PAHs are ubiquitous in the environment and are carcinogenic compounds and tend to accumulate in food chains due to their low bioavailability and poor biodegradability. Cycloclasticus is an obligate marine PAH degrader and is widespread in marine environments, while the PAH degradation pathways remain unclear. In this report, the degradation pathways for naphthalene, phenanthrene, and pyrene were revealed, and an integrated PAH metabolic network covering pyrene, phenanthrene, and naphthalene was constructed in Cycloclasticus. This overlapping network provides streamlined processing of PAHs to intermediates and ultimately to complete mineralization. Furthermore, these results provide an additional context for the prevalence of Cycloclasticus in oil-polluted marine environments and pelagic settings. In conclusion, these analyses provide a useful framework for understanding the cellular processes involved in PAH metabolism in an ecologically important marine bacterium.
KEYWORDS: Cycloclasticus, polycyclic aromatic hydrocarbon, polycyclic aromatic hydrocarbon degradation pathways
ABSTRACT
Bacteria play an important role in the removal of polycyclic aromatic hydrocarbons (PAHs) from polluted environments. In marine environments, Cycloclasticus is one of the most prevalent PAH-degrading bacterial genera. However, little is known regarding the degradation mechanisms for multiple PAHs by Cycloclasticus. Cycloclasticus sp. strain P1 was isolated from deep-sea sediments and is known to degrade naphthalene, phenanthrene, pyrene, and other aromatic hydrocarbons. Here, six ring-hydroxylating dioxygenases (RHDs) were identified in the complete genome of Cycloclasticus sp. P1 and were confirmed to be involved in PAH degradation by enzymatic assays. Further, five gene clusters in its genome were identified to be responsible for PAH degradation. Degradation pathways for naphthalene, phenanthrene, and pyrene were elucidated in Cycloclasticus sp. P1 based on genomic and transcriptomic analysis and characterization of an interconnected metabolic network. The metabolic pathway overlaps in many steps in the degradation of pyrene, phenanthrene, and naphthalene, which were validated by the detection of metabolic intermediates in cultures. This study describes a pyrene degradation pathway for Cycloclasticus. Moreover, the study represents the integration of a PAH metabolic network that comprises pyrene, phenanthrene, and naphthalene degradation pathways. Taken together, these results provide a comprehensive investigation of PAH metabolism in Cycloclasticus.
IMPORTANCE PAHs are ubiquitous in the environment and are carcinogenic compounds and tend to accumulate in food chains due to their low bioavailability and poor biodegradability. Cycloclasticus is an obligate marine PAH degrader and is widespread in marine environments, while the PAH degradation pathways remain unclear. In this report, the degradation pathways for naphthalene, phenanthrene, and pyrene were revealed, and an integrated PAH metabolic network covering pyrene, phenanthrene, and naphthalene was constructed in Cycloclasticus. This overlapping network provides streamlined processing of PAHs to intermediates and ultimately to complete mineralization. Furthermore, these results provide an additional context for the prevalence of Cycloclasticus in oil-polluted marine environments and pelagic settings. In conclusion, these analyses provide a useful framework for understanding the cellular processes involved in PAH metabolism in an ecologically important marine bacterium.
INTRODUCTION
Polycyclic aromatic hydrocarbons (PAHs) are carcinogenic compounds with two or more fused benzene rings that originate from both natural sources and anthropogenic activities (1, 2). PAHs are ubiquitously distributed on Earth (3) in general. Low-molecular-weight (LMW) PAHs are those with two or three aromatic rings, while those with four or more aromatic rings are considered high-molecular-weight (HMW) PAHs. An increase in the number of fused rings leads to higher hydrophobicity and recalcitrance to microbial degradation.
In marine environments, PAHs are widespread and can even be found in remote deep-sea sediments and hydrothermal vent sulfide minerals (1, 4–7). PAHs can be used as both carbon and energy sources by certain oligotrophic bacteria in harsh environments. For instance, Cycloclasticus is the most well-known obligate marine PAH degrader which can also degrade chlorinated derivatives of PAHs (8). Cycloclasticus bacteria are globally distributed and have been detected in estuaries (9–11), coastal areas (12–17), deep-sea sediments (5, 18, 19), and polar oceans (20–22). Further, Cycloclasticus dominated the oil-degrading community in the deep waters (∼1,500 m) of the Gulf of Mexico after the Deepwater Horizon oil spill (23–27). Recently, Cycloclasticus bacteria have been observed as symbionts with mussels and sponges dwelling in deep-sea gas and oil seeps (28). Hence, Cycloclasticus is an ecologically important marine bacterial genus capable of degrading various aromatic hydrocarbons and hydrocarbon gases.
Despite their prevalence and global distribution, Cycloclasticus bacteria are difficult to cultivate due to their unique oligotrophic lifestyles. Their recalcitrance to cultivation has been especially noted on solid medium. Thus, only three species with validly published names have been characterized to date: Cycloclasticus pugetii (9), Cycloclasticus spirillensus (10), and Cycloclasticus oligotrophus (29). Consequently, the physiological mechanisms that enable these bacteria to play a key role in PAH removal remains elusive.
To gain insight into the mechanisms underlying Cycloclasticus PAH metabolism, the degradation pathway needs to be elucidated first. PAH degradation is usually initiated by dihydroxylation mediated by a ring-hydroxylating dioxygenase (RHD). However, little is known regarding the whole pathway or of the commonalities among different PAH degradation pathways ranging from those of pyrene to single-ring compounds. Only a few bacteria have been documented to degrade pyrene, and it is thus an enigmatic degradation pathway. One is pyrene degradation by Mycobacterium vanbaalenii PYR-1, which has been intensively investigated (30–32). However, M. vanvaalenii is a Gram-positive bacterium, and it is unclear whether its degradation pathway represents the corresponding pathways in other PAH degraders, especially marine microorganisms. Further, there is a paucity of knowledge about the regulatory mechanisms involved in different PAH degradation pathways.
To gain new insights into PAH degradation, we investigated the metabolic network of PAH degradation in Cycloclasticus sp. strain P1, for which the range of PAHs that can be degraded and the naphthalene degradation pathway have been previously investigated (18, 33). This study shows that Cycloclasticus sp. P1 is a known pyrene degrader within the genus Cycloclasticus. In addition to pyrene, it can use naphthalene, 2-methylnaphthalene, 2,6-dimethylnaphthalene, biphenyl, fluorene, acenaphthene, dibenzofuran, dibenzothiophene, phenanthrene, and anthracene as its sole carbon and energy sources during growth (18). In our previous announcement of the Cycloclasticus sp. P1 genome sequence, the genome consisted of a circular, closed, 2,363,215-bp chromosome with a G+C content of 42.48%. The chromosome contains 2,248 predicted protein-coding genes (CDS) with an average size of 958 bp, 35 tRNA genes, and one rRNA operon (33). A total of 18 dioxygenase α-subunits were identified in the genome of Cycloclasticus sp. P1 (33).
In the present study, we characterized several RHDs for aromatic hydrocarbon hydroxylation in Cycloclasticus sp. P1 and constructed an integrated PAH metabolic pathway for naphthalene, phenanthrene, and pyrene based on genomic data, transcriptomic data, and PAH metabolic intermediate analysis. Further, we reassessed the capacity of PAH metabolism in this strain.
RESULTS AND DISCUSSION
PAH degradation and growth of Cycloclasticus sp. P1.
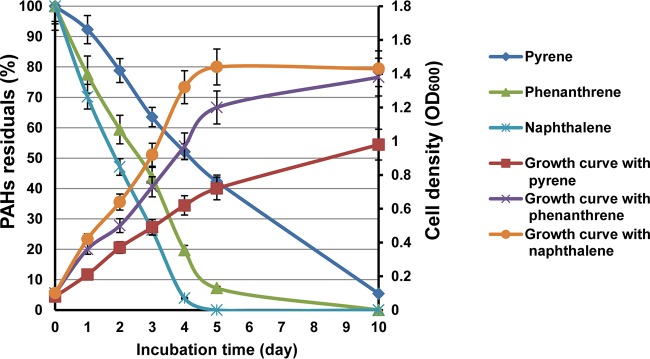
We previously demonstrated that Cycloclasticus sp. P1 can degrade various aromatic hydrocarbons (18). More specifically, naphthalene, phenanthrene, and pyrene were used as representative PAHs with two, three, and four rings, respectively. Here, degradation rates were determined for each PAH (Fig. 1). High-performance liquid chromatography (HPLC) quantification indicated that 98%, 92%, and 57% of the naphthalene, phenanthrene, and pyrene were degraded within 5 days, respectively, and 95% of the pyrene was degraded after 10 days (Fig. 1). Strain P1 grew fastest with naphthalene and slowest with pyrene (Fig. 1). All cultures, regardless of PAH treatment, reached exponential growth phase prior to the fourth day. Naphthalene cultures reached stationary phase first, followed by the cultures grown on phenanthrene and pyrene. After 10 days of growth, naphthalene and phenanthrene cultures exhibited cell yields of 1.4 and 1.3 optical densities at 600 nm (OD600), respectively, while pyrene resulted in only an OD600 of 0.96.
FIG 1.
PAH degradation efficiency and growth of Cycloclasticus sp. P1 with several PAHs as the sole carbon source. The left axis shows PAH degradation efficiency as the residual PAH percentage after growth. The right axis shows cell density (OD600) under optimal growth conditions of 28°C, 3.0% salinity, and pH of 7.0. Vertical bars represent means ± standard deviations (SDs) of results from triplicate treatments.
