Abstract
The gut microbiota shows a wide inter-individual variation, but its within-individual variation is relatively stable over time. A functional core microbiome, provided by abundant bacterial taxa, seems to be common to various human hosts regardless of their gender, geographic location, and age. With advancing chronological age, the gut microbiota becomes more diverse and variable. However, when measures of biological age are used with adjustment for chronological age, overall richness decreases while a certain group of bacteria associated with frailty increases. This highlights the importance of considering biological or functional measures of aging. Studies using model organisms indicate that age-related gut dysbiosis may contribute to unhealthy aging and reduced longevity. The gut microbiome depends on the host nutrient signaling pathways for its beneficial effects on host health and lifespan, and gut dysbiosis disrupting the interdependence may diminish the beneficial effects or even have reverse effects. Gut dysbiosis can trigger the innate immune response and chronic low-grade inflammation, leading to many age-related degenerative pathologies and unhealthy aging. The gut microbiota communicates with the host through various biomolecules, nutrient signaling-independent pathways, and epigenetic mechanisms. Disturbance of these communications by age-related gut dysbiosis can affect the host health and lifespan. This may explain the impact of the gut microbiome on health and aging.
Keywords: Aging, Gut, Health, Intestine, Lifespan, Longevity, microbiome, microbiota
The human gut microbiota
The human digestive tract is inhabited by numerous microorganisms. The total estimated number of gut microorganisms is somewhere between 1013 and 1014, hovering around the total estimated number of human body cells (3~4×1013) [1]. Bacteria outnumber all other domains of gut microbes, and the total number of species found in the gut microbiota is estimated to be about 500 ~1,000 [2]. The most populous bacterial phyla, constituting more than 90% of the gut microbiota, are Bacteriodetes and Firmicutes [3]. The remainder consists of many species in other phyla in lower abundance, some of which may provide important metabolites and functions for healthy aging.
According to the 16S ribosomal DNA sequencing data of fecal samples, individual gut microbiotas show distinct profiles, and this inter-individual variation is greater in older adults [4]. Longitudinally, however, gut microbiotas of healthy adults are relatively stable even for decades [5]. These phylogenetic data are supported by metagenomic analysis of whole shotgun sequencing data: SNP variation patterns show stability over time [6]. Thus, once established early in life (even within three years after birth) [7], the gut microbiota seems to be rather stably maintained. Nevertheless, it is responsive to the host’s dietary and health conditions [8], much as the host’s epigenome is to various environmental cues. In fact, the gut microbiota interfaces the gut environment with the epigenome, but its communication with the host systems involves various signaling networks and their mediators. For instance, the “gut-brain axis” connects the gut microbiome with the central nervous system via neurons, hormones, or cytokines [9].
Despite the marked inter-individual variation in the gut microbiota profile, an array of bacterial genes exists that individual hosts share, as shown by functional metagenomics [10]. This “functional core microbiome” (in this review, microbiome denotes the combined genomes of the constituent microbes in the microbiota) is collectively provided by different microbial taxa, indicating that different microbial species can functionally replace one another. The presence of such a core microbiome makes sense if the core functions concern housekeeping or other important biochemical or physiological pathways [11,12]. In this regard, the core microbiome is likely to consist of omnipresent taxa. Fecal samples collected from different countries over several continents contain three distinct microbial metagenomic clusters, designated as enterotypes [13]. These enterotypes are characterized by the most abundant genera in Bacteroidaceae, Prevotellaceae, and Ruminococcuceae families. The first two belong to the Bacteroidetes phylum and the last to the Firmicutes. The enterotypes are not correlated with host features such as body mass index, gender, and even age, which implies the universality of the enterotypes. A separate phylogenetic study found a core microbiota common to four different age groups: young (22-48), elderly (65-75), centenarian (99-104), and semi-supercentenarian (105-109) [12]. This core microbiota includes Bacteroidaceae, Ruminococcaceae, and Lachnospiraceae families, the last two of which belong to the Firmicutes. Thus, most adult age groups, from young to extremely old, seem to possess a common core function in their microbiomes that is provided by members of abundant taxa. If so, what’s important in the gut microbiota for healthy aging could be a compositional change in the functional core microbiome or an enrichment of non-core functions with advancing age.
Changes in the gut microbiota associated with longevity or healthy aging
One potentially productive approach to the roles of the gut microbiota in human aging is to compile age-related changes in the gut microbiota and examine whether these changes have any biological relevance. Cross-sectional studies of fecal samples from individuals in different age groups suggest age-related changes in the gut microbiota composition and diversity, which concurs with longitudinal study results [4]. In general, the gut microbiota of the elderly becomes more diverse and variable with advancing age [4]. For instance, the three bacterial families in the core microbiota, mentioned in the previous section, become less abundant in older age groups, while certain health-associated species become more abundant in older age groups including centenarians and semi-supercentenarians (aged 105-109) [12]. These changes in composition and diversity are also reflected in age-dependent reshaping of co-abundance networks.
