Abstract
YAP/TAZ are nuclear effectors of the Hippo pathway regulating organ growth and tumorigenesis. Yet, their working as transcriptional regulators remains underinvestigated. By ChIP-seq analyses in breast cancer cells, we discovered that the YAP/TAZ transcriptional response is pervasively mediated by a dual element: TEAD factors, through which YAP/TAZ bind to DNA, co-occupying chromatin with Activator Protein-1 (AP-1, dimer of JUN and FOS proteins) at composite cis-regulatory elements harboring both TEAD and AP-1 motifs. YAP/TAZ/TEAD and AP-1 form a complex that synergistically activates target genes directly involved in the control of S-phase entry and mitosis. This control occurs almost exclusively from distal enhancers that contact target promoters through chromatin looping. YAP/TAZ-induced oncogenic growth is strongly enhanced by gain-of-AP-1 and severely blunted by its loss. Conversely, AP-1-promoted skin tumorigenesis is prevented in YAP/TAZ conditional knockout mice. This work highlights a novel layer of signaling integration, feeding on YAP/TAZ function at the chromatin level.
Introduction
YAP/TAZ1,2 are potent inducers of cell proliferation and main drivers of tumorigenesis in a number of contexts3–6. Yet, the mechanisms underpinning this activity remain enigmatic. Only a handful of direct targets have been described in mammalian cells, leaving largely undefined what are the immediate downstream effectors by which YAP/TAZ exert their biological effects3. Moreover, lack of systematic studies results in just a scattered understanding of the transcriptional partners by which nuclear YAP/TAZ control transcription on a genome-wide scale. Also unknown is whether and how, after binding to DNA, YAP/TAZ achieve combinatorial control of gene expression, for example through cooperation with other nuclear oncogenes during YAP/TAZ-driven oncogenic growth.
Results
A genomic map of YAP/TAZ recruitment to chromatin
To elucidate how YAP/TAZ regulate gene expression in tumor cells we performed chromatin immunoprecipitation assays with YAP and TAZ antibodies followed by next-generation sequencing (ChIP-seq) in MDA-MB-231 breast cancer cells, bearing genetic inactivation of the Hippo pathway (NF2 null)7. 7107 peaks were identified by both antibodies (Fig. 1a). YAP/TAZ-bound regions included the promoters of previously established YAP/TAZ direct targets (CTGF, CYR61, ANKRD1, AXL, AMOTL2, AJUBA and WTIP, Supplementary Figs. 1b-d).
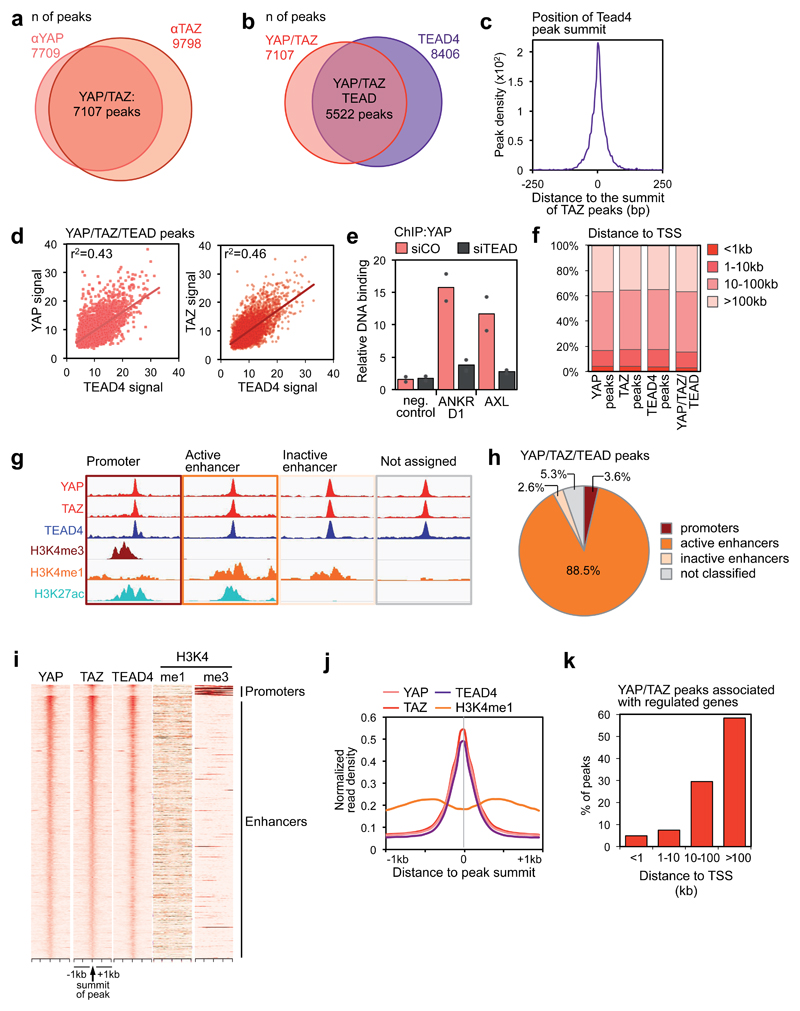
Figure 1. Genome-wide co-localization of YAP, TAZ and TEAD on enhancers.
(a) Overlap of peaks identified with YAP and TAZ antibodies. See Supplementary Figure 1a for specificity controls of the antibodies, and Table S1 for the results of de novo motif finding in YAP/TAZ peaks.
(b) Overlap of YAP/TAZ peaks and TEAD4 peaks. See Supplementary Figure 1g for specificity controls of TEAD4 antibody, and Supplementary Figure 1h for ChIP-seq profiles at positive control loci.
(c) Position of TEAD4 peak summits relative to the summits of the overlapping YAP/TAZ peaks, in a 500 bp window surrounding the summit of YAP/TAZ peaks.
(d) Linear correlation between the signal of YAP or TAZ and TEAD4 peaks in the 5522 shared binding sites. r2 is the coefficients of determination of the two correlations.
(e) ChIP-qPCR showing YAP binding to the indicated sites in MDA-MB-231 cells transfected with control (siCO) or TEAD siRNAs (siTEAD A). Relative DNA binding was calculated as fraction of input and normalized to IgG (IgG bars are omitted); data from 2 biological replicates from one representative experiment are shown.
(f) Absolute distance of YAP peaks (n=7709), TAZ peaks (n=9798), TEAD4 peaks (n=8406) or overlapping YAP/TAZ/TEAD peaks (n=5522) to the nearest TSS.
(g-h) Association of YAP/TAZ/TEAD peaks to promoters and enhancers according to ChIP-seq data for histone modifications. (g) Scheme illustrating peak classification. (h) Fraction of YAP/TAZ/TEAD peaks associated with each category. See Supplementary Figure 1j for validation of the enhancer/promoter status of a set of YAP/TAZ/TEAD-bound regions.
(i) Heatmap representing YAP/TAZ/TEAD binding sites located on promoters (top) and enhancers (bottom). YAP, TAZ and TEAD4 peaks are ranked from the strongest to weakest signal in TAZ ChIP, in a window of ±1kb centered on the summit of TAZ peaks. H3K4me1 and H3K4me3 signal in the corresponding genomic regions is shown on the right.
(j) Bimodal distribution of H3K4me1 signal around the summit of YAP/TAZ and TEAD4 peaks.
(k) Distance between YAP/TAZ/TEAD binding sites and the TSS of the direct target genes they are associated to. Overall, 635 peaks were associated to 379 genes positively regulated by YAP/TAZ.
See Methods for reproducibility of experiments.
YAP/TAZ do not carry a DNA-binding domain, and thus can contact the DNA only indirectly, through transcription factors (TFs) partners. So far, a number of these partners have been described, including TEAD1-4, RUNX, p73, KLF4, TBX5, and others3,8–11. Motif analyses at YAP/TAZ peaks revealed that the main platforms by which YAP/TAZ interact with DNA are TEAD proteins: their consensus sequence was present in at least 75% of YAP/TAZ peaks, typically at the summit of YAP/TAZ peaks (Supplementary Figs. 1e-f). While TEAD factors have been repeatedly associated as mediators of YAP/TAZ transcriptional responses3,9, surprisingly no motifs for the other proposed DNA-binding platforms of YAP/TAZ were significantly enriched (with the exception of RUNX, found in a minority of peaks, see Supplementary Table 1).
Following these results, we performed a ChIP-seq experiment for endogenous TEAD4 and found that 78% (5522) of YAP/TAZ peaks overlapped with TEAD4 peaks (Fig. 1b and Supplementary Table 2); furthermore, the summits of TEAD4 peaks coincide with the summit of the corresponding YAP/TAZ peaks (Fig. 1c), indicating that TEAD factors are indeed the major driver for YAP/TAZ recruitment to chromatin. In support of this notion, the signal of TEAD4 peaks is positively correlated with that one of YAP/TAZ peaks (Fig. 1d), and binding of YAP to chromatin is strongly affected by combined knockdown of TEAD1/2/3/4 as assessed by ChIP-qPCR (Fig. 1e).
Global association of YAP/TAZ/TEAD to enhancer elements
We analyzed the distribution of YAP, TAZ or TEAD binding sites relative to genes annotated in the human genome, and found that only a minute fraction of peaks mapped close (±1 kb) to Transcription Start Sites (TSS) whereas the vast majority of peaks was located farther than 10kb from the closest TSS (Fig. 1f). Analyses of publicly available TEAD ChIP-seq data revealed that this pattern is conserved in several cancer cell types (Supplementary Fig. 1i).
Due to their remote location, we asked whether most YAP/TAZ/TEAD common peaks are located in enhancers. Enhancers can be distinguished from promoters by their epigenetic features, that is, relative enrichment of histone H3 monomethylation (H3K4me1) on lysine 4 at enhancers, and trimethylation (H3K4me3) at promoters12. ChIP-seq data for these epigenetic marks in MDA-MB-231 cells were recently reported13, allowing us to compare this map of promoters and enhancers with the location of YAP/TAZ/TEAD4 peaks (Fig. 1g). Notably, only a very small fraction (3.6%) of YAP/TAZ/TEAD4 peaks are located on promoters; instead, 91% of peaks are located in enhancer regions (Figs. 1h-i). Furthermore, most of these enhancers are in an active state as revealed by H3K27 acetylation and reduced nucleosome occupancy at the peak center, also resulting in a bimodal distribution of H3K4me1 around the peak summit (Figs. 1h-j and Supplementary Fig. 1k).
