Antibiotic resistance is a global problem. This study, conducted in rural western Uganda, describes antibiotic resistance patterns in Escherichia coli bacteria near two forested national parks. Resistance was present not only in people, but also in their livestock and in nearby wild nonhuman primates. Multidrug resistance and class 1 integrons containing genes that confer resistance were common and were similar in people and animals. The percentage of resistant isolates decreased with increasing local price of the antibiotic. Antibiotic resistance in this setting likely reflects environmental diffusion of bacteria or their genes, perhaps facilitated by local ecological and socioeconomic conditions.
KEYWORDS: antibiotic resistance, Escherichia coli, Africa, protected areas, primates, class 1 integrons
ABSTRACT
Antibiotic resistance is a global concern, although it has been studied most extensively in developed countries. We studied Escherichia coli and class 1 integrons in western Uganda by analyzing 1,685 isolates from people, domestic animals, and wild nonhuman primates near two national parks. Overall, 499 isolates (29.6%) were resistant to at least one of 11 antibiotics tested. The frequency of resistance reached 20.3% of isolates for trimethoprim-sulfamethoxazole but was nearly zero for the less commonly available antibiotics ciprofloxacin (0.4%), gentamicin (0.2%), and ceftiofur (0.1%). The frequency of resistance was 57.4% in isolates from people, 19.5% in isolates from domestic animals, and 16.3% in isolates from wild nonhuman primates. Isolates of livestock and primate origin displayed multidrug resistance patterns identical to those of human-origin isolates. The percentage of resistant isolates in people was higher near Kibale National Park (64.3%) than near Bwindi Impenetrable National Park (34.6%), perhaps reflecting local socioeconomic or ecological conditions. Across antibiotics, resistance correlated negatively with the local price of the antibiotic, with the most expensive antibiotics (nalidixic acid and ciprofloxacin) showing near-zero resistance. Among phenotypically resistant isolates, 33.2% harbored class 1 integrons containing 11 common resistance genes arranged into nine distinct gene cassettes, five of which were present in isolates from multiple host species. Overall, these results show that phenotypic resistance and class 1 integrons are distributed broadly among E. coli isolates from different host species in this region, where local socioeconomic and ecological conditions may facilitate widespread diffusion of bacteria or resistance-conferring genetic elements.
IMPORTANCE Antibiotic resistance is a global problem. This study, conducted in rural western Uganda, describes antibiotic resistance patterns in Escherichia coli bacteria near two forested national parks. Resistance was present not only in people, but also in their livestock and in nearby wild nonhuman primates. Multidrug resistance and class 1 integrons containing genes that confer resistance were common and were similar in people and animals. The percentage of resistant isolates decreased with increasing local price of the antibiotic. Antibiotic resistance in this setting likely reflects environmental diffusion of bacteria or their genes, perhaps facilitated by local ecological and socioeconomic conditions.
INTRODUCTION
Antibiotic resistance is ubiquitous in the developing world (1–3). In some African countries, resistance to commonly available antibiotics can exceed 75% (3–9). Cost, ease of use, and time on the market influence the use of antibiotics in Africa and the consequent resistance of bacteria to those antibiotics (10). Antibiotic resistance is also ubiquitous in African livestock, deriving from diverse genetic mechanisms of resistance in common bacteria, such as Escherichia coli (11).
Most reports of antibiotic resistance in Africa are from studies of urban populations, often in settings such as hospitals and schools (3, 9, 12). However, patterns and drivers of resistance may be different in rural settings, where the majority of sub-Saharan Africa's population lives (13) and where people experience higher disease burdens and perhaps increased need for antibiotics (12). Additional factors, such as unregulated use and counterfeit antibiotics, may compound the development and maintenance of resistance in such settings (14, 15). Moreover, studies that consider both animals and humans as contributors to antibiotic resistance in low-resource settings are rare but needed (16).
We conducted a study of antibiotic resistance and class 1 integrons, which are horizontally transmissible genetic elements that may contain resistance-conferring genes (17), in E. coli bacteria from rural western Uganda. We sampled not only people and domestic animals, but also wild nonhuman primates, the habitats of which overlap extensively with that of people and livestock in this region, with documented consequences for environmentally mediated bacterial transmission (18–21). Antibiotic use in the region occurs in people and, to a lesser extent, in domestic animals, but it is almost entirely absent in the region's wild primates (18–21). The setting therefore offers an opportunity to examine resistance and its dissemination in a context where extensive ecological overlap occurs among host species, but where patterns of use differ among these species.
RESULTS
A total of 485 E. coli isolates were collected and analyzed from 202 people in rural western Uganda near two national parks (Fig. 1). The median age of participants was 14 years (range, <1 year to 77 years), with most either 6 to 10 years old (21.8%; n = 44) or 21 to 49 years old (21.8%; n = 44), and the population consisted mainly of subsistence farmers of the Batooro tribe, who keep small herds of domestic livestock (cattle, goats, pigs, and sheep). People in this population report frequent interactions with wild primates, especially in locations where forest habitats occur near agricultural lands (22, 23). A total of 37 (18.3%) participants reported taking an antibiotic within 4 weeks prior to donating a fecal sample, but the identity of the antibiotic taken could be determined in only three (8.1%) of these cases.
FIG 1.
Map of Uganda (in East Africa) showing the locations of Kibale National Park (Kibale) and Bwindi Impenetrable National Park (Bwindi).
