Abstract
The advent of multidrug resistance among pathogenic bacteria is imperiling the worth of antibiotics, which have previously transformed medical sciences. The crisis of antimicrobial resistance has been ascribed to the misuse of these agents and due to unavailability of newer drugs attributable to exigent regulatory requirements and reduced financial inducements. Comprehensive efforts are needed to minimize the pace of resistance by studying emergent microorganisms, resistance mechanisms, and antimicrobial agents. Multidisciplinary approaches are required across health care settings as well as environment and agriculture sectors. Progressive alternate approaches including probiotics, antibodies, and vaccines have shown promising results in trials that suggest the role of these alternatives as preventive or adjunct therapies in future.
Keywords: antibiotics, multidrug resistance, evolution, alternative therapies
Introduction
Antibiotic resistance is ancient and the “resistome” is a dynamic and mounting problem. Causes of the global resistome are overpopulation, enhanced global migration, increased use of antibiotics in clinics and animal production, selection pressure, poor sanitation, wildlife spread, and poor sewerage disposal system.1,2 Antibiotic treatment is one of the main approaches of modern medicine which is used to combat infections. The “golden era” of antibiotics ranged from the 1930s to 1960s which gave rise to many antibiotics.3 Unfortunately, this era ended because researchers were unable to maintain the pace of antibiotic discovery in the face of emerging resistant pathogens. Persistent failure to develop or discover new antibiotics and nonjudicious use of antibiotics are the predisposing factors associated with the emergence of antibiotic resistance.4
Antimicrobial resistance (AMR) poses a serious global threat of growing concern to human, animal, and environment health. This is due to the emergence, spread, and persistence of multidrug-resistant (MDR) bacteria or “superbugs.”5 MDR bacteria exist across the animal, human, and environment triangle or niche and there is interlinked sharing of these pathogens in this triad. The plausible causes of “the global resistome” or AMR include excessive use of antibiotics in animals (food, pets, aquatic) and humans, antibiotics sold over-the-counter, increased international travel, poor sanitation/hygiene, and release of nonmetabolized antibiotics or their residues into the environment through manure/feces. These factors contribute to genetic selection pressure for the emergence of MDR bacterial infections in the community. Recently, the global consumption of antimicrobials in livestock has indicated the hotspots of antibiotics use across the continents that will have economic and public health impacts in the years to come. In food animals, antibiotics are commonly used in cattle, chicken, and pigs and it is projected that in 2030 such use will increase up to 67% in the most populated countries of the world.6
The effective use of an antimicrobial agent is undermined due to the possible tolerance or resistance developed from the very initial time this compound is used. This is true for antimicrobial agents used to treat bacterial, viral, fungal, and parasitic infections. Several physiological and biochemical mechanisms may steer this developing resistance. The intricacy of all the mechanisms associated with the emergence and distribution of the resistance should not be overplayed. Furthermore, lack of elementary data on these specific subjects is a vital concern, which has caused a lack of significant achievements from being made to manage the development of resistance. Worldwide, various institutes and agencies have recognized this serious global public health problem. Many recommendations and resolutions have been proposed, several reports have also been written, but so far little progress has been made. Unfortunately, the increase in antibiotic resistance is a persistent issue.7
The discovery of antibiotics was a defining moment in the history of mankind that revolutionized medicine and saved countless lives. Unfortunately, these “magic bullets” have been accompanied by these emerging resistant strains of pathogens. Currently, medical experts are raising real concern for a return to the preantibiotic age. After the analysis of the available bacterial genomes, it has been concluded that over 20,000 potential resistance genes (r genes) are present, fortunately; however, the functional resistance determinants in various microbes are far less in number.5
During the late 1950s and early 1960s, antibiotic resistance to multiple antimicrobial agents was detected, for the very first time, among enteric bacteria namely Salmonella, Shigella, and Escherichia coli. These resistant strains caused huge clinical, economic losses and loss of life, mainly in the developing world. However, in the developed world it was considered a mild health problem restricted to enteric microbes. This misconception changed in the 1970s when it was observed that Neisseria gonorrhoeae and Haemophilus influenzae are resistant to ampicillin, while in the case of Haemophilus it was further reported to resist tetracycline and chloramphenicol as well. Due to the increasing use of antimicrobials, the incidence of resistance accelerated, particularly in the developing world where these drugs were freely accessible without any prescription. Deprived hygiene settings facilitated the transmission of resistance and insignificant health care funds limited access to novel and effective antibiotics.8
Numerous important organizations, like the Centers for Disease Control and Prevention (CDC), Infectious Diseases Society of America, World Economic Forum, and the World Health Organization (WHO) have declared antibiotic resistance to be a “global public health concern.”9,10 The World Health Assembly requested WHO to propose a global action plan to fight the antibiotic resistance problem.11 Another valuable input to the issue has been the recent publication of a book titled “The evolving threat of antimicrobial resistance – options for action.” In the UK, people voted for a government-sponsored prize of £10 million (Longitude Prize challenge) to create novel solutions in combating antibiotic resistance.12,13 In America, by 2015, President Barack Obama under the recommendations of the US President’s Council of Advisors on Science and Technology ordered the National Security Council to draft a comprehensive national action plan to tackle antibiotic resistance.14,15
Global antibiotic resistance shows no signs of decline, though it may perhaps shift direction. The etiology of antibiotic resistance is multifaceted, and its consequences pose an impact across the globe. Numerous attempts have been made to delineate the diverse aspects of antibiotic resistance and possible solutions required to deal with this global challenge have been tried. However, a principally coordinated campaign is lacking, particularly at the political level worldwide.16 Antibiotics played an unprecedented role in social and medical development, and at present, antibiotics are obligatory in all health care systems. The success of modernized medicine, namely organ transplantation, cancer therapy, management of preterm babies, or a surplus of advanced major surgeries, might not have been possible without effective antibiotic treatment to control for bacterial infections. If effective global action plans are not adopted soon, then we might encounter terrible complications for social, medical, and economic prospects. Here, we try to portray the global magnitude, major etiologies, and consequences and identify key areas that require urgent attention.17
The global economic scenario of antibiotic resistance
An estimation of the exact economic impact of resistant bacterial infections is still a huge global challenge. In this matter, measuring the disease distribution associated with antibiotic resistance is a key prerequisite. Antibiotic resistance is a substantial economic burden to the whole world. In the USA alone, antibiotic-resistant pathogen-associated hospital-acquired infections (HAIs) cause 99,000 deaths annually. In 2006, about 50,000 Americans died due to two common HAIs, namely pneumonia and sepsis, costing about $8 billion to the US economy.18 Patients with antibiotic-resistant bacterial infections need to stay in the hospital for at least 13 days, adding an additional 8 million hospital days annually. Costs of up to $29,000 per patient treated with an antibiotic-resistant bacterial infection have been reported. In total, economic losses of about $20 billion have been recorded in the US, while losses of about $35 billion annually have also been recorded in terms of lost productivity due to antibiotic resistance in health care systems.15
According to the analysts of Research and Development Corporation, a US nonprofit global organization, a worst-case scenario may evolve in the coming future where the world might be left without any potent antimicrobial agent to treat bacterial infections. In this situation, the global economic burden would be about $120 trillion ($3 trillion per annum), which is approximately equal to the total existing annual budget of the US health care. In general, the world population would be hugely affected: as of the year 2050, about 444 million people would succumb to infections and birthrates would rapidly decline in this scenario.19,20 These losses are calamitous, but due to data constraints like the inclusion of overall conditions and diseases susceptible to resistance these figures represent incomplete images of the economic costs of antibiotic resistance. Another very significant trait of AMR that was absent in the investigation is the use of antibiotics in livestock and food industry. It is an active participant in the increasing AMR and possibly brings its own expected economic losses. A malpractice of the use of antimicrobials as growth promoters is also found in many developing countries. Since 2006, this practice has been banned in the European Union.21,22
