Significance
The Mycobacterium tuberculosis (Mtb) ClpB is a ring-shaped, ATP-driven disaggregase. The ability to rescue aggregated proteins is crucial for Mtb to grow and persist in the host. Despite extensive studies in the past two decades, it is still not well understood how a bacterial disaggregase couples ATP binding and hydrolysis to peptide translocation. Our cryo-EM study of the Mtb ClpB in the presence of a peptide substrate and the slowly hydrolysable adenosine 5′-[γ-thio]triphosphate revealed two active conformations in the midst of the substrate-threading process. This, together with the resolved nucleotide state in each of the 12 nucleotide-binding domains of the ClpB hexamer, helps define a detailed atomic trajectory that couples ATP binding and hydrolysis to mechanical protein translocation.
Keywords: Mycobacterium tuberculosis, proteostasis, AAA-ATPase, disaggregase, cryo-EM
Abstract
The protein disaggregase ClpB hexamer is conserved across evolution and has two AAA+-type nucleotide-binding domains, NBD1 and NBD2, in each protomer. In M. tuberculosis (Mtb), ClpB facilitates asymmetric distribution of protein aggregates during cell division to help the pathogen survive and persist within the host, but a mechanistic understanding has been lacking. Here we report cryo-EM structures at 3.8- to 3.9-Å resolution of Mtb ClpB bound to a model substrate, casein, in the presence of the weakly hydrolyzable ATP mimic adenosine 5′-[γ-thio]triphosphate. Mtb ClpB existed in solution in two closed-ring conformations, conformers 1 and 2. In both conformers, the 12 pore-loops on the 12 NTDs of the six protomers (P1–P6) were arranged similarly to a staircase around the bound peptide. Conformer 1 is a low-affinity state in which three of the 12 pore-loops (the protomer P1 NBD1 and NBD2 loops and the protomer P2 NBD1 loop) are not engaged with peptide. Conformer 2 is a high-affinity state because only one pore-loop (the protomer P2 NBD1 loop) is not engaged with the peptide. The resolution of the two conformations, along with their bound substrate peptides and nucleotides, enabled us to propose a nucleotide-driven peptide translocation mechanism of a bacterial ClpB that is largely consistent with several recent unfoldase structures, in particular with the eukaryotic Hsp104. However, whereas Hsp104’s two NBDs move in opposing directions during one step of peptide translocation, in Mtb ClpB the two NBDs move only in the direction of translocation.
Bacterial ClpB and eukaryotic Hsp104 belong to the Hsp100 family of chaperones (1). They cooperate with DnaK/Hsp70 cochaperones and additional accessory proteins to refold and rescue damaged proteins from their aggregated states, thereby helping cells survive under stress conditions (2–4). ClpB/Hsp104 utilize energy from ATP hydrolysis to thread polypeptides through their central pore and therefore are called “protein disaggregases” (1, 5). Structural studies have revealed a hexameric ring architecture with each protomer having three main domains: an N-terminal domain (NTD) for substrate binding and two tandem AAA+-type nucleotide-binding domains, NBD1 and NBD2, responsible for ATP-driven peptide translocation (6–11). ClpB/Hsp104 has a coiled-coil middle domain (MD) inserted in the NBD1 that distinguishes it from other AAA+ ATPases and regulates the disaggregation activity (6–8, 10, 12, 13). Despite their similar activity, there may be functional distinctions between the two homologs. For example, Hsp104, but not ClpB, has the ability to sever amyloid (14). The individual protomers of the Hsp104 hexamer were found to collaborate in a noncooperative manner, whereas those of ClpB function cooperatively (15). Structurally, Hsp104 has an extra C-terminal extension important for its oligomerization, but ClpB lacks such an extension (16). Recently, an elegant cryo-EM study revealed that yeast Hsp104 bound to casein had a closed and an open conformation, and on the basis of these states a ratchet-like substrate translocation mechanism was proposed (6). However, in the cryo-EM study of a ClpB variant of Escherichia coli ClpB with two Walker B mutations, E279A and E678A (called “BAP”), only a single closed conformation with a seam in the NBD1 ring was derived for the asymmetric double-ring structure bound to casein (7). It is currently uncertain if an open state exists in a bacterial disaggregase and whether there is any mechanistic distinction between a bacterial ClpB and eukaryotic Hsp104.
ClpB is important for the survival and persistence of Mycobacterium tuberculosis (Mtb) in the host and has been proposed as a novel target for the development of antituberculosis (anti-TB) drugs (17). ClpB facilitates asymmetrical distribution of protein aggregates within Mtb and between a given Mtb cell’s two progeny as part of its defense of the pathogen against host immunity and chemotherapy (18). In an in vitro system to reconstitute refolding of a model protein aggregate, the full function of Mtb ClpB required several partners, including DnaK, DnaJ1, DnaJ2, GrpE, and Hsp20 (4). Structural studies of Mtb ClpB are important not only to gain a mechanistic understanding but also for the future development of small molecules targeting this essential proteostasis system to treat TB, the leading cause of death due to an infectious disease worldwide. Using cryo-EM, we have captured Mtb ClpB in the midst of threading a peptide in two distinct but largely closed conformations at a resolution of 3.8–3.9 Å. These structures have enabled us to propose a coordinated, sequential ATP hydrolysis-coupled peptide translocation mechanism for this bacterial disaggregase.
Results
Mtb ClpB Is a Left-Handed Open Spiral in Either ADP-Bound or Adenylyl-Imidodiphosphate-Bound States but Switches to a Right-Handed Spiral in the Presence of Adenosine 5′-[γ-thio]triphosphate.
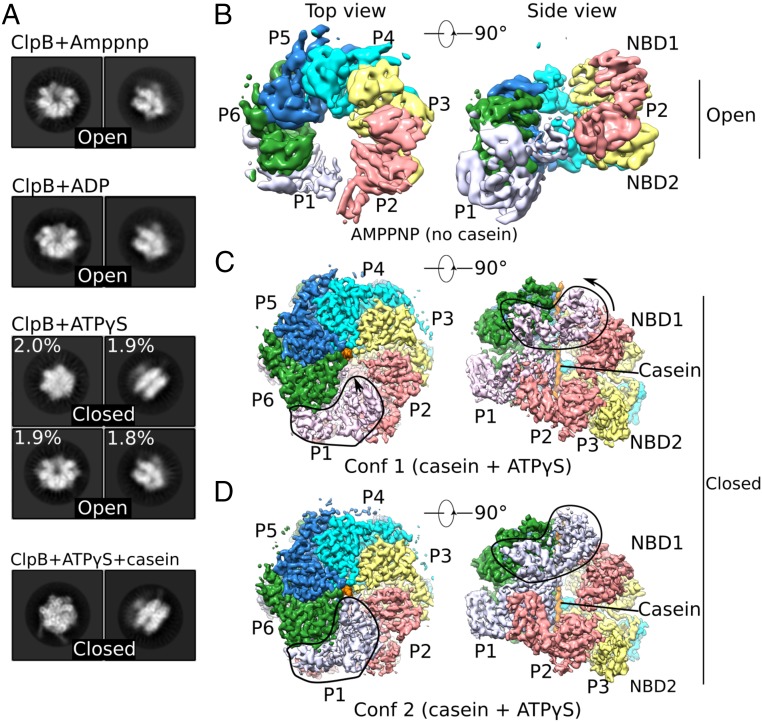
Full-length Mtb ClpB was expressed in E. coli and purified as a hexamer, as reported (4). We examined by cryo-EM the overall architecture of the hexamer in the absence of a peptide substrate but in the presence of different nucleotides by incubating the protein in 2 mM ADP, the nonhydrolysable ATP analog adenylyl-imidodiphosphate (AMP-PNP), or the weakly hydrolysable ATP analog adenosine 5′-[γ-thio]triphosphate (ATPγS). Reference-free 2D image classification revealed that in the presence of either ADP or AMP-PNP, ClpB adopted an open and asymmetric ring (Fig. 1A). 3D reconstruction of the AMP-PNP form allowed us to derive a 6.3-Å resolution map of six ClpB protomers arranged in a left-handed spiral architecture with NBD1 of the bottom end protomer (protomer 1; P1) and NBD2 of the top end protomer (protomer 2; P2) contacting each other (Fig. 1B and SI Appendix, Fig. S1). This left-handed Mtb ClpB open spiral resembled that of yeast Hsp104 bound to either ADP or AMP-PNP (6, 11). It is unlikely that a fully ADP- or ATP-bound state (as mimicked in vitro here by AMP-PNP) could exist under physiological conditions. Nevertheless, such an open state may represent a nonprocessive configuration ready for initial peptide engagement or poised to let go of unfolded peptide (6, 11). In the presence of ATPγS, Mtb ClpB took on two conformations. One represented the open state described above. The other, termed the “closed state,” was more symmetric, with the central chamber of the hexameric ring almost sealed off (Fig. 1A). Interconversion between the open and closed states required a large conformational rearrangement, which might be facilitated by the hydrolysis of ATPγS in some of the six ClpB protomers. These nucleotide-induced conformational changes were consistent with previous studies of E. coli ClpB and yeast Hsp104 (6, 7).