Overall, 155 (76.7%) people harbored E. coli isolates resistant to at least one antibiotic, and 119 (58.9%) E. coli isolates were resistant to at least two antibiotics. The proportion of people harboring at least one resistant isolate was highest for trimethoprim-sulfamethoxazole (61.9%; n = 125), followed by streptomycin (48.5%; n = 98) and tetracycline (47.5%; n = 96). The proportion of individuals harboring at least one resistant isolate was lowest for ciprofloxacin (1.0%; n = 2), nalidixic acid (2.0%; n = 4), and gentamicin (0.5%; n = 1). This measure of resistance was higher in the youngest (0 to 6 years) and oldest (40 to 77 years) age categories than in the intermediate age categories (adjusted odds ratios of 4.37 [1.40 to 13.60] and 7.53 [1.94 to 29.26] and P values of 0.01 and 0.004, respectively) and nearly twice as high in people living near Kibale as in people living near Bwindi (64.3% versus 34.6%, respectively; Fisher's exact test, P < 0.001). Across all antibiotics, there was a statistically significant and nonlinear decrease in the proportion of resistant isolates with increasing local antibiotic prices (r2 = 0.68; t = −3.8; P = 0.007; Fig. 2), with the most expensive antibiotics (nalidixic acid and ciprofloxacin) associated with very low resistance proportions (0.8% and 0.4% of isolates, respectively).
FIG 2.
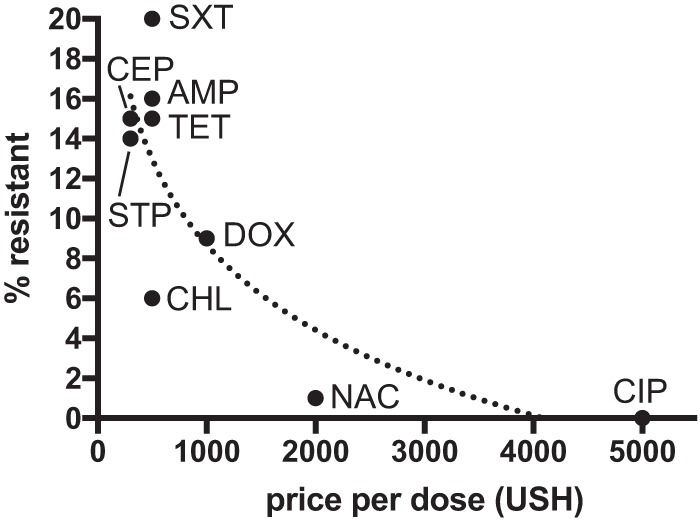
Antibiotic resistance versus price. The plot shows the percentages of 485 human-origin isolates resistant to nine antibiotics available in western Uganda for which local price information (Ugandan shillings [USH] per dose) was also available. Abbreviations for antibiotics are shown beside data points. AMP, ampicillin; CEP, cephalothin; CIP, ciprofloxacin; CHL, chloramphenicol; DOX, doxycycline; NAC, nalidixic acid; STP, streptomycin; SXT, sulfamethoxazole-trimethoprim; and TET, tetracycline.
A total of 1,200 E. coli isolates from 259 animals were analyzed (Table 1). Overall, the proportion of resistant isolates was approximately three times lower in animals than in people (17.7% and 57.4%, respectively; Fisher's exact test, P < 0.001; Table 1). Within and across both locations, isolates from domestic animals and wild primates showed statistically indistinguishable proportions of resistance (19.5% versus 16.3% of isolates, respectively; Fisher's exact test, P = 0.1468). The most frequent patterns of multiple resistance in human-derived isolates also occurred with the highest frequency in animals (Table 2). Among primates, the red-tailed monkey (Cercopithecus ascanius) in Kibale harbored isolates with nearly four times higher resistance rates than those in other primate species (Table 1).
TABLE 1.
Frequency of antibiotic resistance in E. coli isolates from people, livestock, and wild primates in western Ugandaa
| Antibioticb | % of resistant isolates |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bwindi |
Kibale |
All (n = 1,685) | |||||||||||
| HU (n = 112) | CA (n = 12) | GT (n = 127) | GO (n = 252) | HU (n = 373) | CA (n = 38) | GT (n = 318) | DO (n = 17) | BW (n = 124) | RC (n = 134) | RT (n = 61) | CZ (n = 117) | ||
| AMP | 20.1 | 50.0 | 5.5 | 8.7 | 37.1 | 10.5 | 5.3 | 29.4 | 0.0 | 0.8 | 0.0 | 6.8 | 19.2 |
| CHL | 14.5 | 0.0 | 0.8 | 0.0 | 15.5 | 2.6 | 2.2 | 23.5 | 0.8 | 0.0 | 1.6 | 0.0 | 6.4 |
| CIP | 0.0 | 0.0 | 0.0 | 0.0 | 1.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.4 |
| DOX | 7.6 | 33.3 | 2.4 | 0.4 | 24.4 | 0.0 | 2.8 | 0.0 | 0.0 | 0.0 | 3.3 | 0.0 | 8.5 |
| TET | 21.4 | 41.7 | 3.9 | 0.4 | 39.3 | 2.6 | 6.3 | 35.3 | 0.8 | 0.0 | 8.2 | 0.0 | 15.1 |
| GEN | 0.6 | 0.0 | 0.0 | 0.0 | 0.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 |
| STP | 23.3 | 41.7 | 4.7 | 1.2 | 36.3 | 2.6 | 5.3 | 29.4 | 0.0 | 0.0 | 0.0 | 1.7 | 14.2 |
| SXT | 28.9 | 41.7 | 6.3 | 2.0 | 52.9 | 2.6 | 6.3 | 35.3 | 0.8 | 0.0 | 9.8 | 4.3 | 20.3 |
| NAC | 2.5 | 0.0 | 1.0 | 0.0 | 1.7 | 0.0 | 0.0 | 0.0 | 0.8 | 0.0 | 0.0 | 0.0 | 0.8 |
| CEP | 8.8 | 41.7 | 20.0 | 12.7 | 24.8 | 18.4 | 6.6 | 5.9 | 6.5 | 11.9 | 36.1 | 8.6 | 15.3 |
| XNL | 0.6 | 0.0 | 0.0 | 0.4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.1 |
| Anyc | 35.6 | 58.3 | 26.8 | 17.5 | 64.3 | 18.5 | 15.5 | 35.3 | 8.1 | 11.9 | 44.3 | 12.8 | 29.6 |
Values in parentheses indicate sample sizes of isolates from each population. Values in the table indicate percentages of resistant isolates. HU, human; CA, cattle; GT, goat/sheep; GO, mountain gorilla; DO, dog; BW, black-and-white colobus; RC, red colobus; RT, red-tailed guenon; and CZ, chimpanzee.