Current estimates of resistance-related costs have limited scope and do not consider the broader social values of antibiotics. These are predisposing factors which steer inaccuracy in the estimation of the actual economic burden that the world is facing due to this issue. To get the precise estimation of the economic ramifications caused, prospective studies should employ macroeconomic methods, which consider all the effects of escalating antibiotic resistance especially the reduction in effectiveness of various antibiotics in modernized medicine. Until we address these issues, the exact estimate of the global economic burden of antibiotic resistance may not be fully calculated.15,23
Antibiotic discovery and supremacy
In various parts of the world, from Greece to China, control of microbial infection has been well documented in the past. In 1928, the discovery of penicillin opened the door to the modern era of medicine. Ever since these “magic bullets” have transformed medicine and saved countless lives. In the 1940s, the very first prescription of antibiotics was made. During World War II, penicillin was considered as the drug of choice and used to control bacterial infections among the armed forces.24,25 Unfortunately, soon after that resistance against penicillin turned out to be a significant clinical concern. So, most of the advancements made in the 1940s were threatened during the 1950s. However, to maintain the use of antibiotic treatment strategies, scientists soon discovered and developed novel β-lactam antibiotics. Howbeit, the first case of methicillin-resistant Staphylococcus aureus (MRSA) was reported in the UK and the US in 1962 and 1968, respectively, the same decade when new antibiotics were applied.24,26
Regrettably, with the passage of time, resistance has been observed to almost all the antibiotics developed during that period. Later in 1972, an antibiotic called “vancomycin” was presented to the clinicians to treat MRSA.24 During that period, it was believed that development of vancomycin resistance was unlikely to occur in clinical settings. However, during the late 1970s and early 1980s few cases of coagulase-negative vancomycin-resistant staphylococci were reported. Until the early 1980s, the pharmaceutical industry developed and introduced many new antibiotics to resolve the resistance issue, and with the passage of time, the pace of antibiotic development staggered, so very few new antibiotics were introduced. Consequently in 2015, after ~70 years since the first patient was treated with antibiotic, bacterial infection became a serious threat to life once again.24,26
Although at present antibiotic resistance emerges as a global threat, there are certain breakthroughs antibiotics have handed to humankind, especially in medicine and surgery. Antibiotics have treated or prevented bacterial infections that may occur during chemotherapy or major surgeries like joint replacement, organ transplant, or cardiac surgery. These magic bullets have also changed the outcome of bacterial infections which extended the average life expectancy. During 1920, expected life spans were 56.4 years in the US; on the other hand, at present, the average life span in the US is about 80 years. Antibiotics have had a parallel useful impact across the globe, such as in the developing world with deprived public health infrastructure, as antibiotics reduce the morbidity and mortality rate caused by bacterial infections in humans and livestock alike.8,25,27
The emergence of antibiotic resistance
Microorganisms underwent Darwinian selection to develop some stringent mechanisms to escape the lethal effects of antimicrobial substances. Naturally, most of the antibiotics are produced by microbes like saprophytic bacteria or environmental fungi, others are modified synthetic antibiotics, whereas a few are completely synthetic like fluoroquinolones and sulphonamides.28 Various organisms evolved defensive phenomena against them such as alterations in the target site, inhibition of drug entry or distribution, and enzyme production that may degrade the antimicrobials. For that reason, antibiotic resistance may simply depict the Darwinian competition from natural microbial-derived antimicrobial elements.29,30 The functional meta-genomic analysis of soil microbes has revealed an extensive diversity in genetic determinants associated with antibiotic resistance. Although very little has been reported in human pathogens about this aspect, production of enzymes (β-lactamases) is an exceptional example as a naturally present resistance mechanism and has an impact on human health.31,32
It is considered that various antimicrobial molecules produced by saprophytic bacteria impede the growth of other organisms present in that environment, a mutual benefit in such environmental settings, although few findings proposed a more complicated interaction. Primarily, it is suggested that the concentration of antimicrobial substances in the soil appears much lower, and may not be able to inhibit the growth of neighboring bacteria.28,29 Second, available data suggest that antimicrobial substances with sublethal concentrations have a significant impact on microbial physiology, microbial evolution, and they may act as effective signaling molecules which may induce host or microbial gene expression.32–34 Another important query is that few saprophytic bacteria produce a vital type of broad spectrum antibiotic known as carbapenems. Various genes involved in carbapenems synthesis may also play a part in biofilm formation or quorum sensing. These findings introduce more queries about the unexpected impacts of these antibiotics. This emergence of resistance is not limited to natural antimicrobials only, it occurs against synthetic antimicrobials too.33
Causes of antibiotic resistance
At present, the multifaceted etiology of antibiotic resistance has many factors which are at play. These include inadequate regulations and usage imprecisions, awareness deficiency in best practices which steers undue or inept use of antibiotics, use of antibiotics as a poultry and livestock growth promoter rather than to control infection, and online marketing which made the unrestricted availability of low-grade antibiotics very accessible.20,26
Primarily, overuse of antibiotics is the principal cause of resistance evolution, as it was also warned by Sir Alexander Fleming that “public will demand [the drug and] then will begin an era … of abuses.” Antibiotics kill sensitive bacteria but allow resistant pathogens to remain which then reproduce and thrive through natural selection. Although overuse of antibiotics is strongly discouraged, there remains overprescription across the globe. Several studies have revealed that treatment indications, agent choice, and antibiotic therapy duration are inappropriate in 30%–50% of the cases.15,35,36 Globally, antibiotics are used as a growth promoter in livestock. According to an estimate, about 80% of the antibiotics are sold in the US only for use as growth supplements and to control infection in animals. In another study, a global map of 228 countries was drawn which depicted the consumption of antibiotics in livestock; it was estimated that the total antibiotic consumption was 63,151 tons in 2010.6 van Boeckel et al have also projected a 67% rise in antibiotic consumption by 2030 that would approximately double in Brazil, Russia, India, China, and South Africa block of the rapidly developing and highly populated countries of the world.6
Drivers of antibiotic resistance transmission
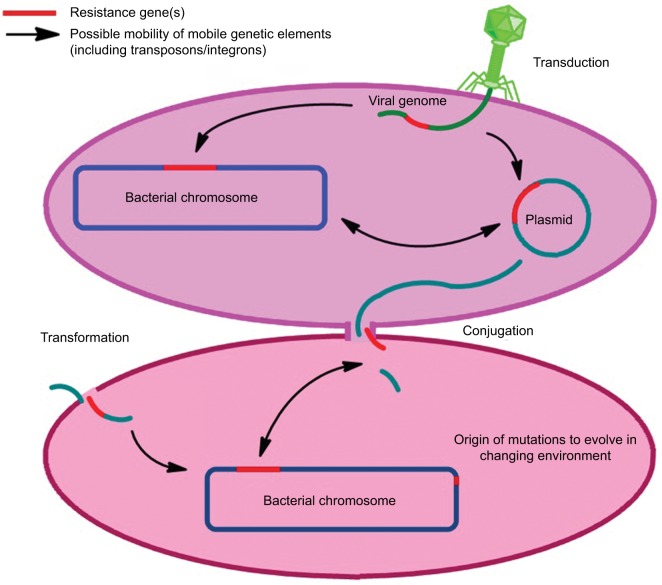
At present, the understanding of various drivers of antibiotic resistance is the key to addressing this global concern. The occurrence of resistance in microbes is a natural process (Figure 1); until now, antibiotic resistance selection is driven using different antibiotics in health care systems, environment, and in agriculture/livestock. Additional important factors which are potent drivers of antibiotic resistance include sanitation settings, infection control standards, water hygiene systems, drug quality, diagnostics and therapeutics, and travel or migration quarantine. In addition to the mutation in various genes residing on the chromosome of the microorganism, exchange of genetic material between organisms plays a vital role in the distribution of antibiotic resistance.28 Plasmid transmission is the most important phenomenon which may transfer genes of antibiotic resistance to the host cell. Antibiotics may influence this process by inducing the transmission of resistance elements; these antimicrobials may additionally exert a selective pressure to the emergence of resistance.37,38 Demonstration of transmission dynamics of resistance has increased the awareness and understanding of how resistant pathogens transmit from human to human.17 At the community level, feco–oral route is the most important route of transmission especially for resistant pathogens of the family Enterobacteriaceae, usually due to sanitation failure. Community-acquired (CA)-MRSA is also a good example to understand the transmission dynamics of resistance at the human–human level, which is usually transmitted due to prolonged hospital stay or unhygienic hospital settings. Sexual contact is also a source of transmission for resistant N. gonorrhoeae.39–41
Figure 1.
Drivers of antibiotic resistance transmission.