Fig. 1.
Cryo-EM analyses of multiple states of Mtb ClpB disaggregase. (A) Representative 2D images of the ClpB asymmetric hexamer (Left: top view; Right: side view) in the presence of different nucleotides with or without casein substrate. (B–D) Cryo-EM maps of different trapped conformations of ClpB, segmented by individual protomers. B shows an open left-handed spiral structure of ClpB in the presence of AMP-PNP. C and D show two closed conformations of ClpB in a complex with casein and ATPγS. Outlined regions exhibit major conformational differences between these two conformations. Conf 1, conformer 1; Conf2, conformer 2.
Mtb ClpB Switches to a Closed State When Bound to Casein.
As noted above, Mtb ClpB in the presence of ATPγS existed in both open and closed states. We found that Mtb ClpB existed primarily in the closed state when bound to casein, which mimicked a disordered protein substrate, and in the presence of ATPγS (Fig. 1A). This observation implies that the open state is converted to the closed state upon ClpB engagement with a protein substrate. We derived a cryo-EM 3D density map of Mtb ClpB-casein-ATPγS to an average resolution of 3.6 Å (SI Appendix, Fig. S2). We noticed that P1 had weak densities, indicative of significant conformational dynamics in this region. Another round of 3D classification led to the identification of two conformations of the closed state, one at 3.8-Å resolution termed “conformer 1” and the other at 3.9-Å resolution termed “conformer 2” (Fig. 1 C and D and SI Appendix, Fig. S2). In both conformers, the NTDs were invisible, likely due to high flexibility, but the NBD1 and NBD2 of each protomer were visualized in sufficient structural detail for atomic modeling (Fig. 2). In the atomic model of each conformer, the six NBD1s and six NBD2s of the ClpB hexamer assembled into two stacked right-handed spirals. However, the ClpB hexamer maintained an overall closed architecture in both conformers because protomer P1 at the beginning of the spiral was topologically connected with protomer P2 at the end of the spiral (Figs. 1 C and D and 2A). Although the overall architecture of Mtb ClpB was similar to that of Hsp104 (6), key differences existed between these two disaggregases. Both Hsp104 and ClpB were in a state where they had casein and ATPγS bound. Hsp104 was found to have a closed state and an open state, and in the open state both NBD1 and NBD2 rings were fully open and topologically discontinuous (6). In Mtb ClpB, however, the two observed conformers were both in the closed state, and the major changes from conformer 1 to conformer 2 occurred when NBD1 of protomer P1 rotated and moved upward to make contact with NBD1 of neighboring P6 and moved inward to bind casein, while the entire NBD2 ring was largely unchanged (Fig. 1 C and D). These differences may underlie a subtle mechanistic distinction between ClpB and Hsp104, discussed below.
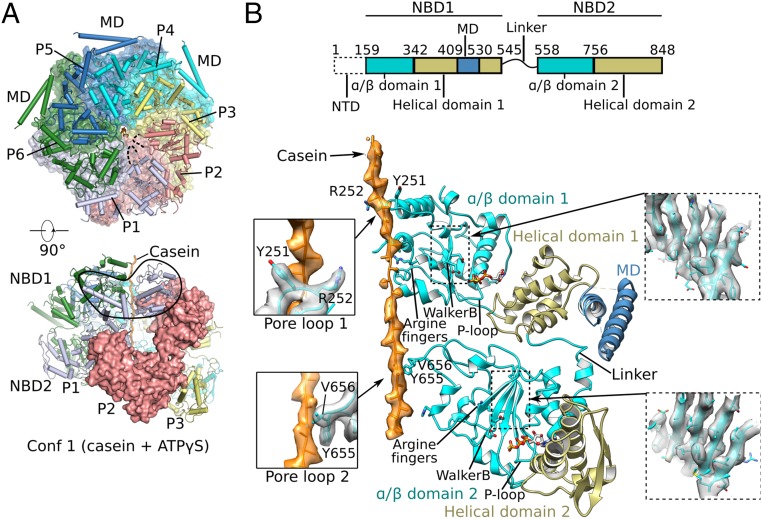
Fig. 2.
Cryo-EM structure of conformer 1 of Mtb ClpB complexed with ATPγS and casein. (A) Two orthogonal views of the atomic model of conformer 1 (corresponding to Fig. 1C). (Upper) The NBD2 ring is shown, overlapped with a semitransparent surface view. (Lower) Side view of conformer 1 with one protomer (P2) shown in surface view. (B, Upper) The domain architecture of ClpB. (Lower) Detailed structural elements of protomer P4 are shown in cartoon representation. The two α/β subdomains are shown in cyan, two helical subdomains in olive, MD domain in dark blue, and casein peptide in orange with electron density superimposed in semitransparent surface. Key features such as arginine fingers, Walker B motifs, P loops, and the linker between NBD1 and NBD2 are labeled. (Left Insets) Enlarged views of the interactions between the casein peptide and pore-loop 1 (Upper) and pore-loop 2 (Lower). (Right Insets) Electron density of the middle β-sheet of NBD1 (Upper) and NBD2 (Lower).
Mtb ClpB Conformers 1 and 2 Engage Casein Differently.
