Significance
Direct interactions between natural killer (NK) cells and bacteria are rarely observed, and the consequences of these interactions not well understood. We show that human NK cells exposed to the bacterium group B Streptococcus (GBS) undergo pyroptosis, a type of inflammatory cell death. By releasing inflammatory mediators, pyroptosis is thought to be a way for sentinels to die and alert the immune system of infection. Interestingly, we also found that GBS has a protein that binds to Siglec-7, an inhibitory molecule on NK cells. Siglec-7 inhibits pyroptosis and silences the sentinel activity of NK cells by preventing the release of inflammatory molecules. These studies suggest that a human pathogen can silence the sentinel by specific interaction with a Siglec protein.
Keywords: natural killer cells, Siglec, group B Streptococcus, pyroptosis, inflammasome
Abstract
Natural killer (NK) cells are innate immune lymphocytes that recognize and destroy abnormal host cells, such as tumor cells or those infected by viral pathogens. To safely accomplish these functions, NK cells display activating receptors that detect stress molecules or viral ligands displayed at the cell surface, balanced by inhibitory receptors that bind to self-molecules. To date, such activating and inhibitory receptors on NK cells are not known to recognize bacterial determinants. Moreover, NK cell responses to direct interactions with extracellular bacteria are poorly explored. In this study, we observed the human neonatal pathogen group B Streptococcus (GBS) can directly engage human NK cells. The interaction was mediated through the B6N segment of streptococcal β-protein, binding to the inhibitory receptor Siglec-7 via its amino-terminal V-set domain. Unlike classical Siglec binding, the interaction is also independent of its sialic acid recognition property. In contrast to WT GBS, mutants lacking β-protein induced efficient pyroptosis of NK cells through the NLRP3 inflammasome, with production and secretion of the proinflammatory cytokine IL-1β and dissemination of the cytotoxic molecule granzyme B. We postulate that GBS evolved β-protein engagement of inhibitory human Siglec-7 to suppress the pyroptotic response of NK cells and thereby block recruitment of a broader innate immune response, i.e., by “silencing the sentinel.”
Natural killer (NK) cells are lymphocytes of the innate immune system that recognize endogenous eukaryotic cells under stress, such as tumor cells or cells infected by intracellular pathogens, modulating this process through an array of activating and inhibitory receptors (1–3). Activating receptors on human NK cells include NKG2D (4–6) and the natural cytotoxicity receptor family consisting of NKp46, NKp44, and NKp30 (7). These receptors bind to a variety of ligands displayed on the surface of eukaryotic cells during infection, or in response to stress or transformation (7–9). To avoid inadvertent destruction of healthy host cells, NK cells also express inhibitory receptors that bind to host molecules recognized as “self” (1), including the KIR (killer-cell Ig-like receptor) family, which recognizes HLA class I molecules expressed on normal autologous cells. The combined landscape of activating and inhibitory ligands on a target’s surface determines whether the NK cell becomes activated, leading to cytokine secretion and release of cytotoxic molecules such as perforin, granulysin, and granzymes (3). These activating and inhibitory receptors are not known to recognize determinants on bacteria, and direct interactions or responses against extracellular bacteria by NK cells are poorly explored (3, 10). Here we report on the unexpected finding that the important human pathogen group B Streptococcus (GBS) engages another known inhibitory receptor on human NK cells, the sialic acid-recognizing Ig-like lectin-7 (Siglec-7).
Siglec-7 is a member of the Siglec subfamily of CD33-related Siglecs (CD33rSiglecs) (11), which are single-pass transmembrane sialic acid-binding Ig-like lectins typically found on the surface of leukocytes (12–14). The cytosolic domains of most CD33-related Siglecs harbor inhibitory intracellular ITIM motifs that induce an immunosuppressive signal, but some can instead recruit DAP-12 with an activating intracellular domain, leading to augmentation of the immune response. Inhibitory Siglecs, which constitute the majority of CD33rSiglecs, can block cytokine secretion induced through Toll-like receptor (TLR) signaling (14–18) and may have evolved as a self-tolerance mechanism in which host leukocytes are inhibited when they recognize “self-associated molecular patterns” (SAMPs) presented by sialic acids abundantly displayed on host cell surfaces (12, 19–23).
Notably, certain bacterial pathogens have convergently evolved diverse mechanisms for displaying Siglec ligands on their cell surface, apparently to inhibit antipathogen immune responses via molecular mimicry (24–26). For example, sialylated polysaccharides of GBS engage inhibitory CD33rSiglecs found on neutrophils and myeloid lineage cells. Most such recognized microbial mimics of SAMPs for CD33rSiglec recognition are glycans. However, in at least one instance, Siglec-5 engagement also occurs through the cell wall-anchored β-protein expressed by certain GBS strains, with a similar suppression of the innate immune response of myeloid lineage cells like neutrophils (27, 28). As Siglec-5 is not prominent on human lymphocytes (29), it is not clear whether GBS β-protein can also inhibit this class of leukocytes.
GBS induces a form of immunogenic cell death called pyroptosis, mediated by an intracellular signaling complex called the inflammasome, which comprises several different signaling domains that multimerize upon binding of key ligands (30–33). Under canonical inflammasome activation, multimerization of the complex processes a proenzyme, such as caspase-1, into its mature form, enabling it to enzymatically cleave proinflammatory cytokines such as IL-1β from its proform into its mature form (34, 35). Cells activated for pyroptotic cell death ultimately swell and burst, allowing dissemination of the enzymatically cleaved proinflammatory cytokines into the extracellular space for the recruitment and activation of nearby leukocytes such as neutrophils and macrophages. Through this inflammatory pathway, pyroptosis acts as an “alarm signal” that promotes downstream host-protective antibacterial activities.
