Abstract
Gas vesicles are a unique class of gas-filled protein nanostructures whose physical properties allow them to serve as highly sensitive imaging agents for ultrasound and magnetic resonance imaging (MRI), detectable at sub-nanomolar concentrations. Here we provide a protocol for isolating gas vesicles from native and heterologous host organisms, functionalizing these nanostructures with moieties for targeting and fluorescence, characterizing their biophysical properties and imaging them using ultrasound and magnetic resonance imaging. Gas vesicles can be isolated from natural cyanobacterial and haloarchaeal host organisms or from E. coli expressing a heterologous gas vesicle gene cluster, and purified using buoyancy-assisted techniques. They can then be modified by replacing surface-bound proteins with engineered, heterologously expressed variants, or through chemical conjugation, resulting in altered mechanical, surface and targeting properties. Pressurized absorbance spectroscopy is used to characterize their mechanical properties, while dynamic light scattering and transmission electron microscopy are used to determine nanoparticle size and morphology, respectively. Gas vesicles can then be imaged with ultrasound in vitro and in vivo using pulse sequences optimized for their detection versus background. They can also be imaged with hyperpolarized xenon MRI using chemical exchange saturation transfer between gas vesicle-bound and dissolved xenon – a technique currently implemented in vitro. Taking 3–8 days to prepare, these genetically encodable nanostructures enable multi-modal, noninvasive biological imaging with high sensitivity and potential for molecular targeting.
Keywords: Gas vesicles, protein nanostructures, noninvasive imaging, contrast agents, molecular reporters, ultrasound, hyperpolarized MRI, magnetic resonance imaging, acoustics
EDITORIAL SUMMARY
The physical properties of a unique class of proteins called gas vesicles allow them to be used as contrast agents for ultrasound and magnetic resonance imaging (MRI). This protocol describes how to isolate gas vesicles, functionalize them with moieties for targeting and fluorescence, and image them in vitro and in vivo.
INTRODUCTION
The study of biological function and disease and the development of clinical diagnostics require technologies for noninvasive imaging of cells and molecules in intact organisms. Ultrasound and magnetic resonance imaging (MRI) are two widely used noninvasive imaging modalities that provide high spatial and temporal resolution but are currently restricted in their molecular imaging capabilities due to limited repertoires of nanoscale contrast agents. For ultrasound, conventional imaging agents based on microbubbles – micron-sized particles of trapped gas stabilized by a lipid or protein shell – are limited by their size and physical instability to labeling the vasculature and endovascular targets1,2. Conventional MRI contrast agents based on superparamagnetic iron oxides3,4 or lanthanide chelates5 are limited by their potential toxicity and the requirement that they be present at relatively high concentrations (typically in the μM range) for detection by MRI .
Development of the Protocol
To develop biomolecular imaging agents addressing the aforementioned limitations, we recently introduced a new class of molecular imaging agents for ultrasound and MRI based on gas vesicles (GVs) – genetically encoded gas-filled protein nanostructures from buoyant photosynthetic microorganisms6,7. GVs comprise a protein shell with spatial dimensions of ~200 nm and thickness of ~2 nm enclosing a hollow gas-filled interior. Gas dissolved in the surrounding media partitions freely in and out of this nanoscale compartment, while liquid water is excluded by the shell’s hydrophobic inner surface. Microbes such as the cyanobaterium Anabaena flosaquae (Ana) and the archaeon Halobacteria salinarum (Halo) form GVs as a means to regulate cellular buoyancy for optimal access to light and nutrients. In these organisms, GVs are encoded by operons of 8–14 genes, which include the primary structural proteins GvpA and GvpC and several minor constituents and chaperones.
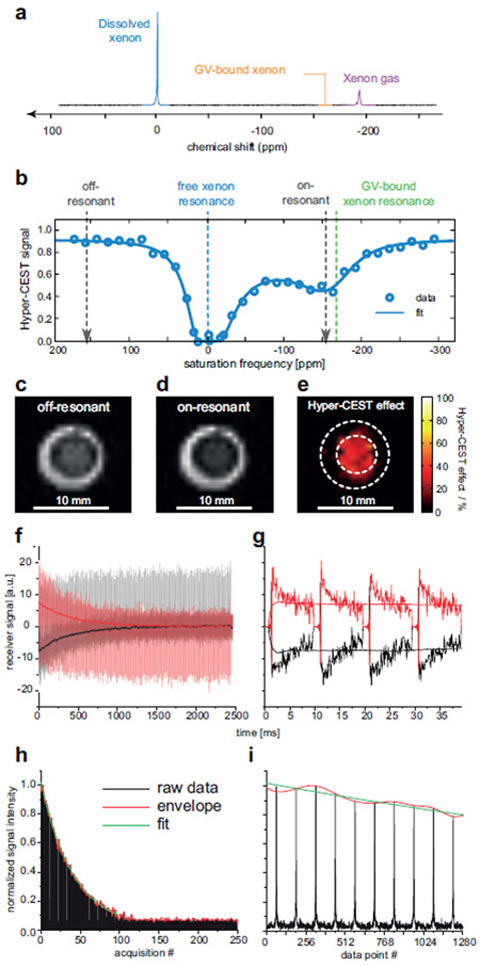
We recently discovered that GVs purified from cyanobacteria and haloarchaea scatter sound waves and thereby produce ultrasound contrast at sub-nanomolar concentrations8. Furthermore, we found that GVs’ unique physical properties enable them to produce harmonic ultrasound signals useful for contrast specificity in vitro and in vivo9,10. We also showed that GVs from different species can be imaged in multiplex based on their differential responses to acoustic pressure, and that conditional GV clustering leads to contrast enhancement, allowing them to be used as molecular sensors8. In parallel, we demonstrated that GVs produce contrast in hyperpolarized xenon MRI11, an emerging form of imaging that takes advantage of non-equilibrium spin polarization to increase molecular sensitivity by factors of up to 105. We discovered that atoms of hyperpolarized 129Xe dissolved in aqueous media exchange in and out of the GV interior, where their distinct chemical shift enables amplified MRI contrast to be produced using chemical exchange saturation transfer (CEST) pulse sequences. Using this hyperpolarized CEST (HyperCEST) technique allows GVs to be detected at picomolar concentrations11. In addition, the distinct chemical shifts of GVs in different microbial species enable multiplexed imaging. HyperCEST is currently performed in vitro, but is advancing toward in vivo use12,13.
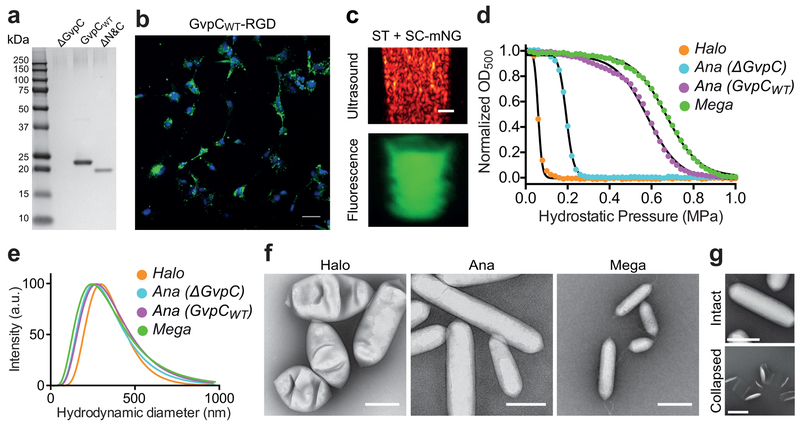
One particularly promising aspect of GVs is the possibility of engineering their physical and biochemical properties at the genetic level. To take advantage of this possibility, we recently developed a convenient genetic engineering platform, in which a single target protein, GvpC, on the surface of GVs purified from Ana cells is replaced by heterologously expressed engineered versions14. This strategy allows the modification of GV mechanical properties, including their harmonic response to sound waves and collapse under pressure for enhanced contrast and multiplexed imaging. In addition, genetic modification enables surface display of peptides such as lysine-rich protein to modify surface charge, CD47 to minimize uptake by macrophages, polyarginine for cellular internalization and RGD for targeting to integrin receptors on tumor cells14.
Overview of the Procedure
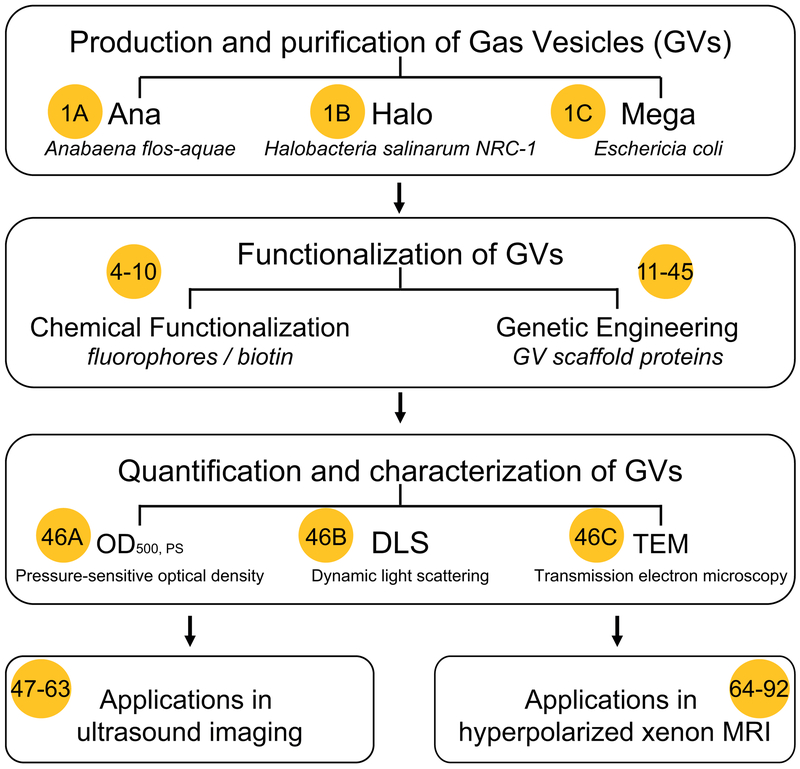
Here, we present a detailed optimized protocol to express and purify GVs (Steps 1|(A-C)) and quantify (Steps 2–3), functionalize and genetically engineer them (Steps 4–45), characterize their size and shape (Steps 46|(A-C)), and use them as contrast agents for non-invasive imaging applications (Steps 47–98). As illustrated in Figure 1, the procedure begins with isolation of GVs from cultures of Ana and Halo, or from E. coli expressing a heterologous GV gene cluster from Bacillus megaterium (Mega)15 (note that the Mega GVs were referred to as E. coli GVs in the cited paper). This results in three nanostructure populations with distinct properties (Table 1). These GVs are then quantified and characterized using pressure-dependent spectroscopy, dynamic light scattering (DLS) and transmission electron microscopy (TEM). Protocols for modification and functionalization using genetic and chemical approaches are provided as an added option based on the desired end application. Finally, the GVs are imaged in vitro and in vivo using ultrasound and in vitro using HyperCEST MRI.
Figure 1. Flowchart illustrating the experimental design and workflow for gas vesicle (GV) production, quantification, characterization, functionalization and imaging applications.
Table 1:
Characteristics of different types of GVs
| Ana GV | Halo GV | Mega GV | |
|---|---|---|---|
| Host/origin | Anabaena flosaquae | Halobacteria salinarum | Heterologous expression of a gene cluster from Bacillus megaterium in E. coli. |
| Shape | Cylindrical | Spindle | Cylindrical |
| Resistance to pressure-induced collapse | Medium (can be tuned) | Low | High |
| Ultrasound contrast | High Nonlinear after engineering |
High Nonlinear |
Low |
| Stability in Xe-MRI phantoms | High | Low | High |
| Ease of genetic modification | High | Low | Not established |
Advantages and Limitations:
Among the properties that make GVs attractive as imaging agents are their nanometer size, their fundamental physical stability, their biocompatibility as particles made entirely of protein, and their lack of requirement for metals or other inorganic cofactors to produce image contrast. In addition, as the first genetically encodable contrast agents for ultrasound and hyperpolarized xenon MRI, they offer the possibility of being developed as reporter genes16, creating new opportunities for non-invasive imaging at the molecular and cellular level.
Among the limitations of GVs in molecular imaging applications are their lower echogenicity per particle compared to conventional microbubble contrast agents for ultrasound and the specialized hyperpolarized Xe approach currently needed to take advantage of their sensitivity as reporters for MRI. In addition, the modular engineering approach presented here is done in vitro on purified GVs, and future efforts will focus on extending this platform to engineering of these nanostructures completely within cells. Furthermore, the use of GVs as dynamic molecular sensors based on conditional clustering requires ascertaining that a given molecular design does not result in non-specific aggregation, which must be done on a case-by-case basis.
EXPERIMENTAL DESIGN:
Here, we describe key experimental parameters for each stage of the protocol. One important initial consideration is the choice of GV-producing species. Although this protocol presents methods for producing three different types of GVs – Ana, Halo and Mega – one of these types may be most appropriate for a given application (see Table 1). For ultrasound, unmodified Halo GVs can be used directly in ultrasound imaging to obtain non-linear signals8,10. Ana GVs are the system of choice if one wishes to genetically tune the properties of GVs for multiplexing, multimodal imaging and targeting applications9,14. Mega GVs produce lower echogenicity under ultrasound compared to Ana and Halo GVs (Figure S1, a-d), but have a higher critical collapse pressure that may make them useful for multiplexing. Halo GVs produce non-linear ultrasound contrast immediately after purification, while Ana GVs require a chemical treatment. With regard to Xe-MRI, Ana and Mega GVs are more stable under pressure and during the bubbling of hyperpolarized xenon (as described below) compared to Halo GVs11. All three species have a unique chemical shift in Xe-MRI, allowing multiplexing.
Production and Purification of Gas Vesicles (GVs):
GVs are obtained from Ana, Halo or heterologously-expressing E. coli. Ana is a green, filamentous cyanobacterium that naturally inhabits fresh water lakes6. Halo is a pink halophilic and thermophilic archaea that grows in salt-water ponds7. Ana is cultured in low-salinity medium supplemented with trace metals and buffering agents, while Halo is cultured in high salinity medium for GV production.
Ana and Halo cultures natively produce ample GVs after a few weeks of growth8,9,14. Ana cultures additionally require a controlled gaseous environment and illumination for optimal growth (Figure 2a-b). A freshly inoculated culture of cells may require several rounds (typically 2–3) of subculture to become strongly proliferative (Figure 2c-d). The confluent culture of microbes is then transferred to a separatory funnel that is left undisturbed for up to a week to allow the buoyant cells producing GVs to float to the top and separate from media (Figure 2e-f). Buoyant cells are then lysed using hyper-osmotic shock for Ana and hypo-osmotic shock for Halo. Subsequently, centrifugally assisted floatation is used to isolate GVs from the cell lysate to yield a concentrated, milky-white solution of GVs in the buffer of choice8,9,14 (Figure 2g-j). Heterologous production of Mega GVs in E. coli is accomplished by expression from a plasmid encoding a Mega GV gene, followed by detergent-mediated lysis11.
Figure 2. Equipment setup and expected results for native production and purification of GVs.
(a-b) Shaker-incubators adapted for the growth of cyanobacteria Anabaena flosaquae providing controlled illumination, temperature, aeration and CO2. (c-d) Confluent green and pink cultures of Anabaena flosaquae and Halobacteria salinarum respectively, just before harvesting. (e-f) Effective separation of buoyant cells from spent media in separatory funnel for isolation and harvesting of Ana and Halo cells producing GVs. Purified Halo (g) and Ana (h) and Mega (i) GVs as a dense milky-white layer post centrifugally assisted floatation. (j) Resuspended milky-white solutions of purified Ana (left), Halo (middle) and Mega (right) GVs in PBS at OD500,ps ~6 prior to use in ultrasound imaging experiments.
The procedures leading from inoculation of GV-producing microbes to harvesting and purification is summarized in the Table 2, along with important parameters that affect processing time, yield and quality.
Table 2:
Experimental Parameters for GV Production, Purification and Storage
| Procedure | Design Parameters |
|---|---|
| Inoculation of starter culture | Type of culture (suspension vs. solid), amount of inoculum, total volume of culture |
| Growth of starter culture | Temperature, rotation speed, duration, illumination |
| Sub-culturing | Number of flasks, volume of culture and media |
| Harvesting of GVs | Composition of lysis buffer and duration of lysis, concentration of cells |
| Purification | Selection of centrifugation speed, type of rotor, tube and syringe needle |
| Storage | Storage temperature, buffer and type of vial/tube |
Growth conditions are chosen to facilitate optimal proliferation of each host strain and GV expression. One unusual variable to keep track of is pressure, since GVs collapse irreversibly at hydrostatic pressures of 50 to 800 kPa, depending on species6. For example, the cultures should be grown under mild agitation, as excessive shaking may lead to GV collapse. During centrifugation steps, it is necessary to calculate the hydrostatic pressure generated for a particular g-force on the liquid column of GVs and ensure that it is well below the GV critical collapse pressure. Long-term storage of purified GV stocks should preferably be done in screw-top vials, as micro-centrifuge tubes with snap-lock caps may cause GV collapse due to pressurization of the sample while opening or closing the tube.
Quantification and Characterization of GVs:
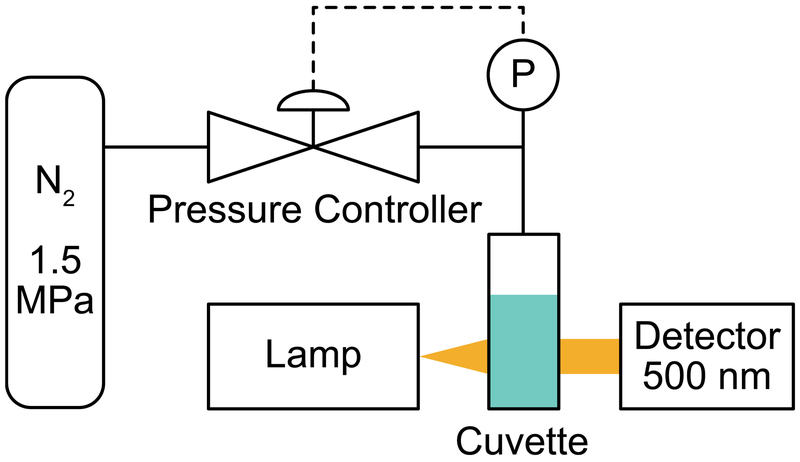
Purified GVs resuspended in the buffer of choice (e.g. phosphate buffered saline, PBS) can be quantified by measuring the optical density at 500 nm, or OD500, since GVs scatter visible light (Step 2). Collapsed GVs (in the same buffer), which do not scatter light, are typically used as the blank control for measurements, yielding a pressure-sensitive OD reading (OD500, ps). It is important to note that clustering of GVs, whether by design or due to functionalization with aggregation-prone moieties can confound OD500 measurements and contribute to errors in calculating concentration from OD500. Pressurized absorbance spectroscopy assays GV mechanical strength by measuring OD500 under increasing hydrostatic pressure using the device described in the Equipment Setup section (Figure 3) and the protocol described in Steps 46|(A)(i-ix).
Figure 3. Collapsometry setup.
Illustration of the collapsometry setup used for determining the critical collapse pressure of GVs.
Dynamic light scattering (DLS) is used to estimate the hydrodynamic size of GVs for routine non-destructive characterization and quality control (Steps 46|(B)(i-ii)). DLS can be used to assess GV clustering. Care should be taken in the interpretation of DLS readings of GVs due to the spherical assumption of the Einstein–Smoluchowski relation and the non-spherical shape of GVs. Negative contrast TEM is used for imaging GV size, shape, texture and integrity following production and physical or biochemical treatments (Steps 46|(C)(i-viii)). Negative staining with uranyl acetate is used to produce contrast, and use of a buffer such as HEPES is preferred over phosphate buffers that may precipitate with the uranyl acetate. The concentration of the GV solution spotted on the grid directly correlates with the density of GV particles on the grid.