AMP, ampicillin; CHL, chloramphenicol; CIP, ciprofloxacin; DOX, doxycycline; TET, tetracycline; GEN, gentamicin; STP, streptomycin; SXT, sulfamethoxazole-trimethoprim; NAC, naladixic acid; CEP, cephalothin; and XNL, ceftiofur.
Frequency of resistance to any of the antibiotics tested.
TABLE 2.
Frequency of multiple antibiotic resistance patterns among E. coli isolates from people, livestock, and wild primates in western Ugandaa
| Patternb | % of isolates with antibiotic resistance pattern |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bwindi |
Kibale |
All (n = 379) | |||||||||||
| HU (n = 43) | CA (n = 5) | GT (n = 8) | GO (n = 17) | HU (n = 259) | DO (n = 6) | CA (n = 4) | GT (n = 24) | BW (n = 1) | RC (n = 1) | RT (n = 5) | CZ (n = 6) | ||
| AMST | 1.06 | 0.26 | 0.26 | 0 | 7.92 | 0.26 | 0 | 1.32 | 0 | 0 | 0 | 0 | 11.08 |
| ADMST | 1.32 | 0 | 0 | 0 | 6.60 | 0 | 0 | 0.26 | 0 | 0 | 0 | 0 | 8.18 |
| AP | 0 | 0 | 0.53 | 3.96 | 0.53 | 0 | 0.79 | 0 | 0 | 0.26 | 0 | 1.06 | 7.12 |
| ACMST | 1.58 | 0 | 0 | 0 | 3.96 | 0.79 | 0 | 0.53 | 0 | 0 | 0 | 0 | 6.86 |
| ACMPST | 1.06 | 0 | 0 | 0 | 4.49 | 0 | 0.26 | 0 | 0 | 0 | 0 | 0 | 5.80 |
| ADMPST | 0.26 | 1.06 | 0 | 0.26 | 3.69 | 0 | 0 | 0.53 | 0 | 0 | 0 | 0 | 5.80 |
| AMPS | 0.53 | 0 | 0 | 0 | 2.90 | 0 | 0 | 0.26 | 0 | 0 | 0 | 0.53 | 4.22 |
| DMT | 0 | 0 | 0 | 0 | 3.17 | 0 | 0 | 0.79 | 0 | 0 | 0.26 | 0 | 4.22 |
| ACDMST | 0.53 | 0 | 0 | 0 | 3.43 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.96 |
| ACDMPST | 0 | 0 | 0 | 0 | 3.69 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.69 |
| DMST | 0.26 | 0 | 0.79 | 0 | 2.64 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.69 |
| DT | 0 | 0 | 0 | 0 | 3.17 | 0 | 0 | 0.26 | 0 | 0 | 0 | 0 | 3.43 |
| ACMPS | 0.26 | 0 | 0 | 0 | 2.90 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.17 |
| MT | 0.26 | 0 | 0.26 | 0 | 2.11 | 0 | 0 | 0 | 0 | 0 | 0.53 | 0 | 3.17 |
| MP | 0.26 | 0 | 0 | 0 | 1.85 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2.11 |
| AMS | 0.26 | 0 | 0 | 0 | 1.06 | 0 | 0 | 0.26 | 0 | 0 | 0 | 0 | 1.58 |
| MS | 0 | 0 | 0 | 0 | 1.58 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.58 |
| ACDMPT | 0 | 0 | 0 | 0 | 1.06 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.06 |
| AMP | 0 | 0 | 0 | 0 | 1.06 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.06 |
| AMT | 0 | 0 | 0 | 0 | 1.06 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.06 |
| Otherc | 3.69 | 0 | 0.26 | 0.26 | 9.50 | 0.53 | 0 | 2.11 | 0.26 | 0 | 0.53 | 0 | 17.15 |
| Anyd | 11.35 | 1.32 | 2.11 | 4.49 | 68.34 | 1.58 | 1.06 | 6.33 | 0.26 | 0.26 | 1.32 | 1.58 | 100 |
Values in parentheses indicate sample sizes of multiply resistant E. coli isolates. Values in the table indicate percentages of isolates showing the specified pattern. Only common patterns (present in 1% or more of 379 multiply resistant isolates) are specified. HU, human; CA, cattle; GT, goat/sheep; GO, mountain gorilla; DO, dog; BW, black-and-white colobus; RC, red colobus; RT, red-tailed guenon; and CZ, chimpanzee.
A, ampicillin; C, chloramphenicol; D, doxycycline; M, sulfamethoxazole-trimethoprim; P, cephalothin; S, streptomycin; and T, tetracycline.
Thirty-seven other unique patterns occurred with a frequency of <1%. Additional antibiotics in these patterns were ciprofloxacin, gentamicin, and naladixic acid.