In contrast to myeloid lineage cells, the predominant inhibitory CD33rSiglec on NK cells is Siglec-7, which engages sialylated ligands on tumor cells to suppress NK cell responses (12, 21, 36–41). However, as this outcome does not favor the host, the natural role of Siglec-7 in controlling NK cell reactivity remains poorly explored. In this study, we report that GBS β-protein binds Siglec-7. As direct interactions between extracellular bacteria and NK cells are rarely observed and incompletely understood (10), we examined the consequence of this engagement upon the NK cell innate immune response. We found that NK cells undergo pyroptosis after exposure to bacteria. However, when Siglec-7 was engaged by β-protein, pyroptosis was inhibited, suggesting that GBS evolved this binding ability to down-regulate inflammasome activation and prevent recruitment of other innate immune cells.
Results
GBS β-Protein Engages NK Cells via Siglec-7.
GBS sialylated polysaccharide capsules of various serotypes engage the extracellular domains of specific recombinant soluble human Siglec receptors, including Siglec-7 (24). We later found that certain GBSs can engage Siglec-5 via their β-protein, but it was unclear whether other Siglec receptors also bind to this protein (27). A subsequent screen for binders to β-protein revealed that Siglec-7 also bound. As Siglec-7 is expressed primarily on human NK cells, we used confocal microscopy to demonstrate direct contact between GBS labeled with FITC and the surface of NK cells (Fig. 1A). Although Siglec-7 is uniformly distributed across the surface of uninfected NK cells, immunofluorescent labeling revealed clear coclustering of receptors toward multiple FITC-labeled WT GBSs on the surface of NK cells. Interestingly, based on these confocal images, there may even be intracellular Siglec-7 receptors residing in endosomal compartments, but further validation by costaining with endosomal markers will be required for a more in-depth understanding.
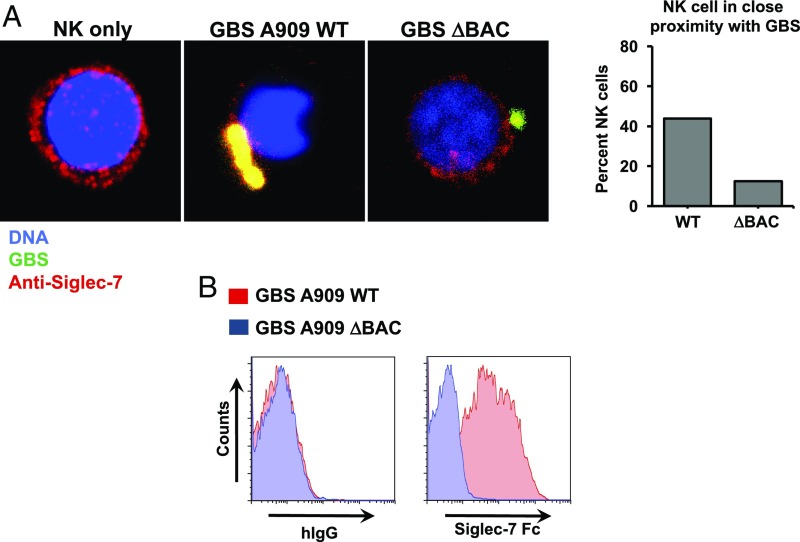
Fig. 1.
GBS strain A909 (GBS A909) interacts with human NK cells through the cell wall-anchored β-protein and Siglec-7 receptor. (A) Images taken by confocal microscopy show even distribution of Siglec-7 (red) across the cell surface of human NK cells under normal conditions. When NK cells are infected with FITC-labeled GBS (green), Siglec-7 on NK cells clusters toward GBS. Infection with β-protein–deficient mutant GBS ΔBAC restores the even distribution of Siglec-7 across the surface of NK cells. (B) Flow cytometry histograms of WT GBS A909 and ΔBAC stained with recombinant chimeric Siglec-7-Fc protein or IgG-Fc control. GBS A909 stained with Siglec-7-Fc showed positive signals above IgG-Fc, but GBS ΔBAC did not display any positive signals.
In contrast to the results with WT GBS, Siglec-7 did not cluster around β-protein–deficient GBS (ΔBAC). It is worth noting that there are no morphological differences between WT GBS vs. ΔBAC, but rather we believe there were more WT GBSs captured in direct contact with the NK cell visualized. To complement these observations, flow cytometric analysis of WT GBS indeed showed a positive signal for binding to a Siglec-7 Fc chimera compared with total human IgG (negative control), whereas virtually no signal was seen for Siglec-7 Fc binding to the ΔBAC mutant (Fig. 1B).
The Siglec-5–binding region (B6N) of GBS β-protein spans amino acid residues 1–153, but the adjacent IgA binding region (IgA-BR) spanning amino acid residues 154–225 is not recognized by Siglec-5 (28). Western blot analysis of purified B6N and IgA-BR peptides probed with Siglec-7 Fc also showed a direct interaction between Siglec-7 and B6N, but not IgA-BR (SI Appendix, Fig. S1A). ELISA analysis with immobilized B6N or IgA-BR probed by Siglec-7 Fc supported these findings (SI Appendix, Fig. S1B). The Siglec-7–B6N interaction appeared to be relatively stronger than the previously recognized Siglec-5–B6N interaction. When Siglec-7 Fc was coincubated with the S7.7 anti–Siglec-7 monoclonal antibody, the binding signal was reduced to background levels (SI Appendix, Fig. S1C), but anti–Siglec-7 antibody 194212 did not reduce the signal in a significant manner. In view of these findings, subsequent experiments used clone S7.7 as a blocking antibody against Siglec-7 and β-protein interaction.
To determine which extracellular domain on Siglec-7 might contribute to B6N recognition, we probed immobilized B6N by using previously constructed Siglec-Fc proteins with the V-set and underlying C2 domain swapped between Siglec-7 and Siglec-9 (26). By ELISA, we found that Siglec-7 V-set + Siglec-9 C2 fusion Fc bound to B6N at nearly equal levels as Siglec-7 Fc (SI Appendix, Fig. S1D). However, the Siglec-9 V-set + Siglec-7 C2 fusion Fc had no detectable binding to B6N, suggesting that the V-set domain of Siglec-7 is primarily responsible for B6N engagement.
NK Cell Responses to GBS Are Suppressed by β-Protein Engagement of Siglec-7.
