Abstract
Genetic variation may explain some of the disparity in prevalence and control of hypertension across sub-Saharan Africa. Twenty-seven blood pressure (BP) related single nucleotide polymorphisms (SNPs) were genotyped among 2881samples from participants in the MEPI-CVD survey. Associations with known BP variants were evaluated for SBP, DBP and PP as continuous variables and for HTN as a binary variable. Eleven SNPS were associated with at least one BP trait (P<0.05). Four SNPs; rs2004776, rs7726475, rs11837544 and rs2681492 whose nearest genes are AGT, NPR3/SUB1, PLXNC1 and ATP2B1 respectively were associated with SBP. Six SNPs: rs2004776, rs11977526, rs11191548, rs381815, rs2681492 and rs1327235 close to AGT, IGFBP3, CYP17A1, PLEKHA7, ATP2B1 and JAG respectively were associated with DBP while two SNPs located within AGT and IGFBP-3 genes associated with HTN. For PP, four variants rs1458038, rs11725861, rs7726475 and rs11953630 whose corresponding genes are FGF5, CHIC2, SUB1/NPR3 and EBF1 reached significance (P<0.05). Eight SNPs were replicated in the same effect direction as the parent studies. Risk scores defined using published effect sizes were significantly associated with both SBP (P = 0.0026) and DBP (P = 0.0214). The replication of multiple BP variants among East Africans suggests that these variants may have universal effects across ethnic populations.
Keywords: diastolic blood pressure, East Africa, genetics, SNPs, systolic blood pressure
Graphical Abstract
INTRODUCTION
Cardiovascular disease is a leading contributor to disease burden in Africa accounting for nearly a half of the mortality due to non-communicable disease in the region(1). High blood pressure is the principal risk factor for cardiovascular disease (CVD). An increasing prevalence of hypertension has been documented in Africa; however detection and treatment rates have been dismal (2). In Uganda, for example, more than a quarter of the population have documented hypertension but less than 15% of all those treated achieve target blood pressure control (3, 4). As a result high blood pressure- related CVD morbidity and mortality occurs at a much younger age in Africa compared to the rest of the world (5–7).
Blood pressure (BP) is a heritable trait with an estimated 39 −50% variation attributed to genetics (8). Further evidence is derived from several twin and family studies wherein greater correlation has been noted between monozygotic twins compared to dizygotic twins and greater resemblance of blood measurements documented within families than between families (9, 10). Hypertension is 3.8 times more frequent among persons with a positive family history of hypertension before the age of 55 years (11). In native West Africans, heritability for both systolic and diastolic blood pressure is estimated at 45% (12). Epidemiological studies in sub-Saharan Africa (SSA) have also been unanimous in their findings of a strong association between family history of hypertension and its incidence (13).
People within the sub-Saharan region are disproportionately affected by hypertension. Numerous prevalence studies have documented a higher prevalence of hypertension and poorer control in the East and Southern African regions than the North African region (2, 14). Differences in environment risks, while important, may not completely explain the disparity in prevalence of hypertension and its control across SSA (15). The other mechanisms responsible for these differences, including variation in genetic predisposition as a result of varying frequencies of common disease susceptibility patterns or the presence of population specific alleles especially in populations with a high level of genetic heterogeneity such as those of SSA (16–18) need to be further examined.
To evaluate the genetic basis of hypertension, both genome-wide association studies (GWAS) and candidate gene approaches have been employed. The strength of GWAS compared to candidate gene studies, lies in the identification of novel hypothesis-free loci and the reproducibility of the findings (19). Since 2009, GWAS has identified multiple blood pressure regulatory loci with many of these loci being replicated in independent population samples (20) . Replication of GWAS findings is required to improve generalizability of study findings (21). The few BP GWAS and replications in Africans have been performed on samples derived from populations of West African origin, including African Americans (22, 23). There is a paucity of genetic studies to delineate the genetic basis of blood pressure in other African populations. It is also not clear whether previously identified blood pressure genetic variants can be generalized to sub-Saharan populations.
We therefore sought to determine whether a set of genetic loci significantly associated with blood pressure- traits could be replicated among samples from the Medical Education Partnership Initiative for Cardiovascular Disease (MEPI-CVD) survey in Uganda, East Africa.
MATERIALS AND METHODS:
The methods of the Medical Education Partnership Initiative (MEPI-CVD) Survey have been described in detail elsewhere (3, 24). The MEPI-CVD Survey was a cross-sectional study conducted between September 2012 and May, 2013 in Wakiso district of central Uganda among men and women aged 18 years and older to describe CVD risk factors in a peri-urban Ugandan population. Data on CVD risk factors was collected by trained research nurses using a World Health Organisation (WHO) modified expanded STEPs questionnaire. The study variables were age, gender, address, dietary habits, tobacco and alcohol consumption, exercise, smoke exposure, socio-economic status (housing characteristics), family history and symptoms of heart disease including angina. Self-reported history of hypertension diagnosis, diabetes, dyslipidaemia and the treatment for these conditions were also recorded. Anthropometric measurements gathered included height, weight and waist circumference. Blood pressure, fasting cholesterol and blood sugar were also measured.
All study data were collected by the study staff within the survey area and entered into a central survey database. All case report forms and laboratory reports were reviewed for data consistency before data entry. Information collected from blood analysis was electronically transferred to the central survey database.
Ethical considerations:
The survey protocol was approved by the Makerere University School of Medicine Research and Ethics Committee and the Uganda National Council of Science and Technology. Written informed consent was obtained from all study participants.