Modification and Functionalization of GVs:
GVs can be modified and functionalized through genetic and chemical methods. Genetic engineering of GV mechanical and surface properties is conveniently performed via exchange of the native outer scaffolding protein GvpC (gas vesicle protein C) with recombinant GvpC variants (Steps 11–41). For example, fusing short peptides to the termini of GvpC allows for cell-specific targeting, and truncated GvpC variants enable acoustic multiplexing14. GvpC tolerates terminal fusions of peptides up to 100 residues. To covalently attach larger proteins, one can use the modular SpyTag-SpyCatcher protein assembly system17. GVs reconstituted with SpyTag-GvpC can be reacted with proteins of interest fused to SpyCatcher, forming a covalent bond (Steps 42–45).
Chemical conjugation to GVs makes use of lysine residues on their protein shells and amine-reactive crosslinkers such as sulfo-N-hydroxysuccinimide esters (Sulfo-NHS). Chemical moieties including polymers (e.g. polyethylene glycol), fluorophores and small molecules (e.g. biotin) can be conjugated using this method (Steps 4–10). Biotinylated GVs can subsequently react with streptavidin or avidinated antibodies8. The Sulfo-NHS coupling reaction can be conducted in PBS (pH 7.4) as a one-pot reaction. Depending on the application, the desired extent of labeling can be tuned by varying the molar ratio of Sulfo-NHS to GVs and by changing the incubation time. Either dialysis or buoyancy purification can be used to separate the labeled GVs from excess reactants.
Ultrasound Imaging of GVs:
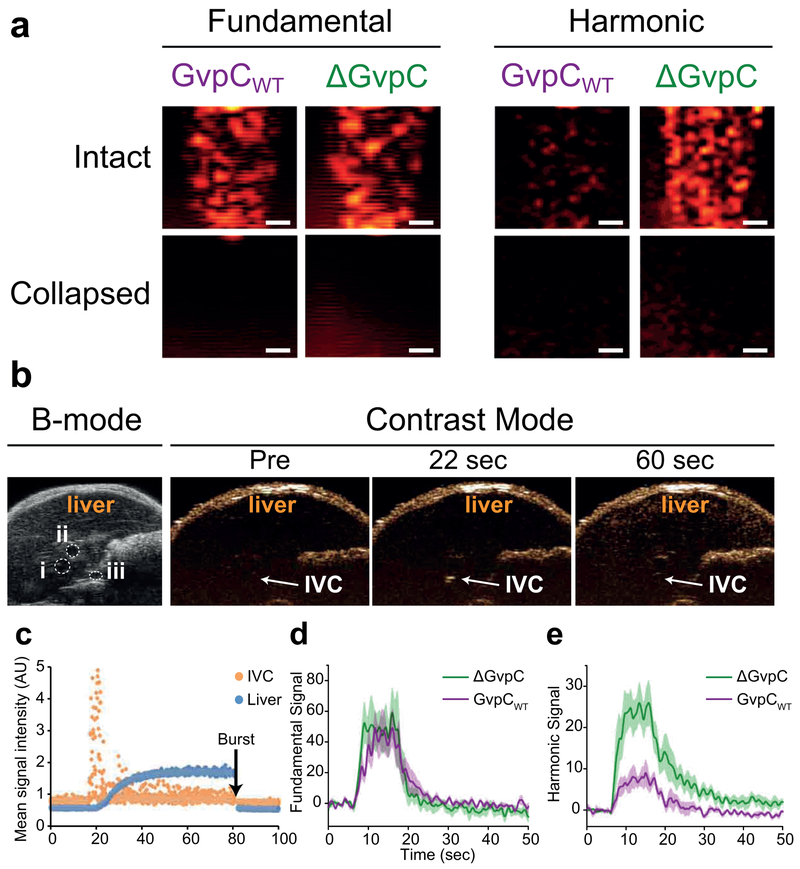
For in vitro imaging, GVs can be embedded and imaged in multi-well agarose phantoms (Figure 4a, Steps 47–62). A typical experimental setup for in vitro and in vivo ultrasound imaging is shown in Figure 4, a-d. Experimental design parameters include phantom composition, (i.e. percentage of agarose, buffer, background scattering particles), concentration and amount of GV sample loaded. Phantom molds can be made using 3-D printing to obtain defined well size, shape and spacing. Imaging parameters include plane of imaging (longitudinal versus transverse cross-section of the phantom wells), mode of ultrasound imaging (conventional or nonlinear imaging), transducer frequency range, transmit waveform characteristics such as pulse envelope shape, number of cycles, amplitude and frequency. Importantly, the amplitude used for imaging must be below the GVs’ acoustic critical collapse pressure. PBS is typically used as a negative control for ultrasound contrast and 5 μm polystyrene beads that scatter linearly at medical ultrasound frequencies are used as a reference sample. All GV samples and controls are mixed with melted agarose solution prior to loading. Solidification of the agarose after loading into the phantom wells ensures that samples are uniformly distributed throughout the well and that GVs remain suspended in the agarose matrix without floating during imaging. Typically, a final OD500 of 2.25 for Ana GVs and 0.4% (wt/vol) for polystyrene beads is used to match echogenicity. For Halo GVs, a final OD of 0.5 gives good signal without attenuation, with 0.83% (wt/vol) polystyrene beads to match echogenicity. In-situ collapse of GVs using pulses with amplitudes above the GVs’ acoustic critical collapse pressures cause GV ultrasound signals to disappear, allowing confirmation of GV-based signals and background subtraction. In addition, differential collapse at multiple pressures and spectral unmixing allow multiplexed imaging of mechanically distinct GV classes14. Quantification of signals in ultrasound images is typically performed using MATLAB or ImageJ software.
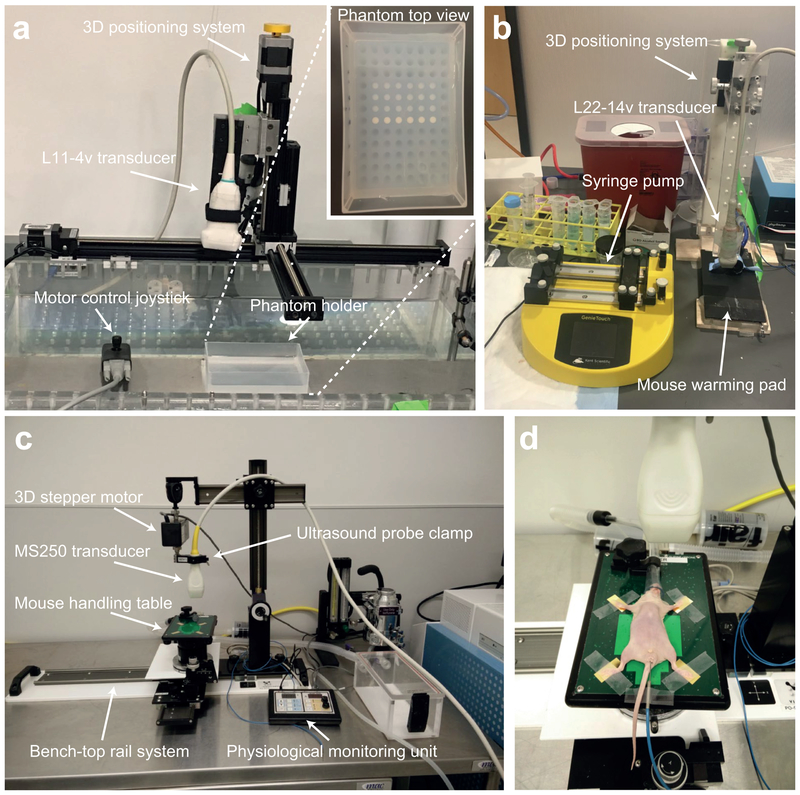
Figure 4. Ultrasound setup for in vitro and in vivo imaging.
(a) Setup of the in vitro imaging system with the Verasonics L11–4v transducer mounted on a 3-D translatable clamp (computer-controlled) and a fixed imaging phantom holder. Inset shows an agarose phantom loaded with GV and polystyrene samples prior to imaging. (b) In vivo imaging setup with the Verasonics L22–14v transducer, mounted on a 3-D positioning system (manual), an animal mounting platform with a heating pad and a syringe pump for controlled sample injections. (c) Setup of the Vevo imaging station includes the ultrasound probe clamp attached to the 3D stepper, the mouse handling table and the physiological monitoring unit. All components except the physiological monitoring unit are attached to the bench-top rail system. (d) Animal positioning with the mouse being laid down in a supine position with the nose and mouth in the nose cone and each paw extending outwards onto the electrodes and secured with surgical tape. A rectal probe is used to monitor the core temperature of the animal.
The in vivo ultrasound imaging protocol has been used to look at GV passage through the inferior vena cava (IVC) and subsequent induced contrast enhancement in the liver after intravenous injection8,14 (Step 63|(A-B)), but can be adapted for other organs or tissues. Depending on the tissue region of interest, some ultrasound imaging parameters, such as image gain and field of view may need to be adjusted accordingly. As in in vitro imaging, it is critical that the transmit power is kept at a value that provides sufficient signal without collapsing the GVs. Further adjustments can be made to GV solution concentration and volume, according to experimental needs. Functionalized GVs can also be imaged using this protocol, as long as appropriate controls are used. When planning an imaging experiment using functionalized GVs, we suggest using native GVs from the same batch as a control. It is possible to administer multiple injections of GVs in the same mouse, as long as the total injection volume does not exceed the limit stated in institutional guidelines. If required, GV solutions can be tested for bacterial endotoxins using quantitative, chromogenic endpoint LAL assays, such as the QCL-1000™ Assay (Lonza). Endotoxins can be removed using commercially available affinity resins such as the ToxinEraser™ Endotoxin Removal Resin (GenScript). Once injected, ensure GVs have cleared completely and tissue contrast signal is back to baseline before a second bolus injection.
129Xe MRI Imaging of GVs:
Hyperpolarized 129Xe-MRI is an active area of molecular imaging research, which is currently mainly performed in vitro with pure contrast agents or labeled cells18,19, while in vivo techniques are being developed12,13. This protocol describes the imaging of GVs using hyperpolarized 129Xe MRI in vitro, as previously done with purified GVs, GV-expressing cells and GV-labeled cells11. This involves preparing the hyperpolarized gas, introducing it into a liquid sample of GVs and applying a HyperCEST pulse sequence11 (Steps 64–92). This requires some specialized equipment, some of which has been described earlier in the context of xenon NMR and MRI applications. The working steps and the equipment setup in particular require people with skills in NMR/MRI and some experience in setting up custom-made electronic devices. The hyperpolarization system is available as a commercial unit (see Equipment section and Figure 5a-e) or can be built according to previous publications20. In either case, the system should be capable of delivering hyperpolarized 129Xe under very stable, continuous flow conditions 21,22.
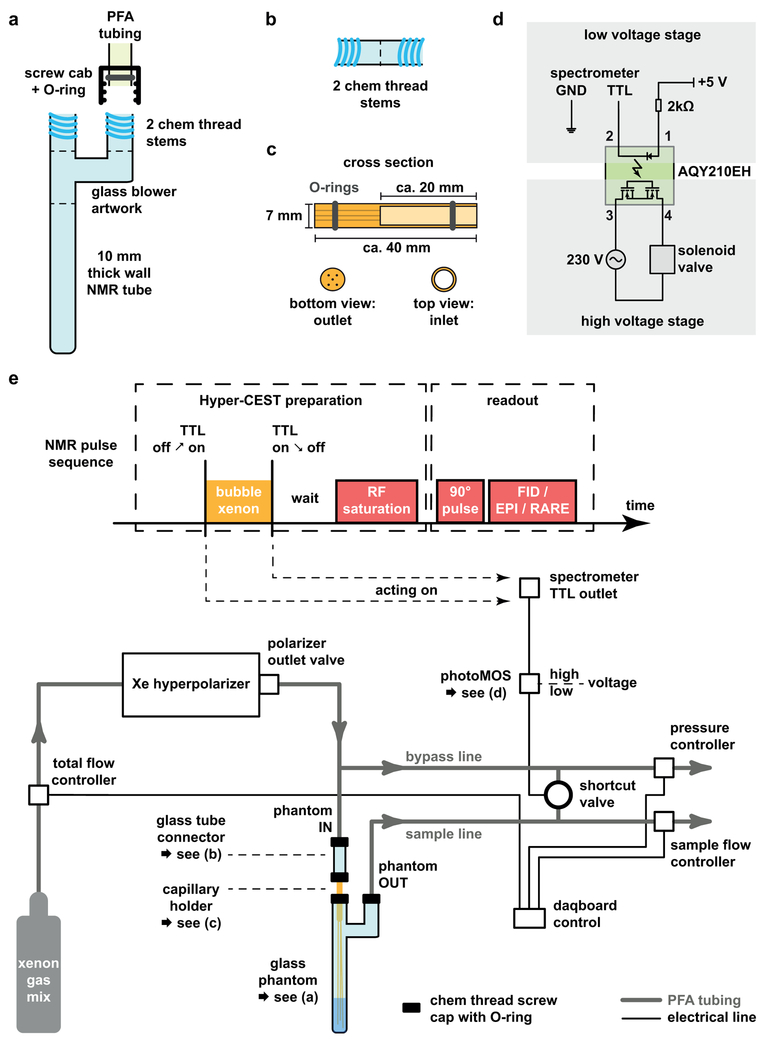
Figure 5. Required elements for assembling the gas delivery manifold for 129Xe-MRI.
(a) Main glass phantom made from 10 mm thick wall NMR tube and chem thread connectors; PFA tubings are connected with a chem thread srew cap and an O‐ring. (b) Glass tube connector. This is also used for step 3, but mainly for connecting the PFA tubing with the capillary holder. (c) Capillary holder. (d) PhotoMOS circuit for controlling the solenoid valve. (e) Schematics of the communication between the NMR pulse sequence and the gas delivery manifold with corresponding tube and electrical connections.
To achieve good Hyper-CEST performance with a particular GV type, it is necessary to optimize the saturation power and duration to match the exchange conditions for Xe between GVs and the surrounding media23,24. In addition, attention must be paid to GV integrity during the bubbling or inflow of xenon into the phantom, since both hydrostatic pressure and bubbles can cause irreversible GV collapse.
MATERIALS:
REAGENTS:
Anabaena flosaquae/Aphanizomenon flosaquae (Culture Collection of Algae and Protozoa, Scottish Marine Institute, cat. no. CCAP 1403/13F)
Halobacterium sp. NRC-1 as brine inclusions (Carolina Biological Supply Company, cat. no. 154777) (OR) Halobaterium sp. plate (Carolina Biological Supply Company, cat. no. 154801)
Rosetta™ 2(DE3) pLysS competent E. coli cells (EMD Millipore, cat. no.71401–3)
BL21 (DE3) competent E. coli cells (NEB, cat. no. C25271)
Cyanobacteria BG-11 Freshwater solution 50x (Sigma, cat. no. C3061)
Phosphate Buffered Saline without calcium and magnesium (PBS, Corning, cat. no. 21–040)
Sodium Nitrate, High Purity Grade (NaNO3, Amresco, cat. no. 0598)
Potassium Phosphate monobasic (KH2PO4, Sigma, cat. no. P5655)
Magnesium Chloride hexahydrate (MgCl2.6H2O, Sigma, cat. no. M9272)
Trisodium Citrate dihydrate (Sigma-Aldrich, cat. no. S1804)
Potassium Chloride (KCl, Sigma, cat. no. 746436)
Casein Hydrosylate (Sigma, cat. no. 22090)
Yeast Extract (BD, cat. no. 212750)
Magnesium sulfate heptahydrate (MgSO4.7H2O, Sigma, cat. no. 230391)
Calcium Chloride dihydrate (CaCl2.2H2O, BDH Chemicals, cat. no. BDH9224)
Sodium metasilicate nonahydrate (Na2SiO3.9H2O, Sigma, cat. no. S4392)
Sodium Carbonate (Na2CO3, Sigma, cat. no. 71345)
Sodium Bicarbonate (NaHCO3, Sigma, cat. no. S5761)
Citric Acid, 1M (BDH Chemicals, cat. no. BDH7397)
Ethylenediaminetetraacetic acid (EDTA), disodium salt dihydrate (Alfa Aesar, cat. no. A15161)
Ferric Ammonium Citrate (Amresco, cat. no. 0846)
Solulyse Bacterial Protein Extraction Reagent, in Tris.HCl Buffer, pH 7.4 (Genlantis, cat. no. L200500)
D-Sorbitol (Sigma, cat. no. S1876)
Deionized (DI) water (tap supply)
Anhydrous dimethyl sulfoxide (Life technologies, cat. no. D12345)
N-Hydroxy-Succinimidyl-Ester moiety such as Alexa-488-NHS (Thermo Fisher, cat. no. A20000)
Tris-HCl buffer (Teknova, cat. no. T1080)
HEPES (Sigma, cat. no. H3375)
Terrific broth (Sigma, cat. no. T0918)
Isopropyl β-D-1-thiogalactopyranoside (IPTG) (Anatrace, cat. no. I1003)
Kanamycin (Sigma-Aldrich, cat. no. K1377)
2% Uranyl Acetate (EMS, cat. no. 22400–2)
Urea (VWR, cat. no. 0568)
Sodium Hydroxide (NaOH, Sigma, cat. no. 221465)
Sodium Chloride (NaCl, Sigma, cat. no. 793566)
Sodium Phosphate Monobasic (NaH2PO4, Amresco, cat. no. 0823)
10X Tris/Glycine/SDS buffer (Bio-Rad, cat. no. 161–0732)
Protein standard (Bio-Rad, cat. no. 161–0375)
Laemmli sample buffer (Bio-Rad, cat. no. 161–0737)
Gel electrophoresis power supply (Bio-Rad, cat. no. 164–5050)
Gel electrophoresis cell (Bio-Rad, cat. no. 165–8004)
Any kD SDS-polyacrylamide gels (Bio-Rad, cat. no. 456–9034)
SimplyBlue™ SafeStain (Invitrogen, cat. no. LC6060)
Bradford Reagent (Sigma-Aldrich, cat no. B6916–500)
Pierce™ 660 nm protein assay reagent (Thermo-Fisher Scientific, cat. no. 22660)
Bovine serum albumin (Thermo-Fisher Scientific, cat. no. 15561020)
Ni-NTA Superflow Resin (Qiagen, cat. no. 30450)
Poly-prep chromatography columns (Bio-Rad, cat. no. 7311550)
Imidazole (Sigma, cat. no. 792527)
2-mercaptoethanol (Bio-Rad, cat. no. 1610710)
DNaseI (NEB, cat. no. M0303S)
Lysozyme (Sigma-Aldrich, cat. no. L6876)
GvpCWT, ΔN&C-GvpC and GvpC-SpyTag genes in pET28a plasmid (Addgene, ID no. 85732, 85736 and 85737 respectively)
SpyCatcher-mNeonGreen in pET26b plasmid (https://www.addgene.org/vector-database/9135 )
pST39 plasmid containing pNL29 Mega GV gene cluster (Addgene ID no. 91696)
SOC Outgrowth Medium (NEB, cat. no. B90205)
Luria-Bertani (LB) Broth base powder (Invitrogen™ by Life Technologies, cat. no.12795–084)
Ampicillin (Sigma, cat. no. A0166–25G)
Chloramphenicol (Sigma-Aldrich, cat. no. C0378)
Glucose (Sigma-Aldrich, cat. no.G8270)
Glycerol (Sigma-Aldrich, cat. no.G5516)
Phosphate buffered saline (PBS), 20x solution (Teknova, cat. no. P1225)
-
SCID Hairless Outbred (SHO) (OR) SCID Hairless Congenic (SHC) mice, female, 4–6 weeks of age (Charles River, cat. no. 474 (OR) 488)
!CAUTION All procedures involving animal use must be performed in accordance to institutional guidelines and regulations and approved by relevant Animal Care and Use Committees.