Certain changes in composition and diversity are associated with biological or functional age, independent of chronological age. Various measures of frailty have been used as indicators of biological age [14], and gut microbiota composition is associated with biological age [8,15-17]. Also, gut microbial diversity inversely correlates with biological age, but not with chronological age [16,17]. Furthermore, a co-abundance module consisting of Ruminococcus, Coprobacillus, and Eggerthella genera becomes abundant with an increase in biological age, independent of chronological age (Maffei 2017). The first two genera of this module belong to the Firmicutes phylum, and the last one to the Actinobacteria. An interpretation of these results is that as biological age increases, overall gut microbiota richness decreases, while some microbial taxa associated with unhealthy aging emerge. Thus, what happens in the gut microbiota with advancing biological age can be very different from what happens with chronological age, which illustrates the importance of using a biological or functional measure in aging studies.
Even with an age-related change that seems biologically meaningful on hand, it is difficult to determine any causal roles of the human microbiota in aging. However, the lack of such information has been partially circumvented by data generated from studies using tractable model organisms.
Animal models of gut microbiota in longevity and healthy aging
Nutrient signaling pathways
Nutrition is the major factor that shapes the host’s gut microbiota. It also affects the host’s epigenome; for example, folate and choline, as dietary methyl-donors, can affect DNA methylation [18]. Furthermore, nutrition is a key environmental factor that interacts with the host genes, especially those in the nutrient-signaling pathways. Thus, nutrition is a common factor that can link the gut microbiome with the host genome.
Dietary or caloric restriction (CR), a moderate reduction in food intake, extends both health and lifespan [19]. It is a widely conserved intervention that engages biological pathways that are evolutionarily conserved from yeast to primates. However, much remains to be learned. Nutrient availability may not be the only input that can affect the pathways that actuate the CR response, because modifications of gustatory or olfactory neurons or even treatment of animals with diet-derived odors can modulate lifespan in Caenorhabditis elegans and Drosophila melanogaster [20,21].
The nutrient signaling pathways include the insulin/insulin-like growth factor-1 signaling (IIS) pathway [19]. Activation of AKT (protein kinase B) by nutrient abundance leads to inactivation of FOXO (a Forkhead box O transcription factor). FOXO is a central factor to longevity as it induces expression of many proteins involved in cell metabolism, autophagy, and stress-response [22]. Thus, reduced nutrient availability or mutations that weaken the AKT activity result in enhanced FOXO activity. FOXO is abundantly expressed in the C. elegans intestine [23], and intestinal FOXO expression alone fully restores the lifespan of FOXO-deficient mutants [24].
Besides FOXO, FKH, a homolog of FOXA (Forhhead box A), is required for the IIS. Gut barrier and nutrient transport functions decline with aging, and dysbiosis weakens the intestinal barrier function, resulting in higher mortality of aged fruit flies [25]. Gut-specific FKH upregulation improves gut barrier function and increases expression of nutrient transporters in aged flies, resulting in lifespan extension. FKH is also required for the mTOR pathway (see below), thus connecting both pathways [26].
Another important nutrient signaling pathway involves mTOR (mechanistic or mammalian target of rapamycin), which responds not only to nutrients but also other signals, such as growth hormones and mitogens. As the name indicates, rapamycin targets mTOR to block its signaling pathway for cell growth and proliferation. A serine/threonine kinase, mTOR exists in two protein complexes, mTORC1 and mTORC2, as a catalytic subunit in each complex. Both complexes regulate many cellular processes important for cell growth, proliferation, and survival, but some differences exist between the two [27]. For example, the effects of mTORC1 seem to be more extensive than those of mTORC2, and the inhibition of mTOR by rapamycin in mTORC1 is more immediate than that in mTORC2. Thus, the anti-aging effects of rapamycin are viewed to be mainly through its effect on mTORC1 [27]. Significantly, the health and lifespan extension by dietary intervention or inhibition of TOR activity by rapamycin involves altered gut microbiota in mice [28,29].
As mentioned above, the gut microbiome and the host nutrient signaling pathways are interconnected in part because nutrition is the major common factor. A series of experiments using C. elegans shows that the relationship between the gut microbiome and the host IIS and TOR signaling pathways goes deep to the functional level to modulate the host’s health and lifespan. These worms are grown on media containing a single bacterial strain as a food source, which greatly facilitates sorting out bacterial genes of interest. Han et al. used a collection of E. coli viable deletion mutants as a food source and found 29 bacterial mutants robustly extending the host lifespan [30]. Some of them also lowered the mortality of worms carrying germline tumors or human amyloid-β. Interestingly, the lifespan extending effects of many of these bacterial mutants disappeared in the host worms containing mutations that disable the IIS/TOR signaling pathways. This suggests that these bacterial mutants in the gut depend on the host IIS/TOR pathways for their lifespan extending effects. Disturbance of the dependence may well diminish or even reverse the beneficial effects.
Gut dysbiosis and innate immunity
Changes in microbial composition or outgrowths of certain taxa lead to gut dysbiosis, which refers to disruption of the commensal homeostasis between the host and the gut microbiota. Gut dysbiosis is associated with many pathological conditions, including inflammatory bowel disease, obesity, diabetes, cardiovascular diseases, and neurodegenerative diseases [9,31]. Furthermore, a study using African turquoise killifish indicates that prevention of age-related gut dysbiosis can be conducive to longevity. Smith et al. found that in the killifish, loss of the gut microbial richness occurs during aging, and middle-aged killifish that were treated with antibiotics and then with intestinal contents from young fish showed significant lifespan extension [32]. The long-lived recipient fish maintained higher mobility and microbial diversity compared with controls.
