Significance
Actin polymerization is a divalent cation-dependent process. Here we identify a cation binding site on the surface of actin in a 2.0-Å resolution X-ray structure of actin and find evidence of three additional sites in published high-resolution structures. These cations are stable in molecular dynamics (MD) simulations of the filament, suggesting a functional role in polymerization or filament rigidity. Polymerization activates the ATPase activity of the incorporating actin protomers. Careful analysis of water molecules that approach the ATP in the MD simulations revealed Gln137-activated water to be in a suitable position in F-actin, to initiate attack for ATP hydrolysis, and its occupancy was dependent on bound cations.
Keywords: X-ray crystallography, molecular dynamics, actin, ATP hydrolysis
Abstract
The structure of the actin filament is known at a resolution that has allowed the architecture of protein components to be unambiguously assigned. However, fully understanding the chemistry of the system requires higher resolution to identify the ions and water molecules involved in polymerization and ATP hydrolysis. Here, we find experimental evidence for the association of cations with the surfaces of G-actin in a 2.0-Å resolution X-ray structure of actin bound to a Cordon-Bleu WH2 motif and in previously determined high-resolution X-ray structures. Three of four reoccurring divalent cation sites were stable during molecular dynamics (MD) simulations of the filament, suggesting that these sites may play a functional role in stabilizing the filament. We modeled the water coordination at the ATP-bound Mg2+, which also proved to be stable during the MD simulations. Using this model of the filament with a hydrated ATP-bound Mg2+, we compared the cumulative probability of an activated hydrolytic water molecule approaching the γ-phosphorous of ATP, in comparison with G-actin, in the MD simulations. The cumulative probability increased in F-actin in line with the activation of actin’s ATPase activity on polymerization. However, inclusion of the cations in the filament lowered cumulative probability, suggesting the rate of hydrolysis may be linked to filament flexibility. Together, these data extend the possible roles of Mg2+ in polymerization and the mechanism of polymerization-induced activation of actin’s ATPase activity.
Actin is highly conserved across the entire range of eukaryotes, from plants to protists and from metazoa to fungi (1). Its ability to form dynamic noncovalent double-stranded helical filaments, F-actin (2, 3), confers the protein the ability to participate in critical cellular processes such as cell motility (4) and cell division (5). The F-actin protomer together with the monomeric form, G-actin, adopt U-shaped structures formed from four subdomains that surround the nucleotide, either ATP or ADP, and a divalent cation (6). Subdomains 2 and 4 are referred to as the pointed face, and 1 and 3, the barbed face of the monomer, which can associate with the barbed end or pointed end of a filament, respectively. Although the structure of the filament has been determined by cryoelectron microscopy (cryoEM) at resolutions that allow interpretation of the architecture (7), understanding the chemistry of the filament, particularly the exact roles of nucleotides, water molecules, and cations in the processes of polymerization and depolymerization, is challenging at limited resolution. For example, even the highest resolution cryoEM filament map does not have sufficient detail to determine the H2O coordination of the ADP-bound Mg2+ (7, 8).
Divalent cations are critical determinants of actin polymerization and ATP hydrolysis. The polymerization and stability of the filament are dependent on the ionic composition of the solution, which affects the critical concentration over which polymerization occurs (9–12). Mg2+- or Ca2+-induced differences in biochemical and structural properties of the actin protomer (13, 14) lead to variations in critical concentrations that have been exploited in many in vitro polymerization assays in which a switch from Ca2+ to Mg2+ initiates polymerization (15). Furthermore, the rate of ATP hydrolysis on forming F-actin is linked to the type of divalent cation bound to ATP, with Mg2+ being six times faster than Ca2+ (16). However, understanding the precise mechanisms by which divalent cations affect actin polymerization is hampered by the limited resolution of the F-actin structure. Recently it has been proposed that there are two key “polymerization” and “stiffness” Mg2+ binding sites at the interface between two adjacent protomers (17).
Cations also influence actin filament stability through associating with ATP. Polymerization switches on the ATPase activity of actin. G-actin ATPase activity is ∼7,000-fold slower than for the protomers in F-actin (18). On removal of the γ-phosphate from the Mg-ATP through hydrolysis, the ADP-Pi–bound sections of actin filaments are relatively stable (19). The release of the phosphate results in a destabilization of the filament. The mechanism of hydrolysis is hypothesized to involve Gln137, which is positioned to allow its side chain oxygen to form a hydrogen bond with a nearby water molecule, activating it for nucleophilic attack on ATP’s γ-phosphate (20). Supporting this mechanism, a Q137A mutant indicated that this residue has a role in both the ATPase activity and in the conformational transition between G- and F-actin (21). Molecular dynamics (MD) simulations on the fiber diffraction structure of F-actin solved at 3.3- to 5.6-Å resolution (22), indicate that Gln137 is closer to ATP’s γ-phosphate than in G-actin (23). Quantum mechanics/molecular mechanics (QM/MM) simulations on this model indicate that the leaving H+ is shuttled via water molecule-mediated proton relay to Asp154 and then to the leaving phosphate (24, 25). Nevertheless, the precise interactions between ATP/ADP, the divalent cation, and surrounding water molecules, remain to be empirically proven.
In this study, we provide experimental evidence for the specific association of cations with the surface of G-actin. MD simulations confirm that, once occupied, these sites are stable in the filament based on the 3.6-Å F-actin cryoEM structure (7). MD simulations of the F-actin cryoEM structure indicate that the Mg2+ at the ATP binding site is coordinated by H2O in a similar way to G-actin. Comparative MD simulations of G-actin and F-actin support the mechanism by which the G- to F-actin transition produces a repositioning of Gln137 and a water molecule to activate ATP hydrolysis (23). Together these data provide insight into the roles of Mg2+ in filament stability and ATP hydrolysis.
Results
Crystal Structure of the Actin/CoblNW Complex.
The X-ray structure of a chemically synthesized Cordon-Bleu WH2 motif (CoblNW) bound to Oryctolagus cuniculus (rabbit) actin was determined at a 2.0-Å resolution (Fig. 1 and SI Appendix, Table S1). The ordered region of the WH2-motif peptide comprises the entire 22-residue long peptide (Ser1184/Val1205). In comparison with a previously published lower resolution Cordon-Bleu WH2 motif/actin crystal (2.91 Å, PDB 4JHD, ref. 26), this 2.0-Å structure allows for the unambiguous tracing of the Cordon-Bleu WH2 motif on the surface of actin and reveals a slightly different route in connecting the N-terminal amphipathic alpha helix to the LKKV motif (SI Appendix, Fig. S1 A and B). Reanalysis of the 4JHD X-ray data (26) indicates that the present structure describes the 4JHD X-ray data more accurately (SI Appendix, Fig. S1 D–G) and highlights other regions of the 4JHD structure that are not supported by sufficient electron density (SI Appendix, Fig. S1 H–K).