Sterile PBS (Life Technologies, cat. no.10010–023)
Isoflurane USP (Fresenius Kabi, cat. no.M60303)
-
Medical air USP cylinder (Praxair)
!CAUTION Gaseous anesthesia must be scavenged properly using a charcoal canister or house vacuum to minimize potential exposure to humans.
Charcoal canister (F-Air, Braintree, cat. no. F-AIR EA)
Eye ointment (Lacri-Lube, Refresh) (OR) Lubricating gel (Aquagel, Parker, cat. no. 57–05)
0.9% Sodium Chloride Injection, (Baxter, cat. no. JB1322 (OR) VWR, cat. no. 47729–565 )
Ultrasound gel (WavelengthCL, National Therapy Production Inc., cat. no. NTPC500X) (OR)
Aquasonic 100 ultrasound transmission gel (Parker Lab Inc. ,VWR, cat. no. 68200–712)
Electrode gel (Signa gel, Parker Lab Inc., cat. no. 15–25)
Nair™ lotion (Church & Dwight co., Inc.)
Vetbond tissue adhesive glue (Fisher Scientific, cat. no. NC9259532)
SPHERO™ polystyrene particles, 4–5-4.9 μm (Spherotech, cat. no.PP-45–10)
Agarose (Biorad, cat. no. 161–3102)
-
Xenon gas mixture: 2–5% (vol/vol) Xe (natural abundance with 26.4% (vol/vol) 129Xe is sufficient), 10% (vol/vol) N2, He balance, grade 5.0 (Praxair))
▴CRITICAL The gas tank should have a pressure regulator for adjustments around 2 bar absolute pressure. The gas amount to be used depends on the flow rate through the system. A typical value would be 0.25 L/min, i.e. 15 L per hour.
EQUIPMENT:
Shaker-Incubator with cooling, CO2 control and illumination (Infors HT, Multitron II) (Exact setup used is shown in Figure 2a-b)
Benchtop centrifuge with swinging bucket rotor (Beckman Coulter, Allegra X-15R)
Benchtop micro-centrifuge (Eppendorf 5424R)
Barnstead™ GenPure™ xCAD Plus UV/UF Water Purification System (Thermo Scientific, cat. no. 50136146)
Pyrex™ 500 mL and 1000 mL Erlenmeyer Flasks (Corning, cat. no.4980, stopper no. 9)
Pyrex™ 500 mL Separatory Funnel (Cat. no.6402–500, stopper no. 24)
Pyrex™ Culture Tubes (Corning, cat. no.9820)
Pyrex™ 250 mL, 500 mL and 1 L media storage bottles (Corning, cat. no. 1395)
4L Beaker (Thermo Scientific Nalgene, cat. no. 1201–4000)
15 and 50 mL centrifuge/falcon tubes
1.5, 1.7 and 2 mL micro-centrifuge tubes
Metal ring stand for separatory funnel
Vacuum driven disposable bottle top filter (Durapore™,0.22 μm, Millpore, cat. no. SCGVT05RE)
Adjustable volume pipets and pipet tips (100–1000 μL, 20–200 μL, 2–20 μL and 0.5–2.5 μL, Eppendorf Research Plus)
Serological pipets (1, 5, 10 mL and 25 mL, VWR)
Syringes, Luer-lok tip, 1mL, 3mL, 10 mL and 30mL (BD, cat. no.309602, 309657, 309604 and 309650)
18G and 1”/1.5” 21.5G flat bottom needles (SAI B18–100 and B21–150/B21–100)
Gel-loading pipet tips (Corning, cat. no. 4884)
Regenerated cellulose dialysis tubing, 6–8kDa cutoff (Spectrapor, cat. no. 1326650)
Formvar/Carbon 200 mesh Cu TEM grids (Ted Pella Inc., cat. no. 01801)
Ultracentrifuge (Beckman-Coulter Avanti J-30I)
Ultracentrifuge bottles (Beckman-Coulter)
PD10 desalting columns (GE Healthcare Life Sciences, cat. no. GE17–0851-01)
1 cm path-length quartz cuvette (Hellma Analytics, cat. no. 176.700-QS)
Pressure controller (Alicat Scientific, cat. no. 73907)
UV-Vis Spectrophotometer (Ocean Optics Inc., STS series microspectrometer, 350–800 nm, 50 μm slit, 400 core/1cm SMA input, cat. no. STS-VIS-L-50–400-SMA) equipped with a 400 μm fiber in the VIS/NIR range (2m, cat. no. P400–2-VIS-NIR) and a Krypton lightsource (2000 hour bulb, ECOVIS) and accompanying Oceanview spectroscopy software.
-
Compressed Nitrogen cylinder with pressure regulator (Air Liquide)
!CAUTION Proper safety techniques should be followed when opening and handling the compressed gas cylinder. Make sure the cylinder is secured to a wall and properly connected to pressure regulator before opening.
ZetaPALS dynamic light scattering instrument (Brookhaven Instruments)
Disposable cuvette (Eppendorf, cat. no. Z605050)
Plate-reader spectrophotometer (Molecular Devices SpectraMax M5)
Gel imaging system (BioRad Chemidoc MP, model no. 170–8280)
Water bath (various sources, e.g. Fisher Scientific, cat. no.15–462-6Q)
Disposable culture tube, 16 × 100mm (VWR, cat. no. 47729–576)
Dialysis clips, weighted/unweighted closures, 55 mm sealing width (Spectrum Labs Inc., cat. no. 132745/132737)
Transmission electron microscope (FEI Tecnai T12 LaB6 120 kV) equipped with a Gatan Ultrascan 2k X 2k CCD and Leginon automated data collection software suite
Glow Discharging System (Emitek K100X)
PELCO reverse, anti-capillary tweezers (TedPella, 5378-NM)
Glass slides (VWR Microslides, cat. no. 48382–171)
Filter paper (Whatman™, cat. no. 1001–090)
Pipet-Aid (Drummond Scientific Co.)
Magnetic Stirring Bars (VWR)
Rotatory Shaker (Rotamix, Appropriate Technical Resources, model RKVS)
Microvolume Spectrophotometer (NanoDrop™ 2000C series, Thermo Scientific, cat. no. ND-2000C)
Tail vein catheter (SAI Infusion Technologies, cat. no. BF-27–01)
Syringe with tip cap, 12 mL (Luer Lock Kendall Monoject, cat. no.1181200777)
Needle, 27G x 1/2 (PrecisionGlide, BD, cat. no. 305109)
Tuberculin Syringe, 1/2cc (BD, cat. no. 305620)
Surgical tape (Transpore, 3M, cat. no. 1527–1)
Alcohol wipes (Webcol, Covidien, cat. no. 63001) (OR) (BD, cat. no. BD326895)
Cotton swabs
Syringe pump (New Era Pump System, cat. no. NE-1000 (OR) Kent Scientific Corporation, model. GenieTouch) (Typical setup shown in Figure 4b).
Heatlamp (Home Hardware)
Ultrasound imaging system (Vevo®2100, VisualSonics, Fujifilm (OR) Verasonics Vantage 256™, Verasonics. Inc.) (Typical setup shown in Figure 4a-d).
Ultrasound transducer (L11–4v Verasonics Inc., (OR) L22–14v Verasonics, Inc., (OR) MS250, VisualSonics, Fujifilm)
Acoustic absorber sheet, 300mm x 300 mm (Precision Acoustics Ltd., cat. no. F28-SMALL)
Multi-well phantom mold (3-D printed)
Plastic reservoir for phantom preparation (Nalgene™ flat-bottom polypropylene robotic reservoirs , cat. no. 1200–1300)
Computer-controlled translation stage (X-Slide, Velmex, Inc.).
MATLAB (Licensed Software, Mathworks, Inc.)
Animal heat pad (Braintree Scientific, Inc. cat. no. HP 1M)
Temperature controller (Braintree Scientific, Inc. cat. no. TCAT 2DF)
Rectal probe for mice (Braintree Scientific, Inc. cat. no. RET 3)
Vevo® Imaging Station (VisualSonics, Fujifilm)
PE10 plastic tubing (Becton Dickinson, cat. no. 427400)
30G needles (Becton Dickinson, cat. no. 305106)
Rodent anesthesia machine (Patterson Veterinary, cat. no. 07–870-3592 (OR) VetEquip, cat. no. 901806)
Weighing Balance (sensitive in the g and mg range)
-
MR imaging system (Bruker); preferably a vertical NMR spectrometer at field strength of 7 T or higher with micro-imaging capability for 1H and 129Xe; a variable temperature unit (VTU) is desirable to keep the sample temperature stable either at room temperature or at physiological body temperature.
▴CRITICAL The operating console needs at least one available trigger signal output (TTL) to be connected to a photoMOS relay for regulating the gas flow
Dual-tuned 1H/129Xe NMR coil with 10 mm inner diameter (Bruker)
Non-selective NMR pulse sequence for acquiring a direct Xe NMR spectrum after fresh Xe delivery. This is implemented by using a simple (pulse – acquire) sequence; the sequence starts with two trigger signals to communicate with the gas delivery manifold and then a block excitation pulse of adjustable amplitude, followed by an FID acquisition
Multi-echo acquisition MRI pulse sequence (echo planar imaging [EPI] or rapid acquisition with relaxation enhancement [RARE], Bruker) with activatable magnetization transfer module (alternative: fat suppression or saturation transfer module) and two triggers.
-
Hyperpolarization system for producing spin-hyperpolarized 129Xe(commercial systems are available through www.xemed.com, www.polarean.com or as an open source design)
! CAUTION The hyperpolarization system contains ~1 g of a highly reactive alkali metal such as Rb. Make sure to be familiar with the procedures for handling the system as provided by the manufacturer
▴CRITICAL The system must be capable of producing hyperpolarized Xe in continuous flow mode for repeated re-delivery into the sample solution.
-
Gas flow controller with a maximum flow rate of100 standard mL/min (SMLM) (Omega Engineering Inc., model no. FMA 5408) and second one of 500 SMLM flow (Omega Engineering Inc., model FMA5412)
▴CRITICAL Such gas flow controllers are typically calibrated for N2 gas; the correction factors for systems based on heat flux measurements applied to the Xe gas mix are: 1.44 for the Xe fraction, 1.454 for the He fraction. Hence the correction factor for a 5% Xe mix is 0.05×1.44 + 0.1×1 + 0.85×1.454 = 1.408.
Pressure controller for up to 6 bar and 1 standard liter per minute flow (Bronkhorst, model no. P-702CV-6K0A-AAD-22-V)
Direct solenoid actuated poppet “shortcut” valve (Norgren, model no. 9502310,)
PhotoMOS relay (Panasonic, model no. AQY210EH,)
PC-operated control interface for the hyperpolarization system and the gas flow controllers based on a data acquisition board (Daq/Board3000, Omega Engineering Inc.) and a graphic user interface (e.g., DASYLab or LabView, National Instruments)
Glass phantom (made from thick wall NMR tube of 10 mm outer diameter, Wilmad) with inlet and outlet adapter with size 7 chem threads (pieces CG-350–01 from ChemGlass Life Sciences), see Figure 5a
5 mm standard NMR tube to be inserted into the glass phantom to create a second compartment inside the 10 mm NMR tube
-
¼ inch PFA flexible tubing (Swagelok)
▴CRITICAL Ensure that the PFA tubing directing the Xe flow from the hyperpolarizer into the sample does not go through any zero-field crossings in the fringe field of the superconducting magnet because this might cause loss of hyperpolarization.
Fused silica glass capillaries for Xe dispersion (e.g five pieces of ca. 15 cm length each and 350 μm outer diameter, e.g. material no. 106815–0030, Polymicro)
Glass connecting tube with size 7 chem threads on either end (Figure 5b)
Home-made capillary holder; ca. 40 mm long, on the inlet side a 20 mm deep, 5mm diameter bore, on the outlet side 5 clear bores matching the outer diameter of the capillaries (like in Figure 5c; Teflon or Delrin material)
pH meter
Magnetic Stir Plate (Thermo Scientific, cat. no. S194925)
Autoclave
Bunsen Burner
Spark Lighter
REAGENT SETUP:
G625 growth medium for Anabaena flos-aquae (Ana) (4 L):
5.84 mM NaNO3, 224 μM KH2PO4, 304 μM MgSO4.7H2O, 208 μM Na2SiO3.9H2O, 189 μM Na2CO3, 10 mM NaHCO3, 245 μM CaCl2, 31 μM citric acid and 3 μM EDTA. Separately, add 24 mg of ferric ammonium citrate to 1 mL of ultrapure water (in a 1.5 mL micro-centrifuge tube). Heat in a 42°C water-bath for 10–15 min to dissolve before adding to the media in the Nalgene 4L beaker. Adjust to pH 8.0 with HCl. Filter sterilize using a 0.22 μm bottle-top filter. G625 growth media prepared using this method is stable for at least 6 months if stored in a cool, dry place at room temperature under sterile conditions. Since the iron sediments, make sure that the bottle is swirled each time before use.
Carolina growth medium for Halobacteria salinarum-NRC1 (Halo) (1 L):
4.278 M NaCl, 81 mM MgSO4.7H2O, 10 mM Trisodium citrate dihydrate, 27 mM KCl, 5g/L casein hydrolysate and 3g/L yeast extract. Adjust pH to 7.2 with NaOH and autoclave at 121˚C and 18 psi for 30 minutes. Autoclaved media can be stored for at least 6 months under sterile conditions at room temperature.
TMC lysis buffer for Halo GVs:
10 mM Tris-HCl, 2.5 mM MgCl2 and 2mM CaCl2, pH 7.5. Store indefinitely.
Sorbitol lysis buffer for Ana GVs:
1M D-Sorbitol in distilled water. Store indefinitely.
Terrific Broth (TB):
Add 47.6 g of TB powder and 8 mL glycerol to 1L of distilled water. Autoclave at 121°C for 15 m. Autoclaved media can be stored for at least 6 months under sterile conditions at room temperature.
Luria-Bertani (LB) Broth:
Add 25 g of LB powder to 1 L of distilled water. Autoclave at 121°C for 15 m. Autoclaved media can be stored for at least 6 months under sterile conditions at room temperature.
Kanamycin stock solution:
50 mg/mL in distilled water.
Ampicillin stock solution:
100 mg/mL in distilled water.
Chloramphenicol stock solution:
25 mg/mL in ethanol.
IPTG inducer stock solution:
1M IPTG in distilled water.
GV stripping buffer (Round 1):
10M urea, 100mM Tris-HCl, pH 8.0–8.5.
GV stripping buffer (Round 2):
6M urea, 60mM Tris-HCl, pH 8.0–8.5.
TGS buffer:
1:9 (v/v) dilution of 10X TGS stock in DI H2O.
Inclusion body solubilization buffer:
6M urea, 20mM Tris-HCl, 500mM NaCl, pH 8.0.
Inclusion body wash buffer:
6M urea, 20mM Tris-HCl, 500mM NaCl, 20mM imidazole, pH 8.0.
Inclusion body elution buffer:
6M urea, 20mM Tris-HCl, 500mM NaCl, 250mM imidazole, pH 8.0.
Soluble protein wash buffer:
50mM NaH2PO4, 300mM NaCl, 20mM imidazole, 1mM 2-mercaptoethanol, pH 8.0.
Soluble protein elution buffer:
50mM NaH2PO4, 300mM NaCl, 250mM imidazole, pH 8.0.
▴CRITICAL Antibiotic stocks and IPTG solution are at 1000x concentration and should be filter-sterilized after preparation and stored at −20°C. IPTG is light-sensitive and should be stored in the dark at −20°C. Lysis buffers and stock solutions for media preparation should be filtered (0.22 μm filter) and stored at room temperature. All protein purification buffers (wash and elution buffers) should be filter-sterilized and stored at 4°C.It is not advisable to use urea buffers more than a week old for protein purification. GV stripping buffers should be freshly prepared before each use.
Ultrasound gel preparation for imaging experiments:
Take a 30 mL luer-lok syringe, remove the piston and place the syringe with the tip facing down in a 50 mL falcon tube. Fill the syringe with ultrasound gel before loading the falcon tube into a table top centrifuge. Spin at 2500g for 30 minutes to ensure that there are no air bubbles in the gel and keep at room temperature. It is advisable to prepare the gel on the same day of the experiment, as longer storage may lead to drying if the syringe is not sealed tightly.
EQUIPMENT SETUP:
Cleaning and sterilization of glassware:
Clean and sterilize all glassware by autoclaving and fully dry them before use.
Setting up the apparatus for pressurized absorbance spectroscopy:
The setup used to conduct pressurized absorbance spectroscopy is illustrated in Figure 3 and includes the following components: (i) computer equipped with MATLAB to run the collapse pressure scripts, (ii) compressed N2 tank with control valves and a regulator, (iii) pressure controller, (iv) flow-through quartz cuvette, (v) UV-Vis spectrometer equipped with a light source and cuvette holder.
Couple the pressure controller to the gas tank via a stainless steel gas connector, to the computer via a USB port, and to the flow-through cuvette via plastic cannulae. Connect the Ecovis Krypton light source of the UV-Vis spectrometer to a power supply, allowing light to pass through the sample in the quartz cuvette. The transmitted light then passes through an optical fiber connector into the OceanOptics STS microspectrometer. The spectrometer is connected to the computer and controlled by a MATLAB script, which also interfaces with the pressure controller. These scripts can be downloaded from http://shapirolab.caltech.edu/?page_id=525. An equilibration time of 7 seconds at each pressure step and a final collapse pressure of 1400 kPa (for blanking) is hard-coded into the script and kept constant for all measurements. Before acquiring sample measurements, the system is calibrated by recording the spectra for full transmission (using the cuvette loaded with PBS or buffer of choice) and zero transmission (by turning the cuvette 90° so that no light passes through).
!CAUTION Ensure that the compressed nitrogen tank is transported and stored safely. Cylinders should be secured and stored upright in a cool dry area protected from combustible materials. The gas valve seal must always be in place. When not in use, keep all the cylinder and regulator valves shut and the pressure controller and spectrometer powered off.
Setting up the Verasonics imaging station and imaging parameters:
The Verasonics imaging system and setup for in vitro and in vivo experiments is shown in Figure 4a-b and comprises the following components: (i) computer, (ii) ultrasound probe, (iii) translatable probe clamp equipped with three independent motors that move in the x, y and z directions with μm-level precision (iv) fixed phantom holder and (v) Verasonics scripts running on MATLAB software. Adapt the setup for in vivo imaging by replacing the phantom holder with a mouse mounting platform equipped with a heating pad, anesthesia equipment and nose-cone for the animal and a manually translatable probe clamp.
CRITICAL Make sure that the probe is well coupled to the sample and that the transducer array of the probe is immersed in liquid or covered with ultrasound gel before transmitting ultrasound.
GVs can be imaged using the Ray-lines Verasonics example script. Imaging is typically performed at around 18 MHz using the L22–14v transducer with an F-number of 2 or 3, focal depth of 8 mm (matching the natural focus of the transducer) and 6 cycle pulses. For these parameters, nondestructive imaging of GVs can be performed at voltages of or below 3V. The onset of GV collapse usually takes place at around 5V. For efficient GV destruction, a 25V pulse is applied for a few seconds.
GVs are imaged at lower frequencies around 6.25 MHz using the L11–4v transducer and the corresponding Ray-lines script. The F-number is set to 2, transmit focus to 20 mm and transmit voltage to 1.6V. Images are saved from the Verasonics graphical user interface.