Another very important facet known as “one health” also plays a significant role in the transmission dynamics of antibiotic resistance.28 Irrational use of antimicrobial growth promoters in farm animals is associated with the transmission of resistance to humans via animal products; important pathogens under consideration in this aspect are Salmonella spp. and Campylobacter spp. Additionally, indistinguishable resistant mechanisms have been found in bacteria isolated from humans or animals. Resistant bacteria and mobile genetic elements (MGEs) may make their way from animals to humans through various means.42–44 The influence of the environment toward antibiotic resistance is also a concern. In the agriculture sector, metals are used as microbicides and may contribute critically to resistance development. The role of sewage systems, pharmaceutical industry pollution, and waste management procedures in human–environment transmission are also well documented. A number of resistant pathogens have been isolated from pre- and posttreatment sewage systems.39,45
Superbugs and superresistance
The word “superbugs” denotes microbes with higher morbidity and mortality rate increased due to several mutations providing resistance to various classes of antibiotics. Therapeutic preferences for these resistant microbes are less, and these are associated with a prolonged stay at hospital and increased economic cost. In certain cases, superresistant strains have developed increased virulence and improved transmissibility as well.5 Due to aberrant use of antibiotics, numerous bacterial human pathogens have evolved into MDR types. MDR Mycobacterium tuberculosis is a prominent example from both the developing and the developed world.46,47 Other severe bacterial infections include Acinetobacter, Burkholderia, Campylobacter, Citrobacter, Clostridium, Enterobacter spp., Enterococcus, E. coli, Haemophilus, Klebsiella, Proteus, Pseudomonas, Salmonella, Serratia, S. aureus, Staphylococcus epidermidis, and Streptococcus pneumoniae.5
S. aureus is considered as the most notorious superbug. It is a nasal commensal of humans and can cause common skin infections. At present, CA-MRSA with increased acquired virulence has emerged as a major community concern. Although most of the CA-MRSA properties are similar to MRSA, CA-MRSA has some additional characteristics as well such as different mec gene clusters and genes encoding the cytotoxic Panton–Valentine leukocidin.48,49
Antibiotic resistance and virulence
Bacteria reside on human skin, mucous membrane, and inside the human body as well. Among these, bacteria include harmless organisms, many of them are useful commensals, and few are even essential. However, some are known pathogens which cause infection; these are competent enough to colonize, invade, and harm the host tissue.50 Pathogenicity is the ability of a bacterium to cause disease, and a pathogen carries a number of factors that facilitate the bacterium to increase their degree of pathogenicity known as virulence. The most important properties which help a pathogen to cause a disease include toxicity and invasiveness. The ultimate sense of balance of a bacterial disease course may be influenced by virulence and the host immune status. Both host and bacteria have coevolved with time, which may be as long as millions of years. Pathogens have amended virulence to get used to the immune system of the host during this time period.51 This is contrary to the evolution of antibiotic resistance which is a relatively recent occurrence mostly in the past five decades after the discovery of antibiotics. It is therefore believed that resistance and virulence have evolved during different eras.52
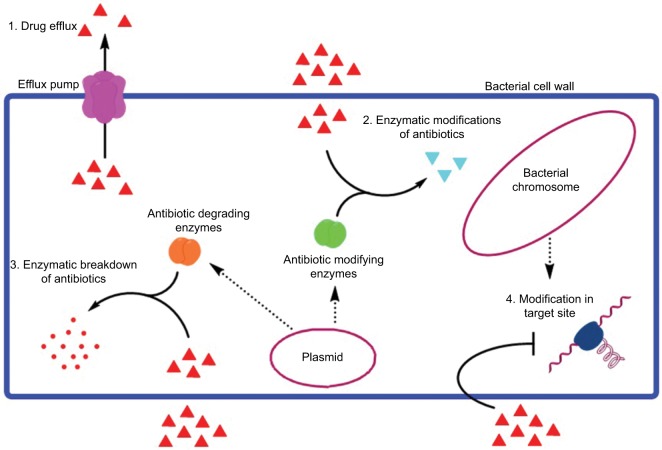
Regardless of evolutionary differences, these processes share common features, and both processes are indispensable for the survival of bacteria under adverse environments. Antibiotic resistance enables bacteria able to overcome therapeutics and virulence is necessary to fight the host immune system.51,53 Second, horizontal gene transfer is the mechanism which both resistance and virulence factors use to transmit the determinants between genera or species. Although other mechanisms like adaptive or compensatory mutations may play their role, the transfer of MGEs might be the key genetic procedure for distribution and coselection of resistance and virulence genes.54,55 Thirdly, in the case of intracellular or biofilm producing bacteria, antibiotic resistance is also linked to virulence.56,57 Additionally, various properties common to resistance and virulence include cell wall modifications, porins (Figure 2), the direct contribution of efflux pumps, and two-component systems that stimulate or inhibit the gene expression involved in virulence and resistance.58–60
Figure 2.
Various mechanisms of antibiotic resistance, including drug efflux with the help of efflux pump, enzymatic modifications of the antibiotic, enzymatic breakdown of the antibiotics, and modification in the target sites.
MDR opportunistic pathogens like Acinetobacter baumannii and Pseudomonas aeruginosa may grow in niches with high antibiotic pressure where several other bacteria may not be able to survive and these pathogens may dislocate the host microbiota as well. This is an example of how antibiotic resistance may improve the virulence or pathogenicity of such pathogens and make these bacteria be able to survive in such niches. Therefore, antibiotic resistance might be considered as a virulence factor, particularly in the case of hospital settings where drug-resistant opportunistic bacteria may cause high morbidity and mortality.61,62
Molecular insights of antibiotic resistance
In the past two decades, acquired MDR infections have increased due to the production of β-lactamases (eg, extended spectrum β-lactamases [ESBLs] enzymes, carbapenemases, and metallo-β-lactamases), leading to third generation cephalosporin and carbapenem resistance.63 Among 21 known AMR genes, the important genes responsible for MDR Salmonella and E. coli are AmpC, bla-TEM-1, bla-CTX-M-15, VIM-1, NDM-1, floR, tetG, and the recent mcr-1 gene coding colistin drug resistance.64–66 Different mechanisms of resistance are described, and novel transmission vectors and genes are identified or characterized on a regular basis. In general, bacteria may use two mechanisms for resistance: 1) intrinsic resistance and 2) acquired resistance.67 The ability of a bacterium to resist the action of a specific antibiotic due to inherent structural or functional properties is known as intrinsic resistance. Pseudomonas displays a very good example of intrinsic resistance mechanism, due to the absence of a susceptible target site for a particular antibiotic. Triclosan which is a broad spectrum biocide especially against Gram-positive bacteria and several Gram-negative bacteria as well is incapable of preventing the growth of Pseudomonas. Initially, it was supposed that active efflux is the reason behind this resistance, but recently the presence of fabI (insensitive allele) encoding an extra enoylacyl carrier protein reductase enzyme, which is a target site for triclosan, has been reported.68,69 Lipopeptide daptomycin, approved in 2003 for clinical practice, is an active drug against Gram-positive bacteria; however, it is ineffective against Gram-negative bacteria. The reason behind this is the intrinsic variation in the cytoplasmic membrane composition. Since the cytoplasmic membrane of Gram-negative bacteria has a lesser proportion of anionic phospholipids as compared to Gram-positive bacteria, the efficacy of the Ca2+-mediated daptomycin insertion into the cytoplasmic membrane, which is necessary for the antibacterial activity of daptomycin, is reduced.70 Additionally, some antibacterial compounds are unable to cross the outer membrane, also considered as a way of intrinsic resistance. In Gram-positive bacteria, vancomycin inhibits peptidoglycan crosslinking by targeting d-Ala-d-Ala peptides while in the case of Gram-negative bacteria, vancomycin cannot pass through the outer membrane. By using high-throughput tools of high-density genome mutant libraries in P. aeruginosa, S. aureus, and E. coli, various genes associated with intrinsic resistance to different classes of antibiotics like β-lactams, aminoglycosides, and fluoroquinolones have been identified in recent times.71,72 As an example, inactivation of redundant E. coli genes identified putative targets for susceptibility phenotyping, like FabI, SapC, RecQ, TrxA (thioredoxin), TrxB (thioredoxin reductase), DacA, and the d-Ala-d-Ala carboxypeptidase. Inhibition of these genes may significantly stimulate drug activity like triclosan, rifampicin, aminoglycosides, nitrofurantoin, and some β-lactams.72 In addition to the intrinsic mechanism of resistance, bacteria may acquire antibiotic resistance as well. Various other mechanisms may also help bacteria to acquire resistance including antibiotic efflux or poor drug penetration resulting in the reduction of the intracellular concentration of antibiotic, modification of the antibiotic target site due to posttranslational target modification or genetic mutation of the target, and inactivation of the antibiotic by modification or hydrolysis.73–76 Plasmid coding colistin-resistant (mcr-1 dependent) E. coli was first isolated from raw meat, animals, and humans in China.77 In Pakistan, mcr-1 harboring E. coli was also reported from migratory birds and from human clinical isolates. MDR (ESBLs, quinolones) E. coli was also reported from poultry farms, wild birds, pigeons, dogs, and humans.78
Therapeutic strategies to combat antibiotic resistance
Control of infection has long been a serious concern of the evolutionary race, regardless of the clinical proof that approaches based on small molecule monotherapy are inadequate in resistance settings. Assessment of various nominees for infection control under developmental process suggests that the focus of research would be on discovery and identification of novels antibiotics. Unfortunately, bio-therapeutics like antibiotics, novel combination remedies, and drug delivery procedures still lag behind the production of novel small molecules that are usually an extension of the present drug classes.79 Approaches of systemic monotherapy where resistance has overtaken drug development are inadequate worldwide. Therefore, instead of combating bacterial evolution, control strategies might serve better by pursuing resistance mechanisms based on genetic inspiration, for example, furanones deployed by red sea algae to disturb the quorum sensing of resistant bacteria.80–82
Though the role of biologics in the control of bacterial infection is in its infancy, its potential to combat MDR cannot be overlooked. Small molecules would always have a key role in the control of infection; however, on the basis of biological inspiration the search for an effective drug candidate may possibly be pursued more logistically.79,83 Various tools used for drug development are essential to handle the shortcomings of biologics, like controlled delivery options, partial in vitro stability, inadequate high-throughput, advanced screening tools, inefficient pharmacokinetics, and comparatively unknown pharmacodynamics, and are not as radical as those accessible for the development of small molecules. By chance, a vast potential subsists between biologics combinations, molecules based on biological inspiration, and drug delivery tools.84,85 Therefore, a paradigm shift like cancer and complex viruses might be adapted to eradicate susceptible bacteria, handle antibiotic resistance, and shield the host microbiota. This type of approach may be crafted through the combination of conventional antibiotics, novel adjuvants, and viable limited delivery strategies. In contrast to various expensive development models which often fail in trials, advanced bioinformatics to determine effective combination delivery and new targets will possibly bring substantial benefits.86
Implementation of various methods of antibiotic resistance is very significant, as it has a substantial impact on humans and livestock. Although the resolutions have been anticipated, with few being thrown to tackle the crisis, to date very little action has been taken toward these solutions.15,87 At present, antibiotics are obligatory in health settings, and the economic, social, and medical costs would be calamitous due to disorganized effort. In the face of rising antibiotic resistance, the antibiotic efficacy looks increasingly bleak. Epidemiological trends in MDR bacteria must be considered in developing treatment strategies for infection control. Optimistically, constant highlighting of the problem would encourage researchers to adopt novel strategies to control bacterial infection with the following criteria: discovery of novel antibiotics, anticipation and prevention of antibiotic resistance, and protection of host microbiota. Preeminent approaches that come across with this criterion would help the formation of novel combination strategies together with smart and local delivery tools, for example, directed nanoparticles, liposomes, and infection-responsive polymer-controlled delivery. Alternative approaches to antibiotics that are presently in clinical trials are listed in Table 1.
