Abstract
Melanoma is one of the most highly mutated malignancies, largely a function of its generation through ultraviolet light and other mutational processes. The wide array of mutations in both “driver” and “passenger” genes can present a confusing array of data for practitioners, particularly in the context of the recent targeted and immune therapy revolutions. While mutations in BRAF V600 clearly confer sensitivity to BRAF and MEK inhibitors, the clinical implications of most other mutations are less often discussed and understood. In this review, we provide an overview of the high-frequency genomic alterations and their prognostic and therapeutic relevance in melanoma.
Keywords: Melanoma, Mutation, BRAF, NRAS, PD-1, KIT, CDKN2A
Introduction
Cancer therapeutics have been dramatically reshaped over the past 15 years by nearly concurrent revolutions in precision medicine-driven targeted therapies and novel cancer immune therapies. The detection of particular actionable mutations driving growth and progression of several cancers has led to the development of small molecule kinase inhibitors and/or monoclonal antibodies, which have in turn improved clinical outcomes.1–5 Coupled with the development of protein-directed antibody therapies (e.g. rituximab, trastuzumab, bretuximab vedotin, cetuximab, etc.), hormone-directed therapies (e.g. tamoxifen, enzalutamide), angiogenesis-targeted therapies (e.g. vascular endothelial growth factor inhibitors), and cell-cycle kinase inhibitors, these precision-medicine type approaches have dramatically improved therapeutic outcomes in many solid and hematologic cancers. Immunotherapies, agents that target programmed death-1 and its ligand (PD-1/PD-L1) and cytotoxic T lymphocyte antigen-4 (CTLA-4) have similarly revolutionized a partially overlapping group of cancers. These agents frequently present superior toxicity profiles, and produce rapid and/or durable responses.
Central to the development of these therapies, particularly mutation-directed approaches, has been the ability to molecularly profile tumors in the clinic. While this type of technology was completely unavailable prior to the human genome project in 2001, 6, 7 rapid advances have permitted the now routine sequencing of large fractions of the cancer genome. As such, determining the presence of relevant therapeutic mutations is now a routine procedure in many cancers, particularly melanoma, non-small cell lung cancer (NSCLC), colon adenocarcinoma, and others. While a few mutations have clear therapeutic implications, most detected mutations have less well-defined clinical relevance. Thus, next generation sequencing platforms that sequence hundreds of genes (e.g. FoundationOne™, Caris™) can provide a figurative mountain of genomic data through which to parse. In particular, a highly mutated tumor like melanoma may yield data that is challenging for the practioner to interpret. Herein, we discuss the clinical and biologic implications of common mutations and other genomic alterations in melanoma.
Melanoma is highly mutated
One consideration is that the mutation rate is widely variable between human cancers. The somatic mutation frequency rate in 3,083 matched tumor-normal pairs from 27 different tumor types including pediatric tumors, hematologic malignancies, and solid tumors, was analyzed for median frequency of non-synonymous mutations. The anlaysis revealed a 1000-fold variation across the different tumor types. In melanoma, the frequency of somatic mutations ranged widely from 0.1–100/Mb, but overall had the highest mutation frequency of all cancers analyzed.8 The variability in the mutation frequency in melanoma may be attributed to the presence or absence of a known carcinogen, such as UV exposure.
The landscape of genomic alterations in cutaneous melanoma is wide-ranging. Data published with The Cancer Genome Atlas (TCGA) Network found that whole exome sequence analysis of 333 primary and/or metastatic melanoma patients, melanomas could be classified into four genomic subtypes: mutant BRAF, mutant NRAS, mutant NF1, and triple-wildtype.9 The different genomic subtypes may be of predictive value given the therapeutic targets currently available. The TCGA database and other key sequencing studies has provided a comprehensive survey of the genetic landscape of melanomas.10
In melanoma, the activated BRAF mutated kinase can be inhibited by BRAF targeting drugs, and its downstream protein MEK kinase can be inhibited by a MEK targeted drug. The combination of targeted inhibitors have had a very significant impact on survival in patients with BRAF mutant melanoma with a median overall survival exceeding 2 years.11 On the other hand, the other common genomic subtypes, including mutant NRAS, mutant NF1, and triple-wildtype have thus far not been effectively targeted.
Mutation patterns: UV is the key
Mutation patterns vary substantially between melanoma subtypes, both in terms of total numbers of mutations (single nucleotide variants) and driver oncogenes (Table).10 Melanomas from chronically sun-exposed skin tend to have the highest numbers of mutations, and often have NF1, NRAS and occasionally BRAF V600K mutations present.12 BRAF V600E mutations, by contrast, are rare in these melanomas. Melanomas from intermittently sun-exposed skin frequently have intermediate numbers of mutations, and have mutations in BRAF V600E (50%) or NRAS (15–20%). Tumors arising from noncutaneous sites (mucosal, acral, uveal) have significantly lower total numbers of mutations by several orders of magnitude, with uveal melanoma being a particularly genomically simple tumor, thus demonstrating that ultraviolet light is not the only driver of melanogenesis. Age is also a factor with younger patients (<40 years) more likely to have BRAF V600E mutations. Acral melanomas arising on the palms, soles, and nailbeds have mutations in BRAF, NRAS, and KIT in approximately 15% each. Mucosal melanomas, by contrast, have only infrequent BRAF or NRAS mutations, but have KIT mutations in about 15% (primarily in genitourinary or anal, rather than sinonasal melanomas). Uveal melanomas have totally distinct genomic patterns, and harbor mutations either in GNAQ or GNA11 in >90%, while, BAP1, SF3B1, and EIFAX are generally distinct subsets and non-overlapping.
Table.