Phenotype (Blood pressure) measurement:
Participants were requested beforehand to refrain from smoking, drinking alcohol or caffeinated beverage half an hour prior to blood pressure measurement. Blood pressure and heart rate were measured with an Omron automated sphygmomanometer model HEM-907. The BP was measured on the left arm after the participant had sat for at least five minutes. The blood pressure was taken in the sitting position, legs uncrossed, the arm resting on a table and the ante-cubital fossa at the level of the lower sternum. Two arm cuffs that fitted arm circumferences 9-13 inches and 13-17 inches were used in the process. Three readings were taken three minutes apart and the mean of the closest two values were used to describe the blood pressure of the subject. Additional measurements included height which was measured to the nearest 0.1 cm as the perpendicular distance between the top of the head (the vertex) and the bottom of the feet by a SECA 214 portable stadiometer. The weight was measured to the nearest 0.1 kg using a SECA 762 weighing scale with the subjects putting on loose clothing. The waist circumference was measured to the nearest 0.1 cm at the level of the midpoint between the inferior margin of the last rib and the crest of the ilium in the mid-axillary plane using a non-stretchable tape measure.
Candidate single nucleotide polymorphism (SNP) selection:
Thirty single nucleotide polymorphisms (SNPs) were selected for genotyping and association analysis with BP. These SNPs were selected based on the previous association evidence with BP from GWASs or admixture mapping analysis (23, 25–29) (Supplementary Table 1).
DNA genotyping and quality control:
Complete phenotype information was obtained in 2,881 participants. Genomic DNA was extracted from whole blood drawn from these participants using Gentra Puregene kits from QIAGEN. Genotyping was performed using the PlexSeq process (Plexseq Diagnostics, Cleveland, OH). Primers were designed to amplify regions surrounding each SNP using a multiplexed approach. Each primer design included additional sequence at the 5’ end that was then used to anneal universal barcoded Illumina primers in a secondary amplification reaction. All samples (including negative and positive controls) were uniquely barcoded and sequenced simultaneously using a MiSeq instrument (Illumina, San Diego, CA). Fastq files were analyzed using Plexcall software (PlexSeq Diagnostics, Cleveland, OH) which provides genotype calls for all SNP’s in each sample.
SNP genotyping quality was evaluated for departures from Hardy-Weinberg equilibrium (HWE) and call rate. One SNP had significant departure from the HWE with P ≤ 0.001 and was excluded from analysis. Two SNPs with a missing rate larger than 90% were also excluded from analysis. Two hundred and fourteen samples with genotype call rate less than 85% were excluded from the analysis. After QCs, 27 SNPs and 2667 individuals were left for further association analysis.
Association analysis
All data management was carried out using R (30). Genotype and phenotype association analysis was carried out using software PLINK (31). Systolic blood pressure (SBP) and diastolic and blood pressure (DBP) values were obtained as an average of the closest two measurements. Pulse pressure (PP) was calculated as the difference of SBP and DBP. . Only 12.5% of individuals with hypertension were treated with pharmacotherapy. However treatment information was sparse and incomplete and therefore BP traits were not imputed for hypertension treatment. Hypertension status (HTN) was defined by either SBP > 140mm Hg or DBP > 90mm Hg. Only individuals with complete phenotype and covariate data were included in analysis. Linear regression was applied for SBP, DBP, MAP and PP while logistic regression was applied for HTN. In all the analysis the covariates: age, age2, gender and BMI were included. The SNP effect was coded additively. Since all the 27 SNPs passed QCs were selected from previous confirmed BP variants, a significance level of 0.05 was considered as replication in this study.
Multi-trait statistical analyses using CPASSOC
We applied the CPASSOC package to combine association evidence of SBP, DBP and HTN. CPASSOC provided two statistics, SHom and SHet as previously described (32). SHom is similar to the fixed effect meta-analysis method (33) but accounts for the correlation of summary statistics of the correlated traits. SHom uses a sample size of a trait as a weight. SHet is an extension of SHom but power can be improved when the effect sizes are different for different traits. The distribution of SHet under the null hypothesis was obtained through an estimated beta distribution. To calculate statistics SHom and SHet, a correlation matrix is required to account for the correlation among traits or induced by overlapped or related samples from different cohorts. In this study, we directly use the correlation matrix calculated from the residual of three blood pressure traits after adjusting for gender, age, age2 and BMI.
RESULTS
Baseline population characteristics
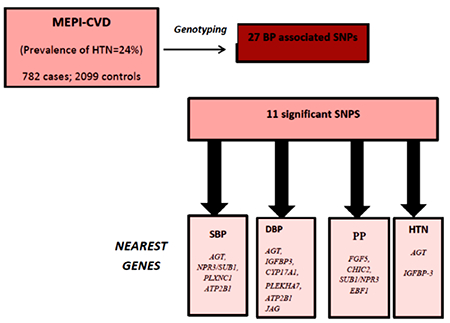
The prevalence of hypertension among the MEPI-CVD study was 1237/5183 (24%). Among these 782 cases and 2099 controls were selected for genotyping). Table 1 presents the demographic characteristics of study participants. Participants with hypertension had a higher BMI and waist hip ratio compared to those with normal blood pressure. Participants with hypertension also had higher total cholesterol and low density lipoprotein cholesterol concentrations compared to participants with normal blood pressure. High density lipoprotein was lower among hypertensive patients.
Table 1.
