Acidithiobacillus ferrooxidans is an iron- and sulfur-oxidizing chemolithoautotroph and is a key member of the microbial consortia used in industrial biomining applications. There is interest in exploiting these cells for other metal recovery applications as well as in developing them as unique nonmodel microbial cell factories. Plasmid-driven expression of exogenous genes has been reported, and homologous recombination has been used to knock out some gene expression. Here, new selection protocols facilitated the development of a transposition method for chromosomal integration of exogenous genes into A. ferrooxidans. This greatly expands the available genetic toolbox, which will open the door to greater metabolic engineering efforts for these cells.
KEYWORDS: Acidithiobacillus ferrooxidans, biofuel, biomining, genetic engineering, sulfur oxidation, transformation, transposons
ABSTRACT
The development of Acidithiobacillus ferrooxidans as a non-model host organism for synthetic biology is hampered by a lack of genetic tools and techniques. New plating and liquid-based selection methods were developed to improve the identification of transformed cell lines. Enabled by these methods, a hyperactive transposase was used to generate mutants with integrated genes for the expression of the superfolder green fluorescent protein (sfGFP) gene or a 2-keto decarboxylase (KDC) gene, which enabled the production and secretion of isobutyric acid (IBA). An inverse PCR method was used to identify the insertion sites of the KDC gene in several mutants, leading to the identification of a region on the chromosome that may be suitable for future genetic insertions. These results demonstrate that functional exogenous metabolic genes have been chromosomally integrated into A. ferrooxidans, and this advance will facilitate the future development of these cells for new biotechnology applications.
IMPORTANCE Acidithiobacillus ferrooxidans is an iron- and sulfur-oxidizing chemolithoautotroph and is a key member of the microbial consortia used in industrial biomining applications. There is interest in exploiting these cells for other metal recovery applications as well as in developing them as unique nonmodel microbial cell factories. Plasmid-driven expression of exogenous genes has been reported, and homologous recombination has been used to knock out some gene expression. Here, new selection protocols facilitated the development of a transposition method for chromosomal integration of exogenous genes into A. ferrooxidans. This greatly expands the available genetic toolbox, which will open the door to greater metabolic engineering efforts for these cells.
INTRODUCTION
Acidithiobacillus ferrooxidans is an acidophilic, chemolithoautophic bacterium that obtains energy from the oxidation of ferrous iron or reduced inorganic sulfur compounds (RISCs) (1, 2). A. ferrooxidans is a key member of microbial consortia used in industrial bioleaching of copper and other metal sulfides, and it has been explored for use in other applications such as sour gas treatment, coal desulfurization, and electronic waste recycling (3–5). Recently, there has been interest in the development of A. ferrooxidans as a biotechnology platform organism for biofuel and biochemical production (6, 7); however, this has been hampered by a lack of tools and techniques to genetically modify the cells.
One significant bottleneck in the development of engineered A. ferrooxidans strains has been transformation with foreign DNA. The methods for conjugating plasmids from Escherichia coli into A. ferrooxidans have been established, and the conjugation conditions for IncQ-type plasmids have been improved by several orders of magnitude by modifying the formulations of media (8, 9). The solid 2:2 medium (0.625% [wt/vol] agar) used for conjugations [containing 0.2% Fe(II), 0.2% (wt/vol) S2O3, 100 µM diaminopimelic (DAP), 40 µg/ml leucine, and 200 µg/ml kanamycin] includes both ferrous iron and thiosulfate, both of which provide energy to A. ferrooxidans. However, it appears that the high concentrations of ferrous iron in the 2:2 medium may prevent kanamycin and streptomycin from providing transformation selection, likely due to the involvement of ferrous iron in degradation of the antibiotics. These antibiotics are found to be ineffective in liquid media at the acidic pH ranges optimal for growth (10). While the use of mercury resistance has been explored as an alternative selectable marker, heavy metals are undesirable for handling in cultures because of their toxicity (11). It has recently been reported that robust growth of engineered A. ferrooxidans cells was possible in liquid media containing ferric iron and sulfur, indicating that ferrous iron is not necessary for A. ferrooxidans cultivation (12). Therefore, it was hypothesized that selection procedures may be improved with the removal of ferrous iron from the solid and liquid selection media.
To date, the genetic engineering of A. ferrooxidans has been limited to the expression of just a few endogenous and exogenous genes. Previous plasmid-based expression studies included the overexpression of native iron oxidation genes rus, cyc2, and tetH (13–15). The superfolder green fluorescent protein (sfGFP) gene has been introduced to the cells, and to explore the biosynthetic capabilities of this organism, the 2-keto decarboxylase (KDC) gene from Lactococcus lactis was expressed to enable the production of isobutyric acid (IBA) from precursors in the valine biosynthesis pathway (6, 16). In addition, two genes (encoding acyl-acyl carrier protein [acyl-ACP] reductase and aldehyde deformylating oxygenase) from Synechococcus elongatus were expressed to enable the production of the long-chain hydrocarbon heptadecane (6). These transient transformations were made using modifications to the broad-host-range pJRD215 plasmid (17). The wild-type (WT) strain and the genetically modified strains have been investigated using a variety of medium compositions, including the addition of metal chelators, to improve growth rates and maximize chemical yields in batch and continuous cultures (16, 18, 19). Although several constitutive promoters of different strengths have been characterized with respect to driving gene expression, the need for inducible control of gene expression within this organism led to the identification of endogenous promoter sequences compatible with the growth conditions for A. ferrooxidans (12, 20). Thus, these developments with plasmid-based systems have greatly expanded the ability to modify A. ferrooxidans cells for different future applications.
Efforts aimed at the introduction of plasmids from various incompatibility groups have shown that plasmids are steadily lost from transformed cells over time. Due to the difficulty in applying selective pressure in liquid media, even the most stable IncQ-type plasmid, pJRD215, had a roughly 70% retention rate in the type strains of A. ferrooxidans (8). Transitioning foreign gene expression from a plasmid to the chromosome would overcome the drawbacks associated with plasmid stability and the metabolic burden of plasmid maintenance from the cells.
Despite the progress made in expanding the available genetic engineering toolbox, permanent genetic manipulation of the chromosome has been difficult, with only a few reports describing the creation of mutant knockout strains. Homologous recombination has been used to knock out the recA, pfkB, and tetH genes to better understand the role of these native genes in A. ferrooxidans (15, 21, 22). Far less has been reported for chromosomal gene integration in these cells, and yet the ability to express heterologous genes from the chromosome is often critical for industrial processes where sustained gene expression over an extended period is required. The recombination strategies used to produce knockouts could be used for chromosomal integration, although this has not yet been reported in the literature. Evidence has been found of the presence of a native insertion sequence in A. ferrooxidans which enables horizontal gene transfer causing phenotypic switching under different growth conditions (23), which indicates active native transposition activity. The Tn5 transposition system has been widely used to study microorganisms through random mutagenesis and for the engineering of bacterial species by inserting recombinant genes into the chromosomes of Gram-negative bacteria (24, 25). In prior work in A. ferrooxidans, a suicide plasmid harboring the Tn5 transposase was used to demonstrate delivery of kanamycin resistance, but that approach had not been further explored and there was no reported attempt to identify the location of the insertion sites (9). Because of the size of the pJRD215 backbone, it has been difficult to develop complex genetic systems (26). The additional utility of transposon insertions arises from the ability to insert genes of any size into A. ferrooxidans. Moving genes onto the chromosome will allow the expression of multiple genes within A. ferrooxidans without being limited by the space available on the pJRD215 vector, which contains genes for plasmid mobilization and replication.