Although NK cells are not typically studied as primary responders to extracellular bacteria, we determined the consequence of exposing NK cells to WT or ΔBAC GBS. We unexpectedly observed through flow cytometry analysis of forward and side scatter that there were major morphological changes in NK cells incubated with the ΔBAC mutant compared with cells incubated with WT GBS (Fig. 2A). Propidium iodide (PI) staining showed a much greater proportion of PI-positive NK cells when exposed to the ΔBAC mutant vs. WT GBS (Fig. 2B). These data indicate that GBS β-protein is cytoprotective for NK cells.
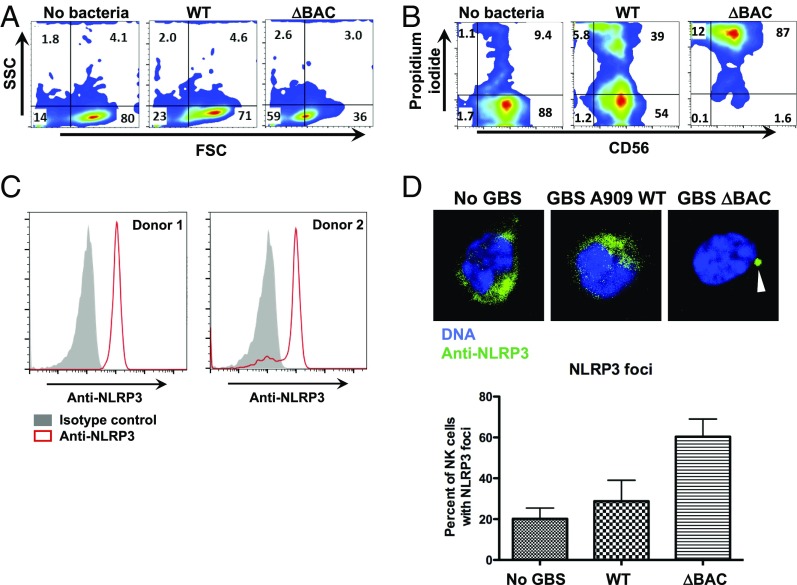
Fig. 2.
Streptococcal β-protein protects NK cells from death after infection. (A) Forward-and side-scatter analysis of primary human NK cells in the uninfected state or infected with GBS WT or GBS ΔBAC. (B) Flow cytometry analysis of primary CD56+ cells stained with PI to determine cell death without infection or with infection by GBS WT or GBS ΔBAC. (C) Flow cytometry histograms of permeabilized primary human NK cells stained with anti-NLRP3 antibody from two donors. (D) Confocal microscopy images of NK cells stained with anti-NLRP3 antibody without infection or with infection by GBS WT or GBS ΔBAC. The graph quantifies the percentage of NK cells with NLRP3 foci per field of view for each incubation condition. Error bars represent SD.
GBS induces pyroptosis in cells that express inflammasome-associated proteins, and the β-hemolysin/cytolysin (β-h/c) toxin has been implicated as the principal activator of the NLRP3 inflammasome complex (30, 31, 33). To explore inflammasome activation in human NK cells, we confirmed expression of NLRP3 in freshly isolated, sterile human NK cells by flow cytometry in peripheral blood from healthy donors with an anti-NLRP3 antibody (Fig. 2C). These results support a previous study that also described NLRP3 expression in human NK cells (42). We next examined inflammasome-induced cell death events such as release of intracellular contents caused by cellular swelling and bursting, dissemination of the proinflammatory cytokine IL-1β, and oligomerization of inflammasome proteins into foci. NK cells were exposed to WT or ΔBAC GBS and then stained by using an anti-NLRP3 antibody after fixation and permeabilization. By using confocal microscopy to visualize signals from the anti-NLRP3 antibody, we found that a greater percentage of NK cells contained NLRP3 foci when exposed to the ΔBAC mutant compared with WT GBS (Fig. 2D and SI Appendix, Fig. S2). Consistent with pyroptosis, release of the intracellular enzyme lactate dehydrogenase (LDH), a classic marker of membrane-disrupted cell death, as well as IL-1β, was much higher in the supernatant of NK cells exposed to ΔBAC vs. WT GBS (Fig. 3 A and B).
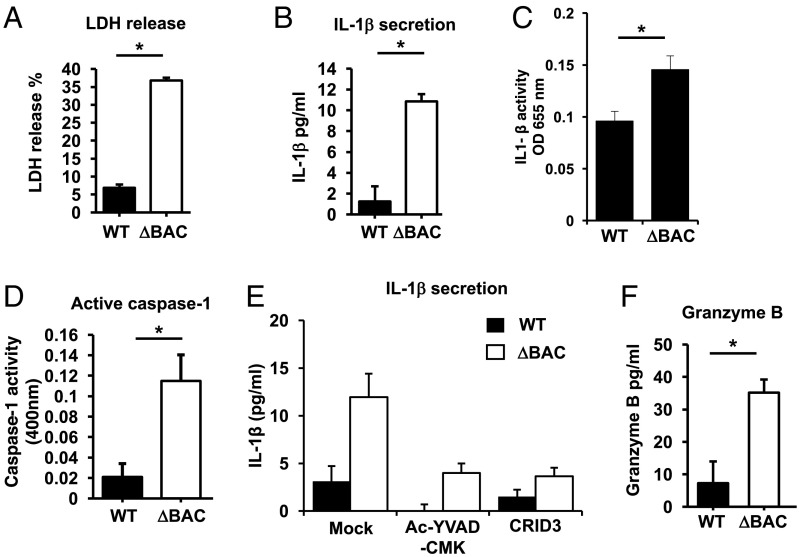
Fig. 3.
