Abstract
Clinically relevant subtypes exist for pancreatic ductal adenocarcinoma (PDAC), but molecular characterization is not yet standard in clinical care. We implemented a biopsy protocol to perform time-sensitive whole exome sequencing and RNA-sequencing for patients with advanced PDAC. Therapeutically relevant genomic alterations were identified in 48% (34/71) and pathogenic/likely pathogenic germline alterations in 18% (13/71) of patients. Overall, 30% (21/71) of enrolled patients experienced a change in clinical management as a result of genomic data. Twenty-six patients had germline and/or somatic alterations in DNA-damage repair genes, and 5 additional patients had mutational signatures of homologous recombination deficiency but no identified causal genomic alteration. Two patients had oncogenic in-frame BRAF deletions, and we report the first clinical evidence that this alteration confers sensitivity to MAP-kinase pathway inhibition. Moreover, we identified tumor/stroma gene expression signatures with clinical relevance. Collectively, these data demonstrate the feasibility and value of real-time genomic characterization of advanced PDAC.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the third-leading cause of cancer-related death in the United States and is projected to become the second leading cause by 2030 (1). Most patients present with advanced disease and die within 12 months of diagnosis (2,3). Recent genomic studies of primary PDAC resection specimens have identified recurrent molecular alterations and genomic subtypes of the disease (4–12). Moreover, RNA analyses of PDAC cohorts have identified gene expression signatures with prognostic and biological relevance (7,12–14). While these molecular subtypes of PDAC may theoretically help guide precision medicine approaches, molecular characterization of PDAC in patients with advanced disease is not yet standard clinical practice. Biopsy-driven genomic studies have been challenging due to rapid disease progression and the small-volume and heterogeneous nature of biopsies that impede deep molecular characterization. Despite the fact that conventional therapies are often ineffective, the rate of enrollment of PDAC patients onto clinical trials is extremely low (15). A proactive, standardized approach to acquire PDAC biopsy tissue and perform rapid turnaround molecular analysis is required to efficiently generate and utilize genomic information in PDAC patients.
Results
Biopsy approach and patient cohort
The PancSeq protocol was developed as an institutional review board (IRB)-approved multi-disciplinary biopsy program to obtain tissue from core needle biopsies or fine needle aspirates in patients with metastatic or locally advanced PDAC for rapid turnaround genomic analysis. Between March 2015 and June 2017, 79 patients underwent biopsy on the PancSeq protocol (Supplementary Table S1, Supplementary Figure S1). An initial pilot phase (n=10 patients) was conducted in patients having clinically indicated biopsies to optimize workflow, tissue processing, nucleic acid extraction, whole exome sequencing (WES) and RNA sequencing (RNA-seq) from small volume needle biopsies (approximately 15–20 mg of tissue per biopsy). Subsequently, 69 additional patients underwent biopsy and WES was performed in a CLIA-certified laboratory, with return of selected somatic and germline variants to referring clinicians. Patients had a median age of 64 years and most had metastatic disease (96%) and no prior therapy (70%) (Supplementary Table S1). Most patients had a diagnosis of PDAC (95%), although several patients with less common histologies were enrolled. Biopsies were obtained from liver (n=63), pancreas (n=7), peritoneum (n=6), lymph nodes (n=2) and ovary (n=1) (Supplementary Table S2). Biopsies were performed percutaneously by interventional radiology (n=72), with endoscopic ultrasound (n=6), or intraoperatively (n=1), and a median of 5 (range, 1–10) cores or biopsy specimens were collected per patient. A low rate of serious complications was observed, with only one patient having a self-limited hepatic subcapsular hematoma in the setting of a clinically indicated liver biopsy and therapeutic anticoagulation with an oral factor Xa inhibitor (rivaroxaban) for prior deep venous thrombosis.
Real-time DNA sequencing and exome analysis
From the FFPE core, tumor content was quantified by histopathology and 92% (73/79) of cases had sufficient tumor content (≥5% tumor nuclei) to proceed with WES. ABSOLUTE purity (16) was examined in WES data and successful mutation and copy number calls could be made for all but two of these samples (Supplementary Table S2, Supplementary Figure S1).
Consistent with prior genome sequencing studies of primary PDAC samples, samples from our cohort of advanced PDAC patients displayed low neoplastic cellularity, with a median cellularity by histologic assessment of 40%, and by WES using the ABSOLUTE algorithm of 34% (Supplementary Figure S2A-B)(16). Neoplastic cellularity estimates by histologic assessment and by the ABSOLUTE algorithm on WES data were significantly correlated (Pearson’s product-moment correlation = 0.386, 95% CI (0.195 – 0.548); P = 0.00016; Supplementary Figure S2A). This level of correlation is consistent with that observed in other studies (12,16) and is impacted by variability between biopsy specimens as well as differential sensitivity between the two methods for identification of neoplastic cellularity. For our biopsy samples, we performed deep WES with a median of mean target coverage of 191X in the tumor and 176X in the normal (Supplementary Table S2). Within the CLIA-certified phase of the study, analyzed results were available within an average of 39 days (range, 16–67) from the date of clinically indicated biopsies and 28 days (range, 15–51) for research-only biopsies (Supplementary Table S2). Additional time for return of results for clinically indicated biopsies was required, as these specimens were held in the pathology department until a formal histologic diagnosis was confirmed. A report of clinically relevant events was returned to the referring clinician detailing somatic mutations, small insertions/deletions, and copy number alterations (CNAs) as well as pathogenic/likely pathogenic germline alterations in a curated list of 81 PDAC-relevant genes (Supplementary Table S3). These genes were chosen based on somatic or germline clinical relevance, therapeutic actionability and/or recurrent mutation across published PDAC genome sequencing cohorts (4–12). Most of these genes (n=69) were associated with clinical trial or off-label FDA-approved targeted therapies. Comprehensive analysis of all genes from WES data was simultaneously performed in the research setting.
Landscape of somatic mutations and copy number alterations (CNAs) in advanced PDAC
Genome sequencing studies have elucidated the molecular landscape of archived primary PDAC tumors and have demonstrated distinct molecular subtypes of disease (6–12). In our cohort of 71 advanced PDAC patients, we identified significantly recurrent mutations in KRAS, TP53, CDKN2A, SMAD4, ARID1A and TGFBR2 – a collection of genes that were also recurrently mutated in primary PDAC tumors (7,10,12) (Figure 1). Moreover, we observed frequent mutations in additional tumor suppressor genes (e.g. RNF43) or oncogenes (e.g. BRAF, GNAS), as well as recurrent mutations in genes involved in DNA-damage repair (DDR) and chromatin modification (Figure 1). Recurrent high-level amplifications were observed in several genomic loci, encompassing genes such as MYC, AKT2 and GATA6 (Figure 1, Supplementary Figure S3). Deletions were identified at numerous loci, including frequent homozygous deletions of CDKN2A and SMAD4 (Figure 1, Supplementary Figure S3).
Figure 1:
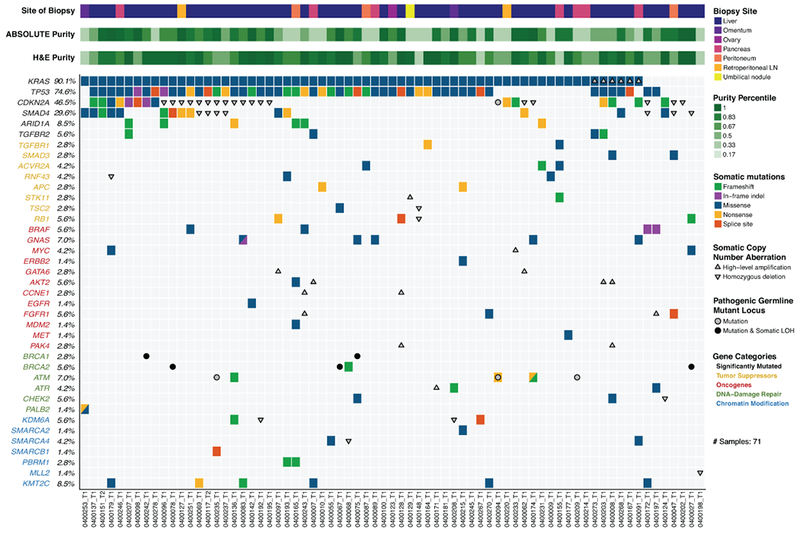
Landscape of genomic alterations identified by whole exome sequencing in biopsies of advanced pancreatic ductal adenocarcinoma (PDAC) patients. Co-mut plot displaying integrated genomic data for 71 samples displayed as columns, including: somatic mutations, high-level amplifications and homozygous deletions, and germline mutations for selected genes. For each sample, the site of biopsy, the ABSOLUTE neoplastic cellularity (purity) from WES data, and the neoplastic cellularity as assessed by histologic evaluation are shown as tracks at the top. Significantly mutated genes with q value ≤ 0.1 that were identified by exome sequencing are listed at the top (black) vertically in order of decreasing significance. Genes from recurrently altered functional classes are also shown, including tumor suppressor genes (yellow), oncogenes (red), DNA damage repair genes (green) and chromatin modification genes (blue). LOH, loss of heterozygosity. LN, lymph node. Indel, insertion/deletion.