Table 1.
Various product types as alternatives to antibiotics for the therapeutic management of bacterial infections
| Target bacteria | Product name | Product type | Company | References |
|---|---|---|---|---|
| Clostridium difficile | AmpliPhage-004 | Bacteriophages | AmpliPhi Biosciences | 113 |
| Phenylbutyrate/vitamin D | Immune stimulator | Akthelia Pharmaceuticals | 114, 115 | |
| C. difficile toxoid vaccine | Vaccine | Sanofi Pasteur | 116 | |
| IC84 | Vaccine | Valneva | 117 | |
| NVB302 | Antimicrobial peptides | Novacta Biosystems | 118 | |
| AP-114 | Antimicrobial peptides | Adenium Biotech | 119 | |
| Bezlotoxumab | Antibodies | Merck | 120, 121 | |
| RBX2660 | Probiotics | Rebiotix | 122 | |
| SER-109 | Probiotics | Seres Therapeutics | 123 | |
| VP20621 | Probiotics | ViroPharma (Shire) | 124 | |
| Staphylococcus aureus | SA4Ag | Vaccines | Pfizer | 125 |
| AP-138 | Antimicrobial peptide | Adenium Biotech | 119 | |
| SAL200 | Lysins | iNtRON Biotechnology | 126 | |
| CF-301 | Lysins | ContraFect | 127 | |
| MEDI4893 | Antibodies | MedImmune | 121, 128 | |
| Salvecin | Antibodies | Aridis Pharmaceuticals | 129 | |
| 514G3 | Antibodies | XBiotech | 130 | |
| Pseudomonas aeruginosa | AmpliPhage-001 | Bacteriophages | AmpliPhi Biosciences | 131 |
| PT-3.1 | Bacteriophages | Phico Therapeutics | 132 | |
| IC43 | Vaccines | Valneva | 133 | |
| Murepavadin | Antimicrobial peptides | Roche | 134 | |
| Aerumab | Antibodies | Aridis Pharmaceuticals | 135 | |
| Aerucin | Antibodies | Aridis Pharmaceuticals | 136 | |
| MEDI3902 | Antibodies | MedImmune | 137 |
The use of nanotechnology is emerging in medicine and it is not astounding to see these technologies being applied for the management of the antibiotic resistance menace. Nanoparticles can be utilized for the therapeutic management of infections in different ways. They can be coupled with existing antimicrobial agents for enhancement of their physiochemical behavior against drug-resistant microbes. Second, the colloidal forms of zinc, silver, copper, and titanium can itself be used as antimicrobial agents. Although the principal targets of antibiotics include the inhibition or disruption of bacterial cell wall, inhibition of proteins, and nucleic acids synthesis, nanoparticles are reported to affect the respiration system and thereby generation of reactive oxygen species that ultimately leads to bacterial death. Nanoparticles also target the bacterial cell wall; therefore, for example, silver nanoparticles can be coupled with the relevant antibiotics to enhance their antibacterial action through synergy.88,89
Antimicrobial peptides (AMPs) are emerging antimicrobial agents found in animals, microorganisms, and plants and have a broader spectrum especially active against different bacteria, fungi, and protozoans. The amphipathic nature of AMPs enables interaction and insertion into the cell wall and cellular membranes of microbes.90 Although AMPs usually demonstrate antimicrobial activity because of damage to the cellular membranes, they can targets other proteins, DNA, RNA, and regulatory enzymes and therefore seem to be a promising substitute for classical antibiotics.91–95 However, the development of resistance to AMPs is expected as soon as AMPs are used in clinical practice; therefore, it is essential to explore molecular mechanisms of their action and better understanding of the resistance against these compounds is necessary for rational planning of the use of AMPs as an alternate to antibiotics.96,97
Global action plan to control the menace of antibiotic resistance
The global burden of AMR has no signs of receding, rather it piles up the pressure on human and veterinary medicine. Akin to global warming, AMR is an ecological calamity of indefinite magnitude and has no apparent way out. Until the first decade of the current century, scientists and clinicians were not up-to-the-minute about AMR, whereas resistant bacteria had been identified before the discovery of penicillin.98 Consequently, WHO made a landmark by declaring and promoting AMR as a global health concern. The agenda for such global health concerns is at the developmental stages; for example, a book named “The evolving threat of antimicrobial resistance – options for action” is a valuable addition to the archive.99
Few countries across the globe reported the control of various resistant pathogens especially MRSA, but reasons behind the decline in the resistance of these microorganisms are disputed. For example, the apparent success to confine MRSA attained in the US by practicing various schemes of hygiene and screenings has been thrown aside because some supplementary factors like overall national decline in the prevalence of MRSA may be a strong reason of MRSA control.100 In Europe, the same actions have also been significantly found at the national level. Scientific fraternity must appreciate such arguments because we are still struggling to recognize the main interventions prerequisite for AMR control. Perhaps, the eradication of AMR is unattainable but its progression may be decelerated.101,102 Different global agencies such as the Global Antimicrobial Resistance Surveillance System under WHO should work for the control of antibiotic resistance; Food and Agriculture Organization, CDC, and Office International des Epizooties are making strong efforts to control antibiotic resistance. Other programs dealing with the global threat of antibiotic resistance include the Global Health Security Agenda (GHSA), Antimicrobial Resistance Action Package (GHSA Action Package Prevent-1).103,104
The fallacy about the enormity and peril of AMR mapped the current epidemic. Additionally, studies showing the burden of AMR influenced the think tanks to peruse the issue with urgency. Countries that crafted inclusive national plans remain successful in controlling AMR. These approaches include cautious use of antibiotics, surveillance of antibiotics by employing the “One Health Approach,” advancement of health care setup, development of health insurance policies, restricted drug promotion, consistent disease control strategies, and stewardship plans in the community.17 On the other hand, these strategies demand patience and time to be organized. Furthermore, these require a comprehensive endorsement from the government authorities with ample funds. In developing countries, progress is at a very low pace, although some Asian countries including India, in particular, have taken some courageous steps toward AMR control in recent times, for example, the Chennai declaration.105
Diagnostics pose a significant impact in the setting up of AMR because it drives clinicians to use antibiotics imprudently. Diagnostics is a pressing issue especially in developing countries because they still rely on conventional microbiological tools to identify bacteria. These gaps could be bridged by developing personalized medicine established based on new and efficient molecular diagnostic tools that would be very helpful to identify patients who actually need antibiotic therapy. One Health Approach could be a very useful tool to study the human–animal interface and guide researchers to design novel screening tools. This subject is of unique importance and must be taken up earnestly because the transmission pathway of AMR between all components (human, animal, and environment) of one health has been established.17,106
Unwise and irrational use of antibiotics is also a salient factor associated with AMR especially in low and middle income countries (LMICs). Antibiotics are used imprudently due to many reasons, such as patient’s contentment over a prescription of the physician, inappropriate information about antibiotics, improper diagnosis, quackery especially in developing countries, and sinfully, temptations of the pharmaceutical industry for the physicians. Having a dearth of new antibiotics, it is quite difficult to confront this portion of the problem.107 Innovation in antibiotic development, combination therapy, and technological development are indispensable.108 Another aspect of colossal importance in this regard is the integration of the health care system and antibiotic stewardships. Data about the impact of AMR on health and economy of developing countries are inadequate. LMICs have a dire situation in terms of clinical outcomes of many fatal bacterial diseases like newborn sepsis, typhoid, etc. So, the task is not simply to use conventional options but some customized control policies should also be applied; for example, resistant pathogens should be prioritized like MDR M. tuberculosis. Up to the tertiary level, WHO recommendations on infection and disease control should be followed. Comprehensive data should be collected by taking all the national stakeholders including government, academia, and industry on board. Additionally, the industry–academia interface should be improved as both academia and industry have made some fundamental mistakes in the past. First, in the late 20th century, academia and researchers had a mistaken belief about the elimination of infectious diseases and industry-produced antibiotics enormously. Second, although financial incentives in the form of research grants were provided to the academia and pharmaceutical industry, an inadequate proportion was allocated to academia and industries for the generation of sufficient novel classes of antimicrobial agents as compared to the surge of antibiotic resistance.109 In atonement, a group of experts named as Infectious Disease Society of America submitted a 10×20 plan; this was about the development of 10 novel safe and effective antibiotics by the end of 2020.110
Use of alternative therapies for the treatment and control of infectious diseases could also be a different and a prolific option to fend off AMR. These therapies include anti-virulence strategies (to play with the virulence factors of bacteria), biological therapies (use of monoclonal antibodies, insulin, erythropoietin, etc), and vaccines (vaccines against MRSA, MDR M. tuberculosis).111,112 Moreover, herbal medicines have elusive properties; there is a strong viewpoint that it could be a practicable alternate option.
