Abstract
2018 marks the 100-year anniversary of the 1918 influenza pandemic, which killed ~50 million people worldwide. The severity of this pandemic resulted from a complex interplay between viral, host, and societal factors. Here, we review the viral, genetic and immune factors that contributed to the severity of the 1918 pandemic and discuss the implications for modern pandemic preparedness. We address unresolved questions of why the 1918 influenza H1N1 virus was more virulent than other influenza pandemics and why some people survived the 1918 pandemic and others succumbed to the infection. While current studies suggest that viral factors such as haemagglutinin and polymerase gene segments most likely contributed to a potent, dysregulated pro-inflammatory cytokine storm in victims of the pandemic, a shift in case-fatality for the 1918 pandemic toward young adults was most likely associated with the host's immune status. Lack of pre-existing virus-specific and/or cross-reactive antibodies and cellular immunity in children and young adults likely contributed to the high attack rate and rapid spread of the 1918 H1N1 virus. In contrast, lower mortality rate in in the older (>30 years) adult population points toward the beneficial effects of pre-existing cross-reactive immunity. In addition to the role of humoral and cellular immunity, there is a growing body of evidence to suggest that individual genetic differences, especially involving single-nucleotide polymorphisms (SNPs), contribute to differences in the severity of influenza virus infections. Co-infections with bacterial pathogens, and possibly measles and malaria, co-morbidities, malnutrition or obesity are also known to affect the severity of influenza disease, and likely influenced 1918 H1N1 disease severity and outcomes. Additionally, we also discuss the new challenges, such as changing population demographics, antibiotic resistance and climate change, which we will face in the context of any future influenza virus pandemic. In the last decade there has been a dramatic increase in the number of severe influenza virus strains entering the human population from animal reservoirs (including highly pathogenic H7N9 and H5N1 viruses). An understanding of past influenza virus pandemics and the lessons that we have learnt from them has therefore never been more pertinent.
Keywords: influenza, pandemic, 1918, host factors, viral factors, external factors, societal factors, prevention
Introduction
In 1918 a mysterious and deadly disease spread around the world in three consecutive waves (spring 1918, autumn 1918, and winter 1918–19). This pandemic infected over one third of the world's population and killed an estimated 50 million people (Johnson and Mueller, 2002; Murray et al., 2006), with unusually severe clinical manifestations in previously healthy young adults (Collins, 1931; Hoffman, 2011). In 1918, the etiological agent that caused this disease was unknown (Hildreth, 1991). However, we now know that these events represented the largest influenza virus pandemic on record: the catastrophic 1918 influenza pandemic. Since 1918, the world has experienced three additional influenza pandemics: the 1957 “Asian” influenza pandemic, the 1968 “Hong Kong” influenza pandemic and the 2009 so-called “swine flu” pandemic. These pandemics, although mild in comparison to that of 1918, highlight the constant threat that influenza virus poses to human health. Given that almost 100 years have passed since 1918, it behooves us to ask: are we truly better prepared for the next influenza virus pandemic or are there still lessons to be learned? This review gives an overview of lessons learned from the 1918 influenza pandemic, highlighting new insights into our understanding of viral pathogenesis and their impact on our preparedness for the next outbreak of influenza.
The origins 1918 influenza virus
The 1918 influenza pandemic is often colloquially referred to as the “Spanish” influenza pandemic. However, it is unlikely that the 1918 influenza virus originated in Spain. Instead, influenza cases were widely reported in Spain due to the fact that, as a neutral country in World War I, Spain did not practice censorship in the press. In contrast, other countries involved in the war, such as Germany, Britain and France, most likely limited the news of this deadly pandemic, so as not to lower the moral of the troops and raise questions about their military readiness (Johnson, 2006). Today, the general consensus is that the 1918 influenza virus originated in the Midwest of the United States of America (Barry, 2004). Medical records reported the first cases of “influenza of a severe type” around March 1918 in military camps in Kansas (Barry, 2004). From here, the virus is thought to have spread throughout the United Stated and then transported by American troop ships to the battlefields of France, where it gradually spread throughout Europe and the rest of the world (Patterson and Pyle, 1991). The spread of the virus beyond port cities was further facilitated by local transport networks, predominately railways (Patterson and Pyle, 1991; Johnson, 2006). However, it is possible that the predecessor of this killer virus first entered human population prior to 1918 and became more virulent and/or more transmissible over time. Unusual influenza activity was already reported in the United States and several European countries before the first (spring) wave of the 1918 influenza outbreak (Frost, 1919; Johnson, 2006; Hoffman, 2011). Military camps in France already reported influenza infections accompanied with high mortality in the winter of 1916–17 (Hammond et al., 1917), which was followed 2 months later by a similar outbreak near London at Aldershot, one of Britain's biggest military camps (Oxford et al., 1999, 2002, 2005). Interestingly, no records of civilian influenza cases around that time exist, possibly because influenza cases were not recorded at the time or because they got lost with time. Alternatively, it is tempting to speculate that military camps, with their high population density, close proximity to livestock, high mobility, and large number of people with pre-existing lung conditions (due to exposure to toxic gasses in the trenches) served as the perfect breeding ground for the emergence of this catastrophic pandemic (Oxford et al., 2005).
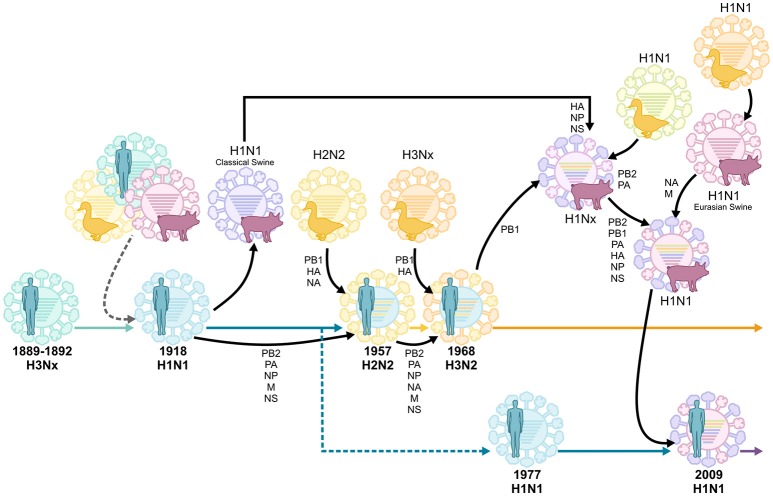
Just as the geographic origins of the 1918 virus remain unclear, the original animal reservoir of the virus also remains controversial. As a segmented virus, influenza virus is capable of undergoing the process of reassortment. Reassortment occurs when two influenza virus strains co-infect the same cell, facilitating the emergence of a new “reassortant” virus which contains a novel constellation of genes. Reassortment between avian and human influenza viruses gave rise to the 1957 and 1968 influenza pandemics (Figure 1; Scholtissek et al., 1978; Kawaoka et al., 1989; Schäfer et al., 1993). In contrast, the 2009 influenza pandemic resulted from a reassortment event between avian, human and swine influenza viruses (Figure 1; Garten et al., 2009; Smith et al., 2009b). Unlike these more recent influenza pandemics, the 1918 virus is thought to have been directly introduced in the human population (i.e., in the absence of reassortment) from a single unidentified host (Taubenberger et al., 2005). This notion is supported by the fact that the 8 individual gene segments of the 1918 virus appear to have co-evolved in the same host. However, the exact identity of this host remains unclear, as the nucleotide sequence of the virus is genetically distinct to all other known avian and mammalian influenza viruses (Reid et al., 2004a,b; Taubenberger et al., 2005). In contrast, others argue that the 1918 influenza virus could have indeed originated from a reassortment event between avian and mammalian, possibly swine and/or human, influenza viruses in the years prior to the 1918 pandemic (Smith et al., 2009a; Worobey et al., 2014). Unfortunately, in the absence of influenza virus sequence data in the years preceding the 1918 pandemic, this question may never be definitively answered.
Figure 1.
Reassortment events of historic pandemic influenza A viruses, adapted from van de Sandt et al. (2015b). Historic serum analysis suggests that the Russian influenza pandemic of 1889–1892 was of the H3Nx subtype and seasonally circulated up to the 1918 influenza pandemic. It remains undefined whether the 1918 H1N1 pandemic virus originated from multiple reassortment events between avian, swine and human influenza viruses, or if it was introduced by a direct zoonotic transmission event of an avian, swine or other influenza virus. The H1N1 virus continued to circulate, causing seasonal epidemics, until 1957 when it reassorted with an avian H2N2 virus. This virus circulated until 1968, when it reassorted again with the avian H3Nx virus, which has caused seasonal epidemics ever since. In 1977 the H1N1 virus was reintroduced in the human population and co-circulated with H3N2 viruses until the influenza pandemic of 2009 when it was replaced by another H1N1 virus which was the result of multiple reassortment events between avian, swine, and human influenza viruses.
A broad spectrum of clinical disease
During the 1918 influenza pandemic, a broad spectrum of clinical illness was observed (Brundage and Shanks, 2008). In the first spring wave of the pandemic, disease was typically mild and mortality rates were not unusually high (Johnson and Mueller, 2002). However, there was a surprisingly large number of young adults who were affected by the outbreak (Ahmed et al., 2007). The second or autumn wave of influenza emerged in late August 1918 and by the end of 1918 almost no country was spared (Patterson and Pyle, 1991; Johnson, 2006). The striking feature of the autumn wave was its unprecedented virulence (Taubenberger et al., 2001). Patients typically suffered from a high fever, cyanosis, and fluid accumulation in the lungs (Johnson, 2006). In ~5% of the fatal cases, death occurred rapidly after the onset of clinical symptoms (i.e., within 3 days), although for the majority of cases the time from clinical symptoms to death was ~7–10 days (Brundage and Shanks, 2008). The third and final wave of influenza emerged at the start of 1919 (Beveridge, 1977). This wave was typically not as virulent as the fall wave and it did not affect every country (Beveridge, 1977; Taubenberger and Morens, 2006). During the course of the pandemic, ~500 million people worldwide were infected, resulting in a case-fatality rate of >2.5% (Johnson and Mueller, 2002; Johnson, 2006; Taubenberger and Morens, 2006). While this fatality rate was certainly higher than other influenza virus pandemics (Taubenberger and Morens, 2006), these data indicate that influenza virus infection was not always fatal and that a large number of people survived the infection. These data raise two intriguing questions: (i) Why was the 1918 influenza pandemic more virulent than other influenza pandemics of the twentieth century; and (ii) Why did some people survive the pandemic and others succumb to the infection?
Viral factors associated with the severity of the 1918 influenza pandemic
It was not until 1933, more than a decade after the devastating pandemic of 1918–19, when the influenza virus was first isolated and demonstrated to be the causative agent of seasonal influenza virus infections (Smith et al., 1933). However, even then, an in-depth understanding of the viral factors that contributed to the severity of the 1918 pandemic was thwarted by the absence of any available biological material from the virus in question. Finally, in the late 1990s the virus' genetic material was successfully isolated from formalin-fixed, paraffin-embedded lung tissue from 1918 influenza victims and from the lungs of a 1918 influenza victim buried in Alaska's permafrost (Taubenberger et al., 1997; Reid et al., 1999). These efforts unraveled a partial viral sequence from four viruses and the complete genomic sequence of one virus (Reid et al., 1999). The fully reconstructed 1918 influenza virus proved to be highly pathogenic in mice (Tumpey et al., 2005), ferrets (Memoli et al., 2009), and macaques (Kobasa et al., 2007). Interestingly, a recent study in ferrets demonstrated that the 1918 influenza virus could spread to, and induce cytokine responses in tissues outside the respiratory tract, which likely contributed to the severity of the infection (de Wit et al., 2018) and could explain the neurological complications observed during the 1918 influenza pandemic (Alexander, 1919; Ravenholt and Foege, 1982). Various reverse genetics experiments suggest that the high pathogenicity exerted by the 1918 influenza virus was most likely an interplay between different virulence factors, in which proteins encoded by the viral haemagglutinin (HA) and polymerase gene segments played a crucial role (Kobasa et al., 2004; Tumpey et al., 2005; Kash et al., 2006; Conenello et al., 2007; Pappas et al., 2008; Watanabe et al., 2009; Jagger et al., 2010).
One of the best-known virulence determinants of influenza virus is the presence of a multibasic cleavage site in the HA (Horimoto and Kawaoka, 1994; Subbarao et al., 1998). In avian species, influenza viruses without a multibasic cleavage site require the HA to be cleaved by host trypsin-like proteases for infection. Trypsin-like proteases are commonly found in the respiratory tract, thus limiting replication of these viruses to these tissues. However, the presence of the multibasic cleavage site means that these viruses can be cleaved by ubiquitously expressed proteases. The presence of a multibasic cleavage site in modern highly pathogenic avian influenza viruses can be associated with increased virulence in mammalian hosts (Schrauwen et al., 2012; Suguitan et al., 2012). However, none of the available 1918 HA sequences contained a multibasic cleavage site (Kawaoka and Webster, 1988; Taubenberger et al., 1997; Reid et al., 1999). Instead, analysis of the HA sequence of the 1918 viruses revealed that these viruses were adapted to bind to human epithelial cells. The influenza A virus HA protein requires only one amino acid change in order to switch binding to α-2,3-linked sialic acids (typically found on avian cells) to binding to the α-2,6-linked sialic acids (typically found on human epithelial cells in the upper respiratory tract) (Glaser et al., 2005). Compared to victims from the spring wave, an increased incidence of this mutation in the HA was observed in viruses isolated from victims of the more severe autumn wave (Reid et al., 2003; Glaser et al., 2005; Sheng et al., 2011). A second mutation, which strengthens the virus binding to the human receptor, could only be found in some of the 1918 HA sequences (Reid et al., 2003; Sheng et al., 2011). These data suggest that at least two H1N1 influenza viruses circulated in 1918, which differed in their binding affinity for the human receptor. Both viruses displayed a similar cell tropism in the respiratory tract of terminal stage human influenza victims (Sheng et al., 2011). However, this secondary adaptation is essential for effective transmission of the 1918 influenza virus between ferrets (Tumpey et al., 2007). Mutations in other gene segments, including PB2, PA, and PB1-F2 of the 1918 influenza virus can also play a role in host adaptation (Dunham et al., 2009; Jagger et al., 2010; Mehle et al., 2012; Mazel-Sanchez et al., 2018). In the absence of additional influenza virus sequences from 1918, it is hard to establish whether these or other mutations contributed to the dramatic increase in case-fatality seen during the autumn wave of the pandemic (Simonsen et al., 2018).
Gain- and loss-of-function experiments, such as those described above, have provided important insights as to how novel influenza viruses adapt to the human population (Subbarao et al., 1993; Mehle and Doudna, 2009; Herfst et al., 2012; Imai et al., 2012; Belser et al., 2013; Richard et al., 2013; Zhu et al., 2013; Watanabe et al., 2014). Specifically, this information is used to evaluate the pandemic potential of novel influenza viruses, including avian H7N9 and H5N1 viruses, which are frequently crossing the species barrier into the human population. The Global Influenza Surveillance and Response System (GISRS), a surveillance program that monitors which influenza virus strains circulate at a given time (Hay and McCauley, 2018; World Health Organization, 2018a), uses this information for a rapid risk assessment when a potentially pandemic virus is reported to circulate in animals (predominately birds or swine) or has crossed the species barrier into the human population. It is hoped that extensive surveillance activities, in combination with rapid clinical diagnosis, will afford us a “head start” in the case of a future influenza pandemic. However, the success of such surveillance programs is contingent upon their geographical breadth (Krammer et al., 2018). International cooperation and support for influenza surveillance will become even more pertinent in the future as climate changes continues to affect animal reservoirs and avian migration patterns, both of which could lead to the spread of influenza viruses to new locations and across a wider range of avian species (Klaassen et al., 2012; Shaman and Lipsitch, 2013; Audubon, 2018).
