Abstract
The liver is the primary organ involved in handling of bile salts, a class of amphipathic molecules with signaling activities as well as desired and detrimental detergent actions. To allow in-depth investigation of functions of bile salts in healthy and diseased liver, the spatial distribution of bile salt species within the liver needs to be studied. Therefore, the aim of our study was to determine hepatic bile salt distribution and identify specific lipid markers that define the structural elements of the liver. Matrix-assisted laser desorption/ionization-mass spectrometry imaging (MALDI-MSI) was used to monitor the spatial distribution of bile salts and lipids in liver sections of rat, dog, and patients with unaffected and cholestatic parenchyma. MALDI-MSI in negative ion mode showed the local presence of a variety of bile salts, predominantly taurine-conjugates, as localized patches of varying sizes (representing the bile ducts) throughout the liver tissue. Specific molecular markers were identified for the connective tissue (phosphatidic acids, e.g., [PA (18:0_18:1)–H]−), the liver parenchyma (phosphatidylinositols, e.g., [PI (18:0_20:4)-H]−), and the bile ducts (hydroxylated-sulfatides, e.g., [ST–OH (18:1_24:0)-H]−). One of these sulfatides (at m/z 906.6339) was found to be uniquely localized in a thin lining on the inside of the bile duct, colocalized with cytokeratins, and encased luminal bile salts. A similar distribution of the aforementioned sulfatide was observed, albeit in constricted ductular structures, in the liver of a patient with a mild clinical phenotype of primary sclerosing cholangitis (PSC). In contrast, sulfatides were virtually absent in the liver of patients with PSC and a severe clinical phenotype, with (atypical) cholanoids (e.g., the bile alcohol 5-cyprinolsulfate) abundant in the extra-ductular space and glyco(cheno)deoxycholic acid-3-sulfate localized to fibrotic connective tissue. The latter two molecular species were able to discriminate between healthy liver tissue (n = 3) and tissue from PSC patients with a severe clinical phenotype (n = 3). In conclusion, the distinct structural elements of the mammalian liver are characterized by specific classes of lipids. We propose that (hydroxylated-)sulfatides are specific molecular markers of the bile duct.
During the past two decades, bile salts have been upgraded from molecules required for absorption of dietary lipids and fat-soluble vitamins, to signaling molecules regulating biological processes as diverse as nutrient metabolism, inflammation, and liver regeneration.1−4 The discovery of bile salt receptors that mediate these signaling activities was key to the renewed interest in bile salts by academic and pharmaceutical communities. In particular, Farnesoid-X Receptor (FXR), a bile salt-activated transcription factor with an essential role in bile salt homeostasis, has been studied extensively. FXR agonism-based therapy has already been approved for treatment of patients with primary biliary cholangitis (PBC) and inadequate response to ursodeoxycholic acid.5 Multiple clinical trials are ongoing to evaluate efficacy of FXR agonists in treatment of metabolic liver disorders like nonalcoholic steatohepatitis (NASH).6,7
The liver is the primary organ involved in the synthesis and handling of bile salts. Being amphipathic molecules, bile salts in the aqueous phase can interact with biological membranes. Consequently, the liver is the prime site where injury occurs when bile formation or flow of bile is impaired, and toxic levels of bile salts build up inside the parenchymal cells. At a certain threshold, intracellular bile salts damage mitochondrial membranes and initiate a sterile inflammatory response that results in immune-mediated tissue injury.8−10 In drug development, drug-induced cholestasis is a common reason for halting further development in preclinical phases of testing.11
Distinct bile salt species arise from different synthetic routes in the host, utilization of either glycine or taurine for formation of N-amidated conjugates, and biotransformation by the gut microbiota.1 Individual bile salt species have distinct signaling properties and toxicity profiles. Mass spectrometry (MS)-based analytical tools have been of great value to determine bile salt composition of (homogenized) biospecimens (e.g., serum, bile, urine, feces, and liver tissue) from healthy and diseased subjects.
Study of hepatic bile salt composition requires the preparation of a homogenate, inevitably resulting in loss of spatial information. As several aspects of hepatic bile salt handling are known to be zonated (e.g., extraction of bile salts from the portal supply in the periportal area, de novo synthesis in the pericentral area),12 information on the local distribution of bile salts within the liver could further advance our understanding of bile salt (patho)physiology.
Matrix-assisted laser desorption/ionization-mass spectrometry imaging (MALDI-MSI) has been extensively used to monitor the distribution of drugs, lipids, peptides, and proteins in tissue sections, making this an ideal tool to determine the molecular profiles of the various compartments of healthy and diseased liver. Here we used MALDI-MSI to study the spatial distribution of bile salts within the liver of animal species commonly used in preclinical drug testing, as well as in noncholestatic and cholestatic human liver parenchyma. Specific aims were (i) to determine whether it is feasible to determine the spatial distribution of distinct bile salt species in the liver, including those with identical molecular mass, (ii) to identify lipid molecular markers that define the structural elements of the liver anatomy, and (iii) determine if there are unique molecular profiles that can distinguish between healthy and diseased human liver and compare with those obtained from preclinical animal species. This spatial molecular information can be employed to address roles of defined molecules in liver (dys)function, both in the context of human pathology and drug screening programs.
Methods
Materials
Cholic acid (CA), deoxycholic acid (DCA), chenodeoxycholic acid (CDCA), lithocholic acid (LCA), taurocholic acid (TCA), taurodeoxycholic acid (TDCA), taurochenodeoxycholic acid (TCDCA), glycocholic acid (GCA), glycochenodeoxycholic acid (GCDCA), glycodeoxycholic acid (GDCA), 2,5-dihydroxybenzoic acid (DHB), 9-aminoacridine (9AA), chloroform (CHCl3), and trifluoroacetic acid (TFA) were purchased from Sigma-Aldrich (Zwijndrecht, The Netherlands). Methanol (MeOH) was purchased from Biosolve (Valkenswaard, The Netherlands).
Liver Specimens
A healthy canine liver specimen (Marshall Beagle, n = 1) was obtained in the framework of an ongoing study at Janssen Pharmaceutica N. V. The study was approved by the local animal ethical committee and conducted in facilities accredited by national institutions adhering to AAALAC guidelines. Human liver specimens were obtained from patients undergoing either resection for hepatobiliary malignancies (nonaffected liver tissue sample distant from the tumor, n = 3) or undergoing liver transplantation for end-stage liver disease (PSC, n = 6). Patients with PSC were classified as severe cases if bilirubin was ≥34 μmol/L along with ≥2 abnormal liver tests, which included aspartate aminotransferase (AST), alanine aminotransferase (ALT), alkaline phosphatase (ALP), gamma-glutamyl transferase (GGT), and international normalized ratio (INR) (see Table S1). Specimens were procured as part of biobank initiatives at the Aachen (Medical Ethical approval no. EK 20609) and Maastricht (Medical Ethical approval no. 14-4-153) locations of the Euregional Surgical Center Aachen Maastricht (ESCAM) or from the Erasmus MC, Rotterdam (Medical Ethical approval no. 167.902/1998/33 and no. 2014-060). Patients gave written informed consent for biobanking and research use of surgical specimens. A rat liver specimen (Wistar Han, n = 1) was obtained from a control group of rats sacrificed for a study on colonic anastomotic healing (approved by the local Animal Ethics Committee of the Radboud University, Nijmegen). Per liver specimen, multiple sections throughout the tissue were analyzed [i.e., dog (n = 7), noncholestatic patient (n = 7), PSC patients (n = 4), and rat (n = 9)].
Tissue Preparation
Liver tissue was sectioned using a Microm HM535 cryo-microtome (Microm International, Walldorf, Germany) at a temperature of −20 °C to produce 10 μm thick sections, which were thaw-mounted onto clean indium tin oxide (ITO)-coated glass slides (4–8 Ω resistance, Delta Technologies, Stillwater, MN, USA). Tissue sections were scanned prior to matrix application using a Nikon Super CoolScan 5000 ED (Nikon Corporation, Tokyo, Japan) to produce high-quality optical images. Consecutive tissue sections were thaw-mounted on regular glass slides (Thermo Scientific) for histological and immunohistochemical staining.
Matrix Application
Tissue sections were coated with 15 mg/mL DHB in 2:1 CHCl3:MeOH with 0.2% TFA using the SunCollect automated pneumatic sprayer equipped with dispenser system (Sunchrom GmbH, Friedrichsdorf, Germany) in a series of 15 layers. The initial seeding layer was performed at 10 μL/min; subsequent layers were performed at 20, 30, and 40 μL/min, respectively (all at speed 450 mm/min, track spacing: 2 mm, height: 23 mm). For negative-ion mode some tissue sections were coated with 10 mg/mL 9AA in 70:30 MeOH:H2O in a series of 10 layers. The initial seeding layer was performed at 10 μL/min; subsequent layers were performed at 15, 20, and 25 μL/min, respectively (all at speed, 450 mm/min; track spacing, 2 mm; height, 25 mm).
