Abstract
Life is built on cooperation between genes, which makes it vulnerable to parasitism. Selfish genetic elements that exploit this cooperation can achieve large fitness gains by increasing their transmission relative to the rest of the genome. This leads to counter-adaptations that generate unique selection pressures on the selfish genetic element. This arms race is similar to host–parasite coevolution, as some multi-host parasites alter the host’s behaviour to increase the chance of transmission to the next host. Here, we ask if, similarly to these parasites, a selfish genetic element in house mice, the t haplotype, also manipulates host behaviour, specifically the host’s migration propensity. Variants of the t that manipulate migration propensity could increase in fitness in a meta-population. We show that juvenile mice carrying the t haplotype were more likely to emigrate from and were more often found as migrants within a long-term free-living house mouse population. This result may have applied relevance as the t has been proposed as a basis for artificial gene drive systems for use in population control.
Keywords: t complex, meiotic drive, natal dispersal, Mus musculus, intra-genomic conflict, arms race
1. Introduction
The genes within a genome must work together to produce a viable organism, but their interests are not identical [1]. This causes conflict because not all genes in an organism will be transmitted equally to the next generation. Consequently, a fair chance of transmission is necessary for cooperation within the genome over evolutionary time. Genes that violate this rule by increasing their chance of transmission can gain large fitness advantages at the cost of those that transmit in a Mendelian fashion [2]. This leads to selection for selfish adaptations and, as a result, counter-adaptations to this selfishness, initiating an arms race between selfish genetic elements and the rest of the genome. This arms race is similar to the one between hosts and parasites, where some parasites even manipulate their hosts. For example, a parasite of the paper wasp Polistes dominula manipulates the behaviour of its host through changes in gene expression [3]. Instead of behaving as a member of the ‘worker’ caste, a parasitized female will behave more like the nest-founding ‘gyne’ caste. However, she will not actually found nests, but will instead transmit the parasite to other nests. Other manipulations have been observed, for example, in fungi-infected ants that climb vegetation and remain latched onto it post-mortem. The fungus will then produce spores, which disperse out of the dead ant’s body [4].
Host defences against parasites and ‘parasitic’ [5,6] selfish genetic elements range from behavioural changes to increased resistance in infected populations. For example, populations of the amphipod Gammarus pulex that are not naturally infected with the parasite Pomphorhynchus laevis are more sensitive to the parasite’s manipulation than naturally infected populations [7]. This is evidence of an arms race. A similar counter-adaptation to selfish genetic elements is the suppression of the drive mechanism. For example, in systems with X chromosome drive in Drosophila, which lead to the killing of Y-carrying sperm, some (Y) chromosomes suppress the drive, restoring production of sons [8–11]. Behavioural adaptations are also evident, especially in mating preferences that reduce transmission of parasites or selfish genetic elements. In the woodlouse Armadillidium vulgare, males discriminate against ‘neo-females’ infected with feminizing Wolbachia bacteria, another type of selfish genetic element [12]. Similarly, females discriminate against individuals carrying a selfish genetic element in stalk-eyed flies (Cyrtodiopsis) [13].
Male meiotic drivers are selfish genetic elements that manipulate spermatogenesis to favour the sperm that carry them by harming the sperm that do not [14,15]. This is expected to decrease the competitiveness of a male carrying the meiotic driver by decreasing the number of viable sperm and potentially damaging the driver-carrying sperm as a by-product [15,16]. In consequence, driver-carrying individuals will perform worse [17,18] in sperm competition, in which sperm of different males compete over fertilization. Additionally, females evolve higher remating rates in response to the presence of a selfish genetic element in Drosophila pseudoobscura, which increases sperm competition and reduces the element’s fitness [19]. Potentially, the driver carriers might not sire a single offspring despite mating [16] and the driver could go locally extinct [20]. Because of this strong disadvantage, females can be selected to increase sperm competition to decrease the risk of transmitting a driver to their offspring [19,21,22]. In response, the driver could manipulate the male host’s reproductive behaviour, as may be the case in Wolbachia-infected Drosophila that show higher mating rates [23]. Not much is otherwise known about how male meiotic drivers respond to this counter-adaptation that increases the risk of their extinction.
The t haplotype (t) is a male meiotic driver in the house mouse Mus musculus. It consists of a set of genes, making up about 1.5% or 40 Mb of the mouse genome, that are linked by inversions [2,24] and distort Mendelian inheritance patterns so that 90–99% of the offspring inherit the t from a heterozygous sire [25,26]. It harms its host in at least two ways. The t carries recessive lethal alleles, so that t/t die prenatally [16,27]. In addition, t heterozygous (+/t) males are very poor sperm competitors, siring only 11%–24% of offspring when mating with a female who also mates with a wild-type male in the same oestrus cycle [16,28]. In house mice, sperm competition intensity varies between populations [29] and is higher in larger populations [30], so that fitness losses of +/t males from sperm competition are likely to vary with population demography. This is consistent with a negative association between population size and t frequency found in a trapping study [31]. In an intensively monitored free-living large house mouse population, the frequency of the t decreased significantly over 6 years until no +/t were left [20] while population size increased [32]. Experimental evidence shows that t frequency decline in this population is not linked to mate choice against the t haplotype [33,34], as found by Lenington et al. [35] in another population, but is influenced by sperm competition [16,20].
The decline of the t in the population was even more rapid than predicted by a model based on sperm competition [20]. One additional contributing factor could be that +/t individuals are more likely to emigrate from the population than +/+. We will use the term ‘emigration’ when we mean leaving the natal population (the first step of dispersal [36]), ‘migration’ when we mean leaving and entering another deme or population [37] and ‘dispersal’ when we mean migrating and then breeding. Early theoretical work predicted that increased dispersal rates should be beneficial for the t haplotype by preventing it from becoming extinct owing to drift and allowing it to increase in frequency rapidly when dispersing to a suitable population [38]. In this view, a suitable population would be one that has no +/t in it, because the fitness of the t is frequency dependent, with lower fitness at high t frequencies [39]. This is owing to negative fitness effects (up to homozygous lethality) of deleterious mutations on the t [25]. Combined with the more recent discovery of low sperm competitiveness, the most suitable population for the t would therefore be one with as few +/t and as little sperm competition as possible, which is expected in smaller populations [30]. A t variant that is more likely to disperse to such a population should therefore be at a selective advantage compared to other variants.