Host factors associated with variations in influenza morbidity and mortality in 1918
The 1918 influenza pandemic is notorious for its high morbidity and mortality rates. However, it is important to recognize that there were substantial variations in mortality, both within and between countries (Mills, 1986; Johnson and Mueller, 2002; Johnson, 2006). General estimations assume an overall death rate of 2.5–5 per 1,000 individuals worldwide. Although this might be an accurate estimate for some countries [e.g., Australia (2.8/1000), Austria (3/1000), Demark (4.1/1000)], it represents an overestimation for some countries [e.g., Argentina (1.2/1000), Uruguay (1.4/1000), American Samoa (0/1000)], and a gross underestimation of others [e.g., Nauru (160/1000), Western Samoa (236/1000), Cameroon (445/1000)] (Johnson and Mueller, 2002; Johnson, 2006). These data indicate that in addition to viral factors, host factors had a major impact on the outcome of infection.
Age
An individual's age played a major role in determining one's risk of death during the 1918 influenza pandemic. Typically, when the mortality rates of seasonal influenza are graphed against the age of the population, a “U” shaped curve is produced, as the highest mortality occurs in the very young and old (Johnson, 2006). In contrast, pandemic outbreaks (to various degrees) are characterized by a shift in case-fatality toward younger age groups (Simonsen et al., 1998; Olson et al., 2005; Ahmed et al., 2007; Georgantopoulos et al., 2009). This was particularly pronounced during 1918 pandemic when young adults (15–30 years) displayed such usually high mortality rate that a “W” shaped mortality curve was produced (Olson et al., 2005; Ahmed et al., 2007; Shanks and Brundage, 2012). The underlying mechanisms driving this mortality shift are not fully understood but are likely to be associated with the host's immune status.
Immunopathology
The high mortality rates observed in young adults during the 1918 pandemic has traditionally been attributed to the induction of an aberrant, dysregulated pro-inflammatory response (often referred to as a “cytokine storm”). This hypothesis is based upon experimental studies in various animal models using the reconstructed 1918 influenza virus. These experimental studies showed that the 1918 influenza virus triggered a potent, dysregulated pro-inflammatory response, which likely contributed to the severe lung lesions observed in victims of the 1918 influenza pandemic (Kobasa et al., 2004, 2007; Kash et al., 2006; Memoli et al., 2009; de Wit et al., 2018). Indeed, this dysregulated immune response has also been observed in natural and experimental infections with both the highly pathogenic avian H5N1 virus and the 2009 pandemic influenza virus (de Jong et al., 2006; To et al., 2010). However, it is important to note that all experimental 1918 influenza virus studies to date have been performed in immunologically-naïve animals. This is not necessarily indicative of the human situation in 1918, as influenza viruses caused epidemics and pandemics prior to 1918 (Dowdle, 1999; Johnson, 2006; Morens and Fauci, 2007; Morens et al., 2009; Valleron et al., 2010). It can therefore be assumed that a large proportion of the human population in 1918, with the possible exception of isolated countries/communities, would have encountered at least one previous influenza virus infection, resulting in pre-existing humoral and cellular immunity. It remains unclear whether such pre-existing immunity would cross-react with the 1918 H1N1 virus, and if so, whether it would enhance or dampen any dysregulated pro-inflammatory response in young adults.
Humoral immune response
In contrast to young adults, older adults (aged 30–60 years) fared significantly better during the 1918 pandemic (Luk et al., 2001). This observation is likely to reflect the beneficial effects of pre-existing humoral immunity. It is theorized that an H1 and/or N1 influenza virus circulated in the human population prior to 1889, when it was replaced by a H3 influenza virus that caused the so-called “Russian” influenza pandemic (1889–1892) (Ahmed et al., 2007). Accordingly, individuals born before 1889 (i.e., those 30 years or older during the 1918 pandemic) would have had cross-protective antibodies, while people born after 1889 would have been immunological naïve to the 1918 H1N1 pandemic virus (Dowdle, 1999; Ahmed et al., 2007). The lack of pre-existing 1918 influenza virus-specific or cross-reactive antibodies in children and young adults likely contributed to the high attack rate and rapid spread of the virus (Ahmed et al., 2007). Only people infected during the first “spring” wave of the pandemic acquired a protective immune response against the second, more virulent, “fall” wave of the 1918–19 pandemic (Gibbon, 1919; Shope, 1958; Palmer and Rice, 1992; Barry et al., 2008; Mathews et al., 2010; Shanks et al., 2010, 2011b; Fraser et al., 2011). Interestingly, unlike the majority of elderly populations worldwide, elderly populations in remote settings, including Indigenous Australians, Alaskan Natives, and Latin Americans, experienced high mortality during the 1918 pandemic. This most likely reflects the fact that these remote populations were not exposed to the previously circulating influenza viruses that conferred cross-protection (Ahmed et al., 2007).
Conclusive evidence that protective influenza virus-specific antibody responses are indeed long-lived came from the 2009 influenza pandemic. Here, elderly people who were exposed to the 1918 influenza virus (or its immediate descendant), 60–90 years prior to the pandemic of 2009, were protected from infection and severe disease, as they maintained the antibody response that cross-reacted with the 2009 pandemic strain (Yu et al., 2008; Hancock et al., 2009; Ikonen et al., 2010; Reed and Katz, 2010).
Interesting, a recent study suggested that individuals medically-treated for influenza-like illness in the years prior to the 1918 pandemic (1916–1918) were actually at an increased risk of having clinically significant respiratory illness during the autumn wave of the 1918 pandemic (Shanks et al., 2016a). Similarly, the presence of cross-reactive but non-neutralizing antibodies, was associated with immune complex deposition and increased disease severity during the 2009 influenza pandemic (Monsalvo et al., 2011). These data suggest that protection against disease is dependent not just upon the presence of pre-existing antibodies, but rather their ability to neutralize the influenza virus strain in question.
Cellular immune response
The issue of prior influenza virus exposure in the general population prior to 1918 raises the question as to why a pre-existing cellular immune response, in particular cross-reactive CD8+ T cells, offered so little protection to young adults during the 1918 influenza pandemic?
A robust CD8+ T cell response plays an important role in protection against novel influenza virus strains and subtypes. Unlike antibodies, CD8+ T cells can recognize the internal proteins of influenza virus. Since these internal proteins do not undergo rapid antigenic change, CD8+ T cells are able to provide cross-protection against a broad range of different influenza virus strains. Accordingly, pre-existing influenza virus specific CD8+ T cells provided protection against severe disease during the influenza pandemics of both 1957 and 2009 (Slepushkin, 1959; McMichael et al., 1983; Epstein, 2006; Sridhar et al., 2013; Hayward et al., 2015). In addition, seasonally induced influenza virus-specific CD8+ T cells can cross-react with novel potentially pandemic avian influenza viruses (Kreijtz et al., 2008; Lee et al., 2008; van de Sandt et al., 2014) and facilitate more rapid recovery in patients following infection with low pathogenic H7N9 avian influenza virus (Wang et al., 2015). The presence of conserved CD8+ T cell peptides in the viral protein sequences of the 1918 influenza virus (Quiñones-Parra et al., 2014) and the ability the of the 2009 H1N1 pandemic influenza virus to recall influenza virus-specific CD8+ T cells, cross-reacting with the 1918 H1N1 influenza virus (Gras et al., 2010) suggest that pre-existing CD8+ T cells should have been protective against severe infection with the 1918 H1N1 influenza virus, especially in the case of young adults. CD8+ T cells may not have been optimal in very young children (age 0–4 years) due lack of exposure to previous influenza viruses (Bodewes et al., 2011a; Sauerbrei et al., 2014). Similarly, immunosenescence may have impaired CD8+ T cell function in the elderly (>65 years) (Goronzy and Weyand, 2013). This may partially explain the high mortality observed in the youngest and oldest age groups during seasonal epidemics (U-shaped curve). However, individuals between 15 and 65 years of age, who suffered the greatest burden of disease during the 1918 pandemic, are thought to display the “gold-standard” immune response, with optimal cross-reactive CD8+ T cell responses. The absence of protective immunity in this age group is unlikely to be due to the fact that heterosubtypic immunity is short-lived (Mathews et al., 2010), as the longevity of influenza virus-specific CD8+ T cells in healthy individuals has recently been demonstrated (van de Sandt et al., 2015a). It is possible that the recall of pre-existing influenza virus-specific CD8+ T cell responses was not rapid enough for the extremely virulent 1918 pandemic virus, causing rapid appearance of clinical disease and death within 3 days (Ahmed et al., 2007). Alternatively, pandemic H1N1 influenza viruses (1918 and 2009) may have suppressed immunogenic RIPK3-driven dendritic cell death needed for the induction of an effective CD8+ T cell response (Hartmann et al., 2017). In addition, it is likely that ethnically defined genetic variations in HLA molecules influenced cross-reactive CD8+ T cell responses in influenza virus infected individuals (Quiñones-Parra et al., 2014). This (combined with other socio-economic factors) would leave some ethnicities, like Alaskan Natives and Indigenous Australians, more susceptible to severe influenza virus infections. Indeed, alarmingly high morbidity and mortality rates were observed amongst these populations during the pandemics of 1918 (Ahmed et al., 2007) and 2009 (La Ruche et al., 2009; Flint et al., 2010). Similarly, it is striking to note that the matrix protein of the 1918 virus already contained extra-epitopic amino acid residues that were associated with evasion from the pre-existing influenza virus CD8+ T cells (van de Sandt et al., 2015b), a phenomenon not observed in the comparatively mild 2009 pandemic influenza virus (van de Sandt et al., 2018a,b).
Finally, it is important to note that the highest influenza virus infection rates in 1918 were observed among children of school age (5–15 years). However, this increased infection rate occurred in the absence of high morbidity (Shanks and Brundage, 2012; Mamelund et al., 2016). Thus, school-aged children are thought to be in a “honeymoon period” of superior immunity, whereby they display increased resistance to a variety of different bacterial and viral pathogens (Ahmed et al., 2007). However, the mechanisms underlying such superior immunity are completely unexplored, despite the fact that it holds key information for inducing effective immune responses to influenza viruses.
In this review, we would like to propose an additional hypothesis that might have influenced the effectiveness of the cross-reactive cellular immune response and possibly contributed to the disproportional mortality amongst young adults in 1918; namely immune suppression as a resulting from recent measles infections (Moss et al., 2004; Griffin, 2010; de Vries et al., 2012; Mina et al., 2015). Measles epidemics were frequently reported at the end of the nineteenth and in the early twentieth century (Cliff et al., 1983; Duncan et al., 1997; Shulman et al., 2009; Shanks et al., 2011a, 2014), including a large measles outbreak in the US military camps in the winter of 1917–1918 (Shanks et al., 2014; Morens and Taubenberger, 2015). The elderly population would have experienced measles in their childhood and their immunity would have protected them from contracting measles in the years prior to the 1918 influenza pandemic. However, children and young adults, without prior measles infections, would have been immunologically susceptible to measles in the years preceding the 1918 influenza pandemic. Recent studies have demonstrated that the measles virus infects memory T lymphocytes, resulting in apoptosis and a prolonged state of immune suppression up to 3 years after the initial measles infection (Moss et al., 2004; Griffin, 2010; de Vries et al., 2012; Mina et al., 2015). Influenza virus-specific CTL responses could have been suppressed in young individuals who had to endure a measles infection in the years prior to the 1918 influenza pandemic, which may have increased their susceptibility to a severe influenza infection. The combination of recovering from immunosuppression and an infection with an unexpectedly highly virulent virus might have contributed to severe inflammatory related pathology in a mechanism better known as the immune reconstitution inflammatory syndrome (IRIS) (Hirsch et al., 2004; Morens and Fauci, 2007; Shulman et al., 2009; Barber et al., 2012). Whether recent measles infections indeed led to immunosuppression of the influenza virus-specific T cell responses, resulting in a higher susceptibility for severe influenza virus infections and potential IRIS or alternatively contributed to dampening CD8+ T cell immunopathology remains an important area of future research. Fortunately, measles vaccines are now widely available and have greatly reduced the prevalence of measles worldwide (Moss and Griffin, 2012; Perry et al., 2014; Mina et al., 2015). However, the increased number of measles outbreaks in recent years and declining vaccination rates represent a key point of concern for future influenza virus pandemics.
Together, these data demonstrate that an individual's age (and the associated differences in their immune response) played an important role in determining disease outcome in the context of pandemic influenza virus infections. In 2009, age and pre-existing humoral immunity were taken into account when identifying priority individuals for vaccination. In 2009, the elderly population were less susceptible to severe influenza (Dawood et al., 2012) as they were protected through cross-reactive antibodies and CD8+ T cells acquired during previous seasonal infections, including the antigenically related A/H1N1 virus that circulated prior to 1957 (Yu et al., 2008; Hancock et al., 2009; Ikonen et al., 2010). Based on these findings, the first limited 2009 influenza vaccine stocks were administered to younger individuals, instead of being misdirected to the traditional high risk group: the elderly (National Center for Respiratory Diseases, CDC, and Centers for Disease and Prevention (CDC), 2009). Improving cross-reactive CD8+ T cell responses to influenza vaccinations and natural infections remains a key research priority for the future (Clemens et al., 2018). This includes an understanding of CD8+ T cell functionality in ethnically diverse populations and different age groups (Clemens et al., 2018).
Genetic differences
In addition to the role of humoral and cellular immunity, there is a growing body of evidence to suggest that individual genetic differences contribute to differences in the severity of influenza virus infections. For example, during the 2009 influenza pandemic several single-nucleotide polymorphisms (SNPs) were strongly associated with severe pneumonia. These included SNPs in the genes for interferon response factor 7 (Ciancanelli et al., 2015), Fc fragment of immunoglobulin G, low-affinity IIA, receptor (Zúñiga et al., 2012), RPA interacting protein (Zúñiga et al., 2012), complement component 1q subcomponent binding protein (Zúñiga et al., 2012), CD55 (Zhou et al., 2012), IL-1α (Liu et al., 2013), IL-1β (Liu et al., 2013), surfactant protein B gene (To et al., 2014) and interferon induced transmembrane protein 3 (Everitt et al., 2012; Zhang et al., 2013), and IRF9 (Hernandez et al., 2018). Unfortunately, there is insufficient information available to conclude whether mortality variations in 1918 were influenced by any of the aforementioned SNPs. Defining which SNPs confer increased susceptibility to severe influenza remains an important aspect of influenza pandemic preparedness, as it will help to inform which populations are most at risk of severe disease.
Malnutrition
Host nutritional status has long been recognized as an important factor in the outcome of a variety of different infectious diseases (Cohen, 2000). In India in 1918, the effects of malnutrition and famine on influenza severity were particularly pronounced. The 1918 influenza pandemic hit India during a widespread drought, which affected the viability of many important food crops (Mills, 1986). Consequently, many Northern-Western, Western and Central Indian provinces experienced a famine during 1918 (Mills, 1986). It was these provinces which also experienced the highest 1918 influenza mortality rates (Mills, 1986). Due to the unusual age distribution of the pandemic, those who succumbed to the disease were typically young adults who formed the majority of the agricultural labor force (Mills, 1986). The resultant labor shortage only served to exacerbate the severity of the influenza pandemic (Mills, 1986). The exact mechanisms by which malnutrition and famine increase the severity of influenza remain to be defined. However, experimental studies suggest that not only does malnutrition suppress the host's immune response to influenza virus, but that it may also facilitate the emergence of novel viral variants, which display increased pathogenicity relative to the original parental strain (Beck et al., 2004).