Instrumentation
MALDI-MSI
High-speed imaging was performed on a Bruker RapifleX MALDI Tissuetyper system (Bruker Daltonik GmbH, Bremen, Germany) equipped with a neodymium-doped yttrium aluminum garnet (Nd:YAG) laser (355 nm, 10 kHz); a more detailed description of this instrument is provided elsewhere.13 The instrument was operated in reflectron mode in both positive- and negative-ion modes in the mass range m/z 400–1000. The instrument was calibrated prior to analysis using red phosphorus clusters in both polarities. The laser power was 80% with an accumulation of 200 shots. Images of the whole tissue sections were acquired using a 50 × 50 μm raster (25 × 25 μm beam scan area). The images were generated using the FlexImaging version 5.0 software (Bruker Daltonik GmbH) and were normalized to the total ion current (TIC).
MALDI-FTICR-MSI
For confirmation of molecular identity, high-mass resolution (100,000 at m/z 500) measurements were performed on a Bruker SolariX FTICR mass spectrometer (Bruker Daltonik GmbH, Bremen, Germany) equipped with a 9.4 T superconducting magnet and Nd:YAG laser. The instrument was operated in both positive- and negative-ion modes in the mass range m/z 100–2000. The instrument was calibrated prior to analysis using red phosphorus clusters in both polarities. The laser power was 30% (2 kHz) with an accumulation of 100 shots. Images were acquired with a pixel size of 15 μm. A 95% data reduced profile spectrum and spectrum peak list were saved for further analysis. The images were generated using the FlexImaging version 4.1 software (Bruker Daltonik GmbH) and were normalized to the total ion current (TIC).
MALDI-MS/MS
For further confirmation of molecular identity, a Waters MALDI HDMS Synapt G2-Si mass spectrometer (Waters Corporation, Manchester, UK) equipped with an Nd:YAG laser was used to perform ion mobility separation (IMS) and tandem mass spectrometry (MS/MS) measurements. The instrument was calibrated using red phosphorus prior to analysis. MS/MS was performed on consecutive tissue sections, the laser power was 150 arbitrary units (1 kHz), and the ions of interest were fragmented by collision induced dissociation (CID) in the trap cell with an isolation window of approximately 3 Da. For ion mobility, the IMS wave velocity was 900 m/s and the wave height was 40 V. For presentation purposes, mass spectra from the MassLynx version 4.1 software (Waters Corporation) were exported in the form of text files and imported into mMass, open-source software used for mass spectral processing.14 The MS/MS spectra from the commercial bile acid standards and tissue sections were compared to those in the literature or databases such as lipid maps (www.lipidmaps.org) and the ALEX123 lipid calculator (www.alex123.info). The fragment ions observed in each MS/MS spectrum were assigned based on the recently proposed nomenclature for lipids.15
TOF-SIMS
High spatial resolution imaging was performed using a PHI nanoTOF II TOF-SIMS instrument (Physical Electronics, Minnesota, MN, USA) equipped with a 30 kV Binq+ cluster liquid metal ion gun (LMIG), which was operated with a Bi3+ cluster ion beam. Images were acquired using a tile size of 500 × 500 μm at 30 frames with an ion dose of 2.59 × 1012 ions/cm2 in negative ion mode; the data was acquired in the mass range m/z 0–1850 and a spatial resolution of ∼1 μm. The instrument was calibrated using a combination of elements and hydrocarbon fragments. The images were generated using software PHI TOF-DR software (Physical Electronics).16
Tissue Staining
Hematoxylin and eosin (H&E) and Masson’s trichrome (MTC) staining was performed on consecutive tissue sections (10 μm) followed by aqueous mounting. Optical images of all stained tissue sections were acquired using a Mirax desk scanner (Zeiss, Gottingen, Germany). Cryosections (5 μm thickness) of liver tissue were used for immunohistochemistry; thinner tissue sections were required to reduce background signal. The bile duct epithelium was stained using a wide spectrum anticytokeratin antibody (Dako/Agilent Pathology Solutions) and secondary detection reagents. Detailed protocols are located in the Supporting Information.
Statistical Analysis
Statistical analyses were performed on the MALDI-FTICR-MSI data using the in-house built ChemomeTricks toolbox for Matlab version 7.0 software (The MathWorks, Natick, MA, USA).17 Briefly, peak picking was performed on an average spectrum (created using the peaks sqlite file) at a 0.001 Da bin size. The center of the peaks and integration ranges were defined, to account for peak shifting from pixel-to-pixel. Principal component analysis (PCA) was then performed on the converted data sets, which were normalized to the total ion current. The peak list from each principal component that highlighted different areas of the liver was exported, and the top ten peaks (based on intensity) were then identified.
Further statistical analysis was performed on the MALDI-FTICR-MS imaging data set from healthy human liver (n = 3) and liver from patients with severe PSC (n = 3), using SCiLS lab version 2016b (SCiLS GmbH, Bremen, Germany). The results were normalized to the total ion current (TIC), and an average spectrum was created from all samples which was imported into the mMass software and subjected to peak picking, to create a peak list which was imported back into SCiLS lab. A receiver operating characteristic (ROC) analysis was performed to determine the power of selected molecular species to discriminate between healthy liver and cases of severe PSC.
Results and Discussion
Identification of Bile Salts
In order to have optimal sensitivity and selectivity of the method applied, the physiologically most relevant (non)conjugated primary and secondary bile salts (Figure S1) were spotted individually on a target plate, coated with 9AA, and analyzed with MALDI-MS, according to previous works.18,19 All bile salts were observed in their deprotonated form ([M – H]−). MS/MS experiments were performed on the standards (Figure S2). The taurine-conjugated bile salts showed characteristic product ions, indicating the fragmentation or loss of taurine such as m/z 80 (SO3–), 107 (C2H3O3S–), and 124 (C2H6NO3S–), whereas the glycine-conjugated bile salts showed the loss of glycine m/z 74 (C2H5NO2–).20,21 The unconjugated bile salts showed fragments resulting from the loss of H2O, CO2, and CH2O2.21 Isomeric species, such as chenodeoxycholic acid (CDCA), deoxycholic acid (DCA), and their conjugated forms, can be separated using chromatographic techniques.22 Here, ion mobility separation (IMS) was evaluated for separation of isomeric bile salts. Standards of the isomeric bile salts were applied as a mixture on a target plate and analyzed. Using this technique, it was possible to differentiate between DCA and CDCA (Figure S3A); however, it was not possible to separate the taurine (Figure S3B) or glycine-conjugated variants thereof (Figure S3C). This is likely due to the limited resolution of current commercial ion mobility instrumentation. Since the bulk of hepatic bile salts are conjugated, IMS was not suitable to analyze isomeric bile salt species in intact tissues.
Spatial Distribution of Bile Salts and Lipids in Healthy Dog Liver
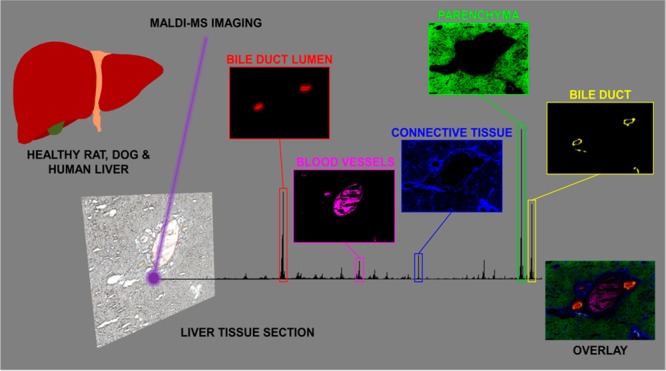
High-speed imaging experiments on whole liver tissue sections (at a spatial resolution of 50 μm) showed the distinct distribution of various molecular markers linked to the different tissue types found in the liver. Regions such as the parenchyma, the bile ducts, the connective tissue, and the bile duct lumen could be clearly distinguished in these MALDI-MS images (Figure 1A and S4A). Following analysis of the whole tissue section, high mass and high spatial resolution MALDI-FTICR-MSI was performed on specific areas, selected based on histological and molecular features (Figure 1B and S4B). These images provided greater spatial detail of the selected molecular species and improved mass accuracy, improving molecular identification.