We hypothesized that a t mutant that increases the migration propensity of its host generally would more often disperse to suitable populations and would thereby be selected. The increase in migration propensity could be a function of population density (i.e. +/t might only emigrate more than +/+ out of dense populations where sperm competition is more common [29,30]). This has not yet been tested, but for parasites, theoretical work has demonstrated that they would benefit in general from manipulating their host’s migration propensity [40,41]. We analysed juvenile disappearances from and juvenile migration within an open population of wild house mice (the same as analysed for t frequency dynamics by Manser et al. [20]) to investigate if +/t individuals are more likely to disappear than +/+. We found that +/t juveniles were more likely to disappear from the population than +/+, particularly when juvenile densities were high. To our knowledge, this is the first evidence of increased migration propensity of carriers of any selfish genetic element in a free-living population. Our research is particularly timely, as the t haplotype is proposed as a basis for artificial gene drive systems to eradicate house mouse populations [42,43] and behavioural differences in migration propensity between +/t and +/+ would need to be considered in modelling and implementing such systems.
2. Methods
(a). The population
We analysed data that were collected between the years 2004 and 2012 in a free-living house mouse Mus musculus domesticus population in an old barn near Zurich, Switzerland [44]. We provided a human-made and provisioned environment similar to that found in barns housing animals, but easier to monitor. We provided food and water regularly ad libitum. The barn is divided into four similarly sized sectors [44]. However, mice can easily travel between these sectors and also freely enter and leave the barn. Thus emigration could not be monitored directly owing to the numerous and unpredictable exit routes that mice use (that were however small enough to exclude predators). Instead, we used an indirect measure of emigration (see §2b). We considered individuals from 1 to 16 days as pups, then (when they began to be weaned) as juveniles before reaching 17.5 g in body mass, which is when we classified them as adults, as females do not breed until they exceed this body mass [32]. The sex ratio of the population was roughly equal (48% female).
(i). Monitoring
When pups reached 13 days of age (allowing for ±2 days of difference from this), they were ear-punched to provide a DNA sample. Every 10–13 days, the barn was searched for new litters. Every seven weeks, on average, every individual in the barn was caught. On this occasion, all individuals above 17.5 g in body mass received a radio-frequency identification (RFID) transponder and were then considered adults. On average in the years studied, 16.1% of the population received a transponder (was newly classified as an adult) on such a capture event. Additionally, we regularly searched the barn visually and with transponder scanners for dead individuals or lost transponders. When found, dead individuals were removed and identified via their transponder or a new genetic sample. Finally, there has been an automatic antenna system in the population since 2007 that tracks exits and entries of transpondered mice into and out of 40 nest boxes [44]. We used these data in addition to data from manual checks to determine when an adult individual was last detected in the population if it was never found dead. This was relevant for the population size calculations; see §2c.
(ii). Identification
We genetically identify each individual as a pup, as a newly classified adult, or as a corpse if found dead without a transponder. We do so based on multi-locus genotypes based on 25 micro-satellite loci [45]. The genotypes allow us to link individuals as pups to their adult transponder ID or to a corpse, allowing for one allelic mismatch using the software CERVUS [46]. We use the micro-satellite locus Hba-ps4 that has a 16-bp t specific insertion [47] to identify the t haplotype. Sexing of individuals was performed by testing for the presence of Y-chromosome-specific micro-satellite markers Y8, Y12 and Y21 [48].
(b). Definitions of migration
(i). Disappearing from the population
Individuals that fulfilled all of the following criteria were classified as juveniles that disappeared from the population: (i) the individual was genotyped as a 13 ± 2 day old pup, (ii) its genotype never matched to an adult’s sample, and (iii) also never to a corpse’s sample. Following this definition, the time at which the individual disappeared must have been between 13 ± 2 days of age and an adult age (defined by body mass as described earlier) and therefore the individual was a juvenile. Consequently, individuals that disappeared from the barn as adults were not classified as disappeared in this analysis, but are instead treated as juveniles that stayed until adulthood. We excluded individuals born in the year 2005 from the analysis because monitoring was considerably less intense in this year and thus there is a larger potential to misclassify individuals that died within the population as ones that disappeared. Therefore, we analysed 7 birthyears (2004 and 2006–2011) in which the t was present in the population (it then went extinct). We also excluded individuals about whom we did not have enough information (such as incomplete genotype or conflicting sex information) from the analysis. Furthermore, we removed those that died as juveniles, because we cannot know whether they might have emigrated later. Following these exclusions, 261 +/t and 2677 +/+ remained for the analysis (see the electronic supplementary material, S1, for an overview).
(ii). Migration within the population
We defined the four distinct sectors within the population described earlier as sub-populations between which mice can migrate. We did so because from earlier analyses [49] we know that the dividing walls between the four sectors are social barriers for the mice. While mice are regularly seen moving within each sector, movements and social interactions between the sectors are less frequent [49]. Furthermore, 61% of adults (in their adult lifetime) that were located at least nine times were found within the same sector every time. Thirty-one per cent were found in two sectors in total, 7% in three and less than 1% in all four. We defined juvenile within-population migrants as individuals that were first found as adults in a different sector from where they were last seen in as pups. Thus, these individuals migrated in the same age range as those that disappeared. The dataset was based on the same restrictions made for the disappearance analysis, except that only those individuals that stayed in the population until adulthood could be analysed.
(c). Controlling variables
Mice were counted towards the population size from birth until death or until they were last seen in the population. When they were last seen was based on both manually locating the animal (in regular population monitoring) or information from our automatic antenna system. A large proportion of the individuals disappeared from our population before they received their RFID transponder (the disappearances analysed in this study). These mice were counted for 30 days from the time of their birth on as part of the population. This cut-off is based on a handful of individuals that reached the body mass we designate as minimum for the transponder (17.5 g) at 35 days of age, reports of an early dispersal phase in 30-day-old juveniles [50], and a weaning age (nutritional independence and end of active maternal care) in mice of about 23 days [51,52]. Therefore, it is a conservative estimate of the minimum amount of time an emigrant would spend in the population after birth. However, the results of this study do not change fundamentally when this time frame is increased (we used 50 days of age as an alternative cut-off; see the electronic supplementary material, S1).
We subdivided the population size into adult and juvenile population sizes. We did so because we do not know how individual mice decide whether they migrate, therefore we wanted to disentangle the current and the future reproductive environment reflected by these two variables. The two population sizes are correlated, but do not explain much of each other's variation (linear model with R2 = 0.08). Individuals that remained in the population until adulthood were counted from age 31 days on as part of the adult population (and before as juveniles), whereas individuals that were never found as adults were only counted for 30 days as juveniles and never as adults. We also considered using local adult population sizes in the four sectors, but overall did not find that to be more informative for the questions asked here (see electronic supplementary material, S2). Similarly, we tested whether controlling for relatedness would influence the results, but concluded that this was not the case (see electronic supplementary material, S3).