Undernutrition (often exacerbated by ongoing civil conflicts) remains a problem for influenza pandemics of the twenty-first century and beyond. Indeed, chronic malnutrition was thought to have contributed to the high morbidity and mortality seen in Guatemalan children during the 2009 influenza pandemic (Reyes et al., 2010). Climate change may result in crop failures and exacerbate any food shortages in the future. However, in any future influenza virus pandemic, we will face a “double burden” of malnutrition whereby a proportion of the world's population will experience severe disease because of undernutrition and a proportion of the world's population will experience severe disease because of overnutrition. Specifically, it is now well accepted that obesity increases one's risk of being hospitalized with, and dying from, an influenza virus infection (Morgan et al., 2010; Louie et al., 2011; Van Kerkhove et al., 2011). Perhaps of even greater concern is the fact that obesity inhibits both virus-specific CD8+ T cell responses and antibody responses to the seasonal influenza vaccine (Sheridan et al., 2012). The challenge for future influenza pandemics is therefore not only to protect those affected by undernutrition (in particularly in light of the growing problem of climate change), but also the growing number of people living with obesity.
Underlying infections
Co-infection with bacterial pathogens
Historical autopsy reports and examination of lung tissue sections from 1918 to 19 influenza case material indicated that for a significant number of patients, the cause of death was not primary viral pneumonia (Brundage and Shanks, 2008; Morens et al., 2008; Chien et al., 2009). Instead, these individuals succumbed to a secondary bacterial infection, most commonly pneumonia caused by bacteria such as Streptococcus pneumoniae, Haemophilus influenzae, Staphylococcus aureus, and Streptococcus pyogenes (Morens et al., 2008). H. influenzae was so frequently observed in influenza patients that it was often cited as the cause of the pandemic (and was thus named accordingly) (Hildreth, 1991). The role of secondary bacterial infections during the 1918 pandemic is consistent with epidemiological observations that while influenza virus attack rates in 1918 were similar among soldiers and civilians, mortality rates were much higher amongst newly arrived soldiers (Shanks et al., 2016b). The unhygienic circumstances in the army camps led to frequent bacterial infections, especially amongst immunologically naïve new army recruits. Thus, following an influenza virus infection, new army recruits were more likely to develop a lethal secondary bacterial pneumonia than civilians or long-serving soldiers (Shanks et al., 2010, 2016b). These observations have been echoed by numerous experimental animal studies, showing that co-infection with influenza virus and bacterial pathogens results in increased disease severity compared to infection with either pathogen alone (Brightman, 1935; Glover, 1941; Francis and de Torregrosa, 1945; Harford et al., 1946; Wilson et al., 1947; Short et al., 2012a, 2013). Different mechanisms have been proposed to explain this viral-bacterial synergism (McCullers, 2006; McAuley et al., 2007; Smith et al., 2013; Hrincius et al., 2015). These include, but are not limited to, reduced mucociliary clearance of inhaled bacteria following influenza virus infection, bacterial adhesion to the basement membrane (Morens et al., 2008; Taubenberger et al., 2012; Chertow and Memoli, 2013) and/or sialic acids exposed by influenza virus (McCullers and Bartmess, 2003; Peltola et al., 2005), viral alterations to the host immune response (Navarini et al., 2006; van der Sluijs et al., 2006; Ballinger and Standiford, 2010; Nakamura et al., 2011; Ellis et al., 2015; Lee et al., 2015) and the bacterial inhibition of epithelial cell repair following initial damage by influenza virus infection (Kash et al., 2011). Importantly, experimental studies suggest that influenza viruses not only increases the severity of secondary bacterial infections, but that it also increases the transmission of S. pneumoniae (Diavatopoulos et al., 2010; Short et al., 2012b).
In addition to co-infections with bacterial pathogens such as S. pneumoniae, chronic bacterial infections, such as those with Mycobacterium tuberculosis, contributed to variations in influenza mortality during the 1918 pandemic. For example, data from a Swiss sanatorium during the 1918 pandemic suggested that the risk of influenza death was higher among tuberculosis (TB) patients than non-TB controls (Oei and Nishiura, 2012). Similarly, individuals with TB were 2.2 times more likely to contract the 1918 influenza virus than non-TB individuals living in the same household (Noymer and Garenne, 2000; Noymer, 2011). A synergistic relationship between M. tuberculosis and influenza viruses has also been supported by experimental studies (Redford et al., 2014). The predominance of TB amongst young adults in 1918 may have contributed to the striking “W shaped” mortality curve associated with the 1918 influenza pandemic (Oei and Nishiura, 2012).
Severe complications and morbidity as a result of bacterial co-infections were not unique to the 1918 influenza pandemic. Rather, bacterial co-infections were also observed in the influenza pandemics of 1957, 1968 and 2009, albeit to a lesser extent than in 1918 (Oswald et al., 1958; Robertson et al., 1958; Louria et al., 1959; Oseasohn et al., 1959; Chertow and Memoli, 2013; Joseph et al., 2013). In the 2009 influenza pandemic, TB was also identified as a risk factor for the development of severe disease (Morales et al., 2017). Thankfully, unlike in 1918, the severity of bacterial infections during these more recent influenza pandemics was likely minimized by the use of antibiotics, advanced medical care and the availability of bacterial vaccines [such as the pneumococcal polysaccharide and H. influenzae type b (Hib) vaccine] (Oswald et al., 1958; Robertson et al., 1958; Louria et al., 1959; Madhi et al., 2004; Wahl et al., 2018). However, as the rate of antibiotic resistance continues to rise and as pathogens such as methicillin-resistant S. aureus (MRSA) (Memoli et al., 2008) and multidrug-resistant M. tuberculosis (Zumla et al., 2013; Millard et al., 2015) become more common, we potentially face a future where antibiotics will be ineffectual in the treatment of bacterial infections. This will have direct and severe implications for any future influenza virus pandemic (Memoli et al., 2008). It must be considered an urgent priority to not only minimize antibiotic resistance, but also to invest in the discovery of new antibiotics and alternative treatment options for bacterial infections.
Malaria
In addition to individuals with bacterial co-infections, mortality during the 1918 influenza pandemic was considerably higher amongst malaria-infected individuals (Langford and Storey, 1992; Afkhami, 2003; Shanks, 2015). Although the underlying mechanism is not fully understood, a malaria-induced procoagulant state could play a role in increasing inflammation and subsequent clinical outcome (Shanks, 2015).
Today, chemopreventive strategies have lowered the disease burden associated with malaria and new eradication strategies are being developed. However, malaria still causes significant worldwide morbidity and mortality, there is ever increasing drug resistance and new malaria vaccines have yet to provide long-lasting benefits at a population level (Ashley et al., 2018). Until effective control measures have been developed and implemented, areas endemic for malaria remain at high risk for increased mortality during the next influenza pandemic.
Non-pharmaceutical interventions
In 1918, a variety of different approaches were employed to limit the spread of influenza virus and treat infected patients. While many of these methods were of little or no avail, they contain important lessons for influenza pandemic preparedness in the twenty-first century.
Maritime quarantine
In 1918, when the severity of the second wave of influenza became apparent, many countries imposed strict quarantine measures on all incoming ships to try and prevent the spread of influenza (Johnson, 2006). For the most part, these attempts were unsuccessful (Johnson, 2006). Quarantine measures were either implemented too late and the virus was already present within the country or quarantine was breached by infected individuals who were not yet symptomatic (Crosby, 1976; Tomkins, 1992). Thus, countries such as the U.K. and South Africa dismissed maritime quarantine as impractical and ineffectual (Blakely, 2006; Johnson, 2006). However, Australia imposed the maritime quarantine before any victims of the second wave were reported. All arriving vessels had to be cleared by Commonwealth Quarantine Officials before disembarking. This quarantine protected Australia from the second wave of the pandemic until December 1918 when the quarantine was finally breached. Maritime quarantine thus helped to protect Australia from the worst of the pandemic (Crosby, 2003; Johnson, 2006) and indirectly contributed to protecting certain Pacific Islands that depended on Australian supply ships (Shanks et al., 2018).
The most striking example of this was the mortality difference between American Samoa and Western Samoa. A strict maritime quarantine was imposed in American Samoa by the U.S. Governor in 1918 (Shanks and Brundage, 2013). This quarantine prevented influenza from entering the country, and no deaths from the 1918 influenza were ever recorded in America Samoa (Johnson, 2006; Shanks and Brundage, 2013). This was in sharp contrast to the nearby Western Samoa (located ~100 km away), which did not practice strict maritime quarantine (Tomkins, 1992; Shanks and Brundage, 2013). As a result, Western Samoa was infected by the New Zealand supply ship, the Talune, and it is estimated that influenza killed more than a quarter of the population (Tomkins, 1992).
Global transportation has experienced a major transformation in the last century, with ships being replaced by the faster and more widely used air travel. The rise of commercial air travel helps explains the rapid global spread of the more modern influenza pandemics of 1957, 1968, and 2009 in the absence of major military troop movements (Rvachev and Longini, 1985; Hufnagel et al., 2004; Khan et al., 2009; Bajardi et al., 2011; Lemey et al., 2014). Accordingly, maritime quarantine is unlikely to play a role in limiting the spread of any future influenza pandemic. However, in 2009 authorities tried to limit the spread of influenza by using the modern-day equivalent of maritime quarantine: airport arrival screening. Unfortunately, analysis of arrival passengers at Sydney airport in 2009 suggested that airport screening had only a sensitivity of 6.67% for detecting influenza-infected patients, while costing ~$50,000 AUD per case detected (Gunaratnam et al., 2014). This limited efficacy likely reflects the fact that individuals infected with influenza virus can be contagious prior to becoming symptomatic (Hollingsworth et al., 2006). Airport arrival screening is therefore unlikely to control the spread of influenza by international air travel. Rather, advancing modeling (e.g., identifying which travel routes are most vulnerable to disease spread) and a variety of different education campaigns (e.g., raising awareness amongst the general public about the risks of traveling if they have been exposed to an infected individual) are likely to play a more significant role in future pandemic preparedness.
Mass gatherings
In addition to limiting maritime travel, in 1918 most cities implemented simple non-pharmaceutical interventions to restrict the viral spread. These included imposing restrictions on social gatherings where person-to-person transmission could occur. As a result, schools, theaters, churches, and dance halls were closed, while mass gatherings such as weddings and funerals were banned in order to prevent overcrowding (Frost, 1919; Johnson, 2006; Bootsma and Ferguson, 2007; Hatchett et al., 2007). The peak death rate was lower in cities that rapidly implemented these non-pharmaceutical interventions within a few days after the first local cases were recorded, compared to those which waited a few weeks to respond (Bootsma and Ferguson, 2007; Hatchett et al., 2007). The timing when these interventions were lifted also affected the overall mortality (Bootsma and Ferguson, 2007; Hatchett et al., 2007). Thus, while restrictions on gatherings of people helped reduce influenza virus transmission, as soon as these restrictions were relaxed (typically within 2–8 weeks of their implementation) efficient viral transmission recommenced (Hatchett et al., 2007).
Following the outbreak of the 2009 pandemic influenza virus in Mexico, an 18-day period of mandatory school closure was implemented in the greater Mexico City area (Chowell et al., 2011). This was associated with a 29–37% reduction in influenza transmission (Chowell et al., 2011). Similarly, in Hong Kong there was approximately a 25% reduction influenza virus transmission following secondary schools closures from June 11 to July 10, 2009 (Wu et al., 2010). However, just as in 1918, the duration of these intervention strategies affected their efficacy, and there was a dramatic increase influenza activity in 32 Mexican states in the autumn of 2009, a period which coincided with schools opening for the autumn term (Chowell et al., 2011).
Facemasks and hygiene
Facemasks were a popular preventative measure employed during the 1918 pandemic. While people were unsure of the etiological agent of the pandemic, the consensus was that it was an airborne disease and wearing a facemask would prevent infections (Crosby, 1976). Accordingly, many cities and regions, including Guatemala City, San Francisco, and certain prefectures of Japan, made wearing a facemask in public places obligatory, and special task forces and education campaigns were established to enforce this regulation (Crosby, 1976; Rice and Palmer, 1993; Rice, 2011). However, in order for a facemask to be at least partially effective against influenza virus it must be (i) worn at all times, (ii) properly made and fitted, and (iii) made of appropriate material. The surgical gauze masks of 1918 often failed to meet these criteria (Crosby, 1976). Thus, the mortality rate of Ontario, Canada (where wearing a mask was voluntary) was not significantly different from Alberta, Canada, (where mask wearing was enforced by law) (MacDougall, 2007). In fact, influenza deaths in Alberta continued to rise even after mask wearing was sanctioned by law, suggesting that in 1918 wearing a facemask was not sufficient to prevent deaths from influenza (World Health Organization Writing Group et al., 2006).
Proper hygiene (e.g., frequent hand washing) would also have helped limiting the spread of the influenza virus during the 1918 pandemic, as influenza viruses are transmissible via hand to face contact (World Health Organization Writing Group et al., 2006; Thomas et al., 2014). Thus, the Japanese traditional attitude to disease and illness might have contributed to a lower national pandemic mortality in 1918–19, as Japanese children are taught to remove their shoes and wash their hands upon re-entering the home (Rice and Palmer, 1993).
In the context of modern influenza pandemics, facemasks and handwashing/hand sanitizers have been used as preventative, non-pharmaceutical interventions. However, during the 2009 influenza pandemic for the most part, the use of facemasks was not obligatory (CDC, 2009). Rather, the CDC only recommended facemasks for individuals at increased risk of severe illness from influenza and/or individuals who were the direct careers of persons infected with the pandemic virus (CDC, 2009). Moreover, the effectiveness of facemasks in preventing the transmission of influenza virus remains unclear (Cowling et al., 2010) and just as was observed in 1918, low public compliance significantly limits the utility of facemasks in a modern pandemic setting (Cowling et al., 2010). Perhaps such interventions will be of greatest relevance to medical personal, who serve in the front line of a pandemic and are at high risk for infection. In contrast, handwashing and the use of hand sanitizers (whether or not in combination with wearing a facemask) had a clear protective effect during the influenza pandemic of 2009 (Larson et al., 2012; Suess et al., 2012; Wong et al., 2014).
These data suggest that non-pharmaceutical interventions such as social distancing, handwashing/hand sanitizers, and facemasks in any future influenza pandemic may buy valuable time before vaccines become widely available. However, the success of these interventions will depend upon their early and continuous implementation and also people's willingness to comply. The 1918 pandemic has shown that measures are most effective when they are voluntary, as people have low tolerance for mandatory health measures (Spinney, 2017). Indeed, a behavioral study showed that individuals were more likely to wear a facemask when they received autonomy-supportive advice as compared to controlled instructions (Chan et al., 2015).
Medical interventions, therapies and vaccines: then and now
The 1918 influenza pandemic occurred during a period in history when controlling infectious diseases had become a realistic goal of the medical profession (Tomkins, 1992; Johnson, 2006). Public health initiatives had already proven successful in limiting the spread of diseases such as cholera and TB (Hildreth, 1991; Tomkins, 1992; Tognotti, 2003). Thus, there was initially little to suggest that an influenza outbreak could not be effectively controlled (Hildreth, 1991; Tomkins, 1992; Tognotti, 2003). However, despite the dramatic advances in microbiology in the previous decades, the etiological agent of the 1918 influenza pandemic remained a mystery. In the absence of clear information about the causative agent of the pandemic, a range of different therapeutic and preventative treatments were attempted. People experimented with medications (including Asprin) and homemade remedies such as mustard poultice, quinine, tobacco, beef tea, the inhalation of zinc sulfate, opium, salt water, and alcohol (Rice and Palmer, 1993; Johnson, 2006; Starko, 2009; Keeling, 2010). Traditional eastern medicine, like the Japanese Kanpo medicine (consisting of herbal remedies accompanied by green tea) may have had some beneficial effect by stimulating perspiration (helping to reduce fever), improving vitamin C levels and replacing lost fluids (Palmer and Rice, 1992; Rice and Palmer, 1993). Similarly, the use of Traditional Chinese Medicine may have reduced the severity of influenza infections in at least some individuals (Kobayashi et al., 1999; Cheng and Leung, 2007; Chen et al., 2011). However, for the most part little was available in terms of effective therapeutic and/or prophylactic treatments. Nursing care actually proved to have contributed to the recovery of patients, especially those suffering from secondary bacterial pneumonia (Robinson, 1990; Rice and Palmer, 1993). In addition, mortality rates were significantly higher in places deprived from nursing care, e.g., mining compounds (Phimister, 1973; Rice and Palmer, 1993). Unfortunately, during the 1918–19 pandemic many of those who typically performed these duties were either serving overseas or were sick themselves (Crosby, 2003; Keeling, 2010; Shanks et al., 2011b).