Setting up the Vevo imaging station and imaging parameters:
The imaging station setup is depicted in Figure 4c-d, and encompasses the following components: (i) bench-top rail system; (ii) 3D stepper motor; (iii) ultrasound probe clamp; (iv) mouse handling table; and (v) physiological monitoring unit. Make sure that the 3D motor is connected to the Vevo®2100 ultrasound system to allow for 3D image acquisition. Also connect the physiological monitoring unit to the Vevo®2100 system to allow on screen tracking of physiological parameters, such as heart rate, respiratory rate and body temperature.
Halo gas vesicles are imaged under the following imaging settings using the Vevo®2100 system: (i) frequency - 21MHz and 18MHz for B-mode and contrast mode respectively; (ii) transmit power - 2%; and (iii) gain - 16dB and 32dB for B-mode and contrast mode respectively. The imaging field of view can be adjusted accordingly to fit the tissue region of interest, by increasing or decreasing the image width and depth.
▴CRITICAL It is crucial to keep the transmit power on the Vevo®2100 system to 2%, to allow for stable imaging of Halo gas vesicles. Higher transmit power will collapse exposed GVs.
Setting up the syringe pump:
Gas vesicle injections are administered as a bolus using a syringe pump (shown in Figure 4b). The following settings are used for each injection: (i) syringe diameter - 3.55 mm (inner diameter of a BD 1/2cc Tuberculin Syringe); (ii) injection rate - 300 μL/min to 500 μL/min; and (iii) injection volume – 50 μL.
Installing the control electronics for the 129Xe MRI:
Set up the TTL output of the spectrometer with the photoMOS relay such that the latter one controls the status of the shortcut valve (see Figure 5d for the circuit). This is used to alternatingly force gas flow through the sample or to bypass the sample. From the Daq/board, use one analog output to operate the set points of the flow and pressure controllers. Connect one analog input each to read the process values of the controllers. Display the corresponding parameters through appropriate elements in the DASYLab/LabView software.
Preparation of the Xe gas delivery manifold:
The timing of Xe bubbling is controlled by the pulse sequence and achieved through a proper gas delivery system by connecting the PFA tubing and controllers according to Figure 5e. Prior to each experiment, insert the (five) capillaries into the capillary holder. The end tips should reach close to the bottom of the glass phantom.
▴CRITICAL This design of the flow and pressure controllers prevents Xe gas from passing through any needle valves or (electro-) magnetic units during its transfer from the hyperpolarizer to the sample, thus minimizing the depolarization of Xe.
▴CRITICAL Gas bubbles can cause serious image artifacts if persistent during image acquisition. Therefore, the bubbling has to be stopped and the sample has to settle down prior to acquisition. Fresh hyperpolarized xenon has to be re-delivered for each subsequent acquisition.
MRI pulse sequence setup:
The non-selective NMR sequence for acquiring direct Xe NMR spectra is adapted from an existing simple FID sequence by implementing the TTL trigger pulses and bubble/wait delays prior to the 90° block pulse (Figure 5e). The FID sampling window is typically 1000 ms and the spectral width to cover is ca. 350 ppm. For Xe MRI imaging sequence, single-shot pulse sequences should be used to make the most efficient use of the non-renewable Xe polarization. A single-shot EPI pulse sequence can be used when long T2* values are expected. Alternatively, a single-shot RARE pulse sequence can be used and is limited by T2. To set up the imaging sequence, take a standard sequence with image encoding of your choice (EPI or RARE) and implement a magnetization transfer or saturation module (Bruker ParaVision terminology) after the TTL trigger pulse for xenon delivery and prior to the k-space encoding. The saturation offsets are defined through a saturation frequency list. This list can be accessed by providing in the ParaVision graphical user interface a menu to enter a fixed number of saturation pulse frequency offsets. Point these entries to the saturation frequency list. Finally, loop the entire xenon delivery, wait time, the saturation pulse and the imaging sequence multiple times to walk through the entire saturation frequency list. Some general parameters when implementing the sequence are: a field-of-view of 20 × 20 mm2; matrix size 32 × 32 (in-plane spatial resolution: 625 μm2); slice thickness: 20 mm; repetition time including saturation time and xenon delivery: ca. 18 s.
Detailed parameters for RARE sequence: 90° Hermite excitation pulse: length = 3.375 ms, bandwidth = 1,600 Hz; 180° Mao refocusing pulse: length = 3.105 ms, bandwidth = 2,000 Hz; short echo time of ca. 10 ms (this is also the effective echo time as centric encoding is used here), echo readout time of 2.66 ms; ensure the RARE factor is set to give a single-shot acquisition, i.e., a RARE factor of 32 is used when the image matrix size is 322.
Detailed parameters for EPI sequence: 90° Gaussian-shaped excitation pulse: length = 1.0 ms, bandwidth = 2,740 Hz; encoding: partial Fourier-acceleration factor = 1.68 (i.e. 19 out of 32 k-space lines) and double sampling; short echo time of ca. 6 ms; echo train readout time of 6.01 ms for 19 k-space lines.
PROCEDURE:
Production and purification of gas vesicles (GVs):
1| Gas vesicles can be produced natively in cyanobacterial (Ana GVs) or haloarchaeal (Halo GVs) hosts. In addition, they can be heterologously expressed in E. coli (Mega GVs). To produce and purify Ana, Halo or Mega GVs, follow steps in Option A, B, or C respectively.
(A). Producing and purifying Ana GVs ● TIMING 4.5 weeks
-
(i)
Making Ana starter cultures (Steps i-iv: ● TIMING 2 wks) Open the seal of the primary culture tube of Anabaena flosaquae (as received from CCAP, UK), and loosen the screw cap to allow aeration of the culture. Let the tube stand upright and undisturbed for 2 days in a cool (room temperature i.e. (18–25 °C)), dry place with access to ambient light. Viable Ana cells producing GVs will float to the top of the tube, forming a dark green layer.
-
(ii)
Take a pre-sterilized Pyrex glass culture tube. Aliquot 9 mL of G625 growth media into the tube in the presence of a flame to ensure sterility. Add 0.2 mL of 50x BG-11 cyanobacteria freshwater solution. Swirl a few times to ensure complete mixing.
-
(iii)
Gently transfer 0.8 mL of the floating green cells from the primary culture to the G625 media in the culture tube to make a total volume of 10 mL. Swirl gently to disperse the cells. The culture should have just a hint of green at this stage.
-
(iv)
Transfer the starter culture to the shaker incubator (shown in Figure 2b) and allow it to grow at 25°C, 100 rpm and 1% CO2 with cycles of 14 hours of light illumination and 10 hours in the dark until the cells form a dense, floating green layer that is clearly visible at the top of the liquid phase. This growth typically takes around 1–2 weeks. Note that the doubling time of Anabaena flosaquae during exponential phase is ~ 56.5 hours25, so depending on the number of viable cells in the starter culture, it will take 1–2 weeks to produce enough floating green cells that have the ability to produce GVs. Viable cells typically form the dense layer at the top and dead cells sink to the bottom of the tube.
?TROUBLESHOOTING
-
(v)
Sub-culturing Ana for GV production (Steps v-vi: ● TIMING 2 wk) Aliquot 200–250 mL of G625 media into an autoclaved 1L Erlenmeyer flask (use a bunsen burner flame or a laminar hood to provide a sterile environment to avoid contamination). Add 1x final concentration of BG-11 freshwater solution (4–5 mL of the 50x stock) just before inoculation with Ana starter culture. Swirl to mix components.
-
(vi)
Inoculate 2 mL of the green, floating layer of the starter culture per flask and grow cells in the incubator at 25°C, 100 rpm, 1% CO2, 14 h light and 10 h dark cycle until they become confluent (as shown in Figure 2b-c). This should take ~ 2 weeks, with the exact time depending on the number of viable cells inoculated.
?TROUBLESHOOTING
-
(vii)
Harvesting Ana GVs (Steps vii-xi: ● TIMING 1 d) Pour the confluent culture from the flask into a separatory funnel (pre-sterilized with stopcork in place), allowing it to stand undisturbed on a metal ring stand for 12–16 hours until the GV-producing Ana cells separate from the spent media to form a compact green layer at the top (Figure 2e).
-
(viii)
Carefully remove most of the spent media by opening the stopcork at the bottom of the funnel and retain only the green fraction at the top along with ~30 ml of media.
-
(ix)
Using a 25 mL serological pipet, gently resuspend the cells and transfer to a 50 mL tube.
-
(x)
Use 10 mL of fresh G625 growth media to collect cells sticking to the walls of the funnel. Each flask should be reconstituted to ~ 50 mL of concentrated cell suspension.
-
(xi)
plit the suspension into two falcon tubes and add equal volume of 1M sorbitol (final concentration is 500 mM) and 10% (vol/vol) of Solulyse (i.e. 5.5 mL in 55 mL) to achieve hypertonic lysis. Place the tubes in a rotatory shaker at 4°C for 6–8 hours. Green lysates will have a milky tinge post-lysis due to the presence of GVs.
CRITICAL STEP We have observed that GVs become structurally compromised by certain detergents and surfactants used routinely for cell lysis such as BugBuster (EMD Millipore), Triton-X and sodium dodecyl sulfate. It is therefore advisable to thoroughly characterize the effect of any new detergent on GV structure and integrity using techniques such as TEM and pressure absorbance spectroscopy before using them for ultrasound and MRI.
?TROUBLESHOOTING
-
(xii)
Isolation and purification of Ana GVs from lysate (Steps xii-xvii: ● TIMING 2–3 d) Transfer 35 mL of cell lysate to a 50 mL tube and centrifuge at 350g, 8°C for ~24 hours in a table-top centrifuge, preferably equipped with a swinging bucket rotor.
?TROUBLESHOOTING
-
(xiii)
Slowly remove as much as possible of the green subnatant liquid and cell pellet using a 25 mL serological pipet, without disturbing the white GV layer on top.
-
(xiv)
For the second and third round of centrifugation (350g, 8°C), resuspend the white supernatant layer containing GVs in 25 mL of sterile 1x PBS (Corning). During each resuspension step, gently wash the inner walls of the falcon tube with fresh PBS to recover GVs sticking to the sides. The green subnatant should progressively become clearer with each spin. The second and third rounds of centrifugation are shorter than the first, with the endpoint being when all the GVs rise to the top of the tube.
-
(xv)
For the final spin, resuspend GVs in 10–12 mL of PBS (total volume can be varied depending on GV yield) and aliquot them into 2 mL tubes. Spin in a microcentrifuge at 350g for 4 hrs at 4–8°C.
CRITICAL STEP Do not fill the tube to the brim as GVs will get stuck to the lid of the tube when they float to the top during centrifugation
?TROUBLESHOOTING
-
(xvi)
Using a 21.5 G flat needle attached to a 3 mL syringe, maintain the angle of the microcentrifuge tube and slowly insert the needle through the GV film to the bottom. Collect most of the subnatant and slowly take out the needle. Some GV loss will occur at this step. This is OK as long as most of the GVs do not get sucked up into the syringe.
-
(xvii)
Resuspend the purified GVs in ~ 6–8 mL of PBS (exact volume of added PBS can be varied and is determined by how concentrated the GV solution needs to be for the end application) and aliquot the milky white GV solution (Figure 2j) into screw-top vials or microcentrifuge tubes.
▪ PAUSE POINT The Ana GVs aliquoted into tubes can be stored for up to a year at 4°C.
CRITICAL STEP Avoid freezing and subjecting the tube to shocks, such as dropping to the ground or snapping the cap, as this may collapse the GVs.
(B). Producing and purifying Halo GVs ● TIMING 3.5 weeks
-
(i)
Growing Halo cultures for GV production (Steps i-iii: ● TIMING 2 wks) Aliquot 250 mL of Carolina growth medium in to an autoclaved 1L Erlenmeyer flask under sterile conditions.
-
(ii)
Inoculate Halo cultures using one of the following methods. (1) Using a sterile pipette tip or toothpick, scrape a small amount of pink culture from the agar plate to add to the flask as inoculum. (2) Use one to two brine crystals containing Halo for inoculation. (3) Inoculate 2.5 mL from a healthy pink liquid starter culture into 250 mL of fresh growth medium (1:100).
-
(iii)
Grow the culture in an incubator at 42˚C with 100 rpm shaking. Depending on the health of the parent culture, it may take ~ 2 weeks for the inoculated culture to become confluent . Confluency is determined by the color and turbidity of cultures as shown in Figure 2d.
?TROUBLESHOOTING
-
(iv)
Harvesting Halo GVs (Steps iv-vi: ● TIMING 1 wk) Gently pour the culture from the flask into a separatory funnel (pre-sterilized with stopcork in place). Allow the culture to remain undisturbed until a visible ring is formed at the top (Figure 2f). This typically takes 4–6 days.
-
(v)
Remove as much of the spent media as possible by opening the stopcork, retaining only the buoyant layer of milky-pink cells for lysis. The retained volume is~10–20 mL , and a lot of cells stick to the sides of the funnel.
-
(vi)
Using equal volume (10–20 mL) of TMC lysis buffer (pH 7.5), gently wash the cells stuck on the sides of the funnel and retrieve as many cells as possible. The volume of TMC buffer used might be varied depending on the cell density to achieve efficient hypo-osmotic lysis. However, note that if too much buffer is used in this step, the number of 2 mL aliquots will proportionately increase for the next step, thus increasing sample processing time.
?TROUBLESHOOTING
-
(vii)
Isolation and purification of Halo GVs from lysate (Steps vii-xi: ● TIMING 2 d) Aliquot ~1.6 mL of cells in 2 mL tubes and spin in a microcentrifuge at 300g for 4 hours at 8˚C.
▴CRITICAL STEP Close the tubes gently; the pressure wave caused from snapping the lid will collapse a large number of Halo GVs.
?TROUBLESHOOTING
-
(viii)
At the top of the tube, a mixed layer of Halo GVs (white) and unlysed Halo cells (milky-pink) will be visible. Using a blunt end 18.5 or 21.5G needle, aspirate the pellet at the bottom of the tubes as well as the pink cell lysate. CRITICAL STEP Take care to limit the amount of floating Halo cells and Halo GVs (white) that are aspirated in to the syringe.
-
(ix)
Transfer the GVs and unlysed Halo cells to fresh tubes and bring to 1.6 mL with 1x PBS. Centrifuge tubes at 300 g for 4 hours at 8˚C.
-
(x)
Repeat steps viii and ix. After each step, the amount of milky-pink buoyant cells will reduce and white Halo GVs will increase. Continue with centrifugally-assisted floatation until all the cells have lysed and there is no evidence of pink cell lysate in the subnatant.
-
(xi)
Resuspend the purified GVs in PBS and aliquot the milky white GV solution (Figure 2j) into screw top vials or microcentrifuge tubes.
▪ PAUSE POINT The aliquoted Halo GVs can be stored for up to one year at 4°C.
CRITICAL STEP Avoid freezing and subjecting the tube to mechanical shocks, such as dropping to the ground or snapping the cap, as this may collapse the GVs.
(C). Producing and purifying Mega GVs ● TIMING 4 d
-
(i)
Heterologous expression of Mega GVs in E. coli (Steps i-iv: ● TIMING 2 d) Transform 50 μL chemically competent Rosetta™ 2(DE3) pLysS cells using > 1 ng of pST39 plasmid containing the pNL29 Mega GV gene cluster11 by mixing the two components in a 1.5 mL tube and incubating on ice for 30 minutes. Heat shock the tube in a 42 °C water bath for 45 seconds, and put the tube back on ice for a minute. Add 500μL of SOC outgrowth medium and incubate in a shaker at 37°C and 250 rpm for 1 hour.
-
(ii)
Prepare 3 mL of LB broth containing 1x Ampicillin (100 μg/mL), 1x Chloramphenicol (25 μg/mL) and 1% (wt/vol) glucose in a glass culture tube. Resuspend 300 μL of the transformed E. coli in the broth. Grow the culture in a shaker-incubator at 37°C and 250 rpm until OD600 reaches 0.4 – 0.6. Make 100 μL aliquots of the culture in sterile tubes, and mix with 100 μL of 50% sterile glycerol. Freeze the tubes at −80°C as E. coli glycerol stocks.
▪ PAUSE POINT The glycerol stocks can be stored at –80°C and used for up to 3 months. Note that while using glycerol stocks is convenient, the GV yield is reduced when using frozen stocks, so we recommend fresh overnight transformations for best results.
-
(iii)
Resuspend a tube of the aliquoted glycerol stock in 3 mL LB broth containing 1x Ampicillin, 1x Chloramphenicol and 1% (wt/vol) glucose. Grow the E. coli culture to saturation (OD600 > 4). For fresh transformations, aliquot 500 uL of the transformed E.Coli from Step (i) into 5 mL of LB broth containing 1x Ampicillin, 1x Chloramphenicol and 1% (wt/vol) glucose. Allow it to grow overnight until the culture reaches saturation (~ 16 hours).
-
(iv)
Prepare 100 mL LB broth containing 1x Ampicillin, 1x Chloramphenicol and 0.2% (wt/vol) glucose, and inoculate 1 mL of the saturated E. coli culture into the broth. Grow at 37°C for ~ 2 hours until OD600 reaches 0.4 to 0.6. Induce the culture by adding 20 μM IPTG (final concentration), and grow at 30°C for an additional 16–24 hours.
-
(v)
Harvesting and purifying Mega GVs from E. coli cultures (Steps v-x: ● TIMING 2 d) Split the culture equally into three 50 ml Falcon tubes and spin for 1 hour at 500g and 25°C.
CRITICAL STEP Avoid higher speeds because they may cause collapse of GVs.
? TROUBLESHOOTING
-
(vi)
Insert a 10mL syringe with needle to > 1 cm below the surface of the solution and withdraw the clear liquid component of the solution. Withdraw the liquid slowly to preserve the thin layer of cells floating at the top of the solution, as well as the pellet at the bottom, both of which contain Mega GVs.
-
(vii)
To lyse the cells, add 4 ml SoluLyse-Tris reagent per 50 ml of E. coli culture, 250 μg/ml lysozyme and 10 μg/ml DNAseI. Rotate the tubes for 10 minutes at room temperature and then aliquot 1.5 mL of the solution to 2 mL tubes. Spin samples for 4 hours at 800 g and 8°C. Mix the floating GV layer gently with supernatant and transfer to a clean tube.
-
(viii)
Spin the samples for 4 h at 800g. Use a 3 mL syringe to remove the bottom fraction, which sometimes includes a small pellet. Gently resuspend GVs in 1 mL of PBS. Repeat the spin and wash steps 3 times.
CRITICAL STEP Be aware that GVs are susceptible to desiccation and resuspend GVs immediately after withdrawing the liquid.
-
(ix)
Mega GVs are natively clustered. To uncluster them, GV-containing solution is mixed with 10 M urea in a 2:3 ratio to achieve 6 M final urea concentration, and the resulting solution is gently rotated for 30 min.
-
(x)
Dialyze GVs overnight in 6–8 kDa MWCO tubing against 4L of PBS. This step can be omitted for experiments with no stringent requirements for buffer conditions. The white layer of unclustered GVs at the top of the liquid phase after buoyancy purification, as well as the re-suspended milky-white solution of Mega GVs in PBS is shown in Figure 2i-j.
▪ PAUSE POINT Mega GVs can be stored for up to one year at 4°C .
CRITICAL STEP Avoid freezing and subjecting the tube to mechanical shocks, such as dropping to the ground or snapping the cap, as this may collapse the GVs.
Quantification of GVs by measuring pressure-sensitive optical density ● TIMING 15 m
2| Determine the concentration of a solution of gas vesicles by measuring its pressure-sensitive optical density at 500 nm (OD500,ps) using a NanoDrop 2000 Spectrophotometer. Load 2 μL of sample on the pedestal for each measurement. Collapsed gas vesicles in the same buffer are used as a blank for measurements. Prepare collapsed GVs by sonication in a water bath until the solution turns completely clear or by manual collapse in a capped syringe. For manual collapse, remove the plunger from a 12 mL Luer-Lock syringe closed with a tip cap and place 5–10 μL of gas vesicle solution at the bottom of the syringe. Making sure that the tip cap is screwed on tight, replace the plunger and push down until there is significant resistance. The increase in pressure will collapse the gas vesicles, turning the milky white solution clear. A shortcut for quick measurements of GV concentration is to blank with the GV resuspension buffer. For most samples, this will give an OD reading that is very close to that measured when using collapsed GVs as a blank. However, for some samples containing GVs that are fluorescent, it is necessary to use the collapsed GVs as the blank.