We defined the months April to September as the main breeding season, because these are the six months with the highest counts of new pups. The remaining months (October to March) were defined as the off-season. Eighty-seven per cent of the birth dates in our dataset fall within the main breeding season. To account for inter-annual variation in the environment (like temperature or noises in the area) that could possibly affect migration propensity, we added the year of birth (N = 7) as a random effect in the disappearance models. Finally, we also controlled for the age when individuals were first sampled (between 11 and 15 days of age, with most being sampled at 13 days). We did so because preliminary data visualizations revealed a relation between this age and disappearances.
(d). Statistical analyses
(i). Disappearing from the population
We used a generalized mixed effect model with a binomial distribution, a logit-link function, and fit by maximum-likelihood. All statistical analyses and figures were done in R 3.4.4 [53] with RStudio [54] and the packages ggplot 2 2.2.1 [55], and lme4 1.1–17 [56], the latter using the function glmer. The dependent variable was binary (1 when the individual disappeared as a juvenile and 0 if it did not). The independent variables were adult and juvenile population size (each standardized and fitted as linear and quadratic terms), the season, the sex and the genotype. The population sizes and the season were taken from 30 days after an individual’s birth to reflect the environment that the juvenile was exposed to around the time when it either did or did not emigrate. The year of birth was used as a random effect. We used predictInterval of merTools 0.3.0 [57] with its integrated bootstrapping method with 10 000 simulations, using the median and a confidence interval of 95% for figure 1.
Figure 1.
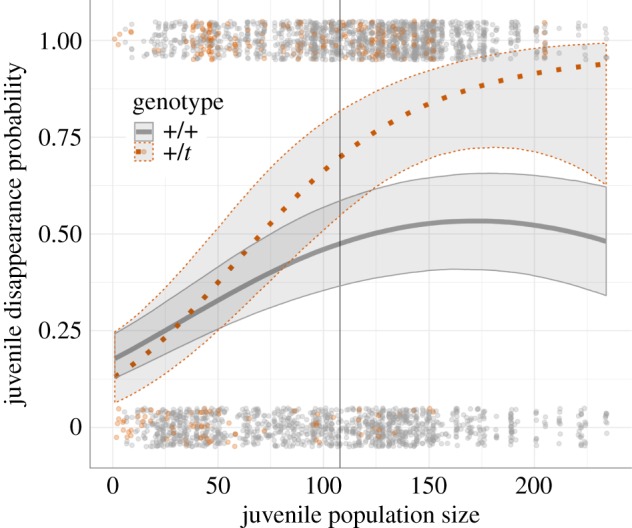
Predicted probabilities of juvenile disappearance out of the study population (lines) with 95% confidence intervals and actual data points (top and bottom, jittered) of +/t (orange, dotted line) and +/+ (grey, solid line) individuals in varying juvenile population sizes (N = 2938). This example plot is based on predictions from the most informative disappearances model (model 2) for a female born in the off-season in average adult population size for no specific birthyear (fixed effects only). The vertical line indicates the mean juvenile population size.
We used pbkrtest 0.4–7 [58] for parametric bootstrapping-based model comparisons with a significance level of 5%. Each dataset was simulated 10 000 times. The p-value is based on the PB statistic provided by the function PBmodcomp. It represents the fraction of likelihood ratio test (LRT) values of the simulated (bootstrapped) datasets that were larger or equal to the observed LRT value. Some of the runs can result in negative values of the LRT statistic. These runs are excluded automatically. We tested the significance of the genotype’s effect and the interaction between genotype and the population sizes by comparing a model with the respective predictors to a model without them (see table 1; electronic supplementary material, S1 for all comparisons). We list ΔAIC values in the table to ease understanding, but did not use them for interpretation. We tested interactions of genotype and season as well as genotype and sex to explore potential relationships that we did not hypothesize (electronic supplementary material, S1).
Table 1.
Excerpt overview of models of juvenile disappearances out of the study population (see electronic supplementary material, S1 for the full table). LRT indicates the likelihood ratio test statistic of the observed dataset. The p-value is the fraction of simulated datasets with LRT larger than the observed LRT. Runs indicate the absolute values on which the p is based. The superscripted ‘2’ in the formula refers to quadratic terms. The ‘x’ indicates model term interactions.
| models | formula | comparison | LRT | p-value | runs | ΔAIC |
|---|---|---|---|---|---|---|
| null model with covariates | ∼ juvenile population size | n.a. | n.a. | n.a. | n.a. | n.a. |
| + juvenile population size2 | ||||||
| + adult population size | ||||||
| + adult population size2 | ||||||
| + season + sex | ||||||
| + age when sampled | ||||||
| model 1 | ∼ genotype | null model | 16.00 | 0.0003 | 1/5869 | −14.0 |
| + null model variables | ||||||
| model 2 | ∼ genotype × juv. pop. size | model 1 | 11.62 | 0.005 | 26/5815 | −7.62 |
| + genotype × juv. pop. size2 | ||||||
| + model 1 variables |
To test whether pup condition differences could be an alternative explanation for the disappearance differences, we used the same environmental variables to set up a linear mixed model that predicts pup body mass and then compared this model to one that also included the genotype as an effect (electronic supplementary material, S4). We then added pup body mass as a predictor to our disappearance null model and our most informative disappearances model (electronic supplementary material, S1) to test whether (a) disappearance is predicted by pup body mass and (b) the genotype explains the same variation as does the pup body mass. All analyses that included body mass are reduced in their sample size by 40 individuals for whom we did not have this information.
(ii). Migration within the population
For this analysis, we have a reduced sample size because only mice that stayed alive and remained within the population until adulthood can be analysed. We also excluded one more birthyear because in 2011 no +/t stayed in the population until adulthood. We analysed 873 mice. The number of +/t in this dataset is small (60), which complicates statistical analyses. We compared the numbers of juvenile migrants between the genotypes with Pearson’s χ2 test using R. We also used generalized linear models to control for the same variables as in the disappearance analysis. The smaller sample size made this approach less informative. These results can be found in electronic supplementary material, S5.
