Abstract
Background
Neuroblastoma is characterized by substantial clinical heterogeneity. Despite intensive treatment, the survival rates of high-risk neuroblastoma patients are still disappointingly low. Somatic chromosomal copy number aberrations have been shown to be associated with patient outcome, particularly in low- and intermediate-risk neuroblastoma patients. To improve outcome prediction in high-risk neuroblastoma, we aimed to design a prognostic classification method based on copy number aberrations.
Methods
In an international collaboration, normalized high-resolution DNA copy number data (arrayCGH and SNP arrays) from 556 high-risk neuroblastomas obtained at diagnosis were collected from nine collaborative groups and segmented using the same method. We applied logistic and Cox proportional hazard regression to identify genomic aberrations associated with poor outcome.
Results
In this study, we identified two types of copy number aberrations that are associated with extremely poor outcome. Distal 6q losses were detected in 5.9% of patients and were associated with a 10-year survival probability of only 3.4% (95% confidence interval [CI] = 0.5% to 23.3%, two-sided P = .002). Amplifications of regions not encompassing the MYCN locus were detected in 18.1% of patients and were associated with a 10-year survival probability of only 5.8% (95% CI = 1.5% to 22.2%, two-sided P < .001).
Conclusions
Using a unique large copy number data set of high-risk neuroblastoma cases, we identified a small subset of high-risk neuroblastoma patients with extremely low survival probability that might be eligible for inclusion in clinical trials of new therapeutics. The amplicons may also nominate alternative treatments that target the amplified genes.
Neuroblastoma is a pediatric tumor of the sympathetic nervous system, affecting mainly children younger than age five years (1). It is the most common extracranial solid cancer, making up 5% of childhood cancer diagnoses, while accounting for approximately 10% of childhood cancer deaths (2,3). Neuroblastoma is characterized by extensive clinical heterogeneity, illustrated by different clinical evolutions ranging from spontaneous regression to aggressive disease. Therefore, prognostic markers that accurately differentiate low- and high-risk patients are essential. Current risk stratification of neuroblastoma patients is mainly performed according to the International Neuroblastoma Risk Group (INRG) classification system and is based on clinical parameters including age and stage of the disease, histopathological parameters including differentiation status of the tumor, and genetic parameters including MYCN amplification, 11q loss, and the global copy number profile (4,5). Despite intensive multimodal treatment, five-year survival probability within the high-risk group remains disappointingly low (4). Therefore, there is a need for more precise biomarkers for risk stratification that can discriminate patients who will benefit from current high-risk treatment protocols from those with very poor prognosis who might benefit from experimental therapy trials.
For several adult cancer types, transcriptome profiling enabled the identification of new prognostic biomarkers, some of which are currently used in clinical practice (6). Also for neuroblastoma, several studies have shown that classifiers based on gene expression data can predict survival outcome (7–9). However, most published classifiers perform well in the global cohort of neuroblastoma patients but have only limited prognostic value within the high-risk patient subgroup. No gene expression classifier is currently in routine clinical use.
While recurrent single nucleotide mutations, such as those targeting ALK and ATRX, occur in up to 10% of patient tumors at diagnosis (10,11), several copy number aberrations occur at much higher frequency and are strongly associated with disease outcome. Large segmental chromosomal imbalances and focal aberrations are abundant in high-stage tumors, while low-stage tumors typically present with whole-chromosome imbalances (12). More specifically, tumors with only numerical aberrations have a favorable prognosis, while any presence of segmental aberrations is indicative of poor survival outcome (13). However, as the majority of high-risk tumors have segmental aberrations, prognostic stratification based on the absence or presence of segmental aberrations is not applicable within high-risk neuroblastoma. Moreover, no studies have been undertaken so far to identify copy number aberrations that are discriminating patients who will die or survive within the high-risk subgroup.
Therefore, the aim of this study is to investigate whether specific (combinations of) segmental DNA copy number aberrations allow better discrimination of high-risk patients with fatal outcomes. To achieve this with sufficient statistical power, we collected the DNA copy number profiles of 556 high-risk neuroblastoma patients within the Ultra-High-Risk (UHR) working group of the International Neuroblastoma Response Criteria (INRC) consortium (5). Using this unique data set, we aimed to identify genomic aberrations associated with survival outcome in high-risk neuroblastoma patients. First, we attempted to create a genome-wide classifier to stratify high-risk patients. Second, we used this unique large data set to identify single events associated with survival.
Methods
DNA Copy Number Data
DNA copy number data were collected from 671 high-risk neuroblastomas enrolled in the SIOPEN (cohort 1), GPOH (cohort 2), COG (cohort 3), and Japanese (cohort 4) treatment protocols. Parts of the SIOPEN cohort (14,15) and the Japanese cohort (10) have been published previously. After quality control analyses (eg, excluding samples with normal cell contamination and/or silent profiles), 556 samples remained (Table 1; Supplementary Table 1, available online). Data were segmented using SegAnnDB (16) and then converted into a regions-by-samples matrix at 1 kb resolution (using R) (see the Supplementary Methods, available online for details). The data have been deposited in GEO (accession number GSE103123).
Table 1.
Summary of characteristics for the 556 samples included in the study
| Patient cohort | No. (%) |
|---|---|
| MYCN status | |
| Nonamplified | 301 (54.1) |
| Amplified | 255 (45.9) |
| Stage | |
| Non–stage 4 | 41 (7.4) |
| Stage 4 | 515 (92.6) |
| Age, y | |
| <1 | 24 (4.3) |
| 1–1.5 | 45 (8.1) |
| >1.5 | 485 (87.2) |
| NA | 2 (0.4) |
| Treatment cohort | |
| 1 | 159 (28.6) |
| 2 | 122 (21.9) |
| 3 | 207 (37.2) |
| 4 | 68 (12.2) |
Copy Number Aberration Calling
SegAnnDB, a webtool that combines mathematical modeling with visual inspection, was used to identify segments with equal copy number status and to call breakpoints from copy number data. We focused on clonal events by setting platform-dependent cutoffs to call aberrations: gains (aCGH: 0.2, SNP: 0.15), losses (aCGH: –0.3, SNP: –0.25), amplifications (Agilent: 2), and homozygous deletions (–2 for all platforms). This computational pipeline generates a good general view of the aberrations present in the study population but occasionally misses some aberrations (see the Supplementary Methods, available online, for details).