! CAUTION Before doing manual collapse, ensure that the tip-cap on the syringe is facing away from you and securely fastened, as pressurization of the syringe may cause ejection of the cap, resulting in potential injury to the user. Wear protective eyewear and clothing.
▴CRITICAL STEP It is important to ensure that the GVs are homogenously re-suspended in solution just before measurements. For each sample, take the average OD500,ps value after multiple measurements (n>=3) to ensure precision and accuracy.
Quantification of GV protein concentration ● TIMING 15 m
3| Measure the protein concentrations using the Pierce 660nm protein assay to obtain relationships between optical density and protein content for the GV solutions. We have established the protein concentrations to OD relationships of our three types of GVs, and the results are as shown in Table 3 below (N = 4, 5, 3 for Mega, Ana and Halo GVs respectively and the errors are in SEM). The molecular weight is derived from the TEM data summarized in Table 4, assuming a spindle shape for Halo GVs, a cylindrical shape for Mega and Ana GVs, a wall thickness of 18 Å and a protein density of 1.4 g/mL.
Table 3.
Quantification and calculation of GV molecular weight and molar concentration.
| Ana | Halo | Mega | |
|---|---|---|---|
| Protein concentration to OD500 ratio ([μg/mL] / OD) | 36.6 ± 2.6 | 13.4 ± 2.2 | 145.5 ± 6.4 |
| Estimated molecular weight (MDa) | 320 | 282 | 71.7 |
| Estimated molar protein concentration to OD500 ratio (pM / OD) | 114 | 47.3 | 2,030 |
| Estimated gas fraction to OD500 ratio (v/v/OD) | 0.000417 | 0.000178 | 0.000794 |
Table 4:
GV Dimensions
| Spatial Dimension | Ana GV | Halo GV | Mega GV |
|---|---|---|---|
| Length (nm) | 519 ± 160 | 400 ± 113 | 249 ± 99 |
| Width (nm) | 136.3 ± 21.0 | 250.8 ± 51.4 | 72.5 ± 13.6 |
Chemical functionalization of GVs ● TIMING 2 d
CRITICAL Purified Ana, Halo and Mega GVs contain lysine residues on the surface that can be used to chemically conjugate a variety of moieties such as polyethylene glycol, fluorophores and biotin using an amine-reactive coupling group such as N-hydroxysuccinimide ester.
4| Measure the concentration of purified GVs using the OD relationships in Table 2.
5| Aliquot the NHS-moiety in anhydrous DMSO at 100x of the required molar concentration for the amine-NHS reaction. For Alexa-488-NHS conjugation to Ana GVs, aliquot 5 μl of the 10 mM stock solution of the dye pre-prepared in anhydrous-DMSO.
▴CRITICAL STEP Ensure that the NHS-moiety solution does not contain detergents or surfactants that might affect the integrity and properties of GVs.
6| Adjust the concentration and volume of GVs to the desired amount and ensure that the buffer is free of amines (avoid Tris buffer). For Alexa-488-NHS conjugation to Ana GVs, bring Ana GVs to OD1 in 1 mL of PBS at pH 7.4.
▴CRITICAL STEP If GVs were previously in a buffer containing free amines or PBS with pH less than 7, ensure complete buffer exchange with PBS at pH 7 – 9 before proceeding with the amine-NHS reaction.
7|Add 105 molar excess of the NHS-moiety to GVs, keeping the DMSO concentration at 0.5% or less of the total reaction volume. For Alexa-488-NHS conjugation to Ana GVs, add 5 μl of the 10 mM Alexa-488-NHS in DMSO to 1 mL of Ana GV solution. Based on the average number of gvpA and gvpC protein monomers that make up Ana GVs, approximately 50,000 lysine residues are present for every Ana GV. One can tune the molar ratio of the two reactants (NHS-moiety:GV) to achieve the desired reaction efficiency.
8| Allow the reaction to proceed for 4 hours at room temperature under gentle rotation.
?TROUBLESHOOTING
9| Quench the unreacted NHS-moieties using Tris-HCl buffer at pH 8 to a final concentration of 10 mM for 20 minutes at room temperature under gentle rotation
10| Add the whole reaction to dialysis tubes (6–8 kDa cutoff) and dialyze against a 4000x volume excess of PBS at 4˚C for 8 hours. Replace the buffer and allow dialysis to continue for an additional 8 hours.
CRITICAL STEP If NHS-moiety is not amenable to dialysis, repeated rounds of centrifugally-assisted purification is an alternative method to remove excess reactants and/or for buffer exchange.
?TROUBLESHOOTING
▪ PAUSE POINT Chemically functionalized GVs can be stored in PBS buffer for up to one year at 4˚C −.
Optional: Genetic modification and functionalization of Ana GVs ● TIMING 3 days
CRITICAL The outer scaffold protein of Ana GVs, gas vesicle protein C (GvpC), can be removed and replaced with genetically modified recombinant versions as a molecular handle for GV mechanical modification and functionalization. This procedure comprises stripping native GvpC off Ana GVs (Steps 11–19), preparing recombinant GvpC (Steps 20–38) and re-adding this GvpC onto ∆GvpC Ana GVs via dialysis (Steps 39–41). In addition, GVs functionalized with GvpC fused to the SpyTag peptide enable convenient downstream covalent functionalization with proteins fused to the SpyCatcher moiety (Steps 42–45).
11| Preparation of ∆GvpC GVs (Steps 11-19: ● TIMING 12 h) Dilute purified Ana GVs in PBS such that OD500,ps <10.
12| Prepare 3:2 (vol/vol) mix of GV stripping buffer (10M urea in 100mM Tris buffer) and GV solution in PBS. Pipet 1.7 mL into 2 mL microcentrifuge tubes.
13| Centrifuge at 300g for 4 hours, or until the subnatant is completely clear. Remove the clear subnatant with a syringe using a 21G flat needle. Retain the milky white supernatant in the tube. Resuspend the GV-containing supernatant in GV stripping Buffer (Round 2 i.e. 6M urea, 60mM Tris-HCl). Repeat this step 1 time.
14| Confirm GvpC removal by performing SDS-PAGE. Incubate a 1:1 (vol/vol) mix of GVs in 2x Laemmli buffer (containing 5% (vol/vol) 2-mercaptoethanol) at 95°C for 5 m. Centrifuge briefly to collect condensate.
15| Assemble the electrophoresis cell with the comb and tape removed from the polyacrylamide gels. Fill the inner chamber completely with 200 mL 1x TGS buffer. Ensure that the cell is not leaking fluid. Fill the outer chamber up to mark with 600 mL 1x TGS buffer.
16| Load the protein ladder and samples in the gel using gel-loading tips. GVs should be at OD500 > 3 prior to the 1:1 dilution. If purified proteins are being run on the same gel for comparison, load > 100 ng.
CRITICAL STEP In order to prevent contamination between wells, do not exceed the maximum recommended volume per well.
17| Connect electrophoresis cell to power supply and run the gel for 55 m at 120V.
18| Recover the gel by disassembling the electrophoresis cell and the gel cassette. Incubate the gel in a holder with DI H2O for 10 m, then stain for 1 h with 10 mL SimplyBlue™ SafeStain. De-stain the gel for at least 1 h with 10 mL DI H2O.
19| Image the gel using a Coomassie imaging protocol using the gel imaging system to visualize protein bands. The GvpC band at approximately 25 kDa should be missing.
▪ PAUSE POINT Store the ΔGvpC GVs in urea buffer at 4°C for no more than 1 week. When preparing ΔGvpC GVs for long term storage without any further genetic functionalization or recombinant GvpC addition (steps 39–45), we recommend dialyzing the GV solution against PBS in order to completely remove the urea.
?TROUBLESHOOTING
20| Preparation of recombinant GvpC (Steps 20-38) :● TIMING 1 d) Transform > 1 ng pure plasmid encoding recombinant GvpC with a C-terminal hexahistidine tag into BL21 (DE3) competent cells and grow culture in terrific broth with 50 μg/ml kanamycin overnight.
21| Dilute 500 μL of the starter culture 1:1 with 50% glycerol in water and store at −80°C. Future starter cultures can be grown from aliquots of this glycerol stock instead of fresh transformations.
22| Dilute starter culture 1:250 in terrific broth with 50 μg/ml kanamycin and grow to OD600~0.4–0.7 with shaking (250 rpm) at 37ºC. Induce at a final concentration of 1mM IPTG. Grow culture for 6–12 h at 30ºC with shaking.
CRITICAL STEP Frozen IPTG stocks should be thawed fully and vortexed to mix contents before use.
23| Pellet the cells in ultracentrifuge tubes at 5,500g for 15 min at 4°C and discard the supernatant.
▪ PAUSE POINT Cell pellets can be stored at −20°C. Protein extraction is typically more effective with frozen cells.
24| Resuspend the pellets in 10 mL Solulyse with 10 μg/mL DNAse. Rotate at room temperature for 10 min.
25| Centrifuge at 20,000g for 15 m at 4°C to clear the lysate and discard the supernatant.
26| Resuspend the pellet in 10mL Solulyse and lysozyme (0.25 mg/mL). Rotate at room temperature for 10 m.
27| Add 5mL Solulyse and vortex. Centrifuge at 20,000g for 20 m at 4°C and discard the supernatant.
28| Thoroughly resuspend the pellet in 5 mL of inclusion body solubilization buffer.
29| Centrifuge at 20,000g for 20 m at 4°C.
30| Add 1.5mL Ni-NTA slurry to the supernatant, incubate at 4°C with shaking (60 rpm) for 2 h or more.
31| Pour into a polyprep column and collect all the flow-through, wash and elutions in the next steps. Collecting all fractions is good practice for troubleshooting and analyzing purification steps using SDS-PAGE.
32| Wash with 10 column volumes of inclusion body wash buffer.
33| Elute with 2 column volumes of inclusion body elution buffer.
34| To quantify the eluted protein using the Bradford assay, prepare a standard curve of bovine serum albumin (BSA) at a final concentrations of 100, 250, 500, 750, 1000 and 1500 μg/mL in 60 μL of 3x diluted inclusion body elution buffer in PBS. Prepare dilutions of eluted protein 1:2 in PBS with a final volume 60 μL. Prepare a negative control of 3x diluted inclusion body elution buffer in PBS.
35| To 25 μL of the sample and BSA standards, add 1 mL of Bradford reagent, vortex and incubate at room temperature for 5–10 m. Prepare all samples in duplicate.
36| Blank the spectrophotometer with a negative control sample and measure the OD595.
37| Measure the OD595 of the standard curve samples. Plot the OD595 versus the concentration, and compute linear regression fit.
38| Measure the OD595 of the eluted protein samples. Use the linear fit from Step 37 to compute the unknown concentrations, and multiply by 3 (dilution factor) to obtain concentration of stock elution solution.
▪ PAUSE POINT Store the elutions separately at 4°C. Elution 1 has > 80% of collected pure protein and is used for the subsequent experiments. Elution 2 is more dilute and is typically stored as backup or for running protein controls for SDS-PAGE.
?TROUBLESHOOTING
39| Preparation of GVs with recombinant GvpC (Steps 39-41● TIMING 1 d) Add recombinant GvpC to ∆GvpC GVs according to the formulation: 2 * OD * 198 nM * volume (in L) of GVs = nmol recombinant GvpC. This provides a 2-fold stoichiometric excess of GvpC relative to binding sites on an average Ana GV, assuming a 1:25 molar ratio of GvpC : GvpA binding based on previous work26. The exact volume of recombinant GvpC to be added is calculated based on the molar mass of the particular variant and the concentration of eluted GvpC (measured by Bradford according to steps 34–38). For truncated GvpC variants with a lower GV binding affinity, a higher stoichiometric excess may be added to promote attachment of GvpC to the GV surface. However, note that adding too much excess of GvpC might lead to protein aggregation during dialysis.
40| Soak the dialysis tubing in PBS for 5 minutes. Add samples (GVs + recombinant GvpC) into dialysis tubing and clip both sides. Dialyze in 4L PBS with stirring on low speed for at least 12 h. CRITICAL STEP The length of dialysis tubing used for each sample depends on the total volume of the dialysate, which is determined by the amount of engineered GVs required for the end application. The type of dialysis tubing used (molecular weight cutoff) can change depending on the GvpC variant, as truncated variants may have a much lower molecular weight.
41| Transfer the dialysate into 2-mL centrifuge tubes and spin at 300g for 3 hours, or until subnatant is clear. Remove subnatant with syringe with a 21.5G flat needle. Retain the milky white supernatant in the tube. Resuspend GVs in PBS. Repeat this centrifugation step one time.
▪ PAUSE POINT Store at 4°C.
?TROUBLESHOOTING
42| Optional: Preparation of SpyCatcher-functionalized GVs (Steps 42-45: ● TIMING 5 h) Prepare Ana GVs with GvpC fused to SpyTag peptide using Steps 39–41.
43| Mix SpyTag-functionalized GVs with SpyCatcher-fused proteins according to the formulation: 2 * OD * 395 nM * volume (in L) of SpyTag GVs= nmol SpyCatcher-fused protein. This results in a 2-fold excess of SpyCatcher to SpyTag in the reaction, based on the stoichiometry described in Step 39. Note that the SpyCatcher-mNeonGreen (SC-mNG) fusion protein used in our published work14 is expressed separately in E.Coli following procedures in Steps 20–23 and using the plasmid containing SC-mNG (details in the reagents section). SC-mNG is expressed as a soluble protein and hexahistidine-tagged, enabling purification using the same Ni-NTA slurry used for recombinant GvpC purification. Unlike GvpC inclusion bodies, soluble proteins are in the supernatant after cell lysis with Solulyse/DNAse (Step 24), allowing direct incubation of the supernatant with the slurry (Step 30). Wash and elution (Steps 31–33) are performed with soluble protein wash buffer and soluble protein elution buffer respectively, and the protein is desalted into PBS using PD10 desalting columns. Protein quantification is done using the Pierce or Bradford assay before use.
44| Incubate 1 hour or more at room temperature.
45| Centrifuge at 300 g for 3 hours or until subnatant is clear. Remove clear subnatant with syringe with 21.5G needle. Retain milky white supernatant in the tube. Resuspend supernatant in PBS. Repeat this centrifugation step one time.
▪ PAUSE POINT Store at 4°C.
?TROUBLESHOOTING
GV Characterization
46| To characterize the purified GVs, follow the procedure in A to determine the critical collapse pressure of GVs using pressurized absorbance spectroscopy. Follow option B for DLS measurements and option C for preparing the GV specimens for TEM.
(A). Determining critical collapse of GVs with pressurized absorbance spectroscopy ● TIMING 40m
-
(i)
Before acquiring measurements, connect the spectrophotometer to a power supply for 30 m to allow it to warm up.
-
(ii)
Run the Alicat_startup script to initialize the pressure controller.
-
(iii)
Blank the spectrophotometer with a cuvette filled with PBS or GV resuspension buffer and run OceanOptics_startup_FullTrans to save the data.
-
(iv)
Establish a zero-transmission baseline with the opaque side of cuvette blocking the light path using the OceanOptics_startup_NoTrans script.
-
(v)
Fill cuvette with intact GV sample (OD500nm = 0.2 in PBS) and fasten the cannulae securely. To assist loading, use elongated gel-loading micropipette tips.
-
(vi)
Open the N2 tank valve, pressure regulator, and pressure controller valve.
-
(vii)
Run the Collapse script to measure the OD500,ps under increasing hydrostatic pressure (0 kPa – 1.4 MPa in 20-kPa increments).
-
(viii)
Between these measurements, rinse the cuvette with DI H2O, 70% ethanol and acetone to ensure that cuvette is completely clean and dry before adding the next sample.
-
(ix)
After measurements, close gas the valves and turn off the spectrophotometer.
?TROUBLESHOOTING
(B). Preparation of GV specimens for dynamic light scattering (DLS) ● TIMING 5 m
-
(i)
Dilute the GV samples to OD500,ps = 0.2 in PBS.
-
(ii)
Measure the particle size on ZetaPALS instrument with an angle of 90° and refractive index 1.33.
(C). Preparation of GV samples for transmission electron-microscopy (TEM) ● TIMING 30 m
-
(i)
Buffer-exchange the purified GVs in 10 mM HEPES buffer with 150 mM NaCl or an alternative non-phosphate containing buffer via centrifugally-assisted floatation (same procedure as used for GV isolation) at 8°C and 300g. Replace the subnatant with equal volume 10 mM HEPES buffer with 150 mM NaCl. Repeat 3 times. The aim of this step is to prevent phosphate in the PBS from causing unwanted precipitation of the uranyl acetate stain used downstream in step (vi). Therefore, if the GV solution is very concentrated, direct dilution of the GV sample into the above-mentioned HEPES buffer to a final OD of 0.2 would be a quicker alternative.
-
(ii)
Dilute the GVs to a final OD of 0.2.
-
(iii)
Spin 2% Uranyl acetate solution in a benchtop centrifuge at 14,000xg for 5 minutes to pellet any precipitate.
-
(iv)
Charge Formvar TEM grids using the glow discharge system with 15 mA current for 1 minute.
▴CRITICAL STEP Gently handle the TEM grids and avoid contact of the tweezer tip with the center of the grid to prevent bending and damage to the grids.
-
(v)
Place 2 μl of well-mixed GV solution on the charged Formvar TEM grids for 3 minutes. The sample should be placed on the carbon side of the grid; avoid placing sample on the copper side. For convenience, we use PELCO reverse, anti-capillary tweezers to hold the TEM grid while adding sample and negative stain.
-
(vi)
Add 5 μl of 2% uranyl acetate to the GV solution on the TEM grid for 30 seconds.
-
(vii)
Using a Whatman filter paper, wick the solution by gently touching the edge of the grid. For consistent results, leave a thin film of sample on the grids and leave to air dry.
-
(viii)
Image the grids using TEM.
?TROUBLESHOOTING
Ultrasound imaging of GVs in vitro in agarose phantoms
47| Agarose phantom preparation and sample loading (Steps 47–55: ● TIMING 3 h) Make a 250 mL solution of 1% (wt/vol) agarose in PBS and microwave for 3 minutes to ensure dissolution. Ensure that the lid is loosely fastened and swirl the bottle at 1 min intervals to prevent the agarose from settling.
48| Fasten the lid and put the bottle in a water bath at 50–55°C for at least 30–45 min to allow the solution to equilibrate to the bath temperature
49| Fill the Nalgene plastic holder (L 12.5 cm x W 8 cm, 300 mL) with 150 mL of molten 1% agarose in PBS. Keep the remaining agarose solution in the water bath for sample loading.
50| Immerse the multi-well phantom mold on top of the agarose solution.
▴CRITICAL STEP Remove any small air bubbles with a syringe needle and lower the mold gently onto the surface of the agarose, maintaining a small angle to allow any air trapped on the interface between the agarose and mold to escape.
51| Let the agarose phantom solidify at room temperature for 1 h.
52| Carefully take out the multi-well phantom mold without disturbing the agarose beneath.
53| Prepare the GV samples and controls at 2x of the final desired concentration.
54| Prepare a 1:1 mixture of the 1% agarose in PBS with the GVs or polystyrene beads.
▴CRITICAL STEP Make sure the 1% agarose in PBS is equilibrated to a temperature of 50–55°C before and during the mixing with GVs or polystyrene beads. Just prior to mixing with the agarose, place the GV or polystyrene sample for 10–15 seconds in a 50°C water bath to warm the sample. This allows proper mixing of agarose with the sample and provides enough time to load the sample into the well before the agarose solidifies.