Mutational signature analysis from WES data
Mutational signature analysis was performed using a Bayesian variant of the non-negative matrix factorization (NMF) approach in a two-stage manner from the set of single nucleotide variants (SNV) in our dataset, as previously described (SignatureAnalyzer, Supplementary Experimental Methods, (17–20)). First, we performed de novo signature discovery and our analytic pipeline identified three primary signatures: SigA that best resembled COSMIC signature 3 with cosine similarity 0.87 (BRCA mutant signature suggestive of homologous recombination deficiency [HRD]); SigB that best resembled COSMIC signature 1 with cosine similarity 0.96 (C>T transitions at CpG dinucleotides, Aging); and SigC that best resembled COSMIC signature 17 with cosine similarity 0.91 (etiology unknown) (Supplementary Figure S4A-B). In addition, we observed a relative elevation of C>G transversions and C>T transitions at TC[A/T] contexts in SigA corresponding to canonical hotspots of APOBEC mutagenesis (COMSIC signature 2 and 13), suggesting that APOBEC signature is possibly operative in this cohort, but not cleanly separable due to a lack of mutations (17). Based on this de novo analysis, we concluded that four main mutational processes were likely active in these data (Aging/COSMIC1; BRCA/HRD/COSMIC3; APOBEC/COSMIC2+13; and COSMIC17). To better evaluate discrete contributions of these mutational processes in our data and to minimize signature contamination, we next performed a projection analysis to infer a signature activity across our biopsy cohort using the signature profiles of the five contributing COSMIC signatures: COSMIC1, 2, 3, 13, 17 (Figure 2A and Supplemental Experimental Methods). Consistent with the de novo analysis, the inferred signature activity with these five COSMIC signatures reveals a clear contribution of the Aging/COSMIC1 signature in almost all patients and a notable activity of the HRD/COSMIC3 signatures in many patients (Figure 2A).
Figure 2:
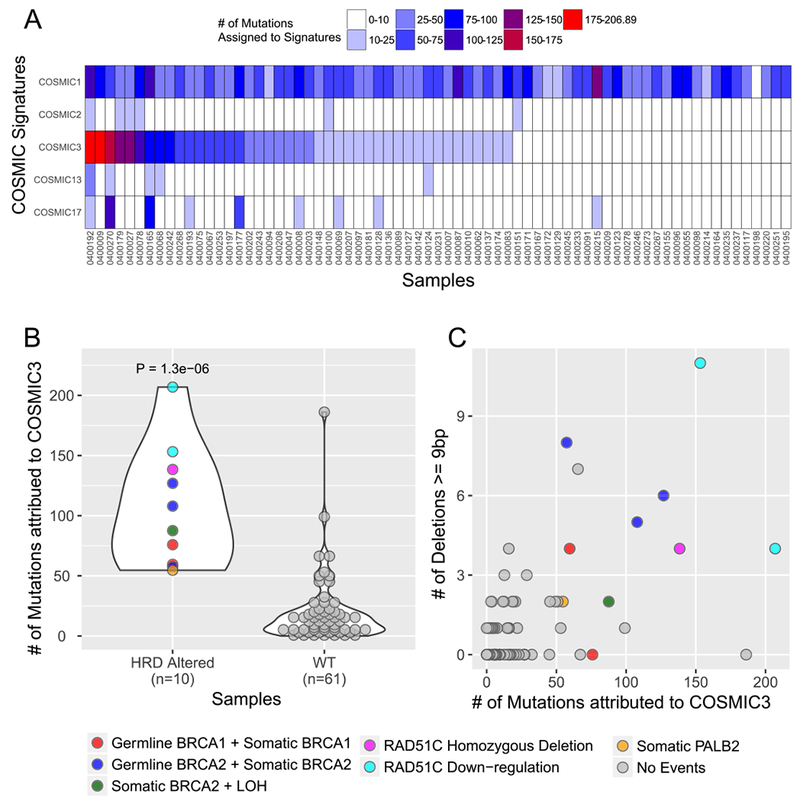
Mutational signature analysis of whole exome sequencing data from advanced PDAC patients. A) Projection of signatures representing four main mutational processes identified in de novo signature analysis. All 71 samples in the cohort are listed as columns. Each row represents a signature as defined in the text: COSMIC1 (C>T transitions at CpG dinucleotides, Aging), COSMIC2 and 13 (APOBEC), COSMIC3 (homologous recombination deficiency [HRD] or BRCA deficient), and COSMIC17 (unknown). B) Samples are shown by the number of HRD/COSMIC3 mutations (y-axis) and binned according to whether they have a mutation or gene expression alteration in the known HR genes BRCA1, BRCA2, PALB2 of RAD51C (HRD Altered) (x-axis). Legend at right indicates coloring based on type of alteration. RAD51C downregulation refers to more than 2-fold down-regulation of mRNA-expression levels below the mean value for the entire cohort. “No events” refers to no detected mutation, copy number alteration or mRNA downregulation in the genes indicated. C) Scatter plot of samples displayed by the number of large (≥ 9 base pairs) deletions on the y-axis and the number of HRD/COSMIC3 mutations on the x-axis. Coloring as shown in the legend at right.
To further investigate the HRD/COSMIC3 signature in these data, we integrated mutation and copy number data from WES and gene expression data from RNA-seq for a core set of known homologous recombination (HR) genes (BRCA1, BRCA2, PALB2 and RAD51C) that are known to be associated with the HRD/COSMIC3 signature (18) and categorized samples as “HRD altered” or “WT.” We defined HRD altered samples based on the presence of damaging germline and somatic mutations (null, truncating, and splice-site variants), homozygous deletions, or more than 2-fold down-regulation of mRNA-expression levels. We observed a significant enrichment of HRD/COSMIC3 signature mutations within HRD altered samples (Figure 2B, P < 0.000002 by one-tailed Wilcoxon rank-sum test). The increased occurrence of large deletions of up to 50 base pairs with overlapping microhomology is another characteristic mark of HR-deficient samples, and we indeed observed an increased incidence of such deletions (≥ 9 base pairs) in our HRD altered samples (Figure 2C, p < 0.00002 by one-tailed Wilcoxon rank-sum test). Eight out of the top fourteen samples with HRD/COSMIC3 signature activity harbored deleterious mutations or homozygous deletions in one of the four core HRD genes. Six out of these eight HRD-altered samples had both germline and somatic events or a somatic alteration with co-existing loss-of-heterozygosity (LOH) in BRCA1 or BRCA2, supporting a “two-hit” hypothesis. Another sample (0400253) had both a p.Q750* nonsense and a p.F1016S missense mutation in PALB2. Furthermore, one sample harbored homozygous deletion of RAD51C. Two additional samples with HRD/COSMIC3 signature activity but without genomic alterations in the four core HRD genes displayed down-regulation of RAD51C expression in the RNA-sequencing data, an event previously associated with HRD/COSMIC3 signature activity (18). Thus, a total of 10 of 14 samples with a high HRD/COSMIC3 signature activity could be explained by genomic alterations or downregulation of gene expression in BRCA1, BRCA2, PALB2 or RAD51C (Figure 2B).
Notably, we also observed 4 samples that did not have clear DNA alterations or mRNA downregulation of BRCA1, BRCA2, PALB2 or RAD51C but nevertheless had enrichment of the HRD/COSMIC3 mutational signature at a level equal to or greater than those samples in the HRD altered class (Figure 2B-C). We conducted a broader examination of genes involved in DDR, including those responsible for HR, non-homologous end joining (NHEJ), base excision repair (BER), nucleotide excision repair (NER) and DDR checkpoint responses. One patient with a high activity of the HRD/COSMIC3 signature had a ERCC2N238S mutation in a conserved helicase domain of this key enzyme involved in NER. This ERCC2N238S mutation corresponds to one of the recurrent mutational hotspots observed in patients with muscle-invasive urothelial carcinoma who achieved a complete response to neoadjuvant cisplatin-based therapy (21). ERCC2 mutations have been associated with a separate mutational signature that has a significant overlap with the HRD/COSMIC3 signature (20); thus, the apparent enrichment of the HRD/COSMIC3 signature for this PDAC patient with an ERCC2N238S mutation may be attributed to the presence of an ERCC2 mutational signature. Notably, this patient experienced a partial response to platinum-based FOLFIRINOX therapy (5-FU, leucovorin, irinotecan and oxaliplatin), but progressed after 4–5 months of treatment. For the remaining three samples with unexplained HRD/COSMIC3 signature enrichment, we did not observe other mutations or copy number events in recognized DDR genes. Thus, our data suggest that the HRD/COSMIC3 signature analysis in WES data may detect additional patients with HRD not identified solely by mutation profiling of specific genes known to be related to HR.
Germline mutations
Identification of germline variants associated with PDAC can have important implications for treatment of the proband as well as for counseling and genetic screening of family members. Thus, in addition to somatic mutation and copy number analysis, we simultaneously interrogated each patient’s germline sequencing data for pathogenic/likely pathogenic variants in 81 genes (Supplementary Table S3). In total, we observed pathogenic/likely pathogenic germline variants in 18% (13/71) of patients (Table 1, Figure 1). These patients were referred for genetic counseling and outreach to family members (Table 1).