In Drosophila intestinal epithelium, deregulated proliferation of intestinal stem cells (ISCs) increases in aging flies, leading to polyploid and poorly differentiated cells [33]. This dysplasia accompanies epithelial malfunctions, such as loss of the intestinal barrier for selective permeability, leading to increased infection and mortality. Curbing the ISC proliferation in old flies extends lifespan [34]. The number of gut microbes increases in older flies [35], and old flies reared axenically (e.g., with antibiotics) show delayed dysplasia [36]. These results suggest that gut dysbiosis most likely underlies the intestine epithelial dysplasia in aging flies.
In Drosophila, which lacks adaptive immune function, the primary immune defense is provided by the innate immune response. Two main innate immune pathways are at work in ISCs. One is through activation of dual oxidase (Duox) that generates reactive oxygen species (ROS) in response to bacterial uracil [37,38]. The other pathway is through activation of the immune deficiency (IMD/Relish) pathway, in response to bacteria-derived peptidoglycan, resulting in increased expression of antimicrobial peptides (AMP). The ROS or AMP initiates an immediate immune response against invading pathogens or gut dysbiosis [37,38].
The gut microbiota of old animals is so dysbiotic that it can initiate an innate immune response. The number of gut microbes increases with age [35], and AMP expression significantly increases in aging flies, likely in response to increasing inflow of microbes. Flies with chronically activated IMD/Relish (NFκB) signaling are prone to bacterial infection and lifespan reduction. Modulation of dysplasia and lifespan can be achieved by regulated expression of peptidoglycan recognition proteins in conventionally grown flies, but not in antibiotics-treated flies [21,36]. Thus, the innate immune response, triggered by gut dysbiosis, can affect gut dysplasia and mortality of the host.
The causative role of gut dysbiosis in innate-immunity-induced inflammation becomes convincing in a study using mice. Transfer of the gut microbiota from old mice to young germ-free mice triggers innate immunity and inflammatory responses mimicking “inflammaging” [39]. These include increased CD4+ T cell differentiation in spleen, inflammation in the intestine and upregulated expression of inflammatory cytokine genes, such as TNF-α, and increased circulation of inflammatory factors of bacterial origin. Circulation of bacterial compounds in the host is probably due to damage of the intestinal epithelium by inflammation. All these results indicate that the elevated innate immune response caused by dysbiosis in the aging gut can provoke chronic inflammation, leading to gut dysplasia. Gut dysplasia in turn can cause defective epithelial functioning, making the host prone to unhealthy aging, infection, and increased mortality.
The gut microbiota in animal models of diseases
Factors that contribute to gut dysbiosis are linked to development of pathological conditions. These factors include unbalanced diet, environmental toxins, drugs, ROS, psychological stressors, and other proinflammatory factors. For instance, gut dysbiosis caused by taking antibiotics or high-fat or carbohydrate diet is associated with obesity and metabolic disorders [40,41]. Causal roles of such dysbiotic factors in pathogenesis can be seen better in animal studies.
Radaura et al. [42] transplanted fecal matter from twins discordant for obesity into germ-free mice and found that the mice’s body composition measurements vary according to the human donor’s body composition features. The differences in body composition between mice with the obese twin’s microbiota and mice with the lean twin’s microbiota are associated with the differences in meta-transcriptome profiles. These results show a causal role of the gut microbiota in obesity as well as the potential use of the gut microbiota as a therapeutic target.
Another approach to causal relationships between microbes and phenotypes was made using gnotobiotic mice carrying or missing distinct microbial groups [43]. These investigators generated groups of gnotobiotic mice that carry defined microbiotas and exhibit differential survival rates for colitis. By creating hybrid mice with mixed microbiotas that display intermediate susceptibility to colitis, they found the Lachnospiraceae family beneficial. By directly administering suspected members of the family into colitis-prone mice, they found Clostridium immunis to be protective against colitis-associated death of the host.
Unlike aged mice fed a low-glycemic diet, those fed a high-glycemic, iso-caloric diet develop many features of age-related macular degeneration (AMD) [44]. Changing from high- to low-glycemic diet, even late in life, can stop or reverse progression of the AMD features. Low glycemic diets seem to suppress accumulation of several modified molecules, including advanced glycation end products. Furthermore, the severity of retina damage is inversely correlated with the abundance of serotonin, suggesting a protective role of serotonin. In fact, selective serotonin reuptake inhibitors (SSRIs), used as antidepressants, increase serotonin levels in the brain, and people using them are less likely to develop diabetic retinopathy [45]. Most of this mono amine neurotransmitter is synthesized by tryptophan hydroxylase 1 (TPH1) in enterochromaffin cells of the digestive tract. Certain spore-forming gut bacteria, whose identity is yet to be determined, stimulate TPH1 expression and serotonin release from enterochromaffin cells, which seems to be mediated by more than a dozen gut metabolites, including short-chain fatty acids [46]. These results suggest that the effects of glycemic diets on AMD features may involve signaling biomolecules along the gut-brain axis.
