Significance
Spontaneous overexpression of endogenous IRIS, an alternatively spliced product of the tumor suppressor gene BRCA1, allows it to function as an oncoprotein that stimulates a potentially lethal outcome, i.e. metastasis of human cancer cells to tissues served, in part, by the arterial circulation. It does so by suppressing phosphatase and tensin homolog (PTEN) mRNA synthesis, thereby stabilizing and activating HIF-1α in normoxic cells. Thus, this study provides a strong rationale for exploring the therapeutic value of interfering with spontaneously overexpressed IRIS function in multiple types of tumors that can naturally overexpress it.
Keywords: BRCA1-IRIS, metastasis, HIF-1α, PTEN
Abstract
BRCA1 is an established breast and ovarian tumor suppressor gene that encodes multiple protein products whose individual contributions to human cancer suppression are poorly understood. BRCA1-IRIS (also known as “IRIS”), an alternatively spliced BRCA1 product and a chromatin-bound replication and transcription regulator, is overexpressed in various primary human cancers, including breast cancer, lung cancer, acute myeloid leukemia, and certain other carcinomas. Its naturally occurring overexpression can promote the metastasis of patient-derived xenograft (PDX) cells and other human cancer cells in mouse models. The IRIS-driven metastatic mechanism results from IRIS-dependent suppression of phosphatase and tensin homolog (PTEN) transcription, which in turn perturbs the PI3K/AKT/GSK-3β pathway leading to prolyl hydroxylase-independent HIF-1α stabilization and activation in a normoxic environment. Thus, despite the tumor-suppressing genetic origin of IRIS, its properties more closely resemble those of an oncoprotein that, when spontaneously overexpressed, can, paradoxically, drive human tumor progression.
Germ-line mutations of the breast cancer susceptibility gene BRCA1 predispose women to early-onset breast and/or ovarian cancers (1). An understanding of the BRCA1 function(s) that suppress cancer development remains limited, and how defective BRCA1 function contributes to tumorigenesis and metastasis is a mystery.
IRIS is an alternatively spliced, nuclear polypeptide product of the BRCA1 gene (2). It controls cell proliferation, at least in part, by binding to and modulating the replication initiation-regulating protein Geminin (2). It also acts as a transcriptional coactivator by associating with selected promoter elements and thereby influencing the transcription of certain genes (3). Others have detected its overexpression in cancer cells and associated it with certain transformed properties, such as an epithelial-to-mesenchymal transition (EMT) and chemotherapy resistance in animal cancer models (4, 5). However, a detailed biochemical role for IRIS in sustaining human cancers has not yet been established, and knowledge of the mechanisms by which it drives certain canonical tumor-associated properties remains limited.
Spontaneous overexpression of endogenous IRIS can now be detected in broad collections of sporadic human cancers. Moreover, we have found that spontaneously overexpressed endogenous IRIS promotes the metastasis of primary and cell line-based human breast cancer cells in mouse models and does so, at least in part, through the arterial circulation.
IRIS is also a transcription cofactor that, we find, operates under normoxic conditions by suppressing phosphatase and tensin homolog (PTEN) mRNA synthesis. This, in turn, activates PI3K signaling and results in AKT activation (6–10). The latter triggers the inhibition of GSK-3β (11), which otherwise catalyzes HIF-1α phosphorylation at distinct sites within its transactivation domain (12, 13). Blockade of HIF-1α phosphorylation stabilizes and activates HIF-1α, even in a normoxic environment, allowing it to manifest its known metastasis-promoting function (14–16).
Thus, IRIS is a product of a classical tumor suppressor gene that, when overexpressed, paradoxically stimulates tumor progression. It does so by perturbing established elements of tumor-suppression signaling at the center of which is its target, the prominent human tumor suppressor PTEN.
Results
Expression of IRIS in Sporadic Human Cancer.
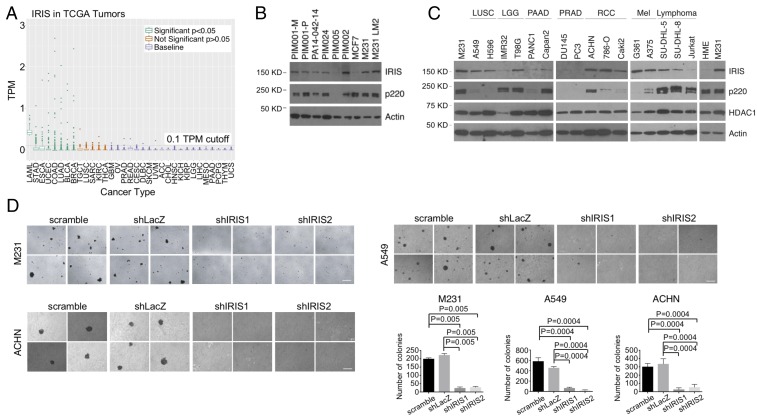
When ectopically overexpressed, IRIS stimulates cellular proliferation. It is also spontaneously overexpressed in certain breast cancer cell lines (2). Hence, we extended IRIS expression analysis to cells from a variety of human tumors, using RNA-sequencing (RNA-Seq) data available in The Cancer Genome Atlas (TCGA) (17).
An alignment-free approach, Kallisto (18), was used to estimate IRIS-specific RNA abundance from FASTQ files in this endeavor. In these datasets, despite the low estimated overall abundance, relatively high levels of IRIS mRNA were detected in breast, stomach, endometrial, bladder, colon, esophageal, and lung carcinoma and in acute myeloid leukemia (Fig. 1A and Datasets S1 and S2). BRCA1-p220 (also known as “p220”) messenger levels were also determined in all tumor cases in TCGA (SI Appendix, Fig. S1A and Dataset S1). Of note, IRIS mRNA expression did not correlate with p220 mRNA abundance in most cancer types (SI Appendix, Fig. S1A and Dataset S3), indicating that expressions of these two isoforms are regulated differently.
Fig. 1.
Expression of IRIS in sporadic human cancer. (A) Dot plot showing IRIS RNA-Seq data in various sporadic tumors in the TCGA data collection. TPM, transcripts per million. Refer to Dataset S2 for a description of abbreviations of each cancer type and P values. (B) Western blot showing IRIS protein levels in samples from multiple human breast cancer PDX models. (C) Western blot showing IRIS protein levels in various human cancer cell lines. LGG, lower grade glioma; LUSC, lung squamous cell carcinoma; Mel, melanoma; PAAD, pancreatic adenocarcinoma; PRAD, prostate adenocarcinoma; RCC, renal clear cell carcinoma. (D) Agar colony formation by M231, A549, and ACHN cells before and after IRIS depletion (n = 6). (Scale bars, 200 μm.)
We also analyzed IRIS expression in primary, triple-negative breast cancer (TNBC) patient-derived xenograft (PDX) cells (19). Although some tumors, such as PA14-0421-14 and PIM005, displayed low or even undetectable levels of IRIS protein, the levels in tumors such as PIM001-M and PIM002 were much higher. Indeed, they were comparable to levels in some IRIS-overexpressing human breast cancer cell lines such as MDA-MB-231 (also known as “M231”) (2) and M231 LM2 (20) (Fig. 1B). Taken together, these results show that endogenous IRIS is abundantly expressed in a variety of sporadic human cancers, including breast cancer, and that both RNA and protein overexpression were detected.
IRIS protein levels were also studied in various cancer cell lines in the Cancer Cell Line Encyclopedia (21). Using the elevated level in M231 breast cancer cells as an example of IRIS overexpression in a cancer cell line (2), IRIS protein overexpression was detected in multiple cell lines of different cancers, e.g., lung, glioma, renal cell, melanoma, and lymphoma (Fig. 1C). In the lines in which p220 was less abundant, IRIS abundance failed to parallel these effects, in keeping with the strict target specificity of our p220 and IRIS antibodies (Fig. 1C).