3. Results
(a). Disappearances from the population
Fifty-six per cent of all individuals born (N = 2938) in the years of this analysis who were alive shortly before weaning disappeared (overview in electronic supplementary material, S1). The most informative disappearance model included the genotype and an interaction between the genotype and the juvenile population size (model 2, see table 1 and electronic supplementary material, S1). This model indicated that +/t were more likely to disappear, particularly with increasing numbers of juveniles in the population (figure 1). At mean juvenile densities, the probability that a +/t juvenile disappears was 47.5% higher than the probability for a +/+ juvenile (based on model predictions used for figure 1). A standard deviation increase in juvenile population size increased this difference by 13.3 percentage points. As can be seen in figure 1, +/t and +/+ were similar in their probability to disappear when there were few juveniles in the population, but then diverged with increasing juvenile density. Disappearance probability decreased with increasing adult population sizes, but was not differently affected in +/+ and +/t. Similarly, being born in the main breeding season and being female increased the probability of disappearance for both genotypes (electronic supplementary material, S1).
To test possible alternative explanations (other than migration propensity) for the disappearance probability of +/t (like a mortality or condition bias), we analysed data on dead juveniles found in the same time frame. We analysed data on 218 dead juveniles. We compared the number of dead juveniles with the number of individuals that were found alive as adults between +/+ and +/t and found no difference (+/t: 17.8% of 90 died as juveniles, +/+: 14.2% of 1424, χ2=0.62, p=0.43). We decided not to conduct a more detailed model for this comparison because of the limited amount of juvenile +/t corpses found (16). For better comparison of this simple mortality analysis with the disappearance model, we used the same simple statistical test for the disappearance data used in the model and again found the difference between +/t and +/+ (71.6% of 261 +/t and 54.4% of 2677 +/+ disappeared as juveniles, χ2 = 28.16, p = 1.1 × 10−7). We also tested whether there were any differences in the individual body mass as a pup (as a measure of the condition of the pup) between +/+ and +/t. We found that +/t pups were slightly heavier than +/+ pups (β = 0.17g, p=0.03, 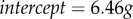 , details in electronic supplementary material, S4), but did not find that the body mass as a pup predicts disappearances, either when the genotype was in the model or when it was absent (models 7 and 8; electronic supplementary material, S1). Thus, we concluded that differences in juvenile disappearances between the genotypes cannot be explained by differences in juvenile mortality or condition.
, details in electronic supplementary material, S4), but did not find that the body mass as a pup predicts disappearances, either when the genotype was in the model or when it was absent (models 7 and 8; electronic supplementary material, S1). Thus, we concluded that differences in juvenile disappearances between the genotypes cannot be explained by differences in juvenile mortality or condition.
(b). Migration within the population
Of the 873 individuals analysed, 9.4% migrated as juveniles within the population, i.e. they were found in a different sub-population as adults from where they were last seen in as pups. Within the population, 16.8% of the 60 +/t migrated as juveniles compared to 8.9% of 813 +/+, a statistically significant difference (χ2 = 4.01, d.f. = 1, p = 0.045). Controlling for other explanatory variables in a generalized linear model (GLM) was more challenging owing to the reduced sample size. We found overall that the genotype remained an informative predictor in interactions with juvenile population size and sex (comparison with null model: p = 0.01, ΔAIC = −7.22, details in electronic supplementary material, S5). Particularly, male +/t had a high migration propensity in the smallest population sizes.
4. Discussion
We provide evidence for a higher migration propensity of +/t juveniles compared to +/+ juveniles. We found that carrying the t haplotype is a strong positive predictor for juvenile disappearances out of our study population. Our hypothesis that +/t should be selected to increase migration propensity was also modestly supported by a +/t bias in migratory movements within the population. Given that variation in behaviours related to dispersal is generally heritable to moderate degrees [59], a manipulation by the t in the t’s favour is a probable explanation. Our results further suggest that the rates of +/t disappearances are increased particularly in denser populations. This is consistent with previous results because the t was found to be less fit in denser populations owing to an increase in sperm competition [20,30]. The +/t that did not disappear from the population were found to be more likely to migrate within the population when juvenile densities were low. A possible explanation for this could be that there was more open habitat available when fewer juveniles were in the population and the migration-prone +/t were able to migrate within the population instead of needing to leave it.
We did not find a different effect of sex between the genotypes in our disappearance analysis, but did find one in the within-population migration analysis. The lack of difference agrees with a theoretical model that showed that t migration propensity manipulation need not be biased towards males (in which t drives), because migration of both male and female +/t was found to be more effective than male-only migration [38]. However, +/t males were more likely than females to migrate within the population as juveniles. The test of this interaction was exploratory and not driven by a hypothesis. The result may reflect sex-specific costs and benefits of within-population migration for +/t mice, which have yet to be fully elucidated. It is interesting, but needs further verification, particularly given that the disappearance analysis with its larger dataset does not show this pattern.
One drawback of our disappearance analysis is that it is at best an indirect measure of emigration, which we expect to be less precise. Despite that, we detected a strong signal. We considered alternative explanations of the strong +/t disappearance bias. We tested for a difference in juvenile mortality but did not find one, which is further supported by a lack of difference in pup survival until weaning from laboratory-bred mice taken from the same population [26]. We found a slightly increased pup body mass for +/t, but showed that this was not predictive of the disappearances (electronic supplementary material, S1) and migration events (electronic supplementary material, S5). Furthermore, there is evidence from another laboratory study that +/t and +/+ from the same study population do not differ in adult body mass (males and females) [16]. Differences in social dominance could be another explanation for disappearance patterns. Studies looking at dominance either found less dominant +/t males [60], more dominant +/t males [61], or no difference in dominance between males and less dominant +/t females [35,62]. However, if dominance differences were the cause of our disappearance results, we might expect to see an informative interaction between sex and genotype. Furthermore, we know from previous analyses that +/t males do not differ in survival from +/+, but +/t females live longer than +/+ in our population [20]. Survival can predict dominance in house mice [62] and thus there is no clear evidence that dominance differs between the genotypes in our population. Finally, the mice in our population could go on exploratory trips outside the barn. Some of the exploring mice could be preyed upon on their trips. In that case, our results would in part reflect differences in exploration propensity. However, studies in mammals indicate that individuals that are more likely to explore are also more likely to migrate [63–65] and if that is true in the study population we would still measure migration propensity indirectly through exploration propensity. Alternatively, if +/t juveniles are somehow more likely to be preyed upon than +/+, it would cause them to disappear more often without necessarily an increased migration or exploration propensity. We cannot test this idea with the data that are available to us. However, we believe that this alternative explanation is weaker than the one we offer. The difference between the genotypes in disappearances is larger in denser populations. This is more consistent with a density-dependent migration propensity than with predation risk. Furthermore, we found evidence that +/t may also migrate differently within the population from +/+, which provides further support for a difference in migration propensity. We cannot completely rule out a difference in predation risk as an explanation, but we argue that it is less likely than differences in migration propensity.