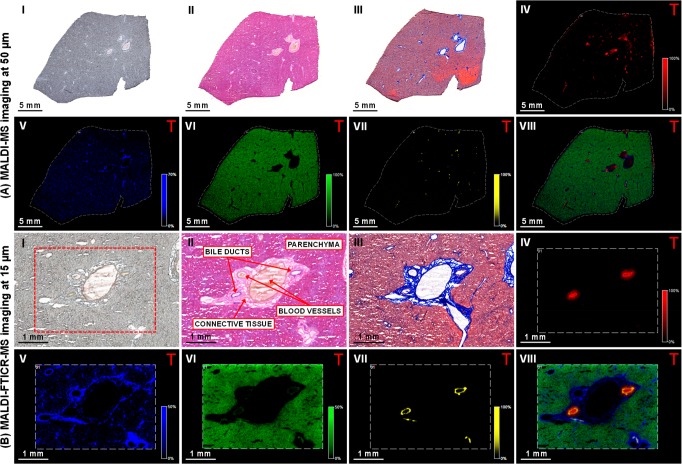
Figure 1.
Multimodal imaging of healthy dog liver tissue. (I) Optical image of the tissue section prior to matrix application or staining. (II) Tissue sections stained with hematoxylin and eosin stain, and (III) Masson’s trichrome. Negative-ion mode (A) MALDI-MS and (B) MALDI-FTICR-MS images showing the distribution of selected molecular species in the (IV) bile duct lumen ([TCA – H]− at m/z 514.2849); (V) connective tissue ([PA (18:0_18:1) – H]− at m/z 701.5129); (VI) parenchyma ([PI (18:0_20:4) – H]− at m/z 885.5504); (VII) bile duct ([ST–OH (18:1_24:0) – H]− at m/z 906.6339); and (VIII) overlay of the selected species. (Designated masses are from MALDI-FTICR-MS imaging measurements, MALDI-MS image field of view 140785 pixels, area 124 mm2, and spatial resolution 50 μm. MALDI-FTICR-MS image field of view 51761 pixels, 11.6 mm2 area, and spatial resolution 15 μm. Images normalized with TIC).
The optical image of the whole liver tissue section prior to analysis (Figure 1, panels A and B, I) shows a homogeneous parenchyma with a cluster of large blood vessels in the center of the tissue. Histological staining of consecutive tissue sections was performed for reference. The hematoxylin and eosin stain (Figure 1, panels A and B, II) differentiates the eosinophilic hepatocellular parenchyma from the brighter (slightly eosinophilic) stained portal areas, whereas the blue Masson’s trichrome stain (Figure 1, panels A and B, III) highlights the presence of collagen, indicating the portal connective tissue. This is abundant around the large blood vessels, with smaller patches located throughout the tissue section.
MALDI-MS imaging in negative-ion mode showed the presence of predominantly taurine-conjugated bile salts, the most abundant were taurocholic acid (TCA) and taurochenodeoxycholic/taurodeoxycholic acid (TCDCA/TDCA). This is to be expected as dogs use primarily taurine for bile acid conjugation.23,24 The distribution of TCA at m/z 514.28 (Figure 1A, IV) showed localized patches of varying sizes throughout the tissue, most likely representing the bile ducts. The larger ones (average diameter 175–250 μm), which are in close proximity to the large blood vessels, are septal bile ducts. Smaller bile ducts (average diameter 20–70 μm) distributed throughout the tissue, are the interlobular bile ducts.25 The identity of TCA was confirmed by MS/MS measurements (Figure S5A). Interestingly, 3-keto-TCA and 3-keto-TDCA/TCDCA were also present in the bile duct lumen. Keto bile salts were previously detected in bear bile.26 Next to this, taurolithocholic acid was also present. The connective tissue showed a strong presence of a lipid at m/z 701.51 (Figure 1A, V), which was identified by MS/MS measurements as a phosphatidic acid [PA (18:0_18:1) – H]− (Figure S5B). This species was distributed throughout the tissue but appeared to be more abundant in the connective tissue, which correlates well with Masson’s trichrome staining (Figure 1, panels A and B, III). The connective tissue was also characterized by other phosphatidic acids, phosphatidylserines (PS), and sphingomyelin (SM) fragments ([M – CH3]−).27 It should be noted that the positive ion counterparts of the SM fragments colocalized with observed species, hence confirming their identification (see Figure S4). In the surrounding liver parenchyma, the most abundant species, which was homogeneously distributed throughout the parenchyma but absent from the other areas of the tissue was identified using MS/MS measurements as a phosphatidylinositol (PI), specifically [PI (18:0_20:4) – H]− at m/z 885.55 (Figure 1A, VI and Figure S5C). Other PIs, PAs, and phosphatidylethanolamines (PE) were predominantly present in this area.
Another lipid species at m/z 906.64 (Figure 1A, VII) was distributed in small localized patches throughout the tissue and colocalized with the distribution of bile salts. The overlay of the selected species (Figure 1A, VIII) shows that selected species colocalize well together and clearly highlight the different boundaries of the molecular components that make up the bile duct and surrounding tissue. Following analysis of the whole tissue section, high mass and high spatial resolution MALDI-FTICR-MS imaging was performed on selected areas, which was selected based on histological and molecular features. This provided far more detail of the selected molecular species, as well as improved mass accuracy enabling better confirmation of their identity (Figure 1B, III–V). Interestingly, MALDI-FTICR-MS analysis revealed that the species at m/z 906.6339 was localized in a thin lining on the inside of the bile duct (Figure 1B, VII). This compound was identified, by MS/MS measurements, as a hydroxylated-sulfatide [ST–OH (18:1_24:0) – H]− (Figure S5D), a ceramide derivative previously identified by MALDI-MS/MS imaging of brain tissue.28 Nonhydroxylated sulfatides (ST) were also present in the bile duct. The overlay of the selected molecular species (Figure 1B, VIII) recapitulated the general histological features of the liver. All peaks identified in the different regions of the healthy dog liver are shown in Tables S2 and S3.
Spatial Distribution of Bile Salts and Lipids in Healthy Human Liver
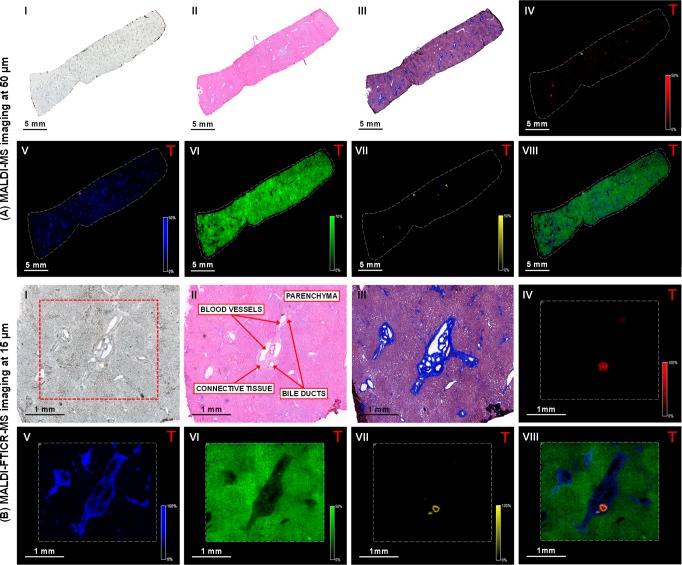
Healthy human liver tissue was analyzed next to study (dis)similarities in spatial distribution of bile salt species and molecular lipid markers of structural elements in the human liver (Figure 2 and S6). Optical and histological images were again acquired for reference (Figure 2, panels A and B, I–III). MALDI-MSI showed high signal intensities of TCA at m/z 514.28 and TCDCA/TDCA at m/z 498.28, whereas lower signal intensities of glyco(cheno)deoxycholic acid (GCDCA/GDCA) at m/z 448.30 and glycocholic acid (GCA) at m/z 464.30 were present (Table S3). These findings were somewhat unexpected since the human bile salt pool consists largely of glycine-conjugated species, with taurine-conjugated bile salts comprising approximately 20% of the biliary bile salt pool.24 This discrepancy is most likely attributed to the fact that the sulfonate group of taurine conjugates is more easily ionized in negative-ion mode than the carboxylic acid group of glycine conjugates.18,19 The distribution of TCA at m/z 514.28 (Figure 2A, IV) showed a few small localized patches throughout the tissue, indicating the presence of bile ducts. Both septal and interlobular bile ducts were apparent in the studied specimen. Taurine-conjugated keto bile salts and lithocholate were also detected in the bile ducts. Also in human liver tissue, the connective tissue was defined by [PA (18:0_18:1) – H]− at m/z 701.51 (Figure 2A, V), and distribution of this lipid species matched the Masson’s trichrome staining (Figure 2, panels A and B, III). Connective tissue was also characterized by SM fragments and PS. Human liver parenchyma (Figure 2A, VI) was characterized by a PI [PI (18:0_20:4) – H]− at m/z 885.55, as well as other PIs, PAs, and PEs. The hydroxylated-sulfatide [ST–OH (18:1_24:0) – H]− at m/z 906.63 (Figure 2A, VII) appeared as small localized patches throughout the tissue. The overlay of the selected species (Figure 2A, VIII) shows how the selected ions colocalize and indicated the best area for high spatial resolution MALDI-FTICR-MS imaging. The images again only showed detailed views of the other selected species (Figures 2B, IV–VI). MALDI-FTICR-MSI analysis showed the hydroxylated-sulfatide [ST–OH (18:1_24:0) – H]− at m/z 906.6340 as a thin band located in the bile duct (Figure 2B, VII). The overlay of the selected species (Figure 2B, VIII) demonstrated the localization of the bile salts within the bile duct lumen, which was bordered by a lining of the aforementioned hydroxylated-sulfatide. Additional hydroxylated and nonhydroxylated sulfatide species were observed in the bile duct wall. The peaks identified in the different regions of the healthy human liver are shown in Tables S2 and S3.