Demographic characteristics of study subjects
| Whole study (n=2881) | Normotensive (n=2099 ) | Hypertensive (n= 782) | p-value | |
|---|---|---|---|---|
| Age (years) | 30 (25-43) | 22 (24-38) | 42 (30-55) | <0.0001 |
| Males n (%) | 831 (28.8) | 598 (28.5) | 233 (29.8) | 0.492 |
| BMI (Kg m−2) | 24.4 ± 5.1 | 23.95 ± 4.7 | 25.59 ± 5.95 | <0.0001 |
| SBP | 126.4 ± 21.1 | 117.0 ± 11.6 | 151.6 ± 20.3 | <0.0001 |
| DBP | 80.0 ± 12.3 | 74.9 ± 7.7 | 93.5 ± 12.4 | <0.0001 |
| Waist | 80.8 ± 12.2 | 78.7 ± 11.5 | 83.6 ± 13.2 | <0.0001 |
| WHR | 0.82 ± 0.07 | 0.82 ± 0.07 | 0.84 ± 0.07 | <0.0001 |
| Total cholesterol (mmol/l) | 2.84 (2.2-3.86) | 2.76 (2.18-3.49) | 2.88 (2.2-4.0) | 0.009 |
| HDL(mmol/l) | 0.84 (0.46-1.71) | 0.79 (0.43-1.71) | 0.99 (0.56-1.73) | 0.0002 |
| LDL(mmol/l) | 1.08 (0.84-1.34) | 1.01 (0.8-1.3) | 1.15 (0.9-1.44) | <0.0001 |
| Triglycerides(mmol/l) | 0.97 (0.66-1.47) | 0.94 (0.63-1.42) | 1.06 (0.74-1.56) | <0.0001 |
Association analysis among Ugandan samples with four blood pressure traits
Thirty SNPs which are known to be associated with blood pressure or hypertension were genotyped (Supplementary Table 1). Three of them failed QCs and were therefore were not further included for analysis. Table 2 lists the 11 SNPS that were significantly associated with at least one BP trait (P<0.05). To control for the effects of multiple testing, we used Li and Ji’s approach which assumes that correlated tests are independent and adjusts them according to an ‘effective number’ (Meff) of independent tests (34). These Meff based tests have been shown to control error rate accurately, resulting in an increase in power. We estimated 56 independent tests for the 27 SNPs and the four BP traits. Under null hypothesis that none of 56 independent tests were significant, we can expect 56×0.05 = 2.7, around 3 SNPs with P < 0.05. We observed 11 significant SNPs, which are substantially higher than expected.
Table 2.
The SNPs whose association with BP traits was significant in the MEPI-CVD samples
| SNP | Chr | Gene | Effect Allele/Other Allele | EAF | Trait | N | BETA | SE | |
|---|---|---|---|---|---|---|---|---|---|
| rs2004776 | 1 | AGT | C/T | 0.46 | SBP | 2659 | 1.38 | 0.53 | 0.01 |
| DBP | 2659 | 0.79 | 0.32 | 0.01 | |||||
| PP | 2659 | 0.59 | 0.37 | 0.12 | |||||
| HTN | 2602 | 0.16 | 0.07 | 0.02 | |||||
| CPASSOC SHom |
2659/ 2659/ 2602 |
NA | NA | 0.0050 | |||||
| CPASSOC SHet |
2659/ 2659/ 2602 |
NA | NA | 0.03 | |||||
| rs1458038 | 4 | FGF5 | T/C | 0.04 | SBP | 2665 | 2.25 | 1.36 | 0.10 |
| DBP | 2665 | 0.31 | 0.83 | 0.71 | |||||
| PP | 2665 | 1.94 | 0.97 | 0.04 | |||||
| HTN | 2601 | 0.17 | 0.17 | 0.33 | |||||
| CPASSOC SHom |
2665/ 2665/ 2601 |
NA | NA | 0.27 | |||||
| CPASSOC SHet |
2665/ 2665/ 2601 |
NA | NA | 0.35 | |||||
| rs11725861 | 4 | CHIC2 | C/T | 0.22 | SBP | 2663 | −0.28 | 0.63 | 0.66 |
| DBP | 2663 | 0.66 | 0.38 | 0.08 | |||||
| PP | 2663 | 0.94 | 0.45 | 0.04 | |||||
| HTN | 2598 | 0.14 | 0.08 | 0.09 | |||||
| CPASSOC SHom |
2663/ 2663/ 2598 |
NA | NA | 0.20 | |||||
| CPASSOC SHet |
2663/ 2663/ 2598 |
NA | NA | 0.01 | |||||
| rs7726475 | 5 | SUB1/NPR3 | A/G | 0.01 | SBP | 2667 | −10.41 | 3.30 | 0.0016 |
| DBP | 2667 | −3.62 | 2.01 | 0.07 | |||||
| PP | 2667 | −6.79 | 2.35 | 0.0039 | |||||
| HTN | 2602 | −0.44 | 0.48 | 0.36 | |||||
| CPASSOC SHom |
2667/ 2667/ 2602 |
NA | NA | 0.03 | |||||
| CPASSOC SHet |
2667/ 2667/ 2602 |
NA | NA | 0.01 | |||||
| rs11953630 | 5 | EBF1 | A/G | 0.12 | SBP | 2665 | 0.80 | 0.79 | 0.31 |
| DBP | 2665 | −0.47 | 0.48 | 0.34 | |||||
| PP | 2665 | 1.27 | 0.56 | 0.02 | |||||
| HTN | 2600 | −0.09 | 0.10 | 0.40 | |||||