Mycobacterium sp. strain PYR-1 degraded 90% of the phenanthrene and 56% of the pyrene after 14 days of incubation (initial concentrations of each were 500 ppm) (30). Previous analyses have shown that 19.8% of fluoranthene (initial concentration, 300 ppm) was degraded by Microbacterium paraoxydans JPM1 after 5 days of incubation (34), 63% of pyrene (initial concentration, 300 ppm) was degraded by Acinetobacter strain USTB-X after 16 days of incubation (35), Achromobacter sp. strain HZ01 degraded 29.8%, 50.6%, and 38.4% of anthracene, phenanthrene, and pyrene, respectively (initial concentrations, 350 ppm) after 30 days of incubation (36), Klebsiella pneumonia PL1 degraded 63.4% of 500 mg liter−1 pyrene and 55.8% of 300 mg liter−1 benzo[a]pyrene after 10 days (37), and Bacillus megaterium YB3 could degrade 72.44% of 500 mg liter−1 pyrene within 7 days (38). The consumption rates of naphthalene, phenanthrene, and pyrene by Cycloclasticus sp. P1 after 5 days suggested that strain P1 was more active in PAH degradation than the above-mentioned strains.
Transcriptomic analysis of Cycloclasticus sp. P1 grown on naphthalene, phenanthrene, and pyrene.
Transcriptomic analyses of strain P1 grown under different conditions were conducted to confirm the relevance of genes with putative annotated functions in PAH degradation based on the genomic analysis. Napththalene, phenanthrene, and pyrene were used to grow P1 cells, along with an acetate control, and all cultures were subjected to transcriptomic analyses. The expression profiles of strain P1, when grown on phenanthrene and pyrene, were similar, while the naphthalene-induced profile was distinct from those of the other two treatments. Growth with each PAH led to hundreds of genes that were significantly differentially expressed relative to acetate-grown cells (Fig. 2). A total of 1,065 genes exhibited at least 2-fold differential expression, among which 522 genes were shared among the three PAH treatments (see Table S1 in the supplemental material). Notably, 199 of the 522 genes belonged to categories that are assigned to PAH degradation pathways (mainly oxidoreductases), including cell motility (flagellar systems), two-component signal transduction systems, and membrane transport systems (Table S1). Thus, many of these genes are likely involved in PAH metabolism. Under pyrene growth, 505 genes were upregulated more than 2-fold and 364 genes were downregulated more than 2-fold (data not shown). Under napththalene and phenanthrene growth, 454 and 527 genes were upregulated more than 2-fold and 401 and 359 genes were downregulated more than 2-fold, respectively (data not shown).
FIG 2.
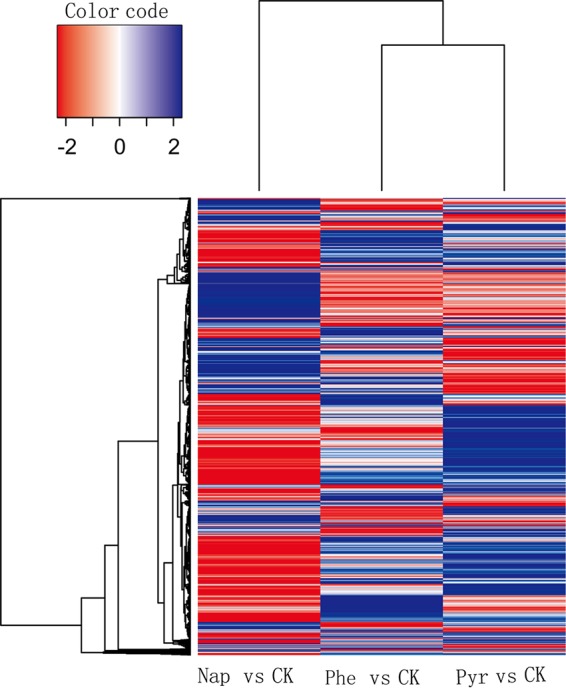
Heat map showing transcriptomic features of Cycloclasticus sp. P1 grown with three different PAHs. Blue indicates upregulated genes, and red indicates downregulated genes. Columns represent different PAH treatment groups, and rows represent different genes. Changes in the PAH treatment groups are shown for genes with significantly different expression levels under PAH treatments compared to those of the control (CK) and includes genes within treatments that were not significantly different from those of the control. Nap, naphthalene; Phe, phenanthrene; Pyr, pyrene; CK, control group (non-PAH treatment).
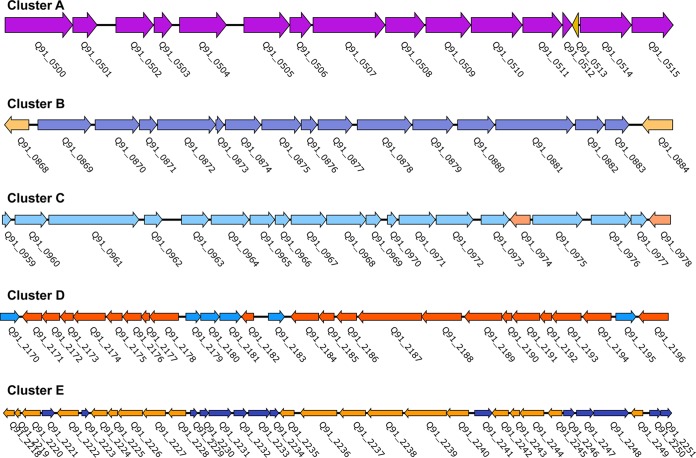
Five gene clusters are involved in PAH biodegradation of strain P1.
To identify genes involved in PAH degradation, the genome of strain P1 was mapped with the transcriptomes generated under different PAH substrates. More than 100 genes that are associated with the metabolism of aromatic hydrocarbons were identified (Table S2). These genes comprised five large gene clusters (annotated as clusters A to E) in the genome of strain P1 (Fig. 3; Table S2). Pyrene treatments resulted in more than 2-fold upregulation of 18 genes involved in PAH degradation pathways (Table S3). Two pairs of colocalized genes, Q91_0875/Q91_0876 and Q91_0870/Q91_0871, which encoded the α- and β-subunits of dioxygenases (termed RHD-1 and RHD-4), respectively, were remarkably upregulated. Interestingly, these four genes were upregulated only in response to pyrene degradation (Table S3). In addition, two genes (Q91_0872 and Q91_1047) that encoded dihydrodiol dehydrogenase and decarboxylase, respectively, were also highly upregulated only under pyrene growth (Table S3).
FIG 3.
Gene clusters involved in the biodegradation of PAHs in Cycloclasticus sp. P1. Detailed information for the ORFs (identified as Q91_XX) for clusters A to E is presented in Table S2 in the supplemental material. The transcriptional directions of ORFs are represented by the directions of the arrows.
Phenanthrene led to the specific upregulation of two subunit-encoding genes, Q91_2243 and Q91_2244, which encoded a dioxygenase (termed RHD-2), in addition to Q91_2235, which encoded a dihydrodiol dehydrogenase (Table S4). Two genes, Q91_0507 and Q91_0344, which encoded an aldehyde dehydrogenase and a hydroxylase, respectively, were upregulated in the phenanthrene and pyrene treatments, respectively (Table S3 and S4).
Naphthalene growth resulted in 10 upregulated genes in one of the degradation pathways described above, as initiated by RHD-3. These genes included the two subunit genes of RHD-3 (Q91_2225 and Q91_2226) and genes coding for a ferredoxin (Q91_2219), a ferredoxin reductase (Q91_2220), a ring cleavage dioxygenase (Q91_2224), a dihydrodiol dehydrogenase (Q91_2228), a salicylaldehyde dehydrogenase (Q91_0488), and a salicylaldehyde hydroxylase (Q91_1959) (Table S5). Naphthalene growth resulted in the specific upregulation of Q91_2225, Q91_2226, and Q91_2228 (Table S5), while the other upregulated genes were all upregulated in the other PAH treatments (Tables S3, S4, and S5).
Only one pair of Fe-S electron transfer components was found in the genome. The pair comprised a ferredoxin and a ferredoxin reductase that were encoded by open reading frames (ORFs) Q91_2219 and Q91_2220, respectively. Intriguingly, these two components were upregulated in all of the PAH treatments (Tables S3, S4, and S5). As in other bacterial species, multiple terminal oxygenases of strain P1 likely share one set of electron transfer components in order to reduce the cost of biosynthesis.
Ring-cleaving dioxygenases are a critical dearomatization step in bacterial degradation pathways of single-ring and multiring aromatic compounds (39). Only two ring cleavage dioxygenases encoded by ORFs Q91_2224 and Q91_0932 were identified in the P1 genome. Q91_2224 was upregulated in all of the PAH treatments (Table S3, S4, and S5), while Q91_0932 was not expressed in any of the treatments, including the control (data not shown). Thus, we deduced that the metabolic intermediates dihydroxypyrene, dihydroxyphenanthrene, and dihydroxynaphthalene were converted to their corresponding ring cleavage products by a ring cleavage dioxygenase encoded by ORF Q91_2224 (see below).
Ring hydroxylation of PAHs by different RHDs.