Figure 2.
Multimodal imaging of healthy human liver tissue. (I) Optical image of tissue section prior to matrix application or staining. (II) Tissue sections stained with hematoxylin and eosin and (III) Masson’s trichrome stain. Negative-ion mode (A) MALDI-MS and (B) MALDI-FTICR-MS images showing the distribution of selected molecular species in the (IV) bile duct lumen ([TCA – H]− at m/z 514.2849); (V) connective tissue ([PA (18:0_18:1) – H]− at m/z 701.5128); (VI) parenchyma ([PI (18:0_20:4) – H]− at m/z 885.5504); and (VII) bile duct ([ST–OH (18:1_24:0) – H]− at m/z 906.6339). (VIII) Overlay of the selected species. (Designated masses are from MALDI-FTICR-MS imaging measurements, MALDI-MS image field of view 78624 pixels, area 110 mm2, and spatial resolution 50 μm. MALDI-FTICR-MS image field of view 24915 pixels, 5.5 mm2 area, and spatial resolution 15 μm. Images normalized with TIC).
Sulfatides are Novel Molecular Markers for the Bile Duct Epithelium
Hydroxylated-sulfatide [ST–OH (18:1_24:0) – H]− was identified in both healthy dog and human liver tissue and specifically localized as a thin band lining the bile duct. This compound was the most prominent of several hydroxylated and nonhydroxylated sulfatides that colocalized in the bile duct (Table S3). In order to determine whether sulfatides are specific markers for the bile duct, immunohistochemical staining for cytokeratins was performed on sections of dog and human liver (Figure 3). This revealed the presence of cytokeratins in the cholangiocytes/bile duct epithelium (Figure 3, panels A and B, II). MALDI-MSI of a consecutive section revealed [ST–OH (18:1_24:0) – H]− at m/z 906.63 distributed as a ring-like structure (Figure 3, panels A and B, III). High spatial resolution TOF-SIMS imaging of consecutive sections of dog and human liver confirmed the confinement of sulfatides to the bile duct epithelium (Figure 3, panels A and B, IV). The molecular images correlate well with the immunohistochemical staining of the bile ducts indicating that these species, as well as the other sulfatide species (Table S3) are molecular markers for the bile duct epithelium.
Figure 3.
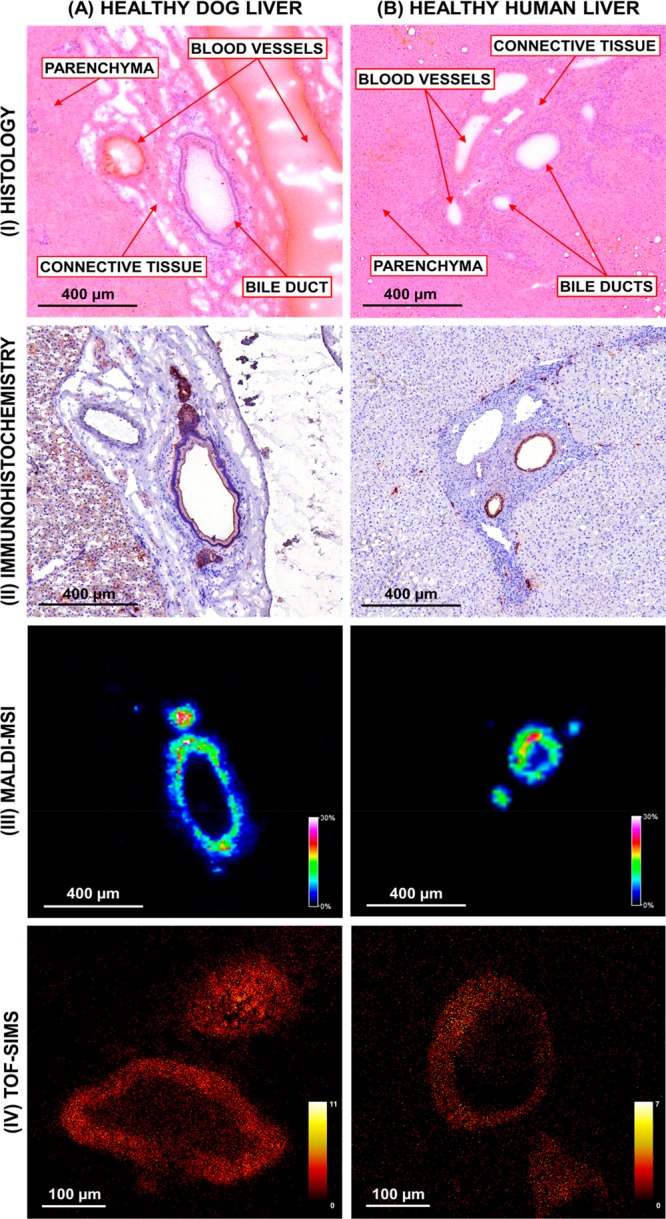
Comparison of immunohistochemical staining, MALDI-MS imaging and TOF-SIMS imaging of (A) healthy human and (B) dog liver. (I) Hematoxylin and eosin stain of tissue section post-MALDI-MSI analysis. (II) Cytokeratin staining shows the bile duct epithelium. Negative-ion mode (III) MALDI-MS imaging and (IV) TOF-SIMS imaging of the bile ducts showing the distribution of [ST–OH (18:1_24:0) – H]− at m/z 906.63. (MALDI-MS image (dog) field of view 15875 pixels, area 1.60 mm2, and spatial resolution 10 μm. MALDI-MS image (human) field of view 30420 pixels, area 3.05 mm2, and spatial resolution 10 μm. TOF-SIMS (dog/human) field of view 250000 pixels, area 0.25 mm2, and spatial resolution 1 μm. Images normalized with TIC).
The unique localization of sulfatides to bile duct epithelium suggests a biologically relevant role. Sulfatides are glycosphingolipids that contain an extra sulfate group and hence are commonly observed in negative-ion mode. Sulfatides are predominantly found in the exoleaflet of the membrane bilayer and have been detected in different tissues, including the brain where they are an abundant component of the myelin sheath, the pancreas (the islets of Langerhans), and the kidney.29 Although sulfatides were previously detected in liver, their presence could not be attributed to a particular region because those studies were performed on tissue extracts.30 Sulfatides are negatively charged and will likely repel negatively charged compounds, such as bile salts, and may accordingly play a role in protecting the bile duct epithelium from detergent action of bile salt anions. Information on specific roles of sulfatides in liver biology is sparse. Sulfatides are self-lipid antigens, and presence of autoantibodies appears to be common in chronic autoimmune liver diseases like autoimmune hepatitis and primary biliary cholangitis (PBC).31 Sulfatides presented by CD1d protein are recognized by type 2 natural killer T (NKT) cells. In the liver, NKT cells are abundant in the sinusoids, with type I and type II NKT cells having a pro- and anti-inflammatory action, respectively.32 Immune-mediated liver injury in an animal model of alcoholic liver disease is dependent on type I NKT cells.33 Interestingly, activation of type II NKT cells by sulfatides prevented inflammation and liver disease in this model and may have a broader role in a tolerogenic response in inflammatory liver disease.34 Given the abundance of this lipid in the bile ducts, a link with inflammatory cholangiopathies like PBC and primary sclerosing cholangitis (PSC) is plausible.