A YAP/TAZ-regulated transcriptional program driving cell proliferation
We then sought to link YAP/TAZ/TEAD4 peaks to corresponding target genes (Supplementary Fig. 1l). All the peaks located in promoter regions (201) were assigned to the nearest genes. However, application of this proximity criterion to the distant enhancers bound by YAP/TAZ was questionable, as enhancers can regulate target genes over long distance, often skipping intervening genes. Instead, the specificity of enhancer-promoter associations is dictated by the 3D higher order chromatin structure, whereby enhancers interact with their target promoters via chromatin looping12. Importantly, a recently reported high-resolution map of chromatin interactions (HiC) has been shown to predict bona-fide enhancer-promoter pairs with great accuracy14. Notably, the large majority of these long-range chromatin interactions are conserved across cell types15. By using the map of enhancer-promoter pairs discovered by Jin et al., 201314, we could associate more than half of YAP/YAZ/TEAD4-bound enhancers to a set of 2957 candidate target genes; considering also the genes with peaks in their promoters, the list extends to 3089 genes. Of these candidates, 379 are in fact expressed in MDA-MB-231 cells, and in a YAP/TAZ-dependent manner, as determined by Affymetrix profiling of control and YAP/TAZ-depleted cells (using two independent combinations of siRNAs) (Supplementary Table 3). Crucially, the vast majority (88.6%) of these bona-fide YAP/TAZ direct targets are controlled only from distal enhancers, mostly located farther than 100,000 bp from the TSS (Fig. 1k); we further note that these genes could not have been identified by assigning the peak to the closest gene.
To identify the main biological processes regulated by YAP/TAZ, we performed Gene Ontology (GO) analyses on the list of YAP/TAZ direct targets. A large fraction of positive targets (135) are linked to processes related to cell cycle progression (Fig. 2a; Supplementary Tables 4 and 5); YAP/TAZ/TEAD binding sites are located exclusively on distal enhancers for 115 of these genes, and both on promoters and enhancers for other 11 genes. Other positive targets (13.5% of the total) are connected to RNA metabolism and RNA transport.
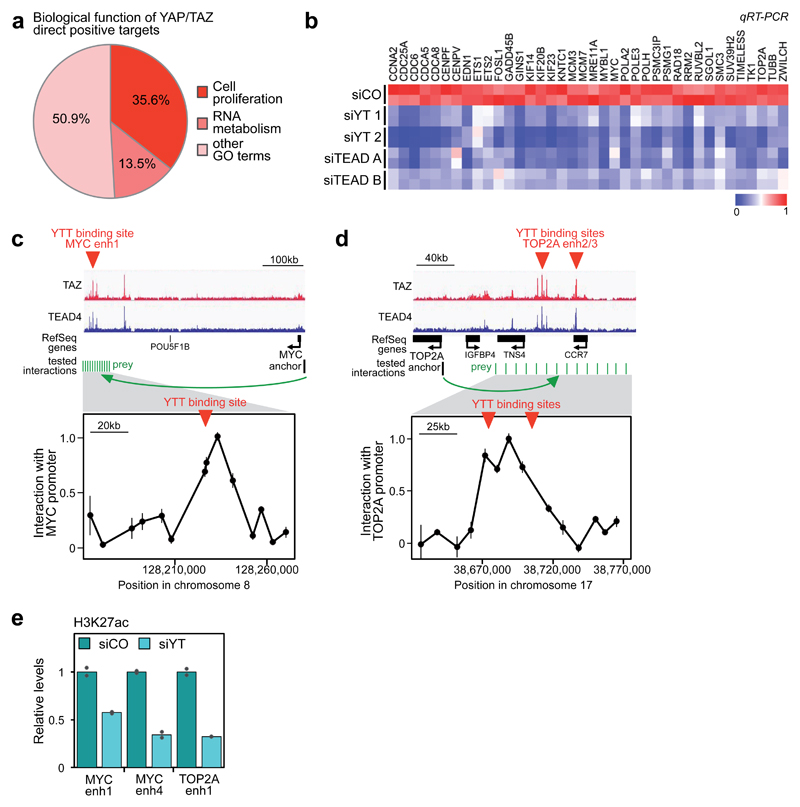
Figure 2. YAP/TAZ/TEAD transcriptional program.
(a) Biological functions associated to YAP/TAZ direct positive targets, identified by GO terms.
(b) YAP/TAZ or TEAD depletion impairs the expression of YAP/TAZ/TEAD direct target genes involved in cell proliferation, as evaluated by qRT-PCR (siCO=control siRNA, siYT=YAP/TAZ siRNA, siTEAD=TEAD siRNA). For a subset of genes, the downregulation of the corresponding proteins was also verified by Western blot (Supplementary Fig. 2b). See Supplementary Figure 2a for validation of TEAD siRNAs.
(c-d) Validation of the long-range interaction between YAP/TAZ-occupied enhancers and the promoters of MYC (c) and TOP2A (d) by DNA looping, using 3C assays in MDA-MB-231 cells. TAZ and TEAD4 ChIP-seq profiles show the position of YAP/TAZ/TEAD binding sites upstream of MYC or TOP2A genes (here named "MYC enhancer 1", “TOP2A enhancer2” “TOP2A enhancer3”), whereas no YAP/TAZ/TEAD binding sites were detected close to their TSS. The chart shows the frequency of interaction (measured as cross-linking frequency) between MYC or TOP2A promoter ("anchor") and the indicated sites surrounding YAP/TAZ/TEAD (YTT) peaks (green lines). Interaction frequency is higher close to YAP/TAZ peak. Data points are mean+SEM from n=3 biological replicates. See also Supplementary Figs. 2d-e for the additional interactions between MYC and TOP2A promoters and a different set of YAP/TAZ/TEAD-bound enhancers.
(e) ChIP-qPCR comparing the levels of H3K27ac (normalized to total H3 levels) in MDA-MB-231 cells transfected with control (siCO) or combined YAP/TAZ siRNAs (siYT1+2). Data from 2 biological replicates from one representative experiment are shown.
See Methods for reproducibility of experiments.
The YAP/TAZ/TEAD cell-proliferation program comprises essential factors involved in replication licensing, DNA synthesis and repair (e.g. CDC6, GINS1, MCM3, MCM7, POLA2, POLE3, TOP2A, RAD18), transcriptional regulators of the cell cycle (e.g. ETS1, MYC, MYBL1), cyclins and their activators (CCNA2 and CDC25A), and factors required for completion of mitosis (e.g. CENPF, CDCA5, KIF23). We selected about 40 of these genes (containing representative genes belonging to each of the above mentioned categories) and confirmed by qRT-PCR that their expression depends on YAP/TAZ as well as on TEAD1-4 (Fig. 2b). All these new YAP/TAZ/TEAD-regulated genes were associated to YAP/TAZ/TEAD peaks located on enhancers (as exemplified in Supplementary Fig. 2c). By chromatin conformation capture (3C) analysis we confirmed that the interaction of YAP/TAZ/TEAD-bound enhancers with MYC or TOP2A promoters occurs via chromatin looping in MDA-MB-231 cells (Figs. 2c-d and Supplementary Figs. 2d-e), validating the procedure here used to associate distant enhancers to target genes by using HiC data. Remarkably, YAP/TAZ are required for the activity of these enhancers, as acetylation of H3K27 in these regions drops in YAP/TAZ-depleted cells (Fig. 2e).
We next aimed to determine the biological validity of these findings. We found that YAP/TAZ-depleted cells stop proliferating and accumulate in the G1 phase (Figs. 3a-b). These effects are phenocopied by TEAD depletion (Supplementary Figs. 3b-c); moreover, the growth of YAP/TAZ-depleted cells can be fully rescued by re-introduction of wild-type YAP or TAZ, but not by their TEAD-binding deficient mutants (Figs. 3c and Supplementary Fig. 3d). Together, these results underline the relevance of TEAD as master determinant of YAP/TAZ-driven proliferation9.
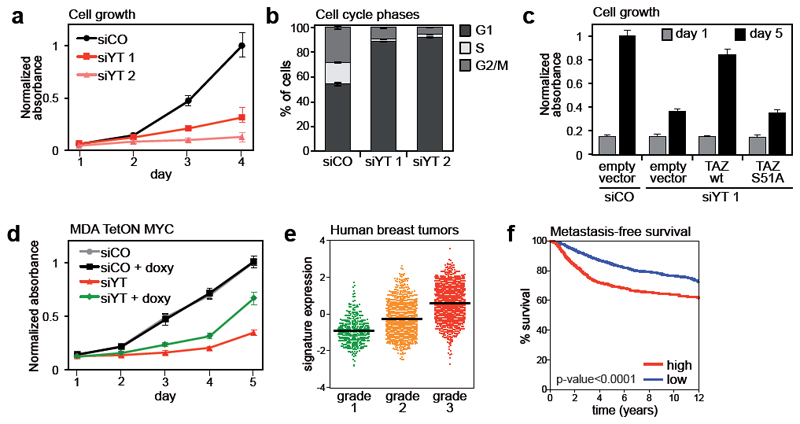
Figure 3. Control of cell proliferation by YAP/TAZ/TEAD.
(a) Growth curve of MDA-MB-231 cells transfected with control siRNA (siCO) or two different combinations of siRNAs targeting YAP and TAZ (siYT). Data are mean+SD of n=8 biological replicates. Individual depletion of YAP or TAZ has no effect on cell growth (Supplementary Fig. 3a).
(b) Percentage of MDA-MB-231 cells in G1, S and G2/M phases of cell cycle, as determined by flow-cytometric analysis of DNA content. Cells were transfected with control (siCO) or YAP/TAZ siRNAs (siYT) 48hr before fixation. Data are mean+SD of n=3 biological replicates.
(c) Sustained expression of TAZ, but not of TEAD-binding deficient TAZS51A, rescues cell proliferation in YAP/TAZ-depleted cells. Empty-vector-, wild-type TAZ- (wt) or TAZS51A-transduced MDA-MB-231 cells were transfected with control (siCO) or YAP/TAZ (siYT) siRNAs, as indicated. Proliferation was evaluated as in (f). Data are mean+SD of n=8 biological replicates.
(d) MDA-MB-231 cells were transduced with lentiviral vectors encoding rtTA and doxycycline-inducible MYC (MDA TetON MYC) and transfected with control or YAP/TAZ siRNAs. Where indicated, MYC expression was induced with 0.1 μg/ml doxycycline at the time of transfection. Cell growth was evaluated as in (f). Data are mean+SD of n=8 biological replicates. A control experiment with doxycycline-inducible EGFP is shown in Supplementary Figure 3h.
(e) Average gene expression values of validated YAP/TAZ/TEAD direct target genes (listed in Fig. 2b) in invasive breast cancer samples, classified according to their histological grade. Individual data points and the mean value (black line) of each group are shown.
(f) Kaplan-Meier graph representing the probability of cumulative metastasis-free survival in breast cancer patients stratified according to the expression of validated YAP/TAZ/TEAD direct target gene signature. High expression of the signature is associated with shorter metastasis-free survival (log-rank p-value < 0.0001).
See Methods for reproducibility of experiments.