CRITICAL STEP Do not heat the GV samples to temperatures above 50°C. GV collapse has been observed even for a 10–20 sec incubation at 65°C or above.
55| Pipette the mixture into the phantom well. The volume of mixture pipetted depends on the dimensions of the well, typically 50–100 μL.
?TROUBLESHOOTING
56| Phantom imaging (Steps 56–62: ● TIMING 1 h) Place the ultrasound acoustic absorber at the bottom of the phantom holder to prevent multiple ultrasound wave reflections during the measurement
57| Immerse the phantom in water or 1X PBS
58| Perform ultrasound imaging on the Verasonics L11–4v or L22–14v ray-line example scripts using the following parameters: set the transmit frequency is to 4.5 MHz, set the number of cycles of the transmitted ultrasound pulse to 3, the F-number to 3, the voltage to 2.5 and the persistence to 90. These parameters can be adjusted by the user in the Verasonics MATLAB script defining the ultrasound pulse sequence, and operated via the VSX GUI. Perform the collapse using a high power pulse at 25V.
59| Perform ultrasound imaging at the fundamental frequency with the L22–14v: the transmit frequency is set to 11.5 MHz, the number of cycles of the transmitted ultrasound pulse is set to 6, the F-number to 2, and the voltage to 3V. Those parameters are adjusted in the script and operated via the VSX GUI.
60| Perform ultrasound imaging at the second harmonic frequency with the L11–4v transducer: echoes at the second harmonic frequency are displayed using a 2 MHz wide band pass filter centered around 9 MHz. Other parameters are identical to the fundamental imaging mode and adjusted in the script. Second harmonic imaging is operated as an option via the VSX GUI. Note that we have recently developed an amplitude modulation strategy that can also be used for highly selective non-linear imaging of native and engineered GVs9.
61| Obtain acoustic multiplexing images by using GVs at OD 1 and a transmit frequency of 6.25 MHz and at 1.6V on the L11–4v, with a 4 cycle pulse, transmit focus of 20 mm, F-number of 2 and persistence of 90. To collapse the GVs, increase the acoustic pressure by lowering the F-number to 0.1 and gradually ramping up the voltage. At each collapse step, the transducer is translated in the y and z planes using the Velmex motors to ensure homogenous collapse over the entire well and field of view.
62| Save the ultrasound images for subsequent processing as .fig images (MATLAB format) directly from the MATLAB figure panel.
?TROUBLESHOOTING
Ultrasound imaging of GVs in vivo
63| GVs can be used as contrast agents for in vivo ultrasound imaging. Option A describes the steps for imaging GVs in mice using the Verasonics system, while Option B describes the procedure for imaging Halo GVs using the Vevo ultrasound system.
CAUTION Any experiments involving live mice must conform to relevant Institutional and National regulations. All animal experiments reported in this article received approval from the Institutional Animal Care and Use Committees of the California Institute of Technology or the Sunnybrook Research Institute.
(A). GV imaging in vivo using the Verasonics ultrasound system ●TIMING 45 m
-
(i)
Place the mouse in the induction chamber with isoflurane 3–4%, room air at 2 liters/min with the vacuum on for isoflurane removal.
-
(ii)
When the animal is unresponsive to toe pinch, move the animal to the heat pad and nose cone for continued anesthesia and place it on the supine position. Reduce the isoflurane to 2% and apply eye lubricant to both eyes to avoid drying. Use NAIR™ and cotton swabs to depilate the abdominal area and wash thoroughly with saline to avoid any burns on the skin.
-
(iii)
Prepare the catheter using 10 cm of PE10 plastic tubing and a 30 G needle. With a razor blade, cut the plastic at the bottom of the needle to allow the blunt end to be free. Push this end into the PE10 plastic tubing.
!CAUTION Follow safe practices and wear proper PPE while handling sharp needles.
-
(iv)
Fill the catheter with saline using a 30 G syringe. Confirm that there is no leaking from the catheter. Remove the saline-filled syringe before the next step.
▴CRITICAL STEP Failure to fill the tubing with liquid to remove the air from the catheter can lead to a gas embolism and death of the animal.
-
(v)
To insert the catheter into the tail-vein, place the mouse on its side for a better view of the lateral tail vein, clean the tail with an alcohol swab. Using the dominant hand to hold the catheter, use the other hand to hold the tail between your thumb and index finger. Insert the bevel needle through the skin and into the vein at a flat angle and advance it for several mm. If the needle is inserted correctly, blood should backflow through the catheter.
-
(vi)
Secure the position of the catheter in the tail vein with tissue adhesive glue and use the 30G syringe filled with saline to flush the blood from the catheter.
CRITICAL STEP The flow of saline should be smooth, confirming proper positioning of the needle tip in the tail vein. Excessive pressure to achieve flow may indicate improper positioning of catheter or clotting of blood.
-
(vii)
To perform ultrasound imaging, slowly adjust the mouse to a supine position and apply ultrasound gel on the depilated area. Place the transducer on the gel.
CRITICAL STEP Make sure that there are no air gaps and that the gel is completely coupled to the transducer.
-
(viii)
Open the Verasonics L11–4v ray-line example script in MATLAB . The transmit frequency is set to 4.5 MHz with the L11–4v transducer. Position the transducer to see the Inferior vena cava (IVC) for vascular imaging. Keep the voltage at 2.5V, 3-cycle pulse, F-number 3 and persistence 20.
▴CRITCAL STEP Ensure that the region of interest in the mouse aligns with the focus of the transducer to acquire optimal images.
-
(ix)
(ix) Remix the gas vesicles thoroughly by gently pipetting up and down, then aliquot 100 μL of solution of OD500,ps ~ 25 into a 1.5 mL micro-centrifuge tube.
▴CRITCAL STEP Gas vesicles must be mixed thoroughly each time before an injection, they are buoyant and will float up to the top of the tube. Make sure to inject immediately after remixing.
-
(x)
Carefully load a 30 G tuberculin syringe with the GV solution, making sure there are no air bubbles. Do not tap as this may collapse the GVs. Replace the saline-filled syringe at the end of the catheter and secure it to the syringe pump.
-
(xi)
Fill the catheter line with GVs at a rate of 5 μL/s.
-
(xii)
Inject 50 μL of GVs into the tail vein at a rate of 5 μL/s and record the ultrasound signal appearing in the IVC and liver.
▴CRITCAL STEP The total amount of injected saline and sample should be monitored and within the approved safety guidelines to prevent adverse effects on the animal caused by blood dilution.
-
(xiii)
Perform imaging using B-mode or harmonic filtering as described in steps 58–60 for in vitro specimens.
-
(xiv)
At the end of the imaging session, turn off the isoflurane and remove the mouse from the nosecone. Wait for the animal to completely recover from the anesthesia before returning it back to the cage.
?TROUBLESHOOTING
(B). Halo gas vesicle imaging in vivo using the Vevo ultrasound system ●TIMING 45 m
-
(i)
Setup the ultrasound mouse imaging station (see ‘ Setting up the Vevo Imaging Station’ in Equipment Setup) and attach the 21 MHz, MS250 MicroScan™ Transducer to the probe clamp. Make sure that the transducer is placed in the active port of the Vevo®2100 system.
-
(ii)
Adjust your ultrasound imaging parameters (see ‘Setting imaging parameters on the Vevo®2100 system’ in Equipment Setup) for both B-mode and nonlinear contrast mode imaging.
▴CRITICAL STEP Ensure that all imaging parameters are kept the same, so data can be compared across animals.
-
(iii)
Warm the mouse for 5 – 10 minutes to dilate its blood vessels. Care should be taken to not over heat the animal.
!CAUTION Heat lamp can get very hot. Handle with care.
-
(iv)
After the animal has been warmed up, anesthetize for tail vein cannulation. Mice are anesthetized using a tabletop animal anesthesia system. Place the mouse in the induction chamber and open the valve of the medical air cylinder. Set the flow meter to 1.5 L/min and vaporizer to 5% isoflurane.
-
(v)
After 1–2 minutes transfer the mouse to the handling table and re-route the anesthesia from the induction chamber to the nosecone. Fit nosecone over the nose and mouth of the mouse. Toe pinch to make sure there is no reflex, then lower the vaporizer to 2% for maintenance.
▴CRITICAL STEP Use medical air instead of oxygen as the carrier for the isoflurane to increase the stability of gas vesicles.
-
(vi)
Fill a 1mL syringe with a 27G x 1/2 needle with 0.9% sodium chloride solution, and attach the end of the catheter tubing to the needle.
-
(vii)
Flush the catheter tubing with the 0.9% sodium chloride solution until the solution leaks out from the catheter needle.
▴CRITICAL STEP Failure to fill the tubing with liquid to remove the air from the catheter can lead to a gas embolism and death of the animal.
-
(viii)
Insert catheter into tail vein as described in step 63A(v).
-
(ix)
Place eye ointment in both eyes of the mouse to prevent drying.
-
(x)
If desired, heart rate and respiratory rate monitoring can be accomplished as follows: place a small amount of electrode gel onto the 4 electrodes located at the corners of the mouse handling table, and secure each paw of the mouse to the electrodes with surgical tape. Make sure the animal is placed in a supine position to allow for ultrasound probe access to the liver. Refer to Figure 4d.
-
(xi)
For body temperature monitoring, lubricate the temperature probe and insert gently into the rectum of the animal. Refer to Figure 4d.
-
(xii)
Apply ultrasound gel to the upper abdominal region of the mouse, right below the rib cage, and place the transducer on top of the gel.
-
(xiii)
Starting with B-mode imaging, adjust the X, Y and Z positioning knobs to locate a slice of the liver where the inferior vena cava (a), portal vein (b) and abdominal aorta (c) can be observed in the same imaging plane. Once an imaging plane has been established, switch the imaging mode over to nonlinear contrast.
-
(xiv)
Resuspend the gas vesicles thoroughly by gently pipetting up and down, then aliquot 60 μL of solution (50 μL for injection + 10 μL to fill catheter tubing) into a 1.5 mL micro-centrifuge tube. We typically use OD500,ps ~ 5 for Halo injections, although one can go higher or lower based on their experimental needs and design (injected concentrations as low as OD500,ps 1.5 can be detected in the IVC).
▴CRITICAL STEP Gas vesicles must be mixed thoroughly each time before an injection, they are buoyant and will float up to the solution surface very quickly. Make sure to inject immediately after remixing.
-
(xv)
Withdraw the aliquoted gas vesicle solution into a 1/2cc tuberculin syringe and secure into the syringe pump. With the needle pointing up, remove any bubbles by gently pulling the plunger down and up. Do not flick the syringe as this will collapse GVs.
-
(xvi)
Imaging under nonlinear contrast mode, acquire ultrasound images before, during and after a bolus injection of gas vesicles, to observe the change in signal from the inflow of gas vesicles. A high power burst sequence can also be applied to collapse any gas vesicles in the imaging plane and diminish the signal.
? TROUBLESHOOTING
-
(xvii)
Steps xiii - xv can be repeated for other tissue regions of interest.
-
(xviii)
At the end of the imaging session, turn off the isoflurane and remove the mouse from the nosecone. Wait for the animal to completely recover from the anesthesia before returning it back to the cage.
GV imaging using 129Xe-MRI in vitro ● TIMING 2–3 h
-
64.
Preparing GV samples for MRI: Dilute the stock solution of GVs in phosphate-buffered saline solution to achieve an OD500, PS of ca. 0.33. For each sample, 2 mL of GVs are needed.
▴CRITICAL STEP While Ana and Mega GVs are stable and survive multiple xenon re-deliveries and data sets, Halo GVs are very fragile. Therefore, use a fresh Halo sample for each measurement.
OD measurements should be performed before and after NMR measurements to ensure GV concentrations stay constant.
-
65.
Starting up the polarizer (Steps 65–72:● TIMING 20 m) Turn on the hyperpolarizer according to the manufacturer’s protocols.
-
66.
Select an absolute gas pressure in the system of ca. 1.2 bar (i.e. 0.2 bar above atmospheric pressure). This pressure is adjusted through the Daq/Board-regulated pressure controller behind the sample and will be reached once the hyperpolarizer outlet valve has been opened. The manual pressure regulator directly at the gas tank should be set to a higher value (1.5 to 2 bar).
▴CRITICAL STEP Since the GVs are pressure sensitive, an excessive pressure set point for the sample could later damage the GVs in solution when performing step 76. On the other hand, the gas pressure determines the amount of dissolved Xe and eventually the image signal intensity. Considerations regarding the expected spin concentrations are described in Box 1.
-
67.
Connect the gas delivery tubing (coming from the hyperpolarizer) with the gas outlet tubing (going into the pressure controller) using glass connecting tube shown in Figure 5b.
-
68.
Slowly open the outlet valve of the hyperpolarizer to pressurize the gas manifold and to achieve the selected total pressure of 1.2 bar in the entire system.
! CAUTION Back-flow of air (oxygen) or humidity into the system and the Rb-containing optical pumping cell in the polarizer setup must be avoided at any time to prevent violent reactions with the reactive Rb.
-
69.
Select a total gas flow rate of 250 SMLM through the system for the flow controller between the gas tank and the hyperpolarizer. The pressure controller will now maintain the 1.2 bar while releasing incoming excess gas at the selected flow rate.
-
70.
Wait until the process temperatures have reached a steady state.
-
71.
Stop the gas flow by setting the flow controller set point to 0 SMLM and isolate the hyperpolarizer by closing its outlet valve.
-
72.
Depressurize the gas manifold by carefully unscrewing the glass tube connector while keeping the polarizer in standby.
-
73.
Checking the gas delivery conditions (Steps 73-82)● TIMING 10 m Decide if the glass phantom should be used with a one compartment for a single sample solution or with two compartments for including a second solution (e.g., a reference buffer solution without GVs); in the latter case, insert a 5mm NMR tube into the glass phantom.
-
74.
Load the sample solution(s) into the glass phantom and screw the capillary holder onto the inlet thread (distribute the capillaries as needed between the two compartments in case the 5mm tube has been inserted). Make sure that the filling level is not too high, so that the NMR coil can also detect some of the gas atmosphere on top of the solution.
-
75.
Connect the gas delivery tube from the hyperpolarizer setup to the capillary holder of the phantom and the gas outlet tube to the other chem thread of the glass phantom.
-
76.
Select a total flow rate through the polarizer of 250 SMLM and activate the flow. This will ensure that gas is flowing into the system once the next step is performed.
-
77.
Carefully pressurize the PFA tubing system by slowly opening the outlet valve of the hyperpolarizer to reach the overpressure of 0.2 bar.
CRITICAL STEP Make sure the shortcut valve is always open for pressurizing or depressurizing the sample; this avoids abrupt pressurization that could push the sample solution out of the glass phantom into the PFA tubing which can later contaminate the flow or pressure controller.
▴CRITICAL STEP When pressuring the GV-containing sample with xenon gas mix, a fast change in pressure has to be avoided and a rate of 0.1 bar/30 seconds should not be exceeded to ensure the GVs remain intact.
-
78.
Once the pressure has stabilized, ensure that the total flow rate of 250 SMLM through the polarizer is still active while the shortcut valve is open; this flow will now go through the bypass without bubbling the sample.
-
79.
To bubble the sample, adjust the sample delivery flow to 40 SMLM.
-
80.
For activating the flow, the pulse sequence needs to be executed. However, all RF pulses should be turned off since there is no sample in the NMR coil that absorbs the transmitted power. To do so, select first the pulse-acquire FID sequence including the TTL triggers and set the amplitude of the excitation pulse to 0.
-
81.
Choose a bubble time of ca. 10–20 sec and a waiting time after bubbling of ca. 1–2 sec.
-
82.
Set the number of repetitions to 5–10 and start the sequence while watching the sample for sufficient gas dispersion during periodic gas delivery and possible unwanted excessive foam formation inside the glass phantom. Ensure that any foam and gas bubbles disappear within the waiting time after bubbling (typically 1–2 sec).
▴CRITICAL STEP Prolonged bubbling could collapse GVs via shear forces. The flow of gas through the sample should be long enough to have decent xenon signal but short enough to not stress the GVs too much. The subsequent waiting time should be as short as possible to minimize the longitudinal relaxation of the xenon while long enough to ensure all bubbles to collapse.
? TROUBLESHOOTING
-
83.
Preparative MRI measurements (Steps 83-91:● TIMING 10 m) Insert the pressurized phantom carefully into the NMR probe that is already mounted inside the magnet in its correct position.
-
84.
If desired, adjust the temperature of the sample with the VTU of the spectrometer. This sample data was acquired at room temperature (T = 298 K).
▴CRITICAL STEP If another set point, e.g. body temperature, is chosen, the sample might need 20–40 min to reach stable temperature. It is possible to continue with the next steps, but step 87 () should not be performed until stable conditions are reached.
-
85.
Tune and match the 1H and 129Xe resonators, perform a global automatic shim on the sample and acquire a proton reference image with automated adjustments for 1H resonance frequency, reference attenuation (flip angle), and receiver gain according to the spectrometer manufacturer’s protocol.
-
86.
Set Xe delivery flow to 40 SMLM, 10 sec bubbling time and 2 sec wait time or as optimized in (c).
-
87.
Take a 1D Xe NMR spectrum with the pulse-acquire FID sequence including the TTL triggers with ca. 350 ppm bandwidth and four signal averages. If the amplitude for a flip angle ~90° is not known, proceed with the steps in Box 2 (and Figure 6f-i) to determine the reference pulse.
-
88.
Perform Fourier transform and automatic phase correction to display the NMR spectrum. Find the resonance frequency of the left intense peak (Figure 6a) and use it as the transmitter frequency for further 129Xe MRI acquisition.
? TROUBLESHOOTING
-
89.
For the imaging experiments to follow, enter the reference pulse power into the pulse sequence control interface (typically the pulse amplitude for a 1 ms block pulse to achieve a 90° flip of the magnetization). Switch to an MRI sequence and choose the same bubbling time, wait time and transmitter frequency.
-
90.
Ensure that the excitation for either the EPI or the RARE sequence is effectively a 90° pulse and that other sequence parameters are set as in the equipment preparation section. The number of acquisitions should be set to one.
-
91.
Run the Xe MRI acquisition without any saturation pulses (deactivate the MT module) and without phase encoding gradient (GSP in Bruker’s ParaVision) and set the receiver gain such that the maximum incoming signal reaches ca. 30% of the full ADC range.
BOX 1 | GENERAL SIGNAL CONSIDERATIONS ● TIMING 5 m.
In comparison to 1H MRI, the concentration of Xe is approximately 5 orders of magnitude lower. This is compensated by its higher polarization, approximately 4 orders of magnitude. Hence, higher Xe partial pressure helps to increase the detectable signal, which is directly proportional to the xenon concentration in solution. The calculation to determine the Xe concentration in solutions can be done as follows using the Ostwald solubility coefficient:
For the phosphate-buffered saline solution, assume an Oswald solubility coefficient similar to that of water: 0.11 L/bar or 4.9 mM/bar (1 mole of an ideal gas corresponds to 22.41 L). The partial pressure of the Xe fraction (5%) for 1.2 bar of gas mixture in the system is then 0.05 × 1.2 bar and the concentration of dissolved Xe is [Xe] = 0.11 L/bar × 0.05 × 1.2 bar × 4.9 mM/bar = 295 μM (keep in mind that only 26.4% is NMR-active isotope 129Xe).
To get an idea how long it takes to saturate the solution with Xe, consider the following: It has been shown that bubbling water with a 2% Xe mix at a flow corresponding to 100 SMLM (calibrated to N2, i.e. ca, 140 SMLM 2% Xe mix) exponentially saturates the solution with Xe with a time constant of ca. 8 sec (see supplementary information of ref. [23]). A 5% Xe mix at 40 SMLM (described in this protocol) delivers ca. 29% less Xe per unit time, and the time constant increases accordingly.