Identifying Prognostic Copy Number Biomarkers
The construction of a genome-wide classifier is described in the Supplementary Methods (available online). To select copy number aberrations associated with overall survival, the R statistical package was used to perform regression analyses for each of the 27 565 regions in the regions-by-samples matrix. The primary end point was binary: death from any cause within 18 months of diagnosis (=case) vs survival with at least five years of follow-up (=control). Logistic regression was performed on 83 case vs 53 control samples from the training set (cohort 1 and 2). A secondary end point was overall survival time, defined as the time from diagnosis until death from any cause, whereby patients who were alive were censored on the date of last contact. Cox proportional hazards regression for overall survival was performed on all 273 training samples (proportionality assumption verified by testing whether Schoenfeld residuals slope equals 0). From logistic and Cox regression analyses, regions with a statistically significant P value (not adjusted for multiple testing) and occurring in at least 5% of the samples were selected (neighboring statistically significant regions were collapsed to one). The selected regions were evaluated in validation cohorts 3 and 4 by assessing survival differences (log-rank test) of patients with and without an aberration in the selected region.
Statistical Analysis and Data Visualization
Kaplan-Meier estimates were calculated with the R package “survival” (default settings). P values reported with Kaplan-Meier plots result from log-rank tests. P values to test co-occurrences result from chi-square tests. All P values are two-sided and tested against an α of .05; multiple testing correction relies on the Benjamini-Hochberg method. More details and additional methods for data visualization are described in the Supplementary Methods (available online).
Results
Exploratory Analysis of Copy Number Aberrations in High-risk Neuroblastoma Patients
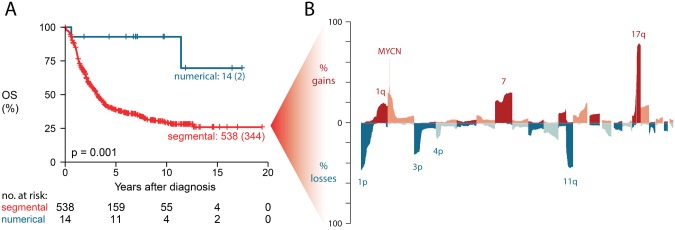
In a first step, we explored the collected data by evaluating copy number aberrations known to be linked to high-risk neuroblastoma disease. Of interest, 2.5% (14/556) of high-risk tumors presented with a copy number profile of only numerical aberrations that is typically observed in low-risk neuroblastoma (12). Kaplan-Meier analysis confirmed a statistically significantly better outcome (P = .001) for this small group of patients, with a 10-year survival probability of 92.9% (95% confidence interval [CI] = 80.3% to 100%) for both overall (Figure 1A) and event-free survival (P < .001) (Supplementary Figure 3, available online). Because these samples represent a different class of genomic aberrations, they were omitted from further analysis. The frequency plot of chromosomal gains and losses in the 542 high-risk tumors with segmental aberrations (Figure 1B) shows recurrent loss of 1p, 3p, 4p, and 11q, gain of 1q, 2p, 7, and 17q, and amplification of MYCN (255/542, 47.0%) at expected frequency levels (12). Comparing the aberration frequencies according to MYCN status confirms known associations (Supplementary Figure 4, available online); that is, MYCN amplification frequently co-occurs with 1p loss (Padjusted < .001), and MYCN-nonamplified cases more frequently present with 3p, 4p, 11q loss and 1q and 17q gain (all Padjusted < .001). Overall, these analyses confirm previous findings on the copy number data of high-risk neuroblastoma tumors (12). A detailed heatmap depicting all aberrations per sample is provided in Supplementary Figure 5 (available online).
Figure 1.
Exploratory analysis of DNA copy number aberrations. A) Overall survival of high-risk patients with numerical DNA copy number profiles compared with patients with segmental profiles, showing two-sided P value of log-rank test. Curve labels represent the number of samples with number of events between brackets. B) Frequency of copy number gains/amplifications (upper part) and losses (lower part) for chromosomes 1 to 22 in 542 high-risk neuroblastoma samples with segmental copy number aberrations. OS = overall survival.
Construction of a Multiregion DNA Copy Number Prognostic Classifier
To construct a prognostic classifier that would discriminate high-risk patients with different outcomes, we compared genomic profiles of high-risk patients of cohorts 1 and 2 with contrasting disease outcome, that is, 83 high-risk patients who died from any cause within 18 months (cases) vs 53 high-risk patients who survived with at least five years of follow-up (controls). From the regions-by-samples matrix, 754 out of 27 565 regions were selected using logistic regression as being associated with survival outcome (Supplementary Table 2, Supplementary Figure 6, available online) and subsequently used to train a classifier with random forests. However, the prognostic value of the classifier could not be validated in cohort 3 (201 samples, P = .66) and cohort 4 (68 samples, P = .63) (Supplementary Figures 7 and 8, available online). The alternative approach, in which one-third of the pooled samples were used for training, also resulted in poor classification of the remaining samples (data not shown). In addition, a classifier built using prognostic regions identified with the random forest algorithm, with Cox regression or a combination of Cox and logistic regression analysis, could not improve the classification performance (data not shown).