Table 1:
Patients with pathogenic/likely pathogenic germline mutations. LOH, loss of heterozygosity. Dx, diagnosis. Family history was obtained by review of the patient’s medical records. The following samples harbor Ashkenazi Jewish founder mutations: 0400027_T1, 0400067_T1 and 0400075_T1.
| Case | Germline Mutation | Somatic Event | Family History of Cancer | Age at Dx |
|---|---|---|---|---|
| 0400094_T2 | ATM (p.D1013fs) CDKN2A (p.G101W) |
Nonsense mutation none |
Mother: breast cancer Father: melanoma |
51 |
| 0400209_T1 | ATM (splice site) | none | No family history | 61 |
| 0400235_T1 | ATM (p.E1978*) | none | Mother: breast cancer Maternal uncle: melanoma |
65 |
| 0400027_T1 | BRCA2 (p.S1982fs) | LOH | Sister: breast cancer | 64 |
| 0400067_T1 | BRCA2 (p.S1982fs) | LOH | Maternal half-brother: melanoma Maternal half-sister: colon cancer Paternal grandfather: unknown primary cancer |
59 |
| 0400078_T1 | BRCA2 (p.W1692Mfs*3) | LOH | Father: melanoma and prostate cancer Paternal aunt 1: breast cancer Paternal aunt 2: brain cancer Paternal grandmother: lung cancer |
39 |
| 0400075_T1 | BRCA1 (p.Q1756fs) | LOH | Mother: ovarian cancer Maternal grandmother: ovarian cancer |
58 |
| 0400242_T1 | BRCA1 (p.T276Afs*14) | LOH | Mother: breast cancer Brother: pancreatic cancer |
63 |
| 0400124_T1 | CHEK2 (Ex2_3del) | LOH | Mother: breast cancer Father: prostate cancer Brother: prostate cancer Paternal grandfather: colon cancer Maternal grandmother: intraadominal/pelvic cancer |
73 |
| 0400215_T1 | BLM (p.P1320fs) | none | Brother: glioblastoma Father: lung cancer Maternal grandmother: brain cancer |
53 |
| 0400214_T1 | FANCA (p.Q343*) | none | Sister: ovarian cancer | 59 |
| 0400164_T1 | FANCL (p.T367fs) | none | No family history | 70 |
| 0400192_T1 | RAD50 (p.S653*) | none | Daughter: lung cancer | 67 |
Pathogenic/likely pathogenic germline variants were observed in several DDR genes, including BRCA1 (n=2), BRCA2 (n=3), and ATM (n=3). An additional pathogenic CHEK2 deletion was observed through commercial testing in a patient who was referred due to a strong family history of malignancy (Table 1). Notably, our CLIA certified WES platform was not validated for detection of exon-level CNAs in the germline at the time of this study. However, re-examination of the germline WES sequencing data confirmed the CHEK2 deletion seen on commercial testing.
All five patients with a germline mutation in BRCA1 or BRCA2 demonstrated evidence of enrichment in the genomic HRD/COSMIC3 signature (Figure 2B), as well as loss of heterozygosity (LOH) through somatic copy number loss of the wild-type allele (Table 1). However, only one of three patients with a germline ATM mutation harbored a somatic alteration in the other allele. A somatic second hit was also not identified for patients with germline pathogenic/likely pathogenic mutations in BLM, FANCA, FANCL, or RAD50. Beyond the BRCA1/2 mutant cases, none of the patients harboring other germline DDR gene mutations showed evidence of HRD/COSMIC3 signature enrichment (Figure 2B). The mean age at diagnosis did not significantly differ between cohorts with or without a pathogenic/likely pathogenic germline mutation (60.2 versus 63.9 years, respectively; two-tailed t-test, p = 0.14).
RNA sequencing of PDAC biopsies
Recent gene expression studies have identified subtypes of PDAC with prognostic and biological relevance (7,12–14). These studies converge on at least two major neoplastic subtypes of PDAC including a squamous/basal-like/quasimesenchymal subtype and a classical/pancreatic progenitor subtype. To investigate these neoplastic PDAC subsets within our metastatic biopsy cohort, we performed RNA-seq on a separate biopsy specimen from that used for WES. We achieved successful RNA extraction and sequencing from 80% (63/79) of patients (Supplementary Figure S1). Consistent with our prior observations in primary PDAC specimens, we observed clear distinctions at the RNA level of the basal-like and classical subtypes (14) (Figure 3A). However, samples with low neoplastic cellularity were more difficult to classify with the basal-like and classical subtype gene sets, and several of these samples showed a strong association with gene expression from normal liver tissue (Figure 3A-B, Supplementary Figure S5A), suggesting that the biopsy may have captured primarily adjacent liver parenchyma rather than the target tumor lesion. Moreover, integrated analysis of tumor and normal gene expression enabled clustering of samples by tumor-specific subtype and site of biopsy as well as identification of outlier samples based on tumor type, atypical genetic lesions or predominant contribution from adjacent normal or stromal gene expression (Supplementary Figure S5B).
Figure 3:
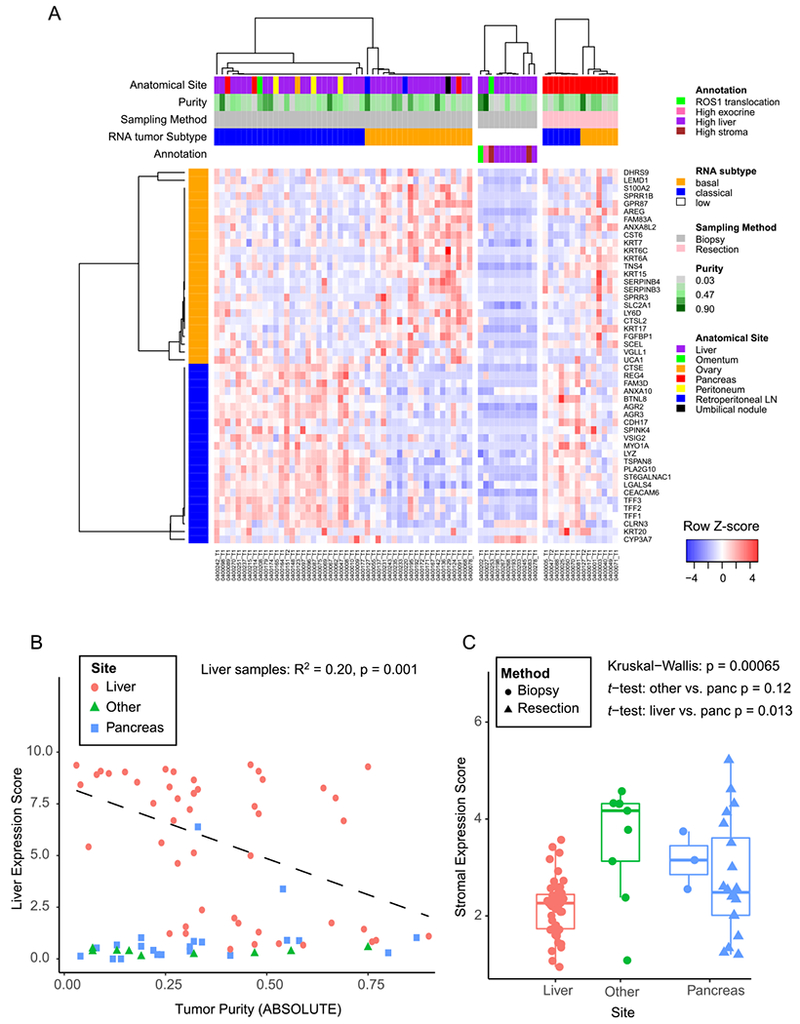
PDAC gene expression signatures in biopsy cohort. A) Heatmap showing each sample as a column, with rows displaying genes sets defining the Moffitt Basal-like (orange bar) and Classical (blue bar) PDAC gene expression programs (14). Tracks at the top also show anatomic site and ABSOLUTE purity by WES of the sample. To the right of the main biopsy heatmap, samples with low tumor content (middle panel), and samples from resected cases (right most panel) are shown. B) Liver gene expression score (y-axis) is plotted versus the ABSOLUTE purity of each sample (x-axis). The linear regression shown includes only samples from the liver. C) A composite stromal score is displayed for each sample, with samples binned according to the biopsy site. Boxplots represent first, second and third quartiles, and whiskers depict the furthest sample from the median which is within 1.5 times the inter-quartile range.
In addition to neoplastic subtypes of PDAC, two stromal subtypes have been identified: “normal” and “activated” stromal subtypes (14). To capture a single composite stroma signature, we generated a merged “stroma score” reflecting the combined total expression of the top 25 activated and top 25 normal expressed stroma genes. Notably, biopsies from metastatic liver lesions on average demonstrated lower stroma scores than those from other sites or compared with primary tumor resection specimens (Figure 3C). Comparison of ABSOLUTE neoplastic cellularity derived from WES revealed a trend toward higher neoplastic cellularity for liver lesions (mean 0.39) versus pancreatic biopsies or resections (mean 0.30; two sample t-test, p = 0.07, Supplementary Figure S2B). Despite liver metastases showing on average lower stroma scores, many samples did show elevated scores at a level similar to those seen in primary resections or biopsies of other sites. Thus, the stroma score shows important differences according to site of biopsy but also significant interpatient variability within each biopsy site.
Clinically relevant genomic events
Genomic analyses of primary specimens have suggested that approximately 40% of patients with PDAC may harbor clinically relevant genomic events that could potentially impact treatment decisions (12). However, most PDAC patients do not undergo timely genomic analysis to identify these events for clinical decision making. Leveraging our rapid turnaround CLIA-certified sequencing program, we performed real time assessment of genomic data for use in clinical decision making (Supplementary Figure S6A)(Supplementary Table S4). Excluding common events in KRAS or CDKN2A, 48% (34/71) of patients within this cohort had cancers with at least one genomic alteration that could potentially confer eligibility for current clinical trials or support off-label usage of an agent approved for another indication (Supplementary Figure S6A). Furthermore, 11% (8/71) of the patients had cancers with two or more such events, suggesting a potential basis for genotype-driven combination therapy trials.