To investigate the newly identified YAP/TAZ direct targets, we focused on MYC, given its established prominence as regulator of cell proliferation. As shown in Supplementary Figs. 3e-g, MYC knockdown caused a minor, but significant reduction of cell proliferation and a substantial increase of cells in the G1 phase, in part phenocopying the requirement of YAP/TAZ. Overexpression of MYC in YAP/TAZ-depleted cells triggered a significant, but only a partial rescue of cell proliferation (Fig. 3d). This indicates that MYC represents an important functional effector of YAP/TAZ in this context; however, MYC alone cannot fully recapitulate the biological effectiveness of YAP/TAZ.
We next sought to determine whether genes identified as YAP/TAZ/TEAD targets in MDA-MB-231 cells are exploited in human breast cancers. YAP/TAZ are required and sufficient to confer malignant traits to more benign tumor cells16–18. In line, elevated levels of YAP/TAZ in human breast cancer specimens identify aggressive tumors (defined as high histopathological grade, or “G3”), and those with worse prognosis16. Direct targets of YAP/TAZ/TEAD should thus similarly earmark aggressive tumors and be prognostic. To test this idea, we used a dataset of >3600 clinically annotated and transcriptionally profiled breast cancer samples (Supplementary Table 8) and confirmed that the signature enlisting the validated YAP/TAZ/TEAD direct targets (see Fig. 2b) was differentially expressed in G3 and G1 tumors, and identified tumors with poor prognosis as determined by Kaplan-Meier analyses (Figs. 3e-f). Moreover, the signature of direct targets statistically correlates with the expression of independent YAP/TAZ signatures, and with the levels of TAZ mRNA, which is amplified in a subset of breast tumors16, 19 (Supplementary Fig. 3i). Thus the expression of direct YAP/TAZ/TEAD target genes here identified correlates with YAP/TAZ activation in human tumors.
Genomic co-occupancy of AP-1 and YAP/TAZ/TEAD
De novo motif analyses in YAP/TAZ/TEAD peaks revealed that, after TEAD consensus, the second most frequent motif corresponded to the consensus for Activator Protein-1 (AP-1) transcription factors (Supplementary Table 1). AP-1 are dimers of JUN (JUN, JUNB, JUND) and FOS (FOS, FOSB, FOSL1 and FOSL2) families of leucine-zipper proteins20. Many of these factors are archetypal oncogenes involved in the control of cellular growth and neoplastic transformation. The majority of YAP/TAZ/TEAD peaks (70%) contained both a TEAD and an AP-1 motif, with a median distance of about 60 bp.
We next verified that AP-1 factors are indeed recruited to the same genomic regions bound by YAP/TAZ/TEAD4. For this, we considered JUN as a surrogate mark for bound AP-1, because it is a common component of JUN/FOS and JUN/JUN dimers20. By ChIP-seq we identified >24,000 JUN binding sites. JUN was present in 78% of YAP/TAZ/TEAD binding sites (4306/5522; Figs. 4a-b), and 93% of these shared binding sites are located on active enhancers. This is in line with the notion that composite TEAD and AP-1 motifs dominate the YAP/TAZ-cistrome.
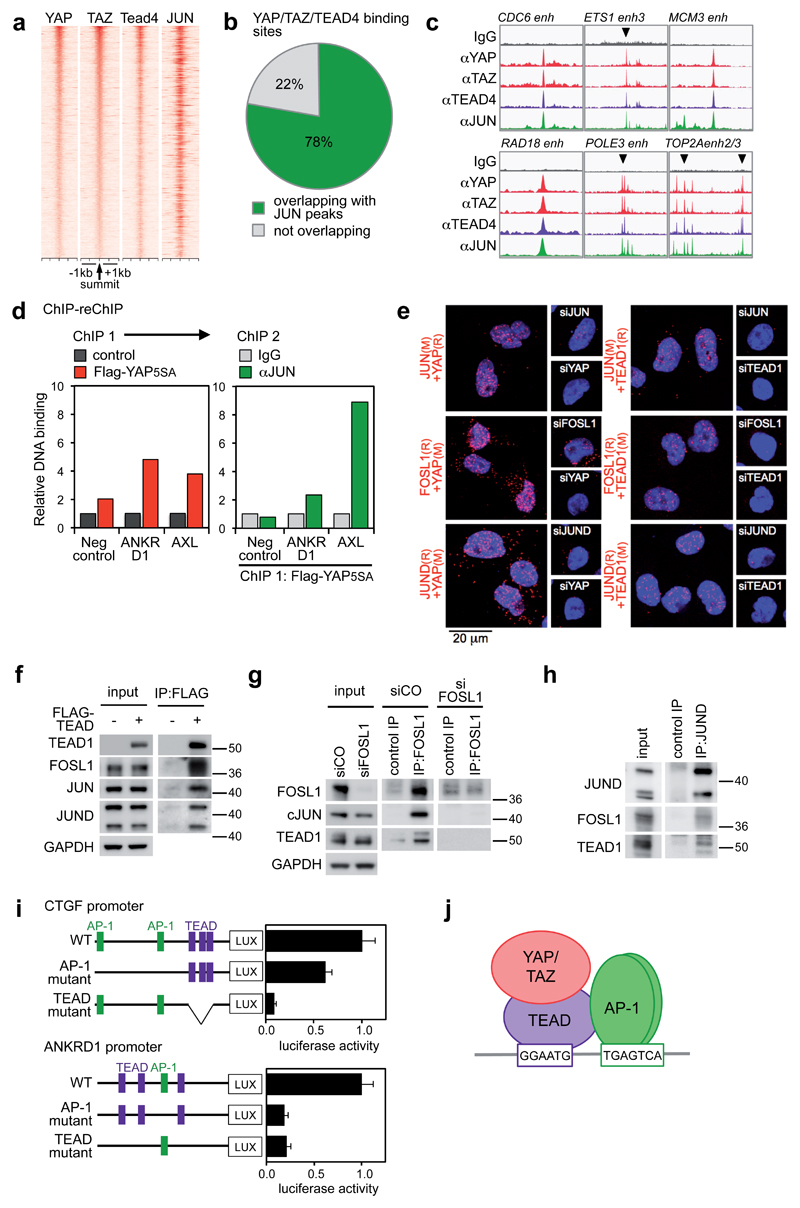
Figure 4. Co-occupancy of YAP/TAZ/TEAD and AP-1 at the same genomic loci.
(a) Density maps of YAP, TAZ, TEAD4 and JUN ChIP-seq reads at YAP/TAZ/TEAD-bound loci. See Supplementary Figures 4a-b for specificity controls of JUN antibody.
(b) Percentage of YAP/TAZ/TEAD4 peaks (n=5522) overlapping with JUN ChIP-seq peaks.
(c) Representative examples of YAP/TAZ/TEAD-bound enhancers co-occupied by JUN in the genome of MDA-MB-231 cells.
(d) Co-presence of YAP and JUN at the same genomic regions. Results are fold enrichment relative to FLAG IP in control (empty vector-transduced) cells (ChIP 1) or relative to IgG negative control (ChIP 2). Data from one representative experiment are shown; experiments were repeated twice with similar results.
(e) In situ PLA detection of endogenous YAP/AP-1 and TEAD1/AP-1 interactions in MDA-MB-231 cells. Nuclei were counterstained with DAPI (blue). The detected dimers are represented by fluorescent dots (red). The specificity of the interactions is revealed by the reduced number of dots detected after depletion of either of the partners with siRNAs. See Supplementary Figure 5a for different combinations of antibodies and Supplementary Table 5 for details about antibodies. Magnification is the same for all pictures.
(f) AP-1 proteins co-precipitate with FLAG-TEAD1 in protein lysates of MDA-MB-231 cells. Input and IP were run on different gels.
(g) TEAD1 co-precipitates with FOSL1 at endogenous protein levels in MDA-MB-231 cells. IP was performed with FOSL1 (N-17) antibodies. All samples were run on the same gel. JUN is a positive control.
(h) TEAD1 co-precipitates with JUND at endogenous protein levels in MDA-MB-231 cells. FOSL1 is a positive control. All samples were run on the same gel.
(i) CTGF and ANKRD1 promoters (either wild-type, or carrying mutations in AP-1 or TEAD binding sites) were cloned upstream of the luciferase coding sequence and their activity was measured in MDA-MB-231 cells. Data are normalized to wild-type promoter sequences. Data are presented as mean+SD of n=4 biological replicates from 2 independent experiments. Binding of TEAD and AP-1 proteins to their respective binding sites was verified by DNA pull down; mutations abolished binding (Supplementary Fig. 5j).
(j) Model of the complex formed by YAP/TAZ/TEAD and AP-1 on DNA.
See Supplementary Figure 7 for uncropped Western blots, and Methods for reproducibility of experiments.
JUN peaks were detected on the regulatory regions of well-established YAP/TAZ/TEAD target genes (CTGF and ANKRD1 - Supplementary Fig. 4c), and on the enhancers of 85% of the new targets defining the YAP/TAZ/TEAD cell-proliferation program (as in Fig. 4c). We indeed re-validated by ChIP-qPCR the presence of both JUN and FOSL1 in all tested binding sites (Supplementary Fig. 4d-e). By considering ChIP-seq data from the ENCODE project21, TEAD4 and AP-1 peaks largely overlap in all examined tumor cell lines (Supplementary Figs. 4f-g), indicating that co-occupancy of TEAD and AP-1 on the same regions is a widespread phenomenon. To test the possibility that AP-1 and YAP/TAZ can simultaneously co-occupy chromatin, we carried out a sequential ChIP for YAP followed by anti-JUN re-ChIP at selected loci. The results indicated that both factors co-occupy the same cis-regulatory elements at the same time (Fig. 4d).
Given their vicinity on DNA, we tested whether YAP/TAZ, TEAD and AP-1 proteins could physically interact. By in situ proximity ligation assay (PLA)22, we found YAP and TEAD1 in close proximity with AP-1 proteins (JUN, FOSL1, JUND) in the nuclei of MDA-MB-231 cells (Fig. 4e). Similar results were obtained in A549 and HCT116 cells (Supplementary Fig. 5b). These interactions were confirmed at the biochemical level: endogenous FOSL1, JUN and JUND robustly co-immunoprecipitated with Flag-tagged TEAD1 (Fig. 4f). Finally, endogenous TEAD1 was co-purified with endogenous FOSL1 and JUND (Figs. 4g-h and Supplementary Fig. 5c).
It has been recently reported that FOS tethers YAP to the promoter of Vimentin, leading to a model in which FOS may directly recruit YAP/TAZ on DNA independently of TEAD23. By co-IP, we have been unable to detect any protein-protein interaction between YAP/TAZ and the main AP-1 factors expressed in MDA-MB-231, that is JUN, JUND and FOSL1 (Supplementary Fig. 5d-f). By ChIP-seq, the signal of YAP/TAZ peaks, while matching with TEAD signal, did not correlate with that one of JUN (Supplementary Fig. 5g). Finally, we monitored the capacity of YAP to activate a luciferase reporter containing polymerized TEAD binding sites (8xGT-lux) or AP-1 binding sites. Despite being artificial, these reporters are highly specific and sensitive, and allow evaluating the contribution of individual TFs in absence of other, and potentially confounding, binding sites (a risk of natural promoters). YAP could activate 8xGT-lux but not the AP-1 sensor (which instead was activated by treatment with the phorbol ester TPA, an established inducer of AP-1; Supplementary Fig. 5h-i). Collectively, our results argue against the possibility that AP-1 mediate YAP/TAZ binding to DNA; that said, it remains possible that, in certain contexts, AP-1 factors may also interact with YAP/TAZ to further enhance the stability of the YAP/TAZ-TEAD and AP-1 complex.