DNA Copy Number Breakpoint Counts
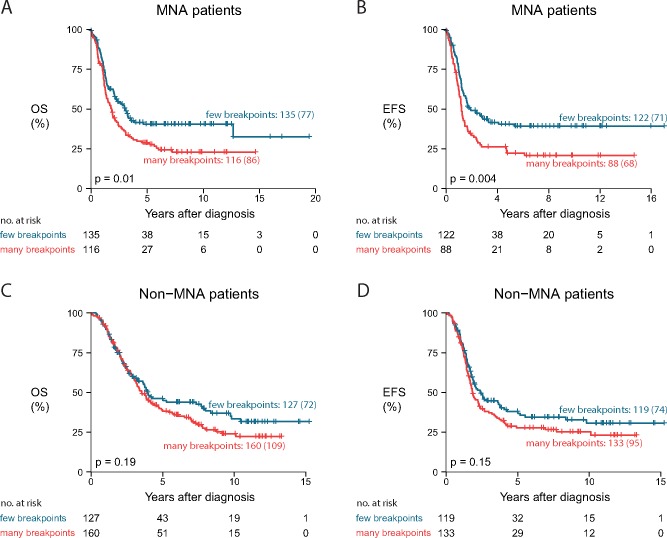
Given previous reports on the association of the number of copy number breakpoints with survival (17), we investigated whether the number of DNA copy number breakpoints is linked to survival outcome in high-risk patients. On average, the copy number profiles in the high-risk cohort contain 10.7 breakpoints, ranging from 1 to 69. A high number of breakpoints (above the median) is associated with worse overall and event-free survival, both in the global cohort (P = .01 for overall survival and P = .005 for event-free survival) (Supplementary Figure 9, available online) and in a subset of MYCN-amplified patients (P = .01 for overall survival and P = .004 for event-free survival) (Figure 2, A and B). Within the subset of MYCN-nonamplified patients, a subtle but statistically nonsignificant difference in survival was observed (P = .19 for overall survival and P = .15 for event-free survival) (Figure 2, C and D).
Figure 2.
Impact of number of breakpoints on patient survival. Comparison of overall survival (A and C) and event-free survival (B and D) of cases with many breakpoints (9–61, above or equal to median) vs cases with a lower number of breakpoints (2–8, below median), within both the subgroup of MYCN-amplified cases (A and B) and the subgroup of MYCN-nonamplified cases (C and D). P values represent two-sided log-rank tests. Curve labels represent the number of samples with the number of events between brackets. EFS = event-free survival; MNA = MYCN-amplified; non-MNA = MYCN-nonamplified; OS = overall survival.
Amplifications in Regions Not Encompassing the MYCN Locus
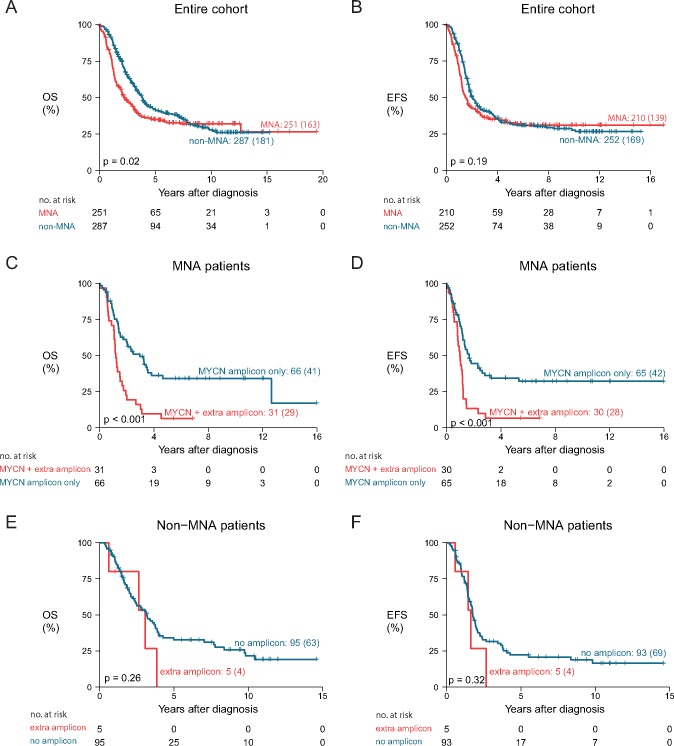
MYCN amplification is an important prognostic biomarker for neuroblastoma patients. In this data set of high-risk patients, the difference in survival of MYCN-amplified vs nonamplified cases is statistically significant (log-rank P = .02), but the proportional hazards assumption is violated, and in the end the survival curves coincide (Figure 3, A and B).
Figure 3.
Association of patient survival with the presence of amplicons (MYCN locus and other loci). A and B) Comparison of survival of patients with and without MYCN amplification, for overall survival (A) and event-free survival (B). C and D) Within the subgroup of patients with MYCN-amplified tumors, comparison of overall (C) and event-free survival (D) of patients with an additional amplicon (not encompassing the MYCN locus) and patients with only the MYCN amplification. E and F) Within the subgroup of patients with MYCN-nonamplified tumors, comparison of overall (E) and event-free (F) survival for patients with an amplicon (not encompassing MYCN) and patients without an amplicon. P values represent two-sided log-rank tests. Curve labels represent the number of samples with the number of events between brackets. EFS = event-free survival; MNA = MYCN-amplified; non-MNA = MYCN-nonamplified; OS = overall survival.
