Abstract
Purpose of Review:
We present study design and methodological suggestions for population-based studies that integrate molecular -omics data and highlight recent studies that have used such data to examine the potential impacts of prenatal environmental exposures on fetal health.
Recent Findings:
Epidemiologic studies have observed numerous relationships between prenatal exposures (smoking, toxic metals, endocrine disruptors), fetal and early-life molecular profiles, though such investigations have so far been dominated by epigenomic association studies. However, recent transcriptomic, proteomic, and metabolomic studies have demonstrated their promise for the identification of exposure and response biomarkers.
Summary:
Molecular -omics have opened new avenues of research in environmental health that can improve our understanding of disease etiology, contribute to the development of exposure and response biomarkers. Studies that incorporate multiple -omics data from different molecular domains in longitudinally collected samples hold particular promise.
Keywords: environmental, prenatal, genomics, epigenomics, proteomics, metabolomics
Introduction:
Environmental health research aims to identify the ways in which environmental exposures contribute to human disease. This is a complex task given that around 140,000 [1] and 85,000 [2] chemicals have been registered for use in Europe and the United States, respectively, demonstrating the vast array of chemicals, pollutants and contaminants that humans may potentially be exposed to, the majority of which lack the substantive data to perform comprehensive risk assessment. Traditional environmental epidemiology has aimed to characterize how individual, or small subsets of exposures, are associated with one or a few health outcomes. Though this hypothesis-driven approach has successfully identified and characterized numerous environmental risk factors for disease, it cannot efficiently search across this vast exposure space to discover or characterize all environmentally-associated risks.
Technological advancements over the last decade have resulted in the improved capacities and precision in the generation of high-dimensional molecular data, measuring the quantities and interactions of numerous small molecules in biological samples, that are informative about internalization of exposures and perturbations to genomic regulation and physiological activity [3]. These types of data have broadened researchers’ capabilities for examining the underlying etiology of environmentally-associated diseases. This approach to studying the environment, when used as a complement to traditional environmental epidemiology and experimental studies, can lead to the breakthroughs in understanding the impacts of chemicals on health, identification of exposure and/or pre-symptomatic biomarkers of health outcomes, and an expanded understanding of disease etiology. In this review, we aim to introduce the role that molecular -omics data currently play in population-based studies of environmental health issues, discuss issues to consider when designing an epidemiologic study utilizing molecular -omics, and highlight recent studies that have incorporated these types of data into investigations of prenatal and early life exposures.
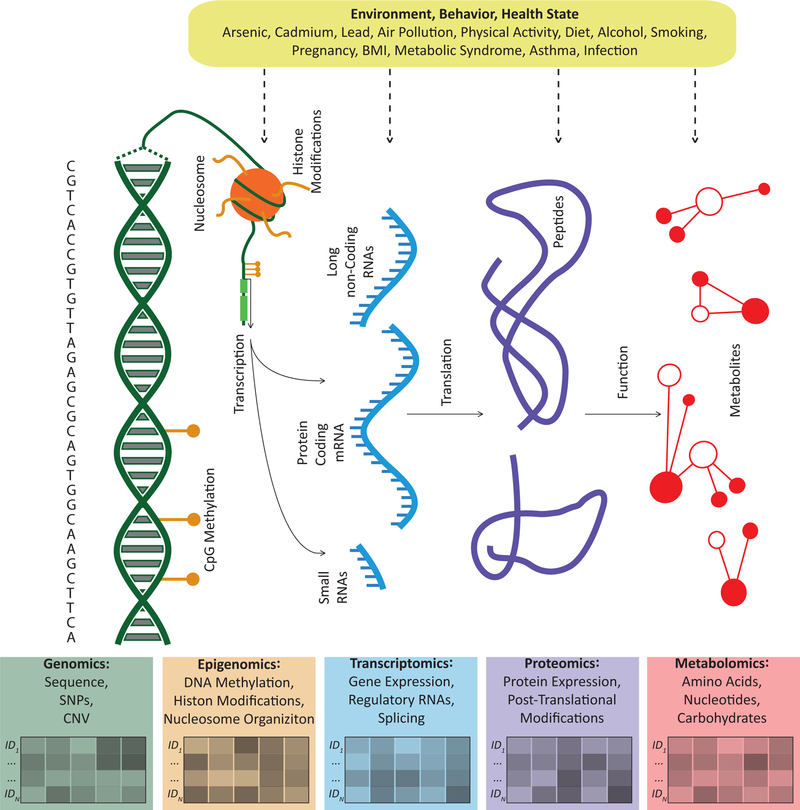
Genomics, an extension of genetics beyond individual candidate loci to encompass the complete sequence of DNA, was the first -omic approach to be utilized in population-based studies. Since publication of the first genome-wide association study (GWAS) in 2005, the scale of testable genetic loci has risen from 116,204 single nucleotide polymorphisms (SNPs) [4] to tens of millions of loci when integrating modern laboratory assays with imputation based on reference genomes [5]. Additionally, other -omics disciplines have emerged and grown at a similar pace, including (1) epigenomics: covalent chemical additions to DNA and histones that regulate gene-expression potential without altering the genetic sequence [6], (2) transcriptomics: RNA expression, which includes protein-coding and non-coding RNAs [7], (3) proteomics: the abundance of and interactions between proteins [8], and (4) metabolomics: the identification, quantification, and profiling of metabolites [9]. While genomics largely represents an individual’s inherited biological blueprint, these other molecular -omic features represent intermediary domains that capture a combination of biological potential, adaptive or reactive responses to stimuli, or internalization of exogenous exposures. The genome being most proximal to the underlying biological potential, the metabolome more proximal to the internalization of exposures and immediate physiological response, with the epigenome, transcriptome and proteome representing intermediate steps linking that potential to an actual response (Figure 1).
Figure 1 Caption:
Environmental exposures, as well as behaviors and disease processes, can influence the activities of multiple molecular domains individually or at the systems-level. Epigenomics includes molecules that interact with DNA to regulate gene expression potential, transcriptomics includes expression of RNAs, proteomics includes the expression levels of peptides, and metabolomics includes the activities and interactions of metabolites. While the genomic sequence is biologically inherited, genotype may influence the biological response to certain exposures across multiple domains. SNP = single nucleotide polymorphisms, CNV = copy number variations, CpG = cytosine-phosphate-guanine.
