taxa may play a role in the pathogenesis of NAFLD, possibly through interactions with bile acid metabolism.(29) Furthermore, our results suggest that in the future, the gut microbiota may offer potential to help identify youth at risk for NAFLD or to identify youth who may be particularly amenable to microbiota-based interventions for NAFLD.
Supplementary Material
Acknowledgments
Statement of Financial support: The EPOCH study was funded by grant R01 DK068001 (PI: Dr. Dabelea) from the National Institute of Diabetes, Digestive and Kidney Diseases (NIDDK)
Footnotes
Disclosure Statement: The authors have nothing to disclose.
References
- 1.Loomba R, Sanyal AJ. The global NAFLD epidemic. Nat Rev Gastroenterol Hepatol. 2013;10:686–90. doi: 10.1038/nrgastro.2013.171. [DOI] [PubMed] [Google Scholar]
- 2.Bellentani S, Marino M. Epidemiology and natural history of non-alcoholic fatty liver disease (NAFLD) Ann Hepatol. 2009;8:S4–S8. [PubMed] [Google Scholar]
- 3.Ali R, Cusi K. New diagnostic and treatment approaches in non-alcoholic fatty liver disease (NAFLD) Ann Med. 2009;41:265–78. doi: 10.1080/07853890802552437. [DOI] [PubMed] [Google Scholar]
- 4.DeVore S, Kohli R, Lake K, et al. A Multidisciplinary Clinical Program is Effective in Stabilizing BMI and Reducing Transaminase Levels in Pediatric Patients with NAFLD. J Pediatr Gastroenterol Nutr. 2013;57:119–23. doi: 10.1097/MPG.0b013e318290d138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boursier J, Diehl AM. Implication of Gut Microbiota in Nonalcoholic Fatty Liver Disease. PLOS Pathog. 2015;11:e1004559. doi: 10.1371/journal.ppat.1004559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leung C, Rivera L, Furness JB, Angus PW. The role of the gut microbiota in NAFLD. Nat Rev Gastroenterol Hepatol. 2016;13:412–25. doi: 10.1038/nrgastro.2016.85. [DOI] [PubMed] [Google Scholar]
- 7.Finelli C, Tarantino G. Is there any consensus as to what diet or lifestyle approach is the right one for NAFLD patients. J Gastrointestin Liver Dis. 2012;21:293–302. [PubMed] [Google Scholar]
- 8.Canfora EE, Jocken JW, Blaak EE. Short-chain fatty acids in control of body weight and insulin sensitivity. Nat Rev Endocrinol. 2015;11:577. doi: 10.1038/nrendo.2015.128. [DOI] [PubMed] [Google Scholar]
- 9.Tarantino G, Finelli C. Systematic review on intervention with prebiotics/probiotics in patients with obesity-related nonalcoholic fatty liver disease. Future Microbiol Lond. 2015;10:889–902. doi: 10.2217/fmb.15.13. [DOI] [PubMed] [Google Scholar]
- 10.Hussain HK, Chenevert TL, Londy FJ, et al. Hepatic Fat Fraction: MR Imaging for Quantitative Measurement and Display—Early Experience. Radiology. 2005;237:1048–55. doi: 10.1148/radiol.2373041639. [DOI] [PubMed] [Google Scholar]
- 11.Hussain HK, Oral EA, Rohrer S, et al. Validation of Hepatic Fat Quantified on 3T MRI via Histopathologic Correlation in Patients with Non-Alcoholic Steatohepatitis. Proc Int Soc Magn Reson Med. 2008;16 [Google Scholar]
- 12.Rastogi R, Gupta S, Garg B, Vohra S, Wadhawan M, Rastogi H. Comparative accuracy of CT, dual-echo MRI and MR spectroscopy for preoperative liver fat quantification in living related liver donors. Indian J Radiol Imaging. 2016;26:5–14. doi: 10.4103/0971-3026.178281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Block G, Murphy M, Roullet J, Wakimoto P. Pilot validation of a FFQ for children 8–10 years; Fourth International Conference on Dietary Assessment Methods; 2000. [Google Scholar]
- 14.Crume TL, Scherzinger A, Stamm E, et al. The Long-term impact of intrauterine growth restriction in a diverse US cohort of children: The EPOCH study. Obesity. 2014;22:608–15. doi: 10.1002/oby.20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Centers for Disease Control and Prevention. SAS Program (ages 0 to < 20 years) | Resources | Growth Chart Training | Nutrition | DNPAO | CDC [Internet] [cited 2017 Jan 6];Growth Chart Train. Available from: https://www.cdc.gov/nccdphp/dnpao/growthcharts/resources/sas.htm.