We then questioned whether the presence of amplifications in regions not encompassing the MYCN locus has prognostic potential in high-risk neuroblastoma. Importantly, not all copy number platforms could accurately identify the presence of MYCN amplification (Supplementary Figure 10, available online), agreeing with published findings (18). Copy number data generated on the Agilent array platform showed a large dynamic range at the MYCN locus. Therefore, only the 199 profiles analyzed on this platform (cohort 2 and part of cohort 1) were selected to evaluate the prognostic power of amplicons. Amplicons (log2 ratio > 2), other than those encompassing MYCN, were observed in 36 of these samples (18.1%). Eight regions were recurrently amplified: that is, 2p25.1 encompassing the ODC1 locus (12 samples), 2p23.2 including ALK (5), 2p25.1 including GREB1/NTSR2 (4), 6q16.3 including LIN28B (3), 12q15 including MDM2 (2), 12q13.3/14.1 including CDK4 (2), 11q13.2/13.3 including MYEOV and CCND1 (2), and 5p15.33 including TERT (2). Several other amplicons, including 8q24.21 encompassing the MYC gene, were detected in only one sample. All identified amplicons are summarized in Supplementary Table 3 (available online).
Remarkably, patients with an amplicon other than the MYCN amplicon have a very low 10-year survival probability of 5.8% (95% CI = 1.5% to 22.2%, P < .001). (Supplementary Figure 11, available online). Most of the cases with these amplicons also presented with MYCN amplification, associated with a 10-year overall survival probability of 6.5% (95% CI = 1.7% to 24.7%) (Figure 3C). Only five cases without MYCN amplification presented with another amplicon. Those patients show a 10-year overall survival probability of 0.0% (95% CI not available) (Figure 3E). Event-free survival is depicted in Figure 3, D and F. The presence of an amplicon other than MYCN thus identifies a small subgroup of patients with extremely low survival probability.
On the other hand, the presence of homozygous deletions (26 samples), such as CDKN2A, could not be linked with survival outcome (deletions are listed in Supplementary Table 4, available online).
Distal 6q Losses
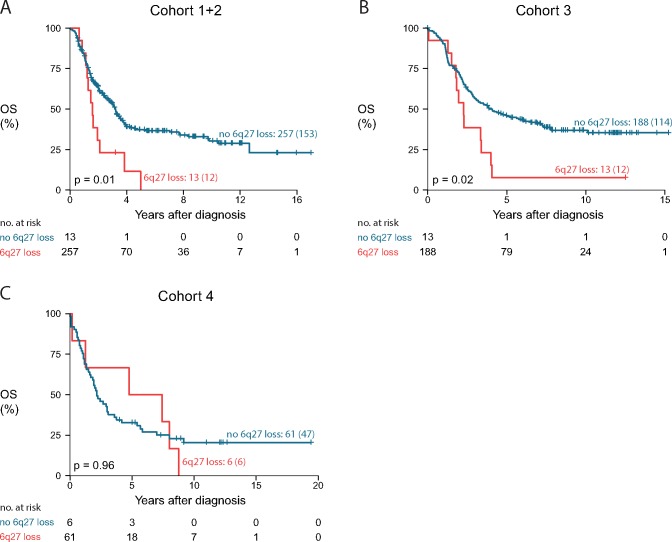
We also investigated whether a single prognostic region identified using Cox or logistic regression analysis could delineate a subgroup of high-risk patients with aggressive disease. Cox regression analysis (on 27 565 genomic regions) in the training set (cohort 1 and 2) identified five genomic aberrations statistically significantly associated with survival outcome including a 7 Mb region at distal 6q (6q27) that could be validated in cohort 3 (P = .02, Padjusted = .09, Benjamini-Hochberg method) (Supplementary Figure 12, available online). While not statistically significant (P = .96), the presence of 6q loss seems also to be associated with survival outcome in cohort 4 as all six cases with a 6q loss died of disease. Thirteen patients out of 273 (4.8%), 13 patients out of 201 (6.5%), and six out of 68 (8.8%) harbored a loss at the distal 6q region in the training cohort, cohort 3, and cohort 4, respectively. These patients showed a 10-year overall survival probability of 0.0% (no 95% CI), 7.7% (95% CI = 1.2% to 50.6%), and 0.0% (no 95% CI), respectively (only two of these patients survived) (Figure 4). Overall survival and event-free survival for combined and individual cohorts are shown in Supplementary Figures 13 and 14 (available online), respectively. Interestingly, two samples with a 6q loss have a focal LIN28B gain or amplification adjacent to the 6q loss, suggesting that a single event generated the amplification and deletion. In addition, we observed that only 25.0% of samples with 6q loss also harbor MYCN amplification and that 6q loss samples have statistically significantly more breakpoints than samples without 6q loss (P = .003) (Supplementary Figure 15, available online). Cox regression analysis with both 6q loss and the number of breakpoints as covariate shows that the presence of 6q loss confers an additional decrease in survival probability, independent from the presence of a high number of breakpoints (above the median; P = .006 and .03 for 6q loss/breakpoints coefficients).
Figure 4.
Association of distal 6q loss with patient survival. Overall survival of patients with a distal 6q loss compared with patients without a distal 6q loss in the training set (cohort 1 + 2) (A) and the two validation sets, cohort 3 (B) and cohort 4 (C). P values represent two-sided log-rank tests. Curve labels represent the number of samples with the number of events between brackets. OS = overall survival.