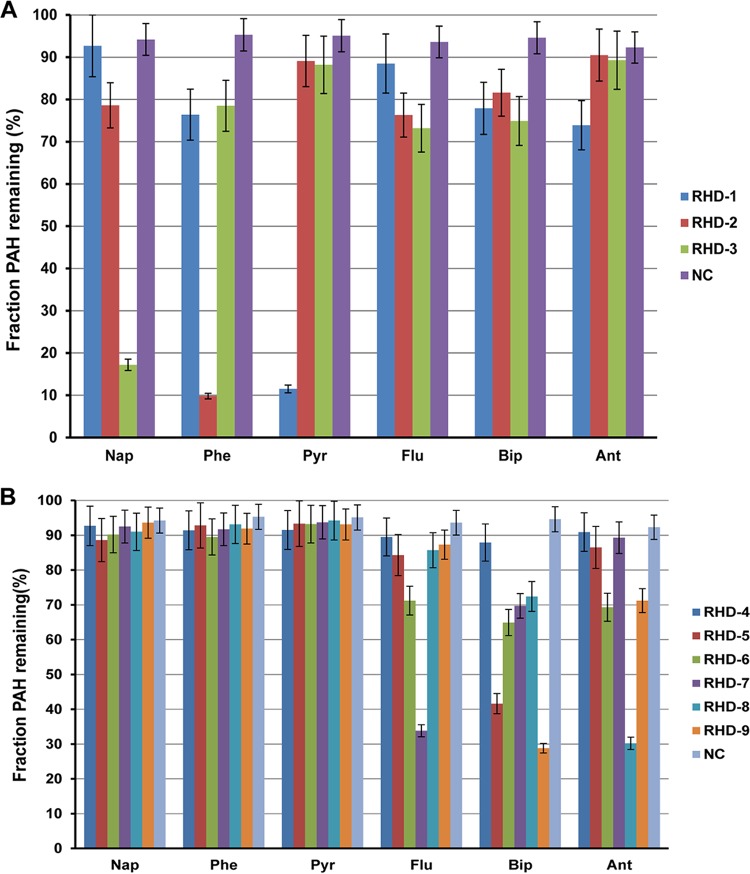
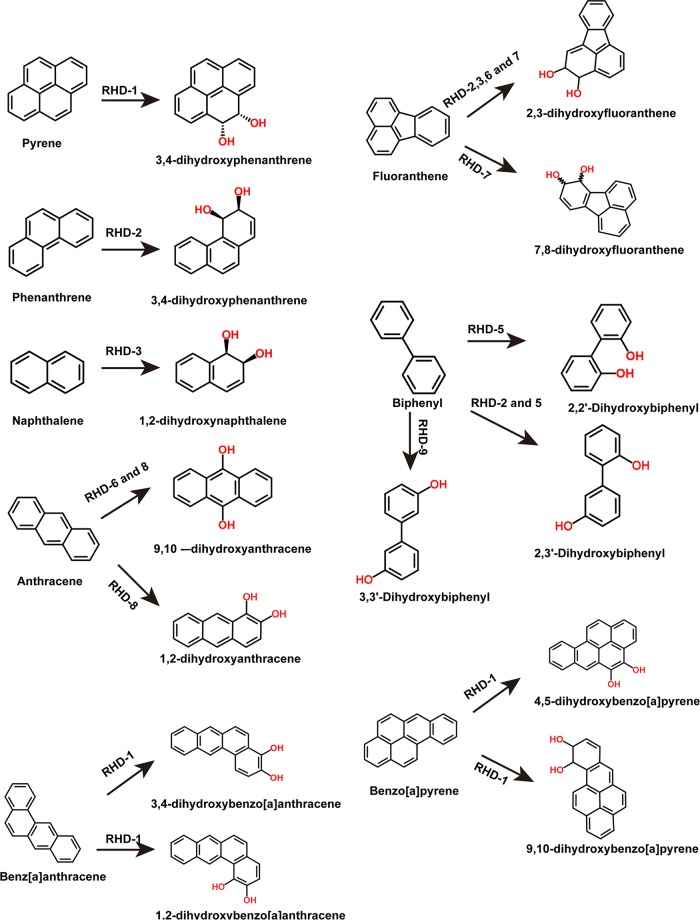
RHDs are the key enzymes of PAH degradation pathways. Nine RHDs were identified in the Cycloclasticus sp. P1 genome, indicated as RHD-1 to -9 (Table S1). All of the RHDs are differentially expressed in the presence of PAHs. To examine their substrate specificity, each pair of RHD genes (α- and β-subunits) were cloned into an expression vector downstream of the ferredoxin and reductase genes Q91_2219 and Q91_2220 and transferred to Escherichia coli (see Materials and Methods). The ring-hydroxylating activity of each RHD was assessed with naphthalene, phenanthrene, pyrene, fluoranthene, anthracene, and biphenyl as the substrate. After incubation of the recombinant E. coli strains with all six PAHs, the remaining PAH was quantified in the culture and the reduction in PAH concentration was used to reflect the enzymatic activity of RHD. Of the nine RHDs, only RHD-1 was capable of hydroxylating pyrene and added two hydroxyl groups at the 4,5 sites of pyrene (Fig. 4A and 5). Notably, Cycloclasticus sp. P1 could not degrade benz[a]anthracene and benzo[a]pyrene (data not shown). RHD-1 converted benzo[a]pyrene to 4,5-dihydroxybenzo[a]pyrene and 9,10-dihydroxybenzo[a]pyrene (Fig. 5). In the benz[a]anthracene experiments, RHD-1 produced traces of 1,2-dihydroxybenz[a]anthracene and 3,4 -dihydroxybenz[a]anthracene (Fig. 5). RHD-2 was capable of hydroxylating naphthalene, phenanthrene, biphenyl, and fluoranthene, although it was most efficient at phenanthrene transformation, wherein it produced 3,4-dihydroxyphenanthrene (Fig. 4A and 5). The enzymatic specificity of RHD-3 indicated that it was capable of transforming naphthalene, fluoranthene, and phenanthrene to 1,2-dihydroxynaphthalene, 2,3-dihydroxyfluoranthene, and 3,4-dihydroxyphenanthrene, respectively. Interestingly, RHD-3 was the only RHD that could efficiently transform naphthalene to the corresponding hydroxylated 1,2-dihydroxynaphthalene (Fig. 4A and 5).
FIG 4.
PAH degradation activity and substrate range for nine RHDs (RHD-1 to -3 [A]; RHD-4 to -9 [B]) within strain P1. The decrease in the concentrations of the PAH parent compounds by E. coli cells expressing RHD genes (and associated ferredoxin and reductase genes) from strain P1 are presented relative to those of E. coli cells expressing only the RHD-associated ferredoxin and reductase genes. Values represent means ± standard deviations, which are given as error bars for triplicate incubations. Nap, naphthalene; Flu, fluoranthene; Ant, anthracene; Bip, biphenyl; Phe, phenanthrene; Pyr, pyrene; RHD-1 to -9, gene pairs corresponding to RHD-1 to -9 and their associated electron transport genes cloned into pRSET-A vectors and transformed into E. coli BL21(DE3) pLyS host cells.
FIG 5.
Metabolite structures produced from aromatic substrates by enzymatic activity of RHD-1 to -9 enzymes.
Two genes, Q91_0870 and Q91_0871, were located upstream of the RHD-1 genes and encoded the α- and β-subunits of RHD-4. These genes were also significantly upregulated in the presence of pyrene alone, which was similar to RHD-1 (Table S3). However, RHD-4 failed to transform any of the six tested PAHs (Fig. 4B). In addition to the activity of the above-mentioned RHDs, RHD-5 and RHD-9 significantly degraded biphenyl (Fig. 4B and 5). RHD-6 was capable of dihydroxylating fluoranthene and anthracene, thereby producing 2,3-dihydroxyfluoranthene and 9,10-dihydroxyanthracene, respectively (Fig. 4B and 5). RHD-7 significantly transformed fluoranthene, producing 2,3-dihydroxyfluoranthene and 7,8-dihydroxyfluoranthene. Lastly, RHD-8 significantly transformed anthracene and produced 1,2-dihydroxyanthracene and 9,10-dihydroxyanthracene (Fig. 4B and 5).
Ring hydroxylation is the initial and rate-limiting step in PAH biodegradation and is carried out by di- or mono-oxygenases that produce metabolites with one or two -OH radicals (40, 41). The α-subunit of RHDs is one such dioxygenase that hydroxylates the aromatic ring by using a nonheme iron catalytic domain (42). Recently, the sequence, structure, and enzyme biochemistry of RHDs have been reviewed as to how these properties relate to biodegradation, biotransformation, and biocatalysis (43). The PhnA dioxygenase of Sphingomonas sp. strain VKMB-2434 is able to oxidize anthracene and phenanthrene, while the ArhA dioxygenase is able to oxidize acenaphthylene (44). Several RHDs have been reported to function in HMW PAH degradation. Two of these, NidAB and NidA3B3 from M. vanbaalenii PYR-1, are involved in the initial oxidation of pyrene and fluoranthene (45, 46). An additional dioxygenase, PdoAB from Mycobacterium sp. strain NJS-P, is capable of oxidizing anthracene, phenanthrene, pyrene, and benzo[a]pyrene but not fluoranthene or benzo[a]anthracene (47). The PahA dioxygenase of Novosphingobium sp. strain HR1a is able to oxidize chrysene, biphenyl, and pyrene (48). Recently, we identified a 7,8-dioxygenase with activity toward fluoranthene in Celeribacter indicus P73T, although the dioxygenase belongs to the toluene/biphenyl-degrading family (49). Similarly, the RHD from Microbacterium paraoxydans JPM1 is capable of oxidizing fluoranthene via the initial dihydroxylation at the C-1 and C-2 positions (34).
