Significance
In this study, we used an inducible cellular model for FUS proteinopathy to demonstrate that mitochondrial dysfunction occurs as the earliest detectable change induced by FUS. In cellular and fly models, FUS interacts with the mitochondrial ATP synthase β-subunit (ATP5B), disrupts ATP synthase complex assembly, suppresses the activity of mitochondrial ATP synthase, and activates the mitochondrial unfolded protein response (UPRmt). ATP5B expression is increased in cells and flies expressing FUS. Down-regulating expression of ATP5B or UPRmt genes ameliorates FUS-induced neurodegeneration. Our data uncover a previously unknown role of FUS in targeting mitochondrial ATP synthesis and activating UPRmt.
Keywords: FUS proteinopathy, mitochondria, ATP synthase, mitochondrial unfolded protein response, frontotemporal lobar degeneration
Abstract
FUS (fused in sarcoma) proteinopathy is a group of neurodegenerative diseases characterized by the formation of inclusion bodies containing the FUS protein, including frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Previous studies show that mitochondrial damage is an important aspect of FUS proteinopathy. However, the molecular mechanisms by which FUS induces mitochondrial damage remain to be elucidated. Our biochemical and genetic experiments demonstrate that FUS interacts with the catalytic subunit of mitochondrial ATP synthase (ATP5B), disrupts the formation of ATP synthase complexes, and inhibits mitochondrial ATP synthesis. FUS expression activates the mitochondrial unfolded protein response (UPRmt). Importantly, down-regulating expression of ATP5B or UPRmt genes in FUS transgenic flies ameliorates neurodegenerative phenotypes. Our data show that mitochondrial impairment is a critical early event in FUS proteinopathy, and provide insights into the pathogenic mechanism of FUS-induced neurodegeneration.
FUS (fused in sarcoma, or translocated in liposarcoma) is a multifunctional DNA/RNA-binding protein associated with neurodegeneration. FUS proteinopathy is a group of neurodegenerative diseases characterized by the presence of protein inclusion bodies containing FUS, including amyotrophic lateral sclerosis (ALS-FUS) and different forms of frontotemporal lobar degeneration with FUS pathology (FTLD-FUS) (1–4). Recent studies have suggested potential mechanisms underlying FUS proteinopathy (reviewed in refs. 5–7), including the DNA damage response (8, 9), stress granule formation (10, 11), dysregulation of RNA metabolism (12), synaptic dysfunction (13), disrupted endoplasmic reticulum–mitochondria association (14), and impaired mitochondrial function or dynamics (15–18).
Our previous work has demonstrated that FUS protein interacts with the mitochondrial chaperone HSP60 to localize into mitochondria, leading to mitochondrial fragmentation, membrane potential loss, increased mitochondrial reactive oxygen species production, and defects in mitochondrial axonal transport (17, 18). Together with studies by other groups (14–16), these data suggest that mitochondrial dysfunction may play an important role in the pathogenesis of FUS proteinopathy.
Mitochondrial ATP synthesis is essential for many aspects of cellular homeostasis, affecting not only metabolic processes but also cell survival and death (19, 20). Mitochondrial ATP synthase (complex V) generates ATP using the electrochemical gradient across the mitochondrial inner membrane as a result of the concerted activities of respiratory chain complexes (complexes I to IV) (21). Proper assembly of complex V is essential for mitochondrial ATP synthesis (reviewed in ref. 22). Down-regulating or inhibiting a single or multiple subunits of the mitochondrial electron transport chain (ETC), encoded by either the nuclear genome (nDNA) or the mitochondrial genome (mtDNA), results in an imbalance between nDNA- versus mtDNA-encoded subunits as well as the accumulation of unassembled subunits in mitochondria. This leads to the activation of the mitochondrial unfolded protein response (UPRmt) (23). UPRmt may play a protective role in neurodegenerative disorders by reducing mitochondrial damage. UPRmt activation has been reported in animal models for Parkinson’s disease (PD) and ALS-SOD1 (24–26). However, the role of UPRmt in FUS proteinopathy has not been reported.
Our data presented here show that when accumulated inside mitochondria, FUS interacts with the mitochondrial ATP synthase catalytic subunit ATP5B and reduces mitochondrial ATP synthesis. This mitochondrial impairment is the earliest effect detected, preceding any detectable signs of cell death. In both cellular and animal models, expression of wild-type (Wt) or an ALS-associated mutant (P525L) FUS disrupts the formation of the mitochondrial ATP synthase supercomplexes and suppresses the activity of ATP synthase, resulting in mitochondrial cristae loss followed by mitochondrial fragmentation. Expression of FUS increases levels of ATP5B and ATP5B monomer not assembled into the ATP synthase supercomplexes. FUS expression activates UPRmt. Importantly, down-regulation of ATP5B or UPRmt genes by RNAi partially rescues neurodegenerative phenotypes. Our data suggest that mitochondrial impairment may be a common pathogenic mechanism underlying FTLD-FUS and ALS-FUS, and that blocking mitochondrial damage may provide a therapeutic approach to these devastating diseases.
Results
Expression of Wt or P525L-Mutant FUS Induces Cell Death.
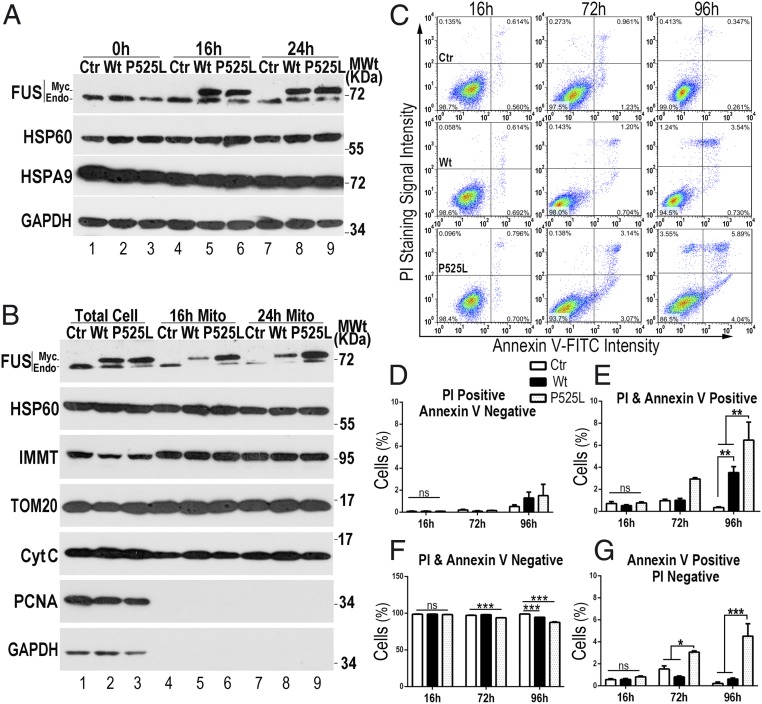
To detect early changes induced by FUS, we generated stable HEK293 cell lines expressing FUS as a myc-tagged protein using a tetracycline (Tet)-inducible system. This system allowed us to examine cellular and molecular changes at different time points following the induction of expression of Wt or P525L-mutant FUS. Both endogenous and exogenous FUS proteins were detected in purified mitochondria by the 16- or 24-h time point after FUS induction (Fig. 1 A and B). Expression of several mitochondrial proteins was not obviously affected by FUS, including HSP60, HSPA9, TOM20, or IMMT (inner-membrane mitochondrial protein) or CytC (cytochrome c) (Fig. 1 and SI Appendix, Fig. S1).