Integrative personal -omics profiling (iPOP), gathering high-throughput data on multiple molecular markers from the same individual, is increasingly recognized for its utility in personalized medicine [10,11]. Initial iPOP studies have demonstrated that personal molecular profiles may be particularly useful for identifying molecular events that mark transitions between healthy and diseased states. The Adverse Outcome Pathway (AOP) framework, characterizing how chemical exposures perturb different aspects of biology and ultimately lead to some adverse biological event, provides a context in which -omics profiling may be particularly useful in broadening environmental health research [12]. This idea has been proposed in the context of epigenetic epidemiology – organizing the findings from these studies into an AOP framework could improve causal reasoning by characterizing biologic plausibility in an evidence-based manner [13]. Incorporating other molecular features, beyond those studied in epigenomics, could help to clarify which biological perturbations represent molecular initiating events (MIE), key events (KE), and key event relationships (KER) on the path to different adverse outcomes (AO). In fact, molecular -omics integration into environmental health studies has been suggested as an approach that would improve health risk assessment by informing the development of AOPs via identifying the molecular targets of environmental exposure and determining whether groups of chemicals and/or stressors impact related biological processes [14]. Multiple exposures that have the same molecular targets or affect similar biological pathways may be more likely to exhibit joint-effects, multiplicative and or additive, on health outcomes and should be studied as mixtures or co-exposures. In addition to furthering our understanding of AOPs, incorporating iPOP approaches into population based studies can distinguish inter- and intra-individual variations in molecular characteristics [10] and help to characterize whether the impacts of an exposure depend on genetics, lifestage, or prior exposure and/or disease status [14].
Design Considerations:
Population-based studies that involve molecular -omics need to consider unique potential sources of bias related to sampling of tissues and analyzing of high-dimensional data, so that appropriate conclusions can be drawn from these studies.
In human environmental health research, -omics data are typically measured in tissues or biological matrices that are easily accessible and may be composed of heterogeneous cell populations. This presents two potential issues: (1) whether the expected exposure-outcome relationships are detectable and meaningful in the accessible tissue, and (2) whether cellular or tissue heterogeneity could confound these exposure-outcome relationships. Whether the available tissue samples are the target tissue or can be an effective surrogate, is largely dependent on the exposure-outcome relationships being studied and whether the specific loci or features demonstrate tissue-specific patterns [15,16]. Thus, in epidemiologic studies, investigators need to consider whether the tissues that will be accessible or available are expected to capture the associations between exposures and the biological responses. Additionally, some epigenetic loci are differentially methylated [17] and certain genes are differentially expressed [18] in highly cell-type specific patterns. Thus, when attempting to investigate differences between groups of samples (exposed vs unexposed, or cases vs controls) it is difficult to disentangle whether the observed differences are capturing an average change across all cell-types, a change in just a subset of cells, or a shift in the distribution or composition of the cell mixture itself.
Some researchers have suggested that epigenomic studies should focus on analyses within sorted subsets of specific cell types [19] and a similar argument could be made for studies of other molecular features including telomere length, transcriptomics, etc. Performing these types of studies within specific subsets of cells should yield some important benefits, including (1) larger magnitudes of associations, (2) improved sensitivity and specificity for classification, (3) a more focused understanding of the biological consequences of those epigenetic changes, and (4) reduced sample size requirements given the potentially larger effect sizes [19]. However, this approach also has some limitations that cannot be ignored. In order to perform an association study within specific cell-types, researchers require a priori knowledge of which cell-types are most likely to be involved in the exposure-response relationship, something that is largely unknown for many prenatal and early-life exposures and outcomes. Additionally, cell-sorting of most tissues can only be performed on biosamples that have been collected, transported and stored in a manner that preserves cellular membranes and epitopes, and the steps to preserve these structures can differ for specific cell-types. Thus, many existing biosamples which were not collected according to such protocols would be inappropriate for cell-type specific studies, and the incorporation of such protocols in future studies would require additional resources that may limit the sample size that can be investigated. Finally, although cell-type specific association studies have the potential to yield larger magnitudes of effect, and possibly improved sensitivity/specificity as biomarkers, the additional steps required (collection, enrichment, purification, sorting) to utilize those biomarkers may also make them prohibitive in some settings. Thus, although these approaches may have utility in directed, hypothesis driven mechanistic studies, they may hold less utility in the broader, discovery-style studies that are the current state of the science for prenatal and early life genomics research.
For studies that aim to perform molecular assessments in heterogenous biosamples that will not be sorted, direct measurements of cell-mixtures or heterogeneity of tissue should be obtained in parallel to the molecular -omic data when possible, or estimated when this is infeasible. Numerous tools have been developed which estimate tissue heterogeneity using epigenomic [20–22] or transcriptomic [23,24] data, so that the mixture can be statistically adjusted for during data analysis. Controlling for cellular heterogeneity in epigenome-wide association studies is often critical to reducing the rate of false-positive findings [17]. Even when the goal of an analysis is solely to detect molecular biomarkers of an exposure or disease, it is important to explore whether those epigenetic markers are acting purely as surrogates for different distributions of cell types, as this can be informative of the toxicant’s mechanism of action or on the potential consequences of these alterations. Though similar tools are not yet available in the context of proteomic or metabolomic data, and it is not as apparent that these molecular features are as susceptible to confounding by tissue heterogeneity, studies incorporating these data-types should consider whether cellular heterogeneity may potentially confound the relationships that the investigators seek to identify.
It is also important to discuss how the tissue of origin can impact findings in molecular epidemiology. The majority of studies with research questions about prenatal and early-life exposures utilize tissues that are most readily accessible, including placenta, blood (peripheral or cord), and buccal cells. In some cases, the accessible tissues are relevant to the expected exposure-outcome relationships. The placenta, for instance, is known to play critical roles in the growth and development of the fetus, and placental activities have been shown to be responsive to multiple maternal exposures and lead to restricted fetal growth [25]. Thus, the placenta is a likely target tissue for environmental exposures that are thought to impair fetal growth. However, the available tissues do not always represent the ideal target tissues for some of health outcomes that are most relevant to the exposures of interest. For instance, neurotoxicants that affect behavior or cognition are most likely to alter the development and/or functions of brain cells, but brain tissue can only be obtained from humans post mortem. Thus, before utilizing a surrogate tissue, such as blood, to study the impacts of neurotoxicants it is critical to consider whether a molecular response in that surrogate tissue is meaningful for the health outcomes that are relevant for that exposure. Bakulski et al (2016) provide a thorough review of human epigenomic studies of neuropsychiatric disorders in both target brain tissues and surrogate tissues such as blood and buccal cells – they found that surrogate tissues could be useful for both biomarker of disorders and to provide insights in disease mechanisms [26]. Molecular epidemiologists need to carefully consider their research questions and examine the existing literature to determine whether the tissues that will be available to them, and the molecular measure they are considering, can capture the expected exposure-response relationships.