Generally, an increased migration propensity of +/t could help to explain why the t continues to exist in nature despite its homozygous and heterozygous fitness costs owing to recessive lethals and low sperm competitiveness. Compared to a t variant that does not influence migration, variants of the t that increase migration propensity could have an increased chance of reaching or founding populations where there are few other +/t and polyandrous matings are less frequent. The t is expected to rapidly increase in frequency given such circumstances [20,31,66–68]. Thus, it would likely out-compete t variants that did not affect migration. Competition between t variants is consistent with genetic evidence that a single t haplotype variant recently replaced previous variants in a sweep [69]. We do not know how an increased migration propensity could be encoded within the t haplotype, but the t comprises several hundred genes that are protected from recombination [25]. Alternatively, instead of manipulation by the t, the increased migration propensity could also be an evolved response by the rest of the genome to the presence of the t, if increasing migration propensity is increasing the fitness of the rest of the genome when t is present. More work is needed to better understand this interesting dynamic.
Emigration is only the first step of successful dispersal. Emigrants also need to breed as an immigrant or founder, which is challenging for mice [70]. Unfortunately, there were too few +/t that migrated within the population for us to analyse their breeding success. However, Anderson et al. [71] were able to ‘infect’ an island population with the t haplotype by manually migrating +/t. Although the t was able to establish itself in the initial area over a period of a few years, it did not spread much across the island. For Pennycuik et al. [72], introducing the t to an enclosure was more difficult. However, they managed to do so when there were open territories in the population. They also reported many of the +/t males and females migrating between sub-populations. However, the t was almost extinct 2 years later, at the end of the study. It is evident from these experiments that there will be many populations to which the t cannot disperse successfully. In our study population, we have no evidence for immigration of any individuals (unpublished). This makes increased migration propensity counterintuitive because the migration will often fail. Still, because not migrating is also not beneficial for the t, it makes migration attempts potentially even more necessary for the t’s fitness.
When house mice invade an island that has evolved without mammalian predators, their presence can be very damaging to the ecosystem [73–75]. Recently, efforts have been made to use a modified t haplotype for potential eradication of such house mouse populations [42,43,76,77]. The tSRY variant is a t haplotype that is synthetically combined with the male-determining gene SRY. Every +/tSRY individual is thus expected to be male. Owing to the t’s transmission advantage, more than 90% of the offspring of a +/tSRY are then male, which could then drive populations extinct via lack of one sex [42,78,79]. So far, only some of the t’s characteristics have been explicitly considered in trying to facilitate the use of tSRY to eradicate wild populations [42]. However, accounting for the entirety of the known attributes of the t is crucial to successfully predict how a synthesized variant works in the field. Increased migration propensity would likely aid in the distribution of +/tSRY mice to target locations, but could also increase the possibility of tSRY reaching populations it was not intended for.
5. Conclusion
We found that juvenile mice carrying the t haplotype were more likely to disappear from the population at high densities and were over-represented in migrants within the population. To our knowledge, this is the first evidence of a change in migration propensity that is linked to a selfish genetic element. Our results should be of broad interest. First, they have implications for research on other selfish genetic elements, considering that low sperm competitiveness is expected in many male meiotic drive systems like the t [14,15,17,18,80]. Recessive deleterious alleles and therefore frequency-dependent fitness would also be expected in other meiotic drivers, because without negative fitness effects the driver would spread to fixation [81,82]. This would provide further advantages for migratory variants of these drivers. Similarly, parasites could also benefit from manipulating dispersal behaviour [40]. Second, the recent work on artificial gene drive systems based on the t haplotype will benefit from incorporating as many traits of the t as are available. A difference in migration propensity could have important implications for such a system. Third, a selfish genetic element affecting migration propensity could be an important finding for research on dispersal and migration in general. Dispersal attempts are risky [83] and the different selective pressures for the t and similar elements could help to explain better when this behaviour—which often results in no fitness gains—is most beneficial. Therefore, arms races like the one studied here could be a causal mechanism driving the evolution of dispersal. We will further investigate this new direction in t haplotype research with theoretical and experimental approaches.
Supplementary Material
Supplementary Material
Supplementary Material
Acknowledgements
We are particularly grateful to Barbara König for her perseverance in keeping this long-term field study going and for her generous support, and to her and all others who have contributed to collection of the data. We also thank Jari Garbely for genetic laboratory work. Additionally, we thank Barbara König, Tom Price, Erik Postma and Andri Manser for comments on an earlier version of this manuscript. Finally, we acknowledge Natalie Wagner Niepoth for her recommendation to consider local population sizes in the bioRxiv comments [84].
Ethics
The data were collected under permits 26/2002, 210/2003, 215/2006, 51/2010 from the Swiss Animal Experimentation Commission.
Data accessibility
The datasets supporting this article have been uploaded as part of the electronic supplementary material, S6 and S7.
Authors' contributions
The study was conceived and the manuscript written by J.-N.R. and A.K.L. The data were collected and the genetic analyses performed by A.K.L. and colleagues. Statistical analyses were performed by J.-N.R.
Competing interests
We declare we have no competing interests.
Funding
This study was funded by the SNF(31003A-120444, 310030M_138389 and 31003A_160328), the University of Zurich, the Promotor Foundation, Julius Klaus Foundation and the Claraz-Stiftung.