Fig. 1.
Expression of Wt or P525L-mutant FUS induces cell death. Stable HEK cell lines expressing either Wt or P525L-mutant FUS as myc-tagged proteins following induction by tetracycline (0.5 μg/mL). (A and B) At different time points following Tet induction of FUS expression, total cell lysates or purified mitochondria were analyzed using Western blotting with the indicated antibodies. Control, Ctr. (C) Representative flow cytometry analysis of cells following staining with an Annexin V-FITC/PI kit to quantify cell death in cells expressing Wt or P525L-mutant FUS. (D–G) Quantification of distinct cell populations at different time points (also see SI Appendix, Fig. S2). Data were analyzed using a two-way ANOVA with Bonferroni post hoc test (representing three independent experiments; *P < 0.05, **P < 0.01, ***P < 0.001; ns, not significant). Error bars represent mean ± SEM.
We next examined cell death by staining cells using an Annexin V-FITC/PI (propidium iodide) kit followed by flow cytometry analyses. Cells expressing P525L-mutant FUS began to show increased cell death by the 72-h time point following FUS induction, whereas no significant cell death was detected at the 16- or 48-h time point in cells expressing either Wt or P525L-mutant FUS (Fig. 1 C–G and SI Appendix, Fig. S2). By the 96-h time point after FUS induction, both Wt and P525L-mutant FUS induced significant cell death compared with control cells (Fig. 1 C–G). Expression of P525L-mutant FUS showed more mitochondrial FUS accumulation (Fig. 1B) and induced more cell death (Fig. 1 C–G) compared with Wt FUS, suggesting that increased mitochondrial FUS is associated with elevated cytotoxicity.
FUS Induces Mitochondrial Damage Before Cell Death.
To investigate whether FUS expression leads to the mitochondrial dysfunction at an early stage, we examined FUS-expressing cells by electron microscopy (EM) between 16 and 72 h after FUS induction. At 16 h, mitochondria in cells expressing Wt or P525L-mutant FUS showed a prominent loss of cristae compared with those in control cells (Fig. 2 A–C). By 72 h, the size of mitochondria was significantly reduced in cells expressing either Wt or P525L-mutant FUS (Fig. 2 A and D). Interestingly, the mitochondrial membrane potential of cells expressing Wt or P525L-mutant FUS exhibited a transient increase at 16 h after FUS induction, and then decreased by 72 h (Fig. 2 E–H and SI Appendix, Fig. S3). Furthermore, mitochondrial cristae loss was also found in fly photoreceptors expressing Wt or P525L-mutant FUS (SI Appendix, Fig. S4), indicating that increased FUS expression disrupts the formation or maintenance of mitochondrial cristae in vivo. It has been reported that an ATP synthase defect may cause mitochondrial hyperpolarization (27) and lead to mitochondrial cristae loss (28, 29). Our results demonstrate that transient hyperpolarization and mitochondrial cristae loss induced by FUS occur at an early stage, preceding mitochondrial fragmentation and cell death.
Fig. 2.
Increased FUS expression induces mitochondrial cristae loss and mitochondrial membrane hyperpolarization before cell death. All assays were done 16 or 72 h postinduction as specified. (A) EM images showing mitochondrial morphology of HEK293 cells expressing the vector control or Wt or P525L-mutant FUS. (B) Quantification of mitochondrial size; >100 mitochondria per group (128, 116, and 150 in Ctr, Wt, and P525L groups, respectively) were measured. (C) Mitochondrial cristae density; >50 mitochondria per group (52, 53, and 54 in Ctr, Wt, and P525L groups, respectively) were quantified. (D) Mitochondrial size at 72 h postinduction; >100 mitochondria per group (115, 117, and 141 in Ctr, Wt, and P525L groups, respectively) were measured. (E–H) FACS analyses of cells expressing Ctr, Wt, or P525L-mutant FUS with quantification of TMRM (tetramethylrhodamine methyl ester) signal intensity at 16 and 72 h. Data were analyzed using a one-way ANOVA with Bonferroni post hoc test (*P < 0.05, **P < 0.01, ***P < 0.001). Error bars represent mean ± SEM.
FUS Suppresses Mitochondrial ATP Synthase Activity and Disrupts the Formation of ATP Synthase Supercomplex.
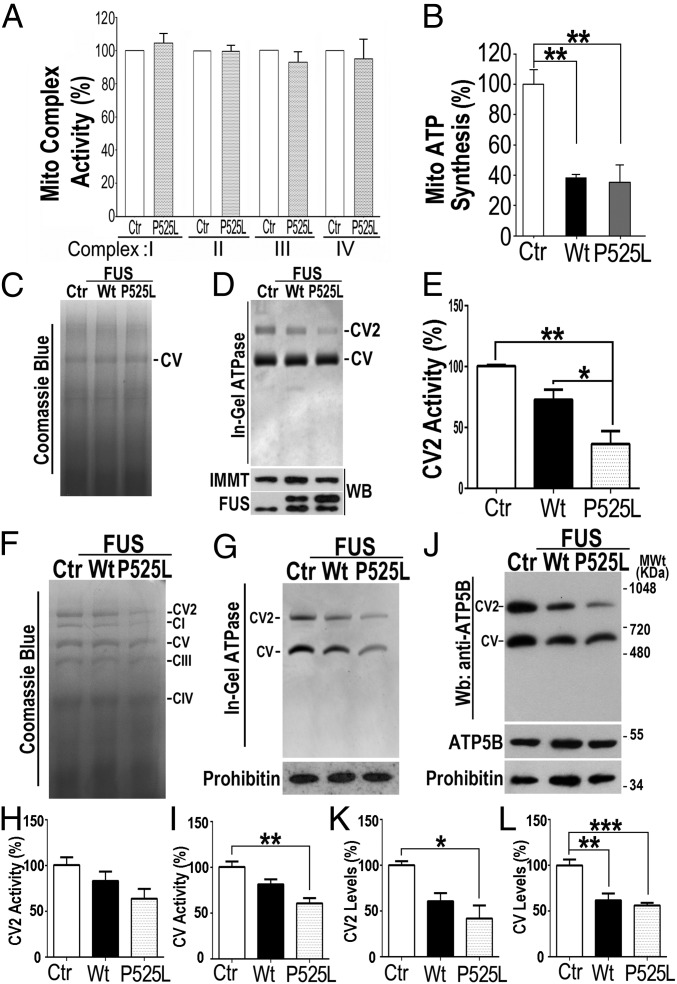
We systematically examined mitochondrial function in these FUS-expressing cells. By the 16 h time point following induction of expression of either Wt or P525L-mutant FUS, mitochondrial ATP synthesis was reduced by more than 50%, reflecting a significant impairment of mitochondrial function. In contrast, the activities of mitochondrial complexes I through IV were unaffected by FUS expression (Fig. 3 A and B). This is consistent with the possibility that the ongoing activity of ETC complexes without proton influx through the dysfunctional ATP synthase led to a transient increase in the membrane potential (Fig. 2 E and F). We then focused analyses on the activity and formation of mitochondrial ATP synthase (complex V) using blue native polyacrylamide gel electrophoresis (BN-PAGE) (Fig. 3C). At 16-h postinduction, an in-gel ATPase assay after BN-PAGE was performed to examine the activities of the complex V monomer and dimer (Fig. 3D). Although ATP synthase complex monomer (CV) activity was not affected by FUS expression, CV dimer (CV2) activity was decreased in cells expressing Wt or P525L-mutant FUS compared with control cells (Fig. 3 D and E). No obvious differences were detected in the level of the ATP synthase CV by Coomassie blue staining (Fig. 3C). Thus, complex V formation was further examined using clear native polyacrylamide gel electrophoresis (CN-PAGE) analyses of purified mitochondria from these cells. Western blot analyses after CN-PAGE using anti-ATP5B antibody detected not only the complex V complexes but also faster-migrating bands with molecular masses between 66 and 150 kDa, corresponding to ATP5B monomers and ATP5B-containing intermediate complexes. Notably, the levels of ATP5B monomers and ATP5B-containing intermediate complexes were increased in purified mitochondria from cells expressing either Wt or P525L-mutant FUS (especially the mutant FUS) compared with the control group (SI Appendix, Fig. S5). Together, these data demonstrate that FUS expression disrupts the formation and suppresses the activity of ATP synthase complexes. It is important to note that FUS-induced disruption of ATP synthase function occurs at an early stage (16 h), before any detectable signs of cell death (72 to 96 h postinduction).