In this study, RHD-1, along with a ferredoxin and a ferredoxin reductase (Q91_2219 and Q91_2220), could oxidize HMW PAHs and exhibited a substrate preference for pyrene. This represents an RHD enzyme system that can oxidize pyrene in the Cycloclasticus genus. The α-subunit of the RHD-1 dioxygenase exhibited only 23.5% and 20.5% amino acid sequence identity to NidA (AF249301) and NidA3 (DQ028634) of M. vanbaalenii PYR-1, respectively. However, homologues of RHD-1 genes from Cycloclasticus sp. P1 are present in the genomes of several other Cycloclasticus species (19, 26, 50, 51), suggesting that RHD-1 plays an important role in the degradation of HMW PAHs in Cycloclasticus.
Biodegradation intermediates of naphthalene, phenanthrene, and pyrene.
To identify the degradation pathways for different PAHs in strain P1, the biodegradation intermediates were then characterized using gas chromatography-mass spectrometry (GC-MS) (see Materials and Methods). The intermediates that were detected are summarized in Table 1. 1,2-Naphthalenediol, salicylaldehyde, salicylic acid, and catechol were the major intermediate products in the degradation pathways of all three PAHs. The occurrence of 1,2-naphthalenediol in the naphthalene-incubated culture reconfirmed that RHD-3 is a naphthalene dioxygenase. In addition, 3,4-dihydroxyphenanthrene and 2-carboxy-1-naphthol were detected in the phenanthrene- and pyrene-grown cultures, which is consistent with intermediates produced by Sphingomonas, Cronobacter, and Enterobacter (52, 53). 3,4-Dihydroxyphenanthrene was generated as an intermediate in phenanthrene degradation, which reconfirmed that RHD-2 was active in phenanthrene hydroxylation. This result is consistent with the transcriptome and enzymatic assay results described above. The occurrence of 3,4-dihydroxyphenanthrene as an intermediate in pyrene degradation supports the hypothesis that pyrene is degraded via the phenanthrene metabolic pathway. In the pyrene culture supernatant extract and resting cells, a metabolite had a molecular ion peak at an m/z of 254, with characteristic fragment ions at m/z values of 236, 221, 202, 167, 149, 128, 113, 96, and 40 and which corresponds to phenanthrene-4,5-dicarboxylic acid (52, 53). In addition, 4,5-dihydroxypyrene was identified as a pyrene-derived intermediate. These results confirm that pyrene is dioxygenated at the 4 and 5 positions in strain P1 and then transformed to a 4,5-dihydrodiol intermediate. Taken together, these results are consistent with RHD-1 functioning as a pyrene dioxygenase.
TABLE 1.
GC-MS data for the metabolites of naphthalene, phenanthrene, and pyrenea
| Substrate | Retention time (min) | m/z of major ion peaks (%) | Suggested metabolite |
|---|---|---|---|
| Pyrene | 44.576 | 236 (M+, 100), 200 (38), 174 (8), 150 (6), 118 (12), 100 (32), 74 (7), 4 (12) | 4,5-Dihydroxypyrene |
| 48.365 | 254 (M+, 38), 236 (21), 221 (100), 202 (31), 167 (45), 149 (27), 128 (33), 113 (18), 96 (54), 40 (70) | Phenanthrene-4,5-dicarboxylic acid | |
| 38.683 | 208 (M+, 100), 193 (86), 176 (23), 164 (24), 150 (26), 132 (19), 118 (22), 105 (44), 79 (62), 51 (29) | 3,4-Dihydroxyphenanthrene | |
| 46.365 | 279 (M+, 23), 167 (37), 149 (100), 132 (13), 113 (15), 71 (35), 57 (34), 43 (28) | 2-Carboxy-1-naphthol | |
| 18.643 | 161 (M+, 100), 146 (65), 120 (36), 94 (53), 69 (31) | 1,2-Naphthalenediol | |
| 7.893 | 122 (M+, 100), 121 (90), 104 (22), 93 (38), 76 (24), 65 (33) | Salicylaldehyde | |
| 9.578 | 138 (M+, 72), 120 (100), 92 (64), 64 (33) | Salicylic acid | |
| 8.751 | 110 (M+, 100), 92 (28), 81 (18), 64 (35), 63 (19) | Catechol | |
| Phenanthrene | 38.175 | 208 (M+, 100), 193 (88), 176 (30), 164 (17), 150 (28), 132 (12), 118 (24), 105 (33), 79 (59), 51 (13) | 3,4-Dihydroxyphenanthrene |
| 47.024 | 279 (M+, 43), 167 (24), 149 (100), 132 (22), 113 (12), 71 (24), 57 (37), 43 (16) | 2-Carboxy-1-naphthol | |
| 18.594 | 161 (M+, 100), 146 (35), 120 (28), 94 (34), 69 (14) | 1,2-Naphthalenediol | |
| 7.605 | 122 (M+, 100), 121 (77), 104 (31), 93 (27), 76 (26), 65 (36) | Salicylaldehyde | |
| 9.604 | 138 (M+, 57), 120 (100), 92 (55), 64 (20) | Salicylic acid | |
| 8.816 | 110 (M+, 100), 92 (41), 81 (25), 64 (17), 63 (20) | Catechol | |
| Naphthalene | 19.034 | 161 (M+, 100), 146 (46), 120 (31), 94 (20), 69 (11) | 1,2-Naphthalenediol |
| 7.506 | 122 (M+, 100), 121 (66), 104 (24), 93 (30), 76 (19), 65 (22) | Salicylaldehyde | |
| 9.742 | 138 (M+, 38), 120 (100), 92 (64), 64 (15) | Salicylic acid | |
| 8.542 | 110 (M+, 100), 92 (50), 81 (10), 64 (28), 63 (37) | Catechol |
The metabolites were obtained from the organic extracts of the cultures and resting cell incubations of Cycloclasticus sp. P1.
The intermediate metabolites 1,2-naphthalenediol, salicylaldehyde, salicylic acid, and catechol were the major products of all three PAH treatments. The consilience in major intermediates among these pathways suggests that the three PAHs share the same downstream pathway through salicylate degradation via catechol. Further, catechol is converted to biological precursor molecules, such as pyruvate, that are funneled into the tricarboxylic acid (TCA) cycle for energy generation (Fig. 6). Thus, strain P1 incorporated degradation pathways of naphthalene and phenanthrene into the pyrene degradation pathway, forming a relay system of ring decomposition.
FIG 6.
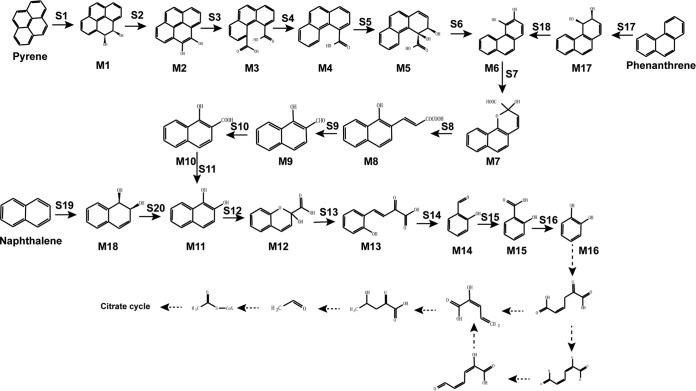
Complete PAH degradation pathway in Cycloclasticus sp. P1 based on genomic, transcriptomic, and other experimental analyses. Chemical designations: M1, pyrene-cis-4,5-dihydrodiol; M2, 4,5-dihydroxypyrene; M3, phenanthrene-4,5-dicarboxylic acid; M4, phenanthrene-4-carboxylate; M5, cis-3,4-dihydroxyphenanthrene-4-carboxylate; M6, 3,4-dihydroxyphenanthrene; M7, 2-hydroxy-2H-benzo[h]chromene-2-carboxylic acid; M8, trans-4-(1′-hydroxynapth-2-yl)-2-oxobut-3-enoic acid; M9, 1-hydroxy-2-naphthaldehyde; M10, 1-hydroxy-2-naphthoic acid; M11, 1,2-dihydroxynaphthalene; M12, 2-hydroxy-2H-chromene-2-carboxylic acid; M13, trans-o-hydroxybenzylidenepyruvic acid; M14, salicylaldehyde; M15, salicylic acid; M16, catechol; M17, cis-3,4-phenanthrenedihydrodiol; M18, (1R,2S)cis-l,2-naphthalenedihydrodiol. Enzyme designations: S1, pyrene dioxygenase (RHD-1); S2 and S6, dihydrodiol dehydrogenase (Q91_0872); S3, S4, S7, and S12, cleavage dioxygenase (Q91_2224); S5, dioxygenase (RHD-4); S8 and S13, isomerase (Q91_2218); S9 and S14, hydratase-aldolase (Q91_0437); S10, aldehyde dehydrogenase (Q91_0507); S11, 1-hydroxy-2-naphthoate hydroxylase (Q91_0344); S15, salicylaldehyde dehydrogenase (Q91_0488); S16, salicylate 1-hydroxylase (Q91_1959); S17, phenanthrene dioxygenase (RHD-2); S18, dihydrodiol dehydrogenase (Q91_2235); S19, naphthalene 1,2-dioxygenase (RHD-3); S20, NAD+-dependent cis-1,2-naphthalenedihydrodiol dehydrogenase (Q91_2228). Known metabolic pathways are represented by solid arrows. Predicted metabolic pathways are represented by dotted arrows.
The degradation network of naphthalene, phenanthrene, and pyrene in strain P1.