Here, chromosomal integration into A. ferrooxidans was investigated with the pBAM1 plasmid, which contains the mini-Tn5 transposase where the transposase has been mutated to be hyperactive. This system allows the suicide delivery of a gene within the transposon to the genome (27, 28). To improve this process, a new suicide plasmid, pBAM2, was created which contains a further mutation to endow increased transposase activity. The transformation of the pBAM2 plasmid derivatives was facilitated by the development of new solid medium and liquid medium formulations that are more suitable for selection with antibiotics. The hyperactive mutant Tn5 transposase was used to integrate a fluorescent marker gene, the sfGFP gene, as well as to introduce a metabolic gene, KDC, into the genome to create strains that would perpetually make IBA. The locations of integration of the KDC gene were identified in the genome by an inverse PCR method. Thus, the transposase-mediated chromosomal integration of functional metabolic genes in the A. ferrooxidans genome is demonstrated.
RESULTS
Construction of plasmids and conjugation donor strains.
Experiments with the mini-Tn5 transposase in the pBAM1 plasmid did not generate large numbers of mutants for subsequent characterization; thus, the hyperactive transposase as described by Martínez-García et al. (29) was modified by inclusion of another mutation, P242G, to endow more hyperactivity. The hyperactive Tn5 transposase was codon optimized to increase gene expression in A. ferrooxidans, and this was used to create the pBAM2 plasmid (see Table S1 and Fig. S1 in the supplemental material). The sfGFP operon and the KDC operon were inserted into pBAM2 to generate the pBAM2-GFP and pBAM2-KDC plasmids (Fig. S2) (6, 12). The plasmids were electroporated into donor E. coli S17-1 λpir lacI cells for conjugation.
Modified solid and liquid media.
Low transformation rates have inhibited the genetic engineering of A. ferrooxidans, and so modified medium formulations were developed to increase the reproducibility and ease of transferring DNA to A. ferrooxidans. By developing solid and liquid variants of the 2:2 solid medium using ferric iron and sulfur, these changes allowed better growth throughout the conjugation process, resulting in faster formation of larger colonies on the solid medium, and this enabled effective screening of colonies with the liquid medium. Transconjugants containing the A. ferrooxidans KDC (AF-KDC) plasmid on S204 solid medium plates (0.625% [wt/vol] agar) containing 0.04% (wt/vol) Fe(III), 0.2% (wt/vol) S2O3, 100 µM diaminopimelic acid (DAP), 40 µg/ml leucine, and 225 µg/ml kanamycin appeared between days 7 and 14, which was a shorter period than that observed for colony formation with the previous solid medium formulation. The use of this solid medium indicated that ferrous sulfate is not necessary for growth of A. ferrooxidans on plates. The cells were able to grow in the presence of thiosulfate as a sole energy source at pH 5.0 with a small amount of ferric sulfate. These results contrast with previous work where no colony formation was observed on solid medium when thiosulfate was used as the sole energy source at pH 4.5 or above (30, 31). Ferric iron may play a role in allowing thiosulfate oxidation at this higher pH.
A liquid selection medium (SM4 medium) made up of AFM basalt salts adjusted to pH 5.0 with NaOH supplemented with 0.0005% (wt/vol) Fe(III), 1 mM citric acid, 100 µM DAP, 40 µg/ml leucine, 0.1% (wt/vol) colloidal sulfur, and 250 µg/ml kanamycin as needed was developed to select transformed colonies. The selection was validated with wild-type cells and cells transformed with the AF-KDC plasmid. The kanamycin resistance on the plasmid allowed mutants to oxidize sulfur to sulfate, gradually acidifying the media to approximately pH 2.0 within 1 to 2 weeks (Fig. 1). Untransformed, wild-type cells were inhibited by kanamycin and thus were unable to oxidize sulfur as demonstrated by the lack of change in the pH in the same time frame.
FIG 1.
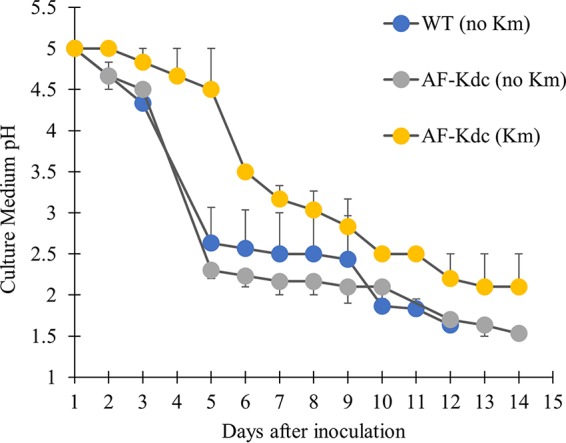
Antibiotic selection of transformed A. ferrooxidans cultures in liquid cultures. Batch cultures of AF-KDC cells with kanamycin (250 μg/ml) (yellow circles) were compared to cultures of wild-type cells (blue circles) and of AF-KDC cells (gray circles) without kanamycin in SM4 media. Cell growth was monitored by analysis of the decrease in culture media pH over time. Wild-type cells without kanamycin resistance did not produce changes in the SM4 medium pH in the presence of kanamycin for the duration of incubation (data not shown). The growth data are presented as means of results from three biological replicates. Cultures were measured either until 14 days or until the growth medium pH reached approximately pH 2. To prevent overlapping, the error bars that appear above the symbols (and which represent standard errors) correspond to AF-KDC cells (with kanamycin [Km]) and WT cells (no Km), and the error bars that appear below the symbols correspond to AF-KDC cells (no Km). Cell growth in each culture was also quantified by a MPN assay as described in the text.
A most-probable-number (MPN) assay (32, 33) was used to quantify the growth of the cultures, as optical methods for determining cell concentrations are unreliable for A. ferrooxidans cultures, especially in the dilute SM4 medium. In kanamycin-free SM4 medium, the wild-type cells grew to a concentration of log10(7.97 ± 0.52) per ml, and AF-KDC cells grew to a concentration of log10(8.69 ± 0.44) per ml. In SM4 with kanamycin, AF-KDC plasmid-transformed cells grew to a concentration of log10(8.26 ± 0.17) per ml.
This liquid selection medium was used to identify mutants that had obtained kanamycin resistance after transformation with the pBAM2 transposase plasmids. Transconjugants obtained from conjugations were put through two passages in SM4 medium to confirm extended kanamycin resistance, a result that suggested that the transposons had moved from the pBAM2 plasmids into the genome.
Construction and characterization of GFP-integrated A. ferrooxidans strains.
When pBAM2-GFP was used to deliver the sfGFP gene, kanamycin-resistant strains were successfully grown in SM4 medium. The portion of the plasmid backbone harboring the ampicillin resistance and the transposase should have been lost, and the DNA fragment containing that region should not have been detectable. PCR analysis using the DNA obtained from strains by the use of a miniprep kit after the two passages in SM4 medium and one passage in F2S medium [containing 0.8 g/liter (NH4)2SO4, 0.1 g/liter HK2PO4, 2.0 g/liter MgSO4·7H2O, 5 ml/liter ATCC MD-TMS, 10 mM citric acid, 100 mM FeSO4·7H2O, and 0.1 (wt/vol) colloidal sulfur] confirmed that only the positive-control sample with the pBAM2-GFP plasmid produced a PCR product using primer pair pBAM2F/pBAM2R. To confirm the presence of GFP within the cells, the GFP-integrated strains were imaged with confocal microscopy. The cells exhibited green fluorescence, and no qualitative differences between the levels of fluorescence of the different strains were noted (Fig. 2). The persistence of green fluorescence after several passages in liquid medium suggested that GFP had been integrated into the chromosome from the transposase, as the plasmid originally containing the gene had apparently been lost.