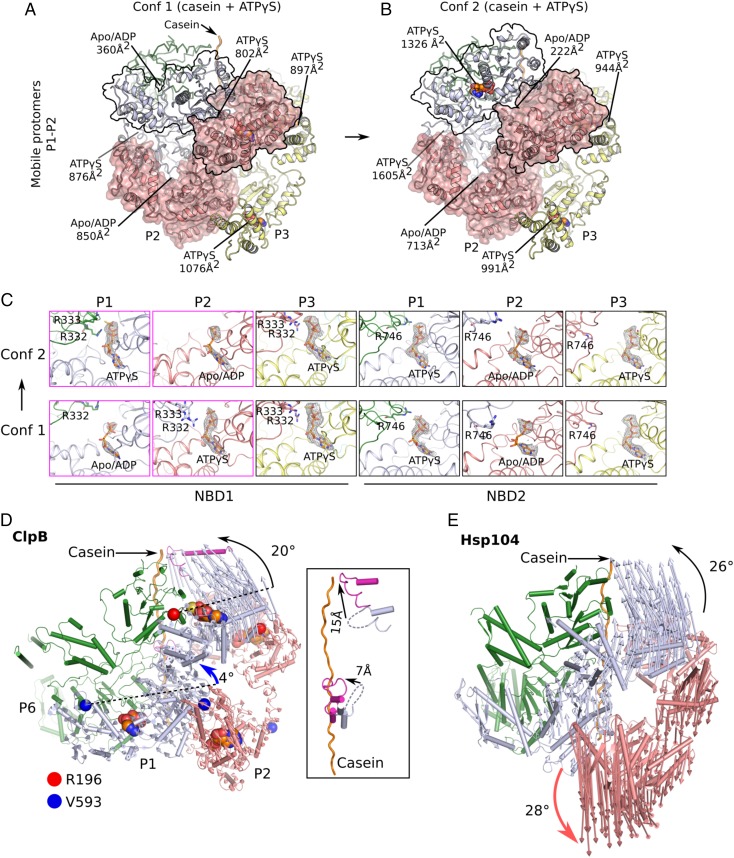
In the two Mtb ClpB conformers, the axial substrate-translocation channels were lined with 12 pore-loops: six pore-loops 1 of the six NBD1s (hereafter “pore-loops 1”) and six pore-loops 2 of the six NBD2s (hereafter “pore-loops 2”) (Figs. 2B and 3 A–C). More specifically, the six Y251 residues of the six pore-loops 1 and six Y655 residues and six V656 residues of the six pore-loops 2 were arranged in a right-handed staircase with a step size of two amino acids, ∼6.6 Å in height. These bulky and hydrophobic residues stacked against the peptide backbone of the bound casein (Fig. 3C). This general mode of substrate engagement was maintained by several intersubunit interactions. Inside the NBD1 ring, the side chain of R252 of one protomer was oriented toward pore-loop 1 of a neighboring protomer to form an H-bond with the carbonyl oxygen of S249 and another H-bond with the side-chain hydroxyl of the same residue (Fig. 3D). Inside the NBD2 ring, the Y658 phenyl ring in pore-loop 2 formed a hydrophobic interaction with V656 (Fig. 3E). These residues are well conserved. Their individual substitution by alanine nearly abolished ClpB-mediated substrate unfolding activity in vitro (Fig. 3F).
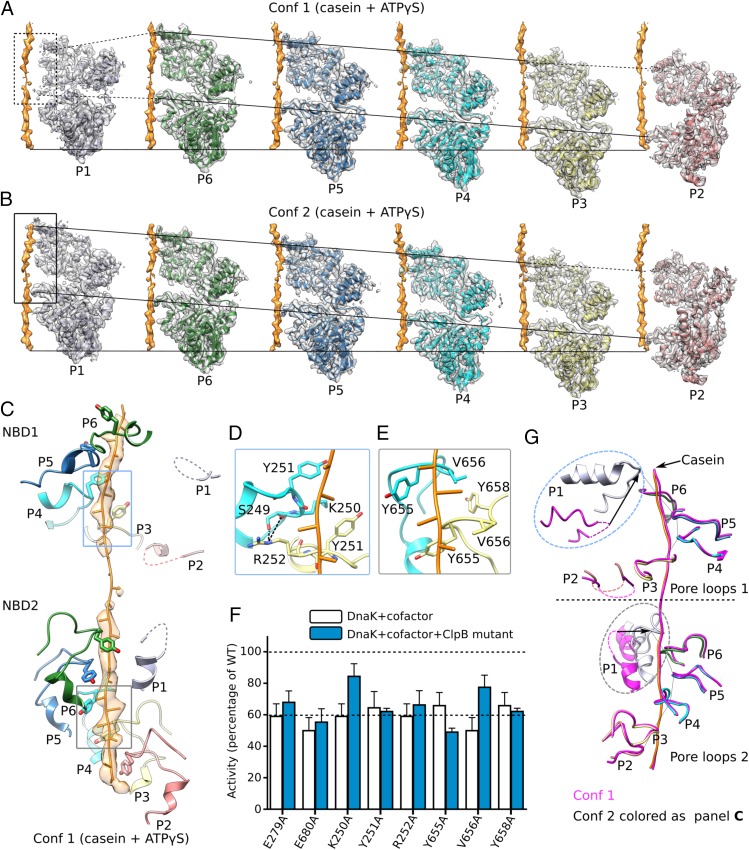
Fig. 3.
Interactions between ClpB and bound substrate mimic casein. (A and B) Individual protomers within conformer 1 (Conf 1) (A) and conformer 2 (Conf 2) (B) are shown separately with a 60° rotation around the bound casein, superposed with their corresponding cryo-EM densities. The casein peptide was aligned across panels (horizontal solid lines) to show the relative height of each protomer in the hexamer. The two slanted lines in each panel connect the pore-loops 1 and 2 across individual protomers, respectively. The dashed lines connect to disordered loops. In both conformations, protomers P3–P6 were fully engaged with the casein through their respective pore-loops 1 and 2, and the tyrosine sidechains of all eight pore-loops had good densities (Fig. 2B). Furthermore, protomer P2 contacted casein via pore-loop 2, but its pore-loop 1 was disordered, so P2 was partially engaged with the peptide. Both pore-loops of P1 were disordered or unengaged in conformer 1 but become fully engaged in conformer 2. (C) In conformer 1, the pore-loops inside the central chamber surrounded and bound the substrate casein in a right-handed spiral mode. This spiral was discontinued at pore-loops 1 of protomers P1 and P2 in the NBD1 ring and at pore-loop 2 of protomer P1 in the NBD2 ring; these pore-loops were flexible and did not contact the substrate. (D) Interactions among neighboring pore-loops 1 and the substrate. Y251 stacked with the casein main chain while R252 H-bonded with S249 of neighboring pore-loops to stabilize the right-handed spiral arrangement. (E) Interactions among neighboring pore-loops 2 and the substrate. Y655 and V656 stacked with the peptide while V656 stacked with Y658 of neighboring loops to stabilize their spiral arrangement. (F) Mutational analysis of the key residues in the pore-loops using a luciferase-based model protein aggregate reactivation assay. The Walker B mutations E279A and E680A were used as negative controls. Data were normalized to reactions containing wild-type ClpB, Hsp20, and DnaK along with cofactors DnaJ1, DnaJ2, and GrpE, which together reactivate denatured luciferase. The dashed line at 60% of wild-type activity defines the yield of reactions lacking ClpB (white bars), indicating the background refolding activity of DnaK and cofactors alone. Hence, reactions with ClpB mutants (blue bars) that show about 60% activity are defined as nonfunctional. (G) A comparison of pore-loops in conformer 1 (magenta) and conformer 2 (colored by protomers as in C). The two conformations were aligned by their respective caseins. Dashed curves mark disordered loops.