The degradation pathways of naphthalene, phenanthrene, and pyrene of strain P1 were constructed based on the above results (Fig. 6). The degradation of pyrene begins with pyrene dioxygenase (RHD-1) attacking the aromatic ring to form pyrene-cis-4,5-dihydrodiol (M1), which is subsequently dehydrogenated to 4,5-dihydroxypyrene (M2) by a dihydrodiol dehydrogenase (Q91_0872). Then, 4,5-dihydroxypyrene is converted to phenanthrene-4,5-dicarboxylic acid (M3) by a ring cleavage dioxygenase (Q91_2224). Phenanthrene-4,5-dicarboxylic acid is decarboxylated to phenanthrene-4-carboxylate (M4), which is subsequently oxidized by a dioxygenase (RHD-4) and then dehydrogenated to 3,4-dihydroxyphenanthrene (M6). Lastly, 3,4-dihydroxyphenanthrene is further transformed to catechol via the phenanthrene degradation pathway.
Strain P1 catabolizes phenanthrene via an initial dioxygenation at the bay region of phenanthrene by a phenanthrene dioxygenase (RHD-2), producing phenanthrene-cis-3,4-dihydrodiol (M17). Phenanthrene-cis-3,4-dihydrodiol is then dehydrogenated to 3,4-dihydroxyphenanthrene (M6) by a dihydrodiol dehydrogenase (Q91_2235). 3,4-Dihydroxyphenanthrene subsequently undergoes meta-cleavage to yield 2-hydroxy-2H-benzo[h]chromene-2-carboxylic acid (M7) by the same ring cleavage dioxygenase (Q91_2224) and is further metabolized by an isomerase (Q91_2218) to form trans-4-(1′-hydroxynapth-2-yl)-2-oxobut-3-enoic acid (M8). The metabolite trans-4-(1′-hydroxynapth-2-yl)-2-oxobut-3-enoic acid (M8) is further metabolized to 1-hydroxy-2-naphthaldehyde (M9) by a hydratase-aldolase (Q91_0437) and 1-hydroxy-2-naphthoic acid (M10) by an aldehyde dehydrogenase (Q91_0507). The 1-hydroxy-2-naphthoic acid (M10) is then hydroxylated to 1,2-dihydroxynaphthalene (M11) by 1-hydroxy-2-naphthoate hydroxylase (Q91_0344). 1,2-Dihydroxynaphthalene is further transformed to catechol via the naphthalene degradation pathway (Fig. 6).
In naphthalene degradation by strain P1, the first catabolic step is the formation of (1R,2S)naphthalene-cis-l,2-dihydrodiol (M18) by naphthalene 1,2-dioxygenase (RHD-3), which is then dehydrogenated to 1,2-dihydroxynaphthalene (M11) by an NAD-dependent cis-1,2-naphthalenedihydrodiol dehydrogenase (Q91_2228). The 1,2-dihydroxynaphthalene is cleaved by a ring cleavage dioxygenase (Q91_2224) that is shared with the degradation pathways of the other two PAHs, to form an unstable ring cleavage product, 2-hydroxy-4-(2′-oxo-3,5-cyclohexadienyl)-buta-2,4-dienoic acid. The unstable ring cleavage product then undergoes spontaneous recyclization to 2-hydroxy-2H-chromene-2-carboxylic acid (M12). Next, a 2-hydroxychromene-2-carboxylate isomerase (Q91_2218) isomerizes 2-hydroxy-2H-chromene-2-carboxylic acid to trans-o-hydroxybenzylidenepyruvic acid (M13). Subsequently, trans-o-hydroxybenzylidenepyruvic acid (M13) is metabolized by a hydratase-aldolase (Q91_0437) to salicylaldehyde (M14), followed by oxidation to salicylic acid (M15) via salicylaldehyde dehydrogenase (Q91_0488). The salicylic acid then undergoes oxidative decarboxylation by salicylate 1-hydroxylase (Q91_1959) to produce catechol (M16). As mentioned above, the catabolic pathway for naphthalene can also be integrated into the phenanthrene catabolism pathway (Fig. 6).
In addition to the degradation pathways described above, a complete catechol degradation pathway was identified in the genome of strain P1 (18). Through this pathway, catechol is further converted to biological precursor molecules, such as pyruvate, which can then be funneled into the TCA cycle (Fig. 6; Table S6).
An increasing number of bacteria have been found to catabolize HMW PAHs with four or more rings over the last decade (35, 43). Among these strains, the soil isolate M. vanbaalenii PYR-1 is the most intensively investigated pyrene-degrading bacterium, and a pyrene degradation pathway has been described for it (30). M. vanbaalenii PYR-1 can oxidize pyrene by two pathways: via initial dioxygenation at the C-1 and C-2 positions to form O-methylated derivatives of pyrene-1,2-diol or via initial dioxygenation at the C-4 and C-5 positions (K region) to generate cis-4,5-dihydroxy-4,5-dihydropyrene (pyrene cis-4,5-dihydrodiol). Frequently, PAHs of three and more benzene rings form sequentially an intermediate dihydriol with n-1 aromatic rings until reaching 1,2-naphthalene dihydriol (54). However, the PAH degradation pathways described above did not form a complete metabolic network.
Here, we show that naphthalene, phenanthrene, and pyrene degradation pathways form an interconnected metabolic network in Cycloclasticus sp. P1. This overlapping network provides streamlined processing of PAHs to intermediates and ultimately to complete mineralization. Further, we describe the complete pathway of naphthalene, phenanthrene, and pyrene degradation in an ecologically important marine bacterium that specializes in PAH metabolism. These results provide additional context for the prevalence of Cycloclasticus in oil-polluted marine environments and pelagic settings. However, it is still unclear how these PAH degradation pathways are coordinated at the transcriptional level and, in particular, how naphthalene and phenanthrene degradation is coordinated with pyrene degradation. The results reported here provide a foundation for revealing the regulatory mechanisms underlying these degradation pathways. Future investigations of these mechanisms will provide a greater understanding of the ecological role of Cycloclasticus in marine environments. Lastly, a number of genes involved in pathway regulation and potential transport of PAHs were also predicted by the transcriptomic data and require further characterization to fully understand the process of PAH degradation by this important marine bacterial genus.
MATERIALS AND METHODS
Bacterial strains, chemicals, and media.
Cycloclasticus sp. P1 (MCCC 1A01040) was originally isolated from deep-sea sediments of the Pacific Ocean (18). Naphthalene, phenanthrene, pyrene, naphthalene-13C6, phenanthrene-13C6, and pyrene-13C6 were purchased from Sigma (Sigma-Aldrich, Shanghai, China).
ML medium contained 30 g/liter NaCl, 1 g/liter NH4NO3, 0.35 g/liter KCl, 1 g/liter KH2PO4, 1 g/liter K2HPO4, 0.08 g/liter KBr, 0.05 g/liter CaCl2, 24 mg/liter SrCl2·6H2O, 1 × 10−4 g/liter ZnSO4·7H2O, 3.5 g/liter MgSO4·7H2O, 0.01 g/liter FeCl3, 3 g/liter sodium pyruvate, 3 g/liter sodium citrate, 0.5 g/liter yeast extract, 1 g/liter tryptone, and 1 liter of H2O at pH 8.0.
Artificial seawater medium (ASM) without a carbon source contained 24 g/liter NaCl, 7.0 g/liter MgSO4·7H2O, 1 g/liter H2O NH4NO3, 0.7 g/liter KCl, 2.0 g/liter KH2PO4, 3.0 g/liter Na2HPO4, and 10 ml of a trace element solution, with 1 liter of H2O at pH 7.5.
Growth assays and degradation capacity of naphthalene, phenanthrene, and pyrene.
Cells were grown at 28°C in a rotary shaker with shaking at 150 rpm in 250-ml Erlenmeyer flasks containing 100 ml ASM that was supplemented with naphthalene, phenanthrene, or pyrene as the sole carbon source (final concentration of 500 ppm; dimethyl sulfoxide [DMSO] at a final concentration of 0.5% was used as a cosolvent to normalize concentrations). Cultures were incubated for between 0 and 10 days. Abiotic control assays without bacterial inoculation were conducted in parallel, and all treatments were performed in triplicate. After induction, cells were harvested by centrifugation and resuspended in 100 ml ASM. Bacterial growth was monitored by measuring culture ODs at 600 nm.
Five milliliters of culture from each treatment was sampled each day starting on the first day. Cell samples were subjected to cell rupture with ultrasonic disruption and centrifugation to remove precipitates. Culture supernatants were then extracted with 3 volumes of ethyl acetate. The concentrations of PAHs in the lipid phase were analyzed by HPLC, as described previously (55). All treatments were performed in triplicate.
GC-MS identification of PAH degradation intermediates.
To identify PAH degradation intermediates, 100 ml of culture was sampled after incubation for 3 days with naphthalene, phenanthrene, and pyrene, which corresponded to cell density OD600 values of ∼0.8 to 1.3. Cells were harvested from cultures and subjected to cell rupture with ultrasonic disruption of cells, followed by centrifugation to remove precipitates. The resultant lysate was extracted with 3 volumes of ethyl acetate. The extraction was then acidified with concentrated HCl to pH 2 and extracted again with 3 volumes of ethyl acetate. The extracts were then dried over anhydrous sodium sulfate and evaporated to 10 ml using a rotatory evaporator at 40°C (56) and were further dried in a vacuum and stored at −20°C. The final extract that was dried in a vacuum was then dissolved in hexane for GC-MS analysis on a gas chromatograph with an HP 5973 mass spectrometer system (Agilent Technologies/Hewlett Packard). For GC-MS, the column was a TR-5MS (5% phenyl polysilphenylene siloxane; 30 mm by 0.25-mm diameter, 0.25-μm film thickness), and helium was used as the carrier gas with a constant flow at 1 ml/min. The column temperature was held at 70°C for 5 min and then increased at a rate of 4°C/min to 290°C, followed by a hold for 10 min. To remove any remaining compounds, a ramp of 20°C/min to 320°C was conducted, followed by a hold for 20 min. The mass spectrometer was operated in electron impact (EI) mode at 70 eV (EV) in the full-scan mode from 85 to 450 m/z over 6.5 to 85 min. Injector and detector temperatures were 270°C and 280°C, respectively (52).