Common Genomic Alterations in Melanoma and Clinical Implications
| Mutation | Percentage (%) | Clinical features or other comments |
|---|---|---|
| BRAF V600 | 40–50 | Confers susceptibility to BRAF/MEK inhibitors, more common on intermittent sun exposed skin |
| NRAS | 15–20 | Poor prognosis, may have higher response to immunotherapy |
| NF1 | 10–15 | More common on sun exposed skin, may have higher response to immunotherapy |
| KIT | 1–2 | Confers susceptibility to KIT inhibitors, more common in mucosal (15–20%) and acral melanomas (15–20%) |
| Atypical BRAF (non-V600) | 4–5 | May confer susceptibility to MEK or RAF inhibitors |
| GNAQ/GNA11 | 80–90 (uveal) | |
| TERT promoter | 40–50 | Poor prognosis, UV-mediated mutation |
| CDKN2A | 25–35 | Deep deletions more common than mutations |
| PTEN | 4–8 | May correlate with immune resistance, deep deletions more common than mutations |
Notwithstanding the less common non-cutaneous melanomas, ultraviolet light is the major driver of mutagenesis in cutaneous melanomas. While it remains challenging to conclusively model the role of UV light in melanoma, a wealth of observational studies have unequivocally demonstrated linked this carcinogen with melanoma generation.13 UV light produces a distinct mutational signature, with C>T and CC>TT transitions. Interestingly, melanoma precursors appear to progress through a stereotypical pattern of mutagenesis in many cases 14. Unequivocally benign lesions (nevi) harbored only BRAF V600E mutations. Intermediate lesions acquired other mutations, such as TERT promoter mutations, or by contrast, had NRAS mutations. Invasive melanomas often acquired CDKN2A loss, PTEN loss, or TP53 mutations. Further, point mutations increased at each stage of evolution. This study reinforced findings from in vitro models; specifically that BRAF V600E mutations alone are insufficient to drive oncogenesis and require additional genetic “hits,” and that ultraviolet light was the major driver of mutagenesis. While ultraviolet light is a major culprit in driving mutagenesis and progression, other pathways, such pheomelanin generated oxidative damage in the red hair/fair skin phenotype, exemplified by patients or mice with MC1R gene polymorphisms.15 Other germline polymorphisms, such as those in CDKN2A are linked with higher incidences of melanoma.16
Lots of mutations: Does it matter?
Until recently, the total number of mutations present in a tumor was not considered of major clinical relevance. At the low end of the spectrum, with <1 mutation/megabase (MB) of DNA are most hematologic malignancies and childhood cancers.8 Many common adult solid tumors, such as breast, colon, prostate, and pancreas cancers, harbor mutation burdens slightly higher than this, but still almost always <5 MB. On the higher end of the spectrum, carcinogen induced cancers, including cancers of the cervix, bladder, head and neck, and lung, typically have a median of 5–10 mutations/MB, although intratumoral variation is quite high. Melanoma is at the highest end of the spectrum, with a median of >10 mutations/MB and many tumors with 10-fold greater mutation numbers. Interestingly, normal human skin has also been found to have approximately 2–6 mutations/MB, similar to many solid tumors.17, 18 On average, the only other cancer subsets more highly mutated than melanoma are those with microsatellite instability (MSI) or POLE mutations where DNA repair is impaired.
One feature of melanoma, that on the surface is seemingly unrelated to passenger mutations, is its high degree of response to immune therapy. Melanoma has historically been thought of as an immune responsive cancer. Indeed, the only effective historical therapies for metastatic disease were interleukin-2 and adoptive cell therapy.19, 20 More recently, immune checkpoint inhibitors have further transformed the treatment of metastatic melanoma. In particular, single-agent nivolumab (anti-programmed death-1; [PD-1]) produces a response rate of approximately 45%, and combination ipilimumab (anti-cytotoxic T lymphocyte antigen-4 [CTLA-4]) and nivolumab causes nearly 60% of patients to respond.21 Reliable biomarkers to predict response, and indeed the causative factors underlying responses in individual patients have been somewhat elusive.
Several studies have demonstrated intriguing predictive and perhaps causative links between mutational burden and response to immunotherapy. The initial assumption in these and other studies referenced below is that putative “passenger” mutations actually function to generate so-called neoantigens, peptides that may be recognized as foreign to the adaptive immune system.22 These neoantigens generate a pre-existing immune response that may be unleashed by checkpoint blockade. This model may be simplistic, as other studies have suggested that mutational burden is not linked with pre-existing immune signatures.23 Regardless of the mechanism, mutation burden has been clearly linked, in multiple tumor types to response to immune therapy.
The first study involved a cohort of patients treated with ipilimumab. In this study, patients who had durable clinical benefit (>6mos of stable or responsive disease) had substantially higher mutation numbers compared with those who failed to respond.24 Initially, a tetrapeptide signature corresponding to viral or bacterial sequences was reported, suggesting that mutations may generate microbial-like sequences susceptible to immune targeting. A subsequent large sequencing study failed to replicate the tetrapeptide signature associations, although it redemonstrated strong associations between ipilimumab benefit and high mutation burdens.25
Anti-PD-1 benefit in melanoma patients has also been linked with high mutation burdens. Two studies that performed whole exome sequencing on pre-treatment melanoma samples, and found that high mutation numbers correlated with improved clinical outcomes after treatment.26, 27 Other studies have also strongly correlated mutation load, as determined by whole exome sequencing, with improved clinical outcomes to anti-PD-1 in non-small cell lung cancer 28, 29 and microsatellite unstable cancers.30, 31
While whole exome sequencing provides comprehensive genomic characterization of tumors, it remains impractical to perform in routine clinical practice, given the requirement for matched germline tissue and extensive informatics support. Our group and others evaluated whether mutation burden in smaller, clinically useful gene panels (comprised of 170–315 genes) could serve as a useful surrogate for total exonic mutation burden. These studies showed that predicted mutation burden correlated strongly with total exonic mutation load and more importantly, with immunotherapy responses. Among 65 patients with melanoma, mutation burden calculated using FoundationOne (315 genes) was strongly associated with response, PFS, and OS, particularly at high (>20 mutations/MB) levels. NF1 mutations, likely as a surrogate for highly mutated melanomas, also correlated with improved clinical outcomes. These findings (using FoundationOne) were redemonstrated in a prospectively accrued cohort of patients with urothelial bladder carcinoma treated with atezolizumab.32 Mutation burden calculated with a platform of 170 genes performed at MD Anderson also correlated with response to ipilimumab and adoptive T cell therapy (in melanoma) and anti-PD-1 (in NSCLC).33 Thus, mutation load using smaller panels may serve as a biomarker of response to immunotherapy. Our group is conducting a clinical trial of nivolumab in highly mutated NSCLC who have not received any prior treatment. The impact of mutation load on combination immunotherapy strategies (e.g. ipilimumab/nivolumab) is not known.