The ability to detect true associations, statistical power, is dependent on the magnitude of effect, the variation in the effect, and noise. Studies with small sample sizes are more likely to be underpowered, which increases the probability of false positives, false negatives, and exaggerated estimated effect sizes [27]. Additionally, variations in levels of DNA methylation [28], transcription [28], and metabolite concentrations [29] can originate from many sources including within- and between-individual variability, as well as technical variability. Thus, when designing environmental health studies that will incorporate molecular -omics data, power calculations must be performed to estimate an adequate sample sizes to identify the estimated effects in the face of these multiple sources of variability. Additionally, all of these molecular data need to undergo appropriate quality control, normalization, processing, and statistical adjustments to correct for batch effects to limit the influence of technical variability on study results [30–33].
A number of experimental considerations also must be contemplated when designing an -omics experiment. The timing of sampling may be critical, on multiple scales of time. In considering disease outcomes, prospective sampling will allow for unbiased assessment of causality, but may be difficult to obtain dependent on the type of outcome being interrogated. On a more immediate scale, time of day can impact certain -omics measures, particularly metabolomics or transcriptomics where circadian patterns or timing specific confounders may alter specific readouts. In addition, timing and protocol from sample collection through processing is also a critical consideration, particularly when the molecular feature may be labile, such as those of RNA and some metabolites.
Carefully addressing the above considerations when designing an -omics based environmental health study can help investigators deal with the potential sources of bias proactively and improve the rigor and reproducibility of those studies. In addition to the above potential issues that are specific to molecular analysis in population-based studies, other traditional epidemiologic concepts should be carefully considered, including confounding, selection bias, effect modification, mediation, and temporality.
We present illustrative examples of how molecular -omics data have been successfully integrated into environmental health studies to date, with a focus on prenatal and/or early life exposures. Prenatal exposures may be particularly important to human health since organs are undergoing development and programming at this time, and alterations to these processes can have acute consequence to the fetus and alter long-term risk of disease later in life [34]. The mechanisms through which prenatal environmental exposures may impact birth outcomes and long-term health in humans have primarily been studied through the lens of fetal epigenetics [34], though transcriptomics, proteomics and metabolomics have begun to provide valuable insights into these exposure-outcome relationships as well.
Highlights of -omics studies of prenatal exposures and early life health
The prenatal exposure that has been most thoroughly studied in relation to fetal molecular -omics is maternal smoking during pregnancy (MSDP), largely because the relationship between smoking and reduced birth size has been observed consistently across numerous independent studies [34] and small size at birth is a risk factor for the development of chronic diseases later in life. Multiple single-cohort epidemiologic studies have examined the relationship between MSDP and fetal epigenetics, as a potential mechanism through which smoking may impact birth weight and long-term health outcomes. To date, the largest of such studies was undertaken by Joubert et al (2016) who performed a meta-analyses of 13 independent cohorts from the Pregnancy and Childhood Epigenetics (PACE) consortium [35], including 6,685 newborns. This meta-analysis identified over 6,000 epigenetic loci that were differentially methylated in relation to MSDP at a 5% FDR threshold. The top differentially methylated loci was at the aryl hydrocarbon receptor repressor (AHRR) gene [36], which had been associated with tobacco smoke exposure in multiple previous studies. Joubert et al (2016) implemented numerous epidemiologic principles that contribute to the strength of evidence surrounding these findings, performing functional network and pathway enrichment analyses, testing the relationships between methylation and expression, and testing for associations between MSDP and DNA methylation in older children, to provide additional evidence for biological plausibility that these epigenetic variations may impact biological function and can persist after birth [36]. This meta-analysis served as a confirmatory study not only for the AHRR gene, which is predictive of smoking-associated morbidity and mortality [37], but for 967 epigenetic loci that had previously been reported as differentially methylated with MSDP, thus identifying numerous candidate loci that may deserve follow-up in future studies. Additionally, Reese et al (2017) have since developed a method for estimating prenatal exposure to maternal smoking from cord blood methylation data, demonstrating the utility of high-throughput epigenomics data for developing biomarkers of environmental exposure [38]. Epigenetic variations in other fetal tissues have also been associated with MSDP, such as placenta [39] and fetal cortex [40]. The top hits from these studies were in different genomic locations than those identified in cord blood and may represent tissue-specific responses to MSDP, which emphasizes the need for additional studies of MSDP in other fetal tissues, and well as validation studies in placenta and brain tissue in independent studies with larger sample sizes.
Studies of the fetal transcriptome in response to MSDP have also been performed, identifying differentially expressed genes involved in xenobiotic metabolism, coagulation, and thrombosis, despite these studies being done in independent samples with relatively small sample sizes [41,42]. Additionally, a study of the placental proteome found that protein networks involved in cellular morphology, organization and compromise, or involved in DNA replication, recombination, and repair, energy production and nucleic acid metabolism may be impacted by MSDP. The authors also examined difference in selected transcript levels of candidate genes, again finding genes involved in xenobiotic metabolism to be differentially expressed [43]. All three of these placental transcriptome/proteome studies have been performed on small sample sizes, but produced some strikingly similar results, warranting more comprehensive examinations in larger studies.
Multiple studies have also examined the relationships between air pollution exposure and molecular profiles measured via -omics technologies in fetal tissues. Small numbers of differentially methylated loci have been identified in placental tissue associated with distance to roadway [44] and in cord blood associated with nitrogen dioxide (NO2) exposure [45], suggesting possible air-pollution-associated differential epigenetic regulation of protein tyrosine phosphatase receptors and mitochondrial activity. Kingsley et al (2016) also identified that prenatal exposure to black carbon and PM2.5 were associated with variations in expression of a large proportion of placental imprinted genes [46], some of which had been previously linked to birthweight in the same cohort and are known to play roles in fetal development [47]. Winckelmans et al. (2017) examined relationships between maternal long-term and short-term PM2.5 exposure and gene expression patterns in cord blood related to immune and DNA damage response, and that many of the observed relationships may be highly dependent on fetal sex with [48]. While Martens et al (2017) studied associations between PM2.5 and cord blood oxylipins, a subset of metabolites generated from fatty-acid oxidation, observing associations with metabolites derived from the lipoxygenase pathway which is involved in inflammatory signaling [49]. These studies have begun to elucidate how maternal air pollution exposure can impact the molecular activities in fetal tissues, but independent studies with large sample sizes are needed to validate many of these findings.
