Abstract
Peptidic-nanodiscs are useful membrane-mimetic tools for structural and functional studies of membrane proteins, and membrane interacting peptides including amyloids. Here, we demonstrate anti-amyloidogenic activities of a nanodisc-forming 18-residue peptide (denoted as 4F), both in lipid-bound and lipid-free states by using Alzheimer’s amyloid-beta (Aβ40) peptide as an example. Fluorescence based amyloid fibrillation kinetic assays showed a significant delay in Aβ40 amyloid aggregation by the 4F peptide. In addition, 4F-encased lipid-nanodiscs, at an optimal concentration of 4F (>20 μM) and nanodisc size (<10 nm), significantly affect amyloid fibrillation. A comparison of experimental results obtained from nanodiscs with that obtained from liposomes revealed a substantial inhibitory efficacy of 4F-lipid-nanodiscs against Aβ40 aggregation and were also found to be suitable to trap Aβ40 intermediates. A combination of atomistic molecular dynamics (MD) simulations with NMR and circular dichroism experimental results exhibited a substantial change in Aβ40 conformation upon 4F binding through electrostatic and π-π interactions. Specifically, the 4F peptide was found to interfere with the central β-sheet-forming residues of Aβ40 through substantial hydrogen, π-π and π-alkyl interactions. Fluorescence experiments and coarse-grained MD simulations showed the formation of a ternary complex, where Aβ40 binds to the proximity of peptidic-belt and membrane surface that deaccelerate amyloid fibrillation. Electron microscopy images revealed short and thick amyloid fibers of Aβ40 formed in presence of 4F or 4F-lipid-nanodsics. These findings could aid in the development of amyloid inhibitors as well as in stabilizing Aβ40 intermediates for high-resolution structural and neurobiological studies.
Keywords: Alzheimer’s disease, Beta-amyloid, Nanodisc, Membrane scaffold protein, Protein aggregation
Graphical abstract
Introduction
Disordered soluble proteins are potentially capable of growing into insoluble aggregates that are implicated in neuropathic disorders like Alzheimer’s disease (AD) and Parkinson’s disease and non-neuropathic disease like type-2 diabetes [1,2]. Increasing studies have been focused on the elucidation of detailed mechanisms of amyloid aggregation in order to aid in the development of amyloid inhibitors for potential medical treatment [3,4]. However, the accumulating discoveries from previous studies do not converge on a single mechanism of protein aggregation [5]. One of the impediments to understand the mechanism is that the disordered proteins are capable of growing into various types of aggregates due to their conformational plasticity [6,7]. According to biophysical and biochemical studies, the aggregates are different in their growth rate, morphology, and toxicity [8,9]. Their final and intermediate states seem to be continuously affected by countless factors such as physical environments and balances with other molecules [10–13]. In addition, pathological studies show the presence of insoluble aggregates of disordered proteins at the cell membrane interface, and the cell membrane has also been shown to play a catalytic role in the formation of toxic amyloid intermediates [14–16]. This evidence indicates that disordered proteins collaborate with cell membrane to progressively deprive cells of physiological functions though their aggregation [17,18].
To address the mechanism of the cooperative relationship between the cell membrane and disordered proteins, different types of membrane-mimetics have been designed. The use of liposomes and bicelles revealed that the peptide aggregation collapses membranes, and the area, hydrophobic thickness and curvature of lipid membrane affect the aggregation of an amyloid peptide or protein [19]. Lipid specificity has also been shown to modulate the conformational plasticity of disordered proteins in the formation of a partial folded structure that is known to be an important initial nucleating step for further aggregation of many different disordered proteins and peptides [20–23]. To avoid the effect of membrane curvature, recent studies utilized nanodiscs as a versatile system for protein aggregation studies [24,25]. Peptidic nanodiscs prepared using an amphipathic α-helical apoA-I mimetic membrane scaffold protein (MSP) or short peptides (for example, the 4F peptide: Ac-D-W-F-K-A-F-Y-D-K-V-A-E-K-F-K-E-A-F-NH2) have recently been used to characterize the aggregation properties of amyloidogenic proteins [24,26]. These nanodiscs are stable for a long time as well as capable of reconstituting various types of lipids that differ in acyl chain length and/or head group chemistry [27]. These unique advantages of nanodiscs have also been well utilized in NMR based structural studies of membrane proteins [28].
The AD associated amyloid-beta (Aβ) peptides have been widely studied to establish mechanisms of amyloid formation and to investigate the roles of cell-membrane in modulating the aggregation behaviour. Different molecular targets such as small molecule compounds [29,30], peptides [31–33], antibodies [34,35] etc. have been discovered to inhibit Aβ aggregation and the progression implicated in AD. Studies have shown that electrostatic interaction between Aβ and target peptide inhibitors is crucial for the development of potent peptide inhibitors [32,33]. In addition, nanoparticles constituting with cationic and anionic surfactants have recently been tested to inhibit Aβ aggregation [36]. An alternative therapeutic approach for AD using high-density lipoprotein nanodiscs in animal model has also been reported recently [37]. Considering the charge interaction for the potential development of Aβ inhibitors, the amphipathic MSP or 4F peptides in solution or lipid nanodiscs with exposed charged residues to solvent could modulate the aggregation behaviour of Aβ. While lipid-nanodisc is becoming an useful membrane mimetic to investigate the roles of lipid membrane on amyloid aggregation and also to stabilize intermediates for structural studies [24,26], it is important to examine if the charged belt of the nanodiscs plays a role in modulating amyloid aggregation. In this study, we reveal a concerted ternary association driven by both nanodiscs scaffold protein and lipids with Aβ40 that significantly delay/abolish protein aggregation. Our results demonstrate that the 4F peptide exhibit significant retardation of Aβ40’s aggregation both in lipid-nanodiscs associated and lipid-free states. The conformational changes induced by the intermolecular interaction between 4F and Aβ40, and the interacting residues, are identified using CD, NMR and MD simulations. Employing atomistic all-atom and coarse-grained (CG) MD simulations, we further reveal the interactions between the 4F-peptidic belt and Aβ40 which are supported by fluorescence quenching measurements.
Results
Aβ40 aggregation is affected by 4F peptide in lipid-free solution or in nanodiscs
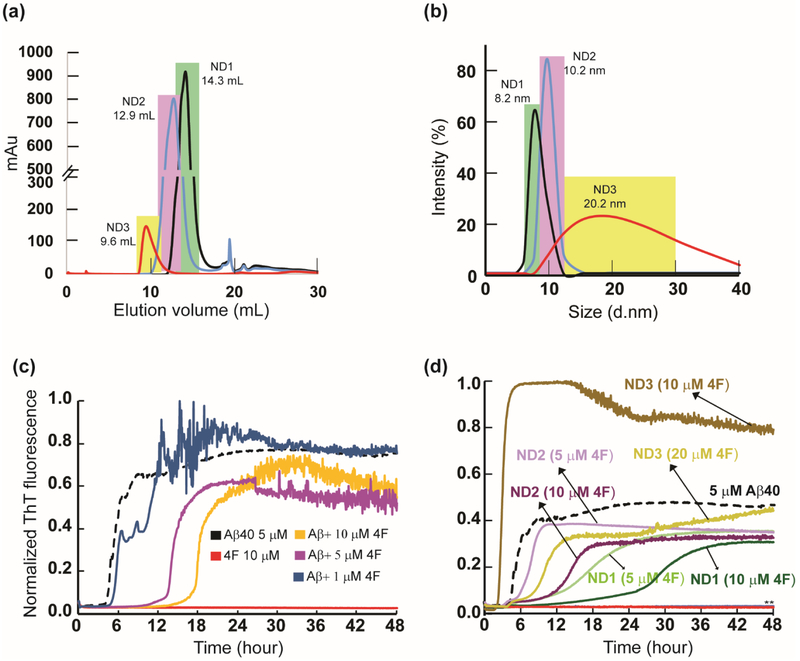
The 4F peptide was used to prepare three differently sized nanodiscs (ND1, ND2 and ND3) at a 4F-peptide/lipid (w/w) ratio of 1, 0.5 and 0.25 (see the Materials and Methods section) and characterized using size-exclusion chromatography (SEC) (Fig. 1a). The dynamic light scattering (DLS) distribution analysis of SEC purified ND1, ND2 and ND3 samples presented an average size distribution with a hydrodynamic diameter of 8.2, 10.2 and 20.2 nm, respectively (Fig. 1b). Thioflavin-T (ThT) based fluorescence assay was used to monitor Aβ40 aggregation both in lipid-free solution containing 4F peptides and in presence of 4F-lipid-nanodiscs, and the results are shown in Fig. 1c and d. The 4F peptide was found to significantly delay Aβ40 aggregation, and the lag-time (Tlag) of Aβ40 fibrillation was found to increase by ~ 3 and 4 folds at equimolar and two times higher molar concentration of 4F, respectively (Fig. 2a). Even for a low concentration (1 μM) of the 4F peptide, we observed a weak retardation of the Aβ40 aggregation process (Fig. 1c).
Fig. 1.