Conclusion
We should seek to increase our knowledge regarding the extent of the AMR issue. Comprehensive, unfailing data collection is crucial in AMR control. Current angst about this issue is due to inadequate information. One cannot predict the future scenario with surety at this stage but with the scarcity of novel antibiotics, the control of AMR seems very difficult. Multipronged strategies should be adopted to confront this issue. Constant and refreshing education of medical students, physicians, and pharmacists is required. Regulations should be implemented with strict monitoring of antibiotic use as part of the policy. A global and interdisciplinary approach must be considered for the development of new screening and diagnostic tools. Ecological and environmental aspects of the issue need not be ignored; all the elements of “one health” should be part and parcel of the control policy. Alternative strategies may also play a fruitful role, especially in developing countries. Current global interest indicates that AMR is not an unheeded issue anymore. Albeit this attention is not itself adequate to combat AMR, a global code of conducts implementing all the options of action against AMR might eliminate AMR in the future. Alternatives to antibiotics such as probiotics and lytic bacteriophages can help to decrease the burden of AMR globally. The spread and sharing of AMR can be contained by the rational use of antibiotics, infection control, immunization, promoting good practices in food supply, and control of person-to-person spread by screening, treatment, awareness, and education. At the national, regional, and global levels, the tracking, bio-surveillance, and response and prevention strategies of AMR and MDR pathogens may help to control the “global resistome.”
Availability of data and materials
The aggregate data supporting findings contained within this manuscript will be shared upon request submitted to the corresponding author. Identifying patient data will not be shared.
Acknowledgments
We thank Dr Barry Bradford and Omar Alfituri of The Roslin Institute, The University of Edinburgh for proof reading, English style editing, and useful suggestions. This study was supported by a grant from the China Postdoc Fund. The funding body had no role in the design of the study, collection, analysis, interpretation of data, or writing of the manuscript.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Singer AC, Shaw H, Rhodes V, Hart A. Review of antimicrobial resistance in the environment and its relevance to environmental regulators. Front Microbiol. 2016;7:1728. doi: 10.3389/fmicb.2016.01728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marshall BM, Levy SB. Food animals and antimicrobials: impacts on human health. Clin Microbiol Rev. 2011;24(4):718–733. doi: 10.1128/CMR.00002-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nathan C, Cars O. Antibiotic resistance-problems, progress, and prospects. N Engl J Med. 2014;371(19):1761–1763. doi: 10.1056/NEJMp1408040. [DOI] [PubMed] [Google Scholar]
- 4.Nathan C. Antibiotics at the crossroads. Nature. 2004;431(7011):899–902. doi: 10.1038/431899a. [DOI] [PubMed] [Google Scholar]
- 5.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74(3):417–433. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Boeckel TP, Brower C, Gilbert M, et al. Global trends in antimicrobial use in food animals. Proc Natl Acad Sci U S A. 2015;112(18):5649–5654. doi: 10.1073/pnas.1503141112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roca I, Akova M, Baquero F, et al. The global threat of antimicrobial resistance: science for intervention. New Microbes New Infect. 2015;6:22–29. doi: 10.1016/j.nmni.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rossolini GM, Arena F, Pecile P, Pollini S. Update on the antibiotic resistance crisis. Curr Opin Pharmacol. 2014;18:56–60. doi: 10.1016/j.coph.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 9.Michael CA, Dominey-Howes D, Labbate M. The antimicrobial resistance crisis: causes, consequences, and management. Front Public Health. 2014;2:145. doi: 10.3389/fpubh.2014.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spellberg B, Srinivasan A, Chambers HF. New societal approaches to empowering antibiotic stewardship. JAMA. 2016;315(12):1229–1230. doi: 10.1001/jama.2016.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffman SJ, Caleo GM, Daulaire N, et al. Strategies for achieving global collective action on antimicrobial resistance. Bull World Health Organ. 2015;93(12):867–876. doi: 10.2471/BLT.15.153171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Payne DJ, Miller LF, Findlay D, Anderson J, Marks L. Time for a change: addressing R&D and commercialization challenges for antibacterials. Philos Trans R Soc Lond B Biol Sci. 2015;370(1670):20140086. doi: 10.1098/rstb.2014.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luepke KH, Mohr JF. The antibiotic pipeline: reviving research and development and speeding drugs to market. Expert Rev Anti Infect Ther. 2017;15(5):425–433. doi: 10.1080/14787210.2017.1308251. [DOI] [PubMed] [Google Scholar]
- 14.Landers T, Kavanagh KT. Is the Presidential Advisory Council on Combating Antibiotic Resistance missing opportunities? Am J Infect Control. 2016;44(11):1356–1359. doi: 10.1016/j.ajic.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 15.Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. P T. 2015;40(4):277–283. [PMC free article] [PubMed] [Google Scholar]
- 16.Prestinaci F, Pezzotti P, Pantosti A. Antimicrobial resistance: a global multifaceted phenomenon. Pathog Glob Health. 2015;109(7):309–318. doi: 10.1179/2047773215Y.0000000030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laxminarayan R, Duse A, Wattal C, et al. Antibiotic resistance-the need for global solutions. Lancet Infect Dis. 2013;13(12):1057–1098. doi: 10.1016/S1473-3099(13)70318-9. [DOI] [PubMed] [Google Scholar]
- 18.Guidos RJ. Combating antimicrobial resistance: policy recommendations to save lives. Clin Infect Dis. 2011;52(Suppl 5):S397–S428. doi: 10.1093/cid/cir153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gould IM, Bal AM. New antibiotic agents in the pipeline and how they can help overcome microbial resistance. Virulence. 2013;4(2):185–191. doi: 10.4161/viru.22507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartlett JG, Gilbert DN, Spellberg B. Seven ways to preserve the miracle of antibiotics. Clin Infect Dis. 2013;56(10):1445–1450. doi: 10.1093/cid/cit070. [DOI] [PubMed] [Google Scholar]
- 21.Economou V, Gousia P. Agriculture and food animals as a source of antimicrobial-resistant bacteria. Infect Drug Resist. 2015;8:49–61. doi: 10.2147/IDR.S55778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maron DF, Smith TJ, Nachman KE. Restrictions on antimicrobial use in food animal production: an international regulatory and economic survey. Global Health. 2013;9:48. doi: 10.1186/1744-8603-9-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Founou LL, Founou RC, Essack SY. Antibiotic resistance in the food chain: a developing country-perspective. Front Microbiol. 2016;7:1881. doi: 10.3389/fmicb.2016.01881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sengupta S, Chattopadhyay MK, Grossart HP. The multifaceted roles of antibiotics and antibiotic resistance in nature. Front Microbiol. 2013;4:47. doi: 10.3389/fmicb.2013.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piddock LJ. The crisis of no new antibiotics – what is the way forward? Lancet Infect Dis. 2012;12(3):249–253. doi: 10.1016/S1473-3099(11)70316-4. [DOI] [PubMed] [Google Scholar]
- 26.Spellberg B, Gilbert DN. The future of antibiotics and resistance: a tribute to a career of leadership by John Bartlett. Clin Infect Dis. 2014;59(Suppl 2):S71–S75. doi: 10.1093/cid/ciu392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wright GD. Something old, something new: revisiting natural products in antibiotic drug discovery. Can J Microbiol. 2014;60(3):147–154. doi: 10.1139/cjm-2014-0063. [DOI] [PubMed] [Google Scholar]
- 28.Holmes AH, Moore LS, Sundsfjord A, et al. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet. 2016;387(10014):176–187. doi: 10.1016/S0140-6736(15)00473-0. [DOI] [PubMed] [Google Scholar]