| CPASSOC SHom |
2665/ 2665/ 2600 |
NA | NA | 0.67 | |||||
| CPASSOC SHet |
2665/ 2665/ 2600 |
NA | NA | 0.02 | |||||
| rs11977526 | 7 | IGFBP-3 | T/C | 0.33 | SBP | 2666 | 1.04 | 0.59 | 0.08 |
| DBP | 2666 | 0.76 | 0.36 | 0.03 | |||||
| PP | 2666 | 0.28 | 0.42 | 0.51 | |||||
| HTN | 2601 | 0.18 | 0.08 | 0.02 | |||||
| CPASSOC SHom |
2666/ 2666/ 2601 |
NA | NA | 0.01 | |||||
| CPASSOC SHet |
2666/ 2666/ 2601 |
NA | NA | 0.06 | |||||
| rs11191548 | 10 | CYP17A1 | G/A | 0.03 | SBP | 1998 | −0.56 | 1.87 | 0.77 |
| DBP | 1998 | −2.47 | 1.13 | 0.03 | |||||
| PP | 1998 | 1.91 | 1.32 | 0.15 | |||||
| HTN | 1984 | −0.11 | 0.25 | 0.67 | |||||
| CPASSOC SHom |
1998/ 1998 1984 |
NA | NA | 0.26 | |||||
| CPASSOC SHet |
1998/ 1998/ 1984 |
NA | NA | 0.13 | |||||
| rs381815 | 11 | PLEKHA7 | T/C | 0.28 | SBP | 2664 | 0.88 | 0.57 | 0.12 |
| DBP | 2664 | 0.70 | 0.35 | 0.04 | |||||
| PP | 2664 | 0.18 | 0.41 | 0.66 | |||||
| HTN | 2600 | 0.00 | 0.09 | 0.99 | |||||
| CPASSOC SHom |
2664/ 2664 2600 |
NA | NA | 0.21 | |||||
| CPASSOC SHet |
2664/ 2664/ 2600 |
NA | NA | 0.18 | |||||
| rs11837544 | 12 | PLXNC1 | T/A | 0.20 | SBP | 2665 | −1.40 | 0.65 | 0.03 |
| DBP | 2665 | −0.68 | 0.39 | 0.09 | |||||
| PP | 2665 | −0.73 | 0.46 | 0.12 | |||||
| HTN | 2601 | −0.16 | 0.09 | 0.07 | |||||
| CPASSOC SHom |
2665/ 2665 2601 |
NA | NA | 0.03 | |||||
| CPASSOC SHet |
2665/ 2665/ 2601 |
NA | NA | 0.12 | |||||
| rs2681492 | 12 | ATP2B1 | G/A | 0.14 | SBP | 2661 | −1.92 | 0.74 | 0.01 |
| DBP | 2661 | −1.34 | 0.45 | 0.0029 | |||||
| PP | 2661 | −0.58 | 0.53 | 0.27 | |||||
| HTN | 2598 | −0.12 | 0.10 | 0.21 | |||||
| CPASSOC SHom |
2661/ 2661/ 2598 |
NA | NA | 0.01 | |||||
| CPASSOC SHet |
2661/ 2661/ 2598 |
NA | NA | 0.02 | |||||
| rs1327235 | 20 | JAG 1 | A/G | 0.42 | SBP | 2666 | −0.82 | 0.52 | 0.12 |
| DBP | 2666 | −0.70 | 0.32 | 0.03 | |||||
| PP | 2666 | −0.12 | 0.37 | 0.75 | |||||
| HTN | 2601 | −0.10 | 0.07 | 0.15 | |||||
| CPASSOC SHom |
2666/ 2666/ 2601 |
NA | NA | 0.05 | |||||
| CPASSOC SHet |
2666/ 2666/ 2601 |
NA | NA | 0.12 |
Among the significant SNPs, four SNPs were associated with SBP: rs2004776, rs7726475, rs11837544 and rs2681492 and the nearest genes are AGT, NPR3/SUB1, PLXNC1 and ATP2B1, respectively. Six SNPS; rs2004776, rs11977526, rs11191548, rs381815, rs2681492 and rs1327235 and the nearest genes are AGT, IGFBP3, CYP17A1, PLEKHA7, ATP2B1 and JAG, respectively, were associated with DBP. Two SNPs located within AGT and IGFBP-3 genes respectively were associated with HTN. For PP, four variants rs1458038, rs11725861, rs7726475 and rs11953630 whose corresponding genes are FGF5, CHIC2, SUB1/NPR3 and EBF1 reached significance (P<0.05). Since combining evidence of the four BP traits will improve the power of association evidence, we performed CPASSOC analysis (32). Eight of the 11 variants were significantly associated with all the BP traits (Table 2).
We next calculated a risk score for each individual byg ∑βigi, where βi and gi are the effect size and genotype value of the ith variant of the 27 variants we selected and tested the association between the risk score and each of BP traits. The effect size βi was obtained from the published data (Supplementary Table 1). Significant linear regression association evidence was observed for SBP (effect size=0.64, P = 0.0026), DBP (effect size=0.45, P = 0.0214). However, there was no significant association for HTN (effect size= 1.28, P= 0.089). We further compared the effect sizes estimated from this study and that from the published data (Supplementary Table 1) and observed that the effect sizes were highly correlated (Figure 1).
Figure 1.