Today, the identification of the etiological agent of influenza has dramatically improved diagnostic speed and accuracy. Rapid and highly accurate molecular diagnostic techniques have largely replaced the labor intensive and time consuming “gold standard” cell culture method for diagnosing influenza virus infections (Ellis and Zambon, 2002), which allows for rapid isolation of infected individuals. Furthermore, risk assessment of potentially pandemic viruses has greatly improved by screening the viral genome of human and animal virus isolates for the presence of mutations that increase human adaptation and/or virulence.
In addition, we are able to deploy both anti-viral drugs and vaccines in the case of an influenza virus pandemic. Antivirals (such as the neuraminidase inhibitors oseltamivir and zanamivir) can be used as a therapeutic in severely ill patients, while also being employed prophylactically in outbreak situations (Cooper et al., 2003; Hayden et al., 2004; De Clercq, 2006; Zambon, 2014; Krammer et al., 2018). At present, potentially pandemic influenza viruses (such as avian H7N9 and H5N1 viruses) are sensitive to both oseltamivir and zanamivir (Herfst et al., 2012). However, acquired resistance to oseltamivir has been observed in several H5N1 isolates (De Clercq, 2006). Similarly, oseltamivir resistance is known to emerge in H7N9 viruses within just 2 days from the start of treatment (Hay and Hayden, 2013). These data suggest that in the case of any future influenza virus pandemic, antivirals should be used judiciously, and the emergence of drug-resistant viral variants closely monitored.
Influenza virus vaccines have also played a major role in reducing the morbidity and mortality associated with seasonal influenza. Unfortunately, antibodies elicited by seasonal influenza vaccines do not provide protection in the case of an antigenically distinct influenza virus of a novel subtype, such as A/H5N1 or A/H7N9 (De Jong et al., 2000). Furthermore, current inactivated seasonal influenza vaccines may even prevent the induction of cross-reactive CD8+ T cell responses, which are our primary protection in case of a pandemic outbreak and may therefore prove to be a double-edged sword (Bodewes et al., 2009a,b, 2011b,c). Rapid vaccine production also remains a challenge for future influenza virus pandemics (World Health Organization, 2005; Rockman and Brown, 2010; Pada and Tambyah, 2011). This was particularly apparent during the 2009 pandemic when sufficient amounts of the vaccine against the pandemic virus were only available in October 2009, well and truly after the pandemic had spread globally (Butler, 2010). Vaccine production in a pandemic scenario may be further complicated by the fact that some avian influenza viruses can kill the embryonated chicken eggs needed for vaccine production (Tumpey et al., 2005). Novel vaccines strategies, in combination with alternative vaccine production platforms are needed to accelerate vaccine production and circumvent such problems (Schotsaert and García-Sastre, 2014). However, an influenza vaccine that offers long-lasting, broad-spectrum immunity remains the gold standard for pandemic preparedness. How basic fundamental humoral and cellular biology and human clinical data can be considered for the implementation of a universal influenza vaccine has recently been reviewed (Clemens et al., 2018).
Concluding remarks
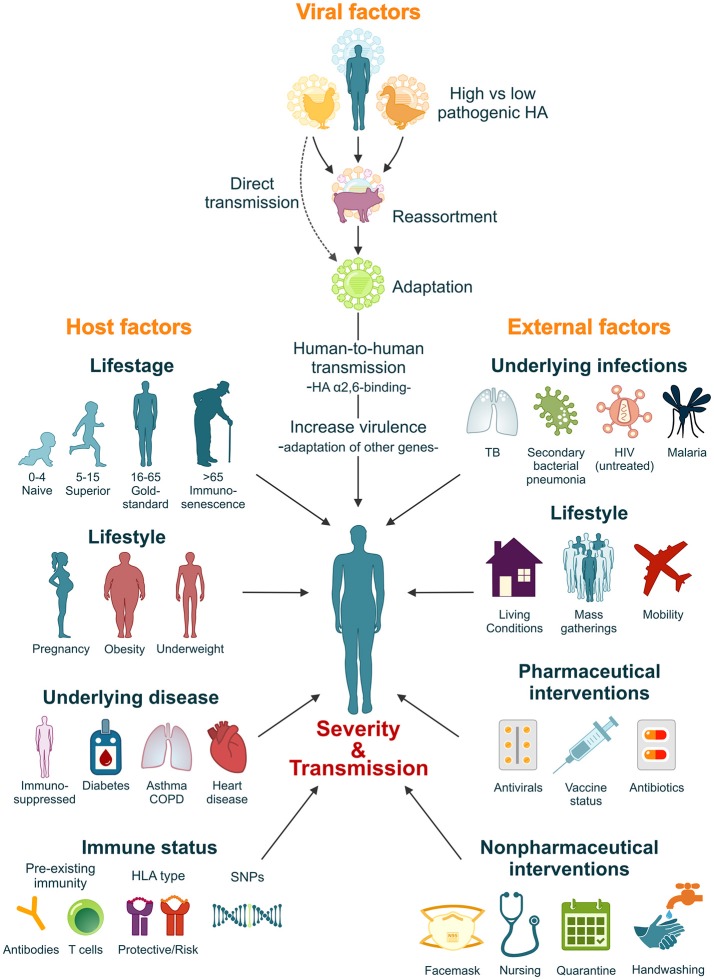
It is estimated that if a pandemic influenza virus were to re-appear today, with a similar virulence and attack-rate as the 1918 influenza virus, mortality could rise to 21–147 million (Murray et al., 2006; Madhav, 2013). However, the high morbidity and mortality rates associated with the 1918 influenza pandemic resulted from a complex interplay between factors intrinsic to the 1918 virus itself, the host's immune response and the social context in which the pandemic struck. It is thus unlikely that this exact combination of factors would repeat itself in the future. Nevertheless, a comprehensive understanding of the factors that contributed to the severity of the 1918 pandemic plays an important role in our preparedness for the next influenza pandemic (Figure 2).
Figure 2.
Factors that influence the severity and transmissibility of a pandemic influenza virus. The severity and transmissibility of pandemic influenza viruses are the result of a complex interplay of viral, host, and external factors. We have come a long way since 1918 and pandemic preparedness programs have learned from the 1918 and later pandemic outbreaks. Although unlikely, we cannot exclude the possibility that an influenza pandemic with similar severity will repeat itself in the future. However, lessons learned from the 1918 influenza pandemic will ensure that we are better prepared.
Today, we are better prepared for the next influenza virus pandemic than we were 100 years ago. Global influenza surveillance programs have been established to constantly monitor whether influenza viruses cross the species barrier into the human population (Hay and McCauley, 2018; World Health Organization, 2018a; Ziegler et al., 2018). This has already resulted in improved management strategies (Krammer et al., 2018) and the preventive slaughter of vast numbers of poultry that were infected with potentially pandemic viruses, such as H5N1 and H7N9 viruses (Oi, 2018). In addition, an improved understanding of the host-adaptation of influenza viruses and the existence of pre-existing immunity are likely to contribute to a more accurate predication of viral severity even before the influenza virus in question becomes established as a pandemic (Kreijtz et al., 2008; Lee et al., 2008; Herfst et al., 2012; Imai et al., 2012; Richard et al., 2013; Quiñones-Parra et al., 2014; van de Sandt et al., 2014; Wang et al., 2015). A better understanding of the human immune response against (pandemic) influenza viruses will eventually aid the development of broad-protective influenza vaccines (Clemens et al., 2018). However, in the interim, the majority of countries have established a pandemic preparedness program, which defines the precautionary measurements needed to be taken in case of an emerging viral pandemic (van Genugten et al., 2001; RIVM, 2018; World Health Organization, 2018b). These programs include surveillance, diagnostics, screening of passengers traveling from a potential outbreak region, quarantine procedures, stockpiling antibiotics, antivirals, bacterial and viral vaccines and the distribution of medical supplies (Brundage, 2006; Memoli et al., 2008; Mossad, 2009; World Health Organization, 2018b). We have also learnt from the 2009 H1N1 pandemic that it is important to have a somewhat flexible approach to pandemic preparedness, which allows countries to develop and implement their own risk assessments based on the global assessments provided by the WHO (Rudenko et al., 2015). However, good communication between countries and the WHO remains essential. In addition, it will be important for governments and those in authority to gain public trust before the next major pandemic outbreak. This will ensure that the public knows what to expect, how to act and is likely to improve compliance with preventative measures in a pandemic scenario. The internet represents a powerful and effective tool to disperse such information (Little et al., 2015).
Despite the advances that we have made in pandemic preparedness over the last 100 years, there are also several new challenges that we face in the context of twenty-first century (and later) influenza pandemics. Today's population demographic is dramatically different to that of 1918. Today, a large percentage of the world's population is either elderly (Morens et al., 2008; Mossad, 2009; Murray and Chotirmall, 2015) and/or living with one or more chronic medical conditions [such as heart disease, obesity, asthma, chronic obstructive pulmonary disease (COPD), and/or diabetes mellitus] (Morens et al., 2008; Jain et al., 2009; La Ruche et al., 2009; Flint et al., 2010; Hulme et al., 2017). The number of immunosuppressed individuals (due to untreated HIV infection, transplantation or/and chemotherapy) is also increasing (Jain et al., 2009; Kunisaki and Janoff, 2009; Sheth et al., 2011). This changing population demographic is of significance as each one of these host factors is known to increase the severity of even mild influenza virus infections. Mitigating the severity of future influenza pandemics will be further complicated by the prevalence of antibacterial resistance (Memoli et al., 2008), an increasing negative attitudes toward vaccination for other infectious diseases (such as measles and the pneumococcal polysaccharide and Hib vaccine) (Moss and Griffin, 2012; Perry et al., 2014) and an increase in seasonal influenza vaccinations of healthy individuals affecting the cross-reactive immune response otherwise induced by natural influenza virus infections (Bodewes et al., 2009b, 2011b). The high prevalence of underlying infections in less economically developed countries (such as TB and HIV), coupled with an underprepared health care system, places less economically developed countries at particularly high risk of severe morbidity during future influenza virus pandemics (Murray et al., 2006). These effects may even be compounded by the impacts of climate change which will lead to food shortages, famine and migration of climate refugees (van Schaik and Bakker, 2017).
At present, it is impossible to predict which influenza virus strain will cause the next pandemic. However, the growing number of human infections with the avian H7N9 virus represents a point of concern (especially in light of the approximate 40% mortality rate of this virus in humans). Like previous influenza pandemic viruses, human H7N9 virus infections have thus far displayed multiple waves of infections and shown signs of adaptation to human hosts (Zhu et al., 2017). Although this virus has yet to display efficient human-to-human transmission (Chen et al., 2013), it serves as a timely reminder that even though it has been 100 years since the 1918 pandemic, influenza pandemic preparedness remains of a paramount importance.
Author contributions
CvdS planned the review. KS, KK, and CvdS wrote the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. CvdS has received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 792532. KS is supported by an Australian Research Council DECRA (DE180100512). KK is supported by a NHMRC Senior Research Fellowship Level B (GNT#1102792).
References
- Afkhami A. (2003). Compromised constitutions: the Iranian experience with the 1918 influenza pandemic. Bull. Hist. Med. 77, 367–392. 10.1353/bhm.2003.0049 [DOI] [PubMed] [Google Scholar]
- Ahmed R., Oldstone M. B., Palese P. (2007). Protective immunity and susceptibility to infectious diseases: lessons from the 1918 influenza pandemic. Nat. Immunol. 8, 1188–1193. 10.1038/ni1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander J. B. (1919). Cases resembling encephalitis lethargica occurring during the influenza epidemic. Br. Med. J. 1, 794–795. 10.1136/bmj.1.3052.794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley E. A., Pyae Phyo A., Woodrow C. J. (2018). Malaria. Lancet 391, 1608–1621. 10.1016/S0140-6736(18)30324-6 [DOI] [PubMed] [Google Scholar]
- Audubon (2018). Audubon's Birds and Climate Change Report. Available online at: http://climate.audubon.org (Accessed May 6, 2018).
- Bajardi P., Poletto C., Ramasco J. J., Tizzoni M., Colizza V., Vespignani A. (2011). Human mobility networks, travel restrictions, and the global spread of 2009 H1N1 pandemic. PLoS ONE 6:e16591. 10.1371/journal.pone.0016591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballinger M. N., Standiford T. J. (2010). Postinfluenza bacterial pneumonia: host defenses gone awry. J. Interferon Cytokine Res. 30, 643–652. 10.1089/jir.2010.0049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber D. L., Andrade B. B., Sereti I., Sher A. (2012). Immune reconstitution inflammatory syndrome: the trouble with immunity when you had none. Nat. Rev. Microbiol. 10, 150–156. 10.1038/nrmicro2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry J. M. (2004). The site of origin of the 1918 influenza pandemic and its public health implications. J. Transl. Med. 2:3. 10.1186/1479-5876-2-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry J. M., Viboud C., Simonsen L. (2008). Cross-protection between successive waves of the 1918-1919 influenza pandemic: epidemiological evidence from US Army camps and from Britain. J. Infect. Dis. 198, 1427–1434. 10.1086/592454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M. A., Handy J., Levander O. A. (2004). Host nutritional status: the neglected virulence factor. Trends Microbiol. 12, 417–423. 10.1016/j.tim.2004.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser J. A., Gustin K. M., Pearce M. B., Maines T. R., Zeng H., Pappas C., et al. (2013). Pathogenesis and transmission of avian influenza A (H7N9) virus in ferrets and mice. Nature 501, 556–559. 10.1038/nature12391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge W. I. (1977). The start of pandemics: site, season and spread. Dev. Biol. Stand. 39, 443–444. [PubMed] [Google Scholar]
- Blakely D. E. (2006). Mass Mediated Disease: A Case Study Analysis of Three Flu Pandemics and Public Health Policy. Oxford: Lexington Books. [Google Scholar]
- Bodewes R., de Mutsert G., van der Klis F. R., Ventresca M., Wilks S., Smith D. J., et al. (2011a). Prevalence of antibodies against seasonal influenza A and B viruses in children in Netherlands. Clin. Vaccine Immunol. 18, 469–476. 10.1128/CVI.00396-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodewes R., Fraaij P. L., Geelhoed-Mieras M. M., van Baalen C. A., Tiddens H. A., van Rossum A. M. (2011b). Annual vaccination against influenza virus hampers development of virus-specific CD8+ T cell immunity in children. J. Virol. 85, 11995–12000. 10.1128/JVI.05213-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodewes R., Kreijtz J. H., Baas C., Geelhoed-Mieras M. M., de Mutsert G., van Amerongen G. (2009a). Vaccination against human influenza A/H3N2 virus prevents the induction of heterosubtypic immunity against lethal infection with avian influenza A/H5N1 virus. PLoS ONE 4:e5538. 10.1371/journal.pone.0005538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodewes R., Kreijtz J. H., Geelhoed-Mieras M. M., van Amerongen G., Verburgh R. J., van Trierum S. E., et al. (2011c). Vaccination against seasonal influenza A/H3N2 virus reduces the induction of heterosubtypic immunity against influenza A/H5N1 virus infection in ferrets. J. Virol. 85, 2695–2702. 10.1128/JVI.02371-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodewes R., Kreijtz J. H., Rimmelzwaan G. F. (2009b). Yearly influenza vaccinations: a double-edged sword? Lancet Infect. Dis. 9, 784–788. 10.1016/S1473-3099(09)70263-4 [DOI] [PubMed] [Google Scholar]
- Bootsma M. C., Ferguson N. M. (2007). The effect of public health measures on the 1918 influenza pandemic in U.S. cities. Proc. Natl. Acad. Sci U.S.A. 104, 7588–7593. 10.1073/pnas.0611071104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brightman I. J. (1935). Streptococcus infection occurring in ferrets inoculated with human influenza virus. Yale J. Biol. Med. 8, 127–135. [PMC free article] [PubMed] [Google Scholar]
- Brundage J. F. (2006). Interactions between influenza and bacterial respiratory pathogens: implications for pandemic preparedness. Lancet Infect. Dis. 6, 303–312. 10.1016/S1473-3099(06)70466-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundage J. F., Shanks G. D. (2008). Deaths from bacterial pneumonia during 1918-19 influenza pandemic. Emerg. Infect. Dis. 14, 1193–1199. 10.3201/eid1408.071313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler D. (2010). Portrait of a year-old pandemic. Nature 464, 1112–1113. 10.1038/4641112a [DOI] [PubMed] [Google Scholar]
- CDC (2009). Interim Recommendations for Facemask and Respirator Use to Reduce 2009 Influenza A (H1N1) Virus Transmission. Available online at: https://www.cdc.gov/h1n1flu/masks.htm (Accessed May 10, 2018).