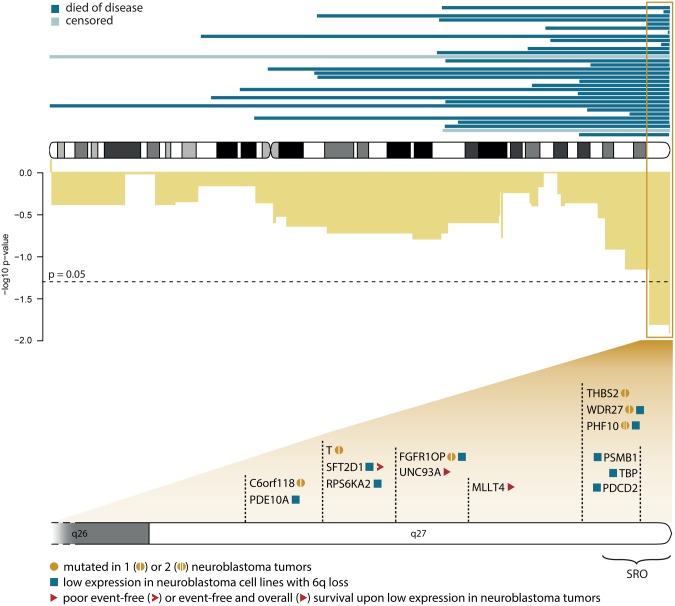
In Figure 5, the 7 Mb region at distal 6q is depicted together with 14 genes that meet at least one of the following criteria: (1) statistically significantly lower expression of the gene in neuroblastoma cell lines with vs without a 6q loss, (2) at least one mutation in the gene as described in 625 primary tumors, and (3) statistically significantly worse survival outcome when gene expression is low (<10th percentile) in 125 high-risk neuroblastoma tumors.
Figure 5.
Detailed view of distal 6q losses. Top: Exact location of the distal 6q deletions found in 32 patients (all cohorts). Censored patients are indicated with a lighter shade. Middle: -log10 of P values (uncorrected) of Cox regression for chromosome 6 in the training set (cohort 1 + 2). Bottom: Genes in the statistically significant region (based on Cox regression) that met at least one of the following criteria: statistically significantly lower expression of the gene in neuroblastoma cell lines with vs without a 6q deletion (squares), at least one mutation in the gene as described in primary tumors (circles), and statistically significantly worse survival outcome when gene expression is low (lowest 10th percentile) in high-risk neuroblastoma tumors (triangles). SRO = smallest region of overlap.
In summary, losses overlapping with the distal 6q region are present in 5.9% of samples and are associated with extremely poor survival outcome (10-year survival probability of 3.4%, 95% CI = 0.5% to 23.3%, P = .002) and pinpoint interesting tumor suppressor candidate genes.
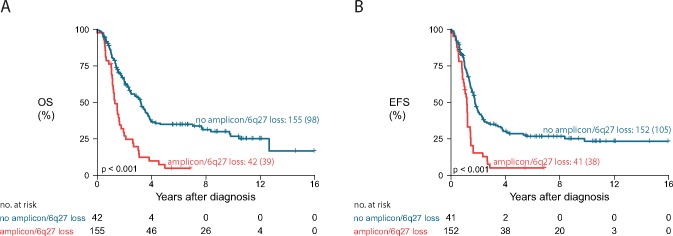
The presence of a distal 6q deletion and/or an amplification distinguish a group of 21.1% of the high-risk patients (within the subgroup tested using Agilent arrays) with poor outcome (Figure 6).
Figure 6.
Survival of a small subset of high-risk patients characterized by the presence of a 6q loss and/or an amplification not encompassing the MYCN locus. Kaplan-Meier and log-rank analyses of the cases with and without these genomic aberrations are depicted for overall survival (A) and event-free survival (B). Only samples analyzed on Agilent arrays are taken into account. P values represent two-sided log-rank tests. Curve labels represent the number of samples with the number of events between brackets. EFS = event-free survival; OS = overall survival.
Discussion
As an international effort of the UHR working group of the INRC consortium, we collected an unprecedentedly large DNA copy number profiling data set of high-risk neuroblastoma tumors. After validation of this unique data set, we investigated different approaches to identify a prognostic multiregion classifier; however, this classifier could not be validated in independent cohorts. As the developed multiregion classifier was biased toward the more abundant aberrations, we shifted our focus to the identification of more infrequent recurrent aberrations that are associated with clinical outcome. Indeed, with this approach, we could distinguish two small subgroups of highly aggressive neuroblastoma disease, that is, patients with tumors that present with an amplicon other than MYCN and patients with tumors with a deletion that encompasses the distal 6q region.
The observation that patients with an amplicon not encompassing MYCN have a 10-year survival probability of only 5.8% might impact therapeutic stratification of neuroblastoma patients in two ways. Not only is the presence of an additional amplicon in the tumor genome a strong prognostic biomarker, it may also predict the sensitivity to small molecule inhibitors that target proteins encoded by genes within the amplified region. Indeed, it has previously been described that ERBB2-amplified breast cancers are more sensitive to trastuzumab (19) and FGFR1-amplified squamous cell lung carcinoma to FGFR1 inhibitors (20). The low survival probability for neuroblastoma patients whose tumors at diagnosis harbor an amplification in a region not encompassing MYCN might justify novel therapeutic approaches such as enrollment of these patients in clinical trials of therapies targeting the amplified genes. Of the 36 patients, 23 present with amplicons encompassing the genes ODC1, ALK, CDK4, MDM2, CCND1, TERT, and MYC, for which inhibitors are currently being tested in clinical trials (21–23). A previous study by Guimier et al. (24) has also investigated the effect of additional amplifications on survival probability, however, in a global cohort of neuroblastoma, also including low-risk cases. The present study only includes high-risk cases and thus underlines the clinical significance of amplifications in this patient subgroup.
A second important observation in this study is the identification of a recurrent but infrequent 6q loss in 32 neuroblastoma tumors collected at diagnosis, associated with a 10-year survival probability of 3.4%. The importance of 6q deletions in aggressive neuroblastoma is further evidenced by a recent study in which five neuroblastoma cases were identified with a distal 6q deletion upon relapse of the tumor (not present in the primary tumor) (25). Moreover, the observation that 22.9% (8/35) of neuroblastoma cell lines (originating from high-risk tumors) harbor a 6q deletion further supports the association of 6q deletion with tumor aggressiveness. Distal 6q deletions have also been described in other diseases such as ovarian cancer and 6q terminal deletion syndrome causing cognitive disorders, both of which suggest important roles for genes localized to 6q27 (26,27).