To assess the role of AP-1 in YAP/TAZ/TEAD-mediated transcription, we generated luciferase reporters containing CTGF and ANKRD1 regulatory sequences; both contain motifs for TEAD and AP-1. Mutation of either TEAD or AP-1 motif reduces luciferase activity (Fig. 4i), indicating that both sites are required to mediate YAP/TAZ-dependent transcription. Taken together, the data indicate that, at YAP/TAZ-bound cis-regulatory elements, YAP/TAZ/TEAD and AP-1 form a TF complex bound to composite regulatory elements harboring both TEAD and AP-1 motifs (Fig. 4j), and jointly regulate gene transcription.
Role of AP-1 in YAP/TAZ-regulated cell growth and tumorigenesis
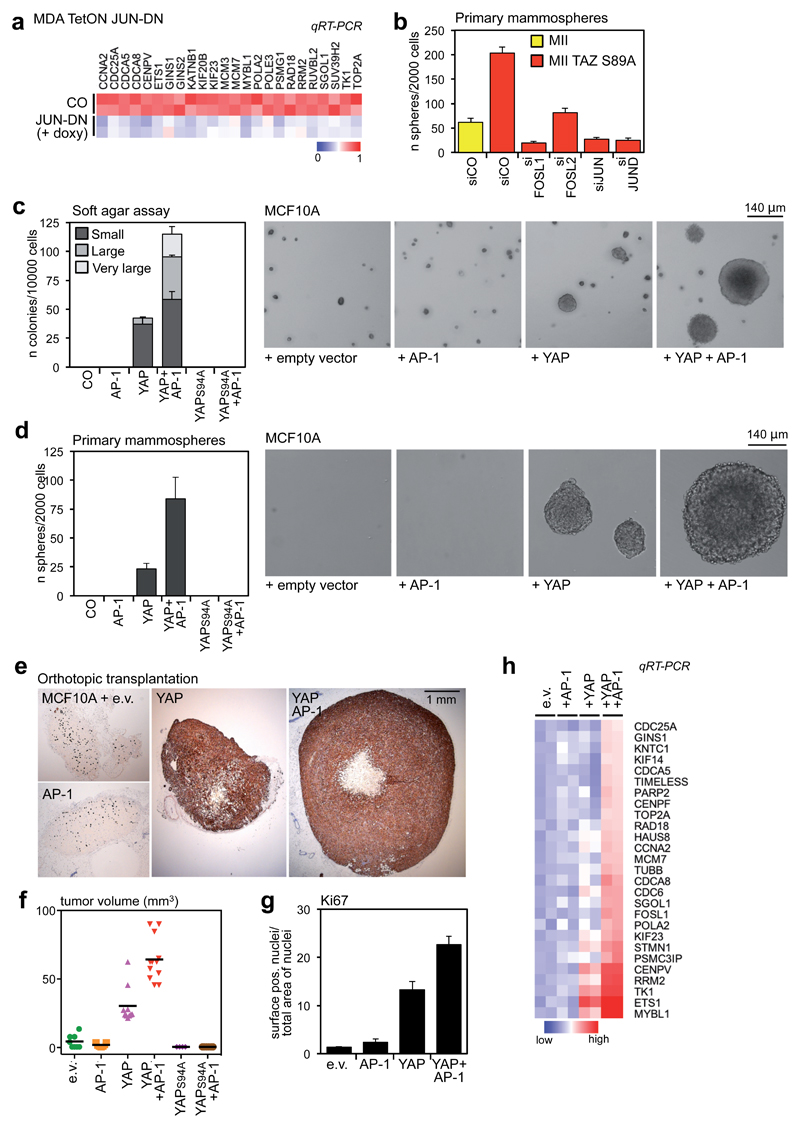
To verify the requirement of AP-1 for endogenous YAP/TAZ/TEAD target genes, we overexpressed a dominant-negative mutant of JUN (JUN-DN), which allows simultaneous repression of all the AP-1 complexes24. Molecularly, expression of JUN-DN reduced transcription of the YAP/TAZ/TEAD cell-proliferation program (Fig. 5a). Functionally, expression of JUN-DN reduced the growth of MDA-MB-231 cells, partially phenocopying the effects of YAP/TAZ depletion or TEAD1/2/3/4 depletion (Supplementary Fig. 6b).
Figure 5. AP-1 factors synergize with YAP/TAZ/TEAD to promote oncogenic growth.
(a) The expression of YAP/TAZ/TEAD target genes involved in cell growth depends on AP-1. MDA-MB-231 cells were transduced with rtTA and doxycycline-inducible JUN-DN. Cells were left untreated (CO) or treated with 1 μg/ml doxycycline for 48h. As control, JUN-DN reduces the expression of FOSL1, CTGF and ANKRD1 (see Supplementary Fig. 6a). All expression levels are normalized to GAPDH.
(b) Control and TAZS89A-overexpressing MII cells were transfected with the indicated siRNAs and tested for mammosphere formation. Data are mean+SD of n=6 biological replicates from a representative experiment. See Supplementary Figure 6c for a comparison with control MII cells.
(c) Quantification and representative pictures of colonies formed by the indicated MCF10A derivatives in soft agar assays. Only background/not growing cell clusters were formed by control and MCF10A+AP-1 cells, and were not counted as colonies. Data are presented as mean+SD of n=3 biological replicates from a representative experiment. Magnification is the same for all pictures.
(d) Quantification and representative pictures of primary mammospheres formed by the indicated MCF10A derivatives. Data are presented as mean+SD of n=6 biological replicates from a representative experiment. Magnification is the same for all pictures.
(e-g) YAP and AP-1 synergize to promote tumor growth. (e) Representative IHC pictures of xenografts formed by the indicated cell lines. MCF10A cells were stained with a human-specific pan-cytokeratin antibody. (f) Tumor volumes at harvesting (individual tumors are plotted, line is the mean). (g) Quantification of Ki67-positive cells in tumor sections; data are mean + SEM of at least n=8 different samples.
(h) YAP5SA and AP-1 cooperate to activate YAP/TAZ/TEAD proliferative program in MCF10A cells. MCF10A derivatives were grown on a thick Matrigel coating for one week before harvesting for RNA extraction. mRNA levels of the indicated genes were evaluated by qRT-PCR and normalized to GAPDH.
See Methods for reproducibility of experiments.
The above findings raise the possibility that the oncogenic properties of YAP/TAZ may require AP-1. It has been previously reported that overexpression of TAZ confers “cancer stem cell” properties to benign MII breast cancer cells16,25. We first investigated whether AP-1 factors are involved in these effects. As shown in Fig. 5b, siRNA-mediated depletion of FOSL1, FOSL2, JUN or JUND severely reduced mammosphere formation by MII-TAZS89A cells, indicating that AP-1 factors are essential for these biological activities of TAZ.
Next, we tested if AP-1 is instrumental to enhance YAP/TAZ activity during transformation and oncogenic growth. For this, we transduced immortalized non-tumorigenic mammary epithelial cells (MCF10A) with the following cDNAs: a JUN~FOSL1 tethered dimer alone26 (+AP-1 in Figs. 5c-h), YAP5SA alone, or the two constructs together. By monitoring anchorage-independent growth in soft-agar and mammosphere assays, we found that concomitant expression of YAP and AP-1 strongly enhanced the number and size of colonies when compared to YAP alone (Figs. 5c-d). No colonies were induced by AP-1 alone. Notably, the synergism between YAP and AP-1 requires YAP binding to TEAD, as no soft-agar colonies or mammospheres formed upon expression of YAP5SA/S94A (which cannot interact with TEAD), irrespectively of the expression of AP-1 (Figs. 5c-d).
To investigate whether the cooperation between YAP and AP-1 is important for induction of tumorigenic properties, we injected the MCF10A variants described above in the fat-pad of immunocompromised mice. As shown in Figures 5e-g, expression of AP-1, while per se insufficient to induce tumors, strongly enhanced the growth of YAP-induced tumors, as indicated by tumor size and Ki67 staining. Mice injected with cells expressing TEAD-binding deficient YAP instead developed no tumors, irrespectively of the expression of AP-1 dimer (Fig. 5f). Importantly, the functional synergism between AP-1 and YAP/TAZ/TEAD on tumor growth is a mirror of their transcriptional cooperation: although AP-1 alone was insufficient to turn on the YAP/TAZ/TEAD proliferative program, it strongly synergized with YAP to activate transcription of these genes (Fig. 5h). Collectively, results of gain- and loss-of-function assays indicate that AP-1 are instrumental for YAP/TAZ transcriptional and biological effects.
Role of YAP/TAZ in AP-1-dependent tumor growth
Having established the relevance of AP-1 for YAP/TAZ-induced cell proliferation and tumor growth, we next asked the converse question, that is, whether YAP/TAZ are important for AP-1-controlled tumorigenesis. To this end we examined skin tumorigenesis induced by chemical carcinogens. In this model, mutation of H-Ras triggered by a mutagen (DMBA) is per se insufficient for tumor development, requiring cooperation with tumor “promoters”, requiring AP-1 activation. The classic tumor promoter is indeed treatment with phorbol-esters (TPA)27,28, well-established triggers of AP-1 activity. In the context of chemical carcinogenesis of the skin, tumor promotion by TPA is blocked by AP-1 inhibition29; conversely, TPA treatment can be substituted by gain-of-AP-1 to fully promote tumor development after DMBA initiation30. In line, skin tumors genetically rely on AP-1 activity11,31,32.