Spatial Distribution of Bile Salts and Sulfatides in Liver of PSC Patients
Liver tissue from six patients undergoing liver transplantation for treatment of PSC was analyzed by MALDI-FTICR-MSI (Figures 4 and S7) to explore whether molecular species are globally similar in diseased liver tissue. The first PSC patient (case 1, Table S1) was noncholestatic and was considered to have a mild clinical phenotype. Histological evaluation of a liver biopsy taken prior to transplantation revealed mild inflammation but no fibrosis. Severe cases of PSC (n = 3) were characterized by cholestasis (bilirubin ≥ 34 μmol/L) and cholestatic liver injury/dysfunction (≥2 liver test abnormalities) (Table S1).
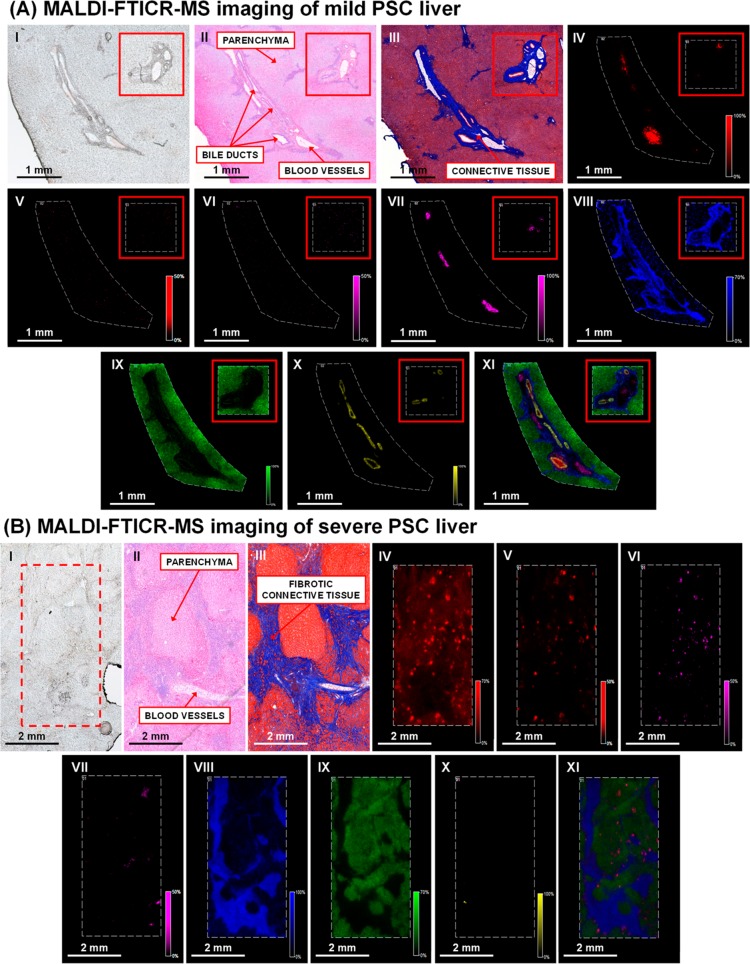
Figure 4.
Multimodal imaging of human liver from patients with (A) mild PSC and (B) severe PSC. (I) Optical image of tissue section prior to matrix application or staining. Consecutive tissue sections stained with (II) hematoxylin and eosin (III) Masson’s trichrome. Negative-ion mode MALDI-FTICR-MS images showing the distribution of (IV) [TCA – H]− at m/z 514.2855; (V) sulfated bile alcohol ([M – H]−) at m/z 531.2999; (VI) bilirubin diglucuronide ([M – H]−) at m/z 935.3218; (VII) heme ([M – H]−) at m/z 615.1704; (VIII) [PA (18:0_18:1) – H]− at m/z 701.5128; (IX) [PI (18:0_20:4) – H]− at m/z 885.5504; and (X) [ST–OH (18:1_24:0) – H]− at m/z 906.6353. (XI) Overlay of the selected species. (Mild PSC image field of view 20326 pixels and area 10 mm2, severe PSC image field of view 63744 pixels, and area 14 mm2. Spatial resolution of both images 15 μm, normalized with TIC).
Mild PSC
The optical image of the tissue section showed an elongated cluster of bile ducts/blood vessels surrounded by connective tissue and a homogeneous parenchyma (Figure 4A, I). Hematoxylin and eosin (Figure 4A, II) and Masson’s trichrome staining of liver sections of the initial patient with mild PSC (Figure 4A, III) revealed concentric periductal fibrosis with little periductal inflammatory infiltrate. In addition, mixed inflammatory cells and bile ductular proliferation were found at the periphery of the portal area extending into the interlobular septa. For both patients (cases 1 and 2), the histological features observed in the hepatic parenchyma, bile ducts, and portal areas are associated with PSC.35 Despite these morphological differences, liver tissue of the patient with mild PSC showed the same distribution of selected molecular species as healthy human liver tissue. Taurine-conjugated species were the most abundant bile salt species and localized to the bile duct lumen (Figure 4A, IV). Blood vessels in the portal area were marked by the presence of heme (Figure 4A, VII). Molecular markers identified in the healthy human liver also defined the connective tissue (Figure 4A, VIII), parenchyma (Figure 4A, IX), and the bile ducts in the patient with mild PSC. The distribution of [ST–OH (18:1_24:0) – H]− at m/z 906.6353 (Figure 4A, X) indicated that the bile ducts were narrowed, a hallmark histological feature observed in PSC.37 The overlay shows that the selected species colocalize well together and clearly highlight the different boundaries of the molecular components (Figure 4A, XI). The mild/severe PSC (case 4) is classed as such due to elevated bilirubin and clotting function, the liver also has slightly above normal aspartate aminotransferase levels (>40 U/L). All other parameters within the normal range (Table S1).
Severe PSC
The optical image shows an inhomogeneous parenchyma with fibrotic connective tissue dispersed throughout the tissue (Figure 4B, I). Histological examination of the liver from the initial patient with severe PSC (case 2, Figure 4B, II and III) showed a diminished number/absence of interlobular bile ducts, portal-to-portal bridging fibrosis, bile ductular proliferation in portal areas, and fibrous septa with minimal mononuclear inflammatory infiltrate. In addition nodular hepatocellular regeneration, ballooning degeneration of hepatocytes, and yellow to brown pigmentation were noted. Taurine-conjugated species were also present in the liver of the patient with severe PSC (Figure 4B, IV); however, they appear to have diffused out of the bile ducts into the surrounding parenchyma. This is reminiscent of regurgitation of bile salts from leaky bile ducts into the portal tracts in an animal model of PSC.36 Moreover, a unique bile salt species at m/z 531.2999 was observed (Figure 4B, V). MS/MS measurements indicated loss of m/z 97 (HSO4–) and m/z 80 (SO3–), leading to its tentative identification as 5-cyprinolsulfate (5-CS). To our knowledge, the presence of 5-CS (a sulfated bile alcohol abundant in cypriniform fishes)37 has not been previously reported in humans and likely relates to cholestasis where elevation of urinary bile alcohols was previously observed.38 In order to confirm molecular identity, a whole-body tissue section of a zebra fish, an authentic source of 5-CS, was analyzed. The MALDI-MS/MS spectrum of 5-CS obtained from the initial patient with severe PSC was comparable to that obtained from the zebra fish (Figure S8A). Glyco(cheno)deoxycholic acid-3-sulfate (G(C)DCA-S) at m/z 528.2639 was also identified in the liver of the initial patient with severe PSC, which was not detected in the mild PSC case. The MALDI-MS/MS spectrum of an authentic standard is shown in Figure S8B. Molecular identity of G(C)DCA-S in liver tissue was further supported by MALDI-FTICR-MS and MS/MS measurements. In general, sulfation of bile salts is an adaptive mechanism to promote elimination of bile salts that have accumulated in the liver.39
Furthermore, both unconjugated and conjugated bilirubin was observed. The MALDI-FTICR-MS images show that the distribution of bilirubin at m/z 583.2569, bilirubin monoglucuronide at m/z 759.2899, and bilirubin diglucuronide at m/z 935.3218 (Figure 4B, VI) coincided with that of bile salts. Of note, 5-CS and bilirubin (conjugated and unconjugated) were not detected in the liver from the patient with mild PSC (Figures 4A, V and VI, respectively) whose serum bilirubin levels were within the normal range. Elevation of bilirubin can be seen following obstruction of bile flow resulting in the release of conjugated bilirubin into the bloodstream. The lipid at m/z 701.5128, identified as [PA (18:0_18:1) – H]−, was abundant in the connective tissue surrounding the blood vessels (Figure 4B, VII) and bile ducts, with a notably thicker layer when compared to liver of the patient with mild PSC (Figure 4A, VIII) and is dispersed throughout the parenchyma (Figure 4B, IX) which is apparent from the overlay of the selected species (Figure 4B, XI). Despite the occurrence of discrete patches of biliary constituents (i.e., bile salts and bilirubin glucuronides) that indicates that bile ducts are present (as confirmed by staining of consecutive sections for cytokeratins), hydroxylated and nonhydroxylated sulfatides were virtually absent in the liver of the initial patient with severe PSC (Figure 4B, X and Figure S9). It is currently undetermined if absence/disappearance of sulfatides from the bile ducts has pathological relevance. The presence of sulfatides in tissue of severe PSC cases (Figure 4B, X) could be attributed to small bile ducts that are typically not affected in PSC.35 In contrast, the larger bile ducts appeared to be generally devoid of sulfatides and may have been damaged, as bile salts were found in abundance into the surrounding parenchyma. Thus, bile salts do not colocalize with the sulfatides that are still present in severe PSC.