These broadly based examples of IRIS overexpression in sporadic human tumor samples and in human cancer cell lines prompted the question of whether overexpressed IRIS contributes to a tumor cell neoplastic phenotype. To test this, we asked whether IRIS is required for anchorage-independent growth of three IRIS protein-overproducing human cancer cell lines, M231, A549 (a human lung cancer cell line), and ACHN (a human renal cancer cell line). All overexpressed IRIS, and all readily formed colonies in semisolid medium (Fig. 1 C and D). By contrast, after shRNA-mediated IRIS depletion these cells formed fewer and/or smaller colonies in semisolid medium than controls (Fig. 1D). Furthermore, IRIS depletion in A549 and ACHN cells led to an induction of E-cadherin expression (SI Appendix, Fig. S1B), suggesting that endogenously overexpressed IRIS gives rise to an EMT phenotype in these cells (22).
Moreover, shIRIS-induced inhibition of M231 cell anchorage-independent growth was rescued by expression of an RNAi-resistant IRIS complementary DNA (cDNA), FL IRIS (SI Appendix, Fig. S1C). By contrast, an RNAi-resistant IRIS mutant that lacked the C-terminal intron 11-encoded IRIS functional domain, Tr IRIS, failed to rescue the growth of IRIS-depleted M231 cells in soft agar when tested in parallel (SI Appendix, Fig. S1C). This reflected the specificity of these results.
IRIS sustains a normal rate of cell proliferation by modulating the rate of DNA replication (2). Therefore, we asked whether decreased levels of anchorage-independent growth of IRIS-depleted cells were a result of persistent growth retardation. Colony-forming assays on plastic surfaces were performed on IRIS-depleted and control hairpin-expressing M231, A549, and ACHN cells. These cells all exhibited similar, robust colony-forming efficiency (SI Appendix, Fig. S1D).
Collectively, these data indicate that overly expressed endogenous IRIS drives anchorage-independent growth, a property of some tumor-initiating cells and other neoplastic phenotype-bearing cells (23–25), without affecting attached cell proliferation and colony formation. Furthermore, although IRIS is a BRCA1 gene product and was first found to be overexpressed in breast cancer cell lines (2), its aberrant expression and its ability to sustain anchorage-independent growth and to suppress E-cadherin expression were also detected in nonmammary cancer lines (Fig. 1 C and D and SI Appendix, Fig. S1B).
IRIS Depletion Attenuates Breast Cancer Cell Metastatic Activity but Not Primary Tumor Formation.
Since IRIS is overexpressed in some of the breast cancer PDX models that were tested, we asked whether this property contributes to mammary tumorigenesis. Control hairpin or specific IRIS hairpin-expressing M231 cells were injected both s.c. and into the mammary fat pads of nude mice, and subsequent tumor growth was analyzed.
Gross tumor masses appeared at both sites after the injection of either control or IRIS-depleted cells (SI Appendix, Fig. S2 A and B). The latter exhibited slightly delayed tumor growth (SI Appendix, Fig. S2A), possibly due to modest proliferation suppression (2). Nonetheless, it appeared that IRIS overexpression was not essential for these xenografts to generate primary breast tumors.
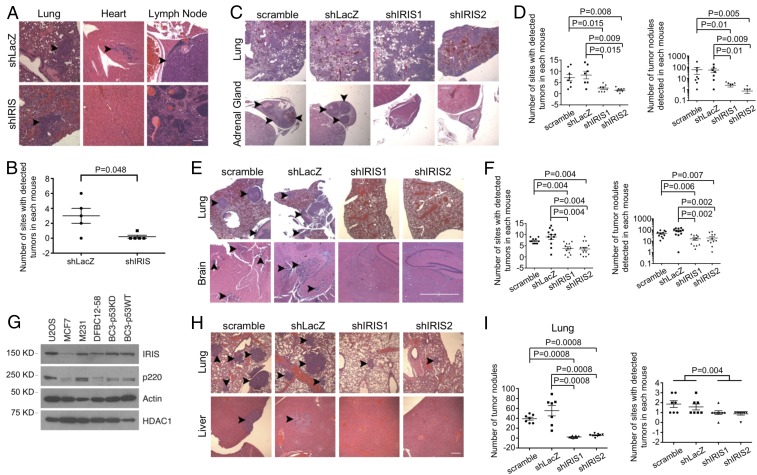
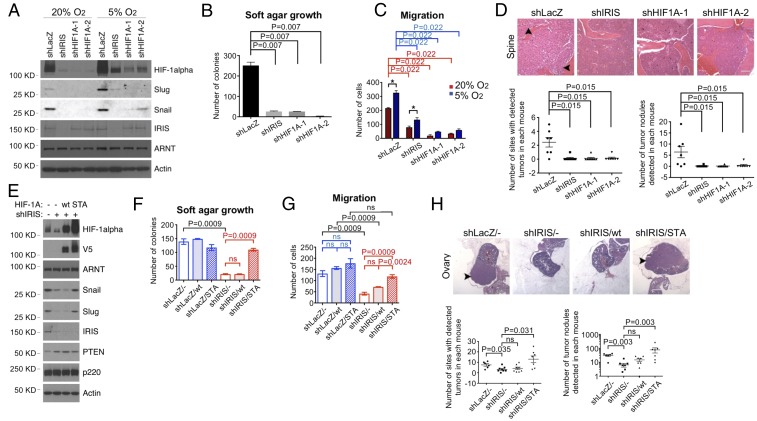
In the same experiments, we also asked whether IRIS depletion affects metastatic M231 tumor growth following mammary fat pad injection. When multiple H&E-stained tissues of these M231-injected animals were analyzed 7 wk after injection, metastatic tumor cells were detected histologically in a variety of organs and tissue sites, including lung, lymph node, abdominal fat collections, cardiac muscle, and brain. By contrast, in mice injected in parallel with IRIS-depleted M231 cells, metastatic tumor deposits were detected primarily in lung and were undetectable elsewhere (Fig. 2 A and B and SI Appendix, Fig. S2C). IRIS depletion did not lead to an obvious decrease in the growth of tumor lung deposits (SI Appendix, Fig. S2C). Thus, high IRIS expression is required for the development of overt metastatic activity following mammary fat pad injection, except in the lung.
Fig. 2.
IRIS depletion attenuates metastasis of breast cancer cells. (A) IRIS depletion reduced metastatic deposits arising from primary tumors of M231 cells. (Scale bar, 80 μm.) Arrowheads pointing at dense, H&E-stained cell collections identify tumor deposits. (B) Quantitation of metastatic deposits detected in the mammary fat pad injection experiments (n = 5). (C) IRIS depletion significantly reduced metastatic deposits of M231 cells in organs and tissue sites other than the lung in the i.v. model. (Scale bar, 400 μm.) (D) Summaries of metastatic deposit quantitation in mice injected and analyzed in the i.v. model (scramble: n = 8; shLacZ: n = 7; shIRIS1: n = 8; shIRIS2: n = 7). (Left) Quantitation of the number of organ sites that contained tumor deposits as reflected in a standard number of sections, excluding lung. (Right) Quantitation of the number of tumor deposits detected in a standard number of sections, excluding lung. (E) Depletion of IRIS significantly depressed M231 cell tumor growth following IC injection. (Scale bar, 200 μm.) (F) Quantitation of metastatic deposits in a standard number of sections (lung included) in the IC injection assays (scramble: n = 11; shLacZ: n = 12; shIRIS1: n = 12; shIRIS2: n = 13). (G) Western blot showing expression levels of certain proteins in various cell strains growing in culture. (H) IRIS depletion attenuated metastasis of BC3-p53KD cells. (Scale bar, 160 μm.) (I) Summaries of metastatic activity of BC3-p53KD cells (scramble: n = 7; shLacZ: n = 7; shIRIS1: n = 8; shIRIS2: n = 8).