Fig. 3.
(A–E) Expression of Wt or P525L-mutant FUS impairs mitochondrial ATP synthesis in inducible HEK293 cells (16 h postinduction). (A) Activities of the mitochondrial complexes I to IV were determined in HEK293 inducible FUS-expressing cells. (B) Mitochondrial ATP synthesis was measured. (C) BN-PAGE analyses of mitochondrial complexes using purified mitochondria from cells expressing Ctr, Wt, or P525L-mutant FUS. (D) The in-gel ATPase assay was performed using BN-PAGE of digitonin-solubilized mitochondria from corresponding groups, showing ATPase activity of monomeric or dimeric complex V (CV or CV2, respectively). The mitochondrial marker IMMT and FUS protein levels were examined by Western blotting (WB). (E) Quantification of mitochondrial ATPase activity from the corresponding groups shown in D, normalized to the protein levels. (F–L) Expression of Wt or P525L-mutant FUS in flies disrupts complex V formation and suppresses ATP synthase activity. (F) BN-PAGE analyses of mitochondrial complexes using purified mitochondria from flies expressing Ctr, Wt, or P525L-mutant FUS at day 15 postinduction. (G) The in-gel ATPase assay was performed as in D. (H and I) Quantification of the mitochondrial ATPase activity shown in G (normalized to the intensity of the corresponding bands in Coomassie blue staining of F). (J) Western blotting analyses following BN-PAGE to show the protein levels of the ATP synthase monomeric or dimeric complexes (CV or CV2), as detected using a specific antibody against ATP5B. Another mitochondrial protein, prohibitin, was used as a loading control. (K and L) Quantification of CV and CV2 protein levels shown in J. Fly genotypes: Ctr: actin5C-Gal4/tubulin-Gal80ts/UAS-RFP; Wt: actin5C-Gal4/tubulin-Gal80ts/UAS-Wt-FUS-RFP; P525L: actin5C-Gal4/tubulin-Gal80ts/UAS-P525L-FUS-RFP. Data were analyzed using a one-way ANOVA with Bonferroni post hoc test (representing three independent experiments; *P < 0.05, **P < 0.01, ***P < 0.001). Error bars represent mean ± SEM.
In our previous work, we had established a Drosophila model of FUS proteinopathy and showed that expression of Wt or ALS-mutant FUS led to mitochondrial damage and neurodegeneration (17, 30). To examine whether FUS expression affected mitochondrial ATP synthase activity and formation in vivo, we used transgenic flies expressing human Wt or P525L-mutant FUS under a temperature-inducible actin5C-Gal4/tubulin-Gal80ts driver (31). FUS expression was induced in adult flies for 15 d before mitochondria were purified from fly heads. BN-PAGE was then performed using purified mitochondria to measure the activity and formation of complex V (Fig. 3F). The in-gel ATPase assay following BN-PAGE revealed that expression of Wt or P525L-mutant FUS decreased the activities of both the monomer CV and the dimer CV2 compared with the control group, with reduction of CV activity in the P525L-mutant group reaching statistical significance (Fig. 3 G–I). The protein levels of the monomer CV and the dimer CV2 were significantly decreased in flies expressing P525L-mutant FUS compared with the control group (Fig. 3 J–L). These results indicate that increased FUS expression in vivo disrupts the formation of mitochondrial ATP synthase supercomplexes and suppresses the ATP synthase activity.
FUS Induces Mitochondrial Unfolded Protein Response.
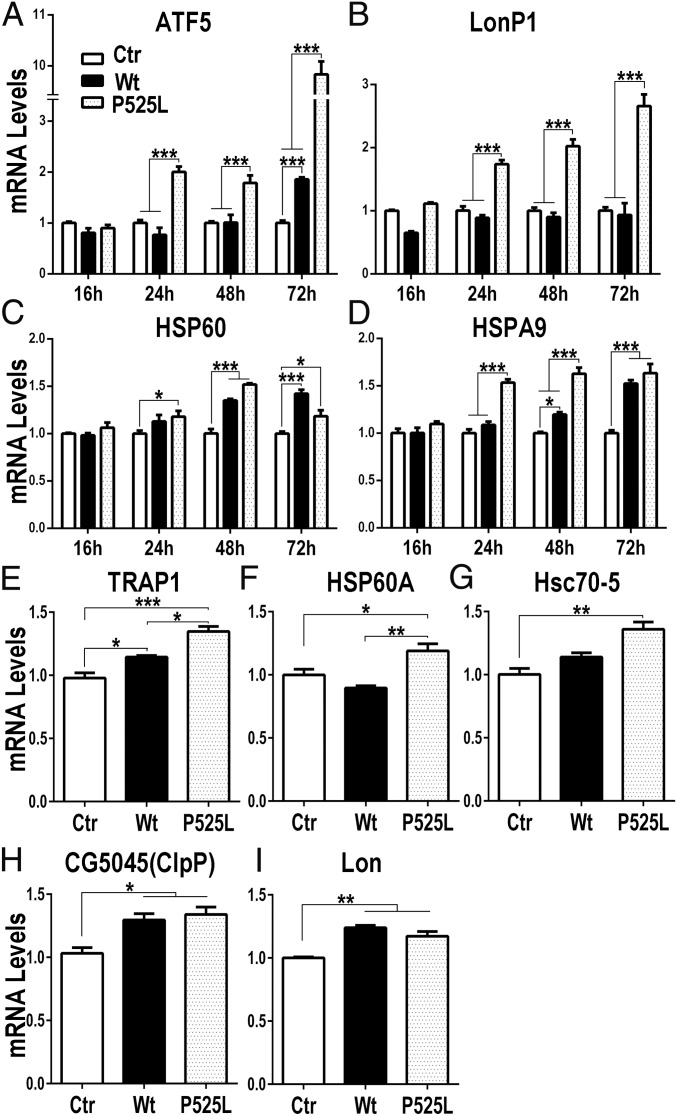
Recent studies have shown that altering the formation or activity of the mitochondrial ETC activates the mitochondrial unfolded protein response; one possible trigger for UPRmt activation is a disruption in the balance of the mtDNA- versus nDNA-encoded components of ETC complexes (23, 32–35). Our data presented above indicate that when accumulated in mitochondria, FUS disrupts the formation and function of the ATP synthase dimer CV2. This prompted us to examine UPRmt in our cellular and animal models of FUS proteinopathy. Using the inducible HEK293 cells expressing FUS, we performed quantitative RT-PCR (qRT-PCR) at different time points following FUS induction using specific primers for genes critical for mammalian UPRmt, including ATF5, LonP1, HSP60, and HSPA9 (also known as mitochondrial HSP70 or mortalin) (36, 37). Quantitative analyses revealed that these UPRmt-associated genes were up-regulated following induction of expression of either Wt or P525L-mutant FUS (Fig. 4 A–D). All four genes were up-regulated 24 h after FUS induction in cells expressing P525L-mutant FUS and remained up-regulated through 72 h. HSP60 and HSPA9 were up-regulated 48 h after FUS induction in cells expressing Wt FUS, and ATF5 was up-regulated after the 72-h time point. These data show that increased expression of FUS induces UPRmt in cultured cells.