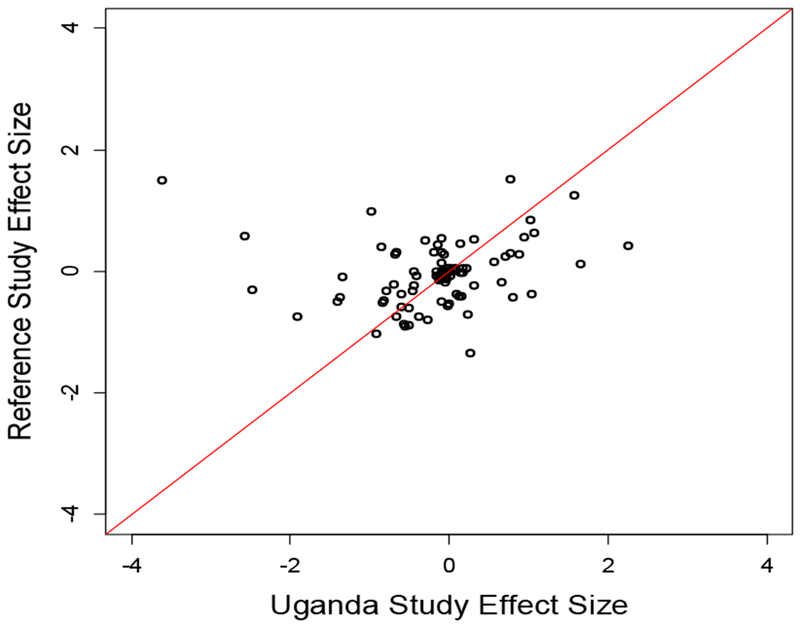
Scatter plots of effects size for Uganda study with the reference studies of the 28 SNPs for SBP, DBP and HTN.
DISCUSSION
Replication of genetic variants identified in BP GWAS in different populations is necessary to improve generalizability of findings to diverse populations. In particular, replication of findings in samples of populations with lower LD, notably African populations, usually leads to more refined estimates of positions of causal variants (18, 35). In this replication study, the first of its kind among East African population, we were able to replicate eight of the selected 30 SNPs for SBP, DBP, PP and HTN status in a direction consistent with the primary studies. These were higher than the three variants expected under the null hypothesis. Another three SNPs were significantly associated with any one of the four blood pressure traits, albeit in an effect direction different to the primary studies. These included rs2681492 and rs7726475, the SNPs with the strongest association evidence to the studied BP traits. However, there were still a substantial number of SNPs which failed to reach the 0.05 significance level. We then performed power analysis with the sample size 2,881. Using the effect size of 0.1% BP variation accounted for by a single BP variant, as suggested by GWASs (36, 37), our study has 31% statistical power to detect a BP variant at the 0.05 significance level, which is consistent with what we observed. The genetic score defined by the 27 variants are also significantly associated with both SBP and DBP. We further demonstrated that the effect sizes estimated in this study are consistent with those obtained from published literature of different ethnic populations (Figure 1). This is consistent with our previous study (37) which shows that BP loci may have universal effects across studied populations, and further demonstrates that multi-ethnic samples are critical for the identification, fine mapping and understanding trait variability.
Among the 8 replicated SNPs, rs2004776 near the AGT had the strongest association evidence with BP traits in this study and was replicated for SBP, DBP and HTN. The AGT gene has been implicated in both the pathogenesis of hypertension and blood pressure response to ACE inhibitors and angiotensin receptor blockers (38). Multiple consortium-based GWAS have confirmed this SNP -BP trait association in samples derived from population of European and East Asian ancestry (38–40). The effect direction of this association in European samples is same to that found in our samples (41) (Supplemental Table 1). Many candidate studies also suggested the variants in AGT are associated with BP traits (42, 43). Previously, other AGT variants associated with hypertension in Caucasians could not be replicated in indigenous Africans(44).
The SNP rs381815 found near the PLEKHA7 gene was replicated for DBP; whereas rs11977526 which is near IGFBP-3 was replicated for DBP and hypertension. These two variants have consistent association evidence in multi-ethnic populations such as Europeans, South East Asians and African Americans and now in East Africans, suggesting that this association may have a broad role in BP regulation across ethnicities (45–47). Post- GWAS evaluation for the mechanistic contribution of PLEKHA7 to BP traits and cardiovascular function has revealed a role in the regulation of ion calcium, nitric oxide bioavailability, and vasculature responsiveness (48). Zhu et al, using the summary statistics from COGENT African ancestry GWAS, identified rs11977526 as the most significant IGFBP3-related SNP in association with multiple BP traits among SBP, DBP and HTN (49). IGFBP-3 binds IGF-1 thereby reducing action at the cellular level which subsequently decreases vascular resistance through stimulation of nitric oxide synthesis (50).
The SNPS rs1458038, rs11725861 and rs11953630 found in the proximity of FGF5, CHIC2 and EBF1 respectively were significantly associated with PP. In addition, rs11725861 and rs11953630 were significant for all the traits in the combined analysis. These SNPs have demonstrated interethnic replication among diverse ethnicities (49, 51, 52). More importantly, the three variants have been associated with blood pressure responsiveness to beta-blocker and thiazide diuretic monotherapy with among Caucasians(53). The SNP rs1458038 near FGF5, for example, was associated with significant BP responses to both atenolol and HCTZ, albeit in opposite directions. Whereas the aim of this study was not to assess variants for pharmacological response to BP medication, the multi-ethnic replication for other BP traits may have similar effects on BP response to pharmacological treatment.