Fig. 1.
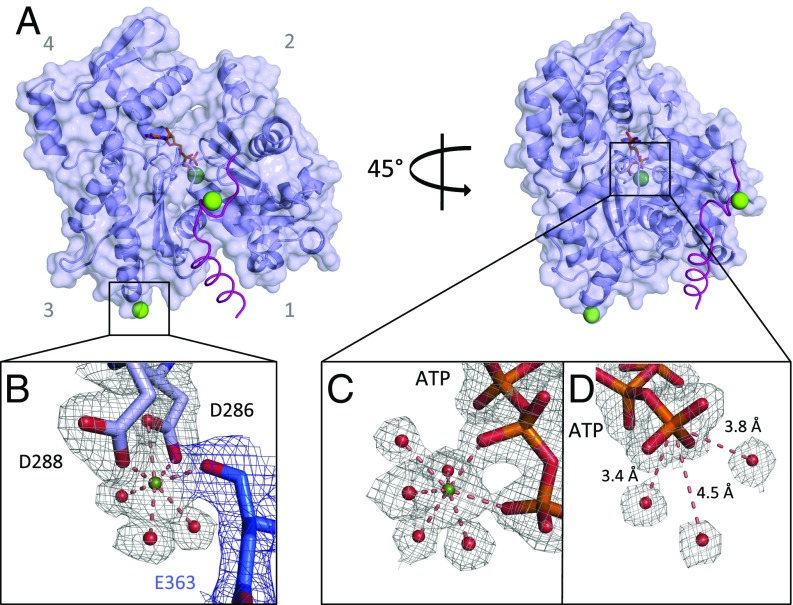
Crystal structure of the actin/CoblNW complex at a 2.0-Å resolution. (A) Actin, light blue cartoon/surface. CoblNW, α-carbons purple trace. Ca2+, green spheres. ATP is shown as orange sticks. (B) Closeup of the D288/D286 Ca2+ site. Residues from the two symmetry-related molecules are shown in light blue and blue, associated with electron density, gray and blue mesh, respectively, contoured at 1 σ. Water molecules are shown as red spheres. (C) The ATP-associated Ca2+ site. (D) Waters close to the ATP γ-phosphate.
As observed in the thymosin β4/actin complex structure (27), the binding of CoblNW narrows the distance between actin subdomains 4 and 2 at the pointed face of the monomer, compared with native actin (PDB 3HBT, ref. 28). This narrowing of the nucleotide binding cleft flattens the structure more substantially than in thymosin β4-actin (27). This flattening of the G-actin (∼5°) is in the same direction, but to a far lesser extent, than observed in F-actin, which undergoes a 20° flattening in the G- to F-actin transition (PDB 5JLF; ref. 7, 22, 29). The CoblNW/actin structure includes an ATP with an associated Ca2+. This Ca2+, chelated by oxygen atoms from the β- and γ-phosphates of ATP and by five water molecules, forms the typical pentagonal bipyramid coordination observed in most high-resolution actin ATP/Ca2+ crystal structures (Fig. 1C). In the CoblNW/actin structure, three water molecules lie within 5 Å of the ATP γ-phosphate (Fig. 1D). These waters are candidates for ATP γ-phosphate nucleophilic attack to initiate hydrolysis.
Two additional Ca2+ were found on the outer surface of the CoblNW/actin complex (Fig. 1A). One bound to Glu1200 of CoblNW and to Asp187 of the symmetry-related actin, and the second Ca2+ is bound to Asp286 and Asp288 of actin and to Glu363 of the symmetry-related actin, again forming a pentagonal bipyramidal coordination (Fig. 1B). This Ca2+ resides at the predicted “polymerization site” (17), providing experimental evidence that the hypothesized binding site is capable of accepting a cation. Thus, the CoblNW/actin structure clarifies the binding mode of this WH2 motif, but importantly presents structural evidence for the cation binding that is involved in actin polymerization.
Published Crystal Structures Reveal G-Actin–Bound Cations.
We next asked whether surface-bound cation binding sites have been observed in previous actin structures. There are more than 100 actin structures that have been solved using X-ray crystallography (73%), EM (25%), or fiber diffraction and electron crystallography (2%) deposited in the Protein Data Bank (www.rcsb.org). We inventoried all of the structures with a resolution of 2.5 Å or better (100% X-ray structures), resolutions at which cation binding can be predicted with some certainty (8), and cataloged the divalent cations bound to the outer surface of actin (SI Appendix, Table S2). Some cation binding sites reoccurred in several structures (Fig. 2). These sites were segregated into two groups: the red-framed cation binding sites refer to polymerization and stiffness sites (17), whereas the green-framed sites are repeatedly found in several crystal structures, but have not been reported to bear any role in polymerization (Fig. 2).
Fig. 2.
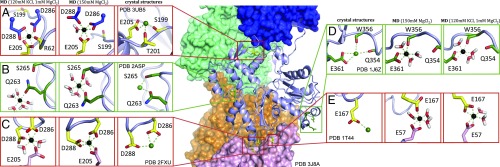
Review of divalent cation binding sites found in the PDB crystal structure library. F-actin is represented in the Middle (PDB 3J8A), in which four actin protomers are illustrated as surfaces (blue, green, orange, and pink), and the central protomer as a light blue cartoon. (A–E “crystal structures”) Enlargements of the cation binding sites from crystal structures are shown as green or yellow sticks, and Mg2+ and Ca2+ as black and green spheres, respectively. Sites that were previously described are boxed by red squares (A, C, and E) and those that were additionally observed in several crystal structures are boxed by green squares (B and D). (A–E “MD”) Mg2+ coordination after 100-ns MD simulation, in respective conditions. A and C are the upper and lower polymerization site, and E is the lower stiffness site (the upper site is identical and omitted for clarity).
MD Simulations to Study Outer Surface Mg2+ Binding Sites.