Protomers P3, P4, P5, and P6 all engaged the substrate in a similar manner in both conformers 1 and 2, with their respective pore-loops 1 and 2 all interacting with the substrate peptide. Therefore, the key differences between the two conformers resided in protomers P1 and P2 (Fig. 3 A and B). In conformer 1, pore-loops 1 and 2 of protomer P1 and pore-loop 1 of protomer P2 were disordered and had little contact with the casein peptide (Fig. 3 A and G). In conformer 2, however, pore-loops 1 and 2 of protomer P1 were ordered, and both were in close contact with the substrate (Fig. 3 B and G). Because more pore-loops in conformer 2 than in conformer 1 are engaged with the peptide, we refer to conformer 2 as the “high-affinity state” and conformer 1 as the “low-affinity state.” The difference in substrate affinity was a result of conformational changes driven by nucleotide hydrolysis, as described next.
The MD-Mediated Interprotomer Interactions Are Essential for Mtb ClpB Activity.
The MD coiled-coil of ClpB/Hsp104 is further divided into a distal motif 1 and a proximal motif 2, and the MD motif 1 of one protomer can latch onto the NBD1 of a neighboring protomer, forming an interprotomer brace (6, 7, 10). In the cryo-EM maps of Mtb ClpB, only the MD motifs 1 of protomers P4–P6 were resolved, including some large amino acid side chains. These densities enabled us to rigid-body fit the crystal structure of the E. coli ClpB MD domains to build an atomic model for the three MD motifs 1 (Fig. 4 A and B and SI Appendix, Fig. S3A). Therefore, the entire MD domains of protomers P1–P3 and the MD motifs 2 of protomers P4–P6 were dynamic. The flexibility of all six MD motifs 2 in Mtb ClpB was notable and in our structure might be due to the absence of a binding partner, such as one of the DnaK cochaperones (19). The three resolved MD motifs 1 of protomers P4–P6 were of a similar configuration in both conformers 1 and 2, stabilized by three salt bridges with their respective neighboring NBD1s: D414 to R188, R418 to D184, and E426 to R352 (Fig. 4B). The bracing interaction of these MD motifs 1 might be important for conformational coupling between the protomers in the ClpB hexamer, because we found that individually breaking up these salt bridges by reversing the charges nearly abolished the peptide-unfolding activity of Mtb ClpB in vitro (Fig. 4C). Although not observed here in Mtb ClpB, it is known that the MD motif 1 can assume an alternative configuration by protruding outwards and dissociating from the neighboring NBD1 (6–8). The unbracing motion of MD motif 1 required a rotation around a highly conserved hydrophobic region comprised of P410, I413, and W463 (Fig. 4B and SI Appendix, Fig. S3 B and C). This region was previously unappreciated but is essential for ClpB activity, because replacement of P410 with an alanine rendered Mtb ClpB virtually nonfunctional (Fig. 4C). Hence, the interconversion of MD motif 1 between the bracing and unbracing configurations is important.
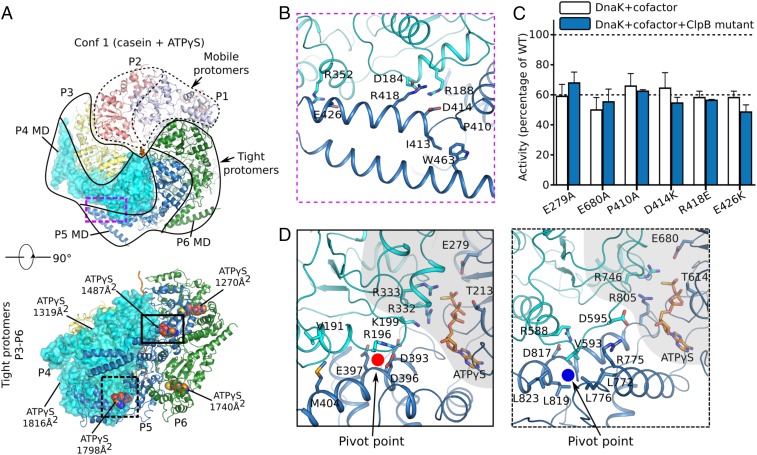
Fig. 4.
Protomer organization in conformer 1 (Conf 1) of Mtb ClpB bound to casein. (A) Mobile (protomers P1 and P2; dashed protomer boundary) vs. tightly packed protomers (protomers P3–P6; solid boundary) in conformer 1. (B) Three pairs of salt bridges between MD motif 1 of protomer P5 and NBD1 of the neighboring protomer P4 in the ClpB region marked by the purple dashed box in A. A hydrophobic region formed by P410, I413, and W463 at the right end of coiled-coil motif 1 might facilitate the relative rotation between motif 1 and motif 2 (not shown) of the MD domain. (C) Impact of the mutation of key residues in the MD domain on ClpB activity. The same Walker B mutations E279A and E680A shown in Fig. 3F were used as negative controls. Refer to Fig. 3F for a description of this assay. (D) Close-up views of the interfaces between the two NBD1 domains (Left) and two NBD2 domains (Right) of protomers P4 and P5 in regions marked by the solid-outlined and dashed-outlined boxes in A, Lower, respectively. The NBD1–NBD1 and NBD2–NBD2 interfaces were similar; both involved the ATP-binding pocket (shaded in gray) that interacted with the arginine finger from a neighboring protomer and a helix of the small helical domain that interacted with a loop from a neighboring protomer, either by salt bridges in NBD1 (R196 vs. D393/D396/E397) or by hydrophobic packing in NBD2 (V593 vs. L772/L776/L819/L823).
Interprotomer Contacts in NDB1 and NBD2 Rings.
Protomers P1 and P2 in conformer 1 were of weaker densities, packed loosely in the ClpB hexamer, and therefore are termed “mobile protomers,” compared with the “tight protomers” P3–P6 with very strong densities (Fig. 4A). The four tight protomers were arranged in a continuous right-handed spiral (Figs. 1 C and D and 2A) and were essentially in the same conformation (SI Appendix, Fig. S4) with extensive interprotomer interactions between their respective NBD1 domains as well as between NBD2 domains (Fig. 4D). Each of the four tight protomers bound two well-resolved ATPγS (SI Appendix, Fig. S5), one in NBD1 and one in NBD2, with their respective nucleotide phosphates binding to the P-loop of a Walker-A motif typical of AAA+ ATPases (Fig. 2B) (20). The conserved arginine fingers of the adjacent protomer, R332 and R333 in NBD1 and R746 in NBD2, interacted with γ-phosphate of the bound ATPγS. In NBD2, R805 in the helical subdomain provided additional contact with the γ-phosphate (Fig. 4D). Additional notable interprotomer contacts included (i) salt-bridges in the NBD1 ring formed between D393, D396, and E397 of one protomer and R196 and K199 of the neighboring protomer; this contact was further strengthened by an interprotomer hydrophobic interaction between V191 and M404; and (ii) hydrophobic stacking of L772, L776, L819, and L823 of one protomer with V593 of the adjacent protomer in the NBD2 ring, which was further strengthened by two salt-bridges, one between R775 and D595 and the other between D817 and R588. These interprotomer interactions in ClpB NBD1 and NBD2 were topologically similar, involving the same α-helices of the small helical domain and the same loop preceding the Walker A motif of the adjacent protomer (Fig. 4D). Similar interprotomer interactions are also seen in other AAA+ unfoldases, such as Hsp104, Vps4, and Yme1 (SI Appendix, Fig. S6) (6, 21, 22), which likely provide the pivot points for nucleotide-dependent domain rotations while maintaining the ring architecture.