FIG 2.
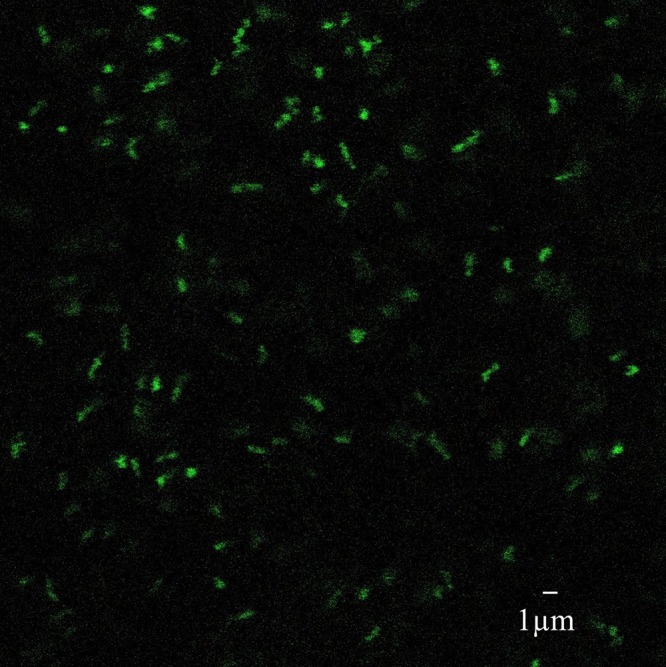
A confocal image of A. ferrooxidans transformed with the pBAM2-GFP transposase plasmid. After the apparent loss of the pBAM2-GFP suicide plasmid was confirmed, GFP fluorescence in the transformed A. ferrooxidans cells was imaged to validate that transgene expression was occurring, indicating likely chromosomal integration.
Construction and characterization of KDC-integrated A. ferrooxidans strains.
After we demonstrated that persistently green, kanamycin-resistant strains could be obtained using the hyperactive transposase, the pBAM2-KDC vector construct was explored, as chromosomal integration of an exogenous metabolic enzyme is more relevant than the use of a reporter gene for future applications. Numerous colonies were obtained on the solid selection plates from the conjugation of the pBAM2-KDC plasmid. Approximately 10 colonies were randomly chosen from each selection plate to be inoculated into the SM4 media for further selection. In every case, some of the transconjugants were not viable in SM4 medium, indicating that the kanamycin resistance had been lost. The strains successfully grown in SM4 medium were expanded in F2S medium to extract both genomic and plasmid DNA and to analyze the production of IBA from each strain using gas chromatography-mass spectrometry (GC/MS) analysis. PCR analysis using the DNA obtained from strains by the use of a miniprep kit after the two passages in SM4 medium and one passage in F2S medium confirmed the apparent loss of the plasmid, as only the positive control with the pBAM2-KDC plasmid produced a PCR product using primer pair pBAM2F/pBAM2R.
Comparison of levels of IBA production in KDC-integrated and plasmid-driven strains in mixed iron sulfur medium.
The production of IBA in the supernatant of cells transformed with pBAM2-KDC was measured by GC/MS and compared to the levels seen with cells harboring the pJRD215 plasmid driving transient KDC expression. The sulfur-supplemented F2S medium, which should have more energy available for growth and product formation, was used. Three IBA-producing mutants (AFKI1, AFKI2, and AFKI3) were characterized, and various amounts of IBA were observed. AFKI1 produced 0.79 ± 0.01 ppm of IBA in its supernatant over 72 h, AFKI2 produced 0.45 ± 0.03 ppm, and AFKI3 produced 0.91 ± 0.02 ppm (Fig. 3). In contrast, the KDC gene expressed from the pJRD-KDC plasmid produced 3.5 ± 0.1 ppm of IBA in the F2S medium, indicating that multicopy expression of KDC from the pJRD vector allows higher production of IBA than single-copy expression from the genome.
FIG 3.
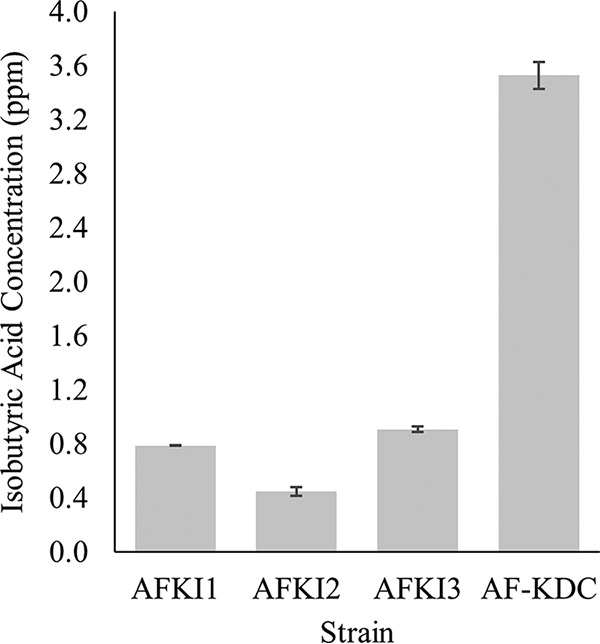
Comparison of the levels of isobutyric acid production of KDC-integrated strains (AFKI1, AFKI2, and AFKI3) and plasmid-driven strain AF-KDC in the sulfur-based F2S medium. Cultures were grown for 72 h, and supernatants were obtained for analysis. Measurements were made in triplicate, and the error bars represent standard deviations from the mean.
Identification and genetic analysis of transposon insertions.
An inverse PCR technique using nested primers was used to distinguish the desired amplicons from nonspecific PCR products in order to characterize the Tn5 transposon insertion sites in the A. ferrooxidans genomes (Fig. S3). EcoRI was used as the restriction enzyme for the genomic digestion as it provided a single cut within the transposon containing the KDC gene. While the primers used produced off-target PCR bands, comparison of the bands corresponding to the PCR products from the KDC-integrated strains with the bands obtained from the control wild-type DNA enabled the identification of a pair of nested bands that could be sequenced for each strain. As the lengths of the transposon fragments on either side of the EcoRI were known to be 2.6 kb and 0.8 kb, amplified bands below these amplicon sizes were ignored even when nested bands were obtained from the amplification of the mutant strain. The transposon integration loci for the three strains whose IBA production was characterized were identified using this technique.
After identifying the integration loci, the validity of the inverse PCR method was confirmed by amplifying the region around each locus using primer pairs AFKI1Fwd/AFKI1Rev, AFKI2Fwd/AFKI2Rev, and AFKI3Fwd/AFKI3Rev. These results demonstrated that the 3.5-kb transposon containing the KDC gene was successfully integrated into the genome as observed by determination of the increase in the length of the PCR amplicon between the wild-type DNA and mutant genomic DNA (Fig. 4).
FIG 4.