Frequency of any multiple-resistance pattern (i.e., resistance to ≥2 antibiotics).
Class 1 integrons were detected in 33.2% of phenotypically resistant E. coli isolates (Table 3). Class 1 integrons were present in 42.9% of isolates from humans but in only 15.4% of isolates from animals (Fisher's exact test, P < 0.001). The proportion of isolates containing class 1 integrons was higher in people living near Bwindi than in people living near Kibale (61.2% versus 39.5%, respectively; Fisher's exact test, P = 0.0073). Class 1 integrons were present in isolates from domestic animals at a frequency of 26.9%, which was 3.5 times higher than the frequency of class 1 integrons in isolates from wild primates (7.7%; Fisher's exact test, P = 0.0007).
TABLE 3.
Frequency of class 1 integrons in phenotypically resistant E. coli isolates from people, livestock, and wild primates in western Ugandaa
| Integronb | Cassette | % of isolates containing integron cassette |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bwindi |
Kibale |
All (n = 499) | ||||||||||||
| HU (n = 48) | CA (n = 3) | GT (n = 27) | GO (n = 37) | HU (n = 276) | DO (n = 0) | CA (n = 8) | GT (n = 40) | BW (n = 11) | RC (n = 11) | RT (n = 25) | CZ (n = 13) | |||
| A | dfrA17 | 33.3 | 66.7 | 14.8 | 0.0 | 24.3 | 0.0 | 12.5 | 10.0 | 0.0 | 0.0 | 0.0 | 0.0 | 18.8 |
| B | dfrA1 aadA1 | 4.2 | 0.0 | 7.4 | 2.7 | 3.6 | 0.0 | 0.0 | 10.0 | 9.1 | 0.0 | 16.0 | 0.0 | 4.8 |
| C | dfrA5 | 8.4 | 0.0 | 0.0 | 0.0 | 2.2 | 0.0 | 0.0 | 5.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.4 |
| D | dfrA17 aadA5 | 8.4 | 0.0 | 0.0 | 0.0 | 2.9 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.4 |
| E | dfrA15 aadA1 | 2.1 | 0.0 | 0.0 | 0.0 | 2.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.6 |
| F | aadA1 | 6.3 | 0.0 | 0.0 | 0.0 | 1.1 | 0.0 | 0.0 | 2.5 | 0.0 | 0.0 | 0.0 | 0.0 | 1.4 |
| G | blaOXA-1 aadA2 | 0.0 | 0.0 | 0.0 | 0.0 | 2.2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.2 |
| H | dfrA1 catB3 aacA4 | 0.0 | 0.0 | 0.0 | 0.0 | 0.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.4 |
| I | dfrA12 aadA2 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.5 | 0.0 | 0.0 | 0.0 | 0.0 | 0.2 |
| Allc | 62.7 | 66.7 | 22.2 | 2.7 | 39.5 | 0.0 | 12.5 | 30.0 | 9.1 | 0.0 | 16.0 | 0.0 | 33.3 | |
Values in parentheses indicate numbers of phenotypically resistant (multiple and single resistance) E. coli isolates tested for class 1 integrons using PCR. Values in the table indicate percentages of such isolates containing the specified integron cassette. HU, human; CA, cattle; GT, goat/sheep; GO, mountain gorilla; DO, dog; BW, black-and-white colobus; RC, red colobus; RT, red-tailed guenon; CZ, chimpanzee. Complete integron cassette sequences are available in GenBank (accession numbers MH560801 to MH560966).
Nine E. coli isolates, all from humans, contained two class 1 integrons, i.e., four isolates containing integrons C and F, two isolates containing integrons A and B, two isolates containing integrons A and D, and one isolate containing integrons A and F.
Frequency of all class 1 integrons combined.
The 166 class 1 integrons detected by PCR contained nine distinct gene cassettes comprised of different combinations of 11 common resistance-conferring genes, ranging in frequency from 0.2% to 18.8% (Table 3; GenBank accession numbers MH560801 to MH560966). Of these 11 genes, five encoded dihydrofolate reductase (dfrA) (24), three encoded aminoglycoside adenyltransferase (aadA) (25), one encoded class D beta-lactamase (blaOXA) (26), one encoded chloramphenicol acetyltransferase (catB) (27), and one encoded aminoglycoside N(6′)-acetyltransferase (aacA) (28). Eight distinct gene cassettes were present in isolates from humans, five were present in isolates from domestic animals, and one was present in isolates from primates (Table 3).
DISCUSSION
Our results highlight the ubiquity of antibiotic resistance in people in the developing world, including in Africa (1–3), and they offer insights into carriage of resistance and resistance-conferring genes in animals. In the human populations in our study, approximately 20% of E. coli isolates were resistant to the commonly available antibiotic combination sulfamethoxazole-trimethoprim, with approximately 15% of isolates resistant to ampicillin, tetracycline, doxycycline, streptomycin, sulfamethoxazole-trimethoprim, and cephalothin. In contrast, the proportion of isolates resistant to ciprofloxacin, gentamicin, and nalidixic acid was less than 1%, and ceftiofur, a veterinary antibiotic not available in Uganda at the time of the study, showed near-zero resistance.