To determine the generality of the above findings, liver samples from four additional patients with PSC were analyzed and confirmed the presence and localization of 5-CS and G(C)DCA-S in each sample, albeit in varying abundance (Figure S10). To determine whether the observed molecular species can differentiate between healthy human liver tissue and severe PSC liver tissue, ROC analysis was performed. For this, three healthy liver specimens were compared to the three cases of severe PSC where cholestatic liver injury was evident (cases 2, 5, and 6). Liver from the three cases of mild PSC (1, 3, and 4) was also compared with healthy liver, revealing that ROC values for 5-CS, G(C)DCA-S, and the other characteristic ions were approximately 0.5. Thus, it was not possible to differentiate between healthy tissue and cases of mild PSC (data not shown).
G(C)DCA-S at m/z 528.2633 (AUC 0.957) and 5-CS at m/z 531.3000 (AUC 0.835) clearly differentiated between healthy liver tissue and severe PSC liver tissue (Figure S11). The atypical shape of the 5-CS ROC plot is due to the peak picking threshold, and the fact that a lot of pixels in the image have an intensity of zero. This is also apparent from the MALDI-FTICR-MS images of 5-CS (Figure S10), which is observed as small localized patches. In contrast, G(C)DCA-S is distributed throughout the tissue, hence the ROC plot has a later plateau due to the high number of pixels. 5-CS and G(C)DCA-S displayed different spatial distributions in severe PSC (Figure S10, panels F and G), with 5-CS colocalizing with TCA and both seeming to spread into the parenchyma. In contrast, G(C)DCA-S appeared to localize to connective tissue. The mechanistic base for these distinct distributions is unclear. The other examined molecular species (e.g., TCA, T(C)DCA and bilirubin) did not have as much discriminating power (AUC 0.626, 0.721, and 0.568, respectively, Table S4). The molecular species identified in the liver of both mild and severe cases of PSC samples are shown in Tables S2 and 3.
Interspecies Comparison of Molecular Profiles of Healthy Livers
Healthy rat liver was analyzed in the same manner, the resulting MALDI-FTICR-MS images are shown in Figures S12 and S13. A distinct feature of the rodent liver is the synthesis of muricholic acids, that along with TCA are major bile salts in rodent bile. These trihydroxy bile salt species (and taurine-conjugated versions thereof) have an identical molecular mass as cholic acid (and taurine-conjugated versions thereof), and these isomeric species cannot be discriminated using current IMS equipment. Hence, it is most likely that the signal observed at m/z 514.28 corresponds to multiple taurine-conjugated trihydroxy bile salts. The most abundant molecular species observed in the connective tissue, parenchyma, and bile duct of healthy dog and human liver were also observed in healthy rat liver (Tables S2 and S3).
PCA was performed on the MALDI-FTICR-MSI data sets of the healthy liver specimens in order to determine the molecular profiles of each liver region in the animal species studied.40 The top 10 of identified lipid classes were determined for the positive and negative ion mode data sets (Tables S2 and S3, respectively) and classified by the following liver tissue structures: bile duct lumen, bile duct, connective tissue, and parenchyma (Table 1). Localization of the most abundant lipid species for each of these regions is also depicted in Figures 1, 2, and 4. The same molecular classes were observed in the examined structures across all species, the only difference was the pattern of bile salt conjugation. In human and mild PSC liver tissue, both glycine- and taurine-conjugated bile salt species were detected, whereas only taurine-conjugated bile salts were apparent in dog and rat liver. Interestingly, in cases of severe PSC, taurine conjugated bile salts, bilirubin (conjugated/unconjugated) and sulfated bile salts/alcohols were present in the liver. Negative ion mode was far more informative than positive ion mode, as the bile salts and a wide range of lipids are more easily detected in negative ion mode. In addition, the mass spectrum is less complicated as only deprotonated ([M – H]−) species are present, whereas in positive ion mode the spectrum is compounded by protonated species ([M + H]+) and adducts of sodium ([M + Na]+) and potassium ([M + K]+). The positive ion MALDI-FTICR-MS images from healthy dog and human liver, as well as diseased human liver are shown in Figures S4, S6, and S7 respectively. In positive ion mode, a number of yet unknown molecular species (ranging from m/z 544.24–592.24) separated by 16 Da were detected in the bile duct lumen (data not shown). No specific molecular species were observed in the bile duct epithelium in positive ion mode.
Table 1. Summary of Lipid Species Detected in Healthy and Cholestatic Liver Tissuea.
| positive | ||||
|---|---|---|---|---|
| sample | bile duct | bile duct lumen | connective tissue | parenchyma |
| rat | NDb | NAc | PC and SM | PC |
| dog | NDb | NAc | PC and SM | PC |
| human | NDb | NAc | PC and SM | PC |
| mild PSC | NDb | NAc | PC and SM | PC |
| severe PSC | NDb | NAc | PC and SM | PC |
| negative | ||||
|---|---|---|---|---|
| sample | bile duct | bile duct lumen | connective tissue | parenchyma |
| rat | ST and ST–OH | BA (T) | PA, PS, and SM | PA, PE, and PI |
| dog | ST and ST–OH | BA (T) | PA, PS, and SM | PA, PE, and PI |
| human | ST and ST–OH | BA (G/T) | PA, PS, and SM | PA, PE, and PI |
| mild PSC | ST and ST–OH | BA (G/T) | PA, PS, and SM | PA, PE, and PI |
| severe PSC | ST and ST–OH | BA (G/T/S) and BIL | PA, PS, and SM | PA, PE, and PI |
Species in positive ion mode were observed as protonated ([M + H]+ molecule, sodium ([M + Na]+) and potassium ([M + K]+) adducts. Species in negative ion mode were observed as deprotonated ([M – H]−). Key: bile acids (BA), bilirubin (BIL), phosphatidic acid (PA), phosphatidylethanolamine (PE), phosphatidylserine (PS), phosphatidylinositol (PI), phosphatidylcholine (PC), sphingomyelin (SM), sulfatide (ST), and hydroxylated-sulfatide (ST–OH). Glycine (G), taurine (T), and sulfate (S).
No species related to the bile duct were detected in the positive ion mode.
Unknown species present in the bile duct lumen (identification in progress).
Conclusions
Using MALDI-MSI, we identified lipid-specific distributions in different compartments of the liver in all three species (rat, dog, and human), in both positive and negative ion mode. In the studied liver specimens, bile salts were largely confined within the biliary lumen. It will be interesting to examine the spatial distribution of the distinct bile salt species in the setting of bile salt retention as a consequence of intra- or extrahepatic bile duct obstruction. (Hydroxylated)-sulfatides were identified as specific molecular marker for the bile ducts and localized uniquely to bile duct epithelium. Detection of 5-CS and G(C)DCA-S allowed the discrimination between healthy human liver and liver from patients with severe PSC, and these molecules could potentially be used as diagnostic markers. Experimental links with inflammatory events warrants exploration of a possible involvement of sulfatides in inflammatory cholangiopathies like PBC and PSC.
Acknowledgments
This work has been made possible with the financial support of the Dutch province of Limburg as part of the “LINK” program and the Integrated Technology Strategy (ITS) program sponsored by Janssen Pharmaceutica NV. The authors thank Dr. Anjali Röth (Uniklinik RWTH Aachen) and Dr. Monique Verstegen (Erasmus MC, Rotterdam) for providing patient liver specimens, Anne Bruinen (Maastricht University) for providing the whole-body zebra fish tissue sections, and Chantal van Heugten (Maastricht University) and Kristel Buyens (Janssen Pharmaceutica NV) for technical assistance. The authors are indebted to Hang Nguyen (Maastricht University) for proofreading the manuscript.