Box 2|FLIP ANGLE CALIBRATION FOR HYPERPOLARIZED XENON ● TIMING 10m
Flip angle calibration is usually done with a series of repetitive excitations and subsequent analysis of the obtained signal intensities. It is used to derive pulse amplitudes for the shaped excitation pulses but also to determine the saturation power in μT. Since hyperpolarization does not self-replenish, it is convenient to use a constant small flip angle excitation and analyze the signal decay after a single Xe delivery as follows:
ADDITIONAL MATERIALS
Non-selective NMR pulse sequence for acquiring a series of Xe solution signals after a single Xe delivery. This is implemented by using a simple (pulse – acquire – crusher) sequence with fast repetition between the excitations. As an example, the sequence can have a 100 μs block excitation pulse of adjustable amplitude, followed by a 9.72 ms FID sampling time (99 data points collected) and crusher gradients to remove all remaining transverse magnetization.
Evaluation tool like a gnuplot fitting routine or Origin to fit the data to a user-defined exponential decay model function.
PROCEDURE
Choose the above-mentioned non-selective NMR sequence with single Xe delivery and multiple acquisitions.
-
Select a relatively small excitation pulse amplitude (assuming α~ 7° – 11°).
▴CRITICAL STEP If the amplitude is too high, the signal decay will be either too fast or not monotonous; if the amplitude is too low, the observed signal is too noisy over the entire series.
Run the sequence with a repetition time of 18 ms and NEX = 250 repetitions; the crusher gradients burn for ca. 1 ms at an amplitude of ca. 90 G/cm.
The incoming time domain signal will show a series of 250 truncated FID signals. The spectrometer software may display this as a series of 250 FT magnitude spectra with decaying signal intensity (Figure 6f-g). For an evaluation without custom-made scripts like gnuplot fitting routine, perform the following steps:
Export the magnitude spectra data as .txt file and load them into Origin.
Perform ParaVision standard post-processing: each spectrum contains 128 data points of which only the peak heights are important.
Plot the concatenated spectra and evaluate the envelope formed by the peak heights. This envelope can be reconstructed with the υAnalysis υSignal Processing υEnvelope tool in Origin with using ca. 100 smoothing points.
Fit the envelope using the function I(x) = M0 + I0 sin α (cos α)(x/p - 1) with α as the desired unknown flip angle, M0 as a constant offset representing the non-vanishing noise level for large x, p the fixed number of displayed data points between two peaks. The fit should be performed for the range where the actual data is still above the noise level (Figure 6h-i). This evaluation neglects T1 relaxation during the short acquisition time. Fitting the observed signal intensity to this function therefore yields the flip angle α that the 100 μs block pulse achieved for the selected pulse amplitude.
The scaling factor for the pulse amplitude of a 10-fold longer standard block pulse (1 ms) is then given by 90°/(10*α). Use this for the reference gain for all derived pulse amplitudes.
Knowing the amplitude for a 90° pulse of 1 ms length, derive the pulse power in μT as follows: a full nutation takes 4 ms, hence the precession about the B1 field occurs at 250 Hz. The gyromagnetic ratio for 129Xe is γ = 11.777 kHz/mT. Hence, the applied field strength is B1 = 250/11,777 mT = 0.0212 mT = 21.23 μT. This can be used to derive the pulse powers for a desired saturation level in μT.
Figure 6. 129Xe HyperCEST MRI.
(a) 1D 129Xe NMR spectrum of a solution containing GVs, illustrating the signal from Xe gas in the space above the sample solution (magenta), the below-the-noise signal from GV-bound Xe (yellow) and the signal from the dissolved Xe in solution (cyan), which is assigned to 0 ppm for referencing. (b) Hyper-CEST z-spectrum of Mega GVs (0.67 nM in PBS, T = 295 K). On-resonant (- 156.5 ppm) and off-resonant (+ 156.5 ppm) saturation (B1 = 15 μT, tsat = 5 s) produced different signal intensity, and the spectrum was fitted to the exponential of two Lorentzian line shapes28,29. Analysis based on Bloch-McConnell equations predicts that the GV-bound Xe resonance frequency (green dashed line) is about −170 ppm. (c-d) Axial off- and on-resonant saturation xenon MR image (16 times averaged). A 5 mm NMR tube containing Mega GVs is nested inside a 10 mm NMR tube containing only PBS. (e) The differential images is normalized by the intensity of the off-resonant image and demonstrates the ~ 50 % Hyper-CEST contrast of GVs. The white dashed circles indicate the glass walls of the double-tube phantom. (f-i) Flip angle calibration. (f) Full time-domain FID signal train of 250 acquisitions (black: real domain, and red: imaginary) showing an overall exponential decay (bold solid lines) that is regularly interrupted by the ringing artefacts of opening the ADC gate. (g) Zoom-in of the first four FIDs. (h) Concatenated magnitude spectra after Fourier transforming the data displayed in (f). The fitting (green line) is based on the first 100 spectra, giving α = 12.6°. (i) Zoom-in for the first 10 spectra.
92| For Hyper-CEST MRI acquisition, follow the steps in option A for acquiring an entire image series with different saturation offsets for obtaining spectral information of the CEST response or option B for acquiring data with only one on-resonant and one off-resonant saturation to obtain a single Hyper-CEST image. Spectral information is obtained when changing the frequency of the saturation pulse (Figure 6b). This results in a so-called z-spectrum whose characteristic shape is influenced by xenon’s exchange properties with the GV. Option C should be performed if the exact resonance frequency of Xe inside the GVs is not known. Images with improved quality can then be obtained subsequently with option F.
(A). Imaging series and z-spectrum ● TIMING 45 m
-
(i)
Select the desired number of frequency offsets and enter the values for the offsets. The example data in Figure 6b were taken with 31 offsets covering a range from – 300 ppm to + 175 ppm with an increment of ca. 16 ppm.
-
(ii)
Select the saturation power B1 and saturation time tsat. The example here used saturation conditions of B1 = 15 μT, and tsat = 5 s.
! CAUTION Ensure that the power limits for the NMR coil are not exceeded as this might permanently damage the coil.
▴CRITICAL STEP It is not useful to choose a saturation time that exceeds the T1 relaxation time of Xe in water; the signal loss is then dominated by intrinsic relaxation and not by the CEST effect24 .
▴CRITICAL STEP The choice of the right saturation conditions is crucial for optimal z-spectra quality and for correct interpretation of saturation transfer in xenon MR images. As a rule of thumb, short but strong saturation is desired first. If multiple agents (different GV types in this context) are present, then the saturation strength should be reduced and the saturation time should be adapted similarly.
-
(iii)
Start the image series acquisition and make sure that the system does not perform any auto-adjustments that overwrite any of the previous settings (GOP command in Bruker’s Paravision).
▴CRITICAL STEP If the images are acquired individually because the operating system does not allow an array of measurements like in Bruker’s ParaVision, it is important that all images are taken with the same receiver gain and that the system does not automatically re-adjust the receiver gain before each acquisition.
-
(iv)
Open the image evaluation tool of the spectrometer software and draw a region of interest (ROI) for signal integration in the xenon MR images that were acquired for different saturation pulse frequencies.
-
(v)
Plot the signal intensity of the ROI versus the saturation frequency offset to obtain the z-spectrum.
? TROUBLESHOOTING
-
(vi)
Analyze the obtained data: the full signal loss for 0 ppm saturation offset is the direct saturation of free Xe in solution. The GV-related response is expected around −170 to −190 ppm offset. This local minimum in signal intensity corresponds to the frequency for the best Hyper-CEST response. Optimum conditions for option B are then obtained for choosing this offset for on-resonant saturation and its negative value for the off-resonant control image.
▴CRITICAL STEP While the true GV-bound xenon resonance frequency in the sample data is at about −170 ppm (green dashed line, Figure 6b), the maximum xenon depolarization spectrally appears at a different frequency which is shifted towards that of free xenon (blue dashed line) at about −156.5 ppm. This difference is caused by so-called spillover between the GV-bound xenon resonance and that of free xenon. This would not occur if the Hyper-CEST signal between both resonances would recover entirely back to the baseline. Two parameters affect the “intensity” of spillover: 1) The exchange regime (given by the ratio of the xenon exchange rate and chemical shift difference in Hz), and 2) The strength of the saturation pulse B1. To avoid spillover effects, one can use GVs with slower Xe exchange or reduce the saturation power.
-
(vii)
An accurate evaluation is based on normalized z-spectra that also accounts for the T1 relaxation. For this purpose, acquire data points with no saturation (i.e. setting the saturation time to zero) and negligible relaxation: set the number of saturation offsets to 5, the saturation frequency offset to +300 ppm, and –most importantly – the saturation power and duration to 0 μT and 0 sec respectively. Start the acquisition and draw an ROI in the xenon MR images for signal integration.
-
(viii)
Plot the signal intensity of the ROI for the 5 acquisitions; they should be almost identical and the average value can be used for normalizing the values of the z-spectrum obtained in (v).
(B). Single Hyper-CEST image from two saturation frequencies ● TIMING 20 m
-
(i)
Set the number of saturation offsets to 2.
-
(ii)
Enter the on-resonant saturation frequency offset and the off-resonant saturation frequency offset according to the z-spectrum interpretation in step |()92|(A)(vi)).
-
(iii)
Select the saturation power B1 and saturation time tsat. The example here used saturation conditions of B1 = 15 μT, and tsat = 5 s.
! CAUTION Ensure that the power limits for the NMR coil are not exceeded as this might permanently damage the coil.
▴CRITICAL STEP It is not useful to choose a saturation time that exceeds the T1 relaxation time of Xe in water24 , the signal loss is then dominated by intrinsic relaxation and not by the CEST effect.
-
(iv)
Enter the desired number of image averages, i. e. the number of excitations NEX, to achieve good signal intensity, e.g. NEX = 16.
-
(v)
Start the sequence which will perform 2 × NEX acquisitions.
-
(vi)
Check the results in the image display software.
-
(vii)
The difference image displaying the areas with the Hyper-CEST response can be obtained by subtracting the image with on-resonant saturation from the image with off-resonant saturation using the image display software; if desired, normalize the difference image by the off-resonant control image .
▴CRITICAL STEP Ensure that the intensity scaling is the same for the two images when performing any difference image calculations.
Shutting down the system ● TIMING 10 m
-
93.
Set the flow into the polarizer to 0 SMLM.
-
94.
Close the polarizer gas outlet and shut down the system according to manufacturer’s SOP.
-
95.
Ensure that the shortcut valve is open.
-
96.
Carefully depressurize the phantom by slowly ramping down the set point of the pressure controller.
▴CRITICAL STEP Fast depressurizing may damage the GVs and render them unusable for future experiments or quality control.
-
97.
Disconnect the sample tube from the PFA tubings and store them for later experiments.
-
98.
Turn off the VTU.
●TIMING
GV production and purification
Media preparation: 1 hour
Steps 1Ai-vi 1Bi-iii: Growing Ana and Halo cultures: 2–4 weeks (depending on the type and confluency of culture)
Steps 1Avii-xi and 1Biv-vi: Harvesting Ana and Halo GVs: 1 day
Steps 1Axii-xvii and 1Bvii-xi: Isolation and purification of GVs from Ana and Halo cultures: 2–3 days
Steps 1Ci-iv: Heterologous expression of Mega GVs in E. coli: 2 days
Steps 1Cv-x: Harvesting and purification of Mega GVs: 2 days
GV quantification
Step 2: Measuring pressure-sensitive OD using Nanodrop: 15 mins
Step 3: Measuring the GV protein concentration: 15 mins
Chemical functionalization of GVs
Steps 4–10: Chemical functionalization of GVs: 2 days (includes 1 day for dialysis)
Steps 11–19: Preparation of ΔGvpC GVs: 12 hours
Steps 20–38: Preparation of recombinant GvpC: 1 day
Steps 39–41: Preparation of GVs with recombinant GvpC: 1 day
Steps 42–45: Functionalization of SpyCatcher-functionalized GVs: 5 hours
GV characterization
Step 46A: Determining critical collapse with pressurized absorbance spectroscopy: 40 mins
Step 46B: GV preparation for DLS: 5 mins
Step 46C: Preparation of GV samples for TEM: 30 mins
Ultrasound imaging of GVs in vitro
Steps 47–55: Agarose phantom preparation and sample loading: 3 hours
Steps 56–62: Phantom Imaging: 1 hour (can be more depending on number of samples and mode of acquisition)
Ultrasound imaging of GVs in vivo
Step 63A: GV imaging in vivo using the Verasonics ultrasound system: 45 mins
Step 63B: Halo gas vesicle imaging in vivo using the Vevo ultrasound system: 45 min (timing may vary based on number of acquisitions)
GV imaging using 129Xe-MRI in vitro
Steps 64–91: Preparing GV sample and MRI equipment for data acquisition: 50 mins
Step 92 Option A: Imaging series and z-spectrum: One experimental session: 1.5 hours
Step 92 Option B: Single Hyper-CEST image from two saturation frequencies: One experimental session: 1.5 hours
Steps 93–98: Shutting down the system: 10 minutes
?TROUBLESHOOTING
Troubleshooting advice can be found in Table 5.
Table 5.
Troubleshooting Table
| Step No. | Problem | Possible Reason | Solution |
|---|---|---|---|
| 1A(iv), 1A(vi) 1B(iii) | Cultures growing too slowly | Not enough viable cells | Increase the amount of starter inoculum. |
| 1A(iv), 1A(vi) 1B(iii) | Cell death | Contamination | Start fresh from primary cultures and follow sterile procedures. |
| Incubator conditions (aeration, pH, temperature and lighting) not optimal | Loosen the cap/foil to allow aeration, monitor CO2 levels. For Ana cultures, ensure temperature inside incubator does not increase during light cycle due to heat from the lamps and confirm proper exposure of the cultures to light. | ||
| 1A(vii)-(xi), 1B(iv)-(vi) | Poor GV yield | Cultures not confluent | Allow cultures to grow for a few more days. Confluency is determined by the color and turbidity of the cultures as described in the procedure and shown in Figure 2 |
| 1A(xi), 1B(vi) | Incomplete lysis | Dilute the concentrated cell suspension with lysis buffer and/or allow lysis to proceed longer. | |
| 1A(vii)-(xi), 1B(iv)-(vi) | Collapse of GVs | Make sure the cultures are not subjected to agitation above 100 rpm and handle flasks gently while transporting and placing on hard surfaces. | |
| 1A(x), 1B(vi) | Inefficient retrieval of buoyant cells from the separatory funnel | Thoroughly wash the inner walls of the funnel with media/buffer to retrieve the buoyant cells sticking to the sides (especially for Halo). | |
| 1A(xii) | Incomplete separation of GVs from lysate/subnatant | Lysate is too dense, requiring longer spins for GVs to float to the top | Increase time of centrifugation and/or dilute lysate with PBS prior to spin |
| 1A(xii), 1C(v) | Collapse of GVs during spins | Hydrostatic Pressure on GVs greater than critical collapse pressure | Aliquot suspensions into more tubes to reduce height of the column (volume in a single falcon tube should not exceed 40 mL). |
| 1A(xv), 1B(vii) | Pressurization of tubes while opening and closing | Ensure microcentrifuge tubes are closed very gently. The caps of such tubes should not make a snap sound when closed. | |
| 8 | NHS-amine reaction did not work | NHS will hydrolyze in the presence of water/moisture. | Ensure DMSO used for storage is anhydrous. Limit exposure to ambient moisture. Store in a desiccated environment. |
| 4-10 | GVs aggregate after reaction with NHS-moiety | Conjugation of moieties on the GVs destabilizes the protein. | Conjugate fewer moieties on the GV surface by reducing the concentration of NHS-moiety and/or reaction time. |
| Presence of surfactants in the reactant sample. | Purify the NHS-moieties using chromatography or other methods. | ||
| 9, 10 | Subsequent reaction using NHS-moiety conjugated GVs does not work. | Presence of excess NHS-moieties in solution. | Repeat dialysis until excess NHS-moieties are below level of detection. |
| 11-19 | Incomplete GvpC stripping | GV concentration is too high for efficient removal of GvpC | Dilute native Ana GVs in PBS to an OD ~ 5 before adding 10 M urea buffer. |
| 20-38 | Low or no GvpC yield | Low expression of GvpC variant | Check plasmid for mutations, prepare fresh transformations and try reducing temperature and increasing duration of expression. |
| Poor binding to the Ni column | Ensure column is charged and increase incubation time with resin or add more resin. | ||
| Inefficient retrieval of inclusion bodies | Ensure that inclusion pellets are not lost during repeated rounds of centrifugation and resuspension | ||
| 39-41 | Incomplete GvpC re-addition | GV concentration is too high for re-addition reaction to go to completion | Reduce GV OD500 during GvpC re-addition. Confirm re-addition by SDS-PAGE and Coomassie staining. |
| GvpC concentration is too low | Inaccurate quantification of GvpC eluate. | ||
| 39-41 | Protein precipitation is seen after re-addition | Excess unbound GvpC forms precipitates in PBS. | Reduce stoichiometric excess factor of GvpC during re-addition. |
| 42-45 | No fluorescence from SpyCatcher-GFP-labeled GVs | Incomplete SpyTag functionalization | Confirm GvpC-SpyTag re-addition using SDS-PAGE. |
| Incomplete SpyTag-SpyCatcher reaction | Increase excess factor of SpyCatcher during reaction and/or increase reaction time. | ||
| 46A | Collapse curves are inconsistent or noisy | Incomplete collapse | Make sure that there are no air leaks in the connectors or bubbles in the sample |
| Presence of detergents and/or incomplete GvpC addition | Always run control GV samples such as ΔGvpC and GvpCWT to ensure that the re-addition was done under proper conditions and corroborate with SDS-PAGE results. | ||
| 46C | Too much black background on TEM grid | GV sample contains contaminants or phosphate-containing molecules | Using dialysis or centrifugally-assisted purification to resuspend GVs in 10 mM HEPES with 150 mM NaCl buffer. |
| Uranyl acetate precipitate present in solution | Avoid extended exposure to ambient light. Uranyl acetate will precipitate when exposed to light and UV. | ||
| 46C | Too many or too few GVs on the TEM grid | Incorrect GV OD measurement | Measure GV OD and double check the dilution calculation. |
| GVs were not given adequate time to adhere on charged grids. | Excessive wicking can cause too many GVs to come off grids. Reduce how much solution is being wicked off. | ||
| 47-62 | Only the top of the GV sample for in vitro ultrasound phantoms shows contrast | The concentration of GVs is too high. | Lower the concentration of GVs. |
| 47-62 | The signal from GVs and polystyrene beads is very different. | The concentration of GVs or polystyrene beads is too high or too low. | Matching of GVs’ concentration to concentration of beads. |
| 63A, 63B | Weak or no signal after injection | IVC not in the image | Move the transducer to find the optimal location in the IVC and try to align the imaging target with the natural focus of the transducer |
| GVs collapsed prior to or during injection | Check that there is no pressure when injecting through the catheter by manually injecting a small volume of saline with a 30G syringe. If you feel pressure, readjust the catheter into the tail vein and ensure that there are no blocks, allowing the saline to flow smoothly | ||
| Transmit power set too high | Ensure that transmit power does not generate an acoustic pressure on the GVs that exceeds the critical collapse pressure | ||
| 82 | Excessive foam formation | The overall protein concentration in the sample is too high | Reduce the protein concentration – or – add an antifoam agent such as pluronic L81 (ca. 0.1% by vol.); however, such an agent can also have an impact on the integrity of the protein structure |
| 88 | The Xe NMR spectrum does not show any signal or very distorted signals | The excitation pulse is too weak | Increase the amplitude for the excitation pulse |
| The field homogeneity is poor | Perform another shim | ||
| There are still gas bubbles in the sample while acquiring the FID signal | Increase the waiting time after bubbling or consider adding an antifoam agent | ||
| The Rb is oxidized and no hyperpolarized Xe is produced | Check the laser absorption profile of the hyperpolarizer and replace the Rb in the pumping cell if necessary | ||
| 92A(v) | No CEST response from the GVs is observed in the z-spectrum, or the image with on-resonant saturation does not really show a signal loss | GVs have collapsed | Measure the OD of the sample and prepare a fresh one from stock solution if necessary |
| GV concentration too low | Add GVs from stock solution | ||
| Saturation power too low or saturation time too short | Increase the saturation power or the saturation time | ||
| 92A(v) | The saturation response from the GVs is too broad and merges into the direct saturation response | Saturation power is too high | Reduce the saturation power |
ANTICIPATED RESULTS:
GV expression and purification
Ana and Halo GV expression and purification:
Healthy, viable Ana and Halo cultures look dark green (Figure 2c) and light pink (Figure 2d) upon reaching confluency. Figure 2e and f show the separation of buoyant Ana and Halo cells from spent media. For Halo cultures, the buoyant cells are visible as a thin layer or ring of milky pink at the top of the liquid phase. After lysis, the lysate will appear milky due to the presence of GVs. After one round of centrifugally-assisted floatation, the GVs will appear as a white layer at the top with a green (Ana) or pink (Halo) liquid subnatant, along with a pellet of cell debris at the bottom. For the Halo, a mixed layer of white (Halo GVs) and milky pink (unlysed Halo cells) may be visible after the initial spins. Figures 2g and h show representative images of the purified Ana and Halo GVs in 2 mL tubes after the final spin, with the white GVs forming a dense layer at the top and a clear subnatant of PBS. Figure 2j shows tubes containing milky-white solutions of purified Ana and Halo GVs (~OD500,ps 6) resuspended in PBS. The typical GV yield is ~ 3 mL of OD500,ps 10 for Ana and Halo GVs per flask of culture.