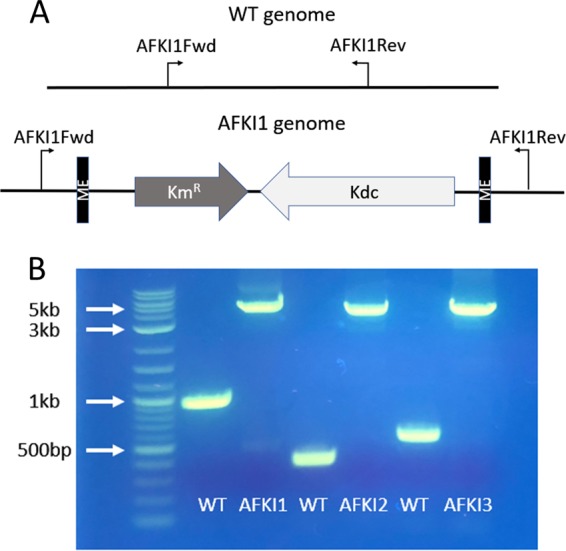
The transposon integration sites were confirmed by amplification of the region of the genome flanking the insertion sites. Panel A shows the genomic PCR amplification scheme for the AFKI1 strain. The primers flanking the insertion site were designed from the published genome sequence for ATCC 23270. In the wild-type genome, the PCR amplicon is of a known size. In the transposon-integrated strains, the same PCR primers were used to amplify the transposon region as well, resulting in a 3.5-kb increase in amplicon size. Panel B shows the results of the method applied to the three transposon locations identified in the mutant strains and visualized on an agarose gel, confirming the successful identification of integration sites.
Using the blastx program at NCBI, the nucleotide sequences of the genes surrounding of the integration loci were compared to sequences in the nonredundant protein sequence database to obtain the putative functions of the disrupted genes for each mutant strain (34). These functions and the locations of the 9-bp sequence repetitions created on both sides of the transposon are shown in Fig. 5. The locations of the insertions for the two higher-IBA producers were separated by only 2 kb.
FIG 5.
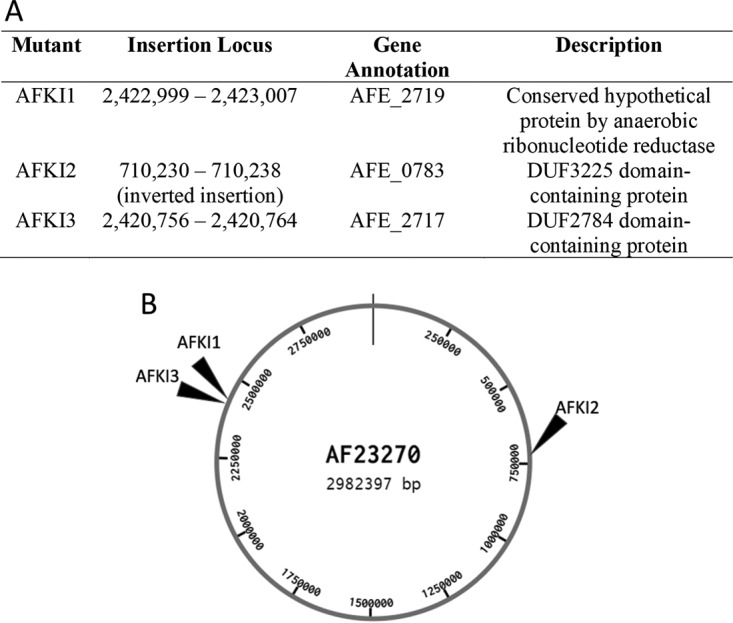
Location of the chromosomal integration sites. Panel A shows the integration loci for KDC-integrated strains. The insertion locus identifies the 9-bp sequence duplicated by the transposase to insert the transposon. Panel B shows the approximate locations of the transposon insertions for the mutant strains in relation to the whole A. ferrooxidans 23270 genome.
DISCUSSION
The exploitation of A. ferrooxidans for various biotechnology applications will be accelerated by the development of new genetic tools and techniques. A critical challenge of working with acidophilic microorganisms is the (lack of) stability of selectable antibiotic markers. Genome-level engineering efforts in A. ferrooxidans are only beginning to be reported in the literature. Here, a combination of solid and liquid formulations of media using sulfur as the main energy source which can improve transformation and selection procedures is described, and these methods were used to demonstrate a transposition technique to integrate exogenous metabolic genes into the genome. This capability will be necessary to further engineer A. ferrooxidans for new biotechnology applications and will enable further investigation of the fundamental physiology of this and related organisms (35, 36).
Other acidophilic cell lines, such as Acidithiobacillus caldus and Sulfolobus species, are capable of oxidizing RISCs, and some sophisticated genetic tools have been developed for these systems (37). A. ferrooxidans cells are especially interesting for their unusual ability to oxidize both iron and sulfur. The cells have been traditionally cultured in ferrous-iron-containing media due to the greater understanding of the iron oxidation pathway and physiological state of A. ferrooxidans than of sulfur oxidation pathways under these conditions (1, 2, 38, 39). However, the use of ferrous iron at low pH presents problems in the selection of transformed cells as the decomposition of aminoglycoside antibiotics, such as kanamycin and streptomycin, is accelerated under these conditions. While these antibiotics have been successfully used in solid formulations of media, successful liquid selection methods have not been reported. The complex interplay between iron activity and sulfur oxidation is still being evaluated, and robust growth on sulfur has been observed in the presence of ferric iron (40, 41). Therefore, new solid and liquid medium selection protocols were developed where sulfur is used as the energy source for the cells and where the lack of ferrous iron presumably reduces the degradation of the antibiotics. This medium formulation has improved the ability to identify transformed colonies on solid selection plates and to ensure their transformation through cultivation in antibiotic-containing liquid medium.
Growth in selective media does not guarantee that functional transposition has taken place, especially since the rate of suicide plasmid loss in A. ferrooxidans has not been evaluated. In this work, all colonies that were able to grow and expand in the SM4 selective media were found to either express sfGFP (when transformed with pBAM2-GFP) or produce IBA (when transformed with pBAM2-KDC). Analysis by PCR was performed to confirm suicide plasmid loss, and the suicide plasmid backbones were not detected, indicating that transposition, as opposed to persistent residence of the plasmid vectors, was likely to have occurred.
In order to definitively confirm that a transposition event has taken place, the location of the transposon in the genome must be determined. An inverse PCR method was used, and 3 different chromosomal insertion sites were identified in the A. ferrooxidans genome. All 3 strains produced IBA at concentrations below 1 ppm, which is lower than the concentration that was observed with cells transiently transformed with a pJRD215 vector containing the KDC gene, which was above 3 ppm. This difference is likely due to a copy number effect; only a single copy of the KDC gene is located in the genome whereas multiple copies of the plasmid are present in the cells.
The two top IBA-producing strains had transposon integration sites very close to each other in the same region of the genome (Fig. 5). This suggests that insertions into this region may be effective to achieve relatively high expression of the KDC-encoding gene and possibly of other genes of interest. For the 3 clones characterized, a range of IBA production levels were obtained between the highest-producing (AFKI3) strain and lowest-producing (AFKI2) strain (Fig. 3). While it is difficult to correlate the activity of a metabolic gene to its expression levels, the difference in the levels of productivity of the strains suggest that the KDC gene was integrated into loci that may attenuate expression levels of the single copy of the gene. Although Tn5 is known to have a slight preference for G/C pairs at the end of the nine-base-pair sequence duplicated by the insertion, it has been shown to be sufficiently random in various species with no significant bias after characterizing numerous transposon insertions (42, 43). Further use of this method to generate and identify transposon insertions will be useful in determining whether Tn5 has a sequence bias within A. ferrooxidans, with GC content of 59% (1), affecting where transposons can be inserted into the genome, or whether the two close insertions producing large amounts of isobutyric acid had occurred by chance.