The gut microbiota is also causally linked to development of neurodegenerative disorders [9]. One of the major risk factors for Parkinson’s disease is aggregation of α-synuclein in brain neurons. Mice overexpressing α-synuclein develop α-synuclein aggregates and defects in motor function and gut motility [47]. However, germ-free mice overexpressing α-synuclein show significantly fewer α-synuclein aggregates, and fecal samples from Parkinson’s patients exacerbate motor dysfunction in mice, indicating the causative role of the gut microbiota in α-synuclein aggregation and the resulting pathology. Furthermore, enteroendocrine cells in the gut epithelium express α-synuclein, and these enteroendocrine cells are physically close to α-synuclein-containing enteric neurons, prompting the hypothesis that α-synuclein originates from the gut and spreads to the central nervous system [48].
Causative roles of the gut microbiota in Alzheimer’s disease were studied similarly. Cerebral deposition of amyloid-β (Aβ) plaques is a critical risk factor for various type of dementia, including Alzheimer’s disease. Transgenic mice expressing Aβ precursor protein start to accumulate cerebral Aβ early on, and their gut microbiota composition differs greatly compared with that of the non-transgenic littermates [49]. Transgenic mice rendered germ free show lower Aβ levels and significantly reduced cerebral Aβ deposition compared with conventionally reared transgenic mice. Furthermore, transfer of the gut microbiota from conventionally-reared transgenic mice to germ-free transgenic mice was more effective than transfer from wild-type mice in the induction of Aβ pathology. All these observations highlight the importance of the gut brain axis in development of neurodegenerative disorders.
Biomolecules in the gut microbiota that influence healthy aging
Many compounds, either endogenous or exogenous, are known to modulate health and lifespan. One such biomolecule of exogenous origin is rapamycin, which is a natural metabolite produced by soil bacteria. Clinically used as an immunosuppressant, rapamycin binds to its immunophilin, an FK binding protein (FKBP12), and the complex targets mTOR to inhibit its function. In mice, rapamycin alters not only the host gene expression profiles but also the gut metagenomes [29,50]. Another example of compound of exogenous origin is the diabetes drug metformin. In C. elegans, excessive folate limits lifespan [51], and metformin extends lifespan through inhibition of bacterial folate and methionine metabolism [52,53]. Interestingly, however, altering folate levels of the host has no effect on its longevity [53]. It could be a secondary metabolite generated from bacterial folate metabolism that is responsible for the altered lifespan. Nitric oxide (NO) is a simple yet versatile signaling molecule involved in many physiological functions [54]. It can be produced by the human gut microbiota, but the contribution of the NO generated by gut flora was unknown [55]. A clue was found in a study using C. elegans, which cannot produce NO due to lack of the NO synthase. NO provided either by gut bacteria or by exogenous supplementation increases lifespan and stress resistance, demonstrating a profound biological function of NO and the gut microbiota [56].
Many biomolecules are produced endogenously by commensal microbes in the digestive tract. These biomolecules include various vitamins, fermentation products, and gut-derived hormones and compounds that are important in neurological health [9]. Colanic acid, either bacteria-derived or exogenously added, extends lifespan in C. elegans and D. melanogaster [30]. Colanic acid is an exopolysaccharide composed of a repeating unit of sugar monomers that is extracellularly secreted by bacteria. The beneficial effects of colanic acid are independent of the CR signaling pathways. Colanic acid promotes mitochondrial fission and enhances the mitochondrial unfolded protein response under stress conditions.
Of the fermentation products, short-chain fatty acids have profound effects on the host [9]. Short-chain fatty acids are products of the breakdown of dietary fibers by the anaerobic gut microbiota. They can easily enter the circulation from the gut and have beneficial roles in energy metabolism. Acetate can reduce serum cholesterol and triglyceride levels, propionate can lower glucose levels, and butyrate can increase insulin sensitivity in mice. On the other hand, short-chain fatty acids can have harmful effects. In the mouse model of Parkinson’s disease mentioned above [47], α-synuclein aggregates activate immune cells, including phagocytic microglial cells in the CNS, in a gut-microbiota dependent manner. Treatment of germ-free mice overexpressing α-synuclein with a mixture of acetate, propionate and butyrate leads to neuroinflammation and motor deficits. These results indicate that short-chain fatty acids constitute the main causal factor for the α-synuclein-related pathology caused by the gut microbiota. Age-related changes in the metagenomics of short-chain fatty acid production have been observed. For instance, frequencies of genes encoding short-chain fatty acid production and those involved in carbohydrate breakdown decrease, while those of genes involved in protein breakdown increase [57]. The reduced frequency of genes for short-chain fatty acid production is also associated with frailty [8]. Thus, the short-chain fatty acids have the potential to modulate healthy aging.
Short-chain fatty acids seem to have both negative and positive effects on health, and to understand this complexity, at least two things need to be considered. Physiological responses to input doses are often nonlinear (e.g., U-shaped or J-shaped). To take extreme examples, inactivation of FOXO shortens lifespan, so does its chronic activation [36]. Similarly, chronic activation of IMD/Relish or inactivation of Duox results in shortened lifespan in flies [36,37]. This indicates that given a dose-response relationship, there is an optimal dose range for the best desired response. The other thing to consider is that the overall effect of a global factor is likely to be the net effect (balance) of all the positive and negative effects combined. Thus, FOXO, a transcription factor regulating expression of many proteins, is an important factor for healthy aging and lifespan of model organisms; given muscle metabolism, however, activation of FOXO by HDACs can cause skeletal muscle atrophy in mice, probably via autophagy [58], and inactivation of HDAC by butyrate can reverse the atrophy (see below) [59].