The two SNPs with the most significant associations had an effect direction opposite to that demonstrated among European and Southeast Asian GWAS (45, 54, 55) The SNP rs2681492 located in ATP2B1 gene was associated with SBP and DBP. The ATP gene encodes the plasma membrane calcium ATPase isoform 1 that plays a critical role in intracellular calcium homeostasis by removing bivalent calcium ions from eukaryotic cells against very large concentration (56) The variant rs7726475 which is located on chromosome 5 between the SUB1 and NPR3 genes was associated with SBP and PP. Among African American samples, using admixture mapping, Zhu et al showed an association between rs7726475 and SBP and DBP (26). The NPR3 gene was reported to be significantly associated with BP traits with another SNP, rs1173771 with same direction, in previous interethnic GWAS in East Asians and Europeans (55, 57). The difference in effect direction of the SNPs compared to the primary populations associations suggests that the same locus may have varying effects among different ethnic groups. True genetic architectural variation among populations and differences in linkage disequilibrium may be responsible for these differential effect directions although the effect of environmental cannot be completely ruled out (58).
The clinical implications of identified variants are limited by available data on cardiovascular outcomes in populations of African ancestry. However, three of the replicated SNPs have demonstrated association with responsiveness to antihypertensive medication. This may have implications for pharmacological control of blood pressure among diverse ancestries. While it is clear that many variants display trans-ethnic replication, the finding that our most significant variants have an opposite effect direction to that found in the primary studies calls for further studies to elucidate the effect of these variants on blood pressure traits in the African.
CONCLUSION:
We were able to replicate multiple BP variants identified from previous BP GWAS of Europeans, African Americans and Eastern Asians or admixture mapping analysis in our Eastern African population, which has not been studied previously. However, we also failed to replicate many variants possibly because of insufficient statistical power of the current sample size. We also observed that some variants have different effect directions in different ethnic populations which may suggest that some genes, while regulating blood pressure across diverse populations, may have a differential effect for various BP traits. Our study also suggests BP variants may have universal effects across ethnic populations, which was suggested by Franceschini et al.(37).
Supplementary Material
Acknowledgements
This work was supported by Grant R24TW008861 (MEPI- Linked: Building Capacity for cardiovascular research and Training in Uganda.) funded by Office of the United States Global Aids Coordinator, National Institutes of Health and Health Resources and Services Administration. The work was partially supported by the National Institutes of Health HG003054 from the National Human Genome Research Institute.
Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the supporting offices. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
The authors would like to thank Evelyn Bakengesa of the Publication and Information Centre, Makerere University College of Health Sciences and Rhoda Namubiru, Administrator of the MEPI- Linked Cardiovascular program for assisting in the editing and submission of the manuscript.
Footnotes
Conflicts of interest
The authors have no conflicts of interest to declare
Contributor Information
James Kayima, Division of Adult Cardiology, Uganda Heart Institute Department of Medicine, Makerere University College of Health Sciences, P.O.Box 7072 Kampala, Uganda.
Jingjing Liang, Department of Epidemiology & Biostatistics, School of Medicine, Case Western Reserve University, Cleveland, OH, USA.
Yanina Natanzon, Department of Epidemiology & Biostatistics, School of Medicine, Case Western Reserve University, Cleveland, OH, USA.
Joaniter Nankabirwa, Clinical Epidemiology Unit, Department of Medicine, School of Medicine, College of Health Sciences, Makerere University.
Isaac Ssinabulya, Division of Adult Cardiology, Uganda Heart Institute, Department of Medicine, Makerere University College of Health Sciences, Kampala, Uganda.
Jane Nakibuuka, Department of Medicine, Makerere University College of Health Sciences, Kampala, Uganda.
Achilles Katamba, Clinical Epidemiology Unit, Department of Medicine, School of Medicine, College of Health Sciences, Makerere University.
Harriet Mayanja-Kizza, Department of Medicine, Makerere University College of Health Sciences, Kampala, Uganda.
Alexander Miron, Department of Genetics, School of Medicine, Case Western Reserve University, Cleveland, OH, USA.
Chun Li, Department of Epidemiology & Biostatistics, School of Medicine, Case Western Reserve University, Cleveland, OH, USA.
Xiaofeng Zhu, Department of Epidemiology & Biostatistics, School of Medicine, Case Western Reserve University, Cleveland, OH, USA.