Fig. 4.
FUS induces mitochondrial unfolded protein response. (A–D) Expression levels of ATF5, LonP1, HSP60, and HSPA9 mRNAs in control group or HEK293 cells expressing Wt or P525L-mutant FUS were measured by qRT-PCR at different time points postinduction. Data were analyzed using a two-way ANOVA with Bonferroni post hoc test. (E–I) Expression levels of TRAP1, HSP60A, Hsc70-5, CG5045 (ClpP), and Lon mRNAs were determined by qRT-PCR in control flies or flies expressing Wt or P525L-mutant FUS at 15 d postinduction. Data were analyzed using a one-way ANOVA with Bonferroni post hoc test. *P < 0.05, **P < 0.01, ***P < 0.001. Error bars represent mean ± SEM. Fly genotypes: Ctr: actin5C-Gal4/tubulin-Gal80ts/UAS-RFP; Wt: actin5C-Gal4/tubulin-Gal80ts/UAS-Wt-FUS-RFP; P525L: actin5C-Gal4/tubulin-Gal80ts/UAS-P525L-FUS-RFP.
We next examined whether FUS expression elicited UPRmt in vivo. The actin5C-Gal4/tubulin-Gal80ts driver was used to induce expression of either Wt or P525L-mutant FUS in adult flies for 15 d, the same time point when the formation of mitochondrial ATP synthase supercomplexes was disrupted by FUS (Fig. 3 G–L). Quantitative RT-PCR analyses revealed that UPRmt-associated genes, including TRAP1, HSP60A, Hsc70-5, CG5045 (ClpP), and Lon, were up-regulated in flies expressing either Wt or P525L-mutant FUS compared with the control flies (Fig. 4 E–I).
To investigate the role of UPRmt in FUS-induced neurodegeneration, we examined whether altering expression of genes critical for UPRmt modified FUS-induced neurotoxicity. Unexpectedly, knocking down UPRmt-associated genes, including TRAP1, Hsc70-5, CG5045 (ClpP), and Lon, reduced retinal degeneration in flies expressing Wt or P525L-mutant FUS, with a recovery of ommatidial organization to various extents, whereas knocking down these genes in control flies did not show any detectable effects (Fig. 5 and SI Appendix, Fig. S6 A and B). While control flies overexpressing Lon showed moderate retinal degeneration (SI Appendix, Fig. S6A), Lon overexpression in FUS transgenic flies exacerbated FUS-induced retinal degeneration, with eye atrophy and a more severe loss of ommatidial organization (Fig. 5A). Overexpression of ClpP, another UPRmt gene, did not show a detectable effect (SI Appendix, Fig. S6C). These data suggest that UPRmt activation by FUS may not protect against neurotoxicity; instead, excessive UPRmt activation may contribute to the pathogenesis of FUS proteinopathy.
Fig. 5.
Down-regulating UPRmt genes partially rescues retinal degeneration phenotype. (A) Microscopic images of fly eyes (day 5 adults) show that RNAi knockdown of UPRmt genes partially rescued retinal degeneration in flies expressing Wt or P525L-mutant FUS. Magnified retinal regions (marked by arrows) are shown (Insets). (B and C) Quantification of the degenerated area (percentage of the entire eye surface) in fly eyes expressing Wt or P525L-FUS, respectively, in the indicated groups shown in A. Data were analyzed by a one-way ANOVA with Bonferroni post hoc test (representing three independent experiments; *P < 0.05, **P < 0.01, ***P < 0.001). Error bars represent mean ± SEM. Fly genotypes: Ctr: GMR-Gal4/UAS-Wt-FUS-RFP or GMR-Gal4/UAS-P525L-FUS-RFP; siTRAP1: GMR-Gal4/UAS-Wt-FUS-RFP/UAS-siTRAP1 or GMR-Gal4/UAS-P525L-FUS-RFP/UAS-siTRAP1; siHsc70-5: GMR-Gal4/UAS-Wt-FUS-RFP/UAS-siHsc70-5 or GMR-Gal4/UAS-P525L-FUS-RFP/UAS-siHsc70-5; siClpP (siCG5045): GMR-Gal4/UAS-Wt-FUS-RFP/UAS-siClpP or GMR-Gal4/UAS-P525L-FUS-RFP/UAS-siClpP; siLon: GMR-Gal4/UAS-Wt-FUS-RFP/UAS-siLon or GMR-Gal4/UAS-P525L-FUS-RFP/UAS-siLon; Lon OE: GMR-Gal4/UAS-Wt-FUS-RFP/UAS-Lon or GMR-Gal4/UAS-P525L-FUS-RFP/UAS-Lon.
FUS Interacts with Mitochondrial ATP Synthase β-Subunit.
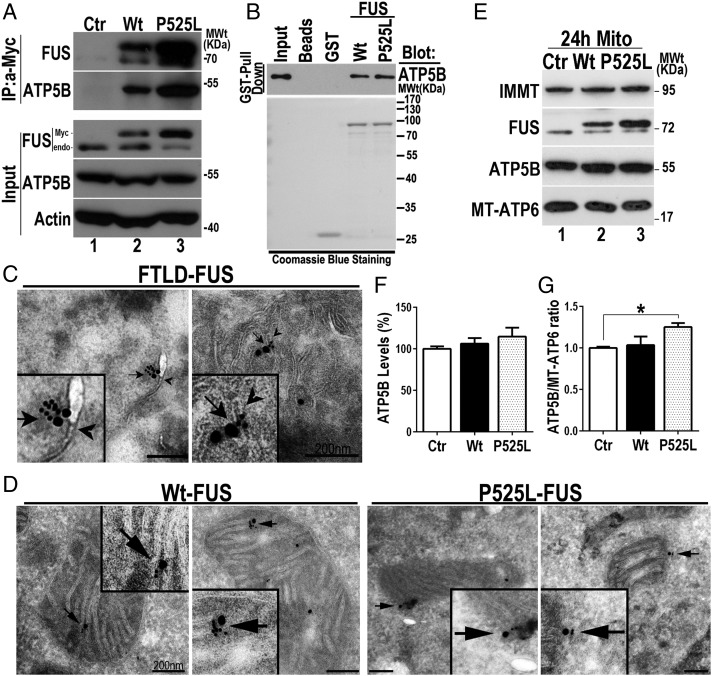
To dissect the mechanism by which FUS affects mitochondrial ATP synthase function, we examined whether FUS interacted with any components of the ATP synthase complex. Our previous study using an immunopurification-coupled mass spectrometry approach identified several mitochondrial proteins interacting with FUS, including ATP5B (17). Coimmunoprecipitation followed by Western blotting with specific antibodies against different subunits of the mitochondrial ATP synthase complexes revealed that both Wt and P525L-mutant FUS specifically interacted with ATP5B, the catalytic subunit of the ATP synthase (Fig. 6A). No interaction was detected between FUS and ATP5A1, ATP5O, or ATP5F1 (SI Appendix, Fig. S7A). To test if FUS directly interacted with ATP5B, we performed GST pull-down experiments using purified Wt or P525L-mutant FUS proteins tagged with GST together with His-tagged ATP5B protein. In the GST pull-down assay, both Wt and P525L-mutant FUS interacted with the purified ATP5B protein (Fig. 6B). The interaction between FUS and ATP5B was not dependent on RNA, because such interaction was not affected by RNase treatment (SI Appendix, Fig. S7B). These data indicate that FUS interacts with ATP5B in an RNA-independent manner.