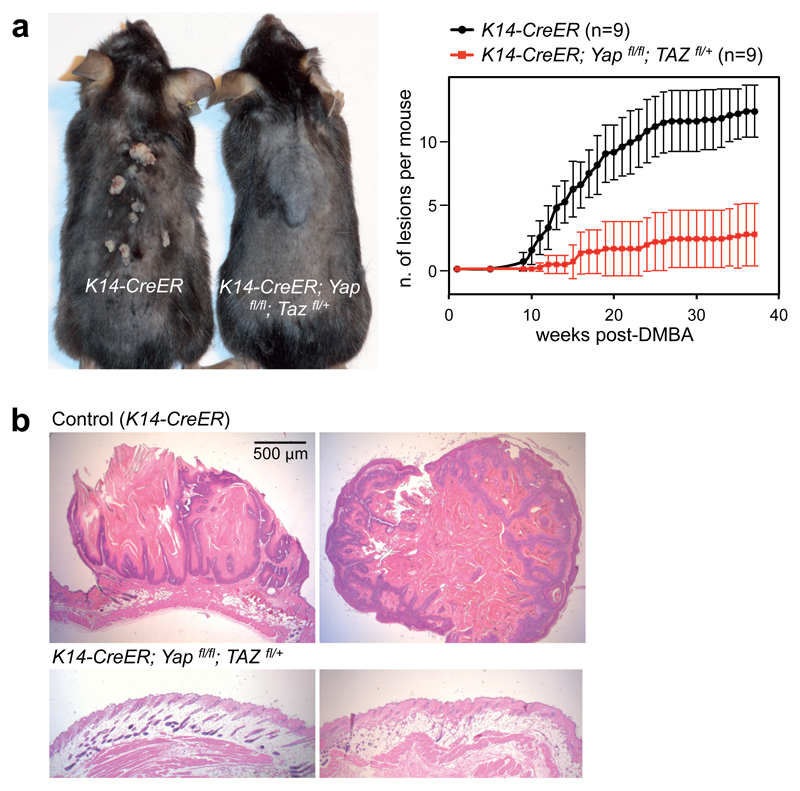
We thus sought to determine whether YAP/TAZ deficiency phenocopies the requirement of AP-1 in tumor promotion. To test this, we treated adult K14-Cre-ERT2;YAPfl/fl;TAZfl/+ mice33 with tamoxifen to inactivate YAP/TAZ in the skin basal layer; these mice were phenotypically normal, and their epidermis was histologically indistinguishable from wild-type controls (Fig. 6b and Supplementary Fig. 6e). Control and YAP/TAZ-deficient mice were subjected to the chemical carcinogenesis protocol. As shown in Figure 6a, at 40 weeks all control mice developed papillomas (average 9.3 tumor/mouse, n=9). Strikingly, YAP/TAZ deficient mice (which still had a normal skin after YAP/TAZ inactivation) only developed rare papillomas (1.5 tumor/mouse, n=9), and 2 mice did not develop any tumor. Histological examination confirmed that control mice mostly developed typical papillomas or, more rarely, foci of squamous cell carcinomas; in contrast, the treated epidermis of K14-Cre-ERT2;YAPfl/fl;TAZfl/+mice retained normal morphology (Fig. 6b and Supplementary Fig. 6e). These results complement the above findings on the role of AP-1 for YAP/TAZ-induced tumor growth in mammary cells, and show that YAP/TAZ play a crucial role in the DMBA/TPA model of skin tumorigenesis, thus phenocopying the requirement of AP-1 in chemical carcinogenesis of the skin.
Figure 6. YAP/TAZ are required for AP-1-driven skin tumorigenesis.
(a) K14-CreER and K14-CreER;Yapfl/fl;Tazfl/+ mice were treated with tamoxifen to activate Cre in the skin basal layer; after two weeks, mice received a single DMBA administration, followed by repeated TPA treatments for 40 weeks. Left: representative picture of control (K14-CreER) and YAP/TAZ deficient mice at time of harvesting (40 weeks after the beginning of the experiment). Right: time course of tumor development in mice with the indicated genotypes (data are mean+SD, n=9 for both experimental groups). See Supplementary Figure 6d for the verification of Cre-mediated recombination of YAP and TAZ alleles.
(b) Representative H&E -stained sections of tumors from control mice, or ostensibly normal skin from DMBA/TPA-treated YAP/TAZ conditional knockout mice. Lesions developed by control mice are characterized by skin folds integrated by a core of connective tissue and lined by an hyperplastic, hyperkeratotic, stratified squamous epithelium; some foci of squamous cell carcinoma were also observed (see Supplementary Fig. 6e). Magnification is the same for all pictures. Scale bar is 1mm.
See Methods for reproducibility of experiments.
Discussion
By carrying out genome-wide analyses of YAP/TAZ binding sites in breast cancer cells through ChIP-seq, we found that the vast majority (91%) of YAP/TAZ-bound cis-regulatory regions coincide with enhancer elements, located distant from TSS. This provides a departure from all previous studies on this topic, so far centered exclusively on promoters9, allowing us to capture new and essential aspects of YAP/TAZ-mediated transcriptional regulation. By using this list of YAP/TAZ genomic interactions, a map of enhancer-promoter pairs and transcriptomic analyses of YAP/TAZ-dependent genes, we unveiled a complex repertoire of direct YAP/TAZ downstream effectors devoted to the control of cell proliferation, a critical process in most YAP/TAZ-dependent biological events. Many YAP/TAZ targets are proteins directly involved in specific steps of the cell cycle. Of these genes, only one (CDC6) was previously proposed as a direct YAP/TAZ target35. The program also includes transcription factors, such as MYC, potentially able to amplify the effects of YAP/TAZ. Overall, our work massively extends the previous knowledge on the transcriptional regulation of cell proliferation by YAP/TAZ and highlights yet unexplored aspects of YAP/TAZ biology, as well as a host of new therapeutic targets. The biological significance of these findings is further emphasized by the clinical validity of a signature of YAP/TAZ target genes as prognostic tool in a large set of breast cancer patients.
The transcriptional responses of YAP/TAZ bear at least superficial similarities with the role of E2F factors: both are required for G1-to-S transition by directly controlling the molecular engines essential for DNA replication. Indeed, by ChIP-PCR experiments mainly using overexpressed YAP and TEAD it has been recently suggested that YAP/TEAD cooperate with E2F in the regulation of some genes from promoters potentially bearing composite TEAD and E2F binding consensuses35. However, at the genome-wide level, our data at endogenous protein level do not favor this model, as the E2F consensus is not enriched at YAP/TAZ/TEAD bound regions (Supplementary Table 1). That said, the E2F motif is present in a number of promoters of genes here identified in the YAP/TAZ-proliferative program (67%, Supplementary Table 5). Collectively, this raises the intriguing hypothesis that YAP/TAZ/TEAD bound to distant enhancers may cooperate with E2F bound to promoters through chromatin looping, thus possibly explaining the requirement of E2F for YAP-mediated cell proliferation35. Future work will be required to dissect this model.
A plethora of DNA-binding platforms have been reported for YAP/TAZ3,8–11. Our ChIP-seq analyses indicate that TEAD factors are the anchors that tether YAP/TAZ to DNA at the genome-wide level; functionally, TEADs are essential for the execution of the entire YAP/TAZ-dependent cell proliferation program. Surprisingly, of the various TF proposed to work as YAP/TAZ DNA-binding platforms, only RUNX1/2 motif displays a low albeit significant enrichment in our context (Supplementary Table 1) but it is not preferentially enriched close to the summit of YAP/TAZ peaks; this suggests that, in general, RUNX factors are unlikely to serve as YAP/TAZ DNA binding platforms.
A key finding of our genome-wide analyses is that AP-1 factors are critical and global regulators of YAP/TAZ/TEAD-dependent gene expression. AP-1 are present in the vast majority of YAP/TAZ/TEAD binding sites, forming a TF complex bound to composite regulatory elements harboring both the TEAD and the AP-1 motifs. AP-1 factors do not mediate YAP/TAZ DNA-recognition, and cannot sustain YAP/TAZ activities in absence of YAP/TAZ binding to TEAD; instead AP-1 strongly support YAP/TAZ/TEAD-dependent gene expression and greatly enhance oncogenic growth triggered by YAP. Conversely, AP-1 inactivation blunts the proliferative and cancer stem cell properties induced by TAZ. It is interesting to note that YAP/TAZ directly activate FOSL1 (Fig. 2b and Supplementary Figs. 2b-c), suggesting a feed-forward/self-enabling loop.
AP-1 factors are classic proto-oncogenes and are important as tumor “promoters”, cooperating with Ras “initiating” mutations in skin chemical carcinogenesis27,29,30. Our findings suggest that part of these attributes could rely on the ability of AP-1 to synergize with YAP/TAZ on chromatin. Supporting this notion, gain-of-AP-1 is per se insufficient to sustain oncogenic growth in absence of YAP overexpression in mammary epithelial cells. In line, YAP/TAZ are genetically required for skin tumorigenesis upon the classical tumor initiation/promotion protocol with DMBA/TPA. That said, further experiments are required to formally demonstrate the transcriptional cooperation between YAP/TAZ/TEAD and AP-1 in skin tumors.
Envisioning AP-1 as general factors in YAP/TAZ-transcriptional regulation adds a number of new modalities to feed information to the YAP/TAZ genetic program. For example, AP-1 are activated by inflammation and stress inputs20, and these may sensitize cells to YAP/TAZ inducing inputs3. Then, there is a widely reported implication of AP-1 in EMT, especially in breast cancer cells; for example, recent reports have linked high FOSL1 expression to EMT and metastasis36,37. Given the established connections between YAP/TAZ and EMT/malignant traits3, it is thus tempting to speculate that AP-1 factors may cooperate with YAP/TAZ for metastatic dissemination or colonization.
It is also notable that AP-1 are dimers of variable composition, and not always behaving as oncogenes20, adding further complexity and potentially cell specificity to the effects of YAP/TAZ. Further studies are required to dissect these regulations in distinct biological contexts in which YAP/TAZ and AP-1 have been so far only independently implicated, including cancer, stem cell biology, regeneration and differentiation.
Methods
Plasmids
pCS2-FLAG-mTAZ, pBABE-hygro-FLAG-mTAZ wt and S89A, pCDNA-FLAG-YAP, FU-tet-o-EGFP-ires-PURO, 8xGTIIC-luc were previously described7,16,33,38. TAZS51A and YAPS94A were generated by PCR-mediated mutagenesis and cloned into pBABE retroviral vectors. pCMV6-FLAG-MYC-TEAD1 was from Origene. FU-tet-o-hc-myc (#19775, Ref 34) and FUdeltaGW-rtTA (#19780, Ref 34) were purchased from Addgene. pBABE-puro-JUN~FOSL1-FLAG was a kind gift of L. Bakiri26. Doxycycline-inducible JUN-DN lentiviral construct was obtained by substituting the Oct4 sequence in FUW-tetO-hOCT4 (Addgene #20726) with JUN-DN cDNA from pMIEG3-JunDN (Addgene #40350). pAP1-luc was from Clontech and pRL-TK from Promega. CTGF and ANKRD1 luciferase reporters were generated by amplifying CTGF (hg19, chr6:132272417-132273191)/ANKRD1(chr10:92680870-92681128) promoters by PCR from genomic DNA and cloning into pGL3 basic; for AP-1 mutants, point mutations were introduced by PCR in AP-1 binding sites; for TEAD mutants, a single deletion comprising the three TEAD binding sites was introduced in CTGF reporter, whereas point mutations were generated in ANKRD1 promoter. All constructs were confirmed by sequencing.