4F peptide (lipid-free or nanodisc-bound) delay amyloid aggregation of Aβ40. (a) Size-exclusion chromatography showing variable size 4F-encased DMPC nanodiscs at peptide:lipid (w/w) ratio of 1:1 (ND1), 1:2 (ND2) and 1:4 (ND3). (b) Size distribution profile of nanodiscs as a function of their hydrodynamic diameter measured using DLS. The highlighted colors in DLS indicate their corresponding SEC fractions. ThT fluorescence monitor the effect of 4F (lipid-free) peptide in solution (c) and 4F-encased lipid-nanodiscs (d) on Aβ40 aggregation (at 5 μM) kinetics at 37 °C in 10 mM sodium phosphate buffer (pH 7.4). The ThT fluorescence of Aβ40 aggregation in the absence of 4F is shown in black and the retardation of aggregation with an increasing concentration of 4F peptide is indicated with different colors. The double asterisks (**) marked in Fig. 1d indicate ND1 and ND2 (containing 20 μM 4F) abolish Aβ40 aggregation. None of the nanodiscs (ND1-3) bind to ThT (see Figure S1a). It should be noted that a change in the 4F-peptide concentration also indicates a corresponding change in the lipid concentration, and therefore an increase in the 4F-peptide concentration indicates an increase in the nanodiscs concentration in the sample.
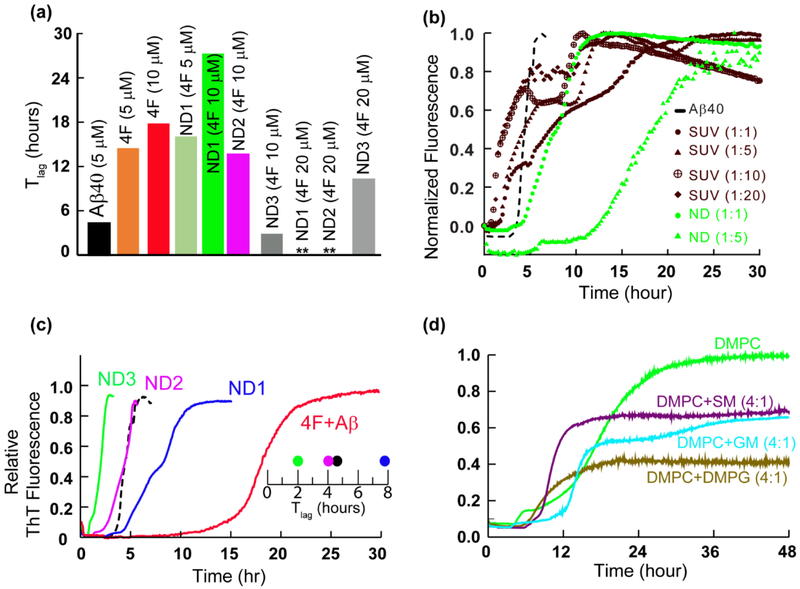
Fig. 2.
Curvature and size of nanodiscs control Aβ40 aggregation. (a) The lag-time of Aβ40 aggregation in 4F and 4F-nanodisc system as a function of time (Tlag) calculated from Fig. 1c and d. (b) ThT assay of Aβ aggregation (5 μM) in presence of nanodiscs (green curves) and DMPC SUVs (brown curves) at the indicated peptide to lipid concentration. NDs at Aβ40 to lipid molar ratio of 1:10 or 1:20 substantially quenches Aβ aggregation and are shown in the figure S1b. (c) ThT fluorescence showing Aβ40 aggregation (5 μM) in absence of nanodiscs and 4F (black) and in presence of nanodiscs (blue, pink and green traces) or 4F (red trace). The Tlag shows a linear correlation between the delay in Aβ40 aggregation and the size of nanodiscs at a minimal DMPC concentration (5 μM) containing 1.5 μM (ND1), 0.75 μM (ND2) and 0.375 μM (ND3) of 4F peptide, (d) Lipid-dependent Aβ aggregation in nanodiscs composed of the indicated lipids. SM and GM denote sphingomyelin and gangliosides, respectively.
Next, we studied the effect of the 4F peptide present as the peptide-belt in all three different 4F-DMPC nanodiscs (ND1, ND2 and ND3) on the aggregation kinetics of Aβ40 (Fig. 1d). ND1 (8.2 nm diameter) containing 5 μM 4F showed a substantial delay in Aβ40 aggregation. Upon increasing ND1 concentration, i.e. containing 10 μM of 4F, delay in Aβ40 fibrillation over a day was observed (Figs. 1d and 2a). Interestingly, a further increase in ND1 concentration (containing 20 or 40 μM of 4F) completely abolished Aβ40 aggregation as indicated by double asterisk in Fig. 1d. Unlikely, ND2 containing 5 μM 4F did not significantly affect the Tlag; but, ND2 containing 10 μM 4F peptide deaccelerated Aβ40 aggregation by increasing the Tlag to ≈13.7 hrs and abolished Aβ40 fibrillation containing 20 μM of 4F (Figs. 1d and 2a). Remarkably, ND3 containing 10 μM 4F peptide accelerated (Tlag≈2.8 hrs), but 20 μM 4F peptide deaccelerated (Tlag≈10.3 hrs) Aβ40 aggregation.
While membrane curvature dependent alteration of Aβ40 aggregation by DMPC vesicles have been reported previously [38], our results show that the presence of nanodiscs alter the lag-time in a size-dependent manner (Figs.1d and 2a). Specifically, the lag-time was increased by small size nanodiscs (ND1 and ND2) and decreased by large size nanodiscs (ND3) at a defined 4F peptide (or nanodisc) concentration (Fig. 1d). Unlike the lipid vesicles, the nanodiscs that are devoid of curvature may be used to trap Aβ40 monomers and/or oligomers on their planar lipid-bilayer surface. To further investigate the ability of 4F peptide nanodiscs to slow down the aggregation kinetics of Aβ40 by trapping intermediates, we compared the aggregation kinetics observed from small unilamellar vesicles (SUVs; Fig. S1b) with that observed from nanodiscs (Figs. 2b and S1c). The presence of SUVs promoted Aβ40 aggregation as shown in Fig.2b (brown traces). ThT fluorescence analysis showed DMPC SUVs with an increasing concentration of lipids (from 1:1 to 1:20 Aβ:DMPC molar ratio) did not significantly alter the lag-time (Fig.2b, brown traces), which is in agreement with previous studies [38]. In contrast, nanodiscs depicted a significant delay in Aβ40 aggregation with an increasing concentration of 4F-DMPC nanodiscs (Fig. 2b, green traces for 1:1 and 1:5Aβ:DMPC molar ratios; and 1:10 and 1:20 Aβ:DMPC shown in Fig.S1c). At Aβ40 to lipid molar ratio of 1:10 or 1:20, nanodiscs substantially abolish Aβ40 aggregation (Figure S1c; lag-time >24 hours), whereas SUVs accelerated its fibrillation (Fig.2b, brown traces). As illustrated in Fig.2b, the notable difference between the lag-times of Aβ40 aggregation observed in SUVs (about 2 hours) and nanodiscs (about 12 hours) for 1:5 Aβ:lipid molar ratio indicates the differential aggregation kinetics influenced by the shape of SUVs or nanodiscs.
While previous studies revealed that a decrease in the size of zwitterionic liposomes accelerated Aβ40 fibrillation due to a high membrane curvature and more water-accessible hydrophobic surface for peptide interactions [38], our results show a counteractive role of 4F-DMPC-nanodiscs size on Aβ40’s aggregation. As shown in Fig 1d, at a defined 4F (10 μM) concentration, an increase in the size of nanodiscs was observed to accelerate Aβ40’s aggregation (Tlag≈27.5, 13.7 and 2.8 hrs for ND1 (8.2 nm diameter), ND2 (10.2 nm diameter) and ND3 (20.2 nm diameter), respectively). It should also be noted that the presence of small size nanodiscs were found to significantly deaccelerate Aβ40’s aggregation as compared to lipid-free Aβ aggregation. Based on the experimental studies using lipid vesicles, previous studies have shown that Aβ40 aggregation is promoted by very low lipid concentration [20,38,39]. However, as shown in Fig.2c, a very low DMPC concentration in nanodiscs (at 1:1 DMPC:Aβ molar ratio) exhibited a linear correlation between 4F concentration and Aβ40 aggregation (Tlag) kinetics indicating its counter protective role against Aβ40 fibrillation.
The role of lipids interaction in nanodiscs with Aβ40 was further studied using Fourier Transform infrared spectroscopy (FTIR). Binding of Aβ40 to 4F-DMPC nanodiscs affected the vibrational bands of both symmetric (~1090 cm−1) and asymmetric (1234 cm−1) stretching modes of the lipid head group (PO2−) (see Fig. S2). This indicated a concerted ternary association of Aβ40 with the lipid and 4F-peptide-belt of nanodiscs. Several other membrane components such as anionic lipids, cholesterol, gangliosides (GM) and sphingomyelin (SM) are connected to the pathology of membrane mediated AD progression [40–42]. To further investigate the role of lipids in nanodiscs in modulating Aβ40 aggregation kinetics, we designed three different 4F-nanodiscs (size <10 nm, Fig.S3a) with membrane compositions DMPC:DMPG (4:1), DMPC:GM (4:1) and DMPC:SM (4:1). For a defined nanodisc (containing 5 μM of 4F) to Aβ40 (5 μM) molar ratio, different membrane compositions of 4F-nanodiscs showed differential behavior of Aβ40 aggregation kinetics (Fig. 2d). For example, GM containing nanodiscs were observed to increase the lag-time compared to SM or anionic lipid containing nanodiscs. These experimental observations further confirm the involvement of lipids in modulating Aβ40 aggregation. A further increase in the nanodisc concentration (containing 10 μM of 4F) substantially quenched the ThT fluorescence for 24 hours (Fig. S3b).