Finally, while overall mutation load has been correlated with immunotherapy response, there is intensive research to identify immunogenic neoantigens. If identified, vaccines or perhaps cellular therapies could be designed against neoantigens of interest. Two recent elegant, proof-of-concept studies were published, showing preliminary clinical activity of this concept.34, 35
Specific mutations
BRAF
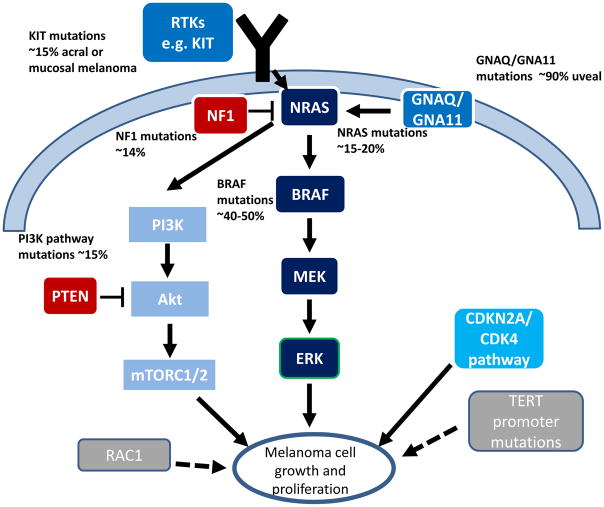
In 2002, BRAF mutations were found to occur frequently in up to 66% of patients with melanoma.36 These activating mutations in BRAF result in constitutive activation of kinase function independent of upstream signaling from RAS. BRAF interacts with MEK resulting in MEK phosphorylation, leading to subsequent activating phosphorylation of ERK, ultimately promoting cellular growth and inhibiting apoptosis.37 Subsequent studies have confirmed the incidence of BRAF mutations as approximately 40–50% in patients with cutaneous melanoma (Figure).38–40 The most common mutation in BRAF is a substitution of glutamic acid for valine at amino acid 600 (V600E) accounting for 70–88% of all BRAF mutations.38, 39, 41 Less common mutations in BRAF include V600K, V600R, and V600M, comprising 11–20%, 2–5%, and 1–4% of BRAF mutations respectively.38–40 Other, non-V600 alterations in BRAF occur in approximately 5% of all melanomas, most commonly at codons 466, 469, 597, and 601, and BRAF fusions.
Figure.
Schematic of relevant cell signaling pathways and frequency of mutations
Melanomas that arise on skin with intermittent sun exposure are more likely to have a BRAF mutation than melanomas on chronically sun-exposed skin, unexposed skin, or mucosal melanomas.42 Other clinicopathologic features that have been associated with BRAF mutant melanoma include younger age, superficial spreading or nodular melanoma, presence of mitoses, occult primary melanoma, and truncal location.39
The relationship between melanocytic nevi and the development of melanoma is incompletely understood. BRAF mutations are an acquired event and found in 70–80% of nevi, and tend to occur more often in melanomas arising from pre-existing nevi. However, the majority of nevi will not progress to melanoma. BRAF mutations are frequent in melanocytic nevi and vertical growth phase melanomas but infrequent in radial growth phase and in situ melanomas.43–45 Thus, while BRAF mutations clearly drive melanoma growth and progression, they are insufficient by themselves to induce melanomas. Melanomagenesis requires cooperation between mutant BRAF and other pathways. Mouse models have demonstrated that concurrent BRAF activating mutations and PTEN inactivating mutations result in melanomagenesis46–48. Also, mice with BRAF V600E mutations as well as CDKN2A loss develop larger melanomcytic nevi with a very small proportion progressing to melanoma. However, when BRAF/CDKN2A mutant mice also have loss of Lkb1, they demonstrate marked activation of mTORC2/Akt resulting in rapidly progressive melanomas46, 49. These findings demonstrate some of the complexities underlying melanomagenesis and the need for further understanding of the relationship between BRAF and other mutations.
The key clinical relevance of BRAF mutations lies in their response to BRAF and/or MEK inhibitors. Available evidence suggests that various BRAF V600 mutations have similar capacity to respond to these therapies, although response rates may be less with V600K mutations compared with V600E.50 Several initial phase III trials demonstrated superiority of single agent BRAF and MEK inhibitors compared with chemotherapy.51–53 Subsequently, combining these agents has demonstrated superiority to single-agent therapy, with progression-free survival (PFS) in the range of 12 months, and nearly 70% response rates.11, 54 Thus, combined BRAF/MEK inhibition is a pillar of standard therapy for metastatic melanoma. More recently, combined dabrafenib and trametinib have demonstrated improved PFS and overall survival (OS) in patients with high-risk, resected melanoma compared with observation alone.55 Importantly, available evidence suggests that BRAF mutations predispose to response to BRAF/MEK inhibition in other cancers, including lung cancer, thyroid cancer, and hematologic malignancies. By contrast, colon cancers appear relatively insensitive to pathway inhibition. This highlights important differences in the tissue type and concurrent genetic makeup, and underscores the importance of dissecting clinical activity data in different cancer types.
A number of extensive studies have assessed the genomic correlates of both acquired and intrinsic resistance to BRAF inhibitors. A variety of mutations appear to arise in the context of acquired resistance, and reignite MAPK signaling (or parallel signaling networks) despite the presence of BRAF inhibition, and include mutations in NRAS, PI3K/AKT pathway members, and amplification and alternative splicing of BRAF.56–58 Unraveling intrinsic resistance has been a greater challenge, although pre-existing mutations in PTEN and MAP2K1 appear to correlate with shorter responses 59, 60. These mutations, however, do not preclude therapeutic responses.