One of the genes in the 7 Mb statistically significant 6q region is PHF10 (BAF45a), which is a member of the npBAF complex, a SWI-SNF complex specifically active in neuron progenitor cells. The expression of this gene is lower in neuroblastoma cell lines with vs without 6q deletion, and two PHF10 mutations (one probably and one possibly damaging) have been described in neuroblastoma tumors (28). Interestingly, the npBAF complex also contains ARID1B, which has been described to be mutated or deleted in a subset of very aggressive neuroblastoma tumors (29). ARID1B is located on 6q slightly proximal to the 7 Mb statistically significant region and is deleted in 27 of the 32 tumors with 6q deletion.
Our observation that tumors with 6q loss, encompassing PHF10 and/or ARID1B, have a more unstable genome, as indicated by the higher number of breakpoints, agrees with the findings of Dykhuizen et al. (30), which showed that mutations in BAF members such as BRG1 lead to anaphase bridge formation that can ultimately lead to chromosome gains and losses, and Watanabe et al. (31), which showed that cancer cells lacking certain SWI/SNF factors, including ARID1A/B, are deficient in DNA repair and vulnerable to DNA damage. Further scrutinizing the potential tumor-suppressive role of PHF10 or other 6q genes in neuroblastoma disease is needed to better understand their role in tumor progression, therapy resistance, and as a potential target.
Of further interest, our extensive study revealed a number of high-risk neuroblastoma tumors (14 or 2.5%) with only numerical aberrations. Despite the high-risk clinical parameters (all are stage 4, with age at diagnosis between one and four years), these patients with only numerical aberrations have statistically significantly better survival outcome (10-year survival probability = 92.9%), confirming published findings (13).
In addition, we demonstrated that a high number of breakpoints is linked to worse outcome within this high-risk cohort. This observation was previously made in a global neuroblastoma population (17) as well as in children older than 1.5 years with localized unresectable neuroblastoma without MYCN amplification (32). The importance of this observation in high-risk neuroblastoma cases, however, should not be overestimated as the difference in survival is not sufficient to influence clinical decisions.
Although the presented approach to design a classifier to predict the outcome of high-risk disease patients was unsuccessful, other classification methods might be more suitable for this type of data, such as methods based on Bayesian models and support vector machines (33–35) or identifying heterogeneous markers (36). Moreover, focusing on extreme outcomes (case–control study) may not be as useful as anticipated. Another approach would be the integration of other omics-level data such as transcriptomics to build a robust prognostic classifier, as shown in other studies (37–39). In addition, interpretation of survival data could benefit from adding minimal follow-up time to sample selection criteria. However, this is difficult without compromising sample size. A minor limitation of this study is that the full potential of this unique large sample set could not be exploited maximally due to the fact that amplifications could only be correctly identified in samples analyzed on the Agilent platform.
In conclusion, we report that the presence of a distal 6q loss and amplicons not encompassing the MYCN region are strongly predictive of poor outcome within high-risk neuroblastoma. These two observations combined distinguish a group of approximately 20% of high-risk patients with poor outcome. These patients may harbor chemotherapy-resistant tumors, and in case of resistance or progression, these alterations should be taken into account when considering biomarker-based early clinical trials.
Funding
This work was supported by the National Cancer Institute Pediatric and Adolescent Solid Tumor Steering Committee for administrative support. PD was supported by the Fund for Scientific Research Flanders (Research project GOD8815N in the context of ERA-NET Transcan-2 cofound JTC 2014). VB was supported by the ATIP-Avenir Program, the ARC Foundation (grant number ARC-RAC16002KSA-R15093KS), Worldwide Cancer Research Foundation (grant number WCR16-1294 R16100KK), and the “Who Am I?” laboratory of excellence ANR-11-LABX-0071, funded by the French Government through its “Investissement d'Avenir” program operated by the French National Research Agency (ANR; grant number ANR-11-IDEX-0005-02). RC was supported by the Fund for Scientific Research Flanders. MF was supported by the Federal German Ministry of Education and Research (BMBF; grant numbers 01ZX1303A, 01ZX1603A, 01ZX1307D, and 01ZX1607D) and the German Cancer Aid (grant number 110122). RN was supported by the National Health Institute Carlos III and European Regional Development Fund (ISCIII&FEDER), Spain (grant numbers PI14/01008, RD12/0036/0020, and CB16/12/00484). JT was supported by the Deutsche Kinderkrebsstiftung (grant number DKS 2006.10). RV was supported by an ERC Advanced grant. GS received support from the Annenberg Foundation, the Nelia and Amadeo Barletta Foundation (FNAB), and the Association Hubert Gouin Enfance et Cancer. This study was also funded by the Associations Enfants et Santé, Les Bagouz à Manon, Les amis de Claire. Funding was also obtained from SiRIC/INCa (grant number INCa-DGOS-4654), from the CEST of Institute Curie, and PHRC (grant number IC2007-09).