Streptococcal β-protein suppresses inflammasome activation of human NK cells. (A) LDH release from NK cells was measured after infection by GBS WT or GBS ΔBAC. (B) ELISA analysis of IL-1β secretion from NK cells after infection by GBS WT or ΔBAC. (C) Relative amounts of active of IL-1β released from NK cells infected by GBS WT or GBS ΔBAC was determined by using IL-1β–sensitive cells. (D) Relative concentrations of active caspase-1 released into the supernatant by NK cells infected by GBS WT or GBS ΔBAC measured by a caspase-1–cleavable reporter peptide. (E) ELISA analysis of IL-1β secreted into the supernatant by NK cells infected with GBS WT or GBS DBAC in the absence or presence of caspase-1 inhibitor Ac-YVAD-CMK or NLRP3 inhibitor CRID3. (F) ELISA analysis of granzyme B released into the supernatant of NK cells infected with GBS WT or GBS ΔBAC.
Pro–IL-1β requires proteolytic cleavage by caspase-1 to become active. To assess whether the IL-1β released from GBS-exposed NK cells is functionally active, supernatants from GBS-exposed NK cells were applied to an IL-1β reporter cell line. Supernatant from NK cells exposed to ΔBAC elicited a stronger signal from the IL-1β reporter cell line compared with supernatants from WT GBS-exposed NK cells (Fig. 3C). Furthermore, we found greater concentrations of active caspase-1 in the supernatant of ΔBAC-infected vs. WT-infected NK cells (Fig. 3D). IL-1β secretion was reduced in the presence of the caspase-1 inhibitor Ac-YVAD-CMK and specific NLRP3 inhibitor CRID3, indicating a crucial role of the NLRP3 inflammasome for NK cell pyroptosis (Fig. 3E).
NK cells possess cytolytic molecules and proteases including granzymes (3). Granzyme B was released into the supernatant with greater concentrations from NK cells exposed to ΔBAC mutant compared with WT GBS (Fig. 3F). Collectively, these data suggest that the GBS β-protein inhibits formation of the inflammasome complex and its downstream effects such as cell lysis and IL-1β secretion in NK cells.
Siglec-7 Engagement by β-Protein Suppresses Inflammasome Responses in NK Cells.
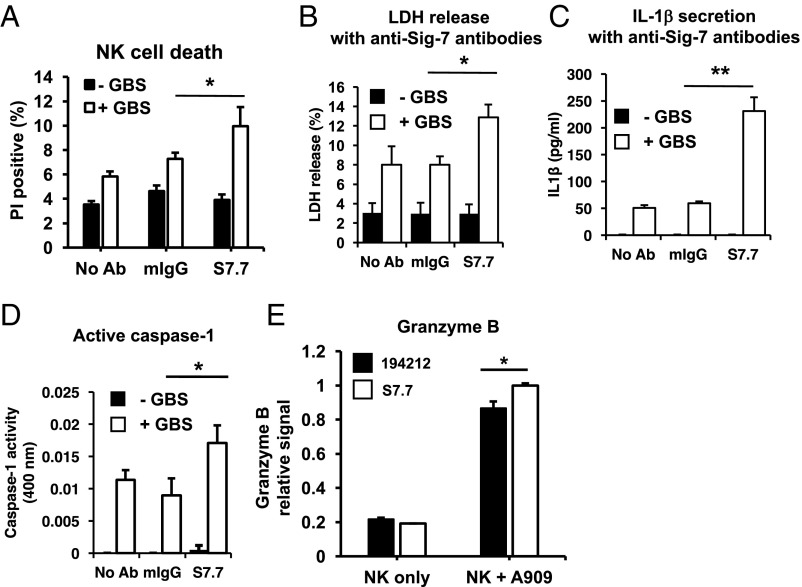
Α greater percentage of NK cells incubated with WT GBS in the presence of anti–Siglec-7 blocking antibodies were PI-positive compared with no antibody or isotype control antibody (Fig. 4A). LDH release was also increased from NK cells exposed to WT GBS in the presence of anti–Siglec-7 antibodies compared with isotype control antibodies (Fig. 4B). These observations were recapitulated upon determination of the relative concentrations of IL-1β, caspase-1, and granzyme B in the NK cell supernatant (Fig. 4 C–E). These data indicate that Siglec-7 can modulate inflammasome formation in NK cells, and our observation that anti–Siglec-7 antibody blocks ligand engagement to enhance inflammasome formation is consistent with the known inhibitory properties of Siglec-7.
Fig. 4.
NK cell inflammasome activation modulated by Siglec-7. (A) Flow cytometry analysis of NK cells stained with PI with or without infection by GBS WT in the presence of isotype control or anti–Siglec-7 antibody clone S7.7. (B) Detection of relative LDH released into the supernatant of NK cells with or without infection by GBS WT in the presence or absence of anti–Siglec-7 antibody. (C) ELISA analysis of IL-1β secreted into the supernatant of NK cells with or without infection by GBS WT in the presence or absence of anti–Siglec-7 antibody. (D) Relative levels of active caspase-1 in the supernatant of NK cells with or without infection in the absence or presence of anti–Siglec-7 antibody was determined by a fluorescent caspase-1 cleavable reporter peptide. (E) Release of granzyme B was determined by ELISA analysis of supernatant of NK cells with or without infection in the absence or presence of anti–Siglec-7 antibody clone 194212 or clone S7.7.
Discussion
In this work, we have explored a rarely studied direct interaction between NK cells and extracellular bacteria. Our experiments uncovered three previously undescribed immune interactions: pyroptosis of NK cells in response to exposure to extracellular bacteria, Siglec-mediated inhibition of such pyroptosis, and NK cell Siglec interactions with β-protein on an important human pathogen, GBS. Based on these findings, we propose that NK cells may function as early sentinels of bacterial infection via their ability to undergo pyroptosis, alerting the other components of the immune response via production of the critical proinflammatory cytokine IL-1β and release of intracellular contents recognized as “danger-associated molecular patterns” (DAMPs). Indeed, a different class of lymphocytes called NKT cells recognize bacterial glycolipids and release cytokines that can act as alarm signals to recruit other innate immune cells (43). From an evolutionary perspective, GBS may derive a survival benefit in the host, as β-protein binding to Siglec-7 suppresses the danger signaling response of the NK cells. This “silencing of the sentinel” is consistent with the otherwise paradoxical “protective” effect of the interaction on the NK cells. Functional consequences of this interaction are supported by the observation that certain other bacteria have evolved molecules to inhibit this pathway (44–46).