KRAS mutations were observed in 90% (64/71) of patients in our cohort. Although this alteration has been used to enroll patients onto clinical trials of MAPK-directed therapy, these trials have demonstrated limited efficacy to date (22). However, in the subset of KRAS wild-type tumors, we observed mutations and CNAs in additional MAPK pathway activating genes, such as BRAF mutations (see below), a ROS1 translocation and high-level FGFR1 amplification (Figure 1, Supplementary Figure S6A). Across the entire cohort, we observed multiple other alterations that could warrant experimental therapies, including RNF43 mutation (e.g. Porcupine inhibitor), AKT2 amplification (e.g. AKT inhibitor), TSC2 mutation (e.g. MTOR inhibitor), MYC amplification (e.g. bromodomain inhibitor), and CDK4 amplification (e.g. CDK4 inhibitor) (Supplementary Figure S6A). Moreover, sequencing identified other lesions that may contraindicate therapy with certain agents, such as RB1 mutations (n=4 patients) and CDK4/6 inhibition.
A total of 24% (17/71) of patients enrolled on the PancSeq study were treated with an experimental agent, either through enrollment onto a clinical trial or through off-label use of an approved agent (Table 2). In particular, genomic information from WES dictated the choice of experimental agent in 15% (11/71) of cases. Moreover, 18% (13/71) of patients had a clinically relevant germline mutation necessitating referral for genetic counseling (Table 1). Thus, accounting for both new therapeutic options and genetic counseling indications, a total of 30% (21/71) of patients enrolled on the PancSeq protocol experienced a change in clinical management as a result of the obtained genomic data (Tables 1 and 2).
Table 2:
Patients on PancSeq protocol who underwent treatment with experimental agents. Bolded genes indicate those genetic alterations that guided choice of experimental therapy. A “g” preceding the gene name refers to “germline.” GA, gemcitabine plus albumin-bound paclitaxel; FOLFIRINOX, 5-FU/Leucovorin/Irinotecan/Oxaliplatin; FOLFOX, 5-FU, Leucovorin, Oxaliplatin; mAb, monoclonal antibody; Nal-Iri, nanoliposomal irinotecan; Cape, capecitabine; CT, clinical trial; OL, off-label; SP-IND, single-patient investigational new drug application.
| Identifier | Gender | Age (yrs) | Stage | Pre-Biopsy Treatments for Advanced Disease | Genomic Features | Post-Biopsy Treatments |
|---|---|---|---|---|---|---|
| 0400068_T1 | M | 44 | Metastatic | No prior chemotherapy | BRCA2, KRAS | FOLFIRINOX -- PARP inhibitor (OL) |
| 0400075_T1 | F | 58 | Metastatic | No prior chemotherapy | CHEK2, KRAS, gBRCA1 | FOLFIRINOX -- PARP inhibitor vs. placebo (CT) |
| 0400078_T1 | F | 39 | Metastatic | GA/JAK inhibitor (CT), FOLFIRINOX | KRAS, CDKN2A, gBRCA2 | UBA1 inhibitor (CT) -- PARP inhibitor (OL) |
| 0400096_T1 | M | 57 | Metastatic | No prior chemotherapy | KRAS, CDKN2A | FOLFIRINOX -- GA/Anti-MUC5AC mAb (CT) |
| 0400097_T1 | M | 48 | Metastatic | No prior chemotherapy | KRAS | FOLFIRINOX -- GA -- CHK1/2 inhibitor (CT) |
| 0400117_T2 | F | 65 | Metastatic | FOLFIRINOX, GA | KRAS, CDKN2A | PI3K/mTOR inhibitor/CDK4/6 inhibitor (CT) |
| 0400127_T1 | F | 60 | Metastatic | No prior chemotherapy | KRAS, CDKN2A | FOLFIRINOX -- GA -- CDK4/6 inhibitor/MEK1/2 inhibitor (CT) |
| 0400151_T2 | F | 61 | Metastatic | FOLFIRINOX, GA | KRAS, CDKN2A | CDK2/5/9 inhibitor (CT) |
| 0400165_T1 | F | 73 | Metastatic | FOLFIRINOX | ERCC2, KRAS | GA -- Prostaglandin E2 receptor EP4 antagonist (CT) |
| 0400172_T1 | M | 58 | Metastatic | No prior chemotherapy | BRAF, CDKN2A | FOLFIRINOX -- MEK1/2 inhibitor (OL) -- GA -- ERK1/2 inhibitor (SP-IND) |
| 0400174_T1 | M | 77 | Metastatic | No prior chemotherapy | ATM (biallelic), KRAS, CDKN2A | GA -- 5FU/LV/Nal-Iri -- PARP inhibitor (OL) |
| 0400177_T1 | F | 64 | Metastatic | FOLFOX, 5FU/LV/Nal-Iri, GA | KRAS, NBN, FANCM | PARP inhibitor/CDK1/2/5/9 inhibitor (CT) |
| 0400197_T1 | F | 65 | Metastatic | FOLFIRINOX, GA | BRAF, FGFR1 (amplification) | MEK1/2 inhibitor (OL) -- ERK1/2 inhibitor (SP-IND) |
| 0400202_T1 | M | 83 | Metastatic | ROS1 inhibitor (CT) | CDKN2A, ROS1 (translocation) | ROS1 inhibitor (OL) -- ROS1 inhibtor (OL) |
| 0400242_T1 | F | 63 | Metastatic | No prior chemotherapy | KRAS, CDKN2A, gBRCA1 | FOLFIRINOX -- PARP inhibitor vs. placebo (CT) |
| 0400245_T1 | M | 57 | Metastatic | No prior chemotherapy | KRAS | FOLFIRINOX -- anti-PD-1 mAb/CXCR4 antagonist (CT) -- GA |
| 0400270_T1 | F | 66 | Metastatic | GA, Cape, 5FU/LV/Nal-Iri, FOLFOX | KRAS | anti-PD-1 mAb/anti-GITR mAB agonist (CT) |
DNA-damage repair mutations and PARP-inhibitor therapy
In 20% of patients, we observed germline and/or somatic alterations in one or more of the following DDR genes: BRCA1, BRCA2, PALB2, ATM and CHEK2 (Figure 1, Supplementary Figure S6B). When considering mutations in a wider spectrum of genes across several DDR classes, as well as specific mutational signatures consistent with HRD, we identified a total of 44% (31/71) of patients displaying genomic evidence of potential DDR deficiency (Table 1, Supplementary Figure S6B). DDR gene mutations may confer increased sensitivity to platinum chemotherapy (9). Moreover, BRCA1 or BRCA2 gene mutations have been reported to confer sensitivity to poly-ADP polymerase (PARP) inhibition in preclinical models and early clinical trials of PDAC (9,23,24). All six patients in the cohort with germline or somatic mutations in the BRCA1 or BRCA2 genes demonstrated evidence of the HRD/COSMIC3 mutational signature (Supplementary Figure S6B). By contrast, none of the patients with mutations in ATM, ATR or CHEK2 demonstrated enrichment of the HRD/COSMIC3 signature. All six patients with BRCA mutations received an oxaliplatin-based chemotherapy regimen (FOLFIRINOX or FOLFOX) in the first or second line setting and all demonstrated some degree of radiographic response to these regimens (Figure 1, Supplementary Table S4). Two of these patients with a germline BRCA1 mutation were subsequently enrolled onto a randomized trial of the PARP inhibitor olaparib versus placebo for maintenance therapy after receipt of 4–6 months of FOLFIRINOX (NCT02184195). Two further patients with a BRCA2 mutation received off-label olaparib, including one patient with a germline mutation and one with a somatic mutation. Besides patients with these BRCA1/2 mutations, two other patients with DDR gene mutations were treated with a PARP inhibitor (Table 2). Eight of ten patients with ATM, ATR or CHEK2 mutations were treated with an oxaliplatin-based chemotherapy regimen and five demonstrated clinical benefit as defined by partial response or stable disease at first radiographic follow-up scans.
As noted above, two patients with BRCA2 alterations were treated with off-label olaparib. The patient harboring a somatic BRCA2 mutation was a 45 year-old man who presented with jaundice and abdominal pain and was diagnosed with metastatic PDAC involving the liver and lymph nodes, with a serum CA19–9 of 3,592 U/mL on presentation. He underwent biopsy of a metastatic liver lesion which confirmed a poorly differentiated PDAC. Genome sequencing on the PancSeq protocol revealed KRAS (p.G12R), TP53 (p.M246fs), and somatic BRCA2 (p.LS298fs) mutations (Figure 4A). He received first line therapy with the platinum-containing regimen FOLFIRINOX and experienced normalization of serum CA19–9 levels (Figure 4B) and a complete radiographic response to therapy within 5 months of treatment (Figure 4C). FOLFIRINOX was discontinued after a total of 13 two-week cycles due to transaminitis and neuropathy. The patient chose to forego further cytotoxic chemotherapy and was initiated on off-label olaparib 300 mg twice daily as maintenance therapy. He remains free of radiographically detectable disease 28 months after diagnosis and 20 months after beginning olaparib (Figure 4C).