In a different tumor cell administration protocol, i.e., i.v. cell injection, we again compared IRIS-regulated metastasis displayed by control vs. IRIS-depleted M231 cells. Here, by 3 wk after tail vein inoculation, large lung tumor deposits were apparent in most mice in all treatment groups (Fig. 2C). The i.v. injection of control hairpin-transduced cells also led to widespread tumor development at other sites, such as adrenal gland, brain, myocardium, muscle, and bone, while mice injected with M231 expressing either of two different IRIS hairpins underwent limited metastasis development except in the lung (Fig. 2C). Quantitations of both metastasis-containing organ/tissue sites and the collective number of tumor deposits detected in each mouse were performed. Standardized histopathology assessment showed that IRIS depletion was associated with markedly reduced metastatic tumor growth other than in the lung (Fig. 2 C and D).
To search for a relationship between lung tumor cell colonization and metastasis to organs fed by the arterial circulation (e.g., adrenal gland, brain, myocardium, muscle, and bone), we repeated these experiments using an intracardiac (IC) tumor cell injection protocol. Unlike the massive pulmonary invasion by IRIS-overexpressing and IRIS-depleted tumor cells following i.v. injection, far fewer and more discrete lung tumor deposits appeared after IC injection of control cells (Fig. 2E and SI Appendix, Fig. S2D). Furthermore, no lung tumor deposits were detected after IC injection of IRIS-depleted cells (Fig. 2E and SI Appendix, Fig. S2D).
Similarly, control hairpin-expressing tumor cells also gave rise to significant numbers of metastases in organ/tissue sites, such as brain, vertebral spine, pancreas, and ovary, all reached by tumor cells in the arterial circulation. By contrast, fewer and smaller metastases were detected in these or any other sites analyzed after IC injection of IRIS hairpin-expressing cells (Fig. 2 E and F). In addition, metastases derived from cells carried to nonlung organ/tissue sites via the arterial circulation were detected at similar frequencies after i.v. and IC injection (Fig. 2 D and F and SI Appendix, Fig. S2D). Lung metastases were much less abundant after IC than after i.v. injection and were even less abundant after IC injection of IRIS-depleted cells vs. controls (Fig. 2 C and E and SI Appendix, Fig. S2D).
Taken together, these results strongly suggest that overexpression of endogenous IRIS enhances arterial M231 metastasis.
IRIS Is Required for Breast Cancer PDX-Derived Cell Metastasis.
In three different human PDX strains (DFBC12-58, BC3-p53KD, and BC3-p53WT) (19, 26) we detected elevated expression levels of IRIS that were comparable to the IRIS level in M231 cells (Fig. 2G). Expression of IRIS hairpins in BC3-p53KD cells led to inhibition of growth in semisolid medium without altering their growth rate when attached (SI Appendix, Fig. S3 A and B). Moreover, BC3-p53KD cells formed primary and secondary mammospheres. This property was suppressed following the expression of IRIS-directed hairpins but not after the expression of irrelevant control hairpins (SI Appendix, Fig. S3C). Anchorage-independent growth and mammosphere formation are manifestations of certain tumor-initiating and stem-like breast cancer cells (23–25, 27, 28).
Distinctive morphological changes also appeared in both BC3-p53KD and BC3-p53WT cells after IRIS depletion. While control cells appeared to be spindle-shaped and fibroblastic, the IRIS-depleted cells displayed a cobblestone-like, epithelial morphology (SI Appendix, Fig. S3D). This implies that IRIS depletion in these cells led to a mesenchymal-to-epithelial transition (MET) (29).
Similarly, in i.v.-injection metastasis experiments, IRIS-depleted cells led to fewer and smaller lung tumor deposits than did control cells (Fig. 2 H and I). The growth of BC3-p53KD tumors was not as aggressive as that of M231 tumors. The experimental animals bearing BC3-p53KD tumors survived longer than animals bearing M231 tumors. This allowed us to observe a defect in the pulmonary tumor deposit formation associated with IRIS-depleted BC3-p53KD cells. Although the metastatic activity in other organs of either control or IRIS-depleted BC3-p53KD cells was relatively low, control cell metastases were nonetheless more abundant than IRIS-depleted ones (Fig. 2 H and I). Thus, IRIS overexpression promoted anchorage-independent growth, mammosphere formation, EMT, and metastasis by primary human breast cancer cells.
Mechanism of IRIS-Driven Metastasis.
IRIS stabilizes and activates HIF-1α.
The state of oxygenation constitutes a major difference between the venous and the arterial circulation. This and the prediction that IRIS-driven metastasizing cells traverse the arterial circulation on their way to their target organs/tissues led us to consider a role for oxygenation-response proteins in the IRIS-driven metastatic process.
The transcription factor HIF-1α mediates adaptive responses to changes in oxygenation (30–32). In the face of tissue, organ, or cellular oxygen deprivation, it becomes stabilized and activated. The opposite is true in normoxic settings. HIF-1α has also been implicated as a metastasis-promoting factor and is often overexpressed in clinical breast cancer (14, 15).
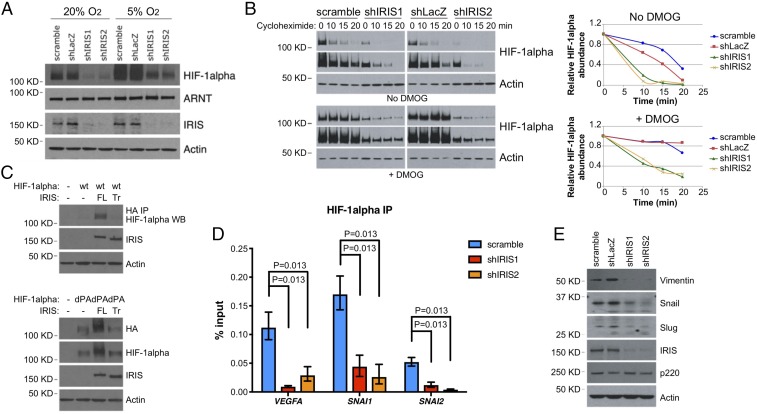
Given the aforementioned observations, we assessed the relative levels of HIF-1α protein in M231 cells before and after IRIS depletion. IRIS depletion resulted in a marked decrease in the steady-state level of HIF-1α in cells cultivated in room air, and the reduction persisted when the cells were switched to a hypoxic environment (Fig. 3A). This suggests that overexpressed IRIS contributes to HIF-1α overexpression in normoxic and hypoxic environments alike.
Fig. 3.
IRIS regulates HIF-1α protein stability. (A) Down-regulation of HIF-1α protein by IRIS depletion under normoxic and hypoxic conditions in M231 cells. (B) Endogenous overexpression of IRIS increased the half-life of HIF-1α protein in the presence and absence of DMOG. (Left) Short and long exposures of HIF-1α and actin Western blots. (Right) Kinetics of HIF-1α protein degradation normalized over actin abundance. (C) The effect of FL or Tr mutant IRIS expression on the abundance of ectopically expressed HIF-1α. (D) IRIS depletion dramatically reduced the binding of HIF-1α to its target gene promoters as reflected by the results of a ChIP assay. Values are normalized mean ± SD (n = 6). (E) Abundance of various proteins in control and IRIS-depleted M231 cells.
Tissue oxygenation-dependent regulation of protein stability underlies the physiological control of HIF-1α abundance in varying states of tissue oxygenation (31, 32). Therefore, we compared the half-life of HIF-1α in IRIS-depleted cells with that detected in control cells. In a cycloheximide-chase assay, the HIF-1α half-life decreased from ∼15 min in control hairpin-expressing cells to ∼5 min in IRIS-depleted cells (Fig. 3B, no DMOG). To mimic a hypoxic condition, we treated the cells with dimethyloxalylglycine (DMOG). DMOG inhibits PHD2, the enzyme that modifies HIF-1α and results in its marked physiological instability in a normoxic environment (33–36). Following DMOG exposure, the half-life of HIF-1α in IRIS-depleted cells was still significantly shorter than that in undepleted cells (10 vs. >20 min) (Fig. 3B, +DMOG). This implies that IRIS stimulates endogenous HIF-1α expression, at least in part, by modulating its stability in a PHD2-independent manner.