Of the primary short-chain fatty acids, butyrate, like trichostatin A, inhibits histone deacetylases (HDACs) and thus has a profound effect on the host epigenome [60]. HDACs catalyze removal of the acetyl group from lysine residues in histones, leading to heterochromatization and transcription repression. Thus, inhibition of HDACs by butyrate promotes histone lysine acetylation, leading to an open chromatin state and transcription activation. Butyrate increases lifespan in Drosophila [61]. In aging mice, butyrate counters muscle atrophy [59], as does activation of FOXO by HDAC1 [58]. Butyrate also enhances memory functioning of aged mice [62]. These results demonstrate that relatively simple organic compounds, such as butyrate, can have far-reaching systemic effects on healthy aging.
Summary
With advancing chronological age, the gut microbiota becomes more diverse, increasing in phylogenetic richness (Fig. 1). However, when biological age is used with adjustment for chronological age, overall richness decreases while certain bacterial taxa associated with unhealthy aging thrive. Thus, as biological age increases, the homeostatic relationship between the gut microbiota and the host deteriorates, while gut dysbiosis increases. These dysbiotic changes in the aging gut can negate the beneficial effects of the gut microbiome on the nutrient signaling pathways, and provoke pro-inflammatory innate immunity and other pathological conditions. Gut dysbiosis can also disturb the communications between the gut microbiota and the host through various biomolecules, CR-independent signaling pathways, and epigenetic mechanisms, affecting host health and longevity.
Fig. 1.
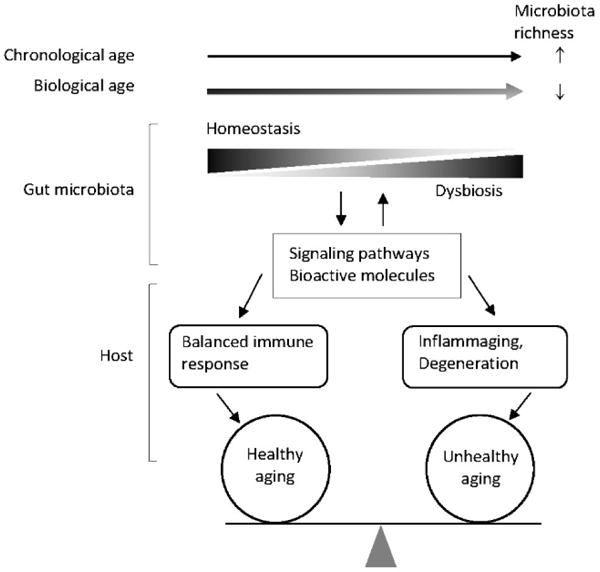
Biological age-dependent gut dysbiosis and unhealthy aging. Biology of aging is better approached using a functional measure of age. An increase in chronological age (in the direction of the arrow) is associated with an increase in phylogenetic richness of the gut microbiota, but an increase in biological age shows an inverse association. As biological age increases, homeostasis of the gut microbiota with the host decreases, while dysbiosis increases. The dysbiotic changes are communicated to the host through various signaling pathways and bioactive molecules, which either delays or promotes proinflammatory immune responses and age-related degenerative pathologies.
Acknowledgments
This work was supported by grants from the National Institute of General Medical Sciences (P20GM103629) and the National Institute on Aging (P01AG022064 and K01AG027905) of the National Institutes of Health.
Footnotes
Conflicts of interests
None declared.
References
- 1.Sender R, Fuchs S, Milo R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016;14:e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sommer F, Backhed F. The gut microbiota–masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 3.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, Mende DR, Li J, Xu J, Li S, Li D, Cao J, Wang B, Liang H, Zheng H, Xie Y, Tap J, Lepage P, Bertalan M, Batto JM, Hansen T, Le Paslier D, Linneberg A, Nielsen HB, Pelletier E, Renault P, Sicheritz-Ponten T, Turner K, Zhu H, Yu C, Li S, Jian M, Zhou Y, Li Y, Zhang X, Li S, Qin N, Yang H, Wang J, Brunak S, Dore J, Guarner F, Kristiansen K, Pedersen O, Parkhill J, Weissenbach J, Meta HITC, Bork P, Ehrlich SD, Wang J. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claesson MJ, Cusack S, O’Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, Marchesi JR, Falush D, Dinan T, Fitzgerald G, Stanton C, van Sinderen D, O’Connor M, Harnedy N, O’Connor K, Henry C, O’Mahony D, Fitzgerald AP, Shanahan F, Twomey C, Hill C, Ross RP, O’Toole PW. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4586–4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, Rosenbaum M, Gordon JI. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schloissnig S, Arumugam M, Sunagawa S, Mitreva M, Tap J, Zhu A, Waller A, Mende DR, Kultima JR, Martin J, Kota K, Sunyaev SR, Weinstock GM, Bork P. Genomic variation landscape of the human gut microbiome. Nature. 2013;493:45–50. doi: 10.1038/nature11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, Heath AC, Warner B, Reeder J, Kuczynski J, Caporaso JG, Lozupone CA, Lauber C, Clemente JC, Knights D, Knight R, Gordon JI. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, Harris HM, Coakley M, Lakshminarayanan B, O’Sullivan O, Fitzgerald GF, Deane J, O’Connor M, Harnedy N, O’Connor K, O’Mahony D, van Sinderen D, Wallace M, Brennan L, Stanton C, Marchesi JR, Fitzgerald AP, Shanahan F, Hill C, Ross RP, O’Toole PW. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 9.Westfall S, Lomis N, Kahouli I, Dia SY, Singh SP, Prakash S. Microbiome, probiotics and neurodegenerative diseases: deciphering the gut brain axis. Cell Mol Life Sci. 2017;74:3769–3787. doi: 10.1007/s00018-017-2550-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lozupone CA, Stombaugh JI, Gordon JI, Jansson JK, Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lloyd-Price J, Abu-Ali G, Huttenhower C. The healthy human microbiome. Genome Med. 2016;8:51. doi: 10.1186/s13073-016-0307-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biagi E, Franceschi C, Rampelli S, Severgnini M, Ostan R, Turroni S, Consolandi C, Quercia S, Scurti M, Monti D, Capri M, Brigidi P, Candela M. Gut Microbiota and Extreme Longevity. Curr Biol. 2016;26:1480–1485. doi: 10.1016/j.cub.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 13.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Dore J, Meta HITC, Antolin M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Foerstner KU, Friss C, van de Guchte M, Guedon E, Haimet F, Huber W, van Hylckama-Vlieg J, Jamet A, Juste C, Kaci G, Knol J, Lakhdari O, Layec S, Le Roux K, Maguin E, Merieux A, Melo Minardi R, M’Rini C, Muller J, Oozeer R, Parkhill J, Renault P, Rescigno M, Sanchez N, Sunagawa S, Torrejon A, Turner K, Vandemeulebrouck G, Varela E, Winogradsky Y, Zeller G, Weissenbach J, Ehrlich SD, Bork P. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S, Jazwinski SM. Quantitative measures of healthy aging and biological age. Healthy Aging Res. 2015;4 doi: 10.12715/har.2015.4.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Tongeren SP, Slaets JP, Harmsen HJ, Welling GW. Fecal microbiota composition and frailty. Appl Environ Microbiol. 2005;71:6438–6442. doi: 10.1128/AEM.71.10.6438-6442.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson MA, Jeffery IB, Beaumont M, Bell JT, Clark AG, Ley RE, O’Toole PW, Spector TD, Steves CJ. Signatures of early frailty in the gut microbiota. Genome Med. 2016;8:8. doi: 10.1186/s13073-016-0262-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maffei VJ, Kim S, Blanchard Et, Luo M, Jazwinski SM, Taylor CM, Welsh DA. Biological Aging and the Human Gut Microbiota. J Gerontol A Biol Sci Med Sci. 2017;72:1474–1482. doi: 10.1093/gerona/glx042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson OS, Sant KE, Dolinoy DC. Nutrition and epigenetics: an interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. J Nutr Biochem. 2012;23:853–859. doi: 10.1016/j.jnutbio.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alic N, Partridge L. Death and dessert: nutrient signalling pathways and ageing. Curr Opin Cell Biol. 2011;23:738–743. doi: 10.1016/j.ceb.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alcedo J, Kenyon C. Regulation of C. elegans Longevity by Specific Gustatory and Olfactory Neurons. Neuron. 2004;41:45–55. doi: 10.1016/s0896-6273(03)00816-x. [DOI] [PubMed] [Google Scholar]
- 21.Libert S, Chao Y, Chu X, Pletcher SD. Trade-offs between longevity and pathogen resistance in Drosophila melanogaster are mediated by NFkappaB signaling. Aging Cell. 2006;5:533–543. doi: 10.1111/j.1474-9726.2006.00251.x. [DOI] [PubMed] [Google Scholar]
- 22.Martins R, Lithgow GJ, Link W. Long live FOXO: unraveling the role of FOXO proteins in aging and longevity. Aging Cell. 2016;15:196–207. doi: 10.1111/acel.12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwon ES, Narasimhan SD, Yen K, Tissenbaum HA. A new DAF-16 isoform regulates longevity. Nature. 2010;466:498–502. doi: 10.1038/nature09184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- 25.Clark RI, Salazar A, Yamada R, Fitz-Gibbon S, Morselli M, Alcaraz J, Rana A, Rera M, Pellegrini M, Ja WW, Walker DW. Distinct Shifts in Microbiota Composition during Drosophila Aging Impair Intestinal Function and Drive Mortality. Cell reports. 2015;12:1656–1667. doi: 10.1016/j.celrep.2015.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolukbasi E, Khericha M, Regan JC, Ivanov DK, Adcott J, Dyson MC, Nespital T, Thornton JM, Alic N, Partridge L. Intestinal Fork Head Regulates Nutrient Absorption and Promotes Longevity. Cell reports. 2017;21:641–653. doi: 10.1016/j.celrep.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017;168:960–976. doi: 10.1016/j.cell.