To better understand the capacity of cations to bind to actin filaments, a 100-ns long MD simulation of F-actin was carried out. The initial F-actin model was built by combining the 3.6-Å F-actin cryoEM structure (PDB 5JLF, ref. 7), with the ATP and bound Mg2+ taken from PDB 2V52 (30). The F-actin model was composed of the protomer to be analyzed surrounded by the other four protomers to complete the entire coordination of a protomer within a filament. This filament model was run in the MD simulation for 100 ns in a water-solvated environment containing either high 150 mM MgCl2, or 1 mM MgCl2 and 120 mM KCl, to mimic physiological cellular conditions (31–33). In these conditions, no Mg2+ was found to be binding in the polymerization or stiffness sites, or any other site identified in the crystal structures. Subsequently, the initial model was modified to contain Mg2+ at each site that was consistently found in crystal structures (Fig. 2 and Dataset S1), as well as earlier predictions and MD simulations (17, 34). In this mode, the Mg2+ binding was stable at each site for the 150 mM MgCl2 simulation. However, some changes in coordination were observed. The polymerization site cation lost interaction with Ser199 and Thr201 (Fig. 2A), and the cation bound to Glu361, Trp356, and Gln354 was no longer bound to Gln354 after 100 ns (Fig. 2D). Interestingly, the polymerization site had slightly different conformations at the pointed and barbed end faces of the central protomer (compare Fig. 2 A and C). At the pointed end, a water molecule was integrated in the Mg2+ chelation sphere, whereas the barbed end integrated the second oxygen of Asp288 in the chelation. In both polymerization sites, the lower protomer contributes Glu205 to the Mg2+ chelation sphere (Fig. 2 A and C). The simulation revealed that the stiffness site Mg2+ is chelated by Glu167 from the upper protomer and Glu57 from the lower protomer (Fig. 2E). Thus, the polymerization site is coordinated by three acid residues, and the stiffness site is coordinated by two acid residues, while the additional sites do not show multiple coordination by acidic residues. For the MD simulation under 1 mM MgCl2 and 120 mM KCl conditions (Datasets S2 and S3), several differences in stability were observed. The polymerization site integrated Arg62 in its coordination at the pointed end, whereas the barbed end released an oxygen from Asp288 to integrate water molecules (Fig. 2 A and C). The stiffness site integrated both Glu57 and Glu167 in the Mg2+ coordination (Fig. 2E). The newly found site Glu361/Tpr356 was retained under these conditions (Fig. 2D), whereas Gln263/Ser265 lost its coordination. An amino acid conservation analysis of actin in eukaryotes shows that the polymerization and stiffness sites, as well as the Glu361/Tpr356 site, are well conserved, even in Asgard archaea, as opposed to Gln363/Ser265, which are poorly conserved (SI Appendix, Fig. S2). Together, these data are consistent with a previously published MD analysis generated from an earlier 6.6-Å resolution cryoEM actin filament model (29) with Mg2+ added to the polymerization and stiffness sites (34).
Nucleotide Mg2+ Coordination.
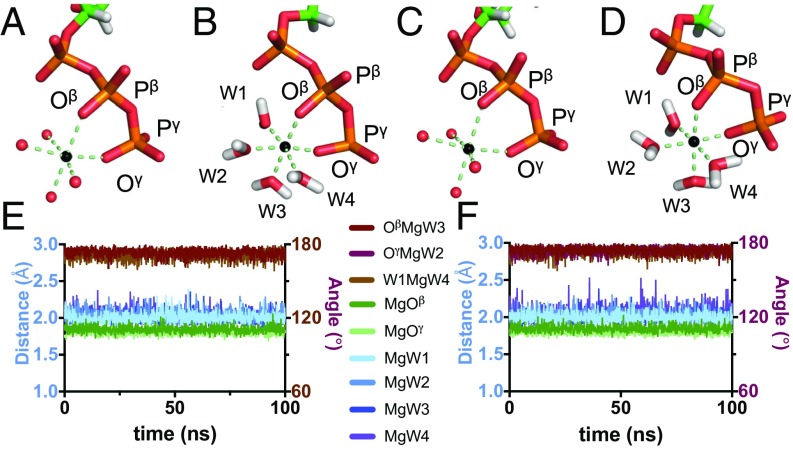
Approximately 70% of the actin X-ray structures have Ca2+ as a nucleotide-bound cation. However, in the cell, filaments are expected to have an Mg2+ bound at the ATP/ADP binding site. We used the MD simulations to assess the stability and the chemical coordination of ATP-bound Mg2+ in G- and F-actin. For G-actin, the initial model was prepared by taking the actin chain from the actin-DNaseI crystal structure (PDB 1ATN, ref. 6) in combination with the ATP and ATP-bound Mg2+ from PDB 2V52 (30). The F-actin model is that presented in the previous paragraph. Both models had Mg2+ added at the surface cation binding sites. These initial models were prepared with the Mg2+ chelated via octahedral coordination to four water molecules and an oxygen atom each from the β- and γ-phosphates of ATP (Oβ and Oγ, Fig. 3 A and C). The distances and angles formed around each Mg2+ were subsequently analyzed over the 100 ns of the simulation, under 1 mM MgCl2 and 120 mM KCl conditions. For both G-actin and F-actin, the octahedral cation coordination remained stable over the simulation. The distances from the Mg2+ to the chelating water molecules were on average 2.11 Å for G-actin and 2.02 Å for F-actin (Fig. 3 E and F, blue lines), and the distances to Oβ and Oγ were 1.85 Å and 1.79 Å for G-actin and 1.84 Å and 1.80 Å for F-actin (Fig. 3 E and F, green lines). The angles OβMgW3, OγMgW2, and W1MgW4 had an average of 173.2° for G-actin and 172.8° for F-actin and were also very stable for the whole simulation (Fig. 3 E and F, brown lines). The conformations after the 100-ns simulation in the presence of 1 mM MgCl2 and 120 mM KCl are presented in Fig. 3 B and D for G- and F-actin, respectively, and are similar in all aspects to the initial models.
Fig. 3.
Symmetry conservation around the ATP-bound Mg2+ in MD. (A and C) Initial MD models of ATP and ATP-bound Mg2+ for F- and G-actin, respectively. (B and D) Final snapshots after a 100-ns long MD simulation of F- and G-actin, respectively. ATP, sticks. Mg2+, black spheres. Water, red spheres. (E and F) Analysis of different angles (brown lines) and distances (blue and green lines) around ATP-bound Mg2+, over time, for F- and G-actin, respectively. Simulations were performed in 120 mM KCl and 1 mM MgCl2.