Figure 4:
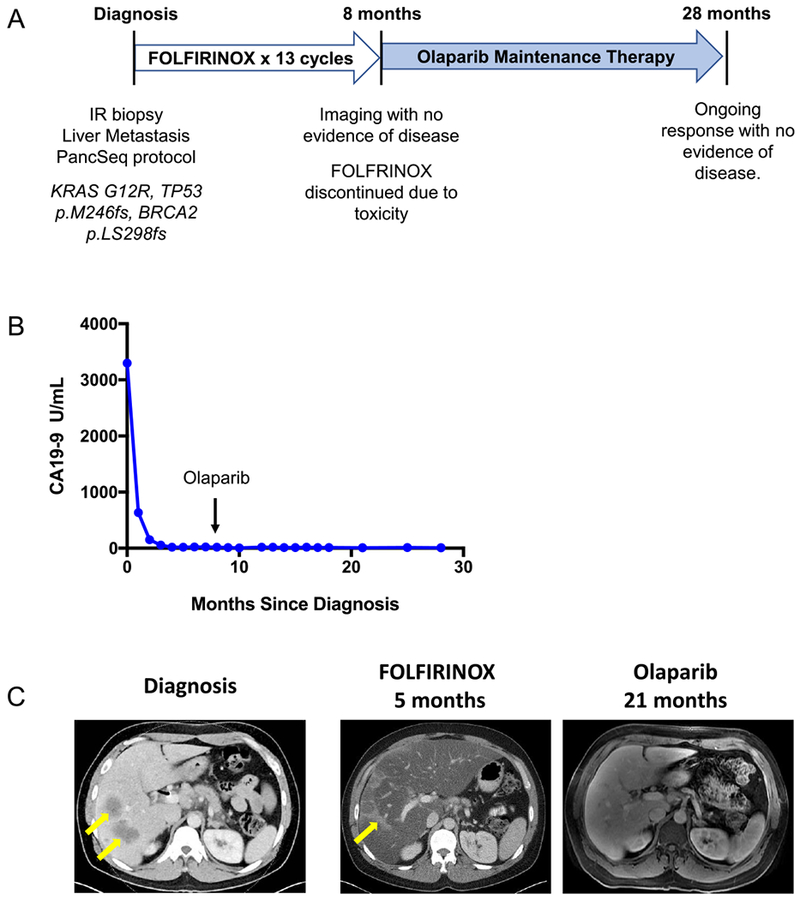
Patient with somatic BRCA2-mutant PDAC demonstrates a radiographic complete response to platinum chemotherapy and subsequent olaparib maintenance therapy. A) 45 year-old man presented with jaundice and abdominal pain and was diagnosed with metastatic PDAC involving the liver and lymph nodes and underwent the depicted treatment course. B) Serum CA19–9 measurements from diagnosis throughout the patient’s treatment course. The arrow indicates transition from FOLFIRINOX chemotherapy to Olaparib (PARP inhibitor) maintenance therapy. C) Computed Tomography (CT) scans are shown at diagnosis demonstrating liver metastases (left panel, yellow arrows) and at the time of cessation of FOLFIRINOX chemotherapy (middle panel) with resolution of liver metastases. Hepatic toxicity of FOLFIRINOX resulted in fatty infiltration of the liver, as noted by severe diffuse attenuation of the liver parenchyma seen in the five month scan. Areas of focal fat sparing in this scan represent treatment effect at the site of liver metastases (middle panel, yellow arrow), denoting a complete response to therapy. The follow-up magnetic resonance imaging (MRI) scan at 21 months after diagnosis, on olaparib maintenance therapy for 13 months, is shown with complete regression of liver metastases (right panel). The patient remains on olaparib therapy without evidence of disease now 28 months after diagnosis.
BRAF mutant PDAC and response to MAP-kinase inhibition
Within our CLIA-certified cohort, we discovered two patients with in-frame deletions in the BRAF oncogene (Figure 5A). This class of deletions near the alphaC-helix region of the kinase domain has recently been shown to occur in KRAS wild-type PDAC and activates the protein to drive MAPK signaling (12,25,26). Furthermore, PDAC cell lines grown in vitro or as in vivo xenografts have been shown to be sensitive to MAPK inhibition, although this approach had not been tested in humans (26). To further define the frequency of this BRAF alteration in PDAC, we investigated a larger collection of samples (N=406) profiled with a targeted genome sequencing panel at Dana-Farber Cancer Institute (27,28) and found four KRAS wild-type tumors that harbored the same in-frame deletion (p.N486_P490del) and one additional KRAS wild-type tumor with a small, likely oncogenic in-frame insertion (p.T599dup). Thus, BRAF in-frame insertions or deletions occurred in approximately 10% of our patients with KRAS wild-type PDAC or 1% of all PDAC patients (12,25,26).
Figure 5:
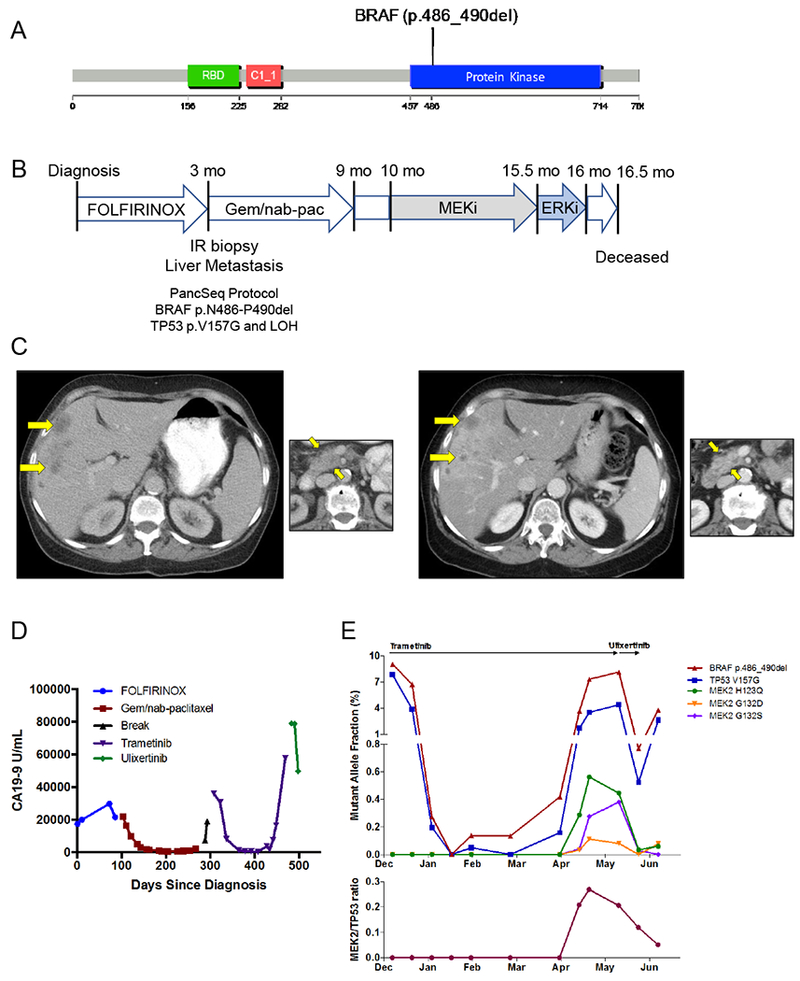
BRAF in-frame deletion confers response to MAPK inhibition. A) An in-frame deletion in BRAF was identified leading to a five amino acid deletion in the kinase domain. B) A 66 year old woman presented with dyspepsia and weight loss and was diagnosed with PDAC and liver metastases and underwent the indicated treatment course. Gem/nab-pac, gemcitabine + nab-paclitaxel. C) CT scans of the liver (large panels) and primary tumor (small panels) at diagnosis (left) and at the time of first restaging scan after 8 weeks of treatment (right) showing partial response to trametinib. D) Serum CA19–9 levels measured throughout the patient’s disease course reflect response and resistance to each therapeutic regimen (color coded by regimen). E) Cell free DNA (cfDNA) measurements for the indicated alleles obtained through droplet digital PCR (ddPCR, BRAF and TP53 alleles) or Guardant360 assay (MEK2 alleles) on plasma collected throughout the patient’s treatment course with trametinib and ulixertinib. Top panel depicts the overall mutant allele fraction of each allele in cfDNA. The bottom panel shows the relative frequency of the MEK2 resistance alleles compared to the overall tumor burden (as measured by the total MEK2/TP53 ratio).
In one case, a 66 year-old woman presented with dyspepsia and weight loss and was diagnosed with PDAC and liver metastases. She underwent first-line therapy with FOLFIRINOX but experienced progression of disease after only five two-week cycles. She underwent a biopsy and WES on the PancSeq protocol and then began second-line therapy with gemcitabine plus nab-paclitaxel that was discontinued after 6 four-week cycles due to disease progression (Figure 5B). Sequencing revealed a BRAF in-frame deletion (p.N486-P490del) and TP53 mutation (p.V157G) (Figure 5A-B). Given that cell lines harboring this mutation were sensitive to MEK inhibitors but resistant to selective BRAF inhibitors in pre-clinical models (26), she was initiated on off-label treatment with the MEK1/2 inhibitor trametinib, which is FDA-approved for use in BRAFV600E mutant melanoma (Figure 5B) (29–31). Within four weeks of initiating therapy, her serum CA19–9 had fallen from 36,000 to 8,100 U/ml, and the first restaging scan done 8 weeks after initiation of trametinib showed a partial response to therapy (Figure 5C-D). Serial plasma samples were collected to measure the fractional abundance of BRAF and TP53 mutant alleles in circulating cell-free DNA (cfDNA) by droplet digital PCR (ddPCR) (32,33). cfDNA measurements for BRAF and TP53 alleles (two clonal alterations in this tumor) revealed a dramatic decline in response to trametinib therapy that mirrored the radiographic findings (Figure 5E). After 5 months on trametinib therapy, a rise in the fractional abundance of the BRAF and TP53 mutant alleles in cfDNA was observed. After six months of therapy, radiographic progression was identified. Evaluation of a more comprehensive panel of genes within cfDNA was pursued through a commercial test (Guardant360®) at the time of progression and demonstrated the emergence of multiple subclonal mutations in the MAP2K2 (MEK2) gene (Figure 5E). Notably, these mutations are homologous to MEK1 mutations that have been previously described to confer resistance to MEK inhibition in vitro in mutagenesis studies in BRAF mutant melanoma (34). Retrospective analysis of the MEK2 mutations in serial plasma samples collected through the patient’s treatment course revealed emergence of these mutant alelles in concert with increases in fractional abundance of mutant BRAF and TP53 alleles (Figure 5E). Thus, these data likely reflect the emergence of heterogeneous polyclonal resistance mechanisms that evolved under the selective pressure of trametinib therapy.