Cell lines and transfections
MDA-MB-231, A549 and HCT116 cells were from ICLC; MCF10A and HEK293 cells were from ATCC. MII cells were a gift of F.R. Miller25. MDA-MB-231 cells were authenticated by DNA profiling of highly polymorphic STR loci. All cell lines were routinely tested for mycoplasma contamination and were negative. None of the cell lines used in this study is present in the database of commonly misidentified cell lines. A549 cells were cultured in DMEM/F12 (Life Technologies) supplemented with 10% FBS, glutamine and antibiotics; culture conditions for all the other cell lines were as previously described16,39.
siRNAs were transfected with Lipofectamine RNAi-MAX (Life Technologies), and DNA transfections were performed with TransitLT1 (Mirus Bio) according to manufacturer instructions. Cells were harvested 48h after transfection, unless differently specified. Sequences of siRNAs are provided in Supplementary Table 9. Retroviral and lentiviral infections were carried out as in Refs. 40–41. MCF10A e.v. cells were infected with pBABE-puro and pBABE-blasti empty vectors; MCF10A+AP-1 were infected with pBABE-puro-JUN~FOSL1-FLAG and pBABE-blasti empty vector; MCF10A+YAP (or 5SA/S94A) were infected with pBABE-puro empty vector and pBABE-blasti-FLAG-YAP5SA (or 5SA/S94A); MCF10A+YAP+AP-1 were infected with pBABE-puro-JUN~FOSL1-FLAG and pBABE-blasti-FLAG-YAP5SA (or 5SA/S94A).
ChIP-seq, ChIP-qPCR and ChIP-reChIP
Chromatin immunoprecipitation was performed as described in Ref. 42. Antibodies are listed in Supplemetary Table 6. Briefly, cells were cross-linked with 1% formaldehyde (Sigma) in culture medium for 10min at room temperature, and chromatin from lysed nuclei was sheared to 200-600bp fragments using a Branson Sonifier 4500D. For ChIP-seq, ~200 μg of chromatin were incubated with 10 μg of antibody overnight at 4°C. Antibody/antigen complexes were recovered with ProteinA-Dynabeads (Invitrogen) for 2h at 4°C. ChIP'd DNA form 3 immunoprecipitations was pooled to generate libraries with the Ovation Ultra Low Library Prep Kit (NuGEN) according to manufacturer's instructions. Sequencing was performed on an Illumina HiSeq 2500 platform.
For ChIP-qPCR, ~100 μg of sheared chromatin and 3-5μg of antibody were used. For ChIPs of modified histones, at least 50 μg of chromatin were incubated with 2 μg of antibody. Quantitative real-time PCR was carried out with a Rotor-Gene Q (Qiagen) thermal cycler; each sample was analyzed in triplicate. The amount of immunoprecipitated DNA in each sample was determined as fraction of input [amplification efficiency^(Ct INPUT-Ct ChIP)], and normalized to IgG control. Primers are listed in Supplementary Table 9.
For ChIP-reChIP, MDA-MB-231 cells stably expressing FLAG-YAP5SA were used. Chromatin was incubated with anti-FLAG magnetic beads (Sigma) for 4h at 4°C; after washing, immunocomplexes were eluted by incubating beads in lysis buffer 3 + 0.5 μg/μl 3x FLAG peptide (Sigma) for 1h at 4°C; two sequential elutions were performed. Eluted chromatin was diluted 1:3 with lysis buffer 3, supplemented with 1% Triton X-100 and incubated with 3 μg of normal mouse IgG or anti-JUN antibody, overnight at 4°C.
Peak Calling and Data Analysis
Raw reads were aligned using Bowtie (version 0.12.7)43 to build version hg19 of the human genome retaining only uniquely mapped reads. Redundant reads were removed using SAMtools. The IDR (Irreproducible Discovery Rate) framework44 was used to assess the consistency of replicate experiments and to obtain a high confidence single set of peak calls for each Transcription Factor as described in the ChIP-seq guidelines of the ENCODE consortium45. MACS2 version 2.0.1046 was used to call peaks in individual replicates using IgG ChIP-seq as control sample and an IDR threshold of 0.01 was applied for all datasets to identify an optimal number of peaks.
Normalized read density (reads per million, rpm) was calculated from pooled replicates using MACS2 callpeak function and displayed using Integrative Genomics Viewer (IGV).
Heatmaps were generated using a custom R script which considers a 2-kb window centered on peak summits and calculates the normalized reads density with a resolution of 50 bp.
The genomic location of the peaks and their distance to TSS of annotated genes were calculated using annotatePeakInBatch function of ChIPpeakanno R package and GENCODE annotation version 1647. Only genes classified as protein coding and with status equal to KNOWN were considered.
The findOverlappingPeaks function of the same package was used with default parameters to identify overlapping peaks and calculate the distance between their summits. TAZ peaks coordinates and summit positions were used to represent common peaks between YAP and TAZ peaks (YAP/TAZ peaks) and were used when comparing YAP/TAZ peaks with other ChIP-seq data.
Definition of MDA-MB-231 promoters and enhancers
Raw reads for ChIP-seq data of histone modifications (H3K4me1, H3K4me3 and H3K27ac) in MDA-MB-23113 were downloaded from SRA (SRP028597) and aligned using Bowtie version 0.12.9 to build version hg19 of the human genome retaining only uniquely mapped reads. Redundant reads were removed using SAMtools. Peak calls and read density tracks were generated using SPP version 1.1148 with default parameters and using as control sample the IgG ChIP-seq data generated in our laboratory because of the low sequencing depth of the Input DNA contained in SRP028597.
The distance between histone modifications peaks and the transcription start sites (TSS) of protein coding genes (GENCODE v. 16 and REFSEQ annotations), and the overlap between histone marks peaks were calculated as previously described for TF peaks.
The presence of H3K4me1 and H3K4me3 peaks, their genomic locations and their overlap were the criteria used to define promoters and enhancers: i) H3K4me3 peaks not overlapping with H3K4me1 peaks and close to a TSS (± 5kb) were defined as promoters, as NA otherwise; ii) H3K4me1 peaks not overlapping with H3K4me3 peaks were defined as enhancers; iii) regions with the co-presence of H3K4me1 and H3K4me3 peaks were visually inspected on IGV and were defined as promoters, enhancers or NA after the evaluation of the proximity to a TSS and the comparison of the enrichment signals. Finally, promoters or enhancers were defined as active if overlapping with H3K27ac peaks.
YAP/TAZ/TEAD peaks annotation
YAP/TAZ/TEAD peaks were annotated as promoters or enhancers if their summit was overlapping with promoter or enhancer regions as defined above. Peaks with the summit falling in regions with no H3K4me1 or H3K4me3 peaks, or in NA regions were defined as "not assigned" and discarded from subsequent analyses.
YAP/TAZ/TEAD peaks summits were compared with FAIRE peaks using the list downloaded from GSE49651.
YAP/TAZ/TEAD peaks falling on promoters were assigned to the closest TSS. YAP/TAZ/TEAD peaks falling on active enhancers were annotated using the chromatin interactions reported in Supplementary data 2 of Ref. 14, derived from a high resolution Hi-C experiment; the data sheets report the genomic locations of all target peaks interacting with more than 10 thousands anchors located at gene promoters. YAP/TAZ peaks overlapping with these target peaks were assigned to the corresponding interacting promoter region. Finally, YAP/TAZ/TEAD peaks falling on inactive enhancers were not assigned to targets.
Motif discovery in ChIP-seq peaks
De novo motifs discovery was performed with findMotifsGenome function of Homer software49. Motifs were searched in 500 bp windows centered at the peak summits. Occurrences of de novo and known motifs inside the peaks were found using annotatePeaks function of the same software. Known motifs were retrieved from Homer motif database and from JASPAR database (http://jaspar.genereg.net).
Gene expression analysis by Affymetrix microarrays
MDA-MB-231 cells were transfected with control or two independent combinations of YAP/TAZ siRNAs for 48h; four biological replicates for each sample were prepared. Transcriptomic data were obtained using Affymetrix GeneChips Human Genome U133 Plus 2.0. Raw data were as in Ref. 50, but analyzed as detailed below.
Microarray analyses were performed in R (version 2.15.1). Probe level signals were converted to expression values using robust multi-array average procedure RMA51 of Bioconductor affy package and a custom chip definition file based on the Entrez gene database (version 17, Ref. 52). Differentially expressed genes were identified using Significance Analysis of Microarray algorithm coded in the samr R package53. In SAM, we estimated the percentage of false positive predictions (i.e., False Discovery Rate, FDR) with 100 permutations. To identify genes regulated by YAP/TAZ, we selected those genes coherently downregulated with a FDR ≤ 0.1% in both silencing experiments, and present in GENCODE v.16 annotation. This selection resulted in 1534 downregulated genes. These genes were compared with the list of candidate YAP/TAZ direct target genes described above. As a result, 379 down-regulated genes and associated with a YAP/TAZ peak were defined as direct positive YAP/TAZ targets.
Gene Ontology analysis
Gene Ontology (GO) analyses were performed using DAVID54. The full list of GO terms of the Biological Process category enriched in direct positive YAP/TAZ targets is presented in Supplementary Table 4. GO terms with a Benjamini-Hochberg FDR ≤ 5% were considered significantly enriched. GO terms significantly enriched among YAP/TAZ/TEAD direct positive target genes could be assigned to two broad categories: “cell proliferation” and “RNA metabolism and transport” (Supplementary Table 4).
Promoters of the genes involved in cell proliferation were defined as 1000 bp windows centered at the TSS. De novo motifs discovery and occurrence of known motifs were performed as described above. Used known motifs are E2F4(E2F) from Homer motif database and (MA0024.1) for E2F1 from JASPAR database.
Analysis of public ChIP-seq data
ChIP-seq datasets for transcription factors and histone modifications that were re-analyzed in this study are listed in Supplementary Table 7.
For data of the ENCODE project21, aligned reads and peak calls were downloaded from the ENCODE project repository. When available, TF peaks uniformly generated by the ENCODE Analysis Working Group (available at http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeAwgTfbsUniform/) were used. Otherwise, aligned reads and peak calls of the first replicate were used. Genomic annotation of TEAD4 or TEAD1 peaks and overlap between peaks were calculated as described for ChIP-seq data of MDA-MB-231 cells.
Gene expression analysis
MDA-MB-231 cells were transfected with siRNAs (or treated with doxycycline) 48h before harvesting. RNA extraction was performed with RNeasy Mini Kit (QIAGEN). MCF10A cells were cultured as spheres on a thick coating of gelled Matrigel for 8 days (described in Ref. 55); cells were recovered with BD Cell Recovery Solution (BD Biosciences), and harvested in Trizol (Invitrogen) for total RNA extraction.