Prenatal exposures to toxic metals have also received substantial attention for their potential impacts on fetal epigenetic patterns. Multiple studies have demonstrated that DNA methylation levels in cord blood and/or placenta are associated with prenatal exposure to metals, including arsenic (As) [50–54], mercury (Hg) [55,56], cadmium (Cd) [57–59], and lead (Pb) [60]. These studies demonstrate how molecular markers may be able to detect functional changes in developing tissues associated with maternal exposures to environmental contaminants, often trace exposure levels, and explore the implications for health and development. For instance, Everson et al (2018) found numerous epigenetic loci associated with Cd in placental tissue that were consistent across two populations, then incorporated transcriptomic data to infer the biological processes in the placenta that may be impacted by these variations, such as inflammatory signaling, cellular growth, and metabolism [59]. Prenatal As exposure has also been studied for associations with fetal metabolomics and proteomics. Prenatal exposure to inorganic arsenic (iAs) has been associated with variations in the cord serum metabolome, potentially related to regulation of the citrate cycle, as well as vitamin and amino acid metabolism [61], and a pilot study implicated that the toxic effects of in-utero iAs on birth weight may be mediated through alterations to metabolic activity in cord blood, specifically laurate, 17-methylstearate, and 4-vinylphenol sulfate [62]. Additionally, prenatal iAs has been associated with the expression of numerous proteins that are involved in Tumor necrosis factor (TNF) mediated immune and/or inflammatory response [63].
Other prenatal environmental exposures have been studied in relation to fetal molecular profiles as well. Using a meet-in-the-middle approach, Pennings et al (2016) found that cord blood expression levels of 52 genes that had been associated with prenatal PFAS exposure were also associated with episodes of the common cold or rubella titers, thus providing supporting evidence that prenatal PFAS exposure may have immunotoxic effects on offspring [64]. Remy et al (2016) examined how the cord blood transcriptome was perturbed by multiple prenatal environmental exposures, including multiple endocrine disruptors, metals, and particulates, and identified p, p’-DDE the most impactful environmental exposure on the fetal transcriptome, and p, p’-DDE was most strongly associated with differential expression of the glucocorticoid receptor (NR3C1) which plays critical roles in fetal development [65]. The approach used by Remy et al (2016) identifies the molecular targets, in this case RNA transcripts, that are associated with environmental exposures, which could be informative for identifying sets of exposures that impact the same molecular features and which exposures have the greatest impact on individual features.
Although metabolomics does have the ability to describe molecular profiles that are informative about physiological activity that may be related to fetal programming, it can also be utilized to characterize internalized exposures or health states during pregnancy. Sulek (2017) compared metabolomic profiles derived from maternal hair samples between cases of fetal growth restriction (FGR) to controls [66]. They identified 32 metabolites that significantly differed between cases and controls, and a multivariate model including just 5 of these metabolites achieved an ROC of 0.998, and speculate that many of these difference may be related to loss of redox control [66]. Maitre et al (2016) examined the relationships between maternal urinary metabolites with FGR and preterm birth in the Rhea cohort, FGR pregnancies were less likely among mother with higher levels of tyrosine, acetate, trimethylamine and formate, while medically induced preterm births were associated with higher levels of N-acetyl glycoproteins and spontaneous preterm birth was associated with high lysine and low formate levels, and posit that these metabolites may be related to maternal metabolic health during pregnancy [67]. Maternal urinary metabolomics was also studied in two independent Spanish birth cohorts, Sabadell and Gipuzka, and the authors observed 10 maternal urinary metabolites that were strongly predictive of birth weight, including branched-chain amino acids (BCAAs) and steroid hormone by-products [68]. Hellmuth et al (2017) performed a metabolomics study of cord blood, and found numerous strong associations with birth weight, but these metabolites were not strongly predictive of weight gain or BMI in adolescences; they propose that this may be due to the metabolome being highly influenced by the immediate environment, and thus may be more sensitive for detecting associations in cross-sectional settings [69]. On the other hand, Isganaitis et al (2015) identified several cord blood metabolites that were strongly associated with postnatal weight gain, and observed that the ratio of glutamine to glutamate was lower, a potential marker of cardiometabolic risk, among those with accelerated weight gain [70]. Prenatal and early-life vitamin D exposure has also been shown to influence metabolomics profiles of children at 3 years of age, which may suggest a potential role for metabolomics in detecting metabolic reprogramming [71]. Maternal smoking during pregnancy has been associated with alterations to both the maternal and fetal metabolome [72], as well as the placental proteome and metabolome and suggest that these differences may be related to increased oxidative stress in placenta of smoking mothers [43]. Though only a few studies have currently been performed, early studies of fetal metabolomics in association with maternal exposures are promising. These studies demonstrate the promising potential of metabolomics for examining biomarkers of maternal exposures, as well as maternal and fetal health states, to more comprehensively assess how the maternal exposome impacts fetal and neonatal health.
Multi-omics Analyses
The above studies highlight many of the successful incorporations of -omics data within environmental health studies of prenatal exposures. However, most of these studies only include a single high-dimensional molecular data set and thus are only capable of identifying associations within the molecular domains they investigated. Multi-omics analyses on the other-hand examine the associations between features across multiple high-throughput data sets. Two approaches for multi-omics analyses include the quantitative trait approach and the multi-omics integration approach.
Quantitative trait loci (QTL) studies seek to identify all associations between features on different molecular datasets, or at least those within close genomic proximity to each other, thus characterizing the functional relationships between molecular -omics domains. Methylation QTLs (mQTLs) are CpG sites whose methylation levels are influenced by genetic variants, while expression QTLs (eQTLs) are genes whose expression levels are influenced by genetic variants, and expression quantitative trait methylations (eQTM) are genes whose expression levels are influenced by DNA methylation [73]. Gutierrez-Arcelus (2013) performed eQTL, mQTL, and eQTM in umbilical cord tissue samples [74], Peng et al (2017) performed an eQTL analysis of placental tissues [75], and Gaunt et al. (2016) have developed a database of mQTLs in human blood at various ages throughout the life-course [76]. These may serve as valuable resources that should be accessed by researchers when interpreting -omics data obtained from fetal tissues. In addition to providing functional context, QTL approaches can be performed within individual cohorts in a reductive way, to only test the features that exhibit QTL associations for additional associations with exposures or outcomes. Teh et al. (2014) did this, identifying mQTLs in fetal tissue, followed by an association study between mQTLs and maternal exposures that likely affect the in-utero environment (smoking, maternal depression, maternal BMI, infant birth weight, gestational age, and birth order). They identified numerous strong exposure-methylation associations via this method, also finding that variations in mQTLs were better explained by gene-by-environment interactions, than by genetics alone at ~75% of the identified mQTLs [77]. In the majority of these G-by-E associated mQTLs, the environment tended to be associated with DNA methylation only among individuals of a particular genotype [77]. An alternative approach can be applied to the loci identified in epigenome-wide association studies, correlating DNA methylation with gene expression, to characterize whether the top hits from an EWAS may be regulators of nearby gene expression. For instance, the expression of the LYRM 2 gene was shown to be inversely correlated with DNA methylation at the loci identified from an EWAS of prenatal As exposure [54], and the expression the TNFAIP2 gene was positively correlated with DNA methylation at the loci identified from an EWAS of placental Cd concentrations [59]. Incorporating gene-expression analyses into EWAS studies such as these, improves the biological plausibility that an environmentally-associated epigenetic variation can influence the functions of the tissue from which it was measured.