Given the emergence of MAP2K2 resistance mutations while on trametinib therapy, the patient was subsequently treated with ulixertinib/BVD-523, an inhibitor of ERK1/2 (which signals downstream of MEK1/2), on a single-patient investigational new drug application. After treatment with ulixertinib/BVD-523 for 17 days, cfDNA analysis by ddPCR demonstrated a rapid decline in the fractional abundance of the BRAF and TP53 mutant alleles (Figure 5E). The MEK2 resistance alleles also rapidly decreased in absolute frequency and in relative abundance with respect to the overall tumor burden (as measured by the total MEK2/TP53 ratio) (Figure 5E). Unfortunately, the patient’s functional status was quickly declining as she initiated therapy with ulixertinib/BVD-523, and this medication was stopped after 3 weeks of therapy. A last blood collection after cessation of ulixertinib/BVD-523 demonstrated rebound of the fractional abdundance of the BRAF and TP53 mutant alleles. The patient expired at home several weeks later with hospice services. This patient’s clinical course suggests that a BRAF in-frame deletion may serve as an important biomarker for response to MAPK inhibition in patients with pancreatic cancer.
Discussion
Many barriers to precision medicine exist for patients with PDAC, including low neoplastic cellularity of tumors and the aggressive nature of the disease that makes timely biopsy and genomic analysis difficult. We have established an integrated biopsy program for advanced PDAC patients that enables CLIA-certified genomic profiling and rapid turnaround of WES data to referring clinicians. To overcome the challenges of low neoplastic cellularity, we have performed deep WES on small volume tumor biopsies and germline DNA and have employed analytic algorithms to accurately quantitate tumor DNA content and to identify mutations and CNAs. In addition, we have identified molecular driver alterations and clinically relevant subsets of disease in real time, during the course of the patient’s illness. In an initial biopsy cohort of 79 patients, we obtained high quality genomic information for 71 patients and identified potentially actionable somatic and germline alterations in 48% of cases. These include 26 patients harboring tumors with DDR gene mutations and 7 with KRAS wild-type tumors, including two with BRAF alterations and one with a ROS1 translocation. We have demonstrated how these data can impact clinical decision making, utilizing molecular information to treat multiple patients on clinical trials or with use of off-label targeted therapies. Nevertheless, further study will be necessary to confirm the rate and therapeutic implications of actionable alterations in a multi-institutional population of patients with PDAC. Overall, 30% (21/71) of enrolled patients experienced a change in clinical management as a result of genomic data, including 15% of patients for which genomic information dictated the choice of an experimental agent and 18% of patients whose germline data warranted referral for genetic counseling. These numbers far exceed the average experience for PDAC patients, and these data support the implementation of genomic evaluation as a standard clinical practice in patients with advanced PDAC.
We identified pathogenic/likely pathogenic germline mutations in 18% of patients in our cohort of advanced PDAC patients. Analysis of germline variants was performed within a set of 81 genes included in our CLIA-certified analysis pipeline (Supplementary Table S3). Thus, this analysis may underestimate the true number of PDAC patients with a hereditary pancreatic cancer predisposition. However, 18% is higher than the approximately 4–12% incidence of pathogenic germline variants that has been reported in recent primary tumor cohorts (12,35,36). This higher rate of germline variants may be due to the larger number of genes examined, different distribution of Ashkenazi Jewish patients (who are known to have higher rates of germline variants), the selected patient population who provided informed consent for research biopsy, and potential enrichment of germline variants in patients with advanced PDAC. Regarding the latter point, a similar observation of enrichment for germline mutations in DNA-damage repair genes has been made in metastatic prostate cancer (37). Nevertheless, the frequency of germline mutations in patients with advanced PDAC will require further validation. Annotation of Ashkenazi Jewish descent was incomplete for patients included in the current study. However, only 3 of the 13 identified germline alterations were known to be Ashkenazi Jewish founder mutations.
While all tumors with germline mutant BRCA1 and BRCA2 genes demonstrated somatic mutation or LOH of the second allele, only 2 of 9 (22%) of the other identified germline mutant genes demonstrated somatic mutation or LOH. The functional implications for loss or retention of the wild-type allele remain an important unanswered question that is worthy of further investigation. Given the prevalence of germline mutations in PDAC patients, several groups now advocate for universal multigene germline testing for all patients, irrespective of family history or age at diagnosis (35,38–40). The most effective approach to implementing universal germline testing as well as the optimal bioinformatic and functional frameworks for interpreting these data remain important to define.
As has been suggested in prior studies of primary PDAC resection specimens (7,9,12), we observed a striking prevalence of germline and somatic mutations in DDR genes in our cohort of advanced PDAC patients. Beyond the 37% (26/71) of patients with DDR gene mutations or CNAs, nine of whom had HRD/COSMIC3 signature enrichment, an additional 7% (5/71) of patients had enrichment of an HRD/COSMIC3 signature but no clear associated HR gene alteration. Two of these cases could be explained by downregulation of mRNA expression of RAD51C; however, three samples had HRD/COSMIC3 signature activity but no clear causal genetic or gene expression feature to explain the signature. While the therapeutic implications of HRD/COSMIC3 signature enrichment without an identifiable mutation need further validation in preclinical models, this result suggests that more patients may have functional DDR deficiency than are detectable by DNA mutational profiling alone, thus arguing in favor of performing more global genomic characterization approaches such as WES in concert with integrative analyses of gene expression in PDAC patients. Notably, whole genome sequencing of PDAC has also suggested that chromosomally unstable tumors with a large number of structural variation events are associated with a BRCA mutational signature (9). We were unable to similarly evaluate the number of structural rearrangements in these tumors using WES, and additional studies will be needed to understand the association of DDR gene mutations, HRD/COSMIC3 mutational signature enrichment and structural variation events.
In this study, six patients with DDR gene mutations were treated off-label or enrolled on clinical trials of PARP inhibitor therapy. Understanding the gene- and allele-specific differences for DDR genes conferring sensitivity to platinum chemotherapy, PARP inhibition or other inhibitors of DDR checkpoints will be critical to enabling effective stratification of patients with PDAC onto efficacious therapies. As noted above, the functional implications of genomic correlates on therapeutic responsiveness, including somatic LOH vs. retention of the wild type allele or COSMIC3/HRD signature activity, also needs further investigation in patient-derived models as well as in human clinical trials.
We and others have noted the importance of alternative oncogenic driver events in KRAS wild-type PDAC (10,12,41). Here, we demonstrate multiple oncogenic and targetable lesions that occur in KRAS wild-type tumors, including alterations in BRAF, ROS1, FGFR1 and other genes. In particular, we have identified in-frame deletions in BRAF in two patients in our cohort. These lesions have been reported to result in oncogenic activation (25,26). We report the first human therapeutic experience with MAPK inhibition in a patient harboring a BRAF in-frame deletion. We demonstrate substantial clinical benefit in a patient who had a partial response to the MEK1/2 inhibitor trametinib (Figure 5). Furthermore, by serial monitoring of driver alterations in plasma cfDNA through multiple lines of treatment, we provide molecular evidence for disease response and progression, as well as the emergence of heterogeneous MEK2 resistance alleles that correspond to radiographic progression of disease on trametinib. This patient was subsequently treated with an ERK inhibitor after progression on trametinib and showed a second decline in mutant BRAF and TP53 alleles within cfDNA and suppression of MAP2K2 resistance alleles, suggesting further evidence of response to MAPK inhibition. A second patient with rapidly progressive BRAF mutant disease was also treated with off-label trametinib but failed to show a response. This heterogeneity of primary and secondary resistance mechanisms will necessitate effective combination therapy strategies with MAPK inhibition. These data suggest that a larger multi-center clinical trial with proper molecular correlates, including cfDNA monitoring of therapeutic response and resistance, should be developed to fully investigate the therapeutic efficacy of MAPK inhibition in patients with activating oncogenic BRAF deletions.