- Chan D. K., Yang S. X., Mullan B., Du X., Zhang X., Chatzisarantis N. L., et al. (2015). Preventing the spread of H1N1 influenza infection during a pandemic: autonomy-supportive advice versus controlling instruction. J. Behav. Med. 38, 416–426. 10.1007/s10865-014-9616-z [DOI] [PubMed] [Google Scholar]
- Chen W., Lim C. E., Kang H. J., Liu J. (2011). Chinese herbal medicines for the treatment of type A H1N1 influenza: a systematic review of randomized controlled trials. PLoS ONE 6:e28093. 10.1371/journal.pone.0028093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liang W., Yang S., Wu N., Gao H., Sheng J., et al. (2013). Human infections with the emerging avian influenza A H7N9 virus from wet market poultry: clinical analysis and characterisation of viral genome. Lancet 381, 1916–1925. 10.1016/S0140-6736(13)60903-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K. F., Leung P. C. (2007). What happened in China during the 1918 influenza pandemic? Int. J. Infect. Dis. 11, 360–364. 10.1016/j.ijid.2006.07.009 [DOI] [PubMed] [Google Scholar]
- Chertow D. S., Memoli M. J. (2013). Bacterial coinfection in influenza: a grand rounds review. J. Am. Med. Assoc. 309, 275–282. 10.1001/jama.2012.194139 [DOI] [PubMed] [Google Scholar]
- Chien Y. W., Klugman K. P., Morens D. M. (2009). Bacterial pathogens and death during the 1918 influenza pandemic. N. Engl. J. Med. 361, 2582–2583. 10.1056/NEJMc0908216 [DOI] [PubMed] [Google Scholar]
- Chowell G., Echevarría-Zuno S., Viboud C., Simonsen L., Tamerius J., Miller M. A., et al. (2011). Characterizing the epidemiology of the 2009 influenza A/H1N1 pandemic in Mexico. PLoS Med. 8:e1000436. 10.1371/journal.pmed.1000436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciancanelli M. J., Huang S. X., Luthra P., Garner H., Itan Y., Volpi S., et al. (2015). Infectious disease. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science 348, 448–453. 10.1126/science.aaa1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens E. B., van de Sandt C., Wong S. S., Wakim L. M., Valkenburg S. A. (2018). Harnessing the power of T cells, the promising hope for a Universal Influenza vaccine. Vaccines 6:E18. 10.3390/vaccines6020018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliff A. D., Haggett P., Graham R. (1983). Reconstruction of diffusion at local scales: the 1846, 1882 and 1904 measles epidemics in northwest Iceland. J. Hist. Geogr. 9, 347–368. 10.1016/0305-7488(83)90254-2 [DOI] [PubMed] [Google Scholar]
- Cohen M. L. (2000). Changing patterns of infectious disease. Nature 406, 762–767. 10.1038/35021206 [DOI] [PubMed] [Google Scholar]
- Collins S. D. (1931). Age and sex incidence of influenza and pneumonia morbidity and mortality in the epidemic of 1928-29 with comparative data for the epidemic of 1918-19: based on surveys of families in certain localities in the united states following the epidemics. Public Health Rep. 46, 1909–1937. 10.2307/458013919315308 [DOI] [Google Scholar]
- Conenello G. M., Zamarin D., Perrone L. A., Tumpey T., Palese P. (2007). A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog. 3, 1414–1421. 10.1371/journal.ppat.0030141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper N. J., Sutton A. J., Abrams K. R., Wailoo A., Turner D., Nicholson K. G. (2003). Effectiveness of neuraminidase inhibitors in treatment and prevention of influenza A and B: systematic review and meta-analyses of randomised controlled trials. BMJ 326:1235. 10.1136/bmj.326.7401.1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling B. J., Zhou Y., Ip D. K., Leung G. M., Aiello A. E. (2010). Face masks to prevent transmission of influenza virus: a systematic review. Epidemiol. Infect. 138, 449–456. 10.1017/S0950268809991658 [DOI] [PubMed] [Google Scholar]
- Crosby A. W. (1976). Epidemic and Peace 1918. Santa Barbara, CA: Abc-Clio. [Google Scholar]
- Crosby A. W. (2003). America's Forgotten Pandemic: the Influenza of 1918. Cambridge: Cambridge University Press. [Google Scholar]
- Dawood F. S., Iuliano A. D., Reed C., Meltzer M. I., Shay D. K, Cheng P. Y., et al. (2012). Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. Lancet Infect. Dis. 12, 687–695. 10.1016/S1473-3099(12)70121-4 [DOI] [PubMed] [Google Scholar]
- De Clercq E. (2006). Antiviral agents active against influenza A viruses. Nat. Rev. Drug Discov. 5, 1015–1025. 10.1038/nrd2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong J. C., Rimmelzwaan G. F., Fouchier R. A., Osterhaus A. D. (2000). Influenza virus: a master of metamorphosis. J. Infect. 40, 218–228. 10.1053/jinf.2000.0652 [DOI] [PubMed] [Google Scholar]
- de Jong M. D., Simmons C. P., Thanh T. T., Hien V. M., Smith G. J., Chau T. N., et al. (2006). Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 12, 1203–1207. 10.1038/nm1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries R. D., McQuaid S., van Amerongen G., Yüksel S., Verburgh R. J., Osterhaus A. D., et al. (2012). Measles immune suppression: lessons from the macaque model. PLoS Pathog. 8:e1002885. 10.1371/journal.ppat.1002885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., Siegers J. Y., Cronin J. M., Weatherman S., van den Brand J. M., Leijten L. M., et al. (2018). 1918 H1N1 influenza virus replicates and induces proinflammatory cytokine responses in extrarespiratory tissues of ferrets. J. Infect. Dis. 217, 1237–1246. 10.1093/infdis/jiy003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diavatopoulos D. A., Short K. R., Price J. T., Wilksch J. J., Brown L. E., Briles D. E., et al. (2010). Influenza A virus facilitates Streptococcus pneumoniae transmission and disease. FASEB J. 24, 1789–1798. 10.1096/fj.09-146779 [DOI] [PubMed] [Google Scholar]
- Dowdle W. R. (1999). Influenza A virus recycling revisited. Bull. World Health Organ. 77, 820–828. [PMC free article] [PubMed] [Google Scholar]
- Duncan C. J., Duncan S. R., Scott S. (1997). The dynamics of measles epidemics. Theor. Popul. Biol. 52, 155–163. 10.1006/tpbi.1997.1326 [DOI] [PubMed] [Google Scholar]
- Dunham E. J., Dugan V. G., Kaser E. K., Perkins S. E., Brown I. H., Holmes E. C., et al. (2009). Different evolutionary trajectories of European avian-like and classical swine H1N1 influenza A viruses. J. Virol. 83, 5485–5494. 10.1128/JVI.02565-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis G. T., Davidson S., Crotta S., Branzk N., Papayannopoulos V., Wack A. (2015). TRAIL+ monocytes and monocyte-related cells cause lung damage and thereby increase susceptibility to influenza-Streptococcus pneumoniae coinfection. EMBO Rep. 16, 1203–1218. 10.15252/embr.201540473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J. S., Zambon M. C. (2002). Molecular diagnosis of influenza. Rev. Med. Virol. 12, 375–389. 10.1002/rmv.370 [DOI] [PubMed] [Google Scholar]
- Epstein S. L. (2006). Prior H1N1 influenza infection and susceptibility of cleveland family study participants during the H2N2 pandemic of 1957: an experiment of nature. J. Infect. Dis. 193, 49–53. 10.1086/498980 [DOI] [PubMed] [Google Scholar]
- Everitt A. R., Clare S., Pertel T., John S. P., Wash R. S., Smith S. E., et al. (2012). IFITM3 restricts the morbidity and mortality associated with influenza. Nature 484, 519–523. 10.1038/nature10921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint S. M., Davis J. S., Su J. Y., Oliver-Landry E. P., Rogers B. A., Goldstein A., et al. (2010). Disproportionate impact of pandemic (H1N1) 2009 influenza on Indigenous people in the Top End of Australia's Northern Territory. Med. J. Aust. 192, 617–622. [DOI] [PubMed] [Google Scholar]
- Francis T., de Torregrosa M. V. (1945). Combined infection of mice with influenzae, H. and influenza virus by the intranasal route. J. Infect. Dis. 76, 70–77. [Google Scholar]
- Fraser C., Cummings D. A., Klinkenberg D., Burke D. S., Ferguson N. M. (2011). Influenza transmission in households during the 1918 pandemic. Am. J. Epidemiol. 174, 505–514. 10.1093/aje/kwr122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost W. H. (1919). Public health weekly reports for August 15, 1919. Public Health Rep. 34, 1823–1926. 10.2307/4575271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garten R. J., Davis C. T., Russell C. A., Shu B., Lindstrom S., Balish A., et al. (2009). Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325, 197–201. 10.1126/science.1176225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgantopoulos P., Bergquist E. P., Knaup R. C., Anthony J. R., Bailey T. C., Williams M. P., et al. (2009). Importance of routine public health influenza surveillance: detection of an unusual W-shaped influenza morbidity curve. Am. J. Epidemiol. 170, 1533–1540. 10.1093/aje/kwp305 [DOI] [PubMed] [Google Scholar]
- Gibbon J. (1919). Acquired immunity in influenza. Lancet 193:583 10.1016/S0140-6736(01)25706-7 [DOI] [Google Scholar]
- Glaser L., Stevens J., Zamarin D., Wilson I. A., García-Sastre A., Tumpey T. M., et al. (2005). A single amino acid substitution in 1918 influenza virus hemagglutinin changes receptor binding specificity. J. Virol. 79:11533–11536. 10.1128/JVI.79.17.11533-11536.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover R. E. (1941). Spread of infection from the respiratory tract of the ferret. II. Association of influenza A virus and streptococcus type C. Br. J. Exp. Pathol. 22, 98–107. [Google Scholar]
- Goronzy J. J., Weyand C. M. (2013). Understanding immunosenescence to improve responses to vaccines. Nat. Immunol. 14, 428–436. 10.1038/ni.2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gras S., Kedzierski L., Valkenburg S. A., Laurie K., Liu Y. C., Denholm J. T., et al. (2010). Cross-reactive CD8+ T-cell immunity between the pandemic H1N1-2009 and H1N1-1918 influenza A viruses. Proc. Natl. Acad. Sci. U.S.A. 107, 12599–12604. 10.1073/pnas.1007270107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin D. E. (2010). Measles virus-induced suppression of immune responses. Immunol. Rev. 236, 176–189. 10.1111/j.1600-065X.2010.00925.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunaratnam P. J., Tobin S., Seale H., Marich A., McAnulty J. (2014). Airport arrivals screening during pandemic (H1N1) 2009 influenza in New South Wales, Australia. Med. J. Aust. 200, 290–292. 10.5694/mja13.10832 [DOI] [PubMed] [Google Scholar]
- Hammond J. A. B., Rolland W., Shore T. H. G. (1917). Purulent bronchiti. Lancet 190, 41–46. 10.1016/S0140-6736(01)56229-7 [DOI] [Google Scholar]
- Hancock K., Veguilla V., Lu X., Zhong W., Butler E. N., Sun H., et al. (2009). Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N. Engl. J. Med. 361, 1945–1952. 10.1056/NEJMoa0906453 [DOI] [PubMed] [Google Scholar]
- Harford C. G., Smith M. R., Wood W. B. (1946). Sulfonamide chemotherapy of combined infection with influenza virus and bacteria. J. Exp. Med. 83, 505–518. 10.1084/jem.83.6.505 [DOI] [PubMed] [Google Scholar]
- Hartmann B. M., Albrecht R. A., Zaslavsky E., Nudelman G., Pincas H., Marjanovic N., et al. (2017). Pandemic H1N1 influenza A viruses suppress immunogenic RIPK3-driven dendritic cell death. Nat. Commun. 8:1931. 10.1038/s41467-017-02035-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatchett R. J., Mecher C. E., Lipsitch M. (2007). Public health interventions and epidemic intensity during the 1918 influenza pandemic. Proc. Natl. Acad. Sci. U.S.A. 104, 7582–7587. 10.1073/pnas.0610941104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay A. J., Hayden F. G. (2013). Oseltamivir resistance during treatment of H7N9 infection. Lancet 381, 2230–2232. 10.1016/S0140-6736(13)61209-X [DOI] [PubMed] [Google Scholar]
- Hay A. J., McCauley J. W. (2018). The WHO global influenza Surveillance and Response System (GISRS) - a future perspective. Influenza Other Respir Viruses. 12, 551–557 10.1111/irv.12565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden F. G., Belshe R., Villanueva C., Lanno R., Hughes C., Small I., et al. (2004). Management of influenza in households: a prospective, randomized comparison of oseltamivir treatment with or without postexposure prophylaxis. J. Infect. Dis. 189, 440–449. 10.1086/381128 [DOI] [PubMed] [Google Scholar]
- Hayward A. C., Wang L., Goonetilleke N., Fragaszy E. B., Bermingham A., Copas A., et al. (2015). Natural T cell-mediated protection against seasonal and pandemic influenza. Results of the Flu Watch Cohort Study. Am. J. Respir. Crit. Care Med. 191, 1422–1431. 10.1164/rccm.201411-1988OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herfst S., Schrauwen E. J., Linster M., Chutinimitkul S., de Wit E., Munster V. J., et al. (2012). Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336, 1534–1541. 10.1126/science.1213362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez N., Melki I., Jing H., Habib T., Huang S. S. Y., Danielson J., et al. (2018). Life-threatening influenza pneumonitis in a child with inherited IRF9 deficiency. J Exp Med. 10.1084/jem.20180628. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildreth M. L. (1991). The influenza epidemic of 1918-1919 in France: contemporary concepts of aetiology, therapy, and prevention. Soc. Hist. Med. 4, 277–294. 10.1093/shm/4.2.277 [DOI] [PubMed] [Google Scholar]
- Hirsch H. H., Kaufmann G., Sendi P., Battegay M. (2004). Immune reconstitution in HIV-infected patients. Clin. Infect. Dis. 38, 1159–1166. 10.1086/383034 [DOI] [PubMed] [Google Scholar]
- Hoffman B. L. (2011). Influenza activity in Saint Joseph, Missouri 1910-1923: evidence for an early wave of the 1918 pandemic. PLoS Curr. 2:RRN1287. 10.1371/currents.RRN1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth T. D., Ferguson N. M., Anderson R. M. (2006). Will travel restrictions control the international spread of pandemic influenza? Nat. Med. 12, 497–499. 10.1038/nm0506-497 [DOI] [PubMed] [Google Scholar]
- Horimoto T., Kawaoka Y. (1994). Reverse genetics provides direct evidence for a correlation of hemagglutinin cleavability and virulence of an avian influenza A virus. J. Virol. 68, 3120–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrincius E. R., Liedmann S., Finkelstein D., Vogel P., Gansebom S., Ehrhardt C., et al. (2015). Nonstructural protein 1 (NS1)-mediated inhibition of c-Abl results in acute lung injury and priming for bacterial co-infections: insights into 1918 H1N1 pandemic? J. Infect. Dis. 211, 1418–1428. 10.1093/infdis/jiu609 [DOI] [PubMed] [Google Scholar]
- Hufnagel L., Brockmann D., Geisel T. (2004). Forecast and control of epidemics in a globalized world. Proc. Natl. Acad. Sci. U.S.A. 101, 15124–15129. 10.1073/pnas.0308344101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulme K. D., Gallo L. A., Short K. R. (2017). Influenza virus and glycemic variability in diabetes: a killer combination? Front. Microbiol. 8:861. 10.3389/fmicb.2017.00861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonen N., Strengell M., Kinnunen L., Osterlund P., Pirhonen J., Broman M., et al. (2010). High frequency of cross-reacting antibodies against 2009 pandemic influenza A(H1N1) virus among the elderly in Finland. Euro Surveill. 15:19478. 10.2807/ese.15.05.19478-en [DOI] [PubMed] [Google Scholar]
- Imai M., Watanabe T., Hatta M., Das S. C., Ozawa M., Shinya K., et al. (2012). Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486, 420–428. 10.1038/nature10831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagger B. W., Memoli M. J., Sheng Z. M., Qi L., Hrabal R. J., Allen G. L., et al. (2010). The PB2-E627K mutation attenuates viruses containing the 2009 H1N1 influenza pandemic polymerase. MBio 1:e00067–10. 10.1128/mBio.00067-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S., Kamimoto L., Bramley A. M., Schmitz A. M., Benoit S. R., Louie J., et al. (2009). Hospitalized patients with 2009 H1N1 influenza in the United States, April-June 2009. N. Engl. J. Med. 361, 1935–1944. 10.1056/NEJMoa0906695 [DOI] [PubMed] [Google Scholar]