Reference
- 1.Frank SA. 2003. Perspective: repression of competition and the evolution of cooperation. Evolution 57, 693–705. ( 10.1554/0014-3820(2003)057%5B0693:PROCAT%5D2.0.CO;2) [DOI] [PubMed] [Google Scholar]
- 2.Burt A, Trivers RL. 2006. Genes in conflict. Cambridge, MA: Harvard University Press. [Google Scholar]
- 3.Geffre AC, Liu R, Manfredini F, Beani L, Kathirithamby J, Grozinger CM, Toth AL. 2017. Transcriptomics of an extended phenotype: parasite manipulation of wasp social behaviour shifts expression of caste-related genes. Proc. R. Soc. B 284, 20170029 ( 10.1098/rspb.2017.0029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Bekker C, Quevillon LE, Smith PB, Fleming KR, Ghosh D, Patterson AD, Hughes DP. 2014. Species-specific ant brain manipulation by a specialized fungal parasite. BMC Evol. Biol. 14, 166 ( 10.1186/s12862-014-0166-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Östergren G. 1945. Parasitic nature of extra fragment chromosomes. Botaniska Notiser 2, 157–163. [Google Scholar]
- 6.Orgel LE, Crick FHC. 1980. Selfish DNA: the ultimate parasite. Nature 284, 604–607. ( 10.1038/284604a0) [DOI] [PubMed] [Google Scholar]
- 7.Franceschi N, Cornet S, Bollache L, Dechaume-Moncharmont FX, Bauer A, Motreuil S, Rigaud T. 2010. Variation between populations and local adaptation in acanthocephalan-induced parasite manipulation. Evolution 64, 2417–2430. ( 10.1111/j.1558-5646.2010.01006.x) [DOI] [PubMed] [Google Scholar]
- 8.Mercot H, Atlan A, Jacques M, Montchamp-Moreau C. 1995. Sex-ratio distortion in Drosophila simulans: co-occurence of a meiotic drive and a suppressor of drive. J. Evol. Biol. 8, 283–300. ( 10.1046/j.1420-9101.1995.8030283.x) [DOI] [Google Scholar]
- 9.Jaenike J. 2001. Sex chromosome meiotic drive. Annu. Rev. Ecol. Syst. 32, 25–49. ( 10.1146/annurev.ecolsys.32.081501.113958) [DOI] [Google Scholar]
- 10.Jaenike J. 1999. Suppression of sex-ratio meiotic drive and the maintenance of Y-chromosome polymorphism in Drosophila. Evolution 53, 164 ( 10.2307/2640929) [DOI] [PubMed] [Google Scholar]
- 11.Hatcher MJ. 2000. Persistence of selfish genetic elements: population structure and conflict. Trends Ecol. Evol. 15, 271–277. ( 10.1016/S0169-5347(00)01875-9) [DOI] [PubMed] [Google Scholar]
- 12.Moreau J, Bertin A, Caubet Y, Rigaud T. 2001. Sexual selection in an isopod with Wolbachia-induced sex reversal: males prefer real females. J. Evol. Biol. 14, 388–394. ( 10.1046/j.1420-9101.2001.00292.x) [DOI] [Google Scholar]
- 13.Wilkinson GS, Presgraves DC, Crymes L. 1998. Male eye span in stalk-eyed flies indicates genetic quality by meiotic drive suppression. Nature 391, 276–279. ( 10.1038/34640) [DOI] [Google Scholar]
- 14.Taylor DR, Ingvarsson PK. 2003. Common features of segregation distortion in plants and animals. Genetica 117, 27–35. ( 10.1023/A:1022308414864) [DOI] [PubMed] [Google Scholar]
- 15.Price TAR, Wedell N. 2008. Selfish genetic elements and sexual selection: their impact on male fertility. Genetica 134, 99–111. ( 10.1007/s10709-008-9253-y) [DOI] [PubMed] [Google Scholar]
- 16.Sutter A, Lindholm AK. 2015. Detrimental effects of an autosomal selfish genetic element on sperm competitiveness in house mice. Proc. R. Soc. B 282, 20150974 ( 10.1098/rspb.2015.0974) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilkinson GS, Fry CL. 2001. Meiotic drive alters sperm competitive ability in stalk-eyed flies. Proc. R. Soc. Lond. B 268, 2559–2564. ( 10.1098/rspb.2001.1831) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Champion de Crespigny FE, Wedell N. 2006. Wolbachia infection reduces sperm competitive ability in an insect. Proc. R. Soc. B 273, 1455–1458. ( 10.1098/rspb.2006.3478) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price TAR, Hodgson DJ, Lewis Z, Hurst GDD, Wedell N. 2008. Selfish genetic elements promote polyandry in a fly. Science 322, 1241–1243. ( 10.1126/science.1163766) [DOI] [PubMed] [Google Scholar]
- 20.Manser A, Lindholm AK, König B, Bagheri HC. 2011. Polyandry and the decrease of a selfish genetic element in a wild house mouse population. Evolution 65, 2435–2447. ( 10.1111/j.1558-5646.2011.01336.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeh JA, Zeh DW. 1996. The evolution of polyandry I: intragenomic conflict and genetic incompatibility. Proc. R. Soc. Lond. B 263, 1711–1717. ( 10.1098/rspb.1996.0250) [DOI] [Google Scholar]
- 22.Wedell N. 2013. The dynamic relationship between polyandry and selfish genetic elements. Phil. Trans. R. Soc. B 368, 20120049 ( 10.1098/rstb.2012.0049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Champion de Crespigny FE, Pitt TD, Wedell N. 2006. Increased male mating rate in Drosophila is associated with Wolbachia infection. J. Evol. Biol. 19, 1964–1972. ( 10.1111/j.1420-9101.2006.01143.x) [DOI] [PubMed] [Google Scholar]
- 24.Kelemen RK, Vicoso B. 2018. Complex history and differentiation patterns of the t-haplotype, a mouse meiotic driver. Genetics 208, 365–375. ( 10.1534/genetics.117.300513) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silver LM. 1985. Mouse t haplotypes. Annu. Rev. Genet. 19, 179–208. ( 10.1146/annurev.ge.19.120185.001143) [DOI] [PubMed] [Google Scholar]
- 26.Lindholm AK, Musolf K, Weidt A, König B. 2013. Mate choice for genetic compatibility in the house mouse. Ecol. Evol. 3, 1231–1247. ( 10.1002/ece3.534) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Safronova LD. 2009. Embryonal effects of t-haplotypes in mice. Russ. J. Dev. Biol. 40, 23–30. ( 10.1134/S1062360409010032) [DOI] [PubMed] [Google Scholar]
- 28.Manser A, Lindholm AK, Simmons LW, Firman RC. 2017. Sperm competition suppresses gene drive among experimentally evolving populations of house mice. Mol. Ecol. 38, 42–49. ( 10.1111/mec.14215) [DOI] [PubMed] [Google Scholar]