Scrutiny of Water Molecules Close to the ATP.
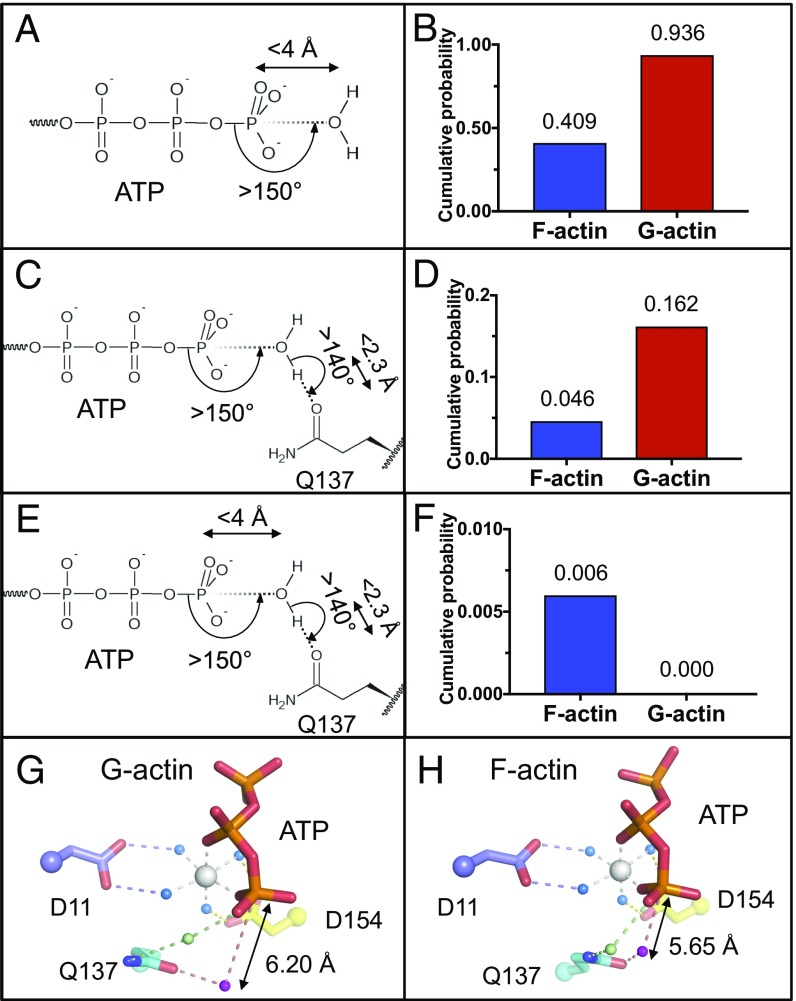
We next assessed the behavior of the H2O close to the ATP γ-phosphate in the MD simulations to better understand actin’s ATPase activity (35, 36). We used the G- and F-actin MD models using positional restraints for the protein atoms to probe the role of H2O in hydrolysis. To assess the conformations of G- and F-actin during the MD simulations, the distance between residues 364 (subdomain 1) and 37 (subdomain 2) was monitored. This distance changes between the two conformations of actin during the flattening in the G-to-F transition. Both G- and F-actin simulations show stability in their conformations after 50-ns equilibration (SI Appendix, Fig. S3 A and B). Thus, the proteins were allowed to equilibrate for 50 ns with restraints. Then analyses were performed on a further 50 ns of MD simulation using the following protocol. First, the number of water molecules that lie within a spherical cone emanating from the Pγ formed by an angle larger than 150°, and also have a distance from the water oxygen to the Pγ of less than 4 Å, was counted to ensure the availability of H2O in positions that may approach the Pγ on a sterically unhindered path (37, 38) (Fig. 4A). The cumulative probability, defined as the fraction of time a water molecule is present in this cone (38), was calculated for G- and F-actin. Under these criteria, water molecules are more often present in the G-actin spherical cone (cumulative probability 0.94) than in F-actin (0.41) (Fig. 4B). To become hydrolytic and start nucleophilic attack toward ATP’s γ-phosphate group, the water needs to form a hydrogen bond with an activating residue, and Gln137 has been reported to bear that role (21, 35, 39). Next, we assessed the numbers of water molecules that form a hydrogen bond with Gln137’s side chain oxygen, with an angle larger than 140° and a distance lower than 2.3 Å (40), and lie within an angle greater than 150° toward Pγ to be in a position enabling nucleophilic attack (Fig. 4C). Under these criteria, waters in the G-actin simulation (0.162) are more numerous than those in F-actin simulation (0.046) (Fig. 4D). The average distances of these water molecules are 6.20 Å and 5.65 Å to the Pγ for G- and F-actin, respectively (Fig. 4 G and H). Finally, a cutoff distance of 4 Å between the Pγ and the activated water molecule was added to the criteria (38), combining the first two analyses (Fig. 4E). No water molecule from the G-actin simulation satisfied these conditions, whereas a cumulative probability of 0.006 was observed for F-actin simulation (Fig. 4F). Thus, 13.0% of the water molecules forming a hydrogen bond with Gln137 in the F-actin simulation are within 4 Å of ATP’s γ-phosphorus and have an appropriate angle for nucleophilic attack, in line with the predictions from the fiber diffraction structure-based MD simulations (23). These analyses were also performed with the model with and without incorporation of the surface Mg2+ found in crystal structures in 150 mM MgCl2 (SI Appendix, Fig. S4). Similar trends were observed, a higher final cumulative probability of 0.096 (27.1% of Gln137 oxygen-bound water molecules) and 0.030 (11.9%) without and with surface Mg2+, whereas no water molecules satisfied all of the conditions for G-actin (SI Appendix, Fig. S4 H and I).
Fig. 4.
MD analysis of H2O close to the γ-phosphate and Gln137. (A, C, and E) Schematic representation of analysis conditions applied around ATP and Gln137. (B, D, and F) Cumulative probability of H2O with applied conditions from A, C, and E. (G) G-actin model, resulting from the combination of actin-DNaseI structure (PDB 1ATN) and ATP and ATP-bound Mg2+ of PDB 2V52. (H) Typical snapshot of MD simulation frame, satisfying all conditions from E. ATP and actin, sticks. Waters coordinated by Mg2+ are shown as blue, green, and purple spheres.