The 5% of melanomas harboring non-V600 mutations have less clear therapeutic relevance. Responses to MEK inhbitors and pan-RAF inhibitors (e.g. sorafenib) have been reported in patients and using in vitro models.61–63 We are currently conducting a clinical trial of trametinib in patients with non-V600 BRAF mutations. In addition, one arm of the NCI MATCH study is testing trametinib in this same population across cancer types.
It remains controversial whether to select targeted therapy (BRAF/MEK inhibitors) or immune therapy as initial therapy in the BRAF mutant population. Ideally, we would be able to determine molecular or clinical biomarkers to help guide these decisions. At this time, it appears most clinical factors that correlate with response to BRAF/MEK inhibitors also occur in patients who have good outcomes with immunotherapy (e.g. low disease burden, low LDH, good functional status).64 One randomized study is now comparing dabrafenib and trametinib with ipilimumab and nivolumab as first-line therapy with planned crossover at progression. Until these data are available, we tend to treat patients with bulky or symptomatic disease with BRAF/MEK inhibitors to induce a rapid response, and most other patients with front-line immunotherapy.
NRAS
The RAS family includes three primary proto-oncogenes: NRAS, KRAS, and HRAS, that regulate cell proliferation and apoptosis. NRAS mutations constitutively activate the MAPK, PI3K, and other cell signaling pathways causing cell growth, proliferation, and cell cycle dysfunction. NRAS mutations occur in >20% of patients with cutaneous melanoma, most commonly in codon 61 and less commonly in codons 12 and 13.38, 65 Melanomas with NRAS mutations are associated with an aggressive clinical course and poor prognosis. They are more common in non-sun exposed skin.66
Treatment for NRAS mutant melanoma has been met with limited success to date. Initial early responses to the MEK inhibitor binimetinib led to a phase III study where binimetinib was compared with chemotherapy in patients with NRAS mutant melanoma. Surprisingly, though response rates (15% vs. 9%, p=0.02) and PFS were modestly improved with binimetinib (median 2.8 vs. 1.5 months, p<0.001), no difference in OS was observed (11 vs. 10.1 months, p=0.5).67 Thus, further development of binimetinib monotherapy in NRAS mutant melanoma is unlikely to be pursued. One study suggested that NRAS mutations may correspond with response to anti-PD-1 but the sample size was small.68 Notably, outcomes to binimetinib in patients with prior immunotherapy appeared to be superior, suggesting that the combination of MEK inhibitors and immunotherapy could be useful in this population. Ongoing studies are evaluating the efficacy of MEK inhibitors combined with CDK4/6 inhibitors.
KIT
KIT is a proto-oncogene receptor tyrosine kinase that is found on the cell membrane and binds to stem cell factor. This activates the KIT protein resulting in the activation of multiple signaling pathways affecting cell growth, proliferation, survival, and migration. Mutations in KIT occur in 1–3% of all melanomas and are most commonly found in exon 11 (L576P) or exon 13 (K642E).69 KIT mutations occur more commonly found in acral or mucosal melanomas (~15% each, in vulvovaginal more often than sinonasal) and in areas of chronic sun damage (~2%).70, 71 Tyrosine kinase inhibitors, particularly imatinib, have demonstrated some activity in the treatment of KIT-mutant melanoma. Responses to treatment in patients with exon 11 mutations occur in the range of 30–50%, although acquired resistance typically occurs within one year.72, 73 KIT amplification, by contrast, does not appear to mediate imatinib sensitivity.
NF1
The neurofibromatosis type 1, NF1, gene product is a GTPase activating protein dampens MAPK signaling by downregulating RAS activity, and mutations and/or loss of NF1 leads to MAPK activation. NF1 mutations define the third most common genomically defined subset of melanoma and occur in 14% of the TCGA melanoma samples, including up to 70% of BRAF/NRAS wildtype samples.9 Most NF1 mutations lead to a loss of function of this tumor suppressor, with about 80% of patients having a nonsense mutation, an insertion, or a deletion that leads to a truncated protein.74
In co-occuring BRAF/NF1 mutant tumors in mice, there is insensitivity to BRAF inhibitor therapy, presumably due to NF1 loss of function leading to the dysregulation of the PI3K/AKT/mTOR pathway.75 There is preclinical data to suggest that NF1 loss may lead to MEK inhibitor sensitivity; however, this has not clinically translated in a meaningful way.76–78 “Pan-RAF” or type II RAF inhibitors in combination with MEK inhibitors, or PI3K/mTOR inhibitors could be considered in NF1 mutated melanomas. NF1 mutations seem to be correlated with the strongest UV signature and a high mutational burden and so there is rationale for using immunotherapeutic agents in this patient population.79
Other genomic subtypes
CDKN2A
The TCGA data showed alterations in the RAS/MAPK/AKT, CDKN2A, and MDM2/TP53 pathways, in 91%, 69%, and 19% of cases, respectively.9 Mutations in CDKN2A specifically occurred in 13% of tumors with another ~30% harboring CKDN2A deletions. Mutations in the p16/CDK4/cyclinD1 pathway have been implicated in melanomagenesis. CDKN2A mutations that result in p16 loss are often seen in familial melanomas.80 Melanoma genome wide-association study (GWAS) have shown this pathway may be important in more than just familial melanomas, as genomewide risk loci have been identified in nearby the CDKN2A and CCND1 loci.81, 82 CDK4 and CCND1 amplifications are seen more frequently in triple-wildtype melanomas. While it has been hypothesized that mutations in this pathway may contribute to sensitivity to CDK4/6 inhibitors, this has yet to be shown conclusively in the clinic.83
P53
TP53 is mutated in about 15% of TCGA melanomas, and appear to occur later during tumor development.9 The TP53 mutations were frequently found in melanomas harboring any of the major subsets of BRAF, NRAS, and NF1 mutated tumors. In comparison, in triple-wildtype tumors, there was an increased prevalence of MDM2 amplifications.9 Currently, agents that inhibit MDM2 (and thus restore wild type p53 activity) are being combined with BRAF and/or MEK inhibitors.