NK cells are most often studied in the context of recognizing aberrant host cells, such as tumor cells or cells infected by intracellular pathogens (2, 3). Indeed, NK cells have evolved recognition receptors to detect when cells are infected or transformed (1). Some of these receptors bind to viral ligands, and others bind to induced “self” molecules. NK cells also express receptors that can detect extracellular bacteria, including pattern recognition receptors. In our study, we define a role for the inhibitory receptor Siglec-7 in blocking GBS-induced pyroptosis of NK cells.
GBS and other similar hemolytic bacteria induce pyroptosis upon infection. In macrophages, pyroptosis can be induced by a GBS β-hemolysin and associated pigment that destabilizes the membrane and induces K+ efflux (31, 33), a known upstream activator of the NLRP3 inflammasome (47). It is likely that this pathway also promotes NK cell pyroptosis, as we have found evidence for NLRP3 oligomerization within NK cells after GBS infection. It is also for this reason that we have focused on NLRP3-mediated pyroptosis by GBS in human NK cells, although a previous study has also demonstrated NOD2 expression in NK cells as well (42). It remains to be seen whether other inflammasome receptors are also expressed in human NK cells.
We found that Siglec-7 engagement can block activation of the inflammasome by GBS in NK cells. Siglec-7 belongs to the immunosuppressive CD33-related primate Siglec family, which harbors intracellular ITIM domains that phosphorylate and recruit SHP2 phosphatase upon ligand binding, a first step of the inhibitory signaling cascade well characterized for Siglec-7 (48). Our results are consistent with other studies demonstrating how pathogens exploit ITIM receptors to suppress inflammasome activation and IL-1β production (49, 50). However, it remains to be understood how the Siglec-7–mediated intracellular signaling cascade can cross-talk to prevent oligomerization of the NLRP3 inflammasome. Siglecs block TLR activation, and a recent model of inflammasome activation places TLR signaling as a “priming” event (51, 52). Indeed, a prior study found that MyD88 deficiency in dendritic cells blocked inflammasome activity induced by GBS (30). Admittedly, the series of experiments determining whether an anti–Siglec-7 blocking antibody negates the suppressive interaction by β-protein did not yield as striking increases in signals as we would have expected (Fig. 4). However, anti–Siglec-7 antibodies have also been observed to cross-link and functionally suppress NK cell activity, so the true difference in signal may have been dampened by this effect (40).
Immunosuppressive Siglec-7 likely evolved as a mechanism for NK cells to recognize “self” antigens such as α2-8–linked disialic acid structures (in cis) or GD3 (in trans) and prevent NK cell attack on healthy endogenous cells (22, 53). The cell wall-anchored β-protein on GBS effectively hijacks Siglec-7 to suppress activation of the NLRP3 inflammasome. Even though this proinflammatory function is mechanistically aligned with the immunosuppressive properties of Siglec-7, Siglecs modulating pyroptotic cell death have not previously been reported to our awareness. Notably, unlike the case with Siglec-5/14 (15) and Siglec-11/16 in myeloid lineage cells (17), there is no known paired activating receptor counterpart to Siglec-7 on NK cells.
A major barrier preventing deeper understanding of NK cell pyroptosis in response to GBS infections is the lack of an animal model. CD33-related Siglecs are rapidly evolving, and murine counterparts are only definable as functionally equivalent orthologs or paralogs of primate CD33-related Siglecs. Although the murine Siglec counterparts provide a conceptual understanding of Siglec-based immunomodulation, it is not possible to directly translate murine Siglec studies to human Siglec biology, and expression of Siglecs on murine NK cells have not yet been described (13).
We observed release of granzyme B into the extracellular space after pyroptosis of NK cells. Recent studies have shown that granzyme B, in the presence of granulysin, can directly kill bacteria and have suggested a role for these molecules in the eradication of intracellular bacteria (54, 55). These studies suggest that cytotoxic lymphocytes recognize infected host cells and use cytotoxic granules to kill the host cell and the intracellular pathogen. Our results are different from these previous studies because we demonstrate a direct interaction between NK cells and extracellular bacteria. Although we have not shown a functional role for extracellular granzyme B in the context of a pyroptotic response, it is known that granzyme B can remodel the ECM by cleaving architectural matrix proteins, perhaps increasing trafficking of professional bacteria-killing leukocytes such as neutrophils and macrophages to the infected tissue (56). The release of granzyme B, cleaved caspase-1, and other intracellular contents indicate that the NK cell response to GBS involved cell death. This contrasts with a recent study showing that dendritic cells could activate the inflammasome via caspase-11 without inducing pyroptotic cell death, leading to a “hyperactive” state (57). In the case of granzyme B and IL-1β, it is difficult to conclude whether the dissemination of these molecules through pyroptosis of NK cells will have a significant physiological impact without an appropriate animal model, but the same hurdles for using an animal model applies as we had discussed earlier. However, as the IL-1β released from pyroptotic NK cells was applied to a reporter cell line with a detectable signal (Fig. 4C), we are confident that the cytokine was cleaved into its active form at a concentration detectable by the assay. We emphasize that the pyroptotic response of NK cells to GBS contrasts with “classic” NK cell activities such as IFN-γ production and cytotoxicity. Although GBS–Siglec-7 interactions can suppress pyroptosis, it is not known whether this would also suppress IFN-γ production and cytotoxicity. However, it is difficult for us to envision a natural and physiological situation in which NK cells are stimulated to produce IFN-γ but are in direct contact with GBS to suppress its production, as GBS is not known to have ligands to activate NKG2D or other NK cell-activating receptors. Nevertheless, it is worth noting that previous studies have demonstrated that functional engagement of Siglec-7 can suppress production of IFN-γ by NK cells stimulated by pharmacologic reagents (40).