In normoxic cells, PHD2-mediated hydroxylation of HIF-1α on two conserved proline residues (Pro402 and Pro564) plays a key role in promoting its degradation (33–36). To further assess the contribution of IRIS to HIF-1α protein stability, we transfected, in parallel, 293T cells with FL IRIS- or Tr IRIS-encoding cDNA. Alternatively, we cotransfected these cells with WT HIF-1α or with the double proline-to-alanine HIF-1α mutant (Pro402Ala and Pro564Ala, dPA), the latter being resistant to PHD2-mediated hydroxylation.
Ectopic expression of FL but not Tr IRIS resulted in elevated steady-state levels of both WT and dPA HIF-1α protein (Fig. 3C). This implies that IRIS overexpression stabilizes HIF-1α protein via an O2/PHD2-independent mechanism. It also indicates that the intron 11-encoded segment of IRIS is required for this effect.
When it accumulates to a sufficient level, HIF-1α activates the transcription of numerous target genes, including VEGFA and SNAI1 (37, 38). To test the outcome of IRIS-driven HIF-1α stabilization, we asked whether it influences the DNA-binding activity of HIF-1α at target gene promoters.
ChIP assays using a HIF-1α antibody were performed, and enrichment of antibody reactivity at the VEGFA, SNAI1, and SNAI2 promoter regions flanking their hypoxia-response elements (HRE) was tested. In control cells, binding of HIF-1α to these promoters was readily detected, and it fell significantly after IRIS depletion (Fig. 3D). These reductions of HIF-1α binding translated into diminished Snail and Slug protein expression (Fig. 3E). Therefore, these results imply that in a normoxic environment IRIS promotes HIF-1α binding to certain hypoxia-responsive promoters by increasing its stability.
IRIS protects HIF-1α from GSK-3β phosphorylation-driven degradation.
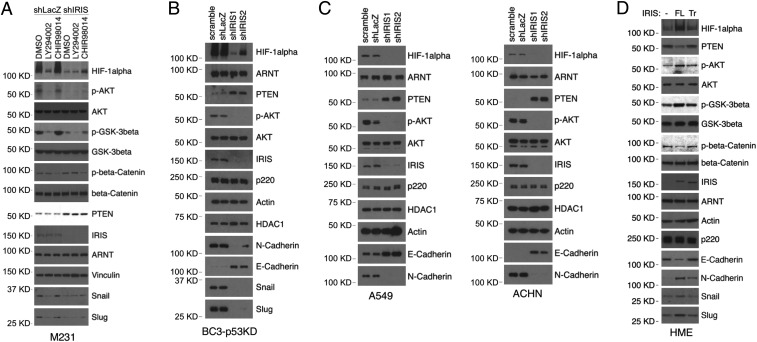
The serine/threonine protein kinases PI3K, AKT, and GSK-3β have been implicated in the PHD2-independent modulation of HIF-1α stability (11–13). Therefore, to gain insight into the mechanism of IRIS-mediated stabilization of HIF-1α protein, we explored the potential roles of PI3K, AKT, and GSK-3β in this process. Exposure of M231 cells to LY294002, a specific PI3K inhibitor, led to AKT inhibition (reflected by a reduction in AKT phosphorylation), hypophosphorylation of GSK-3β, and depletion of HIF-1α (Fig. 4A, compare lanes 2 and 1). The change in the HIF-1α protein level correlated with expression of the HIF-1α transcriptional targets Snail and Slug (Fig. 4A). These findings are consistent with the reported roles of AKT and GSK-3β in the regulation of HIF-1α protein stability in a normoxic environment (12). Intriguingly, hairpin-driven IRIS depletion triggered AKT, GSK-3β, and HIF-1α outcomes similar to those elicited by LY294002 (Fig. 4A, compare lanes 1, 2, and 4).
Fig. 4.
IRIS inhibits HIF-1α degradation via the AKT/GSK-3β pathway. (A) Effects of various kinase inhibitors on HIF-1α protein expression in M231 cells. LY294002 is a PI3K inhibitor; CHIR98014 is a GSK-3β inhibitor. (B) Western blot showing expression levels of various proteins in BC3-p53KD cells before and after IRIS depletion. (C) Effects of IRIS expression on levels of HIF-1α, PTEN, and phospho-AKT in A549 and ACHN cells. (D) Effects of IRIS expression on the expression of the downstream PI3K pathway and certain other targets in HME cells.
Similar effects were detected not only in IRIS-depleted BC3-p53KD, BC3-p53WT, and Hs578t cells (a human TNBC cell line) but also in IRIS-depleted A549 and ACHN cells (Fig. 4 B and C and SI Appendix, Fig. S4 A and B). These hairpin-driven IRIS depletion-mediated effects on the HIF-1α protein level as well as on AKT/GSK-3β signaling were overridden by ectopic overexpression of RNAi-resistant FL IRIS in IRIS-depleted M231 cells (SI Appendix, Fig. S4C). These rescue effects reflect the specificity of the hairpin targeting of endogenous IRIS. They also suggest that, at a minimum, the PI3K/AKT/GSK-3β pathway can operate under IRIS control both in breast cancer cells and in cell lines derived from other cancer types.
In keeping with this hypothesis, exposure to the GSK-3β inhibitor CHIR98014 led to a partial recovery of the HIF-1α protein depletion effect as well as that affecting its targets Snail and Slug that occurred in IRIS-depleted M231 cells (Fig. 4A, compare lanes 6 and 4). This implies that its phosphorylation by GSK-3β is, at least in part, responsible for accelerated HIF-1α degradation in IRIS-depleted cells.
To further explore the role of IRIS in regulating AKT and GSK-3β phosphorylation and function, we analyzed signaling changes in IRIS-transduced, telomerase-immortalized, primary human mammary epithelial (HME) cells. Overexpression of FL but not Tr IRIS resulted in higher levels of phospho-AKT (activated enzyme), phospho-GSK-3β (inactivated enzyme), and HIF-1α (Fig. 4D). Tr IRIS was significantly less active in this regard.
Notably, FL IRIS-overexpressing HME cells exhibited mesenchymal morphology unlike control HME cells, which revealed a typical epithelial phenotype. Cells overexpressing Tr IRIS lacked this phenotypic change (SI Appendix, Fig. S4E). In addition, FL IRIS-overexpressing HME cells grew in semisolid medium and formed mammosphere structures (SI Appendix, Fig. S4 F and G). Both anchorage-independent growth and mammosphere-forming potential have been shown to accompany EMT development in mammary epithelial cells (27, 39, 40).
These data again show that IRIS, when overproduced, stabilizes HIF-1α by differentially modulating AKT and GSK-3β activities. They also reinforce the notion that the unique intron 11-encoded C terminus of IRIS participates actively in this process.
HIF-1α overexpression is required for IRIS-induced neoplastic phenotypes.
Next, we asked whether IRIS not only promotes HIF-1α stability but also sustains a sufficiently high abundance of functional HIF-1α to induce EMT and promote metastasis. HIF-1α is a known metastasis driver, and metastasizing epithelial cells frequently display an EMT phenotype. The latter is strongly suspected of abetting the metastatic process (16, 22, 41).
Although they retained high endogenous IRIS expression when HIF-1α protein was depleted, M231 cells lost Snail and Slug expression, failed to proliferate in soft agar, experienced reduced migratory activity, and reacquired epithelial morphology. The opposite of each of these phenotypes persisted in undepleted cells (Fig. 5 A–C and SI Appendix, Fig. S5 A–C). Furthermore, HIF-1α–depleted cells were unable to metastasize to arterial circulation-fed organs/tissues by comparison with control cells, thereby phenocopying IRIS-depleted cells (Fig. 5D). In light of the established association of deregulated Snail/Slug and HIF-1α with metastasis (14, 42–44), these results strongly imply that HIF-1α is a necessary participant in IRIS-dependent metastasis.