Notes
Affiliations of authors: Center for Medical Genetics, Ghent University, Ghent, Belgium (PD, RC, FS, KDP); Cancer Research Institute Ghent, Ghent, Belgium (PD, RC, FS, KDP); Institut Cochin, Inserm U1016, CNRS UMR 8104, Université Paris Descartes UMR-S1016, Paris, France (VB); Institut Curie, Inserm U900, Mines ParisTech, PSL Research University, Paris, France (VB); Department of Human Genetics, McGill University, Montreal, Quebec, Canada (TDH); Data Mining and Modelling for Biomedicine Group, VIB Center for Inflammation Research, Ghent, Belgium (RC); Children's Cancer Research Institute, Vienna, Austria (IMA, PFA, RL, UP); Department of Pediatrics, Medical University of Vienna, Vienna, Austria (IMA, PFA, RL); Division of Hematology/Oncology, Children's Hospital Los Angeles, University of Southern California Keck School of Medicine, Los Angeles, CA (SA); Division of Oncology (EFA, MDH, JMM) and Center for Childhood Cancer Research (EFA, JMM), Children's Hospital of Philadelphia, Philadelphia, PA; Department of Pediatrics (EFA, JMM), Perelman School of Medicine (MDH), University of Pennsylvania, Philadelphia, PA; Centre Léon-Bérard, Laboratoire de Recherche Translationnelle, Lyon, France (VC); Department of Pathology (RD, KM) and Laboratory of Molecular Biology (LV), Istituto Giannina Gaslini, Genova, Italy; Department of Experimental Pediatric Oncology (MF), Department of Pediatric Oncology and Hematology (BH), and Department of Experimental Pediatric Oncology (JT), University Children's Hospital Cologne, Medical Faculty, and Center for Molecular Medicine Cologne (MF), University of Cologne, Cologne, Germany; Division of Hematology-Oncology, Hospital for Sick Children, University of Toronto, Toronto, ON, Canada (MSI); Department of Oncogenomics, Academic Medical Center, University of Amsterdam, Amsterdam, the Netherlands (JK, RV); Department of Pediatrics, Duke University School of Medicine, Durham, NC (SK); Genetic Somatic Unit (EL) and U830 INSERM, Recherche Translationnelle en Oncologie Pédiatrique (RTOP) and Department of Pediatric Oncology (GS), Institut Curie, Paris, France (EL); Department of Pediatric Hematology and Oncology, Ghent University Hospital, De Pintelaan, Ghent, Belgium (GL); Dana-Farber/Boston Children's Cancer and Blood Disorders Center, Harvard Medical School, Boston, MA (WBL); Saga Medical Center KOSEIKAN, Saga, Japan (AN); Pathology Department, Medical School, University of Valencia, Valencia, Spain (RN); Medical Research Foundation INCLIVA, Valencia, Spain (RN); CIBERONC, Madrid, Spain (RN); Research Institute for Clinical Oncology Saitama Cancer Center, Saitama, Japan (MO); Seattle Children's Hospital and University of Washington, Seattle, WA (JRP); Laboratory of Neuroblastoma, Onco/Haematology Laboratory, University of Padua, Pediatric Research Institute (IRP)-Città della Speranza, Padova, Italy (GPT); Institut Gustave Roussy, Université Paris Sud, Paris, France (DVC); Department of Pediatrics, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA (JMM); Abramson Family Cancer Research Institute, Philadelphia, PA (JMM).
The funding sources had no role in the design of the study, the collection, analysis, or interpretation of the data, the writing of the manuscript, or the decision to submit the manuscript.
EFA reports a grant from National Cancer Institute/National Institutes of Health and employment at Janssen R&D. MF reports honoraria at Novartis. BH reports grants from German Cancer Aid. JRP reports honoraria from Bristol Myer Squibb. The other authors have no disclosures.
Supplementary Material
References
- 1. Westermann F, Schwab M.. Genetic parameters of neuroblastomas. Cancer Lett. 2002;1842:127–147. [DOI] [PubMed] [Google Scholar]
- 2. Smith MA, Seibel NL, Altekruse SF, et al. Outcomes for children and adolescents with cancer: challenges for the twenty-first century. J Clin Oncol. 2010;2815:2625–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bosse KR, Maris JM.. Advances in the translational genomics of neuroblastoma: From improving risk stratification and revealing novel biology to identifying actionable genomic alterations. Cancer. 2016;1221:20–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cohn SL, Pearson AD, London WB, et al. The International Neuroblastoma Risk Group (INRG) classification system: An INRG Task Force report. J Clin Oncol. 2009;272:289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Park JR, Bagatell R, Cohn SL, et al. Revisions to the International neuroblastoma response criteria: A consensus statement from the National Cancer Institute clinical trials planning meeting. J Clin Oncol. 2017;3522:2580–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nalejska E, Mączyńska E, Lewandowska MA.. Prognostic and predictive biomarkers: Tools in personalized oncology. Mol Diagn Ther. 2014;183:273–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Oberthuer A, Berthold F, Warnat P, et al. Customized oligonucleotide microarray gene expression–based classification of neuroblastoma patients outperforms current clinical risk stratification. J Clin Oncol. 2006;2431:5070–5078. [DOI] [PubMed] [Google Scholar]
- 8. Vermeulen J, De Preter K, Naranjo A, Vercruysse L.. Predicting outcomes for children with neuroblastoma using a multigene-expression signature: A retrospective SIOPEN/COG/GPOH study. Lancet Oncol. 2009;107:663–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. De Preter K, Vermeulen J, Brors B, Delattre O.. Accurate outcome prediction in neuroblastoma across independent data sets using a multigene signature. Clin Cancer Res. 2010;165:1532–1541. [DOI] [PubMed] [Google Scholar]
- 10. Chen Y, Takita J, Choi Y, et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;4557215:971–974. [DOI] [PubMed] [Google Scholar]
- 11. Cheung N-KVK, Zhang J, Lu C, et al. Association of age at diagnosis and genetic mutations in patients with neuroblastoma. JAMA. 2012;30710:1062–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vandesompele J, Baudis M, De Preter K, et al. Unequivocal delineation of clinicogenetic subgroups and development of a new model for improved outcome prediction in neuroblastoma. J Clin Oncol. 2005;2310:2280–2299. [DOI] [PubMed] [Google Scholar]