The other multi-omic approach involves -omics integration, similar to iPOP, to examine interrelationships between multiple different -omics data simultaneously, and ideally longitudinally. To date the iPOP framework has primarily been applied to small sample sizes and focused on changes in health states rather than environmental exposures [10,11]. Chen et al. (2012) followed one individual over a 14-month period while examining changes in molecular profiles over time and speculated about indicators for the onset risk for type II diabetes, recognizing that risk cannot be ascertained on a study of one individual [11], while Piening et al. (2018) extended this iPOP framework to a sample of 23 individuals, examining how molecular profiles change with weight gain and weight loss, compared profiles between insulin-sensitive and insulin-resistant persons, and evaluated inter- and intra-individual variations in those profiles [10]. The iPOP framework has primarily been studied through a lens of personalized medicine and the possibility of improved clinical outcomes. However, the clinical utility of the vast majority of detectable molecular markers is currently unknown and requires additional study. We posit that incorporating longitudinal multi-omic profiling in human population-based studies, particularly in birth cohort studies, can help to characterize how molecular profiles change throughout the life-course, in response to environmental exposures, and in association with various health outcomes. Some epidemiologic investigations have utilized multiple integrated -omics data in larger sample sizes. Inouye et al (2010) examined interrelationships between genomics, transcriptomics, and metabolomics in relations to immune response, metabolism and adiposity [78], then performed a multi-sample study that incorporated longitudinal follow-up to validate the original findings and more comprehensively characterize immuno-metabolic cross-talk [79]. These illustrative examples demonstrate the abilities of multi-omics data integration to comprehensively characterize physiological activity, but such integrative approaches have only been applied to environmental health research in a limited capacity.
Conclusions and Future Directions
The pathophysiologies of environmentally induced diseases are complex and often will not be centered around one individual molecular target. Instead, environmental exposures may influence the activity of multiple biological factors, and combinations of these factors may lead to disease development. The emergence and rapid growth of -omics technologies are opening avenues of research for environmental health, that will allow for better detection and characterization of intermediate molecular biomarkers along these exposure-response relationships, and provide novel opportunities to develop preventive strategies or identification of therapeutic targets. Epigenomics has become a common component to investigating the mechanisms underlying how maternal exposures may alter fetal programming and thus impact birth outcomes and fetal health, and has demonstrated substantial utility for this task. However, studies focused on transcriptomic, proteomic, and metabolomic data have also yielded promising results, particularly for identifying biomarkers of exposure and predictors of outcomes. As -omics technologies continue to improve and become more affordable, epidemiologic studies need obtain multiple -omics measures from the same individuals, and from multiple tissues, ideally in a longitudinal manner so that inter- and intra-individual variations can be more well characterized, and targets identified as possible biomarkers to be validated in independent populations. The issues and challenges associated with molecular -omics data require that investigators consider traditional epidemiologic sources of bias, such as confounding, selection bias, measurement error, and reverse causation, but also include unique source of bias which could be biological confounders or technical variability. To ensure that environmental health research benefits from these advancements, environmental health researchers should receive cross training in molecular epidemiology and -omics and seek collaborations with researchers that have expertise in the use of these tools.
References:
- [1].ECHA. Guidance for identification and naming of substances under REACH and CLP. Version 2.1, May 2017. [2018. doi: 10.2823/538683. [DOI] [Google Scholar]
- [2].USEPA. About the TSCA Chemical Substance Inventory 2018. https://www.epa.gov/tsca-inventory/about-tsca-chemical-substance-inventory (accessed February 2, 2018).
- [3].Kyrtopoulos SA. Making sense of OMICS data in population-based environmental health studies. Environ Mol Mutagen 2013;54:468–79. doi: 10.1002/em.21778. [DOI] [PubMed] [Google Scholar]
- [4].Klein RJ, Zeiss C, Chew EY, Tsai J-Y, Sackler RS, Haynes C, et al. Complement factor H polymorphism in age-related macular degeneration. Science (80-) 2005;308:385–9. doi: 10.1126/science.1109557.Complement. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Auton A, Abecasis GR, Altshuler DM, Durbin RM, Bentley DR, Chakravarti A, et al. A global reference for human genetic variation. Nature 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Langevin SM, Kelsey KT. The fate is not always written in the genes: Epigenomics in epidemiologic studies. Environ Mol Mutagen 2013;54:533–41. doi: 10.1002/em.21762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].McHale CM, Zhang L, Thomas R, Smith MT. Analysis of the Transcriptome in Molecular Epidemiology Studies. Env Mol Mutagen 2013;54:500–17. doi: 10.1002/em. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Breker M, Schuldiner M. The emergence of proteome-wide technologies: Systematic analysis of proteins comes of age. Nat Rev Mol Cell Biol 2014;15:453–64. doi: 10.1038/nrm3821. [DOI] [PubMed] [Google Scholar]