- 29.Forsberg KJ, Patel S, Gibson MK, et al. Bacterial phylogeny structures soil resistomes across habitats. Nature. 2014;509(7502):612–616. doi: 10.1038/nature13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aminov RI. The role of antibiotics and antibiotic resistance in nature. Environ Microbiol. 2009;11(12):2970–2988. doi: 10.1111/j.1462-2920.2009.01972.x. [DOI] [PubMed] [Google Scholar]
- 31.Martinez JL. The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proc Biol Sci. 2009;276(1667):2521–2530. doi: 10.1098/rspb.2009.0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Andersson DI, Hughes D. Microbiological effects of sublethal levels of antibiotics. Nat Rev Microbiol. 2014;12(7):465–478. doi: 10.1038/nrmicro3270. [DOI] [PubMed] [Google Scholar]
- 33.Morita Y, Tomida J, Kawamura Y. Responses of Pseudomonas aeruginosa to antimicrobials. Front Microbiol. 2014;4:422. doi: 10.3389/fmicb.2013.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khurshid M, Rasool MH, Ashfaq UA, Aslam B, Waseem M. Emergence of ISAba1 harboring carbapenem-resistant Acinetobacter baumannii isolates in Pakistan. Future Microbiol. 2017;12:1261–1269. doi: 10.2217/fmb-2017-0080. [DOI] [PubMed] [Google Scholar]
- 35.Read AF, Woods RJ. Antibiotic resistance management. Evol Med Public Health. 2014;147(1) doi: 10.1093/emph/eou024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lushniak BD. Antibiotic resistance: a public health crisis. Public Health Rep. 2014;129(4):314–316. doi: 10.1177/003335491412900402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munita JM, Arias CA. Mechanisms of antibiotic resistance. Microbiol Spectr. 2016;4(2) doi: 10.1128/microbiolspec.VMBF-0016-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stokes HW, Gillings MR. Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram-negative pathogens. FEMS Microbiol Rev. 2011;35(5):790–819. doi: 10.1111/j.1574-6976.2011.00273.x. [DOI] [PubMed] [Google Scholar]
- 39.Wellington EM, Boxall AB, Cross P, et al. The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect Dis. 2013;13(2):155–165. doi: 10.1016/S1473-3099(12)70317-1. [DOI] [PubMed] [Google Scholar]
- 40.Lewis DA. The role of core groups in the emergence and dissemination of antimicrobial-resistant N. gonorrhoeae. Sex Transm Infect. 2013;89(Suppl 4):47–51. doi: 10.1136/sextrans-2013-051020. [DOI] [PubMed] [Google Scholar]
- 41.Chamchod F, Ruan S. Modeling methicillin-resistant Staphylococcus aureus in hospitals: transmission dynamics, antibiotic usage and its history. Theor Biol Med Model. 2012;9:25. doi: 10.1186/1742-4682-9-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anderson ES, Lewis MJ. Drug resistance and its transfer in Salmonella typhimurium. Nature. 1965;206(984):579–583. doi: 10.1038/206579a0. [DOI] [PubMed] [Google Scholar]
- 43.Molbak K. Spread of resistant bacteria and resistance genes from animals to humans – the public health consequences. J Vet Med B Infect Dis Vet Public Health. 2004;51(8–9):364–369. doi: 10.1111/j.1439-0450.2004.00788.x. [DOI] [PubMed] [Google Scholar]
- 44.Humphrey TJ, Jorgensen F, Frost JA, et al. Prevalence and subtypes of ciprofloxacin-resistant Campylobacter spp. in commercial poultry flocks before, during, and after treatment with fluoroquinolones. Antimicrob Agents Chemother. 2005;49(2):690–698. doi: 10.1128/AAC.49.2.690-698.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kristiansson E, Fick J, Janzon A, et al. Pyrosequencing of antibiotic-contaminated river sediments reveals high levels of resistance and gene transfer elements. PLoS One. 2011;6(2):e17038. doi: 10.1371/journal.pone.0017038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shah NS, Wright A, Bai GH, et al. Worldwide emergence of extensively drug-resistant tuberculosis. Emerg Infect Dis. 2007;13(3):380–387. doi: 10.3201/eid1303.061400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sotgiu G, Ferrara G, Matteelli A, et al. Epidemiology and clinical management of XDR-TB: a systematic review by TBNET. Eur Respir J. 2009;33(4):871–881. doi: 10.1183/09031936.00168008. [DOI] [PubMed] [Google Scholar]
- 48.Deleo FR, Chambers HF. Reemergence of antibiotic-resistant Staphylococcus aureus in the genomics era. J Clin Invest. 2009;119(9):2464–2474. doi: 10.1172/JCI38226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watkins RR, David MZ, Salata RA. Current concepts on the virulence mechanisms of methicillin-resistant Staphylococcus aureus. J Med Microbiol. 2012;61(Pt 9):1179–1193. doi: 10.1099/jmm.0.043513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beceiro A, Tomas M, Bou G. Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin Microbiol Rev. 2013;26(2):185–230. doi: 10.1128/CMR.00059-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martinez JL, Baquero F. Interactions among strategies associated with bacterial infection: pathogenicity, epidemicity, and antibiotic resistance. Clin Microbiol Rev. 2002;15(4):647–679. doi: 10.1128/CMR.15.4.647-679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fair RJ, Tor Y. Antibiotics and bacterial resistance in the 21st century. Perspect Medicin Chem. 2014;6:25–64. doi: 10.4137/PMC.S14459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feldman MW, Laland KN. Gene-culture coevolutionary theory. Trends Ecol Evol. 1996;11(11):453–457. doi: 10.1016/0169-5347(96)10052-5. [DOI] [PubMed] [Google Scholar]
- 54.Burrus V, Waldor MK. Shaping bacterial genomes with integrative and conjugative elements. Res Microbiol. 2004;155(5):376–386. doi: 10.1016/j.resmic.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 55.Handel A, Regoes RR, Antia R. The role of compensatory mutations in the emergence of drug resistance. PLoS Comput Biol. 2006;2(10):e137. doi: 10.1371/journal.pcbi.0020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patel R. Biofilms and antimicrobial resistance. Clin Orthop Relat Res. 2005;437:41–47. doi: 10.1097/01.blo.0000175714.68624.74. [DOI] [PubMed] [Google Scholar]
- 57.Seral C, van Bambeke F, Tulkens PM. Quantitative analysis of gentamicin, azithromycin, telithromycin, ciprofloxacin, moxifloxacin, and oritavancin (LY333328) activities against intracellular Staphylococcus aureus in mouse J774 macrophages. Antimicrob Agents Chemother. 2003;47(7):2283–2292. doi: 10.1128/AAC.47.7.2283-2292.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsai YK, Fung CP, Lin JC, et al. Klebsiella pneumoniae outer membrane porins OmpK35 and OmpK36 play roles in both antimicrobial resistance and virulence. Antimicrob Agents Chemother. 2011;55(4):1485–1493. doi: 10.1128/AAC.01275-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Moya B, Juan C, Alberti S, Perez JL, Oliver A. Benefit of having multiple ampD genes for acquiring beta-lactam resistance without losing fitness and virulence in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2008;52(10):3694–3700. doi: 10.1128/AAC.00172-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yeung AT, Bains M, Hancock RE. The sensor kinase CbrA is a global regulator that modulates metabolism, virulence, and antibiotic resistance in Pseudomonas aeruginosa. J Bacteriol. 2011;193(4):918–931. doi: 10.1128/JB.00911-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lye DC, Earnest A, Ling ML, et al. The impact of multidrug resistance in healthcare-associated and nosocomial Gram-negative bacteraemia on mortality and length of stay: cohort study. Clin Microbiol Infect. 2012;18(5):502–508. doi: 10.1111/j.1469-0691.2011.03606.x. [DOI] [PubMed] [Google Scholar]
- 62.Sohail M, Rashid A, Aslam B, et al. Antimicrobial susceptibility of Acinetobacter clinical isolates and emerging antibiogram trends for nosocomial infection management. Rev Soc Bras Med Trop. 2016;49(3):300–304. doi: 10.1590/0037-8682-0111-2016. [DOI] [PubMed] [Google Scholar]
- 63.Blair JM, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJ. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 2015;13(1):42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- 64.Voulgari E, Poulou A, Koumaki V, Tsakris A. Carbapenemase-producing Enterobacteriaceae: now that the storm is finally here, how will timely detection help us fight back? Future Microbiol. 2013;8(1):27–39. doi: 10.2217/fmb.12.130. [DOI] [PubMed] [Google Scholar]
- 65.Johnson AP, Woodford N. Global spread of antibiotic resistance: the example of New Delhi metallo-beta-lactamase (NDM)-mediated carbapenem resistance. J Med Microbiol. 2013;62(Pt 4):499–513. doi: 10.1099/jmm.0.052555-0. [DOI] [PubMed] [Google Scholar]