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hurez V, Dao V, Liu A, Pandeswara S, Gelfond J, Sun L, Bergman M, Orihuela CJ, Galvan V, Padron A, Drerup J, Liu Y, Hasty P, Sharp ZD, Curiel TJ. Chronic mTOR inhibition in mice with rapamycin alters T, B, myeloid, and innate lymphoid cells and gut flora and prolongs life of immune-deficient mice. Aging Cell. 2015;14:945–956. doi: 10.1111/acel.12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bitto A, Ito TK, Pineda VV, LeTexier NJ, Huang HZ, Sutlief E, Tung H, Vizzini N, Chen B, Smith K, Meza D, Yajima M, Beyer RP, Kerr KF, Davis DJ, Gillespie CH, Snyder JM, Treuting PM, Kaeberlein M. Transient rapamycin treatment can increase lifespan and healthspan in middle-aged mice. Elife. 2016;5 doi: 10.7554/eLife.16351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han B, Sivaramakrishnan P, Lin CJ, Neve IAA, He J, Tay LWR, Sowa JN, Sizovs A, Du G, Wang J, Herman C, Wang MC. Microbial Genetic Composition Tunes Host Longevity. Cell. 2017;169:1249–1262. e1213. doi: 10.1016/j.cell.2017.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lau K, Srivatsav V, Rizwan A, Nashed A, Liu R, Shen R, Akhtar M. Bridging the Gap between Gut Microbial Dysbiosis and Cardiovascular Diseases. Nutrients. 2017;9 doi: 10.3390/nu9080859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith P, Willemsen D, Popkes M, Metge F, Gandiwa E, Reichard M, Valenzano DR. Regulation of life span by the gut microbiota in the short-lived African turquoise killifish. Elife. 2017;6 doi: 10.7554/eLife.27014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jasper H. Exploring the physiology and pathology of aging in the intestine of Drosophila melanogaster. Invertebr Reprod Dev. 2015;59:51–58. doi: 10.1080/07924259.2014.963713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biteau B, Karpac J, Supoyo S, Degennaro M, Lehmann R, Jasper H. Lifespan extension by preserving proliferative homeostasis in Drosophila. PLoS Genet. 2010;6:e1001159. doi: 10.1371/journal.pgen.1001159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren C, Webster P, Finkel SE, Tower J. Increased internal and external bacterial load during Drosophila aging without life-span trade-off. Cell Metab. 2007;6:144–152. doi: 10.1016/j.cmet.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 36.Guo L, Karpac J, Tran SL, Jasper H. PGRP-SC2 promotes gut immune homeostasis to limit commensal dysbiosis and extend lifespan. Cell. 2014;156:109–122. doi: 10.1016/j.cell.2013.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ha E-M, Lee K-A, Park SH, Kim S-H, Nam H-J, Lee H-Y, Kang D, Lee W-J. Regulation of DUOX by the Gαq-Phospholipase Cβ-Ca2+ Pathway in Drosophila Gut Immunity. Developmental Cell. 2009;16:386–397. doi: 10.1016/j.devcel.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 38.Leulier F, Royet J. Maintaining immune homeostasis in fly gut. Nat Immunol. 2009;10:936–938. doi: 10.1038/ni0909-936. [DOI] [PubMed] [Google Scholar]
- 39.Fransen F, van Beek AA, Borghuis T, Aidy SE, Hugenholtz F, van der Gaast-de Jongh C, Savelkoul HFJ, De Jonge MI, Boekschoten MV, Smidt H, Faas MM, de Vos P. Aged Gut Microbiota Contributes to Systemical Inflammaging after Transfer to Germ-Free Mice. Front Immunol. 2017;8:1385. doi: 10.3389/fimmu.2017.01385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turta O, Rautava S. Antibiotics, obesity and the link to microbes - what are we doing to our children? BMC Med. 2016;14:57. doi: 10.1186/s12916-016-0605-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan YK, Estaki M, Gibson DL. Clinical consequences of diet-induced dysbiosis. Ann Nutr Metab. 2013;63(Suppl 2):28–40. doi: 10.1159/000354902. [DOI] [PubMed] [Google Scholar]
- 42.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, Muehlbauer MJ, Ilkayeva O, Semenkovich CF, Funai K, Hayashi DK, Lyle BJ, Martini MC, Ursell LK, Clemente JC, Van Treuren W, Walters WA, Knight R, Newgard CB, Heath AC, Gordon JI. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Surana NK, Kasper DL. Moving beyond microbiome-wide associations to causal microbe identification. Nature. 2017;552:244–247. doi: 10.1038/nature25019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rowan S, Jiang S, Korem T, Szymanski J, Chang ML, Szelog J, Cassalman C, Dasuri K, McGuire C, Nagai R, Du XL, Brownlee M, Rabbani N, Thornalley PJ, Baleja JD, Deik AA, Pierce KA, Scott JM, Clish CB, Smith DE, Weinberger A, Avnit-Sagi T, Lotan-Pompan M, Segal E, Taylor A. Involvement of a gut-retina axis in protection against dietary glycemia-induced age-related macular degeneration. Proc Natl Acad Sci U S A. 2017;114:E4472–E4481. doi: 10.1073/pnas.1702302114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yekta Z, Xie D, Bogner HR, Weber DR, Zhang X, Harhay M, Reese PP. The association of antidepressant medications and diabetic retinopathy among people with diabetes. J Diabetes Complications. 2015;29:1077–1084. doi: 10.1016/j.jdiacomp.