PTEN
As mentioned, a host of other low-frequency mutations occur in melanoma and are difficult to interpret clinically. PTEN mutations or deep deletions occur in <10% of melanomas and activate P13K/AKT signaling. As mentioned, these may limit response to BRAF inhibition, but have also been implicated in exclusion of T cells from the tumor microenvironment and lack of response to immunotherapy.84
RAC1
Mutations in RAC1 occur in approximately 10% of sun-exposed melanomas and tend to co-occur with BRAF or NRAS mutations. In vitro studies suggest that the primary “hotspot” mutation (RAC1 P29S) activates downstream signaling pathways, thus promoting proliferation and migration.12 Subsequent studies have suggested that this mutation may regulate PD-L1 expression and mediate resistance to BRAF and MEK inhibitors, although this mutation does not preclude clinical responses.85, 86
TERT
Mutations in TERT (telomerase reverse transcriptase) are somewhat unique in that they usually occur in the promoter, rather than the coding region, leading to increased gene expression. These mutations occur in most melanomas, including 69% of all melanomas, and 86% of cutaneous melanomas.87 They are UV-light related (C>T or CC>TT), confer a poor prognosis in one study (median OS 80 vs. 291 months), although were not associated with telomere length.10
GNAQ/GNA11
Mutations in these g-proteins occur in nearly 90% of uveal melanomas and occasionally non-uveal melanomas (but extremely rarely outside of the melanocytic tumor family).88, 89 These activating mutations trigger MAPK and PI3K/AKT signaling, likely through RasGRP3 and protein kinase C.90 The importance of MAPK signaling has led to trials of MEK inhibitors in uveal melanoma, although the results have largely been disappointing.91 Notably, several other genomic events correlate with prognosis in uveal melanoma, including SF3B1 mutations (good), BAP1 loss (poor), monosomy 3 (poor), and disomy 3 (good).88
Other mutations
MYC amplifications occur in up to 8% of melanomas and correlate with poor prognosis and lack of pigmentation 92. Other in vitro studies have suggested that MYC amplifications lead to immune exclusion and T cell dysfunction in the tumor microenvironment 93. Mutations in the WNT/CTNNB1 pathway (primarily APC and CTNNB1) occur in approximately 10% of melanomas. While these have less clear clinical implications, CTNNB1 signaling has also been implicated in immune cell exclusion 94. Finally, low frequency mutations in ARID2, PPP6C, MAP2K1, IDH1, RB1, and many others have been described 38, 95. The clinical implications of these mutations remain unclear.
Conclusions
Deciphering the clinical and biologic implications of molecular features, including mutations and other genomic alterations remains in its early stages. Despite the lack of clear clinical relevance of most mutations, a few salient exceptions exist. BRAF V600 mutations clearly predict sensitivity to inhibitors of BRAF and MEK, as do KIT exon 11 mutations to KIT inhibitors. Further, high mutation burden correlates with response to single-agent anti-PD-1 therapy. In addition, other mutations such as NRAS or atypical BRAF mutations and others, may enable enrollment in clinical trials. These remain the primary clinically actionable findings at this time.
Melanoma has been at the center of both the precision medicine/targeted therapy and immune therapy revolutions. A key challenge for the future includes developing more effective targeted therapies for BRAF wild type patients; promising combinations include those targeting MEK and CDK4/6. Further, overcoming resistance to BRAF/MEK inhibitors has been a formidable obstacle that could be targeted by ERK inhibitors or next generation RAF inhibitors. Unraveling the mutational complexities of melanoma continues to be a compelling challenge as we work to improve care for patients with this disease.
Acknowledgments
Funding: NIH/NCI K23 CA204726 (DBJ), James C. Bradford Jr. melanoma fund (DBJ)
Footnotes
Conflicts of interest: DBJ: Advisory boards for BMS, Incyte, Novartis, Merck. JAS: Advisory boards for Array, Incyte, Merck.
References
- 1.Sosman JA, Kim KB, Schuchter L, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366:707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitsudomi T, Kosaka T, Endoh H, et al. Mutations of the epidermal growth factor receptor gene predict prolonged survival after gefitinib treatment in patients with non-small-cell lung cancer with postoperative recurrence. J Clin Oncol. 2005;23:2513–2520. doi: 10.1200/JCO.2005.00.992. [DOI] [PubMed] [Google Scholar]
- 3.Shaw AT, Ou SH, Bang YJ, et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med. 2014;371:1963–1971. doi: 10.1056/NEJMoa1406766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaw AT, Kim DW, Nakagawa K, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368:2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 5.Verweij J, Casali PG, Zalcberg J, et al. Progression-free survival in gastrointestinal stromal tumours with high-dose imatinib: randomised trial. Lancet. 2004;364:1127–1134. doi: 10.1016/S0140-6736(04)17098-0. [DOI] [PubMed] [Google Scholar]
- 6.Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 7.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 8.Alexandrov LB, Nik-Zainal S, Wedge DC, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Genomic Classification of Cutaneous Melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayward NK, Wilmott JS, Waddell N, et al. Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545:175–180. doi: 10.1038/nature22071. [DOI] [PubMed] [Google Scholar]
- 11.Long GV, Weber JS, Infante JR, et al. Overall Survival and Durable Responses in Patients With BRAF V600-Mutant Metastatic Melanoma Receiving Dabrafenib Combined With Trametinib. J Clin Oncol. 2016;34:871–878. doi: 10.1200/JCO.2015.62.9345. [DOI] [PubMed] [Google Scholar]