Reverse transcription and qPCR were performed as described in Ref. 7. Expression levels are normalized to GAPDH. Primers are listed in Supplementary Table 9. For experiments in Figures 2b, 5a and 5h cDNA was synthesized with High Capacity RNA-to-cDNA Kit (Invitrogen) and target genes were quantified with custom TaqMan Low Density Arrays on a 7900HT Fast Real-Time PCR System (Applied Biosystems), using TaqMan Universal PCR Master Mix (Applied Biosystems). TaqMan assays included in the array are listed in Supplementary Table 9. Expression levels are normalized to GAPDH.
Chromatin conformation capture (3C)
3C was performed as described in Ref. 56 with some modifications. Adherent cells were incubated with a solution containing 1.5% formaldehyde for 10min at RT, followed by 5min treatment with 0.125M Glycine/PBS. Cells were incubated with 0.05% trypsin for 10 min at 37°C, before completely detaching them with a cell scraper. Harvested cells were pelletted, washed in PBS and incubated with Lysis buffer for 15 min at 4°C. Nuclei were digested with HindIII restriction enzyme (NEB), and highly diluted digested chromatin was ligated with T4 DNA ligase (NEB). De-crosslinked DNA fragments were purified by phenol-chloroform extraction and ethanol precipitation. A reference template was generated by digesting, mixing and ligating Bacterial Artificial Chromosomes spanning the genomic regions of interest. 3C templates and the reference template were used to perform semi-quantitative PCR with GO Taq G2 Flexi DNA Polymerase (Promega). Primers flanked the HindIII restriction sites located close to MYC and TOP2A promoters (anchors) and enhancers (primer sequences are provided in Supplementary Table 9). Data are presented as the ratio of amplification obtained with 3C templates from MDA-MB-231 cells and with the reference template. PCR performed on control 3C template obtained from not-crosslinked cells never yielded any product. PCR products were verified by sequencing.
Collection and processing of breast cancer gene expression data
We started from a collection of 4640 samples from 27 major datasets comprising microarray data of breast cancer samples annotated with histological tumor grade and clinical outcome (Supplementary Table 8). The collection was normalized and annotated with clinical information as described in Ref. 50. This resulted in a compendium (meta-dataset) comprising 3661 unique samples from 25 independent cohorts (Supplementary Table 8).
Survival analysis
To identify two groups of tumors with either high or low YAP/TAZ/TEAD direct target genes signature we used the classifier described in Ref 55, i.e. a classification rule based on the YAP/TAZ/TEAD direct target genes signature score. Tumors were classified as "YAP/TAZ/TEAD direct target genes signature" Low if the combined score was negative and as "YAP/TAZ/TEAD direct target genes signature" High if the combined score was positive. To evaluate the prognostic value of the signature, we estimated, using the Kaplan-Meier method, the probabilities that patients would remain free of metastasis. The Kaplan-Meier curves were compared using the log-rank (Mantel-Cox) test. P-values were calculated according to the standard normal asymptotic distribution. Survival analysis was performed in GraphPad Prism.
Average signature expression and correlation
Average signature expression has been calculated as the standardized average expression of all signature genes in sample subgroups. To test the association between YAP/TAZ/TEAD direct target genes signature, TAZ (WWTR1) expression level, and YAP/TAZ signatures38,57, we calculated the Pearson's product moment correlation coefficient using the cor.test function of the stat R package.
Co-imunoprecipitation assays and DNA pull down
For immuniprecipitation of FLAG-tagged proteins, MDA-MB-231 cells were transfected with 80 ng/cm2 of FLAG-TEAD1, FLAG-YAP or FLAG-TAZ plasmids. DNA amount was adjusted to 160 ng/cm2 with pBluescript. For immunoprecipitation of endogenous proteins, MDA-MB-231 and HCT116 cells were transfected with siRNAs as indicated. Cells were harvested 48h after transfection. Cell lysis and immunoprecipitation were performed as in Ref. 33. DNA pull down was performed as in Ref. 16.
Western blot
Whole cell lysates were obtained by sonication in lysis buffer (20 mM HEPES [pH 7.8], 100 mM NaCl, 5% glycerol, 5 mM EDTA, 0.5% Np40, and protease and phosphatase inhibitors). The Western blot procedure was carried out as described in Ref. 39. Primary antibodies are listed in Supplementary Table 6.
In situ proximity ligation assay
In situ PLA was performed with DuoLink In Situ Reagents from Olink Bioscience (Sigma). MDA-MB-231 cells were transfected with siRNAs in standard cell culture dishes, seeded in fibronectin-coated glass chamber slides 24h later, and fixed in 4% PFA for 10 min at RT 48h after transfection. Proximity ligation assays were performed as indicated by the provider's protocol, after an overnight incubation with primary antibodies following the immunofluorescence protocol described in Ref. 38. Images were acquired with Leica SP5 confocal microscope equipped with a CDD camera; for each field, a Z-stack was acquired; images were processed using Volocity software (PerkinElmer). Primary antibodies are listed in Supplementary Table 6.
Luciferase reporter assays
TEAD reporter (8xGTIIC-Lux38) and AP-1 reporter (pAP1-luc, Clontech) (25 ng/cm2) were transfected in HEK293 cells, together with increasing doses of pCDNA-FLAG-YAPwt (1.25, 2.5, 6.25 and 12.5 ng/cm2) and TK-Renilla (25 ng/cm2) to normalize for transfection efficiency. CTGF and ANKRD1 luciferase constructs were co-transfected with pRL-TK in MDA-MB-231 cells (75 ng/cm2). DNA content in all samples was kept uniform by adding pBluescript plasmid up to 250 ng/cm2. Where indicated, 2nM TPA was added 24h after DNA transfection, and cells were harvested 48h after DNA transfection. Firefly and Renilla luciferase activity was measured with an Infinite F200PRO plate reader (TECAN). Data are presented as firefly/Renilla luciferase activity.
Growth assays and cell cycle analysis
For growth assays, cells were seeded in 96-well plates (4000 cells/well, 8 wells/sample) and fixed at the indicated time points with a crystal violet solution (0.05% w/v Crystal violet, 1% formaldehyde, 1% methanol in PBS) for 20 min at RT; stained cells were washed and air-dried. Crystal violet was extracted with 1% SDS (w/v in ddH2O, 100 μl/well) and absorbance at λ=595 nm was measured.
To determine the fraction of cells in each phase of the cell cycle, subconfluent cells were trypsinized 48 hours after siRNA transfection, fixed in cold 70% ethanol and stained with 0.02 mg/ml Propidium Iodide + 0.2 mg/ml RNase A in PBS. Stained cells were analyzed on a FC500 cytofluorimeter (Beckman Coulter).
Soft agar and mammosphere assays
For soft agar assay, 104 MCF10A cells in complete growth medium with 0.35% agar were layered onto 0.5% agar beds in six-well plates; complete medium was added on top of cells and was replaced with fresh medium twice a week for 15 days. Colonies larger than 100 μm in diameter were counted as positive for growth. Thresholds were arbitrary set to classify colonies according to their size.
For mammosphere formation assay, 1000 cells/cm2 were seeded on ultra-low attachment plates (Costar), in DMEM/F12 supplemented with 10 ng/ml EGF, 5 μg/ml insulin, 0.5 μg/ml hydrocortisone, 52 μg/ml bovine pituitary extract and B27 supplement. Mammospheres were counted after one week.
Mice
Animal experiments were performed adhering to our institutional guidelines (CEASA, Comitato Etico per la Sperimentazione Animale, that is our Animal Welfare Body).
For orthotopic transplantation experiments, MCF10A cells were injected in the abdominal mammary glands of 6-10 week-old RAG-/- female mice. For each injection, 106 cells were mixed 1:1 with Matrigel, in a final volume of 100 μl. Tumor growth in the injected site was monitored by repeated caliper measurements. Mice were sacrificed after one month and tumors were explanted, fixed in 4% PFA and embedded in paraffin for histological analyses. The number of injections were: control (e.v.) n=8, +AP-1 n=10, +YAP n=10, +YAP+AP-1 n=12, +YAPS94A n=6, +YAPS94A+AP-1 n=6.
Yap1fl/fl mice were gently provided by Duojia Pan58. K14-CreERT2 mice were kindly provided by Pierre Chambon34. Tazfl/fl mice were described in Ref. 33. Animals were genotyped with standard procedures59, and with the recommended set of primers. To selectively ablate YAP/TAZ in epidermal keratinocytes of adult mice, mice carrying LoxP-sites-containing Yap and Taz alleles were bred with hemizygous K14-CreERT2tg/0 transgenic mice, to produce K14-CreERT2tg/0; Yapfl/fl; Tazfl/+. 6-8 week-old mice (4 females and 5 males in each experimental group) received 3 intraperitoneal injection of tamoxifen (1 mg in 100 μl corn oil) to produce control (from K14-CreERT2tg/0) or conditional Yap/Taz KO mice (from K14-CreERT2tg/0; Yapfl/fl; Tazfl/+). 2 weeks after tamoxifen injection, mice were shaved to synchronize the hair cycle and treated the day after with a single dose of DMBA (Sigma, 100 μg in 100 μl acetone). One week after DMBA application, TPA (Sigma, 5 μg in 100 μl acetone) was applied topically twice a week for up to 40 weeks. The number of tumors per mouse was recorded weekly. Tumors or skin explants were excised at the end of the DMBA/TPA treatment, fixed in 4% PFA and embedded in paraffin for histological sections. DNA was extracted from skin and genotyped with the recommended set of primers for Yap null allele (as in Ref. 58), Taz null allele (as in Ref. 33) and K14-CreERT2 (for: TGGGAAAGTGTAGCCTGCAG; rev: TCCCCTTGGCTTTCATCACC; 182 bp).
All tested animals were included; no statistical method was used to predetermine sample size; no randomization or blinding was used.
Immunohistochemistry
Immunohistochemical staining was performed as described in Ref. 60. Primary antibodies were anti-human Ki67 (Dako, M7240) and anti-human cytokeratin (Dako, M0821).
Reproducibility of experiments
ChIP seq experiments contained two biological replicates, obtained from two independent experiments. For ChIP-qPCR, at least two independent experiments (each containing 2 biological replicates) were performed with similar results. For gene-expression analysis with Affymetrix Microarrays four biological replicates were analyzed for each sample. For qRT-PCR, 3 independent experiments (each with 2 biological replicates) were performed, with similar results. 3C experiments contained n=3 biological replicates and were repeated twice with similar results. Co-immunoprecipitation assays, DNA-pull down, Western blots and PLA assays were performed three times with similar results. For luciferase assays, each experiment contained 2 biological replicates and was repeated at least three times independently with similar results. For growth assays, 8 independent replicate wells were analyzed for each sample; each experiment was performed at least twice, with similar results. For cell cycle analysis, experiments contained n=3 biological replicates for each condition, and were repeated at least twice with similar results. Soft agar assays contained n=3 biological replicates for each condition and were performed three times, with similar results. Mammosphere assays contained n=6 biological replicates for each condition and were performed three times, with similar results.