Nucleotide Hydrolysis-Dependent Conformational Changes from Conformers 1 and 2.
In conformer 1 of Mtb ClpB, the tight protomers P3–P6 formed a strictly right-handed open spiral, which was bridged by the two mobile protomers, P1 and P2 (Fig. 2A). The relative positions of NBD1 and NBD2 of the mobile protomers were different from those of the tight protomers (SI Appendix, Fig. S4A), leading to the weak interactions between the mobile protomers (Fig. 5A). Importantly, protomers P1 and P2 were not in the expected uniform ATPγS-bound state. Instead, NBD1 of protomer P1 and NBD2 of protomer P2 had little to very low density for nucleotides and therefore were tentatively assigned as apo/ADP-like states. However, NBD2 of protomer P1 and NBD1 of protomer P2 remained in the ATP-like state with well-defined ATPγS density (Fig. 5C). Consistent with the disrupted interactions between the nucleotide and arginine fingers, the two apo/ADP sites had the smallest interprotomer contact areas (Fig. 5A). In conformer 2 of Mtb ClpB both NBD1 and NBD2 of protomer P2 were in the apo/ADP-like state, while the other 10 nucleotide pockets were occupied by ATPγS (Fig. 5C). The ring architecture remained nearly intact, and structural changes compared with conformer 1 were limited to protomers P1 and P2, with NBD1 of protomer P1 rotating up ∼20° toward the casein, generating a small seam with the protomer P2 NBD1, and the two NBD2s of protomers P1 and P2 together rotating ∼4° upwards (Fig. 5 A, B, and D). Both NBD1 and NBD2 rotations were around their respective pivot points described above (Fig. 4D). These changes in Mtb ClpB were very different from those in yeast Hsp104, in which the protomer equivalent to ClpB protomer P1 was found to move upwards, and the adjacent protomer corresponding to ClpB P2 moved downward, leading to a gap in the hexamer ring (Fig. 5E) (6). The changes in ClpB converted protomer P1 into the same configuration as protomers P3–P6 (SI Appendix, Fig. S4B). As a result, protomers P6 and P1 then formed a tight interface, with pore-loops 1 and 2 of protomer P1 moving up by 15 Å and 7 Å, respectively, to make contacts with the casein (Fig. 5D, Inset). Therefore, conformer 2 was a higher affinity state for the substrate than conformer 1. From conformer 1 to conformer 2, only the two NBD1 domains of protomers P1 and P2 switched their nucleotide states: NBD1 of protomer P1 switched from an apo/ADP-like to an ATP-like state, and NBD1 of protomer P2 switched from an ATP-like state to an apo/ADP-like state. Therefore, ATP hydrolysis by protomer P2 NBD1 reduced the interprotomer contact interface between protomers P1 and P2 from 1,652 Å2 to 935 Å2, while switching from an Apo/ADP-like to an ATP-like state at protomer P1 NBD1 strengthened the interprotomer interaction between protomers P6 and P1 from 1,236 Å2 to 2,931 Å2. These nucleotide-binding/hydrolysis events drove protomer P1 NBD1 away from protomer P2 NBD1 and toward protomer P6 NBD1, leading to the tight interface between protomers P1 and P6 in conformer 2 (Fig. 5D).
Fig. 5.
Nucleotide states and structural changes between conformers 1 and 2 of Mtb ClpB bound to casein. (A and B) The different arrangement of the mobile protomers P1 and P2 in conformer 1 (Conf 1) (A) and conformer 2 (Conf 2) (B). The contact areas of NBD1–NBD1 and NBD2–NBD2 and the bound-nucleotide states are labeled. (C) A comparison of the bound nucleotides in NBD1 and NBD2 of each protomer between conformers 1 and 2, with all of the densities displayed at the same threshold. The nucleotide states in protomers P4–P6 which bound ATPγS in both NBD1 and NBD2 are shown in SI Appendix, Fig. S5. (D) Structural changes from conformer 1 (cartoon view) to conformer 2 (hidden) are revealed by aligning their respective bound peptides. Vectors connect the same structural elements in the two conformers. Changes occurred primarily in protomer P1 (light blue), much less in protomer P2 (salmon), and were negligible in protomers P3–P6. Changes in protomers P1 and P2 between conformers 1 and 2 were described as rigid-body rotations of NBD1 and NBD2 around their respective pivotal points. The pivot point residues are marked by a red or a blue circle, respectively, as in Fig. 4D. The Inset shows the pore-loop positions in the two conformers. (E) Structural changes of the eukaryotic Hsp104 from the closed state (PDB ID code 5VJH; shown in cartoon) to the extended state (PDB ID code 5VYA) are illustrated by the vectors connecting the same residues in the two states. The bidirectional movement of the two neighboring protomers, as highlighted by the curved black and red arrows, creates a transient gap stage in both the NBD1 and NBD2 rings in Hsp104.
A Potential ATP-Binding and Hydrolysis-Coupled Peptide Translocation Mechanism of Mtb ClpB.
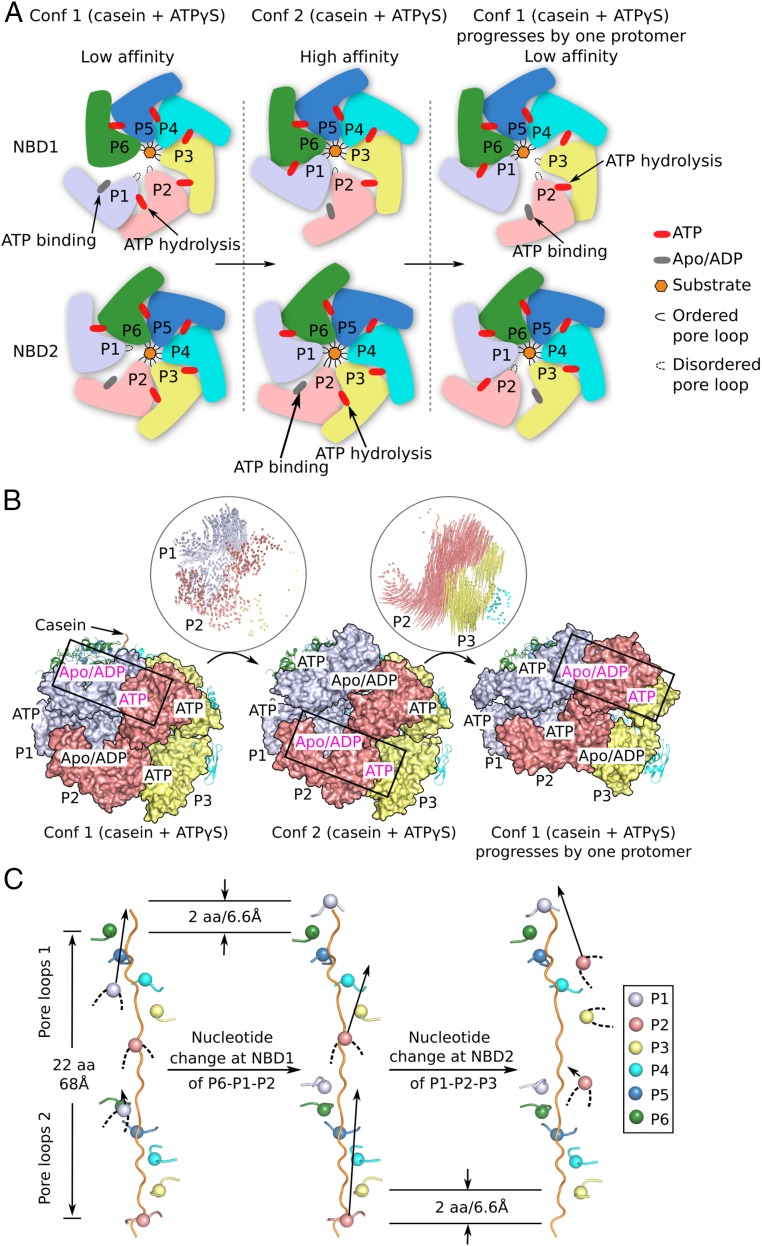
The two conformers of Mtb ClpB bound to casein as described above likely represent two distinct steps during the process of protein substrate disaggregation. Because of the relatively good resolution, we were able to assign the nucleotide state of each of the 12 NBDs in both conformers. By alternating the two conformers, we propose the following atomic model for an ATP-binding and hydrolysis-coupled three-stage peptide translocation mechanism (Fig. 6 and Movie S1). In the first stage, the Mtb ClpB hexamer in conformer 1 is in a low-affinity state, because three of the 12 pore-loops (i.e., both pore-loops of protomer P1 and pore-loop 1 of protomer P2) do not contact casein (Fig. 6). In the second stage, ATP binding to NBD1 of protomer P1 and ATP hydrolysis by NBD1 of protomer P2 convert the ClpB hexamer to conformer 2, in which both pore-loops of protomer P1 now bind to casein by moving upward by two amino acids on the substrate peptide, equivalent to a step size of ∼6.6 Å (Fig. 6B). ClpB had high affinity for substrate in conformer 2, because pore-loop 1 of protomer P2 was the only one of the 12 pore-loops that did not bind casein (Fig. 6A). The transition from conformer 1 to conformer 2 created two apo/ADP sites in protomer P2 and a small seam between protomers P1 and P2 (Fig. 5 A–D). In the third stage, ATP binding to NBD2 of protomer P2 and ATP hydrolysis by NBD2 of protomer P3 propagate, in a counterclockwise direction, the substrate-binding pattern of protomers P1 and P2 in stage 1 to protomers P2 and P3, in which the two protomer P2 pore-loops move upward to contact the substrate and the protomer P3 pore-loop 1 releases casein, thereby driving ClpB back to the low-affinity state (Fig. 6). Continued propagation of the low- and high-affinity states across the hexameric ring would generate a unidirectional force by the shifting pore-loops, powered by ATP binding and hydrolysis in individual protomers, to provide the power stroke that threads the substrate through the hexameric chamber.