Supporting Information Available
Figure S1. Information on bile acids. (A) Base structure of bile acids and (B) positions of the hydroxyl groups and site of conjugation. (C) Observed masses of bile acid standards in negative ion mode. Figure S2. MALDI-MS/MS spectra of deprotonated ([M-H]−) bile acid standards in negative ion mode. (A) cholic acid (m/z 407.26), (B) chenodeoxycholic acid (m/z 391.27), (C) deoxycholic acid (m/z 391.27), (D) lithocholic acid (m/z 375.27), (E) taurocholic acid (m/z 514.28), (F) glycocholic acid (m/z 464.30), (G) taurodeoxycholic acid (m/z 498.30), (H) glycodeoxycholic acid (m/z 448.30), (I) taurochenodeoxycholic acid (m/z 498.30), (J) glycochenodeoxycholic acid (m/z 448.30). Figure S3: MALDI-IMS-MS of isomeric bile acids in negative ion mode. Mobilograms of (A) DCA/CDCA (m/z 391.27), (B) TDCA/TCDCA (m/z 498.28) and (C) GDCA/GCDCA (m/z 448.30). Figure S4. Multimodal imaging of healthy dog liver tissue. (I) Optical image of the tissue section prior to matrix application. Positive-ion mode (A) MALDI-MS and (B) MALDI-FTICR-MS images showing the distribution of selected molecular species in the (II) bile duct lumen (unknown at m/z 576.2418), (III) blood vessels (heme [M]+ at m/z 616.1804), (IV) connective tissue ([PC (32:0)+K]+ at m/z 772.5254), (V) parenchyma ([PC (38:4)+K]+ at m/z 848.5564) and (VI) overlay of the selected species. (Designated masses are from MALDI-FTICR-MS imaging measurements, MALDI-MS image field of view 141828 pixels, area 124 mm2 and spatial resolution 50 μm, MALDI-FTICR-MS image field of view 41700 pixels, 9.3 mm2 area and spatial resolution 15 μm. Both images normalized with TIC). Figure S5. MALDI-MS/MS spectra obtained from the most abundant species. MALDI-MS/MS spectra obtained from (A) bile duct lumen, (B) connective tissue, (C) parenchyma and (D) bile duct. Spectra obtained from healthy dog liver tissue. Figure S6. Multimodal imaging of healthy human liver tissue. (I) Optical image of tissue section prior to matrix application. Positive-ion mode (A) MALDI-MS and (B) MALDI-FTICR-MS images showing the distribution of selected molecular species in the (II) bile duct lumen (unknown at m/z 560.2427), (III) blood vessels (heme [M]+ at m/z 616.1782), (IV) connective tissue ([PC (32:0)+K]+ at m/z 772.5254), (V) parenchyma ([PC (34:2)+K]+ at m/z 796.5252) and (VI) overlay of the selected species. (Designated masses are from MALDI-FTICR-MS imaging measurements, MALDI-MS image field of view 72058 pixels, area 110 mm2 and spatial resolution 50 μm, MALDI-FTICR-MS image field of view 22338 pixels, 5 mm2 area and spatial resolution 15 μm. Images normalized with TIC). Figure S7. Multimodal imaging of liver from a patient with mild PSC (case 1). (A) Optical image of tissue section prior to matrix application. Positive-ion mode MALDI-FTICR-MS images showing the distribution of selected molecular species in the (B) bile duct lumen (unknown at m/z 560.2434), (C) heme ([M]+) at m/z 616.1804, (D) connective tissue ([PC (32:0)+K]+ at m/z 772.5251), (E) parenchyma ([PC (34:2)+K]+ at m/z 796.5254) and (F) overlay of the selected species. (Designated masses are from MALDI-FTICR-MS imaging measurements, field of view 20238 pixels, area 10 mm2 and spatial resolution 15 μm. Images normalized with TIC). Figure S8. MALDI-MS/MS spectra of 5-CS and GCDCA-S in negative ion mode. (A) MALDI-MS/MS spectra of 5-CS at m/z 531.28 obtained from severe PSC liver (blue) and whole-body zebrafish (red) tissue sections. (B) MALDI-MS/MS spectra of G(C)DCA-S at m/z 528.24 obtained from pure standard (blue) and severe PSC liver tissue section (red). * indicate peaks coming from unidentified endogenous species tentatively identified as fatty acids. Figure S9. Comparison of immunohistochemical stain and MALDI-MS images from initial patient with severe PSC (case 2). (A) Optical image of tissue section prior to matrix application and (B) cytokeratin staining of a consecutive tissue section. MALDI-MS images showing the distribution of (C) [TCA-H]− at m/z 514.28, (D) 5-cyprinolsulfate ([M-H]−) at m/z 531.30, (E) heme ([M-H]−) at m/z 615.17, (F) [PA (18:0_18:1)-H]− at m/z 701.51, (G) [PI (18:0_20:4)-H]− at m/z 885.55, (H) [ST–OH (18:1_24:0)-H]− at m/z 906.63 and (I) bilirubin diglucuronide ([M-H]−) at m/z 935.32. (J) Overlay of the selected species. (Field of view 81186 pixels, area 8 mm2 and spatial resolution 10 μm. Images normalized with TIC). Figure S10. Multimodal imaging of liver tissue from patients with severe PSC (cases 3–6). (A) Optical image of tissue section prior to matrix application, (B) H&E staining of tissue sections post imaging and (C) cytokeratin staining of a consecutive tissue section. Negative-ion mode MALDI-FTICR-MS images showing the distribution of (D) [TCDCA/TDCA-H]− at m/z 498.2896, (E) [TCA-H]− at m/z 514.2844, (F) [G(C)DCAS-H]− at m/z 528.2639, (G) [5-CS-H]− at m/z 531.2998, (H) [PS (18:0_18:1)-H]− at m/z 788.5449, (I) [PI (18:0_18:2)-H]− at m/z 861.5502, (J) [ST–OH (18:1_24:0)-H]− at m/z 906.6352 and (K) overlay of the selected species. (PSC cases 3−6 fields of view 10356, 25482, 18987, and 21872 pixels and areas 2.3, 5.7, 4.2, and 4.8 mm2 respectively. Spatial resolution of all images 15 μm, normalized with TIC). Figure S11. Receiver operating characteristic (ROC) analysis of (A) G(C)DCA-S at m/z 528.2642 and (B) 5-CS at m/z 531.2999 from healthy liver tissue and tissue from severe PSC cases. Inserts show the relative ion intensity of the displayed ions. Figure S12. Multimodal imaging of healthy rat liver tissue. (A) Optical image of tissue section prior to matrix application. Negative-ion mode MALDI-FTICR-MS images showing the distribution of selected molecular species in the (B) bile duct lumen ([TCA-H]− at m/z 514.2845), (C) connective tissue ([PA (18:0_18:1)-H]− at m/z 701.5129), (D) parenchyma ([PI (18:0_20:4)-H]− at m/z 885.5499), (E) bile duct ([ST–OH (18:1_24:0)-H]− at m/z 906.6348) and (F) overlay of the selected species. (Designated masses are from MALDI-FTICR-MS imaging measurements, field of view 4352 pixels, area 1 mm2 and spatial resolution 15 μm. Images normalized with TIC). Figure S13. Multimodal imaging of healthy rat liver tissue. (A) Optical image of tissue section prior to matrix application. Positive-ion mode MALDI-FTICR-MS images showing the distribution of selected molecular species in the (B) bile duct lumen (unknown at m/z 560.2414), (C) heme ([M]+) at m/z 616.1775, (D) connective tissue ([PC (32:0)+K]+ at m/z 772.5254), (E) parenchyma ([PC (38:4)+K]+ at m/z 848.5566) and (F) overlay of the selected species. (Designated masses are from MALDI-FTICR-MS imaging measurements, field of view 400 pixels, area 3.8 mm2 and spatial resolution 100 μm. Images normalized with TIC). Table S1. Serum biochemistry from all PSC patients in this study. Table S2. Comparison of lipids present in the different areas of the liver across different species in positive ion mode. Table S3. Comparison of lipids present in the different areas of the liver across different species in negative ion mode. Table S4. ROC values for other molecular markers. The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.8b01378.
(PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Hofmann A. F. Ann. Hepatol. 2007, 6, 15–27. [PubMed] [Google Scholar]
- Schaap F. G.; Trauner M.; Jansen P. L. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 55–67. 10.1038/nrgastro.2013.151. [DOI] [PubMed] [Google Scholar]
- Kuipers F.; Bloks V. W.; Groen A. K. Nat. Rev. Endocrinol. 2014, 10, 488–498. 10.1038/nrendo.2014.60. [DOI] [PubMed] [Google Scholar]
- Marin J. J. G.; Macias R. I. R.; Briz O.; Banales J. M.; Monte M. J. Curr. Drug Metab. 2015, 17, 4–29. 10.2174/1389200216666151103115454. [DOI] [PubMed] [Google Scholar]
- Nevens F.; Andreone P.; Mazzella G.; Strasser S. I.; Bowlus C.; Invernizzi P.; Drenth J. P. H.; Pockros P. J.; Regula J.; Beuers U.; Trauner M.; Jones D. E.; Floreani A.; Hohenester S.; Luketic V.; Shiffman M.; Van Erpecum K. J.; Vargas V.; Vincent C.; Hirschfield G. M.; Shah H.; Hansen B.; Lindor K. D.; Marschall H. U.; Kowdley K. V.; Hooshmand-Rad R.; Marmon T.; Sheeron S.; Pencek R.; MacConell L.; Pruzanski M.; Shapiro D. N. Engl. J. Med. 2016, 375, 631–643. 10.1056/NEJMoa1509840. [DOI] [PubMed] [Google Scholar]
- Neuschwander-Tetri B. A.; Loomba R.; Sanyal A. J.; Lavine J. E.; Van Natta M. L.; Abdelmalek M. F.; Chalasani N.; Dasarathy S.; Diehl A. M.; Hameed B.; Kowdley K. V.; McCullough A.; Terrault N.; Clark J. M.; Tonascia J.; Brunt E. M.; Kleiner D. E.; Doo E. Lancet 2015, 385, 956–965. 10.1016/S0140-6736(14)61933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend S. A.; Newsome P. N. Aliment. Pharmacol. Ther. 2017, 46, 494–507. 10.1111/apt.14210. [DOI] [PubMed] [Google Scholar]
- Jansen P. L.; Ghallab A.; Vartak N.; Reif R.; Schaap F. G.; Hampe J.; Hengstler J. G. Hepatology 2017, 65, 722–738. 10.1002/hep.28965. [DOI] [PubMed] [Google Scholar]
- Perez M. J.; Briz O. World J. Gastroenterol. 2009, 15, 1677–1689. 10.3748/wjg.15.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S. Y.; Ouyang X.; Chen Y.; Soroka C. J.; Wang J.; Mennone A.; Wang Y.; Mehal W. Z.; Jain D.; Boyer J. L. JCI Insight 2017, 2, e90780 10.1172/jci.insight.90780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinken M. Chem. Res. Toxicol. 2015, 28, 1391–1397. 10.1021/acs.chemrestox.5b00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt R.; Matz-Soja M. World J. Gastroenterol. 2014, 20, 8491–8504. 10.3748/wjg.v20.i26.8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogrinc-Potocnik N.; Porta T.; Becker M.; Heeren R. M. A.; Ellis S. R. Rapid Commun. Mass Spectrom. 2015, 29, 2195–2203. 10.1002/rcm.7379. [DOI] [PubMed] [Google Scholar]
- Strohalm M.; Kavan D.; Novak P.; Volny M.; Havlicek V. Anal. Chem. 2010, 82, 4648–4651. 10.1021/ac100818g. [DOI] [PubMed] [Google Scholar]
- Pauling J. K.; Hermansson M.; Hartler J.; Christiansen K.; Gallego S. F.; Peng B.; Ahrends R.; Ejsing C. S. PLoS One 2017, 12, e0188394. 10.1371/journal.pone.0188394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher G. L.; Bruinen A. L.; Ogrinc Potocnik N.; Hammond J. S.; Bryan S. R.; Larson P. E.; Heeren R. M. A. Anal. Chem. 2016, 88, 6433–6440. 10.1021/acs.analchem.6b01022. [DOI] [PubMed] [Google Scholar]
- Eijkel G. B.; Kükrer Kaletaş B.; Van der Wiel I. M.; Kros J. M.; Luider T. M.; Heeren R. M. A. Surf. Interface Anal. 2009, 41, 675–685. 10.1002/sia.3088. [DOI] [Google Scholar]
- Mims D.; Hercules D. Anal. Bioanal. Chem. 2003, 375, 609–616. 10.1007/s00216-003-1771-y. [DOI] [PubMed] [Google Scholar]
- Mims D.; Hercules D. Anal. Bioanal. Chem. 2004, 378, 1322–1326. 10.1007/s00216-003-2475-z. [DOI] [PubMed] [Google Scholar]
- Stroobant V.; de Hoffmann E.; Libert R.; Van Hoof F. J. Am. Soc. Mass Spectrom. 1995, 6, 588–596. 10.1016/1044-0305(95)00203-P. [DOI] [PubMed] [Google Scholar]
- Qiao X.; Ye M.; Liu C. F.; Yang W. Z.; Miao W. J.; Dong J.; Guo D. A. Steroids 2012, 77, 204–211. 10.1016/j.steroids.2011.11.008. [DOI] [PubMed] [Google Scholar]
- Scherer M.; Gnewuch C.; Schmitz G.; Liebisch G. J. Chromatogr. B: Anal. Technol. Biomed. Life Sci. 2009, 877, 3920–3925. 10.1016/j.jchromb.2009.09.038. [DOI] [PubMed] [Google Scholar]
- Schadt H. S.; Wolf A.; Pognan F.; Chibout S. D.; Merz M.; Kullak-Ublick G. A. Clin. Res. Hepatol. Gastroenterol. 2016, 40, 257–266. 10.1016/j.clinre.2015.12.017. [DOI] [PubMed] [Google Scholar]
- Wildgrube H. J.; Stockhausen H.; Petri J.; Füssel U.; Lauer H. J. Chromatogr. 1986, 353, 207–213. 10.1016/S0021-9673(01)87090-4. [DOI] [PubMed] [Google Scholar]
- Strazzabosco M.; Fabris L. Anat. Rec. 2008, 291, 653–660. 10.1002/ar.20664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao X.; Song W.; Lin X. H.; Wang Q.; Bo T.; Guo D. A.; Liu J.; Ye M. Anal. Methods 2014, 6, 596–601. 10.1039/C3AY41605D. [DOI] [Google Scholar]
- Löhmann C.; Schachmann E.; Dandekar T.; Villmann C.; Becker C. M. J. Neurochem. 2010, 114, 1119–1134. 10.1111/j.1471-4159.2010.06836.x. [DOI] [PubMed] [Google Scholar]
- Cerruti C. D.; Benabdellah F.; Lapŕevote O.; Touboul D.; Brunelle A. Anal. Chem. 2012, 84, 2164–2171. 10.1021/ac2025317. [DOI] [PubMed] [Google Scholar]
- Takahashi T.; Suzuki T. J. Lipid Res. 2012, 53, 1437–1450. 10.1194/jlr.R026682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H.; Sun G.; Yang K.; Gross R. G.; Han X. J. Lipid Res. 2010, 51, 1599–1609. 10.1194/jlr.D004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda G.; Ikeda Y.; Kashiwagi M.; Iwamori M.; Oka H. Hepatology 1990, 12, 664–670. 10.1002/hep.1840120408. [DOI] [PubMed] [Google Scholar]
- Kumar V. J. Hepatol. 2013, 59, 618–620. 10.1016/j.jhep.2013.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maricic I.; Sheng H.; Marrero I.; Seki E.; Kisseleva T.; Chaturvedi S.; Molle N.; Mathews S. A.; Gao B.; Kumar V. Hepatology 2015, 61, 1357–1369. 10.1002/hep.27632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder R. C.; Aguilera C.; Maricic I.; Kumar V. J. Clin. Invest. 2007, 117, 2302–2312. 10.1172/JCI31602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschfield G. M.; Karlsen T. H.; Lindor K. D.; Adams D. H. Lancet 2013, 382, 1587–1599. 10.1016/S0140-6736(13)60096-3. [DOI] [PubMed] [Google Scholar]
- Fickert P.; Fuchsbichler A.; Wagner M.; Zollner G.; Kaser A.; Tilg H.; Krause R.; Lammert F.; Langner C.; Zatloukal K.; Marschall H. U.; Denk H.; Trauner M. Gastroenterology 2004, 127, 261–274. 10.1053/j.gastro.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Hofmann A. F.; Hagey L. R.; Krasowski M. D. J. Lipid Res. 2010, 51, 226–246. 10.1194/jlr.R000042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M.; Une M.; Takenaka S.; Tazawa Y.; Nozaki S.; Imanaka T.; Kuramoto T. Clin. Chim. Acta 2001, 314, 101–106. 10.1016/S0009-8981(01)00636-2. [DOI] [PubMed] [Google Scholar]
- Alnouti Y. Toxicol. Sci. 2009, 108, 225–246. 10.1093/toxsci/kfn268. [DOI] [PubMed] [Google Scholar]
- Klerk L. A.; Broersen A.; Fletcher I. W.; van Liere R.; Heeren R. M. A. Int. J. Mass Spectrom. 2007, 260, 222–236. 10.1016/j.ijms.2006.11.014. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.