In parallel to WES of DNA alterations, we also performed RNA sequencing and demonstrated that RNA signatures of neoplastic PDAC subtypes are readily discernible from small volume metastatic biopsies. The two primary neoplastic subtypes of PDAC have been shown to have prognostic importance and may correlate with chemosensitivity (14,42); thus, identifying these subtypes and proactively incorporating this stratification into clinical trials remains an important translational priority. Detection of a high stromal signature was consistent across primary tumors and several metastatic sites but more variable in liver biopsy specimens, suggesting that the stromal make-up of liver lesions may be distinct from that of primary tumors. This observation has been made recently using an automated histological approach (43) and will need rigorous follow-up with histologic and molecular approaches. Stromal fibroblasts have been proposed to play an important role in promoting PDAC progression and in blunting chemotherapeutic response (42,44,45); however, attempts to target fibroblasts in mouse models as well as clinical trials have yielded conflicting results (46–48). Additional trials of stroma-directed therapies are underway, including vitamin D receptor agonists to modulate stromal gene expression and improve chemosensitivity (42). Our data highlight the importance of biopsy and RNA-seq analysis to identify patients and even specific metastatic lesions that may respond to stroma-directed therapy.
Together with recent work from other PDAC referral centers (41,49), this study demonstrates the feasibility and value of real-time genomic characterization of advanced PDAC and provides a path forward for treatment of PDAC patients with molecularly defined therapy. To harness all potential therapeutic opportunities in this highly aggressive disease, we propose that biopsy-based genomic analysis early in a patient’s treament course should become standard of care for all PDAC patients.
Methods
Investigators obtained informed, written consent for each patient enrolled to the PancSeq protocol (DF/HCC #14–408), and this study was performed in accordance with standard ethical guidelines approved by the Dana-Farber/Harvard Cancer Center IRB. DNA and RNA were extracted from tumor samples (and normal whole blood for germline DNA control), and whole exome sequencing (WES) was performed in a laboratory certified by the Clinical Laboratory Improvement Amendments (CLIA, #22D2055652)(12,50). WES data was processed through the Broad Institute “Picard” pipeline (http://picard.sourceforge.net/), generating a BAM file for each sample. Mutation calling was performed using the MuTect algorithm (51). MutSigCV2 was used to determine significantly mutated genes (52). GISTIC2.0 was used to identify recurrent deletions and amplifications (53). ABSOLUTE (16) was used to determine purity, ploidy, and whole genome doubling status using allelic copy number data along with the allelic fraction of all somatic mutations as input. Annotated WES data were cross-referenced with a curated list of 81 PDAC-relevant genes and variants in these genes were reviewed by a certified clinical geneticist. A report of clinically relevant germline and somatic events was returned to the referring clinician detailing somatic mutations, small insertions/deletions, and CNAs as well as pathogenic/likely pathogenic germline alterations. Only germline variants with population frequency of <1% upon comparison with the ExAC database (http://exac.broadinstitute.org/) were retained for review as pathogenic/likely pathogenic. Mutational signature analysis was performed on the set of single nucleotide variants (SNV) in our dataset using SignatureAnalyzer, as previously described (17–20). RNA sequencing was performed on poly-A selected mRNA at the Broad Institute, and gene expression signatures were derived from Moffitt et al. (14). cfDNA analysis was performed by ddPCR as previously described (32,33), or through a commercial assay (Guardant360®)(54). See also the Supplementary Experimental Methods for further description of experimental procedures. Data in this study have been deposited in dbGaP under accession number phs001652.v1.p1.
Supplementary Material
Statement of Significance.
Molecular analyses of metastatic PDAC tumors are challenging due to the heterogeneous cellular composition of biopsy specimens and rapid progression of the disease. Using an integrated multi-disciplinary biopsy program, we demonstrate that real-time genomic characterization of advanced PDAC can identify clinically relevant alterations that inform management of this difficult disease.
Acknowledgements
We dedicate this work to the memory of our patients, including Dr. Andrew Tager, a beloved colleague, mentor and friend whose courageous battle with pancreatic cancer continues to inspire our pursuit of improved diagnosis and treatment for this difficult disease.
Financial Support
We acknowledge primary research support from Lustgarten Foundation (AJ Aguirre, S Raghavan, WC Hahn, DA Tuveson, BM Wolpin), and Dana-Farber Cancer Institute Hale Center for Pancreatic Cancer Research (AJ Aguirre, S Raghavan, WC Hahn, BM Wolpin), with additional support from Hope Funds for Cancer Research (AJ Aguirre, S Raghavan), the Doris Duke Charitable Foundation (AJ Aguirre), Conquer Cancer Foundation of ASCO Young Investigator Award (AJ Aguirre, S Raghavan), Broman Fund for Pancreatic Cancer Research (K Ng), National Institutes of Health National Cancer Institute K08 CA218420–01 (AJ Aguirre), P50CA127003 (AJ Aguirre, MB Yurgelun, WC Hahn, RB Corcoran, BM Wolpin), U01 CA224146 (AJ Aguirre, WC Hahn), U01 CA210171 (BM Wolpin), R01 CA199064 (JJ Yeh), The Harvard Clinical and Translational Science Center UL1 TR001102 (AJ Aguirre, S Raghavan), DFCI Medical Oncology Translational Research Project Award (MB Yurgelun), Pancreatic Cancer Action Network (AJ Aguirre, BM Wolpin), Noble Effort Fund (BM Wolpin), Peter R. Leavitt Family Fund (BM Wolpin), Wexler Family Fund (BM Wolpin), and Promises for Purple (BM Wolpin).
Footnotes
Conflict of Interest Disclosure: Matthew B. Yurgelun acknowledges research funding from Myriad Genetic Laboratories, Inc. James M. Cleary reports research funding from Merck. Geoffrey I. Shapiro reports research funding from Lilly, Merck, EMD Serono, Sierra Oncology and Pfizer and is a consultant to Pfizer, G1 Therapeutics, Lilly, Roche and Merck/EMD Serono. Charles S. Fuchs is a consultant to CytomX, Sanofi, Eli Lilly, Merck and Entrinsic Health. Levi A. Garraway is an employee of Eli Lilly and Company and has ownership interest in Tango Therapeutic and Foundation Medicine. Bruce E. Johnson reports research funding from Toshiba and Novartis. Nikhil Wagle reports ownership interest in Foundation Medicine. Ryan B. Corcoran reports research funding from AstraZeneca and Sanofi, and is a consultant to Astex, Amgen, Avidity Biosciences, BMS, FOG Pharma, Genentech, LOXO Oncology, Merrimack, N-of-one, Roche, Roivant, Shire, Symphogen, Taiho and WarpDrive Bio. Richard B. Lanham is an employee of and has ownership interest in Guardant Health, Inc. Dean J. Welsch is an employee of BioMed Valley Discoveries. Brian M. Wolpin acknowledges research funding from Celgene. The remaining authors report no potential conflicts of interest.
References
- 1.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer research 2014;74(11):2913–21 doi 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 2.Wolfgang CL, Herman JM, Laheru DA, Klein AP, Erdek MA, Fishman EK, et al. Recent progress in pancreatic cancer. CA Cancer J Clin 2013;63(5):318–48 doi 10.3322/caac.21190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ryan DP, Hong TS, Bardeesy N. Pancreatic adenocarcinoma. N Engl J Med 2014;371(11):1039–49 doi 10.1056/NEJMra1404198. [DOI] [PubMed] [Google Scholar]
- 4.Roberts NJ, Norris AL, Petersen GM, Bondy ML, Brand R, Gallinger S, et al. Whole Genome Sequencing Defines the Genetic Heterogeneity of Familial Pancreatic Cancer. Cancer discovery 2016;6(2):166–75 doi 10.1158/2159-8290.CD-15-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sahin IH, Iacobuzio-Donahue CA, O’Reilly EM. Molecular signature of pancreatic adenocarcinoma: an insight from genotype to phenotype and challenges for targeted therapy. Expert Opin Ther Targets 2016;20(3):341–59 doi 10.1517/14728222.2016.1094057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008;321(5897):1801–6 doi 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531(7592):47–52 doi 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 8.Biankin AV, Waddell N, Kassahn KS, Gingras MC, Muthuswamy LB, Johns AL, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature 2012;491(7424):399–405 doi 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015;518(7540):495–501 doi 10.1038/nature14169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Witkiewicz AK, McMillan EA, Balaji U, Baek G, Lin WC, Mansour J, et al. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nature communications 2015;6:6744 doi 10.1038/ncomms7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Notta F, Chan-Seng-Yue M, Lemire M, Li Y, Wilson GW, Connor AA, et al. A renewed model of pancreatic cancer evolution based on genomic rearrangement patterns. Nature 2016;538(7625):378–82 doi 10.1038/nature19823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Research Network aadhe. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer cell 2017;32(2):185–203 e13 doi 10.1016/j.ccell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med 2011;17(4):500–3 doi 10.1038/nm.2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SG, Hoadley KA, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nature genetics 2015;47(10):1168–78 doi 10.1038/ng.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoos WA, James PM, Rahib L, Talley AW, Fleshman JM, Matrisian LM. Pancreatic cancer clinical trials and accrual in the United States. J Clin Oncol 2013;31(27):3432–8 doi 10.1200/JCO.2013.49.4823. [DOI] [PubMed] [Google Scholar]