- 13. Janoueix-Lerosey I, Schleiermacher G, Michels E, et al. Overall genomic pattern is a predictor of outcome in neuroblastoma. J Clin Oncol. 2009;277:1026–1033. [DOI] [PubMed] [Google Scholar]
- 14. Coco S, Theissen J, Scaruffi P, et al. Age-dependent accumulation of genomic aberrations and deregulation of cell cycle and telomerase genes in metastatic neuroblastoma. Int J Cancer. 2012;1317:1591–1600. [DOI] [PubMed] [Google Scholar]
- 15. Fieuw A, Kumps C, Schramm A, et al. Identification of a novel recurrent 1q42.2-1qter deletion in high risk MYCN single copy 11q deleted neuroblastomas. Int J Cancer. 2012;13011:2599–2606. [DOI] [PubMed] [Google Scholar]
- 16. Hocking TD, Boeva V, Rigaill G, et al. SegAnnDB: Interactive Web-based genomic segmentation. Bioinformatics. 2014;3011:1539–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schleiermacher G, Janoueix-Lerosey I, Ribeiro A, et al. Accumulation of segmental alterations determines progression in neuroblastoma. J Clin Oncol. 2010;2819:3122–3130. [DOI] [PubMed] [Google Scholar]
- 18. Curtis C, Lynch AG, Dunning MJ, et al. The pitfalls of platform comparison: DNA copy number array technologies assessed. BMC Genomics. 2009;10:588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mass RD, Press MF, Anderson S, et al. Evaluation of clinical outcomes according to HER2 detection by fluorescence in situ hybridization in women with metastatic breast cancer treated with trastuzumab. Clin Breast Cancer. 2005;63:240–246. [DOI] [PubMed] [Google Scholar]
- 20. Weiss J, Sos ML, Seidel D, et al. Frequent and focal FGFR1 amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;262:62–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wagner AH, Coffman AC, Ainscough BJ, et al. DGIdb 2.0: Mining clinically relevant drug-gene interactions. Nucleic Acids Res. 2016;44(D1):D1036–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sholler GL, Gerner EW, Bergendahl G, et al. A phase I trial of DFMO targeting polyamine addiction in patients with relapsed/refractory neuroblastoma. PLoS One. 2015;105:e0127246 http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0127246. Accessed March 2, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Burgess A, Chia KM, Haupt S, Thomas D, Haupt Y, Lim E.. Clinical overview of MDM2/X-targeted therapies. Front Oncol. 2016;6:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guimier A, Ferrand S, Pierron G, et al. Clinical characteristics and outcome of patients with neuroblastoma presenting genomic amplification of loci other than MYCN. PLoS One. 2014;97:e101990 http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0101990. Accessed September 19, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Eleveld TF, Oldridge DA, Bernard V, Koster J.. Relapsed neuroblastomas show frequent RAS-MAPK pathway mutations. Nat Genet. 2015;478:864–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cooke IE, Shelling AN, Meuth LV.. Allele loss on chromosome arm 6q and fine mapping of the region at 6q27 in epithelial ovarian cancer. Genes Chromosomes Cancer. 1996;154:223–233. [DOI] [PubMed] [Google Scholar]
- 27. Peddibhotla S, Nagamani SC, Erez A, et al. Delineation of candidate genes responsible for structural brain abnormalities in patients with terminal deletions of chromosome 6q27. Eur J Hum Genet. 2015;231:54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S.. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;453:279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sausen M, Leary R, Jones S, et al. Integrated genomic analyses identify ARID1A and ARID1B alterations in the childhood cancer neuroblastoma. Nat Genet. 2012;451:12–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dykhuizen EC, Hargreaves DC, Miller EL, et al. BAF complexes facilitate decatenation of DNA by topoisomerase IIα. Nature. 2013;4977451:624–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Watanabe R, Ui A, Kanno S, Ogiwara H, Nagase T.. SWI/SNF factors required for cellular resistance to DNA damage include ARID1A and ARID1B and show interdependent protein stability. Cancer Res. 2014;749:2465–2475. [DOI] [PubMed] [Google Scholar]
- 32. Defferrari R, Mazzocco K, Ambros IM, et al. Influence of segmental chromosome abnormalities on survival in children over the age of 12 months with unresectable localised peripheral neuroblastic tumours without MYCN amplification. Br J Cancer. 2015;1122:290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guha S, Ji Y, Baladandayuthapani V.. Bayesian disease classification using copy number data. Cancer Inform. 2014;13(Suppl 2):83–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Riccadonna S, Jurman G, Merler S, Paoli S, Quattrone A, Furlanello C.. Supervised classification of combined copy number and gene expression data. J Integrat Bioinformatics. 2007;43:168–185. [Google Scholar]
- 35. Carter L, Rothwell DG, Mesquita B, et al. Molecular analysis of circulating tumor cells identifies distinct copy-number profiles in patients with chemosensitive and chemorefractory small-cell lung cancer. Nat Med. 2017;231:114–119. [DOI] [PubMed] [Google Scholar]
- 36. Ronde JJ, Rigaill G, Rottenberg S, Rodenhuis S, Wessels LF.. Identifying subgroup markers in heterogeneous populations. Nucleic Acids Res. 2013;4121:e200 https://academic.oup.com/nar/article/41/21/e200/1283469. Accessed February 22, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Horlings HM, Lai C, Nuyten D, Halfwerk H.. Integration of DNA copy number alterations and prognostic gene expression signatures in breast cancer patients. Clin Cancer Res. 2010;162:651–663. [DOI] [PubMed] [Google Scholar]
- 38. Ross-Adams H, Lamb AD, Dunning MJ, Halim S.. Integration of copy number and transcriptomics provides risk stratification in prostate cancer: A discovery and validation cohort study. EBioMedicine. 2015;29:1133–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Taskesen E, Babaei S.. Integration of gene expression and DNA-methylation profiles improves molecular subtype classification in acute myeloid leukemia. BMC Bioinformatics. 2015;16(Suppl 4):S5 https://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-16-S4-S5. Accessed February 21, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.