Fig. 6.
FUS interacts with ATP5B and colocalizes with ATP5B in mitochondria. (A) Interaction of FUS with ATP5B was detected in coimmunoprecipitation experiments. A specific anti-Myc monoclonal antibody was used to immunoprecipitate FUS protein in HEK293 cells expressing Wt or P525L-mutant FUS as Myc-tagged proteins. ATP5B was detected among proteins immunoprecipitated by anti-Myc in cells expressing Wt or P525L-mutant FUS, but not in cells expressing the vector control. IP, immunoprecipitation. (B) GST pull-down assay using purified FUS and ATP5B proteins indicates that Wt or P525L-mutant FUS interacts with ATP5B. (Top) Detection of ATP5B by Western blotting in the GST pull-down assay using GST-tagged Wt or P525L-mutant FUS. (Bottom) Coomassie blue staining of purified GST-tagged Wt or P525L-mutant FUS protein preparations as well as the control GST protein used in the GST pull-down experiments. Approximately 1/40 of the total input (20 ng ATP5B-His) was loaded in lane 1 to avoid excessively strong WB signals; 50 ng of Wt or P525L-mutant FUS-GST together with 20 ng ATP5B-His was used for GST pull-down in the last two lanes. (C) IEM of FTLD-FUS brain samples. Cryosections were immunostained with a specific murine anti-FUS antibody and a secondary anti-murine IgG conjugated to 10-nm gold particles. Mitochondrial ATP5B was labeled with rabbit anti-ATP5B and anti-rabbit IgG conjugated to 25-nm gold particles. Because the majority of mitochondria in FTLD-FUS brain samples exhibited a loss of mitochondrial membrane or damaged cristae (17), only the remaining mitochondrial cristae are shown, rather than typical healthy mitochondria. Arrows mark colocalization of FUS IEM signals with ATP5B IEM signals. Arrowheads mark mitochondrial cristae. Magnified areas are shown (Insets). (D) IEM of HEK293 stable cells expressing Wt or P525L-mutant FUS. FUS was labeled with 10-nm gold particles, and ATP5B was labeled with 25-nm gold particles. Arrows mark colocalization of FUS with ATP5B in the mitochondria. Magnified areas are shown (Insets). (E) Mitochondria were purified from HEK293 cells expressing control, Wt, or P525L-FUS 24 h postinduction and analyzed by Western blotting with the indicated antibodies to show the levels of nuclear-encoded ATP5B and mitochondrial genome-encoded MT-ATP6 proteins. IMMT was used as an internal control. (F and G) Quantification of data in B to show the ATP5B protein levels or the ATP5B:MT-ATP6 ratio in Ctr, Wt, or P525L-FUS groups. Data were analyzed using a one-way ANOVA with Bonferroni post hoc test (representing three independent experiments; *P < 0.05). Error bars represent mean ± SEM.
To examine if FUS protein colocalized with ATP5B inside mitochondria, we performed immunoelectron microscopy (IEM) using FTLD-FUS brain samples as reported in our previous study (17). In FTLD-FUS brain samples, FUS-immunostaining signals (10-nm gold particles) were detected in close proximity to ATP5B-immunostaining signals (25-nm gold particles) (marked by arrows in Fig. 6C), and both were in close association with mitochondrial cristae (arrowheads in Fig. 6C). Similar IEM results were obtained in cultured HEK293 cells stably expressing the Wt or P525L-mutant FUS. ATP5B colocalized with either Wt or P525L-mutant FUS on mitochondrial cristae (marked by arrows in Fig. 6D). Further examination of purified mitochondria at 24 h postinduction showed that the ATP5B protein levels were moderately increased in cells expressing Wt or P525L-mutant FUS (not reaching statistical significance; Fig. 6 E and F). Interestingly, the ratio of the nuclear-encoded ATP5B to the mitochondrial-encoded MT-ATP6 increased in cells expressing P525L-mutant FUS at 24 h postinduction (Fig. 6G), consistent with UPRmt activation at this time point in these cells (Fig. 4 A–D). FUS knockout did not affect the mitochondrial ATP5B protein level (SI Appendix, Fig. S8A), suggesting that the increase in AT5B protein level is not due to stabilization by FUS. On the other hand, ATP5B mRNA levels increased in cells expressing Wt or P525L-mutant FUS (SI Appendix, Fig. S8B).
Knocking Down ATP5B Rescues Mitochondrial and Retinal Degeneration Phenotypes in FUS Transgenic Flies.
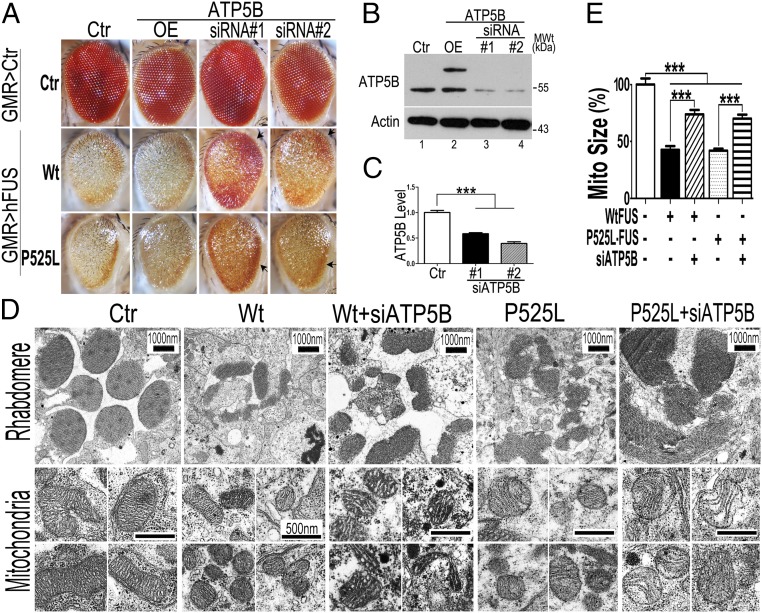
To investigate if FUS interacts with ATP5B in vivo, we performed genetic experiments by overexpressing or down-regulating ATP5B in FUS transgenic flies. Flies expressing Wt or P525L-mutant FUS in photoreceptors exhibited retinal degeneration with a loss of ommatidia, as reported previously (17, 18, 30). Western blotting showed that ATP5B was increased in fly eyes expressing Wt or P525L-mutant FUS (SI Appendix, Fig. S9 A and B). Overexpressing ATP5B in FUS flies exacerbated the retinal degeneration phenotype, with a more severe loss of ommatidia (Fig. 7A), and also further reduced locomotor function of larvae expressing FUS (SI Appendix, Fig. S9C). On the other hand, down-regulating ATP5B suppressed FUS-induced retinal degeneration. Two independent RNAi fly lines, siATP5B 1 and 2, were examined in which the ATP5B protein level was significantly reduced (Fig. 7 B and C). Both siATP5B lines showed suppression of the retinal degeneration phenotypes in fly eyes expressing either Wt or P525L-mutant FUS, with a partial recovery of ommatidial organization (Fig. 7A). These results indicate that down-regulating ATP5B partially rescued the neurodegeneration phenotype in vivo. As siATP5B lines 1 and 2 showed a similar rescue effect, siATP5B line 1 was used for subsequent experiments. EM revealed a loss of rhabdomeres and mitochondrial damage in photoreceptors expressing Wt or P525L-mutant FUS. Although down-regulating ATP5B did not restore the rhabdomeres to completely normal morphology, it significantly increased the mitochondrial size in photoreceptors (Fig. 7 D and E).