To better understand how the structural differences between G- and F-actin affect the presence of water at the hydrolytic position, we compared the transition by morphing between the two conformations. The morph highlights that in forming the “closed” conformation of F-actin, Gln137 rotates closer to the γ-phosphate group. Indeed, Gln137, located in the cleft between actin subdomains 1 and 3, pivots toward ATP, as a result of a rotation of subdomains 1 and 2 around the nucleotide cleft, which is tightly constrained within subdomains 3 and 4 (Movies S1 and S2). Further analyses were performed on the residues lining the ATP cleft that point toward the γ-phosphorus. Strikingly, residues 11, 13, and 137 were found to be on average closer to the Pγ, whereas residues 74, 154, 159, and 161 were farther away, thus changing the cleft to allow hydrolysis as observed in the fiber diffraction structure and MD simulations (22, 24) (SI Appendix, Fig. S3C and Table S3). Moreover, Gln137 is closer to the γ-phosphorus in the F-actin simulation by 1.39 Å for the Cα and 0.97 Å for the oxygen (OE) that is responsible for the hydrogen bond to the hydrolytic water (SI Appendix, Fig. S3C and Table S3). The distance to the second H2O coordinated by Gln137 (via the nitrogen) remained constant. These and previous simulations (24) predict that the approach of the Gln137 oxygen-bound hydrolytic water molecule toward ATP’s γ-phosphorus, mediated by the Gln137 nitrogen-bound water molecule, to be a structural consequence of the G- to F-actin transition that triggers ATP hydrolysis (Fig. 4 G and H and Movie S3). His161 has recently been reported to be a secondary activator of the Gln137-bound water for ATP hydrolysis (41). In these MD simulations, we found no correlation between a water molecule being in a position to initiate hydrolysis and the position of His161.
Discussion
The identification of a cation bound to the surface of actin in the crystal structure of CoblNW/actin structure prompted us to reevaluate actin X-ray structures for further experimental evidence of divalent cation-binding sites (Fig. 2 and SI Appendix, Table S2). Two of these sites were repeatedly observed that had been previously described as stiffness and polymerization sites, which have a bearing on the dynamics of the actin filament (Fig. 2 A, C, and E) (17). These two sites were stable in the MD simulations. Two additional recurring binding sites were identified, one of which was also stable in the MD simulations, thus possibly having a role to play in modulating the dynamics of actin (Fig. 2 B and D). Despite the stability of the three cation-binding sites, F-actin constructed without the cations in the initial model did not accept Mg2+ from solution in the MD simulations. This suggests that these cation-binding sites may be incorporated in the polymerization process and/or in the bending of longer filaments than those used in the MD simulations. Alignment of diverse eukaryotic actin protein sequences (SI Appendix, Fig. S2B) showed that the polymerization and stiffness sites are highly conserved (SI Appendix, Fig. S2A). Interestingly, residues that are part of a cation chelation in crystal structures, but not after MD simulations—Ser199, Thr201, and Gln354—showed lower conservation. Residues of the newly found MD unstable site Ser265 and Gln263 (Fig. 2B), even though found in several different crystal structures (SI Appendix, Table S2), are poorly conserved (SI Appendix, Fig. S2A), whereas residues in the new MD stable site Trp356 and Glu361 are highly conserved across actin sequences.
The 3.6-Å resolution cryoEM F-actin structure (7) allowed for the modeling of the Mg2+ bound waters at the ATP in the F-actin state. The octahedral coordination of the Mg2+ remained stable during the MD simulations for G- and F-actin (100 ns, Fig. 3 E and F), which had not been observed in earlier MD simulations (23). This strongly suggests that this Mg2+ retains its water coordination during the G- to F-actin structural transition. Thus, the two halves of actin swivel around the Mg2+ via water molecules bound to Asp11 (N-terminal half) and Asp154 (C-terminal half) while Mg2+ coordinates the ATP (Fig. 4 G and H and Movie S3). Since the precise geometry of the residues around ATP’s γ-phosphate is coordinated by the bound cation, differences in cation will affect hydrolysis, as observed for Ca2+ (coordination sphere 7), which is six times slower than Mg2+ (coordination sphere 6) (16). Incorporation of actin subunits into a filament activates the ATP hydrolysis, and Gln137 has been widely reported to bear a key role in this process (20–22, 24, 42–45). The MD simulations presented here support the mechanism that a Gln137-bound water molecule (pink in Fig. 4 G and H) becomes positioned for nucleophilic attack on ATP’s γ-phosphate group, agreeing with the earlier analysis of MD simulations on the fiber diffraction structure of F-actin (22, 23). Gln137 moves into position while retaining association with the ATP’s γ-phosphate group via hydrogen bonds through a second water molecule (light green in Fig. 4 G and H). Other residues, Asp11, Asp154, and His161, have been proposed to have a role in the hydrolysis activity (20, 21, 46). We observed no change in the distances between Asp11 or Asp154 side chains with the water molecules interacting with ATP-bound Mg2+ (Movie S3). Thus, these residues appear to hold the Mg2+-ATP in place while allowing for the G- to F-actin conformational change, which results in a slight distortion of the ATP cleft in the MD simulations (SI Appendix, Fig. S3C and Table S3). We performed further analyses on the openness of the ATP cleft (distance between subdomains 1 and 3), and the distance between subdomains 2 and 4, but found no significant correlation between these and the presence of a water molecule in an ATP-hydrolysis position. Thus, the positioning of the water for nucleophilic attack on ATP is dominated by the pivoting of subdomains 1 and 2 (Movie S2). During the G- to F-actin transition, Gln137 (cyan) slightly rotates toward ATP (Movie S1). This change in conformation results in an increase in the probability of water being located in the vicinity of the γ-phosphate of ATP. Thus, the MD simulations based on the latest cryoEM filament structure reinforce the mechanism determined from the fiber diffraction-based MD simulations (24). An aspect of the present study was that the inclusion of surface Mg2+ in MD simulation led to a lower probability of water being located in the vicinity of the γ-phosphate of ATP. This indicates that the hydrolytic water position occupancy and thus the rate of hydrolysis may be linked to flexibility in the filament structure. These data provide insight into the roles of water molecules and Mg2+ in filament stability and in activating the ATPase activity during the G-to-F transition.
Materials and Methods
Actin Preparation.
The O. cuniculus (rabbit) alpha skeletal muscle actin (UniProt P68135) was purified as previously described (47). The protein was further purified by size exclusion chromatography on a HiLoad 26/60 Superdex 200 (GE Healthcare) in 2.0 mM Tris⋅HCl, 0.2 mM ATP, 0.2 mM CaCl2, and 0.2 mM DDT, pH 7.2 (Buffer A). The highly pure G-actin was verified by SDS/PAGE and its concentration assessed by measuring OD at 290 nm.