Fig. 6.
A putative three-stage, rotary sequential nucleotide-driven substrate translocation mechanism of Mtb ClpB. (A) A sketch of the proposed mechanism in which the hexamer progresses from stage 1 (conformer 1) to stage 2 (conformer 2) and finally to stage 3. Stage progression is driven by ATP binding and hydrolysis in NBD1 and NBD2 of the two mobile protomers. (B) Atomic trajectories of the progressive conformational changes between stage 1 and 2 and between stage 2 and 3 as illustrated in A. To generate the model for stage 3, conformer 1 was superposed with conformer 2 by aligning protomer P5 in conformer 1 onto protomer P6 in conformer 2. (C) Unidirectional and progressive rearrangements of the 12 pore-loops as the ClpB hexamer goes through the three stages. From stage 1 (Left) to stage 2 (Middle), protomer P1 transits from the unengaged state with none of pore-loops 1 and 2 contacting casein to the fully engaged state with both pore-loops 1 and 2 contacting casein. This is caused by the ATP rebinding to NBD1 of protomer P1 and ATP hydrolysis by NBD1 of protomer P2. From stage 2 (Middle) to stage 3 (Right), protomer P2 transits from a partially engaged state with only pore-loop 2 contacting casein to an unengaged state with neither pore-loop 1 nor 2 contacting casein, and protomer P3 transits from a fully engaged state to a partially engaged state. This step may be caused by ATP rebinding to NBD2 of P2 and ATP hydrolysis by NBD2 of P3. The net effect of going through these three stages is a counterclockwise propagation of conformer 1 by one protomer. Continued cycling through the three stages leads to a rotary sequential model for substrate translocation in Mtb ClpB. Conf 1, conformer 1; Conf 2, conformer 2.
Discussion
Previous structural studies have revealed common features among several peptide-translocating AAA+ machines, including the asymmetric hexameric architecture and the sequential mode for substrate threading (6, 7, 21–23). The structure of human p97 was determined to a high resolution of 2.3–3.3 Å in multiple conformations, but the structure was symmetric, likely due to the absence of a protein substrate (24). The structure of VAT, an archaeal homolog of eukaryotic p97, bound to a peptide, was determined to 4.8-Å resolution and in a single conformation (23). Single conformations were also determined for the yeast unfoldase Vps4 bound to an ESCRT-III peptide substrate at 3.2-Å resolution (22), the mitochondrial protease ATPase YME1 bound to a peptide at 3.4-Å resolution (21), and E. coli ClpB bound to casein at 4.6-Å resolution (7). The structure of yeast Hsp104 was determined in two conformations in complex with casein at a resolution of 4.0–4.1 Å, just shy of resolving the nucleotides, so that all sites were assigned as ATPγS (6). In the current study, the Mtb ClpB was determined in two conformations at a slightly improved resolution of 3.8–3.9 Å, enabling assignment of the nucleotide states of all 12 NBDs. This has led us to propose a rotary sequential mechanism for Mtb ClpB that is largely consistent with the previously studied unfoldases and especially with the evolutionarily conserved Hsp104 (6).
However, we also noted a significant difference between Mtb ClpB and Hsp104 in the direction of movement of the NBD domains. In the Mtb ClpB structures, NBD1 and NBD2 always move in one direction: upwards. However, in Hsp104 NBD1 was found to move upwards, whereas NBD2 moves downward (6). The bidirectional movement leads to the formation of a transient gap stage in the Hsp104 hexamer ring that is not observed in Mtb ClpB. Further studies are needed to investigate if the observed structural differences are related to their different substrate preferences (14) and to their potentially different ATPase activities, i.e., the reported cooperative ATP hydrolysis of ClpB vs. the probabilistic ATP hydrolysis of Hsp104 (15). Finally, because Mtb is a major pathogen, our detailed structure and a mechanistic understanding could help in antimycobacterial drug design.
Materials and Methods
Overexpression and Purification of MtbClpB.
The construct of Mtb ClpB with an N-terminal His-SUMO tag was from a previous study (4). A similar sample preparation procedure was used with some modifications. Briefly, E. coli Rosetta2 cells (Novagen) transformed with the N-terminal His-SUMO–tagged Mtb ClpB were cultured in LB medium supplemented with 100 μg/mL ampicillin and 34 μg/mL chloramphenicol at 37°C and were grown to OD600 ∼0.5–0.6 before cooling to 25 °C. Protein expression was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside for 5 h. Collected frozen cells were lysed in buffer containing 20 mM Tris (pH 8.0) and 300 mM NaCl. After centrifugation, the protein was purified from the supernatant using nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography. During dialysis into buffer A [20 mM Hepes (pH 7.5), 40 mM NaCl, and 10 mM MgCl2], the His-SUMO tag was cleaved by incubation with His-tagged protease Ulp1 at a 10:1 ClpB:Ulp1 weight:weight ratio. The protease and cleaved His-SUMO tag were removed by Ni-NTA affinity chromatography, and the flow-through was concentrated and subjected to a gel-filtration purification step (Superose 6; GE Healthcare) in buffer A. The fractions corresponding to the estimated hexamer elution peak were pooled for further use. The protein concentration at this stage was ∼1 mg/mL.