Fig. 5.
HIF-1α is necessary and sufficient for IRIS neoplastic functions. (A) Immunoblots reflecting the expression of various proteins synthesized in M231 cells following the expression of various hairpins. (B) HIF-1α depletion inhibited the growth of M231 cells in soft agar (n = 6). (C) Changes in the migration of M231 cells incubated in two different oxygen environments after transduction with the indicated hairpins (n = 6). *P < 0.05. (D) HIF-1α depletion suppressed i.v.-injected M231 cell metastasis to sites and organs other than lung (shLacZ: n = 7; shIRIS: n = 8; shHIF1A-1: n = 8; shHIF1A-2: n = 7). (Scale bar, 160 μm.) (E) Immunoblot-based expression analysis of various proteins in M231 cells after transduction of shIRIS and, where indicated, transduction of WT vs. STA HIF-1α. (F) STA HIF-1α stimulated the growth of IRIS-depleted M231 cells in soft agar (n = 9). ns, not significant. (G) STA HIF-1α restored the high migratory activity of IRIS-depleted M231 cells (n = 9). ns, not significant. (H) STA HIF-1α rescued metastasis of IRIS-depleted M231 cells (shLacZ/-: n = 6; shIRIS/-: n = 8; shIRIS/wt: n = 8; shIRIS/STA: n = 7). (Scale bar, 400 μm.) ns, not significant.
Stabilized GSK-3β phosphorylation-resistant mutant HIF-1α rescues the defects associated with IRIS depletion.
To probe further the relevance of the AKT/GSK-3β phosphorylation cascade to IRIS/HIF-1α–promoted metastasis, we generated a GSK-3β phosphorylation-resistant mutant of HIF-1α (Ser551Ala, Thr555Ala, and Ser589Ala, STA HIF-1α) (12, 13). Given the aforementioned signaling analysis, we predicted that this mutant HIF-1α species would be innately more stable than the WT protein and therefore might drive metastasis in the absence of IRIS coexpression.
After transduction, the STA HIF-1α protein proved to be more abundant than WT HIF-1α when the overexpressing cells were cultivated in room air (SI Appendix, Fig. S5D). Moreover, STA HIF-1α expression was unaffected by IRIS depletion, unlike WT HIF-1α, whose expression repeatedly decreased in the absence of IRIS overexpression (SI Appendix, Fig. S5D). To directly compare the stabilities of WT and STA HIF-1α, the half-lives of these proteins were examined. In both the presence and absence of DMOG the half-life of ectopically expressed WT HIF-1α was shorter than that of STA HIF-1α (SI Appendix, Fig. S5E).
In this setting, overexpression of the HIF-1α STA mutant restored Snail and Slug expression, rescued the anchorage-independent growth defect, enhanced cellular migration, and led to the reappearance of mesenchymal morphology in IRIS-depleted cells (Fig. 5 E–G and SI Appendix, Fig. S5 F–H). More to the point, IRIS-depleted, STA-overexpressing cells exhibited widespread metastatic tumor growth (Fig. 5H). By contrast, despite reaching higher expression levels than did endogenous HIF-1α in control M231 cells, exogenous WT HIF-1α failed to exhibit dramatic rescue effects like those generated by STA (Fig. 5 E–H and SI Appendix, Fig. S5 F–H).
We further examined the requirement for IRIS to support local cell invasion, a phenotypic characteristic of EMT (22). By comparison with undepleted control cells, IRIS-depleted M231 cells exhibited a markedly reduced ability to penetrate Matrigel-coated membranes that mimic extracellular matrix (SI Appendix, Fig. S5I). Thus, in keeping with the codevelopment of an MET phenotype, IRIS depletion inhibited matrix invasion by these endogenous IRIS-overexpressing cells.
Together, these data reveal that IRIS-driven EMT/metastasis is a product of sufficiently activated and stabilized HIF-1α. In this context, IRIS not only stabilized HIF-1α but also activated it by indirectly inhibiting its phosphorylation by GSK-3β (12, 13).
IRIS expression and its effect on PTEN.
The PTEN tumor suppressor is a major antagonist of AKT activation by the PI3K pathway (6–10). Loss of PTEN function is one of the most common known human cancer cell phenotypes (45). Moreover, the PI3K pathway and AKT are strongly activated by IRIS, as documented above. We therefore analyzed PTEN expression in relation to that of spontaneously overexpressed IRIS.
In several spontaneous IRIS-overproducing cell lines, e.g., M231, Hs578t, A549, and ACHN, as well as in PDX-derived human breast cancer cells, PTEN protein levels were higher in IRIS-depleted cells than in IRIS-overexpressing control cells. Moreover, in IRIS-depleted cells, PTEN levels correlated inversely with reduced levels of AKT phosphorylation (Fig. 4 A–C and SI Appendix, Fig. S4 A–D).
By contrast, when FL IRIS was ectopically overexpressed, the PTEN protein levels dropped, unlike the case of Tr IRIS-overexpressing HME cells (Fig. 4D). Thus, by downregulating PTEN protein abundance, IRIS activates the PI3K pathway and AKT function.
To further examine the role of PTEN in IRIS-mediated control of HIF-1α stability, we assessed HIF-1α expression in response to IRIS depletion in PTEN-mutated cells. MDA-MB-468 (also known as “M468”) and BT549 cells both harbor deleterious mutations in the PTEN gene (46). Therefore, no PTEN expression was detected in these cells (SI Appendix, Fig. S4D). IRIS depletion had only small effects on the levels of HIF-1α protein. In contrast, IRIS depletion in two WT PTEN-expressing cell lines, M231 and Hs578t, resulted in major decreases in HIF-1α levels and increases in PTEN expression (SI Appendix, Fig. S4D). Although these cell lines are not isogenic, these data imply that PTEN is an important target in connecting sufficient IRIS expression to an increase in HIF-1α stability.
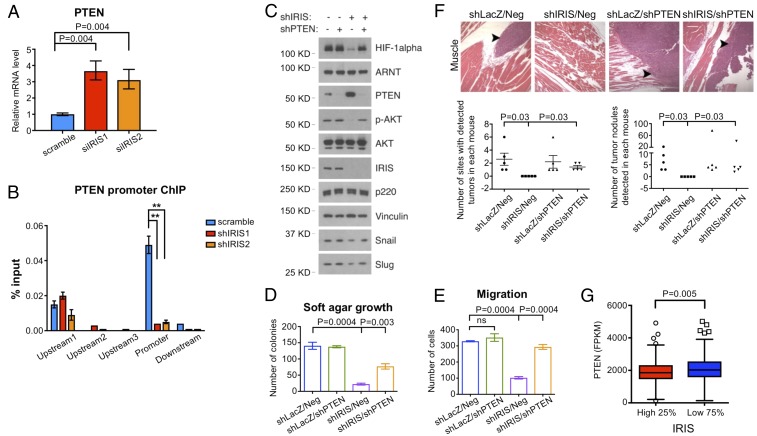
In search of the source of the elevated PTEN protein levels in IRIS-depleted M231 cells, we measured their PTEN mRNA levels. They were two- to threefold higher in cells transfected with two different IRIS siRNAs than in control cells (Fig. 6A). A similar effect was observed in Hs578t cells (SI Appendix, Fig. S6A). These data imply that IRIS overexpression represses PTEN mRNA synthesis.
Fig. 6.
PTEN depletion rescues phenotypic changes in IRIS-depleted cells. (A) PTEN mRNA levels increased after IRIS depletion in M231 cells. Values are normalized mean ± SD (n = 6). (B) Relative abundance of endogenous IRIS associated with the PTEN promoter in M231 cells as assayed by ChIP. Values are normalized mean ± SD (n = 6). **P < 0.01. (C) Western blot showing various protein levels in M231 cells under each knockdown condition. (D) PTEN depletion rescued the growth of IRIS-depleted M231 cells in soft agar (n = 9). Neg, negative control hairpin. (E) PTEN depletion restored the migratory activity of IRIS-depleted M231 cells (n = 12). ns, not significant. (F) IRIS-depleted M231 cells regained the ability to metastasize after codepletion of PTEN (n = 5). (Scale bar, 160 μm.) (G) Low PTEN expression correlated with relatively high IRIS messenger levels in the TCGA breast cancer dataset. High 25%: cases with IRIS mRNA levels in the top quartile. Low 75%: the remaining cases.