- 16.Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 2012;30(5):413–21 doi 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature 2013;500(7463):415–21 doi 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polak P, Kim J, Braunstein LZ, Karlic R, Haradhavala NJ, Tiao G, et al. A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nature genetics 2017;49(10):1476–86 doi 10.1038/ng.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasar S, Kim J, Improgo R, Tiao G, Polak P, Haradhvala N, et al. Whole-genome sequencing reveals activation-induced cytidine deaminase signatures during indolent chronic lymphocytic leukaemia evolution. Nature communications 2015;6:8866 doi 10.1038/ncomms9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim J, Mouw KW, Polak P, Braunstein LZ, Kamburov A, Kwiatkowski DJ, et al. Somatic ERCC2 mutations are associated with a distinct genomic signature in urothelial tumors. Nature genetics 2016;48(6):600–6 doi 10.1038/ng.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Allen EM, Mouw KW, Kim P, Iyer G, Wagle N, Al-Ahmadie H, et al. Somatic ERCC2 mutations correlate with cisplatin sensitivity in muscle-invasive urothelial carcinoma. Cancer discovery 2014;4(10):1140–53 doi 10.1158/2159-8290.CD-14-0623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papke B, Der CJ. Drugging RAS: Know the enemy. Science 2017;355(6330):1158–63 doi 10.1126/science.aam7622. [DOI] [PubMed] [Google Scholar]
- 23.Kaufman B, Shapira-Frommer R, Schmutzler RK, Audeh MW, Friedlander M, Balmana J, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol 2015;33(3):244–50 doi 10.1200/JCO.2014.56.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Domchek SM, Hendifar AE, McWilliams RR, Geva R, Epelbaum R, Biankin A, et al. RUCAPANC: An open-label, phase 2 trial of the PARP inhibitor rucaparib in patients (pts) with pancreatic cancer (PC) and a known deleterious germline or somatic BRCA mutation. Journal of Clinical Oncology 2016;34(15_suppl):4110– doi 10.1200/JCO.2016.34.15_suppl.4110.27863191 [DOI] [Google Scholar]
- 25.Foster SA, Whalen DM, Ozen A, Wongchenko MJ, Yin J, Yen I, et al. Activation Mechanism of Oncogenic Deletion Mutations in BRAF, EGFR, and HER2. Cancer cell 2016;29(4):477–93 doi 10.1016/j.ccell.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 26.Chen SH, Zhang Y, Van Horn RD, Yin T, Buchanan S, Yadav V, et al. Oncogenic BRAF Deletions That Function as Homodimers and Are Sensitive to Inhibition by RAF Dimer Inhibitor LY3009120. Cancer discovery 2016;6(3):300–15 doi 10.1158/2159-8290.CD-15-0896. [DOI] [PubMed] [Google Scholar]
- 27.Sholl LM, Do K, Shivdasani P, Cerami E, Dubuc AM, Kuo FC, et al. Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight 2016;1(19):e87062 doi 10.1172/jci.insight.87062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garcia EP, Minkovsky A, Jia Y, Ducar MD, Shivdasani P, Gong X, et al. Validation of OncoPanel: A Targeted Next-Generation Sequencing Assay for the Detection of Somatic Variants in Cancer. Arch Pathol Lab Med 2017;141(6):751–8 doi 10.5858/arpa.2016-0527-OA. [DOI] [PubMed] [Google Scholar]
- 29.Falchook GS, Lewis KD, Infante JR, Gordon MS, Vogelzang NJ, DeMarini DJ, et al. Activity of the oral MEK inhibitor trametinib in patients with advanced melanoma: a phase 1 dose-escalation trial. The Lancet Oncology 2012;13(8):782–9 doi 10.1016/S1470-2045(12)70269-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flaherty KT, Infante JR, Daud A, Gonzalez R, Kefford RF, Sosman J, et al. Combined BRAF and MEK inhibition in melanoma with BRAF V600 mutations. N Engl J Med 2012;367(18):1694–703 doi 10.1056/NEJMoa1210093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flaherty KT, Robert C, Hersey P, Nathan P, Garbe C, Milhem M, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med 2012;367(2):107–14 doi 10.1056/NEJMoa1203421. [DOI] [PubMed] [Google Scholar]
- 32.Russo M, Siravegna G, Blaszkowsky LS, Corti G, Crisafulli G, Ahronian LG, et al. Tumor Heterogeneity and Lesion-Specific Response to Targeted Therapy in Colorectal Cancer. Cancer discovery 2016;6(2):147–53 doi 10.1158/2159-8290.CD-15-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goyal L, Saha SK, Liu LY, Siravegna G, Leshchiner I, Ahronian LG, et al. Polyclonal Secondary FGFR2 Mutations Drive Acquired Resistance to FGFR Inhibition in Patients with FGFR2 Fusion-Positive Cholangiocarcinoma. Cancer discovery 2017;7(3):252–63 doi 10.1158/2159-8290.CD-16-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emery CM, Vijayendran KG, Zipser MC, Sawyer AM, Niu L, Kim JJ, et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proceedings of the National Academy of Sciences of the United States of America 2009;106(48):20411–6 doi 10.1073/pnas.0905833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shindo K, Yu J, Suenaga M, Fesharakizadeh S, Cho C, Macgregor-Das A, et al. Deleterious Germline Mutations in Patients With Apparently Sporadic Pancreatic Adenocarcinoma. J Clin Oncol 2017;35(30):3382–90 doi 10.1200/JCO.2017.72.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaffee KG, Oberg AL, McWilliams RR, Majithia N, Allen BA, Kidd J, et al. Prevalence of germ-line mutations in cancer genes among pancreatic cancer patients with a positive family history. Genet Med 2018;20(1):119–27 doi 10.1038/gim.2017.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pritchard CC, Mateo J, Walsh MF, De Sarkar N, Abida W, Beltran H, et al. Inherited DNA-Repair Gene Mutations in Men with Metastatic Prostate Cancer. N Engl J Med 2016;375(5):443–53 doi 10.1056/NEJMoa1603144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klein AP. Genetic susceptibility to pancreatic cancer. Mol Carcinog 2012;51(1):14–24 doi 10.1002/mc.20855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hiripi E, Lorenzo Bermejo J, Li X, Sundquist J, Hemminki K. Familial association of pancreatic cancer with other malignancies in Swedish families. Br J Cancer 2009;101(10):1792–7 doi 10.1038/sj.bjc.6605363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yurgelun MB. Germline Testing for Individuals With Pancreatic Cancer: The Benefits and Challenges to Casting a Wider Net. J Clin Oncol 2017;35(30):3375–7 doi 10.1200/JCO.2017.74.7535. [DOI] [PubMed] [Google Scholar]
- 41.Lowery MA, Jordan EJ, Basturk O, Ptashkin RN, Zehir A, Berger MF, et al. Real-Time Genomic Profiling of Pancreatic Ductal Adenocarcinoma: Potential Actionability and Correlation with Clinical Phenotype. Clinical cancer research : an official journal of the American Association for Cancer Research 2017;23(20):6094–100 doi 10.1158/1078-0432.CCR-17-0899. [DOI] [PubMed] [Google Scholar]
- 42.Sherman MH, Yu RT, Engle DD, Ding N, Atkins AR, Tiriac H, et al. Vitamin D receptor-mediated stromal reprogramming suppresses pancreatitis and enhances pancreatic cancer therapy. Cell 2014;159(1):80–93 doi 10.1016/j.cell.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Torphy RJ, Wang Z, True-Yasaki A, Volmar KE, Rashid N, Yeh B, et al. Stromal Content Is Correlated With Tissue Site, Contrast Retention, and Survival in Pancreatic Adenocarcinoma. JCO Precision Oncology 2018(2):1–12 doi 10.1200/po.17.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohlund D, Handly-Santana A, Biffi G, Elyada E, Almeida AS, Ponz-Sarvise M, et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. The Journal of experimental medicine 2017;214(3):579–96 doi 10.1084/jem.20162024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Olive KP, Jacobetz MA, Davidson CJ, Gopinathan A, McIntyre D, Honess D, et al. Inhibition of Hedgehog signaling enhances delivery of chemotherapy in a mouse model of pancreatic cancer. Science 2009;324(5933):1457–61 doi 10.1126/science.1171362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olive KP, Tuveson DA. The use of targeted mouse models for preclinical testing of novel cancer therapeutics. Clinical cancer research : an official journal of the American Association for Cancer Research 2006;12(18):5277–87 doi 10.1158/1078-0432.CCR-06-0436. [DOI] [PubMed] [Google Scholar]
- 47.Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer cell 2014;25(6):735–47 doi 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ozdemir BC, Pentcheva-Hoang T, Carstens JL, Zheng X, Wu CC, Simpson TR, et al. Depletion of carcinoma-associated fibroblasts and fibrosis induces immunosuppression and accelerates pancreas cancer with reduced survival. Cancer cell 2014;25(6):719–34 doi 10.1016/j.ccr.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aung KL, Fischer SE, Denroche RE, Jang GH, Dodd A, Creighton S, et al. Genomics-Driven Precision Medicine for Advanced Pancreatic Cancer: Early Results from the COMPASS Trial. Clinical cancer research : an official journal of the American Association for Cancer Research 2017. doi 10.1158/1078-0432.CCR-17-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghazani AA, Oliver NM, St Pierre JP, Garofalo A, Rainville IR, Hiller E, et al. Assigning clinical meaning to somatic and germ-line whole-exome sequencing data in a prospective cancer precision medicine study. Genet Med 2017;19(7):787–95 doi 10.1038/gim.2016.191. [DOI] [PubMed] [Google Scholar]
- 51.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31(3):213–9 doi 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013;499(7457):214–8 doi 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome biology 2011;12(4):R41 doi 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zill OA, Greene C, Sebisanovic D, Siew LM, Leng J, Vu M, et al. Cell-Free DNA Next-Generation Sequencing in Pancreatobiliary Carcinomas. Cancer discovery 2015;5(10):1040–8 doi 10.1158/2159-8290.CD-15-0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