- 16.CDC. [cited 2017 Mar 14];Defining Childhood Overweight & Obesity [Internet] Available from: https://www.cdc.gov/obesity/childhood/defining.html.
- 17.Centers for Disease Control and Prevention. [cited 2017 Dec 8];National Health and Nutrition Examination Survey (NHANES) Anthropometry procedures manual [Internet] 2007 Available from: https://www.cdc.gov/nchs/data/nhanes/nhanes_07_08/manual_an.pdf.
- 18.Crume TL, Ogden L, Daniels S, Hamman RF, Norris JM, Dabelea D. The Impact of In Utero Exposure to Diabetes on Childhood Body Mass Index Growth Trajectories: The EPOCH Study. J Pediatr. 2011;158:941–6. doi: 10.1016/j.jpeds.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.R Core Team. R: A language and environment for statistical computing [Internet] Vienna, Austria: R Foundation for Statistical Computing; 2016. Available from: https://www.R-project.org/ [Google Scholar]
- 20.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl Environ Microbiol. 2006;72:5069–72. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 23.Lozupone C, Lladser ME, Knights D, Stombaugh J, Knight R. UniFrac: an effective distance metric for microbial community comparison. ISME J. 2011;5:169. doi: 10.1038/ismej.2010.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friedman J, Hastie T, Tibshirani R. Sparse inverse covariance estimation with the graphical lasso. Biostatistics. 2008;9:432–41. doi: 10.1093/biostatistics/kxm045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu L, Baker SS, Gill C, et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: A connection between endogenous alcohol and NASH. Hepatology. 2013;57:601–9. doi: 10.1002/hep.26093. [DOI] [PubMed] [Google Scholar]
- 27.Del Chierico F, Nobili V, Vernocchi P, et al. Gut microbiota profiling of pediatric NAFLD and obese patients unveiled by an integrated meta-omics based approach. Hepatology. 2016 doi: 10.1002/hep.28572. n/a-n/a. [DOI] [PubMed] [Google Scholar]
- 28.Sze MA, Schloss PD. Looking for a Signal in the Noise: Revisiting Obesity and the Microbiome. mBio. 2016;7:e01018–16. doi: 10.1128/mBio.01018-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poeta M, Pierri L, Vajro P. Gut–Liver Axis Derangement in Non-Alcoholic Fatty Liver Disease. Children. 2017;4:66. doi: 10.3390/children4080066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ridlon JM, Kang DJ, Hylemon PB, Bajaj JS. Bile Acids and the Gut Microbiome. Curr Opin Gastroenterol. 2014;30:332–8. doi: 10.1097/MOG.0000000000000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hardison WG. Hepatic taurine concentration and dietary taurine as regulators of bile acid conjugation with taurine. Gastroenterology. 1978;75:71–5. [PubMed] [Google Scholar]
- 32.Devkota S, Wang Y, Musch M, et al. Dietary fat-induced taurocholic acid production promotes pathobiont and colitis in IL-10−/− mice. Nature. 2012;487:104–8. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Keefe SJD, Li JV, Lahti L, et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun. 2015;6:6342. doi: 10.1038/ncomms7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu GD, Chen J, Hoffmann C, et al. Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes. Science. 2011;334:105–8. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–63. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Konikoff T, Gophna U. Oscillospira: a Central, Enigmatic Component of the Human Gut Microbiota. Trends Microbiol. 2016;24:523–4. doi: 10.1016/j.tim.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 37.Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. [cited 2014 Apr 14];Nature [Internet] 2012 doi: 10.1038/nature11053. Available from: http://www.nature.com/doifinder/10.1038/nature11053. [DOI] [PMC free article] [PubMed]
- 38.Walters WA, Xu Z, Knight R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 2014;588:4223–33. doi: 10.1016/j.febslet.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Byers T. Food Frequency Dietary Assessment: How Bad Is Good Enough? Am J Epidemiol. 2001;154:1087–8. doi: 10.1093/aje/154.12.1087. [DOI] [PubMed] [Google Scholar]
- 40.Maier TV, Lucio M, Lee LH, et al. Impact of Dietary Resistant Starch on the Human Gut Microbiome, Metaproteome, and Metabolome. mBio. 2017;8:e01343–17. doi: 10.1128/mBio.01343-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.