Mega GV expression and purification in E. coli:
While native expression of Ana and Halo GVs usually results in majority of the cells being buoyant, most of the E. coli cells heterologously expressing Mega GVs reside in the pellet after centrifugation. In our experience, the typical yield of Mega GVs is approximately 1.5 mg/L cells per unit OD600. Figure 2i shows a representative image of purified and unclustered Mega GVs forming dense white layer at the top, with a clear subnatant of PBS. Figure 2j shows a milky-white solution of purified and unclustered Mega GVs resuspended in PBS (~ OD500,ps 6).
GV functionalization
Ana GvpC is purified from inclusion bodies in E.coli. The recombinant GvpC can be harvested with yields of up to 8 mg protein / L culture. The inclusion bodies yield >95% pure protein (Figure 7a). GvpC-based functionalization relies upon complete removal of the native Ana GvpC layer before reconstitution with recombinant protein. Urea treatment results in nearly complete (>95%) removal of native GvpC, indicated by absence of GvpC on SDS-PAGE (Figure 7a) and a lowered critical collapse pressure (shown in the following section on GV characterization).. GVs retain their milky white appearance throughout the functionalization process. The binding of recombinant GvpC variants to GVs can be assayed by SDS-PAGE and Coomassie staining. ΔGvpC Ana GVs do not exhibit a band around 25 kDa (molecular weight of GvpC) while wild type and truncated GvpC variants bind to GVs (Figure 7a).
Figure 7. Anticipated results for GV characterization and in vitro ultrasound imaging.
(a) Coomassie stained SDS-PAGE gel indicating complete removal of native GvpC (ΔGvpC, lane 2) and re-addition of two recombinant GvpC variants, GvpCWT and ΔN&C GvpC (lanes 3 and 4). The smeared bands around 250 kD represent GvpA polymers forming the inner GV scaffold whereas GvpC bands are visible at lower molecular weights. (b) Confocal fluorescence image showing dual-functionalized Ana GVs with RGD peptide and Alexa Fluor-488 (green) after a 24 h incubation with U87 glioblastoma cells (DAPI-stained nuclei, blue). Scale bars are 50 μm. (c) Dual-mode imaging of GVs functionalized with SpyCatcher-mNeonGreen with ultrasound (top panel) and fluorescence (ex 506 nm, em 550 nm) (bottom panel). Scale bars are 1 mm. Figures (b) and (c) adapted with permission from Lakshmanan, A. et al. Molecular Engineering of Acoustic Protein Nanostructures, ACS Nano, Copyright © 2016, American Chemical Society. (d) Collapse pressure profiles for GV variants (Halo, Mega, Ana ΔGvpC and Ana GvpCWT). (e) DLS measurements showing typical size distributions for purified Halo, Mega, ΔGvpC and GvpCWT Ana GVs (f) Representative TEM images for purified Halo, Ana and Mega GVs. Scale bars are 200 nm. (g) TEM showing intact and collapsed Ana GVs. Scale bars are 200 nm. Figure (g) adaptedwith permission from Shapiro, M.G. et al. Biogenic gas nanostructures as ultrasonic molecular reporters, Nature Nanotechnology, Copyright © 2014, Nature Research Group.
Figure 7b shows an application using dual-functionalized GVs for targeted cell-specific imaging. In this example, Ana GVs functionalized with GvpC fused to peptide RGD are chemically conjugated to Alexa Fluor 488. The functionalized GVs bind to integrin receptors on glioblastoma cells via the RGD motif and the fluorescent labeling of GVs enables rapid assessment of targeting efficacy using confocal microscopy. SpyTag-GvpC binds to GVs with similar productivity and stoichiometry as wild-type GvpC. We have previously shown that each modified GV has an average of 1000 SpyTag functionalities14. The collapse pressure of SpyTag GVs is not altered upon SpyCatcher binding, indicating that attachment of a large 37.9 kD protein to the GV surface does not adversely affect GvpC strengthening of the GV wall14.
Functionalization of SpyTag-GVs with SpyCatcher-GFP enables multi-modal imaging of GVs with ultrasound and fluorescence. Ultrasound images were acquired in an agarose phantom with a Verasonics L22–14V transducer at 19 MHz, 5.0 V and F-number 3. The ultrasound images show similar echogenicity between fluorescently labeled GVs and wild-type and unreacted controls (Figure 7c). Optical images taken through the green channel of a BioRad Chemidoc MP system indicate that SpyTag-SpyCatcher-GFP labeled GVs are fluorescent (Figure 7c). Fluorescence quantification of Halo GVs after chemical conjugation to Alexa Fluor 488 indicates that functionalized GVs are around 40-fold more fluorescent than native GVs.
GV characterization
Critical collapse pressure of GVs:
The collapse pressure profiles of purified Ana, Halo and Mega GVs as well as functionalized GVs are obtained by measuring change in OD500nm of GV samples under increasing hydrostatic pressure. The mean collapse pressures are 59 kPa (Halo), 587 kPa (Ana GvpCWT) and 750 kPa (Mega) (Figure 7d). Ana GVs without GvpC have a significantly lower critical collapse pressure of 193 kPa. Collapse pressure measurements also indicate the degree of mechanical strengthening by recombinant GvpC variants.
DLS Measurements:
DLS values for hydrodynamic diameters exhibit variability due to the biogenic nature of GVs. Representative DLS profiles are given in Figure 7e. Hydrodynamic diameters of Halo GVs range from 260 nm – 320 nm (polydispersity 0.15 – 0.21), ΔGvpC Ana GVs range from 250 nm – 360 nm (polydispersity 0.21 – 0.24), and GvpCWT Ana GVs range from 240 nm – 340 nm (polydispersity 0.17 – 0.26), Mega GVs from 200–380 nm (polydispersity 0.23 – 0.34).
TEM visualization of GVs:
GVs appear as bright particles on a dark background under negative staining with uranyl acetate. Typically, TEM allows visualization of the morphology, size, shape and overall surface features of GVs. Figure 7f shows representative TEM images for purified wild-type Halo, Ana and Mega GVs. While Halo GVs appear spindle shaped, Ana and Mega GVs are more cylindrical with conical tips in the longitudinal dimension. Although the size and shape of GVs are determined primarily by the genotype, each type possesses a certain degree of heterogeneity. For example, Ana GVs have length distribution with a standard deviation of 35% of the mean6. It is also important to note that not all Halo GVs have a spindle morphology, and a small subpopulation may appear more cylindrical with biconical ends27. Ana GVs that are produced in their native host are usually longer and wider than heterologously expressed Mega GVs. Typical GV dimensions obtained from TEM images summarized in Table 4 (N=107, 125 and 61 for Ana Halo, and Mega respectively, errors in standard deviation).
The high resolution provided by TEM allows the visualization of ribs on the surface of the gas vesicles at higher magnifications. Figure 7g shows intact and collapsed Ana GVs, demonstrating that collapse causes complete rupture and opening of the protein shell, leading to a flattened pancake-like structure. GVs chemically functionalized with biotin can be coated with streptavidin and visualized by TEM8.
In vitro ultrasound imaging:
Typical ultrasound images acquired in the fundamental mode and after second harmonic filtering show contrast from intact wild-type (WT) and ΔGvpC Ana GVs (Figure 8a, top panel). Acoustic collapse of GVs leads to elimination of ultrasound signal (Figure 8a, bottom panel), confirming that contrast arises from the intact GVs. Lowering of collapse pressure is typically associated with higher non-linear signals from GV samples, allowing tuning of GV harmonic response by modification or removal of GvpC. The ultrasound contrast from Halo GVs and their utility for nonlinear imaging in vitro and in vivo has been demonstrated in our previous work8. The acoustic behavior of Halo GVs at ultrasound frequencies of 12.5–27.5 MHz has also been investigated through modeling and experiments, suggesting acoustic buckling as the mechanism underlying generation of non-linear signals10. In parallel, we recently developed an amplitude modulation scheme taking advantage of the nonlinear pressure dependence of backscattered signals in engineered Ana GVs, allowing selective imaging of these nanostructures9.
Figure 8. Anticipated results for in vitro and in vivo ultrasound imaging.
(a) Fundamental and second harmonic images for intact ΔGvpC and GvpCWT Ana GVs (top panel) acquired using the Verasonics L11–4v transducer with a 4.46 MHz transmission and band-pass filtered around 4.46 and 8.92 MHz respectively. A high power burst from the transducer is used to collapse the GVs (bottom panel). Scale bars are 1 mm. (b) B-mode image of the liver showing the (i) inferior vena cava (IVC), (ii) portal vein and (iii) abdominal aorta. Contrast mode images acquired using high-frequency ultrasound show the liver and IVC with the baseline contrast image before injection of Halo gas vesicles. Following a bolus injection of gas vesicles, contrast enhancement is seen in the IVC followed by the liver. (c) Time intensity curve depicting the nonlinear contrast signal change over time for the IVC (orange) and the liver (blue) before, during, and after an intravenous administration of Halo gas vesicles. Time intensity curve in the (d) fundamental and (e) second harmonic mode depicting the change in acoustic signal in the IVC before, during and after steady infusion with ΔGvpC and GvpCWT Ana GVs. Shaded regions represent SEM (N=6).Figures (a, d-e) adapted with permission from Lakshmanan, A. et al. Molecular Engineering of Acoustic Protein Nanostructures, ACS Nano, Copyright © 2016, American Chemical Society.
In vivo ultrasound imaging:
When imaging gas vesicles in vivo, we suggest starting with a vascularized organ such as the liver. A normal liver will look homogeneous under B-mode ultrasound with vessel cross-sections appearing as hypoechogenic regions (Figure 8b, left panel). When imaging a mouse in the transverse plane, it is possible to locate a region of the liver containing the inferior vena cava (IVC).
After gas vesicles are administered intravenously via the lateral tail vein, nonlinear contrast enhancement will be observed first in the IVC, followed by the liver (Figure 8b). Using the Vevo®LAB software, regions of interests (ROIs) encompassing the IVC and liver can be drawn, and a time intensity curve of the mean contrast signal change over time within each ROI can be plotted.
As shown in Figure 8c, once a 50μL bolus of gas vesicles has been injected, an initial bulk signal is observed in the IVC, which will decrease over times as the gas vesicles are dispersed into the circulation. Signal enhancement in the liver will follow a few seconds after with a steady signal increase and then a plateau. The signal plateau in the liver is likely due to the uptake of non-functionalized gas vesicles by the Kupffer cells, similar to many other nano-sized particles. This contrast signal can then be eliminated by employing a high power burst that will collapse all the gas vesicles in the imaging plane.
It is possible to engineer Ana GVs to produce enhanced non-linear signals in vivo (Figure 8d-e). After bolus injection of GVs, ultrasound imaging of the IVC is performed in fundamental and harmonic modes for up to a minute (transmission at 4.46 MHz and reception filtered around 4.46 and 8.9 MHz center frequencies, respectively). The mean signal intensity plotted as a function of time (Figure 8d-e) show that ΔGvpC Ana GVs produce higher harmonic signals compared to Ana GvpCWT in vivo, while their fundamental signals are comparable. This observation is also consistent with the in vitro data in Figure 8a.
Hyperpolarized Xe MRI imaging of GVs:
Typical GV concentrations in the nano- to picomolar range will not allow the observation of the GV-bound Xe directly. A 129Xe NMR spectrum therefore only shows the signal from dissolved xenon gas (assigned to 0 ppm) and the resonance from Xe gas on top of the solution (appearing ca. 190 ppm right of the solution peak) (Figure 6a). Data obtained in step 92|(A)(v) should look similar to the spectrum in Figure 6b, except that it is not yet normalized. Taking the relaxation and saturation free reference signal from step 92|(A)(viii) into account, the spectrum should yield intensities between 0 and 1 like Figure 6b. It should be noted that the longer the saturation time, the more the baseline has dropped below 1.
Data from option 92|B can be displayed as the (normalized) Hyper-CEST effect map that gives a signal intensity in %. The original data is plotted in Figure 6c-d and the Hyper-CEST effect map in Figure 6e. This Hyper-CEST effect map is obtained by post-processing the off- and on-resonant xenon MR images according to: Hyper-CEST effect map = (off-resonant – on-resonant)/off-resonant × 100
Supplementary Material
ACKNOWLEDGEMENTS:
This research was supported by the NIH (R01-EB018975 to MGS), DARPA (W911NF-14–1-0111 to MGS), CIHR (Grant # to FSF) and HFSP (RGP0050 to MGS and LS). A.L. is supported by the NSF graduate research fellowship (award number 1144469). A.F. is supported by the NSERC graduate fellowship. S.P.N. is supported by the Caltech Summer Undergraduate Research Fellowship. D.M. is supported by the HFSP Cross-Disciplinary Postdoctoral Fellowship. Research in the Shapiro laboratory is also supported by the Heritage Medical Research Institute, the Burroughs Wellcome Fund, the Pew Charitable Trust, the Sontag Foundation and the David and Lucile Packard Foundation. Research in the Schröder laboratory is also supported by the Michael J. Fox Foundation for Parkinson’s (grant agreement no. 12549) and a Koselleck Grant by the German Research Foundation (DFG; project SCHR 995/5–1).
Footnotes
COMPETING FINANCIAL INTERESTS:
The authors declare no competing financial interests.
REFERENCES:
- 1.Ferrara K, Pollard R & Borden M Ultrasound microbubble contrast agents: fundamentals and application to gene and drug delivery. Annu. Rev. Biomed. Eng 9, 415–47 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Kaufmann BA & Lindner JR Molecular imaging with targeted contrast ultrasound. Curr. Opin. Biotechnol 18, 11–6 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Weissleder R et al. Ultrasmall superparamagnetic iron oxide: characterization of a new class of contrast agents for MR imaging. Radiology 175, 489–493 (1990). [DOI] [PubMed] [Google Scholar]
- 4.Lee J-H et al. Artificially engineered magnetic nanoparticles for ultra-sensitive molecular imaging. Nat. Med 13, 95–99 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Caravan P, Ellison JJ, McMurry TJ & Lauffer RB Gadolinium (III) chelates as MRI contrast agents: structure, dynamics, and applications. Chem. Rev 99, 2293–2352 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Walsby AE Gas vesicles. Microbiol. Rev 58, 94–144 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfeifer F Distribution, formation and regulation of gas vesicles. Nat. Rev. Microbiol 10, 705–15 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Shapiro MG et al. Biogenic gas nanostructures as ultrasonic molecular reporters. Nat. Nanotechnol 9, 311–316 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maresca D et al. Nonlinear ultrasound imaging of nanoscale acoustic biomolecules. Appl. Phys. Lett 110, 073704 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cherin E et al. Acoustic Behavior of Halobacterium salinarum Gas Vesicles in the High-Frequency Range: Experiments and Modeling. Ultrasound Med. Biol 43, 1016–1030 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shapiro MG et al. Genetically encoded reporters for hyperpolarized xenon magnetic resonance imaging. Nat. Chem 6, 629–634 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Hane FT et al. In vivo detection of cucurbit [6] uril, a hyperpolarized xenon contrast agent for a xenon magnetic resonance imaging biosensor. Sci. Rep 7, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barskiy DA et al. NMR Hyperpolarization Techniques of Gases. Chem. Eur. J, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lakshmanan A et al. Molecular Engineering of Acoustic Protein Nanostructures. ACS Nano 10, 7314–7322 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li N & Cannon MC Gas vesicle genes identified in Bacillus megaterium and functional expression in Escherichia coli. J. Bacteriol 180, 2450–8 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilad AA & Shapiro MG Molecular Imaging in Synthetic Biology, and Synthetic Biology in Molecular Imaging. Mol. Imaging Biol, 1–6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zakeri B et al. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. PNAS 109, E690–E697 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schröder L, Chapter 8 HyperCEST Imaging In Chemical Exchange Saturation Transfer Imaging: Advances and Applications, Pan Stanford Publishing: 2017; pp 121–158. [Google Scholar]
- 19.Schröder L, Chapter 17 - Xenon Biosensor HyperCEST MRI A2 - Albert, Mitchell S In Hyperpolarized and Inert Gas MRI, Hane FT, Ed. Academic Press: Boston, 2017; pp 263–277. [Google Scholar]
- 20.Nikolaou P et al. Near-unity nuclear polarization with an open-source 129Xe hyperpolarizer for NMR and MRI. PNAS 110, 14150–14155 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Witte C, Kunth M, Rossella F & Schröder L Observing and preventing rubidium runaway in a direct-infusion xenon-spin hyperpolarizer optimized for high-resolution hyper-CEST (chemical exchange saturation transfer using hyperpolarized nuclei) NMR. J. Chem. Phys 140, 084203 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Witte C et al. Hyperpolarized xenon for NMR and MRI applications. JoVE, e4268–e4268 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kunth M, Witte C, Hennig A & Schröder L Identification, classification, and signal amplification capabilities of high-turnover gas binding hosts in ultra-sensitive NMR. Chem. Sci 6, 6069–6075 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kunth M, Witte C & Schröder L Continuous‐wave saturation considerations for efficient xenon depolarization. NMR Biomed. 28, 601–606 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Smith R & Peat A Growth and gas-vacuole development in vegetative cells of Anabaena flosaquae. Arch. Microbiol 58, 117–126 (1967). [Google Scholar]
- 26.Buchholz B, Hayes P & Walsby A The distribution of the outer gas vesicle protein, GvpC, on the Anabaena gas vesicle, and its ratio to GvpA. Microbiology 139, 2353–2363 (1993). [DOI] [PubMed] [Google Scholar]
- 27.Simon RD Morphology and protein composition of gas vesicles from wild type and gas vacuole defective strains of Halobacterium salinarium strain 5. Microbiology 125, 103–111 (1981). [Google Scholar]
- 28.Kunth M, Witte C & Schröder L Quantitative chemical exchange saturation transfer with hyperpolarized nuclei (qHyper-CEST): Sensing xenon-host exchange dynamics and binding affinities by NMR. J. Chem. Phys 141, 194202 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Zaiss M, Schnurr M & Bachert P Analytical solution for the depolarization of hyperpolarized nuclei by chemical exchange saturation transfer between free and encapsulated xenon (HyperCEST). J. Chem. Phys 136, 144106 (2012). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.