CoblNW Preparation.
The peptide CoblNW was designed from Mus musculus (mouse) Cordon-Bleu protein (residues Ser1184/Thr1205, UniProt Q5NBX1). The sequence of the WH2 motif, SLHSALMEAIHSSGGREKLRKT, was modified to obtain the peptide CoblNW of sequence SLHSALNleEAIHSSGGREKLRKV. The peptide was synthesized manually, using solid phase and Fmoc chemistry at the 0.1-mmol scale using H-Ramage-ChemMatrix resin (0.53 mmol⋅g−1). l-amino acids were obtained from Advanced Chemtech. Fmoc-threonine, serine, glutamic acid, and tyrosine were t-butyl protected. All other solvents and reagents were obtained from Sigma-Aldrich. The 1,2-dichloroethane (DCE) was dried overnight over molecular sieves and purged with argon for 30 min before use. The dry resin was swollen with N-methyl-2-pyrrolidone (NMP). The Fmoc-protecting group was removed with 20% piperidine in dimethylformamide (DMF) (15 min). The Fmoc-protected amino acids [5 equivalent (equiv.)] were coupled using O-(7-azabenzotriazol-1-yl)-1,1,3,3-tetramethyluronium hyxafluorophosphate (HATU) as the activating agent (4.9 equiv.) and N,N-diisopropylethylamine (DIPEA) as the base (5 equiv.) in NMP (0.23 M). The coupling time was 60 min for all amino acids except for (S)-N-Fmoc-2-(4′-pentenyl)alanine and (R)-N-Fmoc-2-(7′-octenyl)alanine, which were preactivated and coupled (4 equiv.) for 2 h. The amino acids immediately following the α,α-disubstituted amino acids were double coupled. Following deprotection of the final Fmoc group, the peptides were acetylated using a mixture of acetic anhydride:DIPEA:DMF (2:2:1) for 1 h. After each coupling, deprotection and acetylation reaction, the resin was thoroughly washed with NMP. Cleavage of the peptide from the resin was achieved using a trifluoroacetic acid (TFA) mixture consisting of TFA:triisopropylsilane:water (95:2.5:2.5, 8 mL) for 2 h, followed by filtration and precipitation with diethyl ether. The precipitate was collected by centrifugation and dried. The peptide was purified by reversed-phase chromatography on a GX-271 (Gilson), using Phenomenex Jupiter C12, 4 μm Proteo 90A LC column, 250 × 10 mm (flow rate 5 mL⋅min−1), with a gradient of acetonitrile/0.1% TFA. Peptide molecular weight was confirmed by LCMS, and purity was assessed by HPLC analysis on an Agilent 1260, using Phenomenex Jupiter C12, μm Proteo 90A LC column, 150 × 4.6 mm (flow rate 1 mL⋅min−1).
Crystallization and Structure Solution.
The complex was prepared by incubating CoblNW and rabbit actin (at a 1.5:1 ratio) on ice in buffer A for 1 h and concentrated (10 kDa cutoff Sartorius Vivaspin) to 9.7 mg/mL. No additional agents were added to keep actin monomeric besides CoblNW. Crystals were obtained at 25 °C in 20 mM Mes pH 4.9, 0.2 mM CaCl2 dihydrate, 20% (wt/vol) PEG-3350 using the hanging-drop method in 1.0-μL drops containing a 2:1 (vol/vol) mixture of protein and precipitant. Crystals were frozen in liquid nitrogen, before X-ray diffraction data collection on beamline BL13B1 on an ADSC Quantum-315 CDD detector at the National Synchrotron Radiation Research Center, Taiwan (NSRRC). HKL2000 (48) was used to index, scale, and merge diffraction data. The rabbit actin/CoblNW structure was solved by molecular replacement using the Drosophila melanogaster actin bound to Cordon-Bleu protein complex structure as a model (PDB 4JHD) in MOLREP (49). Clear electron density for the peptide was observed into which the peptide was built (SI Appendix, Fig. S1A, OMIT map). The structure was then refined several times using PHENIX (50), REFMAC (51), and manual building in Coot (52). Quality of the structure was assessed using MolProbity (53). Refinement statistics are found in SI Appendix, Table S1. Structure figures were generated using PyMOL Molecular Graphics System, version 1.8 (Schrödinger, LLC).
Molecular Dynamics Simulations.
MD simulations were performed with the GROMACS 5.1.2 package (54), using the CHARMM36 force field (55–57) and the TIP3P water model. The 4-methyl histidine (Hic73) was treated as a histidine, since it is not methylated in all organisms (58) and not of bonding distance of ATP-hydrolyzing water (59). The models were placed in a dodecahedron box, subsequently filled with 3-point model water molecules (60). A 10.0-Å distance was imposed between the outer surface of the proteins and the edges of the box. Mg2+ and K+ were added to the desired concentrations (1 mM Mg2+ and 120 mM K+ or 150 mM Mg2+), and Cl− was added to reach neutrality. Energy minimization was conducted using the steepest descent minimization for 1,000 steps, and potential monitored at Ep < 1000.0 kJ⋅mol−1⋅nm−1. Next equilibration of the system was performed in two stages: 100 ps canonical ensemble [constant number of particles, volume, and temperature (NVT)] and 100 ps isothermal-isobaric ensemble [constant number of particles, pressure, and temperature (NPT)] simulation with harmonic restraints (force constant: 1 kJ⋅mol−1⋅K−1) applied to the backbone atoms of the proteins. The resulting thermodynamically equilibrated systems were used for further MD simulations of 100 ns at a temperature of 310 K and a pressure of 1 atm. During all simulations, a 2-fs integration time step was used. The temperature was maintained at 310 K with the V-rescale algorithm and the pressure was coupled at 1 bar by isotropic pressure coupling utilizing the Parrinello-Rahman algorithm (time constant 10 ps, isothermal compressibility of water: 4.5e-5 bar−1). Long-range electrostatics interactions were calculated by fourth order particle mesh Ewald (PME) algorithm with a grid spacing of 0.16 nm. All bonds were constrained using a fourth order P-LINCS algorithm. Electrostatic and van der Waals interactions were cut off at 1 nm. Dispersion corrections for the energy and pressure were applied to account for long-range van der Waals interactions. Periodic boundary conditions were implemented in three dimensions. Initial atomic velocities, before NVT equilibration, were obtained from Maxwell’s distribution at 310 K. Monitoring of potential, pressure, temperature, and density, as well as trajectory analysis, was performed using GROMACS routines, and all other analyses were performed using object-oriented python toolkit MDAnalysis (61), Pandas, (62) and NumPy (63).