Fig. 7.
Down-regulating ATP5B ameliorates retinal degeneration and mitochondrial fragmentation in flies expressing Wt or P525L-mutant FUS. (A) Microscopic images of fly eyes (5-d-old adult) show that RNAi knocking down of ATP5B mitigates retinal degeneration in flies expressing Wt or P525L-mutant FUS, whereas overexpression (OE) of ATP5B exacerbates the neurodegenerative phenotypes. (B) Western blotting analyses revealing ATP5B protein levels in the fly eyes of the corresponding groups. (C) Quantification of ATP5B protein levels in two ATP5B RNAi lines. (D) Transmission electron microscopy images of fly retinae at day 5. Knocking down ATP5B expression by siRNA partially rescues mitochondrial fragmentation in flies expressing Wt or P525L-mutant FUS. (E) Mitochondrial size was quantified using ImageJ (NIH). More than 100 mitochondria per group (140, 129, 125, 139, and 129 in Ctr, Wt-FUS, Wt-FUS+siATP5B, P525L-FUS, and P525L-FUS+siATP5B groups, respectively) were quantified. Data were analyzed using one-way ANOVA with Bonferroni post hoc test (***P < 0.001). Error bars represent mean ± SEM. Fly genotypes: GMR>Ctr group: Ctr: GMR-Gal4/UAS-RFP; ATP5B OE: GMR-Gal4/UAS-ATP5B-Myc; ATP5B siRNA#1: GMR-Gal4/UAS-siATP5B#1; ATP5B siRNA#2: GMR-Gal4/UAS-siATP5B#2; GMR>hFUS group (Wt or P525L-mutant): Ctr: GMR-Gal4/UAS-Wt-FUS-RFP or GMR-Gal4/UAS-P525L-FUS-RFP; ATP5B OE: GMR-Gal4/UAS-Wt-FUS-RFP/UAS-ATP5B-Myc or GMR-Gal4/UAS-P525L-FUS-RFP/UAS-ATP5B-Myc; ATP5B siRNA#1: GMR-Gal4/UAS-Wt-FUS-RFP/UAS-siATP5B#1 or GMR-Gal4/UAS-P525L-FUS-RFP/UAS-siATP5B#1; ATP5B siRNA#2: GMR-Gal4/UAS-Wt-FUS-RFP/UAS-siATP5B#2 or GMR-Gal4/UAS-P525L-FUS-RFP/UAS-siATP5B#2.
Discussion
A number of mechanisms have been proposed for FUS proteinopathy, including the DNA damage repair defect, dysregulation of transcription and/or pre-mRNA splicing, oxidative stress, aberrant stress granule formation, mitochondrial impairment, and axonal transport defects (for recent reviews, see refs. 6 and 7). A previous report suggests that FUS may affect ATP synthesis (14). However, the underlying mechanisms were not understood.
Our data here demonstrate that expression of Wt or ALS-mutant FUS leads to mitochondrial dysfunction that precedes cell death (Figs. 1 and 2 and SI Appendix, Figs. S2 and S3). Further, our work demonstrates that FUS interacts with ATP5B, disrupts assembly of the ATP synthase supercomplex(es), and suppresses mitochondrial ATP synthesis (Figs. 3 and 6). ATP synthase complex assembly is critical for efficient ATP synthesis and is closely associated with mitochondrial cristae formation (38). ATP synthase mutants show disorganized cristae in yeast (28, 38, 39). ATP5B, the β-subunit, is the essential catalytic subunit of the mitochondrial ATP synthase (40). Our data show that expression of Wt or ALS-mutant FUS impairs the formation and activity of ATP synthase complexes (complex V) without affecting the other four ETC complexes (Fig. 3). Our study demonstrates that FUS directly targets mitochondrial complex V.
Consistent with the association between complex V assembly and cristae formation, our EM data show that expression of Wt or P525L-mutant FUS leads to a mitochondrial cristae loss in both cellular and animal models (Fig. 2 and SI Appendix, Fig. S4). Remarkably, similar mitochondrial damage, including cristae loss, was recently reported as the earliest pathological changes, preceding synaptic or neuronal loss, in transgenic mice expressing wild-type human FUS protein (41). Similar cristae changes were also observed in FTLD-FUS patient brains (17), indicating that mitochondrial impairment is an intrinsic pathological feature of FUS proteinopathy.
Genetically altering the level of ATP5B not only modifies FUS-induced mitochondrial damage but also modulates FUS neurotoxicity (Fig. 7), suggesting that the interaction of FUS with ATP5B is functionally significant. By interacting with ATP5B, FUS may disrupt the assembly of ATP synthase supercomplexes, resulting in mitochondrial dysfunction and damage (see the working model depicted in Fig. 8). The mitochondrial ATP5B protein level was increased in FUS-expressing HEK293 cells (Fig. 6 E and F) and flies (SI Appendix, Fig. S9 A and B), whereas ATP synthase complex activities and formation were decreased (Fig. 3 C–E and G–L). Our data suggest that there would be an accumulation of unassembled or free ATP synthase subunits (ATP5B or another subunits; ATPx). This may induce an imbalance in the nDNA- versus mtDNA-encoded complex components and thereby activate UPRmt, similar to a previous study (23). Consistent with this notion, FUS expression increases unassembled ATP5B levels in purified mitochondria and increases the mitochondrial ATP5B:MT-ATP6 ratio (Fig. 6 E and G and SI Appendix, Fig. S5).
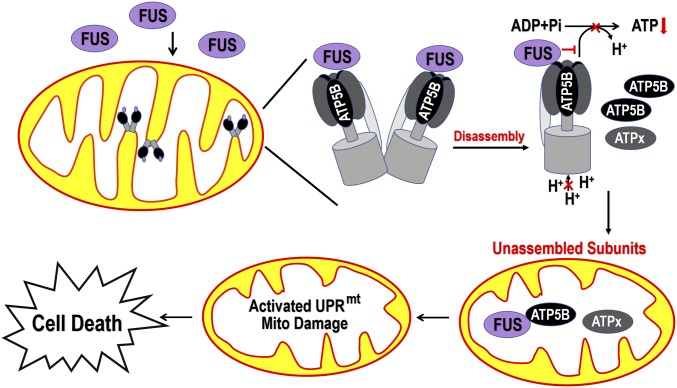
Fig. 8.