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 46.Yano JM, Yu K, Donaldson GP, Shastri GG, Ann P, Ma L, Nagler CR, Ismagilov RF, Mazmanian SK, Hsiao EY. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, Challis C, Schretter CE, Rocha S, Gradinaru V, Chesselet MF, Keshavarzian A, Shannon KM, Krajmalnik-Brown R, Wittung-Stafshede P, Knight R, Mazmanian SK. Gut Microbiota Regulate Motor Deficits and Neuroinflammation in a Model of Parkinson’s Disease. Cell. 2016;167:1469–1480. e1412. doi: 10.1016/j.cell.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chandra R, Hiniker A, Kuo YM, Nussbaum RL, Liddle RA. alpha-Synuclein in gut endocrine cells and its implications for Parkinson’s disease. JCI Insight. 2017;2 doi: 10.1172/jci.insight.92295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harach T, Marungruang N, Duthilleul N, Cheatham V, Mc Coy KD, Frisoni G, Neher JJ, Fak F, Jucker M, Lasser T, Bolmont T. Reduction of Abeta amyloid pathology in APPPS1 transgenic mice in the absence of gut microbiota. Sci Rep. 2017;7:41802. doi: 10.1038/srep41802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jung MJ, Lee J, Shin NR, Kim MS, Hyun DW, Yun JH, Kim PS, Whon TW, Bae JW. Chronic Repression of mTOR Complex 2 Induces Changes in the Gut Microbiota of Diet-induced Obese Mice. Sci Rep. 2016;6:30887. doi: 10.1038/srep30887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Virk B, Correia G, Dixon DP, Feyst I, Jia J, Oberleitner N, Briggs Z, Hodge E, Edwards R, Ward J, Gems D, Weinkove D. Excessive folate synthesis limits lifespan in the C. elegans: E. coli aging model. BMC Biol. 2012;10:67. doi: 10.1186/1741-7007-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cabreiro F, Au C, Leung KY, Vergara-Irigaray N, Cocheme HM, Noori T, Weinkove D, Schuster E, Greene ND, Gems D. Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell. 2013;153:228–239. doi: 10.1016/j.cell.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Virk B, Jia J, Maynard CA, Raimundo A, Lefebvre J, Richards SA, Chetina N, Liang Y, Helliwell N, Cipinska M, Weinkove D. Folate Acts in E. coli to Accelerate C. elegans Aging Independently of Bacterial Biosynthesis. Cell reports. 2016;14:1611–1620. doi: 10.1016/j.celrep.2016.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Snyder SH, Bredt DS. Biological roles of nitric oxide. Sci Am. 1992;266:68–71. 74–67. doi: 10.1038/scientificamerican0592-68. [DOI] [PubMed] [Google Scholar]
- 55.Vermeiren J, Van de Wiele T, Verstraete W, Boeckx P, Boon N. Nitric oxide production by the human intestinal microbiota by dissimilatory nitrate reduction to ammonium. J Biomed Biotechnol. 2009;2009:284718. doi: 10.1155/2009/284718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gusarov I, Gautier L, Smolentseva O, Shamovsky I, Eremina S, Mironov A, Nudler E. Bacterial nitric oxide extends the lifespan of C. elegans. Cell. 2013;152:818–830. doi: 10.1016/j.cell.2012.12.043. [DOI] [PubMed] [Google Scholar]
- 57.Rampelli S, Candela M, Turroni S, Biagi E, Collino S, Franceschi C, O’Toole PW, Brigidi P. Functional metagenomic profiling of intestinal microbiome in extreme ageing. Aging (Albany NY) 2013;5:902–912. doi: 10.18632/aging.100623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Beharry AW, Sandesara PB, Roberts BM, Ferreira LF, Senf SM, Judge AR. HDAC1 activates FoxO and is both sufficient and required for skeletal muscle atrophy. J Cell Sci. 2014;127:1441–1453. doi: 10.1242/jcs.136390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walsh ME, Bhattacharya A, Sataranatarajan K, Qaisar R, Sloane L, Rahman MM, Kinter M, Van Remmen H. The histone deacetylase inhibitor butyrate improves metabolism and reduces muscle atrophy during aging. Aging Cell. 2015;14:957–970. doi: 10.1111/acel.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rahman MM, Kukita A, Kukita T, Shobuike T, Nakamura T, Kohashi O. Two histone deacetylase inhibitors, trichostatin A and sodium butyrate, suppress differentiation into osteoclasts but not into macrophages. Blood. 2003;101:3451–3459. doi: 10.1182/blood-2002-08-2622. [DOI] [PubMed] [Google Scholar]
- 61.Vaiserman AM, Kolyada AK, Koshel NM, Simonenko AV, Pasyukova EG. Effect of histone deacetylase inhibitor sodium butyrate on viability and life span in Drosophila melanogaster. Advances in Gerontology. 2013;3:30–34. [PubMed] [Google Scholar]
- 62.Blank M, Werenicz A, Velho LA, Pinto DF, Fedi AC, Lopes MW, Peres TV, Leal RB, Dornelles AS, Roesler R. Enhancement of memory consolidation by the histone deacetylase inhibitor sodium butyrate in aged rats. Neurosci Lett. 2015;594:76–81. doi: 10.1016/j.neulet.2015.03.059. [DOI] [PubMed] [Google Scholar]