- 29.Firman RC, Simmons LW. 2008. Polyandry, sperm competition, and reproductive success in mice. Behav. Ecol. 19, 695–702. ( 10.1093/beheco/arm158) [DOI] [Google Scholar]
- 30.Dean MD, Ardlie KG, Nachman MW. 2006. The frequency of multiple paternity suggests that sperm competition is common in house mice (Mus domesticus). Mol. Ecol. 15, 4141–4151. ( 10.1111/j.1365-294X.2006.03068.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ardlie KG, Silver LM. 1998. Low frequency of t haplotypes in natural populations of house mice (Mus musculus domesticus). Evolution 52, 1185–1196. ( 10.2307/2411247) [DOI] [PubMed] [Google Scholar]
- 32.König B, Lindholm AK. 2012. The complex social environment of female house mice (Mus domesticus). In Evolution of the house mouse (eds M Macholán, SJE Baird, P Munclinger, J Piálek), pp. 114–134. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 33.Sutter A, Lindholm AK. 2016. No evidence for female discrimination against male house mice carrying a selfish genetic element. Cur. Zool. 62, 675–685. ( 10.1093/cz/zow063) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manser A, König B, Lindholm AK. 2015. Female house mice avoid fertilization by t haplotype incompatible males in a mate choice experiment. J. Evol. Biol. 28, 54–64. ( 10.1111/jeb.12525) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lenington S, Coopersmith C, Williams J. 1992. Genetic basis of mating preferences in wild house mice. Am. Zool. 32, 40–47. ( 10.1093/icb/32.1.40) [DOI] [Google Scholar]
- 36.Matthysen E. 2012. Multicausality of dispersal: a review. In Dispersal ecology and evolution (eds J Clobert, M Baguette, TG Benton, JM Bullock), pp. 3–18. Oxford, UK: Oxford University Press. ( ) [DOI] [Google Scholar]
- 37.Baker RR. 1978. The evolutionary ecology of animal migration. New York, NY: Holmes & Meier Publishers Inc. [Google Scholar]
- 38.Levin BR, Petras ML, Rasmussen DI. 1969. The effect of migration on the maintenance of a lethal polymorphism in the house mouse. Am. Nat. 103, 647–661. ( 10.1086/282631) [DOI] [Google Scholar]
- 39.van Boven M, Weissing FJ. 2001. Competition at the mouse t complex: rare alleles are inherently favored. Theor. Popul. Biol. 60, 343–358. ( 10.1006/tpbi.2001.1551) [DOI] [PubMed] [Google Scholar]
- 40.Lion S, van Baalen M, Wilson WG. 2006. The evolution of parasite manipulation of host dispersal. Proc. R. Soc. B 273, 1063–1071. ( 10.1098/rspb.2005.3412) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boulinier T, McCoy KD, Sorci G. 2001. Dispersal and parasitism. In Dispersal (eds J Clobert, E Danchin, AA Dhondt, JD Nichols), pp. 169–179. New York, NY: Oxford University Press. [Google Scholar]
- 42.Backus GA, Gross K. 2016. Genetic engineering to eradicate invasive mice on islands: modeling the efficiency and ecological impacts. Ecosphere 7, e01589 ( 10.1002/ecs2.1589) [DOI] [Google Scholar]
- 43.Piaggio AJ. et al. 2017. Is it time for synthetic biodiversity conservation? Trends Ecol. Evol. 32, 97–107. ( 10.1016/j.tree.2016.10.016) [DOI] [PubMed] [Google Scholar]
- 44.König B, Lindholm AK, Lopes PC, Dobay A, Steinert S, Buschmann FJ-U. 2015. A system for automatic recording of social behavior in a free-living wild house mouse population. Animal Biotelemetry 3, 39 ( 10.1186/s40317-015-0069-0) [DOI] [Google Scholar]
- 45.Ferrari M, Lindholm AK, König B. 2018. Fitness consequences of female alternative reproductive tactics in house mice (Mus musculus domesticus). Am. Nat. Forthcoming ( 10.1086/700567) [DOI] [PubMed] [Google Scholar]
- 46.Kalinowski ST, Taper ML, Marshall TC. 2007. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 16, 1099–1106. ( 10.1111/j.1365-294X.2007.03089.x) [DOI] [PubMed] [Google Scholar]
- 47.Hammer MF, Schimenti J, Silver LM. 1989. Evolution of mouse chromosome 17 and the origin of inversions associated with t haplotypes. Proc. Natl Acad. Sci. USA 86, 3261–3265. ( 10.1073/pnas.86.9.3261) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hardouin EA, Chapuis J-L, Stevens MI, van Vuuren J, Quillfeldt P, Scavetta RJ, Teschke M, Tautz D. 2010. House mouse colonization patterns on the sub-Antarctic Kerguelen Archipelago suggest singular primary invasions and resilience against re-invasion. BMC Evol. Biol. 10, 325 ( 10.1186/1471-2148-10-325) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perony N, Tessone CJ, König B, Schweitzer F. 2012. How random is social behaviour? Disentangling social complexity through the study of a wild house mouse population. PLoS Comput. Biol. 8, e1002786 ( 10.1371/journal.pcbi.1002786) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lidicker WZ., Jr 1976. Social behaviour and density regulation in house mice living in large enclosures. J. Animal Ecol. 45, 677–697. ( 10.2307/3575) [DOI] [Google Scholar]
- 51.Bronson FH. 1979. The reproductive ecology of the house mouse. Quat. Rev. Biol. 54, 265–299. ( 10.1086/411295) [DOI] [PubMed] [Google Scholar]
- 52.König B, Markl H. 1987. Maternal care in house mice. Behav. Ecol. Sociobiol. 20, 1–9. ( 10.1007/BF00292161) [DOI] [Google Scholar]
- 53.R Core Team. 2017. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. https://www.R-project.org/.
- 54.RStudio Team. 2016. RStudio: integrated development environment for R.
- 55.Wickham H. 2009. ggplot2. New York, NY: Springer New York; ( 10.1007/978-0-387-98141-3) [DOI] [Google Scholar]
- 56.Bates D, Mächler M, Bolker B, Walker S. 2015. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48. (doi:10.18637/jss.v067.i01) [Google Scholar]
- 57.Knowles JE, Frederick C.2016. merTools: tools for analyzing mixed effect regression models. https://CRAN.R-project.org/package=merTools .