Cryo-EM Data Acquisition.
Purified ClpB diluted to ∼0.8 mg/mL was saturated with excessive substrates including 2 mM ADP, 2 mM AMPNP, 2 mM ATPγS, or 2 mM ATPγS, and κ-casein (catalog no. C0406; Sigma) in twofold molar excess to ClpB hexamer. The mixtures were subsequently incubated at 4 °C for 1 h. Three-microliter aliquots of the incubated sample were applied to glow-discharged holey carbon grids (C-Flat Cu CF-1.2/1.3, 300 or 400 mesh). The grids were blotted for 4 s at 4 °C with 95% humidity and were flash-frozen in liquid ethane using an FEI Vitrobot Mark IV device. For initial evaluation, cryo-EM data were recorded on a Falcon II detector operated in linear mode in a 200-kV FEI Arctica Talos electron microscope. Automated data acquisition was performed with the FEI EPU software package. For each sample, 300–400 movies were recorded at a nominal magnification of 120,000×, in a physical pixel size of 1.21 Å per pixel. Defocus values varied from −1.5 μm to −3 μm.
Two samples, ClpB + AMP-PNP and ClpB + ATPγS + casein, were subjected to high-resolution cryo-EM data acquisition in a 300-kV FEI Titan Krios electron microscope with a K2 camera positioned after a GIF quantum energy filter (Gatan), yielding 2,535 and 3,537 movies, respectively. The FEI EPU package was used for automated data acquisition. Micrographs were collected in the superresolution counting mode at a nominal magnification of 130,000×, resulting in a physical pixel size of 1.07 Å. Defocus values varied from −1.1 μm to −3.0 μm. The dose rate was 10.0 electrons per pixel per second. A total exposure of 6 s was dose-fractionated into 30 frames, resulting in a total accumulated dose of 52 electrons/Å2.
Image Processing and 3D Reconstruction.
The movies collected on Arctica Talos were motion-corrected with MotionCor2 (25). Dose-fractionated movies collected on Titan Krios were also motion-corrected, dose-weighted, and binned by a factor of 2 with MotionCor2 (25, 26), resulting in summed micrographs with a physical pixel size of 1.07 Å. CTFFIND4 was used to estimate the contrast transfer function (CTF) parameters for individual micrographs (27). Subsequent image-processing steps were performed using RELION 2.0 (28). For each dataset, a set of manually picked particles was first subjected to 2D classification to generate a set of templates. These templates were used for reference-based automatic particle picking. The picked particles were subject to particle sorting and subsequent reference-free 2D classification.
For the two high-resolution datasets collected in Titan Krios, we first removed contaminants and noisy particles by 2D classification as described above and carried out further processing on the cleaned-up datasets. For the ClpB+AMP-PNP data from 2,535 micrographs, 3D classification was performed using a map generated by cryoSPARC as the initial reference model (29). The dominant 3D class with 112,043 particles was subjected to the final 3D autorefinement with a soft mask. This generated a 3D reconstruction at an overall resolution of 6.3 Å, suggesting substantial flexibility of this structure. For the ClpB+ATPγS+casein dataset from 3,537 micrographs, a map generated by cryoSPARC was used as the initial model for 3D classification. The two most populated 3D classes were combined. 3D autorefinement of this dataset of 354,656 particles with a soft mask resulted in a 3D map with an overall resolution of 3.8 Å. We found that the density for protomer P1 was very weak in this map, indicating substantial flexibility in the region. Therefore, a second round of 3D classification with six classes was carried out, resulting in two different conformations in the protomer P1 region. The most populated 3D class of each conformation was subject to another round of 3D autorefinement, generating two 3D reconstructions, conformer 1 at 3.8-Å resolution, from 108,111 selected particles, and conformer 2 at 3.9-Å resolution, from 76,666 particles. Finally, the particles corresponding to conformers 1 and 2 were combined, and another 3D autorefinement was performed, resulting in a combined map at 3.6-Å overall resolution. The respective resolutions of the three 3D reconstructions were estimated based on the gold standard Fourier shell correlation 0.143 criterion (30). The final maps were corrected for the modulation transfer function (MTF) of the detector and were sharpened by applying a negative B-factor, estimated by the postprocessing procedure in RELION 2.0. Local resolution distribution was estimated using ResMap (31).
Atomic Model Building.
Modeling of conformers 1 and 2 (ClpB+ATPγS+Casein) was based on the crystal structure of E. coli ClpB [Protein Data Bank (PDB) ID code 4CIU] (8). Using this structure, an initial Mtb ClpB protomer model was generated with the SWISS-MODEL server (32). The protomer model was split into two separate domains: NBD1 (amino acids 159–545) and NBD2 (amino acids 546–848). We rigid-body docked NBD1 and NBD2 domains into the 3.6-Å average 3D map using the Fit-in-map function in Chimera (University of California, San Francisco) (33). The docked models were improved by manual adjustments and rebuilding in Coot (34). The rebuilding process was guided by visible densities of many bulky residues such as Trp, Tyr, Arg, and Phe. For the modeling of conformer 1 and conformer 2, six copies of the built models of NBD1 and NBD2 domains were then docked into their corresponding maps, at 3.8-Å resolution for conformer 1 and 3.9-Å resolution for conformer 2. After slight manual adjustments and rebuilding in Coot, the real-space refinement of the atomic models of conformers 1 and 2 against their respective cryo-EM 3D maps was done using the phenix.real_space_refine in PHENIX (35). In the last step, to model the MD domain in a lower-density threshold, the initial motif 1 atomic model was obtained from the crystal structure of E. coli ClpB (PDB ID code 4CIU) and was directly rigid body-docked into the EM densities without further refinement.
The model of the tight protomer P5 of conformer 1 was used for modeling the ClpB-AMP-PNP structure. After removal of the side-chain information, six NBD1 and NBD2 domains were rigid-body fitted into the EM map using Chimera. Then some manual adjustments were performed in Coot to correct the large model deviations from the densities. Because of the low resolution, this model was not subjected to further refinement.
MolProbity (Duke University) was used to assess the final models (36). PyMOL (Schrödinger, LLC.) and Chimera were used to prepare the figures. Statistics of the 3D reconstruction and model refinement are provided in SI Appendix, Table S1.
Protein Reactivation Assay Using Denatured Firefly Luciferase.
This protocol was modified from other reports (37–39) and has been described with M. tuberculosis chaperones (4), which were purified as detailed in that report. Luciferase (100 nM) was heated with Hsp20 (400 nM) in 5-μL aliquots for 5 min at 42 °C in 50 mM Tris(hydroxymethyl)aminomethane (pH 7.5), 150 mM KCl, 20 mM MgCl2, and 2 mM DTT (buffer B) in nonstick tubes and then was cooled on ice for 5 min (to give 2–4% luciferase activity relative to native). Protein disaggregation/refolding reactions were performed by adding the indicated amounts of chaperones and/or cofactors (DnaK: 6 μM; remaining chaperone/cofactor: 2 μM each) in buffer B plus 1 mg/mL BSA and were initiated by adding 2 mM ATP to a final reaction volume of 20 μL and placing tubes at 25 °C for 30 min. Luminescence was measured by placing 2-μL samples into black 96-well polystyrene plates (Costar) and adding 100 μL of luciferase reagent (Promega). Light emission was measured using a SpectraMax L microplate reader luminometer (Molecular Devices) within the linear range of the instrument, and analysis was performed with SoftMax Pro software (Molecular Devices). An integration time of 10 s was used for all measurements. One hundred percent native luciferase activity was measured using luciferase + Hsp20 prepared in the same manner without the denaturation step, diluted in buffer B plus 1 mg/mL BSA, and incubated for the same period of time at the same temperature. Reactions with ClpB and mutants were done in triplicate in two independent experiments and are shown compared with the same reactions lacking ClpB done in at least triplicate. Percent luciferase activity was determined using Prism software as percent of reaction luminescence relative to native luminescence for each and then normalized relative to reactions containing wild-type ClpB measured in the same experiment.