4F-peptide forms a complex with Aβ40 and adopts an α-helical conformation
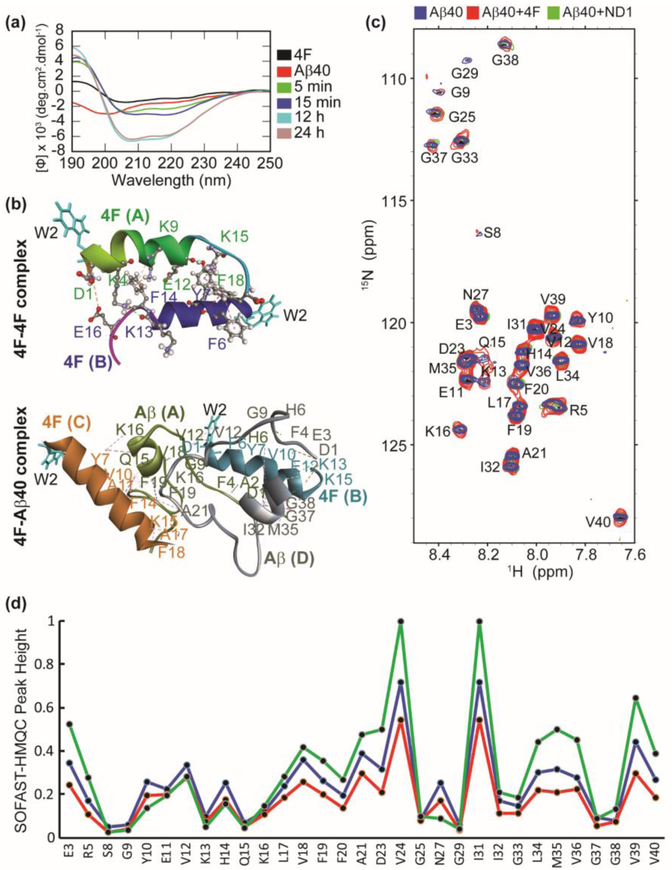
We next studied the binding mechanism of 4F with Aβ40 using CD, NMR and MD simulations. Far-UV CD measurements showed an increase in molar ellipticity at 208 and 222 nm indicating α-helical conformation in 4F-Aβ40 mixture solution within few minutes (Fig. 3a). It should be noted that a favorable helix conformation of 4F peptide has been observed in nanodiscs where it tightly binds to lipids [43]. The time-dependent CD analysis showed a further increase in the helical content and molar ellipticity of the 4F-Aβ40 complex. In contrast, the time-dependent CD spectral analysis in absence of 4F showed a decrease in the molar ellipticity due to the self-assembling properties of Aβ40 (Fig. S4). The increase in the molar ellipticity over time and slow aggregation as revealed from ThT assay (Fig. 1c) indicate a nonfibrillar Aβ40 conformational state induced by the interaction with the 4F-peptide. Structural studies of 4F peptide (Fig S5) in solution using MD simulations showed the formation of 4F dimers (initially 4F monomers were placed ≈ 1 nm away from each other) through symmetric packing along the horizontal axis through electrostatic (D1-E16), hydrogen bonding, and π-π stacking (F6-F18; Y7-F18) interactions (Fig. 3b, Table S1). Similarly, substantial intermolecular interactions between 4F and Aβ40 peptides were observed within a 100 ns MD simulation. Electrostatic (between Aβ:K13/K15 and 4F:D1, and Aβ:D1 with 4F:E12), π-π stacking (between Aβ:F4 and 4F:F14, and Aβ:F19 and 4F:F14), and a substantial number of hydrogen bonds and hydrophobic interactions between 4F and Aβ40 peptides were identified (Fig. 3b, Table S2). The hetero-tetramer 4F-Aβ40 complex showed an unfolded Aβ40 conformation (for both Aβ40 peptide molecules in the complex) during a 100 ns MD simulation; whereas a well-defined helical conformation was identified for 4F in comparison to the dimeric structure obtained in solution, which correlates well with CD results (Fig. 3a and b). The role of central aromatic residues F19 and F20 in modulating the aggregation kinetics through intermolecular π-stacking interactions have been reported for Aβ [44]. Aromatic inhibitors and F19 mutation have been shown to significantly slow down Aβ aggregation [41,42,43]. Thus, the protective role of 4F peptide on Aβ40 aggregation (Fig. 3b) could be explained in terms of π-π and π-alkyl interactions (Table S2) that energetically disfavour Aβ40’s ability to self-assemble by interfering with the β-sheet forming central and C-terminal domains. 2D SOFAST-HMQC NMR experiments showed that the addition of 4F peptide affected the 2D 1H/15N correlation spectrum of Aβ40 (Fig. 3c). The 4F-Aβ40 complex presented a uniform reduction in NMR signal intensities for both N and C-terminal residues (Fig. 3d). On the other hand, the Aβ40 N and C-termini residues depicted a decrease and increase in NMR signal intensities in presence of nanodiscs, respectively. This indicated a coupling between the charged N-terminus of Aβ40 and nanodisc could control the folding and aggregation propensity of Aβ40 as seen in ThT assays (Fig. 1d).
Fig. 3.
Atomistic insights into 4F Aβ40 interactions. (a) Far-UV CD spectra of 4F peptide (20 μM) titrated with Aβ40 (20 μM) measured at the indicated time-intervals. (b) MD snapshots showing the formation of 4F-homodimer and 4F-Aβ40 hetero tetramer. The solvent exposed Trp2 residues are shown in cyan color. The peptide chains are shown in cartoon and indicated in bold. The interacting amino acid residues are labeled and intermolecular hydrogen bonds are shown in dashed lines (see Tables S1 and S2). (c) 2D 1H/15N SOFAST-HMQC spectra of 50 μM Aβ40 (blue), 50 μM Aβ40 mixed with 50 μM 4F (red), and 50 μM Aβ40 mixed with 50 μM 4F of ND2 (green). The NMR spectra were recorded on a 600 MHz Bruker NMR spectrometer at 37°C and the samples were prepared in 10 mM sodium phosphate buffer, pH 7.4 containing 10% D2O. (d) Relative NMR signal intensities of Aβ40 residues measured from Fig. 3c.
Atomistic insights into the interaction between 4F-nanodisc’s belt with Aβ40
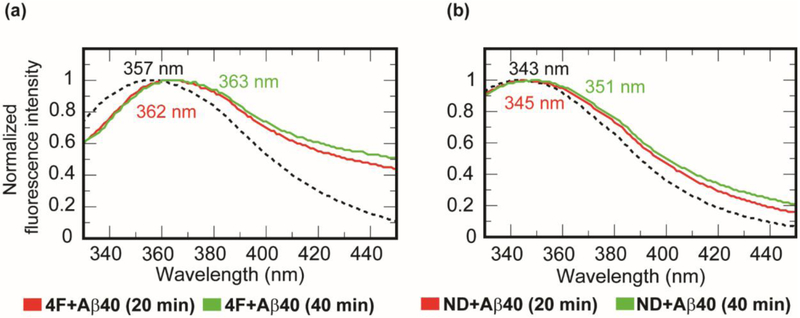
Interaction of the peptidic belt of nanodiscs with Aβ40 was studied by monitoring tryptophan fluorescence using the Trp2 residue in the 4F peptide. A 50 μM of 4F peptide solution (10 mM sodium phosphate buffer, pH 7.4) showed a tryptophan emission spectrum at 357 nm when excited at 295 nm (Fig. 4a) [43]. When 25 μM of Aβ40 peptide was titrated with 50 μM of 4F peptide solution, a small red shift (from 357 to 363 nm) in Trp2 fluorescence was observed over time. This observation suggests the solvent exposure of Trp2 residue of the 4F peptide upon binding with Aβ40 (Fig.4a). This is in agreement with the intermolecular interaction observed between 4F and Aβ40 peptides in all-atom MD simulations (Fig. 3b). To investigate if the presence of lipid-nanodiscs would affect the above-mentioned interaction between 4F and Aβ40 peptides, we carried out Trp2 fluorescence experiments and MD simulations as explained below. In presence of DMPC nanodiscs (containing 50 μM of 4F peptide), a significant blue shift (from 357 to 343 nm) was observed in Trp2 fluorescence indicating Trp2 is oriented inside the hydrophobic-core of the lipid-bilayer in agreement with the amphipathic helical structure of the 4F peptide (Fig. 4b). Interestingly, a titration of 25 μM Aβ40 with 4F-DMPC nanodiscs exhibited a gradual red shift in tryptophan fluorescence (≈ 343 to 351 nm) within 40 min (Fig. 4b). This observed change in Trp2 fluorescence due to the addition of Aβ40 occurred within a time scale (~40 minutes) where majority of the Aβ40 populations are either in monomeric and/or lower order oligomeric states (Tlag≈ 13.7 hrs as shown in Fig. 2a). Therefore, the red shift (343 to 351 nm) indicating the solvent exposure of Trp2 residue of the 4F peptide could be interpreted as a consequence of a direct interaction between 4F and Aβ40 or, it could be due to the lipid bilayer interaction of Aβ40 (monomer and/or oligomers) which can cause membrane thinning and/or induce changes in the lipid packing and therefore the shape of the 4F belt [16]. Since fluorescence experiments can only be used to observe the orientation of the Trp2 residue but not on the size of the 4F-belt, we performed DLS experiments to monitor the change in the size of nanodiscs by adding Aβ40 monomers to ND1 (size of 8.2 nm diameter). After a 1-hour incubation, ND1 showed a ≈ 2 nm increase in the absolute size (as shown in Fig. S6). Thus, the combination of fluorescence and DLS experimental results confirmed the change in the size of the 4F-belt as well as the change in the Trp2 orientation are due to Aβ40 interaction with nanodiscs. However, the direct versus lipid mediated 4F and Aβ40 interactions cannot be differentiated experimentally. Therefore, to gain further insights into the mechanism of Aβ40 interaction with 4F-nanodiscs, we performed both all-atom and coarse-grained (CG) MD simulations as explained below.