- [9].Fearnley LG, Inouye M. Metabolomics in epidemiology: from metabolite concentrations to integrative reaction networks. Int J Epidemiol 2016;45:1319–28. doi: 10.1093/ije/dyw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Piening BD, Zhou W, Contrepois K, Rost H, Gu Urban GJ, Mishra T, et al. Integrative Personal Omics Profiles during Periods of Weight Gain and Loss. Cell Syst 2018;6:1–14. doi: 10.1016/j.cels.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Chen R, Mias GI, Li-Pook-Than J, Jiang L, Lam HYK, Chen R, et al. Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 2012;148:1293–307. doi: 10.1016/j.cell.2012.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12]**.Brockmeier EK, Hodges G, Hutchinson TH, Butler E, Hecker M, Tollefsen KE, et al. The role of omics in the application of adverse outcome pathways for chemical risk assessment. Toxicol Sci 2017;158:252–62. doi: 10.1093/toxsci/kfx097. Emphasizes the ways in which molecular -omics can be used to inform AOPs and improve chemical risk assessment. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Angrish MM, Allard P, McCullough SD, Druwe IL, Helbling Chadwick L, Hines E, et al. Epigenetic Applications in Adverse Outcome Pathways and Environmental Risk Evaluation. Environ Health Perspect 2018;126:1–12. doi: 10.1289/EHP2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cote I, Andersen ME, Ankley GT, Barone S, Birnbaum LS, Boekelheide K, et al. The next generation of risk assessment multi-year study - highlights of findings, applications to risk assessment, and future directions. Environ Health Perspect 2016;124:1671–82. doi: 10.1289/EHP233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rockett JC, Burczynski ME, Fornace AJ, Herrmann PC, Krawetz SA, Dix DJ. Surrogate tissue analysis: Monitoring toxicant exposure and health status of inaccessible tissues through the analysis of accessible tissues and cells. Toxicol Appl Pharmacol 2004;194:189–99. doi: 10.1016/j.taap.2003.09.005. [DOI] [PubMed] [Google Scholar]
- [16].Huang YT, Chu S, Loucks EB, Lin CL, Eaton CB, Buka SL, et al. Epigenome-wide profiling of DNA methylation in paired samples of adipose tissue and blood. Epigenetics 2016;11:227–36. doi: 10.1080/15592294.2016.1146853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17]*.Jaffe AE, Irizarry RA. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol 2014;15. doi: 10.1186/gb-2014-15-2-r31. Addresses the issue of performing epigenome-wide association studies in mixed cell populations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gren ST, Rasmussen TB, Janciauskiene S, Hakansson K, Gerwien JG, Grip O. A single-cell gene-expression profile reveals inter-cellular heterogeneity within human monocyte subsets. PLoS One 2015;10:1–20. doi: 10.1371/journal.pone.0144351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bauer M Cell-type-specific disturbance of DNA methylation pattern: a chance to get more benefit from and to minimize cohorts for epigenome-wide association studies. Int J Epidemiol 2018:1–11. doi: 10.1093/ije/dyy029. [DOI] [PubMed] [Google Scholar]
- [20].Teschendorff AE, Breeze CE, Zheng SC, Beck S. A comparison of reference-based algorithms for correcting cell-type heterogeneity in Epigenome-Wide Association Studies. BMC Bioinformatics 2017;18:1–14. doi: 10.1186/s12859-017-1511-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Rahmani E, Zaitlen N, Baran Y, Eng C, Hu D, Galanter J, et al. Sparse PCA corrects for cell type heterogeneity in epigenome-wide association studies. Nat Methods 2016;13:443–5. doi: 10.1038/nmeth.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Houseman EA, Kile ML, Christiani DC, Ince TA, Kelsey KT, Marsit CJ. Reference-free deconvolution of DNA methylation data and mediation by cell composition effects. BMC Bioinformatics 2016;17:259. doi: 10.1186/s12859-016-1140-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453–7. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Li Y, Xie X. A mixture model for expression deconvolution from RNA-seq in heterogeneous tissues. BMC Bioinformatics 2013;14. doi: 10.1186/1471-2105-14-S5-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Dimasuay KG, Boeuf P, Powell TL, Jansson T. Placental responses to changes in the maternal environment determine fetal growth. Front Physiol 2016;7:1–9. doi: 10.3389/fphys.2016.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bakulski KM, Halladay A, Hu VW, Mill J, Fallin MD. Epigenetic Research in Neuropsychiatric Disorders: the “Tissue Issue “. Curr Behav Neurosci Rep 2016;3:264–74. doi: 10.1007/s40473-016-0083-4.Epigenetic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Christley RM. Power and Error: Increased Risk of False Positive Results in Underpowered Studies. Open Epidemiol J 2010;3:16–9. doi: 10.2174/1874297101003010016. [DOI] [Google Scholar]
- [28].Shvetsov UB, Song M-A, Cai Q, Tiirikainen M, Xiang Y-B, Shu X-O, et al. Intra-individual variation and short-term temporal trend in DNA methylation of human blood. Cancer Epidemiol Biomarkers Prev 2015;24:490–7. doi: 10.1158/1055-9965.EPI-14-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Xiao Q, Moore SC, Boca SM, Matthews CE, Rothman N, Stolzenberg-Solomon RZ, et al. Sources of variability in metabolite measurements from urinary samples. PLoS One 2014;9:e95749. doi: 10.1371/journal.pone.0095749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wehrens R, Hageman JA, van Eeuwijk F, Kooke R, Flood PJ, Wijnker E, et al. Improved batch correction in untargeted MS-based metabolomics. Metabolomics 2016;12. doi: 10.1007/s11306-016-1015-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Leek JT, Scharpf RB, Bravo HC, Simcha D, Langmead B, Johnson WE, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet 2010;11:733–9. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sun Z, Chai HS, Wu Y, White WM, Donkena KV., Klein CJ, et al. Batch effect correction for genome-wide methylation data with Illumina Infinium platform. BMC Med Genomics 2011;4. doi: 10.1186/1755-8794-4-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Peixoto L, Risso D, Poplawski SG, Wimmer ME, Speed TP, Wood MA, et al. How data analysis affects power, reproducibility and biological insight of RNA-seq studies in complex datasets. Nucleic Acids Res 2015;43:7664–74. doi: 10.1093/nar/gkv736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Barker DJP. The Developmental Origins of Adult Disease. J Am Coll Nutr 2004;23:588S–595S. doi: 10.1080/07315724.2004.10719428. [DOI] [PubMed] [Google Scholar]