Transcriptome analysis. (i) RNA extraction.
For transcriptomic analyses, a 100-ml culture of strain P1 was grown in a flask in ML medium at 28°C with shaking at 150 rpm until an OD600 of 0.9 to 1.1 was reached. Cultures were subsampled equally into four sterile 250-ml flasks. Three flasks contained 20 ppm naphthalene, phenanthrene, or pyrene (with DMSO as the cosolvent), and the fourth flask was used as the control. PAH degradation was then measured over a 5-day incubation at 28°C with shaking at 150 rpm. After induction, cells were harvested by centrifugation. RNA was then extracted from harvested cells using an RNA extraction kit according to the manufacturer's protocol (Tiangen, Beijing, China). RNA extracts were digested with DNase (Promega, Madison, WI) to remove DNA contamination. Each RNA sample (0.5 μl) was subjected to PCR with 16S rRNA gene primers to determine if the DNA had been completely digested.
(ii) mRNA purification and cDNA synthesis.
mRNA was enriched in each RNA sample using the MICROBExpress enrichment kit (Ambion) according to the manufacturer's instructions. mRNA-enriched samples were then resuspended in 15 μl of RNase-free water. cDNA was generated from the mRNA enrichments using a SuperScript double-stranded cDNA synthesis kit (Invitrogen) according to the manufacturer's instructions. Briefly, each mRNA sample was mixed with 100 pmol of random hexamer primers (Integrated DNA Technologies) and then incubated at 70°C for 10 min, followed by chilling on ice. A reaction mixture comprising 5 μl of First-Strand reaction buffer (Invitrogen), 2.5 μl of 0.1 M dithiothreitol (DTT), 1.25 μl of 10 mM RNase-free deoxynucleoside triphosphate (dNTP) mix, and 3 μl of SuperScript III reverse transcriptase was then mixed with the sample-primer mixture and incubated at 50°C for 2 h. To generate second-strand cDNA, a second reaction mixture that comprised 86 μl of RNase-free water, 30 μl of second-strand reaction buffer, 3 μl of 10 mM RNase-free dNTP mix, 10 U of E. coli DNA ligase, 40 U of E. coli DNA polymerase, and 2 U of E. coli RNase H was constructed and then incubated at 16°C for 2 h. Following incubation, 10 U of T4 DNA polymerase (Invitrogen) was added, and the reaction mixtures were incubated for an additional 5 min at 16°C. The reactions were quenched by adding 10 μl of 0.5 M RNase-free EDTA and then purified by phenol-chloroform extraction and ethanol precipitation. Residual RNA was removed by treatment with RNase H and RNase A, followed by a phenol-chloroform extraction and ethanol precipitation.
(iii) Illumina sequencing and analysis.
cDNA samples were submitted to the Joint Genome Institute for sequencing on the Illumina genome analyzer platform. Illumina libraries were prepared for sequencing according to the manufacturer's instructions. Sequencing reads were mapped to the complete genome using BLASTn and an E value threshold of 0.0001 and the “_F F” parameter, which allowed reads to be mapped with up to five base mismatches. Reads that mapped to rRNA or that did not map using the aforementioned parameters were removed from further analyses. The number of reads that mapped to each annotated gene was then recorded. Reads from replicate samples were pooled, and the numbers of reads per gene were normalized based on the total number of reads in each library in addition to the gene sizes. The resulting value represented the gene expression index (GEI), which is expressed as normalized reads per kilobase pair.
(iv) Characterization of differentially expressed genes.
χ2 tests were used to identify GEIs representing genes with differential expression between conditions or between strains. Bonferroni's multiple test corrections were applied to the χ2 values, and genes with adjusted P values of <0.05 and with GEI ratios of ≥2 were considered differentially expressed. Descriptive statistical analyses and Kolmogorov-Smirnov tests were performed on all log2-transformed GEI ratios using the Microsoft Excel add-in XLStat (Addinsoft) with the default settings. The distributions of overexpressed genes were analyzed with GenomeViz software v2 or by two-factor analysis of variance (ANOVA) tests with replicates in Microsoft Excel. Overexpressed genes were assigned to clusters of orthologous genes (COG) protein families based on NCBI classifications and then plotted as percentages of the total induced genes in a given gene list. For genes induced under naphthalene, phenanthrene, or pyrene growth conditions, the amino acid sequences of putative ORF products were analyzed using the SignalP and TatP signal prediction servers. SignalP-NN or TatP-NN (neural network) D scores (the average of the S-mean and Y-max scores) were used as indicators for the presence or absence of signal sequences.
Plasmid construction. (i) Construction of recombinant plasmids for RHD gene expression.
Putative gene subunits of PAH-related RHD and electron transport systems involved in PAH metabolism were first identified in strain P1. The genes were then PCR amplified from strain P1 template DNA. To promote expression of the genes in Escherichia coli, PCR primers were designed to include restriction sites for cloning and ribosome-binding sites (RBSs) (see Table S7 in the supplemental material). Forward primers for the first cloned gene also incorporated a stop codon before the RBS in order to prevent the addition of a polyhistidine tag to the protein. Putative RHD α- and β-subunit pairs were amplified together as a single product and therefore possessed only a single RBS that was upstream of the α-subunit. Cloning was performed using previously described methods (57). The vectors and host strains that were used for cloning are provided in Table S5 in the supplemental material. Candidate genes were first PCR amplified and then cloned into pCR4-TOPO (Invitrogen) cells. Clone inserts were then sequenced using 5′ and 3′ sequencing with primers targeting the M13 vector sites and then validated against the P1 genome. Genes were then excised from the cloning vector using restriction endonucleases (New England Biolabs) that targeted sites incorporated into the initial PCR primers. Excised genes were then cloned into pRSET-A vectors (Invitrogen). Restriction sites within some of the cloned dioxygenase gene pairs required partial digestion and gel extraction using a QIAquick gel extraction kit (Qiagen) prior to cloning into the expression vector. Before transformation into the host strain for expression, the presence of four cloned genes in the final plasmid constructs, in the correct orientation and of the correct size, were confirmed by PCR with primers targeting the 5′ end of the ferredoxin gene and the 3′ end of the RHD β-subunit gene.
(ii) RHD heterologous expression and PAH degradation activity assays.
The pRSET-A vectors containing the cloned RHDs, and their associated electron transport genes, were transformed into E. coli host BL21(DE3) pLyS cells for expression by following previously described methods (45). Colonies were grown overnight on LB plates containing ampicillin (50 μg/ml) and chloramphenicol (35 μg/ml). Single colonies were picked and inoculated into 5 ml of LB broth (with antibiotics) and incubated overnight at 37°C with shaking at 200 rpm. A 2-ml aliquot of the culture grown overnight was used to inoculate 200 ml of LB broth that contained antibiotics, and the new cultures were incubated at 37°C with shaking at 200 rpm until the OD600 of the culture reached 0.5. Then, isopropyl-β-d-1-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM to induce plasmid expression, and the flask was returned to the shaker for 2 h. A 90-ml culture subsample was centrifuged to pellet the cells. The resulting cells were then washed with M9 minimal medium containing glucose (M9-glucose) and antibiotics before being resuspended to 90 ml in fresh M9-glucose medium containing antibiotics. A 3-ml culture subsample was added to each of 27 screw-cap glass tubes to test activity for the six PAHs in triplicate. PAHs in DMSO were added to the cell suspensions to achieve final concentrations of 1 μg/ml. The cultures were then incubated in the dark at 37°C with shaking at 200 rpm. Incubations were conducted for 72 to 96 h, with the exception of the no-inoculum controls, which were incubated for 24 h. PAHs were extracted from the cultures by the addition of 7 ml of ethyl acetate, followed by vortexing for 10 s and quantification by HPLC as previously described (58). Abiotic controls comprising medium with PAH additions, but without inoculation, were also analyzed in triplicate. The analysis and identification of metabolites using HPLC and GC-MS were performed as described previously (45).
Accession number(s).
The data supporting this study are available in the NCBI Sequence Read Archive (SRA) under accession numbers SRR926181, SRR926183, SRR926186, and SRR926223.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Science Foundation of China (41876143), the COMRA Program (no. DY135-B-01), the Natural Science Fund for Distinguished Young Scholars of Fujian Province of China (2017J06012), and the National Infrastructure of Natural Resources for Science and Technology Program of China (NIMR-2017-9).
We thank LetPub for providing linguistic assistance during the preparation of the manuscript.
Z.S. fostered ideas and chaired the projects. W.W. and Z.S. designed the research, W.W. and L.W. were associated with genome and transcriptome analysis, W.W. performed the other experiments, and W.W. and Z.S. wrote the manuscript.
The authors have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01261-18.