Fig. 4.
Probing 4F-Aβ40 interactions by tryptophan fluorescence quenching. Monitoring tryptophan (Trp2 residue of the 4F peptide) fluorescence quenching in (a) 50 μM 4F peptide solution and (b) ND1 containing 50 μM 4F peptides in presence (red and green traces) and absence (dashed black traces) of 25 μM Aβ40 at 37 °C at the indicated time intervals (0 min in dashed lines, 20 min in red, and 40 min in green).
We used MSP (a diameter of ~98 Å) encased zwitterionic/anionic atomic lipid bilayer models (see Materials and methods), instead of the 4F-nanodiscs, due to the availability of parametrized all-atom and CG nanodisc model systems at CHARMM-GUI [47] (Fig. S5). We were able to observe intermolecular interactions between Aβ40 and MSP-belt within several nanoseconds in all-atom MD simulations. Our all-atom MD 100 ns simulations showed interactions between Aβ40 and MSP-belt of 4:1 DMPC:DMPS nanodiscs. While majority of Aβ40 molecules were found to be unfolded and aggregated in the aqueous phase, a partially folded Aβ40 was observed to interact with MSP-belt within the defined simulation time length (100 ns) (Fig. S7a). Aβ40 exhibited significant electrostatic and hydrogen bonding interactions through its charged N-terminus with MSP’s charged residues (Arg/Lys/Glu) located in the 120-133 region (see Table S3, Fig. S7a).
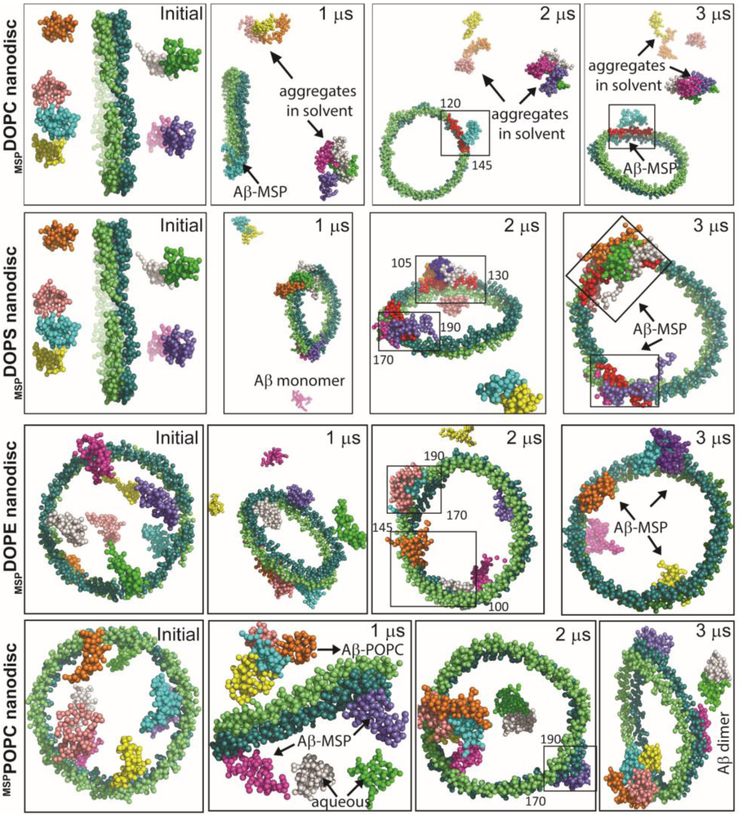
Analysis of hydrogen bond formation between Aβ40 and individual molecules of targeted MSP-encased lipid nanodisc (denoted as mspDMPC/DMPS4:1) showed the formation of a comparatively consistent and higher number of hydrogen bonds between Aβ40 and MSP-belt after 20 ns MD simulation. On the contrary, unstable and relatively small number of hydrogen bonds were identified between Aβ40 and lipids present within the nanodisc (Fig. S8). Within the limited time scale of all-atom MD simulations, we could only observe a single Aβ40 out of total of eight Aβ40 molecules used in the simulations, interacting with the MSP-belt of the nanodisc. To overcome this limitation due to the restricted time length for all-atom MD simulations, we then investigated Aβ40 interaction with MSP-nanodiscs at microsecond time scale using CG model systems of MSP encased DOPC, DOPS, DOPE, POPC or a mixed lipid nanodiscs (see Figs. 5 and S9). Interestingly, several Aβ40 molecules were found to be located close to MSP-belt and in the vicinity of lipids located close to the MSP-belt (see video SV included in the Supporting Information). The Aβ40 molecules were initially observed to aggregate in the aqueous phase followed by a slow transition to water-lipid/MSP interface within microseconds of MD simulation time (Fig. S7b). Surprisingly, Aβ40 molecules were observed to bind directly to the MSP-belt in the case of 100% anionic DOPS lipids within a time scale of ≈ 300 ns (see the video SV included in the Supporting Information). A major population of the well-dispersed Aβ40 molecules in aqueous phase were identified to interact with MSP-belt in all the chosen MSP-nanodisc systems (listed in Table 1) irrespective of their lipid composition. As illustrated in Fig. 5, at the end of 3 μs MD simulations, Aβ40 molecules were localized on the lipid bilayer surface but restricted to interact with the MSP-belt. Similarly, MSP-nanodiscs comprising of mixed lipids mimicking mitochondrial membrane also presented both lipid-bilayer-bound but with a restriction of Aβ40 population to interact with MSP during 3 μs MD simulations (Fig. S9) suggesting that lipid composition has a minimal effect on the Aβ40-MSP interaction, which could potentially delay Aβ40 aggregation. Taken together, both all-atom and CG-MD simulations indicated that the binding efficacy of MSP-belt to Aβ40 molecules could modulate the conformational plasticity of the amyloidogenic peptide to self-assemble in a lipid membrane environment.
Fig. 5.
MD simulations reveal Aβ40-MSP interactions. Illustration of Aβ40 interaction with MSP in nanodiscs at the indicated MD simulation times. MD snapshots are retrieved after every 1 μs from zwitterionic (DOPC/DOPE/POPC) or anionic (DOPS) MSP-based lipid-nanodiscs. The MSP protein chains (ring shape) and Aβ molecules (eight) are represented in vdw using VMD and drawn in different colors. The lipid molecules in the center and water inside the box are not shown for visual clarity. MSP domains binding with Aβ40 are covered in a rectangle and the amino acid residues spanning the binding domains are indicated. All Aβ40 molecules are initially placed ≈ 10 Å away from the lipid bilayer surface and a minimum distance of « 5 Å is maintained between Aβ molecules in aqueous phase.
Table 1.
List of parameters and simulated systems used in molecular dynamics simulations.
| All-atom MD systems | Phospholipid type | Total atoms | Simulation length |
|---|---|---|---|
| 4F-4F (2 molecules) | N/A | 5597 | 100 ns |
| Aβ40 (2 mols)-4F (2 mols) | N/A | 13147 | 100 ns |
| Aβ40-MSPDMPC/DMPS4:1 | 8 Aβ40; 152 DMPC; 38 DMPS | 117079 | 100 ns |
| Coarse-grained MD systems | |||
| Aβ40-MSPDOPC | 8 Aβ40; 172 DOPC | 29648 | 3 μs |
| Aβ40-MSPDOPS | 8 Aβ40; 167 DOPS | 29602 | 3 μs |
| Aβ40-MSPPOPC | 8 Aβ40; 173 POPC | 27215 | 3 μs |
| Aβ40-MSPDOPE | 8 Aβ40; 188 DOPE | 29824 | 3 μs |
| Aβ40-MSPMitochondriaInner | DOPC:DOPE:Cholesterol:Cardiolipin (40.8:26.5:12.2:20.4 %); 8 Aβ40 |
38080 | 3 μs |
| Aβ40-MSPMitochondriaOuter | DOPC:DOPE:Cholesterol:Cardiolipin (58:29:13:0%); 8 Aβ40 |
37420 | 3 μs |
Aβ40 forms short and thick fibers in presence of 4F peptide or 4F-nanodiscs fibers
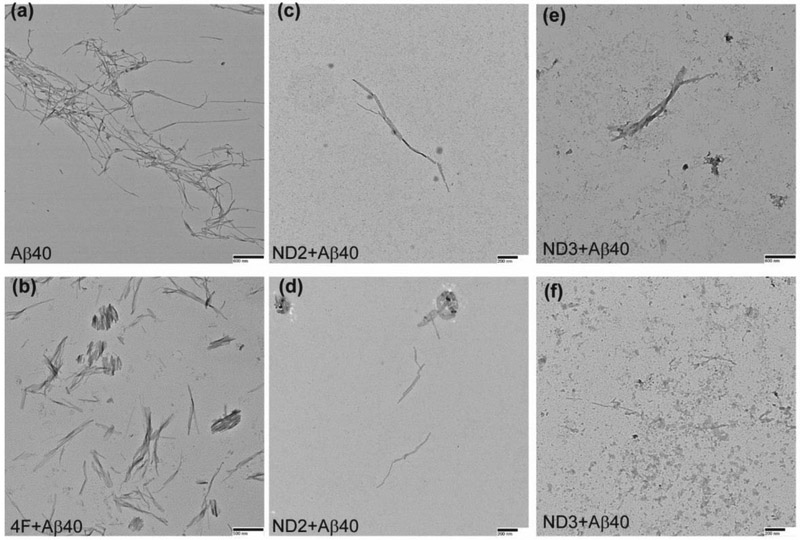
Structural polymorphism are associated with amyloids and studies have focused on understanding the mechanism of amyloid fibril formation and their neurobiological significance [48]. Here, we observed two distinct conformational states of Aβ40 fibers in presence of 4F or 4F-encased lipid-nanodiscs (Fig. S10) using transmission electron microscopy (TEM). The amyloid fibers obtained from 1:1 molar ratio of 4F:Aβ40 (Fig. 6b) or 4F-nanodisc:Aβ40 (Fig. 6c-f) were found to be thicker and shorter than Aβ40 fibers prepared in absence of 4F (Fig. 6a). In addition, we observed relatively small population of short fibers in all nanodisc systems (Fig. 6c-f) that contain equivalent amount of Aβ40 (5 μM) in aqueous solution (Fig. 6a) and in 4F peptide mixed solution (Fig. 6b). Overall, the TEM analysis indicated 4F or 4F-nanodiscs generate short Aβ40 fibers that may exhibit potentially distinct neurobiological activities, which would be worth investigating in the future.