- Johnson N. (2006). Britain and the 1918-19 Influenza Pandemic: A Dark Epilogue. Abingdon: Taylor & Francis Ltd. [Google Scholar]
- Johnson N. P., Mueller J. (2002). Updating the accounts: global mortality of the 1918-1920 “Spanish” influenza pandemic. Bull. Hist. Med. 76, 105–115. 10.1353/bhm.2002.0022 [DOI] [PubMed] [Google Scholar]
- Joseph C., Togawa Y., Shindo N. (2013). Bacterial and viral infections associated with influenza. Influenza Other Respir Viruses 7(Suppl. 2), 105–113. 10.1111/irv.12089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kash J. C., Tumpey T. M., Proll S. C., Carter V., Perwitasari O., Thomas M. J., et al. (2006). Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature 443, 578–581. 10.1038/nature05181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kash J. C., Walters K. A., Davis A. S., Sandouk A., Schwartzman L. M., Jagger B.W., et al. (2011). Lethal synergism of 2009 pandemic H1N1 influenza virus and Streptococcus pneumoniae coinfection is associated with loss of murine lung repair responses. MBio 2:e00172–11. 10.1128/mBio.00172-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka Y., Krauss S., Webster R. G. (1989). Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J. Virol. 63, 4603–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka Y., Webster R. G. (1988). Sequence requirements for cleavage activation of influenza virus hemagglutinin expressed in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 85, 324–328. 10.1073/pnas.85.2.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling A. W. (2010). Alert to the necessities of the emergency: U.S. nursing during the 1918 influenza pandemic. Publ. Health Rep. 125(Suppl. 3), 105–12. 10.1177/00333549101250S313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan K., Arino J., Hu W., Raposo P., Sears J., Calderon F., et al. (2009). Spread of a novel influenza A (H1N1) virus via global airline transportation. N. Engl. J. Med. 361, 212–214. 10.1056/NEJMc0904559 [DOI] [PubMed] [Google Scholar]
- Klaassen M., Hoye B. J., Nolet B. A., Buttemer W. A. (2012). Ecophysiology of avian migration in the face of current global hazards. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 367, 1719–1732. 10.1098/rstb.2012.0008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobasa D., Jones S. M., Shinya K., Kash J. C., Copps J., Ebihara H., et al. (2007). Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 445, 319–323. 10.1038/nature05495 [DOI] [PubMed] [Google Scholar]
- Kobasa D., Takada A., Shinya K., Hatta M., Halfmann P., Theriault S., et al. (2004). Enhanced virulence of influenza A viruses with the haemagglutinin of the 1918 pandemic virus. Nature 431, 703–707. 10.1038/nature02951 [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Davis S. M., Utsunomiya T., Pollard R. B., Suzuki F. (1999). Antiviral effect of gingyo-san, a traditional Chinese herbal medicine, on influenza A2 virus infection in mice. Am. J. Chin. Med. 27, 53–62. 10.1142/S0192415X99000082 [DOI] [PubMed] [Google Scholar]
- Krammer F., Smith G. J. D., Fouchier R. A. M., Peiris M., Kedzierska K., Doherty P. C., et al. (2018). Influenza. Nat. Rev. Dis Prim. 4:3 10.1038/s41572-018-0002-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreijtz J. H., de Mutsert G., van Baalen C. A., Fouchier R. A., Osterhaus A. D., Rimmelzwaan G. F. (2008). Cross-recognition of avian H5N1 influenza virus by human cytotoxic T-lymphocyte populations directed to human influenza A virus. J. Virol. 82, 5161–5166. 10.1128/JVI.02694-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunisaki K. M., Janoff E. N. (2009). Influenza in immunosuppressed populations: a review of infection frequency, morbidity, mortality, and vaccine responses. Lancet Infect. Dis. 9, 493–504. 10.1016/S1473-3099(09)70175-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Ruche G., Tarantola A., Barboza P., Vaillant L., Gueguen J., Gastellu-Etchegorry M., et al. (2009). The 2009 pandemic H1N1 influenza and indigenous populations of the Americas and the Pacific. Euro Surveill. 14:19366 10.2807/ese.14.42.19366-en [DOI] [PubMed] [Google Scholar]
- Langford C. M., Storey P. (1992). Influenza in Sri Lanka, 1918–1919: the impact of a new disease in a premodern third world setting. Health Transition Rev. 2, 97–123. [Google Scholar]
- Larson E. L., Cohen B., Baxter K. A. (2012). Analysis of alcohol-based hand sanitizer delivery systems: efficacy of foam, gel, and wipes against influenza A (H1N1) virus on hands. Am. J. Infect. Control 40, 806–809. 10.1016/j.ajic.2011.10.016 [DOI] [PubMed] [Google Scholar]
- Lee B., Robinson K. M., McHugh K. J., Scheller E. V., Mandalapu S., Chen C., et al. (2015). Influenza-induced type I interferon enhances susceptibility to gram-negative and gram-positive bacterial pneumonia in mice. Am. J. Physiol. Lung Cell. Mol. Physiol. 309, L158–L167. 10.1152/ajplung.00338.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee L. Y., Ha do L. A., Simmons C., de Jong M. D., Chau N. V., Schumacher R., et al. (2008). Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J. Clin. Invest. 118, 3478–3490. 10.1172/JCI32460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemey P., Rambaut A., Bedford T., Faria N., Bielejec F., Baele G., et al. (2014). Unifying viral genetics and human transportation data to predict the global transmission dynamics of human influenza H3N2. PLoS Pathog. 10:e1003932. 10.1371/journal.ppat.1003932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little P., Stuart B., Hobbs F. D., Moore M., Barnett J., Popoola D., et al. (2015). An internet-delivered handwashing intervention to modify influenza-like illness and respiratory infection transmission (PRIMIT): a primary care randomised trial. Lancet 386, 1631–1639. 10.1016/S0140-6736(15)60127-1 [DOI] [PubMed] [Google Scholar]
- Liu Y., Li S., Zhang G., Nie G., Meng Z., Mao D., et al. (2013). Genetic variants in IL1A and IL1B contribute to the susceptibility to 2009 pandemic H1N1 influenza A virus. (2014). BMC Immunol. 14:37 10.1186/1471-2172-14-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie J. K., Acosta M., Samuel M. C., Schechter R., Vugia D. J., Harriman K., et al. (2011). California Pandemic Working, A novel risk factor for a novel virus: obesity and 2009 pandemic influenza A (H1N1). Clin. Infect. Dis. 52, 301–312. 10.1093/cid/ciq152 [DOI] [PubMed] [Google Scholar]
- Louria D. B., Blumenfeld H. L., Ellis J. T., Kilbourne E. D., Rogers D. E. (1959). Studies on influenza in the pandemic of 1957–1958. II. Pulmonary complications of influenza. J. Clin. Invest. 38(1 Part 2), 213–265. 10.1172/JCI103791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luk J., Gross P., Thompson W. W. (2001). Observations on mortality during the 1918 influenza pandemic. Clin. Infect. Dis. 33, 1375–1378. 10.1086/322662 [DOI] [PubMed] [Google Scholar]
- MacDougall H. (2007). Toronto's health department in action: influenza in 1918 and SARS in 2003. J. Hist. Med. Allied Sci. 62, 56–89 10.1093/jhmas/jrl042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhav N. (2013). Modeling a modern-day spanish flu pandemic, in Aircurrents, ed Markey M. J. Available online at: www.air-worldwide.com/Publications/AIR-Currents/2013/Attachments/Modeling-a-Modern-Day-Spanish-Flu-Pandemic (Accessed May 10, 2018).
- Madhi S. A., Klugman K. P., Vaccine Trialist G. (2004). A role for Streptococcus pneumoniae in virus-associated pneumonia. Nat. Med. 10, 811–813. 10.1038/nm1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamelund S. E., Haneberg B., Mjaaland S. (2016). A missed summer wave of the 1918-1919 influenza pandemic, evidence from household surveys in the United States and Norway. Open Forum Infect. Dis. 3:ofw040. 10.1093/ofid/ofw040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews J. D., McBryde E. S., McVernon J., Pallaghy P. K., McCaw J. M. (2010). Prior immunity helps to explain wave-like behaviour of pandemic influenza in 1918-9. BMC Infect. Dis. 10:128. 10.1186/1471-2334-10-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazel-Sanchez B., Boal Carvalho I., Silva F., Dijkman R., Schmolke M. (2018). H5N1 influenza A virus PB1-F2 relieves HAX-1-mediated restriction of avian virus polymerase PA in human lung cells. J. Virol. 92. e00425–18 10.1128/JVI.00425-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuley J. L., Hornung F., Boyd K. L., Smith A. M., McKeon R., Bennink J., et al. (2007). Expression of the 1918 influenza A virus PB1-F2 enhances the pathogenesis of viral and secondary bacterial pneumonia. Cell Host Microbe 2, 240–249. 10.1016/j.chom.2007.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers J. A. (2006). Insights into the interaction between influenza virus and pneumococcus. Clin. Microbiol. Rev. 19, 571–582. 10.1128/CMR.00058-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers J. A., Bartmess K. C. (2003). Role of neuraminidase in lethal synergism between influenza virus and Streptococcus pneumoniae. J. Infect. Dis. 187, 1000–1009. 10.1086/368163 [DOI] [PubMed] [Google Scholar]
- McMichael A. J., Gotch F. M., Noble G. R., Beare P. (1983). Cytotoxic T-cell immunity to influenza. N. Engl. J. Med. 309, 13–17. 10.1056/NEJM198307073090103 [DOI] [PubMed] [Google Scholar]
- Mehle A., Doudna J. A. (2009). Adaptive strategies of the influenza virus polymerase for replication in humans. Proc. Natl. Acad. Sci. U.S.A. 106, 21312–21316. 10.1073/pnas.0911915106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehle A., Dugan V. G., Taubenberger J. K., Doudna J. A. (2012). Reassortment and mutation of the avian influenza virus polymerase PA subunit overcome species barriers. J. Virol. 86, 1750–1757. 10.1128/JVI.06203-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memoli M. J., Morens D. M., Taubenberger J. K. (2008). Pandemic and seasonal influenza: therapeutic challenges. Drug Discov. Today 13, 590–595. 10.1016/j.drudis.2008.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memoli M. J., Tumpey T. M., Jagger B. W., Dugan V. G., Sheng Z. M., Qi L., et al. (2009). An early 'classical' swine H1N1 influenza virus shows similar pathogenicity to the 1918 pandemic virus in ferrets and mice. Virology 393, 338–345. 10.1016/j.virol.2009.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard J., Ugarte-Gil C., Moore D. A. (2015). Multidrug resistant tuberculosis. BMJ 350:h882. 10.1136/bmj.h882 [DOI] [PubMed] [Google Scholar]
- Mills I. D. (1986). The 1918-1919 influenza pandemic–the Indian experience. Indian Econ. Soc. Hist. Rev. 23, 1–40. 10.1177/001946468602300102 [DOI] [PubMed] [Google Scholar]
- Mina M. J., Metcalf C. J., de Swart R. L., Osterhaus A. D., Grenfell B. T. (2015). Long-term measles-induced immunomodulation increases overall childhood infectious disease mortality. Science 348, 694–699. 10.1126/science.aaa3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsalvo A. C., Batalle J. P., Lopez M. F., Krause J. C., Klemenc J., Hernandez J. Z., et al. (2011). Severe pandemic 2009 H1N1 influenza disease due to pathogenic immune complexes. Nat. Med. 17, 195–199. 10.1038/nm.2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales K. F., Paget J., Spreeuwenberg P. (2017). Possible explanations for why some countries were harder hit by the pandemic influenza virus in 2009 - a global mortality impact modeling study. BMC Infect. Dis. 17:642. 10.1186/s12879-017-2730-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens D. M., Fauci A. S. (2007). The 1918 influenza pandemic: insights for the 21st century. J. Infect. Dis. 195, 1018–1028. 10.1086/511989 [DOI] [PubMed] [Google Scholar]
- Morens D. M., Taubenberger J. K. (2015). A forgotten epidemic that changed medicine: measles in the US Army, 1917-18. Lancet Infect. Dis. 15, 852–861. 10.1016/S1473-3099(15)00109-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens D. M., Taubenberger J. K., Fauci A. S. (2008). Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J. Infect. Dis. 198, 962–970. 10.1086/591708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens D. M., Taubenberger J. K., Fauci A. S. (2009). The persistent legacy of the 1918 influenza virus. N. Engl. J. Med. 361, 225–229. 10.1056/NEJMp0904819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan O. W., Bramley A., Fowlkes A., Freedman D. S., Taylor T. H., Gargiullo P., et al. (2010). Morbid obesity as a risk factor for hospitalization and death due to 2009 pandemic influenza A(H1N1) disease. PLoS ONE 5:e9694. 10.1371/journal.pone.0009694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss W. J., Griffin D. E. (2012). Measles. Lancet 379, 153–164. 10.1016/S0140-6736(10)62352-5 [DOI] [PubMed] [Google Scholar]
- Moss W. J., Ota M. O., Griffin D. E. (2004). Measles: immune suppression and immune responses. Int. J. Biochem. Cell Biol. 36, 1380–1385. 10.1016/j.biocel.2004.01.019 [DOI] [PubMed] [Google Scholar]
- Mossad S. B. (2009). Influenza in long-term care facilities: preventable, detectable, treatable. Cleve. Clin. J. Med. 76, 513–521. 10.3949/ccjm.76a.09022 [DOI] [PubMed] [Google Scholar]
- Murray C. J., Lopez A. D., Chin B., Feehan D., Hill K. (2006). Estimation of potential global pandemic influenza mortality on the basis of vital registry data from the 1918-20 pandemic: a quantitative analysis. Lancet 368, 2211–2218. 10.1016/S0140-6736(06)69895-4 [DOI] [PubMed] [Google Scholar]
- Murray M. A., Chotirmall S. H. (2015). The Impact of Immunosenescence on pulmonary disease. Mediators Inflamm. 2015:692546 10.1155/2015/692546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S., Davis K. M., Weiser J. N. (2011). Synergistic stimulation of type I interferons during influenza virus coinfection promotes Streptococcus pneumoniae colonization in mice. J. Clin. Invest. 121, 3657–3665. 10.1172/JCI57762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Center for Respiratory Diseases CDC, and Centers for Disease and Prevention (CDC) (2009). Use of influenza A (H1N1) 2009 monovalent vaccine: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2009. MMWR Recomm Rep. 58, 1–8. [PubMed] [Google Scholar]
- Navarini A. A., Recher M., Lang K. S., Georgiev P., Meury S., Bergthaler A., et al. (2006). Increased susceptibility to bacterial superinfection as a consequence of innate antiviral responses. Proc. Natl. Acad. Sci. U.S.A. 103, 15535–15539. 10.1073/pnas.0607325103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noymer A. (2011). The 1918 influenza pandemic hastened the decline of tuberculosis in the United States: an age, period, cohort analysis. Vaccine 29(Suppl. 2), B38–B41. 10.1016/j.vaccine.2011.02.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noymer A., Garenne M. (2000). The 1918 influenza epidemic's effects on sex differentials in mortality in the United States. Popul. Dev. Rev. 26, 565–581. 10.1111/j.1728-4457.2000.00565.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oei W., Nishiura H. (2012). The relationship between tuberculosis and influenza death during the influenza (H1N1) pandemic from 1918–19. Comput. Math. Methods Med. 2012:124861. 10.1155/2012/124861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oi E. (2018). Avian Influenza Portal, Update On Avian Influenza In Animals (types H5 and H7). World Organization for Animal Health. Available online at: http://www.oie.int/animal-health-in-the-world/update-on-avian-influenza/2018/ (Accessed May 12, 2018).