IRIS can function as a transcriptional regulator in association with other cofactors (3). Therefore, we asked whether endogenous IRIS binds specifically to the PTEN promoter. In ChIP experiments IRIS signals were readily detected at a specific location near the PTEN promoter in both M231 and Hs578t cells. These specific signals were nearly abolished by IRIS depletion (Fig. 6B and SI Appendix, Fig. S6 B and C), in keeping with the increase in PTEN mRNA abundance that accompanied IRIS depletion (Fig. 6A and SI Appendix, Fig. S6A). These results imply that IRIS exerts a negative effect on PTEN mRNA synthesis, at least in part, by repressing PTEN promoter function.
PTEN depletion reestablishes neoplastic phenotypes in IRIS-depleted cells.
To determine whether PTEN plays a role in IRIS-regulated HIF-1α stability, we asked whether PTEN depletion alters downstream signaling and neoplastic outcomes in IRIS-depleted cells. PTEN depletion in shIRIS-expressing M231 cells led to reactivation of AKT, elevation of HIF-1α protein levels, and restoration of Snail and Slug protein expression compared with those in control, IRIS-only–depleted cells (Fig. 6C). These PTEN/IRIS double-knockdown cells displayed anchorage-independent growth, reacquired transmembrane migratory activity, and regained mesenchymal morphology (Fig. 6 D and E and SI Appendix, Fig. S6 D–F). PTEN depletion also led to metastatic tumor development of IRIS-depleted cells not only in lung but also in muscle, bone, and lymph nodes (Fig. 6F).
Thus, it appears that PTEN is an indispensable IRIS target in the perturbed signaling pathway in which spontaneously overexpressed IRIS stabilizes HIF-1α that, in turn, promotes EMT and metastasis.
An inverse relationship between IRIS and PTEN expression.
These data strongly suggest that IRIS participates in the repression of PTEN transcription. This results in the stabilization and activation of HIF-1α, which in turn triggers the development of a multicomponent neoplastic phenotype. The latter includes metastatic activity, in keeping with the view that loss of PTEN expression correlates with human breast cancer metastatic activity (47, 48).
In this context, we asked whether IRIS overexpression correlates with low PTEN expression in clinical samples. Indeed, we detected lower PTEN mRNA levels in the primary breast cancer cells of patients with high IRIS mRNA levels (in the top quartile) than in the breast cancer cells of patients with lower IRIS mRNA levels (below the third quartile) in all WT PTEN-associated TCGA breast cancer cases (Fig. 6G and Dataset S4). Thus, the IRIS/PTEN relationship, first identified in cultured cells, is also valid clinically.
Discussion
By analyzing TCGA RNA-Seq data, we documented spontaneous IRIS RNA overexpression in numerous human cancers derived from various organs. Where studied, multiple IRIS-overproducing human cancer cell lines and two IRIS-overproducing human PDX primary breast cancer strains revealed IRIS overexpression-dependent neoplastic properties. They included anchorage-independent growth, an EMT phenotype, increased cell migration, and, where tested, metastatic behavior in a PDX model. Thus, it appears that, when spontaneously overexpressed, IRIS acts as a tumor-progression driver in multiple common cancers.
Although IRIS-depleted cells displayed drastic suppression of anchorage-independent growth and mammosphere formation in vitro, their potential to develop primary tumors was similar to that in control cells (SI Appendix, Fig. S2 A and B). The cells tested in in vitro assays were cultivated in a normoxic atmosphere in which IRIS depletion led to a marked reduction in HIF-1α level. This is the underlying mechanism whereby IRIS is required for anchorage-independent growth and mammosphere formation. By contrast, in the tumorigenicity assays millions of cells were mixed with Matrigel and injected. These cells may well encounter hypoxia shortly after inoculation, in which case HIF-1α protein would be expected to accumulate even in IRIS-depleted cells. This could contribute to IRIS-independent primary tumor growth.
A key step in the mechanism underlying IRIS-driven metastasis is its suppression of PTEN expression, apparently by participating in the inhibition of PTEN mRNA transcription. This was accompanied by PI3K pathway and AKT activation. Activated AKT inhibits GSK-3β activity by phosphorylating GSK-3β Ser9 (11). GSK-3β inactivation inhibits the phosphorylation of three specific HIF-1α transactivation domain residues, which, in turn, renders HIF-1α resistant to ubiquitylation and subsequent proteasome degradation (12, 13). Thus, a nonphosphorylatable HIF-1α mutant targeting these three residues was exceedingly stable in a normoxic environment, accumulated to abnormally high levels, and substituted effectively for overexpressed IRIS in metastasis and other neoplastic phenotype components.
HIF-1α phosphorylation has been implicated in the negative regulation of its DNA-binding activity and its transcriptional activation of target genes (49, 50). For example, HIF-1α exposed to the serine/threonine phosphatase inhibitor NaF failed to bind to HRE-harboring DNA probes in EMSAs (51). This implied that certain HIF-1α phosphorylation events block its DNA binding and potentially inhibit its transcriptional activation of certain target genes. Thus, interference with HIF-1α phosphorylation, e.g., after endogenous IRIS overexpression leading to GSK-3β inhibition, likely translates into HIF-1α hyperactivity, an event with potential neoplastic consequences.
Given the location of the serine/threonine residues Ser551, Thr555, and Ser589 within the HIF-1α N-terminal transactivation domain, we speculate that GSK-3β–mediated phosphorylation normally facilitates HIF-1α degradation and inhibits its transcriptional activity. This would potentially explain our observation that, despite HIF-1α expression being much higher than in control cells, ectopic overexpression of WT phosphorylatable HIF-1α failed to reverse the inhibited expression of SNAI1 and SNAI2 in IRIS-depleted cells (Fig. 5E). It also failed to reverse the other outcomes arising after IRIS depletion, including failed metastasis (Fig. 5 F–H and SI Appendix, Fig. S5 F–H).
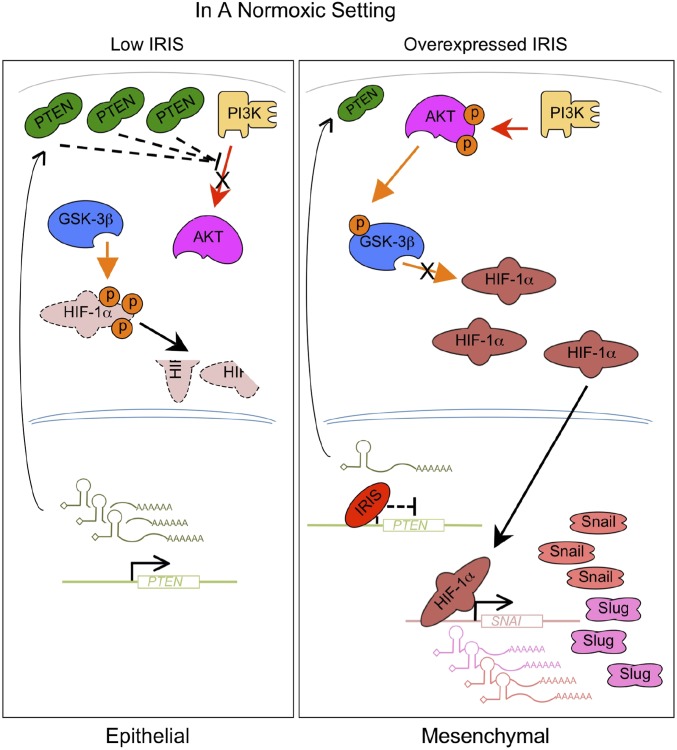
Our findings also reveal that spontaneous overexpression of endogenous IRIS, which appears to be a not uncommon clinical event in multiple, albeit not in all, tumor types, initiates its metastasis-promoting activity by inhibiting PTEN expression. This, in turn, results in HIF-1α stabilization, accumulation, and activation (Fig. 7), which collectively trigger metastasis (16).