Fig. 6.
TEM images showing morphology of Aβ40 fibers in presence and absence of 4F peptide or 4F-peptide-lipid-nanodiscs. The samples analyzed by ThT based experiments (see Figure 1) are used for or TEM imaging. Aβ40 samples in absence of 4F or 4F-nanodiscs (a) and in presence of 4F (b), ND2 (c and d) and ND3 (e and f) were adsorbed on carbon-coated copper grids and negatively stained with 2% uranyl acetate for TEM imaging. Scale bars are shown on the bottom right. TEM image of nanodiscs is shown in Figure S10.
Discussion
Structural characterization of amyloid proteins using lipid nanodiscs is emerging out to be an important approach to better understand the membrane-assisted amyloid aggregation process and to potentially develop therapeutic strategies [24,26,49,50]. While the membrane composition (Gangliosides/Sphingomyelin/Cholesterol) has been shown to modulate amyloid aggregation [51] and the pathological state of Aβ, the findings reported in this study further highlights the protective role of apolipoprotein (MSP) or MSP-derived peptide (i.e., 4F in this study) on amyloidosis. In this study, we revealed that the 4F peptide mimicking a short segment of apo-lipoproteins interacts with Aβ40 and substantially delays the aggregation (Fig. 1c). Importantly, the 4F peptide was not only found to retard the aggregation kinetics of Aβ40, but also alters the fiber morphology by generating short and thick fibers (Fig. 6b). Our structural investigation showed electrostatic and π-π interactions drive 4F-Aβ40 complex formation which is in agreement with the NMR results (Fig. 3c and d). The amyloid core residues including F19, F20, and L34-V36 residues, that drive beta-sheet formation, were significantly affected by the 4F peptide (Table S2) both in aqueous and in nanodisc solutions (Fig. 3d). The therapeutic significance of apolipoprotein A-I mimetic peptides (such as D-4F) in preventing atherosclerosis have been tested in animal models [52,53]. Here, we revealed their potential anti-amyloidogenic activities in the form of 4F-nanodiscs. The 4F-nanodiscs at an optimal concentration (containing > 20 μM 4F) trap Aβ40 intermediates as revealed from the significant ThT fluorescence quenching as shown in Fig. 1d. By controlling the concentration of 4F-nanodiscs, we observed its linear correlation with the delay in Aβ40 aggregation kinetics, and extended the complete abolishment of Aβ40 fibrillation. In reference to a previous study that showed Aβ binding to nanodisc using fluorescence titration experiments [26], it is possible that the Aβ40 aggregation could be modulated by varying membrane lipid composition of 4F-nanodiscs as illustrated in Fig. 2d. Hence, the membrane lipids in association with 4F-belt of the nanodisc most likely to involve in the modulation of Aβ40 aggregation through a concerted mechanism of action (Fig. 2b and 2d). Taken together, the potential anti-amyloidogenic activity of 4F peptide highlighted its importance for Aβ therapy and could be potential nanoparticles to isolate pathologically distinct Aβ40 intermediates for neurobiological functional studies.
Previous studies successfully carried out NMR experimental studies on two different amyloidogenic proteins (such as islet amyloid polypeptide (IAPP) and Aβ42) by using MSP based lipid-nanodiscs [24,26]. In this study, we have successfully demonstrated the use of 4F peptide-based lipid-nanodiscs for NMR experimental investigation of Aβ40. As recently demonstrated peptide-based nanodiscs have additional advantages over MSP-nanodiscs [54]. While Aβ interaction with cell membrane accelerates its aggregation [20,38,39], here we show the solvent exposed charged residues in the peptide belt associated with lipids in the nanodisc play a protective role by delaying Aβ40 aggregation (Fig. 5). The application of apolipoprotein encapsulated lipid-nanodiscs has been tested in-vitro and in-vivo for successful drug-delivery of several poorly soluble drugs including polyphenolic amyloid inhibitors [55]. In addition, synthetic high-density lipoprotein nanoparticles have recently been tested in-vivo that reduce the amount of cerebral amyloid-beta in a transgenic mouse [37]. Considering the emerging application of lipid-nanodiscs, the amyloid inhibiting efficacy of 4F-nanodiscs reported in this study could be useful for potential therapeutic advancement for amyloidosis. It could also be worthwhile to test the bioavailability of amyloid inhibitors by incorporating them into 4F-nanodiscs.
Conclusions
In this study, we have successfully demonstrated the existence of a crosstalk between the amphipathic peptidic belt of lipid-nanodiscs and amyloid aggregation by using a combination of fluorescence, CD, NMR and TEM experiments and MD simulations. This study also enabled us to identify the formation of several off-pathway amyloid aggregates (including the polymorphic fibers) that could be therapeutically important [48], and their high-resolution structures could be worth investigating. Therefore, the ternary association of Aβ, lipid-membrane and peptidic belt must be considered to obtain accurate and complete information from biophysical/biochemical analysis and drug discovery. Based on the ability of nanodisc-belt to inhibit amyloid aggregation reported in this study, screening of more potent anti-amyloidogenic peptides derived from apolipoproteins could further assist in designing novel therapeutic targets to control AD progression. Although the influence of peptide-nanodiscs on Aβ’s aggregation to form fibers is quite complex, and as shown by the results reported in this study that both the peptide-belt and the lipid composition play significant roles, more studies utilizing a variety of lipid composition to trap intermediates of aggregates of several different amyloid proteins/peptides would be useful. Such studies would enable the application of sophisticated biophysical and NMR experiments to obtain high-resolution structures, and could provide valuable insights into the mechanisms of aggregation to form toxic species in a membrane environment and to develop compounds to inhibit toxicity.
Materials and methods
Chemicals
Thioflavin T (ThT), uranyl acetate and all other salts were purchased from Sigma-Aldrich (St. Louis, MO). 1,2-dimyristoyl-sn-glycero-3-phosphatidylcholine (DMPC), 1,2-dimyristoyl-sn-glycero-3-phosphoglycerol (DMPG), ganglioside (GM1), sphingomyelin (SM) were purchased from Avanti Polar Lipids, Inc® (Alabaster, AL). Chemical reagents were purchased from commercial suppliers and used as received.
Aβ40 expression, purification and sample preparation
Unlabeled and uniformly 15N isotope labelled Aβ40 peptides were recombinantly expressed in E. coli BL21 (DE3). The Aβ40 plasmid was a generous gift from Professor Bernd Reif (Technical University of Munich, Germany). The expression and purification procedures of Aβ40 were as reported elsewhere [56,57]. The purified Aβ40 peptide was dissolved in 5% (v/v) NH4OH and lyophilized at a concentration of 0.1 mg/ml. The peptide powder was then dissolved in buffer (10 mM sodium phosphate, pH 7.4) and sonicated for 15 s followed by centrifugation at 14,000 × g for 15 min at 4 °C to remove any small aggregates. The protein concentration was measured using NanoDrop spectrophotometer, and 100 μM stock solutions of Aβ were prepared for experiments. All experiments were performed using 10 mM sodium phosphate buffer, pH 7.4.
Nanodiscs sample preparation
Large unilamellar vesicles (LUVs) were prepared as described elsewhere [27,38]. Briefly, lipid powders were dissolved (10 mg/ml) in HPLC-grade 1:1 chloroform and methanol followed by evaporation under a continuous steam of nitrogen gas. The lipid film (DMPC or DMPC mixed DMPG or GM or SM at 4:1 molar ratio) was kept under vacuum for 4 hours to completely remove any residual solvents. The dehydrated lipid film was hydrated in 10 mM sodium phosphate buffer (pH 7.4) followed by 5 minutes of vortex mixing. The lipid mixture was mixed with the 4F peptide to make a solution with DMPC:4F (w/w) ratios of 1:1 (ND1), 2:1 (ND2) and 4:1 (ND3), followed by vortex mixing for 5 minutes and then incubating overnight at 37 °C under gentle agitation. Small unilamellar vesicles (SUVs) were prepared by dissolving DMPC LUVs in 10 mM sodium phosphate buffer (pH 7.4). The hydrated lipid films were subjected to 30 minutes sonication in a water bath at 37 °C followed by 10 freeze-thaw cycles. Thereafter, the DMPC suspension was extruded 20 times through 30 nm polycarbonate membranes using a mini extruder (purchased from Avanti Polar Lipids, Inc., Alabaster, Alabama). The formation of nanodiscs and SUVs were confirmed by size distribution analysis using DLS experiments.