- 58.Halekoh U, Højsgaard S. 2014. A Kenward-Roger approximation and parametric bootstrap methods for tests in linear mixed models—the R package pbkrtest. J. Stat. Softw. 59, 1–30. (doi:10.18637/jss.v059.i09)26917999 [Google Scholar]
- 59.Saastamoinen M. et al. 2018. Genetics of dispersal. Biol. Rev. 93, 574–599. ( 10.1111/brv.12356) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Carroll LS, Meagher S, Morrison L, Penn DJ, Potts WK. 2004. Fitness effects of a selfish gene (the Mus t complex) are revealed in an ecological context. Evolution 58, 1318–1328. ( 10.1111/j.0014-3820.2004.tb01710.x) [DOI] [PubMed] [Google Scholar]
- 61.Lenington S, Drickamer LC, Robinson AS, Erhart M. 1996. Genetic basis for male aggression and survivorship in wild house mice (Mus domesticus). Aggress. Behav. 22, 135–145. ( 10.1002/(SICI)1098-2337(1996)22:2%3C135::AID-AB6%3E3.0.CO;2-N) [DOI] [Google Scholar]
- 62.Franks P, Lenington S. 1986. Dominance and reproductive behavior of wild house mice in a seminatural environment correlated with T-locus genotype. Behav. Ecol. Sociobiol. 18, 395–404. ( 10.1007/BF00300513) [DOI] [Google Scholar]
- 63.Krackow S. 2003. Motivational and heritable determinants of dispersal latency in wild male house mice (Mus musculus musculus). Ethology 109, 671–689. ( 10.1046/j.1439-0310.2003.00913.x) [DOI] [Google Scholar]
- 64.Hoset KS, Ferchaud A-L, Dufour F, Mersch D, Cote J, Le Galliard J-F. 2011. Natal dispersal correlates with behavioral traits that are not consistent across early life stages. Behav. Ecol. 22, 176–183. ( 10.1093/beheco/arq188) [DOI] [Google Scholar]
- 65.Debeffe L, Morellet N, Cargnelutti B, Lourtet B, Coulon A, Gaillard J, Bon R, Hewison A. 2013. Exploration as a key component of natal dispersal: dispersers explore more than philopatric individuals in roe deer. Anim. Behav. 86, 143–151. ( 10.1016/j.anbehav.2013.05.005) [DOI] [Google Scholar]
- 66.Lewontin RC, Dunn LC. 1960. The evolutionary dynamics of a polymorphism in the house mouse. Genetics 45, 705–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Durand D, Ardlie KG, Buttel L, Levin SA, Silver LM. 1997. Impact of migration and fitness on the stability of lethal t-haplotype polymorphism in Mus musculus: a computer study. Genetics 145, 1093–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Boven M, Weissing FJ. 1999. Segregation distortion in a deme-structured population: opposing demands of gene, individual and group selection. J. Evol. Biol. 12, 80–93. ( 10.1046/j.1420-9101.1999.00011.x) [DOI] [Google Scholar]
- 69.Hammer MF, Silver LM. 1993. Phylogenetic analysis of the alpha-globin pseudogene-4 (Hba-ps4) locus in the house mouse species complex reveals a stepwise evolution of t haplotypes. Mol. Biol. Evol. 10, 971–1001. [DOI] [PubMed] [Google Scholar]
- 70.Pocock MJO, Hauffe HC, Searle JB. 2005. Dispersal in house mice. Biol. J. Linnean Soc. 84, 565–583. ( 10.1111/j.1095-8312.2005.00455.x) [DOI] [Google Scholar]
- 71.Anderson PK, Dunn LC, Beasley AB. 1964. Introduction of a lethal allele into a feral house mouse population. Am. Nat. 98, 57–64. [Google Scholar]
- 72.Pennycuik PR, Johnston PG, Lidicker WZ Jr, Westwood NH. 1978. Introduction of a male sterile allele (tw2) into a population of house mice housed in a large outdoor enclosure. Aust. J. Zool. 26, 69–81. ( 10.1071/ZO9780069) [DOI] [Google Scholar]
- 73.Wanless RM, Angel A, Cuthbert RJ, Hilton GM, Ryan PG. 2007. Can predation by invasive mice drive seabird extinctions? Biol. Lett. 3, 241–244. ( 10.1098/rsbl.2007.0120) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Angel A, Wanless RM, Cooper J. 2009. Review of impacts of the introduced house mouse on islands in the Southern Ocean: are mice equivalent to rats? Biol. Invasions 11, 1743–1754. ( 10.1007/s10530-008-9401-4) [DOI] [Google Scholar]
- 75.St Clair JJ. 2011. The impacts of invasive rodents on island invertebrates. Biol. Conserv. 144, 68–81. ( 10.1016/j.biocon.2010.10.006) [DOI] [Google Scholar]
- 76.Kanavy D, Serr M. 2017. Sry gene grive for rodent control: reply to Gemmell and Tompkins. Trends Ecol. Evol. 32, 315–316. ( 10.1016/j.tree.2017.03.006) [DOI] [PubMed] [Google Scholar]
- 77.Gemmell NJ, Tompkins DM. 2017. Gene drives and rodent control: response to Piaggio et al. Trends Ecol. Evol. 32, 314–315. ( 10.1016/j.tree.2017.03.005) [DOI] [PubMed] [Google Scholar]
- 78.Hamilton WD. 1967. Extraordinary sex ratios. Science 156, 477–488. ( 10.1126/science.156.3774.477) [DOI] [PubMed] [Google Scholar]
- 79.Price TAR, Hurst GDD, Wedell N. 2010. Polyandry prevents extinction. Curr. Biol. 20, 471–475. ( 10.1016/j.cub.2010.01.050) [DOI] [PubMed] [Google Scholar]
- 80.Atlan A, Joly D, Capillon C, Montchamp-Moreau C. 2004. Sex-ratio distorter of Drosophila simulans reduces male productivity and sperm competition ability. J. Evol. Biol. 17, 744–751. ( 10.1111/j.1420-9101.2004.00737.x) [DOI] [PubMed] [Google Scholar]
- 81.Hurst LD, Atlan A, Bengtsson BO. 1996. Genetic conflicts. Q Rev. Biol. 71, 317–364. ( 10.1086/419442) [DOI] [PubMed] [Google Scholar]
- 82.Lindholm AK. et al. 2016. The ecology and evolutionary dynamics of meiotic drive. Trends Ecol. Evol. 31, 315–326. ( 10.1016/j.tree.2016.02.001) [DOI] [PubMed] [Google Scholar]
- 83.Bonte D. et al. 2012. Costs of dispersal. Biol. Rev. 87, 290–312. ( 10.1111/j.1469-185X.2011.00201.x) [DOI] [PubMed] [Google Scholar]
- 84.Runge J-N, Lindholm AK. 2018. The t haplotype, a selfish genetic element, manipulates migration propensity in free-living wild house mice Mus musculus domesticus. bioRxiv. ( 10.1101/271247) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting this article have been uploaded as part of the electronic supplementary material, S6 and S7.