Fig. 7.
A schematic model illustrating the function of overexpressed IRIS in human breast cancer cells. In a normoxic setting, overexpressed IRIS represses PTEN gene transcription. The resulting low PTEN protein levels lead to the activation of AKT and inhibition of HIF-1α phosphorylation mediated by GSK-3β. HIF-1α becomes stabilized and accumulates to activate the transcription of its target genes, such as SNAI1 and SNAI2. Elevated Snail and Slug expression induces an EMT and promotes metastasis.
In addition to HIF-1α stabilization and inhibition of its phosphorylation, another potential route to HIF-1α overexpression and excessive HIF-1α function would be PTEN inhibition-driven mTOR activation followed by enhanced HIF-1α translation (52). Conceivably, this effect, too, operates in IRIS-overexpressing cells.
Our results also show that IRIS-driven metastasis occurs, at least in part, in organs/tissues fed by the arterial circulation and is dependent upon an ability of IRIS to stabilize HIF-1α in what is presumed to be a normoxic setting. High levels of HIF-1α protein are associated with tumor progression and a poor prognosis in breast cancer patients (14, 15). Although they also correlate with tumor size, tumor stage, and histological grade in a variety of human cancers, including advanced breast cancer (14), HIF-1α overexpression has been detected in preneoplastic lesions in breast, colon, and prostate cancer (14) and in early-stage (pT1b) cervical cancer, where it was associated with an unfavorable prognosis (53). Therefore, it is conceivable that an accumulation of activated HIF-1α in normoxic cells, following endogenous IRIS overexpression, can promote cellular transformation and/or stimulate the dissemination of early-stage tumor cells to multiple tissues and organs.
HIF-1α has also been implicated in still other events associated with tumor progression, e.g., the activation of EMT-inducing transcription factors such as SNAI1 and TWIST1 (38, 54). We found that depletion of overexpressed IRIS in multiple human cancer cell lines led to the induction of E-cadherin and reduction of N-cadherin expression which signal MET development (Fig. 4 B and C and SI Appendix, Fig. S4 A and B). By contrast, overexpression of FL IRIS in HME and MCF7 cells, which express low levels of IRIS, resulted in diminished E-cadherin and elevated N-cadherin signals (Fig. 4D and SI Appendix, Fig. S4H). EMT induction is strongly associated with tumor cell invasion and metastasis (22, 41).
A recent report suggested that higher levels of HIF-1α protein expression correlate with IRIS overexpression in certain established human breast cancer cell lines and that IRIS depletion in these cells was accompanied by reduced HIF-1α abundance (55). However, the target specificities of the relevant shRNAs and HIF-1α antibody species were not documented, nor was the molecular connection underlying these phenomena investigated.
Finally, our attempts to analyze the effects of IRIS expression upon survival in the large patient study (TCGA) we consulted were inconclusive. This was a result of (i) the relatively short median follow-up of the TCGA patients; (ii) the limited reporting of survival data and, therefore, the limited ability to discern its relationship to the IRIS concentration in individual tumors; and (iii) the relatively small number of documented survival events and of tissue metastasis patterns that were reported. Thus, an ability to accurately predict the effect of IRIS expression on the clinical course of breast or any of the other IRIS-overproducing cancers has not yet been achieved.
Whatever the case, given its ability to promote the metastasis of primary human breast cancer cells in mouse models, its contribution to anchorage-independent growth, EMT development, and enhanced cell migration behavior, and its naturally occurring overexpression in various primary human cancers, it would not be surprising, if endogenous IRIS overexpression were a negative clinical outcome-inducing factor in certain cancers.
Materials and Methods
Please refer to SI Appendix, Supplementary Materials and Methods for other materials and methods, including cell culture, RNAi, qRT-PCR, ChIP, anchorage-independent growth, cell migration and invasion, mammosphere formation, colony formation, protein half-life estimates, Western blotting, and immunoprecipitation.
Xenograft Studies and Histological Analysis.
All xenograft studies were approved by the Institutional Animal Care and Use Committee at the Dana-Farber Cancer Institute (DFCI) and were conducted under the guidelines of the DFCI Animal Research Facility. All inoculations were carried out blindly, and animals were randomly grouped for inoculation. Please see SI Appendix, Supplementary Materials and Methods for technical details.
Analysis of RNA-Seq Data.
An alignment-free approach, Kallisto (18), was used to estimate p220 and IRIS isoform abundance from the FASTQ files in the TCGA cases (56). Kallisto calculates sets of probabilities for each RNA-Seq read that could have originated from various known transcripts encoded by a given gene. Estimates of the abundance of each transcript are based on the assembly of the probabilities associated with all possible reads derived from a given transcript. To distinguish between p220 and IRIS (ENST00000357654 and ENST00000354071, respectively, from the Ensembl genome browser, https://www.ensembl.org), the specificity was determined by comparing RNA-Seq reads from exons 12–24 with reads that included the exon 11/intron 11 junction as well as reads derived from intron 11. Additional details regarding RNA-Seq data extraction and analysis are presented in SI Appendix, Supplementary Materials and Methods.
Statistical Analysis.
Data are represented as mean ± SEM except when indicated otherwise. The P values shown, except those associated with RNA-Seq analysis, were calculated using a Wilcoxon rank-sum test (two-tailed) and used the Holm–Bonferroni adjustment for multiple comparisons. P < 0.05 is considered significant. No animals were excluded from analysis.
Data Availability.
All the data supporting the findings of this study are available within the article and its supporting information files.
Supplementary Material
Acknowledgments
We thank Y. Li and X. Liu for technical assistance; members of the D.M.L. laboratory for many helpful discussions and valuable comments on the manuscript; R. M. Li for his endless inspiration and tremendous encouragement; and the Rodent Histopathology Core of the Dana-Farber/Harvard Cancer Center, which is supported in part by National Cancer Institute (NCI) Cancer Center Support Grant NIH 5P30CA06516. This body of work was supported by NCI Grant 2P01CA080111, the DFCI/Novartis Program in Drug Discovery, the Breast Cancer Research Foundation, the Susan G. Komen Foundation for the Cure, and the BRCA Foundation. W.G.K. is a Howard Hughes Medical Institute investigator.
Footnotes
Conflict of interest statement: D.M.L. is a grantee of and consultant to the Novartis Institute of Biomedical Research. W.G.K. owns equity in and consults for Lilly. A.L.K. was, during the course of some of this work, a grantee of and consultant to the Novartis Institute of Biomedical Research.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807112115/-/DCSupplemental.