Nanodisc purification and size-distribution analysis
The nanodiscs were purified by passing them through SEC using a Superdex 200 Increase 300/10 GL column operated on an AKTA purifier (GE Healthcare, Freiburg, Germany). DLS (Wyatt Technology Corporation, Goleta, CA) measurements were performed to check the size distribution of all SEC purified nanodiscs using a 1 μl quartz cuvette. The concentration of nanodiscs were calculated by measuring the concentration of 4F peptides using NanoDrop spectrophotometer. All 4F peptide based lipid-nanodiscs (namely, ND1, ND2 and ND3 or mixed lipid nanodiscs) were diluted to a 4F peptide concentration of 200 μM and aliquots of samples were prepared as required for subsequent experiments. All DLS measurements were performed at 37 °C and the scattering results were averaged over 20 scans. Correlation functions were fitted using the isotropic sphere model and fittings were performed using the software provided by the supplier (Wyatt Technology Co., Goleta, CA).
ThT Fluorescence assay
Thioflavin T (ThT) dye based fluorescence experiments were performed to monitor the aggregation kinetics of Aβ40 under various conditions. Fisher 96-well polystyrene plates with a sample (5μM Aβ40 and 10μM ThT, and with a variable concentration of lipid-nanodiscs or SUVs) volume of 100 μl/well were used for fluorescence measurements. The kinetics of amyloid formation was monitored at 3-min intervals using a microplate reader (BioTek Synergy 2, Winooski, VT) with an excitation and emission wavelengths of 440 and 485 nm, respectively, at 37 °C under continuous and medium orbital shaking. The kinetic parameters were obtained by fitting fibrillation curves to the following sigmoid equation [58].
where y0 is the pre-transition baseline, k is the apparent growth rate constant and t0.5 is the half-time when ThT fluorescence reaches half of its maximum intensity. The lag time (Tlag) is defined as Tlag=t0.5– 1/2/k. Fourier transform infrared (FTIR) spectra were obtained for Aβ40 alone (5 μM) and mixed with 4F-DMPC nanodiscs containing 5 μM of 4F in transmission mode within a range of 4000–400 cm−1 using a Thermos scientific ATR-FTIR instrument.
Transmission electron microscopy
Transmission electron microscopy (TEM) images were measured using a HITACHI H-7650 transmission microscope (Hitachi, Tokyo, Japan) at 25 °C following the protocols described elsewhere [20]. The Aβ40 peptide samples used in the ThT assay measurements (see methods) was used for TEM analysis. After ThT measurements, the Aβ40 fiber samples were further incubated at room temperature for 7 days and their TEM images were collected. A 10 μl sample volume of 5 μM of Aβ40 mixed with 10 μM of 4F or 4F-nanodiscs containing 10 μM of 4F (see Fig. 1d) was loaded on a collodion-coated copper grid. The spotted samples were incubated for 2 minutes followed by three times rinsing with Milli-Q water. The copper grid was next stained with 5 μl of 2% (w/w) uranyl acetate for 1 minute followed by three times rinsing with Milli-Q water. The sample loaded grids were incubated overnight at room temperature and electron micrographs were collected.
Tryptophan fluorescence quenching assay
The tryptophan (Trp2) residue in 4F peptide was used for fluorescence measurement. 4F peptide of concentration 50 μM dissolved in 10 mM sodium phosphate buffer (pH 7.4) or 4F-DMPC (ND1) nanodiscs containing 50 μM 4F in presence or absence of 25 μM Aβ40 were prepared for Trp2 fluorescence quenching measurements. Fluorescence emission was measured at 330 nm following an excitation at 295 nm (with a 5 nm bandwidth). The fluorescence measurement was recorded in a continuous mode using a FluoroMax 4® from Horiba Scientific® with a delay time of 1 minute per 5 scans using a 200 μL cuvette at 37 °C. The spectra were normalized and the changes in emission spectral peak were analyzed.
Molecular dynamics simulations
The 3D model structure of 4F peptide (Ac-DWFKAFYDKVAEKFKEAF-NH2) was built using I-TASSER [59]. I-TASSER modelled α-helical 4F peptide was further refined using Discovery studio visualizer. The solution NMR structure of Aβ40 determined in an aqueous environment [60] (PDB: 2LFM) was used for MD simulations. The MSP-encased nanodiscs (all-atom and coarse-grained) comprising of two molecules of MSP (a diameter of ~98 Å) and a variable number of phospholipids (see Table 1) were built using CHARMM-GUI [47]. MD systems comprising of eight molecules of Aβ40 and MSP-nanodiscs of variable lipids built using CHARMM-GUI were generated using GROMACS 5.0.7 [61]. The Aβ40 molecules were separated from one another by a distance of at least ≈ 5 Å (placed in random orientations) and were positioned ≈ 20 Å away from the nanodisc’s lipid bilayer surface. All MD systems were neutralized by adding counter ions followed by energy-minimization using the steepest-descent method prior to carrying out simulations.
For all-atom MD simulations of 4F-4F interactions, two molecules of 4F (separated by ≈ 10 Å) were solvated in a cubic box (size ≈ 20 Å) with random orientations. Similarly, for Aβ40-4F interaction analysis, two molecules of Aβ40 and 4F were suspended in a cubic box. All atom MD simulations were performed using CHARMM36 [62] force field at 37 °C using GROMACS [61] software package, version 5.0.7. The pressure and temperature equilibrations were applied to each complex system as instructed in CHARMM-GUI. A final production MD run of 100 ns was performed for all three all-atom systems (Table 1). The coarse-grained (CG) systems comprising of eight molecules of Aβ40 and nanodiscs (containing DOPC, DOPS, DOPE, POPC or mitochondrial inner and outer membrane mixed lipids) were simulated using Martini (v2.2) force field [63] in GROMACS.
The topology files for all lipids were obtained from CHARMM-GUI and Aβ40 topology was generated using martinize python program [64]. A production MD simulation run of 3 μs was carried out for all systems at 37 °C. MD simulation trajectories were interpreted using visual molecular dynamics [65]. Molecular visualization was carried out using Discovery studio visualizer 3.5 [66] and PyMOL. The final images were built using Adobe illustrator (version 16.0.3). Complex structures retrieved from CG-MD simulations were converted to all-atom structures for structural analysis using CHARMM-GUI.
NMR experiments
2D 1H/15N SOFAST-HMQC [67] NMR spectra were recorded on a 600 MHz Bruker Avance III spectrometer equipped with a z-axis gradient cryogenic probe. Three types of samples were prepared in 10 mM sodium phosphate (pH 7.4) buffer containing 90% H2O/10% 2H2O at 25 °C for NMR measurements: Samples-1: 50 μM of uniformly 15N-labeled Aβ40 in solution; Sample-2: 50 μM of uniformly 15N-labeled Aβ40 mixed with 50 μM 4F peptide and incubated for 1 hour at room temperature; Sample-3: 50 μM of uniformly 15N-labeled Aβ40 mixed with 4F-DMPC nanodisc (ND2) containing 50 μM 4F and incubated for 1 hour at room temperature. All NMR spectra were obtained using 200 t1 increments and with 64 scans. NMR data were processed using TopSpin 3.5 (Bruker) and analyzed using Sparky [68].
Supplementary Material
Highlights.
-
➢
Apolipoprotein mimetic 4F peptide retards beta-amyloid aggregation.
-
➢
4F-nanodiscs substantially inhibit beta-amyloid fibrillation growth.
-
➢
Beta-amyloid forms short and thick fibers in presence of 4F or 4F-nanodiscs.
-
➢
Structural study reveals a ternary association between Aβ40 and 4F-nanodiscs.
Acknowledgement
This study was supported by funds from the National Institutes of Health (AG048934 to A.R.). This work was (in part) performed under the International Collaborative Research Program of Institute for Protein Research, Osaka University, ICR-18-02. We thank Professor Toshimichi Fujiwara in the Institute for Protein Research, Osaka University, for providing parallel computing facility on SGI UV 3000. We thank Professor Bernd Reif for providing us the recombinant expression system and protocol for the production of amyloid-beta-1-40 peptide.