In conclusion, a method to integrate exogenous genes into A. ferrooxidans using a hyperactive Tn5 transposase has been demonstrated. The new selection methods introduced here facilitated the selection of transformed cells, and the results demonstrate that transposition should become a new reliable genetic tool for engineering new A. ferrooxidans strains and open the possibility of using Tn5 for other applications that have been explored in other bacterial strains.
MATERIALS AND METHODS
Chemicals, reagents, bacterial strains, and plasmids.
Unless otherwise noted, all chemicals were sourced from Sigma-Aldrich (St. Louis, MO), and enzymes and reagents for DNA manipulation were purchased from NEB (Ipswich, MA). In-Fusion enzyme and a NucleoSpin tissue kit were purchased from TaKaRa Bio USA (Mountain View, CA). All primers used in this study were obtained from Integrated DNA Technologies (Coralville, IA). All codon-optimized genes were chemically synthesized by ATUM (Newark, CA), and the pBAM1 and pBB528 plasmids were obtained from Addgene (Cambridge, MA). Acidithiobacillus ferrooxidans (ATCC 23270) and the pJRD215 plasmid were obtained as described previously (12). Lambda pir E. coli EC100D cells were sourced from Lucigen (Middleton, WI). All DNA sequencing was performed by Genewiz (South Plainfield, NJ). The bacterial strains and plasmids used in this study are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| E. coli EC100D | F− mcrA Δ(mrr-hsdRMS-mcrBC) Φ80dlacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ− rpsL (Strr) nupG pir+ (DHFR gene) | Lucigen |
| E. coli EC100D lacI | EC100D strain with pBB528 | This study |
| E. coli S17-1 λpir | Tpr Smr recA thi pro hsdR hsdM+ RP4::2-Tc::Mu::Km Tn7 λpir integrated into the chromosome | 22 |
| E. coli S17-1 λpir lacI | S17-1 λpir strain with pBB528 | This study |
| A. ferrooxidans ATCC 23270 | Type strain | ATCC |
| A. ferrooxidans AF-KDC | ATCC 23270 with AF-KDC plasmid | 14 |
| A. ferrooxidans AFGI1 | ATCC 23270 with integrated Tn5-GFP | This study |
| A. ferrooxidans AFKI1 | ATCC 23270 with integrated Tn5-KDC | This study |
| A. ferrooxidans AFKI2 | ATCC 23270 with integrated Tn5-KDC | This study |
| A. ferrooxidans AFKI3 | ATCC 23270 with integrated Tn5-KDC | This study |
| Plasmids | ||
| AF-GFP | pJRD215 containing sfGFP gene with tac promoter | 12 |
| AF-KDC | pJRD215 containing KDC gene with tac promoter | 14 |
| pBAM1 | Ampr, Kmr, R6K replicon, Tnp, Tn5 | 18 |
| pBB528 | Cmr, lacIq | 25 |
| pBAM2 | Codon-optimized transposase gene containing the P242G mutation with tac promoter replacing Tnp in pBAM1 | This study |
| pBAM2-GFP | pBAM2 with codon-optimized sfGFP gene | This study |
| pBAM2-KDC | pBAM2 with codon-optimized KDC gene | This study |
KDC, 2-keto decarboxylase; Tn5-KDC, Tn5 transposon containing KDC; DHFR, dihydrofolate reductase; Km, kanamycin; Amp, ampicillin; Cm, chloramphenicol; sfGFP, superfolder GFP; Tnp, transposase.
Genetic manipulations. (i) pBAM2.
The hyperactive Tn5 transposase in the pBAM1 plasmid was modified by adding an additional point mutation, P242G, to endow more hyperactivity (27, 44). This modified transposase gene was codon optimized for A. ferrooxidans expression (see Fig. S1 and Table S1 in the supplemental material). The hyperactive transposase was flanked by the tac promoter and bla terminator sequences. The purchased operon was PCR amplified from the plasmid obtained from ATUM using primers HypTnpFwd and HypTnpRev. The pBAM1 plasmid was PCR amplified with exclusion of the original transposase operon using primers pBAMFwd and pBAMRev. The vector and insertion were combined with the In-Fusion enzyme, generating SphI and XbaI sites on the ends of the transposase operon to generate pBAM2. The E. coli EC100D strain was transformed with the pBB528 plasmid containing the lacI gene (made in-house) and then used for the transformation of the pBAM2 plasmid (45).
(ii) pBAM2-GFP and pBAM2-KDC.
The superfolder GFP (sfGFP) operon and the KDC operon were PCR amplified from AF-GFP and AF-KDC plasmids using primers Tn5Fwd and Tn5Rev and were digested with NotI as described previously (6, 12). The amplicon was ligated into the pBAM2 plasmid digested with NotI to generate pBAM2-GFP and pBAM2-KDC (Fig. S2). E. coli EC100D lacI was used for the transformation of the ligated plasmids.
Media and culturing of Acidithiobacillus ferrooxidans.
A. ferrooxidans cultures were grown in either ferrous-iron-containing AFM3 medium [0.8 g/liter (NH4)2SO4, and 0.1 g/liter HK2PO4, 2.0 g/liter MgSO4·7H2O, 5 ml/liter ATCC MD-TMS, 10 mM citric acid, 100 mM FeSO4·7H2O] or sulfur-containing F2S medium (12). Specific medium compositions are listed in the supplemental material (Table S2). The growth media were sterilized with 0.2-μm-pore-size filters (Thermo Fisher Scientific, Waltham, MA), and sulfur was added to growth media after filtration. Cells were maintained for use in experiments by weekly subculture into 100 ml AFM3 medium at 30°C.
All cultures were initiated with a starting optical density at 600 nm (OD600) of 0.001 unless otherwise noted. Cells were harvested by centrifugation at 5,000 × g for 7 min. Harvested cells were kept in 10 ml of AFM3 medium and maintained viability for 1 to 2 weeks when stored at 4°C. A. ferrooxidans cultures were cultivated in F2S medium for the characterization of strains and isolation of genomic and plasmid DNA from cells.
The MPN assay was conducted with a 10× serial dilution from SM4 cultures which had reached a pH of approximately 2.0 (6). The serial dilutions were inoculated into AFM medium and were allowed to grow. The change in medium color was used to determine that the dilution had contained viable cells which caused the oxidation of ferrous iron. The assay was continued until no further color change was observed at any further dilutions for at least 48 h.
Plasmid conjugations. (i) Mating and conjugation of plasmid DNA.