REFERENCES
- 1.Louvado A, Gomes NC, Simoes MM, Almeida A, Cleary DF, Cunha A. 2015. Polycyclic aromatic hydrocarbons in deep sea sediments: microbe-pollutant interactions in a remote environment. Sci Total Environ 526:312–328. doi: 10.1016/j.scitotenv.2015.04.048. [DOI] [PubMed] [Google Scholar]
- 2.Duran R, Cravo-Laureau C. 2016. Role of environmental factors and microorganisms in determining the fate of polycyclic aromatic hydrocarbons in the marine environment. FEMS Microbiol Rev 40:814–830. doi: 10.1093/femsre/fuw031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vila J, Tauler M, Grifoll M. 2015. Bacterial PAH degradation in marine and terrestrial habitats. Curr Opin Biotechnol 33:95–102. doi: 10.1016/j.copbio.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Bouloubassi I, Mejanelle L, Pete R, Fillaux J, Lorre A, Point V. 2006. PAH transport by sinking particles in the open Mediterranean Sea: a 1 year sediment trap study. Mar Pollut Bull 52:560–571. doi: 10.1016/j.marpolbul.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Shao Z, Cui Z, Dong C, Lai Q, Chen L. 2010. Analysis of a PAH-degrading bacterial population in subsurface sediments on the Mid-Atlantic Ridge. Deep Sea Res Part I Oceanogr Res Pap 57:724–730. doi: 10.1016/j.dsr.2010.02.001. [DOI] [Google Scholar]
- 6.Mandalakis M, Polymenakou PN, Tselepides A, Lampadariou N. 2014. Distribution of aliphatic hydrocarbons, polycyclic aromatic hydrocarbons and organochlorinated pollutants in deep-sea sediments of the Southern Cretan margin, Eastern Mediterranean Sea: a baseline assessment. Chemosphere 106:28–35. doi: 10.1016/j.chemosphere.2013.12.081. [DOI] [PubMed] [Google Scholar]
- 7.Konn C, Charlou JL, Holm NG, Mousis O. 2015. The production of methane, hydrogen, and organic compounds in ultramafic-hosted hydrothermal vents of the Mid-Atlantic Ridge. Astrobiology 15:381–399. doi: 10.1089/ast.2014.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yakimov MM, Timmis KN, Golyshin PN. 2007. Obligate oil-degrading marine bacteria. Curr Opin Biotechnol 18:257–266. doi: 10.1016/j.copbio.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 9.Dyksterhouse SE, Gray JP, Herwig RP, Lara JC, Staley JT. 1995. Cycloclasticus pugetii gen. nov., sp. nov., an aromatic hydrocarbon-degrading bacterium from marine sediments. Int J Syst Bacteriol 45:116–123. doi: 10.1099/00207713-45-1-116. [DOI] [PubMed] [Google Scholar]
- 10.Chung WK, King GM. 2001. Isolation, characterization, and polyaromatic hydrocarbon degradation potential of aerobic bacteria from marine macrofaunal burrow sediments and description of Lutibacterium anuloederans gen. nov., sp. nov., and Cycloclasticus spirillensus sp. nov. Appl Environ Microbiol 67:5585–5592. doi: 10.1128/AEM.67.12.5585-5592.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niepceron M, Portet-Koltalo F, Merlin C, Motelay-Massei A, Barray S, Bodilis J. 2010. Both Cycloclasticus spp. and Pseudomonas spp. as PAH-degrading bacteria in the Seine estuary (France). FEMS Microbiol Ecol 71:137–147. doi: 10.1111/j.1574-6941.2009.00788.x. [DOI] [PubMed] [Google Scholar]
- 12.Teira E, Lekunberri I, Gasol JM, Nieto-Cid M, Alvarez-Salgado XA, Figueiras FG. 2007. Dynamics of the hydrocarbon-degrading Cycloclasticus bacteria during mesocosm-simulated oil spills. Environ Microbiol 9:2551–2562. doi: 10.1111/j.1462-2920.2007.01373.x. [DOI] [PubMed] [Google Scholar]
- 13.Marcos MS, Lozada M, Di Marzio WD, Dionisi HM. 2012. Abundance, dynamics, and biogeographic distribution of seven polycyclic aromatic hydrocarbon dioxygenase gene variants in coastal sediments of Patagonia. Appl Environ Microbiol 78:1589–1592. doi: 10.1128/AEM.06929-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sauret C, Severin T, Vetion G, Guigue C, Goutx M, Pujo-Pay M, Conan P, Fagervold SK, Ghiglione JF. 2014. ‘Rare biosphere’ bacteria as key phenanthrene degraders in coastal seawaters. Environ Pollut 194:246–253. doi: 10.1016/j.envpol.2014.07.024. [DOI] [PubMed] [Google Scholar]
- 15.Sanni GO, Coulon F, McGenity TJ. 2015. Dynamics and distribution of bacterial and archaeal communities in oil-contaminated temperate coastal mudflat mesocosms. Environ Sci Pollut Res Int 22:15230–15247. doi: 10.1007/s11356-015-4313-1. [DOI] [PubMed] [Google Scholar]
- 16.Messina E, Denaro R, Crisafi F, Smedile F, Cappello S, Genovese M, Genovese L, Giuliano L, Russo D, Ferrer M, Golyshin P, Yakimov MM. 2016. Genome sequence of obligate marine polycyclic aromatic hydrocarbons-degrading bacterium Cycloclasticus sp. 78-ME, isolated from petroleum deposits of the sunken tanker Amoco Milford Haven, Mediterranean Sea. Mar Genomics 25:11–13. doi: 10.1016/j.margen.2015.10.006. [DOI] [PubMed] [Google Scholar]
- 17.Ding Q, Huang X, Hu H, Hong M, Zhang D, Wang K. 2017. Impact of pyrene and cadmium co-contamination on prokaryotic community in coastal sediment microcosms. Chemosphere 188:320–328. doi: 10.1016/j.chemosphere.2017.08.124. [DOI] [PubMed] [Google Scholar]
- 18.Wang B, Lai Q, Cui Z, Tan T, Shao Z. 2008. A pyrene-degrading consortium from deep-sea sediment of the West Pacific and its key member Cycloclasticus sp. P1. Environ Microbiol 10:1948–1963. doi: 10.1111/j.1462-2920.2008.01611.x. [DOI] [PubMed] [Google Scholar]
- 19.Gutierrez T, Biddle JF, Teske A, Aitken MD. 2015. Cultivation-dependent and cultivation-independent characterization of hydrocarbon-degrading bacteria in Guaymas Basin sediments. Front Microbiol 6:695. doi: 10.3389/fmicb.2015.00695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coulon F, McKew BA, Osborn AM, McGenity TJ, Timmis KN. 2007. Effects of temperature and biostimulation on oil-degrading microbial communities in temperate estuarine waters. Environ Microbiol 9:177–186. doi: 10.1111/j.1462-2920.2006.01126.x. [DOI] [PubMed] [Google Scholar]
- 21.Dong C, Bai X, Sheng H, Jiao L, Zhou H, Shao Z. 2015. Distribution of PAHs and the PAH-degrading bacteria in the deep-sea sediments of the high-latitude Arctic Ocean. Biogeosciences 12:2163–2177. doi: 10.5194/bg-12-2163-2015. [DOI] [Google Scholar]
- 22.Brakstad OG, Throne-Holst M, Netzer R, Stoeckel DM, Atlas RM. 2015. Microbial communities related to biodegradation of dispersed Macondo oil at low seawater temperature with Norwegian coastal seawater. Microb Biotechnol 8:989–998. doi: 10.1111/1751-7915.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hazen TC, Dubinsky EA, DeSantis TZ, Andersen GL, Piceno YM, Singh N, Jansson JK, Probst A, Borglin SE, Fortney JL, Stringfellow WT, Bill M, Conrad ME, Tom LM, Chavarria KL, Alusi TR, Lamendella R, Joyner DC, Spier C, Baelum J, Auer M, Zemla ML, Chakraborty R, Sonnenthal EL, D'Haeseleer P, Holman HY, Osman S, Lu Z, Van Nostrand JD, Deng Y, Zhou J, Mason OU. 2010. Deep-sea oil plume enriches indigenous oil-degrading bacteria. Science 330:204–208. doi: 10.1126/science.1195979. [DOI] [PubMed] [Google Scholar]
- 24.Redmond MC, Valentine DL. 2012. Natural gas and temperature structured a microbial community response to the Deepwater Horizon oil spill. Proc Natl Acad Sci U S A 109:20292. doi: 10.1073/pnas.1108756108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gutierrez T, Singleton DR, Berry D, Yang T, Aitken MD, Teske A. 2013. Hydrocarbon-degrading bacteria enriched by the Deepwater Horizon oil spill identified by cultivation and DNA-SIP. ISME J 7:2091–2104. doi: 10.1038/ismej.2013.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kleindienst S, Grim S, Sogin M, Bracco A, Crespo-Medina M, Joye SB. 2016. Diverse, rare microbial taxa responded to the Deepwater Horizon deep-sea hydrocarbon plume. ISME J 10:400–415. doi: 10.1038/ismej.2015.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu P, Dubinsky EA, Probst AJ, Wang J, Cmk S, Tom LM, Gardinali PR, Banfield JF, Atlas RM, Andersen GL. 2017. Simulation of Deepwater Horizon oil plume reveals substrate specialization within a complex community of hydrocarbon degraders. Proc Natl Acad Sci U S A 114:7432–7437. doi: 10.1073/pnas.1703424114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubin-Blum M, Antony CP, Borowski C, Sayavedra L, Pape T, Sahling H, Bohrmann G, Kleiner M, Redmond MC, Valentine DL, Dubilier N. 2017. Short-chain alkanes fuel mussel and sponge Cycloclasticus symbionts from deep-sea gas and oil seeps. Nat Microbiol 2:17093. doi: 10.1038/nmicrobiol.2017.