A role for NK cells in response to extracellular bacteria has been proposed (10), but whether NK cells protect or further exacerbate bacterial infection is not clear. Notably, multiple bacterial toxins can activate NK cells, including streptococcal pyrogenic exotoxin A, staphylococcal enterotoxin B, and listeriolysin O (58–60). At least for the latter toxin, the effect on NK cells was indirect and required recognition of the toxin by antigen-presenting cells. Nevertheless, direct interaction between NK cells and PAMPs (pathogen-associated molecular patterns) has been documented, although typically the expression of receptors for PAMPs (such as TLRs) on NK cells is low and the response of purified NK cells to bacterial ligands is not robust. When they have been activated, NK cells augment innate immune responses via cross-talk with other cell types. This positive role for NK cells in initiating immune responses is supported by findings that other bacterial toxins inhibit NK cells, including leukotoxin derived from Actinobacillus actinomycetemcomitans and exotoxin A from Pseudomonas aeruginosa (61, 62). We also suggest a positive role for NK cells in alerting the immune system to infection, but our observation stands distinct from these prior studies in that we show that NK cells undergo pyroptosis. Moreover, we show direct and strong interaction between a bacterial ligand and Siglec-7, a highly expressed receptor on NK cells. Other groups have found that Siglec-7 also recognizes surface lipooligosaccharides of Campylobacter jejuni that can be modified with ganglioside-like sialylated terminal structures in the core oligosaccharides, potentially influencing host–pathogen interactions (63).
In summary, we found that NK cells undergo pyroptosis in response to bacterial infection, but GBS β-protein mediated binding to the Siglec-7 receptor blunts the pyroptotic phenotype, proteolytic processing and secretion of IL-1β, and release of intracellular contents such as granzyme B. Human pathogens such as GBS may evolve mechanisms such as β-protein engagement of Siglec-7 on NK cells to evade innate immunity.
Materials and Methods
All studies were approved by the University of California, San Diego Institutional Review Board. Consent was obtained for experiments involving human subjects. Human NK cells were derived from different donors in this study. Peripheral blood mononuclear cells were isolated by Ficoll-Paque–based centrifugation, and NK cells were isolated by the Human NK Cell Enrichment Kit (Miltenyi) per manufacturer’s instructions. Streptococcus agalactiae (group B streptococcal type Ia strain A909, or GBS) and its isogenic ΔBAC mutants have been described previously (15). Human NK cell exposure to GBS was performed by coincubation of GBS to NK cells at multiplicities of infection of 10:1 or 1:1 for 10 min or 30 min before NK cells were subjected to further analysis. Further details for the materials and methods used to conduct these studies are provided in SI Appendix, Supplemental Materials and Methods. SI Appendix, SI Materials and Methods also includes details on GBS strains and growth conditions, human NK cell isolation and exposure to GBS methods, ELISA conditions for binding assays and cytokine secretion analysis, flow cytometry procedures, confocal microscopy procedures, and all relevant antibody clones and other reagent details required to execute the experimental procedures referred in the text.
Supplementary Material
Acknowledgments
We thank members of the laboratories of J.D.B., A.V., and V.N. for helpful discussions and suggestions. This study was supported by a grant from The Hartwell Foundation (to J.D.B.), National Institutes of Health (NIH) Grant CA157885 (to J.D.B.), NIH Grant 1P01HL107150/National Heart, Lung, and Blood Institute Program of Excellence in Glycosciences (to A.V. and V.N.), and NIH Grant R01GM032373 (to A.V.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804108115/-/DCSupplemental.
References
- 1.Pegram HJ, Andrews DM, Smyth MJ, Darcy PK, Kershaw MH. Activating and inhibitory receptors of natural killer cells. Immunol Cell Biol. 2011;89:216–224. doi: 10.1038/icb.2010.78. [DOI] [PubMed] [Google Scholar]
- 2.Guillerey C, Huntington ND, Smyth MJ. Targeting natural killer cells in cancer immunotherapy. Nat Immunol. 2016;17:1025–1036. doi: 10.1038/ni.3518. [DOI] [PubMed] [Google Scholar]
- 3.Morvan MG, Lanier LL. NK cells and cancer: You can teach innate cells new tricks. Nat Rev Cancer. 2016;16:7–19. doi: 10.1038/nrc.2015.5. [DOI] [PubMed] [Google Scholar]
- 4.Bauer S, et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285:727–729. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- 5.Wu J, et al. An activating immunoreceptor complex formed by NKG2D and DAP10. Science. 1999;285:730–732. doi: 10.1126/science.285.5428.730. [DOI] [PubMed] [Google Scholar]
- 6.Jamieson AM, et al. The role of the NKG2D immunoreceptor in immune cell activation and natural killing. Immunity. 2002;17:19–29. doi: 10.1016/s1074-7613(02)00333-3. [DOI] [PubMed] [Google Scholar]
- 7.Biassoni R, Bottino C, Cantoni C, Moretta A. Human natural killer receptors and their ligands. Curr Protoc Immunol. 2002;Chapter 14:Unit 14.10. doi: 10.1002/0471142735.im1410s46. [DOI] [PubMed] [Google Scholar]
- 8.Lanier LL. NKG2D receptor and its ligands in host defense. Cancer Immunol Res. 2015;3:575–582. doi: 10.1158/2326-6066.CIR-15-0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Groh V, Wu J, Yee C, Spies T. Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature. 2002;419:734–738. doi: 10.1038/nature01112. [DOI] [PubMed] [Google Scholar]