Luciferase data are presented as mean + SD of n=4 biological replicates from 2 experiments.
For all the other experiments, results from one representative experiment are shown.
Accession numbers
Primary accessions: all data from this study have been deposited in the GEO database under accession number GSE66083.
Referenced accessions: accession numbers for public ChIP-seq and gene expression datasets used in this study are reported in Supplementary Table 7 and 8, respectively.
Supplementary Material
Acknowledgments
We thank O. Wessely, S. Dupont and G. Martello for comments; M. Morgante and E. Aleo for deep-sequencing (IGA, Udine); V. Guzzardo for histology; D.J. Pan for gifts of Yap fl/fl mice; D. Metzger and P. Chambon for K14-Cre-ERT2 transgenics; L. Naldini for lentiviral plasmids and L. Bakiri for AP-1 expression constructs. We are grateful to C. Frasson for flow cytometry analyses. We thank a donation in memoriam of Liana Simonutti. FZ and LA are supported by a fellowship from Italian Association for Cancer Research (AIRC). MF is supported by an assistant professorship from FIRB Accordi di Programma 2011 RBAP11T3WB. This work is supported by AIRC-MFAG to M.C; from AIRC Special Program Molecular Clinical Oncology “5 per mille” to S.P. and S.B.; Epigenetics Flagship project CNR-MIUR grants to S.B. and S.P; AIRC-IG Grant to S.P. Work in SP lab is also supported by ERC-2014-ADG.
Footnotes
Author contributions. FZ and GB performed experiments, MF, MC and SB performed bioinformatic analysis, EQ helped with molecular biology, LA and AR performed in vivo experiments, BB helped with ChIP and 3C protocols, MC and SP planned, discussed and organized the work and wrote the MS.
Author information. Gene expression omnibus accession number is GSE66081 for ChIP-seq data and GSE66082 for gene expression data. (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=idktgqkknnyfpsz&acc=GSE66083). See also "Accession numbers" section in Methods.
The authors declare no competing financial interest.
References
- 1.Sudol M. Yes-associated protein (YAP65) is a proline-rich phosphoprotein that binds to the SH3 domain of the Yes proto-oncogene product. Oncogene. 1994;9:2145–2152. [PubMed] [Google Scholar]
- 2.Kanai F, et al. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. The EMBO journal. 2000;19:6778–6791. doi: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Piccolo S, Dupont S, Cordenonsi M. The biology of YAP/TAZ: hippo signaling and beyond. Physiological reviews. 2014;94:1287–1312. doi: 10.1152/physrev.00005.2014. [DOI] [PubMed] [Google Scholar]
- 4.Ramos A, Camargo FD. The Hippo signaling pathway and stem cell biology. Trends in cell biology. 2012 doi: 10.1016/j.tcb.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harvey KF, Zhang X, Thomas DM. The Hippo pathway and human cancer. Nat Rev Cancer. 2013;13:246–257. doi: 10.1038/nrc3458. [DOI] [PubMed] [Google Scholar]
- 6.Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aragona M, et al. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 8.Strano S, et al. The transcriptional coactivator Yes-associated protein drives p73 gene-target specificity in response to DNA Damage. Mol Cell. 2005;18:447–459. doi: 10.1016/j.molcel.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 9.Zhao B, et al. TEAD mediates YAP-dependent gene induction and growth control. Genes & development. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rosenbluh J, et al. beta-Catenin-driven cancers require a YAP1 transcriptional complex for survival and tumorigenesis. Cell. 2012;151:1457–1473. doi: 10.1016/j.cell.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yagi R, Chen LF, Shigesada K, Murakami Y, Ito Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. The EMBO journal. 1999;18:2551–2562. doi: 10.1093/emboj/18.9.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calo E, Wysocka J. Modification of enhancer chromatin: what, how, and why? Mol Cell. 2013;49:825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhie S, et al. Nucleosome positioning and histone modifications define relationships between regulatory elements and nearby gene expression in breast epithelial cells. BMC Genomics. 2014;15:331. doi: 10.1186/1471-2164-15-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin F, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503:290–294. doi: 10.1038/nature12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rao SS, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cordenonsi M, et al. The Hippo transducer TAZ confers cancer stem cell-related traits on breast cancer cells. Cell. 2011;147:759–772. doi: 10.1016/j.cell.2011.09.048. [DOI] [PubMed] [Google Scholar]
- 17.Bhat KP, et al. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes & development. 2011;25:2594–2609. doi: 10.1101/gad.176800.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamar JM, et al. The Hippo pathway target, YAP, promotes metastasis through its TEAD-interaction domain. Proc Natl Acad Sci U S A. 2012;109:E2441–2450. doi: 10.1073/pnas.1212021109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skibinski A, et al. The Hippo transducer TAZ interacts with the SWI/SNF complex to regulate breast epithelial lineage commitment. Cell reports. 2014;6:1059–1072. doi: 10.1016/j.celrep.2014.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 21.Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koos B, et al. Analysis of protein interactions in situ by proximity ligation assays. Current topics in microbiology and immunology. 2014;377:111–126. doi: 10.1007/82_2013_334. [DOI] [PubMed] [Google Scholar]
- 23.Shao DD, et al. KRAS and YAP1 converge to regulate EMT and tumor survival. Cell. 2014;158:171–184. doi: 10.1016/j.cell.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown PH, Alani R, Preis LH, Szabo E, Birrer MJ. Suppression of oncogene-induced transformation by a deletion mutant of c-jun. Oncogene. 1993;8:877–886. [PubMed] [Google Scholar]
- 25.Santner SJ, et al. Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Research and Treatment. 2001;65:101–110. doi: 10.1023/a:1006461422273. [DOI] [PubMed] [Google Scholar]
- 26.Bakiri L, Matsuo K, Wisniewska M, Wagner EF, Yaniv M. Promoter specificity and biological activity of tethered AP-1 dimers. Mol Cell Biol. 2002;22:4952–4964. doi: 10.1128/MCB.22.13.4952-4964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balmain A, Yuspa SH. Milestones in skin carcinogenesis: the biology of multistage carcinogenesis. The Journal of investigative dermatology. 2014;134:E2–7. doi: 10.1038/skinbio.2014.2. [DOI] [PubMed] [Google Scholar]
- 28.Bailleul B, et al. Skin hyperkeratosis and papilloma formation in transgenic mice expressing a ras oncogene from a suprabasal keratin promoter. Cell. 1990;62:697–708. doi: 10.1016/0092-8674(90)90115-u. [DOI] [PubMed] [Google Scholar]
- 29.Young MR, et al. Transgenic mice demonstrate AP-1 (activator protein-1) transactivation is required for tumor promotion. Proc Natl Acad Sci U S A. 1999;96:9827–9832. doi: 10.1073/pnas.96.17.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Briso EM, et al. Inflammation-mediated skin tumorigenesis induced by epidermal c-Fos. Genes & development. 2013;27:1959–1973. doi: 10.1101/gad.223339.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zenz R, et al. c-Jun regulates eyelid closure and skin tumor development through EGFR signaling. Dev Cell. 2003;4:879–889. doi: 10.1016/s1534-5807(03)00161-8. [DOI] [PubMed] [Google Scholar]
- 32.Chen Y, Lai MZ. c-Jun NH2-terminal kinase activation leads to a FADD-dependent but Fas ligand-independent cell death in Jurkat T cells. J Biol Chem. 2001;276:8350–8357. doi: 10.1074/jbc.M008431200. [DOI] [PubMed] [Google Scholar]
- 33.Azzolin L, et al. YAP/TAZ incorporation in the beta-catenin destruction complex orchestrates the Wnt response. Cell. 2014;158:157–170. doi: 10.1016/j.cell.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 34.Li M, et al. Skin abnormalities generated by temporally controlled RXRalpha mutations in mouse epidermis. Nature. 2000;407:633–636. doi: 10.1038/35036595. [DOI] [PubMed] [Google Scholar]
- 35.Kapoor A, et al. Yap1 activation enables bypass of oncogenic Kras addiction in pancreatic cancer. Cell. 2014;158:185–197. doi: 10.1016/j.cell.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tam WL, et al. Protein kinase C alpha is a central signaling node and therapeutic target for breast cancer stem cells. Cancer Cell. 2013;24:347–364. doi: 10.1016/j.ccr.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bakiri L, et al. Fra-1/AP-1 induces EMT in mammary epithelial cells by modulating Zeb1/2 and TGFbeta expression. Cell death and differentiation. 2015;22:336–350. doi: 10.1038/cdd.2014.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dupont S, et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 39.Dupont S, et al. FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell. 2009;136:123–135. doi: 10.1016/j.cell.2008.10.051. [DOI] [PubMed] [Google Scholar]
- 40.Martello G, et al. A MicroRNA Targeting Dicer for Metastasis Control. Cell. 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 41.Azzolin L, et al. Role of TAZ as mediator of Wnt signaling. Cell. 2012;151:1443–1456. doi: 10.1016/j.cell.2012.11.027. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt D, et al. ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods. 2009;48:240–248. doi: 10.1016/j.ymeth.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Q, Brown JB, Huang H, Bickel PJ. Measuring reproducibility of high-throughput experiments. The Annals of Applied Statistics. 2011;5:1752–1779. [Google Scholar]
- 45.Landt SG, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Research. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Research. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nature Biotechnology. 2008;26:1351–1359. doi: 10.1038/nbt.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Enzo E, et al. Aerobic glycolysis tunes YAP/TAZ transcriptional activity. The EMBO journal. 2015;34:1349–1370. doi: 10.15252/embj.201490379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics (Oxford, England) 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 52.Dai M, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic acids research. 2005;33:e175–e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang F, He J, Zhang Y, Guo Y. Synthesis of biotin-AMP conjugate for 5' biotin labeling of RNA through one-step in vitro transcription. Nat Protoc. 2008;3:1848–1861. doi: 10.1038/nprot.2008.185. [DOI] [PubMed] [Google Scholar]
- 55.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 56.Bodega B, et al. Remodeling of the chromatin structure of the facioscapulohumeral muscular dystrophy (FSHD) locus and upregulation of FSHD-related gene 1 (FRG1) expression during human myogenic differentiation. BMC Biol. 2009;7:41. doi: 10.1186/1741-7007-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang H, et al. TEAD transcription factors mediate the function of TAZ in cell growth and epithelial-mesenchymal transition. J Biol Chem. 2009;284:13355–13362. doi: 10.1074/jbc.M900843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang N, et al. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev Cell. 2010;19:27–38. doi: 10.1016/j.devcel.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Morsut L, et al. Negative control of Smad activity by ectodermin/Tif1gamma patterns the mammalian embryo. Development. 2010;137:2571–2578. doi: 10.1242/dev.053801. [DOI] [PubMed] [Google Scholar]
- 60.Adorno M, et al. A Mutant-p53/Smad Complex Opposes p63 to Empower TGFβ-Induced Metastasis. Cell. 2009;137:87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.