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Button DK, Robertson BR, Lepp PW, Schmidt TM. 1998. A small, dilute-cytoplasm, high-affinity, novel bacterium isolated by extinction culture and having kinetic constants compatible with growth at ambient concentrations of dissolved nutrients in seawater. Appl Environ Microbiol 64:4467–4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kweon O, Kim SJ, Holland RD, Chen H, Kim DW, Gao Y, Yu LR, Baek S, Baek DH, Ahn H, Cerniglia CE. 2011. Polycyclic aromatic hydrocarbon metabolic network in Mycobacterium vanbaalenii PYR-1. J Bacteriol 193:4326–4337. doi: 10.1128/JB.00215-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kweon O, Kim SJ, Kim DW, Kim JM, Kim HL, Ahn Y, Sutherland JB, Cerniglia CE. 2014. Pleiotropic and epistatic behavior of a ring-hydroxylating oxygenase system in the polycyclic aromatic hydrocarbon metabolic network from Mycobacterium vanbaalenii PYR-1. J Bacteriol 196:3503–3515. doi: 10.1128/JB.01945-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim SJ, Kweon O, Sutherland JB, Kim HL, Jones RC, Burback BL, Graves SW, Psurny E, Cerniglia CE. 2015. Dynamic response of Mycobacterium vanbaalenii PYR-1 to BP Deepwater Horizon crude oil. Appl Environ Microbiol 81:4263–4276. doi: 10.1128/AEM.00730-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lai Q, Li W, Wang B, Yu Z, Shao Z. 2012. Complete genome sequence of the pyrene-degrading bacterium Cycloclasticus sp. strain P1. J Bacteriol 194:6677. doi: 10.1128/JB.01837-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin J, Yao J, Liu W, Zhang Q, Liu J. 2017. Fluoranthene degradation and binding mechanism study based on the active-site structure of ring-hydroxylating dioxygenase in Microbacterium paraoxydans JPM1. Environ Sci Pollut Res Int 24:363–371. doi: 10.1007/s11356-016-7809-4. [DOI] [PubMed] [Google Scholar]
- 35.Yuan H, Yao J, Masakorala K, Wang F, Cai M, Yu C. 2014. Isolation and characterization of a newly isolated pyrene-degrading Acinetobacter strain USTB-X. Environ Sci Pollut Res Int 21:2724–2732. doi: 10.1007/s11356-013-2221-9. [DOI] [PubMed] [Google Scholar]
- 36.Deng MC, Li J, Liang FR, Yi M, Xu XM, Yuan JP, Peng J, Wu CF, Wang JH. 2014. Isolation and characterization of a novel hydrocarbon-degrading bacterium Achromobacter sp. HZ01 from the crude oil-contaminated seawater at the Daya Bay, southern China. Mar Pollut Bull 83:79–86. [DOI] [PubMed] [Google Scholar]
- 37.Ping L, Zhang C, Zhang C, Zhu Y, He H, Wu M, Tang T, Li Z, Zhao H. 2014. Isolation and characterization of pyrene and benzo[a]pyrene-degrading Klebsiella pneumonia PL1 and its potential use in bioremediation. Appl Microbiol Biotechnol 98:3819–3828. doi: 10.1007/s00253-013-5469-6. [DOI] [PubMed] [Google Scholar]
- 38.Meena SS, Sharma RS, Gupta P, Karmakar S, Aggarwal KK. 2016. Isolation and identification of Bacillus megaterium YB3 from an effluent contaminated site efficiently degrades pyrene. J Basic Microbiol 56:369–378. doi: 10.1002/jobm.201500533. [DOI] [PubMed] [Google Scholar]
- 39.Fetzner S. 2012. Ring-cleaving dioxygenases with a cupin fold. Appl Environ Microbiol 78:2505–2514. doi: 10.1128/AEM.07651-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kanaly RA, Harayama S. 2000. Biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons by bacteria. J Bacteriol 182:2059. doi: 10.1128/JB.182.8.2059-2067.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seo JS, Keum YS, Li QX. 2009. Bacterial degradation of aromatic compounds. Int J Environ Res Public Health 6:278–309. doi: 10.3390/ijerph6010278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Demaneche S, Meyer C, Micoud J, Louwagie M, Willison JC, Jouanneau Y. 2004. Identification and functional analysis of two aromatic-ring-hydroxylating dioxygenases from a Sphingomonas strain that degrades various polycyclic aromatic hydrocarbons. Appl Environ Microbiol 70:6714–6725. doi: 10.1128/AEM.70.11.6714-6725.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghosal D, Ghosh S, Dutta TK, Ahn Y. 2016. Current state of knowledge in microbial degradation of polycyclic aromatic hydrocarbons (PAHs): a review. Front Microbiol 7:1369. doi: 10.3389/fmicb.2016.01369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baboshin M, Ivashina T, Chernykh A, Golovleva L. 2014. Comparison of the substrate specificity of two ring-hydroxylating dioxygenases from Sphingomonas sp. VKM B-2434 to polycyclic aromatic hydrocarbons. Biodegradation 25:693–703. doi: 10.1007/s10532-014-9692-3. [DOI] [PubMed] [Google Scholar]
- 45.Kim SJ, Kweon O, Freeman JP, Jones RC, Adjei MD, Jhoo JW, Edmondson RD, Cerniglia CE. 2006. Molecular cloning and expression of genes encoding a novel dioxygenase involved in low- and high-molecular-weight polycyclic aromatic hydrocarbon degradation in Mycobacterium vanbaalenii PYR-1. Appl Environ Microbiol 72:1045–1054. doi: 10.1128/AEM.72.2.1045-1054.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kweon O, Kim SJ, Freeman JP, Song J, Baek S, Cerniglia CE. 2010. Substrate specificity and structural characteristics of the novel Rieske nonheme iron aromatic ring-hydroxylating oxygenases NidAB and NidA3B3 from Mycobacterium vanbaalenii PYR-1. mBio 1:e00135-10. doi: 10.1128/mBio.00135-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeng J, Zhu Q, Wu Y, Chen H, Lin X. 2017. Characterization of a polycyclic aromatic ring-hydroxylation dioxygenase from Mycobacterium sp. NJS-P. Chemosphere 185:67–74. doi: 10.1016/j.chemosphere.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 48.Segura A, Hernandez-Sanchez V, Marques S, Molina L. 2017. Insights in the regulation of the degradation of PAHs in Novosphingobium sp. HR1a and utilization of this regulatory system as a tool for the detection of PAHs. Sci Total Environ 590-591:381–393. doi: 10.1016/j.scitotenv.2017.02.180. [DOI] [PubMed] [Google Scholar]
- 49.Cao J, Lai Q, Yuan J, Shao Z. 2015. Genomic and metabolic analysis of fluoranthene degradation pathway in Celeribacter indicus P73T. Sci Rep 5:7741. doi: 10.1038/srep07741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cui Z, Xu G, Gao W, Li Q, Yang B, Yang G, Zheng L. 2014. Isolation and characterization of Cycloclasticus strains from Yellow Sea sediments and biodegradation of pyrene and fluoranthene by their syntrophic association with Marinobacter strains. Int Biodeterior Biodegradation 91:45–51. doi: 10.1016/j.ibiod.2014.03.005. [DOI] [Google Scholar]
- 51.Singh AK, Sherry A, Gray ND, Jones DM, Bowler BF, Head IM. 2014. Kinetic parameters for nutrient enhanced crude oil biodegradation in intertidal marine sediments. Front Microbiol 5:160. doi: 10.3389/fmicb.2014.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hesham Ael-L, Mawad AM, Mostafa YM, Shoreit A. 2014. Biodegradation ability and catabolic genes of petroleum-degrading Sphingomonas koreensis strain ASU-06 isolated from Egyptian oily soil. Biomed Res Int 2014:127674. doi: 10.1155/2014/127674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Umar ZD, Aziz NAA, Zulkifli SZ, Mustafa M. 2017. Rapid biodegradation of polycyclic aromatic hydrocarbons (PAHs) using effective Cronobacter sakazakii MM045 (KT933253). MethodsX 4:104–117. doi: 10.1016/j.mex.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nzila A. 2013. Update on the cometabolism of organic pollutants by bacteria. Environ Pollut 178:474–482. doi: 10.1016/j.envpol.2013.03.042. [DOI] [PubMed] [Google Scholar]
- 55.Bishnoi K, Kumar R, Bishnoi NR. 2008. Biodegradation of polycyclic aromatic hydrocarbons by white rot fungi Phanerochaete chrysosporium in sterile and unsterile soil. J Sci Ind Res 67:538–542. https://pdfs.semanticscholar.org/81f4/4000b0fb76c8fd59cdd7dab5e6d314a10596.pdf. [Google Scholar]
- 56.Zeinali M, Vossoughi M, Ardestani SK. 2008. Degradation of phenanthrene and anthracene by Nocardia otitidiscaviarum strain TSH1, a moderately thermophilic bacterium. J Appl Microbiol 105:398–406. doi: 10.1111/j.1365-2672.2008.03753.x. [DOI] [PubMed] [Google Scholar]
- 57.Sambrook JR, Russell DW (ed). 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 58.Singleton DR, Richardson SD, Aitken MD. 2008. Effects of enrichment with phthalate on polycyclic aromatic hydrocarbon biodegradation in contaminated soil. Biodegradation 19:577–587. doi: 10.1007/s10532-007-9163-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.