- [35].Felix JF, Joubert BR, Baccarelli AA, Sharp GC, Almqvist C, Annesi-Maesano I, et al. Cohort profile: Pregnancy and childhood epigenetics (PACE) consortium. Int J Epidemiol 2018;47. doi: 10.1093/ije/dyx190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36]**.Joubert BR, Felix JF, Yousefi P, Bakulski KM, Just AC, London SJ, et al. DNA Methylation in Newborns and Maternal Smoking in Pregnancy: Genome-wide Consortium Meta-analysis. Am J Hum Genet 2016;98:680–96. doi: 10.1016/j.ajhg.2016.02.019. Large meta-analyses examining the relationship between prenatal smoke exposure and newborn epigenomics. Similar approaches should be encouraged for other molecular omics studies to improve robustness and reproducibility of findings. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Bojesen SE, Timpson N, Relton C, Smith GD, Nordestgaard BG. AHRR (cg05575921) hypomethylation marks smoking behaviour, morbidity and mortality. Thorax 2017:646–53. doi: 10.1136/thoraxjnl-2016-208789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Reese SE, Zhao S, W u MC, Joubert BR, Parr CL, Håberg SE, et al. DNA methylation score as a biomarker in newborns for sustained maternal smoking during pregnancy. Environ Health Perspect 2017;125:760–6. doi: 10.1289/EHP333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Morales E, Vilahur N, Salas LA, Motta V, Fernandez MF, Murcia M, et al. Genome-wide DNA methylation study in human placenta identifies novel loci associated with maternal smoking during pregnancy. Int J Epidemiol 2016;45:1644–55. doi: 10.1093/ije/dyw196. [DOI] [PubMed] [Google Scholar]
- [40].Chatterton Z, Hartley BJ, Seok MH, Mendelev N, Chen S, Milekic M, et al. In utero exposure to maternal smoking is associated with DNA methylation alterations and reduced neuronal content in the developing fetal brain. Epigenetics and Chromatin 2017;10:1–11. doi: 10.1186/s13072-017-0111-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Votavova H, Dostalova Merkerova M, Fejglova K, Vasikova A, Krejcik Z, Pastorkova A, et al. Transcriptome alterations in maternal and fetal cells induced by tobacco smoke. Placenta 2011;32:763–70. doi: 10.1016/j.placenta.2011.06.022. [DOI] [PubMed] [Google Scholar]
- [42].Bruchova H, Vasikova A, Merkerova M, Milcova A, Topinka J, Balascak I, et al. Effect of Maternal Tobacco Smoke Exposure on the Placental Transcriptome. Placenta 2010;31:186–91. doi: 10.1016/j.placenta.2009.12.016. [DOI] [PubMed] [Google Scholar]
- [43].Huuskonen P, Amezaga MR, Bellingham M, Jones LH, Storvik M, Hakkinen M, et al. The human placental proteome is affected by maternal smoking. Reprod Toxicol 2016;63:22–31. doi: 10.1016/j.reprotox.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kingsley SL, Eliot MN, Whitsel EA, Huang YT, Kelsey KT, Marsit CJ, et al. Maternal residential proximity to major roadways, birth weight, and placental DNA methylation. Environ Int 2016;92–93:43–9. doi: 10.1016/j.envint.2016.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Gruzieva O, Xu CJ, Breton CV., Annesi-Maesano I, Anto JM, Auffray C, et al. Epigenome-wide meta-analysis of methylation in children related to prenatal NO2air pollution exposure. Environ Health Perspect 2017;125:104–10. doi: 10.1289/EHP36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kingsley SL, Deyssenroth MA, Kelsey KT, Awad YA, Kloog I, Schwartz JD, et al. Maternal residential air pollution and placental imprinted gene expression. Environ Int 2017;108:204–11. doi: 10.1016/j.envint.2017.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Kappil MA, Green BB, Armstrong DA, Sharp AJ, Lambertini L, Marsit CJ, et al. Placental expression profile of imprinted genes impacts birth weight. Epigenetics 2015;10:842–9. doi: 10.1080/15592294.2015.1073881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Winckelmans E, Vrijens K, Tsamou M, Janssen BG, Saenen ND, Roels HA, et al. Newborn sex-specific transcriptome signatures and gestational exposure to fine particles: findings from the ENVIRONAGE birth cohort. Environ Heal A Glob Access Sci Source 2017;16:1–17. doi: 10.1186/s12940-017-0264-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Martens DS, Gouveia S, Madhloum N, Janssen BG, Plusquin M, Vanpoucke C, et al. Early-Life Environment: An ENVIRONAGE Birth Cohort Study. Environ Health Perspect 2017;125:691–8. doi: 10.1289/EHP291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Cardenas A, Houseman EA, Baccarelli AA, Quamruzzaman Q, Rahman M, Mostofa G, et al. In utero arsenic exposure and epigenome-wide associations in placenta, umbilical artery, and human umbilical vein endothelial cells. Epigenetics 2015;10:1054–63. doi: 10.1080/15592294.2015.1105424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Koestler DC, Avissar-Whiting M, Andres Houseman E, Karagas MR, Marsit CJ. Differential DNA methylation in umbilical cord blood of infants exposed to low levels of arsenic in utero. Environ Health Perspect 2013;121:971–7. doi: 10.1289/ehp.1205925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Rojas D, Rager JE, Smeester L, Bailey KA, Drobna Z, Rubio-Andrade M, et al. Prenatal arsenic exposure and the epigenome: identifying sites of 5-methylcytosine alterations that predict functional changes in gene expression in newborn cord blood and subsequent birth outcomes.Toxicol Sci 2015;143:97–106. doi: 10.1093/toxsci/kfu210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kaushal A, Zhang H, Karmaus WJJ, Everson TM, Marsit CJ, Karagas MR, et al. Genome-wide DNA methylation at birth in relation to in utero arsenic exposure and the associated health in later life. Environ Heal 2017;16. doi: 10.1186/s12940-017-0262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Green BB, Karagas MR, Punshon T, Jackson BP, Robbins DJ, Houseman EA, et al. Epigenome-wide assessment of DNA methylation in the placenta and arsenic exposure in the New Hampshire Birth Cohort Study (USA). Environ Health Perspect 2016;124:1253–60. doi: 10.1289/ehp.1510437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Cardenas A, Koestler DC, Houseman EA, Jackson BP, Kile ML, Karagas MR, et al. Differential DNA methylation in umbilical cord blood of infants exposed to mercury and arsenic in utero. Epigenetics 2015;10:508–15. doi: 10.1080/15592294.2015.1046026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Maccani JZJ, Koestler DC, Lester B, Andrés Houseman E, Armstrong DA, Kelsey KT, et al. Placental dna methylation related to both infant toenail mercury and adverse neurobehavioral outcomes. Environ Health Perspect 2015;123:723–9. doi: 10.1289/ehp.1408561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Kippler M, Engstrom K, Jurkovic Mlakar S, Bottai M, Ahmed S, Hossain MB, et al. Sex-specific effects of early life cadmium exposure on DNA methylation and implications for birth weight. Epigenetics 2013;8:494–503. doi: 10.4161/epi.24401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Sanders