- 66.Woodford N, Turton JF, Livermore DM. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev. 2011;35(5):736–755. doi: 10.1111/j.1574-6976.2011.00268.x. [DOI] [PubMed] [Google Scholar]
- 67.Lynch JP, 3rd, Clark NM, Zhanel GG. Evolution of antimicrobial resistance among Enterobacteriaceae (focus on extended spectrum beta-lactamases and carbapenemases) Expert Opin Pharmacother. 2013;14(2):199–210. doi: 10.1517/14656566.2013.763030. [DOI] [PubMed] [Google Scholar]
- 68.Chuanchuen R, Karkhoff-Schweizer RR, Schweizer HP. High-level triclosan resistance in Pseudomonas aeruginosa is solely a result of efflux. Am J Infect Control. 2003;31(2):124–127. doi: 10.1067/mic.2003.11. [DOI] [PubMed] [Google Scholar]
- 69.Zhu L, Lin J, Ma J, Cronan JE, Wang H. Triclosan resistance of Pseudomonas aeruginosa PAO1 is due to FabV, a triclosan-resistant enoylacyl carrier protein reductase. Antimicrob Agents Chemother. 2010;54(2):689–698. doi: 10.1128/AAC.01152-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Randall CP, Mariner KR, Chopra I, O’Neill AJ. The target of daptomycin is absent from Escherichia coli and other Gram-negative pathogens. Antimicrob Agents Chemother. 2013;57(1):637–639. doi: 10.1128/AAC.02005-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Blake KL, O’Neill AJ. Transposon library screening for identification of genetic loci participating in intrinsic susceptibility and acquired resistance to antistaphylococcal agents. J Antimicrob Chemother. 2013;68(1):12–16. doi: 10.1093/jac/dks373. [DOI] [PubMed] [Google Scholar]
- 72.Liu A, Tran L, Becket E, et al. Antibiotic sensitivity profiles determined with an Escherichia coli gene knockout collection: generating an antibiotic bar code. Antimicrob Agents Chemother. 2010;54(4):1393–1403. doi: 10.1128/AAC.00906-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Floyd JL, Smith KP, Kumar SH, Floyd JT, Varela MF. LmrS is a multidrug efflux pump of the major facilitator superfamily from Staphylococcus aureus. Antimicrob Agents Chemother. 2010;54(12):5406–5412. doi: 10.1128/AAC.00580-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rm H, Liao ST, Huang CC, Huang YW, Yang TC. An inducible fusaric acid tripartite efflux pump contributes to the fusaric acid resistance in Stenotrophomonas maltophilia. PLoS One. 2012;7(12):e51053. doi: 10.1371/journal.pone.0051053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim C, Mwangi M, Chung M, Milheirico C, de Lencastre H, Tomasz A. The mechanism of heterogeneous beta-lactam resistance in MRSA: key role of the stringent stress response. PLoS One. 2013;8(12):e82814. doi: 10.1371/journal.pone.0082814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ogawa W, Onishi M, Ni R, Tsuchiya T, Kuroda T. Functional study of the novel multidrug efflux pump KexD from Klebsiella pneumoniae. Gene. 2012;498(2):177–182. doi: 10.1016/j.gene.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 77.Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 78.Mohsin M, Raza S, Roschanski N, Guenther S, Ali A, Schierack P. Description of the first Escherichia coli clinical isolate harboring the colistin resistance gene mcr-1 from the Indian subcontinent. Antimicrob Agents Chemother. 2017;61(1) doi: 10.1128/AAC.01945-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Brooks BD, Brooks AE. Therapeutic strategies to combat antibiotic resistance. Adv Drug Deliv Rev. 2014;78:14–27. doi: 10.1016/j.addr.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 80.Pena-Miller R, Laehnemann D, Jansen G, et al. When the most potent combination of antibiotics selects for the greatest bacterial load: the smile-frown transition. PLoS Biol. 2013;11(4):e1001540. doi: 10.1371/journal.pbio.1001540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Payne DJ, Gwynn MN, Holmes DJ, Pompliano DL. Drugs for bad bugs: confronting the challenges of antibacterial discovery. Nat Rev Drug Discov. 2007;6(1):29–40. doi: 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- 82.Kalia VC, Wood TK, Kumar P. Evolution of resistance to quorum-sensing inhibitors. Microb Ecol. 2014;68(1):13–23. doi: 10.1007/s00248-013-0316-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Baveja JK, Li G, Nordon RE, et al. Biological performance of a novel synthetic furanone-based antimicrobial. Biomaterials. 2004;25(20):5013–5021. doi: 10.1016/j.biomaterials.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 84.Choi S, Isaacs A, Clements D, et al. De novo design and in vivo activity of conformationally restrained antimicrobial arylamide foldamers. Proc Natl Acad Sci U S A. 2009;106(17):6968–6973. doi: 10.1073/pnas.0811818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Scott RW, Degrado WF, Tew GN. De novo designed synthetic mimics of antimicrobial peptides. Curr Opin Biotechnol. 2008;19(6):620–627. doi: 10.1016/j.copbio.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Weber T. In silico tools for the analysis of antibiotic biosynthetic pathways. Int J Med Microbiol. 2014;304(3–4):230–235. doi: 10.1016/j.ijmm.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 87.Littmann J, Viens AM. The ethical significance of antimicrobial resistance. Public Health Ethics. 2015;8(3):209–224. doi: 10.1093/phe/phv025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kumar M, Curtis A, Hoskins C. Application of nanoparticle technologies in the combat against anti-microbial resistance. Pharmaceutics. 2018;10(1) doi: 10.3390/pharmaceutics10010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang L, Hu C, Shao L. The antimicrobial activity of nanoparticles: present situation and prospects for the future. Int J Nanomedicine. 2017;12:1227–1249. doi: 10.2147/IJN.S121956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brown KL, Hancock RE. Cationic host defense (antimicrobial) peptides. Curr Opin Immunol. 2006;18(1):24–30. doi: 10.1016/j.coi.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 91.Forde E, Devocelle M. Pro-moieties of antimicrobial peptide prodrugs. Molecules. 2015;20(1):1210–1227. doi: 10.3390/molecules20011210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nguyen LT, Haney EF, Vogel HJ. The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 2011;29(9):464–472. doi: 10.1016/j.tibtech.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 93.Dangel A, Ackermann N, Abdel-Hadi O, et al. A de novo-designed antimicrobial peptide with activity against multiresistant Staphylococcus aureus acting on RsbW kinase. FASEB J. 2013;27(11):4476–4488. doi: 10.1096/fj.13-234575. [DOI] [PubMed] [Google Scholar]
- 94.Hao G, Shi YH, Tang YL, Gw L. The intracellular mechanism of action on Escherichia coli of BF2-A/C, two analogues of the antimicrobial peptide Buforin 2. J Microbiol. 2013;51(2):200–206. doi: 10.1007/s12275-013-2441-1. [DOI] [PubMed] [Google Scholar]
- 95.Ghosh A, Kar RK, Jana J, et al. Indolicidin targets duplex DNA: structural and mechanistic insight through a combination of spectroscopy and microscopy. ChemMedChem. 2014;9(9):2052–2058. doi: 10.1002/cmdc.201402215. [DOI] [PubMed] [Google Scholar]
- 96.Maria-Neto S, de Almeida KC, Macedo ML, Franco OL. Understanding bacterial resistance to antimicrobial peptides: from the surface to deep inside. Biochim Biophys Acta. 2015;1848(11 Pt B):3078–3088. doi: 10.1016/j.bbamem.2015.02.017. [DOI] [PubMed] [Google Scholar]
- 97.Parachin NS, Franco OL. New edge of antibiotic development: antimicrobial peptides and corresponding resistance. Front Microbiol. 2014;5:147. doi: 10.3389/fmicb.2014.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Abraham EP, Chain E. An enzyme from bacteria able to destroy penicillin. 1940. Rev Infect Dis. 1988;10(4):677–678. [PubMed] [Google Scholar]
- 99.Gould IM. Antibiotic resistance: understanding how to control it. Int J Antimicrob Agents. 2012;40(3):193–195. doi: 10.1016/j.ijantimicag.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 100.Jain R, Kralovic SM, Evans ME, et al. Veterans Affairs initiative to prevent methicillin-resistant Staphylococcus aureus infections. N Engl J Med. 2011;364(15):1419–1430. doi: 10.1056/NEJMoa1007474. [DOI] [PubMed] [Google Scholar]
- 101.Gurieva T, Bootsma MC, Bonten MJ. Successful Veterans Affairs initiative to prevent methicillin-resistant Staphylococcus aureus infections revisited. Clin Infect Dis. 2012;54(11):1618–1620. doi: 10.1093/cid/cis272. [DOI] [PubMed] [Google Scholar]