- 12.Krauthammer M, Kong Y, Ha BH, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sample A, He YY. Mechanisms and prevention of UV-induced melanoma. Photodermatol Photoimmunol Photomed. 2017 doi: 10.1111/phpp.12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shain AH, Yeh I, Kovalyshyn I, et al. The Genetic Evolution of Melanoma from Precursor Lesions. N Engl J Med. 2015;373:1926–1936. doi: 10.1056/NEJMoa1502583. [DOI] [PubMed] [Google Scholar]
- 15.Mitra D, Luo X, Morgan A, et al. An ultraviolet-radiation-independent pathway to melanoma carcinogenesis in the red hair/fair skin background. Nature. 2012;491:449–453. doi: 10.1038/nature11624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goldstein AM, Chan M, Harland M, et al. Features associated with germline CDKN2A mutations: a GenoMEL study of melanoma-prone families from three continents. J Med Genet. 2007;44:99–106. doi: 10.1136/jmg.2006.043802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martincorena I, Roshan A, Gerstung M, et al. Tumor evolution. High burden and pervasive positive selection of somatic mutations in normal human skin. Science. 2015;348:880–886. doi: 10.1126/science.aaa6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chalmers ZR, Connelly CF, Fabrizio D, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9:34. doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atkins MB, Lotze MT, Dutcher JP, et al. High-dose recombinant interleukin 2 therapy for patients with metastatic melanoma: analysis of 270 patients treated between 1985 and 1993. J Clin Oncol. 1999;17:2105–2116. doi: 10.1200/JCO.1999.17.7.2105. [DOI] [PubMed] [Google Scholar]
- 20.Rosenberg SA, Packard BS, Aebersold PM, et al. Use of tumor-infiltrating lymphocytes and interleukin-2 in the immunotherapy of patients with metastatic melanoma. A preliminary report. N Engl J Med. 1988;319:1676–1680. doi: 10.1056/NEJM198812223192527. [DOI] [PubMed] [Google Scholar]
- 21.Larkin J, Chiarion-Sileni V, Gonzalez R, et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N Engl J Med. 2015 doi: 10.1056/NEJMoa1504030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gubin MM, Schreiber RD. CANCER. The odds of immunotherapy success. Science. 2015;350:158–159. doi: 10.1126/science.aad4140. [DOI] [PubMed] [Google Scholar]
- 23.Spranger S, Luke JJ, Bao R, et al. Density of immunogenic antigens does not explain the presence or absence of the T-cell-inflamed tumor microenvironment in melanoma. Proc Natl Acad Sci U S A. 2016;113:E7759–E7768. doi: 10.1073/pnas.1609376113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snyder A, Makarov V, Merghoub T, et al. Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma. N Engl J Med. 2014 doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350:207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hugo W, Zaretsky JM, Sun L, et al. Genomic and Transcriptomic Features of Response to Anti-PD-1 Therapy in Metastatic Melanoma. Cell. 2016;165:35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riaz N, Havel JJ, Makarov V, et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell. 2017;171:934–949. e915. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rizvi NA, Hellmann MD, Snyder A, et al. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015 doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carbone DP, Reck M, Paz-Ares L, et al. First-Line Nivolumab in Stage IV or Recurrent Non-Small-Cell Lung Cancer. N Engl J Med. 2017;376:2415–2426. doi: 10.1056/NEJMoa1613493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenberg JE, Hoffman-Censits J, Powles T, et al. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016 doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roszik J, Haydu LE, Hess KR, et al. Novel algorithmic approach predicts tumor mutation load and correlates with immunotherapy clinical outcomes using a defined gene mutation set. BMC Med. 2016;14:168. doi: 10.1186/s12916-016-0705-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ott PA, Hu Z, Keskin DB, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547:217–221. doi: 10.1038/nature22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sahin U, Derhovanessian E, Miller M, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547:222–226. doi: 10.1038/nature23003. [DOI] [PubMed] [Google Scholar]
- 36.Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 37.Sharma A, Trivedi NR, Zimmerman MA, Tuveson DA, Smith CD, Robertson GP. Mutant V599EB-Raf regulates growth and vascular development of malignant melanoma tumors. Cancer Res. 2005;65:2412–2421. doi: 10.1158/0008-5472.CAN-04-2423. [DOI] [PubMed] [Google Scholar]
- 38.Cancer Genome Atlas Network. Electronic address imo, Cancer Genome Atlas N. Genomic Classification of Cutaneous Melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Long GV, Menzies AM, Nagrial AM, et al. Prognostic and clinicopathologic associations of oncogenic BRAF in metastatic melanoma. J Clin Oncol. 2011;29:1239–1246. doi: 10.1200/JCO.2010.32.4327. [DOI] [PubMed] [Google Scholar]
- 40.Lovly CM, Dahlman KB, Fohn LE, et al. Routine multiplex mutational profiling of melanomas enables enrollment in genotype-driven therapeutic trials. PLoS One. 2012;7:e35309. doi: 10.1371/journal.pone.0035309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Menzies AM, Haydu LE, Visintin L, et al. Distinguishing clinicopathologic features of patients with V600E and V600K BRAF-mutant metastatic melanoma. Clin Cancer Res. 2012;18:3242–3249. doi: 10.1158/1078-0432.CCR-12-0052. [DOI] [PubMed] [Google Scholar]
- 42.Maldonado JL, Fridlyand J, Patel H, et al. Determinants of BRAF mutations in primary melanomas. J Natl Cancer Inst. 2003;95:1878–1890. doi: 10.1093/jnci/djg123. [DOI] [PubMed] [Google Scholar]
- 43.Pollock PM, Harper UL, Hansen KS, et al. High frequency of BRAF mutations in nevi. Nat Genet. 2003;33:19–20. doi: 10.1038/ng1054. [DOI] [PubMed] [Google Scholar]