REFERENCES
- 1.WHO. Global status report on noncommunicable diseases 2010 2011. April 2011
- 2.Kayima J, Wanyenze RK, Katamba A, Leontsini E, Nuwaha F. Hypertension awareness, treatment and control in Africa: a systematic review. BMC Cardiovasc Disord. 2013;13(54):1471–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kayima J, Nankabirwa J, Sinabulya I, Nakibuuka J, Zhu X, Rahman M, et al. Determinants of hypertension in a young adult Ugandan population in epidemiological transition—the MEPI-CVD survey. BMC Public Health. 2015;15(1):830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayega RW, Makumbi F, Rutebemberwa E, Peterson S, Ostenson CG, Tomson G, et al. Modifiable socio-behavioural factors associated with overweight and hypertension among persons aged 35 to 60 years in eastern Uganda. PLoS One. 2012;7(10):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Donnell MJ, Xavier D, Liu L, Zhang H, Chin SL, Rao-Melacini P, et al. Risk factors for ischaemic and intracerebral haemorrhagic stroke in 22 countries (the INTERSTROKE study): a case-control study. Lancet. 2010;376(9735):112–23. Epub 2010/06/22. [DOI] [PubMed] [Google Scholar]
- 6.Steyn K, Sliwa K, Hawken S, Commerford P, Onen C, Damasceno A, et al. Risk Factors Associated With Myocardial Infarction in Africa. Circulation. 2005;112(23):3554–61. [DOI] [PubMed] [Google Scholar]
- 7.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. The Lancet. 2013;380(9859):2095–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salfati E, Morrison AC, Boerwinkle E, Chakravarti A. Direct estimates of the genomic contributions to blood pressure heritability within a population-based cohort (ARIC). PLoS One. 2015;10(7):e0133031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Longini IM Jr. , Higgins MW, Hinton PC, Moll PP, Keller JB. Environmental and genetic sources of familial aggregation of blood pressure in Tecumseh, Michigan. Am J Epidemiol. 1984;120(1):131–44. [DOI] [PubMed] [Google Scholar]
- 10.Williams RR, Hunt S, Hasstedt S, Hopkins P, Wu L, Berry T, et al. Are there interactions and relations between genetic and environmental factors predisposing to high blood pressure? Hypertension. 1991;18(3 Suppl):I29. [DOI] [PubMed] [Google Scholar]
- 11.Hunt SC, Williams RR, Barlow GK. A comparison of positive family history definitions for defining risk of future disease. Journal of chronic diseases. 1986;39(10):809–21. [DOI] [PubMed] [Google Scholar]
- 12.Rotimi CN, Cooper RS, Cao G, Ogunbiyi O, Ladipo M, Owoaje E, et al. Maximum-likelihood generalized heritability estimate for blood pressure in Nigerian families. Hypertension. 1999;33(3):874–8. [DOI] [PubMed] [Google Scholar]
- 13.van der Sande MA, Walraven GE, Milligan PJ, Banya WA, Ceesay SM, Nyan OA, et al. Family history: an opportunity for early interventions and improved control of hypertension, obesity and diabetes. Bull World Health Organ. 2001;79(4):321–8. [PMC free article] [PubMed] [Google Scholar]
- 14.Addo J, Smeeth L, Leon DA. Hypertension In Sub-Saharan Africa. Hypertension. 2007;50(6):1012–8. [DOI] [PubMed] [Google Scholar]
- 15.Opie LH, Seedat YK. Hypertension in sub-Saharan African populations. Circulation. 2005;112(23):3562–8. [DOI] [PubMed] [Google Scholar]
- 16.Campbell MC, Tishkoff SA. African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annual review of genomics and human genetics. 2008;9:403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gurdasani D, Carstensen T, Tekola-Ayele F, Pagani L, Tachmazidou I, Hatzikotoulas K, et al. The African genome variation project shapes medical genetics in Africa. Nature. 2015;517(7534):327–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324(5930):1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Turner S, Armstrong LL, Bradford Y, Carlson CS, Crawford DC, Crenshaw AT, et al. Quality control procedures for genome-wide association studies. Current protocols in human genetics. 2011:119 1–1.. 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Franceschini N, Reiner AP, Heiss G. Recent findings in the genetics of blood pressure and hypertension traits. American journal of hypertension. 2011;24(4):392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ioannidis JP. Population-wide generalizability of genome-wide discovered associations. Journal of the National Cancer Institute. 2009;101(19):1297–9. [DOI] [PubMed] [Google Scholar]
- 22.Adeyemo A, Gerry N, Chen G, Herbert A, Doumatey A, Huang H, et al. A genome-wide association study of hypertension and blood pressure in African Americans. PLoS genetics. 2009;5(7):e1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franceschini N, Fox E, Zhang Z, Edwards TL, Nalls MA, Sung YJ, et al. Genome-wide association analysis of blood-pressure traits in African-ancestry individuals reveals common associated genes in African and non-African populations. The American Journal of Human Genetics. 2013;93(3):545–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakibuuka J, Sajatovic M, Nankabirwa J, Furlan AJ, Kayima J, Ddumba E, et al. Stroke-Risk Factors Differ between Rural and Urban Communities: Population Survey in Central Uganda. Neuroepidemiology. 2015;44(3):156–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu X, Young JH, Fox E, Keating BJ, Franceschini N, Kang S, et al. Combined admixture mapping and association analysis identifies a novel blood pressure genetic locus on 5p13: contributions from the CARe consortium. Hum Mol Genet. 2011;20(11):2285–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu X, Cooper RS. Admixture mapping provides evidence of association of the VNN1 gene with hypertension. PLoS One. 2007;2(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu X, Luke A, Cooper RS, Quertermous T, Hanis C, Mosley T, et al. Admixture mapping for hypertension loci with genome-scan markers. Nat Genet. 2005;37(2):177–81. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y-J, Tayo BO, Bandyopadhyay A, Wang H, Feng T, Franceschini N, et al. The association of the vanin-1 N131S variant with blood pressure is mediated by endoplasmic reticulum-associated degradation and loss of function. PLoS Genet. 2014;10(9):e1004641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Team RC, . R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: 2015. Available from: http://www.R-project.org/. [Google Scholar]