Working model for FUS-induced mitochondrial damage and neurotoxicity. Under certain stress conditions, either aberrantly increased expression of Wt FUS or mutations in the FUS gene that result in increased cytoplasmic FUS distribution lead to an increase in mitochondrial accumulation of the FUS protein. Excessive mitochondrial FUS protein interacts with ATP5B, disrupts the assembly of the ATP synthase supercomplexes, and impairs ATP synthase activity. This may lead to an accumulation of unassembled components of the ATP synthase complexes, ATP5B, or other components (“ATPx”) and mitochondrial cristae damage. Decreased mitochondrial ATP synthesis together with accumulation of unassembled components of complex V, in turn, may induce the mitochondrial unfolded protein response and further exacerbate mitochondrial damage, ultimately leading to neuronal death. This is supported by the observation that down-regulation of UPRmt-associated genes or ATP synthase subunit ATP5B ameliorates mitochondrial damage and cell death. Our model suggests that disruption of the ATP synthase complexes and excessive UPRmt activation may play a critical role in the pathogenesis of FUS proteinopathy.
UPRmt is an adaptive mechanism to ensure proper communication between mitochondria and the nucleus. Accumulation of mitochondrial unfolded proteins or an imbalance of nDNA- versus mtDNA-encoded mitochondrial proteins may trigger UPRmt to regulate genes crucial for mitochondrial proteostasis and quality control (42–44). However, UPRmt activation is not always beneficial. Under severe or extended mitochondrial stresses, excessive activation of UPRmt may have detrimental effects (24, 45). For example, it was reported that prolonged UPRmt activation induced neurodegeneration in PD models (46). Our data show that expression of Wt or ALS-mutant FUS proteins activates UPRmt (Fig. 4), and that down-regulating UPRmt genes ameliorates FUS-induced neurodegeneration in vivo (Fig. 5 and SI Appendix, Fig. S6). We hypothesize that UPRmt is activated by FUS-induced disruption of mitochondrial ATP synthesis and by an imbalance in nuclear-encoded ATP5B versus mitochondrial-encoded subunits of ATP synthase. This represents an interesting avenue for future work.
Taken together, our data show that FUS interacts with the catalytic subunit of mitochondrial ATP synthase and disrupts assembly and function of the mitochondrial ATP synthase complex(es). This likely leads to an accumulation of unassembled complex V subunits in mitochondria, and possibly thereby activates UPRmt. Excessive or prolonged UPRmt may result in irreparable mitochondrial damage and neuronal death (Fig. 8). Our results suggest that blocking or reversing mitochondrial impairment may provide therapeutic benefits for FUS proteinopathy patients.
Materials and Methods
Fly Strains and Antibodies.
Fly strains and antibodies are described in detail in SI Appendix. The specificity of all antibodies used in this study has been confirmed by published studies.
Ethics Statement.
Deidentified postmortem brain samples were obtained from the Cognitive Neurology and Alzheimer’s Disease Center at Northwestern University following institutional and NIH guidelines. All samples were previously published (17) and collected with informed consent at the Cognitive Neurology & Alzheimer’s Disease Center at Northwestern University with the study approved by the institutional review board. All samples were from deidentified post-mortem autopsy.
Cell Death Detection Assay.
Cell death was measured using Annexin V-FITC Apoptosis Detection Kit I (BD) according to the manufacturer’s instructions. Briefly, cells at different induction time points were detached by trypsin-EDTA, rinsed in cold PBS, and then stained with Annexin V-FITC and PI followed by immediate analysis (within 1 h) using flow cytometry (BD FACSCalibur).
Mitochondrial Purification.
Mitochondria were purified following published protocols (17, 47). Fly mitochondria were purified following a published protocol (48).
Procedures for cell cultures, electron microscopy, mitochondrial purification, measurement of mitochondrial complex activities, mitochondrial ATP synthesis, mitochondrial membrane potential, BN-PAGE and CN-PAGE, in-gel ATPase assay together with qRT-PCR, and larvae movement assays are described in SI Appendix.
Statistical Analyses.
Differences between two groups were analyzed using Student’s t test. Multiple group comparisons were performed using one- or two-way analysis of variance (ANOVA) followed by Bonferroni post hoc test. The bar graphs represent mean ± SEM. Significance is indicated by asterisks: *P < 0.05, **P < 0.01, ***P < 0.001.
Supplementary Material
Acknowledgments
We thank the anonymous reviewers whose constructive comments helped us improve the paper, and Dr. E. Bigio, W. McGee, D. Kuo, and other members of the J.Y.W. lab for helpful suggestions and critical reading of the manuscript. We thank the Center for Biological Imaging (Institute of Biophysics) and S. Sun, L. Sun, L. Wang, and C. Peng for EM work. J.D., P.W., J.L., and L.Z. are supported by grants from the National Natural Science Foundation of China (31671174, 31501133, 31671452, 31701004). J.D. is supported by the China Postdoctoral Science Foundation (2016M600137). J.Y.W. is supported by the NIH (R01CA175360 and RO1AG054008).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806655115/-/DCSupplemental.
References
- 1.Kwiatkowski TJ, Jr, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 2.Vance C, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neumann M, et al. A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain. 2009;132:2922–2931. doi: 10.1093/brain/awp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mackenzie IR, et al. Distinct pathological subtypes of FTLD-FUS. Acta Neuropathol. 2011;121:207–218. doi: 10.1007/s00401-010-0764-0. [DOI] [PubMed] [Google Scholar]
- 5.Ratti A, Buratti E. Physiological functions and pathobiology of TDP-43 and FUS/TLS proteins. J Neurochem. 2016;138:95–111. doi: 10.1111/jnc.13625. [DOI] [PubMed] [Google Scholar]
- 6.Shang Y, Huang EJ. Mechanisms of FUS mutations in familial amyotrophic lateral sclerosis. Brain Res. 2016;1647:65–78. doi: 10.1016/j.brainres.2016.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao FB, Almeida S, Lopez-Gonzalez R. Dysregulated molecular pathways in amyotrophic lateral sclerosis-frontotemporal dementia spectrum disorder. EMBO J. 2017;36:2931–2950. doi: 10.15252/embj.201797568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mastrocola AS, Kim SH, Trinh AT, Rodenkirch LA, Tibbetts RS. The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. J Biol Chem. 2013;288:24731–24741. doi: 10.1074/jbc.M113.497974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiu H, et al. ALS-associated mutation FUS-R521C causes DNA damage and RNA splicing defects. J Clin Invest. 2014;124:981–999. doi: 10.1172/JCI72723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baron DM, et al. Amyotrophic lateral sclerosis-linked FUS/TLS alters stress granule assembly and dynamics. Mol Neurodegener. 2013;8:30. doi: 10.1186/1750-1326-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bentmann E, et al. Requirements for stress granule recruitment of fused in sarcoma (FUS) and TAR DNA-binding protein of 43 kDa (TDP-43) J Biol Chem. 2012;287:23079–23094. doi: 10.1074/jbc.M111.328757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Y, Liu S, Oztürk A, Hicks GG. FUS-regulated RNA metabolism and DNA damage repair: Implications for amyotrophic lateral sclerosis and frontotemporal dementia pathogenesis. Rare Dis. 2014;2:e29515. doi: 10.4161/rdis.29515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sephton CF, et al. Activity-dependent FUS dysregulation disrupts synaptic homeostasis. Proc Natl Acad Sci USA. 2014;111:E4769–E4778. doi: 10.1073/pnas.1406162111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stoica R, et al. ALS/FTD-associated FUS activates GSK-3β to disrupt the VAPB-PTPIP51 interaction and ER-mitochondria associations. EMBO Rep. 2016;17:1326–1342. doi: 10.15252/embr.201541726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang EJ, et al. Extensive FUS-immunoreactive pathology in juvenile amyotrophic lateral sclerosis with basophilic inclusions. Brain Pathol. 2010;20:1069–1076. doi: 10.1111/j.1750-3639.2010.00413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tradewell ML, et al. Arginine methylation by PRMT1 regulates nuclear-cytoplasmic localization and toxicity of FUS/TLS harbouring ALS-linked mutations. Hum Mol Genet. 2012;21:136–149. doi: 10.1093/hmg/ddr448. [DOI] [PubMed] [Google Scholar]