Supplementary Material
Acknowledgments
We thank A*STAR for support and are grateful for the technical services provided by the Synchrotron Radiation Protein Crystallography of the National Core Facility Program for Biotechnology, Ministry of Science and Technology and the National Synchrotron Radiation Research Center, a national user facility supported by the Ministry of Science and Technology of Taiwan, Republic of China.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 5YPU).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1806394115/-/DCSupplemental.
References
- 1.Gunning PW, Ghoshdastider U, Whitaker S, Popp D, Robinson RC. The evolution of compositionally and functionally distinct actin filaments. J Cell Sci. 2015;128:2009–2019. doi: 10.1242/jcs.165563. [DOI] [PubMed] [Google Scholar]
- 2.Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 3.Pollard TD. What we know and do not know about actin. Handb Exp Pharmacol. 2017;235:331–347. doi: 10.1007/164_2016_44. [DOI] [PubMed] [Google Scholar]
- 4.Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J. Actin dynamics, architecture, and mechanics in cell motility. Physiol Rev. 2014;94:235–263. doi: 10.1152/physrev.00018.2013. [DOI] [PubMed] [Google Scholar]
- 5.Pollard TD, Wu JQ. Understanding cytokinesis: Lessons from fission yeast. Nat Rev Mol Cell Biol. 2010;11:149–155. doi: 10.1038/nrm2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Atomic structure of the actin:DNase I complex. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- 7.von der Ecken J, Heissler SM, Pathan-Chhatbar S, Manstein DJ, Raunser S. Cryo-EM structure of a human cytoplasmic actomyosin complex at near-atomic resolution. Nature. 2016;534:724–728. doi: 10.1038/nature18295. [DOI] [PubMed] [Google Scholar]
- 8.Popp D, et al. Advances in structural biology and the application to biological filament systems. BioEssays. 2018;40:e1700213. doi: 10.1002/bies.201700213. [DOI] [PubMed] [Google Scholar]
- 9.Martonosii A, Molino CM, Gergely J. The binding of divalent cations to actin. J Biol Chem. 1964;239:1057–1064. [PubMed] [Google Scholar]
- 10.Strzelecka-Gołaszewska H, Pròchniewicz E, Drabikowski W. Interaction of actin with divalent cations. 1. The effect of various cations on the physical state of actin. Eur J Biochem. 1978;88:219–227. doi: 10.1111/j.1432-1033.1978.tb12441.x. [DOI] [PubMed] [Google Scholar]
- 11.Estes JE, Selden LA, Kinosian HJ, Gershman LC. Tightly-bound divalent cation of actin. J Muscle Res Cell Motil. 1992;13:272–284. doi: 10.1007/BF01766455. [DOI] [PubMed] [Google Scholar]
- 12.Strzelecka-Golaszewska H, Wozniak A, Hult T, Lindberg U. Effects of the type of divalent cation, Ca2+ or Mg2+, bound at the high-affinity site and of the ionic composition of the solution on the structure of F-actin. Biochem J. 1996;316:713–721. doi: 10.1042/bj3160713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rich SA, Estes JE. Detection of conformational changes in actin by proteolytic digestion: Evidence for a new monomeric species. J Mol Biol. 1976;104:777–792. doi: 10.1016/0022-2836(76)90181-9. [DOI] [PubMed] [Google Scholar]
- 14.Frieden C. Polymerization of actin: Mechanism of the Mg2+-induced process at pH 8 and 20 degrees C. Proc Natl Acad Sci USA. 1983;80:6513–6517. doi: 10.1073/pnas.80.21.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooper JA, Buhle EL, Jr, Walker SB, Tsong TY, Pollard TD. Kinetic evidence for a monomer activation step in actin polymerization. Biochem. 1983;22:2193–2202. doi: 10.1021/bi00278a021. [DOI] [PubMed] [Google Scholar]
- 16.Blanchoin L, Pollard TD. Hydrolysis of ATP by polymerized actin depends on the bound divalent cation but not profilin. Biochemistry. 2002;41:597–602. doi: 10.1021/bi011214b. [DOI] [PubMed] [Google Scholar]
- 17.Kang H, et al. Identification of cation-binding sites on actin that drive polymerization and modulate bending stiffness. Proc Natl Acad Sci USA. 2012;109:16923–16927. doi: 10.1073/pnas.1211078109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pollard TD, Weeds AG. The rate constant for ATP hydrolysis by polymerized actin. FEBS Lett. 1984;170:94–98. doi: 10.1016/0014-5793(84)81376-9. [DOI] [PubMed] [Google Scholar]
- 19.Korn ED, Carlier MF, Pantaloni D. Actin polymerization and ATP hydrolysis. Science. 1987;238:638–644. doi: 10.1126/science.3672117. [DOI] [PubMed] [Google Scholar]
- 20.Vorobiev S, et al. The structure of nonvertebrate actin: Implications for the ATP hydrolytic mechanism. Proc Natl Acad Sci USA. 2003;100:5760–5765. doi: 10.1073/pnas.0832273100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwasa M, Maeda K, Narita A, Maéda Y, Oda T. Dual roles of Gln137 of actin revealed by recombinant human cardiac muscle alpha-actin mutants. J Biol Chem. 2008;283:21045–21053. doi: 10.1074/jbc.M800570200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oda T, Iwasa M, Aihara T, Maéda Y, Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457:441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- 23.Saunders MG, Voth GA. Water molecules in the nucleotide binding cleft of actin: Effects on subunit conformation and implications for ATP hydrolysis. J Mol Biol. 2011;413:279–291. doi: 10.1016/j.jmb.2011.07.068. [DOI] [PubMed] [Google Scholar]
- 24.McCullagh M, Saunders MG, Voth GA. Unraveling the mystery of ATP hydrolysis in actin filaments. J Am Chem Soc. 2014;136:13053–13058. doi: 10.1021/ja507169f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun R, Sode O, Dama JF, Voth GA. Simulating protein mediated hydrolysis of ATP and other nucleoside triphosphates by combining QM/MM molecular dynamics with advances in metadynamics. J Chem Theory Comput. 2017;13:2332–2341. doi: 10.1021/acs.jctc.7b00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X, et al. Structural basis of actin filament nucleation by tandem W domains. Cell Reports. 2013;3:1910–1920. doi: 10.1016/j.celrep.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue B, Leyrat C, Grimes JM, Robinson RC. Structural basis of thymosin-β4/profilin exchange leading to actin filament polymerization. Proc Natl Acad Sci USA. 2014;111:E4596–E4605. doi: 10.1073/pnas.1412271111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H, Robinson RC, Burtnick LD. The structure of native G-actin. Cytoskeleton (Hoboken) 2010;67:456–465. doi: 10.1002/cm.20458. [DOI] [PubMed] [Google Scholar]