- 10.Souza-Fonseca-Guimaraes F, Adib-Conquy M, Cavaillon JM. Natural killer (NK) cells in antibacterial innate immunity: Angels or devils? Mol Med. 2012;18:270–285. doi: 10.2119/molmed.2011.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Angata T, Margulies EH, Green ED, Varki A. Large-scale sequencing of the CD33-related Siglec gene cluster in five mammalian species reveals rapid evolution by multiple mechanisms. Proc Natl Acad Sci USA. 2004;101:13251–13256. doi: 10.1073/pnas.0404833101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraschilla I, Pillai S. Viewing Siglecs through the lens of tumor immunology. Immunol Rev. 2017;276:178–191. doi: 10.1111/imr.12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwarz F, Fong JJ, Varki A. Human-specific evolutionary changes in the biology of Siglecs. Adv Exp Med Biol. 2015;842:1–16. doi: 10.1007/978-3-319-11280-0_1. [DOI] [PubMed] [Google Scholar]
- 14.Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7:255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 15.Ali SR, et al. Siglec-5 and Siglec-14 are polymorphic paired receptors that modulate neutrophil and amnion signaling responses to group B Streptococcus. J Exp Med. 2014;211:1231–1242. doi: 10.1084/jem.20131853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fong JJ, et al. Immunomodulatory activity of extracellular Hsp70 mediated via paired receptors Siglec-5 and Siglec-14. EMBO J. 2015;34:2775–2788. doi: 10.15252/embj.201591407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwarz F, et al. Paired Siglec receptors generate opposite inflammatory responses to a human-specific pathogen. EMBO J. 2017;36:751–760. doi: 10.15252/embj.201695581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwarz F, et al. Siglec receptors impact mammalian lifespan by modulating oxidative stress. eLife. 2015;4:e06184. doi: 10.7554/eLife.06184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varki A. Since there are PAMPs and DAMPs, there must be SAMPs? Glycan “self-associated molecular patterns” dampen innate immunity, but pathogens can mimic them. Glycobiology. 2011;21:1121–1124. doi: 10.1093/glycob/cwr087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lizcano A, et al. Erythrocyte sialoglycoproteins engage Siglec-9 on neutrophils to suppress activation. Blood. 2017;129:3100–3110. doi: 10.1182/blood-2016-11-751636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hudak JE, Canham SM, Bertozzi CR. Glycocalyx engineering reveals a Siglec-based mechanism for NK cell immunoevasion. Nat Chem Biol. 2014;10:69–75. doi: 10.1038/nchembio.1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nicoll G, et al. Ganglioside GD3 expression on target cells can modulate NK cell cytotoxicity via Siglec-7-dependent and -independent mechanisms. Eur J Immunol. 2003;33:1642–1648. doi: 10.1002/eji.200323693. [DOI] [PubMed] [Google Scholar]
- 23.Xiao H, Woods EC, Vukojicic P, Bertozzi CR. Precision glycocalyx editing as a strategy for cancer immunotherapy. Proc Natl Acad Sci USA. 2016;113:10304–10309. doi: 10.1073/pnas.1608069113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlin AF, Lewis AL, Varki A, Nizet V. Group B streptococcal capsular sialic acids interact with Siglecs (immunoglobulin-like lectins) on human leukocytes. J Bacteriol. 2007;189:1231–1237. doi: 10.1128/JB.01155-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carlin AF, et al. Molecular mimicry of host sialylated glycans allows a bacterial pathogen to engage neutrophil Siglec-9 and dampen the innate immune response. Blood. 2009;113:3333–3336. doi: 10.1182/blood-2008-11-187302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Secundino I, et al. Host and pathogen hyaluronan signal through human Siglec-9 to suppress neutrophil activation. J Mol Med (Berl) 2016;94:219–233. doi: 10.1007/s00109-015-1341-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carlin AF, et al. Group B Streptococcus suppression of phagocyte functions by protein-mediated engagement of human Siglec-5. J Exp Med. 2009;206:1691–1699. doi: 10.1084/jem.20090691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nordström T, et al. Human Siglec-5 inhibitory receptor and immunoglobulin A (IgA) have separate binding sites in streptococcal beta protein. J Biol Chem. 2011;286:33981–33991. doi: 10.1074/jbc.M111.251728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen DH, Hurtado-Ziola N, Gagneux P, Varki A. Loss of Siglec expression on T lymphocytes during human evolution. Proc Natl Acad Sci USA. 2006;103:7765–7770. doi: 10.1073/pnas.0510484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Costa A, et al. Activation of the NLRP3 inflammasome by group B streptococci. J Immunol. 2012;188:1953–1960. doi: 10.4049/jimmunol.1102543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gupta R, et al. RNA and β-hemolysin of group B Streptococcus induce interleukin-1β (IL-1β) by activating NLRP3 inflammasomes in mouse macrophages. J Biol Chem. 2014;289:13701–13705. doi: 10.1074/jbc.C114.548982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mohammadi N, et al. Neutrophils directly recognize group B streptococci and contribute to interleukin-1β production during infection. PLoS One. 2016;11:e0160249. doi: 10.1371/journal.pone.0160249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whidbey C, et al. A streptococcal lipid toxin induces membrane permeabilization and pyroptosis leading to fetal injury. EMBO Mol Med. 2015;7:488–505. doi: 10.15252/emmm.201404883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Broz P, Dixit VM. Inflammasomes: Mechanism of assembly, regulation and signalling. Nat Rev Immunol. 2016;16:407–420. doi: 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- 35.Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157:1013–1022. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 36.Nicoll G, et al. Identification and characterization of a novel Siglec, Siglec-7, expressed by human natural killer cells and monocytes. J Biol Chem. 1999;274:34089–34095. doi: 10.1074/jbc.274.48.34089. [DOI] [PubMed] [Google Scholar]
- 37.Angata T, Varki A. Siglec-7: A sialic acid-binding lectin of the immunoglobulin superfamily. Glycobiology. 2000;10:431–438. doi: 10.1093/glycob/10.4.431. [DOI] [PubMed] [Google Scholar]
- 38.Jandus C, et al. Interactions between Siglec-7/9 receptors and ligands influence NK cell-dependent tumor immunosurveillance. J Clin Invest. 2014;124:1810–1820. doi: 10.1172/JCI65899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawasaki Y, et al. Ganglioside DSGb5, preferred ligand for Siglec-7, inhibits NK cell cytotoxicity against renal cell carcinoma cells. Glycobiology. 2010;20:1373–1379. doi: 10.1093/glycob/cwq116. [DOI] [PubMed] [Google Scholar]