- 31.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics. 2007;81(3):559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu X, Feng T, Tayo BO, Liang J, Young JH, Franceschini N, et al. Meta-analysis of correlated traits via summary statistics from GWASs with an application in hypertension. American journal of human genetics. 2015;96(1):21–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95(3):221–7. [DOI] [PubMed] [Google Scholar]
- 35.Saccone NL, Saccone SF, Goate AM, Grucza RA, Hinrichs AL, Rice JP, et al. In search of causal variants: refining disease association signals using cross-population contrasts. BMC genetics. 2008;9(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.International Consortium for Blood Pressure Genome-Wide Association S, Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478(7367):103–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franceschini N, Fox E, Zhang Z, Edwards TL, Nalls MA, Sung YJ, et al. Genome-wide association analysis of blood-pressure traits in African-ancestry individuals reveals common associated genes in African and non-African populations. American journal of human genetics. 2013;93(3):545–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson AD, Newton-Cheh C, Chasman DI, Ehret GB, Johnson T, Rose L, et al. Association of hypertension drug target genes with blood pressure and hypertension in 86 588 individuals. Hypertension. 2011;57(5):903–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simino J, Shi G, Bis JC, Chasman DI, Ehret GB, Gu X, et al. Gene-age interactions in blood pressure regulation: a large-scale investigation with the CHARGE, Global BPgen, and ICBP Consortia. The American Journal of Human Genetics. 2014;95(1):24–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeuchi F, Yamamoto K, Katsuya T, Sugiyama T, Nabika T, Ohnaka K, et al. Reevaluation of the association of seven candidate genes with blood pressure and hypertension: a replication study and meta-analysis with a larger sample size. Hypertension Research. 2012;35(8):825–31. [DOI] [PubMed] [Google Scholar]
- 41.Johnson AD, Newton-Cheh C, Chasman DI, Ehret GB, Johnson T, Rose L, et al. Association of hypertension drug target genes with blood pressure and hypertension in 86,588 individuals. Hypertension. 2011;57(5):903–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu X, Chang YP, Yan D, Weder A, Cooper R, Luke A, et al. Associations between hypertension and genes in the renin-angiotensin system. Hypertension. 2003;41(5):1027–34. [DOI] [PubMed] [Google Scholar]
- 43.Zhu X, Fejerman L, Luke A, Adeyemo A, Cooper RS. Haplotypes produced from rare variants in the promoter and coding regions of angiotensinogen contribute to variation in angiotensinogen levels. Human molecular genetics. 2005;14(5):639–43. [DOI] [PubMed] [Google Scholar]
- 44.Tiago AD, Nkeh B, Candy GP, Badenhorst D, Defterios D, Brooksbank R, et al. Association study of eight candidate genes with renin status in mild-to-moderate hypertension in patients of African ancestry. Cardiovascular journal of South Africa: official journal for Southern Africa Cardiac Society [and] South African Society of Cardiac Practitioners. 2000;12(2):75–80. [PubMed] [Google Scholar]
- 45.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, et al. Genome-wide association study of blood pressure and hypertension. Nature genetics. 2009;41(6):677–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fox ER, Young JH, Li Y, Dreisbach AW, Keating BJ, Musani SK, et al. Association of genetic variation with systolic and diastolic blood pressure among African Americans: the Candidate Gene Association Resource study. Human molecular genetics. 2011;20(11):2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hong K-W, Jin H-S, Lim J-E, Kim S, Go MJ, Oh B. Recapitulation of two genomewide association studies on blood pressure and essential hypertension in the Korean population. Journal of human genetics. 2010;55(6):336–41. [DOI] [PubMed] [Google Scholar]
- 48.Endres BT, Priestley JR, Palygin O, Flister MJ, Hoffman MJ, Weinberg BD, et al. Mutation of Plekha7 attenuates salt-sensitive hypertension in the rat. Proceedings of the National Academy of Sciences. 2014;111(35):12817–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu X, Feng T, Tayo BO, Liang J, Young JH, Franceschini N, et al. Meta-analysis of correlated traits via summary statistics from GWASs with an application in hypertension. The American Journal of Human Genetics. 2015;96(1):21–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang L, Curhan GC, Forman JP. Plasma Insulin-Like Growth Factor-1 Level and Risk of Incident Hypertension in Non-Diabetic Women. Journal of hypertension. 2011;29(2):229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Franceschini N, Carty CL, Lu Y, Tao R, Sung YJ, Manichaikul A, et al. Variant Discovery and Fine Mapping of Genetic Loci Associated with Blood Pressure Traits in Hispanics and African Americans. PLoS One. 2016;11(10):e0164132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J, Shi J, Huang W, Sun J, Wu Y, Duan Q, et al. Variant Near FGF5 Has Stronger Effects on Blood Pressure in Chinese With a Higher Body Mass Index. American journal of hypertension. 2015;28(8):1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gong Y, McDonough CW, Wang Z, Hou W, Cooper-DeHoff RM, Langaee TY, et al. Hypertension Susceptibility Loci and Blood Pressure Response to Antihypertensives-Results from the Pharmacogenomic Evaluation of Antihypertensive Responses (PEAR) Study. Circulation: Cardiovascular Genetics. 2012:CIRCGENETICS. 112.964080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, et al. Genome-wide association study identifies eight loci associated with blood pressure. Nature genetics. 2009;41(6):666–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kelly TN, Takeuchi F, Tabara Y, Edwards TL, Kim YJ, Chen P, et al. Genome-wide association study meta-analysis reveals transethnic replication of mean arterial and pulse pressure loci. Hypertension. 2013;62(5):853–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hirawa N, Fujiwara A, Umemura S. ATP2B1 and blood pressure: from associations to pathophysiology. Curr Opin Nephrol Hypertens. 2013;22(2):177–84. [DOI] [PubMed] [Google Scholar]
- 57.Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478(7367):103–9. Epub 2011/09/13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Haga SB. Impact of limited population diversity of genome-wide association studies. Genetics in Medicine. 2010;12(2):81–4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