Abbreviations:
- Aβ
amyloid-beta
- NMR
nuclear magnetic resonance
- MSP
membrane scaffold protein
- CD
circular dichroism
- ThT
Thioflavin-T
- SOFAST-HMQC
band-selective optimized flip-angle short transient heteronuclear multiple quantum coherence
- SEC
size-exclusion chromatography
- DLS
dynamic light scattering
- ND
nanodisc
- SUVs
small-unilamellar vesicles
- LUV
large-unilamellar vesicles
- MD
molecular dynamics
- CG
coarse-grained
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Knowles TPJ, Vendruscolo M, Dobson CM, The amyloid state and its association with protein misfolding diseases, Nat. Rev. Mol. Cell Biol 15 (2014) 384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- [2].Jaikaran ETAS, Clark A, Islet amyloid and type 2 diabetes: From molecular misfolding to islet pathophysiology, Biochim. Biophys. Acta - Mol. Basis Dis 1537 (2001) 179–203. doi: 10.1016/S0925-4439(01)00078-3. [DOI] [PubMed] [Google Scholar]
- [3].Eisele YS, Monteiro C, Fearns C, Encalada SE, Wiseman RL, Powers ET, Kelly JW, Targeting protein aggregation for the treatment of degenerative diseases, Nat. Rev. Drug Discov 14 (2015) 759–780. doi: 10.1038/nrd4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Stefani M, Protein misfolding and aggregation: New examples in medicine and biology of the dark side of the protein world, Biochim. Biophys. Acta - Mol. Basis Dis 1739 (2004) 5–25. doi: 10.1016/j.bbadis.2004.08.004. [DOI] [PubMed] [Google Scholar]
- [5].Invernizzi G, Papaleo E, Sabate R, Ventura S, Protein aggregation: Mechanisms and functional consequences, Int. J. Biochem. Cell Biol 44 (2012) 1541–1554. doi: 10.1016/j.biocel.2012.05.023. [DOI] [PubMed] [Google Scholar]
- [6].Fink AL, Protein aggregation: Folding aggregates, inclusion bodies and amyloid, Fold. Des 3 (1998). doi: 10.1016/S1359-0278(98)00002-9. [DOI] [PubMed] [Google Scholar]
- [7].Uversky VN, Mysterious oligomerization of the amyloidogenic proteins, FEBS J 277 (2010) 2940–2953. doi: 10.1111/j.1742-4658.2010.07721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Paravastu AK, Qahwash I, Leapman RD, Meredith SC, Tycko R, Seeded growth of beta-amyloid fibrils from Alzheimer’s brain-derived fibrils produces a distinct fibril structure., Proc. Natl. Acad. Sci. U. S. A 106 (2009) 7443–7448. doi: 10.1073/pnas.0812033106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wu JW, Breydo L, Isas JM, Lee J, Kuznetsov YG, Langen R, Glabe C, Fibrillar oligomers nucleate the oligomerization of monomeric amyloid β but do not seed fibril formation, J. Biol. Chem 285 (2010) 6071–6079. doi: 10.1074/jbc.M109.069542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Stefani M, Biochemical and biophysical features of both oligomer/fibril and cell membrane in amyloid cytotoxicity, FEBS J 277 (2010) 4602–4613. doi: 10.1111/j.1742-4658.2010.07889.x. [DOI] [PubMed] [Google Scholar]
- [11].Khondker A, Alsop RJ, Rheinstädter MC, Membrane-accelerated Amyloid-β aggregation and formation of cross-β sheets, Membranes (Basel). 7 (2017). doi: 10.3390/membranes7030049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gursky O, Aleshkov S, Temperature-dependent beta-sheet formation in beta-amyloid Abeta(1-40) peptide in water: uncoupling beta-structure folding from aggregation., Biochim. Biophys. Acta 1476 (2000) 93–102. doi: 10.1016/s0167-4838(99)00228-9. [DOI] [PubMed] [Google Scholar]
- [13].Morel B, Varela L, Azuaga AI, Conejero-Lara F, Environmental conditions affect the kinetics of nucleation of amyloid fibrils and determine their morphology, Biophys. J 99 (2010) 3801–3810. doi: 10.1016/j.bpj.2010.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Engel MFM, Membrane permeabilization by Islet Amyloid Polypeptide, Chem. Phys. Lipids 160 (2009) 1–10. doi: 10.1016/j.chemphyslip.2009.03.008. [DOI] [PubMed] [Google Scholar]
- [15].a Jayasinghe S, Langen R, Membrane interaction of islet amyloid polypeptide., Biochim. Biophys. Acta 1768 (2007) 2002–9. doi: 10.1016/j.bbamem.2007.01.022. [DOI] [PubMed] [Google Scholar]
- [16].Williams TL, Serpell LC, Membrane and surface interactions of Alzheimer’s A?? peptide - Insights into the mechanism of cytotoxicity, in: FEBS J., 2011: pp. 3905–3917. doi: 10.1111/j.1742-4658.2011.08228.x. [DOI] [PubMed] [Google Scholar]
- [17].Verdier Y, Zarándi M, Penke B, Amyloid beta-peptide interactions with neuronal and glial cell plasma membrane: binding sites and implications for Alzheimer’s disease., J. Pept. Sci 10 (2004) 229–48. doi: 10.1002/psc.573. [DOI] [PubMed] [Google Scholar]
- [18].Khemtémourian L, Killian JA, Höppener JWM, Engel MFM, Recent insights in islet amyloid polypeptide-induced membrane disruption and its role in beta-cell death in type 2 diabetes mellitus., Exp. Diabetes Res 2008 (2008) 421287. doi: 10.1155/2008/421287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Widenbrant MJO, Rajadas J, Sutardja C, Fuller GG, Lipid-induced β-amyloid peptide assemblage fragmentation, Biophys. J 91 (2006) 4071–4080. doi: 10.1529/biophysj.106.085944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Korshavn KJ, Satriano C, Lin Y, Zhang R, Dulchavsky M, Bhunia A, Ivanova MI, Lee YH, La Rosa C, Lim MH, Ramamoorthy A, Reduced lipid bilayer thickness regulates the aggregation and cytotoxicity of amyloid-β, J. Biol. Chem 292 (2017) 4638–4650. doi: 10.1074/jbc.M116.764092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Askarova S, Yang X, Lee JC-M, Impacts of membrane biophysics in Alzheimer’s disease: from amyloid precursor protein processing to aβ Peptide-induced membrane changes., Int. J. Alzheimers. Dis 2011 (2011) 134971. doi: 10.4061/2011/134971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Brender JR, Salamekh S, Ramamoorthy A, Membrane disruption and early events in the aggregation of the diabetes related peptide IAPP from a molecular perspective, Acc. Chem. Res 45 (2012) 454–462. doi: 10.1021/ar200189b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Korshavn KJ, Bhunia A, Lim MH, Ramamoorthy A, Amyloid-β adopts a conserved, partially folded structure upon binding to zwitterionic lipid bilayers prior to amyloid formation, Chem. Commun 52 (2016) 882–885. doi: 10.1039/C5CC08634E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Rodriguez Camargo DC, Korshavn KJ, Jussupow A, Raltchev K, Goricanec D, Fleisch M, Sarkar R, Xue K, Aichler M, Mettenleiter G, Walch AK, Camilloni C, Hagn F, Reif B, Ramamoorthy A, Stabilization and structural analysis of a membrane-associated hIAPP aggregation intermediate, Elife. 6 (2017). doi: 10.7554/eLife.31226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Borch J, Hamann T, The nanodisc: A novel tool for membrane protein studies, Biol. Chem 390 (2009) 805–814. doi: 10.1515/BC.2009.091. [DOI] [PubMed] [Google Scholar]
- [26].Thomaier M, Gremer L, Dammers C, Fabig J, Neudecker P, Willbold D, High-Affinity Binding of Monomeric but Not Oligomeric Amyloid-β to Ganglioside GM1 Containing Nanodiscs, Biochemistry. 55 (2016) 6662–6672. doi: 10.1021/acs.biochem.6b00829. [DOI] [PubMed] [Google Scholar]
- [27].Barnaba C, Sahoo BR, Ravula T, Medina-Meza IG, Im S-C, Anantharamaiah GM, Waskell L, Ramamoorthy A, Cytochrome-P450-Induced Ordering of Microsomal Membranes Modulates Affinity for Drugs, Angew. Chemie - Int. Ed 35294 (2018) 3391–3395. doi: 10.1002/anie.201713167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hagn F, Etzkorn M, Raschle T, Wagner G, Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins, J. Am. Chem. Soc 135 (2013) 1919–1925. doi: 10.1021/ja310901f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Necula M, Kayed R, Milton S, Glabe CG, Small molecule inhibitors of aggregation indicate that amyloid ?? oligomerization and fibrillization pathways are independent and distinct, J. Biol. Chem 282 (2007) 10311–10324. doi: 10.1074/jbc.M608207200. [DOI] [PubMed] [Google Scholar]
- [30].Churches QI, Caine J, Cavanagh K, Epa VC, Waddington L, Tranberg CE, Meyer AG, Varghese JN, Streltsov V, Duggan PJ, Naturally occurring polyphenolic inhibitors of amyloid beta aggregation, Bioorganic Med. Chem. Lett 24 (2014) 3108–3112. doi: 10.1016/j.bmcl.2014.05.008. [DOI] [PubMed] [Google Scholar]
- [31].Osada Y, Hashimoto T, Nishimura A, Matsuo Y, Wakabayashi T, Iwatsubo T, CLAC binds to amyloid beta peptides through the positively charged amino acid cluster within the collagenous domain 1 and inhibits formation of amyloid fibrils., J. Biol. Chem 280 (2005) 8596–605. doi: 10.1074/jbc.M413340200. [DOI] [PubMed] [Google Scholar]