A modified filter-mating conjugation protocol was used to transfer plasmids pBAM2-GFP and pBAM2-KDC from E. coli to A. ferrooxidans. Plasmids were electroporated into E. coli S17-1 λpir lacI. The S17-1 λpir lacI strains were grown at 37°C in 5 ml of 2:2 basalt salt media supplemented with 5 g/liter yeast extract at 37°C until the late-exponential-growth phase (8). Wild-type A. ferrooxidans cells were cultivated in F2S medium at 30°C until the late-exponential-growth phase. A. ferrooxidans cells were harvested by centrifugation and washed three times with AFM basal salts (12). The washed A. ferrooxidans cells and E. coli cells were then washed three times with 2:2 basal salt medium, and washed cells of both species were adjusted to an OD600 of 10 for mating. The donor and recipient cells were mixed in a donor-to-recipient ratio of 1:1/1:4, and 50 μl of cell suspension were spotted and incubated at 30°C for 5 days on filter paper placed on mating medium plates (0.625% [wt/vol] agar) containing 0.0005% (wt/vol) Fe(III) using Fe2(SO4)3 · 7H2O, 0.05% (wt/vol) S2O3 using sodium thiosulfate, 100 μM diaminopimelic acid (DAP), 40 μg/ml leucine, and 0.05% (wt/vol) yeast extract. After 5 days of incubation, cells were resuspended in 500 μl of 2:2 basal salt medium and 50 μl of the cell suspensions was plated on S204 solid medium plates (0.625% [wt/vol] agar) containing 0.04% (wt/vol) Fe(III), 0.2% (wt/vol) S2O3, 100 μM DAP, 40 μg/ml leucine, and 225 μg/ml kanamycin and incubated at 30°C until colonies appeared. Agar was sterilized by autoclaving, and basal salt medium and other additives described above were sterilized using 0.2-μm-pore-size filters.
(ii) Liquid selection medium for verification of mutants.
Single colonies of A. ferrooxidans transconjugants were inoculated in 10 ml of SM4 medium, which was made up of AFM basalt salts adjusted to pH 5.0 with NaOH supplemented with 0.0005% (wt/vol) Fe(III), 1 mM citric acid, 100 μM DAP, 40 μg/ml leucine, 0.1% (wt/vol) colloidal sulfur, and 250 μg/ml kanamycin as needed. Colloidal sulfur was added after filtration of the other components. Cultures were incubated at 30°C, and the pH of the medium was measured daily over 14 days of incubation or until the medium pH dropped to roughly 2.0, measured using pH strips. A 1-ml volume of each of the cultures exhibiting growth was transferred into new cultures with 100 ml of SM4 medium until the pH reached roughly 2.0.
Screening of suicide plasmid.
A 1-ml sample from the 100-ml SM4 medium cultures was used to inoculate cultures in 100 ml of F2S medium and incubated at 30°C until the concentration of Fe(II) was <5 mM. Cells were harvested by centrifugation, genomic DNAs were isolated using a NucleoSpin tissue kit, and plasmid DNAs were isolated using miniprep kits (Qiagen, Germantown, MD). DNAs from the miniprep kits were screened by PCR performed with pBAM1F and pBAM1R primers to explore the presence of the suicide plasmid. The respective pBAM2 derivatives that were conjugated into A. ferrooxidans were used as positive controls, and a negative-control experiment was conducted without the introduction of template DNA.
Characterization of GFP-integrated strains.
GFP-integrated strains were harvested, washed 3 times with AFM basal salt solution and 3 times with 2:2 basal salt solution, and resuspended in a 2:2 basal salt solution containing 4% glycerol. After cells were adjusted to an OD600 of 10, 5 μl of this suspension was spotted onto a glass microscope slide, covered with a glass coverslip, and viewed using a TCS SP5 multiphoton confocal microscope (Leica, Buffalo Grove, IL) with a 100× oil objective. Cells were imaged using a 488-nm-wavelength argon laser for excitation and a 525/50 bandpass filter for emission.
Characterization of KDC-integrated strains. (i) Measurement of isobutyric acid production by GC/MS.
Supernatants of F2S cultures with KDC-integrated strains were reserved for analysis of IBA production by GC/MS. The F2S cultures were harvested at 72 h, when the remaining ferrous concentration was <5 mM. A solution of 6 M NaOH was added to 1 ml of the reserved supernatant from the F2S cultures of KDC-integrated strains until the iron was fully precipitated from the solution (approximately pH 9.0). Solutions were centrifuged for 1 min at 17,000 × g to separate insoluble particles. The clarified supernatant was returned to approximately pH 3.0 with 1 M H2SO4 and mixed with a 3 M glycine-HCl buffer adjusted to pH 3.0 in a 9:1 sample-to-buffer ratio. The solution was analyzed using a Shimadzu GCMS-QP2010 Ultra GC/MS system and a Restek Stabilwax column (length, 30 m; inner diameter [I.D.], 0.25 mm; film thickness, 0.25 μm). A 0.3-μl sample was injected using the splitless mode with the inlet temperature held at 220°C. The GC oven temperature was initially held at 80°C for 3 min, and then the temperature was ramped up to 120°C at a rate of 40°C/min, then to 200°C at a rate of 20°C/min, and then to 240°C at a rate of 40°C/min and was held at 240°C for 3 min. The IBA peak was identified at 7.0 min by comparisons to the fragmentation pattern and retention time of a standard reference, and quantitative analysis was performed using a calibration curve from the standard reference and concentrations of 10.00 ppm, 5.00 ppm, 3.00 ppm, 1.00 ppm, and 0.10 ppm.
(ii) Identification of integration loci.
An inverse PCR method was adapted and used for the identification of integration sites (46) (Fig. S3). Approximately 100 ng of extracted genomic DNA from the KDC-integrated strains or wild-type A. ferrooxidans was digested with EcoRI overnight. Digested DNA was circularized with T4 DNA ligase using a DNA concentration of 1 ng/μl to favor intramolecular circularization. Using 25 ng of DNA as the template for each strain, 2 nested PCRs for each end of the transposon were performed with the following sets of primers: tn1RFwd/tn1RRev, tn2RFwd/tn1RRev, tn1LFwd/tn1LRev, and tn1LFwd/tn2LRev (Table 2). Bands that did not appear in both PCRs with the nested primers and bands less than the size of the digested transposon were ruled out as representing spurious PCR products. The nested reactions from wild-type DNA were used as a further control to rule out off-target PCR products. The remaining potential bands observed were subjected to gel purification using a PCR cleanup and gel extraction kit from TaKaRa Bio USA (Mountain View, CA) and were sequenced. Primers tnFSeq and tnRSeq were selected at each end of the transposon as sequencing primers. Using the NCBI Nucleotide blastx program, sequences were compared against the published ATCC 23270 genome to identify integration loci.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′–3′) |
|---|---|
| Tn5Fwd | TAT TAT CTG CGG CCG CCA TCG ACT GCA CGG TGC AC |
| Tn5Rev | AGA TAT CTG CGG CCG CTG TCA CTT T |
| Tn5Rev2 | AGA TAT CTC GCG GCC GCA AAA AGG CCA TCC GTC AGG ATG |
| HypTnpFwd | TAC ACA AGT AGC GTC GCA TGC CAT CGA CTG CAC |
| HypTnpRev | TTA GGC GGG CTA CTA TCT AGA TGT CAC TTT GCT TGA TAT ATG AGA ATT ATT TAA C |
| pBAMFwd | GAC GCT ACT TGT GTA CTG TCT CTT ATA CAC ATC TGA CGT CTT GTG T |
| pBAMRev | TAG TAG CCC GCC TAA TGA GCG |
| pBAM2F | AAG CGG GGT AAG CGC AAG AAT |
| pBAM2R | ATC GCC CAT GTT ATG CAG AAA |
| tn1RFwd | CCA CTA CCG GCA AGT TCT CCG |
| tn2RFwd | CAG TTC ACC GAC ACC AAA GGT G |
| tn1RRev | TAT GAA GAT GCA TGA GCC GGT C |
| tn1LFwd | TCG TCG ACC GAG CTT TTG C |
| tn1LRev | GAA AGA GGA TGC GCC GAA AGT G |
| tn2LRev | GGG AAA GCT CTT CGC CGA AC |
| tnFSeq | TGC ACA GCC ATA CCA CAG CTT C |
| tnRSeq | GGC TAC AGC TCG TTT CAC GCT G |
| AFKI1Fwd | TCG CCG TTC GTT TTC TCG |
| AFKI1Rev | GCC ACC GCA TCC AGT AAT C |
| AFKI2Fwd | ATG GTT CAC ACC GAA ATC AAT GC |
| AFKI2Rev | CAT CCA TGC TAC AGC CTA AGT TGC C |
| AFKI3Fwd | CCT GAT GTA GTC GTT GGC GTC C |
| AFKI3Rev | GTT CGT CAA CAG CAA AGT GGA AC |
(iii) Confirmation of integration loci.