- 44.Dong J, Phelps RG, Qiao R, et al. BRAF oncogenic mutations correlate with progression rather than initiation of human melanoma. Cancer Res. 2003;63:3883–3885. [PubMed] [Google Scholar]
- 45.Poynter JN, Elder JT, Fullen DR, et al. BRAF and NRAS mutations in melanoma and melanocytic nevi. Melanoma Res. 2006;16:267–273. doi: 10.1097/01.cmr.0000222600.73179.f3. [DOI] [PubMed] [Google Scholar]
- 46.Damsky WE, Bosenberg M. Melanocytic nevi and melanoma: unraveling a complex relationship. Oncogene. 2017;36:5771–5792. doi: 10.1038/onc.2017.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dankort D, Curley DP, Cartlidge RA, et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41:544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsao H, Goel V, Wu H, Yang G, Haluska FG. Genetic interaction between NRAS and BRAF mutations and PTEN/MMAC1 inactivation in melanoma. J Invest Dermatol. 2004;122:337–341. doi: 10.1046/j.0022-202X.2004.22243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Damsky W, Micevic G, Meeth K, et al. mTORC1 activation blocks BrafV600E-induced growth arrest but is insufficient for melanoma formation. Cancer Cell. 2015;27:41–56. doi: 10.1016/j.ccell.2014.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klein O, Clements A, Menzies AM, O’Toole S, Kefford RF, Long GV. BRAF inhibitor activity in V600R metastatic melanoma. Eur J Cancer. 2012 doi: 10.1016/j.ejca.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 51.Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hauschild A, Grob JJ, Demidov LV, et al. Dabrafenib in BRAF-mutated metastatic melanoma: a multicentre, open-label, phase 3 randomised controlled trial. Lancet. 2012;380:358–365. doi: 10.1016/S0140-6736(12)60868-X. [DOI] [PubMed] [Google Scholar]
- 53.Flaherty KT, Robert C, Hersey P, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med. 2012;367:107–114. doi: 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- 54.Ascierto PA, McArthur GA, Dreno B, et al. Cobimetinib combined with vemurafenib in advanced BRAF(V600)-mutant melanoma (coBRIM): updated efficacy results from a randomised, double-blind, phase 3 trial. Lancet Oncol. 2016;17:1248–1260. doi: 10.1016/S1470-2045(16)30122-X. [DOI] [PubMed] [Google Scholar]
- 55.Long GV, Hauschild A, Santinami M, et al. Adjuvant Dabrafenib plus Trametinib in Stage III BRAF-Mutated Melanoma. N Engl J Med. 2017;377:1813–1823. doi: 10.1056/NEJMoa1708539. [DOI] [PubMed] [Google Scholar]
- 56.Johnson DB, Menzies AM, Zimmer L, Eroglu ZE, Ye F, et al. BRAF inhibitor acquired resistance: A multicenter meta-analysis of the spectrum and clinical implications of resistance mechanisms. J Clin Oncol. 2015;33 doi: 10.1016/j.ejca.2015.08.022. Abstr 9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moriceau G, Hugo W, Hong A, et al. Tunable-Combinatorial Mechanisms of Acquired Resistance Limit the Efficacy of BRAF/MEK Cotargeting but Result in Melanoma Drug Addiction. Cancer Cell. 2015;27:240–256. doi: 10.1016/j.ccell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi H, Hugo W, Kong X, et al. Acquired Resistance and Clonal Evolution in Melanoma during BRAF Inhibitor Therapy. Cancer Discov. 2014;4:80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carlino MS, Fung C, Shahheydari H, et al. Preexisting MEK1P124 mutations diminish response to BRAF inhibitors in metastatic melanoma patients. Clin Cancer Res. 2015;21:98–105. doi: 10.1158/1078-0432.CCR-14-0759. [DOI] [PubMed] [Google Scholar]
- 60.Catalanotti F, Cheng DT, Shoushtari AN, Johnson DB, Panageas KS, et al. PTEN Loss-of-Function Alterations Are Associated With Intrinsic Resistance to BRAF Inhibitors in Metastatic Melanoma. J Clin Oncol Prec Onc. 2017 doi: 10.1200/PO.16.00054. Epub June 23, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dahlman KB, Xia J, Hutchinson K, et al. BRAF L597 mutations in melanoma are associated with sensitivity to MEK inhibitors. Cancer Discov. 2012 doi: 10.1158/2159-8290.CD-12-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Botton T, Yeh I, Nelson T, et al. Recurrent BRAF kinase fusions in melanocytic tumors offer an opportunity for targeted therapy. Pigment Cell Melanoma Res. 2013 doi: 10.1111/pcmr.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Menzies AM, Yeh I, Botton T, Bastian BC, Scolyer RA, Long GV. Clinical activity of the MEK inhibitor trametinib in metastatic melanoma containing BRAF kinase fusion. Pigment Cell Melanoma Res. 2015 doi: 10.1111/pcmr.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Long GV, Grob JJ, Nathan P, et al. Factors predictive of response, disease progression, and overall survival after dabrafenib and trametinib combination treatment: a pooled analysis of individual patient data from randomised trials. Lancet Oncol. 2016;17:1743–1754. doi: 10.1016/S1470-2045(16)30578-2. [DOI] [PubMed] [Google Scholar]
- 65.Krauthammer M, Kong Y, Bacchiocchi A, et al. Exome sequencing identifies recurrent mutations in NF1 and RASopathy genes in sun-exposed melanomas. Nat Genet. 2015;47:996–1002. doi: 10.1038/ng.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Curtin JA, Fridlyand J, Kageshita T, et al. Distinct sets of genetic alterations in melanoma. N Engl J Med. 2005;353:2135–2147. doi: 10.1056/NEJMoa050092. [DOI] [PubMed] [Google Scholar]
- 67.Dummer R, Schadendorf D, Ascierto PA, et al. Binimetinib versus dacarbazine in patients with advanced NRAS-mutant melanoma (NEMO): a multicentre, open-label, randomised, phase 3 trial. Lancet Oncol. 2017;18:435–445. doi: 10.1016/S1470-2045(17)30180-8. [DOI] [PubMed] [Google Scholar]
- 68.Johnson DB, Lovly CM, Flavin M, et al. NRAS mutation: A potential biomarker of clinical response to immune-based therapies in metastatic melanoma (MM) J Clin Oncol. 2013;31:9019. [Google Scholar]