- 17.Deng J, et al. FUS interacts with HSP60 to promote mitochondrial damage. PLoS Genet. 2015;11:e1005357. doi: 10.1371/journal.pgen.1005357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y, et al. PINK1 and Parkin are genetic modifiers for FUS-induced neurodegeneration. Hum Mol Genet. 2016;25:5059–5068. doi: 10.1093/hmg/ddw310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dillin A, Gottschling DE, Nyström T. The good and the bad of being connected: The integrons of aging. Curr Opin Cell Biol. 2014;26:107–112. doi: 10.1016/j.ceb.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Houtkooper RH, Williams RW, Auwerx J. Metabolic networks of longevity. Cell. 2010;142:9–14. doi: 10.1016/j.cell.2010.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Okuno D, Iino R, Noji H. Rotation and structure of FoF1-ATP synthase. J Biochem. 2011;149:655–664. doi: 10.1093/jb/mvr049. [DOI] [PubMed] [Google Scholar]
- 22.Jonckheere AI, Smeitink JAM, Rodenburg RJT. Mitochondrial ATP synthase: Architecture, function and pathology. J Inherit Metab Dis. 2012;35:211–225. doi: 10.1007/s10545-011-9382-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Houtkooper RH, et al. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pimenta de Castro I, et al. Genetic analysis of mitochondrial protein misfolding in Drosophila melanogaster. Cell Death Differ. 2012;19:1308–1316. doi: 10.1038/cdd.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riar AK, et al. Sex specific activation of the ERα axis of the mitochondrial UPR (UPRmt) in the G93A-SOD1 mouse model of familial ALS. Hum Mol Genet. 2017;26:1318–1327. doi: 10.1093/hmg/ddx049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper JF, et al. Activation of the mitochondrial unfolded protein response promotes longevity and dopamine neuron survival in Parkinson’s disease models. Sci Rep. 2017;7:16441. doi: 10.1038/s41598-017-16637-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mourier A, Ruzzenente B, Brandt T, Kühlbrandt W, Larsson NG. Loss of LRPPRC causes ATP synthase deficiency. Hum Mol Genet. 2014;23:2580–2592. doi: 10.1093/hmg/ddt652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strauss M, Hofhaus G, Schröder RR, Kühlbrandt W. Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J. 2008;27:1154–1160. doi: 10.1038/emboj.2008.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giraud MF, et al. Is there a relationship between the supramolecular organization of the mitochondrial ATP synthase and the formation of cristae? Biochim Biophys Acta. 2002;1555:174–180. doi: 10.1016/s0005-2728(02)00274-8. [DOI] [PubMed] [Google Scholar]
- 30.Chen Y, et al. Expression of human FUS protein in Drosophila leads to progressive neurodegeneration. Protein Cell. 2011;2:477–486. doi: 10.1007/s13238-011-1065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL. Spatiotemporal rescue of memory dysfunction in Drosophila. Science. 2003;302:1765–1768. doi: 10.1126/science.1089035. [DOI] [PubMed] [Google Scholar]
- 32.Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rath E, et al. Induction of dsRNA-activated protein kinase links mitochondrial unfolded protein response to the pathogenesis of intestinal inflammation. Gut. 2012;61:1269–1278. doi: 10.1136/gutjnl-2011-300767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Runkel ED, Liu S, Baumeister R, Schulze E. Surveillance-activated defenses block the ROS-induced mitochondrial unfolded protein response. PLoS Genet. 2013;9:e1003346. doi: 10.1371/journal.pgen.1003346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Samuel BS, Breen PC, Ruvkun G. Caenorhabditis elegans pathways that surveil and defend mitochondria. Nature. 2014;508:406–410. doi: 10.1038/nature13204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fiorese CJ, Haynes CM. Integrating the UPRmt into the mitochondrial maintenance network. Crit Rev Biochem Mol Biol. 2017;52:304–313. doi: 10.1080/10409238.2017.1291577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fiorese CJ, et al. The transcription factor ATF5 mediates a mammalian mitochondrial UPR. Curr Biol. 2016;26:2037–2043. doi: 10.1016/j.cub.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paumard P, et al. The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J. 2002;21:221–230. doi: 10.1093/emboj/21.3.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davies KM, Anselmi C, Wittig I, Faraldo-Gómez JD, Kühlbrandt W. Structure of the yeast F1Fo-ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc Natl Acad Sci USA. 2012;109:13602–13607. doi: 10.1073/pnas.1204593109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H, Oster G. Energy transduction in the F1 motor of ATP synthase. Nature. 1998;396:279–282. doi: 10.1038/24409. [DOI] [PubMed] [Google Scholar]
- 41.So E, et al. Mitochondrial abnormalities and disruption of the neuromuscular junction precede the clinical phenotype and motor neuron loss in hFUSWT transgenic mice. Hum Mol Genet. 2018;27:463–474. doi: 10.1093/hmg/ddx415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jensen MB, Jasper H. Mitochondrial proteostasis in the control of aging and longevity. Cell Metab. 2014;20:214–225. doi: 10.1016/j.cmet.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haynes CM, Ron D. The mitochondrial UPR—Protecting organelle protein homeostasis. J Cell Sci. 2010;123:3849–3855. doi: 10.1242/jcs.075119. [DOI] [PubMed] [Google Scholar]
- 44.Jovaisaite V, Mouchiroud L, Auwerx J. The mitochondrial unfolded protein response, a conserved stress response pathway with implications in health and disease. J Exp Biol. 2014;217:137–143. doi: 10.1242/jeb.090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Papa L, Germain D. SirT3 regulates the mitochondrial unfolded protein response. Mol Cell Biol. 2014;34:699–710, and erratum (2014) 34:1378. doi: 10.1128/MCB.01337-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martinez BA, et al. Dysregulation of the mitochondrial unfolded protein response induces non-apoptotic dopaminergic neurodegeneration in C. elegans models of Parkinson’s disease. J Neurosci. 2017;37:11085–11100. doi: 10.1523/JNEUROSCI.1294-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Frezza C, Cipolat S, Scorrano L. Organelle isolation: Functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat Protoc. 2007;2:287–295. doi: 10.1038/nprot.2006.478. [DOI] [PubMed] [Google Scholar]
- 48.Villa-Cuesta E, Rand DM. Preparation of mitochondrial enriched fractions for metabolic analysis in Drosophila. J Vis Exp. 2015;(103):e53149. doi: 10.3791/53149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.