Because accurate data on antibiotic use were not available, we cannot attribute differences in resistance directly to differences in use of antibiotics. However, our observation that the proportion of resistant isolates decreases with increasing local antibiotic price per dose is suggestive. In western Uganda, as in many similar settings, some antibiotics are available over the counter from local clinics and dispensaries, often without the oversight of a physician (29, 30). Compounding this problem are incomplete dosing and a ubiquity of counterfeit, substandard, and degraded drugs (14). We speculate that such local economic conditions influence antibiotic use, which might in turn drive resistance. We caution that route of administration, mechanism of action, availability, clinical effects/side effects, and many other factors may have contributed to the trend observed and that antibiotic consumption is increasing globally (31). We encourage further research into the economic drivers of antibiotic use in developing countries, such as Uganda and elsewhere, and the associated cofactors and confounders that may contribute to the phenomenon.
Our results confirm previous findings that, within the commonly accessible human antibiotics assessed, people and animals harbor bacteria with phenotypically similar (often identical) antibiotic resistance profiles (18–20). In animals, the proportion of resistant isolates was approximately three times lower than that in humans, with isolates from domestic animals and wild primates showing comparable proportions of resistant isolates. The most frequent patterns of multiple resistance in human-derived E. coli isolates were also the most frequent patterns of multiple resistance in E. coli isolates from both livestock and wild primates. Five of the nine class 1 integron gene cassettes present in phenotypically resistant E. coli isolates were present in isolates from either livestock or wild primates (Table 3). These results point to a ubiquity of resistant bacteria and resistance-conferring genes in this setting, regardless of the host species from which the bacteria were derived. Our results show that such patterns vary geographically. The frequency of resistant isolates was lower in people living near Bwindi than in people living near Kibale, with the frequency of class 1 integrons showing the reverse pattern. The reasons for these differences are not clear but could reflect local variation in ecology or socioeconomics.
The mechanisms by which resistant bacteria or resistance-conferring genes diffuse across the landscape and among species in western Uganda are not clear. Our previous work has documented that rates of enteric bacterial transmission between species in western Uganda are enhanced where ecological overlap is high because of such factors as ecotourism (21, 32), forest fragmentation (20), or animal husbandry practices (19). In these cases, environmental contamination appears to be the likeliest mode of transmission. For example, water sources in our study communities tended to be unprotected wells used by both people and livestock, situated at the edges of forest habitats containing primates. Similarly, livestock tended to be grazed in pastures near agricultural fields frequented by people, where primates often raid crops. Fecal contamination of soil, vegetation, and water as a result of shared activity spaces might therefore explain the patterns observed (22, 23). Such effects could also explain the higher proportions of resistant isolates in red-tailed guenons than in other sympatric primates; red-tailed guenons are notorious for entering agricultural lands to raid crops (33).
Even though we studied commensal E. coli, horizontal gene transfer might reduce the efficacy of antibiotics for pathogenic bacteria. We note that the antibiotic for which we found the highest resistance, sulfamethoxazole-trimethoprim, has been routinely administered prophylactically to patients infected with human immunodeficiency virus (HIV), which substantially burdens western Uganda (34, 35). Our previous studies (21, 32) show that ape populations used for tourism are at particular risk for the acquisition of antibiotic-resistant E. coli. In the present study, we found only two E. coli isolates that were resistant to ceftiofur, an extended-spectrum veterinary cephalosporin not available in Uganda at the time of the study; these isolates were from an adult male silverback gorilla in Bwindi and an adult male human who was employed to guide tourists to these gorillas. Such observations, although anecdotal, draw attention to potential ecological pathways of transmission.
Overall, our results demonstrate broad diffusion of antibiotic-resistant bacteria and resistance-conferring genetic elements among locations and host species in rural Uganda. For class 1 integrons (which are only one of many types of genetic elements that often, but not always, contain resistance-conferring genes [17]), our results show their composition to be entirely similar to what has been documented worldwide, suggesting either widespread dispersal of such genes or an inherently limited repertoire of class 1 integron cassettes in E. coli. Our results also suggest that future research into reservoirs of resistance, including water, crops, soil, food, and other features of the physical environment where gastrointestinal bacteria and their genes are deposited and persist, would be fruitful (36). Similarly, our results suggest that local economic factors may influence patterns of resistance, and this too merits further study, especially as agriculture in Uganda and other African countries intensifies (37). To the extent that our findings can be generalized, they illustrate the environmental ubiquity of resistant bacteria and their genes in the developing world, including how ecological and socioeconomic conditions might influence spread and persistence.
MATERIALS AND METHODS
The study took place in rural western and southwestern Uganda in two national parks and the communities surrounding them, Kibale National Park and Bwindi Impenetrable National Park (Fig. 1). Kibale is a 795-km2 forested park near the foothills of the Rwenzori Mountains (0°13′ to 0°41′ N, 30°19′ to 30°32′ E) (38) containing a high diversity and density of primates, including Uganda's largest population of chimpanzees (Pan troglodytes schweinfurthii), which form the focus of a thriving ecotourism industry (39). Bwindi (0° 53′ to 1° 08′ N, 29° 35′ to 29° 50′ E) encompasses 331 km2 of predominantly montane forest and is home to approximately 45% of the world's remaining mountain gorillas (Gorilla beringei beringei), also representing a popular tourist destination (40).
We enrolled volunteers from March through December 2005 in communities surrounding each park. All volunteers were asked to provide a fecal sample in a sterile cup, which was collected within 24 h. From May to August 2005, we also collected fecal samples from domestic animals and wild nonhuman primates in and near the parks, following methodologies described elsewhere (19, 20). At the time of sample collection, we recorded local antibiotic prices per dose during visits to local pharmacies. Survey data about health, medication, livelihoods, and contact with animals were collected and have been reported previously (18–21, 32).