- 69.Shtivelman E, Davies MQ, Hwu P, et al. Pathways and therapeutic targets in melanoma. Oncotarget. 2014;5:1701–1752. doi: 10.18632/oncotarget.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Curtin JA, Busam K, Pinkel D, Bastian BC. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol. 2006;24:4340–4346. doi: 10.1200/JCO.2006.06.2984. [DOI] [PubMed] [Google Scholar]
- 71.Jin SA, Chun SM, Choi YD, et al. BRAF mutations and KIT aberrations and their clinicopathological correlation in 202 Korean melanomas. J Invest Dermatol. 2013;133:579–582. doi: 10.1038/jid.2012.338. [DOI] [PubMed] [Google Scholar]
- 72.Guo J, Si L, Kong Y, et al. Phase II, open-label, single-arm trial of imatinib mesylate in patients with metastatic melanoma harboring c-Kit mutation or amplification. J Clin Oncol. 2011;29:2904–2909. doi: 10.1200/JCO.2010.33.9275. [DOI] [PubMed] [Google Scholar]
- 73.Hodi FS, Corless CL, Giobbie-Hurder A, et al. Imatinib for melanomas harboring mutationally activated or amplified KIT arising on mucosal, acral, and chronically sun-damaged skin. J Clin Oncol. 2013;31:3182–3190. doi: 10.1200/JCO.2012.47.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shen MH, Harper PS, Upadhyaya M. Molecular genetics of neurofibromatosis type 1 (NF1) J Med Genet. 1996;33:2–17. doi: 10.1136/jmg.33.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maertens O, Johnson B, Hollstein P, et al. Elucidating distinct roles for NF1 in melanomagenesis. Cancer Discov. 2013;3:338–349. doi: 10.1158/2159-8290.CD-12-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nissan MH, Pratilas CA, Jones AM, et al. Loss of NF1 in Cutaneous Melanoma Is Associated with RAS Activation and MEK Dependence. Cancer Res. 2014;74:2340–2350. doi: 10.1158/0008-5472.CAN-13-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ranzani M, Alifrangis C, Perna D, et al. BRAF/NRAS wild-type melanoma, NF1 status and sensitivity to trametinib. Pigment Cell Melanoma Res. 2015;28:117–119. doi: 10.1111/pcmr.12316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Catalanotti F, Solit DB, Pulitzer MP, et al. Phase II trial of MEK inhibitor selumetinib (AZD6244, ARRY-142886) in patients with BRAFV600E/K-mutated melanoma. Clin Cancer Res. 2013;19:2257–2264. doi: 10.1158/1078-0432.CCR-12-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cirenajwis H, Lauss M, Ekedahl H, et al. NF1-mutated melanoma tumors harbor distinct clinical and biological characteristics. Mol Oncol. 2017;11:438–451. doi: 10.1002/1878-0261.12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hussussian CJ, Struewing JP, Goldstein AM, et al. Germline p16 mutations in familial melanoma. Nat Genet. 1994;8:15–21. doi: 10.1038/ng0994-15. [DOI] [PubMed] [Google Scholar]
- 81.Bishop DT, Demenais F, Iles MM, et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat Genet. 2009;41:920–925. doi: 10.1038/ng.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Barrett JH, Taylor JC, Bright C, et al. Fine mapping of genetic susceptibility loci for melanoma reveals a mixture of single variant and multiple variant regions. Int J Cancer. 2015;136:1351–1360. doi: 10.1002/ijc.29099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Young RJ, Waldeck K, Martin C, et al. Loss of CDKN2A expression is a frequent event in primary invasive melanoma and correlates with sensitivity to the CDK4/6 inhibitor PD0332991 in melanoma cell lines. Pigment Cell Melanoma Res. 2014 doi: 10.1111/pcmr.12228. [DOI] [PubMed] [Google Scholar]
- 84.Peng W, Chen JQ, Liu C, et al. Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discov. 2015 doi: 10.1158/2159-8290.CD-15-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Watson IR, Li L, Cabeceiras PK, et al. The RAC1 P29S hotspot mutation in melanoma confers resistance to pharmacological inhibition of RAF. Cancer Res. 2014;74:4845–4852. doi: 10.1158/0008-5472.CAN-14-1232-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vu HL, Rosenbaum S, Purwin TJ, Davies MA, Aplin AE. RAC1 P29S regulates PD-L1 expression in melanoma. Pigment Cell Melanoma Res. 2015;28:590–598. doi: 10.1111/pcmr.12392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Griewank KG, Murali R, Puig-Butille JA, et al. TERT promoter mutation status as an independent prognostic factor in cutaneous melanoma. J Natl Cancer Inst. 2014:106. doi: 10.1093/jnci/dju246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Robertson AG, Shih J, Yau C, et al. Integrative Analysis Identifies Four Molecular and Clinical Subsets in Uveal Melanoma. Cancer Cell. 2017;32:204–220. e215. doi: 10.1016/j.ccell.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Johnson DB, Roszik J, Shoushtari AN, et al. Comparative analysis of the GNAQ, GNA11, SF3B1, and EIF1AX driver mutations in melanoma and across the cancer spectrum. Pigment Cell Melanoma Res. 2016 doi: 10.1111/pcmr.12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen X, Wu Q, Depeille P, et al. RasGRP3 Mediates MAPK Pathway Activation in GNAQ Mutant Uveal Melanoma. Cancer Cell. 2017;31:685–696. e686. doi: 10.1016/j.ccell.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Carvajal RD, Sosman JA, Quevedo JF, et al. Effect of selumetinib vs chemotherapy on progression-free survival in uveal melanoma: a randomized clinical trial. JAMA. 2014;311:2397–2405. doi: 10.1001/jama.2014.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pouryazdanparast P, Brenner A, Haghighat Z, Guitart J, Rademaker A, Gerami P. The role of 8q24 copy number gains and c-MYC expression in amelanotic cutaneous melanoma. Mod Pathol. 2012;25:1221–1226. doi: 10.1038/modpathol.2012.75. [DOI] [PubMed] [Google Scholar]
- 93.Casey SC, Tong L, Li Y, et al. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352:227–231. doi: 10.1126/science.aac9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015 doi: 10.1038/nature14404. [DOI] [PubMed] [Google Scholar]
- 95.Hodis E, Watson IR, Kryukov GV, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]