- 29.Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724–728. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- 30.Mouilleron S, Guettler S, Langer CA, Treisman R, McDonald NQ. Molecular basis for G-actin binding to RPEL motifs from the serum response factor coactivator MAL. EMBO J. 2008;27:3198–3208. doi: 10.1038/emboj.2008.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valberg LS, Holt JM, Paulson E, Szivek J. Spectrochemical analysis of sodium, potassium, calcium, magnesium, copper, and zinc in normal human erythrocytes. J Clin Invest. 1965;44:379–389. doi: 10.1172/JCI105151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kasner SE, Ganz MB. Regulation of intracellular potassium in mesangial cells: A fluorescence analysis using the dye, PBFI. Am J Physiol. 1992;262:F462–F467. doi: 10.1152/ajprenal.1992.262.3.F462. [DOI] [PubMed] [Google Scholar]
- 33.Wald T, et al. Quantification of potassium levels in cells treated with Bordetella adenylate cyclase toxin. Anal Biochem. 2014;450:57–62. doi: 10.1016/j.ab.2013.10.039. [DOI] [PubMed] [Google Scholar]
- 34.Hocky GM, et al. Cations stiffen actin filaments by adhering a key structural element to adjacent subunits. J Phys Chem B. 2016;120:4558–4567. doi: 10.1021/acs.jpcb.6b02741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reisler E, Egelman EH. Actin structure and function: What we still do not understand. J Biol Chem. 2007;282:36133–36137. doi: 10.1074/jbc.R700030200. [DOI] [PubMed] [Google Scholar]
- 36.Dominguez R, Holmes KC. Actin structure and function. Annu Rev Biophys. 2011;40:169–186. doi: 10.1146/annurev-biophys-042910-155359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tolman CA. Phosphorus ligand exchange equilibriums on zerovalent nickel. Dominant role for steric effects. J Am Chem Soc. 1970;92:2956–2965. [Google Scholar]
- 38.Afanasyeva A, et al. Lytic water dynamics reveal evolutionarily conserved mechanisms of ATP hydrolysis by TIP49 AAA+ ATPases. Structure. 2014;22:549–559. doi: 10.1016/j.str.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boczkowska M, Rebowski G, Kremneva E, Lappalainen P, Dominguez R. How Leiomodin and Tropomodulin use a common fold for different actin assembly functions. Nat Commun. 2015;6:8314. doi: 10.1038/ncomms9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Legon AC, Millen DJ. Angular geometries and other properties of hydrogen-bonded dimers: A simple electrostatic interpretation of the success of the electron-pair model. Chem Soc Rev. 1987;16:467–498. [Google Scholar]
- 41.Merino F, et al. Structural transitions of F-actin upon ATP hydrolysis at near-atomic resolution revealed by cryo-EM. Nat Struct Mol Biol. 2018;25:528–537. doi: 10.1038/s41594-018-0074-0. [DOI] [PubMed] [Google Scholar]
- 42.Matsuura Y, et al. Structural basis for the higher Ca(2+)-activation of the regulated actin-activated myosin ATPase observed with Dictyostelium/Tetrahymena actin chimeras. J Mol Biol. 2000;296:579–595. doi: 10.1006/jmbi.1999.3467. [DOI] [PubMed] [Google Scholar]
- 43.Pfaendtner J, Lyman E, Pollard TD, Voth GA. Structure and dynamics of the actin filament. J Mol Biol. 2010;396:252–263. doi: 10.1016/j.jmb.2009.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Splettstoesser T, Holmes KC, Noé F, Smith JC. Structural modeling and molecular dynamics simulation of the actin filament. Proteins. 2011;79:2033–2043. doi: 10.1002/prot.23017. [DOI] [PubMed] [Google Scholar]
- 45.Freedman H, Laino T, Curioni A. Reaction dynamics of ATP hydrolysis in actin determined by ab initio molecular dynamics simulations. J Chem Theory Comput. 2012;8:3373–3383. doi: 10.1021/ct3003282. [DOI] [PubMed] [Google Scholar]
- 46.Schüler H. ATPase activity and conformational changes in the regulation of actin. Biochim Biophys Acta. 2001;1549:137–147. doi: 10.1016/s0167-4838(01)00255-2. [DOI] [PubMed] [Google Scholar]
- 47.Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 48.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 49.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 50.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 52.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 53.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abraham MJ, et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- 55.MacKerell AD, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 56.Mackerell AD, Jr, Feig M, Brooks CL., 3rd Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 57.Best RB, et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ(1) and χ(2) dihedral angles. J Chem Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yao X, Grade S, Wriggers W, Rubenstein PA. His(73), often methylated, is an important structural determinant for actin. A mutagenic analysis of HIS(73) of yeast actin. J Biol Chem. 1999;274:37443–37449. doi: 10.1074/jbc.274.52.37443. [DOI] [PubMed] [Google Scholar]
- 59.Nyman T, et al. The role of MeH73 in actin polymerization and ATP hydrolysis. J Mol Biol. 2002;317:577–589. doi: 10.1006/jmbi.2002.5436. [DOI] [PubMed] [Google Scholar]
- 60.Bondi A. van der Waals volumes and radii. J Phys Chem. 1964;68:441–451. [Google Scholar]
- 61.Michaud-Agrawal N, Denning EJ, Woolf TB, Beckstein O. MDAnalysis: A toolkit for the analysis of molecular dynamics simulations. J Comput Chem. 2011;32:2319–2327. doi: 10.1002/jcc.21787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McKinney W. 2011. pandas: A Foundational Python Library for Data Analysis and Statistics. Python High Perform Sci Comput 1–9.
- 63.Oliphant TE. Guide to NumPy. Methods. 2010;1:378. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.