E. coli was cultured from fecal samples, and up to five isolates per individual were tested for resistance to 11 different antibiotics using the disk diffusion method, following previously described methods (20, 21). Briefly, fecal samples were streaked for isolation on MacConkey agar, putative E. coli isolates were collected and confirmed using standard biochemical tests, and confirmed E. coli isolates were reisolated and stored frozen in glycerol stocks for further testing. Resistance to antibiotics was assessed using the disk diffusion method following Clinical and Laboratory Standards Institute (CLSI) protocols (41), as previously described (20, 21), with resistance cutoffs following CLSI guidelines (41). Antibiotics included in the analysis were as follows: ampicillin (AMP), chloramphenicol (CHL), ciprofloxacin (CIP), doxycycline (DOX), tetracycline (TET), gentamicin (GEN), streptomycin (STP), trimethoprim sulfate (SXT), nalidixic acid (NAC), cephalothin (CEP), and ceftiofur (XNL).
PCR was used to detect class 1 integrons using published methods (42). Specifically, reactions were performed in 25-μl volumes containing 2 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate, 0.4 μM primer 5′-CS (5′-GGCATCCAAGCAGCAAGC-3′) and primer 3′-CS (5′-AAGCAGACTTGACCTGAT-3′), and 0.25 U DyNAzyme EXT DNA polymerase (Finnzymes; Espoo, Finland) with 1× DyNAzyme EXT buffer (without MgCl2) and 2.5 μl template. Reactions were cycled in an iCycler thermocycler (Bio-Rad; Hercules, CA) at 94°C for 3 min, followed by 30 cycles at 94°C for 30 s, 62°C for 30 s, and 72°C for 5 min, and ending with a 72°C final extension step for 7 min and an indefinite 4°C soak. To control for PCR failure, a separate positive-control PCR was performed on each isolate using primers complementary to positions 9 to 27 of the 16S rRNA gene (16S rRNA Forward 5′-GAGTTTGATCCTGGCTCA-3′) and positions 2669 to 2654 of the 23S rRNA gene (23S rRNA Reverse 5′-CCGGTCCTCTCGTACT-3′) under the same conditions as the class 1 integron PCR, except with an annealing temperature of 60°C (43). Isolates were reextracted and retested if the positive control PCR failed. Isolates were considered to be negative for a class 1 integron only if they yielded a 16S rRNA amplicon but did not yield an integron amplicon.
PCR products were electrophoresed at 100 V for 30 to 45 min on 1% agarose gels containing 0.5 μg/ml ethidium bromide in Tris-acetate-EDTA (TAE) buffer. Gels were visualized under UV light, and all putative class 1 integron bands were excised for DNA extraction. DNA was recovered using the Zymoclean gel DNA recovery kit (Zymo Research; Orange, CA). Purified amplicons were sequenced directly on ABI 3730XL capillary sequencers. Sequences were edited by hand and PCR products were resequenced to resolve any ambiguous bases. Nucleotide BLAST searches (44) were performed to determine the gene(s) present in each integron cassette based on identity to known sequences. Frequencies of resistance, multiple resistance, and class 1 integrons were compared among populations using statistical methods implemented in the computer programs SPSS version 17.0 (IBM Corp., Armonk, NY) and SAS version 9.3 (SAS Institute, Cary, NC) with a Bonferroni correction implemented to account for multiple comparisons. Frequencies of antibiotic resistance were compared to local antibiotic prices using nonlinear regression.
All protocols were reviewed and approved by the Uganda National Council for Science and Technology, the Uganda Wildlife Authority, and the Institutional Review Board and Institutional Animal Care and Use Committees of the University of Illinois at Urbana-Champaign and the University of Wisconsin-Madison.
Accession number(s).
Sequences of the integron casettes described here have been deposited in GenBank under the accession numbers MH560801 to MH560966.
ACKNOWLEDGMENTS
We gratefully acknowledge the Uganda Wildlife Authority, the Uganda National Council for Science and Technology, and the local government councils for granting us permission to conduct this research. We also thank J. Byaruhanga, P. Katurama, A. Mbabazi, A. Nyamwija, and J. Rusoke for providing assistance in the field, and E. Estoff, M. Lee, M. Lesosky, S. Rao, T. Tranby, and K. Vashisht for providing assistance in the laboratory and with data analyses. We also thank Krista Christensen for providing assistance with data analysis, as well as Kris Bisgard, Jon Meiman, and Carrie Tomasallo for their help reviewing earlier versions of the manuscript.
This material is based upon work conducted as part of the Kibale EcoHealth Project and was supported by the Morris Animal Foundation under award number D07Z0-024 and by the Earth and Society Initiative of the University of Illinois, Urbana-Champaign.