The integration of the transposons was confirmed by designing primers for the genome surrounding the integration loci for each identified strain (Table 2). WT genomic DNA and mutant genomic DNA were PCR amplified using the respective forward and reverse primers for each strain. The bands obtained from the region amplified from wild-type genomic DNA and the genomic DNA from integrated strains were visualized by agarose gel electrophoresis and compared by amplicon size (Fig. 4).
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge funding from the National Science Foundation (CBET-1438263). T.K. and S.B. have a financial interest in Ironic Chemicals, LLC, which is negotiating a license for this technology from Columbia University.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01381-18.
REFERENCES
- 1.Valdés J, Pedroso I, Quatrini R, Dodson RJ, Tettelin H, Blake R II, Eisen JA, Holmes DS. 2008. Acidithiobacillus ferrooxidans metabolism: from genome sequence to industrial applications. BMC Genomics 9:597. doi: 10.1186/1471-2164-9-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quatrini R, Appia-Ayme C, Denis Y, Jedlicki E, Holmes DS, Bonnefoy V. 2009. Extending the models for iron and sulfur oxidation in the extreme acidophile Acidithiobacillus ferrooxidans. BMC Genomics 10:394. doi: 10.1186/1471-2164-10-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rawlings DE. 2002. Heavy metal mining using microbes. Annu Rev Microbiol 56:65–91. doi: 10.1146/annurev.micro.56.012302.161052. [DOI] [PubMed] [Google Scholar]
- 4.Yang Y, Chen S, Li S, Chen M, Chen H, Liu B. 2014. Bioleaching waste printed circuit boards by Acidithiobacillus ferrooxidans and its kinetics aspect. J Biotechnol 173:24–30. doi: 10.1016/j.jbiotec.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Nemati M, Harrison STL, Hansford GS, Webb C. 1998. Biological oxidation of ferrous sulphate by Thiobacillus ferrooxidans: a review on the kinetic aspects. Biochem Eng J 1:171–190. doi: 10.1016/S1369-703X(98)00006-0. [DOI] [Google Scholar]
- 6.Kernan T, Majumdar S, Li X, Guan J, West AC, Banta S. 2016. Engineering the iron-oxidizing chemolithoautotroph Acidithiobacillus ferrooxidans for biochemical production. Biotechnol Bioeng 113:189–197. doi: 10.1002/bit.25703. [DOI] [PubMed] [Google Scholar]
- 7.Banerjee I, Burrell B, Reed C, West AC, Banta S. 2017. Metals and minerals as a biotechnology feedstock: engineering biomining microbiology for bioenergy applications. Curr Opin Biotechnol 45:144–155. doi: 10.1016/j.copbio.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 8.Liu Z, Borne F, Ratouchniak J, Bonnefoy V. 1999. Genetic transfer of IncP, IncQ, IncW plasmids to four Thiobacillus ferrooxidans strains by conjugation, p 39–49. In Amils R, Ballester A (ed), Process metallurgy, vol 9 Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 9.Peng JB, Yan WM, Bao XZ. 1994. Plasmid and transposon transfer to Thiobacillus ferrooxidans. J Bacteriol 176:2892–2897. doi: 10.1128/jb.176.10.2892-2897.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang R, Lin C, Lin J, Pang X, Liu X, Zhang C, Lin J, Chen L. 2017. Construction of novel pJRD215-derived plasmids using chloramphenicol acetyltransferase (cat) gene as a selection marker for Acidithiobacillus caldus. PLoS One 12:e0183307. doi: 10.1371/journal.pone.0183307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kusano T, Sugawara K, Inoue C, Takeshima T, Numata M, Shiratori T. 1992. Electrotransformation of Thiobacillus ferrooxidans with plasmids containing a mer determinant. J Bacteriol 174:6617–6623. doi: 10.1128/jb.174.20.6617-6623.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kernan T, West AC, Banta S. 2017. Characterization of endogenous promoters for control of recombinant gene expression in Acidithiobacillus ferrooxidans. Biotechnol Appl Biochem 64:793–802. doi: 10.1002/bab.1546. [DOI] [PubMed] [Google Scholar]
- 13.Liu W, Lin J, Pang X, Cui S, Mi S, Lin J. 2011. Overexpression of rusticyanin in Acidithiobacillus ferrooxidans ATCC19859 increased Fe(II) oxidation activity. Curr Microbiol 62:320–324. doi: 10.1007/s00284-010-9708-0. [DOI] [PubMed] [Google Scholar]
- 14.Liu W, Lin J, Pang X, Mi S, Cui S, Lin J. 2013. Increases of ferrous iron oxidation activity and arsenic stressed cell growth by overexpression of Cyc2 in Acidithiobacillus ferrooxidans ATCC19859. Biotechnol Appl Biochem 60:623–628. doi: 10.1002/bab.1110. [DOI] [PubMed] [Google Scholar]
- 15.Yu Y, Liu X, Wang H, Li X, Lin J. 2014. Construction and characterization of tetH overexpression and knockout strains of Acidithiobacillus ferrooxidans. J Bacteriol 196:2255–2264. doi: 10.1128/JB.01472-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X, West AC, Banta S. 2016. Enhancing isobutyric acid production from engineered Acidithiobacillus ferrooxidans cells via media optimization. Biotechnol Bioeng 113:790–796. doi: 10.1002/bit.25837. [DOI] [PubMed] [Google Scholar]
- 17.Davison J, Heusterspreute M, Chevalier N, Ha-Thi V, Brunel F. 1987. Vectors with restriction site banks. V. pJRD215, a wide-host-range cosmid vector with multiple cloning sites. Gene 51:275–280. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Mercado R, Berlinger S, Banta S, West AC. 2014. Engineering Acidithiobacillus ferrooxidans growth media for enhanced electrochemical processing. AIChE J 60:4008–4013. doi: 10.1002/aic.14628. [DOI] [Google Scholar]
- 19.Li X, Mercado R, Kernan T, West AC, Banta S. 2014. Addition of citrate to Acidithiobacillus ferrooxidans cultures enables precipitate-free growth at elevated pH and reduces ferric inhibition. Biotechnol Bioeng 111:1940–1948. doi: 10.1002/bit.25268. [DOI] [PubMed] [Google Scholar]
- 20.Wang H, Fang L, Wen Q, Lin J, Liu X. 2017. Application of beta-glucuronidase (GusA) as an effective reporter for extremely acidophilic Acidithiobacillus ferrooxidans. Appl Microbiol Biotechnol 101:3283–3294. doi: 10.1007/s00253-017-8116-9. [DOI] [PubMed] [Google Scholar]