- Olson D. R., Simonsen L., Edelson P. J., Morse S. S. (2005). Epidemiological evidence of an early wave of the 1918 influenza pandemic in New York City. Proc. Natl. Acad. Sci. U.S.A. 102, 11059–11063. 10.1073/pnas.0408290102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oseasohn R., Adelson L., Kaji M. (1959). Clinicopathologic study of thirty-three fatal cases of Asian influenza. N. Engl. J. Med. 260, 509–518. 10.1056/NEJM195903122601101 [DOI] [PubMed] [Google Scholar]
- Oswald N. C., Shooter R. A., Curwen M. P. (1958). Pneumonia complicating Asian influenza. Br. Med. J. 2, 1305–1311. 10.1136/bmj.2.5108.1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxford J. S., Lambkin R., Sefton A., Daniels R., Elliot A., Brown R., et al. (2005). A hypothesis: the conjunction of soldiers, gas, pigs, ducks, geese and horses in northern France during the Great War provided the conditions for the emergence of the “Spanish” influenza pandemic of 1918-1919. Vaccine 23, 940–945. 10.1016/j.vaccine.2004.06.035 [DOI] [PubMed] [Google Scholar]
- Oxford J. S., Sefton A., Jackson R., Innes W., Daniels R. S., Johnson N. P. (2002). World War I may have allowed the emergence of “Spanish” influenza. Lancet Infect. Dis. 2, 111–114. 10.1016/S1473-3099(02)00185-8 [DOI] [PubMed] [Google Scholar]
- Oxford J. S., Sefton A., Jackson R., Johnson N. P, Daniels R. S. (1999). Who's that lady? Nat. Med. 5, 1351–1352. [DOI] [PubMed] [Google Scholar]
- Pada S., Tambyah P. A. (2011). Overview/reflections on the 2009 H1N1 pandemic. Microbes Infect. 13, 470–478. 10.1016/j.micinf.2011.01.009 [DOI] [PubMed] [Google Scholar]
- Palmer E., Rice G. W. (1992). A Japanese physician's response to pandemic influenza, Ijiro Gomibuchi and the “Spanish flu” in Yaita-Cho, 1918-1919. Bull. Hist. Med. 66, 560–577. [PubMed] [Google Scholar]
- Pappas C., Aguilar P. V., Basler C. F., Solórzano A., Zeng H., Perrone L. A., et al. (2008). Single gene reassortants identify a critical role for PB1, HA, and NA in the high virulence of the 1918 pandemic influenza virus. Proc. Natl. Acad. Sci. U. S. A. 105, 3064–3069. 10.1073/pnas.0711815105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson K. D., Pyle G. F. (1991). The geography and mortality of the 1918 influenza pandemic. Bull. Hist. Med. 65, 4–21. [PubMed] [Google Scholar]
- Peltola V. T., Murti K. G., McCullers J. A. (2005). Influenza virus neuraminidase contributes to secondary bacterial pneumonia. J. Infect. Dis. 192, 249–257. 10.1086/430954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R. T., Gacic-Dobo M., Dabbagh A., Mulders M. N., Strebel P. M., Okwo-Bele J. M., et al. (2014). Centers for disease and prevention, global control and regional elimination of measles, 2000-2012. MMWR Morb. Mortal. Wkly. Rep. 63, 103–107. [PMC free article] [PubMed] [Google Scholar]
- Phimister I. R. (1973). The “Spanish” influenza pandemic of 1918 and its impact on the Southern Rhodesian mining industry. Cent. Afr. J. Med. 19, 143–148. [PubMed] [Google Scholar]
- Quiñones-Parra S., Grant E., Loh L., Nguyen T. H., Campbell K. A., Tong S. Y, et al. (2014). Preexisting CD8+ T-cell immunity to the H7N9 influenza A virus varies across ethnicities. Proc Natl Acad Sci U.S.A. 111, 1049–1054 10.1073/pnas.1322229111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravenholt R. T., Foege W. H. (1982). 1918 influenza, encephalitis lethargica, parkinsonism. Lancet 2, 860–864. 10.1016/S0140-6736(82)90820-0 [DOI] [PubMed] [Google Scholar]
- Redford P. S., Mayer-Barber K. D., McNab F. W., Stavropoulos E., Wack A., Sher A., et al. (2014). Influenza A virus impairs control of Mycobacterium tuberculosis coinfection through a type I interferon receptor-dependent pathway. J. Infect. Dis. 209, 270–274. 10.1093/infdis/jit424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed C., Katz J. M. (2010). Serological surveys for 2009 pandemic influenza A H1N1. Lancet 375, 1062–1063. 10.1016/S0140-6736(09)62194-2 [DOI] [PubMed] [Google Scholar]
- Reid A. H., Fanning T. G., Hultin J. V., Taubenberger J. K. (1999). Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc. Natl. Acad. Sci. U.S.A. 96, 1651–1656. 10.1073/pnas.96.4.1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid A. H., Fanning T. G., Janczewski T. A., Lourens R. M., Taubenberger J. K. (2004a). Novel origin of the 1918 pandemic influenza virus nucleoprotein gene. J. Virol. 78, 12462–12470. 10.1128/JVI.78.22.12462-12470.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid A. H., Janczewski T. A., Lourens R. M., Elliot A. J., Daniels R. S., Berry C. L., et al. (2003). 1918 influenza pandemic caused by highly conserved viruses with two receptor-binding variants. Emerging Infect. Dis. 9, 1249–1253. 10.3201/eid0910.020789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid A. H., Taubenberger J. K., Fanning T. G. (2004b). Evidence of an absence: the genetic origins of the 1918 pandemic influenza virus. Nat. Rev. Microbiol. 2, 909–914. 10.1038/nrmicro1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes L., Arvelo W., Estevez A., Gray J., Moir J. C., Gordillo B., et al. (2010). Population-based surveillance for 2009 pandemic influenza A (H1N1) virus in Guatemala, 2009. Influenza Other Respir. Viruses 4, 129–140. 10.1111/j.1750-2659.2010.00138.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G. W. (2011). Japan and New Zealand in the 1918 influenza pandemic: comparative perspectives on official responses and crisis management, in Spanish Influenza Pandemic of 1918–1919: New Perspectives, ed Killingray D. (Melbourne, VIC: Routledge; ). [Google Scholar]
- Rice G. W., Palmer E. (1993). Pandemic influenza in Japan, 1918–19, mortality patterns and official responses. J. Japan. Stud. 19, 389–420. 10.2307/132645 [DOI] [Google Scholar]
- Richard M., Schrauwen E. J., de Graaf M., Bestebroer T. M., Spronken M. I., van Boheemen S., et al. (2013). Limited airborne transmission of H7N9 influenza A virus between ferrets. Nature 501, 560–563. 10.1038/nature12476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- RIVM (2018). De Lci-Richtlijnen, Stappenplannen En Draaiboeken Zijn Er Voor En Door Professionals in De Infectieziektebestrijding, Influenza. RIVM; Available online at: https://lci.rivm.nl/richtlijnen/influenza (Accessed May 05, 2018). [Google Scholar]
- Robertson L., Caley J. P., Moore J. (1958). Importance of Staphylococcus aureus in pneumonia in the 1957 epidemic of influenza A. Lancet 2, 233–236. 10.1016/S0140-6736(58)90060-6 [DOI] [PubMed] [Google Scholar]
- Robinson K. R. (1990). The role of nursing in the influenza epidemic of 1918–1919. Nurs. Forum 25, 19–26. 10.1111/j.1744-6198.1990.tb00845.x [DOI] [PubMed] [Google Scholar]
- Rockman S., Brown L. (2010). Pre-pandemic and pandemic influenza vaccines. Hum. Vaccin. 6, 792–801. 10.4161/hv.6.10.12915 [DOI] [PubMed] [Google Scholar]
- Rudenko L., Sellwood C., Russell C., Herfst S., Gross D., Dingwall R. (2015). Will there ever be a new influenza pandemic and are we prepared? Vaccine 33, 7037–7040. 10.1016/j.vaccine.2015.08.045 [DOI] [PubMed] [Google Scholar]
- Rvachev L. A., Longini I. M. (1985). A mathematical-model for the global spread of influenza. Math. Biosci. 75, 3–22. 10.1016/0025-5564(85)90064-1 [DOI] [Google Scholar]
- Sauerbrei A., Langenhan T., Brandstadt A., Schmidt-Ott R., Krumbholz A., Girschick H., et al. (2014). Prevalence of antibodies against influenza A and B viruses in children in Germany, 2008 to 2010. Euro Surveill. 19. [DOI] [PubMed] [Google Scholar]
- Schäfer J. R., Kawaoka Y., Bean W. J., Süss J., Senne D., Webster R. G. (1993). Origin of the pandemic 1957 H2 influenza A virus and the persistence of its possible progenitors in the avian reservoir. Virology 194, 781–788. 10.1006/viro.1993.1319 [DOI] [PubMed] [Google Scholar]
- Scholtissek C., Rohde W., Von Hoyningen V., Rott R. (1978). On the origin of the human influenza virus subtypes H2N2 and H3N2. Virology 87, 13–20. 10.1016/0042-6822(78)90153-8 [DOI] [PubMed] [Google Scholar]
- Schotsaert M., García-Sastre A. (2014). Influenza vaccines: a moving interdisciplinary field. Viruses 6, 3809–3826. 10.3390/v6103809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrauwen E. J., Herfst S., Leijten L. M., van Run P., Bestebroer T. M., Linster M., et al. (2012). The multibasic cleavage site in H5N1 virus is critical for systemic spread along the olfactory and hematogenous routes in ferrets. J. Virol. 86, 3975–3984. 10.1128/JVI.06828-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaman J., Lipsitch M. (2013). The El Nino-Southern Oscillation (ENSO)-pandemic influenza connection: coincident or causal? Proc. Natl. Acad. Sci. U.S.A. 110(Suppl. 1), 3689–3691. 10.1073/pnas.1107485109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks G. D. (2015). Synergistic mortality caused by Plasmodium falciparum during the 1918 influenza pandemic. Am. J. Trop. Med. Hyg. 92, 941–942. 10.4269/ajtmh.14-0792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks G. D., Brundage J. F. (2012). Pathogenic responses among young adults during the 1918 influenza pandemic. Emerging Infect. Dis. 18, 201–207. 10.3201/eid1802.102042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks G. D., Brundage J. F. (2013). Pacific islands which escaped the 1918–1919 influenza pandemic and their subsequent mortality experiences. Epidemiol. Infect. 141, 353–356. 10.1017/S0950268812000866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks G. D., Burroughs S., Sohn J. D., Waters N. C., Smith V. F., Waller M., et al. (2016b). Variable mortality from the 1918–1919 influenza pandemic during military training. Mil. Med. 181, 878–882. 10.7205/MILMED-D-15-00124 [DOI] [PubMed] [Google Scholar]
- Shanks G. D., Burroughs S. A., Sohn J. D., Waters N. C., Smith V. F., Waller M., et al. (2016a). Enhanced risk of illness during the 1918 influenza pandemic after previous influenza-like illnesses in three military populations. Epidemiol. Infect. 144, 2043–2048. 10.1017/S0950268816000479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks G. D., Hu Z., Waller M., Lee S. E., Terfa D., Howard A., et al. (2014). Measles epidemics of variable lethality in the early 20th century. Am. J. Epidemiol. 179, 413–422. 10.1093/aje/kwt282 [DOI] [PubMed] [Google Scholar]
- Shanks G. D., Lee S. E., Howard A., Brundage J. F. (2011a). Extreme mortality after first introduction of measles virus to the polynesian island of Rotuma, 1911. Am. J. Epidemiol. 173, 1211–1222. 10.1093/aje/kwq504 [DOI] [PubMed] [Google Scholar]
- Shanks G. D., Mackenzie A., McLaughlin R., Waller M., Dennis P., Lee S. E., et al. (2010). Mortality risk factors during the 1918-1919 influenza pandemic in the Australian army. J. Infect. Dis. 201, 1880–1889. 10.1086/652868 [DOI] [PubMed] [Google Scholar]
- Shanks G. D., MacKenzie A., Waller M., Brundage J. F. (2011b). Low but highly variable mortality among nurses and physicians during the influenza pandemic of 1918-1919. Influenza Other Respir. Viruses 5, 213–219. 10.1111/j.1750-2659.2010.00195.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks G. D., Wilson N., Kippen R., Brundage J. F. (2018). The unusually diverse mortality patterns in the Pacific region during the 1918–21 influenza pandemic: reflections at the pandemic's centenary. Lancet Infect Dis. 10.1016/S1473-3099(18)30178-6 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Sheng Z. M., Chertow D. S., Ambroggio X., McCall S., Przygodzki R. M., Cunningham R. E., et al. (2011). Autopsy series of 68 cases dying before and during the 1918 influenza pandemic peak. Proc. Natl. Acad. Sci. U.S.A. 108, 16416–16421. 10.1073/pnas.1111179108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheridan P. A., Paich H. A., Handy J., Karlsson E. A., Hudgens M. G., Sammon A. B., et al. (2012). Obesity is associated with impaired immune response to influenza vaccination in humans. Int. J. Obes. 36, 1072–1077. 10.1038/ijo.2011.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth A. N., Patel P., Peters P. J. (2011). Influenza and HIV: lessons from the 2009 H1N1 influenza pandemic. Curr. HIV/AIDS Rep. 8, 181–191. 10.1007/s11904-011-0086-4 [DOI] [PubMed] [Google Scholar]
- Shope R. E. (1958). Influenza: history, epidemiology, and speculation. Public Health Rep. 73, 165–178. 10.2307/4590072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short K. R., Habets M. N., Hermans P. W., Diavatopoulos D. A. (2012a). Interactions between Streptococcus pneumoniae and influenza virus: a mutually beneficial relationship? Future Microbiol. 7, 609–624. 10.2217/fmb.12.29 [DOI] [PubMed] [Google Scholar]
- Short K. R., Reading P. C., Brown L. E., Pedersen J., Gilbertson B., Job E. R., et al. (2013). Influenza-induced inflammation drives pneumococcal otitis media. Infect. Immun. 81, 645–652. 10.1128/IAI.01278-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short K. R., Reading P. C., Wang N., Diavatopoulos D. A., Wijburg O. L. (2012b). Increased nasopharyngeal bacterial titers and local inflammation facilitate transmission of Streptococcus pneumoniae. MBio 3:e00255-12. 10.1128/mBio.00255-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman S. T., Shulman D. L., Sims R. H. (2009). The tragic 1824 journey of the Hawaiian king and queen to London: history of measles in Hawaii. Pediatr. Infect. Dis. J. 28, 728–733. 10.1097/INF.0b013e31819c9720 [DOI] [PubMed] [Google Scholar]
- Simonsen L., Chowell G., Andreasen V., Gaffey R., Barry J., Olson D., et al. (2018). A review of the 1918 herald pandemic wave: importance for contemporary pandemic response strategies. Ann. Epidemiol. 28, 281–288. 10.1016/j.annepidem.2018.02.013 [DOI] [PubMed] [Google Scholar]
- Simonsen L., Clarke M. J., Schonberger L. B., Arden N. H., Cox N. J., Fukuda K. (1998). Pandemic versus epidemic influenza mortality: a pattern of changing age distribution. J. Infect. Dis. 178, 53–60. 10.1086/515616 [DOI] [PubMed] [Google Scholar]