Supplementary Material
Acknowledgments
Cryo-EM images were collected at the David Van Andel Advanced Cryo-Electron Microscopy Suite in the Van Andel Research Institute. H.L. was supported by NIH Grant R01 AI070285. C.F.N. was supported by NIH Grant U19 AI111143.
Footnotes
The authors declare no conflict of interest.
Data deposition: Cryo-EM maps and models were deposited in the Electron Microscopy Data Bank (EMDB) and the Protein Data Bank (PDB) [EMDB accession no. 7492, and PDB ID code 6DJU (Mtb ClpB-ATPS-casein conformer 1); EMDB accession no. 7493 and PDB ID code 6DJV (Mtb ClpB-ATPS-casein conformer 2); EMDB accession no. 9027 (Mtb ClpB-ATPS-casein with two conformations combined); and EMDB accession no. 9028 and PDB ID code 6ED3 (Mtb ClpB-AMP-PNP)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810648115/-/DCSupplemental.
References
- 1.Doyle SM, Genest O, Wickner S. Protein rescue from aggregates by powerful molecular chaperone machines. Nat Rev Mol Cell Biol. 2013;14:617–629. doi: 10.1038/nrm3660. [DOI] [PubMed] [Google Scholar]
- 2.Mogk A, et al. Identification of thermolabile Escherichia coli proteins: Prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 1999;18:6934–6949. doi: 10.1093/emboj/18.24.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doyle SM, Wickner S. Hsp104 and ClpB: Protein disaggregating machines. Trends Biochem Sci. 2009;34:40–48. doi: 10.1016/j.tibs.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 4.Lupoli TJ, Fay A, Adura C, Glickman MS, Nathan CF. Reconstitution of a Mycobacterium tuberculosis proteostasis network highlights essential cofactor interactions with chaperone DnaK. Proc Natl Acad Sci USA. 2016;113:E7947–E7956. doi: 10.1073/pnas.1617644113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weibezahn J, et al. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119:653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 6.Gates SN, et al. Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104. Science. 2017;357:273–279. doi: 10.1126/science.aan1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deville C, et al. Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase. Sci Adv. 2017;3:e1701726. doi: 10.1126/sciadv.1701726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carroni M, et al. Head-to-tail interactions of the coiled-coil domains regulate ClpB activity and cooperation with Hsp70 in protein disaggregation. eLife. 2014;3:e02481. doi: 10.7554/eLife.02481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee S, et al. The structure of ClpB: A molecular chaperone that rescues proteins from an aggregated state. Cell. 2003;115:229–240. doi: 10.1016/s0092-8674(03)00807-9. [DOI] [PubMed] [Google Scholar]
- 10.Heuck A, et al. Structural basis for the disaggregase activity and regulation of Hsp104. eLife. 2016;5:e21516. doi: 10.7554/eLife.21516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yokom AL, et al. Spiral architecture of the Hsp104 disaggregase reveals the basis for polypeptide translocation. Nat Struct Mol Biol. 2016;23:830–837. doi: 10.1038/nsmb.3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oguchi Y, et al. A tightly regulated molecular toggle controls AAA+ disaggregase. Nat Struct Mol Biol. 2012;19:1338–1346. doi: 10.1038/nsmb.2441. [DOI] [PubMed] [Google Scholar]
- 13.Wendler P, et al. Atypical AAA+ subunit packing creates an expanded cavity for disaggregation by the protein-remodeling factor Hsp104. Cell. 2007;131:1366–1377. doi: 10.1016/j.cell.2007.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shorter J, Lindquist S. Hsp104 catalyzes formation and elimination of self-replicating Sup35 prion conformers. Science. 2004;304:1793–1797. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- 15.DeSantis ME, et al. Operational plasticity enables hsp104 to disaggregate diverse amyloid and nonamyloid clients. Cell. 2012;151:778–793. doi: 10.1016/j.cell.2012.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mackay RG, Helsen CW, Tkach JM, Glover JR. The C-terminal extension of Saccharomyces cerevisiae Hsp104 plays a role in oligomer assembly. Biochemistry. 2008;47:1918–1927. doi: 10.1021/bi701714s. [DOI] [PubMed] [Google Scholar]
- 17.Lupoli TJ, Vaubourgeix J, Burns-Huang K, Gold B. Targeting the proteostasis network for mycobacterial drug discovery. ACS Infect Dis. 2018;4:478–498. doi: 10.1021/acsinfecdis.7b00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaubourgeix J, et al. Stressed mycobacteria use the chaperone ClpB to sequester irreversibly oxidized proteins asymmetrically within and between cells. Cell Host Microbe. 2015;17:178–190. doi: 10.1016/j.chom.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenzweig R, Moradi S, Zarrine-Afsar A, Glover JR, Kay LE. Unraveling the mechanism of protein disaggregation through a ClpB-DnaK interaction. Science. 2013;339:1080–1083. doi: 10.1126/science.1233066. [DOI] [PubMed] [Google Scholar]
- 20.Hanson PI, Whiteheart SW. AAA+ proteins: Have engine, will work. Nat Rev Mol Cell Biol. 2005;6:519–529. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 21.Puchades C, et al. Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing. Science. 2017;358:eaao0464. doi: 10.1126/science.aao0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han H, Monroe N, Sundquist WI, Shen PS, Hill CP. The AAA ATPase Vps4 binds ESCRT-III substrates through a repeating array of dipeptide-binding pockets. eLife. 2017;6:e31324. doi: 10.7554/eLife.31324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ripstein ZA, Huang R, Augustyniak R, Kay LE, Rubinstein JL. Structure of a AAA+ unfoldase in the process of unfolding substrate. eLife. 2017;6:e25754. doi: 10.7554/eLife.25754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banerjee S, et al. 2.3 Å resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science. 2016;351:871–875. doi: 10.1126/science.aad7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng SQ, et al. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grant T, Grigorieff N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 Å reconstruction of rotavirus VP6. eLife. 2015;4:e06980. doi: 10.7554/eLife.06980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kimanius D, Forsberg BO, Scheres SH, Lindahl E. Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2. eLife. 2016;5:e18722. doi: 10.7554/eLife.18722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 30.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–745. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 31.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–65. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 33.Pettersen EF, et al. UCSF Chimera–A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 34.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mogk A, Deuerling E, Vorderwülbecke S, Vierling E, Bukau B. Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol Microbiol. 2003;50:585–595. doi: 10.1046/j.1365-2958.2003.03710.x. [DOI] [PubMed] [Google Scholar]
- 38.Nillegoda NB, et al. Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation. Nature. 2015;524:247–251. doi: 10.1038/nature14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94:73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.