The findings and conclusions in the article are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
REFERENCES
- 1.World Health Organization. 2014. Antimicrobial resistance: global report on surveillance 2014. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Founou LL, Founou RC, Essack SY. 2016. Antibiotic resistance in the food chain: a developing country-perspective. Front Microbiol 7:1881. doi: 10.3389/fmicb.2016.01881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Founou RC, Founou LL, Essack SY. 2017. Clinical and economic impact of antibiotic resistance in developing countries: a systematic review and meta-analysis. PLoS One 12:e0189621. doi: 10.1371/journal.pone.0189621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kisakye A, Makumbi I, Nansera D, Lewis R, Braka F, Wobudeya E, Chaplain D, Nalumansi E, Mbabazi W, Gessner BD. 2009. Surveillance for Streptococcus pneumoniae meningitis in children aged <5 years: implications for immunization in Uganda. Clin Infect Dis 48(Suppl 2):S153–S161. doi: 10.1086/596495. [DOI] [PubMed] [Google Scholar]
- 5.Joloba ML, Bajaksouzian S, Palavecino E, Whalen C, Jacobs MR. 2001. High prevalence of carriage of antibiotic-resistant Streptococcus pneumoniae in children in Kampala Uganda. Int J Antimicrob Agents 17:395–400. doi: 10.1016/S0924-8579(00)00345-9. [DOI] [PubMed] [Google Scholar]
- 6.Medina MJ, Greene CM, Gertz RE, Facklam RR, Jagero G, Hamel M, Shi YP, Slutsker L, Feikin DR, Beall B. 2005. Novel antibiotic-resistant pneumococcal strains recovered from the upper respiratory tracts of HIV-infected adults and their children in Kisumu, Kenya. Microb Drug Resist 11:9–17. doi: 10.1089/mdr.2005.11.9. [DOI] [PubMed] [Google Scholar]
- 7.Kateete DP, Namazzi S, Okee M, Okeng A, Baluku H, Musisi NL, Katabazi FA, Joloba ML, Ssentongo R, Najjuka FC. 2011. High prevalence of methicillin resistant Staphylococcus aureus in the surgical units of Mulago Hospital in Kampala, Uganda. BMC Res Notes 4:326. doi: 10.1186/1756-0500-4-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rutebemberwa E, Mpeka B, Pariyo G, Peterson S, Mworozi E, Bwanga F, Kallander K. 2015. High prevalence of antibiotic resistance in nasopharyngeal bacterial isolates from healthy children in rural Uganda: a cross-sectional study. Ups J Med Sci 120:249–256. doi: 10.3109/03009734.2015.1072606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernabe KJ, Langendorf C, Ford N, Ronat JB, Murphy RA. 2017. Antimicrobial resistance in West Africa: a systematic review and meta-analysis. Int J Antimicrob Agents 50:629–639. doi: 10.1016/j.ijantimicag.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 10.Seni J, Najjuka CF, Kateete DP, Makobore P, Joloba ML, Kajumbula H, Kapesa A, Bwanga F. 2013. Antimicrobial resistance in hospitalized surgical patients: a silently emerging public health concern in Uganda. BMC Res Notes 6:298. doi: 10.1186/1756-0500-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alonso CA, Zarazaga M, Ben Sallem R, Jouini A, Ben Slama K, Torres C. 2017. Antibiotic resistance in Escherichia coli in husbandry animals: the African perspective. Lett Appl Microbiol 64:318–334. doi: 10.1111/lam.12724. [DOI] [PubMed] [Google Scholar]
- 12.Omulo S, Thumbi SM, Njenga MK, Call DR. 2015. A review of 40 years of enteric antimicrobial resistance research in Eastern Africa: what can be done better? Antimicrob Resist Infect Control 4:1. doi: 10.1186/s13756-014-0041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.United Nations Department of Economic and Social Affairs. 2015. World urbanization prospects: the 2014 revision (ST/ESA/SER.A/366). United Nations Department of Economic and Social Affairs, Population Division, New York, NY. [Google Scholar]
- 14.Johnston A, Holt DW. 2014. Substandard drugs: a potential crisis for public health. Br J Clin Pharmacol 78:218–243. doi: 10.1111/bcp.12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vasoo S, Barreto JN, Tosh PK. 2015. Emerging issues in gram-negative bacterial resistance: an update for the practicing clinician. Mayo Clin Proc 90:395–403. doi: 10.1016/j.mayocp.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 16.Rousham EK, Unicomb L, Islam MA. 2018. Human, animal and environmental contributors to antibiotic resistance in low-resource settings: integrating behavioural, epidemiological and One Health approaches. Proc Biol Sci 285:20180332. doi: 10.1098/rspb.2018.0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Domingues S, da Silva GJ, Nielsen KM. 2012. Integrons: vehicles and pathways for horizontal dissemination in bacteria. Mob Genet Elements 2:211–223. doi: 10.4161/mge.22967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rwego IB, Isabirye-Basuta G, Gillespie TR, Goldberg TL. 2008. Gastrointestinal bacterial transmission among humans, mountain gorillas, and livestock in Bwindi Impenetrable National Park, Uganda. Conserv Biol 22:1600–1607. doi: 10.1111/j.1523-1739.2008.01018.x. [DOI] [PubMed] [Google Scholar]
- 19.Rwego IB, Gillespie TR, Isabirye-Basuta G, Goldberg TL. 2008. High rates of Escherichia coli transmission between livestock and humans in rural Uganda. J Clin Microbiol 46:3187–3191. doi: 10.1128/JCM.00285-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goldberg TL, Gillespie TR, Rwego IB, Estoff EE, Chapman CA. 2008. Forest fragmentation as cause of bacterial transmission among primates, humans, and livestock, Uganda. Emerg Infect Dis 14:1375–1382. doi: 10.3201/eid1409.071196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldberg TL, Gillespie TR, Rwego IB, Wheeler ER, Estoff EE, Chapman CA. 2007. Patterns of gastrointestinal bacterial exchange between chimpanzees and humans involved in research and tourism in western Uganda. Biol Conserv 135:511–517. doi: 10.1016/j.biocon.2006.10.048. [DOI] [Google Scholar]