- [32].Assarsson A, Hellstrand E, Cabaleiro-Lago C, Linse S, Charge dependent retardation of amyloid β aggregation by hydrophilic proteins, ACS Chem. Neurosci 5 (2014) 266–274. doi: 10.1021/cn400124r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Webster S, Bonnell B, Rogers J, Charge-based binding of complement component C1q to the Alzheimer amyloid beta-peptide., Am. J. Pathol 150 (1997) 1531–6. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1858209&tool=pmcentrez&rendertype=abstract. [PMC free article] [PubMed] [Google Scholar]
- [34].Sevigny J, Chiao P, Bussière T, Weinreb PH, Williams L, Maier M, Dunstan R, Salloway S, Chen T, Ling Y, O’Gorman J, Qian F, Arastu M, Li M, Chollate S, Brennan MS, Quintero-Monzon O, Scannevin RH, Arnold HM, Engber T, Rhodes K, Ferrero J, Hang Y, Mikulskis A, Grimm J, Hock C, Nitsch RM, Sandrock A, The antibody aducanumab reduces Aβ plaques in Alzheimer’s disease, Nature. 537 (2016) 50–56. doi: 10.1038/nature19323. [DOI] [PubMed] [Google Scholar]
- [35].Du Y, Wei X, Dodel R, Sommer N, Hampel H, Gao F, Ma Z, Zhao L, Oertel WH, Farlow M, Human anti-β-amyloid antibodies block β-amyloid fibril formation and prevent β-amyloid-induced neurotoxicity, Brain. 126 (2003) 1935–1939. doi: 10.1093/brain/awg191. [DOI] [PubMed] [Google Scholar]
- [36].Sudhakar S, Kalipillai P, Santhosh PB, Mani E, Role of Surface Charge of Inhibitors on Amyloid Beta Fibrillation, J. Phys. Chem. C 121 (2017) 6339–6348. doi: 10.1021/acs.jpcc.6b12307. [DOI] [Google Scholar]
- [37].Fernández-De-Retana S, Cano-Sarabia M, Marazuela P, Sánchez-Quesada JL, Garcia-Leon A, Montañola A, Montaner J, Maspoch D, Hernández-Guillamon M, Characterization of ApoJ-reconstituted high-density lipoprotein (rHDL) nanodisc for the potential treatment of cerebral β-amyloidosis, Sci. Rep 7 (2017) 1–13. doi: 10.1038/s41598-017-15215-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Terakawa MS, Yagi H, Adachi M, Lee YH, Goto Y, Small liposomes accelerate the fibrillation of amyloid β(1- 40), J. Biol. Chem 290 (2015) 815–826. doi: 10.1074/jbc.M114.592527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kinoshita M, Kakimoto E, Terakawa MS, Lin Y, Ikenoue T, So M, Sugiki T, Ramamoorthy A, Goto Y, Lee Y-H, Model membrane size-dependent amyloidogenesis of Alzheimer’s amyloid-β peptides, Phys. Chem. Chem. Phys 19 (2017) 16257–16266. doi: 10.1039/C6CP07774A. [DOI] [PubMed] [Google Scholar]
- [40].Amaro M, Šachl R, Aydogan G, Mikhalyov II, Vácha R, Hof M, GM1Ganglioside Inhibits β-Amyloid Oligomerization Induced by Sphingomyelin, Angew. Chemie - Int. Ed 55 (2016) 9411–9415. doi: 10.1002/anie.201603178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Matsuzaki K, How do membranes initiate alzheimers disease? Formation of toxic amyloid fibrils by the amyloid β-protein on ganglioside clusters, Acc. Chem. Res 47 (2014) 2397–2404. doi: 10.1021/ar500127z. [DOI] [PubMed] [Google Scholar]
- [42].Habchi J, Chia S, Galvagnion C, Michaels TCT, Bellaiche MMJ, Ruggeri FS, Sanguanini M, Idini I, Kumita JR, Sparr E, Linse S, Dobson CM, Knowles TPJ, Vendruscolo M, Cholesterol catalyses Aβ42 aggregation through a heterogeneous nucleation pathway in the presence of lipid membranes, Nat. Chem (2018). doi: 10.1038/s41557-018-0031-x. [DOI] [PubMed] [Google Scholar]
- [43].Epand RM, Epand RF, Sayer BG, Melacini G, Palgulachari MN, Segrest JP, Anantharamaiah GM, An Apolipoprotein AI Mimetic Peptide: Membrane Interactions and the Role of Cholesterol, Biochemistry. 43 (2004) 5073–5083. doi: 10.1021/bi049786u. [DOI] [PubMed] [Google Scholar]
- [44].Cukalevski R, Boland B, Frohm B, Thulin E, Walsh D, Linse S, Role of aromatic side chains in amyloid β-protein aggregation, ACS Chem. Neurosci 3 (2012) 1008–1016. doi: 10.1021/cn300073s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Porat Y, Abramowitz A, Gazit E, Inhibition of amyloid fibril formation by polyphenols: Structural similarity and aromatic interactions as a common inhibition mechanism, Chem. Biol. Drug Des 67 (2006) 27–37. doi: 10.1111/j.1747-0285.2005.00318.x. [DOI] [PubMed] [Google Scholar]
- [46].Wang J, Yamamoto T, Bai J, Cox SJ, Korshavn KJ, Monette M, Ramamoorthy A, Real-time monitoring of the aggregation of Alzheimer’s amyloid-β via 1 H magic angle spinning NMR spectroscopy, Chem. Commun 54 (2018) 2000–2003. doi: 10.1039/C8CC00167G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Jo S, Kim T, Iyer VG, Im W, CHARMM-GUI: A web-based graphical user interface for CHARMM, J. Comput. Chem 29 (2008) 1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- [48].Tycko R, Amyloid Polymorphism: Structural Basis and Neurobiological Relevance, Neuron. 86 (2015) 632–645. doi: 10.1016/j.neuron.2015.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Yasuhara K, Arakida J, Ravula T, Ramadugu SK, Sahoo B, Kikuchi JI, Ramamoorthy A, Spontaneous Lipid Nanodisc Fomation by Amphiphilic Polymethacrylate Copolymers, J. Am. Chem. Soc 139 (2017) 18657–18663. doi: 10.1021/jacs.7b10591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Nath A, Trexler AJ, Koo P, Miranker AD, Atkins WM, Rhoades E, Single-Molecule Fluorescence Spectroscopy Using Phospholipid Bilayer Nanodiscs, 2010. doi: 10.1016/S0076-6879(10)72014-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zinser EG, Hartmann T, Grimm MOW, Amyloid beta-protein and lipid metabolism, Biochim. Biophys. Acta - Biomembr. 1768 (2007) 1991–2001. doi: 10.1016/j.bbamem.2007.02.014. [DOI] [PubMed] [Google Scholar]
- [52].Morgantini C, Imaizumi S, Grijalva V, Navab M, Fogelman AM, Reddy ST, Apolipoprotein A-I Mimetic Peptides Prevent Inflammation in a Murine Model of Diabetes, 59 (2010) 3223–3228. doi: 10.2337/db10-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Sherman CB, Peterson SJ, Frishman WH, Apolipoprotein A-I mimetic peptides: A potential new therapy for the prevention of atherosclerosis, Cardiol. Rev 18 (2010) 141–147. doi: 10.1097/CRD.0b013e3181c4b508. [DOI] [PubMed] [Google Scholar]
- [54].Ravula T, Barnaba C, Mahajan M, Anantharamaiah GM, Im S-C, Waskell L, Ramamoorthy A, Membrane environment drives cytochrome P450’s spin transition and its interaction with cytochrome b 5, Chem. Commun 53 (2017) 12798–12801. doi: 10.1039/C7CC07520K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Murakami T, Phospholipid nanodisc engineering for drug delivery systems, Biotechnol. J 7 (2012) 762–767. doi: 10.1002/biot.201100508. [DOI] [PubMed] [Google Scholar]
- [56].Garai K, Crick SL, Mustafi SM, Frieden C, Expression and purification of amyloid-?? peptides from Escherichia coli, Protein Expr. Purif 66 (2009) 107–112. doi: 10.1016/j.pep.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Dasari M, Espargaro A, Sabate R, Lopez Del Amo JM, Fink U, Grelle G, Bieschke J, Ventura S, Reif B, Bacterial Inclusion Bodies of Alzheimer’s Disease β-Amyloid Peptides Can Be Employed To Study Native-Like Aggregation Intermediate States, ChemBioChem. 12 (2011) 407–423. doi: 10.1002/cbic.201000602. [DOI] [PubMed] [Google Scholar]
- [58].Arosio P, Knowles TPJ, Linse S, On the lag phase in amyloid fibril formation, Phys. Chem. Chem. Phys 17 (2015) 7606–7618. doi: 10.1039/C4CP05563B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Zhang Y, I-TASSER server for protein 3D structure prediction, BMC Bioinformatics. 9 (2008). doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Vivekanandan S, Brender JR, Lee SY, Ramamoorthy A, A partially folded structure of amyloid-beta(1-40) in an aqueous environment, Biochem. Biophys. Res. Commun 411 (2011) 312–316. doi: 10.1016/j.bbrc.2011.06.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC, GROMACS: Fast, flexible, and free, J. Comput. Chem 26 (2005) 1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- [62].Huang J, Mackerell AD, CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data, J. Comput. Chem 34 (2013) 2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, De Vries AH, The MARTINI force field: Coarse grained model for biomolecular simulations, J. Phys. Chem. B 111 (2007) 7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- [64].De Jong DH, Singh G, Bennett WFD, Arnarez C, Wassenaar TA, Schäfer LV, Periole X, Tieleman DP, Marrink SJ, Improved parameters for the martini coarse-grained protein force field, J. Chem. Theory Comput 9 (2013) 687–697. doi: 10.1021/ct300646g. [DOI] [PubMed] [Google Scholar]
- [65].Humphrey W, Dalke A, Schulten K, VMD: Visual molecular dynamics, J. Mol. Graph 14 (1996) 33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- [66].San Diego: Accelrys Software Inc., Discovery Studio Modeling Environment, Release 3.5,Accelrys Softw. Inc. (2012). [Google Scholar]
- [67].Schanda P, Brutscher B, Very fast two-dimensional NMR spectroscopy for real-time investigation of dynamic events in proteins on the time scale of seconds, J. Am. Chem. Soc 127 (2005) 8014–8015. doi: 10.1021/ja051306e. [DOI] [PubMed] [Google Scholar]
- [68].Goddard Td., Kneller DG, Sparky 3, Univ. California, San Fr. 14 (2004) 15. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.