- 40.Shao JY, et al. Siglec-7 defines a highly functional natural killer cell subset and inhibits cell-mediated activities. Scand J Immunol. 2016;84:182–190. doi: 10.1111/sji.12455. [DOI] [PubMed] [Google Scholar]
- 41.Mikulak J, Di Vito C, Zaghi E, Mavilio D. Host immune responses in HIV-1 infection: The emerging pathogenic role of Siglecs and their clinical correlates. Front Immunol. 2017;8:314. doi: 10.3389/fimmu.2017.00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiu F, Maniar A, Diaz MQ, Chapoval AI, Medvedev AE. Activation of cytokine-producing and antitumor activities of natural killer cells and macrophages by engagement of Toll-like and NOD-like receptors. Innate Immun. 2011;17:375–387. doi: 10.1177/1753425910372000. [DOI] [PubMed] [Google Scholar]
- 43.Kinjo Y, et al. Invariant natural killer T cells recognize glycolipids from pathogenic Gram-positive bacteria. Nat Immunol. 2011;12:966–974. doi: 10.1038/ni.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.LaRock CN, Cookson BT. The Yersinia virulence effector YopM binds caspase-1 to arrest inflammasome assembly and processing. Cell Host Microbe. 2012;12:799–805. doi: 10.1016/j.chom.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bergsbaken T, Cookson BT. Macrophage activation redirects yersinia-infected host cell death from apoptosis to caspase-1-dependent pyroptosis. PLoS Pathog. 2007;3:e161. doi: 10.1371/journal.ppat.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: Host cell death and inflammation. Nat Rev Microbiol. 2009;7:99–109. doi: 10.1038/nrmicro2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muñoz-Planillo R, et al. K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity. 2013;38:1142–1153. doi: 10.1016/j.immuni.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamaji T, Mitsuki M, Teranishi T, Hashimoto Y. Characterization of inhibitory signaling motifs of the natural killer cell receptor Siglec-7: Attenuated recruitment of phosphatases by the receptor is attributed to two amino acids in the motifs. Glycobiology. 2005;15:667–676. doi: 10.1093/glycob/cwi048. [DOI] [PubMed] [Google Scholar]
- 49.Nakayama M, et al. Inhibitory receptor paired Ig-like receptor B is exploited by Staphylococcus aureus for virulence. J Immunol. 2012;189:5903–5911. doi: 10.4049/jimmunol.1201940. [DOI] [PubMed] [Google Scholar]
- 50.Lu R, Pan H, Shively JE. CEACAM1 negatively regulates IL-1β production in LPS activated neutrophils by recruiting SHP-1 to a SYK-TLR4-CEACAM1 complex. PLoS Pathog. 2012;8:e1002597. doi: 10.1371/journal.ppat.1002597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen GY, et al. Broad and direct interaction between TLR and Siglec families of pattern recognition receptors and its regulation by Neu1. eLife. 2014;3:e04066. doi: 10.7554/eLife.04066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nat Rev Immunol. 2013;13:397–411. doi: 10.1038/nri3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Avril T, North SJ, Haslam SM, Willison HJ, Crocker PR. Probing the cis interactions of the inhibitory receptor Siglec-7 with alpha2,8-disialylated ligands on natural killer cells and other leukocytes using glycan-specific antibodies and by analysis of alpha2,8-sialyltransferase gene expression. J Leukoc Biol. 2006;80:787–796. doi: 10.1189/jlb.1005559. [DOI] [PubMed] [Google Scholar]
- 54.Dotiwala F, et al. Granzyme B disrupts central metabolism and protein synthesis in bacteria to promote an immune cell death program. Cell. 2017;171:1125–1137.e11. doi: 10.1016/j.cell.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dotiwala F, et al. Killer lymphocytes use granulysin, perforin and granzymes to kill intracellular parasites. Nat Med. 2016;22:210–216. doi: 10.1038/nm.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buzza MS, et al. Extracellular matrix remodeling by human granzyme B via cleavage of vitronectin, fibronectin, and laminin. J Biol Chem. 2005;280:23549–23558. doi: 10.1074/jbc.M412001200. [DOI] [PubMed] [Google Scholar]
- 57.Aakhus AM, Stavem P, Hovig T, Pedersen TM, Solum NO. Studies on a patient with thrombocytopenia, giant platelets and a platelet membrane glycoprotein Ib with reduced amount of sialic acid. Br J Haematol. 1990;74:320–329. doi: 10.1111/j.1365-2141.1990.tb02590.x. [DOI] [PubMed] [Google Scholar]
- 58.Sacks LV, et al. A streptococcal erythrogenic toxin preparation augments natural killer activity of peripheral blood mononuclear cells. J Infect Dis. 1991;164:522–526. doi: 10.1093/infdis/164.3.522. [DOI] [PubMed] [Google Scholar]
- 59.D’Orazio JA, Burke GW, Stein-Streilein J. Staphylococcal enterotoxin B activates purified NK cells to secrete IFN-gamma but requires T lymphocytes to augment NK cytotoxicity. J Immunol. 1995;154:1014–1023. [PubMed] [Google Scholar]
- 60.Nomura T, et al. Essential role of interleukin-12 (IL-12) and IL-18 for gamma interferon production induced by listeriolysin O in mouse spleen cells. Infect Immun. 2002;70:1049–1055. doi: 10.1128/IAI.70.3.1049-1055.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shenker BJ, et al. Flow cytometric analysis of the cytotoxic effects of Actinobacillus actinomycetemcomitans leukotoxin on human natural killer cells. J Leukoc Biol. 1994;55:153–160. doi: 10.1002/jlb.55.2.153. [DOI] [PubMed] [Google Scholar]
- 62.Michaels JE, Garfield SA, Hung JT, Cardell RRJ., Jr Labeling of hepatic glycogen after short- and long-term stimulation of glycogen synthesis in rats injected with 3H-galactose. Am J Anat. 1990;188:419–428. doi: 10.1002/aja.1001880410. [DOI] [PubMed] [Google Scholar]
- 63.Avril T, Wagner ER, Willison HJ, Crocker PR. Sialic acid-binding immunoglobulin-like lectin 7 mediates selective recognition of sialylated glycans expressed on Campylobacter jejuni lipooligosaccharides. Infect Immun. 2006;74:4133–4141. doi: 10.1128/IAI.02094-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.