- 22.Paige SB, Bleecker J, Mayer J, Goldberg T. 2017. Spatial overlap between people and non-human primates in a fragmented landscape. Ecohealth 14:88–99. doi: 10.1007/s10393-016-1194-9. [DOI] [PubMed] [Google Scholar]
- 23.Paige SB, Frost SD, Gibson MA, Jones JH, Shankar A, Switzer WM, Ting N, Goldberg TL. 2014. Beyond bushmeat: animal contact, injury, and zoonotic disease risk in Western Uganda. Ecohealth 11:534–543. doi: 10.1007/s10393-014-0942-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alonso H, Gready JE. 2006. Integron-sequestered dihydrofolate reductase: a recently redeployed enzyme. Trends Microbiol 14:236–242. doi: 10.1016/j.tim.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 25.Wright GD. 1999. Aminoglycoside-modifying enzymes. Curr Opin Microbiol 2:499–503. doi: 10.1016/S1369-5274(99)00007-7. [DOI] [PubMed] [Google Scholar]
- 26.Pfeifer Y, Cullik A, Witte W. 2010. Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. Int J Med Microbiol 300:371–379. doi: 10.1016/j.ijmm.2010.04.005. [DOI] [PubMed] [Google Scholar]
- 27.Schwarz S, Kehrenberg C, Doublet B, Cloeckaert A. 2004. Molecular basis of bacterial resistance to chloramphenicol and florfenicol. FEMS Microbiol Rev 28:519–542. doi: 10.1016/j.femsre.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 28.Tran van Nhieu G, Collatz E. 1987. Primary structure of an aminoglycoside 6′-N-acetyltransferase AAC(6′)-4, fused in vivo with the signal peptide of the Tn3-encoded beta-lactamase. J Bacteriol 169:5708–5714. doi: 10.1128/jb.169.12.5708-5714.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okeke IN, Lamikanra A, Edelman R. 1999. Socioeconomic and behavioral factors leading to acquired bacterial resistance to antibiotics in developing countries. Emerg Infect Dis 5:18–27. doi: 10.3201/eid0501.990103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitema ES. 2010. The role of unregulated sale and dispensing of antimicrobial agents on the development of antimicrobial resistance in developing countries, p 403–411. In Sosa ADJ, Byarugaba DK, Amábile-Cuevas CF, Hsueh P-R, Kariuki S, Okeke IN (ed), Antimicrobial resistance in developing countries. Springer, New York, NY. [Google Scholar]
- 31.Klein EY, Van Boeckel TP, Martinez EM, Pant S, Gandra S, Levin SA, Goossens H, Laxminarayan R. 2018. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc Natl Acad Sci U S A 115:E3463–E3470. doi: 10.1073/pnas.1717295115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rwego IB, Isabirye-Basuta G, Gillespie TR, Goldberg TL. 2009. Bacterial exchange between gorillas, humans, and livestock in Bwindi. Gorilla J 38:16–18. [Google Scholar]
- 33.Naughton-Treves L, Treves A, Chapman C, Wrangham R. 1998. Temporal patterns of crop-raiding by primates: linking food availability in croplands and adjacent forest. J Appl Ecol 35:596–606. doi: 10.1046/j.1365-2664.1998.3540596.x. [DOI] [Google Scholar]
- 34.Gasasira AF, Kamya MR, Ochong EO, Vora N, Achan J, Charlebois E, Ruel T, Kateera F, Meya DN, Havlir D, Rosenthal PJ, Dorsey G. 2010. Effect of trimethoprim-sulphamethoxazole on the risk of malaria in HIV-infected Ugandan children living in an area of widespread antifolate resistance. Malar J 9:177. doi: 10.1186/1475-2875-9-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shafer LA, Biraro S, Nakiyingi-Miiro J, Kamali A, Ssematimba D, Ouma J, Ojwiya A, Hughes P, Van der Paal L, Whitworth J, Opio A, Grosskurth H. 2008. HIV prevalence and incidence are no longer falling in southwest Uganda: evidence from a rural population cohort 1989–2005. AIDS 22:1641–1649. doi: 10.1097/QAD.0b013e32830a7502. [DOI] [PubMed] [Google Scholar]
- 36.Braykov NP, Eisenberg JNS, Grossman M, Zhang L, Vasco K, Cevallos W, Muñoz D, Acevedo A, Moser KA, Marrs CF, Foxman B, Trostle J, Trueba G, Levy K. 2016. Antibiotic resistance in animal and environmental samples associated with small-scale poultry farming in northwestern Ecuador. mSphere 1(1):e00021-15. doi: 10.1128/mSphere.00021-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Binswanger-Mkhize HP, Savastano S. 2017. Agricultural intensification: the status in six African countries. Food Policy 67:26–40. doi: 10.1016/j.foodpol.2016.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chapman CA, Struhsaker TT, Lambert JE. 2005. Thirty years of research in Kibale National Park, Uganda, reveals a complex picture for conservation. Int J Primatol 26:539–555. doi: 10.1007/s10764-005-4365-z. [DOI] [Google Scholar]
- 39.Archabald K, Naughton-Treves L. 2001. Tourism revenue sharing around national parks in western Uganda: early efforts to identify and reward local communities. Environ Conserv 23:135–149. [Google Scholar]
- 40.Blomley T, Namara A, McNeilage A, Franks P, Rainer H, Donaldson A, Malpas R, Olupot W, Baker J, Sandbrook C, Bitariho R, Infield M. 2010. Development and gorillas? Assessing fifteen years of integrated conservation and development in south-western Uganda. Natural Resource Issues no. 23. IIED, London, United Kingdom. [Google Scholar]
- 41.CLSI. 2015. Performance standards for antimicrobial susceptibility testing; twenty-fifth informational supplement. CLSI, Wayne, PA. [Google Scholar]
- 42.Levesque C, Piche L, Larose C, Roy PH. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother 39:185–191. doi: 10.1128/AAC.39.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ibrahim A, Gerner-Smidt P, Sjostedt A. 1996. Amplification and restriction endonuclease digestion of a large fragment of genes coding for rRNA as a rapid method for discrimination of closely related pathogenic bacteria. J Clin Microbiol 34:2894–2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]