- Slepushkin A. N. (1959). The effect of a previous attack of A1 influenza on susceptibility to A2 virus during the 1957 outbreak. Bull. World Health Organ. 20, 297–301. [PMC free article] [PubMed] [Google Scholar]
- Smith A. M., Adler F. R., Ribeiro R. M., Gutenkunst R. N., McAuley J. L., McCullers J.A., et al. (2013). Kinetics of coinfection with influenza A virus and Streptococcus pneumoniae. PLoS Pathog. 9:e1003238. 10.1371/journal.ppat.1003238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. J., Bahl J., Vijaykrishna D., Zhang J., Poon L. L., Chen H., et al. (2009a). Dating the emergence of pandemic influenza viruses. Proc. Natl. Acad. Sci. U.S.A. 106, 11709–11712. 10.1073/pnas.0904991106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. J., Vijaykrishna D., Bahl J., Lycett S. J., Worobey M., Pybus O. G., et al. (2009b). Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459, 1122–1125. 10.1038/nature08182 [DOI] [PubMed] [Google Scholar]
- Smith W., Andrewes C. H., Laidlaw P. P. (1933). A virus obtained from influenza patients. Lancet 222, 66–68. 10.1016/S0140-6736(00)78541-2 [DOI] [Google Scholar]
- Spinney L. (2017). Pale Rider: The Spanish Flu of 1918 and How It Changed the World. New York, NY: Public Affairs. [Google Scholar]
- Sridhar S., Begom S., Bermingham A., Hoschler K., Adamson W., Carman W., et al. (2013). Cellular immune correlates of protection against symptomatic pandemic influenza. Nat. Med. 19, 1305–1312. 10.1038/nm.3350 [DOI] [PubMed] [Google Scholar]
- Starko K. M. (2009). Salicylates and pandemic influenza mortality, 1918–1919 pharmacology, pathology, and historic evidence. Clin. Infect. Dis. 49, 1405–1410. 10.1086/606060 [DOI] [PubMed] [Google Scholar]
- Subbarao E. K., London W., Murphy B. R. (1993). A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 67, 1761–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbarao K., Klimov A., Katz J., Regnery H., Lim W., Hall H., et al. (1998). Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 279, 393–396. 10.1126/science.279.5349.393 [DOI] [PubMed] [Google Scholar]
- Suess T., Remschmidt C., Schink S. B., Schweiger B., Nitsche A., Schroeder K., et al. (2012). The role of facemasks and hand hygiene in the prevention of influenza transmission in households: results from a cluster randomised trial; Berlin, Germany, 2009–2011. BMC Infect. Dis. 12:26. 10.1186/1471-2334-12-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suguitan A. L., Jr, Matsuoka Y., Lau Y. F., Santos C. P., Vogel L., Cheng L. I., et al. (2012). The multibasic cleavage site of the hemagglutinin of highly pathogenic A/Vietnam/1203/2004 (H5N1) avian influenza virus acts as a virulence factor in a host-specific manner in mammals. J. Virol. 86, 2706–2714. 10.1128/JVI.05546-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger J. K., Baltimore D., Doherty P. C., Markel H., Morens D. M., Webster R. G., et al. (2012). Reconstruction of the 1918 influenza virus: unexpected rewards from the past. MBio 3:e00201–12. 10.1128/mBio.00201-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger J. K., Morens D. M. (2006). 1918 Influenza: the mother of all pandemics. Emerging Infect. Dis. 12, 15–22. 10.3201/eid1209.05-0979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger J. K., Reid A. H., Janczewski T. A., Fanning T. G. (2001). Integrating historical, clinical and molecular genetic data in order to explain the origin and virulence of the 1918 Spanish influenza virus. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 356, 1829–1839. 10.1098/rstb.2001.1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger J. K., Reid A. H., Krafft A. E., Bijwaard K. E., Fanning T. G. (1997). Initial genetic characterization of the 1918 “Spanish” influenza virus. Science 275, 1793–1796. 10.1126/science.275.5307.1793 [DOI] [PubMed] [Google Scholar]
- Taubenberger J. K., Reid A. H., Lourens R. M., Wang R., Jin G., Fanning T. G. (2005). Characterization of the 1918 influenza virus polymerase genes. Nature 437, 889–893. 10.1038/nature04230 [DOI] [PubMed] [Google Scholar]
- Thomas Y., Boquete-Suter P., Koch D., Pittet D., Kaiser L. (2014). Survival of influenza virus on human fingers. Clin. Microbiol. Infect. 20, O58–O64. 10.1111/1469-0691.12324 [DOI] [PubMed] [Google Scholar]
- To K. K., Hung I. F., Li I. W., Lee K. L., Koo C. K., Yan W. W., et al. (2010). Delayed clearance of viral load and marked cytokine activation in severe cases of pandemic H1N1 2009 influenza virus infection. Clin. Infect. Dis. 50, 850–859. 10.1086/650581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K. K. W., Zhou J., Song Y. Q., Hung I. F. N., Ip W. C. T., Cheng Z. S., et al. (2014). Surfactant protein B gene polymorphism is associated with severe influenza. Chest 145, 1237–1243. 10.1378/chest.13-1651 [DOI] [PubMed] [Google Scholar]
- Tognotti E. (2003). Scientific triumphalism and learning from facts: bacteriology and the “Spanish flu” challenge of 1918. Soc. Hist. Med. 16, 97–110. 10.1093/shm/16.1.97 [DOI] [PubMed] [Google Scholar]
- Tomkins S. M. (1992). The influenza epidemic of 1918–19 in Western Samoa. J. Pac. Hist. 27, 181–197. 10.1080/00223349208572706 [DOI] [Google Scholar]
- Tumpey T. M., Basler C. F., Aguilar P. V., Zeng H., Solórzano A., Swayne D. E., et al. (2005). Characterization of the reconstructed 1918 Spanish influenza pandemic virus. Science 310, 77–80. 10.1126/science.1119392 [DOI] [PubMed] [Google Scholar]
- Tumpey T. M., Maines T. R., Van Hoeven N., Glaser L., Solorzano A., Pappas C., et al. (2007). A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science 315, 655–659. 10.1126/science.1136212 [DOI] [PubMed] [Google Scholar]
- Valleron A. J., Cori A., Valtat S., Meurisse S., Carrat F., Boëlle P. Y. (2010). Transmissibility and geographic spread of the 1889 influenza pandemic. Proc. Natl. Acad. Sci. U.S.A. 107, 8778–8781. 10.1073/pnas.1000886107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Sandt C. E, Sagong K. A., Pronk M. R., Bestebroer T. M., Spronken M. I., Koopmans M. P. G., et al. (2018b). H1N1pdm09 influenza virus and its descendants lack extra-epitopic amino acid residues associated with reduced recognition by M158-66-specific CD8+ T-cells. J Infect Dis. 218, 581–585. 10.1093/infdis/jiy218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Sandt C. E., Hillaire M. L., Geelhoed-Mieras M. M., Osterhaus A. D., Fouchier R. A., Rimmelzwaan G. F. (2015a). Human influenza A virus-specific CD8+ T cell response is long-lived. J. Infect. Dis. 212, 81–5. 10.1093/infdis/jiv018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Sandt C. E., Kreijtz J. H., de Mutsert G., Geelhoed-Mieras M. M., Hillaire M. L., Vogelzang-van Trierum S. E., et al. (2014). Human cytotoxic T lymphocytes directed to seasonal influenza A viruses cross-react with the newly emerging H7N9 virus. J. Virol. 88, 1684–1693. 10.1128/JVI.02843-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Sandt C. E., Kreijtz J. H., Geelhoed-Mieras N. J., Nieuwkoop M. I., Spronken M., van de Vijver D. A., et al. (2015b). Differential recognition of influenza A viruses by M158-66 epitope-specific CD8+ T cells is determined by extraepitopic amino acid residues. J. Virol. 90, 1009–1022. 10.1128/JVI.02439-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Sandt C. E., Pronk M. R., van Baalen C. A., Fouchier R. A. M., Rimmelzwaan G. F. (2018a). Variation at extra-epitopic amino acid residues influences suppression of influenza virus replication by M158-66 epitope-specific CD8+ T lymphocytes. J Virol. 92:e00232–18. 10.1128/JVI.00232-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sluijs K. F., Nijhuis M., Levels J. H., Florquin S., Mellor A. L., Jansen H. M., et al. (2006). Influenza-induced expression of indoleamine 2,3-dioxygenase enhances interleukin-10 production and bacterial outgrowth during secondary pneumococcal pneumonia. J. Infect. Dis. 193, 214–222. 10.1086/498911 [DOI] [PubMed] [Google Scholar]
- van Genugten M. L. L., Heijnen M. L. A., Jager J. C. (2001). Scenario-ontwikkeling zorgvraag bij een influenza pandemie, in RIVM-Rapport. RIVM. [Google Scholar]
- Van Kerkhove M. D., Vandemaele K. A., Shinde V., Jaramillo-Gutierrez G., Koukounari A., Donnelly C. A., et al. (2011). Infection, risk factors for severe outcomes following 2009 influenza A (H1N1) infection: a global pooled analysis. PLoS Med. 8:e1001053. 10.1371/journal.pmed.1001053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Schaik L., Bakker T. (2017). Climate-migration-security: Making the most of a contested relationship, in Clingendael Policy Brief (Den Haag: Planetary Security Initiative; ). [Google Scholar]
- Wahl B., O'Brien K. L., Greenbaum A., Majumder A., Liu L., Chu Y., et al. (2018). Burden of Streptococcus pneumoniae and Haemophilus influenzae type b disease in children in the era of conjugate vaccines: global, regional, and national estimates for 2000–15. Lancet Glob. Health 6, e744–e757. 10.1016/S2214-109X(18)30247-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wan Y., Qiu C., Quinones-Parra S., Zhu Z., Loh L., et al. (2015). Recovery from severe H7N9 disease is associated with diverse response mechanisms dominated by CD8+ T cells. (2010). Nat. Commun. 6:6833 10.1038/ncomms7833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Watanabe S., Shinya K., Kim J. H., Hatta M., Kawaoka Y. (2009). Viral RNA polymerase complex promotes optimal growth of 1918 virus in the lower respiratory tract of ferrets. Proc. Natl. Acad. Sci. U.S.A. 106, 588–592. 10.1073/pnas.0806959106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Zhong G., Russell C. A., Nakajima N., Hatta M., Hanson A., et al. (2014). Circulating avian influenza viruses closely related to the 1918 virus have pandemic potential. Cell Host Microbe 15, 692–705. 10.1016/j.chom.2014.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson H. E., Saslaw S., Doan C. A., Woolpert O. C., Schwab J. L. (1947). Reactions of monkeys to experimental mixed influenza and streptococcus infections: an analysis of the relative roles of humoral and cellular immunity, with the description of an intercurrent nephritic syndrome. J. Exp. Med. 85, 199–215. 10.1084/jem.85.2.199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong V. W., Cowling B. J., Aiello A. E. (2014). Hand hygiene and risk of influenza virus infections in the community: a systematic review and meta-analysis. Epidemiol. Infect. 142, 922–932. 10.1017/S095026881400003X [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2005). Evolution of H5N1 avian influenza viruses in Asia. Emerging Infect. Dis. 11, 1515–21. 10.3201/eid1110.050644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization (2018a). Global Influenza Virological Surveillance. Available online at: http://www.who.int/gho/epidemic_diseases/influenza/virological_surveillance/en/ (Accessed May 6, 2018). [Google Scholar]
- World Health Organization (2018b). Pandemic Preparedness. Available online at: http://www.who.int/influenza/preparedness/pandemic/en/ (Accessed May 12, 2018). [Google Scholar]
- World Health Organization Writing Group. Bell D., Nicoll A., Fukuda K., Horby P., Monto A., et al. (2006). Non-pharmaceutical interventions for pandemic influenza, national and community measures. Emerging Infect. Dis. 12, 88–94. 10.3201/eid1201.051371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worobey M., Han G. Z., Rambaut A. (2014). Genesis and pathogenesis of the 1918 pandemic H1N1 influenza A virus. Proc Natl Acad Sci U.S.A. 111, 8107–8112. 10.1073/pnas.1324197111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J. T., Cowling B. J., Lau E. H., Ip D. K., Ho L. M., Tsang T., et al. (2010). School closure and mitigation of pandemic (H1N1) 2009, Hong Kong. Emerging Infect. Dis. 16, 538–541. 10.3201/eid1603.091216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Tsibane T., McGraw P. A., House F. S., Keefer C. J., Hicar M. D., et al. (2008). Neutralizing antibodies derived from the B cells of 1918 influenza pandemic survivors. Nature 455, 532–536. 10.1038/nature07231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambon M. (2014). Developments in the treatment of severe influenza: lessons from the pandemic of 2009 and new prospects for therapy. Curr. Opin. Infect. Dis. 27, 560–565. 10.1097/QCO.0000000000000113 [DOI] [PubMed] [Google Scholar]
- Zhang Y. H., Zhao Y., Li N., Peng Y. C., Giannoulatou E., Jin R. H., et al. (2013). Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals. Nat. Commun. 4:1418. 10.1038/ncomms2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., To K. K., Dong H., Cheng Z. S., Lau C. C., Poon V. K., et al. (2012). A functional variation in CD55 increases the severity of 2009 pandemic H1N1 influenza A virus infection. J. Infect. Dis. 206, 495–503. 10.1093/infdis/jis378 [DOI] [PubMed] [Google Scholar]
- Zhu H., Wang D., Kelvin D. J., Li L., Zheng Z., Yoon S. W., et al. (2013). Infectivity, transmission, and pathology of human H7N9 influenza in ferrets and pigs. Science 341:183–186. 10.1126/science.1239844. [DOI] [PubMed] [Google Scholar]
- Zhu W., Zhou J., Li Z., Yang L., Li X., Huang W., et al. (2017). Biological characterisation of the emerged highly pathogenic avian influenza (HPAI) A(H7N9) viruses in humans, in mainland China, 2016 to 2017. Euro Surveill. 22:30533. 10.2807/1560-7917.ES.2017.22.19.30533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler T., Mamahit A., Cox N. J. (2018). 65 Years of influenza surveillance by a WHO-coordinated global network. Influenza Other Respir Viruses (2018). 12 558–565 10.1111/irv.12570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A., Raviglione M., Hafner R., von Reyn C. F. (2013). Tuberculosis. N. Engl. J. Med. 368, 745–755. 10.1056/NEJMra1200894 [DOI] [PubMed] [Google Scholar]
- Zúñiga J., Buendía-Roldán I., Zhao Y., Jiménez L., Torres D., Romo J., et al. (2012). Genetic variants associated with severe pneumonia in A/H1N1 influenza infection. Eur. Respir. J. 39, 604–610. 10.1183/09031936.00020611 [DOI] [PMC free article] [PubMed] [Google Scholar]