References
- 1.Miki Y, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 2.ElShamy WM, Livingston DM. Identification of BRCA1-IRIS, a BRCA1 locus product. Nat Cell Biol. 2004;6:954–967. doi: 10.1038/ncb1171. [DOI] [PubMed] [Google Scholar]
- 3.Nakuci E, Mahner S, Direnzo J, ElShamy WM. BRCA1-IRIS regulates cyclin D1 expression in breast cancer cells. Exp Cell Res. 2006;312:3120–3131. doi: 10.1016/j.yexcr.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 4.Paul BT, Blanchard Z, Ridgway M, ElShamy WM. BRCA1-IRIS inactivation sensitizes ovarian tumors to cisplatin. Oncogene. 2015;34:3036–3052. doi: 10.1038/onc.2014.237. [DOI] [PubMed] [Google Scholar]
- 5.Blanchard Z, Paul BT, Craft B, ElShamy WM. BRCA1-IRIS inactivation overcomes paclitaxel resistance in triple negative breast cancers. Breast Cancer Res. 2015;17:5. doi: 10.1186/s13058-014-0512-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maehama T, Dixon JE. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1998;273:13375–13378. doi: 10.1074/jbc.273.22.13375. [DOI] [PubMed] [Google Scholar]
- 7.Myers MP, et al. The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc Natl Acad Sci USA. 1998;95:13513–13518. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stambolic V, et al. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 9.Lee JO, et al. Crystal structure of the PTEN tumor suppressor: Implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 10.Sun H, et al. PTEN modulates cell cycle progression and cell survival by regulating phosphatidylinositol 3,4,5,-trisphosphate and Akt/protein kinase B signaling pathway. Proc Natl Acad Sci USA. 1999;96:6199–6204. doi: 10.1073/pnas.96.11.6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 12.Flügel D, Görlach A, Michiels C, Kietzmann T. Glycogen synthase kinase 3 phosphorylates hypoxia-inducible factor 1alpha and mediates its destabilization in a VHL-independent manner. Mol Cell Biol. 2007;27:3253–3265. doi: 10.1128/MCB.00015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flügel D, Görlach A, Kietzmann T. GSK-3β regulates cell growth, migration, and angiogenesis via Fbw7 and USP28-dependent degradation of HIF-1α. Blood. 2012;119:1292–1301. doi: 10.1182/blood-2011-08-375014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhong H, et al. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 1999;59:5830–5835. [PubMed] [Google Scholar]
- 15.Dales JP, et al. Overexpression of hypoxia-inducible factor HIF-1alpha predicts early relapse in breast cancer: Retrospective study in a series of 745 patients. Int J Cancer. 2005;116:734–739. doi: 10.1002/ijc.20984. [DOI] [PubMed] [Google Scholar]
- 16.Schito L, Semenza GL. Hypoxia-inducible factors: Master regulators of cancer progression. Trends Cancer. 2016;2:758–770. doi: 10.1016/j.trecan.2016.10.016. [DOI] [PubMed] [Google Scholar]
- 17.Ciriello G, et al. TCGA Research Network Comprehensive molecular portraits of invasive lobular breast cancer. Cell. 2015;163:506–519. doi: 10.1016/j.cell.2015.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 19.Powell E, et al. p53 deficiency linked to B cell translocation gene 2 (BTG2) loss enhances metastatic potential by promoting tumor growth in primary and metastatic sites in patient-derived xenograft (PDX) models of triple-negative breast cancer. Breast Cancer Res. 2016;18:13. doi: 10.1186/s13058-016-0673-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minn AJ, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 23.Freedman VH, Shin SI. Cellular tumorigenicity in nude mice: Correlation with cell growth in semi-solid medium. Cell. 1974;3:355–359. doi: 10.1016/0092-8674(74)90050-6. [DOI] [PubMed] [Google Scholar]
- 24.Shin SI, Freedman VH, Risser R, Pollack R. Tumorigenicity of virus-transformed cells in nude mice is correlated specifically with anchorage independent growth in vitro. Proc Natl Acad Sci USA. 1975;72:4435–4439. doi: 10.1073/pnas.72.11.4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallimore PH, McDougall JK, Chen LB. In vitro traits of adenovirus-transformed cell lines and their relevance to tumorigenicity in nude mice. Cell. 1977;10:669–678. doi: 10.1016/0092-8674(77)90100-3. [DOI] [PubMed] [Google Scholar]
- 26.Wang Y, et al. CDK7-dependent transcriptional addiction in triple-negative breast cancer. Cell. 2015;163:174–186. doi: 10.1016/j.cell.2015.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dontu G, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nieto MA. Epithelial plasticity: A common theme in embryonic and cancer cells. Science. 2013;342:1234850. doi: 10.1126/science.1234850. [DOI] [PubMed] [Google Scholar]
- 30.Semenza GL, Wang GL. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol. 1992;12:5447–5454. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: The central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 32.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maxwell PH, et al. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- 34.Ohh M, et al. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat Cell Biol. 2000;2:423–427. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- 35.Ivan M, et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: Implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- 36.Jaakkola P, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- 37.Forsythe JA, et al. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol. 1996;16:4604–4613. doi: 10.1128/mcb.16.9.4604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo D, Wang J, Li J, Post M. Mouse snail is a target gene for HIF. Mol Cancer Res. 2011;9:234–245. doi: 10.1158/1541-7786.MCR-10-0214. [DOI] [PubMed] [Google Scholar]
- 39.Morel AP, et al. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One. 2008;3:e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sorokin AV, Chen J. MEMO1, a new IRS1-interacting protein, induces epithelial-mesenchymal transition in mammary epithelial cells. Oncogene. 2013;32:3130–3138. doi: 10.1038/onc.2012.327. [DOI] [PubMed] [Google Scholar]
- 41.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 42.Blanco MJ, et al. Correlation of Snail expression with histological grade and lymph node status in breast carcinomas. Oncogene. 2002;21:3241–3246. doi: 10.1038/sj.onc.1205416. [DOI] [PubMed] [Google Scholar]
- 43.Martin TA, Goyal A, Watkins G, Jiang WG. Expression of the transcription factors snail, slug, and twist and their clinical significance in human breast cancer. Ann Surg Oncol. 2005;12:488–496. doi: 10.1245/ASO.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 44.Moody SE, et al. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell. 2005;8:197–209. doi: 10.1016/j.ccr.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 45.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 47.Depowski PL, Rosenthal SI, Ross JS. Loss of expression of the PTEN gene protein product is associated with poor outcome in breast cancer. Mod Pathol. 2001;14:672–676. doi: 10.1038/modpathol.3880371. [DOI] [PubMed] [Google Scholar]
- 48.Tsutsui S, et al. Reduced expression of PTEN protein and its prognostic implications in invasive ductal carcinoma of the breast. Oncology. 2005;68:398–404. doi: 10.1159/000086981. [DOI] [PubMed] [Google Scholar]
- 49.Minet E, Michel G, Mottet D, Raes M, Michiels C. Transduction pathways involved in hypoxia-inducible factor-1 phosphorylation and activation. Free Radic Biol Med. 2001;31:847–855. doi: 10.1016/s0891-5849(01)00657-8. [DOI] [PubMed] [Google Scholar]
- 50.Brahimi-Horn C, Mazure N, Pouysségur J. Signalling via the hypoxia-inducible factor-1alpha requires multiple posttranslational modifications. Cell Signal. 2005;17:1–9. doi: 10.1016/j.cellsig.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 51.Wang GL, Jiang BH, Semenza GL. Effect of protein kinase and phosphatase inhibitors on expression of hypoxia-inducible factor 1. Biochem Biophys Res Commun. 1995;216:669–675. doi: 10.1006/bbrc.1995.2674. [DOI] [PubMed] [Google Scholar]
- 52.Zhong H, et al. Modulation of hypoxia-inducible factor 1alpha expression by the epidermal growth factor/phosphatidylinositol 3-kinase/PTEN/AKT/FRAP pathway in human prostate cancer cells: Implications for tumor angiogenesis and therapeutics. Cancer Res. 2000;60:1541–1545. [PubMed] [Google Scholar]
- 53.Birner P, et al. Overexpression of hypoxia-inducible factor 1alpha is a marker for an unfavorable prognosis in early-stage invasive cervical cancer. Cancer Res. 2000;60:4693–4696. [PubMed] [Google Scholar]
- 54.Yang MH, et al. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol. 2008;10:295–305. doi: 10.1038/ncb1691. [DOI] [PubMed] [Google Scholar]
- 55.Ryan D, et al. Correction: A niche that triggers aggressiveness within BRCA1-IRIS overexpressing triple negative tumors is supported by reciprocal interactions with the microenvironment. Oncotarget. 2017;8:113294. doi: 10.18632/oncotarget.23573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tatlow PJ, Piccolo SR. A cloud-based workflow to quantify transcript-expression levels in public cancer compendia. Sci Rep. 2016;6:39259. doi: 10.1038/srep39259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data supporting the findings of this study are available within the article and its supporting information files.