- 21.Liu Z, Guiliani N, Appia-Ayme C, Borne F, Ratouchniak J, Bonnefoy V. 2000. Construction and characterization of a recA mutant of Thiobacillus ferrooxidans by marker exchange mutagenesis. J Bacteriol 182:2269–2276. doi: 10.1128/JB.182.8.2269-2276.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang H, Liu X, Liu S, Yu Y, Lin J, Lin J, Pang X, Zhao J. 2012. Development of a markerless gene replacement system for Acidithiobacillus ferrooxidans and construction of a pfkB mutant. Appl Environ Microbiol 78:1826–1835. doi: 10.1128/AEM.07230-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cabrejos M-E, Zhao H-L, Guacucano M, Bueno S, Levican G, Garcia E, Jedlicki E, Holmes DS. 1999. IST1 insertional inactivation of the resB gene: implications for phenotypic switching in Thiobacillus ferrooxidans. FEMS Microbiol Lett 175:223–229. doi: 10.1111/j.1574-6968.1999.tb13624.x. [DOI] [PubMed] [Google Scholar]
- 24.Reznikoff WS. 2008. Transposon Tn5. Annu Rev Genet 42:269–286. doi: 10.1146/annurev.genet.42.110807.091656. [DOI] [PubMed] [Google Scholar]
- 25.Kang Y, Durfee T, Glasner JD, Qiu Y, Frisch D, Winterberg KM, Blattner FR. 2004. Systematic mutagenesis of the Escherichia coli genome. J Bacteriol 186:4921–4930. doi: 10.1128/JB.186.15.4921-4930.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loftie-Eaton W, Rawlings DE. 2012. Diversity, biology and evolution of IncQ-family plasmids. Plasmid 67:15–34. doi: 10.1016/j.plasmid.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 27.Martínez-García E, Calles B, Arévalo-Rodríguez M, de Lorenzo V. 2011. pBAM1: an all-synthetic genetic tool for analysis and construction of complex bacterial phenotypes. BMC Microbiol 11:38. doi: 10.1186/1471-2180-11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goryshin IY, Reznikoff WS. 1998. Tn5 in vitro transposition. J Biol Chem 273:7367–7374. doi: 10.1074/jbc.273.13.7367. [DOI] [PubMed] [Google Scholar]
- 29.Martínez-García E, Aparicio T, de Lorenzo V, Nikel PI. 2014. New transposon tools tailored for metabolic engineering of gram-negative microbial cell factories. Front Bioeng Biotechnol 2:46. doi: 10.3389/fbioe.2014.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng J-B, Yan W-M, Bao X-Z. 1994. Solid medium for the genetic manipulation of Thiobacillus ferrooxidans. J Gen Appl Microbiol 40:243–253. doi: 10.2323/jgam.40.243. [DOI] [Google Scholar]
- 31.Tuovinen OH, Kelly DP. 1974. Studies on the growth of Thiobacillus ferrooxidans. Arch Microbiol 98:351–364. doi: 10.1007/BF00425295. [DOI] [PubMed] [Google Scholar]
- 32.Haldane JBS. 1939. Sampling errors in the determination of bacterial or virus density by the dilution method. J Hyg (Lond) 39:289–293. doi: 10.1017/S002217240001192X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas HA. 1942. Bacterial densities from fermentation tube tests. J Am Water Works Assoc 34:572–576. [Google Scholar]
- 34.Gish W, States DJ. 1993. Identification of protein coding regions by database similarity search. Nat Genet 3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 35.Gerdes SY, Scholle MD, Campbell JW, Balázsi G, Ravasz E, Daugherty MD, Somera AL, Kyrpides NC, Anderson I, Gelfand MS, Bhattacharya A, Kapatral V, D'Souza M, Baev MV, Grechkin Y, Mseeh F, Fonstein MY, Overbeek R, Barabási AL, Oltvai ZN, Osterman AL. 2003. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J Bacteriol 185:5673–5684. doi: 10.1128/JB.185.19.5673-5684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu BJ, Sung BH, Koob MD, Lee CH, Lee JH, Lee WS, Kim MS, Kim SC. 2002. Minimization of the Escherichia coli genome using a Tn5-targeted Cre/loxP excision system. Nat Biotechnol 20:1018–1023. doi: 10.1038/nbt740. [DOI] [PubMed] [Google Scholar]
- 37.Gumulya Y, Boxall NJ, Khaleque HN, Santala V, Carlson RP, Kaksonen AH. 2018. In a quest for engineering acidophiles for biomining applications: challenges and opportunities. Genes 9:E116. doi: 10.3390/genes9020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quatrini R, Appia-Ayme C, Denis Y, Ratouchniak J, Veloso F, Valdés J, Lefimil C, Silver S, Roberto F, Orellana O, Denizot F, Jedlicki E, Holmes D, Bonnefoy V. 2006. Insights into the iron and sulfur energetic metabolism of Acidithiobacillus ferrooxidans by microarray transcriptome profiling. Hydrometallurgy 83:263–272. doi: 10.1016/j.hydromet.2006.03.030. [DOI] [Google Scholar]
- 39.Bonnefoy V, Holmes DS. 2012. Genomic insights into microbial iron oxidation and iron uptake strategies in extremely acidic environments. Environ Microbiol 14:1597–1611. doi: 10.1111/j.1462-2920.2011.02626.x. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki I, Takeuchi TL, Yuthasastrakosol TD, Oh JK. 1990. Ferrous iron and sulfur oxidation and ferric iron reduction activities of Thiobacillus ferrooxidans are affected by growth on ferrous iron, sulfur, or a sulfide ore. Appl Environ Microbiol 56:1620–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sand W. 1989. Ferric iron reduction by Thiobacillus ferrooxidans at extremely low pH-values. Biogeochemistry 7:195–201. doi: 10.1007/BF00004217. [DOI] [Google Scholar]
- 42.Goryshin IY, Miller JA, Kil YV, Lanzov VA, Reznikoff WS. 1998. Tn5/IS50 target recognition. Proc Natl Acad Sci U S A 95:10716–10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Green B, Bouchier C, Fairhead C, Craig NL, Cormack BP. 2012. Insertion site preference of Mu, Tn5, and Tn7 transposons. Mob DNA 3:3. doi: 10.1186/1759-8753-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steiniger M, Metzler J, Reznikoff WS. 2006. Mutation of Tn5 transposase β-loop residues affects all steps of Tn5 transposition: the role of conformational changes in Tn5 transposition. Biochemistry 45:15552–15562. doi: 10.1021/bi061227v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Marco A, Deuerling E, Mogk A, Tomoyasu T, Bukau B. 2007. Chaperone-based procedure to increase yields of soluble recombinant proteins produced in E. coli. BMC Biotechnol 7:32. doi: 10.1186/1472-6750-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huang G, Zhang L, Birch RG. 2000. Rapid amplification and cloning of Tn5 flanking fragments by inverse PCR. Lett Appl Microbiol 31:149–153. doi: 10.1046/j.1365-2672.2000.00781.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