- 102.Wyllie DH, Walker AS, Miller R, et al. Decline of methicillin-resistant Staphylococcus aureus in Oxfordshire hospitals is strain-specific and preceded infection-control intensification. BMJ Open. 2011;1(1):e000160. doi: 10.1136/bmjopen-2011-000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sirijatuphat R, Sripanidkulchai K, Boonyasiri A, et al. Implementation of global antimicrobial resistance surveillance system (GLASS) in patients with bacteremia. PLoS One. 2018;13(1):e0190132. doi: 10.1371/journal.pone.0190132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Belay ED, Kile JC, Hall AJ, et al. Zoonotic disease programs for enhancing global health security. Emerg Infect Dis. 2017;23(13) doi: 10.3201/eid2313.170544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ghafur A, Mathai D, Muruganathan A, et al. The Chennai Declaration: a roadmap to tackle the challenge of antimicrobial resistance. Indian J Cancer. 2013;50(1):71–73. doi: 10.4103/0019-509X.104065. [DOI] [PubMed] [Google Scholar]
- 106.Cantón R, Akóva M, Carmeli Y, et al. Rapid evolution and spread of carbapenemases among Enterobacteriaceae in Europe. Clin Microbiol Infect. 2012;18(5):413–431. doi: 10.1111/j.1469-0691.2012.03821.x. [DOI] [PubMed] [Google Scholar]
- 107.Bigdeli M, Jacobs B, Tomson G, et al. Access to medicines from a health system perspective. Health Policy Plan. 2013;28(7):692–704. doi: 10.1093/heapol/czs108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fischbach MA. Combination therapies for combating antimicrobial resistance. Curr Opin Microbiol. 2011;14(5):519–523. doi: 10.1016/j.mib.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Coates AR, Halls G, Hu Y. Novel classes of antibiotics or more of the same? Br J Pharmacol. 2011;163(1):184–194. doi: 10.1111/j.1476-5381.2011.01250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Infectious Diseases Society of America. The 10×’20 initiative: pursuing a global commitment to develop 10 new antibacterial drugs by 2020. Clin Infect Dis. 2010;50(8):1081–1083. doi: 10.1086/652237. [DOI] [PubMed] [Google Scholar]
- 111.Escaich S. Antivirulence as a new antibacterial approach for chemotherapy. Curr Opin Chem Biol. 2008;12(4):400–408. doi: 10.1016/j.cbpa.2008.06.022. [DOI] [PubMed] [Google Scholar]
- 112.Rex JH, Eisenstein BI, Alder J, et al. A comprehensive regulatory framework to address the unmet need for new antibacterial treatments. Lancet Infect Dis. 2013;13(3):269–275. doi: 10.1016/S1473-3099(12)70293-1. [DOI] [PubMed] [Google Scholar]
- 113.Research programme: bacteriophage therapeutics – AmpliPhi BioSciences/Leicester and Glasgow Universities. [Accessed May 11, 2018]. Available from: https://adisinsight.springer.com/drugs/800039161.
- 114.Raqib R, Sarker P, Bergman P, et al. Improved outcome in shigellosis associated with butyrate induction of an endogenous peptide antibiotic. Proc Natl Acad Sci U S A. 2006;103(24):9178–9183. doi: 10.1073/pnas.0602888103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bergman P, Norlin AC, Hansen S, Björkhem-Bergman L. Vitamin D supplementation to patients with frequent respiratory tract infections: a post hoc analysis of a randomized and placebo-controlled trial. BMC Res Notes. 2015;8:391. doi: 10.1186/s13104-015-1378-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Matsuoka O, Patel DM, Sasaki S, et al. Safety and immunogenicity of Clostridium difficile toxoid vaccine in Japanese adults. Hum Vaccin Immunother. 2018;14(2):322–328. doi: 10.1080/21645515.2017.1395538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Peng Z, Ling L, Stratton CW, et al. Advances in the diagnosis and treatment of Clostridium difficile infections. Emerg Microbes Infect. 2018;7(1):15. doi: 10.1038/s41426-017-0019-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.NVB 302. [Accessed February 1, 2018]. Available from: https://adisinsight.springer.com/drugs/800030483.
- 119.Research programme – plectasin antibacterials – Adenium Biotech. [Accessed February 2, 2018]. Available from: https://adisinsight.springer.com/drugs/800039698.
- 120.Hernandez LD, Racine F, Xiao L, et al. Broad coverage of genetically diverse strains of Clostridium difficile by actoxumab and bezlotoxumab predicted by in vitro neutralization and epitope modeling. Antimicrob Agents Chemother. 2015;59(2):1052–1060. doi: 10.1128/AAC.04433-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Morrison C. Antibacterial antibodies gain traction. Nat Rev Drug Discov. 2015;14(11):737–738. doi: 10.1038/nrd4770. [DOI] [PubMed] [Google Scholar]
- 122.Dubberke ER, Lee CH, Orenstein R, Khanna S, Hecht G, Gerding DN. Results from a randomized placebo-controlled clinical trial of a RBX2660 – a microbiota-based drug for the prevention of recurrent Clostridium difficile infection. Clin Infect Dis. 2018 Mar 29; doi: 10.1093/cid/ciy259. Epub. [DOI] [PubMed] [Google Scholar]
- 123.Seres Therapeutics. SER-109. [Accessed May 11, 2018]. Available from: http://www.seresthera-peutics.com/clinical-trials/ecosportrial.
- 124.Gerding DN, Meyer T, Lee C, et al. Administration of spores of nontoxigenic Clostridium difficile strain M3 for prevention of recurrent C. difficile infection: a randomized clinical trial. JAMA. 2015;313(17):1719–1727. doi: 10.1001/jama.2015.3725. [DOI] [PubMed] [Google Scholar]
- 125.Begier E, Seiden DJ, Patton M, et al. SA4Ag, a 4-antigen Staphylococcus aureus vaccine, rapidly induces high levels of bacteria-killing antibodies. Vaccine. 2017;35(8):1132–1139. doi: 10.1016/j.vaccine.2017.01.024. [DOI] [PubMed] [Google Scholar]
- 126.Jun SY, Jung GM, Yoon SJ, et al. Preclinical safety evaluation of intravenously administered SAL200 containing the recombinant phage endolysin SAL-1 as a pharmaceutical ingredient. Antimicrob Agents Chemother. 2014;58(4):2084–2088. doi: 10.1128/AAC.02232-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schuch R, Lee HM, Schneider BC, et al. Combination therapy with lysin CF-301 and antibiotic is superior to antibiotic alone for treating methicillin-resistant Staphylococcus aureus-induced murine bacteremia. J Infect Dis. 2014;209(9):1469–1478. doi: 10.1093/infdis/jit637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hua L, Hilliard JJ, Shi Y, et al. Assessment of an anti-alpha-toxin monoclonal antibody for prevention and treatment of Staphylococcus aureus-induced pneumonia. Antimicrob Agents Chemother. 2014;58(2):1108–1117. doi: 10.1128/AAC.02190-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Aridis Pharmaceuticals. AR-301 (Salvecin®) [Accessed May 11, 2018]. Available from: https://aridispharma.com/ar-301/
- 130.Varshney AK, Kuzmicheva GA, Lin J, et al. A natural human monoclonal antibody targeting staphylococcus protein A protects against Staphylococcus aureus bacteremia. PLoS One. 2018;13(1):e0190537. doi: 10.1371/journal.pone.0190537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Phase I/II trial of AmpliPhage-00 in patients with cystic fibrosis-associated Pseudomonas aeruginosa infections. [Accessed May 11, 2018]. Available from: https://adisinsight.springer.com/trials/700252954.
- 132.Research programme: antibacterial protein-based therapeutics – Phico Therapeutics. [Accessed August 4, 2016]. Available from: https://adisinsight.springer.com/drugs/800027172.
- 133.Westritschnig K, Hochreiter R, Wallner G, Firbas C, Schwameis M, Jilma B. A randomized, placebo-controlled phase I study assessing the safety and immunogenicity of a Pseudomonas aeruginosa hybrid outer membrane protein OprF/I vaccine (IC43) in healthy volunteers. Hum Vaccin Immunother. 2014;10(1):170–183. doi: 10.4161/hv.26565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Labiotech. Swiss pharma clears €37M funding to develop a new class of antibiotics. [Accessed May 11, 2018]. Available from: https://labiotech.eu/polyphor-murepavadin-pol6014-funding/
- 135.Aridis Pharmaceuticals. AR-101 (Aerumab™) [Accessed May 11, 2018]. Available from: https://aridispharma.com/ar-101/
- 136.Aridis Pharmaceuticals. AR-105 (Aerucin®) [Accessed May 11, 2018]. Available from: https://aridispharma.com/ar-105/
- 137.Digiandomenico A, Keller AE, Gao C, et al. A multifunctional bispecific antibody protects against Pseudomonas aeruginosa. Sci Transl Med. 2014;6262(262):ra155. doi: 10.1126/scitranslmed.3009655. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The aggregate data supporting findings contained within this manuscript will be shared upon request submitted to the corresponding author. Identifying patient data will not be shared.