AP, Smeester L, Rojas D, DeBussycher T, W u MC, Wright FA, et al. Cadmium exposure and the epigenome: Exposure-associated patterns of DNA methylation in leukocytes from mother-baby pairs. Epigenetics 2014;9:212–21. doi: 10.4161/epi.26798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Everson TM, Punshon T, Jackson BP, Hao K, Lambertini L, Chen J, et al. Cadmium-Associated Differential Methylation throughout the Placental Genome: Epigenome-Wide Association Study of Two U.S. Birth Cohorts. Environ Health Perspect 2018;126:1–13. doi: 10.1289/EHP2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Wu S, Hivert MF, Cardenas A, Zhong J, Rifas-Shiman SL, Agha G, et al. Exposure to low levels of lead in utero and umbilical cord blood DNA methylation in project viva: An epigenome-wide association study. Environ Health Perspect 2017;125:1–10. doi: 10.1289/EHP1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Laine JE, Bailey KA, Olshan AF, Smeester L, Drobna Z, Styblo M, et al. Neonatal Metabolomic Profiles Related to Prenatal Arsenic Exposure. Env Sci Technol 2017;51:625–33. doi: 10.1021/acs.est.6b04374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Wei Y, Shi Q, Wang Z, Zhang R, Su L, Quamruzzaman Q, et al. Maternal/fetal metabolomes appear to mediate the impact of arsenic exposure on birth weight: A pilot study. J Expo Sci Environ Epidemiol 2017;27:313–9. doi: 10.1038/jes.2016.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Bailey KA, Laine J, Rager JE, Sebastian E, Olshan A, Smeester L, et al. Prenatal arsenic exposure and shifts in the newborn proteome: Interindividual differences in tumor necrosis factor (TNF)-responsive signaling. Toxicol Sci 2014;139:328–37. doi: 10.1093/toxsci/kfu053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Pennings JLA, Jennen DGJ, Nygaard UC, Namork E, Haug LS, Van Loveren H, et al. Cord blood gene expression supports that prenatal exposure to perfluoroalkyl substances causes depressed immune functionality in early childhood. J Immunotoxicol 2016;13:173–80. doi: 10.3109/1547691X.2015.1029147. [DOI] [PubMed] [Google Scholar]
- [65].Remy S, Govarts E, Wens B, De Boever P, Den Hond E, Croes K, et al. Metabolic targets of endocrine disrupting chemicals assessed by cord blood transcriptome profiling. Reprod Toxicol 2016;65:307–20. doi: 10.1016/j.reprotox.2016.08.018. [DOI] [PubMed] [Google Scholar]
- [66].Sulek K, Han TL, Villas-Boas SG ranat., Wishart DS cot., Soh SE, Kwek K, et al. Hair metabolomics: identification of fetal compromise provides proof of concept for biomarker discovery. Theranostics 2014;4:953–9. doi: 10.7150/thno.9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Maitre L, Fthenou E, Athersuch T, Coen M, Toledano MB, Holmes E, et al. Urinary metabolomic profiles in early pregnancy are associated with preterm birth and fetal growth restriction in the Rhea mother-child cohort study. BMC Med 2014; 12:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Maitre L, Villanueva CM, Lewis MR, Ibarluzea J, Santa-Marina L, Vrijheid M, et al. Maternal urinary metabolic signatures of fetal growth and associated clinical and environmental factors in the INMA study. BMC Med 2016;14:177. doi: 10.1186/s12916-016-0706-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Hellmuth C, Uhl O, Standl M, Demmelmair H, Heinrich J, Koletzko B, et al. Cord Blood Metabolome Is Highly Associated with Birth Weight, but Less Predictive for Later Weight Development. Obes Facts 2017;10:85–100. doi: 10.1159/000453001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Isganaitis E, Rifas-Shiman SL, Oken E, Dreyfuss JM, Gall W, Gillman MW, et al. Associations of cord blood metabolites with early childhood obesity risk. Int J Obes 2015;39:1041–8. doi: 10.1038/ijo.2015.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Blighe K, Chawes BL, Kelly RS, Mirzakhani H, McGeachie M, Litonjua AA, et al. Vitamin D prenatal programming of childhood metabolomics profiles at age 3 y. Am J Clin Nutr 2017;106:1092–9. doi: 10.3945/ajcn.117.158220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Rolle-Kampczyk UE, Krumsiek J, Otto W, Roder SW, Kohajda T, Borte M, et al. Metabolomics reveals effects of maternal smoking on endogenous metabolites from lipid metabolism in cord blood of newborns. Metabolomics 2016;12:76. doi: 10.1007/s11306-016-0983-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Jones MJ, Fejes AP, Kobor MS. DNA methylation, genotype and gene expression: Who is driving and who is along for the ride? Genome Biol 2013;14. doi: 10.1186/gb-2013-14-7-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74]*.Gutierrez-Arcelus M, Lappalainen T, Montgomery SB, Buil A, Ongen H, Yurovsky A, et al. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. Elife 2013;2:e00523. doi: 10.7554/eLife.00523. Demonstration of the complex interplay between genetics and epigenetics, in their regulatory roles on gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Peng S, Deyssenroth MA, Di Narzo AF, Lambertini L, Marsit CJ, Chen J, et al. Expression quantitative trait loci (eQTLs) in human placentas suggest developmental origins of complex diseases. Hum Mol Genet 2017;26:3432–41. doi: 10.1093/hmg/ddx265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Gaunt TR, Shihab HA, Hemani G, Min JL, Woodward G, Lyttleton O, et al. Systematic identification of genetic influences on methylation across the human life course. Genome Biol 2016;17:61. doi: 10.1186/s13059-016-0926-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Teh AL, Pan H, Chen L, Ong ML, Dogra S, Wong J, et al. The effect of genotype and in utero environment on interindividual variation in neonate DNA methylomes. Genome Res 2014;24:1064–74. doi: 10.1101/gr.171439.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Inouye M, Kettunen J, Soininen P, Silander K, Ripatti S, Kumpula LS, et al. Metabonomic, transcriptomic, and genomic variation of a population cohort. Mol Syst Biol 2010;6. doi: 10.1038/msb.2010.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79]**.Nath AP, Ritchie SC, Byars SG, Fearnley LG, Havulinna AS, Joensuu A, et al. An interaction map of circulating metabolites, immune gene networks, and their genetic regulation. Genome Biol 2017;18:1–15. doi: 10.1186/s13059-017-1279-y. Multi-omics approach was utilized to demonstrate the interplay between genomics, transcriptomics, and metabolomics in immune modulation. [DOI] [PMC free article] [PubMed] [Google Scholar]